Abstract
Mcm10 (Dna43), first identified in Saccharomyces cerevisiae, is an essential protein which functions in the initiation of DNA synthesis. Mcm10 is a nuclear protein that is localized to replication origins and mediates the interaction of the Mcm2–7 complex with replication origins. We identified and cloned a human cDNA whose product was structurally homologous to the yeast Mcm10 protein. Human Mcm10 (HsMcm10) is a 98-kDa protein of 874 amino acids which shows 23 and 21% overall similarity to Schizosaccharomyces pombe Cdc23 and S.cerevisiae Mcm10, respectively. The messenger RNA level of HsMcm10 increased at the G1/S-boundary when quiescent human NB1–RGB cells were induced to proliferate as is the case of many replication factors. HsMcm10 associated with nuclease-resistant nuclear structures throughout S phase and dissociated from it in G2 phase. HsMcm10 associated with human Orc2 protein when overexpressed in COS-1 cells. HsMcm10 also interacted with Orc2, Mcm2 and Mcm6 proteins in the yeast two-hybrid system. These results suggest that HsMcm10 may function in DNA replication through the interaction with Orc and Mcm2–7 complexes.
INTRODUCTION
The replication of eukaryotic chromosome is highly regulated process so that it occurs in S phase only once per cell cycle. Present understanding of the initiation step of DNA replication in eukaryotic cells is mainly based on experiments conducted with Saccharomyces cerevisiae and Xenopus cell-free system. In S.cerevisiae, the origin recognition complex (ORC), a complex of six proteins (Orc1–6), binds to replication origins throughout the cell cycle (1,2), whereas the ORC seems to dissociate from chromatin during mitosis in Xenopus (3–5). After mitosis, the ORC forms a pre-replication complex with Cdc6 and Mcm2–7 complex (3,6–9). Recruitment of the Mcm2–7 complex, which may be the putative replicative helicase (10), is dependent on Cdc6 (3,11–13). Cdc6 is a short-lived protein in yeast and is degraded during S phase through the Cdc4/34/53 pathway (14). Cdc6 is also regulated by subcellular localization in mammalian cells (15). DNA replication initiates upon the activation of the pre-replication complex by S phase cyclin-dependent kinases (S-Cdks) and Cdc7/Dbf4 kinase in S phase (16–18). The activation of both S-Cdks and Cdc7/Dbf4 kinase is required for efficient loading of Cdc45 to a preformed pre-replication complex at each origin with programmed timing (18–21). Cdc45 facilitates assembly of replication forks by loading replication protein A (RPA) and DNA polymerases (20,22,23). After DNA replication initiates, the Mcm2–7 complex and Cdc45 dissociate from origin DNA (19).
Mcm10 (Dna43) was originally found in S.cerevisiae as an essential protein which is involved in the initiation of DNA replication (24,25). The mcm10 mutant shows a drastic reduction of DNA replication initiation at replication origins under the non-permissive temperature (25). The mcm10 mutant also causes the stalling of replication forks at replication origins which did not fire (25). These results suggest that Mcm10 is necessary for the activation of pre-replication complex and/or in the elongation step. Recently, Mcm10 was shown to be localized to replication origins (26). It was also shown that chromatin binding of the Mcm2–7 complex is dependent on Mcm10 and that Mcm10 genetically interacts with Mcm7 (26). These results suggest that Mcm10 is a component of the pre-replication complex and may be required for the release of the Mcm2–7 complex from origins.
MCM10 homologs have so far been identified in Schizosaccharomyces pombe, Candida albicans and Caenorhabditis elegans (26,27). Although the nature of DNA sequences defining an origin of replication has not yet been resolved in mammalian cells, many replication proteins are conserved from yeast to mammal, suggesting that the order of the events that leads to initiation of DNA replication is basically common in eukaryotes. We think characterization of replication factors and their interaction with DNA will be helpful to understand how DNA replication is regulated in mammalian cells. Toward this goal, we have isolated the human MCM10 cDNA based on the sequence homology. The human MCM10 transcript increased at the G1–S transition, implying the involvement of cell growth regulation in the expression of MCM10. Human Mcm10 protein bound to the nuclease-resistant nuclear structures throughout S phase. We also examined whether human Mcm10 protein interacted with other replication factors. We found Mcm10 interacted with Orc2 in mammalian expression system as well as in the yeast two-hybrid system, which suggests a new Mcm10 function.
MATERIALS AND METHODS
Nucleotide sequence in database
The nucleotide sequence of human Mcm10 cDNA was designated as HsMCM10 and deposited in the DDJB /EMBL/GenBank databases with the accession number AB042719. HsMCM10 gene was on the clone RP11-24J20 of chromosome 10, which is in the DDJB/EMBL/GenBank database with the accession number AL138764.
cDNA cloning of human MCM10 homolog
In the Drosophila melanogaster genome database (National Center for Biotechnology Information), a partial sequence (AC006574) was first found with significant homology to a portion of the S.pombe cdc23 gene, which is a homolog of S.cerevisiae MCM10 (27). A BLAST search of the genome database with the AC006574 sequence revealed a recently reported C.elegans MCM10 homolog (AB011244) (26), and a part of the Arabidopsis thaliana MCM10 homolog (AC006234). A BLAST search of the human expressed sequence tag database with the AC006574 sequence also revealed a partial human MCM10 cDNA fragment (AA312197). This fragment was amplified by RT–PCR from HeLa RNA and then used as a probe to screen a HeLa cDNA library as described (28). Thirteen overlapping cDNA clones were obtained from a screen of approximately 3 million plaques. The largest insert (3.7 kb) was subcloned into the pBluescript II and sequenced on both strands.
Northern blot analysis
Human normal fibroblast NB1–RGB cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum and non-essential amino acids. For synchronization, cells were cultured in DMEM supplemented with 0.4% fetal calf serum and non-essential amino acids for 72 h, and then released by changing the medium to DMEM containing 15% fetal calf serum and non-essential amino acids. Poly(A)+ RNA was isolated every 4 h and used for northern blotting as described (29). To monitor DNA synthesis, cells were pulse-labeled with BrdU for 20 min, and incorporated BrdU was quantified by enzyme-linked immunosorbent assay using the BrdU labeling detection kit III (Boehringer Mannheim).
Antibodies and immunoblotting
A 1.1-kb cDNA fragment encoding amino acids 127–512 of HsMcm10 was cloned into the NotI–SalI sites of pET24a (Novagen). A 42-kDa fragment of HsMcm10 was produced in Escherichia coli BL21(DE3) strain, extracted with 8 M urea, and excised from sodium dodecyl sulfate (SDS)–polyacrylamide gel. Purified protein was used to raise an antibody in a rabbit.
For immunoblotting, samples were separated by 8% SDS–polyacrylamide gel and transferred electrophoretically onto 0.45-µm PVDF membranes (Millipore). After incubation of the membranes with first antibodies in TBS (Tris-buffered saline; 50 mM Tris–HCl, pH 7.5 and 150 mM NaCl) containing 5% (w/v) dried milk for 1 h at room temperature, the membranes were washed three times with TBS containing 0.05% Tween 20. The membranes were then incubated for 1 h at room temperature with horseradish peroxidase-conjugated secondary antibodies (Vector Inc.) in TBS containing 5% dried milk and washed again. Detection of the protein bands was performed using the enhanced chemiluminescent reagent SuperSignal (PIERCE) according to the manufacturer’s instruction. Kaleidoscope prestained standards (Bio-Rad) were used as molecular weight standards. The antibodies against human Mcm7 and human lamin B1 were purchased from Santa Cruz. The polyclonal antibody against HA-epitope tag was obtained from MBL. The monoclonal antibody against HA-epitope tag was obtained from Roche.
Fractionations of cellular proteins
HeLa S3 cells were maintained in DMEM supplemented with 10% calf serum. For synchronization at M phase, exponentially growing HeLa S3 cells were treated with 50 ng/ml of nocodazole (Aldorich) for 4 h and collected by mitotic shake-off. For synchronization at the G1/S boundary, mitotic cells were collected as described above, replated, and cultured in the presence of 15 µM aphidicolin (Sigma) for 12–14 h. To monitor DNA synthesis, cells were pulse-labeled with BrdU for 10 min, and incorporated BrdU was detected by immunofluorescent staining as described previously (30).
HeLa cell extracts were prepared according to the procedure described by Fujita et al. (31). HeLa cells were washed three times with ice-cold phosphate-buffered saline (PBS) and scraped into PBS. Aliquots of 2 × 106 cells were lysed in 250 µl of cytoskeleton buffer (CSK buffer: 10 mM PIPES, pH 6.8, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 1 mM EGTA, 1 mM DTT, 0.1 mM ATP, 1 mM Na3VO4, 10 mM NaF, 0.2 mM phenylmethylsulfonyl fluoride, 0.4 µg/ml aprotinin, 0.4 µg/ml leupeptin, 0.2 µg/ml antipain and 0.2 µg/ml pepstatin A) containing 0.1% Triton X-100 at 4°C for 15 min and subjected to low speed centrifugation (3000 r.p.m. for 3 min). The supernatants were further clarified by centrifugation at 15 000 r.p.m. for 10 min to obtain Triton X-100-soluble fractions. After Triton-extracted nuclei were washed once with the 500 µl of CSK containing 0.1% Triton X-100, chromatin was digested in 300 µl of CSK-buffer containing 0.1% Triton X-100 and 1000 U/ml of RNase-free DNaseI (Roche) at 25°C for 30 min. After digestion, soluble and insoluble materials were separated by low speed centrifugation.
Construction of expression vectors for mammalian cells
To make ptetHA, two oligonucleotides, 5′-CCGGT CGCCA CCATG GTGTA CCCAT ACGAC GTCCC AGACT ACGCT A-3′ and 5′-GATCT AGCGT AGTCT GGGAC GTCGT ATGGG TACAC CATGG TGGCG A-3′, were annealed and cloned into the AgeI–BglII sites of ptetGFP (32). HsMcm10 was amplified by PCR using the primers 5′-TAATA GATCT TGGAT GAGGA GGAAG ACAAT CTGTC-3′ and 5′-ATTAG GTACC CTTTA AGGCT GTTCA GAAAT TTAGC-3′, digested with BglII and KpnI, and then subcloned into the compatible sites of pSRhisA (33) to construct pSRhisA/HsMcm10. HsMcm10 was also amplified using the primers 5′-ATTAG TCGAC GATGA GGAGG AAGAC AATCT GTC-3′ and 5′-ATTAC CGCGG TTTTA AGGCT GTTCA GAAAT TTAGC ATG-3′, digested with SalI and SacII, and subcloned into the compatible sites of ptetHA. Sequence analysis confirmed that no mutations had been introduced in the amplified sequence during PCR.
Transfection and immunofluorescent staining
COS-1 cells, which are derived from the African green monkey kidney cell line CV-1 by transformation with an origin-defective SV40 virus, were cultured in DMEM supplemented with 10% fetal calf serum. Transfection was performed using the liposome-mediated method as described (32). Twenty-four hours after transfection, cells were fixed with ethanol:acetic acid (19:1) at –20°C for 10 min. In experiments where cells were first extracted, cells were washed twice with ice-cold CSK buffer, and then incubated with CSK buffer containing 0.5% Triton X-100 for 10 min at 4°C before fixation. Slides were processed for immunofluorescence staining as described previously (34). Anti-six-His monoclonal antibody was purchased from Novagen. FITC or Texas-red conjugated secondary antibodies were purchased from Vector Inc. Cells were visualized using the Olympus IX70 fluorescent microscope equipped with the 100× 1.35NA UPlanApo oil-immersion objective. Images were taken by a CCD camera and assembled using IP Lab software.
Yeast two-hybrid system
The yeast two-hybrid system was performed with the MATCHMAKER Two-Hybrid System (Clontech) according to the manufacturer’s protocol. To make the plasmids for the two-hybrid system, HsMcm10 cDNA was amplified with the primers 5′-GGAGA TCTCA ATTGA TGGAT GAGGA GGAAG ACAA-3′ and 5′-CCGGT ACCGT CGACT TTAAG GCTGT TCAGA AATT-3′. The product was digested with BglII, filled-in with Klenow, digested with SalI, and then subcloned into the EcoRI (also filled-in with Klenow) and XhoI sites of pB42AD. Human Orc2 cDNA (35) was amplified with the primers 5′-CCCAA TTGAT GAGTA AACCA GAATT AAA-3′ and 5′-GGCTC GAGCC TCCTC TTCTT CCTTT T-3′, digested with MunI and XhoI, and then subcloned into the EcoRI and XhoI sites of pLexA. Mouse Mcm2 cDNA (36) was amplified using the primers 5′-CCGGA TCCTG GCGGA GTCTT CTGAG TCT-3′ and 5′-GGCTC GAGGT ACCTA GAACT GCTGT AGGCT CAGT-3′, digested with BamHI and XhoI, and subcloned into the compatible sites of pLexA. Mouse Mcm3 cDNA (37) was amplified with the primers 5′-CCGCT AGCGA ATTCA TGGCG GGCAC AGTAG TGCTG-3′ and 5′-GGCTC GAGGT ACCTA GATAA GGAAG ACGAT GCCC-3′, digested with EcoRI and XhoI, and subcloned into the compatible sites of pLexA. Mouse Mcm4 cDNA (38) was amplified with the primers 5′-CCGCT AGCGA ATTCA TGTCG TCCCC GGCAT CCACC-3′ and 5′-GGCTC GAGGT ACCTA GAGCA GGCGG ACAGT CTTCC-3′, digested with EcoRI and XhoI, and subcloned into the compatible sites of pLexA. Mouse Mcm5 cDNA (38) was amplified with the primers 5′-CCAGA TCTTT CGAAT TCATG TCGGG CTTCG ACGAC C-3′ and 5′-CCCTC GAGAT CTACT TGAGG CGATA GAGCA CCTT-3′, digested with EcoRI and XhoI, and subcloned into the compatible sites of pLexA. Mouse Mcm6 cDNA (36) was amplified with the primers 5′-CCGGA TCCTG GACCT CGCAG CGGCC GCG-3′ and 5′-GGTGT ACAGT CGACT ATTGA CAGAG TGCCC AAG-3′, digested with BamHI and SalI, and subcloned into the BamHI and XhoI sites of pLexA. Mouse Mcm7 cDNA (39) was amplified with the primers 5′-CCGGA TCCGC GAATT CATGG CGCTT AAGGA CTACG CG-3′ and 5′-CCGGA TCCAT GGTAC CTAGA CAAAG GTGAT CCGTG TCC-3′, digested with EcoRI and BamHI, and subcloned into the compatible sites of pLexA.
Co-immunoprecipitation analysis
The expression plasmids pSRhisA/HsMcm10 and pSRα/HA–Orc2 were transfected into COS-1 cells by electroporation as described (40). After transfection, COS-1 cells were washed with PBS, scraped from the plates in PBS, centrifuged for 5 min, and resuspended in CSK buffer containing 0.1% Triton X-100 at 4°C for 30 min. Cells were centrifuged for 3 min at 3000 r.p.m. at 4°C and washed with CSK buffer containing 0.1% Triton X-100 six times. Chromatin was removed by digestion with 500 U/ml of RNase-free DNaseI (Roche) in CSK buffer containing 0.1% Triton X-100 at 25°C for 30 min.
Fifty micrograms of chromatin-bound fractions containing His–HsMcm10, HA–Orc2, or co-expressed His–HsMcm10 and HA–Orc2 were immunoprecipitated with 1 µl of anti-Mcm10 or anti-HA antibody which had been pre-absorbed to protein G–Sepharose (Pharmacia) for 2 h at 4°C in 200 µl of CSK buffer containing 0.1% Triton X-100. After washing with PK buffer (20 mM potassium phosphate, pH 7.5, 100 mM KCl, 0.1% NP-40, 1 mM EDTA, 10% glycerol), precipitates were dissolved with 15 µl of 2× Laemmli sample buffer, subjected to SDS–polyacrylamide gel electrophoresis (PAGE), and then transferred electrophoretically onto PVDF membranes. HsMcm10 and HA–Orc2 were detected by immunoblotting.
RESULTS AND DISCUSSION
Isolation of the cDNA of human MCM10 homolog
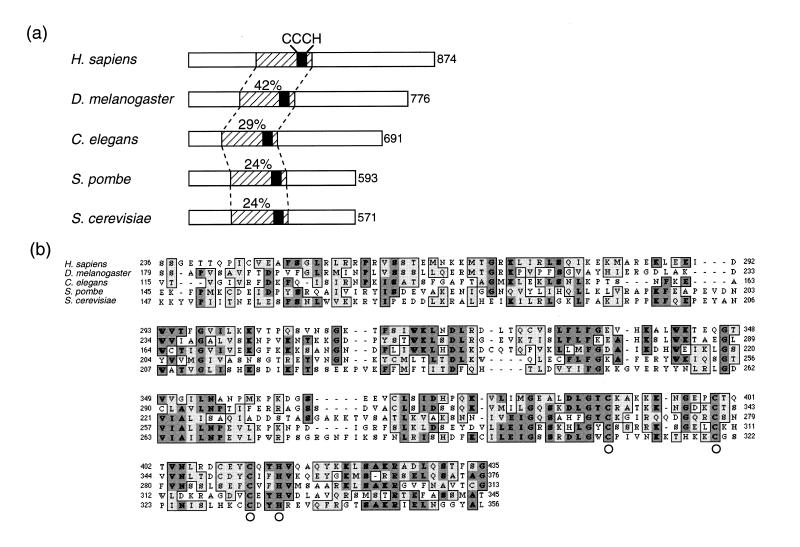
A database search identified a human EST clone (AA312197) as a potential homolog of the S.cerevisiae MCM10. This cDNA fragment was obtained by RT–PCR and used for screening a HeLa cDNA library. The longest human cDNA clone obtained (3721 bp) encoded a predicted protein of 874 amino acids with a calculated molecular mass of 98 kDa (Fig. 1a). The initiator methionine codon was preceded by an untranslated leader sequence that contained a stop codon in the same reading frame. The sequence surrounding the initiation codon matched with the Kozak consensus for eukaryotic translation initiation (41). The cDNA contained an AluI repetitive sequence as well as poly(A)+ signal in the 3′ non-coding region. Human Mcm10 (HsMcm10) was 28% identical and 42% similar to D.melanogaster Mcm10, 19% identical and 33% similar to C.elegans Mcm10, 12% identical and 23% similar to S.pombe Cdc23, and 11% identical and 21% similar to S.cerevisiae Mcm10. The central region of HsMcm10 (from amino acids 236 to 435) showed a high level of identity with the D.melanogaster homolog (42%), C.elegans homolog (29%), S.pombe homolog (24%) and S.cerevisiae homolog (24%) (Fig. 1b). This central homologous region, which contains the sequence resembling a zinc-finger motif (CX9-10-CX11-CX2H), previously noted in both S.cerevisiae MCM10 and S.pombe cdc23 gene products (27), appears conserved among all five species. In S.cerevisiae, the substitution of the second cysteine residue by proline [in the mcm10-43 (dna 43-1) mutant] suggests an important role of this motif in the biological function of Mcm10 (26).
Figure 1.
Comparison of Mcm10 proteins of different species. (a) Schematic diagram of the similarities among human (Homo sapiens), D.melanogaster, C.elegans, S.pombe and S.cerevisiae Mcm10 proteins. Striped boxes indicate the conserved central domain. Black boxes and CCCH indicate the zinc finger-like motif (CX9-10-CX11-CX2H). The amino acid sequence of the human Mcm10 central domain (200 residues between 236 and 435) showed 42% identity to the D.melanogaster protein, 29% identity to the C.elegans protein, and 24% identity to the central domains of both S.pombe and S.cerevisiae proteins, respectively. Numbers at the right of the open boxes represent the amino acid residue number. The DNA sequence of the human Mcm10 gene is available from the DDBJ/GenBank/EMBL database with the accession number AB042719. (b) Multiple sequence alignment of the central domain of the above sequences using the ClustalW program. Identical residues are indicated by dark gray boxes and bold type, and similar residues are indicated by light gray boxes. The zinc finger-like motif is indicated by circles. Numbers indicate the amino acid residues.
Schizosaccharomyces pombe cdc23 was able to complement both S.cerevisiae mcm10-43 (dna43-1) mutant (27) and mcm10-1 mutant (data not shown). As an initial characterization of HsMcm10, we tested whether HsMcm10 could complement a temperature-sensitive allele of Mcm10. Full-length HsMcm10 was subcloned into an expression vector and introduced into the temperature-sensitive mcm10-1, mcm10-43 (dna43-1) and cdc23–M36 strains. HsMcm10 did not functionally replace mutant Mcm10 either in budding yeast or in fission yeast (data not shown).
HsMcm10 mRNA expression during the cell cycle
In both HeLa and NB1–RGB cells, northern blot analysis revealed the presence of two HsMcm10 transcripts of 5.0 and 3.7 kb in a ratio of 2:1. Genomic Southern blot analysis demonstrated that the human genome contained a single copy of the HsMcm10 gene (data not shown). The different mRNA species may result from an alternative splicing event in the 3′ non-coding region. We were not successful in isolating the cDNA clone which contained the entire 3′ non-coding region. However, nine of the 13 isolated cDNA clones, which contained the entire open reading frame, did not contain any alternatively spliced form, although the probe used for library screening recognized two kinds of mRNA species. We also made five probes of ~200 bp which hybridized to the coding region at every 500 bp. All five probes detected two mRNA species by northern blot analysis (data not shown). Therefore, it is unlikely that the two different mRNA species result from alternative splicing within the coding region.
We examined whether mRNA expression of HsMcm10 was growth dependent. The mRNA levels of most proteins involved in DNA replication increase at the G1/S boundary after quiescent mammalian cells are stimulated by serum. NB1–RGB cells were arrested at the G0 phase by serum starvation and stimulated to proliferate by serum addition. The mRNA level was measured by northern blot hybridization at the indicated times after serum addition (Fig. 2). The mRNA level increased from 8 h after serum stimulation, reached maximum at 16 h and subsequently decreased. The ratio of the two mRNA species did not change throughout the cell cycle. This fluctuation is typical for DNA replication proteins, in accord with the notion that Mcm10 is necessary for DNA replication. As a positive control for the effect of serum stimulation, the mRNA level of PCNA, an auxiliary protein of DNA polymerase δ, was also examined and shown to increase before the synthesis of DNA.
Figure 2.
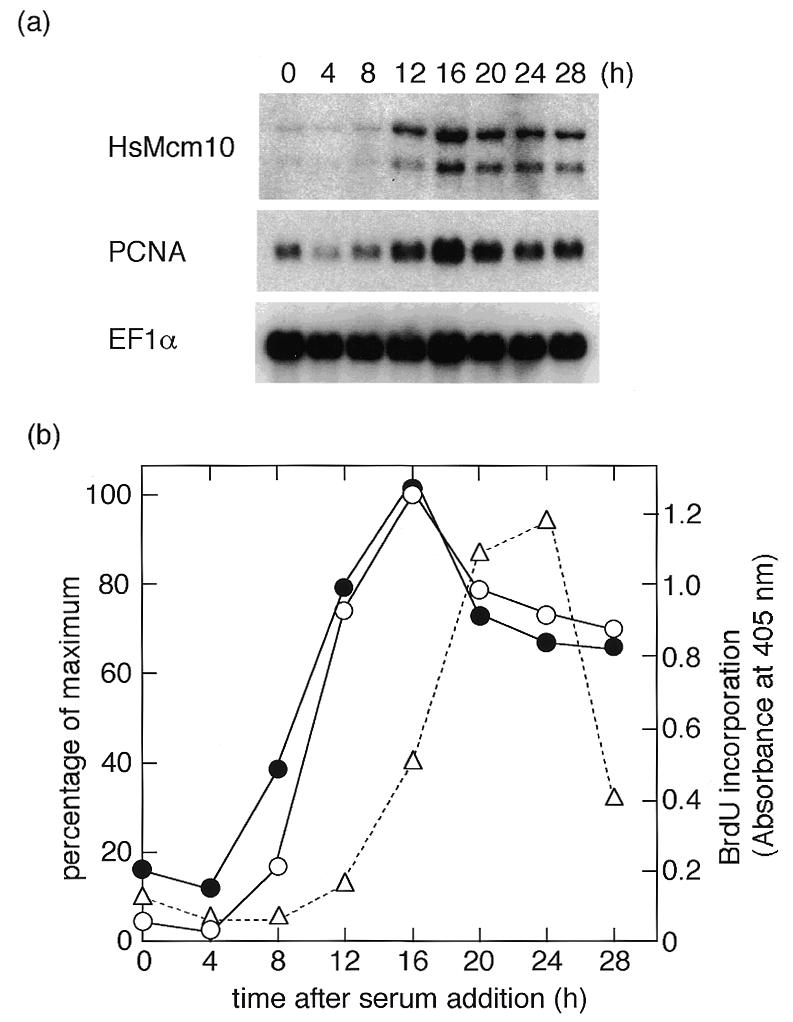
Expression of human Mcm10 mRNA after the growth stimulation. After cultured in DMEM containing 0.4% fetal calf serum for 72 h, NB1–RGB cells were released by changing the medium to DMEM containing 15% fetal calf serum. Two micrograms of poly(A)+ RNA was isolated at the indicated times and used for northern blotting. (a) The membrane blot was probed with human Mcm10 (HsMcm10), PCNA and EF1α cDNA probes. (b) The intensity of radioactivity was measured by BAS2500, and the results are presented as percentage of maximum. DNA synthesis was monitored by BrdU incorporation. Open and closed circles represent HsMcm10 and PCNA mRNA, respectively, and triangles represent the amount of incorporated BrdU. EF1α was used as a reference of the amount of RNA loaded.
HsMCM10 gene was found in the human genome database (accession number AL138764) and mapped on chromosome 10. The upstream sequence of HsMcm10 contained several potential E2F-binding motifs. The transcription of HsMcm10 may be regulated by E2F transcription factors at the G1/S boundary as shown for many other DNA replication proteins including dihydrofolate reductase, DNA polymerase α, Orc1, Mcm4 and Cdc6 (42).
Subcellular localization of the HsMcm10 in mammalian cells
To determine the subcellular localization of HsMcm10, we made the expression vector for HsMcm10. The entire coding region except for the first methionine codon was subcloned into the multiple cloning sites of pSRhisA, which is a mammalian expression vector carrying the SRα promoter as well as six-His-tag in the upstream region of the multicloning sites (33). COS-1 cells were transiently transfected with pSRhisA/HsMcm10, and 24 h after transfection, cells were fixed and stained with monoclonal antibody specific to the six-His-tag and FITC-conjugated secondary antibody. Ectopically expressed HsMcm10 protein was localized in the nuclei and formed a fine array of foci which were excluded from the nucleoli (Fig. 3a). These foci were resistant to detergent extraction as shown in Figure 3b. HsMcm10 was also subcloned into ptetHA, which contains the tetracycline-regulated promoter as well as HA epitope-tag upstream of the multicloning sites, and co-transfected with a tTA-expressing plasmid. HsMcm10 expressed by the ptetHA vector gave the same results (data not shown).
Figure 3.
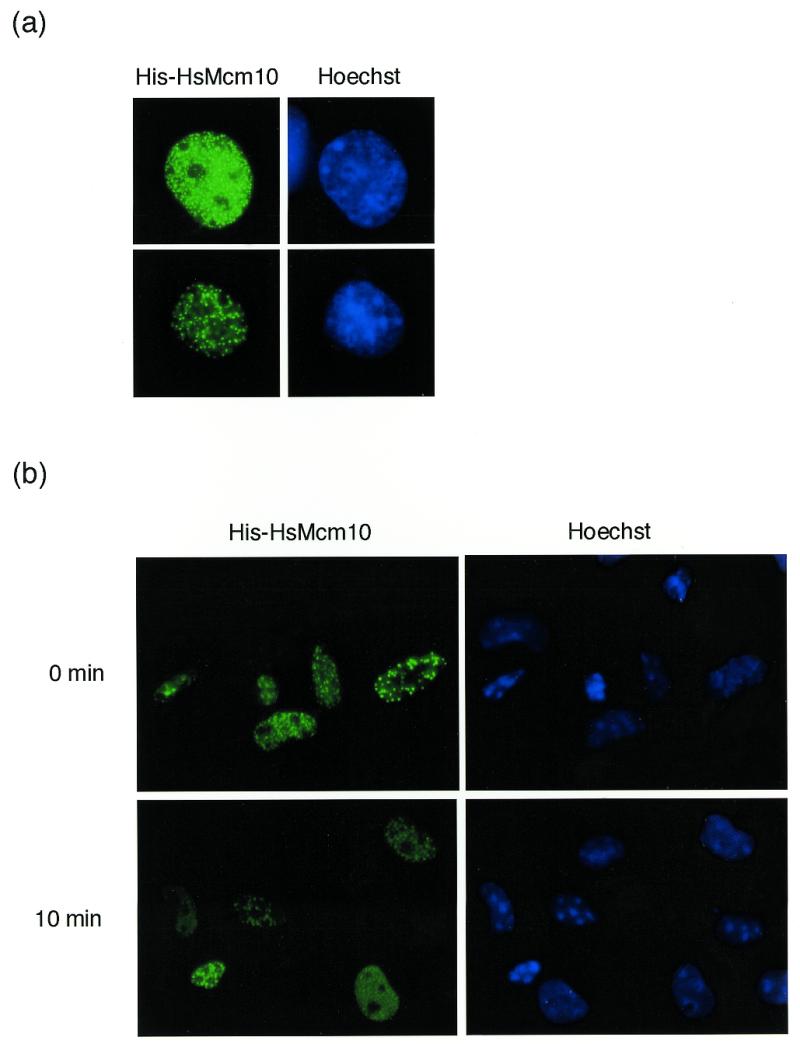
Localization of ectopically expressed HsMcm10 in COS-1 cells. (a) COS-1 cells were transiently transfected with the expression vector encoding the six-His-tagged HsMcm10 protein. Cells were fixed at 24 h after transfection, and the expression was detected by immunostaining with the monoclonal anti-six-His antibody and FITC-conjugated secondary antibody. Nuclear DNA was stained with 1 µg/ml of Hoechst 33258. (b) Twenty-four hours after transfection, cells were either fixed immediately or extracted with 0.5% Triton X-100 at 4°C for 10 min prior to fixation.
The resistance of immunostaining against detergent extraction suggests that HsMcm10 may be associated with chromatin. For further analysis of subcellular localization of HsMcm10, we raised a polyclonal rabbit antibody against the recombinant fragment of HsMcm10. The major protein band of 105 kDa, which was slightly larger than the predicted molecular mass, was detected by immunoblotting of HeLa total cell lysate (Fig. 4a, lane 1, indicated by an arrow). To confirm the specificity of the antibody, HA-tagged HsMcm10 was ectopically expressed by transient transfection in HeLa cells and detected by immunoblotting. Addition of HA-epitope to HsMcm10 produced a 120-kDa protein which was detected by the anti-HsMcm10 antibody (Fig. 4a, lanes 2 and 3, indicated by an arrowhead). The 120-kDa protein was also detected by the anti-HA antibody (data not shown). Therefore, we concluded that the antibody specifically detected HsMcm10 in human cell extract. The minor protein band of 86 kDa which was detected in HeLa total cell lysate (Fig. 4a, lane 1, indicated by an asterisk) seems to be a degradation product of HsMcm10. We observed similar degradation product of overexpressed HA-tagged HsMcm10 as shown in Figure 4a, lane 3 (indicated by a dot).
Figure 4.
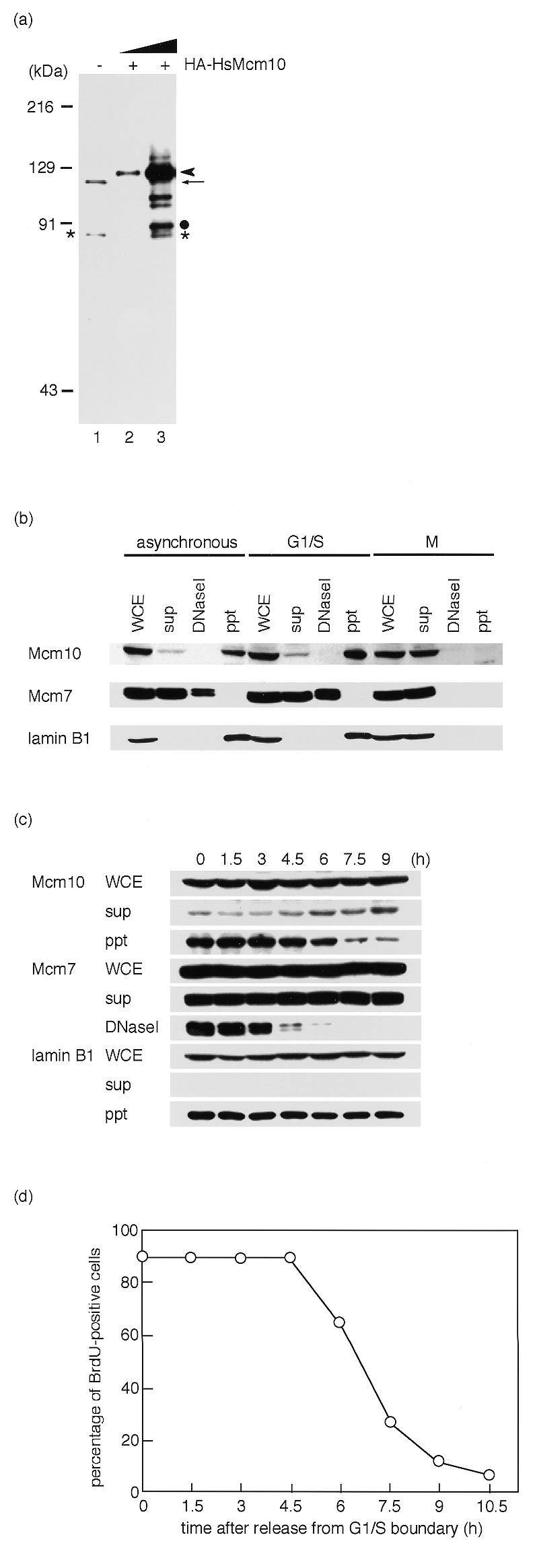
Subcellular distribution of endogenous HsMcm10 in HeLa cells. (a) Detection of HsMcm10 protein with a polyclonal antibody. Whole cell lysates from 2 × 104 HeLa cells were subjected to immunoblotting (lane 1). The arrow indicates the endogenous HsMcm10. The minor additional band positioned at 88 kDa indicated by an asterisk seems to be a degradation product of HsMcm10 protein. Whole cell lysates from 2 × 103 or 2 × 104 HeLa cells overexpressing HA-epitope tagged HsMcm10 were subjected to immunoblotting (lanes 2 and 3). The arrowhead indicates the HA–HsMcm10. The minor band positioned at 91 kDa indicated by a dot seems to be a degradation product of HA–HsMcm10. (b) Whole cell extracts (WCE), Triton X-100-soluble fractions (sup), chromatin-bound fractions solubilized with DNaseI digestion (DNaseI) and DNaseI-unextractable fractions (ppt) were prepared. HeLa cells were synchronized at M phase with nocodazole treatment. HeLa cells were arrested at G1/S boundary with aphidicolin treatment after replating mitotic cells. Each fraction from 1 × 105 cells was analyzed by immunoblotting. (c) HeLa cells were released from G1/S boundary. Cell extracts were prepared at indicated times and subjected to immunoblotting. (d) To monitor S phase, HeLa cells were pulse-labeled at the indicated times with BrdU, which was detected by fluorescent immunostaining.
HeLa whole cell extracts were fractionated into Triton X-100-soluble fractions, chromatin-bound fractions solubilized with DNaseI digestion, and DNaseI-unextractable fractions, and subcellular localization of HsMcm10 was investigated by immunoblotting (Fig. 4b). When extracts were prepared from asynchronously growing HeLa cells, HsMcm10 proteins were mainly localized in DNaseI-resistant fraction. About 10% of HsMcm10 was localized in Triton X-100-extractable fraction. To confirm that fractionation was performed properly, localization of Mcm7 and lamin B1 were investigated. As demonstrated previously, Mcm7 resolved into a doublet on SDS–PAGE. More than half of the Mcm7 proteins were free in soluble fractions in HeLa cells, while the rest of Mcm7 was bound to chromatin, which is consistent with the previous report by Fujita et al. (31). On the other hand, lamin B1 was localized in DNaseI-resistant structure.
Next we fractionated the lysates of HeLa cells arrested at the G1/S boundary or at M phase. Most HsMcm10 proteins were localized in DNaseI-resistant fraction at G1/S boundary, whereas HsMcm10 dissociated from DNaseI-resistant structure and became soluble at M phase (Fig. 4b). To determine whether HsMcm10 dissociates from nuclear structures during the progression of S phase in the way that Mcm2–7 complex does, HeLa cells were arrested at the G1/S boundary by aphidicolin treatment following mitotic shake-off, and then released in aphidicolin-free medium. HsMcm10 remained in DNaseI-resistant structure during S phase, whereas Mcm7 dissociated from chromatin during S phase progression (Fig. 4c and d). HsMcm10 started to dissociate from DNaseI-resistant structure 6 h after release. At 6 h after release, G2 phase cells, which had intact lamina structure and no BrdU incorporation, began to increase. M phase cells began to appear at 9 h after release. Therefore, HsMcm10 seems to dissociate from DNaseI-resistant nuclear structure in G2 phase.
HsMcm10 associated with other replication factors
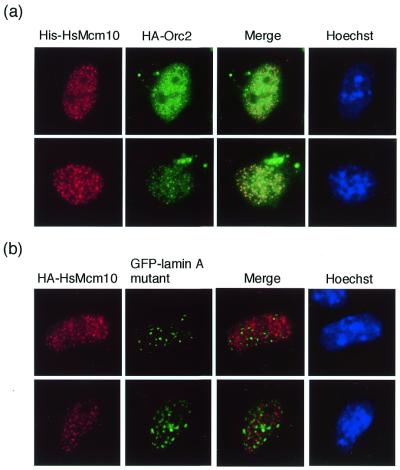
We speculated that Mcm10 might interact with other initiation factors, since the previous report demonstrated that Mcm10 is involved in initiating DNA replication in budding yeast (25). Therefore, the cDNAs for DNA replication factors were subcloned into the expression vector described above and co-transfected with pSRhisA/HsMcm10 or ptetHA/HsMcm10. When HsMcm10 was co-expressed with human Orc2, HsMcm10 was found to co-localize with not all, but some Orc2 in the foci (Fig. 5a), whereas HsMcm10 was not co-localized with PCNA, DNA polymerase α or p21 (data not shown). To confirm that the co-localization between HsMcm10 and Orc2 did not result from the non-specific aggregation from overexpression, the expression vector for GFP-tagged human lamin A mutant, which lacks the C-terminal CaaX sequence, was also introduced with ptetHA/HsMcm10 as a negative control. The CaaX motif is necessary for the efficient integration of lamin A protein into the nuclear lamina, whereas lamin A without the CaaX motif accumulates within nuclei as multiple foci when overexpressed (30,43). HsMcm10 was not co-localized with GFP-tagged lamin A mutant as shown in Figure 5b, suggesting that co-localization of HsMcm10 and Orc2 did not result from the non-specific aggregation caused by overexpression. Co-localization of Mcm10 and Orc was also observed in budding yeast (44).
Figure 5.
Co-localization of ectopically expressed HsMcm10 and Orc2 in COS-1 cells. The expression vector encoding the His-tagged HsMcm10 protein was co-transfected with either the vector expressing the HA-tagged Orc2 (a) or that expressing the GFP-tagged lamin A mutant (b). Cells were fixed at 24 h after transfection, and the six-His-tagged HsMcm10 protein was detected using the monoclonal anti-six-His antibody and the Texas red-conjugated secondary antibody. HA-tagged HsMcm10 and Orc2 were detected with the polyclonal anti-HA antibody and the FITC- or Texas red-conjugated secondary antibody.
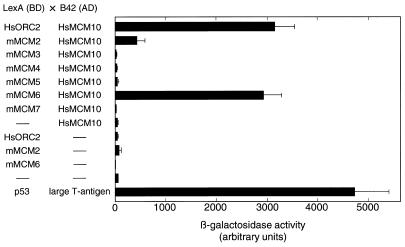
As an independent method for detection of the physical interaction between Mcm10 and Orc2, we used the two-hybrid system to examine whether similar interactions could occur in yeast cells. Full-length cDNAs encoding the HsMcm10 and human Orc2 genes were fused to the B42 transactivation domain (AD) and the N-terminal LexA DNA-binding domain (BD), respectively. These constructs were introduced in the same cell and β-galactosidase activity was measured. As shown in Figure 6, positive interaction was detected in cells co-expressing the HsMcm10 and Orc2 proteins. The β-galactosidase activity caused by HsMcm10 and Orc2 interaction was strong and was ~70% of the activity observed with control cells harboring simian virus 40 large T antigen in the AD vector and p53 protein in the BD vector. When the HsMcm10 and Orc2 cDNAs were switched between BD vector and AD vector, a similar strong signal was obtained (data not shown).
Figure 6.
Two-hybrid interactions between human Mcm10 (HsMCM10), human Orc2 (HsORC), and mouse Mcm2–7 (mMCM2–7). Interactions between pairs of fusion proteins containing either an N-terminal LexA DNA-binding domain (BD) or a B42 transactivation domain (AD) were tested in the yeast two-hybrid assay using the lacZ reporter gene. The reporter β-galactosidase activity was measured using chlorophenol red-β-d-galactopyranoside (CPRG) as the substrate. Each bar represents three independent measurements. Error bars indicate standard deviation. As a positive control, plasmids containing simian virus 40 large T antigen and p53 were used. As a negative control, BD and AD vectors containing no inserts were transfected.
The interaction between Mcm10 and five of the six Mcm2–7 proteins (Mcm2–4, 6 and 7) has been reported in budding yeast (26). The cDNAs for mouse Mcm2–7 were subcloned into the BD vector and cotransfected with HsMcm10 in the AD vector. Mcm2–7 proteins are well conserved between human and mouse, and their proteins are functionally equivalent and interchangeable when the Mcm2–7 complex was reconstituted in vitro (Z.You, personal communication). As shown in Figure 6, interactions between HsMcm10 and mouse Mcm2 and Mcm6 were observed. A similar interaction was observed when the cDNAs were switched between BD and AD vectors (data not shown). None of the other Mcm proteins interacted with HsMcm10. The β-galactosidase activity observed from the interaction between HsMcm10 and Mcm6 was strong and almost identical to the activity caused by HsMcm10 and Orc2. The β-galactosidase activity observed from the interaction between HsMcm10 and Mcm2 was only 10% of the activity detected in cells with simian virus 40 large T antigen and p53 protein (as the positive control), suggesting that the interaction was unstable in yeast. We did not detect an interaction between HsMcm10 and Mcm3, 4 or 7 in the two-hybrid system that was reported to occur in budding yeast (25). However, the inability to detect interactions in the two-hybrid system does not exclude the possibility that these proteins do not interact in vivo. The interaction between HsMcm10 and Mcm2, 3, 4 and 7 may require post-translational modification, for example.
To obtain further evidence of the association of HsMcm10 with replication proteins in vivo, immunoprecipitation was carried out. The plasmids expressing His–HsMcm10 and HA–Orc2 were transiently transfected into COS-1 cells, and HsMcm10 or Orc2 proteins were precipitated. Since HsMcm10 in the nuclease-resistant structure was insoluble and not suitable for immunoprecipitation, HsMcm10 in the Triton-extractable fraction was used for immunoprecipitation at first. However, both His–HsMcm10 and HA–Orc2 in the soluble fraction non-specifically bound to protein G–Sepharose beads, even in the absence of antibodies. Therefore, we used a minor fraction of His–HsMcm10 and HA–Orc2 which was released from DNaseI treatment. HA–Orc2 was co-precipitated with His–HsMcm10 when anti-HsMcm10 antibody was used (Fig. 7, lane 7). Because anti-HsMcm10 antibody reacted with human, mouse and monkey Mcm10 proteins (data not shown), a small amount of HA–Orc2 was precipitated by anti-HsMcm10 antibody via endogenous monkey MCM10 (Fig. 7, lane 6). Furthermore, His–HsMcm10 was co-precipitated with HA–Orc2 when anti-HA antibody was used (Fig. 7, lane 11), confirming that HsMcm10 and Orc2 associated each other in mammalian cells. On the other hand, the Mcm2–7 complex was not present in the precipitates in both cases, although there was a large amount of Mcm2–7 complex in the DNaseI-extractable fraction (data not shown). However, this result does not exclude the possibility that HsMcm10 interacts with the Mcm2–7 complex in vivo. The interaction with the Mcm2–7 complex may be unstable and/or transient. It is also possible that only a small fraction of Mcm2–7, which are close to the replication origins, for example, interacts with HsMcm10.
Figure 7.
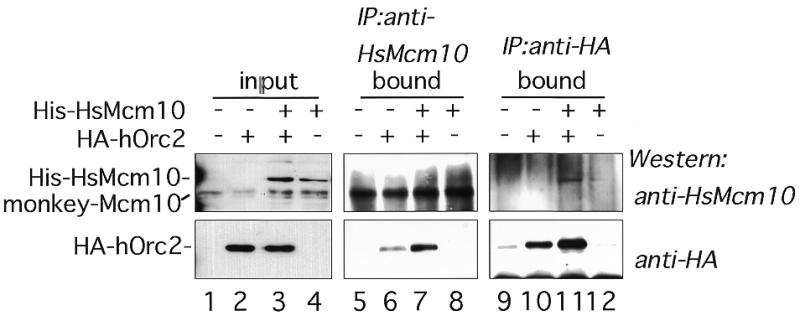
Immunoprecipitation assay with His–HsMcm10 and HA–Orc2. Fifty micrograms of the chromatin-bound fraction containing vector control (lanes 5 and 9), HA–Orc2 (lanes 6 and 10), coexpressed His–HsMcm10 and HA–Orc2 (lanes 7 and 11), or His–HsMcm10 (lanes 8 and 12) were immunoprecipitated with anti-HsMcm10 antibody (lanes 5–8) or anti-HA antibody (lanes 9–12). Precipitates were subjected to western blot analysis using anti-HsMcm10 (upper panels) and anti-HA (lower panels) antibodies. Lanes 1–4 contain 10 µg of the chromatin-bound fractions as input proteins.
CONCLUSION
We report here the identification of a human protein which is structurally homologous to S.cerevisiae Mcm10, after which we named this protein HsMcm10. The homology among the sequences of five species, the regulation of mRNA level, the subcellular localization, and the protein–protein interaction reported here strongly suggest that the protein we have identified is human Mcm10 protein. Several independent experiments reported here suggest that HsMcm10 may interact with Orc2 in vivo. Recently, Orc1 was also reported to be localized in nuclease-resistant nuclear structure (45). Therefore, HsMcm10 may have some role in the initiation step of DNA replication through the interaction with Orc. Since HsMcm10 dissociated from nuclear structure after the completion of DNA replication, HsMcm10 may be involved in the re-setting of pre-replicative complex. Several lines of evidence suggest that Cdc6 and Cdt1 lead to the formation of the pre-replication complex by supporting the loading of the Mcm2–7 complex onto chromatin associated with the Orc proteins (8,12,46,47). Except for an interaction between Orc1 and Cdc6 (15), direct interaction between Orc, Mcm2–7 and Cdc6 has not been reported. The possible interaction of HsMcm10 with the Mcm2–7 complex could provide an attractive hypothesis that Mcm10 might be another factor that recruits the Mcm2–7 complex onto the chromatin-bound Orc.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Toshihiko Eki for the expression vector of human Orc2 cDNA, Dr Hiroshi Kimura for the mouse Mcm2–7 cDNA, and Dr Shigeo Ohno for plasmid pSRhisA. This work was supported by grants from the Ministry of Education, Science, Sports and Culture of Japan.
DDBJ/EMBL/GenBank accession no. AB042719
REFERENCES
- 1.Bell S.P. and Stillman,B. (1992) Nature, 357, 128–134. [DOI] [PubMed] [Google Scholar]
- 2.Diffley J.F.X. and Cocker,J.H. (1992) Nature, 357, 169–172. [DOI] [PubMed] [Google Scholar]
- 3.Coleman T.R., Carpenter,P.B. and Dunphy,W.G. (1996) Cell, 87, 53–63. [DOI] [PubMed] [Google Scholar]
- 4.Romanowski P., Madine,M.A., Rowles,A., Blow,J.J. and Laskey,R.A. (1996) Curr. Biol., 6, 1416–1425. [DOI] [PubMed] [Google Scholar]
- 5.Rowles A., Tada,S. and Blow,J.J. (1999) J. Cell Sci., 112, 2011–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diffley J.F.X., Cocker,J.H., Dowell,S.J. and Rowley,A. (1994) Cell, 78, 303–316. [DOI] [PubMed] [Google Scholar]
- 7.Liang C., Weinreich,M. and Stillman,B. (1995) Cell, 81, 667–676. [DOI] [PubMed] [Google Scholar]
- 8.Cocker J.H., Piatti,S., Santocanale,C., Nasmyth,K. and Diffley,J.F.X. (1996) Nature, 379, 180–182. [DOI] [PubMed] [Google Scholar]
- 9.Rowles A., Chong,J.P.J., Brown,L., Howell,M., Evan,G.I. and Blow,J.J. (1996) Cell, 87, 287–296. [DOI] [PubMed] [Google Scholar]
- 10.Ishimi Y. (1997) J. Biol. Chem., 272, 24508–24513. [DOI] [PubMed] [Google Scholar]
- 11.Donovan S., Harwood,J., Drury,L.S. and Diffley,J.F.X. (1997) Proc. Natl Acad. Sci. USA, 94, 5611–5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tanaka T., Knapp,D. and Nasmyth,K. (1997) Cell, 90, 649–660. [DOI] [PubMed] [Google Scholar]
- 13.Aparicio O.M., Weinstein,D.M. and Bell,S.P. (1997) Cell, 91, 59–69. [DOI] [PubMed] [Google Scholar]
- 14.Drury L.S., Perkins,G. and Diffley,J.F.X. (1997) EMBO J., 16, 5966–5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saha P., Chen,J., Thome,K.C., Lawlis,S.J., How,Z., Hendricks,M., Parvin,J.D. and Dutta,A. (1998) Mol. Cell. Biol., 18, 2758–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bousset K. and Diffley,J.F.X. (1998) Genes Dev., 12, 480–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donaldson A.D., Fangman,W.L. and Brewer,B.J. (1998) Genes Dev., 12, 491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zou L. and Stillman,B. (1998) Science, 280, 593–596. [DOI] [PubMed] [Google Scholar]
- 19.Aparicio O.M., Stout,A.M. and Bell,S.P. (1999) Proc. Natl Acad. Sci. USA, 96, 9130–9135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zou L. and Stillman,B. (2000) Mol. Cell. Biol., 20, 3086–3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jares P. and Blow,J.J. (2000) Genes Dev., 14, 1528–1540. [PMC free article] [PubMed] [Google Scholar]
- 22.Mimura S. and Takisawa,H. (1998) EMBO J., 17, 5699–5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walter J. and Newport,J. (2000) Mol. Cell, 5, 617–627. [DOI] [PubMed] [Google Scholar]
- 24.Solomon N.A., Wright,M.B., Chang,S., Buckley,A.M., Dumas,L.B. and Gaber,R.F. (1992) Yeast, 8, 273–289. [DOI] [PubMed] [Google Scholar]
- 25.Merchant A.M., Kawasaki,Y., Chen,Y., Lei,M. and Tye,B.K. (1997) Mol. Cell. Biol., 17, 3261–3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Homesley L., Lei,M., Kawasaki,Y., Sawyer,S., Christensen,T. and Tye,B.K. (2000) Genes Dev., 14, 913–926. [PMC free article] [PubMed] [Google Scholar]
- 27.Aves S.J., Tongue,N., Foster,A.J. and Hart,E.A. (1998) Curr. Genet., 34, 164–171. [DOI] [PubMed] [Google Scholar]
- 28.Masutani C., Sugasawa,K., Yanagisawa,J., Sonoyama,T., Ui,M., Enomoto,T., Takio,K., Tanaka,K., van der Spek,P.J., Bootsma,D. et al. (1994) EMBO J., 13, 1831–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyazawa H., Izumi,M., Tada,S., Takada,R., Masutani,M., Ui,M. and Hanaoka,F. (1993) J. Biol. Chem., 268, 8111–8122. [PubMed] [Google Scholar]
- 30.Izumi M., Vaughn,A., Hutchison,C. and Gilbert,D.M. (2000) Mol. Biol. Cell, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fujita M., Yamada,C., Tsurumi,T., Hanaoka,F., Matsuzawa,K. and Inagaki,M. (1998) J. Biol. Chem., 273, 17095–17101. [DOI] [PubMed] [Google Scholar]
- 32.Izumi M. and Gilbert,D.M. (1999) J. Cell. Biochem., 76, 280–289. [DOI] [PubMed] [Google Scholar]
- 33.Akimoto K., Takahashi,R., Moriya,S., Nishioka,N., Takayanagi,J., Kimura,K., Fukui,Y., Osada,S., Mizuno,K., Hirai,S. et al. (1996) EMBO J., 15, 788–798. [PMC free article] [PubMed] [Google Scholar]
- 34.Mizuno T., Yamagishi,K., Miyazawa,H. and Hanaoka,F. (1999) Mol. Cell. Biol., 19, 7886–7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takahara K., Bong,M., Brevard,R., Eddy,R.L., Haley,L.L., Sait,S.J., Shows,T.B., Hoffman,G.G. and Greenspan,D.S. (1996) Genomics, 31, 119–122. [DOI] [PubMed] [Google Scholar]
- 36.Kimura H., Ohtomo,T., Yamaguchi,M., Ishii,A. and Sugimoto,K. (1996) Genes Cells, 1, 977–993. [DOI] [PubMed] [Google Scholar]
- 37.Thommes P., Fett,R., Schray,B., Burkhart,R., Barnes,M., Kennedy,C., Brown,N.C. and Knippers,R. (1992) Nucleic Acids Res., 20, 1069–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kimura H., Takizawa,N., Nozaki,N. and Sugimoto,K. (1995) Nucleic Acids Res., 23, 2097–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takizawa N., Kimura,H. and Sugimoto,K. (1995) Gene, 167, 343–344. [DOI] [PubMed] [Google Scholar]
- 40.Sorimachi H., Toyama-Sorimachi,N., Saido,T.C., Kawasaki,H., Sugita,H., Miyasaka,M., Arahata,K., Ishiura,S. and Suzuki,K. (1993) J. Biol. Chem., 268, 10593–10605. [PubMed] [Google Scholar]
- 41.Kozak M. (1996) Mamm. Genome, 7, 563–574. [DOI] [PubMed] [Google Scholar]
- 42.Dyson N. (1998) Genes Dev., 12, 2245–2262. [DOI] [PubMed] [Google Scholar]
- 43.Mical T.I. and Monteiro,M.J. (1998) J. Cell Sci., 111, 3471–3485. [DOI] [PubMed] [Google Scholar]
- 44.Kawasaki Y., Hiraga,S. and Sugino,A. (2000) Genes Cells, in press. [Google Scholar]
- 45.Tatsumi Y., Tsurimoto,T., Shirahige,K., Yoshikawa,H. and Obuse,C. (2000) J. Biol. Chem., 275, 5904–5910. [DOI] [PubMed] [Google Scholar]
- 46.Maiorano D., Moreau,J. and Mechali,M. (2000) Nature, 404, 622–625. [DOI] [PubMed] [Google Scholar]
- 47.Nishitani H., Lygerou,Z., Nishimoto,T. and Nurse,P. (2000) Nature, 404, 625–628. [DOI] [PubMed] [Google Scholar]