Abstract
Activation of the CiaRH two-component signaling system prevents the development of competence for genetic transformation in Streptococcus pneumoniae through a previously unknown mechanism. Earlier studies have shown that CiaRH controls the expression of htrA, which we show encodes a surface-expressed serine protease. We found that mutagenesis of the putative catalytic serine of HtrA, while not impacting the competence of a ciaRH+ strain, restored a normal competence profile to a strain having a mutation that constitutively activates the CiaH histidine kinase. This result implies that activity of HtrA is necessary for the CiaRH system to inhibit competence. Consistent with this finding, recombinant HtrA (rHtrA) decreased the competence of pneumococcal cultures. The rHtrA-mediated decline in transformation efficiency could not be corrected with excess competence-stimulating peptide (CSP), suggesting that HtrA does not act through degradation of this signaling molecule. The inhibitory effects of rHtrA and activated CiaH, however, were largely overcome in a strain having constitutive activation of the competence pathway through a mutation in the cytoplasmic domain of the ComD histidine kinase. Although these results suggested that HtrA might act through degradation of the extracellular portion of the ComD receptor, Western immunoblots for ComD did not reveal changes in protein levels attributable to HtrA. We therefore postulate that HtrA may act on an unknown protein target that potentiates the activation of the ComDE system by CSP. These findings suggest a novel regulatory role for pneumococcal HtrA in modulating the activity of a two-component signaling system that controls the development of genetic competence.
Competence for genetic transformation in the gram-positive human pathogen Streptococcus pneumoniae is a tightly regulated process, influenced by factors including bacterial cell density, pH, and temperature (5, 57-60). Much has been learned recently about the density-dependent initiation of competence orchestrated by the peptide quorum-sensing system encoded by comCDE and comAB (reviewed in reference 7). The product of comC is a small peptide precursor that is exported from the bacterium and thought to be concurrently processed to form the mature 17-amino-acid competence-stimulating peptide (CSP) by the ComAB transporter (18, 19, 22). The transmembrane ComD histidine kinase is then stimulated by extracellular CSP and activates its cognate ComE response regulator, which in turn initiates a transcriptional cascade resulting in DNA uptake and recombination (20, 43). ComE also promotes the transcription of comCDE and comAB (62), resulting in positive feedback that amplifies signaling through the system. Less is understood, however, about the mechanisms that then shut off competence (7)—even as the bacterial density continues to increase—or the adaptive function of limiting competence expression to such a narrow window of growth. A second pneumococcal two-component system, CiaRH, is known to exert an inhibitory influence on the competence system when activated (12, 14, 17, 39). Of note, the CiaRH system shows a sustained pattern of transcriptional activation following the initiation of competence that persists after most genes involved in the development of the competence phenotype have returned to basal levels of expression (45). Although these features suggest that CiaRH may function to terminate competence, DNA binding studies have not revealed a direct interaction between the CiaR response regulator and any known pneumococcal competence genes (39).
The pneumococcal gene htrA encodes a putative serine protease that is predicted to be localized to the surface of the organism (49). The expression of htrA, together with the downstream gene spoJ, is controlled by the CiaRH two-component system (39, 49). Although this gene has been shown to contribute to the fitness of the organism in both nasopharyngeal colonization (49) and invasive disease (23, 24), the protein target(s) on which HtrA acts in S. pneumoniae is not known. Homologues of HtrA are found in many other bacteria, where they have been shown to participate in responses to a variety of stress conditions (reviewed in reference 41). Intriguingly, the stress responses in which HtrA has been implicated vary among bacteria and include resistance to elevated temperature, oxidizing agents, osmotic pressure, and acidic conditions (2, 9, 15, 25, 27, 33, 35, 42, 52, 63). In Escherichia coli, HtrA (also known as DegP) is found in the periplasm (48) and is thought to alleviate stresses through dual activities, both acting as a chaperone for periplasmic proteins at low temperatures and degrading denatured proteins at higher temperatures (6, 50). The function of HtrA in gram-positive organisms lacking a periplasm has been less well studied.
We sought to investigate whether pneumococcal HtrA might mediate the inhibitory effect of CiaRH on the competence system. This hypothesis appeared plausible because the htrA-spoJ locus was one of only seven loci identified by the combination of solid-phase DNA binding studies and microarray transcriptional analysis as being directly regulated by CiaR, none of which were known to participate in competence regulation (39). Furthermore, the presence of an N-terminal signal peptide in HtrA predicted by the PSORT-B program (Brinkman laboratory, Simon Frasier University [http://www.psort.org/psortb/]) suggested that this protease is likely to be surface associated or secreted, either of which could position it ideally to interact with the competence system. This idea was supported by the proximity of htrA to comCDE on the pneumococcal chromosome, where these divergently transcribed loci are separated by 1 kilobase (21, 54). We therefore examined the effects of genetic manipulation of htrA on the expression of competence.
MATERIALS AND METHODS
Bacterial growth conditions.
S. pneumoniae was grown in C+Y medium (29) at pH 7.4, plated on TS agar plates spread with 5,000 U catalase (Worthington Biochemicals, Freehold, NJ), and incubated in candle extinction jars at 37°C unless otherwise noted. Antibiotics were used at the following concentrations for S. pneumoniae: streptomycin, 200 μg/ml; kanamycin, 500 μg/ml; erythromycin, 1 μg/ml; and novobiocin, 5 μg/ml.
Construction of mutant strains.
Pneumococcal strains utilized in this study are listed in Table 1. A type 3 pneumococcal strain with an erythromycin resistance cassette replacing nucleotides 93 to 971 of htrA has been described elsewhere (49). DNA from this strain was back-transformed into the highly transformable strain R6x (56) to generate the htrA::ermAM strain used in this study.
TABLE 1.
Pneumococcal strains used in this study
| Strain | Characteristic(s)a | Reference or source |
|---|---|---|
| R6x | hex laboratory strain | 56 |
| D39 | Type 2 strain | 1 |
| P143 | R6x, but spontaneous Smr mutant | Masureb |
| 0100993 | Type 3 strain, clinical isolate | 55 |
| P1139 | 0100993, but ciaRH::ermAM, Emr | 55 |
| P1140 | 0100993, but htrA::ermAM, Emr | 49 |
| P1149 | 0100993, but ciaRH::ermAM, by transformation with P1139 DNA, Emr | 49 |
| P1178 | R6x, but htrA::ermAM by transformation with P1140 DNA, Emr | This work |
| P1180 | R6x, but htrA::ermAM by transformation with P1178 DNA, Emr | This work |
| 706 | Novr | 30 |
| 772 | trt | 30 |
| P1301 | D39, but Novr by transformation with 706 DNA | This work |
| P1359 | R6x, but comAB::ermAM, Emr | This work |
| CP1296 | cbp3::Janus, Kmr, Sms | 53 |
| P1372 | P143, but htrA::Janus, Kmr, Sms | This work |
| P1373 | P1372, but htrAS234A, Kms, Smr | This work |
| P1374 | P1372, but wild-type htrA, Kms, Smr | This work |
| P1378 | P1372, but htrAΔ298-1152, Kms, Smr | This work |
| P1382 | P1373, but ciaH::Janus, Kmr, Sms | This work |
| P1384 | P1374, but ciaH::Janus, Kmr, Sms | This work |
| P1386 | P1382, but ciaHT230P, Kms, Smr | This work |
| P1387 | P1382, but wild-type ciaH, Kms, Smr | This work |
| P1388 | P1384, but ciaHT230P, Kms, Smr | This work |
| P1389 | P1384, but wild-type ciaH, Kms, Smr | This work |
| P1458 | R6x, but comDD299N, Smr | This work |
| P1461 | P1458, but ciaH::Janus, Kmr, Sms | This work |
| P1462 | P1462, but ciaHT230P, Kms, Smr | This work |
Emr, erythromycin resistant; Kmr, kanamycin resistant; Kms, kanamycin sensitive; Novr, novobiocin resistant; Smr, streptomycin resistant; Sms, streptomycin sensitive.
From the collection of R. Masure, Rockefeller University.
Unmarked mutations in htrA and ciaH were generated using the counterselectable Janus cassette amplified from genomic DNA of strain CP1296 (53). A spontaneous streptomycin-resistant derivative of R6x from the collection of Robert Masure, designated P143, was used as the parent strain for these manipulations. The cassette was inserted into htrA and ciaH by PCR ligation mutagenesis (32) using primer pairs listed in Table 2. Insertion of the cassette conferred kanamycin resistance and caused a dominant return to streptomycin sensitivity. The cassette was then replaced with a fragment of DNA carrying the desired unmarked mutation, causing reacquisition of streptomycin resistance and loss of kanamycin resistance. Overlap-extension PCR with primer pairs listed in Table 2 was used to generate DNA fragments carrying the unmarked mutations for recombination into the pneumococcal chromosome at the site of the Janus cassette. Isogenic control strains were constructed by replacing the Janus cassette with PCR products amplified from pneumococcal DNA containing the unmodified gene sequences. The presence of the intended point mutations was confirmed by sequencing.
TABLE 2.
PCR primers used in this study
| Usage | Primer name | Primer sequence |
|---|---|---|
| PCR ligation mutagenesis | ||
| htrA 5′ flanking region | FPHTRASPOJ | 5′-GATGATAATCCTAAACTTCCCCCAA-3′ |
| HTRA2093(BamHI) | 5′-ACGAGGATCCTGAAAAACTACCCAAGGCTCC-3′ | |
| htrA 3′ flanking region | HTRA3048(ApaI) | 5′-AGCAGGGCCCTCATCAACAGACTTACAAAGTGCTC-3′ |
| HTRA3903R28 | 5′-CATCTAATCCGAGTAGTTTTCTTAACTG-3′ | |
| ciaH 5′ flanking region | CIARH1430F21 | 5′-TTGATTTTGCTGGATTTGATG-3′ |
| CIARH2139(BamHI) | 5′-ACGAGGATCCTAACTGCTTGAGGATTTTCACTC-3′ | |
| ciaH 3′ flanking region | CIARH3254(ApaI) | 5′-AGCAGGGCCCGATTGCCATTCAGACACCATC-3′ |
| CIARH3912R21 | 5′-AACCAACTTAGCGATTTCACG-3′ | |
| Overlap-extension PCR | ||
| htrAS234A 5′ region | FPHTRASPOJ | 5′-GATGATAATCCTAAACTTCCCCCAA-3′ |
| HTRA4716R35SA | 5′-ATCAGTGGGCCGCCGGCGTTACCTGGGTTAATAGC-3′ | |
| htrAS234A 3′ region | HTRA4682F35SA | 5′-GCTATTAACCCAGGTAACGCCGGCGGCCCACTGAT-3′ |
| RPHTRASPOJ | 5′-CTTCAGAGACAGCTAAAGCGGCT-3′ | |
| htrAΔ298-1152 5′ region | HTRA1418F20 | 5′-TGGGGTAAGGTTAGGCGACC-3′ |
| HTRA2286del3140 | 5′-CCTGAACTCTTGTTAAGTTTGATAGAGATTCCCAAAGCTGGACG-3′ | |
| htrAΔ298-1152 3′ region | HTRA3140del2880 | 5′-CGTCCAGCTTTGGGAATCTCTATCAAACTTAACAAGAGTTCAGG-3′ |
| HTRA3829R23 | 5′-TGTTTTTTCTCTGTCAGAAGAGC-3′ | |
| ciaHT230P 5′ region | CIARH1430F21 | 5′-TTGATTTTGCTGGATTTGATG-3′ |
| CIARH2668R44TP | 5′-CGATTTTGCAAAACTGCGAGTGGAGGTCGTAACTCATGACTGGC-3′ | |
| ciaHT230P 3′ region | CIARH2625F44TP | 5′-GCCAGTCATGAGTTACGACCTCCACTCGCAGTTTTGCAAAATCG-3′ |
| CIARH3912R21 | 5′-AACCAACTTAGCGATTTCACG-3′ | |
| Amplification of other loci | ||
| Janus cassette | JANUS765F32 | 5′-GATCGGATCCGTTTGATTTTTAATGGATAATG-3′ |
| JANUS2109R30 | 5′-ACCTGGGCCCCTTTCCTTATGCTTTTGGAC-3′ | |
| rpsL locus | RPSL871F25 | 5′-CGGTACTTTTTACTTTTGGTCTCTC-3′ |
| RPSL1430R22 | 5′-TCTTTATCCCCTTTCCTTATGC-3′ | |
| comD trt allele | COMCDE1093F26 | 5′-CATTTTCAAGTAACATACTCTTCGTG-3′ |
| COMCDE3191R20 | 5′-AGCTGGATAGAGCATTCGCC-3′ | |
| Insertion-deletion mutagenesis | ||
| comAB amplification | COMAPR2 | 5′-CAGGATTAAGATGAAGCACCC-3′ |
| COMAPR9 | 5′-TGATTGGGGTAAGATAGTTGTTATC-3′ | |
| comAB inverse PCR | COMAPR11 | 5′-ACGCGTCGACGGAGAACCACTTTCCAGAGGAG-3′ |
| COMAPR12 | 5′-TCCCCGCGGATCCTTATCCTGCCCAGTCACC-3′ | |
| Recombinant protein expression | ||
| htrA ORF | HTRA(NdeI)fp | 5′-GGGAATTCCATATGAAACATCTAAAAACATTTTACAAA-3′ |
| HTRA(XhoI)rp | 5′-GTACCGCTCGAGAGATTCTAAATCACCTGAACTCTTG-3′ | |
| comD 3′ region | COMCDE2184(NdeI) | 5′-GGGAATTCCATATGGAGAAAGAGATTGCTTTGAAG-3′ |
| COMCDE1516(XhoI) | 5′-GTACCGCTCGAGATTATCTAATGATTTGTCTAAATGTACTGCC-3′ |
The comAB::ermAM deletion strain (P1359) was generated by insertion-deletion mutagenesis using primers COMAPR9 and COMAPR2 to amplify a 3.3-kilobase fragment of the comAB region. The ermAM cassette then was ligated into SacII and SalI restriction sites introduced into the cloned product by inverse PCR with primers COMAPR11 and COMAPR12. The entire insert was excised and used to transform the pneumococcal R6x strain. The length of a PCR product amplified from the comAB region of strain P1359 was consistent with proper replacement of the targeted portion of this gene with the ermAM cassette. Transformation assays showed that P1359 displayed competence only when supplemented with exogenous CSP, as predicted for a strain lacking the ComAB transporter (data not shown).
The trt (transformation-resistant-to-trypsin) phenotype was transferred to strain R6x using a two-step transformation protocol essentially as described by Lacks and Greenberg (30). In the first step, the parental R6x strain was allowed to undergo spontaneous development of competence, mixed with a PCR product amplified from the comD locus of the trt strain 772 (30) using primers COMCDE1093F26 and COMCDE3191R20, and incubated for 1 h at 30°C to allow DNA uptake. Trypsin (10 μg/ml; Invitrogen, Carlsbad, CA) was then added to inhibit competence in bacteria that had not acquired the trt phenotype, and the culture was incubated for 2 h at 37°C. An additional 10 μg/ml of trypsin was added at the midpoint of the incubation. The culture was then diluted 10-fold into fresh medium also containing 10 μg/ml trypsin and incubated for 15 min at 30°C before the addition of a PCR product encoding streptomycin resistance, which was amplified from the rpsL locus of strain P143 with the primers RPSL871F25 and RPSL1430R22. After 30 min at 30°C, DNase I (1 μg/ml; Roche, Indianapolis, IN) was added to digest unincorporated DNA. The culture was then incubated for 90 min at 37°C, and streptomycin-resistant colonies were selected. The resulting strain P1458 was confirmed to show spontaneous competence in the presence of trypsin, and sequencing verified the presence of the comDD299N point mutation previously described for strain 772 (30, 38).
Recombinant HtrA protein production.
The pneumococcal htrA gene from strain 0100993 was amplified using primers HTRA(NdeI)fp and HTRA(XhoI)rp and cloned into the Novagen (Madison, WI) pET-28a expression vector. The predicted amino acid sequences of strains R6, TIGR4, and 0100993 are identical with the exception of the assignment of alternative start codons in the published annotations of the R6 and TIGR4 genomes, resulting in a 4-residue difference at the N terminus. Recombinant protein expression was induced in E. coli BL21(DE3)pLysS cells (Novagen) grown in 4 liters of Luria-Bertani medium supplemented with kanamycin (30 μg/ml), chloramphenicol (34 μg/ml), and 1% glucose by the addition of 2 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Bacterial pellets were resuspended in a 1/25 volume of 5 mM imidazole, 500 mM NaCl, 20 mM Tris-Cl, pH 7.9, and treated with DNase I (37 U/ml; Roche) and RNase A (0.25 mg/ml; Roche). Cells were lysed by passage through a French press and centrifuged for 1 h at 75,000 × g. The supernatants were then loaded onto His-Bind resin (Novagen) columns charged with 50 mM NiSO4 and equilibrated with 5 mM imidazole, 500 mM NaCl, 20 mM Tris-Cl, pH 7.9. Columns were washed with 60 mM imidazole, 500 mM NaCl, 20 mM Tris-Cl, pH 7.9, and samples were then eluted in five 3-ml fractions with 1 M imidazole, 500 mM NaCl, 20 mM Tris-Cl, pH 7.9. Centricon Plus-20 centrifugal concentration tubes (NMWL 10,000; Millipore, Bedford, MA) were used to exchange the buffer of the eluted fractions to phosphate-buffered saline (PBS) with 10% glycerol without changing the final concentrations of larger-molecular-weight species in each sample. Aliquots of recombinant HtrA (rHtrA) were stored at −20°C. A control vector carrying the htrAS234A gene with a mutation at the putative catalytic site was also constructed, although expression from this plasmid yielded a product that further analysis suggested not to be consistent with the intended, full-length recombinant protein, as it did not react with an anti-His6 antibody (data not shown).
Recombinant ComD protein production.
The region of comD predicted to encode the C-terminal cytoplasmic kinase domain of the protein was amplified using the primers COMCDE2184(NdeI) and COMCDE1516(XhoI) and cloned into the Novagen pET-28a expression vector. Recombinant protein expression was induced in E. coli BL21(DE3)pLysS cells (Novagen) grown in 350 ml Luria-Bertani medium supplemented with kanamycin (30 μg/ml), chloramphenicol (34 μg/ml), and 1% glucose by the addition of 2 mM IPTG. The bacterial pellet was resuspended in 14 ml BugBuster solution (Novagen) and sonicated to shear nucleic acids. Samples were then centrifuged at 21,900 × g to remove cellular debris, passed through a 0.22-μm filter (Millipore), and treated with lysozyme (200 μg/ml). Inclusion bodies containing the recombinant protein were then washed by centrifugation at 5,000 × g and resuspended in 5 mM imidazole, 500 mM NaCl, 20 mM Tris-Cl, pH 7.9. After repeated centrifugation, the inclusion bodies were solubilized in 6 M urea, 5 mM imidazole, 500 mM NaCl, 20 mM Tris-Cl, pH 7.9, by sonication. Samples were centrifuged at 14,000 × g and passed through a 0.45-μm filter (Millipore) before loading onto His-Bind resin (Novagen) columns charged with 50 mM NiSO4 and equilibrated with 6 M urea, 5 mM imidazole, 500 mM NaCl, 20 mM Tris-Cl, pH 7.9. Columns were washed with 6 M urea, 20 mM imidazole, 500 mM NaCl, 20 mM Tris-Cl, pH 7.9, and samples were then eluted with 6 M urea, 1 M imidazole, 500 mM NaCl, 20 mM Tris-Cl, pH 7.9. Centricon Plus-20 centrifugal concentration tubes (NMWL 10,000; Millipore) were used to concentrate the recombinant ComD (rComD) protein without buffer exchange. Aliquots of rComD were stored at −20°C.
Zymography and in vitro proteolytic assay.
rHtrA and control eluate samples were prepared in a nonreducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer consisting of 50 mM Tris-Cl, pH 6.8, 2% SDS, 0.1% bromophenol blue, and 10% glycerol and separated by electrophoresis through 10% acrylamide gels containing β-casein (1 mg/ml) within the resolving gel matrix. Protein refolding was facilitated by washing the gels twice for 10 min each in 2.5% Triton X-100, then in 2.5% Triton X-100, 50 mM Tris-Cl, pH 7.4, and finally in 50 mM Tris-Cl, pH 7.4. The gels were then incubated overnight at 37°C in 50 mM Tris-Cl, pH 7.4, 1 mM MgCl2, 1 mM CaCl2. The resulting zymograms were fixed in 10% acetic acid, 10% methanol, stained with Coomassie brilliant blue, and destained in 10% acetic acid, 10% methanol.
For the in vitro assay of rHtrA activity, 4-μl aliquots of rHtrA eluate or a control eluate from a culture having the pET-28a vector encoding pneumococcal htrA but not induced with IPTG were mixed with 4 μg of β-casein. These samples were incubated at 37°C for 20 h in a buffer consisting of 150 mM NaCl, 1 mM CaCl2, 1 mM MgCl2, 50 mM Tris-Cl, pH 7.4. These digests were then analyzed by SDS-PAGE and stained with Coomassie brilliant blue.
HtrA and ComD antisera production and immunoblotting.
Rabbit polyclonal antisera against rHtrA and rComD were generated by Rockland Immunochemicals Inc. (Gilbertville, PA). Rabbit polyclonal antiserum against pneumolysin was obtained from Tim Mitchell (Glasgow, United Kingdom). For Western immunoblotting, samples were separated by SDS-PAGE and transferred to Immobilon-P membranes. Membranes were incubated in Tris-saline blotting buffer (10 mM Tris-Cl, pH 8.0, 0.5 M NaCl, 0.5% Tween 20, 0.02% sodium azide) with primary antisera (anti-rHtrA, 1:500; anti-rComD, 1:250; antipneumolysin, 1:5,000) followed by a monoclonal anti-rabbit immunoglobulin alkaline phosphatase conjugate (1:5,000; Sigma-Aldrich, St. Louis, MO). Five percent nonfat dry milk was used to reduce nonspecific binding for ComD assays, whereas the lysate of an isogenic pneumococcal strain lacking htrA was used to reduce nonspecific binding for HtrA assays. Alkaline phosphatase activity was detected by reaction with p-nitroblue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate. For preparation of bacterial lysates, pneumococci were resuspended in PBS and sonicated. Culture supernatants were filtered through 0.45-μm-pore-size filters (Millipore) and concentrated approximately 100-fold using Centricon Plus-20 centrifugal concentration tubes (NMWL 10,000; Millipore).
Pneumococcal fractionation.
Bacteria from 48-ml cultures of S. pneumoniae were harvested by centrifugation at 1,500 × g, resuspended in 1 ml PBS supplemented with 15% glycerol, and stored at −80°C to await fractionation. The pellet was thawed, washed twice in PBS, and incubated for 10 min in 0.5 ml PBS supplemented with 2% choline chloride. The sample was centrifuged at 1,500 × g and the pellet resuspended in fresh PBS with choline chloride and incubated as before. The choline washes were pooled and passed through a 0.22-μm filter (Millipore). The pellet was then washed twice in protoplast buffer (PBS supplemented with 1 M sucrose) and incubated in 1 ml protoplast buffer containing 500 μg Cpl-1 phage lysozyme (36, 47) for 1 h. The formation of protoplasts was confirmed by demonstrating susceptibility to osmotic lysis upon 20-fold dilution in distilled water (data not shown). Following digestion, 4 μl 0.5 M EDTA was added per ml of sample. Protoplasts were collected by centrifugation at 12,000 × g for 10 min. The supernatant was collected, filtered through a 0.22-μm-pore-size filter, and saved as the cell wall fraction. The protoplast pellet was then washed twice in PBS supplemented with 1 M sucrose and 1 mM EDTA before being resuspended in 50 μl of the same buffer. Osmotic lysis was induced by dilution into 950 μl of 0.1 M Tris-Cl, pH 7.6, 1 mM EDTA and incubation for 15 min. Unlysed and clumped cellular materials were removed by centrifugation at 5,000 × g for 10 min. The supernatant was then centrifuged at 100,000 × g for 30 min to collect membranes. The supernatant remaining after ultracentrifugation was filtered through a 0.22-μm-pore-size filter and saved as the cytoplasmic fraction. The pellet was washed twice in 0.1 M Tris-Cl, pH 7.6, 1 mM EDTA, resuspended in 50 mM Tris-Cl, pH 7.9, 5 mM NaCl, 1 mM EDTA, 0.4% Triton X-100, 0.5% Sarkosyl, and saved as the membrane fraction. All manipulations, including the Cpl-1 digestion, were carried out at 4°C or on ice.
Transformation assays.
S. pneumoniae strains to be tested were grown in C+Y medium (29), pH 7.4, for 2 h and then diluted into fresh medium to achieve an optical density at 620 nm (OD620) reading of approximately 0.005 to start cultures used for transformation assays. At the desired growth densities, 100-μl aliquots were removed and mixed with 200 ng transforming DNA (from strain P143 for streptomycin resistance or strain P1301 for novobiocin resistance). Samples were incubated for 40 min at 30°C before adding DNase I (20 U; Roche) and then for 90 min at 37°C. Transformation efficiency was calculated as the ratio of the number of colonies seen on serial dilutions plated in the presence of selective antibiotics to that in the absence of antibiotics. Data are presented as mean values ± standard deviation (SD), and differences were compared using two-tailed t tests. When applicable, exogenous CSP-1 was added to culture aliquots and samples were incubated at 30°C for 10 min prior to the addition of transforming DNA.
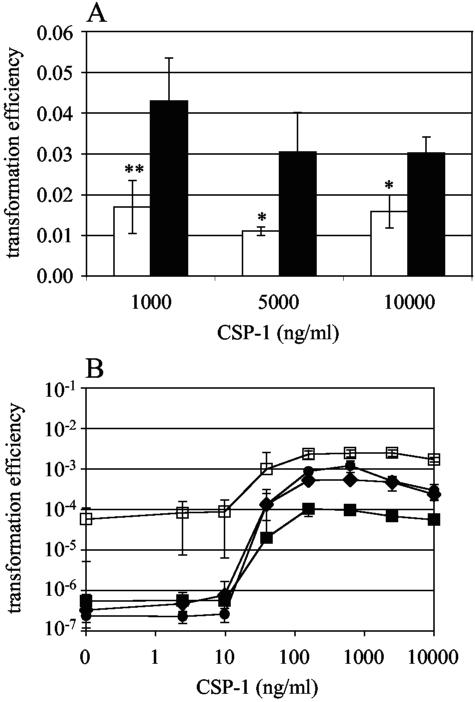
For the transformation assays shown in Fig. 5A, CSP-1 (0.1, 0.5, or 1.0 μg) was incubated with 4-μl aliquots of rHtrA (elution fraction 2) or PBS plus 10% glycerol in C+Y medium (29), pH 7.4, for 1 h at 37°C in order to allow potential digestion of the peptide by rHtrA to occur. These mixtures were then added to 100-μl aliquots of the ΔcomAB strain P1359 taken at an OD620 of 0.110 to 0.154 and incubated for 10 min at 30°C before the addition of 200 ng P143 DNA. Transformation assays were then completed as described above.
FIG. 5.
(A) CSP-1 dose response in strain P1359 (ΔcomAB) treated concurrently with rHtrA (white bars) eluates or PBS plus glycerol (black bars). For comparisons with PBS controls, * indicates a P value of <0.05 and ** indicates a P value of <0.01. Values represent the means of three independent experiments ± 1 SD. (B) CSP-1 dose response in strains R6x (wild type, P3, filled diamonds), P1359 (ΔcomAB, filled circles), P1386 (ciaHT230P htrAS234A, open squares), and P1388 (ciaHT230P, filled squares), n = 3. Values represent the means of three independent experiments ± 1 SD.
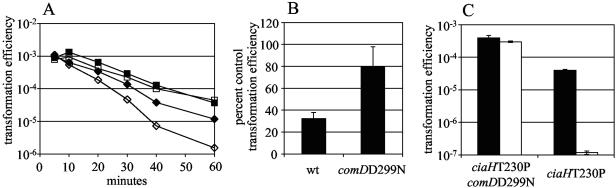
For the transformation assays shown in Fig. 6A and B, strains R6x and P1458 were grown to an OD620 of 0.098 to 0.117, and 100-μl aliquots were mixed with 4 μl rHtrA (elution fraction 2) or an equal volume of PBS plus 10% glycerol. After incubation at 30°C for the stated time period, 200 ng P143 DNA was added, and transformation assays were completed as described above.
FIG. 6.
(A) Time course of decline in transformation efficiency seen with strains R6x (diamonds) and P1458 (comDD299N, squares) after application of rHtrA (open symbols) or PBS plus glycerol (filled symbols). (B) Effect of rHtrA on competence of strains R6x and P1458 40 min after application of recombinant proteins, expressed as the percentage of transformation efficiency of control samples treated with PBS plus glycerol. Values represent the means of three independent measurements ± 1 SD. (C) Transformation of strains P1462 (ciaHT230P comDD299N) and P1388 (ciaHT230P) with (black bars) and without (white bars) the application of exogenous CSP-1. Values represent the means of three independent measurements ± 1 SD.
RESULTS
Protease activity and expression of pneumococcal HtrA.
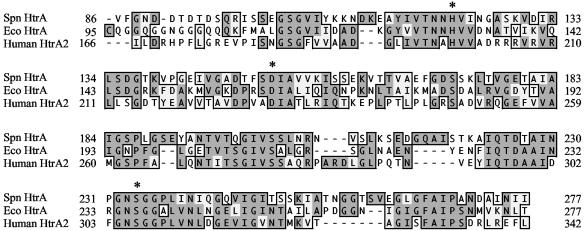
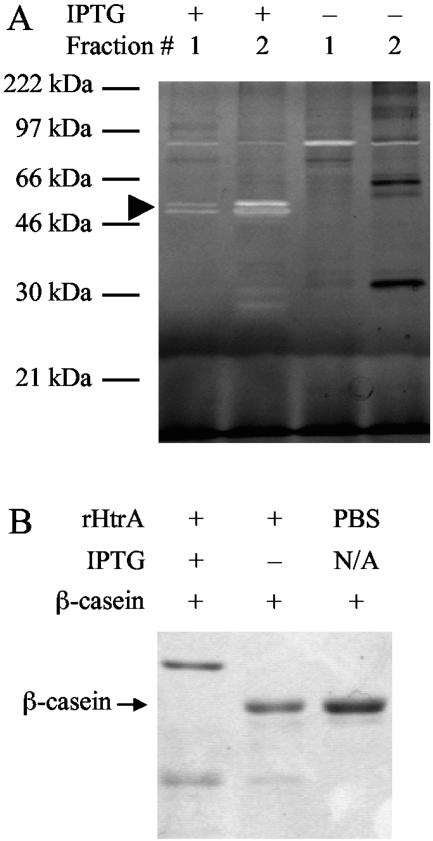
Comparison of the sequence of pneumococcal HtrA to the related serine proteases HtrA/DegP and HtrA2, from E. coli and humans, respectively, for which crystal structures are available (28, 34) revealed conservation of the trypsin-family serine protease domain, including the catalytic triad residues (Fig. 1). In order to assess its proteolytic activity and potential impact on the competence system, pneumococcal HtrA was expressed in E. coli as an IPTG-inducible, recombinant protein (rHtrA) having both N-terminal and C-terminal His6 tags and purified by Ni affinity chromatography. Zymography using β-casein as an artificial substrate showed bands of clearing in the size range expected for rHtrA in column elution fractions derived from cultures in which recombinant protein expression had been induced by IPTG but not in fractions from uninduced cultures (Fig. 2A). The strongest activity in the β-casein zymography was seen in elution fraction 2, which was therefore selected for use in later assays involving pneumococcal competence. The purity of the active protein is difficult to assess due to apparent autoproteolysis within the sample. Analysis of the rHtrA preparation by SDS-PAGE with Coomassie staining revealed an IPTG-induced band consistent with the 45-kilodalton size predicted for the His6-tagged recombinant protein, as well as several smaller, IPTG-induced bands that may represent autoproteolytic digestion products, as both these bands and the full-length band were seen to react with an anti-His6 antibody (data not shown). Two smaller, faint bands of clearing were also seen on the zymogram for fraction 2 of the rHtrA preparation. Comparable bands were not seen in the fractions lacking IPTG induction, supporting the idea that these smaller bands likely represented autodigestion products. Effects of protease activity during purification were also evident in the presence of several protein bands seen in the uninduced sample (Fig. 2A, lane 4) that were not seen in the induced sample.
FIG. 1.
Alignment of the serine protease domains of pneumococcal HtrA (Spn), E. coli HtrA/DegP (Eco), and human HtrA2 generated by the ClustalW algorithm. Asterisks indicate the catalytic triad residues as defined in crystal structures of the E. coli and human proteins (28, 34).
FIG. 2.
(A) β-Casein zymography of recombinant HtrA preparations. Samples were derived from lysates of E. coli expressing pneumococcal HtrA in the presence or absence of IPTG induction. Fractions 1 and 2 of the His-Bind column eluates are shown. The arrowhead indicates full-length rHtrA. (B) In vitro digestion of β-casein by rHtrA from cultures with and without IPTG induction.
Because the protocol for zymography included a refolding step in the matrix of the gel after electrophoresis, aliquots of β-casein were incubated in vitro with rHtrA and assayed for degradation directly by SDS-PAGE (Fig. 2B). The observation of β-casein digestion under these conditions confirmed that activity was present in the eluted samples to be used in later functional assays of pneumococcal competence without requiring refolding during zymography. A lesser amount of activity was also noted using this assay in the control eluate prepared without IPTG induction.
HtrA localization.
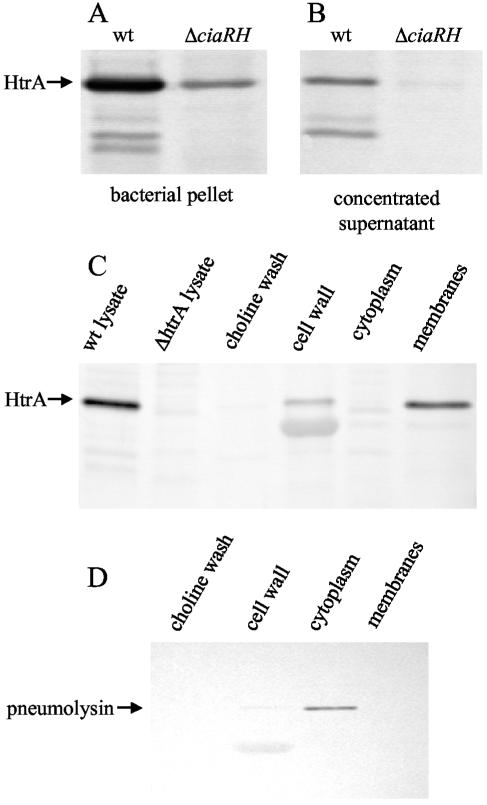
Polyclonal antisera were raised against the recombinant HtrA protein and used in Western blot assays. HtrA was found both in the cellular pellet and in the concentrated supernatant fractions of in vitro pneumococcal cultures, although the amount of protein detected was markedly higher in the bacterial lysate than in the supernatant (Fig. 3). Smaller, immunoreactive bands that may represent HtrA degradation products were also observed. Levels of HtrA expression were substantially lower in strain P1149 containing an insertion-deletion mutation in the ciaRH locus, consistent with the recent report by Ibrahim et al. (24). These assays were performed in the type 3 background in which our previous microarray data (49) had shown decreased expression of htrA mRNA associated with deletion of ciaRH. A similar pattern of HtrA expression was also seen in experiments using the R6x parent strain and an isogenic ciaRH insertion-deletion mutant, although the lower bands were not apparent in this background (data not shown).
FIG. 3.
Western immunoblots for HtrA. (A and B) HtrA expression in bacterial lysates (A) and concentrated supernatants (B) from the wild type (0100993) and ΔciaRH (P1149) (concentration factor, 4.3× relative to the culture volumes from which lysates were harvested). (C and D) Expression of HtrA (C) and pneumolysin (D) in subcellular fractions of S. pneumoniae (P1389, htrA+ ciaH+). Fractionation samples derived from 4.3 ml of original culture were loaded in each lane. Sonicates of whole bacteria containing 2.5 μg of protein from strains P143 (wt) and P1378 (htrAΔ298-1152) are shown as a control for specificity of the HtrA antibody.
The subcellular localization of pneumococcal HtrA was investigated by fractionation of cultures of P1389 (wild-type htrA and ciaH) grown to an OD620 of 0.14 in C+Y medium, pH 7.4 (Fig. 3C). The majority of the HtrA signal was detected in the membrane fraction, although a smaller amount was also seen with the cell wall and culture supernatant. Because the formation of protoplasts during cell wall digestion was monitored by susceptibility to osmotic lysis, the degree to which cell wall-associated proteins were completely disassociated from the underlying membrane is uncertain. That the membrane and cell wall fractions did not contain appreciable amounts of cytoplasmic material was confirmed by probing the samples for pneumolysin (Fig. 3D), an intracellular protein toxin (26). These results demonstrate that pneumococcal HtrA is associated with the surface components of the bacterium, although it remains to be determined whether the protein is linked directly to the cell wall or to the membrane. The finding of a surface localization for pneumococcal HtrA is consistent with the presence of a predicted N-terminal signal peptide in its sequence and with the periplasmic location of HtrA in E. coli (48). A faint, lower band was noted in the cell wall fractions incubated with both the HtrA and pneumolysin antibodies. This band may represent nonspecific hybridization to the 39-kDa Cpl-1 phage lysozyme that was used to digest the bacterial cell wall.
Impact of htrA mutations on competence.
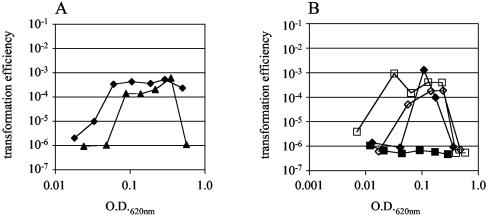
The role of htrA in pneumococcal competence was tested by comparing the development of spontaneous competence during the in vitro growth of strains bearing different targeted mutations of the htrA locus. A strain in which htrA was inactivated by double homologous recombination (P1180), replacing the majority of the gene with an erythromycinresistance cassette, showed decreased transformation compared to the wild type (data not shown). Since the direction of this effect was opposite the augmentation of competence that had been expected based on the phenotype of strains with ciaRH deletions, nonpolar mutations in htrA were constructed using the counterselectable Janus cassette (53). Neither an in-frame deletion of the gene (P1378, htrAΔ298-1152) nor a point mutation inactivating the predicted HtrA catalytic site (P1373, htrAS234A) resulted in competence profiles substantially different from an isogenic control strain with the native htrA sequence (P1374) (Fig. 4). However, an effect of htrA on the induction of competence was seen when examined in the background of a strain bearing the ciaHT230P mutation, which is thought to prevent competence through constitutive activation of the CiaH histidine kinase (17). In the background of CiaH activation, the htrAS234A mutation (P1386) (Fig. 4B) resulted in release of the competence inhibition seen when the ciaHT230P mutation is present alone (P1388). This finding suggests that catalytic activity of HtrA is necessary for the CiaRH two-component system to exert its inhibitory effect on the competence system.
FIG. 4.
(A) Cell density-dependent development of spontaneous competence in strain P1378 (htrAΔ298-1152, triangles) compared to the isogenic strain P1374 (diamonds) having the wild-type htrA sequence. (B) Cell density-dependent development of spontaneous competence in strains P1386 (ciaHT230P htrAS234A, open squares), P1387 (htrAS234A, open diamonds), P1388 (ciaHT230P, filled squares), and P1389 (wild-type ciaH and htrA, filled diamonds).
Effects of exogenous CSP on HtrA-mediated competence inhibition.
Because HtrA is localized to the surface of the bacterium, we investigated whether it acts through degradation of either CSP-1 or the ComD receptor, two potential targets that would likely be accessible to the protease. To test where HtrA interacts with the competence system relative to the accumulation of extracellular CSP, rHtrA was mixed with increasing concentrations of synthetic CSP-1, incubated at 37°C for 1 h, and then added to the comAB deletion strain P1359 that is unable to export the competence-stimulating peptide and therefore requires exogenous CSP for transformation. This strain was used to assess the ability of synthetic CSP-1 to overcome the rHtrA-induced competence defect because it allowed the concentration of CSP-1 to be controlled precisely without variability introduced by endogenous peptide production. Even when CSP-1 was added at concentrations up to 10 μg/ml, however, rHtrA still reduced transformation efficiency (Fig. 5A). This result suggested that the target through which rHtrA caused a decrease in competence was downstream of the accumulation of CSP in the extracellular environment.
Our observation that exogenous CSP-1 cannot reverse the effect of rHtrA on competence is consistent with the report of Guenzi et al. (17) that the transformation deficit of the C306 strain containing the ciaHT230P activating mutation could not be restored by addition of competence factor derived from competent pneumococci. Because synthetic CSP was not available at the time these earlier experiments were performed, we measured the impact of the ciaHT230P mutation on responsiveness to increasing concentrations of synthetic CSP-1 (Fig. 5B). In order to measure the response to CSP-1 of the wild type, samples for these experiments were harvested from early-stage cultures (OD620, 0.02 to 0.03) before the peak of spontaneous competence. The comAB deletion strain P1359 was also tested as a comparison strain that, like P1388 (ciaHT230P), does not develop spontaneous competence. Although transformation was induced in strain P1388 with the application of exogenous CSP-1, the competence response of this strain became saturated at a level nearly 10-fold lower than that of the wild type. In contrast, the P1359 comAB deletion strain showed a response curve nearly identical to the wild type. Inactivation of the expected HtrA catalytic site in the CiaH-activated background (P1386) restored the rate of transformation to a level above that of the wild type at each concentration of CSP-1 tested, even when saturating doses of peptide were applied. These observations suggest that HtrA is responsible for the decreased CSP-1 responsiveness of strain P1388 but that this phenotype is not the result of CSP degradation.
Effects of ComD activation on HtrA-mediated competence inhibition.
The hypothesis was then considered that HtrA might inhibit competence through cleavage of the ComD histidine kinase that acts as a receptor for CSP. The surface location of the HtrA protease could position it to interact with the sensor domain of ComD. Moreover, the degradation of a portion—but not all—of the receptor pool could explain the incomplete loss of CSP responsiveness in the ciaHT230P strain. This model would imply that enhanced activity of the receptor histidine kinase conferred by the comDD299N mutation in the cytoplasmic portion of ComD (30, 38) might bypass the competence-inhibitory effects of CiaH activation and of HtrA. In keeping with this prediction, rHtrA was found to decrease the transformation efficiency of the parent strain R6x but not the comDD299N strain P1458 in both time course (Fig. 6A) and single time point (Fig. 6B) experiments. While there was a small decrease in transformation efficiency for strain P1458 when treated with rHtrA, this difference did not reach statistical significance (P = 0.06). Next, the ability of ComD activation to overcome the competence inhibition caused by the ciaHT230P mutation was tested by constructing a strain (P1462) containing both the comDD299N and ciaHT230P mutations. This strain exhibited competence without requiring the addition of CSP-1 and did not increase its competence significantly in response to synthetic CSP-1 (Fig. 6C) (P = 0.1). Of note, the comDD299N mutation was able to correct the competence defect associated with CiaH activation to a significantly higher level of transformation than was the application of exogenous CSP-1 (Fig. 6C) (P < 0.0001).
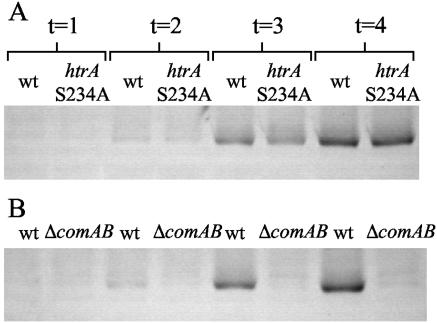
Western blotting was then performed to determine whether mutation of the predicted catalytic site for HtrA would affect ComD (Fig. 7). As anticipated from previous observations for comD RNA levels (12, 13), ComD expression in the control strain P1374 increased as the culture density rose. Strain P1373 (htrAS234A) showed similar expression levels at each time point assayed, and no shift in protein size was apparent. In contrast, strain P1359 (ΔcomAB) did not show induction of ComD, consistent with the failure of this mutant to become competent.
FIG. 7.
Western immunoblots showing ComD expression in whole-cell bacterial lysates taken from cultures of increasing cell density for (A) strain P1374 (wt) versus strain P1373 (htrAS234A) and (B) strain P1374 versus strain P1359 (ΔcomAB). Each lane was loaded with 8 μg total protein. Samples were taken when the culture reached an OD620 of 0.020 to 0.025 (t = 1), 0.070 to 0.074 (t = 2), 0.137 to 0.160 (t = 3), and 0.540 to 0.558 (t = 4). The cultures of P1373 and P1374 displayed spontaneous competence at the second and third time points, whereas that of P1359 did not exhibit competence (data not shown).
DISCUSSION
The impact of HtrA on the development of competence for genetic transformation in S. pneumoniae was evaluated through both site-directed mutagenesis of htrA and studies using recombinant HtrA protease. Although in-frame deletion of htrA and point mutation of the expected HtrA catalytic site did not appear to affect the development of competence in wild-type bacteria, the loss of HtrA activity in a strain with constitutive activation of the CiaRH two-component system restored the development of spontaneous competence. This observation provides evidence that the HtrA protease mediates the inhibitory effects of CiaRH on the competence system. Such a role for HtrA in repressing the development of competence is supported by experiments in which recombinant HtrA caused a decrease in the transformation efficiencies of both wild-type and CSP-stimulated ΔcomAB bacteria.
While the magnitude of the effects on competence seen with recombinant HtrA was much smaller than those due to either genetic inactivation of htrA or activation of ciaH, the pattern of effects from rHtrA was consistently in agreement with the results of these genetic manipulations. Factors that may have contributed to the lesser impact of recombinant HtrA include the unknown level of activity of the recombinant protein preparation relative to native pneumococcal HtrA and the local concentration of the recombinant protein compared to that of the native protein, which is concentrated at the surface of the bacterium. Because in-frame mutations in htrA were not found to impact the spontaneous development of competence in a ciaRH+ background, the question also arises as to whether HtrA plays a role in competence regulation in the wild-type background. Our experiments using the recombinant HtrA protein with wild-type and ΔcomAB strains, however, suggest that HtrA is capable of modulating competence in these contexts as well.
A wide range of molecular targets through which HtrA might interact with the pneumococcal competence system is potentially available, from the proteins responsible for the export and sensing of the competence-stimulating peptide to the surface effector proteins that are directly responsible for DNA uptake. Microarray data presented by Mascher et al. (39), however, show that the expression of late competence genes encoding these effectors is low in the R6ciaHC306 strain that has constitutive activation of the CiaH sensor kinase and therefore imply that the inhibition is likely to take place before these downstream genes are activated rather than through degradation of their products. A second factor that limits the range of candidate targets for HtrA is its surface localization, with the majority of the protein being detected in the bacterial membrane fraction and a smaller amount in the culture supernatant and cell wall fractions.
We therefore chose to investigate components of the CSP export and sensing system as potential targets for HtrA activity. Our finding that exogenous CSP cannot overcome the competence deficit caused by recombinant HtrA indicates that the target is downstream of both the quorum-sensing peptide and the ComAB transporter that exports it. The inability of saturating amounts of synthetic CSP to correct fully the transformation deficit of the CiaH-activated strain is in agreement with this conclusion. The previous description of a point mutation in the intracellular kinase domain of the ComD receptor (30, 38) that enhances its activity provided a means to further define the level at which HtrA interacts with the competence system. Because the activation of ComD substantially reduced the effects of recombinant HtrA and overcame the competence inhibition caused by CiaH activation more completely than did the application of exogenous CSP, we infer that HtrA acts upstream of the intracellular kinase domain of ComD.
Given these results suggesting that HtrA exerts its inhibitory effect on the competence system between the accumulation of CSP and the activation of the ComD kinase domain, we looked for evidence of digestion of ComD by HtrA. Inactivation of HtrA, however, did not alter ComD receptor levels, and consequently the natural substrate for HtrA through which it influences the competence system remains to be determined. Taken together, these data suggest that HtrA may function via the degradation of an unknown protein factor that enhances the activation the ComDE two-component system in response to CSP. We cannot exclude, however, the possibility that HtrA directly inactivates ComD through the removal of a peptide fragment too small to have given rise to a measurable shift in protein size on SDS-PAGE. An increasing number of gene products have now been shown to affect the expression of competence in S. pneumoniae, including the RegR regulator (3), the StkP serine/threonine kinase (11), the ClpP protease (4, 46), and the VicRK two-component system (13, 40, 61), through mechanisms that have not yet been elucidated. Such accessory pathways influencing the development of competence may serve as targets through which HtrA modulates its expression.
Alternatively, it has been suggested that competence represents a part of a broader, generalized stress response in S. pneumoniae (7, 10, 38), and it is possible that HtrA acts to decrease the signal induced by this stress and consequently the underlying stimulus for competence development. The role of HtrA in E. coli in relieving stress resulting from denatured periplasmic proteins (6, 31) is consistent with this model. To be consistent with our results, however, the site at which such a stress signal potentially intersects with the competence pathway would need to be between the accumulation of extracellular CSP and the cytoplasmic activation of ComD. Under this scenario, the putative stress signal would be downstream and independent of CSP, with the competence peptide acting as the postulated “alarmone” (7, 38) only via the secondary feedback amplification of its production through the core competence pathway. Regardless of whether a stress indicator serves as an intermediary, the ultimate result of HtrA activation in S. pneumoniae appears to be a downward modulation of the core signaling system controlling the development of genetic competence.
Ibrahim et al. (24) have recently published data suggesting a stimulatory rather than inhibitory role for HtrA in competence. Their experiments showed that a polar insertion-deletion mutation in htrA was associated with a decrease in competence, which was corrected by complementation with htrA on a plasmid. While these results are similar to our observations with the htrA insertion-deletion mutation containing an erythromycin resistance cassette, the complementation result points to a direct role for HtrA in this effect, whereas our studies using an in-frame deletion of htrA suggest a polar effect of the resistance cassette insertion. The apparent discrepancy in these observations may result from the fact that while our assays measured the development of spontaneous competence in growing pneumococcal cultures, the assays of Ibrahim et al. (24) measured CSP responsiveness after treating the bacteria with synthetic peptide. Interpretation of these experiments is also complicated by the use of the constitutive ami promoter to drive the expression of htrA on the complementing plasmid rather than the native htrA promoter, which is characterized by differential activity under the control of CiaRH. The potential importance of this difference was suggested by our attempt to complement the polar htrA::ermAM mutation with a plasmid expressing htrA from its own 5′ regulatory sequences yielding a strain with a severe growth deficit (data not shown) rather than the restoration of competence observed by Ibrahim et al. (24).
While our findings implicate HtrA in the repression of competence caused by CiaRH, they do not by themselves account for the enhancement of competence that is seen with loss-of-function mutations in ciaRH. Such strains show competence under conditions where it is not normally seen, such as at low culture densities, at decreased pH, and during growth in competence nonpermissive Todd-Hewitt broth (12, 14, 39). In agreement with the report of Mascher et al. (39) using a polar inactivation of htrA, we found that neither the htrAΔ298-1152 in-frame deletion nor the htrAS234A mutation of the putative catalytic site resulted in the expression of competence in Todd-Hewitt broth similar to that seen with a ΔciaRH strain (data not shown). Other components of the CiaRH regulon are therefore likely also to participate in the regulation of competence. The apparent polar effect of the htrA::ermAM mutation suggests that such an additional competence regulator may lie in the vicinity of htrA. Although the downstream gene spoJ would appear to be the most likely candidate for this role, our preliminary experiments have not shown that the deletion of spoJ either affects competence in a wild-type background or restores competence to a CiaH-activated strain (data not shown). While we cannot exclude the possibility that spoJ may affect the development of competence under other conditions not tested, it is also notable that the htrA-spoJ locus lies adjacent to the putative origin of replication of the pneumococcal chromosome (16, 21, 54). A polar effect related to the initiation of DNA replication, similar to that suggested by Claverys et al. (8), might be postulated to result from changes in transcriptional activity in this region.
Pneumococcal htrA has been implicated recently as contributing to the virulence of the organism in models of pneumonia, bacteremia, and nasopharyngeal colonization (23, 24, 49). Our study raises the question of whether these deficits result from the altered regulation of the competence system rather than from direct effects of the protease in relieving environmental stress. More generally, the inhibition of competence by pneumococcal HtrA may represent a novel regulatory role for this protease that has previously been implicated in stress responses (2, 9, 15, 25, 27, 33, 35, 42, 52, 63), the degradation and refolding of denatured proteins (6, 50), and the maturation of exported proteins (37, 44, 51). As the HtrA family of proteases is widely distributed among bacteria, further investigation will be required to determine whether these proteins also regulate signaling through other peptide quorum-sensing pathways or two-component systems. If modulation of signaling by HtrA is found to be a more general phenomenon, it may provide insight into the diverse phenotypes that have been linked to htrA in the different bacteria in which it has been studied.
Acknowledgments
This work was supported by Public Health Service grants AI052129 (M.E.S.), AI38446 (J.N.W.), and AI44231 (J.N.W.) from the National Institute of Allergy and Infectious Diseases.
We thank Donald Morrison for strain CP1296 containing the Janus cassette, Sanford Lacks for the novobiocin-resistant strain 706 and the trt strain 772, David Holmes for the htrA::ermAM and ciaRH::ermAM strains P1140 and P1139, Tim Mitchell for the pneumolysin antiserum, and Vincent Fischetti for the Cpl-1 phage lysozyme. We are grateful to Lin-Sheng Li for his excellent technical assistance during the purification of recombinant HtrA.
REFERENCES
- 1.Avery, O. T., C. M. MacLeod, and M. McCarty. 1944. Studies on the chemical nature of the substance inducing transformation of pneumococcal types. J. Exp. Med. 79:137-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boucher, J. C., J. Martinez-Salazar, M. J. Schurr, M. H. Hudd, H. Yu, and V. Deretic. 1996. Two distinct loci affecting conversion to mucoidy in Pseudomonas aeruginosa in cystic fibrosis encode homologs of the serine protease HtrA. J. Bacteriol. 178:511-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chapuy-Regaud, S., A. D. Ogunniyi, N. Diallo, Y. Huet, J. F. Desnottes, J. C. Paton, S. Escaich, and M. C. Trombe. 2003. RegR, a global LacI/GalR family regulator, modulates virulence and competence in Streptococcus pneumoniae. Infect. Immun. 71:2615-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chastanet, A., M. Prudhomme, J. P. Claverys, and T. Msadek. 2001. Regulation of Streptococcus pneumoniae clp genes and their role in competence development and stress survival. J. Bacteriol. 183:7295-7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, J. D., and D. A. Morrison. 1987. Modulation of competence for genetic transformation in Streptococcus pneumoniae. J. Gen. Microbiol. 133:1959-1967. [DOI] [PubMed] [Google Scholar]
- 6.Clausen, T., C. Southan, and M. Ehrmann. 2002. The HtrA family of proteases: implications for protein composition and cell fate. Mol. Cell 10:443-455. [DOI] [PubMed] [Google Scholar]
- 7.Claverys, J.-P., and L. S. Håvarstein. 2002. Extracellular-peptide control of competence for genetic transformation in Streptococcus pneumoniae. Front. Biosci. 7:1798-1814. [DOI] [PubMed] [Google Scholar]
- 8.Claverys, J.-P., M. Prudhomme, I. Mortier-Barrière, and B. Martin. 2000. Adaptation to the environment: Streptococcus pneumoniae, a paradigm for recombination-mediated genetic plasticity? Mol. Microbiol. 35:251-259. [DOI] [PubMed] [Google Scholar]
- 9.Cortés, G., B. de Astorza, V. J. Benedí, and S. Albertí. 2002. Role of the htrA gene in Klebsiella pneumoniae virulence. Infect. Immun. 70:4772-4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dagkessamanskaia, A., M. Moscoso, V. Hénard, S. Gurial, K. Overweg, M. Reuter, B. Martin, J. Wells, and J. P. Claverys. 2004. Interconnection of competence, stress and CiaR regulons in Streptococcus pneumoniae: competence triggers stationary phase autolysis of ciaR mutant cells. Mol. Microbiol. 51:1071-1086. [DOI] [PubMed] [Google Scholar]
- 11.Echenique, J., A. Kadioglu, S. Romao, P. W. Andrew, and M. C. Trombe. 2004. Protein serine/threonine kinase StkP positively controls virulence and competence in Streptococcus pneumoniae. Infect. Immun. 72:2434-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Echenique, J. R., C.-R. Sabine, and M.-C. Trombe. 2000. Competence regulation by oxygen in Streptococcus pneumoniae: involvement of ciaRH and comCDE. Mol. Microbiol. 36:688-696. [DOI] [PubMed] [Google Scholar]
- 13.Echenique, J. R., and M.-C. Trombe. 2001. Competence repression under oxygen limitation through the two-component MicAB signal-transducing system in Streptococcus pneumoniae and involvement of the PAS domain of MicB. J. Bacteriol. 183:4599-4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Echenique, J. R., and M. C. Trombe. 2001. Competence modulation by the NADH oxidase of Streptococcus pneumoniae involves signal transduction. J. Bacteriol. 183:768-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foucaud-Scheunemann, C., and I. Poquet. 2003. HtrA is a key factor in the response to specific stress conditions in Lactococcus lactis. FEMS Microbiol. Lett. 224:53-59. [DOI] [PubMed] [Google Scholar]
- 16.Gasc, A.-M., P. Giammarinaro, S. Richter, and M. Sicard. 1998. Organization around the dnaA gene of Streptococcus pneumoniae. Microbiology 144:433-439. [DOI] [PubMed] [Google Scholar]
- 17.Guenzi, E., A. M. Gasc, M. A. Sicard, and R. Hakenbeck. 1994. A two-component signal-transducing system is involved in competence and penicillin susceptibility in laboratory mutants of Streptococcus pneumoniae. Mol. Microbiol. 12:505-515. [DOI] [PubMed] [Google Scholar]
- 18.Håvarstein, L. S., G. Coomaraswamy, and D. A. Morrison. 1995. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. USA 92:11140-11144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Håvarstein, L. S., D. B. Diep, and I. F. Nes. 1995. A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates concomitant with export. Mol. Microbiol. 16:229-240. [DOI] [PubMed] [Google Scholar]
- 20.Håvarstein, L. S., P. Gaustad, I. F. Nes, and D. A. Morrison. 1996. Identification of the streptococcal competence-pheromone receptor. Mol. Microbiol. 21:863-869. [DOI] [PubMed] [Google Scholar]
- 21.Hoskins, J., W. E. Alborn, Jr., J. Arnold, L. C. Blaszczak, S. Burgett, B. S. DeHoff, S. T. Estrem, L. Fritz, D.-J. Fu, W. Fuller, C. Geringer, R. Gilmour, J. S. Glass, H. Khoja, A. R. Kraft, R. E. Lagace, D. J. LeBlanc, L. N. Lee, E. J. Lefkowitz, J. Lu, P. Matsushima, S. M. McAhren, M. McHenney, K. McLeaster, C. W. Mundy, T. I. Nicas, F. H. Norris, M. O'Gara, R. B. Peery, G. T. Robertson, P. Rockey, P.-M. Sun, M. E. Winkler, Y. Yang, M. Young-Bellido, G. Zhao, C. A. Zook, R. H. Baltz, S. R. Jaskunas, P. R. Rosteck, Jr., P. L. Skatrud, and J. I. Glass. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 183:5709-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hui, F. M., L. Zhou, and D. A. Morrison. 1995. Competence for genetic transformation in Streptococcus pneumoniae: organization of a regulatory locus with homology to two lactococcin A secretion genes. Gene 153:25-31. [DOI] [PubMed] [Google Scholar]
- 23.Ibrahim, Y. M., A. R. Kerr, J. McCluskey, and T. J. Mitchell. 2004. Control of virulence by the two-component system CiaR/H is mediated via HtrA, a major virulence factor of Streptococcus pneumoniae. Infect. Immun. 186:5258-5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ibrahim, Y. M., A. R. Kerr, J. McCluskey, and T. J. Mitchell. 2004. Role of HtrA in the virulence and competence of Streptococcus pneumoniae. Infect. Immun. 72:3584-3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson, K., I. Charles, G. Dougan, D. Pickard, P. O'Gaora, G. Costa, T. Ali, I. Miller, and C. Hormaeche. 1991. The role of a stress-response protein in Salmonella typhimurium virulence. Mol. Microbiol. 5:401-407. [DOI] [PubMed] [Google Scholar]
- 26.Johnson, M. K. 1977. Cellular location of pneumolysin. FEMS Microbiol. Lett. 2:243-245. [Google Scholar]
- 27.Jones, C. H., T. C. Bolken, K. F. Jones, G. O. Zeller, and D. E. Hruby. 2001. Conserved DegP protease in gram-positive bacteria is essential for thermal and oxidative tolerance and full virulence in Streptococcus pyogenes. Infect. Immun. 69:5538-5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krojer, T., M. Garrido-Franco, R. Huber, M. Ehrmann, and T. Clausen. 2002. Crystal structure of DegP (HtrA) reveals a new protease-chaperone machine. Nature 416:455-459. [DOI] [PubMed] [Google Scholar]
- 29.Lacks, S., and R. D. Hotchkiss. 1960. A study of the genetic material determining an enzyme activity in pneumococcus. Biochim. Biophys. Acta 39:508-517. [DOI] [PubMed] [Google Scholar]
- 30.Lacks, S. A., and B. Greenberg. 2001. Constitutive competence for genetic transformation in Streptococcus pneumoniae caused by mutation of a transmembrane histidine kinase. Mol. Microbiol. 42:1035-1045. [DOI] [PubMed] [Google Scholar]
- 31.Laskowska, E., D. Kuczynska-Wisnik, J. Skórko-Glonek, and A. Taylor. 1996. Degradation by proteases Lon, Clp and HtrA, of Escherichia coli proteins aggregated in vivo by heat shock; HtrA protease action in vivo and in vitro. Mol. Microbiol. 22:555-571. [DOI] [PubMed] [Google Scholar]
- 32.Lau, P. C. Y., C. K. Sung, J. H. Lee, D. A. Morrison, and D. G. Cvitkovitch. 2002. PCR ligation mutagenesis in transformable streptococci: application and efficiency. J. Microbiol. Methods 49:193-205. [DOI] [PubMed] [Google Scholar]
- 33.Li, S.-R., N. Dorrell, P. H. Everest, G. Dougan, and B. W. Wren. 1996. Construction and characterization of a Yersinia enterocolitica O:8 high-temperature requirement (htrA) isogenic mutant. Infect. Immun. 64:2088-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, W., S. M. Srinivasula, J. Chai, P. Li, J. W. Wu, Z. Zhang, E. S. Alnemri, and Y. Shi. 2002. Structural insights into the pro-apoptotic function of mitochondrial serine protease HtrA2/Omi. Nat. Struct. Biol. 9:436-441. [DOI] [PubMed] [Google Scholar]
- 35.Lipinska, B., O. Fayet, L. Baird, and C. Georgopoulos. 1989. Identification, characterization, and mapping of the Escherichia coli htrA gene, whose product is essential for bacterial growth only at high temperatures. J. Bacteriol. 171:1574-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loeffler, J. M., and V. A. Fischetti. 2003. Synergistic lethal effect of a combination of phage lytic enzymes with different activities on penicillin-sensitive and -resistant Streptococcus pneumoniae strains. Antimicrob. Agents Chemother. 47:375-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lyon, W. R., and M. G. Caparon. 2004. Role for serine protease HtrA (DegP) of Streptococcus pyogenes in the biogenesis of virulence factors SpeB and the hemolysin streptolysin S. Infect. Immun. 72:1618-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin, B., M. Prudhomme, G. Alloing, C. Granadel, and J.-P. Claverys. 2000. Cross-regulation of competence pheromone production and export in the early control of transformation in Streptococcus pneumoniae. Mol. Microbiol. 38:867-878. [DOI] [PubMed] [Google Scholar]
- 39.Mascher, T., D. Zähner, M. Merai, N. Balmelle, A. B. de Saizieu, and R. Hakenbeck. 2003. The Streptococcus pneumoniae cia regulon: CiaR target sites and transcription profile analysis. J. Bacteriol. 185:60-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ng, W.-L., G. T. Robertson, K. M. Kazmierczak, J. Zhao, R. Gilmour, and M. E. Winkler. 2003. Constitutive expression of PcsB suppresses the requirement for the essential VicR (YycF) response regulator in Streptococcus pneumoniae R6. Mol. Microbiol. 50:1647-1663. [DOI] [PubMed] [Google Scholar]
- 41.Pallen, M. J., and B. W. Wren. 1997. The HtrA family of serine proteases. Mol. Microbiol. 26:209-221. [DOI] [PubMed] [Google Scholar]
- 42.Pedersen, L. L., M. Radulic, M. Doric, and Y. Abu Kwaik. 2001. HtrA homologue of Legionella pneumophila: an indispensable element for intracellular infection of mammalian but not protozoan cells. Infect. Immun. 69:2569-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pestova, E. V., L. S. Håvarstein, and D. A. Morrison. 1996. Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol. Microbiol. 21:853-862. [DOI] [PubMed] [Google Scholar]
- 44.Poquet, I., V. Saint, E. Seznec, N. Simoes, A. Bolotin, and A. Gruss. 2000. HtrA is the unique surface housekeeping protease in Lactococcus lactis and is required for natural protein processing. Mol. Microbiol. 35:1042-1051. [DOI] [PubMed] [Google Scholar]
- 45.Rimini, R., B. Jansson, G. Feger, T. C. Roberts, M. de Francesco, A. Gozzi, F. Faggioni, E. Domenici, D. M. Wallace, N. Frandsen, and A. Polissi. 2000. Global analysis of transcription kinetics during competence development in Streptococcus pneumoniae using high density DNA arrays. Mol. Microbiol. 36:1279-1292. [DOI] [PubMed] [Google Scholar]
- 46.Robertson, G. T., W.-L. Ng, J. Foley, R. Gilmour, and M. E. Winkler. 2002. Global transcriptional analysis of clpP mutations of type 2 Streptococcus pneumoniae and their effects on physiology and virulence. J. Bacteriol. 184:3508-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanz, J. M., and J. L. Garcia. 1990. Structural studies of the lysozyme coded by the pneumococcal phage Cp-1. Conformational changes induced by choline. Eur. J. Biochem. 187:409-416. [DOI] [PubMed] [Google Scholar]
- 48.Sassoon, N., J. P. Arié, and J. M. Betton. 1999. PDZ domains determine the native oligomeric structure of the DegP (HtrA) protease. Mol. Microbiol. 33:583-589. [DOI] [PubMed] [Google Scholar]
- 49.Sebert, M. E., L. M. Palmer, M. Rosenberg, and J. N. Weiser. 2002. Microarray-based identification of htrA, a Streptococcus pneumoniae gene that is regulated by the CiaRH two-component system and contributes to nasopharyngeal colonization. Infect. Immun. 70:4059-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spiess, C., A. Beil, and M. Ehrmann. 1999. A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell 97:339-347. [DOI] [PubMed] [Google Scholar]
- 51.St Geme, J. W., III, and S. Grass. 1998. Secretion of the Haemophilus influenzae HMW1 and HMW2 adhesins involves a periplasmic intermediate and requires the HMWB and HMWC proteins. Mol. Microbiol. 27:617-630. [DOI] [PubMed] [Google Scholar]
- 52.Strauch, K. L., K. Johnson, and J. Beckwith. 1989. Characterization of degP, a gene required for proteolysis in the cell envelope and essential for growth of Escherichia coli at high temperature. J. Bacteriol. 171:2689-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sung, C. K., H. Li, J. P. Claverys, and D. A. Morrison. 2001. An rpsL cassette, Janus, for gene replacement through negative selection in Streptococcus pneumoniae. Appl. Environ. Microbiol. 67:5190-5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]
- 55.Throup, J. P., K. K. Koretke, A. P. Bryant, K. A. Ingraham, A. F. Chalker, Y. Ge, A. Marra, N. G. Wallis, J. R. Brown, D. J. Holmes, M. Rosenberg, and M. K. Burnham. 2000. A genomic analysis of two-component signal transduction in Streptococcus pneumoniae. Mol. Microbiol. 35:566-576. [DOI] [PubMed] [Google Scholar]
- 56.Tiraby, J. G., and M. S. Fox. 1973. Marker discrimination in transformation and mutation of pneumococcus. Proc. Natl. Acad. Sci. USA 70:3541-3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tomasz, A. 1965. Control of the competent state in Pneumococcus by a hormone-like cell product: an example for a new type of regulatory mechanism in bacteria. Nature 208:155-159. [DOI] [PubMed] [Google Scholar]
- 58.Tomasz, A. 1966. Model for the mechanism controlling the expression of competent state in pneumococcus cultures. J. Bacteriol. 91:1050-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tomasz, A., and R. D. Hotchkiss. 1964. Regulation of the transformability of pneumococcal cultures by macromolecular cell products. Proc. Natl. Acad. Sci. USA 51:480-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tomasz, A., and J. L. Mosser. 1966. On the nature of the pneumococcal activator substance. Proc. Natl. Acad. Sci. USA 55:58-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wagner, C., A. de Saizieu, H. J. Schönfeld, M. Kamber, R. Lange, C. J. Thompson, and M. G. Page. 2002. Genetic analysis and functional characterization of the Streptococcus pneumoniae vic operon. Infect. Immun. 70:6121-6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ween, O., P. Gaustad, and L. S. Håvarstein. 1999. Identification of DNA binding sites for ComE, a key regulator of natural competence in Streptococcus pneumoniae. Mol. Microbiol. 33:817-827. [DOI] [PubMed] [Google Scholar]
- 63.Williams, K., P. C. F. Oyston, N. Dorrell, S. R. Li, R. W. Titball, and B. W. Wren. 2000. Investigation into the role of the serine protease HtrA in Yersinia pestis pathogenesis. FEMS Microbiol. Lett. 186:281-286. [DOI] [PubMed] [Google Scholar]