Abstract
Export of DsbA, a protein disulfide bond-introducing enzyme, across the Escherichia coli cytoplasmic membrane was studied with special reference to the effects of various mutations affecting translocation factors. It was noted that both the internalized precursor retaining the signal peptide and the periplasmic mature product fold rapidly into a protease-resistant structure and they exhibited anomalies in sodium dodecyl sulfate-polyacrylamide gel electrophoresis in that the former migrated faster than the latter. The precursor, once accumulated, was not exported posttranslationally. DsbA export depended on the SecY translocon, the SecA ATPase, and Ffh (signal recognition particle), but not on SecB. SecY mutations, such as secY39 and secY205, that severely impair translocation of a number of secretory substrates by interfering with SecA actions only insignificantly impaired the DsbA export. In contrast, secY125, affecting a periplasmic domain and impairing a late step of translocation, exerted strong export inhibition of both classes of proteins. These results suggest that DsbA uses not only the signal recognition particle targeting pathway but also a special route of translocation through the translocon, which is hence suggested to actively discriminate preproteins.
Protein export from the cytosol to the periplasm or the outer membrane in E. coli is mediated by Sec translocase, which is also used for integration of a number of membrane proteins (for reviews, see references 9, 29, and 51). Integral membrane Sec components SecY, SecE, and SecG constitute the core of the polypeptide-conducting channel (50) and a SecDF-YajC complex cooperates with SecYEG for full translocation activity (51). Another membrane protein, YidC, also functions in conjunction with SecYEG or by itself for the correct integration of membrane proteins (10). The translocation process is driven by the SecA ATPase as well as by the proton motive force across the membrane (29, 30, 51), although some membrane protein segments may translocate/integrate in SecA-independent manners (21, 33). It is known that a number of, but not all, presecretory proteins are chaperoned by SecB to the SecA/SecYEG components on the membrane (40). It is obscure, however, what might target SecB-independent preproteins to the translocon, except that membrane proteins are considered to be targeted cotranslationally by the signal recognition particle (SRP) system (9, 23, 51).
OmpA, an outer membrane protein, has most commonly been used as a substrate for the in vivo and in vitro studies of protein translocation. We have also been using maltose-binding protein (MBP), a periplasmic protein, for in vivo analyses of the translocase functions. Generally, these two cell envelope proteins respond similarly, although not identically, to a variety of physiological or mutational alterations of the translocase. However, there is no reason to assume that all secretory proteins depend equally on different subreactivities of the translocase and its constituents. If they indeed respond differently to a particular perturbation of the translocation machinery, such differences could provide valuable information about the functional differentiation of the machinery.
We describe here what is unusual about the export of DsbA, a periplasmic protein having a thioredoxin fold with a Cys31-Cys33 disulfide bond acting as an oxidant for a dithiol in substrate proteins (for a review, see reference 31). This pair of cysteines are kept oxidized by the action of a membrane protein, DsbB. During the course of characterization of the secY mutations we isolated earlier, we used DsbA as a translocase substrate and found that it behaved anomalously compared with other “conventional” substrates, OmpA and MBP. While this work was in progress, Schierle et al. (44) reported that the signal sequence of DsbA is special in that it directs a normally cytosolic and rapidly folding protein, thioredoxin, to the SRP-dependent pathway of protein export. However, the export properties of DsbA itself have been insufficiently studied. In this work, we systematically examined the dependence of DsbA export on various elements of the Sec translocation machinery, in particular, different mutational lesions of SecY.
MATERIALS AND METHODS
Media.
Minimal medium M9 (27) and complete nutrient medium L (containing 10 g of Bacto Tryptone and 5 g of yeast extract and 5 g of NaCl per liter; pH adjusted to 7.2 by NaOH) were used. For growing plasmid-carrying strains, ampicillin (50 μg/ml) and/or chloramphenicol (20 μg/ml) was added to the medium.
Bacterial strains and plasmids.
The E. coli strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristics | Reference |
|---|---|---|
| E. coli | ||
| MC4100 | araD139 Δ(argF-lac)U169 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR | 8 |
| AD202 | MC4100, ompT::kan | 1 |
| AD208 | AD202, secY39 zhd33::Tn10 | 49 |
| IQ85 | MC4100, secY24 zhd33::Tn10 rpsE | 45 |
| IQ86 | MC4100, secY+zhd33::Tn10 rpsE | 45 |
| SS141 | MC4100, dsbB::kan5/F′ lacIqlacPL8 LacZ+Y+A+pro+ | 19 |
| CK1953 | MC4100, secB::Tn5 | 22 |
| IT41 | lep9(Ts) era::Tn10 | 13 |
| MM66 | MC4100, secA(Am) Tn10 su3(Ts) trp(Am) | 35 |
| PR520 | MC4100, secE501(Cs) argE::Tn10 | 41 |
| WAM113 | ΔlacU169 araD139 rpsL thi ffh1::kan λ(Para-ffh+ Ampr) Ara+ | 38 |
| KI297 | MC4100, rpsE secY24 zhd33::Tn10/F′ lacIqlacPL8 lacZ+lacY+lacA+ | 15 |
| TH521 | AD202, secD1 zaj::Tn10 | 17 |
| TY0 | AD202, rpsE secY+zhd33::Tn10 | 49 |
| TY1 | AD202, secY205 zhd33::Tn10 | 49 |
| TY8 | AD202, rpsE secY125 zhd33::Tn10 | 49 |
| TY22 | AD202, rpsE secY115 zhd33::Tn10 | 49 |
| Plasmids | ||
| pSK220 | pHSG399 derivative carrying dsbA under the lac promoter | 16 |
| pST30 | pSTV29 derivative carrying syd under the lac promoter | 46 |
| pKJ10 | pUC119 derivative carrying dsbA under the lac promoter | K. Inaba |
| pNH5 | pSTV28 derivative carrying secM-met6 under the lac promoter | 32 |
Pulse-labeling analyses of protein export.
Cells were grown in M9 medium containing 0.4% glycerol and 0.4% maltose, 0.4% glucose, or 0.2% arabinose as carbon sources, 2 μg/ml thiamine, and 18 amino acids (20 μg/ml each; other than methionine and cysteine) as well as appropriate antibiotics when required. Cells were pulse labeled with [35S]methionine (43 TBq/mmol, obtained from American Radiolabeled Chemicals Inc.) for 30 or 90 seconds at the specified temperature, and, when indicated, chased with unlabeled methionine (200 μg of l-methionine per ml). The labeled culture was directly treated with trichloroacetic acid (final concentration, 5%) to precipitate cellular proteins, which were then solubilized and processed for immunoprecipitation with appropriate antibodies (3). Antisera against MBP, OmpA, DsbA, and SecM were described previously (2, 32, 46).
To examine the redox states of DsbA, the acid-precipitated proteins were solubilized in buffered sodium dodecyl sulfate (SDS) solution containing 4-acetoamido-4′-maleimidylstilbene-2,2′-disulfonate (AMS) as described previously (20), except that the concentration of AMS was 20 mM, before immunoprecipitation of DsbA. Radioactive proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) (3) and each protein band was visualized and quantified by a Fuji BAS1800 phosphorimager. The export efficiency of the labeled protein was estimated from the proportion of the signal sequence-processed mature form (for OmpA and MBP) or from the proportion of the oxidized form (for DsbA).
Proteinase K accessibility of reduced and oxidized DsbA species.
Pulse-labeled culture was mixed with sodium azide (0.02%) and chloramphenicol (100 μg/ml) and placed in an ice bath. Cells were collected by centrifugation in the cold and resuspended in 30 mM Tris-HCl (pH 8.1)-20% sucrose. The suspension was then mixed with 1/10 volume of 1 mg/ml lysozyme dissolved in 0.1 M EDTA (pH 8.0) and incubated at 0°C for 30 min followed by centrifugation (10 min in a microcentrifuge at 4°C) to obtain supernatant (periplasmic fraction) and precipitates (spheroplasts). The spheroplasts were finally resuspended in 30 mM Tris-HCl (pH 8.1)-20% sucrose. Samples were treated with proteinase K (100 μg/ml) at 0°C for 60 min. The proteolytic reaction was terminated by addition of 1 mM phenylmethylsulfonyl fluoride, and proteins were precipitated with trichloroacetic acid. When indicated, spheroplasts were disrupted by sonication (three 20-second bursts using a Heat Systems sonicator) before the protease treatment. Subsequently, proteins were subjected to AMS modification and gel electrophoresis, as described above.
Immunoblotting.
Immunological detection of cellular accumulated proteins was carried out as described previously (46). SDS-solubilized cellular proteins equivalent to 107 cells were separated by SDS-PAGE and decorated with appropriate antibodies. Protein images were quantified by a Fuji LAS1000 lumino-imager.
Accumulation of the precursor form of DsbA.
We followed the previously published procedure (55) to accumulate a preprotein in the cell. Plasmid pKJ10 (dsbA) was introduced into cells of KI297 (secY24) harboring pST30 (syd). Cells were grown at 30°C in LB medium containing 0.4% glucose. The overexpression of Syd and DsbA was simultaneously induced with isopropyl-β-d-galactoside (1 mM) and cyclic AMP (5 mM); Syd severely impairs the function of translocon with the SecY24 altered subunit (46). After 1 h, total cellular proteins were directly precipitated with 5% trichloroacetic acid and separated by SDS-PAGE. The precursor form of DsbA was identified as a prominently induced band that migrated ahead of a purified preparation of DsbA (a gift from Kenji Inaba). For N-terminal sequencing, proteins were electro-blotted onto a polyvinylidene difluoride membrane filter (Millipore) and stained with Coomassie brilliant blue.
In vitro translation of DsbA.
An S140 extract (56) was prepared from AD202 as described previously (3). Inverted cytosolic membrane vesicles were prepared from the same strain after French press disruption (54). Coupled transcription/translation of dsbA was directed in the presence of [35S]methionine by plasmid pSK220 (dsbA) DNA in the presence or absence of membranes (3) at 37°C for 40 min.
RESULTS
Anomalous electrophoretic behavior of the DsbA secretory precursor.
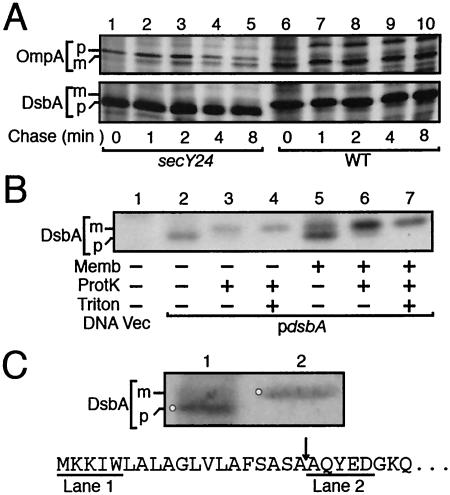
Biosynthesis of DsbA was studied by pulse-chase experiments using wild-type and secY24 mutant cells that had been exposed to 42°C for 2 h, during which the SecY24 mutant protein should have been degraded substantially (4). Whereas OmpA in the wild-type cells was labeled almost exclusively as the mature form (Fig. 1A, upper panel; lanes 6 to 10), that in the mutant cells was initially present as the slow-migrating precursor form, which was gradually converted into the mature form during the chase (Fig. 1A, upper panel; lanes 1 to 5). DsbA in the same cells behaved anomalously in that its electrophoretic mobility was faster in the mutant cells than in the wild-type cells (Fig. 1A, lower panel; compare lanes 1 to 5 and lanes 6 to 10). Unlike OmpA, DsbA did not show any significant mobility change in the mutant during the chase period.
FIG. 1.
Unusual electrophoretic mobility shift of DsbA upon proteolytic processing. (A) In vivo-produced precursor and mature forms. Cells of IQ85 (secY24; lanes 1 to 5) and IQ86 (wild-type; lanes 6 to 10), each carrying pSK220 (encoding DsbA), were grown first at 30°C and then at 42°C for 2 h. Cells were pulse-labeled with [35S]methionine for 30 seconds and chased for 0 (lanes 1 and 6), 1 (lanes 2 and 7), 2 (lanes 3 and 8), 4 (lanes 4 and 9), and 8 (lanes 5 and 10) min. Proteins precipitated by trichloroacetic acid (5%) treatment were directly subjected to SDS-PAGE and autoradiography. Abundant bands of OmpA and overproduced DsbA were visible without immunoprecipitation in this particular experiment. Precursor and mature forms are indicated by p and m, respectively. (B) Primary translation product of DsbA produced in vitro and its protease-cleaved form. In vitro transcription/translation was directed by pHSG399 (vector; lane 1) or pSK220 (DsbA; lanes 2 to 7) as a template in the presence of [35S]methionine and in the presence (lanes 5 to 7) or the absence (lanes 1 to 4) of inverted membrane vesicles. After reaction at 37°C for 40 min, samples were incubated at 0°C for 30 min in the presence or absence of 100 μg/ml of proteinase K (ProtK) and 1% Triton X-100 (Triton) as indicated. Labeled proteins were analyzed by SDS-PAGE and visualized by autoradiography. (C) Confirmation of the anomalous electrophoretic mobility of the DsbA precursor by N-terminal sequencing. DsbA was induced in the SecY-compromised secY24 strain, and total cellular proteins were electrophoresed in lane 1. Lane 2 received purified DsbA. After SDS-PAGE, the electrophoresed proteins were blotted onto a polyvinylidene difluoride membrane filter and stained with Coomassie brilliant blue. The lane 1 band indicated by the circle was barely seen in the uninduced sample (not shown) and should have consisted mainly of pre-DsbA. This band and the DsbA mature band in lane 2 were excised and subjected to N-terminal sequence determination for five residues (APRO Life Science). The N-terminal region of the primary translation product of dsbA is shown below, and the sequences obtained experimentally are underlined. The arrow indicates the signal sequence processing site.
We also encountered unexpected electrophoretic behaviors with in vitro-synthesized DsbA, the primary translation product. The mature DsbA product in vivo exhibited considerable protease resistance, due presumably to its tight folding (see Fig. 5). We found that a main body of the in vitro translation product of dsbA was also resistant to the proteinase K action. The mobility of the in vitro translation product of DsbA (Fig. 1B, lane 2) was shifted to the “higher molecular mass” side upon treatment with proteinase K (Fig. 1B, lanes 3 and 4). It is possible that proteinase K removed the signal peptide, if not precisely, from pre-DsbA and that the truncated form of DsbA migrated slower than the primary translation product. Consistent with this notion, when membrane vesicles were present during in vitro translation, a slow-migrating species identical in mobility to the proteinase K-cleaved DsbA was also produced (Fig. 1B, lane 5). We believe that the membrane vesicles allowed translocation of pre-DsbA and signal peptide cleavage by the leader peptidase in the membrane. Although the intrinsic partial protease resistance of DsbA (Fig. 1B, lanes 3 and 4) complicated the situation, the decreased intensity of the lower-mobility materials in the presence of both proteinase K and Triton X-100 (Fig. 1B, compare lanes 6 and 7) suggests that the slow-migrating species had been protected from protease digestion by the membrane vesicles in the absence of detergent.
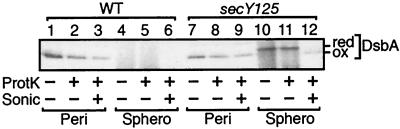
FIG. 5.
Protease accessibility of DsbA species produced in secY125 cell. Cells of TY0 (wild-type; lanes 1 to 6) and TY8 (secY125; lanes 7 to 12) were grown at 30°C and pulse-labeled with [35S]methionine for 30 seconds. Labeled cells were fractionated into periplasmic (lanes 1 to 3 and 7 to 9) and spheroplast (lanes 4 to 6 and 10 to 12) fractions as described in Materials and Methods. Samples were then subjected to sonication (Sonic) and/or proteinase K (ProtK) treatment as indicated. Finally, each sample was treated with trichloroacetic acid, subjected to AMS modification, and immunoprecipitated.
The above results suggest that the precursor form of DsbA migrates faster than the mature form. This was supported further by the following observations. First, DsbA synthesized in the presence of NaN3, an inhibitor of SecA, exhibited faster mobility compared with that produced in the absence of the drug (Fig. 2A, compare lanes 1 and 2). Second, the faster-migrating DsbA species was produced when it was synthesized in the lep9 mutant cells with a temperature-sensitive leader peptidase (Fig. 2B, compare lane 3 with lanes 1 and 2). Finally, the unusual electrophoretic behavior of the DsbA precursor was established by direct determination of the N-terminal sequences of purified mature DsbA (Fig. 1C, lane 2) and the faster-migrating species accumulated in SecY-compromised cells (see Materials and Methods; Fig. 1C, lane 1).
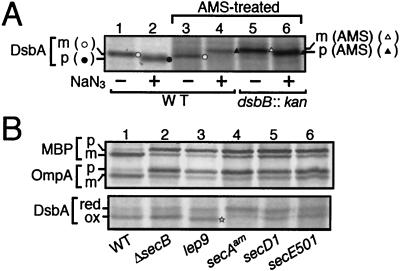
FIG. 2.
Redox states of newly synthesized DsbA as indications of its localization. (A) Migration of internalized and exported DsbA after AMS treatment. Cells of MC4100 (wild-type; lanes 1 to 4), and SS141 (dsbB::kan; lanes 5 and 6) were grown at 30°C and pulse-labeled with [35S]methionine for 60 seconds with (lanes 2, 4, and 6) or without (lanes 1, 3, and 5) prior 60-second exposure to 0.02% NaN3. Samples for lanes 3 to 6 were treated with AMS. Labeled proteins were precipitated with trichloroacetic acid, immunoprecipitated with anti-DsbA, and analyzed by SDS-PAGE. Open circles, unmodified mature form; solid circles, unmodified (oxidized) precursor form; solid triangles, AMS-modified (reduced) precursor form; open triangles, AMS-modified (reduced) and signal sequence-processed form. (B) DsbA synthesized in different mutant cells. Cells of TY0 (wild type; lane 1), CK1953 (ΔsecB; lane 2), IT41 (lep9; lane 3), MM66 [secA(Am); lane 4], THE521 (secD1; lane 5), and PR520 (secE501; lane 6) were grown at 30°C and pulse-labeled with [35S]methionine for 60 seconds. Samples for the upper panel were directly immunoprecipitated with anti-MBP and anti-OmpA, whereas those for the lower panel were first treated with AMS and then precipitated with anti-DsbA. The precursor and mature forms of MBP and OmpA are indicated by p and m, respectively. The reduced and oxidized forms of DsbA are indicated by red and ox, respectively. The oxidized but signal sequence-retaining DsbA species in the lep9 mutant is marked by a star.
Oxidation of DsbA as an indication of its export.
DsbA contains a pair of cysteines that constitute the active site for its activity to introduce disulfide bonds into other proteins. This pair of cysteine residues are kept oxidized by the action of DsbB, a membrane protein. Like other exported proteins in E. coli (7), DsbA should only be oxidized after export to the periplasm, because the oxidation-essential cysteine residues of DsbB are all facing the periplasm (31). We assessed the redox states of newly synthesized DsbA by examining whether pulse-labeled DsbA was modifiable with AMS, a thiol-alkylating agent that causes mobility retardation of the modified protein in SDS-PAGE (20). As expected, export inhibition by NaN3 resulted in production of the reduced and AMS-modifiable form of DsbA (Fig. 2A, compare lanes 3 and 4). The AMS-modified form of DsbA in the NaN3-treated wild-type cells (presumably retaining the signal sequence) migrated slightly faster than the signal sequence-processed but reduced form that was produced in the dsbB cells in the absence of NaN3 (Fig. 2A, compare lanes 4 and 5). Addition of NaN3 to the latter cells resulted in a slight mobility increase of the AMS-modified DsbA (Fig. 2A, compare lanes 5 and 6).
These results are consistent with our finding that signal peptide-unprocessed precursor migrates faster than the processed form; this effect of the signal peptide was observed even when the protein was modified with AMS. As already stated, DsbA synthesized in the leader peptidase-defective lep9 cells, which was supposed to be oxidized but retain the signal peptide, gave a band that migrated faster than the oxidized and signal-processed form (Fig. 2B, lower panel; lane 3). These results confirm that DsbA is oxidized immediately upon export to the periplasm and that signal peptide accelerates the electrophoretic mobility of DsbA.
DsbA export is SecB independent.
The mobility difference between the precursor and mature forms of DsbA was sometimes too subtle to resolve unequivocally. Therefore, we used AMS modifiability to assess the translocation states of DsbA under different in vivo conditions. Whereas export/oxidation of DsbA was retarded in mutant cells having a defect in either secA (Fig. 2B, lane 4), secY (Fig. 1A), secE (Fig. 2B, lane 6), or secD (Fig. 2B, lane 5), it was unaffected by the deletion of secB (Fig. 2B, lane 2). The weak effect of the secE501 mutation was due to the fact that it reduces SecE expression by only about 50% (43).
DsbA export depends on SRP.
Scierle et al. (44) reported that the signal sequence of DsbA can direct thioredoxin, a normally cytosolic protein, to the SRP-dependent export pathway. However, the SRP dependence of DsbA itself was not studied by these authors. We examined the effects of ffh shutdown on export of DsbA, using an engineered strain in which the ffh gene had been placed under arabinose promoter control (38). Additionally, a plasmid encoding SecM-Met6, an SRP-dependent secretory protein (32), was introduced into the strain. Cells were grown first in the presence of arabinose and then in arabinose-free glucose medium to deplete Ffh.
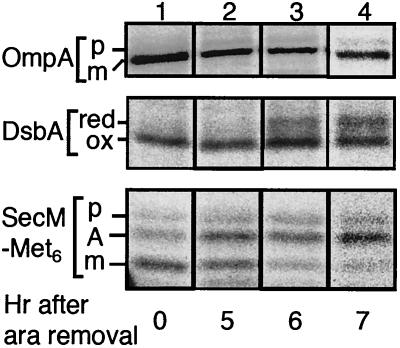
Pulse-labeling and immunoprecipitation experiments revealed that DsbA became labeled as the precursor (reduced) form at about 6 h and thereafter (Fig. 3, middle panel, lanes 3 and 4). Export of OmpA, an SRP-independent protein, was only negligibly retarded by the medium change (Fig. 3, top panel). Thus, secondary inhibition of general protein export was not apparent under these experimental conditions. These results indicate that export of DsbA depends on the SRP function. The SRP depletion effect was observed at ≈5 h for export of SecM-Met6, increasing fractions of which were then labeled as the elongation-arrested intracellular form (32) (Fig. 3, bottom panel, lanes 2 to 4). The earlier onset of the export defect for SecM-Met6 than for DsbA suggests that DsbA may depend on SRP less strongly than SecM-Met6 does. Consistent with this notion, a mild SRP defect caused by the ffh10(Ts) mutation at the permissive temperature (37) retarded integration of a model membrane protein but not export of DsbA (N. Shimohata, unpublished results). In any case, our results indicate that DsbA does depend on SRP for its translocation across the cytoplasmic membrane, consistent with the conclusion of Shierle et al. (44) that its signal sequence is recognized by the SRP targeting pathway.
FIG. 3.
Effects of Ffh depletion on export of DsbA. Plasmid pNH5 (encoding SecM-Met6) was introduced into WAM113 (Para-ffh) cells, which were grown first in M9-amino acids-0.2% arabinose medium, washed three times with arabinose-free medium, and grown further in the same arabinose-free medium supplemented with 0.4% glucose. After 0 (lane 1), 5 (lane 2), 6 (lane 3), and 7 (lane 4) hours in the absence of arabinose, SecM-Met6 was induced with 5 mM cyclic AMP for 15 min and cells were pulse-labeled with [35S]methionine for 30 seconds. Samples were processed for OmpA, DsbA, and SecM immunoprecipitation as indicated. The DsbA sample was AMS modified before immunoprecipitation. SecM-Met6 band A represents the elongation-arrested translation product, the predominant cytosolic form of this protein (32).
Export of DsbA responds to SecY alterations differently from that of previously studied outer membrane and periplasmic proteins.
We studied the export of DsbA in a series of cold-sensitive secY mutants that we had characterized previously (3, 49). The results of pulse-labeling experiments at 20°C with five representative mutants are presented in Fig. 4. The mutation secY39 affects a conserved and essential Arg357 residue and impairs the ability of SecY to interact productively with SecA (28). The secY205 alteration also compromises SecA actions, especially the “membrane insertion” reaction (26). The secY104 mutation may affect some aspect of the SecY-SecG interaction (42). The secY125 mutation affects primarily some late step of protein translocation, such as substrate release from the translocation channel to the periplasmic milieu (24). The secY40 (secY115) mutation is known to be almost silent with respect to protein export (3), and its defect was proposed to lie in the membrane protein integration processes (34).
FIG. 4.
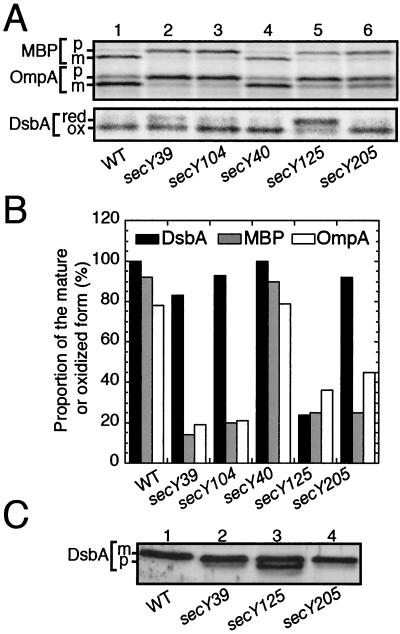
DsbA export in different secY(Cs) mutants. (A) Pulse-labeling at the nonpermissive temperature. Cells of TY0 (wild type; lane 1), AD208 (secY39; lane 2), TY15 (secY104; lane 3), TY22 (secY40; lane 4), TY8 (secY125; lane 5), and TY1 (secY205; lane 6) were grown first at 37°C and then shifted to 20°C for 30 min. Cells were pulse-labeled with [35S]methionine for 90 seconds. Samples for the upper panel were directly immunoprecipitated with anti-MBP and anti-OmpA, whereas those for the lower panel were first treated with AMS and then precipitated with anti-DsbA. (B) Graphic representations of the export efficiencies. The results in A were quantified, and the proportions of exported molecules based on signal sequence processing, for OmpA (open columns) and MBP (shaded columns), or oxidation, for DsbA (solid columns), are shown. (C) Accumulation of DsbA precursor in the secY125 mutant. Total proteins of TY0 (wild type; lane 1), AD208 (secY39; lane 2), TY8 (secY125; lane 3), and TY1 (secY205; lane 4) grown at 30°C were analyzed directly by SDS-PAGE and anti-DsbA immunoblotting.
Export of two commonly used cell surface proteins, OmpA and MBP, was severely retarded in the secY39, secY104, secY125, and secY205 mutants but not significantly in the secY40 mutant, confirming the earlier results (Fig. 4A, upper panel). In contrast, DsbA exhibited nearly normal export, as indicated by its rapid oxidation, in all the mutants other than secY125 (Fig. 4A, lower panel; Fig. 4B). It is remarkable that its export is nearly normal in the secY39 and the secY205 mutants (Fig. 4A, lanes 2 and 6; Fig. 4B).
In the above experiments, the oxidized and reduced forms of DsbA were taken as representing exported and precursor species of this protein, respectively. The validity of this assignment was confirmed by cell fractionation and protease accessibility tests. Pulse-labeling secY125 cells at 30°C, an intermediate temperature, gave both the oxidized and reduced forms of DsbA. The former was fractionated into the soluble periplasmic fraction, whereas the latter was spheroplast associated (Fig. 5). The intensity of the reduced form in the spheroplasts was unchanged by proteinase K treatment (Fig. 5, lanes 10 and 11) but was decreased appreciably when spheroplasts were sonicated before the protease treatment (Fig. 5, lane 12). We believe that DsbA underwent air oxidation after sonication, explaining the increased electrophoretic mobility of the DsbA species that remained after the protease treatment (Fig. 5, lane 12). The results of the experiment shown in Fig. 4 also indicate that DsbA is rapidly folded into a protease-resistant conformation upon export to the periplasm (Fig. 5, lanes 2 and 8).
Direct gel electrophoresis and immunoblotting also revealed that the faster-migrating precursor species accumulated in the 30°C-grown secY125 mutant cells but only insignificantly in the secY39 and secY205 mutant cells (Fig. 4C). The results of the experiment shown in Fig. 4C exclude the possibility that the unusual secY mutational effects on DsbA were only apparent due to the use of oxidation as an indication of export.
DsbA precursor does not undergo posttranslational export.
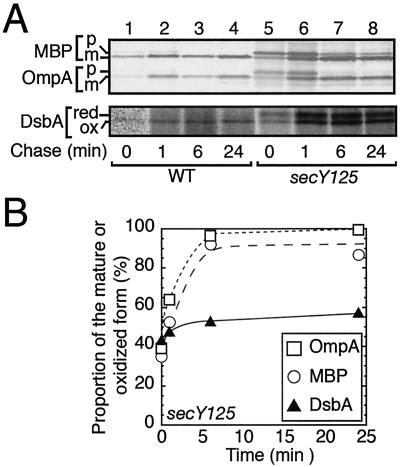
The marked accumulation of the precursor form of DsbA in the secY125 mutant even at 30°C (Fig. 4C) suggests that the precursor form, once produced, does not effectively undergo subsequent processing/export. Whereas roughly a half of the DsbA molecules were labeled as the precursor (reduced) form in secY125 mutant cells at 30°C, they were not chased into the mature (oxidized) form (Fig. 6A, lower panel; Fig. 6B, triangles). OmpA and MBP, on the other hand, exhibited posttranslational processing/translocation, albeit slowly, in secY mutant cells (Fig. 6A, upper panel; Fig. 6B, circles and squares). Similar differences in posttranslational processing were also observed for DsbA and OmpA in secY24 mutant cells (Fig. 1A). Finally, precursor DsbA synthesized in the SRP-depleted cells also remained stable and unprocessed during the chase (data not shown).
FIG. 6.
DsbA precursor, once produced in secY125 cells, is not posttranslationally exported. Cells of TY0 (wild type; lanes 1 to 4) and TY8 (secY125; lanes 5 to 8) were grown at 30°C and pulse-labeled with [35S]methionine for 60 seconds, followed by chase with unlabeled methionine for 0 (lanes 1 and 5), 1 (lanes 2 and 6), 3 (lanes 3 and 7), and 24 (lanes 4 and 8) min. Samples for the upper panel were directly immunoprecipitated with anti-MBP and anti-OmpA, whereas those for the lower panel were first treated with AMS and then precipitated with anti-DsbA. (B) Graphic representations of the chase kinetics. The results for secY125 in A were quantified, and the proportions of exported molecules based on signal sequence processing, for OmpA (open squares) and MBP (open circles), or oxidation, for DsbA (solid circles), are shown.
DISCUSSION
In this paper, we showed that DsbA exhibits several unusual properties in its localization to the periplasm. The precursor form of DsbA proved to migrate faster than the signal sequence-cleaved mature form in SDS-PAGE. Aberrant SDS-PAGE mobility is known for some proteins. Some hydrophilic proteins migrate slower (11), whereas hydrophobic membrane proteins, including SecY (14) and LacY (5), migrate faster than expected from their molecular masses. Waters et al. (52) reported that prepro-α-factor of Saccharomyces cerevisiae runs faster than the signal sequence-processed form in SDS-PAGE.
Two factors are conceivable for these unusual behaviors, SDS binding and residual structure. As pointed out by Schierle et al. (44), the signal sequence of DsbA is more hydrophobic than that of MBP. Using the SOSUI program (12), we also confirmed that the DsbA signal sequence is more hydrophobic than that of OmpA. It may be possible that the signal sequence region of DsbA binds an excess of SDS molecules, which will confer increased negative charges per polypeptide molecular mass. Alternatively, the mature part of DsbA may bind less SDS, which is compensated for by the hydrophobic signal sequence. Still another possibility is that, unlike signal peptides of some other proteins (36), the DsbA signal may induce tighter folding of the protein, which somehow survives the denaturing of SDS.
The E. coli SRP prefers a hydrophobic targeting signal (6, 23). Schierle et al. (44) argued that higher hydrophobicity of the DsbA signal sequence is responsible for the recognition of the DsbA-TrxA fusion protein by SRP. Consistent with this reasoning, we showed that DsbA export depends on Ffh (SRP) but not on SecB. Thus, DsbA, along with SecM (32) and hemoglobin protease (47), belongs to a special class of cell envelope proteins that are targeted by SRP for export across the membrane rather than for integration into it (18).
Our results, that even the in vitro-translated DsbA molecules largely resisted the proteinase K action, suggest that the DsbA precursor can be folded efficiently in spite of the presence of the signal peptide. It is known that the reduced form of DsbA is thermodynamically more stable than the oxidized form (53). Thus, not only the periplasmically exported DsbA molecules but also those retained in the cytosol may be able to fold rapidly. The robust inability of cytosolic pre-DsbA, once accumulated, to undergo posttranslational translocation is consistent with its tight folding. As discussed previously (44), SRP-mediated cotranslational targeting will be particularly beneficial for such rapidly folding precursor proteins. Earlier studies also revealed an inverse correlation between the folding efficiency and export competence of MBP in the cytoplasm (39). Whereas MBP is chaperoned by SecB for its normal export (40), SecB appears to be unable to handle DsbA, due perhaps to its rapid folding potency, which might be to such an extent that it must be captured cotranslationally by SRP.
Among the six secY alleles we examined, only the secY24 and secY125 alterations markedly impaired the export of DsbA. The SecY24 effect can be understood in terms of the proteolytic degradation of this mutant protein (4). What really surprised us was that DsbA export was only insignificantly affected by three well-characterized secY mutations, secY39, secY104, and secY205. Residue Arg357, affected by secY39, is extremely sensitive to substitutions, many of which exhibit partial dominance with respect to the export/growth defect (28), suggesting that this residue plays a crucial role in export of growth-essential E. coli proteins. The mutated translocase requires higher ATP concentration and the proton motive force in vitro (30). Taken together with the fact that the mutational defect can be suppressed by the “superactive” forms of SecA variants (30), the secY39 defects appear to lie in the efficient utilization of the SecA ATPase as the translocation motor.
The secY205 alteration also appears to impair the SecA-SecY interaction, probably at the step of SecA “insertion” into the translocation channel (26). In the in vitro characterization of translocation and integration, Koch and Müller (21) regarded the defect seen with SecY205 membrane vesicles as almost synonymous with the requirement for the SecA function. The secY104 mutation could also affect SecA function because it impairs some aspect of the SecY-SecG interaction (42), while SecG is considered to assist in the SecA reaction cycles (25, 48). In these contexts, the lack of the secY39, secY205, and secY104 effects on export of DsbA could be taken to mean that DsbA is SecA-independently exported. Clearly, this is not the case, however, as our results showed that DsbA export was severely defective in the presence of NaN3 or a SecA-abolishing mutation. It is possible that DsbA and OmpA/MBP utilize different reactivities of SecA, in which this ATPase drives preprotein entry into the translocon.
According to the three-dimensional structure determined for an archaeic SecYEβ complex, the residue of the SecY125 alteration, Ser76 in E. coli SecY, is located within the short α helix, termed TM2a, which was proposed to act as the plug to occlude the hourglass-shaped translocation pore from the periplasmic side (50). The mutational replacement of this residue by bulkier phenylalanine might interfere with the proposed conformational transition of the plug to open the pore. In addition, our previous observation that the secY125 mutant has a protein export defect at some late step of translocation, such as substrate release to the periplasm (24), might suggest that the plug region is also involved in a process of substrate release from the translocon to the periplasm. DsbA and other “conventional” substrates were equally affected by the SecY125 periplasmic alteration but differentially by a number of mutations that affect the cytoplasmic side of SecY. Thus, these substrates may use different routes for the initial targeting and entry steps, and they may subsequently merge at later processes of translocation, in which SecY elements such as the plug will play crucial roles.
Remarkably, we observed that the spectra of secY mutational effects on the integration processes of some model membrane proteins are similar to what we observed here for DsbA export (N. Shimohata, Y. Akiyama, and K. Ito, submitted for publication). These results point to the existence of common translocon elements that DsbA and membrane proteins utilize for their export/integration. Our results suggest that the SecYEG channel is not a passive conduit for polypeptide movement but is able to modulate the mode of its functioning either within the membrane interior or at the initial phase of translocation in a fashion dependent on the mode of targeting of the precursor to the SecA-translocon complex.
Acknowledgments
We thank Kenji Inaba for providing the purified DsbA preparation and DsbA-overproducing plasmid and discussion and Tom Silhavy for the Ffh depletion strain. Thanks are also due to Hiroyuki Mori for discussion and Kiyoko Mochizuki, Michiyo Sano, Mikihiko Yamada, Yasuhide Yoshioka, Kunihito Yoshikaie, and Takuhiko Adachi for technical assistance.
This work was supported by CREST, JST (Japan Science and Technology Agency) (to K.I.), by grants from the Ministry of Education, Culture, Sports, Science and Technology, Japan (to Y.A. and K.I.), and by the National Project on Protein Structural and Functional Analyses of the Ministry of Education, Culture, Sports, Science and Technology, Japan.
REFERENCES
- 1.Akiyama, Y., and K. Ito. 1990. SecY protein, a membrane-embedded secretion factor of E. coli, is cleaved by the OmpT protease in vitro. Biochem. Biophys. Res. Commun. 167:711-715. [DOI] [PubMed] [Google Scholar]
- 2.Akiyama, Y., S. Kamitani, N. Kusukawa, and K. Ito. 1992. In vitro catalysis of oxidative folding of disulfide-bonded proteins by Escherichia coli dsbA (ppfA) gene product. J. Biol. Chem. 267:22440-22445. [PubMed] [Google Scholar]
- 3.Baba, T., A. Jacq, E. Brickman, J. Beckwith, T. Taura, C. Ueguchi, Y. Akiyama, and K. Ito. 1990. Characterization of cold-sensitive secY mutants of Escherichia coli. J. Bacteriol. 172:7005-7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baba, T., T. Taura, T. Shimoike, Y. Akiyama, T. Yoshihisa, and K. Ito. 1994. A cytoplasmic domain is important for the formation of a SecY-SecE translocator complex. Proc. Natl. Acad. Sci. USA 91:4539-4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beyreuther, K., B. Bieseler, R. Ehring, H. W. Griesser, M. Mieschendahl, B. Muller-Hill, and I. Triesch. 1980. Investigation of structure and function of lactose permease of Escherichia coli. Biochem. Soc. Trans. 8:675-676. [DOI] [PubMed] [Google Scholar]
- 6.Bowers, C. W., F. Lau, and T. J. Silhavy. 2003. Secretion of LamB-LacZ by the signal recognition particle pathway of Escherichia coli. J. Bacteriol. 185:5697-5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyd, D., C.-D. Guan, S. Willard, W. Wright, K. Strauch, and J. Beckwith. 1987. Enzymatic activity of alkaline phosphatase precursor depends on its cellular location, p. 89-83. In A. Torriani-Gorini, F. G. Rothman, S. Silver, A. Wright, and E. Yagil (ed.), Phosphate metabolism and cellular regulation in microorganisms. American Society for Microbiology, Washington, D.C.
- 8.Casadaban, M. 1976. Transposition and fusion of the lac operon to selected promoters in E. coli using bacteriophages λ and μ. J. Mol. Biol. 104:541-555. [DOI] [PubMed] [Google Scholar]
- 9.Dalbey, R. E., and M. Chen. 2004. Sec-translocase mediated membrane protein biogenesis. Biochim. Biophys. Acta 1694:37-53. [DOI] [PubMed] [Google Scholar]
- 10.Dalbey, R. E., and A. Kuhn. 2004. YidC family members are involved in the membrane insertion, lateral integration, folding, and assembly of membrane proteins. J. Cell Biol. 166:769-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gill, D. R., and G. P. Salmond. 1990. The identification of the Escherichia coli ftsY gene product: an unusual protein. Mol. Microbiol. 4:575-583. [DOI] [PubMed] [Google Scholar]
- 12.Hirokawa, T., S. Boon-Chieng, and S. Mitaku. 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14:378-379. [DOI] [PubMed] [Google Scholar]
- 13.Inada, T., D. L. Court, K. Ito, and Y. Nakamura. 1989. Conditionally lethal amber mutations in the leader peptidase gene of Escherichia coli. J. Bacteriol. 171:585-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ito, K. 1984. Identification of the secY (prlA) gene product involved in protein export in Escherichia coli. Mol. Gen. Genet. 197:204-208. [DOI] [PubMed] [Google Scholar]
- 15.Ito, K., and Y. Akiyama. 1991. In vivo analysis of integration of membrane proteins in Escherichia coli. Mol. Microbiol. 5:2243-2253. [DOI] [PubMed] [Google Scholar]
- 16.Kamitani, S., Y. Akiyama, and K. Ito. 1992. Identification and characterization of an Escherichia coli gene required for the formation of correctly folded alkaline phosphatase, a periplasmic enzyme. EMBO J. 11:57-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kihara, A., and K. Ito. 1998. Translocation, folding and stability of the HflKC complex with signal-anchor topogenic sequence. J. Biol. Chem. 273:29770-29775. [DOI] [PubMed] [Google Scholar]
- 18.Kim, J., S. Rusch, J. Luirink, and D. A. Kendall. 2001. Is Ffh required for export of secretory proteins? FEBS Lett. 505:245-248. [DOI] [PubMed] [Google Scholar]
- 19.Kishigami, S., E. Kanaya, M. Kikuchi, and K. Ito. 1995. DsbA-DsbB interaction through their active site cysteines. Evidence from an odd cysteine mutant of DsbA. J. Biol. Chem. 270:17072-17074. [DOI] [PubMed] [Google Scholar]
- 20.Kobayashi, T., S. Kishigami, M. Sone, H. Inokuchi, T. Mogi, and K. Ito. 1997. Respiratory chain is required to maintain oxidized states of the DsbA-DsbB disulfide bond formation system in aerobically growing Escherichia coli cells. Proc. Natl. Acad. Sci. USA 94:11857-11862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koch, H. G., and M. Müller. 2000. Dissecting the translocase and integrase functions of the Escherichia coli SecYEG translocon. J. Cell Biol. 150:689-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumamoto, C. A., and J. Beckwith. 1985. Evidence for specificity at an early step in protein export in Escherichia coli. J. Bacteriol. 163:267-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, H. C., and H. D. Bernstein. 2001. The targeting pathway of Escherichia coli presecretory and integral membrane proteins is specified by the hydrophobicity of the targeting signal. Proc. Natl. Acad. Sci. USA 98:3471-3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsumoto, G., T. Homma, H. Mori, and K. Ito. 2000. A mutation in secY that causes enhanced SecA insertion and impaired late functions in protein translocation. J. Bacteriol. 182:3377-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsumoto, G., H. Mori, and K. Ito. 1998. Roles of SecG in ATP- and SecA-dependent protein translocation. Proc. Natl. Acad. Sci. USA 95:13567-13572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsumoto, G., T. Yoshihisa, and K. Ito. 1997. SecY and SecA interact to allow SecA insertion and protein translocation across the Eschrichia coli plasma membrane. EMBO J. 16:6384-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 28.Mori, H., and K. Ito. 2001. An essential amino acid residue in the protein translocation channel revealed by targeted random mutagenesis of SecY. Proc. Natl. Acad. Sci. USA 24:5128-5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mori, H., and K. Ito. 2001. The Sec protein-translocation pathway. Trends Microbiol. 9:494-500. [DOI] [PubMed] [Google Scholar]
- 30.Mori, H., and K. Ito. 2003. Biochemical characterization of a mutationally altered protein translocase: proton-motive force stimulation of the initiation phase of translocation. J. Bacteriol. 185:405-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakamoto, H., and J. C. Bardwell. 2004. Catalysis of disulfide bond formation and isomerization in the Escherichia coli periplasm. Biochim. Biophys. Acta 1694:111-119. [DOI] [PubMed] [Google Scholar]
- 32.Nakatogawa, H., and K. Ito. 2001. Secretion monitor, SecM, undergoes self translation arrest in the cytosol. Mol. Cell 7:185-192. [DOI] [PubMed] [Google Scholar]
- 33.Neumann-Haefelin, C., U. Schafer, M. Müller, and H. G. Koch. 2000. SRP-dependent co-translational targeting and SecA-dependent translocation analyzed as individual steps in the export of a bacterial protein. EMBO J. 19:6419-6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Newitt, J. A., and H. D. Bernstein. 1998. A mutation in the Escherichia coli secY gene that produces distinct effects on inner membrane protein insertion and protein export. J. Biol. Chem. 273:12451-12456. [DOI] [PubMed] [Google Scholar]
- 35.Oliver, D. B., and J. Beckwith. 1982. Regulation of a membrane component required for protein secretion in Escherichia coli. Cell 30:311-319. [DOI] [PubMed] [Google Scholar]
- 36.Park, S., G. Liu, T. B. Topping, W. H. Cover, and L. L. Randall. 1988. Modulation of folding pathways of exported proteins by the leader sequence. Science 239:1033-1035. [DOI] [PubMed] [Google Scholar]
- 37.Park, S. K., P. Jiang, R. E. Dalbey, and G. J. Phillips. 2002. Functional analysis of the signal recognition particle in Escherichia coli by characterization of a temperature-sensitive ffh mutant. J. Bacteriol. 184:2642-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phillips, G. J., and T. J. Silhavy. 1992. The E. coli ffh gene is necessary for viability and efficient protein export. Nature 359:744-746. [DOI] [PubMed] [Google Scholar]
- 39.Randall, L. L., and S. J. Hardy. 1986. Correlation of competence for export with lack of tertiary structure of the mature species: a study in vivo of maltose-binding protein in E. coli. Cell 46:921-928. [DOI] [PubMed] [Google Scholar]
- 40.Randall, L. L., and S. J. Hardy. 2002. SecB, one small chaperone in the complex milieu of the cell. Cell. Mol. Life Sci. 59:1617-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riggs, P. D., A. I. Derman, and J. Beckwith. 1988. A mutation affecting the regulation of a secA-lacZ fusion defines a new sec gene. Genetics 118:571-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Satoh, Y., G. Matsumoto, H. Mori, and K. Ito. 2003. Nearest neighbor analysis of the SecYEG complex. 1. Identification of a SecY-SecG interface. Biochemistry 42:7434-7441. [DOI] [PubMed] [Google Scholar]
- 43.Schatz, P. J., K. L. Bieker, K. M. Ottemann, T. J. Silhavy, and J. Beckwith. 1991. One of three transmembrane stretches is sufficient for the functioning of the SecE protein, a membrane component of the E. coli secretion machinery. EMBO J. 10:1749-1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schierle, C. F., M. Berkmen, D. Huber, C. Kumamoto, D. Boyd, and J. Beckwith. 2003. The DsbA signal sequence directs efficient, cotranslational export of passenger proteins to the Escherichia coli periplasm via the signal recognition particle pathway. J. Bacteriol. 185:5706-5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shiba, K., K. Ito, T. Yura, and D. P. Cerretti. 1984. A defined mutation in the protein export gene within the spc ribosomal protein operon of Escherichia coli: isolation and characterization of a new temperature-sensitive secY mutant. EMBO J. 3:631-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shimoike, T., T. Taura, A. Kihara, T. Yoshihisa, Y. Akiyama, K. Cannon, and K. Ito. 1995. Product of a new gene, syd, functionally interacts with SecY when overproduced in Escherichia coli. J. Biol. Chem. 270:5519-5526. [DOI] [PubMed] [Google Scholar]
- 47.Sijbrandi, R., M. L. Urbanus, C. M. ten Hagen-Jongman, H. D. Bernstein, B. Oudega, B. R. Otto, and J. Luirink. 2003. Signal recognition particle (SRP)-mediated targeting and Sec-dependent translocation of an extracellular Escherichia coli protein. J. Biol. Chem. 14:4654-4659. [DOI] [PubMed] [Google Scholar]
- 48.Suzuki, H., K. Nishiyama, and H. Tokuda. 1998. Coupled structure changes of SecA and SecG revealed by the synthetic lethality of the secAcsR11 and ΔsecG::kan double mutant. Mol. Microbiol. 29:331-341. [DOI] [PubMed] [Google Scholar]
- 49.Taura, T., Y. Akiyama, and K. Ito. 1994. Genetic analysis of SecY: additional export-defective mutations and factors affecting their phenotypes. Mol. Gen. Genet. 243:261-269. [DOI] [PubMed] [Google Scholar]
- 50.van den Berg, B., W. M. Clemons Jr., I. Collinson, Y. Modis, E. Hartmann, S. C. Harrison, and T. A. Rapoport. 2004. X-ray structure of a protein-conducting channel. Nature 427:36-44. [DOI] [PubMed] [Google Scholar]
- 51.Veenendaal, A. K., C. van der Does, and A. J. Driessen. 2004. The protein-conducting channel SecYEG. Biochim. Biophys. Acta 1694:81-95. [DOI] [PubMed] [Google Scholar]
- 52.Waters, M. G., E. A. Evans, and G. Blobel. 1988. Prepro-α-factor has a cleavable signal sequence. J. Biol. Chem. 263:6209-6214. [PubMed] [Google Scholar]
- 53.Wunderlich, M., R. Jaenicke, and R. Glockshuber. 1988. The redox properties of protein disulfide isomerase (DsbA) of Escherichia coli result from a tense conformation of its oxidized form. J. Mol. Biol. 233:559-566. [DOI] [PubMed] [Google Scholar]
- 54.Yamato, I., Y. Anraku, and K. Hirosawa. 1975. Cytoplasmic membrane vesicles of Escherichia coli. A simple method for preparing the cytoplasmic and outer membranes. J. Biochem. (Tokyo) 77:705-718. [DOI] [PubMed] [Google Scholar]
- 55.Yoshihisa, T., and K. Ito. 1996. Pro-OmpA derivatives with a His6-tag in their N-terminal “translocation initiation domain” are arrested by Ni2+ at an early posttargeting stage of translocation. J. Biol. Chem. 271:9429-9436. [DOI] [PubMed] [Google Scholar]
- 56.Zubay, G. 1973. In vitro synthesis of protein in microbial systems. Annu. Rev. Genet. 7:267-287. [DOI] [PubMed] [Google Scholar]