Abstract
Background
Blood-derived mitochondrial DNA copy number (mtDNA-CN) is a proxy measurement of mitochondrial function in the peripheral and central systems. Abnormal mtDNA-CN not only indicates impaired mtDNA replication and transcription machinery but also dysregulated biological processes such as energy and lipid metabolism. However, the relationship between mtDNA-CN and Alzheimer disease (AD) is unclear.
Methods
We performed two-sample Mendelian randomization (MR) using publicly available summary statistics from GWAS for mtDNA-CN and AD to investigate the causal relationship between mtDNA-CN and AD. We estimated mtDNA-CN using whole-genome sequence data from blood and brain samples of 13,799 individuals from the Alzheimer’s Disease Sequencing Project. Linear and Cox proportional hazards models adjusting for age, sex, and study phase were used to assess the association of mtDNA-CN with AD. The association of AD biomarkers and serum metabolites with mtDNA-CN in blood was evaluated in Alzheimer’s Disease Neuroimaging Initiative using linear regression. We conducted a causal mediation analysis to test the natural indirect effects of mtDNA-CN change on AD risk through the significantly associated biomarkers and metabolites.
Results
MR analysis suggested a causal relationship between decreased blood-derived mtDNA-CN and increased risk of AD (OR = 0.68; P = 0.013). Survival analysis showed that decreased mtDNA-CN was significantly associated with higher risk of conversion from mild cognitive impairment to AD (HR = 0.80; P = 0.002). We also identified significant associations of mtDNA-CN with brain FDG-PET (β = 0.103; P = 0.022), amyloid-PET (β = 0.117; P = 0.034), CSF amyloid-β (Aβ) 42/40 (β=-0.124; P = 0.017), CSF t-Tau (β = 0.128; P = 0.015), p-Tau (β = 0.140; P = 0.008), and plasma NFL (β=-0.124; P = 0.004) in females. Several lipid species, amino acids, biogenic amines in serum were also significantly associated with mtDNA-CN. Causal mediation analyses showed that about a third of the effect of mtDNA-CN on AD risk was mediated by plasma NFL (P = 0.009), and this effect was more significant in females (P < 0.005).
Conclusions
Our study indicates that mtDNA-CN measured in blood is predictive of AD and is associated with AD biomarkers including plasma NFL particularly in females. Further, we illustrate that decreased mtDNA-CN possibly increases AD risk through dysregulation of mitochondrial lipid metabolism and inflammation.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13195-024-01601-w.
Keywords: Mitochondria DNA copy number, Biomarkers, Whole genome sequencing, Alzheimer’s disease, Serum metabolites, Mendelian randomization, Causal mediation
Background
Mitochondria are double-membrane organelles and the site of many vital cellular processes, including the generation of ATP through oxidative phosphorylation (OXPHOS), apoptosis, lipid metabolism, and maintaining calcium homeostasis [1, 2]. Human mitochondria have ~ 16.6 kb-long circular DNA (mtDNA) encoding 2 rRNAs, 22 tRNAs, and 13 OXPHOS proteins. A human cell can contain 0–600,000 mitochondria and each mitochondrion contains 1–10 copies of mtDNA [3, 4]. mtDNA copy number (mtDNA-CN) is cell-type-specific and developmental-stage-specific, depending on cellular energy demand. Thus, mtDNA-CN is also a proxy measurement of mitochondrial function. Abnormal mtDNA-CN not only indicates impaired mtDNA replication and transcription machinery [5, 6], but also suggests dysregulated biological processes, leading to mitochondrial dysfunction. Low blood-derived mtDNA-CN is a risk factor for several peripheral diseases, including cardiometabolic disease, type 2 diabetes, chronic kidney disease, and dementia [5, 7–10].
Late-onset Alzheimer disease (AD) is the most common cause of dementia. It is estimated that 6.5 million people aged 65 and older have AD in the United States, and this number will increase to 13.8 million by 2060 [11]. The role of β-amyloid (Aβ) and tau proteins in AD pathogenesis have been intensively studied in the past few decades, but their underlying mechanisms remain unclear [12]. Mitochondrial dysfunction has also been extensively studied and may become a novel therapeutic target for AD [13, 14]. One feature of mitochondrial dysfunction is disturbed mitochondrial genome homeostasis, which is in part reflected by abnormal mtDNA levels [6, 13]. Decreased mtDNA-CN in pyramidal neurons from AD cases compared to cognitively normal (CN) individuals has been observed [15]. Similarly, lower mtDNA-CN was found in peripheral mononuclear blood cells from AD and mild cognitive impairment (MCI) patients compared to CN subjects [16]. These findings suggest that mtDNA-CN may be a marker of mitochondrial dysfunction in a preclinical stage of AD. However, the metabolic mediators of mitochondria and mechanistic connection between blood mtDNA-CN and AD remain unknown. Yang et al. observed that genes whose expression was significantly associated with blood mtDNA-CN were enriched in neurodegenerative disease pathways in multiple tissues [5]. A direct connection of mtDNA-CN to AD was established by a genome-wide association study (GWAS) of blood mtDNA-CN which identified significant associations with genes involved in Aβ clearance [17].
In this study, we employed multiple analytical approaches to evaluate the association of AD with mtDNA-CN estimated from whole genome sequence (WGS) data from more than 13,000 participants of the Alzheimer’s Disease Sequencing Project (ADSP) and to determine the causal relationship between mtDNA-CN and AD using publicly available GWAS summary statistics. We also examined the association of mtDNA-CN with a panel of proteins and metabolites measured in the Alzheimer Disease Neuroimaging Initiative (ADNI) participants. Finally, we investigated possible mediation by proteins and metabolites of the association between mtDNA-CN and AD.
Methods
Overview of analysis design
As depicted in Fig. 1, we assessed associations of mtDNA-CN with AD and pertinent covariates including age, sex, APOE4 status, and ancestry in WGS data derived from brain and blood tissue of ADSP participants. Analyses were performed separately for each ancestry group because the heritability of mtDNA-CN varies among populations and associations of mtDNA-CN with other traits is influenced by environmental factors [18]. The effect of mtDNA-CN level on conversion of CN to MCI and MCI to AD was assessed by survival analysis. Next, we tested the hypothesis that low blood mtDNA-CN is a causal factor to AD using Mendelian Randomization (MR). We also evaluated associations of blood mtDNA-CN with AD biomarkers and metabolites in ADNI datasets using linear regression models. AD status-stratified and sex-stratified analyses were performed to investigate how these associations change across AD stages and between genders. Causal mediation analysis was also conducted to identify which biomarkers and metabolites mediate the association between blood mtDNA-CN and AD.
Fig. 1.
Analysis design. mtDNA-CN = mitochondria DNA copy number; ADSP = Alzheimer’s Disease Sequencing Project; WGS = whole genome sequencing; AD = Alzheimer disease; EA = European Ancestry, AA = African American, CH = Caribbean Hispanic; ADNI = Alzheimer’s Disease Neuroimaging Initiative; MCI = mild cognitive impairment; GWAS = genome-wide association study; * Covariates include age, sex, genetic ancestry group, APOE ε2 and ε4 dosage, sequencing center, and PCR. ** Covariates include age, sex, and original study phase; *** Covariates include age, sex, original study phase, and AD diagnosis; # Cox proportional hazards regression model:
Subjects and whole genome sequencing
Study participants were selected from cohorts assembled by the ADSP for whole genome sequencing. Details of subject ascertainment, diagnostic procedures, and WGS methods using DNA obtained from blood or brain tissue for the ADSP Release 3 (R3) dataset containing 16,774 individuals are described elsewhere [19–21]. The Genome Center for Alzheimer’s Disease (GCAD) quality control (QC) working group performed quality checks of variants and genotypes and samples [19]. Individuals with unknown AD status (n = 2,774) and unexpected genetic duplicate records (n = 286) were excluded, yielding a sample of 13,799 individuals for subsequent analyses (Additional file 2: Table S1). Individuals in the pre-filtered R3 dataset were clustered into population ancestry groups using a Gaussian Mixture model applied to principal component analysis performed with the GENESIS R package [22]. 2,930, 3,027, and 7,842 individuals were identified to be genetically similar to African Americans (AA), Caribbean Hispanics (CH), and Europeans (EA) respectively. Some of the subjects in this dataset were members of multiplex families (comprising two or more individuals affected with AD). The R3 dataset included 1,575 participants in multiple phases of ADNI (ADNI-1, ADNI-GO, ADNI-2) which is a multicenter longitudinal study of MCI and AD that performs cognitive and neuroimaging exams and measures AD biomarkers derived from CSF and blood [23]. WGS for the ADNI samples was performed in two batches several years apart using different sequencing platforms. The distribution of ADNI subjects according to diagnosis and study phase is shown in Table 1.
Table 1.
Demographic characteristics of unrelated ADNI participants with European ancestry
| Cohorts | ADNI-1 | ADNI-GO | ADNI-2 |
|---|---|---|---|
| Total, No. | 585 | 109 | 661 |
| Age, mean (SD), y | 74.8 (6.8) | 71.5 (7.6) | 72.4 (7.1) |
| Female, No. (%) | 233 (39.8%) | 47 (43.1%) | 307 (46.4%) |
| Baseline Diagnosis, No. (%) | |||
| CN | 161 (27.5%) | - | 238 (36.0%) |
| MCI | 301 (51.5%) | 109 (100%) | 300 (45.4%) |
| AD | 123 (21.0%) | - | 123 (18.6%) |
| Progression from CN to MCI, No. (%) | |||
| Yes | 38 (26.2%) | - | 25 (11.7%) |
| No | 107 (73.8%) | - | 189 (88.3%) |
| Progression from MCI to AD, No. (%) | |||
| Yes | 155 (57.2%) | 12 (14.6%) | 77 (30.8%) |
| No | 116 (42.8%) | 70 (85.4%) | 173 (69.2%) |
SD Standard deviation, CN Cognitively normal, MCI Mild cognitive impairment, AD Alzheimer’s Disease, ADNI the Alzheimer’s Disease Neuroimaging Initiative
Estimation of mtDNA-CN
We developed a custom pipeline to estimate mtDNA-CN from WGS data in CRAM files. Genome Analysis Toolkit version 4.0 was used to separate mtDNA read alignments from nuclear read alignments in each CRAM file (Additional file 1: Fig. S1). Paired-end reads that mapped to two different chromosomes were discarded. To efficiently estimate autosomal DNA coverage, we randomly sampled 3,000 1,000-bp regions that meet the following criteria for estimation: (1) no overlap with telomeres, centromeres, gaps, and low complexity regions, and (2) matched average GC content with mtDNA. The mitochondrial control region and both ends with an average read length of the non-control region were excluded from mtDNA coverage estimation for each sequencing library due to their low mapping quality. Mosdepth software was used to count base coverage for both autosomal DNA and mtDNA [24]. mtDNA-CN was estimated as
We compared our customized pipeline with fastMitoCalc [25] by leveraging the 39 technical replicates derived from blood samples. These replicates included three subjects sequenced 9 times (n = 27) and two subjects sequenced 6 times (n = 12). As shown in Fig. S2 (Additional file 1), the mean mtDNA-CN that was estimated using our pipeline (mean = 220.3) was significantly less compared to the mean estimated using fastMitoCalc (mean = 231.2; ANOVA P = 9.61 × 10−9).
Preprocessing/Normalization of blood mtDNA-CN and calculation of polygenic risk scores
Previous studies reported that leukocytes and platelets counts are major confounding factors in the estimation of mitochondrial genome copy number per cell [5, 17, 26, 27]. To adjust for the effect of blood cell type composition on blood mtDNA-CN level estimated in the ADNI dataset, PRSice-2 was used [28] to calculate polygenic risk scores (PRS) by leveraging summary statistics of blood cell count GWAS recently published by Chen et al. [29]. Variants with association p-values < 5 × 10−5 and minor allele frequency > 0.01 from each blood cell count trait GWAS for individuals with European ancestry were selected and then LD-clumped (r2 < 0.1) for the PRS calculation. PRS of neutrophils (P = 1.53 × 10−6), lymphocytes (P = 0.15), and platelets (P = 0.007) and a binary variable of whether the sequencing library was PCR amplified (P = 7.96 × 10−65) were selected using stepwise regression in both directions and included as covariates in a linear regression model with mtDNA-CN as the outcome. Standardized and inverse normal transformed residuals from linear regression were used as adjusted mtDNA-CN for all subsequent association analyses using ADNI data.
AD biomarker, serum metabolite and plasma protein measurements
Baseline measurements of AD biomarkers including fluorodeoxyglucose (FDG) and amyloid-β (Aβ) measured by positron emission tomography (PET) scan and plasma and CSF measures of Aβ42, Aβ40, total tau (t-tau), phosphorylated-tau181 (p-tau), and neurofilament light (NFL) for ADNI participants were downloaded from the Laboratory of Neuro Imaging (LONI) website (https://ida.loni.usc.edu). Measurement protocols are documented in the LONI website and were previously described [30, 31]. Data for a panel of 187 metabolites consisting of lipids, amino acids, and biogenic amines were quantified in serum samples collected from ADNI-1/GO/2 participants at the baseline visit by the Alzheimer’s Disease Metabolomics Consortium (ADMC) [32], as well as for another panel of 190 proteins that were measured in plasma samples collected at baseline from ADNI-1 participants by the Biomarkers Consortium Plasma Proteomics Project were downloaded from the LONI website.
Statistical analysis methods
Two-sample mendelian randomization analysis
To test the hypothesis that lower mtDNA-CN in blood is causally related to increased risk of AD, we performed two-sample MR analyses using summary statistics from two independent GWAS because data for multiple traits were unavailable for the same cohort. We selected 26 single nucleotide polymorphisms (SNPs) as instrument variables from a blood mtDNA-CN GWAS focused on mitochondria disorder-related traits [10]. Summarized results were also obtained for 24 of these SNPs from a large AD GWAS [33]. Data from the two GWAS were harmonized to align the effect allele for each SNP. A synonymous variant, rs62641680, in the DGUOK loci was found to largely influence causal effect estimate in the leave-one-out analysis using MR-Egger (Additional file 2: Table S2) and therefore was excluded, which resulted in 23 SNPs in the final analysis. MR analysis was performed using several approaches including inverse variance weighted (IVW), median weighted, MR-Egger, and robust adjusted profile score (RAPS), which models pleiotropy to overcome the bias caused by violation of the exclusion restriction assumption [34–37], that are implemented in the R package “TwoSampleMR” (version 0.5.6) [38]. MR-PRESSO and HEIDI methods, implemented in R packages “MRPRESSO” and “gsmr” respectively, were used to detect global heterogeneity and pleiotropic outliers [39, 40]. The intercept term of MR-Egger regression was used to determine directional horizontal pleiotropy. The MR Steiger method in “TwoSampleMR” package was used to evaluate the directionality of causality [41].
Association analyses
We evaluated the association of inverse-normal-transformed mtDNA-CN with age, sex, AD status, genetic ancestry, and APOE ε2 and ε4 dosage, using a linear regression model adjusting for sequencing centers and a binary term of whether the sequencing library was PCR-amplified. Analyses included only unrelated individuals and were performed separately for mtDNA-CN derived from brain and blood. Additional models were tested for mtDNA-CN measured in blood for each ancestry group. A sensitivity analysis including terms for AD diagnosis and covariates measured at baseline in ADNI participants was also performed to overcome the confounding effects from time interval between age when the blood sample was drawn for WGS and age at AD diagnosis and blood cells composition. The effect of blood-derived mtDNA-CN on conversion from CN to MCI and conversion from MCI to AD in the ADNI cohort was assessed using a Cox proportional hazards model including covariates for age at baseline, sex, and study phase implemented in the R package “survival” (version 3.3-1, https://cran.r-project.org/web/packages/survival/index.html). Comparison of the rates of conversion from CN to MCI and from MCI to AD between low and high mtDNA-CN levels defined by the median of adjusted mtDNA-CN was evaluated by Kaplan-Meier survival analysis using R package “survminer” (version 0.4.9). We investigated the associations between mtDNA-CN and AD brain imaging markers measured by PET and established AD biomarkers measured in CSF and plasma. Associations of inverse normal-transformed biomarkers, metabolites, and proteins levels with the adjusted mtDNA-CN was evaluated using multiple linear regression models including covariates for age at baseline, sex, original ADNI study phase, and diagnosis at baseline. Analyses were also performed in subgroups stratified by sex or diagnosis at baseline.
Causal mediation analysis
We performed causal mediation analysis to measure the potential role of biomarkers and metabolites as mediators of the association of mtDNA-CN (i.e. exposure variable) with AD or both AD and MCI considered jointly (i.e. outcome variable) using a natural effect model that included covariates for age, sex, and study phase. This approach models the unobservable counterfactual diagnostic outcome if one was exposed to a mtDNA-CN level that is different than his/her real-world measurement and enables decomposition of the total effects of mtDNA-CN on AD risk into natural direct effect (NDE) and natural indirect effect (NIE). The model is implemented as follows. Let denote the AD diagnosis outcome observed when the mtDNA-CN is at level and biomarker/metabolite is at level . In this situation, NDE captures the effect of one standard deviation increase in mtDNA-CN on AD or MCI risk with covariates at baseline levels if one intervenes to set a biomarker or metabolite to a fixed level it would have been as if mtDNA-CN level is unchanged, which can be expressed as an odds ratio using counterfactual notation as
By contrast, NIE captures the effect of mtDNA-CN on AD or MCI risk only due to the effect of one standard deviation increase in mtDNA-CN on the biomarker or metabolite, which can be expressed as
Data were analyzed using a logistic regression model including both exposure, mediator, and covariates as predictors to obtain estimates for the outcome model. Next, 10 hypothetical exposure levels were randomly drawn for each individual from a linear model for exposure, conditioned on all covariates, and were fit in the outcome model along with the baseline levels of mediator and all covariates to impute the unobserved counterfactual outcome. Finally, this ten-fold expanded dataset was fit in the natural effect model to obtain the estimates of natural direct, indirect, and total effects using the R package medflex (version 0.6-7) [42].
Results
mtDNA-CN in brain and blood are associated with AD
In the ADSP WGS sample, we observed an average mtDNA-CN of 2,152.0 ± 1249.0 in DNA derived from brain (Additional file 1: Fig. S3A). Participants diagnosed with AD had a significantly lower brain mtDNA-CN than cognitively normal controls (β = -0.189, P = 0.002, Additional file 1: Fig. S3B). Brain mtDNA-CN was positively associated with age (β = 0.020, P = 2.20 × 10−8, Additional file 1: Fig. S3C) and APOE ε2 dosage (β = 0.265, P = 3.88 × 10−4, Additional file 1: Fig. S3F), but not with ε4 dosage (P = 0.46) or sex (P = 0.76, Additional file 1: Fig. S3D). EA individuals had a significantly greater brain mtDNA-CN compared to AAs (P = 0.006, Additional file 1: Fig. S3E).
The average mtDNA-CN is 193.0 ± 107.4 in DNA derived from blood (Additional file 1: Fig. S4A), much lower than brain mtDNA-CN. Similar to the association findings in brain samples, blood mtDNA-CN was significantly lower in AD cases than controls (β = -0.130, P = 1.33 × 10−9, Additional file 1: Fig. S4B) and positively associated with APOE ε2 dosage (β = 0.072, P = 0.029, Additional file 1: Fig. S4F). However, different from associations with brain mtDNA-CN, blood mtDNA-CN was negatively associated with age (β = -0.008, P = 1.81 × 10−11, Additional file 1: Fig. S4C) and significantly higher in females than males (β = 0.212, P = 2.45 × 10−24, Additional file 1: Fig. S4D). Additionally, AA individuals had significantly more copies of mtDNA than EAs (β = 0.170, P = 1.80 × 10−9) and significantly fewer copies of mtDNA than CH individuals (β = -0.252, P = 7.77 × 10−14, Additional file 1: Fig. S4E). In ancestry-specific analyses, mtDNA-CN measured in blood was associated with lower age in EA (β = -0.011, P = 4.16 × 10−12) and CH (β = -0.012, P = 1.40 × 10−4) individuals and was significantly higher in females than in males in all three ancestry groups (Additional file 2: Table S3). mtDNA-CN was significantly smaller in AD cases compared to controls in the EA (β = -0.211, P = 3.58 × 10−12) and CH (β = -0.263, P = 1.70 × 10−6) groups.
Sensitivity analyses conducted to consider confounding effects from blood cells composition and WGS technical differences revealed that AD cases had significantly lower mtDNA-CN than MCI cases (β = -0.202, P = 0.002) and controls (β = -0.257, P = 2.72 × 10−4). However, mtDNA-CN was not different between MCI cases and controls (Fig. 2A). Lower blood mtDNA-CN was inversely associated with age (β = -0.018, P = 1.34 × 10−7) (Fig. 2B) and mtDNA-CN level was higher in females than males (β = 0.426, P = 2.04 × 10−18) (Fig. 2C). These results are consistent with findings from our analysis of all blood samples (Additional file 1: Fig. S4) and other studies [43].
Fig. 2.
Partial residuals plots showing the relationships between blood mtDNA-CN from ADNI and AD diagnosis (A), age (B), sex (C). Residuals were calculated after fitting a liner model where mtDNA-CN was the dependent variable and age, sex, cohort, and AD diagnosis were the independent variables. Kaplan-Meier plot to visualize the probability of (D) MCI participants at ADNI baseline free from converting to AD and (E) CN patients free from converting to MCI for low mtDNA-CN group and high mtDNA-CN group. mtDNA-CN was dichotomized by the median
Blood mtDNA-CN is associated with conversion from MCI to AD
Among 1,355 genetically unrelated ADNI participants of European ancestry, 63 CN individuals and 244 MCI cases converted to MCI and AD, respectively, during the follow-up period. The mean follow-up time was 5.2 years among those who converted from CN to MCI, and 3.6 years for those who converted from MCI to AD. We found that lower blood mtDNA-CN estimated at baseline visit was significantly associated with a higher risk of conversion from MCI to AD (HR = 0.80, 95% CI = 0.69–0.92, P = 0.002). However, mtDNA-CN was not associated with conversion from CN to MCI. These observations are consistent with findings from survival analysis showing a higher probability of MCI cases with low mtDNA-CN to develop AD (P < 0.0001; Fig. 2D), but no effect of mtDNA-CN on the conversion from CN to MCI (P = 0.28; Fig. 2E).
Decreased blood mtDNA-CN is genetically causal to higher AD risk
After initial analyses using four two-sample MR methods, a synonymous variant, rs62641680, in the DGUOK loci was found to largely influence causal effect estimate in the leave-one-out analysis (Additional file 2: Table S2) and therefore was excluded, which resulted in 23 SNPs in the final analysis. Results obtained using three of the four methods indicate that decreased blood mtDNA-CN is causal to higher AD risk (IVW: β= -0.356, P = 0.044; MR-Egger: β = -0.738, P = 0.041; RAPS: β = -0.390, P = 0.012; Table 2). There was no evidence of heterogeneity assessed using Cochran’s Q test for MR-Egger (Q = 28.1, P = 0.14) or IVW (Q = 30.4, P = 0.11), additional outliers, global horizontal pleiotropy (P = 0.13), or directional horizontal pleiotropy assessed using intercept of MR-Egger regression (P = 0.20). The MR Steiger test of directionality indicated a valid causal direction from mtDNA-CN to AD (P = 1.93 × 10−45).
Table 2.
Estimation of causal effect of blood mtDNA-CN on AD using two-sample mendelian randomization methods
| Method | OR | 95% CI | P-value |
|---|---|---|---|
| Robust adjusted profile score | 0.677 | 0.498–0.920 | 0.013 |
| MR Egger | 0.478 | 0.246–0.928 | 0.041 |
| Weighted median | 0.746 | 0.494–1.125 | 0.162 |
| Inverse variance weighted | 0.700 | 0.495–0.991 | 0.044 |
SE Standard error, 95% CI 95% confidence interval
Blood mtDNA-CN is associated with AD biomarkers in females and serum metabolites
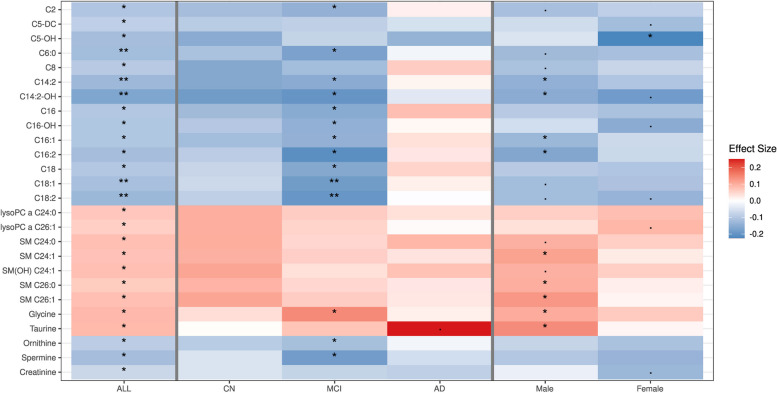
In the total ADNI dataset, mtDNA-CN was nominally associated with CSF Aβ42 level (P = 0.048), but not with any other AD biomarkers (Fig. 3, Additional file 2: Table S4). Amyloid-PET level was positively associated with mtDNA-CN in controls only (β = 0.199, P = 0.002). mtDNA-CN was inversely associated with lower CSF Aβ42 in individuals with MCI (β = -0.102, P = 0.038) and with higher CSF NFL level in AD cases (β = 0.281, P = 0.015). Among females, mtDNA-CN was associated with FDG-PET, amyloid-PET, CSF t-tau and p-tau, and plasma NFL levels, whereas no significant associations were observed in males. In the total sample, mtDNA-CN was significantly associated (Padj < 0.05) with 26 lipid, amino acid, and biogenic amine species (Fig. 4, Additional file 2: Table S5). Significant associations were found for 14 of these metabolites, 11 of which are acylcarnitines, among individuals with MCI, but not in controls or AD cases. Among males, mtDNA-CN was significantly associated with long-chain acylcarnitines, sphingomyelins, glycine, and taurine, whereas only a negative association with hydroxyvalerylcarnitine (C5-OH, β = -0.225, Padj = 0.013) was identified in females. Analyses of plasma protein levels revealed negative associations with matrix metalloproteinase 10 (MMP10) in MCI cases (β = -0.170, P = 0.044) and matrix metalloproteinase 9 (MMP9) in males (β = -0.201, P = 0.009) (Additional file 2: Table S6).
Fig. 3.
P-value and effect size plot to visualize the association between blood mtDNA-CN and classic AD biomarkers for all ADNI participants, each sex and AD diagnosis strata. * P < 0.05; ** P < 0.01. CN = cognitively normal; MCI = mild cognitive impairment; AD = Alzheimer disease
Fig. 4.
Adjusted P-value and effect size plot to visualize the association between blood mtDNA-CN and serum metabolites for all ADNI participants, each sex and AD diagnosis strata. P adj< 0.1; * P adj < 0.05; ** P adj <0.01
AD biomarkers and metabolites mediate the association of blood mtDNA-CN with AD
We found a significant natural indirect effect (NIE) of mtDNA-CN on risk of MCI or AD mediated by FDG-PET, amyloid-PET, and plasma NFL (P < 0.05 for all). However, among these three biomarkers, a significant total effect of mtDNA-CN was observed only for plasma NFL (P = 0.039). A natural direct effect (NDE) of mtDNA-CN on risk of MCI or AD was not evident for any mediators. In a model including only CN and AD participants, we observed very similar patterns of NIE of mtDNA-CN on AD risk mediated by FDG-PET and amyloid-PET, but the significance of NIE was attenuated. On the contrary, the NIE mediated through plasma NFL became more significant (OR = 0.89, 95% CI = 0.82–0.96, P = 0.002). No significant indirect effect of mtDNA-CN was observed in a model including only MCI and AD participants in the analysis. The same results were also observed in the female subgroup, but the NIEs was larger than the total sample analysis (Table 3, Additional file 2: Table S7).
Table 3.
Mediating effects of AD biomarkers and metabolites on the association of mtDNA-CN with AD. (To be placed at line 374, page 18)
| NDE | NIE | TE | ||||||
| Group | Mediator | N | OR (95% CI) | P-value | OR (95% CI) | P -value | OR (95% CI) | P -value |
| (MCI + AD) vs CN | ||||||||
| Total | Plasma NFL | 1157 | 0.901 (0.784, 1.039) | 0.145 | 0.950 (0.914, 0.987) | 0.009 | 0.856 (0.740, 0.994) | 0.039 |
| Hydroxyvalerylcarnitine | 963 | 0.969 (0.824, 1.137) | 0.698 | 0.950 (0.917, 0.985) | 0.005 | 0.920 (0.785, 1.079) | 0.306 | |
| Octadecadienylcarnitine | 1313 | 0.875 (0.753, 1.018) | 0.082 | 0.980 (0.960, 0.999) | 0.044 | 0.857 (0.738, 0.996) | 0.044 | |
| Glycine | 1313 | 0.841 (0.728, 0.966) | 0.017 | 1.018 (1.001, 1.037) | 0.042 | 0.857 (0.741, 0.984) | 0.033 | |
| Serotonin | 1268 | 0.874 (0.756, 1.009) | 0.066 | 0.983 (0.965, 1.002) | 0.073 | 0.859 (0.743, 0.993) | 0.040 | |
| Male | Plasma NFL | 642 | 0.860 (0.705, 1.047) | 0.135 | 0.980 (0.934, 1.029) | 0.411 | 0.843 (0.689, 1.031) | 0.097 |
| Hydroxyvalerylcarnitine | 746 | 0.881 (0.725, 1.072) | 0.204 | 0.967 (0.934, 1.001) | 0.054 | 0.852 (0.699, 1.038) | 0.111 | |
| Octadecadienylcarnitine | 746 | 0.881 (0.717, 1.080) | 0.225 | 0.967 (0.935, 0.999) | 0.045 | 0.852 (0.693, 1.045) | 0.125 | |
| Glycine | 746 | 0.824 (0.681, 1.005) | 0.051 | 1.032 (1.001, 1.065) | 0.042 | 0.850 (0.704, 1.037) | 0.099 | |
| Serotonin | 722 | 0.882 (0.731, 1.064) | 0.189 | 0.991 (0.969, 1.015) | 0.456 | 0.874 (0.723, 1.056) | 0.163 | |
| Female | Plasma NFL | 515 | 0.954 (0.772, 1.183) | 0.666 | 0.919 (0.868, 0.977) | 0.005 | 0.877 (0.705, 1.099) | 0.246 |
| Hydroxyvalerylcarnitine | 410 | 1.076 (0.848, 1.367) | 0.547 | 0.897 (0.830, 0.970) | 0.007 | 0.966 (0.760, 1.229) | 0.775 | |
| Octadecadienylcarnitine | 567 | 0.871 (0.703, 1.082) | 0.211 | 0.997 (0.970, 1.028) | 0.859 | 0.869 (0.702, 1.076) | 0.198 | |
| Glycine | 567 | 0.862 (0.694, 1.067) | 0.176 | 1.008 (0.985, 1.031) | 0.476 | 0.869 (0.700, 1.079) | 0.205 | |
| Serotonin | 546 | 0.864 (0.700, 1.091) | 0.198 | 0.979 (0.949, 1.009) | 0.165 | 0.846 (0.676, 1.057) | 0.141 | |
| AD vs CN | ||||||||
| Total | Plasma NFL | 609 | 0.848 (0.715, 1.008) | 0.059 | 0.886 (0.822, 0.960) | 0.002 | 0.751 (0.625, 0.911) | 0.003 |
| Hydroxyvalerylcarnitine | 457 | 0.855 (0.683, 1.076) | 0.177 | 0.975 (0.942, 1.007) | 0.135 | 0.834 (0.666, 1.046) | 0.114 | |
| Octadecadienylcarnitine | 625 | 0.722 (0.595, 0.871) | 0.001 | 0.992 (0.976, 1.010) | 0.373 | 0.716 (0.591, 0.866) | 0.001 | |
| Glycine | 625 | 0.713 (0.597, 0.862) | 3.25×10-4 | 1.004 (0.982, 1.026) | 0.709 | 0.716 (0.598, 0.866) | 4.16×10-4 | |
| Serotonin | 611 | 0.744 (0.617, 0.905) | 0.002 | 0.970 (0.941, 1.000) | 0.048 | 0.722 (0.599, 0.879) | 0.001 | |
| Male | Plasma NFL | 322 | 0.826 (0.662, 1.043) | 0.099 | 0.958 (0.860, 1.065) | 0.431 | 0.791 (0.613, 1.021) | 0.072 |
| Hydroxyvalerylcarnitine | 331 | 0.767 (0.602, 0.996) | 0.039 | 0.986 (0.954, 1.018) | 0.393 | 0.756 (0.587, 0.974) | 0.030 | |
| Octadecadienylcarnitine | 331 | 0.767 (0.598, 0.983) | 0.037 | 0.986 (0.955, 1.018) | 0.382 | 0.756 (0.589, 0.970) | 0.028 | |
| Glycine | 331 | 0.751 (0.590, 0.965) | 0.022 | 1.009 (0.974, 1.047) | 0.623 | 0.757 (0.593, 0.979) | 0.030 | |
| Serotonin | 325 | 0.802 (0.625, 1.046) | 0.094 | 0.979 (0.944, 1.016) | 0.255 | 0.786 (0.607, 1.017) | 0.067 | |
| Female | Plasma NFL | 287 | 0.882 (0.676, 1.139) | 0.344 | 0.813 (0.724, 0.920) | 0.001 | 0.717 (0.540, 0.950) | 0.021 |
| Hydroxyvalerylcarnitine | 211 | 0.869 (0.607, 1.240) | 0.440 | 0.936 (0.861, 1.024) | 0.135 | 0.813 (0.573, 1.158) | 0.249 | |
| Octadecadienylcarnitine | 294 | 0.680 (0.504, 0.925) | 0.013 | 0.998 (0.979, 1.019) | 0.818 | 0.679 (0.502, 0.917) | 0.012 | |
| Glycine | 294 | 0.678 (0.513, 0.910) | 0.008 | 1.001 (0.968, 1.037) | 0.945 | 0.679 (0.510, 0.903) | 0.008 | |
| Serotonin | 286 | 0.683 (0.508, 0.935) | 0.015 | 0.964 (0.913, 1.016) | 0.173 | 0.658 (0.485, 0.893) | 0.007 | |
NIE Natural indirect effect, NDE Natural direct effect, TE Total effect, CN Cognitively normal, MCI Mild cognitive impairment, AD Alzheimer disease, OR Odds ratio, CI Confidence interval, NFL Neurofilament light
Among serum metabolites, we identified nominal NIE of mtDNA-CN on risk of MCI or AD mediated by two acylcarnitine species, C5-OH (OR = 0.95, 95% CI = 0.92–0.99, P = 0.005) and octadecadienylcarnitine (C18:2: OR = 0.98, 95% CI = 0.96-1.00, P = 0.044), and glycine (OR = 1.02, 95% CI = 1.00-1.04, P = 0.042). The NIE of mtDNA-CN was mediated by serotonin (OR = 0.97; 95% CI = 0.94-1.00, P = 0.048) in a model including only CN and AD participants. Among females, the NIE of mtDNA-CN on risk of MCI or AD was mediated only by C5-OH (P = 0.007), whereas nominally significant NIE through C18:2 (P = 0.045) and glycine (P = 0.042) was observed in males (Table 3, Additional file 2: Table S8).
Discussion
This study leveraged whole genome sequence data obtained from a large sample of ADSP participants to measure mtDNA-CN that is relatively difficult to quantify by traditional genotyping methods [44, 45]. We observed that participants with AD diagnoses had lower mtDNA-CN levels in both brain and blood samples compared to cognitively normal individuals, which is consistent with previous findings [46, 47]. This finding was evident in the EA and CH groups, but not in AAs. MR analysis results suggest that decreased blood mtDNA-CN is causal to AD (OR = 0.68; P = 0.012). We also identified associations of AD biomarkers and metabolites with mtDNA-CN some of which showed sex- and AD status-specific patterns. Our study further characterized the mediators in the association of mtDNA-CN with AD/MCI, indicating the dysfunction of mitochondria for the disease.
In the total sample, mtDNA-CN was positively associated with the APOE ε2 allele in both brain and blood samples. Longchamps et al. also identified a positive association between blood-derived mtDNA-CN and the ε2 SNP (rs7412) [17]. This finding suggests a protective effect of APOE ε2 against mitochondrial dysfunction. Although we did not observe a significant association of mtDNA-CN with the APOE ε4 allele, another study of AD cases reported evidence of impaired mitochondria and oxidative stress in brain tissue from ε4 carriers [48, 49].
We identified female-specific associations of blood mtDNA-CN with several AD biomarkers, including FDG-PET, amyloid-PET, CSF Aβ42/40, CSF t-Tau, CSF p-Tau, and plasma NFL. Previous studies reported sex differences in the association of AD with these biomarkers, except plasma NFL, and related metabolic pathways [50–53], suggesting that unidentified sex-specific factors modify the effect of mtDNA-CN on AD-related processes. However, in the causal mediation analysis, the natural indirect effect and total effect of mtDNA-CN on AD were simultaneously significant when testing for plasma NFL, but not the other AD biomarkers, as a mediator. Plasma NFL has been proposed as a prognostic biomarker for AD because of its association with AD risk, cognitive decline, and chronic inflammation [54–56]. It is shown that immune dysfunction, energy demands, inflammation and altered cell signaling link mitochondrial dysfunction in whole blood to chronic diseases especially cardiovascular diseases [57]. In addition, previous study of ADNI participants showed higher plasma NFL was predictive of brain glucose hypometabolism, which is associated with mitochondrial dysfunction [58], in cognitively impaired and normal subjects [59]. These observations combined with our findings suggest low numbers of mitochondria probably do not respond well to the stressors of inflammation and vascular diseases, leading to inefficient oxidative phosphorylation in mitochondria and AD pathogenesis.
The association findings for mtDNA-CN with amyloid are seemingly inconsistent. Whereas mtDNA-CN was nominally associated with lower CSF Aβ42 in the total sample, it was more significantly associated with increased amyloid-PET level in controls, but not in MCI or AD participants. The evidence for a relationship between amyloid deposition and mtDNA-CN in brain is controversial. Decreased Aβ42 level along with reduced mtDNA-CN was observed in the cortex and hippocampus of an AD transgenic mice that express mitochondrial-targeted endonuclease Mito-PstI [60, 61], whereas another study reported a nominally significant negative association in the dorsolateral prefrontal cortex from post-mortem normal ageing and AD brains [46]. Because blood mtDNA-CN is not necessarily a proxy for brain mtDNA-CN, further investigation is needed to fully understand the interaction between amyloid and mtDNA.
Several studies have identified associations of protein and metabolite levels with AD and cognitive decline in the ADNI cohort [32, 62, 63]. For example, it has been suggested that a combination of markers other than β-amyloid and tau measured in plasma and CSF could prove useful in predicting progression from MCI to AD [51]. Recently, Horgusluoglu et al. showed that short-chain acylcarnitines/amino acids and medium/long-chain acylcarnitines are associated with episodic memory scores and disease severity [63]. Our study, which compared mtDNA-CN with metabolites measured in the same blood sample, identified associations with several lipid species, amino acids, and biogenic amines. Most notably, lower mtDNA-CN was associated with levels of acylcarnitines (ACs). ACs have essential roles in mitochondrial metabolism because they transport fatty acids through mitochondrial membranes for β-oxidation. They shuttle across mitochondrial membranes and are recycled primarily by long-chain acyl-coenzyme A synthetase, carnitine/acylcarnitine translocase, and carnitine palmitoyl-transferase 1 and 2 under normal physiological conditions. We observed that AD cases had elevated levels of blood medium/long-chain ACs, which may be attributed to damaged mitochondria with reduced fatty acid oxidation rate [63]. Curiously, most of our observed associations of mtDNA-CN with ACs were evident primarily in MCI participants. Lack of association of mtDNA-CN with AC levels among AD cases combined with results from a previous study showing that AC levels progressively decline with transitions from cognitively normal to AD [64] suggest there is a floor effect for the impact of the interaction of blood mtDNA-CN with AC levels on AD progression when cognitively declining individuals reach the AD stage. Our findings that mtDNA-CN was associated with levels of four long-chain ACs in males and reduced level of the short-chain acylcarnitine C5-OH was associated with mtDNA-CN in females were supported by mediation analyses which showed that the indirect natural effects mediated by the C18:2 and C5-OH ACs were significant only in males and females, respectively. Although sex differences in AC metabolism have been previously reported [65, 66], mechanisms underlying sex-specific relationships between AC levels and mtDNA-CN are unclear and require further investigation.
Although the association between blood mtDNA-CN and serotonin was not significant after FDR correction, the NIE of mtDNA-CN on AD mediated by serotonin was significant. Serotonin is a key neurotransmitter and its concentration in serum and urine is significantly lower in AD cases compared to controls. It has been suggested that the disruption of serotonergic signaling pathway is linked to enhanced amyloid pathology and, thus, serotonin may be a potential therapeutic target [67]. Because mitochondria provide ATP which is essential to serotonin signaling, mitochondrial dysfunction could result in serotonin dysregulation and subsequent neurodegeneration, an idea that is consistent with our causal mediation analysis findings [68].
Sphingomyelin (SM) is the major type of sphingolipid in blood and has important biological functions including cellular signaling and membrane stabilization. The hydrolysis of SM is the major source of ceramide, which can act as a second messenger and trigger mitochondrial apoptosis. Studies have indicated that Aβ induces apoptosis through SM/ceramide pathway and reduced SM levels have been observed in AD brains and plasma [69–71]. The positive associations we observed between a few SM species and mtDNA-CN, might suggest the degradation of SM results in mitochondrial dysfunction. In addition, there is evidence that plasma SM concentrations are higher and increase faster with age in older females than males [72]. Mielke et al. reported that high plasma SM level is associated with increased AD risk in men, but is protective against AD in women [73]. Interestingly, we observed associations of mtDNA-CN with SMs in males only, suggesting the interaction between SM and mitochondria, especially SM hydrolysis-induced apoptosis pathway, differs between genders.
We also identified associations of blood mtDNA-CN with serum levels of ornithine, spermine, creatinine, and 2 lysophosphatidylcholine (lysoPC) species in the total sample, and with glycine and taurine in males only. Although altered levels of these metabolites have been implicated in mitochondrial dysfunction [74–80], there is little evidence linking them to AD.
Our study has several limitations. First, association analyses of ADSP data included age at last exam for controls and age at symptom onset for AD cases, which are both disconnected from the age when the tissue specimens were collected and, hence, for the calculation of mtDNA-CN. Moreover, age at onset for AD cases is less than the age ascribed to mtDNA-CN derived from brain, but in most instances greater than the age ascribed to mtDNA-CN derived from blood. Second, we lacked information about and thus could not incorporate in our analyses the proportions of brain or blood cell types for all ADSP participants which is an important source of confounding. Power for detecting association of AD with mtDNA-CN was considerably lower in the CH and AA groups compared to the much larger EA group. Complete blood count information was not available for ADNI participants which made it difficult to adjust mtDNA-CN estimates for blood cell composition, which has been recognized as an important confounder in large-scale mtDNA-CN GWAS [10, 17, 81]. Blood cell type proportions can be estimated from bulk RNA-seq data using cell-type enrichment analysis [82] or a deconvolution approach [83], but these data were available for less than half of the ADNI participants. To address this problem, we leveraged ADNI WGS data to calculate a PRS for selected blood cell types as a proxy. Third, we evaluated the association of mtDNA-CN with biomarkers and metabolite levels at baseline only and thus could not assess association patterns over the course of cognitive decline leading to AD. Lastly, the interpretation of the causal mediation analysis results may be imbalanced because data for some biomarkers and metabolites were missing for varying proportions of the sample. Moreover, these analyses considered only the scenario where mtDNA-CN is the exposure and biomarkers and metabolites are mediators, but not vice versa. Elucidating the role of mitochondrial dysfunction in each metabolic pathway that is involved in AD pathogenesis will require future studies.
In conclusion, we demonstrated that lower blood mtDNA-CN estimated from WGS data is causally associated with greater AD risk. In addition, we identified association of mtDNA-CN with several AD biomarkers and serum metabolites, many of which are sex-specific. Causal mediation analyses revealed mediating effects of acylcarnitine, serotonin, and plasma NFL on the influence of blood mtDNA-CN on AD risk, suggesting mitochondrial dysfunction affects multiple metabolic pathways associated with AD and mtDNA-CN is a potential blood-based biomarker. Future analyses incorporating hematology data and longitudinal mtDNA-CN measurements may yield more robust and informative findings about the role of mitochondrial dysfunction in AD.
Supplementary Information
Additional file 1: Fig. S1 Computational pipeline and equation used for estimating mtDNA-CN from WGS data. Fig. S2 Comparison of mtDNA-CN estimated by fastMitoCalc and our pipeline for each subject with WGS technical duplicates. Fig. S3 Multiple linear regression analysis for mtDNA-CN in ADSP R3 brain samples. Fig. S4 Multiple linear regression analysis for mtDNA-CN in ADSP R3 blood samples.
Additional file 2: Table S1. Demographic characteristics of ADSP R3 WGS (17 K). Table S2. Effects of non-genetic factors on blood mtDNA-CN stratified by ancestry. Table S3. Results of single SNP Ward test and leave-one-out test for 24 SNPs from Chong et al. Table S4. Associations between blood-derived mtDNA-CN and AD biomarkers in ADNI participants. Table S5. Associations between blood-derived mtDNA-CN and serum metabolites in ADNI participants. Table S6. Associations between blood-derived mtDNA-CN and plasma proteins in ADNI participants. Table S7. AD biomarkers that mediate the association between blood-derived mtDNA-CN and AD in ADNI participants. Table S8. Serum metabolites that mediate the association between blood-derived mtDNA-CN and AD in ADNI participants.
Acknowledgements
This study was supported by National Institute on Aging grants U01-AG058654, U54-AG052427, U19-AG068753, U01-AG072577, R01AG048927, U01AG062602, U01AG081230, P30AG072978, U19AG068753, and R01AG080810. Some of the data used in the preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). Thus, the investigators within the ADNI contributed to the design and implementation of the ADNI and/or provided data but did not participate in the analysis or writing of this report. A complete listing of ADNI investigators is available at http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf. The Alzheimer’s Disease Sequencing Project (ADSP) is comprised of two Alzheimer’s Disease (AD) genetics consortia and three National Human Genome Research Institute (NHGRI) funded Large Scale Sequencing and Analysis Centers (LSAC). The two AD genetics consortia are the Alzheimer’s Disease Genetics Consortium (ADGC) funded by NIA (U01 AG032984), and the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) funded by NIA (R01 AG033193), the National Heart, Lung, and Blood Institute (NHLBI), other National Institute of Health (NIH) institutes and other foreign governmental and non-governmental organizations. The Discovery Phase analysis of sequence data is supported through UF1AG047133 (to Drs. Schellenberg, Farrer, Pericak-Vance, Mayeux, and Haines); U01AG049505 to Dr. Seshadri; U01AG049506 to Dr. Boerwinkle; U01AG049507 to Dr. Wijsman; and U01AG049508 to Dr. Goate and the Discovery Extension Phase analysis is supported through U01AG052411 to Dr. Goate, U01AG052410 to Dr. Pericak-Vance and U01 AG052409 to Drs. Seshadri and Fornage. Sequencing for the Follow Up Study (FUS) is supported through U01AG057659 (to Drs. PericakVance, Mayeux, and Vardarajan) and U01AG062943 (to Drs. Pericak-Vance and Mayeux). Data generation and harmonization in the Follow-up Phase is supported by U54AG052427 (to Drs. Schellenberg and Wang). The FUS Phase analysis of sequence data is supported through U01AG058589 (to Drs. Destefano, Boerwinkle, De Jager, Fornage, Seshadri, and Wijsman), U01AG058654 (to Drs. Haines, Bush, Farrer, Martin, and Pericak-Vance), U01AG058635 (to Dr. Goate), RF1AG058066 (to Drs. Haines, Pericak-Vance, and Scott), RF1AG057519 (to Drs. Farrer and Jun), R01AG048927 (to Dr. Farrer), and RF1AG054074 (to Drs. Pericak-Vance and Beecham). The ADGC cohorts include: Adult Changes in Thought (ACT) (U01 AG006781, U19 AG066567), the Alzheimer’s Disease Research Centers (ADRC) (P30 AG062429, P30 AG066468, P30 AG062421, P30 AG066509, P30 AG066514, P30 AG066530, P30 AG066507, P30 AG066444, P30 AG066518, P30 AG066512, P30 AG066462, P30 AG072979, P30 AG072972, P30 AG072976, P30 AG072975, P30 AG072978, P30 AG072977, P30 AG066519, P30 AG062677, P30 AG079280, P30 AG062422, P30 AG066511, P30 AG072946, P30 AG062715, P30 AG072973, P30 AG066506, P30 AG066508, P30 AG066515, P30 AG072947, P30 AG072931, P30 AG066546, P20 AG068024, P20 AG068053, P20 AG068077, P20 AG068082, P30 AG072958, P30 AG072959), the Chicago Health and Aging Project (CHAP) (R01 AG11101, RC4 AG039085, K23 AG030944), Indiana Memory and Aging Study (IMAS) (R01 AG019771), Indianapolis Ibadan (R01 AG009956, P30 AG010133), the Memory and Aging Project (MAP) ( R01 AG17917), Mayo Clinic (MAYO) (R01 AG032990, U01 AG046139, R01 NS080820, RF1 AG051504, P50 AG016574), Mayo Parkinson’s Disease controls (NS039764, NS071674, 5RC2HG005605), University of Miami (R01 AG027944, R01 AG028786, R01 AG019085, IIRG09133827, A2011048), the Multi-Institutional Research in Alzheimer’s Genetic Epidemiology Study (MIRAGE) (R01 AG09029, R01 AG025259), the National Centralized Repository for Alzheimer’s Disease and Related Dementias (NCRAD) (U24 AG021886), the National Institute on Aging Late Onset Alzheimer’s Disease Family Study (NIA- LOAD) (U24 AG056270), the Religious Orders Study (ROS) (P30 AG10161, R01 AG15819), the Texas Alzheimer’s Research and Care Consortium (TARCC) (funded by the Darrell K Royal Texas Alzheimer’s Initiative), Vanderbilt University/Case Western Reserve University (VAN/CWRU) (R01 AG019757, R01 AG021547, R01 AG027944, R01 AG028786, P01 NS026630, and Alzheimer’s Association), the Washington Heights-Inwood Columbia Aging Project (WHICAP) (RF1 AG054023), the University of Washington Families (VA Research Merit Grant, NIA: P50AG005136, R01AG041797, NINDS: R01NS069719), the Columbia University Hispanic Estudio Familiar de Influencia Genetica de Alzheimer (EFIGA) (RF1 AG015473), the University of Toronto (UT) (funded by Wellcome Trust, Medical Research Council, Canadian Institutes of Health Research), and Genetic Differences (GD) (R01 AG007584). The CHARGE cohorts are supported in part by National Heart, Lung, and Blood Institute (NHLBI) infrastructure grant HL105756 (Psaty), RC2HL102419 (Boerwinkle) and the neurology working group is supported by the National Institute on Aging (NIA) R01 grant AG033193. The CHARGE cohorts participating in the ADSP include the following: Austrian Stroke Prevention Study (ASPS), ASPS-Family study, and the Prospective Dementia Registry-Austria (ASPS/PRODEM-Aus), the Atherosclerosis Risk in Communities (ARIC) Study, the Cardiovascular Health Study (CHS), the Erasmus Rucphen Family Study (ERF), the Framingham Heart Study (FHS), and the Rotterdam Study (RS). ASPS is funded by the Austrian Science Fond (FWF) grant number P20545-P05 and P13180 and the Medical University of Graz. The ASPS-Fam is funded by the Austrian Science Fund (FWF) project I904), the EU Joint Programme – Neurodegenerative Disease Research (JPND) in frame of the BRIDGET project (Austria, Ministry of Science) and the Medical University of Graz and the Steiermärkische Krankenanstalten Gesellschaft. PRODEM-Austria is supported by the Austrian Research Promotion agency (FFG) (Project No. 827462) and by the Austrian National Bank (Anniversary Fund, project 15435. ARIC research is carried out as a collaborative study supported by NHLBI contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C). Neurocognitive data in ARIC is collected by U01 2U01HL096812, 2U01HL096814, 2U01HL096899, 2U01HL096902, 2U01HL096917 from the NIH (NHLBI, NINDS, NIA and NIDCD), and with previous brain MRI examinations funded by R01-HL70825 from the NHLBI. CHS research was supported by contracts HHSN268201200036C, HHSN268200800007C, N01HC55222, N01HC85079, N01HC85080, N01HC85081, N01HC85082, N01HC85083, N01HC85086, and grants U01HL080295 and U01HL130114 from the NHLBI with additional contribution from the National Institute of Neurological Disorders and Stroke (NINDS). Additional support was provided by R01AG023629, R01AG15928, and R01AG20098 from the NIA. FHS research is supported by NHLBI contracts N01-HC-25195 and HHSN268201500001I. This study was also supported by additional grants from the NIA (R01s AG054076, AG049607 and AG033040 and NINDS (R01 NS017950). The ERF study as a part of EUROSPAN (European Special Populations Research Network) was supported by European Commission FP6 STRP grant number 018947 (LSHG-CT-2006-01947) and also received funding from the European Community’s Seventh Framework Programme (FP7/2007-2013)/grant agreement HEALTH-F4- 2007-201413 by the European Commission under the programme “Quality of Life and Management of the Living Resources” of 5th Framework Programme (no. QLG2-CT-2002- 01254). High-throughput analysis of the ERF data was supported by a joint grant from the Netherlands Organization for Scientific Research and the Russian Foundation for Basic Research (NWO-RFBR 047.017.043). The Rotterdam Study is funded by Erasmus Medical Center and Erasmus University, Rotterdam, the Netherlands Organization for Health Research and Development (ZonMw), the Research Institute for Diseases in the Elderly (RIDE), the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports, the European Commission (DG XII), and the municipality of Rotterdam. Genetic data sets are also supported by the Netherlands Organization of Scientific Research NWO Investments (175.010.2005.011, 911-03-012), the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, the Research Institute for Diseases in the Elderly (014-93-015; RIDE2), and the Netherlands Genomics Initiative (NGI)/Netherlands Organization for Scientific Research (NWO) Netherlands Consortium for Healthy Aging (NCHA), project 050-060-810. All studies are grateful to their participants, faculty and staff. The content of these manuscripts is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the U.S. Department of Health and Human Services. The FUS cohorts include: the Alzheimer’s Disease Research Centers (ADRC) (P30 AG062429, P30 AG066468, P30 AG062421, P30 AG066509, P30 AG066514, P30 AG066530, P30 AG066507, P30 AG066444, P30 AG066518, P30 AG066512, P30 AG066462, P30 AG072979, P30 AG072972, P30 AG072976, P30 AG072975, P30 AG072978, P30 AG072977, P30 AG066519, P30 AG062677, P30 AG079280, P30 AG062422, P30 AG066511, P30 AG072946, P30 AG062715, P30 AG072973, P30 AG066506, P30 AG066508, P30 AG066515, P30 AG072947, P30 AG072931, P30 AG066546, P20 AG068024, P20 AG068053, P20 AG068077, P20 AG068082, P30 AG072958, P30 AG072959), Alzheimer’s Disease Neuroimaging Initiative (ADNI) (U19AG024904), Amish Protective Variant Study (RF1AG058066), Cache County Study (R01AG11380, R01AG031272, R01AG21136, RF1AG054052), Case Western Reserve University Brain Bank (CWRUBB) (P50AG008012), Case Western Reserve University Rapid Decline (CWRURD) (RF1AG058267, NU38CK000480), CubanAmerican Alzheimer’s Disease Initiative (CuAADI) (3U01AG052410), Estudio Familiar de Influencia Genetica en Alzheimer (EFIGA) (5R37AG015473, RF1AG015473, R56AG051876), Genetic and Environmental Risk Factors for Alzheimer Disease Among African Americans Study (GenerAAtions) (2R01AG09029, R01AG025259, 2R01AG048927), Gwangju Alzheimer and Related Dementias Study (GARD) (U01AG062602), Hillblom Aging Network (2014-A-004-NET, R01AG032289, R01AG048234), Hussman Institute for Human Genomics Brain Bank (HIHGBB) (R01AG027944, Alzheimer’s Association “Identification of Rare Variants in Alzheimer Disease”), Ibadan Study of Aging (IBADAN) (5R01AG009956), Longevity Genes Project (LGP) and LonGenity (R01AG042188, R01AG044829, R01AG046949, R01AG057909, R01AG061155, P30AG038072), Mexican Health and Aging Study (MHAS) (R01AG018016), Multi-Institutional Research in Alzheimer’s Genetic Epidemiology (MIRAGE) (2R01AG09029, R01AG025259, 2R01AG048927), Northern Manhattan Study (NOMAS) (R01NS29993), Peru Alzheimer’s Disease Initiative (PeADI) (RF1AG054074), Puerto Rican 1066 (PR1066) (Wellcome Trust (GR066133/GR080002), European Research Council (340755)), Puerto Rican Alzheimer Disease Initiative (PRADI) (RF1AG054074), Reasons for Geographic and Racial Differences in Stroke (REGARDS) (U01NS041588), Research in African American Alzheimer Disease Initiative (REAAADI) (U01AG052410), the Religious Orders Study (ROS) (P30 AG10161, P30 AG72975, R01 AG15819, R01 AG42210), the RUSH Memory and Aging Project (MAP) (R01 AG017917, R01 AG42210Stanford Extreme Phenotypes in AD (R01AG060747), University of Miami Brain Endowment Bank (MBB), University of Miami/Case Western/North Carolina A&T African American (UM/CASE/NCAT) (U01AG052410, R01AG028786), and Wisconsin Registry for Alzheimer’s Prevention (WRAP) (R01AG027161 and R01AG054047). The four LSACs are: the Human Genome Sequencing Center at the Baylor College of Medicine (U54 HG003273), the Broad Institute Genome Center (U54HG003067), The American Genome Center at the Uniformed Services University of the Health Sciences (U01AG057659), and the Washington University Genome Institute (U54HG003079). Genotyping and sequencing for the ADSP FUS is also conducted at John P. Hussman Institute for Human Genomics (HIHG) Center for Genome Technology (CGT). Biological samples and associated phenotypic data used in primary data analyses were stored at Study Investigators institutions, and at the National Centralized Repository for Alzheimer’s Disease and Related Dementias (NCRAD, U24AG021886) at Indiana University funded by NIA. Associated Phenotypic Data used in primary and secondary data analyses were provided by Study Investigators, the NIA funded Alzheimer’s Disease Centers (ADCs), and the National Alzheimer’s Coordinating Center (NACC, U24AG072122) and the National Institute on Aging Genetics of Alzheimer’s Disease Data Storage Site (NIAGADS, U24AG041689) at the University of Pennsylvania, funded by NIA. Harmonized phenotypes were provided by the ADSP Phenotype Harmonization Consortium (ADSP-PHC), funded by NIA (U24 AG074855, U01 AG068057 and R01 AG059716) and Ultrascale Machine Learning to Empower Discovery in Alzheimer’s Disease Biobanks (AI4AD, U01 AG068057). This research was supported in part by the Intramural Research Program of the National Institutes of health, National Library of Medicine. Contributors to the Genetic Analysis Data included Study Investigators on projects that were individually funded by NIA, and other NIH institutes, and by private U.S. organizations, or foreign governmental or nongovernmental organizations. The ADSP Phenotype Harmonization Consortium (ADSP-PHC) is funded by NIA (U24 AG074855, U01 AG068057 and R01 AG059716). The harmonized cohorts within the ADSP-PHC include: the Anti-Amyloid Treatment in Asymptomatic Alzheimer’s study (A4 Study), a secondary prevention trial in preclinical Alzheimer's disease, aiming to slow cognitive decline associated with brain amyloid accumulation in clinically normal older individuals. The A4 Study is funded by a public-private-philanthropic partnership, including funding from the National Institutes of Health-National Institute on Aging, Eli Lilly and Company, Alzheimer's Association, Accelerating Medicines Partnership, GHR Foundation, an anonymous foundation and additional private donors, with in-kind support from Avid and Cogstate. The companion observational Longitudinal Evaluation of Amyloid Risk and Neurodegeneration (LEARN) Study is funded by the Alzheimer's Association and GHR Foundation. The A4 and LEARN Studies are led by Dr. Reisa Sperling at Brigham and Women's Hospital, Harvard Medical School and Dr. Paul Aisen at the Alzheimer's Therapeutic Research Institute (ATRI), University of Southern California. The A4 and LEARN Studies are coordinated by ATRI at the University of Southern California, and the data are made available through the Laboratory for Neuro Imaging at the University of Southern California. The participants screening for the A4 Study provided permission to share their de-identified data in order to advance the quest to find a successful treatment for Alzheimer's disease. We would like to acknowledge the dedication of all the participants, the site personnel, and all of the partnership team members who continue to make the A4 and LEARN Studies possible. The complete A4 Study Team list is available on: a4study.org/a4-study-team.; the Adult Changes in Thought study (ACT), U01 AG006781, U19 AG066567; Alzheimer’s Disease Neuroimaging Initiative (ADNI): Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer's Association; Alzheimer's Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.;Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.;Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California; Estudio Familiar de Influencia Genetica en Alzheimer (EFIGA): 5R37AG015473, RF1AG015473, R56AG051876; Memory & Aging Project at Knight Alzheimer’s Disease Research Center (MAP at Knight ADRC): The Memory and Aging Project at the Knight-ADRC (Knight-ADRC). This work was supported by the National Institutes of Health (NIH) grants R01AG064614, R01AG044546, RF1AG053303, RF1AG058501, U01AG058922 and R01AG064877 to Carlos Cruchaga. The recruitment and clinical characterization of research participants at Washington University was supported by NIH grants P30AG066444, P01AG03991, and P01AG026276. Data collection and sharing for this project was supported by NIH grants RF1AG054080, P30AG066462, R01AG064614 and U01AG052410. We thank the contributors who collected samples used in this study, as well as patients and their families, whose help and participation made this work possible. This work was supported by access to equipment made possible by the Hope Center for Neurological Disorders, the Neurogenomics and Informatics Center (NGI: https://neurogenomics.wustl.edu/) and the Departments of Neurology and Psychiatry at Washington University School of Medicine; National Alzheimer’s Coordinating Center (NACC): The NACC database is funded by NIA/NIH Grant U24 AG072122. NACC data are contributed by the NIA-funded ADRCs: P30 AG062429 (PI James Brewer, MD, PhD), P30 AG066468 (PI Oscar Lopez, MD), P30 AG062421 (PI Bradley Hyman, MD, PhD), P30 AG066509 (PI Thomas Grabowski, MD), P30 AG066514 (PI Mary Sano, PhD), P30 AG066530 (PI Helena Chui, MD), P30 AG066507 (PI Marilyn Albert, PhD), P30 AG066444 (PI John Morris, MD), P30 AG066518 (PI Jeffrey Kaye, MD), P30 AG066512 (PI Thomas Wisniewski, MD), P30 AG066462 (PI Scott Small, MD), P30 AG072979 (PI David Wolk, MD), P30 AG072972 (PI Charles DeCarli, MD), P30 AG072976 (PI Andrew Saykin, PsyD), P30 AG072975 (PI David Bennett, MD), P30 AG072978 (PI Neil Kowall, MD), P30 AG072977 (PI Robert Vassar, PhD), P30 AG066519 (PI Frank LaFerla, PhD), P30 AG062677 (PI Ronald Petersen, MD, PhD), P30 AG079280 (PI Eric Reiman, MD), P30 AG062422 (PI Gil Rabinovici, MD), P30 AG066511 (PI Allan Levey, MD, PhD), P30 AG072946 (PI Linda Van Eldik, PhD), P30 AG062715 (PI Sanjay Asthana, MD, FRCP), P30 AG072973 (PI Russell Swerdlow, MD), P30 AG066506 (PI Todd Golde, MD, PhD), P30 AG066508 (PI Stephen Strittmatter, MD, PhD), P30 AG066515 (PI Victor Henderson, MD, MS), P30 AG072947 (PI Suzanne Craft, PhD), P30 AG072931 (PI Henry Paulson, MD, PhD), P30 AG066546 (PI Sudha Seshadri, MD), P20 AG068024 (PI Erik Roberson, MD, PhD), P20 AG068053 (PI Justin Miller, PhD), P20 AG068077 (PI Gary Rosenberg, MD), P20 AG068082 (PI Angela Jefferson, PhD), P30 AG072958 (PI Heather Whitson, MD), P30 AG072959 (PI James Leverenz, MD); National Institute on Aging Alzheimer’s Disease Family Based Study (NIA-AD FBS): U24 AG056270; Religious Orders Study (ROS): P30AG10161,R01AG15819, R01AG42210; Memory and Aging Project (MAP - Rush): R01AG017917, R01AG42210; Minority Aging Research Study (MARS): R01AG22018, R01AG42210; Washington Heights/Inwood Columbia Aging Project (WHICAP): RF1 AG054023;and Wisconsin Registry for Alzheimer’s Prevention (WRAP): R01AG027161 and R01AG054047. Additional acknowledgments include the National Institute on Aging Genetics of Alzheimer’s Disease Data Storage Site (NIAGADS, U24AG041689) at the University of Pennsylvania, funded by NIA.
Abbreviations
- AA
African Americans
- Aβ
β-amyloid
- AC
Acylcarnitine
- AD
Alzheimer’s disease
- ADMC
Alzheimer’s Disease Metabolomics Consortium
- ADNI
Alzheimer’s Disease Neuroimaging Initiative
- ADSP R3
Alzheimer’s Disease Sequencing Project Release 3
- C5-OH
hydroxyvalerylcarnitine
- C18:2
octadecadienylcarnitine
- CH
Caribbean Hispanics
- CN
Cognitively normal
- CSF
Cerebrospinal fluid
- EA
Europeans
- FDG
Fluorodeoxyglucose
- GCAD
Genome Center for Alzheimer’s Disease
- GWAS
Genome-wide association study
- IVW
Inverse variance weighted
- LD
Linkage disequilibrium
- LONI
Laboratory of Neuro Imaging
- MCI
Mild cognitive impairment
- MR
Mendelian randomization
- mtDNA-CN
mitochondrial DNA copy number
- NDE
Natural direct effect
- NIE
Natural indirect effect
- NFL
Neurofilament light
- OXPHOS
Oxidative phosphorylation
- p-Tau
phosphorylated-tau181
- PET
Positron emission tomography
- PRS
Polygenic risk score
- QC
Quality control
- RAPS
Robust adjusted profile score
- SNPs
Single nucleotide polymorphisms
- SM
Sphingomyelin
- t-Tau
total tau
- WGS
Whole-genome sequence
Authors’ contributions
TT and XZ designed the framework of this study. TT, CZ, ZK, and JJF pre-processed the ADSP R3 WGS data. TT performed the analyses and prepared the tables and figures. XZ, KLL, and LAF supervised the analysis. TT, XZ, LAF, KLL, WQQ, and JLH wrote the manuscript. ERM, WSB, MAP-V, L-SW, GDS, JLH and LAF obtained funding for this study. All authors edited and approved the final manuscript.
Funding
This study was supported in part by National Institute on Aging grants U01-AG062602, U01-AG058654, U54-AG052427, U01-AG032984, R01-AG048927, U01-AG081230, U01-AG072577, R01-AG080810, P30-AG072978 and U19-AG068753, and National Science Foundation grant DMS/NIGMS-2347698.
Data availability
Data for this study are archived and distributed by the National Institute on Aging Alzheimer’s Disease Data Storage Site (NIAGADS) at the University of Pennsylvania (Accession number: NG00067), funded by the National Institute on Aging (U24-AG041689). The AD biomarkers dataset, serum metabolites dataset generated by Alzheimer’s Disease Metabolomics Consortium, and plasma proteomics dataset generated by Biomarkers Consortium Plasma Proteomics Project analyzed during the current study are available in the Laboratory of Neuro Imaging (LONI) Image and Data Archive (IDA) repository on reasonable request, https://ida.loni.usc.edu.
Declarations
Ethics approval and consent to participate
This study was performed in accordance with the Declaration of Helsinki and approved by the.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Lindsay A. Farrer, Email: farrer@bu.edu
Xiaoling Zhang, Email: zhangxl@bu.edu.
References
- 1.Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443(7113):787–95. [DOI] [PubMed] [Google Scholar]
- 2.Tang YG, Zucker RS. Mitochondrial involvement in post-tetanic potentiation of synaptic transmission. Neuron. 1997;18(3):483–91. [DOI] [PubMed] [Google Scholar]
- 3.Babayev E, Seli E. Oocyte mitochondrial function and reproduction. Curr Opin Obstet Gynecol. 2015;27(3):175–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fazzini F, Schopf B, Blatzer M, Coassin S, Hicks AA, Kronenberg F, Fendt L. Plasmid-normalized quantification of relative mitochondrial DNA copy number. Sci Rep. 2018;8:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang SY, Castellani CA, Longchamps RJ, Pillalamarri VK, O’Rourke B, Guallar E, Arking DE. Blood-derived mitochondrial DNA copy number is associated with gene expression across multiple tissues and is predictive for incident neurodegenerative disease. Genome Res. 2021;31(3):349–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Filograna R, Mennuni M, Alsina D, Larsson NG. Mitochondrial DNA copy number in human disease: the more the better? FEBS Lett. 2021;595(8):976–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu X, Longchamps RJ, Wiggins KL, Raffield LM, Bielak LF, Zhao W, Pitsillides A, Blackwell TW, Yao J, Guo X, et al. Association of mitochondrial DNA copy number with cardiometabolic diseases. Cell Genom. 2021;1(1):100006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fazzini F, Lamina C, Raftopoulou A, Koller A, Fuchsberger C, Pattaro C, Del Greco FM, Dottelmayer P, Fendt L, Fritz J, et al. Association of mitochondrial DNA copy number with metabolic syndrome and type 2 diabetes in 14 176 individuals. J Intern Med. 2021;290(1):190–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin LN, Yu BF, Armando I, Han F. Mitochondrial DNA-mediated inflammation in acute kidney injury and chronic kidney disease. Oxidative Med Cell Longev. 2021;2021:9985603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chong M, Mohammadi-Shemirani P, Perrot N, Nelson W, Morton R, Narula S, Lali R, Khan I, Khan M, Judge C, et al. GWAS and ExWAS of blood mitochondrial DNA copy number identifies 71 loci and highlights a potential causal role in dementia. Elife. 2022;11:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alzheimer’s disease facts and figures. Alzheimers Dement. 2022;18(4):700–89. [DOI] [PubMed] [Google Scholar]
- 12.Zhang X, Li YF, Xu HX, Zhang YW. The gamma-secretase complex: from structure to function. Front Cell Neurosci. 2014;8:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang WZ, Zhao FP, Ma XP, Perry G, Zhu XW. Mitochondria dysfunction in the pathogenesis of Alzheimer’s disease: recent advances. Mol Neurodegener. 2020;15(1):22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Swerdlow RH. Mitochondria and mitochondrial cascades in Alzheimer’s Disease. J Alzheimers Dis. 2018;62(3):1403–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rice AC, Keeney PM, Algarzae NK, Ladd AC, Thomas RR, Bennett JP. Mitochondrial DNA copy numbers in pyramidal neurons are decreased and mitochondrial Biogenesis Transcriptome Signaling is disrupted in Alzheimer’s Disease Hippocampi. J Alzheimers Dis. 2014;40(2):319–30. [DOI] [PubMed] [Google Scholar]
- 16.Delbarba A, Abate G, Prandelli C, Marziano M, Buizza L, Varas NA, Novelli A, Cuetos F, Martinez C, Lanni C, et al. Mitochondrial alterations in peripheral mononuclear blood cells from Alzheimer’s disease and mild cognitive impairment patients. Oxidative Med Cell Longev. 2016;2016:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Longchamps RJ, Yang SY, Castellani CA, Shi W, Lane J, Grove ML, Bartz TM, Sarnowski C, Liu C, Burrows K, et al. Genome-wide analysis of mitochondrial DNA copy number reveals loci implicated in nucleotide metabolism, platelet activation, and megakaryocyte proliferation. Hum Genet. 2022;141(1):127–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zaidi A, Verma A, Morse C, BioBank PM, Ritchie M, Mathieson I. The genetic and phenotypic correlates of mtDNA copy number in a multi-ancestry cohort. Hum Genet GENOMICS Adv. 2023;4:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leung YY, Valladares O, Chou YF, Lin HJ, Kuzma AB, Cantwell L, Qu LM, Gangadharan P, Salerno WJ, Schellenberg GD, et al. VCPA: genomic variant calling pipeline and data management tool for Alzheimer’s disease sequencing project. Bioinformatics. 2019;35(10):1768–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Naj AC, Lin HH, Vardarajan BN, White S, Lancour D, Ma YY, Schmidt M, Sun FG, Butkiewicz M, Bush WS, et al. Quality control and integration of genotypes from two calling pipelines for whole genome sequence data in the Alzheimer’s disease sequencing project. Genomics. 2019;111(4):808–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee WP, Choi SH, Shea MG, Cheng PL, Dombroski BA, Pitsillides AN, Heard-Costa NL, Wang H, Bulekova K, Kuzma AB, et al. Association of common and rare variants with alzheimer's disease in over 13,000 diverse individuals with whole-genome sequencing from the alzheimer's disease sequencing project. 2023. Preprint at https://www.medrxiv.org/content/10.1101/2023.09.01.23294953v1. [DOI] [PMC free article] [PubMed]
- 22.Gogarten SM, Sofer T, Chen H, Yu C, Brody JA, Thornton TA, Rice KM, Conomos MP. Genetic association testing using the GENESIS R/Bioconductor package. Bioinformatics. 2019;35(24):5346–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saykin AJ, Shen L, Yao X, Kim S, Nho K, Risacher SL, Ramanan VK, Foroud TM, Faber KM, Sarwar N, et al. Genetic studies of quantitative MCI and AD phenotypes in ADNI: Progress, opportunities, and plans. Alzheimers Dement. 2015;11(7):792–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pedersen BS, Quinlan AR. Mosdepth: quick coverage calculation for genomes and exomes. Bioinformatics. 2018;34(5):867–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qian Y, Butler TJ, Opsahl-Ong K, Giroux NS, Sidore C, Nagaraja R, Cucca F, Ferrucci L, Abecasis GR, Schlessinger D, et al. fastMitoCalc: an ultra-fast program to estimate mitochondrial DNA copy number from whole-genome sequences. Bioinformatics. 2017;33(9):1399–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Picard M. Blood mitochondrial DNA copy number: what are we counting? Mitochondrion. 2021;60:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ganel L, Chen L, Christ R, Vangipurapu J, Young E, Das I, Kanchi K, Larson D, Regier A, Abel H, et al. Mitochondrial genome copy number measured by DNA sequencing in human blood is strongly associated with metabolic traits via cell-type composition differences. Hum Genom. 2021;15(1):34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choi SW, O’Reilly PF. PRSice-2: polygenic risk score software for biobank-scale data. Gigascience. 2019;8(7):giz082. [DOI] [PMC free article] [PubMed]
- 29.Chen MH, Raffield LM, Mousas A, Sakaue S, Huffman JE, Moscati A, Trivedi B, Jiang T, Akbari P, Vuckovic D, et al. Trans-ethnic and ancestry-specific blood-cell Genetics in 746,667 individuals from 5 global populations. Cell. 2020;182(5):1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jagust WJ, Bandy D, Chen KW, Foster NL, Landau SM, Mathis CA, Price JC, Reiman EM, Skovronsky D, Koeppe RA, et al. The Alzheimer’s Disease Neuroimaging Initiative Positron emission tomography core. Alzheimers Dement. 2010;6(3):221–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jagust WJ, Landau SM, Koeppe RA, Reiman EM, Chen KW, Mathis CA, Price JC, Foster NL, Wang AY. The Alzheimer’s Disease Neuroimaging Initiative 2 PET core: 2015. Alzheimers Dement. 2015;11(7):757–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toledo JB, Arnold M, Kastenmuller G, Chang R, Baillie RA, Han XL, Thambisetty M, Tenenbaum JD, Suhre K, Thompson JW, et al. Metabolic network failures in Alzheimer’s disease: a biochemical road map. Alzheimers Dement. 2017;13(9):965–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kunkle BW, Grenier-Boley B, Sims R, Bis JC, Damotte V, Naj AC, Boland A, Vronskaya M, van der Lee SJ, Amlie-Wolf A, et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates a beta, tau, immunity and lipid processing. Nat Genet. 2019;51(3):414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using Summarized Data. Genet Epidemiol. 2013;37(7):658–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bowden J, Smith GD, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44(2):512–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bowden J, Smith GD, Haycock PC, Burgess S. Consistent estimation in mendelian randomization with some Invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40(4):304–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao QY, Wang JS, Hemani G, Bowden J, Small DS. Statistical inference in two-sample summary-data mendelian randomization using robust adjusted profile score. Ann Stat. 2020;48(3):1742–69. [Google Scholar]
- 38.Hemani G, Zhengn J, Elsworth B, Wade KH, Haberland V, Baird D, Laurin C, Burgess S, Bowden J, Langdon R, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018;7:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from mendelian randomization between complex traits and diseases. Nat Genet. 2018;50(5):693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu ZH, Zhang FT, Hu H, Bakshi A, Robinson MR, Powell JE, Montgomery GW, Goddard ME, Wray NR, Visscher PM, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet. 2016;48(5):481. [DOI] [PubMed] [Google Scholar]
- 41.Hemani G, Tilling K, Smith GD. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet. 2017;13(11):e1007081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steen J, Loeys T, Moerkerke B, Vansteelandt S. medflex: An R package for flexible mediation analysis using natural effect models. J Stat Softw. 2017;76(11):1.36568334 [Google Scholar]
- 43.Ding J, Sidore C, Butler TJ, Wing MK, Qian Y, Meirelles O, Busonero F, Tsoi LC, Maschio A, Angius A, et al. Assessing mitochondrial DNA variation and copy number in lymphocytes of similar to 2,000 sardinians using tailored sequencing analysis tools. PLoS Genet. 2015;11(7):e1005306. [DOI] [PMC free article] [PubMed]
- 44.Harerimana NV, Paliwali D, Romero-Molina C, Bennett DA, Pa J, Goate A, Swerdlow RH, Andrews SJ. The role of mitochondrial genome abundance in Alzheimer’s disease. Alzheimers Dement 2022. [DOI] [PubMed]
- 45.Gupta R, Kanai M, Durham TJ, Tsuo K, McCoy JG, Kotrys AV, Zhou W, Chinnery PF, Karczewski KJ, Calvo SE et al. Nuclear genetic control of mtDNA copy number and heteroplasmy in humans. Nature. 2023. [DOI] [PMC free article] [PubMed]
- 46.Klein HU, Trumpff C, Yang HS, Lee AJ, Picard M, Bennett DA, De Jager PL. Characterization of mitochondrial DNA quantity and quality in the human aged and Alzheimer’s disease brain. Mol Neurodegener. 2021;16(1):17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao RN, Ma SL. Is Mitochondria DNA Variation a Biomarker for AD? Genes. 2022;13(10):1789. [DOI] [PMC free article] [PubMed]
- 48.Qin LX, Zhu XW, Friedland RP. ApoE and mitochondrial dysfunction. Neurology. 2020;94(23):1009–10. [DOI] [PubMed] [Google Scholar]
- 49.Yin JX, Reiman EM, Beach TG, Serrano GE, Sabbagh MN, Nielsen M, Caselli RJ, Shi J. Effect of ApoE isoforms on mitochondria in Alzheimer disease. Neurology. 2020;94(23):E2404-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park JC, Lim H, Byun MS, Yi D, Byeon G, Jung G, Kim YK, Lee DY, Han SH, Mook-Jung I. Sex differences in the progression of glucose metabolism dysfunction in Alzheimer’s disease. Experimental Mol Med. 2023;55(5):1023–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buckley RF, Mormino EC, Rabin JS, Hohman TJ, Landau S, Hanseeuw BJ, Jacobs HIL, Papp KV, Amariglio RE, Properzi MJ, et al. Sex differences in the Association of Global Amyloid and Regional Tau Deposition measured by Positron Emission Tomography in clinically normal older adults. Jama Neurol. 2019;76(5):542–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koran MEI, Wagener M, Hohman TJ, Alzheimer’s Neuroimaging I. Sex differences in the association between AD biomarkers and cognitive decline. Brain Imaging Behav. 2017;11(1):205–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mielke MM. Consideration of sex differences in the measurement and interpretation of Alzheimer Disease-Related Biofluid-based biomarkers. J Appl Lab Med. 2020;5(1):158–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Beydoun MA, Noren Hooten N, Beydoun HA, Maldonado AI, Weiss J, Evans MK, Zonderman AB. Plasma neurofilament light as a potential biomarker for cognitive decline in a longitudinal study of middle-aged urban adults. Transl Psychiatry. 2021;11(1):436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Giacomucci G, Mazzeo S, Bagnoli S, Ingannato A, Leccese D, Berti V, Padiglioni S, Galdo G, Ferrari C, Sorbi S, et al. Plasma neurofilament light chain as a biomarker of Alzheimer’s disease in Subjective Cognitive decline and mild cognitive impairment. J Neurol. 2022;269(8):4270–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yao WN, Zhou HJ, Zhang X, Chen HF, Bai F. Alzheimers Dis Neuroimaging I. Inflammation affects dynamic functional network connectivity pattern changes via plasma NFL in cognitive impairment patients. CNS Neurosci Ther. 2024;30(2):e14391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Castellani CA, Longchamps RJ, Sun J, Guallar E, Arking DE. Thinking outside the nucleus: mitochondrial DNA copy number in health and disease. Mitochondrion. 2020;53:214–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dewanjee S, Chakraborty P, Bhattacharya H, Chacko L, Singh B, Chaudhary A, Javvaji K, Pradhan SR, Vallamkondu J, Dey A, et al. Altered glucose metabolism in Alzheimer’s disease: role of mitochondrial dysfunction and oxidative stress. Free Radic Biol Med. 2022;193(Pt 1):134–57. [DOI] [PubMed] [Google Scholar]
- 59.Jung YJ, Damoiseaux JS. The potential of blood neurofilament light as a marker of neurodegeneration for Alzheimer’s disease. Brain. 2024;147(1):12–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilkins HM. Interactions between amyloid, amyloid precursor protein, and mitochondria. Biochem Soc Trans. 2023;51(1):173–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pinto M, Pickrell AM, Fukui H, Moraes CT. Mitochondrial DNA damage in a mouse model of Alzheimer’s disease decreases amyloid beta plaque formation. Neurobiol Aging. 2013;34(10):2399–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lehallier B, Essioux L, Gayan J, Alexandridis R, Nikolcheva T, Wyss-Coray T, Britschgi M. Alzheimers dis neuroimaging I: combined plasma and cerebrospinal fluid signature for the prediction of Midterm Progression from mild cognitive impairment to Alzheimer Disease. Jama Neurol. 2016;73(2):203–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Horgusluoglu E, Neff R, Song WM, Wang MH, Wang Q, Arnold M, Krumsiek J, Galindo-Prieto B, Ming C, Nho K, et al. Integrative metabolomics-genomics approach reveals key metabolic pathways and regulators of Alzheimer’s disease. Alzheimers Dement. 2022;18(6):1260–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cristofano A, Sapere N, La Marca G, Angiolillo A, Vitale M, Corbi G, Scapagnini G, Intrieri M, Russo C, Corso G, et al. Serum Levels of Acyl-Carnitines along the Continuum from Normal to Alzheimer’s Dementia. PLoS ONE. 2016;11(5):e0155694. [DOI] [PMC free article] [PubMed]
- 65.Darst BF, Koscik RL, Hogan KJ, Johnson SC, Engelman CD. Longitudinal plasma metabolomics of aging and sex. Aging-Us. 2019;11(4):1262–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tian Q, Mitchell BA, Erus G, Davatzikos C, Moaddel R, Resnick SM, Ferrucci L. Sex differences in plasma lipid profiles of accelerated brain aging. Neurobiol Aging. 2023;129:178–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Whiley L, Chappell KE, D’Hondt E, Lewis MR, Jiménez B, Snowden SG, Soininen H, Kloszewska I, Mecocci P, Tsolaki M, et al. Metabolic phenotyping reveals a reduction in the bioavailability of serotonin and kynurenine pathway metabolites in both the urine and serum of individuals living with Alzheimer’s disease. Alzheimers Res Ther. 2021;13(1):20. [DOI] [PMC free article] [PubMed]
- 68.Tian J, Du E, Guo L. Mitochondrial Interaction with Serotonin in Neurobiology and its implication in Alzheimer’s Disease. J Alzheimers Disease Rep. 2023;7(1):1165–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hanamatsu H, Ohnishi S, Sakai S, Yuyama K, Mitsutake S, Takeda H, Hashino S, Igarashi Y. Altered levels of serum sphingomyelin and ceramide containing distinct acyl chains in young obese adults. Nutr Diab. 2014;4:e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.He XX, Huang Y, Li B, Gong CX, Schuchman EH. Deregulation of sphingolipid metabolism in Alzheimer’s disease. Neurobiol Aging. 2010;31(3):398–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Costa AC, Joaquim HPG, Forlenza OV, Gattaz WF, Talib LL. Three plasma metabolites in elderly patients differentiate mild cognitive impairment and Alzheimer’s disease: a pilot study. Eur Arch Psych Clin Neurosci. 2020;270(4):483–8. [DOI] [PubMed] [Google Scholar]
- 72.Muilwijk M, Callender N, Goorden S, Vaz FM, van Valkengoed IGM. Sex differences in the association of sphingolipids with age in Dutch and south-asian Surinamese living in Amsterdam, the Netherlands. Biol Sex Differ. 2021;12(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mielke MM, Haughey NJ, Han DF, An Y, Bandaru VVR, Lyketsos CG, Ferrucci L, Resnick SM. The Association between plasma ceramides and sphingomyelins and risk of Alzheimer’s Disease differs by Sex and APOE in the Baltimore Longitudinal Study of Aging. J Alzheimers Dis. 2017;60(3):819–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Law SH, Chan ML, Marathe GK, Parveen F, Chen CH, Ke LY. An updated review of lysophosphatidylcholine metabolism in human diseases. Int J Mol Sci. 2019;20(5):1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zanatta A, Rodrigues MDN, Amaral AU, Souza DG, Quincozes-Santos A, Wajner M. Ornithine and Homocitrulline Impair mitochondrial function, decrease antioxidant defenses and induce cell death in Menadione-stressed rat cortical astrocytes: potential mechanisms of neurological dysfunction in HHH Syndrome. Neurochem Res. 2016;41(9):2190–8. [DOI] [PubMed] [Google Scholar]
- 76.Hofer SJ, Simon AK, Bergmann M, Eisenberg T, Kroemer G, Madeo F. Mechanisms of spermidine-induced autophagy and geroprotection. Nat Aging. 2022;2(12):1112–29. [DOI] [PubMed] [Google Scholar]
- 77.Soda K. Changes in whole blood polyamine levels and their background in age-related diseases and healthy longevity. Biomedicines. 2023;11(10):2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li XH, Sun L, Zhang WD, Li HX, Wang SM, Mu HN, Zhou Q, Zhang Y, Tang YM, Wang Y, et al. Association of serum glycine levels with metabolic syndrome in an elderly Chinese population. Nutr Metab. 2018;15:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ullah R, Jo MH, Riaz M, Alam SI, Saeed K, Ali W, Rehman IU, Ikram M, Kim MO. Glycine, the smallest amino acid, confers neuroprotection againstd-galactose-induced neurodegeneration and memory impairment by regulating c-Jun N-terminal kinase in the mouse brain. J Neuroinflamm. 2020;17(1). [DOI] [PMC free article] [PubMed]
- 80.Oh SJ, Lee HJ, Jeong YJ, Nam KR, Kang KJ, Han SJ, Lee KC, Lee YJ, Choi JY. Evaluation of the neuroprotective effect of taurine in Alzheimer’s disease using functional molecular imaging. Sci Rep. 2020;10(1):15551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Guyatt AL, Brennan RR, Burrows K, Guthrie PAI, Ascione R, Ring SM, Gaunt TR, Pyle A, Cordell HJ, Lawlor DA, et al. A genome-wide association study of mitochondrial DNA copy number in two population-based cohorts. Hum Genom. 2019;13:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aran D, Hu ZC, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18(1):220. [DOI] [PMC free article] [PubMed]
- 83.O’Neill NK, Stein TD, Hu JM, Rehman H, Campbell JD, Yajima M, Zhang XL, Farrer LA. Bulk brain tissue cell-type deconvolution with bias correction for single-nuclei RNA sequencing data using DeTREM. BMC Bioinformatics. 2023;24(1):349. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Fig. S1 Computational pipeline and equation used for estimating mtDNA-CN from WGS data. Fig. S2 Comparison of mtDNA-CN estimated by fastMitoCalc and our pipeline for each subject with WGS technical duplicates. Fig. S3 Multiple linear regression analysis for mtDNA-CN in ADSP R3 brain samples. Fig. S4 Multiple linear regression analysis for mtDNA-CN in ADSP R3 blood samples.
Additional file 2: Table S1. Demographic characteristics of ADSP R3 WGS (17 K). Table S2. Effects of non-genetic factors on blood mtDNA-CN stratified by ancestry. Table S3. Results of single SNP Ward test and leave-one-out test for 24 SNPs from Chong et al. Table S4. Associations between blood-derived mtDNA-CN and AD biomarkers in ADNI participants. Table S5. Associations between blood-derived mtDNA-CN and serum metabolites in ADNI participants. Table S6. Associations between blood-derived mtDNA-CN and plasma proteins in ADNI participants. Table S7. AD biomarkers that mediate the association between blood-derived mtDNA-CN and AD in ADNI participants. Table S8. Serum metabolites that mediate the association between blood-derived mtDNA-CN and AD in ADNI participants.
Data Availability Statement
Data for this study are archived and distributed by the National Institute on Aging Alzheimer’s Disease Data Storage Site (NIAGADS) at the University of Pennsylvania (Accession number: NG00067), funded by the National Institute on Aging (U24-AG041689). The AD biomarkers dataset, serum metabolites dataset generated by Alzheimer’s Disease Metabolomics Consortium, and plasma proteomics dataset generated by Biomarkers Consortium Plasma Proteomics Project analyzed during the current study are available in the Laboratory of Neuro Imaging (LONI) Image and Data Archive (IDA) repository on reasonable request, https://ida.loni.usc.edu.