SUMMARY
Microrchidia 3 (MORC3), a human ATPase linked to several autoimmune disorders, has been characterized both as a negative and positive regulator of influenza A virus. Here, we report that the CW domain of MORC3 (MORC3-CW) is targeted by the C-terminal tail of the influenza H3N2 protein NS1. The crystal structure of the MORC3-CW:NS1 complex shows that NS1 occupies the same binding site in CW that is normally occupied by histone H3, a physiological ligand of MORC3-CW. Comparable binding affinities of MORC3-CW to H3 and NS1 peptides and to the adjacent catalytic ATPase domain suggest that the viral protein can compete with the host histone for the association with CW, releasing MORC3 autoinhibition and activating the catalytic function of MORC3. Our structural, biochemical, and cellular analyses suggest that MORC3 might affect the infectivity of influenza virus and therefore has a role in cell immune response.
Graphical Abstract
In Brief
Influenza virus employs multiple mechanisms to hijack host cellular pathways and evade innate immune responses. Zhang et al. identified MORC3 ATPase as a new target of H3N2-subtype influenza A protein NS1.
INTRODUCTION
Influenza A virus (IAV) is a common human pathogen that causes flu, a contagious respiratory disease. The virally encoded nonstructural protein 1 (NS1) is highly expressed during infection and suppresses antiviral host responses by interacting with multiple binding partners at different stages of infection (Klemm et al., 2018). NS1 consists of the RNA-binding domain, an effector domain, and the C-terminal tail that varies in the amino acid composition and length in different IAV subtypes. Specifically, the C-terminal tail of NS1 derived from the IAV H3N2 strain contains a sequence that mimics the amino-terminal sequence of histone H3 tail. This similarity, first identified by Marazzi et al. (2012), facilitated the idea of virus-mediated hijacking of the histone H3-binding components to compromise gene transcription and other vital programs in the host cells. It was indeed found that the interaction of NS1 with the PAF1 subunit of the human complex hPAF1C leads to suppression of hPAF1C-mediated transcriptional elongation (Marazzi et al., 2012). The same NS1 sequence, but methylated at lysine 229, was also shown to target the CHD1′ double chromodomain, an established reader of methylated lysine 4 of histone H3 (H3K4me) (Marazzi et al., 2012; Qin et al., 2014).
The initial immune response involves activation of several host programs that restrict viral gene replication, including activation of promyelocytic leukemia nuclear bodies (PML-NBs) and associated complexes. Microrchidia 3 (MORC3), a member of the novel family of human ATPases which localizes to PML-NBs, has been described both as a negative and positive regulator of IAV H1N1 and H5N1 subtypes (Bortz et al., 2011; Ver et al., 2015). An antiviral activity of MORC3 toward human herpes simplex virus 1 and human cytomegalovirus has also been observed (Sloan et al., 2016). At the molecular level, the MORC3 protein contains a gyrase, Hsp90, histidine kinase, and MutL (GHKL)-type ATPase domain, a CW-type zinc finger, and a coiled-coil region (Dutta and Inouye, 2000; Li et al., 2013). The CW domain of MORC3 binds to histone H3 tail, exhibiting selectivity to methylated lysine 4 (H3K4me) (Andrews et al., 2016; Li et al., 2012, 2016; Liu et al., 2016). In addition, the CW domain associates with the adjacent ATPase domain and impedes its binding to DNA, which is required for the catalytic activity of MORC3 (Andrews et al., 2016; Zhang et al., 2019). The CW-ATPase association results in MORC3 autoinhibition that is released through binding of CW to the H3 tail (Zhang et al., 2019). In the cell, functional CW has been shown to be essential for subcellular localization of MORC3 and for the recruitment of MORC3 to chromatin and accumulation in NBs (Andrews et al., 2016; Mimura et al., 2010).
In this work, we have identified MORC3 as a new target of the viral protein NS1. We show that the C-terminal tail of NS1 from H3N2-subtype influenza virus has the ability to interact with the CW domain of MORC3. NS1 occupies the H3-binding site of CW and can compete with the histone peptide to alleviate MORC3 autoinhibition. Overexpression of MORC3 affects the infectivity of the H3N2-subtype influenza virus, suggesting a role of MORC3 in immune response.
RESULTS AND DISCUSSION
We initially explored the capability of the viral protein NS1 to recognize the CW domain of MORC3 (MORC3-CW) by NMR spectroscopy. We produced MORC3-CW as a uniformly 15N-labeled protein and collected its 1H,15N heteronuclear single quantum coherence (HSQC) spectra while unlabeled methylated and unmodified NS1 peptides (residues 222–230 of NS1) were gradually added to the protein (Figure 1B). Addition of the NS1 peptide trimethylated at K229 (NS1K229me3) induced large chemical shift perturbations (CSPs) in MORC3-CW, indicating direct binding, and similar in direction but smaller in magnitude CSPs were caused by the unmodified NS1 peptide. These data suggested that both trimethylated and unmodified NS1 peptides occupy the same binding site in MORC3-CW, although the NS1K229me3 peptide binds tighter.
Figure 1. MORC3-CW Is a Target of NS1.
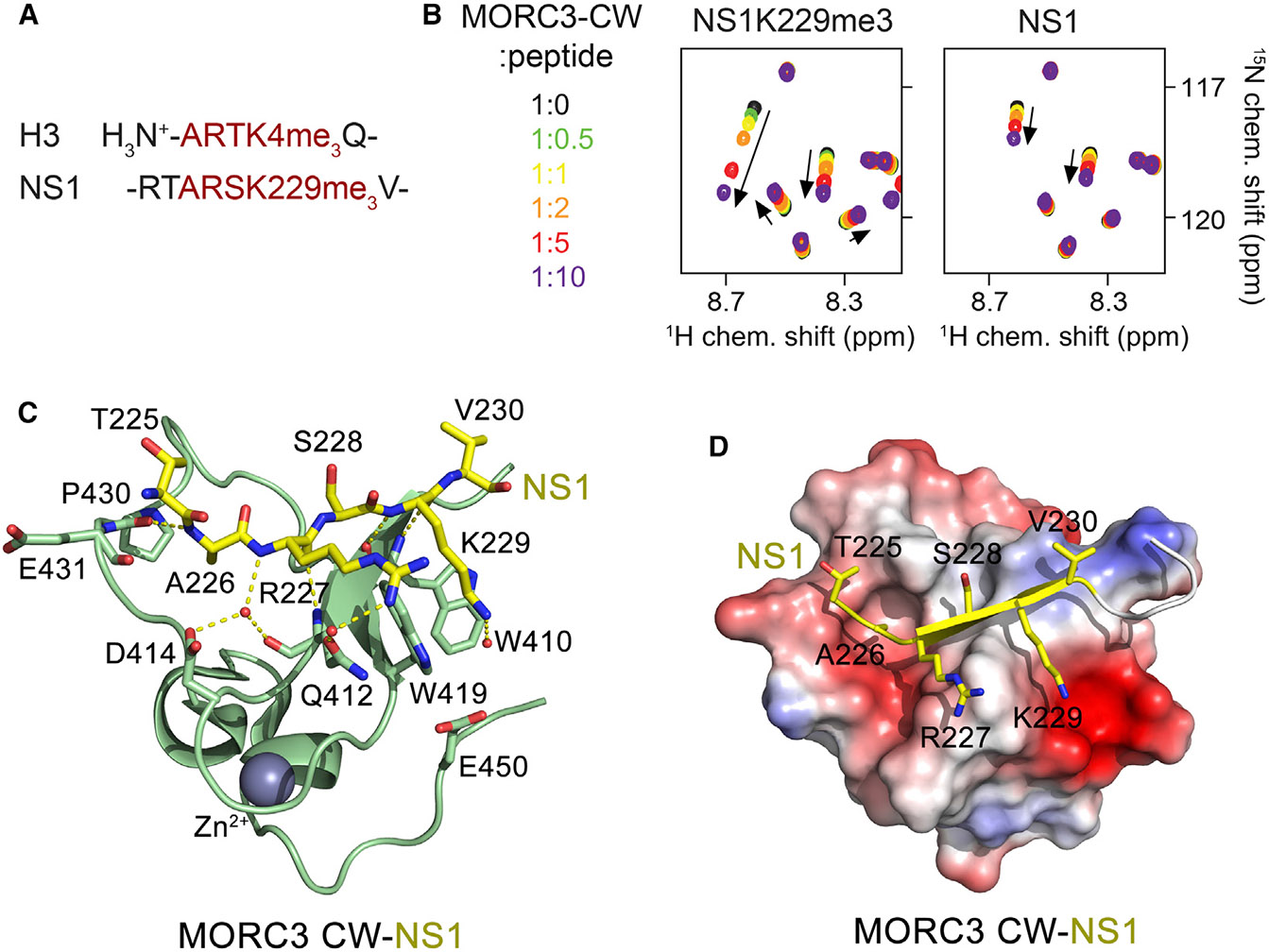
(A) The amino-terminal sequence of H3 (trimethylated at lysine 4) and the C-terminal sequence of NS1 (trimethylated at lysine 229) are shown.
(B) Superimposed 1H,15N HSQC spectra of 15N-labeled MORC3-CW collected upon titration with the methylated and unmodified NS1 peptides. Spectra are color coded according to the protein-to-peptide molar ratio.
(C) The crystal structure of the MORC3-CW:NS1 complex. The CW domain is shown in a ribbon diagram (green), and NS1 is shown as sticks (yellow). The zinc (gray) atom is shown as a sphere.
(D) Electrostatic surface potential of the MORC3-CW domain colored blue and red for positive and negative charges, respectively. The bound NS1 peptide is yellow.
See also Figure S1.
To determine the molecular mechanism for the recognition of NS1K229me3 by the MORC3-CW domain, we generated a chimeric construct that contains residues 225–230 of NS1 linked to residues 407–455 of MORC3 via a GGSG linker. The 1H,15N HSQC spectrum of the linked NS1-CW construct overlaid well with the spectrum of the isolated CW domain in the presence of a 10-fold excess of NS1K229me3 peptide, therefore confirming that the linked and unlinked complexes adopt essentially identical structures in solution (Figure S1). Using this fusion protein, we crystallized the MORC3-CW:NS1 complex and refined its structure to 1.41 Å resolution (Figure 1C; Table 1). The entire sequence of the fusion protein, including the linker region, showed well-defined electron density and therefore was unambiguously traced.
Table 1.
Data Collection and Refinement Statistics for the MORC3-CW:NS1 Complex
| NS1-Linked MORC3-CW | |
|---|---|
| Data collection | |
| Wavelength (Å) | 1.27818 |
| Space group | P212121 |
| Resolution (Å) | 50.00–1.41 |
| Cell dimensions | |
| a, b, c (Å) | 30.91, 37.42, 50.31 |
| α, β, γ (°) | 90.00, 90.00, 90.00 |
| No. of measured reflections | 128,942 |
| No. of unique reflections | 11,669 |
| Completeness (%) | 99.2 (96.3) |
| Redundancy | 11.0 (6.9) |
| I/σ(I) | 92.4 (59.5) |
| Rmerge (%) | 8.0 (10.3) |
| Refinement | |
| Resolution (Å) | 26.34–1.41 |
| No. of reflections | 21,392 |
| R factor (%) | 15.28 (14.8) |
| Rfree (%) | 16.87 (15.1) |
| No. of protein atoms | 479 |
| No. of zinc | 1 |
| No. of water molecules | 135 |
| RMSD from ideal values | |
| Bond lengths (Å) | 0.005 |
| Bond angles (°) | 0.95 |
| Average B values (Å2) | 8.73 |
| Protein | 12.1 |
| Zinc | 8.3 |
| Water | 25.1 |
| Ramachandran plot analysis (%) | |
| Favored | 100 |
| Allowed | 0 |
| Outliers | 0 |
Values in parentheses refer to data in the highest-resolution shell. Datasets collected from a single crystal. RMSD, root-mean-square deviation.
In the complex, the MORC3-CW domain folds into a compact globular structure consisting of a double-stranded antiparallel β sheet and a couple of helical turns stabilized by a zinc-binding cluster (Figure 1C). The R227-V230 region of NS1 adopts an extended conformation and forms the third antiparallel β strand, pairing with the β sheet of MORC3-CW (Figures 1C and 1D). Characteristic β sheet hydrogen bonds are formed between residues R227 and K229 of NS1 and residues W410 and Q412 of CW. In addition, the amide group of R227 forms water-mediated hydrogen bonds with the carbonyl group of Q412 and the carboxyl group of D414, whereas the amide group of A226 donates a hydrogen bond to the carbonyl group of P430. The intermolecular contacts involving the side chains of the NS1 residues also contribute to the interaction. The guanidino moiety of R227 is restrained through a water-mediated hydrogen bond with the side chain carbonyl group of Q412, whereas the K229 amino group is in transient water-mediated contact with the carboxyl group of E450. The hydrophobic part of the K229 side chain lies between the aromatic moieties of W410 and W419, and the methyl group of A226 resides in a well-defined pocket of CW (Figure 1D).
To characterize the interaction in detail, we measured binding affinities of MORC3-CW to methylated and unmodified NS1 peptides using tryptophan fluorescence (Figures 2A, 2B, and S2). We found that MORC3-CW interacts with the unmodified NS1 peptide (residues 222–230) with a dissociation constant (Kd) of 157 μM. In agreement with NMR titration experiments, the binding affinity of MORC3-CW toward NS1K229me3 peptide was increased ~11-fold (Kd = 14 μM). This binding affinity is comparable to the binding affinity of MORC3-CW for histone H3 peptide (Kd = 7 μM), but is weaker than the binding affinity of this protein for H3K4me3 (Kd = 0.6 μM) (Andrews et al., 2016). Notably, similar binding affinities of MORC3-CW for NS1K229me3, and for the ATPase domain (Kd = 9 μM), suggest that binding of the viral protein can alleviate MORC3 autoinhibition and stimulate ATP hydrolysis. Mutation of either W410 or W419 in the CW domain to alanine abrogated binding of NS1K229me3, pointing to the critical role of K229me3 caging (Figures 2A, 2C, and 2D).
Figure 2. Mechanistic Insight into H3 Mimicry by NS1.
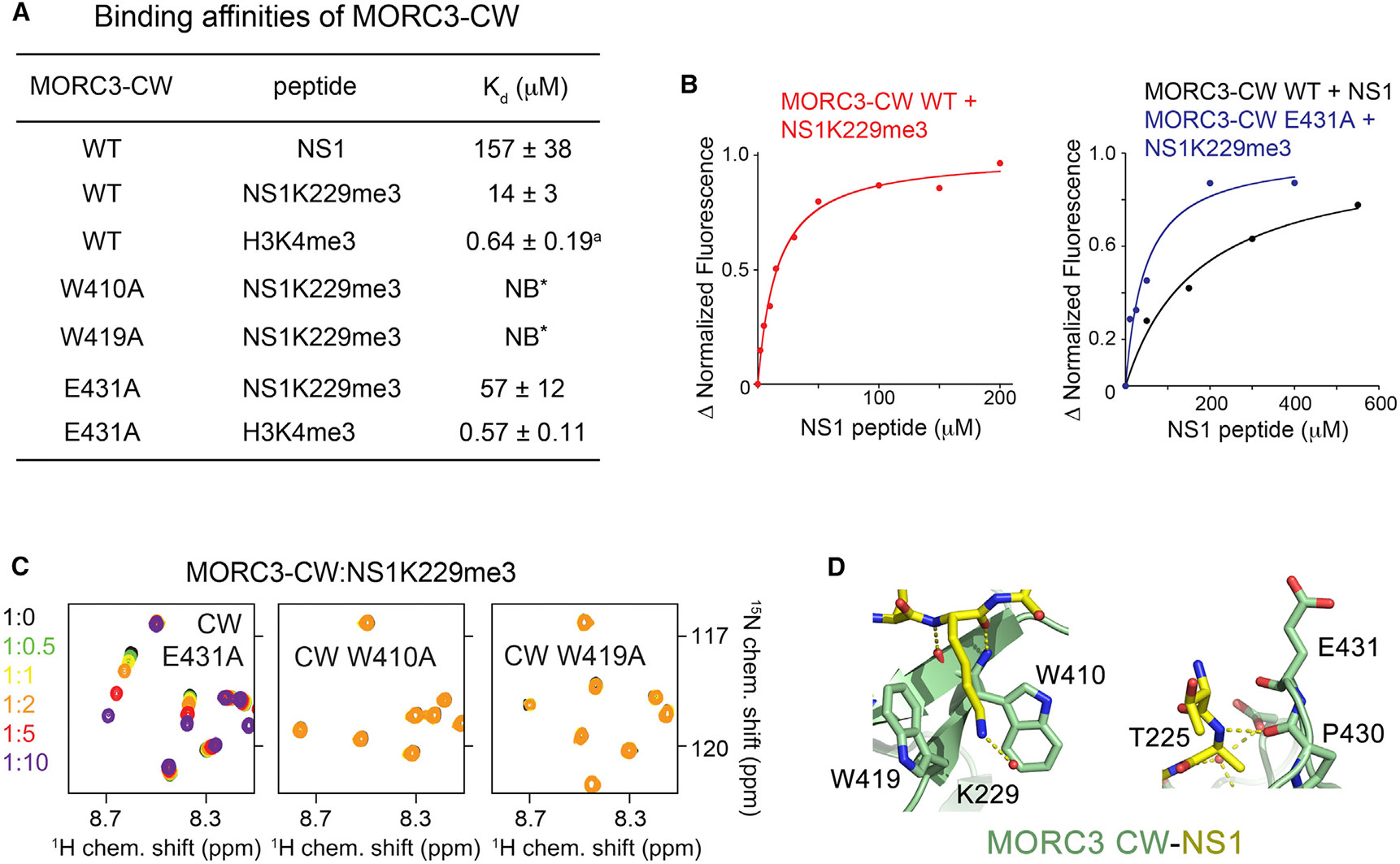
(A) Binding affinities of WT and mutated MORC3-CW to indicated peptides as measured by tryptophan fluorescence or NMR (*). The binding affinity of the CW domain to H3K4me3 peptide was determined previously (a) (Andrews et al., 2016). The experiments were carried out in triplicate (in duplicate for the CW E431A-H3K4me3 interaction). Error bars denote SD.
(B) Representative binding curves used to determine the Kd values by tryptophan fluorescence in (A).
(C) Superimposed 1H,15N HSQC spectra of the 15N-labeled MORC3-CW mutants collected upon titration with the NS1K229me3 peptide. Spectra are color coded according to the protein-to-peptide molar ratio.
(D) Zoom-in views of the NS1 K229- and T225-binding sites of MORC3-CW.
See also Figure S2.
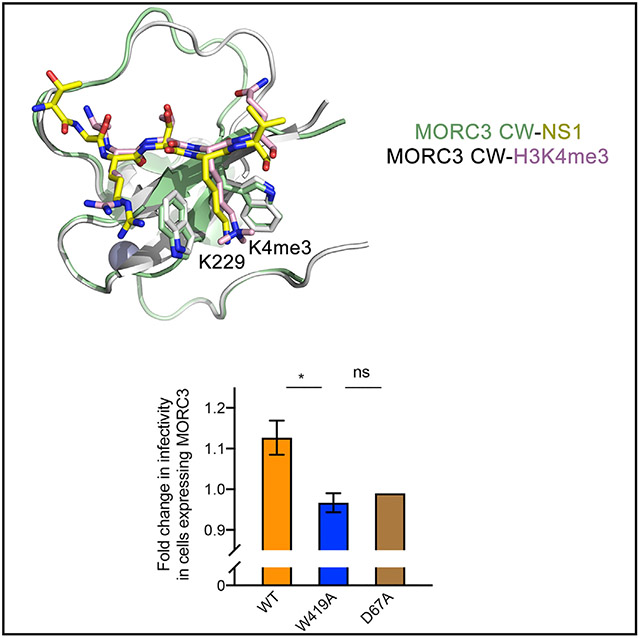
Structural overlay of the CW domain in complex with H3K4me3 and in complex with NS1 reveals that the secondary structure elements and the zinc-binding cluster superimpose well, with a root-mean-square deviation of 0.2 Å (Figure 3A). However, significant conformation variability in the long loop connecting β2 and an α helical turn (residues 421–433 of CW) suggested that this loop may be somewhat flexible in solution. Residues R227-K229 of NS1 and residues R2-K4 in H3 overlay very well, and A226 and V230 in NS1 occupy positions of A1 and Q5 in H3, respectively. The NH3+ group of A1 in the H3K4me3-bound MORC3-CW forms two hydrogen bonds with the backbone carbonyl groups of P430 and E431, and A226 in NS1 retains one hydrogen bond with the carbonyl group of P4300.
Figure 3. Structural Similarity of the MORC3-CW Complexes.
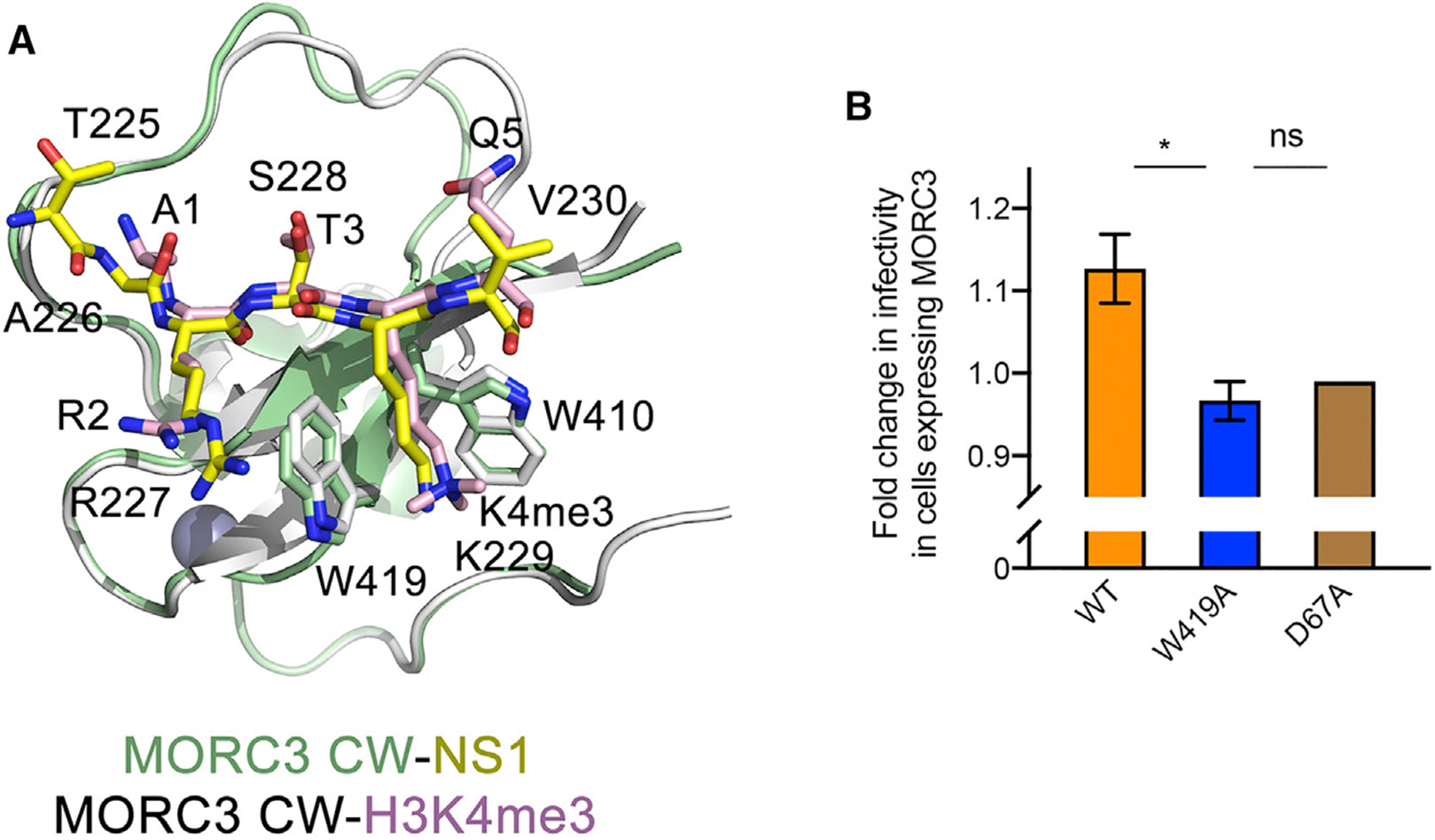
(A) Structural overlay of the MORC3-CW in complex with NS1 peptide (yellow) and H3K4me3 peptide (pink) (PDB: 5SVX).
(B) The fold change in H3N2 influenza virus infectivity in a single round of infection in mCherry-MORC3-positive cells compared with mCherry-negative cells from the same sample. Infectivity measured as a percentage of cells positive of expression of the viral NP protein. The results presented are representative of two independent experiments. The experiments were carried out in triplicate (in duplicate for the D67A mutant). Data are represented as means ± SEM. The statistical significance was determined using the unpaired t test, *p < 0.05.
See also Figures S3 and S4.
In search of differences in histone and NS1 binding mechanisms, we further analyzed the NS1 sequence. Based on the MORC3-CW:NS1 structure, the NS1 residue preceding T225 (R224), which was absent in the crystallized construct, and E431 of CW, should be in close proximity and therefore might electrostatically interact (Figure 2D, right panel). To test this idea, we generated an E431A mutant of MORC3-CW and assessed its interaction with NS1K229me3 (residues 222–230 of NS1) by NMR and fluorescence spectroscopy (Figures 2A-2C). Although the MORC3-CW E431A mutant was folded and capable of binding to the NS1K229me3 peptide, its binding activity was substantially decreased (Kd = 57 μM), indicating that E431 is essential for the interaction with NS1K229me3. Importantly, the E431A mutation did not affect binding of MORC3-CW to H3K4me3 (Kd = 0.6 μM) (Figure 2A). These data imply that the interactions of MORC3-CW with its biological ligand H3K4me3 and the viral protein NS1K229me3 could be differentiated via the E431A mutation. These results also suggest that other H3K4me3-specific readers containing a glutamic acid in the position of E431 of MORC3 might also interact with NS1K229me3, as long as their histone A1-binding pockets are flexible.
MORC3 was shown to interact with the PB1 and PA components of influenza viral polymerase complex during both H1N1- and H3N2-subtype infection and positively regulate H1N1-subtype influenza virus multiplication (Ver et al., 2015). Given that the “ARSK”-motif is uniquely present in NS1 protein of H3N2-subtype influenza virus (Marazzi et al., 2012), we examined the role of MORC3 during H3N2 influenza viral infection. We transfected HEK293T cells with mCherry-MORC3 wild-type (WT), infected with H3N2 influenza virus, and then sorted cells into mCherry-MORC3-positive and -negative populations using flow cytometry. We compared total infectivity and relative viral protein expression levels between MORC3-expressing cells (mCherry+) and control cells (mCherry−). Overexpression of WT MORC3 resulted in a slight increase in viral infectivity (Figure 3B), consistent with previous reports that MORC3 accumulates and positively regulates influenza virus production during infection (Ver et al., 2015). Importantly, the upregulation of infectivity depends on both the functionality of CW domain and the ATPase activity of MORC3, as overexpression of neither the loss-of-function CW W419A mutant nor the loss-of-function ATPase D67A mutant showed similar effects (Figure 3B). These results suggest a CW-dependent role of MORC3 in H3N2 influenza viral infection, possibly because of the competition between the NS1 protein of H3N2 influenza virus and host histone H3.
Influenza virus employs multiple mechanisms to hijack host cellular pathways and evade innate immune responses. The viral NS1 protein can be rapidly expressed to high levels and plays a vital role in influencing infection outcome through modulation of host cell and subversion of innate immunity (Hsu, 2018; Klemm et al., 2018). One of the mechanisms depends on the ability of NS1 from H3N2 virus to disrupt histone H3-binding activities of host nuclear proteins, resulting in altered gene expression programs and related signaling events (Marazzi et al., 2012). Human PAF1 and CHD1 have been shown to be targeted by H3N2 NS1 (Marazzi et al., 2012; Qin et al., 2014), and the MORC3 ATPase represents yet another target of this viral protein. The carboxyl tail of NS1 competes with histone H3 for the CW domain of MORC3 and thus alter chromatin-related functions of MORC3. Our data indicate that increased WT MORC3 level leads to a slight upregulation of H3N2 infectivity, which depends on the functional CW and ATPase domains. Furthermore, MORC3 is highly expressed in immune cells and its expression level is frequently perturbed during influenza infection, also pointing to a role of MORC3 in cellular immune response (Figures S3 and S4). Further studies informed by the results presented here are required to fully understand the MORC3 contribution to these processes.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Tatiana Kutateladze (tatiana.kutateladze@ucdenver.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
MORC3-CW was expressed in Rosetta2 (DE3) pLysS or BL21 (DE3) RIL grown in LB or minimal media. Expression was induced with 0.2~0.5 mM IPTG at A600=0.6 for 16 h at 16°C.
Human embryonic kidney HEK293T (293T) cells were obtained from Dr. Joanna Shisler and were maintained in Gibco’s minimal essential medium with GlutaMax (Life Technologies) supplemented with 8.3% fetal bovine serum (Seradigm) at 37°C and 5% CO2. Recombinant A/Udorn/72 (H3N2) virus was generated using the 8-plasmid rescue system.
METHOD DETAILS
Protein Expression and Purification
The human MORC3 CW domain (aa 401-455) was cloned into a pGEX 6p-1 vector. The NS1-GGSG-MORC3-CW construct (aa 225-230 of NS1, a GGSG linker, aa 407-455 of MORC3-CW) was cloned into a pCIOX vector with N-terminal His-tag and Ulp1 cleavage site. Proteins were expressed in Rosetta2 (DE3) pLysS or BL21 (DE3) RIL in LB or minimal media supplemented with 15NH4Cl and 0.05 mM ZnCl2. Protein expression was induced with 0.2-0.5 mM IPTG for 16 h at 16°C. The His-tag proteins were purified on Ni-NTA beads (Qiagen) in 20 mM Tris-HCl (pH 7.5) buffer, supplemented with 300 mM NaCl and 10 mM βME. The His-tag was cleaved overnight at 4 C with PreScission or Ulp1 protease. The GST-tagged proteins were purified on glutathione Sepharose 4B beads (GE Healthcare) in 20 mM Tris-HCl (pH 7.0) buffer, supplemented with 100 mM NaCl and 5 mM DTT. The GST tag was cleaved overnight at 4°C with PreScission protease. Unlabeled proteins were further purified by size exclusion chromatography and concentrated in Millipore concentrators. All mutants were generated by site-directed mutagenesis using the Stratagene QuikChange mutagenesis protocol, grown and purified as wild-type proteins.
NMR Experiments
NMR experiments were carried out at 298K on Varian INOVA 600 and 900 MHz spectrometers as described (Gatchalian et al., 2017). NMR samples contained 0.1 mM uniformly 15N-labeled WT or mutated CW in 20 mM Tris (pH 6.8) buffer supplemented with 150 mM NaCl, 3 mM DTT, and 8% D2O. Binding was characterized by monitoring chemical shift changes in the proteins induced by NS1 peptides (synthesized by SynPeptide).
X-Ray Crystallography
The purified NS1-MORC3-CW was concentrated to 20 mg/ml in a buffer containing 20 mM Tris-HCl (pH 7.2), 100 mM NaCl, and 1 mM TCEP. Good quality diffracting crystals were obtained at 16°C by hanging drop vapor diffusion in 0.1 M HEPES (7.5) and 1.4 M sodium citrate. Crystals were cryoprotected with the addition of 30% glycerol and the X-ray diffraction data were collected at the Advanced Light Source beamline 4.2.2 administrated by the Molecular Biology Consortium. HKL3000 was used for indexing, scaling, and data reduction. The crystal structure was solved by single-wavelength anomalous-dispersion (SAD) using data collected at a determined wavelength (1.2782 Å). Experimental phases were obtained from experimentally determined f′ and f″ values by using AUTOSOL in PHENIX suite. Model building was performed using Coot (Emsley et al., 2010), and the structure was refined using Phoenix Refine (Adams et al., 2010). The final structure was verified by MOLProbity (Chen et al., 2010). The X-ray diffraction and structure refinement statistics are summarized in Table 1.
Fluorescence Spectroscopy
Spectra were recorded at 25°C on a Fluoromax-3 spectrofluorometer (HORIBA). The samples containing 1.0 μM MORC3 CW fragment (aa 401-455) and progressively increasing concentrations of the peptide were excited at 295 nm. Experiments were performed in buffer containing 20 mM Tris-HCl (pH 6.8), 150 mM NaCl, and 3 mM DTT. Emission spectra were recorded over a range of wavelengths between 330 nm and 360 nm with a 0.5 nm step size and a 1 s integration time and averaged over 3 scans. The KD values were determined using a nonlinear least-squares analysis and the equation:
where is the concentration of the peptide, is the concentration of CW domain, is the observed change of signal intensity, and is the difference in signal intensity of the free and bound states of the CW domain. The Kd value was averaged over three separate experiments (two for the CW E431A-H3K4me3 interaction), with error calculated as the standard deviation between the runs.
Cells, Viruses, DNA Constructs, and Transfection
Human embryonic kidney HEK293T (293T) cells were obtained from Dr. Joanna Shisler and were maintained in Gibco’s minimal essential medium with GlutaMax (Life Technologies) supplemented with 8.3% fetal bovine serum (Seradigm) at 37°C and 5% CO2. Recombinant A/Udorn/72 (H3N2) virus was generated using the 8-plasmid rescue system. Briefly, virus was rescued by transfecting sub-confluent 293T cells with 500ng of each of the appropriate reverse genetics plasmids using JetPRIME (Polyplus) according to the manufacturer’s instructions. Plaque isolates derived from rescue supernatants were amplified into seed stocks in MDCK cells. Working stocks were generated by infecting MDCK cells at an MOI of 0.0001 TCID50/cell with seed stock and collecting and clarifying supernatants at 48 hpi. All viral growth was carried out in MEM with 1 μg/mL trypsin treated with L-(tosylamido-2-phenyl) ethyl chloromethyl ketone (TPCK-treated trypsin; Worthington), 1mM HEPES, and 100 μg/mL gentamicin. Virus stocks were titered via standard tissue culture infectious dose 50 (TCID50) assay.
Virus Infectivity Assays
Transient transfections with N-terminal mCherry-fused MORC3 were performed in sub-confluent 293T cells with 4000ng of the indicated expression plasmid using JetPRIME (Polyplus) according to the manufacturer’s instructions. 24 hours post transfection, we infected cells with A/Udorn/72 at a multiplicity of infection (MOI) of 0.1 TCID50/cell. 3 hours post-infection media containing 50 mM HEPEs and 20 mM NH4Cl was added to block secondary spread of virus. 18 hours post-infection, cells were harvested, fixed and permeabilized using foxP3 fix/perm buffer (eBioscience), and stained for expression of NP and NS1 using the directly conjugated mAbs HB65-AF488 and 1A7-Pacific Blue, respectively. After staining, cells were washed, run on a BD LSR II, and analyzed using FlowJo version 10.1 (Tree Star, Inc.). Data was generated by gating live cells (based on forward and side scatter profiles) into mCherry positive and negative populations. The percentage of NP+ and NS1+ cells were then determined and compared between mCherry positive and negative populations.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistics generated from X-ray crystallography data processing, refinement, and structure validation are displayed in Table 1. For the affinity measurements by fluorescence spectroscopy, emission spectra were recorded over a range of wavelengths between 330 nm and 360 nm and averaged over 3 scans. The Kd values were determined using the change of signal intensity. Data for affinities are shown as mean ± SD based on 3 experiments (2 experiments for the CW E431A-H3K4me3 interaction). For the H3N2 influenza virus infectivity assay, infectivity was measured as % of cells positive of expression of the viral NP protein. The experiments were carried out in triplicates (in duplicates for D67A mutant). Data are represented as mean ± SEM. The statistical significance was determined using unpaired t test.
DATA AND SOFTWARE AVAILABILITY
Software
Software used in this this study has been previously published as detailed in the Key Resources Table.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| Escherichia coli BL21 (DE3) RIL | Andrews et al., 2016 | N/A |
| Escherichia coli Rosetta-2 (DE3) pLysS | Ali et al., 2012 | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Dithiothreitol | Gold Biotechnology | 27565-41-9 |
| NS1(222-230), NS1K229me3(222-230), H3(1-12) and H3K4me3(1-12) | Synpeptide | N/A |
| 15NH4Cl | Sigma-Aldrich | 299251 |
| IPTG | Gold biotechnology | I2481C100 |
| Ni-NTA beads | Thermo Fisher Sci | 88223 |
| Glutathione Sepharose 4B beads | Thermo Fisher Sci | 16101 |
| PreScission and Ulp1 proteases | Home expressed | N/A |
| Deposited Data | ||
| MORC3-CW:NS1 complex | This study | PDB: 6O5W |
| MORC3-CW:H3K4me3 complex | Andrews et al., 2016 | PDB: 5SVX |
| Experimental Models: Cell Lines | ||
| Human: HEK293T | ATCC | CRL-11268 |
| Recombinant DNA | ||
| Plasmid: modified pGEX6p-1 | Andrews et al., 2016 | N/A |
| Software and Algorithms | ||
| Phenix | Adams et al., 2010 | N/A |
| Coot | Emsley et al., 2010 | N/A |
| MOLProbity | Chen et al., 2010 | N/A |
| Genevestigator | https://www.genevestigator.ethz.ch/ | |
Data Resources
Coordinates and structure factors have been deposited in the Protein Data Bank under ID code 6O5W.
Supplementary Material
Highlights.
MORC3 ATPase is a new target of NS1 from H3N2-subtype influenza virus
NS1 interacts with the CW domain of MORC3
NS1 occupies the histone-binding site of CW in the MORC3-CW:NS1 complex structure
Overexpression of MORC3 affects the infectivity of the H3N2-subtype influenza virus
ACKNOWLEDGMENTS
This work was supported by NIH grants GM125195, GM106416, and GM100907 to T.G.K., and a Roy J. Carver Charitable Trust 17-4905 to C.B.B.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.str.2019.03.015.
Data Resources
6O5W
REFERENCES
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews FH, Tong Q, Sullivan KD, Cornett EM, Zhang Y, Ali M, Ahn J, Pandey A, Guo AH, Strahl BD, et al. (2016). Multivalent chromatin engagement and inter-domain crosstalk regulate MORC3 ATPase. Cell Rep. 16, 3195–3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali M, Yan K, Lalonde ME, Degerny C, Rothbart SB, Strahl BD, Côté J, Yang XJ, and Kutateladze TG (2012). Tandem PHD fingers of MORF/ MOZ acetyltransferases display selectivity for acetylated histone H3 and are required for the association with chromatin. J. Mol. Biol 424, 328–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortz E, Westera L, Maamary J, Steel J, Albrecht RA, Manicassamy B, Chase G, Martinez-Sobrido L, Schwemmle M, and Garcia-Sastre A (2011). Host- and strain-specific regulation of influenza virus polymerase activity by interacting cellular proteins. mBio 2, 10.1128/mBio.00151-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, and Richardson DC (2010). MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr 66, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta R, and Inouye M (2000). GHKL, an emergent ATPase/kinase superfamily. Trends Biochem. Sci 25, 24–28. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatchalian J, Wang X, Ikebe J, Cox KL, Tencer AH, Zhang Y, Burge NL, Di L, Gibson MD, Musselman CA, et al. (2017). Accessibility of the histone H3 tail in the nucleosome for binding of paired readers. Nat. Commun 8, 1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu AC (2018). Influenza virus: a master tactician in innate immune evasion and novel therapeutic interventions. Front. Immunol 9, 743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm C, Boergeling Y, Ludwig S, and Ehrhardt C (2018). Immunomodulatory nonstructural proteins of Influenza A viruses. Trends Microbiol. 26, 624–636. [DOI] [PubMed] [Google Scholar]
- Li DQ, Nair SS, and Kumar R (2013). The MORC family: new epigenetic regulators of transcription and DNA damage response. Epigenetics 8, 685–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Yen L, Pastor WA, Johnston JB, Du J, Shew CJ, Liu W, Ho J, Stender B, Clark AT, et al. (2016). Mouse MORC3 is a GHKL ATPase that localizes to H3K4me3 marked chromatin. Proc. Natl. Acad. Sci. U S A 113, E5108–E5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Foley EA, Molloy KR, Li Y, Chait BT, and Kapoor TM (2012). Quantitative chemical proteomics approach to identify post-translational modification-mediated protein-protein interactions. J. Am. Chem. Soc 134, 1982–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Tempel W, Zhang Q, Liang X, Loppnau P, Qin S, and Min J (2016). Family-wide characterization of histone binding abilities of human CW domain containing proteins. J. Biol. Chem 291, 9000–9013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marazzi I, Ho JS, Kim J, Manicassamy B, Dewell S, Albrecht RA, Seibert CW, Schaefer U, Jeffrey KL, Prinjha RK, et al. (2012). Suppression of the antiviral response by an influenza histone mimic. Nature 483, 428–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura Y, Takahashi K, Kawata K, Akazawa T, and Inoue N (2010). Two-step colocalization of MORC3 with PML nuclear bodies. J. Cell Sci 123, 2014–2024. [DOI] [PubMed] [Google Scholar]
- Qin S, Liu Y, Tempel W, Eram MS, Bian C, Liu K, Senisterra G, Crombet L, Vedadi M, and Min J (2014). Structural basis for histone mimicry and hijacking of host proteins by influenza virus protein NS1. Nat. Commun 5, 3952. [DOI] [PubMed] [Google Scholar]
- Sloan E, Orr A, and Everett RD (2016). MORC3, a component of PML nuclear bodies, has a role in restricting herpes simplex virus 1 and human cytomegalovirus. J. Virol 90, 8621–8633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ver LS, Marcos-Villar L, Landeras-Bueno S, Nieto A, and Ortin J (2015). The cellular factor NXP2/MORC3 is a positive regulator of influenza virus multiplication. J. Virol 89, 10023–10030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Klein BJ, Cox KL, Bertulat B, Tencer AH, Holden MR, Wright GM, Black J, Cardoso MC, Poirier MG, and Kutateladze TG (2019). Mechanism for autoinhibition and activation of the MORC3 ATPase. Proc. Natl. Acad. Sci. U S A 10.1073/pnas.1819524116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Software
Software used in this this study has been previously published as detailed in the Key Resources Table.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| Escherichia coli BL21 (DE3) RIL | Andrews et al., 2016 | N/A |
| Escherichia coli Rosetta-2 (DE3) pLysS | Ali et al., 2012 | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Dithiothreitol | Gold Biotechnology | 27565-41-9 |
| NS1(222-230), NS1K229me3(222-230), H3(1-12) and H3K4me3(1-12) | Synpeptide | N/A |
| 15NH4Cl | Sigma-Aldrich | 299251 |
| IPTG | Gold biotechnology | I2481C100 |
| Ni-NTA beads | Thermo Fisher Sci | 88223 |
| Glutathione Sepharose 4B beads | Thermo Fisher Sci | 16101 |
| PreScission and Ulp1 proteases | Home expressed | N/A |
| Deposited Data | ||
| MORC3-CW:NS1 complex | This study | PDB: 6O5W |
| MORC3-CW:H3K4me3 complex | Andrews et al., 2016 | PDB: 5SVX |
| Experimental Models: Cell Lines | ||
| Human: HEK293T | ATCC | CRL-11268 |
| Recombinant DNA | ||
| Plasmid: modified pGEX6p-1 | Andrews et al., 2016 | N/A |
| Software and Algorithms | ||
| Phenix | Adams et al., 2010 | N/A |
| Coot | Emsley et al., 2010 | N/A |
| MOLProbity | Chen et al., 2010 | N/A |
| Genevestigator | https://www.genevestigator.ethz.ch/ | |
Data Resources
Coordinates and structure factors have been deposited in the Protein Data Bank under ID code 6O5W.


