Abstract
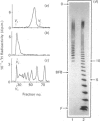
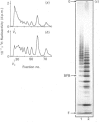
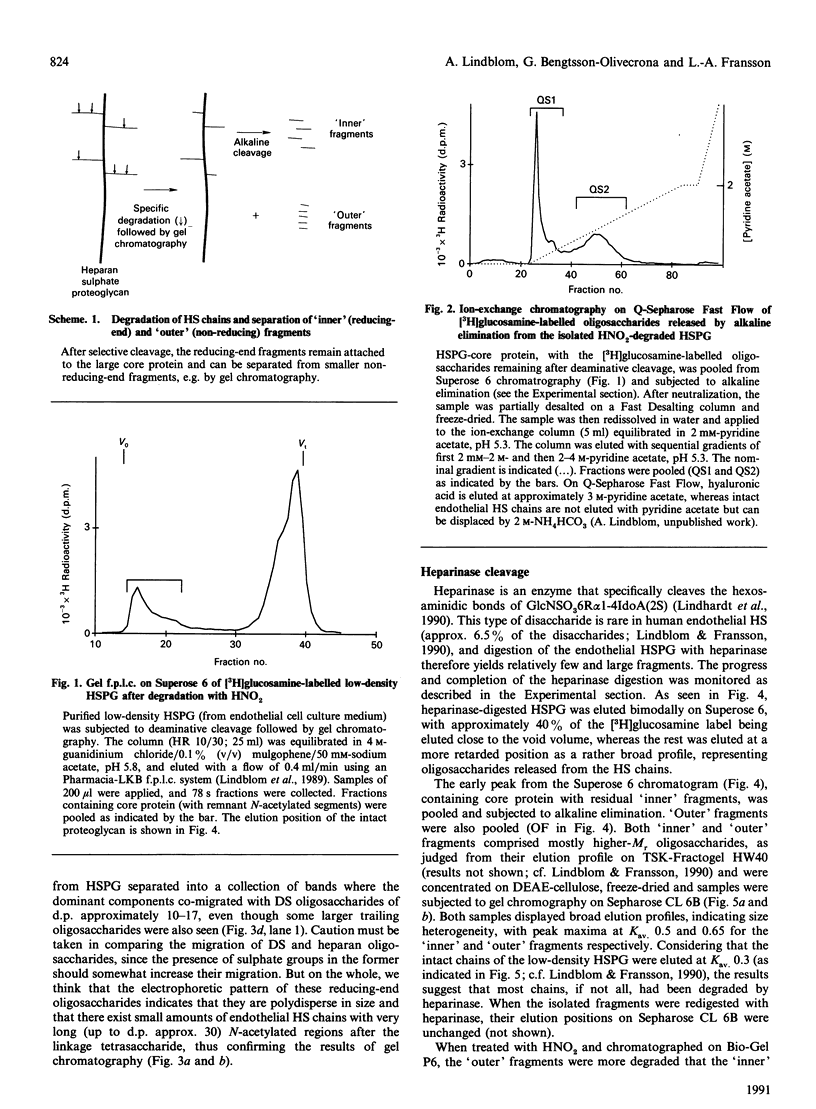
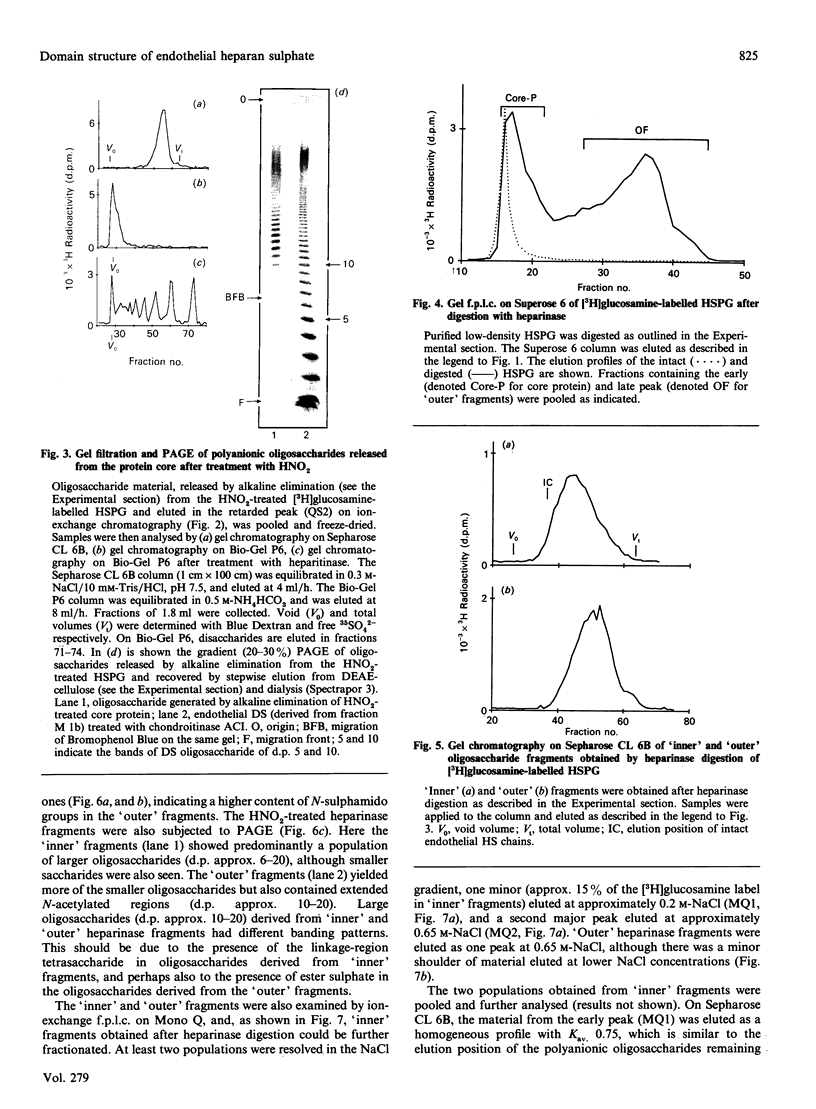
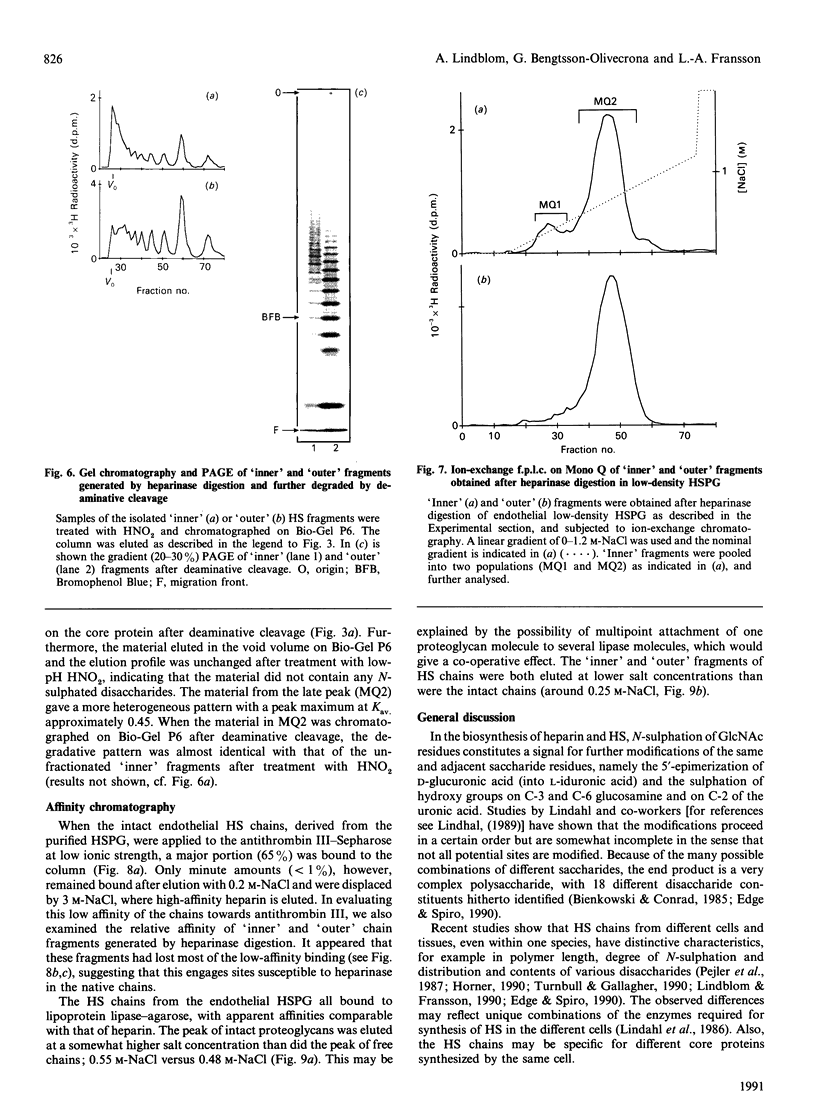
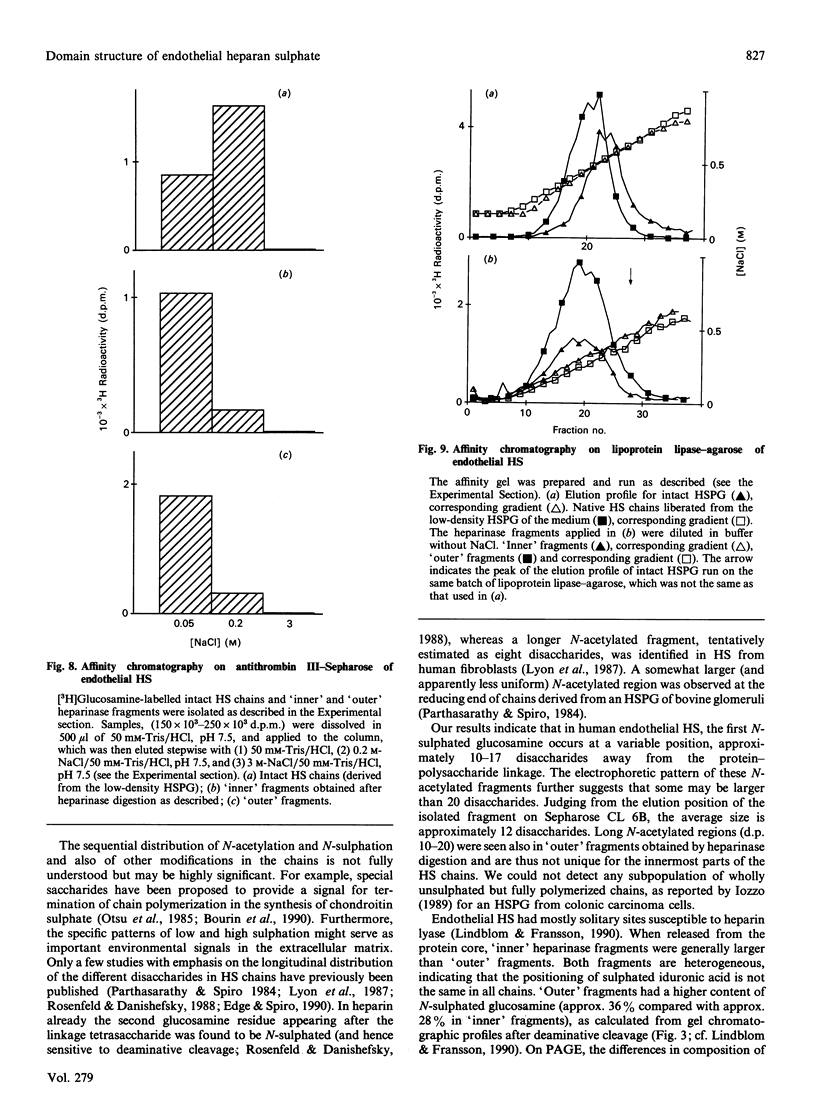
The domain structure of heparan sulphate chains from an endothelial low-density proteoglycan was examined using specific degradations of the chains while attached to the intact proteoglycan. 'Inner' chain fragments, remaining on the protein core, were separated from 'outer' fragments by gel chromatography, and were subsequently released from the protein core by alkaline cleavage. The structure of 'inner' and 'outer' chain fragments was then examined and compared. Using deaminative cleavage we obtained evidence that the first N-sulphated glucosamine residue is variably positioned some 10-17 disaccharides from the xylose-serine linkage of the proteoglycan. Digestion with heparinase yielded 'inner' and 'outer' fragments covering a broad range of different sizes, indicating a scarce and variable distribution of sulphated iduronic acid in the native chains. N-sulphated glucosamine occurred more frequently in the 'outer' fragments. We also studied the affinity of the endothelial heparan sulphate chains towards two presumptive biological ligands, namely antithrombin III and lipoprotein lipase. A major part of the endothelial heparan sulphate chains showed a weak affinity for antithrombin III and the affinity was essentially lost on heparinase digestion. On lipoprotein lipase-agarose the endothelial heparan sulphate chains were eluted at the same salt concentration as heparin, and the binding persisted, although with decreased strength, after digestion with heparinase.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bengtsson G., Olivecrona T., Hök M., Riesenfeld J., Lindahl U. Interaction of lipoprotein lipase with native and modified heparin-like polysaccharides. Biochem J. 1980 Sep 1;189(3):625–633. doi: 10.1042/bj1890625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson G., Olivecrona T. Interaction of heparin with proteins. Demonstration of different binding sites for antithrombin and lipoprotein lipase. FEBS Lett. 1977 Jul 1;79(1):59–63. doi: 10.1016/0014-5793(77)80350-5. [DOI] [PubMed] [Google Scholar]
- Bienkowski M. J., Conrad H. E. Structural characterization of the oligosaccharides formed by depolymerization of heparin with nitrous acid. J Biol Chem. 1985 Jan 10;260(1):356–365. [PubMed] [Google Scholar]
- Bourin M. C., Lundgren-Akerlund E., Lindahl U. Isolation and characterization of the glycosaminoglycan component of rabbit thrombomodulin proteoglycan. J Biol Chem. 1990 Sep 15;265(26):15424–15431. [PubMed] [Google Scholar]
- Damus P. S., Hicks M., Rosenberg R. D. Anticoagulant action of heparin. Nature. 1973 Dec 7;246(5432):355–357. doi: 10.1038/246355a0. [DOI] [PubMed] [Google Scholar]
- Edge A. S., Spiro R. G. Characterization of novel sequences containing 3-O-sulfated glucosamine in glomerular basement membrane heparan sulfate and localization of sulfated disaccharides to a peripheral domain. J Biol Chem. 1990 Sep 15;265(26):15874–15881. [PubMed] [Google Scholar]
- Farndale R. W., Sayers C. A., Barrett A. J. A direct spectrophotometric microassay for sulfated glycosaminoglycans in cartilage cultures. Connect Tissue Res. 1982;9(4):247–248. doi: 10.3109/03008208209160269. [DOI] [PubMed] [Google Scholar]
- Fransson L. A., Havsmark B., Silverberg I. A method for the sequence analysis of dermatan sulphate. Biochem J. 1990 Jul 15;269(2):381–388. doi: 10.1042/bj2690381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher J. T. The extended family of proteoglycans: social residents of the pericellular zone. Curr Opin Cell Biol. 1989 Dec;1(6):1201–1218. doi: 10.1016/s0955-0674(89)80072-9. [DOI] [PubMed] [Google Scholar]
- Heremans A., van der Schueren B., de Cock B., Paulsson M., Cassiman J. J., van den Berghe H., David G. Matrix-associated heparan sulfate proteoglycan: core protein-specific monoclonal antibodies decorate the pericellular matrix of connective tissue cells and the stromal side of basement membranes. J Cell Biol. 1989 Dec;109(6 Pt 1):3199–3211. doi: 10.1083/jcb.109.6.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner A. A. Rat heparan sulphates. A study of the antithrombin-binding properties of heparan sulphate chains from rat adipose tissue, brain, carcase, heart, intestine, kidneys, liver, lungs, skin and spleen. Biochem J. 1990 Mar 1;266(2):553–559. [PMC free article] [PubMed] [Google Scholar]
- Hök M., Björk I., Hopwood J., Lindahl U. Anticoagulant activity of heparin: separation of high-activity and low-activity heparin species by affinity chromatography on immobilized antithrombin. FEBS Lett. 1976 Jul 1;66(1):90–93. doi: 10.1016/0014-5793(76)80592-3. [DOI] [PubMed] [Google Scholar]
- Iozzo R. V. Presence of unsulfated heparan chains on the heparan sulfate proteoglycan of human colon carcinoma cells. Implications for heparan sulfate proteoglycan biosynthesis. J Biol Chem. 1989 Feb 15;264(5):2690–2699. [PubMed] [Google Scholar]
- Kanwar Y. S., Linker A., Farquhar M. G. Increased permeability of the glomerular basement membrane to ferritin after removal of glycosaminoglycans (heparan sulfate) by enzyme digestion. J Cell Biol. 1980 Aug;86(2):688–693. doi: 10.1083/jcb.86.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblom A., Carlstedt I., Fransson L. A. Identification of the core proteins in proteoglycans synthesized by vascular endothelial cells. Biochem J. 1989 Jul 1;261(1):145–153. doi: 10.1042/bj2610145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblom A., Fransson L. A. Endothelial heparan sulphate: compositional analysis and comparison of chains from different proteoglycan populations. Glycoconj J. 1990;7(6):545–562. doi: 10.1007/BF01189076. [DOI] [PubMed] [Google Scholar]
- Linhardt R. J., Turnbull J. E., Wang H. M., Loganathan D., Gallagher J. T. Examination of the substrate specificity of heparin and heparan sulfate lyases. Biochemistry. 1990 Mar 13;29(10):2611–2617. doi: 10.1021/bi00462a026. [DOI] [PubMed] [Google Scholar]
- Liu G. Q., Bengtsson-Olivecrona G., Ostergaard P., Olivecrona T. Low-Mr heparin is as potent as conventional heparin in releasing lipoprotein lipase, but is less effective in preventing hepatic clearance of the enzyme. Biochem J. 1991 Feb 1;273(Pt 3):747–752. doi: 10.1042/bj2730747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lories V., Cassiman J. J., Van den Berghe H., David G. Multiple distinct membrane heparan sulfate proteoglycans in human lung fibroblasts. J Biol Chem. 1989 Apr 25;264(12):7009–7016. [PubMed] [Google Scholar]
- Lyon M., Steward W. P., Hampson I. N., Gallagher J. T. Identification of an extended N-acetylated sequence adjacent to the protein-linkage region of fibroblast heparan sulphate. Biochem J. 1987 Mar 1;242(2):493–498. doi: 10.1042/bj2420493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcum J. A., Rosenberg R. D. Anticoagulantly active heparan sulfate proteoglycan and the vascular endothelium. Semin Thromb Hemost. 1987 Oct;13(4):464–474. doi: 10.1055/s-2007-1003523. [DOI] [PubMed] [Google Scholar]
- Otsu K., Inoue H., Tsuzuki Y., Yonekura H., Nakanishi Y., Suzuki S. A distinct terminal structure in newly synthesized chondroitin sulphate chains. Biochem J. 1985 Apr 1;227(1):37–48. doi: 10.1042/bj2270037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthasarathy N., Spiro R. G. Isolation and characterization of the heparan sulfate proteoglycan of the bovine glomerular basement membrane. J Biol Chem. 1984 Oct 25;259(20):12749–12755. [PubMed] [Google Scholar]
- Pejler G., Bäckström G., Lindahl U., Paulsson M., Dziadek M., Fujiwara S., Timpl R. Structure and affinity for antithrombin of heparan sulfate chains derived from basement membrane proteoglycans. J Biol Chem. 1987 Apr 15;262(11):5036–5043. [PubMed] [Google Scholar]
- Rosenfeld L., Danishefsky I. Location of specific oligosaccharides in heparin in terms of their distance from the protein linkage region in the native proteoglycan. J Biol Chem. 1988 Jan 5;263(1):262–266. [PubMed] [Google Scholar]
- Saksela O., Rifkin D. B. Release of basic fibroblast growth factor-heparan sulfate complexes from endothelial cells by plasminogen activator-mediated proteolytic activity. J Cell Biol. 1990 Mar;110(3):767–775. doi: 10.1083/jcb.110.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shively J. E., Conrad H. E. Formation of anhydrosugars in the chemical depolymerization of heparin. Biochemistry. 1976 Sep 7;15(18):3932–3942. doi: 10.1021/bi00663a005. [DOI] [PubMed] [Google Scholar]
- Simionescu M., Simionescu N., Silbert J. E., Palade G. E. Differentiated microdomains on the luminal surface of the capillary endothelium. II. Partial characterization of their anionic sites. J Cell Biol. 1981 Sep;90(3):614–621. doi: 10.1083/jcb.90.3.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thunberg L., Bäckström G., Lindahl U. Further characterization of the antithrombin-binding sequence in heparin. Carbohydr Res. 1982 Mar 1;100:393–410. doi: 10.1016/s0008-6215(00)81050-2. [DOI] [PubMed] [Google Scholar]
- Turnbull J. E., Gallagher J. T. Molecular organization of heparan sulphate from human skin fibroblasts. Biochem J. 1990 Feb 1;265(3):715–724. doi: 10.1042/bj2650715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasteson A. A method for the determination of the molecular weight and molecular-weight distribution of chondroitin sulphate. J Chromatogr. 1971 Jul 8;59(1):87–97. doi: 10.1016/s0021-9673(01)80009-1. [DOI] [PubMed] [Google Scholar]
- de Agostini A. I., Watkins S. C., Slayter H. S., Youssoufian H., Rosenberg R. D. Localization of anticoagulantly active heparan sulfate proteoglycans in vascular endothelium: antithrombin binding on cultured endothelial cells and perfused rat aorta. J Cell Biol. 1990 Sep;111(3):1293–1304. doi: 10.1083/jcb.111.3.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]