Abstract
We had recently reported that the mymA operon (Rv3083 to Rv3089) of Mycobacterium tuberculosis is regulated by AraC/XylS transcriptional regulator VirS (Rv3082c) and is important for the cell envelope of M. tuberculosis. In this study, we further show that a virS mutant (MtbΔvirS) and a mymA mutant (Mtbmym::hyg) of M. tuberculosis exhibit reduced contents and altered composition of mycolic acids along with the accumulation of saturated C24 and C26 fatty acids compared to the parental strain. These mutants were markedly more susceptible to major antitubercular drugs at acidic pH and also showed increased sensitivity to detergent (sodium dodecyl sulfate) and to acidic stress than the parental strain. We show that disruption of virS and mymA genes impairs the ability of M. tuberculosis to survive in activated macrophages, but not in resting macrophages, suggesting the importance of the mymA operon in protecting the bacterium against harsher conditions. Infection of guinea pigs with MtbΔvirS, Mtbmym::hyg, and the parental strain resulted in an ∼800-fold-reduced bacillary load of the mutant strains compared with the parental strain in spleens, but not in the lungs, of animals at 20 weeks postinfection. Phenotypic traits were fully complemented upon reintroduction of the virS gene into MtbΔvirS. These observations show the important role of the mymA operon in the pathogenesis of M. tuberculosis at later stages of the disease.
Mycobacterium tuberculosis, the etiological agent of extremely serious human infections, is a highly successful intracellular pathogen because it can adapt itself to various hostile environments. The cell envelope of mycobacteria is known to play a major role in their virulence and resistance to hostile environments. Besides, interaction of the mycobacterial cell envelope with host cell receptors facilitates uptake of the bacterium and modulation of host immune responses (12). Mycobacterium tuberculosis has approximately 250 genes involved in its lipid metabolism, and the lipid contents of the pathogen contribute to 60% of the cell dry weight (10, 12). A number of genes involved in the synthesis of essential components of the cell envelope for maintaining appropriate cell wall architecture of M. tuberculosis have been identified, and their requirement for the virulence of M. tuberculosis has been established, suggesting their importance as targets for the development of new antitubercular drugs (4, 8, 19, 22, 57). Genes responsible for the biosynthesis of oxygenated mycolic acids and cyclopropanated mycolic acids were shown to be important for the in vivo growth and persistence of M. tuberculosis (19, 22, 57). The gene cluster involved in the biosynthesis of major complex lipids phthiocerol and phenolphthiocerol dimycocerosates of M. tuberculosis have been shown to play an important role in its virulence (11). Induction of various genes involved in fatty acid metabolism in caseous granuloma (9) or human macrophages (20) or after exposure to sodium dodecyl sulfate (SDS) (37, 57) or acidic stress (21) suggests the importance of fatty acids in maintaining appropriate cell envelope structure for the survival of M. tuberculosis upon its exposure to various stressful conditions encountered in the host. Various reports have suggested that the macrophage environment damages the lipid-rich cell surface of M. tuberculosis (37, 52). However, the survival of M. tuberculosis inside the phagosome and upregulation of a number of genes involved in the modification of the cell envelope clearly suggest the importance of remodeling the cell envelope in the intracellular adaptation of the pathogen (57). The down regulation of the FAS II operon of M. tuberculosis at acidic pH (21), alterations in the mycolic acid composition of Mycobacterium smegmatis upon exposure to environmental stresses like temperature (33), and the requirement of cyclopropanated mycolic acids to counter oxidative stress (67) further suggest the importance of remodeling the cell envelope for adaptation of the pathogen under stressful conditions.
We had recently identified mymA operon (named after the first gene of the operon, Rv3083, which is a homologue of several monooxygenases present in the M. tuberculosis genome and thus designated mymA, i.e., mycobacterial monooxygenase) of M. tuberculosis, which is organized divergently to virS, which acts as a transcriptional regulator of this operon. We had shown that the promoter of mymA operon is induced in macrophages and upon exposure to acidic pH (58). Also, induction of the mymA operon by VirS is important for maintaining appropriate cell envelope structure of M. tuberculosis (58). In this study, we show that the mymA operon is required for maintaining the appropriate mycolic acid composition and permeability of the M. tuberculosis envelope on its exposure to acidic pH. The functional loss of the mymA operon of M. tuberculosis resulted in increased drug sensitivity and killing of the pathogen by activated macrophages and its reduced ability to persist specifically in the spleen of infected guinea pigs. Complementation of the virS mutant of M. tuberculosis with a functional copy of the virS gene resulted in restoration of wild-type phenotype.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
M. tuberculosis Erdman, the parental strain, mutant strains (MtbΔvirS and Mtbmym::hyg) and a virS-complemented strain (MtbΔvirS_virS) were grown in Middlebrook (MB) 7H9 broth (Difco Laboratories) supplemented with 0.5% glycerol, 0.2% Tween 80, and 1× ADC (albumin-dextrose complex; Difco Laboratories) or MB 7H10 medium (Difco Laboratories) supplemented with 1× OADC (oleic acid-albumin-dextrose complex; Difco Laboratories). Escherichia coli DH5 α and HB101 strains were grown in Luria-Bertani (LB) broth. Bacteria were cultured at 37°C with shaking at 200 rpm, and, whenever appropriate, antibiotics were added at the following concentrations: ampicillin, 50 μg/ml for E. coli; kanamycin, 25 μg/ml for E. coli and M. tuberculosis; hygromycin B, 150 μg/ml for E. coli and 50 μg/ml for M. tuberculosis.
Recombinant DNA techniques.
Mycobacterial genomic DNA was extracted as described previously (32). Molecular cloning, digestion with restriction endonucleases, and Southern blot hybridization were carried out by standard techniques (54). Restriction endonucleases and other DNA-modifying enzymes (New England Biolabs Inc.) were used according to manufacturer's recommendations.
Disruption of the mymA gene of M. tuberculosis.
The parent plasmid pSG10 (24), which carries the entire open reading frame of mymA along with upstream and downstream flanking sequences, was employed for the construction of nonreplicative vector p10mym:hk. Plasmid pSG10 was digested with BglII, which cleaves the mymA open reading frame at a unique site at its center. This BglII site was used to clone the hygromycin resistance gene in the mymA open reading frame. The entire hyg resistance cassette for this purpose was excised as a 1.9-kb BamHI fragment from the plasmid p10ΔvirShyg (58) and ligated with BglII-digested pSG10 vector to generate p10mym:hyg. This construct was further modified by cloning the kanamycin resistance gene in the vector backbone. The kanamycin resistance gene was excised out from pSD5 (15) as an NheI-BstEII fragment, end repaired, and cloned into DraI-digested p10mym:hyg, resulting in p10mym:hk.
This plasmid was pretreated with UV radiation (26, 47) and electroporated into M. tuberculosis. The hygromycin-resistant and kanamycin-sensitive transformants, resulting from double crossover, were selected on MB 7H10 agar plates. Loss of mymA (Rv3083) was confirmed by Southern blot and immunoblot analysis using genomic DNA and cell extracts prepared from the wild-type and mymA mutant of M. tuberculosis (Mtbmym::hyg).
Construction of the virS complementation strain.
For complementation studies, the complete virS gene along with its upstream sequences containing virS promoter region was obtained from pSG10 (24) by digesting it with EcoRV-SmaI. This fragment was cloned into the EcoRV site of mycobacteria-E. coli shuttle vector pSD5 (15). The resulting plasmid, pSD5virS, was electroporated into the virS mutant strain of M. tuberculosis (MtbΔvirS) to generate MtbΔvirS_virS. Expression of virS in the MtbΔvirS_virS strain was confirmed by immunoblot analysis using polyclonal antibodies raised against VirS in rabbit.
Electron microscopy.
Transmission electron microscopy was performed as described previously (58).
Isolation, derivatization, and analysis of fatty acids.
The total fatty acids were extracted from various strains of M. tuberculosis grown to an A600 of 1.5 and derivatized to UV-absorbing p-bromophenacyl esters as described previously (7). The separation of fatty acid derivatives was accomplished on a C18 octadecylsilyl reversed-phase cartridge column (250 by 10 mm, with 5-μm-diameter spherical particles). A solvent gradient of increasing concentrations of p-dioxane in acetonitrile, comprising five linear segments (0 to 10 min, 0% p-dioxane; 10 to 20 min, 0 to 5% p-dioxane; 20 to 40 min, 5 to 50% p-dioxane; 40 to 90 min, 50 to 70% p-dioxane; 90 to 100 min, 70 to 50% p-dioxane and constantly at 50% p-dioxane) was run with a flow rate of 1 ml/min. Detection was carried out by monitoring absorbance at 260 nm. The peak produced by the high-molecular-weight standard (Ribi ImmunoChem Research) was used as a reference peak to adjust and calculate the relative retention time (RRT). Percent peak height for each mycolic acid peak was determined as described previously (51). Individual mycolic acid peaks were labeled according to their RRT as determined from multiple runs of mycolic acid samples.
Mycolic acid and fatty acid profiles of M. tuberculosis strains grown at acidic pH.
The total fatty acids of various strains of M. tuberculosis growing at either neutral (7.0) or acidic (5.0) pH were radiolabeled using 1 μCi/ml of 1,2-14C acetate (50 to 62 mCi/mmol [1.85 to 2.29 GBq/mmol]; Amersham). The radiolabeled fatty acids and mycolic acids were extracted, methylated, and analyzed by thin-layer chromatography (TLC) using silica gel plates (Silica gel 60F254; Merck) as described previously (31). The intensities of radioactive fatty acid methyl esters (FAMEs) and mycolic acid methyl esters (MAMEs) were detected and compared using the phosphorimager (Fuji BAS5000). Radioactive FAMEs and MAMEs were also recovered by chloroform from the TLC plate as described previously (30) and subjected to a β scintillation counter for determining the radioactive counts per minute.
Miscellaneous analytical techniques.
For matrix-assisted desorption ionization-time of flight (MALDI-TOF) analysis, MAMEs were separated from FAMEs on preparative silica gel-coated TLC plates (20 cm by 20 cm) by developing in hexane ethyl acetate (95:5) followed by visualization using iodine vapors. MALDI-TOF spectra (in the positive mode) of purified MAMEs were acquired on a Bruker ultraflex MALDI-TOF equipped with a pulsed nitrogen laser emitting at 337 nm in the Reflectron mode as described previously (18).
For gas chromatography, equivalent amounts of FAMEs or MAMEs derived from various strains of M. tuberculosis were injected into a Varian 3800 apparatus equipped with an HP-5 column (30 m by 0.25 mm by 0.25 μm) using nitrogen as carrier gas (flow rate, 1 ml/min). The temperature separation program involved an increase from 150°C to 310°C at the rate of 5°C/min, followed by 20 min at 310°C. The injector temperature for analysis was kept at 250°C for FAMEs and 350°C for MAMEs (for thermal cleavage of MAMEs; pyrolytic gas chromatography [GC]). The fatty acid peaks were identified by comparing retention times with authentic fatty acid methyl ester standards (Sigma). The relative percentages of fatty acid peaks were determined by measuring total peak height, and the percentage of each peak was calculated relative to total peak height.
Drug, detergent, and pH susceptibility/sensitivity assays.
The MIC99s of various antitubercular drugs were determined in triplicate by the serial dilution plate method as described previously (68). Similarly, the antibiotic sensitivities of various strains at acidic pH were determined by using modified MB 7H10 medium adjusted to pH 5.0 as described previously (25). The concentration of antitubercular drugs that resulted in three or fewer colonies was taken as MIC99 and was determined by conducting three independent experiments.
Sensitivity of various M. tuberculosis strains to SDS was determined as described by Camacho et al. (8).
To study the effect of acidic pH, cultures were exposed to pH 4.0 either directly (without preadaptation) or after adaptation at pH 5.0 for 16 to 18 h and percent survival of bacilli was calculated after exposure to pH 4.0 for various time points.
Preparation of M. tuberculosis cells for virulence studies.
Various strains of M. tuberculosis were processed for virulence studies as described previously (59).
In vitro infection of mouse macrophages by M. tuberculosis.
Survival of mycobacterial strains in J774A.1 mouse macrophage cells resting or activated with recombinant gamma interferon (50 U/ml, 16 h) was studied as described earlier (59).
Studies of virulence in guinea pigs.
Virulence of various strains of M. tuberculosis was evaluated in guinea pigs as described earlier (44, 59).
Statistical methods.
The Student's unpaired t test was applied for statistical evaluation of the data assuming normal distribution. A P value of ≤0.05 was considered as a statistically significant observation for a parameter.
RESULTS
Disruption of the mymA gene of M. tuberculosis.
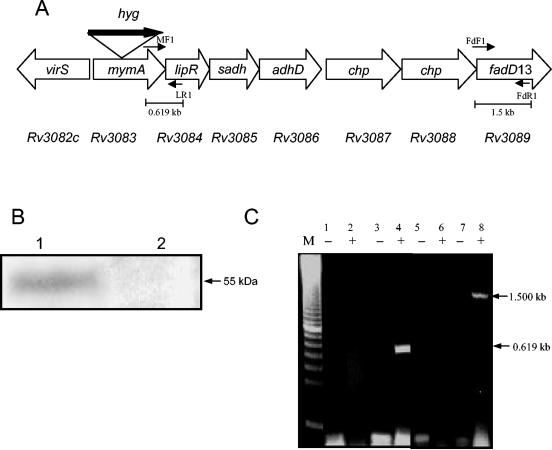
We have carried out disruption of mymA (Rv3083), which is the first open reading frame of the mymA operon, by homologous recombination using nonreplicative vector p10mym:hk (Fig. 1). Electroporation of UV-pretreated p10mym:hk into M. tuberculosis resulted in 130 hygromycin-resistant colonies. Allelic exchange by double crossover should specifically result in the integration of a disrupted mymA allele (mymA::hyg) and not the vector backbone (containing a kanamycin resistance cassette). Further screening of hygromycin-resistant colonies (Hygr) for the kanamycin sensitivity (Kans) phenotype resulted in isolation of two colonies with the desired Hygr Kans phenotype. The disruption of mymA was confirmed in one Hygr Kans colony by Southern blot analysis using the genomic DNA from M. tuberculosis and its mymA mutant strain (Mtbmym::hyg) (data not shown) and by immunoblot analysis using polyclonal antibodies raised in rabbit against MymA. The wild-type strain showed a specific protein band of 55 kDa corresponding to MymA (Fig. 1B, lane 1), which was absent in the mutant strain (Fig. 1B, lane 2). mymA represents the first gene of the mymA operon (Rv3083 to Rv3089). The disruption of the mymA gene by insertion of the hygromycin resistance (Hygr) cassette resulted in the termination of transcription at the end of the hygromycin resistance gene (due to the presence of the trancriptional terminator). Hence, the downstream genes of the mymA operon Rv3084 to Rv3089 were not transcribed. This was confirmed by reverse transcription (RT) analysis. No amplified products were obtained by RT-PCR with primers spanning the intergenic region of mymA and lipR (Rv3084) or fadD13 (Rv3089) when the RNA isolated from Mtbmym::hyg was used, whereas both the primer pairs yielded an RT-PCR product of the predicted size when RNA from M. tuberculosis was employed (Fig. 1C). These results confirmed that the disruption of mymA resulted in functional inactivation of the mymA operon.
FIG. 1.
(A) Disruption of the mymA (Rv3083) gene of M. tuberculosis. Shown is the schematic organization of the mymA operon of M. tuberculosis with a disrupted mymA (Rv3083) gene. Thick arrow, hygromycin resistance gene, which was used both as a disruption element and as a selection marker. MF1-LR1 and FdF1-FdR1 represent primer pairs used to confirm the effect of disruption on the transcription of downstream genes. Predicted sizes of the amplicons using the primer pairs MF1-LR1 and FdF1-FdR1 are shown. (B) Expression of mymA in M. tuberculosis and Mtbmym::hyg. The cell extract (50 μg protein) from M. tuberculosis (lane 1) and Mtbmym::hyg (lane 2) was resolved on a 10% polyacrylamide gel containing 0.1% SDS and transferred to a Hybond C Extra membrane. The blot was probed using polyclonal antibodies raised in rabbit against MymA. (C) Influence of disruption of mymA on the expression of downstream genes of the mymA operon. RT-PCR products corresponding to mymA-lipR intergenic region and the fadD13 gene were amplified using primer sets MF1-LR1 (lanes 1 to 4) and FdF1-FdR1 (lanes 5 to 8), respectively. In the negative control (−), the PCRs were performed directly on the total RNA isolated from Mtbmym::hyg (lanes 1 and 5) and M. tuberculosis (lanes 3 and 7) to confirm the absence of DNA contamination. RT-PCR was performed on the RNA obtained from Mtbmym::hyg (MF1-LR1, lane 2; FdF1-FdR1, lane 6) and M. tuberculosis (MF1-LR1, lane 4; FdF1-FdR1, lane 8). The size of the PCR product is consistent with the predicted size shown in the figure. M, 100-bp marker (U.S. Biochemicals).
Cell wall ultrastructure of the mymA mutant of M. tuberculosis.
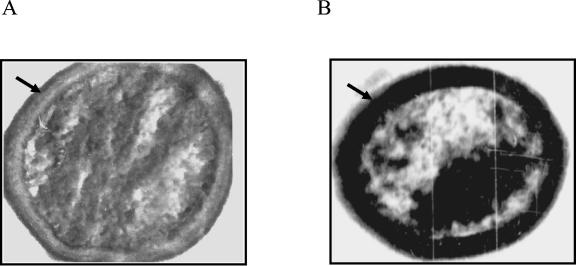
We have previously shown that expression of the mymA operon requires the presence of VirS and that the virS mutant of M. tuberculosis (MtbΔvirS) exhibits altered cell wall ultrastructure (58). Thus, to confirm that the observed phenotype of MtbΔvirS was due to abolished expression of mymA operon, the cell wall of an mymA mutant (Mtbmym::hyg) was analyzed by transmission electron microscopy (TEM) as described previously (58). We observed significant alterations in the cell wall of the Mtbmym::hyg strain, which were identical to the alterations observed in the case of the virS mutant of M. tuberculosis (MtbΔvirS) (Fig. 2) (58). This confirmed that the disruption of mymA or virS results in similar phenotypes. A dense staining of the electron-transparent zone (ETZ) observed in the case of MtbΔvirS and Mtbmym::hyg cells due to compromised permeability of the cell wall is suggestive of alterations in the mycolic acids present in the lipid bilayer (65).
FIG. 2.
Effect of mymA disruption on the cell wall ultrastructure of M. tuberculosis. The cell wall of M. tuberculosis and Mtbmym::hyg was examined by transmission electron microscopy. Cells were cultured in MB 7H9 medium to an A600 of 1.5, harvested, washed with phosphate buffer, and fixed in paraformaldehyde solution for 4 to 8 h. Cells were prepared for electron microscopy as described earlier (58). Shown is electron microscopic analysis (×44,000) of (A) M. tuberculosis and (B) Mtbmym::hyg. The photomicrographs shown are typical of the population as a whole, as judged by viewing many independent fields.
Influence of virS and mymA disruption on the mycolic acid profile of M. tuberculosis.
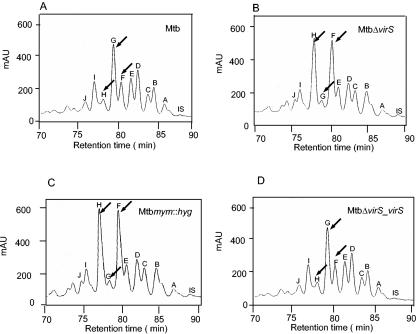
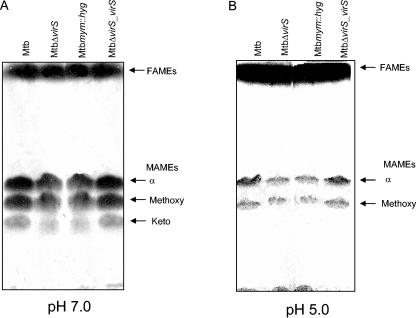
To investigate if the observed changes in the staining pattern of the ETZ were associated with alterations in the mycolic acids, we compared the profiles of mycolic acids extracted from the MtbΔvirS, Mtbmym::hyg, virS-complemented (MtbΔvirS_virS), and parental strains of M. tuberculosis by high-pressure liquid chromatography (HPLC). Strains were grown in 7H9 medium, and extreme care was taken to maintain the medium composition and culture conditions exactly the same for culturing various strains. As shown in Fig. 3, there were clear differences in the profiles of mycolic acids isolated from the MtbΔvirS and Mtbmym::hyg strains when compared with the parental strain of M. tuberculosis. While the parental strain showed peak G as the most prominent peak, in the case of the MtbΔvirS and Mtbmym::hyg strains, peaks H and F were most prominent, as determined by calculating relative percent peak height (Fig. 3). Reintroduction of wild-type virS gene into the MtbΔvirS strain fully restored its mycolic acid profile to the normal profile as exhibited by the parental strain (Fig. 3D). Furthermore, TLC analysis followed by radioactive quantification revealed alterations in the FAMEs and MAMEs of the mutants compared to parental strain of M. tuberculosis. Approximately 30% ± 2.9% reduction and 10% ± 2% accumulation of radioactivity associated with the MAME and FAME fractions of MtbΔvirS and Mtbmym::hyg, respectively, compared to the parental strain were found (Fig. 4A). These differences in the MAMEs and FAMEs isolated from MtbΔvirS and Mtbmym::hyg strains were fully complemented in the MtbΔvirS_virS strain of M. tuberculosis.
FIG. 3.
Representative HPLC chromatograms of mycolic acids from various strains of M. tuberculosis. M. tuberculosis MtbΔvirS, Mtbmym::hyg, and MtbΔvirS_virS strains were inoculated separately in MB 7H9 medium and grown to an A600 of 1.5. Total fatty acids were extracted, derivatized to UV-absorbing p-bromophenacyl esters, and separated by HPLC as described in Materials and Methods. Shown are mycolic acid profiles of (A) M. tuberculosis, (B) MtbΔvirS, (C) Mtbmym::hyg, and (D) MtbΔvirS_virS from HPLC analysis. Peaks were labeled according to their RRTs. IS, internal high-molecular-weight standard. Arrows indicate the significant differences in the mycolic peaks between various strains.
FIG. 4.
Thin-layer chromatography of FAMEs and MAMEs from various strains of M. tuberculosis. M. tuberculosis, MtbΔvirS, Mtbmym::hyg, and MtbΔvirS_virS strains were grown to an A600 of 1.5 in MB 7H9 medium, adjusted either to pH 7.0 or to pH 5.0, and labeled with 1 μCi/ml [1,2-14C]acetate for 12 h. Labeled lipids were extracted, derivatized to methyl esters, and resolved by normal phase TLC using petroleum ether/diethyl ether (17:3). Equal counts (200,000 cpm) were loaded in each lane, and a phosphorimager was used to quantify the proportion of FAMEs and MAMEs derived from various strains of M. tuberculosis. Shown are TLC profiles of total fatty acid esters (FAMEs and MAMEs) extracted from M. tuberculosis, MtbΔvirS, Mtbmym::hyg, and MtbΔvirS_virS strains growing at (A) neutral pH (7.0) or (B) acidic pH (5.0). These experiments were repeated four times, and similar results were obtained.
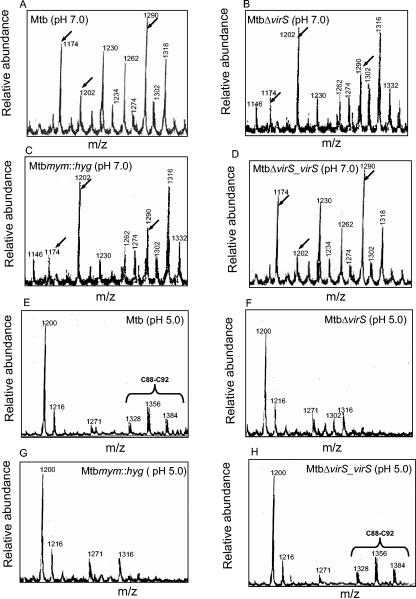
MALDI-TOF analysis of MAMEs from various strains of M. tuberculosis growing at pH 7.0 showed no qualitative difference in the mycolic acids; however, there were quantitative differences in mycolic acid species produced by parental and mutant strains. While, in the case of mutant strains MtbΔvirS and Mtbmym::hyg, the most prominent mycolic acid peaks were represented by masses 1202 and 1316, in the case of parental strain the most prominent mycolic acid peaks were represented by masses 1174, 1230, 1262, 1290, and 1318 (Fig. 5). The virS-complemented strain (MtbΔvirS_virS) showed a distribution of mass intensities indistinguishable from the pattern observed in the parental strain (Fig. 5D). Thus, the data from four independent analytical techniques (TEM, HPLC, TLC, and MALDI-TOF) confirm the role of the mymA operon in maintaining appropriate cell wall structure and mycolic acid composition of M. tuberculosis.
FIG. 5.
MALDI-TOF mass spectra of MAMEs from various strains of M. tuberculosis. M. tuberculosis, MtbΔvirS, Mtbmym::hyg, and MtbΔvirS_virS strains were grown in MB 7H9 medium at pH 7.0 or exposed to pH 5.0 for 12 h. MAMEs were extracted and subjected to MALDI-TOF analysis as described in Materials and Methods. MALDI-TOF mass spectra of MAMEs from M. tuberculosis (A and E), MtbΔvirS (B and F), Mtbmym::hyg (C and G), and MtbΔvirS_virS (D and H) are shown. The psuedomolecular masses of mycolates are indicated on each mass spectrum. Arrows and brackets indicate the major differences observed in MALDI-TOF analysis of MAMEs from various strains.
Influence of acidic stress on the mycolic acid profile of M. tuberculosis.
In our earlier study, based on the up regulation of the mymA operon at acidic pH, we had suggested its role in the modification of the cell envelope of M. tuberculosis in response to acidic stress. Since disruption of the mymA operon resulted in alterations in the mycolic acids of M. tuberculosis, we further analyzed the role of this operon in the modification of mycolic acids upon exposure to acidic stress. Alteration of growth conditions from pH 7.0 to pH 5.0 resulted in a general decline in MAMEs and consequent increase in FAMEs in both the mutants as well as the parental strains (Fig. 4B). However, the amount of MAMEs in the mutant strains (MtbΔvirS and Mtbmym::hyg) was 50% ± 6.4% lower compared to the parental strain. In contrast, the amount of FAMEs in these mutant strains was 20% ± 3.1% higher compared to the parental strain (Fig. 4B).
Further, we carried out MALDI-TOF analysis of the MAMEs extracted from various strains of M. tuberculosis exposed to acidic pH. As shown in Fig. 5, exposure to acidic pH resulted in general suppression of most of the psuedomolecular ion [M+Na+] peaks observed at neutral pH. In addition to the common peaks observed in all strains of M. tuberculosis, in the parental and MtbΔvirS_virS strains, few psuedomolecular masses unique to acidic pH were observed; these included m/z 1271, 1328, 1356, and 1384 (Fig. 5E and H). The latter three mass intensities (1328 to 1384) differed from each other by 28 amu and represent homologous series of mycolic acids differing by a C2 carbon unit (approximately C88 to C92). More importantly, these three mass intensities were not observed in the MALDI-TOF chromatogram of MAMEs extracted from MtbΔvirS and Mtbmym::hyg strains growing at acidic pH (Fig. 5F and G). These results clearly demonstrate the involvement of the mymA operon in the modification of mycolic acids of M. tuberculosis upon exposure to acidic stress.
Inactivation of mymA or virS leads to accumulation of C24:0 and C26:0 fatty acids.
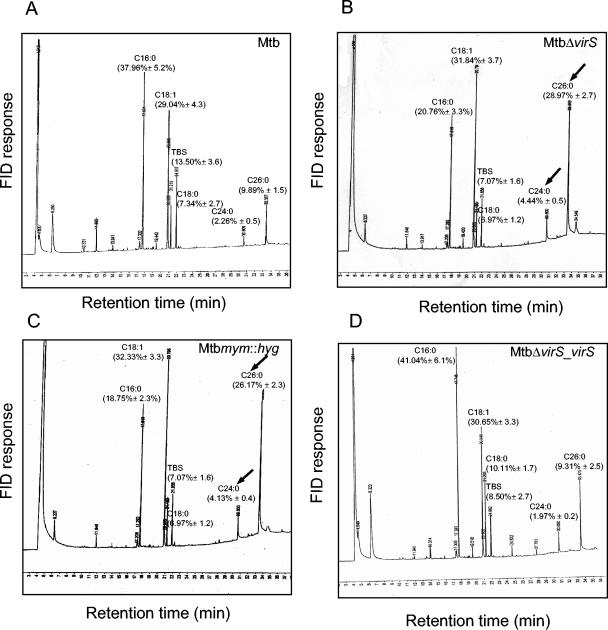
Our TLC experiments showed a consistent increase of 10% to 20% in the radioactivity associated with FAMEs extracted from MtbΔvirS and Mtbmym::hyg compared to the parental strain. To identify the fatty acids responsible for this increase in the mutant strains, the FAMEs from various strains were extracted and subjected to GC. Our GC data showed alterations in the amount of C26:0, C24:0, C16:0, and C18:3 (tuberculostearic acid [TBS]) in the mutant strains when compared with the parental and MtbΔvirS_virS strains. While the relative percentage of other detectable fatty acids C18:1 and C18:0 remained unaltered, we observed a twofold decrease in the levels of C16:0 and TBS and threefold and twofold increases in the levels of C26:0 and C24:0 fatty acids, respectively, in MtbΔvirS and Mtbmym::hyg compared to the parental and MtbΔvirS_virS strains (Fig. 6). Similar alterations in the fatty acid profiles of MtbΔvirS and Mtbmym::hyg were observed by HPLC analysis (data not shown). The identities of the accumulated fatty acids were confirmed by using authentic fatty acid standards for GC and by electrospray-mass spectrometry of accumulated fatty acids collected from HPLC (data not shown). These data strongly suggest that the accumulation of C26:0 and C24:0 fatty acids in the mutants might lead to the reduced synthesis of mycolic acids in these strains.
FIG. 6.
Gas chromatograms of FAMEs from various strains of M. tuberculosis. FAMEs were extracted from various strains of M. tuberculosis and analyzed by gas chromatography as described in the Materials and Methods. The identity of fatty acid peaks was confirmed by using authentic fatty acid standards. Relative percentages of fatty acids were calculated and are indicated on each chromatogram. FID and TBS represent flame ionization detector and tuberculostearic acid, respectively. The values shown are averages of three independent experiments ± standard deviations. (A) M. tuberculosis; (B) MtbΔvirS; (C) Mtbmym::hyg, (D) MtbΔvirS_virS. Accumulation of C26:0 and C24:0 in the mutant strains is indicated by arrows. These experiments were repeated three times with similar results.
Influence of virS and mymA disruption on the mycolic acid cleavage products of M. tuberculosis.
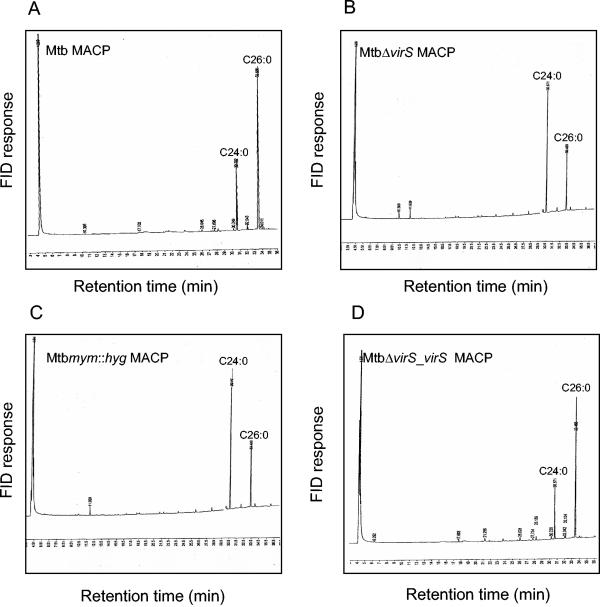
Association between disruption of the mymA operon, reduced synthesis of mycolic acids, and accumulation of C24:0 and C26:0 fatty acids suggested utilization of these fatty acids in the synthesis of mycolic acids by the mymA operon. Thus, we examined the role of this operon, if any, in condensing saturated C24 and C26 fatty acids with meromycolic acid. It has been shown in several studies that increasing the injector temperature of GC to 350°C (pyrolytic GC) results in complete thermal cleavage of C24:0 and C26:0 fatty acids condensed to form mycolic acids of M. tuberculosis (23, 29, 30).
We observed that the thermal cleavage of mycolic acids released an approximately twofold-higher proportion of C26:0 fatty acids compared to C24:0 fatty acids in the case of the parental strain of M. tuberculosis (Fig. 7A). This observation is consistent with the various reports demonstrating a relatively higher release of C26:0 fatty acids in comparison to C24:0 fatty acids from mycolic acids of M. tuberculosis as a result of thermal cleavage (29, 36). However, pyrolysis of mycolic acids from the mutant strains revealed a strikingly reverse pattern of C26:0 and C24:0 fatty acids, where there was an approximately twofold-higher proportion of C24:0 fatty acids released in comparison to C26:0 fatty acids (Fig. 7B and C). These alterations in the pyrolysis of mycolic acids were fully complemented in the case of MtbΔvirS_virS (Fig. 7D).
FIG. 7.
Pyrolytic gas chromatography of MAMEs from various strains of M. tuberculosis. The MAMEs from various strains of M. tuberculosis were subjected to thermal cleavage by increasing the injector temperature to 350°C, and the identity of mycolic acid cleavage product (MACP) was confirmed by using an authentic fatty acid methyl ester standard. (A) M. tuberculosis; (B) MtbΔvirS; (C) Mtbmym::hyg; (D) MtbΔvirS_virS. FID and TBS represent flame ionization detector and tuberculostearic acid, respectively.
Simultaneously, we also extracted and analyzed all the polar and nonpolar lipids as described previously (3, 31), however, we observed no change in the profiles of these lipids from the mutants and parental strain of M. tuberculosis (data not shown). The accumulation of C26:0 and C24:0 fatty acids along with the altered mycolic acid profile in the mutants indicates the role of mymA operon in the synthesis of mycolic acids by utilizing saturated C26 and C24 fatty acids as potential precursors.
Disruption of virS and mymA results in enhanced susceptibility of M. tuberculosis to antibiotics, detergents, and acidic pH.
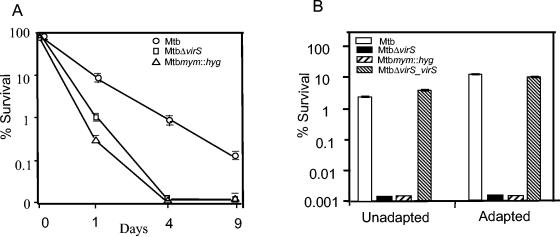
Mycolic acids are essential components of the mycobacterial cell wall; thus we reasoned that alterations in the mycolic acids of mutants might affect their ability to resist drugs, detergent, or stress like acidic pH. We show that MtbΔvirS and Mtbmym::hyg exhibit a fourfold-higher sensitivity towards rifampin and ciprofloxacin and twofold-higher sensitivity towards isoniazid (INH) in comparison to the parental strain (Table 1). Furthermore, these mutants showed a significantly enhanced susceptibility towards the antibiotics at acidic pH. At pH 5.0, MtbΔvirS and Mtbmym::hyg strains were fourfold more sensitive to INH and eightfold more sensitive to rifampin and ciprofloxacin compared to the parental strain. However, the most striking difference was observed in case of susceptibility of the mutant strains towards pyrazinamide (PZA), an antitubercular drug that is active specifically at acidic pH (38, 69). A 50-fold-lower concentration of PZA was sufficient to inhibit the growth of mutant strains in comparison to the concentration required to inhibit the growth of the parental strain (Table 1). These mutants also exhibited greater sensitivity to SDS compared to the parental strain (Fig. 8A). MtbΔvirS and Mtbmym::hyg were also drastically sensitive to acidic pH (pH 4.0) when compared with the parental strain (Fig. 8B). More importantly, prior adaptation of the parental strain to pH 5.0 significantly increased its survival at pH 4.0 (by fourfold [P < 0.02]), although no such influence of adaptation was observed in the case of mutant strains (Fig. 8B). The phenotypes exhibited by the mutants were fully complemented in the MtbΔvirS_virS strain of M. tuberculosis.
TABLE 1.
Influence of virS and mymA disruption on the susceptibility of M. tuberculosis to antibioticsa
| Strain | MIC99 (μg/ml) at indicated pH of:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Rifampin
|
Ciprofloxacin
|
Isoniazid
|
Ethambutol
|
Pyrazinamide | |||||
| 7.0 | 5.0 | 7.0 | 5.0 | 7.0 | 5.0 | 7.0 | 5.0 | 5.0 | |
| M. tuberculosis Erdman | 0.50 | 0.50 | 1.0 | 1.0 | 0.10 | 0.10 | 0.10 | 0.10 | 50 |
| MtbΔvirS | 0.125 | 0.0625 | 0.25 | 0.125 | 0.05 | 0.025 | 0.10 | 0.10 | 1 |
| Mtbmym::hyg | 0.125 | 0.0625 | 0.25 | 0.125 | 0.05 | 0.025 | 0.10 | 0.10 | 1 |
| MtbΔvirS_virS | 0.50 | 0.50 | 1.0 | 1.0 | 0.10 | 0.10 | 0.10 | 0.10 | 50 |
The MIC99 values were determined as described in Materials and Methods. Three independent experiments were carried out, and the same MIC99 values were obtained.
FIG. 8.
(A) Susceptibility of various strains of M. tuberculosis to SDS. M. tuberculosis, MtbΔvirS, and Mtbmym::hyg strains were inoculated separately in MB 7H9 medium and grown to an A600 of 1.5. Cultures were exposed to 0.01%, 0.04%, and 0.1% SDS for the indicated periods of time, and the number of viable bacilli was evaluated by plating on MB 7H10 medium. CFU obtained immediately after the exposure to SDS (∼1 × 108 to 2 × 108 cells per ml) were taken as 100 percent viability. Percent survival for each strain was calculated, and the figure represents percent survival upon exposure to 0.1% SDS. The experiment was carried out thrice in triplicate, and the data are depicted as the mean ± standard errors (SE). (B) Influence of disruption of virS and mymA on the acid tolerance response of M. tuberculosis. M. tuberculosis, MtbΔvirS, Mtbmym::hyg, and MtbΔvirS_virS were grown in MB 7H9 medium (pH 7.0) to an A600 of 1.5. Cultures were exposed either directly to pH 4.0 or after preadaptation at pH 5.0 for 16 to 18 h. Viable counts were taken at 12 h and 24 h after exposure to pH 4.0. Percent survival was calculated as described in Materials and Methods. Viable counts obtained immediately after exposure to pH 4 (∼4 × 107 cells per ml) were taken as 100 percent viability, and percent survival was calculated at various time points. The figure represents percent survival at 24 h postexposure. The experiment was carried out twice in triplicate, and the data are depicted as the means ± SE.
Role of the mymA operon in the in vivo survival of M. tuberculosis in macrophages and guinea pigs.
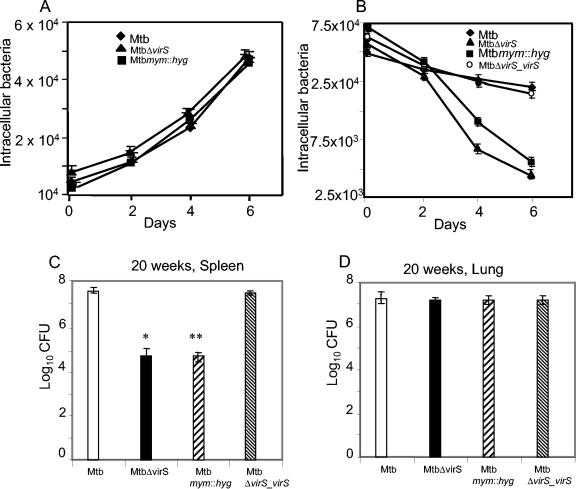
Resting and activated murine macrophage cells were infected with MtbΔvirS, Mtbmym::hyg, MtbΔvirS_virS, and the parental strain of M. tuberculosis, and the survival of intracellular bacteria was determined on days 0, 2, 4, and 6 postinfection. All strains grew at comparable rates inside the resting macrophages (Fig. 9A); however, there were four- and eightfold reductions in the number of intracellular mutant bacilli compared to parental bacilli in activated macrophages at 4 and 6 days, postinfection (Fig. 9B). We further extended our study to investigate the role of the mymA operon in the survival of M. tuberculosis in guinea pigs.
FIG. 9.
Effect of virS and mymA disruption on the in vivo survival of M. tuberculosis in macrophages and guinea pigs. The mouse macrophage cells (J774A.1) were infected with M. tuberculosis, MtbΔvirS, Mtbmym::hyg, and MtbΔvirS_virS strains separately at an multiplicity of infection of 1:10 (macrophage:bacilli). At different time points postinfection (days 0, 2, 4, and 6), macrophages were lysed and the number of intracellular bacilli was assessed by plating on MB 7H10 (A, resting macrophages; B, activated macrophages). The experiment was carried out twice in triplicate, and data depict the means of all six values ± standard errors (SE). Guinea pigs were infected with 5 × 107 CFU of different strains of M. tuberculosis, subcutaneously. Infected guinea pigs (five animals per group at each time point) were euthanized at 10 weeks and 20 weeks postinfection. Spleens and lungs were homogenized in 5 ml of distilled water and 10-fold serial dilutions of the homogenates were plated in duplicate on MB 7H10 medium. Bacillary load in the spleens and lung of guinea pigs euthanized at 20 weeks (C and D, respectively) postinfection is depicted as mean ± SE.
In a preliminary study, we observed that 6 weeks after infection with either MtbΔvirS or the parental strain, the animals showed similar patterns of infection, with no differences in the pathophysiology as well as bacillary load in spleen and lung (data not shown). Thus, we further extended our study to determine the role of the mymA operon in the later stages of the disease. For this, guinea pigs were infected subcutaneously with MtbΔvirS, Mtbmym::hyg, MtbΔvirS_virS, and the parental strain of M. tuberculosis and the animals were euthanized at 10 and 20 weeks postinfection. It was observed that there was no significant difference in the scores and the lung and splenic CFU obtained for various groups of animals at 10 weeks post infection (data not shown). However, at 20 weeks postinfection, the total scores of the animals infected with MtbΔvirS and Mtbmym::hyg strains were significantly lower (17.50 and 24.0, respectively) in comparison to the total scores of animals infected with the parental strain (57.25, P < 0.02) and MtbΔvirS_virS (56.50, P < 0.02). These results were further strengthened by a significant reduction of bacillary load in the spleen of animals infected with the MtbΔvirS (log10, 4.74) and Mtbmym::hyg (log10, 4.71) compared to the bacillary load in the spleen of animals infected with the parental strain (log10, 7.72, P < 0.003) and the MtbΔvirS_virS strain (log10, 7.63, P < 0.002) (Fig. 9C). Thus, an approximately 800-fold (2.8 log)-reduced bacillary load was observed in spleens of animals infected with the mutant strains in comparison to the animals infected with the parental strain. However, the bacillary load in lungs of animals infected with MtbΔvirS (log10, 7.19) and Mtbmym::hyg (log10, 7.25) was comparable to the bacillary load in the lungs of animals infected with the parental strain (log10, 7.25) and MtbΔvirS_virS strain (log10, 7.21) (Fig. 9D). These observations were further substantiated by significant differences in the percent spleen weight per total body weight in case of animals infected with MtbΔvirS (0.16%) and Mtbmym::hyg (0.18%) in comparison to the animals infected with the parental strain (0.31%, P < 0.05) and MtbΔvirS_virS strain (0.36%, P < 0.05); however, we observed no difference in the percent lung weight per total body weight of animals infected with either the mutants or parental strain. These observations strongly suggest the role of the mymA operon in the survival of M. tuberculosis against potentially harsher conditions faced by the pathogen in activated macrophages and also at later stages of progression of disease in guinea pigs.
DISCUSSION
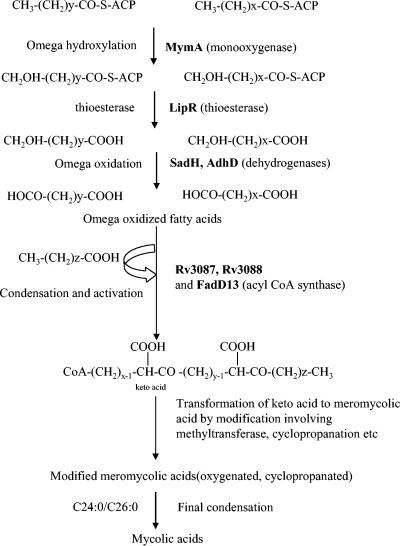
In an earlier study, we had reported that the mymA operon of M. tuberculosis is induced at acidic pH in macrophages and is transcriptionally regulated by VirS (58). We had also shown that MtbΔvirS has an altered cell wall structure and proposed that these alterations were due to abolished expression of the mymA operon. More direct proof of this observation emerged upon disruption of mymA (Rv3083), the first gene in the mymA operon, which resulted in the functional inactivation of the mymA operon and similar alterations in the cell wall ultrastructure phenotype as were observed earlier for MtbΔvirS (58). The Mtbmym::hyg strain exhibited a much denser and darker staining of cell surface, indicating an alteration in the ETZ, which is thought to be composed primarily of mycolic acids arranged perpendicular to the plane of the cell surface (5, 33, 34, 35, 48). Such dense staining of the cell wall has also been observed after treatment of Mycobacterium avium with isoniazid resulting from the inhibition of mycolic acid synthesis by the drug (42). The alterations in the cell surface of MtbΔvirS and Mtbmym::hyg strains were further substantiated by the HPLC profiles of mycolic acids from the mutants and the parental strains. Furthermore, both mutants produced less mycolic acids in comparison to the parental strain as analyzed by TLC. These findings suggest that the observed alterations in the cell wall ultrastructure result from the altered mycolic acid composition although the effect of latter on the arrangement of other cell surface lipids and proteins and their consequent contribution on the observed phenotype cannot be completely ruled out. On exposure to acidic pH, the reduction in mycolic acid synthesis was markedly more prominent in the MtbΔvirS and Mtbmym::hyg strains in comparison to the parental strain. The accumulation of fatty acids (C24:0/C26:0) at acidic pH was also observed to be higher in the mutants compared to the parental strain. Although, a general reduction in the synthesis of mycolic acids at acidic pH can be expected to stem from the repression of the Fas II operon (21), a much sharper decline in mycolic acid synthesis in case of both the mutant strains implicates the mymA operon in the synthesis of mycolic acids on exposure of the pathogen to acidic pH. The emergence of new mass peaks corresponding to the C88 to C92 chain length of mycolic acids (1328, 1356, and 1384) in the parental strain, but not in the mutants, clearly suggested the role of the mymA operon in the synthesis of these mycolic acids at acidic pH. Such modifications in the chain length of mycolic acids in mycobacteria on exposure to environmental stresses have been reported in several studies (2, 31, 33, 63, 64), emphasizing the requirement of the appropriate repertoire of mycolic acids to respond to environmental stresses. Further, the enhanced accumulation of C24:0/C26:0 fatty acids in the mutant strains substantiates their role in the synthesis of mycolic acids by the mymA operon. Conventionally mycolic acids are believed to be synthesized by elongating long chain fatty acids (C16 to C26) to meromycolic acids by the Fas II operon of M. tuberculosis, and the final Claisen type condensation of the C24:0/C26:0 fatty acid with meromycolates results in the production of full-length mycolic acids (6, 16, 33, 35, 41, 43). However, Asselineau et al. (1) have thoroughly reexamined the pathways of mycolic acid synthesis and suggested an alternate approach of mycolic acid synthesis by “head-to-tail” condensation of long chain fatty acids. The synthesis of mycolic acids by this approach involves the condensation of three common fatty acids. First, two of these are subjected to omega oxidation followed by condensation to produce meromycolic acids, which in turn condense with C24:0/C26:0 fatty acids to produce mycolic acids. This approach of mycolic acid synthesis requires enzymes that can carry out omega oxidation of fatty acids and their subsequent condensation (1). Interestingly, analysis of gene products of the mymA operon revealed that Rv3083 (mymA) is a homologue of flavin-containing monooxygenases (58), which can carry out omega hydroxylation of fatty acids (39, 45), the first step in omega oxidation of fatty acids, while Rv3085 and Rv3086 show homologies with dehydrogenases and could possibly carry out subsequent steps to convert terminal methyl groups of fatty acids to carboxylic groups for condensation as described previously (1). Release of acyl carrier protein esterified to the fatty acids by thioesetrase LipR (Rv3084) leads to generation of diacids for the condensation. Rv3087 and Rv3088 contain an HHXXXDG motif required for the thioesterification or Claisen type condensation of fatty acids (28, 46); the last gene, Rv3089, is an acyl-CoA synthase and can activate the fatty acids. Thus, Rv3087 and Rv3088 can carry out “head-to-tail” condensation of fatty acids which were previously omega oxidized by gene products of Rv3083 to Rv3086 and further activation of the condensed fatty acids by Rv3089 can yield long chain fatty acids (keto acids). These keto acids can then be subjected to functional group modification like methylation, decarboxylation, or cyclopropanation to generate meromycolic acids as suggested previously (1). The condensation process described above can produce long chain fatty acids that are indistinguishable from mycolic acids (1). Thus, the genes present in the mymA operon can assemble meromycolic acids beginning from the omega oxidation of fatty acids followed by their condensation with fatty acids (C24:0/C26:0) to produce mycolic acids as shown in Fig. 10.
FIG. 10.
Hypothetical scheme showing the involvement of the mymA operon in the biosynthesis of mycolic acids via “head-to-tail” condensation. Biosynthesis of mycolic acids via “head-to-tail” condensation initiates with the omega hydroxylation of the terminal methyl group of two fatty acid molecules (C24:0/C26:0) by MymA (Rv3083), which are converted into a carboxyl group by the dehydrogenases SadH (Rv3085) and AdhD (Rv3085). Release of acyl carrier protein (ACP) esterified to the fatty acid by thioesterase LipR (Rv3084) leads to generation of diacids for the condensation. Head-to-tail condensation by Rv3087 and/or Rv3088 gene products between three fatty acids, two of which are omega oxidized and activation by FadD13 (Rv3089) produce keto acid. The keto acid can then be subjected to functional modification like methylation, cyclopropanation, etc., to produce meromycolic acids which can be condensed to C24:0/C26:0 to produce full-length mycolic acids.
Both the mutants showed increased sensitivity to major antitubercular drugs along with enhanced susceptibility to SDS and acidic pH. The enhanced susceptibility of M. tuberculosis to antibiotics, detergents, and environmental stresses has been shown to be associated with alterations in the mycolic acid contents and composition (27, 67, 68). A mutant of Mycobacterium smegmatis devoid of mycolic acids has been shown to be hypersusceptible to antibiotics and was unable to grow at 37°C (33), while inactivation of the arylamine N-acetyltransferase gene of Mycobacterium bovis BCG perturbed biosynthesis of mycolic acids, with the consequent increase in the sensitivity to antibiotics (4). Interestingly, these mutants were susceptible to INH, which is known to target mycolic acid biosynthesis (53, 60, 61, 62, 66). The precise target of INH in M. tuberculosis is still debatable, and various pieces of evidence suggest that there might be several targets of INH (60). However, INH-treated M. tuberculosis accumulates C26:0 fatty acid (41), suggesting that INH targets enzymes which utilize C26:0 for generation of mycolic acids. These observations, together with the fact that the mymA operon might utilize C26:0 fatty acids as precursors for mycolic acid biosynthesis, suggest the possibility that the proteins present in this operon are potential targets of INH. This possibility is further strengthened by the presence of a NADH binding site in MymA (58), which has been shown as the target of the active form of INH (iso-NAD) in the case of InhA (17).
The induction of the mymA operon at acidic pH and a significantly reduced ability of MtbΔvirS and Mtbmym::hyg to survive in the activated macrophages compared to the parental strain supports the hypothesis that the mymA operon may play an important role in the survival of M. tuberculosis upon exposure to severely acidic conditions in activated macrophages (56) or caseating granuloma in the later stages of infection (13, 14). This was substantiated by a drastic reduction (∼2.8 log) observed in the ability of the mutant strains to specifically survive in spleen compared to the parental strain at 20 weeks postinfection. The involvement of the mymA operon in persistence during the chronic stage of disease and also the tropism shown by the pathogen are not surprising. Several genes involved in fatty acid metabolism such as icl, encoding isocitrate lyase (catabolizes fatty acids via the glyoxylate cycle) and pcaA, encoding cyclopropane synthase (required for cyclopropanation of mycolic acids), and mutations affecting the lipid-rich cell envelope are all known to reduce the survival of M. tuberculosis in mice, only at the later stages of disease progression (22, 40, 55). Besides, several gene disruption studies have led to the exhibition of tropism by the mutant bacilli; for example, FadD28, FadD33, Pps, MmpL7, and HBHA mutants all showed tissue-specific growth rates (11, 49, 50). This tropism could be due to different physiological conditions or immune responses which the pathogen encounters in different organs. The genes present in the mymA operon apparently may be involved in maintaining the cell wall integrity required for the persistence of M. tuberculosis in spleen.
The involvement of the mymA operon in the persistence of M. tuberculosis together with its role in maintaining appropriate mycolic acid composition to resist antitubercular drugs at acidic pH indicate that precise targeting of the mymA operon gene products may increase the effectiveness of combination chemotherapy and impede the mechanisms involved in the persistence of M. tuberculosis.
Acknowledgments
This work was supported by financial grants received from the Indian Council of Medical Research and Department of Biotechnology, Government of India. A.S. and R.G. are grateful to the Council of Scientific and Industrial Research, India, for research fellowships.
Jaya Gopinath, S. Nambirajan, Lucas, K. Chandran, M. Asokan (TRC, Chennai, India) and Archana (NII, New Delhi, India) are acknowledged for assistance in animal experiments and fatty acid analysis, respectively. We acknowledge T. K. Das, Incharge EM facility, Department of Anatomy, AIIMS, New Delhi, for transmission electron microscopy and D. Chatterji and P. D. Deepalakshmi, Incharge Proteomics facility, IISC, Bangalore, for MALDI-TOF analysis. We thank Bindu Nair and Bahadur Singh for their technical assistance. Rajiv Chawla is thankfully acknowledged for excellent secretarial help.
REFERENCES
- 1.Asselineau, C., J. Asselineau, G. Laneelle, and M.-A. Laneelle. 2002. The biosynthesis of mycolic acids by Mycobacteria: current and alternative hypothesis. Prog. Lipid Res. 41:501-523. [DOI] [PubMed] [Google Scholar]
- 2.Baba, T., K. Kaneda, E. Kusunose, M. Kusunose, and I. Yano. 1989. Thermally adaptive changes of mycolic acids in Mycobacterium smegmatis. J. Biochem. (Tokyo) 106:81-86. [DOI] [PubMed] [Google Scholar]
- 3.Besra, G. S. 1998. Preparation of cell wall fractions from mycobacteria. Methods Mol. Biol. 101:91-107. [DOI] [PubMed] [Google Scholar]
- 4.Bhakta, S., G. S. Besra, A. M. Upton, T. Parish, C. Sholto-Douglas-Vernon, K. J. C. Gibson, S. Knutton, S. Gordon, R. P. daSilva, M. C. Anderton, and E. Sim. 2004. Arylamine N-acetyltransferase is required for synthesis of mycolic acids and complex lipids in Mycobacterium bovis BCG and represents a novel drug target. J. Exp. Med. 199:1191-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brennan, P. J., and H. Nikaido. 1995. The envelope of mycobacteria. Annu. Rev. Biochem. 64:29-61. [DOI] [PubMed] [Google Scholar]
- 6.Brindley, D. N., S. Matsumura, and K. Bloch. 1969. Mycobacterium phlei fatty acid synthase-a bacterial multienzyme complex. Nature 224:666-669. [Google Scholar]
- 7.Cage, H. 1992. High performance liquid chromatography patterns of Mycobacterium gordonae mycolic acids. J. Clin. Microbiol. 30:2402-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Camacho, L. R., P. Constant, C. Raynaud, M.-A. Laneelle, J. A. Triccas, B. Gicquel, M. Daffe, and C. Guilhot. 2001. Identification of a virulence gene cluster of Mycobacterium tuberculosis by signature-tagged mutagenesis. J. Biol. Chem. 276:19845-19854. [DOI] [PubMed] [Google Scholar]
- 9.Chan, K., T. Knaak, L. Satkamp, O. Humbert, S. Falkow, and L. Ramakrishnan. 2002. Complex pattern of Mycobacterium marinum gene expression during long term granulomatous infection. Proc. Natl. Acad. Sci. USA 99:3920-3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V Gordon, K. Eiglmeier, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 11.Cox, J. S., B. Chen, M. McNeil, and W. R. Jacobs, Jr. 1999. Complex lipid determines tissue specific replication of Mycobacterium tuberculosis in mice. Nature 402:79-83. [DOI] [PubMed] [Google Scholar]
- 12.Daffe, M., and P. Draper. 1998. The envelope layers of mycobacteria with reference to their pathogenicity. Adv. Microbiol. Phys. 39:131-203. [DOI] [PubMed] [Google Scholar]
- 14.Dannenberg, A. M., and J. F. Tomashefski. 1991. Pathogenesis of pulmonary tuberculosis. McGraw-Hill, New York, N.Y.
- 13.Dannenberg, A. M., Jr. 1993. Immunopathogenesis of pulmonary tuberculosis. Hosp. Pract. 28:51-58. [DOI] [PubMed] [Google Scholar]
- 15.DasGupta, S. K., S. Jain, D. Kaushal, and A. K. Tyagi. 1998. Expression systems for study of mycobacterial gene regulation and development of recombinant BCG vaccines. Biochem. Biophys. Res. Commun. 246:797-804. [DOI] [PubMed] [Google Scholar]
- 16.Davidson, L. A., and K. Takayama. 1979. Isoniazid inhibition of the synthesis of monounsaturated long-chain fatty acids in Mycobacterium tuberculosis H37Ra. Antimicrob. Agents Chemother. 16:104-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dessen, A., A. Quemard, J. S. Blanchard, W. R. Jacobs, Jr., and J. C. Sacchettini. 1995. Crystal structure and function of the isoniazid target of Mycobacterium tuberculosis. Science 267:1638-1641. [DOI] [PubMed] [Google Scholar]
- 18.Dinadayala, P., F. Laval, C. Raynuad, A. Lemassu, M. A. Laneelle, G. Laneelle, and M. Daffe. 2003. Tracking the putative biosynthetic precursors of oxygenated mycolates of Mycobacterium tuberculosis. J. Biol. Chem. 278:7310-7319. [DOI] [PubMed] [Google Scholar]
- 19.Dubnau, E., J. Chan, C. Raynaud, V. P. Mohan, M. A. Yu, K. Laneelle, A. Quemard, I. Smith, and M. Daffe. 2000. Oxygenated mycolic acids are necessary for virulence of Mycobacterium tuberculosis in mice. Mol. Microbiol. 36:630-637. [DOI] [PubMed] [Google Scholar]
- 20.Dubnau, E., P. Fontan, R. Manganelli, S. Soares-Appel, and I. Smith. 2002. Mycobacterium tuberculosis genes induced during infection of human macrophages. Infect. Immun. 70:2787-2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fisher, M. A., B. B. Plikaytis, and T. M. Shinnick. 2002. Microarray analysis of the Mycobacterium tuberculosis transcriptional response to acidic conditions found in phagosomes. J. Bacteriol. 184:4025-4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glickman, M. S., J. S. Cox, and W. R. Jacobs, Jr. 2000. A novel mycolic acid cyclopropane synthetase is required for cording, persistence and virulence of M. tuberculosis. Mol. Cell 5:717-727. [DOI] [PubMed] [Google Scholar]
- 23.Guerrant, G. O., M. A. Lambert, and C. W. Moss. 1981. Gas-chromatographic analysis of mycolic acid cleavage products in mycobacteria. J. Clin. Microbiol. 13:899-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gupta, S., and A. K. Tyagi. 1993. Sequence of newly identified Mycobacterium tuberculosis gene encoding a protein with sequence homology to virulence regulating proteins. Gene 126:157-158. [DOI] [PubMed] [Google Scholar]
- 25.Heifets, L., and T. Sanchez. 2000. New agar medium for testing susceptibility of Mycobacterium tuberculosis to pyrazinamide. J. Clin. Microbiol. 38:1498-1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hinds, J., E. Mahenthiralingam, K. E. Kempsell, K. Duncan, R. W. Stokes, T. Parish, and N. G. Stoker. 1999. Enhanced gene replacement in mycobacteria. Microbiology 145:519-527. [DOI] [PubMed] [Google Scholar]
- 27.Jackson, M., C. Raynaud, M. Laneelle, C. Guilhot, C. Laurent-Winter, D. Ensergueix, B. Gicquel, and M. Daffe. 1999. Inactivation of the antigen 85C gene profoundly affects the mycolate content and alters the permeability of the Mycobacterium tuberculosis cell envelope. Mol. Microbiol. 31:1573-1587. [DOI] [PubMed] [Google Scholar]
- 28.Kalscheuer, R., and A. Steinbuchel. 2003. A novel bifunctional wax ester/ acyl-CoA:diacylglycerol acyltransferase mediates wax ester and triacylglycerol biosynthesis in Acinetobacter calcoaceticus ADP1. J. Biol. Chem. 278:8075-8082. [DOI] [PubMed] [Google Scholar]
- 29.Kaneda, K., S. Imaizumi, and I. Yano. 1995. Distribution of C22, C24 and C26-α unit-containing mycolic acid homologues in mycobacteria. Microbiol. Immunol. 39:563-570. [DOI] [PubMed] [Google Scholar]
- 30.Kaneda, K., S. Naito, S. Imaizumi, I. Yano, S. Mizuno, I. Tomiyasu, T. Baba, E. Kusunose, and M. Kusunose. 1986. Determination of molecular species composition of C80 or longer chain α-mycolic acids in Mycobacterium spp. by gas chromatography-mass spectrometry and mass chromatography. J. Clin. Microbiol. 24:1060-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kremer, L., Y. Guerardel, S. S. Gurcha, C. Locht, and G. S. Besra. 2002. Temperature-induced changes in the cell-wall components of Mycobacterium thermoresistibile. Microbiology 148:3145-3154. [DOI] [PubMed] [Google Scholar]
- 32.Larsen, M. H. 2000. Some common methods in mycobacterial genetics, p. 313-320. In G. F. Hatfull and W. R. Jacobs, Jr. (ed.), Molecular genetics of mycobacteria. ASM Press, Washington, D.C.
- 33.Liu, J., and H. Nikaido. 1999. A mutant of Mycobacterium smegmatis defective in biosynthesis of mycolic acids accumulates meromycolic acids. Proc. Natl. Acad. Sci. USA 96:4011-4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu, J., C. E. Barry III, G. S. Besra, and H. Nikaido. 1996. Mycolic acid structure determines the fluidity of the mycobacterial cell wall. J. Biol. Chem. 271:29545-29551. [DOI] [PubMed] [Google Scholar]
- 35.Liu, J., C. E. Barry III, and H. Nikaido. 1998. Cell wall—physical structure and permeability, p. 1-44. In C. Ratledge and J. W. Dale (ed.), Mycobacteria: molecular biology and virulence. Chapman & Hall, Philadelphia, Pa.
- 36.Luquin, M., V. Ausina, F. L. Calahorra, F. Belda, M. G. Barcelo, C. Celma, and G. Prats. 1991. Evaluation of practical chromatographic procedures for identification of clinical isolates of mycobacteria. J. Clin. Microbiol. 29:120-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manganelli, R., M. I. Voskuil, G. K. Schoolnik, E. Dubnau, M. Gomez, and I. Smith. 2001. The Mycobacterium tuberculosis ECF sigma factor σE: role in global gene expression and survival in macrophages. Mol. Microbiol. 41:423-437. [DOI] [PubMed] [Google Scholar]
- 38.McDermott, W., and R. Tompsett. 1954. Activation of pyrazinamide and nicotinamide in acidic environments in vitro. Am. Rev. Tuber. 70:748-754. [DOI] [PubMed] [Google Scholar]
- 39.McKenna, E. J., and M. J. Coon. 1970. Enzymatic ω-oxidation. J. Biol. Chem. 245:3882-3889. [PubMed] [Google Scholar]
- 40.McKinney, J. D., K. Honer zu Bentrup, E. J. Munooz-Elias, A. Miczak, B. Chen, W. T. Chan, et al. 2000. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt isocitrate lyase. Nature 406:735-738. [DOI] [PubMed] [Google Scholar]
- 41.Mdluli, K., A. R. Slayden, Y. Zhu, S. Ramaswamy, X. Pan, D. Mead, D. D. Crane, J. M. Musser, and C. E. Barry III. 1998. Inhibition of Mycobacterium tuberculosis β-ketoacyl ACP synthase by isoniazid. Science 280:1607-1610. [DOI] [PubMed] [Google Scholar]
- 42.Mdluli, K., J. Swanson, E. Fischer, R. E. Lee, and C. E. Barry III. 1998. Mechanisms involved in the intrinsic isoniazid resistance of Mycobacterium avium. Mol Microbiol. 27:1223-1233. [DOI] [PubMed] [Google Scholar]
- 43.Minnikin, D. E. 1982. Lipids: complex lipids, their chemistry, biosynthesis and roles, p. 95-184. In C. Ratledge and J. Stanford (ed.), The biology of the mycobacteria: physiology, identification and classification, vol. 1. Academic Press, London, United Kingdom.
- 44.Mitchison, D. A. 1964. The virulence of tubercle bacilli from patients with pulmonary tuberculosis in India and other countries. Bull. Int. Union Tuberculosis 35:287. [PubMed] [Google Scholar]
- 45.Miura, Y., and A. J. Fulco. 1974. ω-2 hydroxylation of fatty acids by a soluble system in Bacillus megaterium. J. Biol. Chem. 249:1880-1888. [PubMed] [Google Scholar]
- 46.Onwueme, K. C., J. A. Ferreras, J. Buglino, C. D. Lima, and L. E. Quadri. 2004. Mycobacterial polyketide-associated proteins are acyltransferases: proof of principle with Mycobacterium tuberculosis PapA5. Proc. Natl. Acad. Sci. USA 101:4608-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parish, T., B. G. Gordhan, R. A. McAdam, K. Duncan, V. Mizrahi, and N. G. Stoker. 1999. Production of mutants in amino acid biosynthesis genes of Mycobacterium tuberculosis by homologous recombination. Microbiology 145:3497-3503. [DOI] [PubMed] [Google Scholar]
- 48.Paul, T. R., and T. J. Beveridge. 1992. Reevaluation of envelope profile and cytoplasmic ultrastructure of mycobacteria processed by conventional embedding and freeze-substitution protocols. J. Bacteriol. 174:6508-6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pethe, K., S. Alonso, F. Biet, G. Delogu, M. J. Brennan, C. Locht, and F. D. Menozzi. 2001. The heparin binding haemagglutinin of M. tuberculosis is required for extrapulmonary dissemination. Nature 412:190-194. [DOI] [PubMed] [Google Scholar]
- 50.Rindi, L., L. Fattorini, B. Daniela, I. Elisabetta, G. Freer, D. Tan, G. Deho, G. Orefici, and C. Garzelli. 2002. Involvement of the fadD33 gene in the growth of Mycobacterium tuberculosis in the liver of BALB/c mice. Microbiology 148:3873-3880. [DOI] [PubMed] [Google Scholar]
- 51.Ritter, D., L. D. Carlson, B. K. Logan, L. S. Ramos, J. O. Kilburn, and M. B. Coyle. 1996. Differentiation of Mycobacterium genavense and Mycobacterium simiae by automated mycolic acid analysis with high-performance liquid chromatography. J. Clin. Microbiol. 34:2004-2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Russell, D. G., H. C. Mwandumba, and E. E. Rhoades. 2002. Mycobacterium and the coat of many lipids. J. Cell Biol. 158:421-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sacchettini, J. C., and J. S. Blanchard. 1996. The structure and function of the isoniazid target in M. tuberculosis. Res. Microbiol. 147:36-43. [DOI] [PubMed] [Google Scholar]
- 54.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 55.Sassetti, C. M., and E. J. Rubin. 2003. Genetic requirements for mycobacterial survival during infection. Proc. Natl. Acad. Sci. USA 100:12989-12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schaible, U. E., S. Sturgill-Koszycki, P. H. Schlesinger, and D. G. Russell. 1998. Cytokine activation leads to acidification and increases maturation of Mycobacterium avium containing phagosomes in murine macrophages. J. Immunol. 160:1290-1296. [PubMed] [Google Scholar]
- 57.Schnappinger, D., S. Ehrt, M. I. Voskuil, Y. Liu, J. A. Mangan, I. M. Monahan, G. Dolganov, B. Efron, P. D. Butcher, C. Nathan, and G. K. Schoolnik. 2003. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insight into the phagosomal environment. J. Exp. Med. 198:693-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Singh, A., S. Jain, S. Gupta, T. Das, and A. K. Tyagi. 2003. mymA operon of Mycobacterium tuberculosis: its regulation and importance in the cell envelope FEMS. Microbiol. Lett. 227:53-63. [DOI] [PubMed] [Google Scholar]
- 59.Singh, R., V. Rao, H. Shakila, R. Gupta, A. Khera, N. Dhar, A. Singh, A. Koul, Y. Singh, M. Naseema, P. R. Narayanan, C. N. Paramasivan, V. D. Ramanathan, and A. K. Tyagi. 2003. Disruption of mptpB impairs the ability of Mycobacterium tuberculosis to survive in guinea pigs. Mol. Microbiol. 50:751-762. [DOI] [PubMed] [Google Scholar]
- 60.Slayden, R. A., and C. E. Barry III. 2000. The genetics and biochemistry of isoniazid resistance in Mycobacterium tuberculosis. Microbes Infect. 2:659-669. [DOI] [PubMed] [Google Scholar]
- 61.Slayden, R. A., and C. E. Barry III. 2002. The role of KasA and KasB in the biosynthesis of meromycolic acids and isoniazid resistance in Mycobacterium tuberculosis. Tuberculosis 82:149-160. [DOI] [PubMed] [Google Scholar]
- 62.Takayama, K., H. K. Schones, E. L. Armstrong, and R. W. Boyle. 1975. Site of inhibitory action of isoniazid in the synthesis of mycolic acids in Mycobacterium tuberculosis. J. Lipid Res. 16:308-317. [PubMed] [Google Scholar]
- 63.Toriyama, S., I. Yano, M. Masui, E. Kusunose, M. Kusunose, and N. Akimori. 1980. Regulation of cell wall mycolic acid biosynthesis in acid-fast bacteria. I. Temperature-induced changes in mycolic acid molecular species and related compounds in Mycobacterium phlei. J. Biochem. 88:211-221. [PubMed] [Google Scholar]
- 64.Toriyama, S., I. Yano, M. Masui, M. Kusunose, and E. Kusunose. 1978. Separation of C50-60 and C70-80 mycolic acid molecular species and their changes by growth temperatures in Mycobacterium phlei. FEBS Lett. 95:111-115. [DOI] [PubMed] [Google Scholar]
- 65.Wang, L., R. A. Slayden, C. E. Barry III, and J. Liu. 2000. Cell wall structure of mutant Mycobacterium smegmatis defective in the biosynthesis of mycolic acids. J. Biol. Chem. 275:7224-7229. [DOI] [PubMed] [Google Scholar]
- 66.Winder, F. G., and P. B. Collins. 1970. Inhibition by isoniazid of synthesis of mycolic acids in Mycobacterium tuberculosis. J. Gen. Microbiol. 63:41-48. [DOI] [PubMed] [Google Scholar]
- 67.Yuan, Y., R. E. Lee, G. S. Besra, J. T. Belisle, and C. E. Barry III. 1995. Identification of a gene involved in the biosynthesis of cyclopropanated mycolic acids in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 92:6630-6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yuan, Y., Y. Zhu, D. D. Crane, and C. E. Barry III. 1998. The effect of oxygenated mycolic acid composition on cell wall function and macrophage growth in Mycobacterium tuberculosis. Mol. Microbiol. 29:1449-1458. [DOI] [PubMed] [Google Scholar]
- 69.Zhang, Y., A. Scorpio, H. Nikaido, and Z. Sun. 1999. Role of acid pH and deficient efflux of pyrazinoic acid in unique susceptibility of Mycobacterium tuberculosis to pyrazinamide. J. Bacteriol. 181:2044-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]