Abstract
DNA regulatory motifs reflect the direct transcriptional interactions between regulators and their target genes and contain important information regarding transcriptional networks. In silico motif detection strategies search for DNA patterns that are present more frequently in a set of related sequences than in a set of unrelated sequences. Related sequences could be genes that are coexpressed and are therefore expected to share similar conserved regulatory motifs. We identified coexpressed genes by carrying out microarray-based transcript profiling of Salmonella enterica serovar Typhimurium in response to the spent culture supernatant of the probiotic strain Lactobacillus rhamnosus GG. Probiotics are live microorganisms which, when administered in adequate amounts, confer a health benefit on the host. They are known to antagonize intestinal pathogens in vivo, including salmonellae. S. enterica serovar Typhimurium causes human gastroenteritis. Infection is initiated by entry of salmonellae into intestinal epithelial cells. The expression of invasion genes is tightly regulated by environmental conditions, as well as by many bacterial factors including the key regulator HilA. One mechanism by which probiotics may antagonize intestinal pathogens is by influencing invasion gene expression. Our microarray experiment yielded a cluster of coexpressed Salmonella genes that are predicted to be down-regulated by spent culture supernatant. This cluster was enriched for genes known to be HilA dependent. In silico motif detection revealed a motif that overlaps the previously described HilA box in the promoter region of three of these genes, spi4_H, sicA, and hilA. Site-directed mutagenesis, β-galactosidase reporter assays, and gel mobility shift experiments indicated that sicA expression requires HilA and that hilA is negatively autoregulated.
Infections with Salmonella serotypes are a major cause of food-borne diseases worldwide (89). Salmonella enterica serovar Typhimurium usually causes gastroenteritis. Although this is often a self-limiting disease marked by diarrhea and abdominal cramps, the infection can be more severe, resulting in bacteremia, fever, or even death (72). Salmonellosis is initiated when S. enterica serovar Typhimurium crosses the intestinal mucosa of a host (41). Many of the genes required for Salmonella epithelial cell invasion are encoded on Salmonella pathogenicity island 1 (SPI1) (23, 101, 105, 106). The invasive phenotype varies greatly in response to growth under different environmental conditions (e.g., osmolarity, oxygen tension, pH) (63, 65).
An intricate regulatory network is responsible for transmitting environmental signals into appropriate gene expression. HilA, a member of the ToxR/OmpR-like family of transcriptional regulators, is a major player in this network. Its expression is dependent upon several transcription factors that are important for virulence, including PhoP, RtsA, SirA, HilC, HilD, and Fis (9, 10, 35, 61, 63, 78, 90-92, 97). HilA in turn activates genes encoding the SPI1 type III secretion machinery and also InvF, which induces the expression of SPI1 secreted effectors (2, 7, 8, 81). Activation of these effectors by InvF requires SicA, a type III secretion system chaperone (22, 24, 32), which has been suggested to stabilize a complex between InvF, RNA polymerase, and DNA (25). HilA also regulates genes in the pathogenicity island SPI4, which is required for the enteric phase of pathogenesis (74, 103).
The SPI1 regulatory cascade is believed to be induced in the small intestine (16, 23, 50), where salmonellae encounter multiple and diverse bacterial species belonging to the endogenous intestinal microbiota (44, 46, 70). There has been recent interest in using some of these intestinal bacterial species as probiotics for the prevention and treatment of food-borne infectious diseases, including salmonellosis (62). Probiotics are live microorganisms which, when administered in adequate amounts, confer a health benefit on the host (39, 40).
We examined gene expression profiles of S. enterica serovar Typhimurium in spent culture supernatant (SCS) of the probiotic Lactobacillus rhamnosus GG (L. rhamnosus GG) (94), which has been reported to antagonize Salmonella infection (47, 57), to define coexpressed genes. Data analysis unveiled a cluster of genes with an expression profile corresponding to genes repressed by L. rhamnosus GG SCS. This cluster was enriched for genes known to be HilA regulated. Motif discovery revealed the presence of a conserved box overlapping with the previously described HilA box (59) in the promoter region of invF and prgH and of spi4_H, sicA and hilA. Using site directed mutagenesis, reporter constructs, and gel mobility shift assays, we confirmed that these latter genes are three additional targets of the master regulator HilA and we could further link the repression of Salmonella's invasion regulatory system to a probiotic effect.
MATERIALS AND METHODS
Bacterial strains, plasmids and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. All strains were grown at 37°C. Lactobacillus rhamnosus GG (ATCC 53103) was inoculated from a glycerol stock (−80°C) in Man-Rogosa-Sharpe medium (MRS, Difco) (27). S. enterica serovar Typhimurium and Escherichia coli were grown in Luria-Bertani (LB) broth (87). L. rhamnosus GG was grown in nonshaking conditions. Except for common cloning procedures and as otherwise stated, salmonellae were cultured under high-osmolarity and limited-aeration conditions, previously shown to promote the induction of SPI1 genes and to induce adherence and invasiveness (8, 56, 63). For agar plates, 15 g/liter agar was added. If appropriate, antibiotics were added at following final concentrations: ampicillin, 100 μg/ml; streptomycin, 25 μg/ml; and tetracycline, 10 μg/ml or 30 μg/ml (when growing plasmid containing strains for β-galactosidase assays).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| E. coli DH5α | F− φ80ΔlacZM15 Δ(lacZYA argF)U169 deoP recA1 endA1 hsdR17 (rK− mK−) | Gibco BRL |
| E. coli TOP10F′ | F′ [lacIq Tn10(TetR)] mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| L. rhamnosus GG (LGG) | Wild type; human isolate | ATCC 53103 |
| S. enterica serovar Typhimurium SL1344 | xyl hisG rpsL; virulent; Smr | 45 |
| S. enterica serovar Typhimurium VV302 | SL1344 ΔhilA-523; hilA mutant | 7 |
| S. enterica serovar Typhimurium LB5010 | LT2 derivative; restriction negative, modification positive (r− m+) for hsdLT, hsdSA, and hsdSB; galE strain sensitive to phage P1; metA22 metE551 ilv-452 leu-3121 trpΔ2 xyl-404 galE856 hsdLT6 hsdSA29 hsdSB121 rpsL120; noninvasive | 14 |
| Plasmids | ||
| pCMPG5321 | 917-bp fragment of pLS31, containing promoter of hilA (PhilA), cloned into pUC19 (EcoRI-BamHI); Ampr | This work |
| pCMPG5322 | 404 bp fragment of pHD11, containing promoter of sicA (PsicA), cloned into pUC19 (EcoRI-BamHI); Ampr | This work |
| pCMPG5324 | Point-mutated pCMPG5321, i.e. C→T at position +78 of hilA; Ampr | This work |
| pCMPG5325 | Point-mutated pCMPG5322, i.e. T→C at position +2 of sicA; Ampr | This work |
| pCMPG5401 | Point-mutated PhilA as an EcoRI-BamHI fragment of pCMPG5324 cloned into pRW50; Tcr | This work |
| pCMPG5402 | Point-mutated PsicA as an EcoRI-BamHI fragment of pCMPG5325 cloned into pRW50; Tcr | This work |
| pFAJ1932 | 1,520-bp PCR fragment containing part of the spi4 H promoter (Pspi4_H) (8821 → 10340 of GenBank entry AF060869) cloned into pUC18 (EcoRI-PstI); Ampr | This work |
| pHD11 | pRW50 containing 404 bp fragment carrying promoter region of sicA (EcoRI/BamHI) (intergenic sequence between spaS and sicA (137 bp) along with 192 bp of the 3′ end of spaS and 76 bp of sicA); Tcr | 22 |
| pLS31 | pRW50 containing −497 to +420 of hilA (EcoRI/BamHI); Tcr | 90 |
| pRW50 | Low-copy-number transcriptional reporter fusion vector (lacZY; 1-2 copies per cell); Tcr | 58 |
| pBAD/Myc-His | Cloning vector to make C-terminal Myc- and His-tagged proteins expressed under arabinose control; Ampr | Invitrogen |
| pCMPG5338 | hilA ORF cloned in pBAD/Myc-His | This work (59) |
| pUC18 | 2.7-kb cloning vector; Ampr | 104 |
| pUC19 | 2.7-kb cloning vector; Ampr | 104 |
Strain and plasmid construction.
Standard protocols were used for buffer preparation, cloning, plasmid isolation, and E. coli competent cell preparation and transformation (87). Salmonellae were transformed as previously described (82). Plasmids isolated from SL1344 were back transferred to E. coli and reisolated prior to restriction analysis. Restriction enzymes were used according to the manufacturer's instructions. DNA fragments were agarose-purified using the QIAquick Gel Extraction kit (Qiagen).
The primers used for PCR (purchased from Eurogentec) are listed in Table 2. PCR was carried out in a Personal Mastercycler (Eppendorf). PCR amplification of the spi4_H putative promoter region was done with the proofreading Pfx enzyme (construction of pFAJ1932), according to the manufacturer's instructions.
TABLE 2.
Primer sequences used for PCRa
| Name | Sequence (from 5′ to 3′) | Description |
|---|---|---|
| M13 reverse | CAGGAAACAGCTATGACC | Sequencing |
| M13 universal | GTTGTAAAACGACGGCCAGT | Sequencing |
| RHI-168 | CCGAATTCAGGGCGCCTATGATATTGAAATC | Putative spi4_H promoter region |
| RHI-169 | GGCTGCAGTTAACGTGTAGCTGCCATCCGCC | Putative spi4_H promoter region |
| RHI-184 | CTGACTCTCTCTGCATCAGGATATACGGCAG | Site-directed mutagenesis hilA |
| RHI-185 | CTGCCGTATATCCTGATGCAGAGAGAGTCAG | Site-directed mutagenesis hilA |
| RHI-186 | GGGTTTAATAACTGCACCAGATAAACGCAGTCG | Site-directed mutagenesis sicA |
| RHI-187 | CGACTGCGTTTATCTGGTGCAGTTATTAAACCC | Site-directed mutagenesis sicA |
| PRO-407 | TTAACCATGGCTCATTTTAATCCTGTTCC | Forward hilA in pCMPG5338 (59) |
| PRO-408 | TTGTTCTAGAATTAATTTAATCAAGCGGGG | Reverse hilA in pCMPG5338 (59) |
Restriction sites are indicated in bold; mutated positions are underscored.
Primers RHI-168 and RHI-169 were used to amplify a 1,520-bp DNA fragment upstream of the spi4_H gene from the SL1344 chromosomal DNA. The 1,520-bp PCR fragment was digested with EcoRI and PstI and cloned into pUC18 that had been digested with EcoRI and PstI, yielding pFAJ1932. Restriction and sequence analysis of pFAJ1932 confirmed the directional insertion of the putative spi4_H promoter in pUC18 (data not shown).
The construction of the hilA-lacZY (pLS31) and sicA-lacZY (pHD11) reporter fusions has been described previously (22, 90). pLS31 and pHD11 were electroporated into SL1344 and VV302 after propagation through LB5010. Cloning steps were performed in E. coli DH5α and TOP10F′.
Single-base-pair substitutions in the putative HilA box occurring in the hilA and sicA promoter sequences were introduced via a PCR approach using the QuickChange site-directed mutagenesis kit (Stratagene), according to the manufacturer's instructions. Since the pLS31 and pHD11 plasmids were too large to obtain a successful point mutation, the promoter containing fragments of both reporter plasmids were subcloned as EcoRI/BamHI fragments into the corresponding sites of pUC19, yielding pCMPG5321 and pCMPG5322, respectively. The primers applied in the mutagenesis protocol are displayed in Table 2. As a result of the engineered mutation, the unique SfaNI site in the sicA promoter fragment disappeared. In the hilA promoter fragment a unique SfaNI site was created and the unique BstNI site disappeared. This information combined with sequence analysis allowed us to confirm the single-base-pair substitutions. The point-mutated hilA and sicA promoter fragments were subcloned into pRW50, resulting in pCMPG5401 and pCMPG5402, respectively. These reporter plasmids were electroporated into SL1344 and VV302.
The construction of pCMPG5338 is based on the design of pCH112 (59). Briefly, primers PRO-407 and PRO-408 (Table 2) were used to amplify hilA with S. enterica serovar Typhimurium SL1344 genomic DNA as a template, while simultaneously introducing restriction sites. The PCR fragment was cloned into Invitrogen's pBAD/Myc-His plasmid, creating an in-frame fusion with the Myc-His C-terminal tag. To this end, both the PCR product and the vector were digested with NcoI and XbaI and subsequently ligated using T4 DNA ligase, yielding pCMPG5338. The tagged HilA is functional and able to activate invasion gene promoters (data not shown).
All constructs were confirmed by sequencing. Sequences were determined by the chain termination dideoxynucleoside triphosphate method (88) either with the AutoRead sequencing kit (Pharmacia-LBK) on an automated sequencer (ALX; Pharmacia-LBK) or via cycle sequencing using the BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems), ethanol/EDTA/sodium acetate precipitation, and subsequent separation of DNA fragments on an ABI 3100-Avant DNA analyzer (Applied Biosystems), according to the manufacturer's instructions. The Cy5-labeled M13 reverse and forward primers were used for sequence confirmation of spi4_H putative promoter region (pFAJ1932) and mutated hilA and sicA promoter sequences (pCMPG5324 and pCMPG5325). The hilA overexpression construct (pCMPG5338) was sequenced using primers PRO-407 and PRO-408 (Table 2). Sequence data banks were screened for similarities by using the BLAST program (4, 5).
Lactobacillus rhamnosus GG spent culture supernatant.
Lactobacillus rhamnosus GG was grown in 10 ml of MRS broth at 37°C overnight without shaking. The culture was inoculated from a −80°C glycerol stock. This L. rhamnosus GG overnight culture was used to inoculate (1:100) fresh MRS broth (350 ml in a 500-ml Erlenmeyer). SCS was obtained from a 24-h culture (37°C, without agitation) by centrifugation for 30 min at 10,000 × g at 4°C, followed by filter sterilization (0.22 μm; Millipore).
Microarray printing.
We used a dedicated array consisting of approximately 500 genes. These genes were hand-picked with a bias towards known virulence determinants and genes we thought may play undiscovered roles in virulence. All steps of PCR product precipitation, resuspension and dilution, glass slide preparation, printing, and processing after printing were performed as previously described (17, 34).
Sample preparation.
Salmonella strain SL1344 was grown overnight in nonaerated culture at 37°C. Overnight cultures were 1:50 diluted into fresh LB medium and incubated for another 3 to 7 h in the same conditions. Cells reached the mid-log phase and were used for induction with SCS. To this end, 109 S. enterica serovar Typhimurium SL1344 cells were centrifuged 5 min at 6,000 × g and 5 ml of the mixture of LB and L. rhamnosus GG SCS (at a 1:12 ratio; pH 5) was used to resuspend the cell pellet. As controls, the following induction media were used in a similar experiment: MRS in fresh LB broth and brought to pH 5 (with HCl), MRS in fresh LB broth (at a 1:12 ratio) (pH = 6.8), and L. rhamnosus GG SCS in fresh LB broth (at a 1:12 ratio) and brought to pH 6.8 (with NaOH). All induction media were filter sterilized (0.22 μm) prior to use. After 1 and 5.5 h of induction, ≈109 CFU of each condition were used for RNA isolation. The cultures were centrifuged (1 min at 14,000 × g) in the presence of Bacterial Protect reagent (Qiagen) according to the manufacturer's instructions for RNA stabilization. No antibiotics were added to the media.
RNA isolation, labeling, and slide hybridization.
Total RNA was isolated with the Qiagen RNeasy mini kit according to the manufacturer's protocol. Contaminating genomic DNA was removed from the RNA samples on-column with Qiagen RNase-free DNase. Removal of DNA was checked by PCR. Prior to labeling, the concentration of total RNA was determined by measuring the absorbance at 260 nm (UV/VIS Lambda 2, Perkin Elmer). cDNA was synthesized from 50 μg RNA with pd(N)6 random hexamer (Amersham Biosciences) and labeled as previously described (34). Genomic Salmonella DNA was used as a labeling and hybridization reference. For each condition, the Cy3 and Cy5 reactions were combined and further handled as described (86). Hybridization took place overnight under a glass coverslip in a humidified slide chamber submerged in a 62°C water bath. The hybridized slides were washed (34), dried, and scanned for fluorescence with a commercial laser scanner (GenePix Scanner 4000A; Axon Instruments Inc., Foster City, CA). Signal intensities and background measurements were obtained for each spot on the array by using the GenePixPro 3.0 software program (Axon Instruments, Foster City, CA).
Data analysis.
Background corrected median values were used for further analysis. Data were normalized using analysis of variance (MatLab script provided by Kerr et al.) (54). This reference design model includes an array main factor, a variety main factor, factors compensating for dye and condition related variation, a gene main factor and the factor of interest, i.e., the variety gene interaction factor, which reflects differences in gene expression level that are not explained by the factor levels (effects) of the main variety and gene factors (36, 52-54). Each of the conditions tested corresponded to a separate variety effect. The independent genomic reference was considered as an additional variety effect.
Normalized values (VG effects) of the nonreference samples were used to cluster the data. Genes with similar expression profiles across the different conditions were grouped by means of the adaptive quality-based cluster algorithm (28) with 0.85% as quality criterion. Subsequently, we searched for statistically overrepresented motifs in the intergenic regions of the coexpressed genes (genes within a cluster). Intergenic regions in this study are defined as a region that contains the noncoding region between two coding regions and are extracted from GenBank files (11) using the modules of INCLusive (19). For the genes in the clusters that are known to belong to an operon, the intergenic region upstream the first gene of the operon was selected. We used Motif Sampler (67, 99), a motif detection procedure based on Gibbs sampling. This algorithm identifies conserved patterns based solely on statistical properties and no prior information on what the motif should look like is required (55). For each data set (cluster) the algorithm was run 100 times using the following parameter settings: motif length 8 to 12 and background order 3.
β-Galactosidase activity assays.
The lacZY fusion strains in either a wild-type SL1344 (45) or hilA deletion background VV302 (7) were grown under derepressing conditions (i.e., high osmolarity [LB, 10 g/liter NaCl], oxygen-limiting) (56) (Fig. 3A and 4A) and repressing conditions (low osmolarity [LB, 0 g/liter NaCl], aeration) (Fig. 4B). Expression of lacZY fusions was assessed using β-galactosidase assays as previously described (71), with minor modifications, resulting in the following optimized microtiter plate-based protocol.
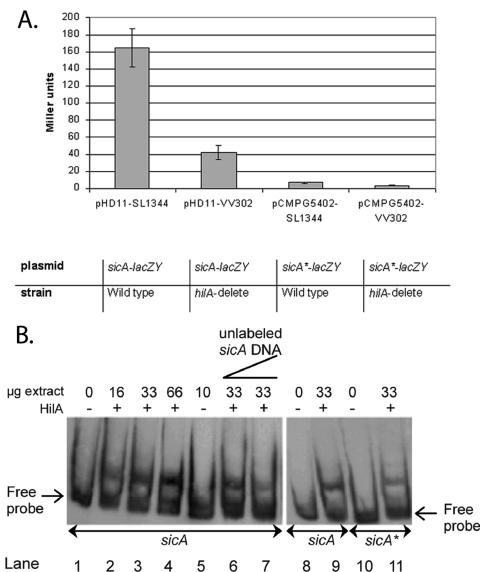
FIG. 3.
HilA box in the promoter region of sicA is important for its HilA regulation. A. sicA reporter gene fusion assays. A single-base-pair substitution was introduced into the promoter sequence of sicA present in pHD11 by site-directed mutagenesis, giving rise to pCMPG5402 (sicA*-lacZY). The lacZY fusion strains in either a wild-type (SL1344) (45) or hilA deletion background (VV302) (7) were grown under derepressing conditions (i.e., high osmolarity [10 g/liter NaCl], oxygen-limiting) (56) and assayed for β-galactosidase activity (71). Values are expressed in Miller units and represent the mean of eight independent experiments. Miller unit values of strains containing the vector pRW50 (58) were zero (data not shown). Error bars indicate standard deviations. B. HilA+ extract alters the gel mobility of the sicA promoter DNA fragment. Gel mobility shift assays were performed with the sicA (lanes 1 to 9) and sicA* (lanes 10 and 11) probe as outlined in Materials and Methods. Lanes 1, 8, and 10 are controls showing the migration of the probe in the absence of any added protein. Lanes 2 to 4 contain increasing amounts of HilA+ extract (i.e., 16, 33, and 66 μg). Lane 5 contains 10 μg of the HilA− extract. Lanes 6 and 7 contain increasing amounts of unlabeled sicA promoter fragment as a specific competitor (i.e., 10 and 50 ng). Lane 11 contains 33 μg of the HilA+ extract.
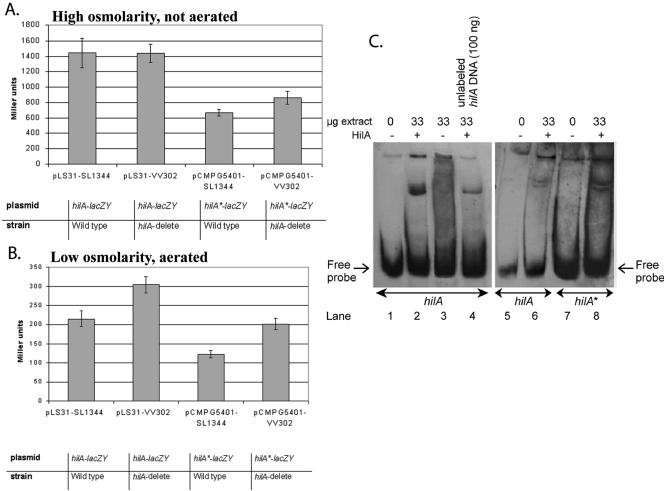
FIG. 4.
Identified HilA box in the hilA promoter is important for hilA regulation. A and B. hilA reporter gene fusion assays. A single-base-pair substitution was introduced into the promoter sequence of hilA present in pLS31 by site-directed mutagenesis, giving rise to pCMPG5401 (hilA*-lacZY). The lacZY fusion strains in either a wild-type (SL1344) (45) or hilA deletion background (VV302) (7) were grown under derepressing conditions (i.e., high osmolarity [10 g/liter NaCl], oxygen-limiting) (56) (A) and repressing conditions (low osmolarity [0 g/liter NaCl], aeration) (B) and assayed for β-galactosidase activity (71). Values are expressed in Miller units and represent the mean of eight independent experiments. Miller unit values of strains containing the vector pRW50 (58) were zero (data not shown). Error bars indicate standard deviations. C. HilA+ extract alters the gel mobility of the hilA promoter DNA fragment. Gel mobility shift assays were performed with the hilA (lanes 1 to 6) and hilA* (lanes 7 and 8) probe as outlined in Materials and Methods. Lanes 1, 5, and 7 are controls showing the migration of the probe in the absence of any added protein. Lanes 2, 6, and 8 contain 33 μg of HilA+ extract. Lane 3 contains 33 μg of the HilA− extract. Lane 4 contains 100 ng unlabeled hilA promoter fragment as a specific competitor.
Single colonies were inoculated into 96-well plates containing 300 μl medium, supplemented with the appropriate antibiotics, and incubated in either derepressing or repressing conditions. Overnight cultures were diluted 1:50 into fresh medium (with the appropriate antibiotics) and cultured for another 4 to 5 h in the same conditions. These cultures were used when assessing the effect of hilA deletion background and point mutations in the putative HilA box on the reporter plasmids. To 10 μl of cell suspensions (optical density at 595 nm [OD-595] of ≈0.3), 90 μl of LacZ buffer (50 mM NaHPO4 [pH 7.0], 14.3 mM β-mercaptoethanol, 1 mM Na2EDTA, 0.1% Triton X-100, 0.1% Na-laurylsarcosine, 25 mM ortho-nitrophenylgalactopyranoside [ONPG, Sigma]) was added. The cultures were diluted 10-fold prior to β-galactosidase activity measurement when pLS31-containing strains were used. This dilution was taken into consideration when calculating the Miller units.
The mixture was incubated at 30°C and the reaction was stopped by adding 35 μl of a 1 M Na2CO3 solution once sufficient yellow color had developed. The reaction was stopped at at least three different time points (replicates) to ascertain that enzyme activity was still linearly increasing with incubation time. The time of reaction was recorded. Optical density was measured at both 420 and 550 nm (OD420 and OD550). Identical treatments were performed with LacZ buffer without cells as control to correct measured sample values. Miller units of β-galactosidase activity were calculated as 1,000 times the increase in absorbance at 420 nm per minute per unit of optical density at 550 nm of the cell suspension: Miller units = 1,000 ×{[(OD420, ONPG − 1.75 × OD550, ONPG) × v1]/(t × vt × OD595)} where t is the time of the reaction in minutes; OD595 reflects the cell density just before the assay; OD420, ONPG reflects absorbance by ONPG, measured after reaction; OD550, ONPG reflects cell density measured after reaction, used as correction for light scattering by cell debris; 1.75 is the corresponding correction factor; v1 is the volume (μl) of cells used in the reaction mixture; and vt is the total volume (μl) of the reaction mixture.
Gel mobility shift assays.
The promoter regions upstream of hilA, sicA, and spi4_H were obtained by digestion of plasmids pLS31 (90) and pHD11 (22) with EcoRI and BamHI and pFAJ1932 with EcoRI and PstI. The point mutated promoter fragments were isolated by digestion of pCMPG5324 and pCMPG5325 with EcoRI and BamHI, respectively. The DNA fragments were purified by agarose gel electrophoresis followed by gel extraction with a QIAquick gel extraction kit; 100 ng of each fragment was end-labeled at 37°C for 15 min with digoxigenin-11-ddUTP using a digoxigenin gel shift kit (Roche Applied Science, Penzberg, Germany), according to the manufacturer's instructions. Labeling efficiency was checked by comparing spotted dilution series of labeling reaction to a labeled control-fragment in a direct detection assay, as outlined in the protocol of the kit.
HilA+ and HilA− extracts were prepared by ultracentrifugation of, respectively, sonicated arabinose-treated (0.02%) and untreated TOP10 cells carrying pCMPG5338, as previously described (59). Since it was reported that as an artifact of overproduction of the protein, HilA is membrane associated (59, 84), only membrane-associated fractions of the extracts were used, i.e., the pellet of the ultracentrifuged extracts. Protein concentration was determined by the Bio-Rad protein assay (13), according to the manufacturer's instructions. Western blotting (data not shown), using anti-c-Myc antibodies (M4439, Sigma), confirmed the presence of HilA in the membrane fraction of HilA+ extracts. No HilA could be detected in the HilA− extract. This extract was used as a negative control.
DNA binding reactions were carried out as previously described (97) in a total volume of 15 μl containing 5 μl of 3x DNA binding buffer (129 mM Tris-HCl, 90 mM potassium acetate, 24 mM MgSO4, 81 mM ammonium acetate, 3 mM dithiothreitol, 240 mM KCl, 30% glycerol) and different concentrations of the HilA+ extract in 5 μl total volume (2.2 μg/μl to 81 ng/μl), 2 μl of labeled DNA fragment (≈0.4 ng/μl, as recommended by the manufacturer of the applied digoxigenin kit), 2 μl of poly(dI-dC) (1 μg/μl), 1 μl of bovine serum albumin (1 μg/μl), and 0.5 μl of 0.5 M EDTA. HilA− extract was used at a concentration of 666 ng/μl. Nonspecific competitor DNA [poly(dI-dC)] and protein (bovine serum albumin) were added to all reactions to minimize nonspecific interactions of the labeled DNA fragments with the proteins. DNA binding reactions were carried out at room temperature for 25 min. Reactions were separated on native Tris-borate-EDTA (TBE)-polyacrylamide gels (5%) prepared and run at 8 mA for 1 h with 20 μl of freshly made 5% thioglycolate in the cold room (59). After 2 to 5 h of electrophoresis (depending on the size of the probe) at 8 V cm−1 in 0.5× TBE buffer, gels were electroblotted (40 min, 300 mA; LKB Bromma 2117 Multiphor II electrophoresis unit) and further handled for chemiluminescent detection as outlined by the manufacturer of the digoxigenin gel shift kit (Roche Applied Science, Penzberg, Germany).
RESULTS AND DISCUSSION
Identification of clusters containing genes with similar expression profiles.
To identify coexpressed genes, we brought salmonellae into contact with different conditions related to the probiotic lactic acid bacterium L. rhamnosus GG and performed S. enterica serovar Typhimurium cDNA microarray experiments. RNA was extracted at 1.0 and 5.5 h after exposure to SCS. It has been reported that the promoter of an important Salmonella SPI-1 invasion gene, sicA (51), is activated in the intestinal lumen by 1 h after infection and in the Peyer's patches by 5 h, but is repressed after 24 h of infection (15). The different experimental conditions included: L. rhamnosus GG spent culture supernatant (SCS) and sterile MRS medium, both at neutral pH and at pH 5.0. However, neutralizing the pH of the SCS eliminates its growth-inhibitory effect on salmonellae (95), so these data should be interpreted with caution. In total, we performed microarray experiments with RNA isolated from bacteria exposed to seven different conditions, described as follows: SCS pH 5.0 (1 h), SCS pH 5.0 (5.5 h), sterile MRS medium pH 6.8 (1 h), MRS pH 6.8 (5.5 h), MRS pH 5.0 (1 h), MRS pH 5.0 (5.5 h), and SCS pH 6.8 (1 h).
Genes with similar expression profiles over the different conditions, i.e., genes that are coexpressed, were grouped by cluster analysis. The expression pattern of one cluster indicated that it contains genes repressed by Lactobacillus SCS. This cluster was enriched for genes important for cell invasion by salmonellae, including hilA, invA, invF, invI, prgH, prgJ, sicA, sigD (sopB), sipB, sopE, spaO, spaQ, spaR, spi4_C, spi4_F, spi4_H, spi4_O, spi4_P, spi4_R, sptP, and yjbA. One of these genes, hilA, is a key virulence regulator that responds to several environmental signals (8, 61) and is potentially a target for therapeutics, including probiotics. Repression of hilA results in the down-regulation of multiple genes important for invasion, including many of the genes that had lower RNA levels in SCS. Moreover, PhoP is a postulated repressor of hilA (8, 10, 42, 81), and RNA levels of both phoP and genes belonging to the PhoP regulon (e.g., pagM, mgtB, marB) (43) increased upon exposure to SCS. These data suggest that L. rhamnosus GG exerts its antagonistic effect on salmonellae in part by repressing Salmonella invasion genes.
While the effect of Lactobacillus acidophilus supernatant on the cell entry of salmonellae has been described before (18), putative Salmonella target genes were not identified. The observation could be partly explained by the effect of low pH on the expression of virulence genes such as hilA (8, 30). This was also observed in our MRS at pH 5. However, in SCS pH 5, the observed repression was more severe, i.e., eightfold difference for hilA (data not shown). This could be due to lactic acid, a major compound present in Lactobacillus SCS. It has been suggested that lactic acid inhibits hilA expression (30, 31), but follow-up experiments were not performed. We found that exposure of salmonellae to L. rhamnosus GG SCS reduces the RNA levels of multiple invasion genes. This microarray experiment was applied to generate clusters of coexpressed genes to focus further experiments on the regulation of these genes.
Validation of clusters through motif detection: a shifted putative HilA box.
Coexpressed genes may have similar transcriptional regulatory mechanisms and their promoter regions may contain common motifs or regulatory elements that bind transcription factors (73, 98, 100). Motif detection strategies involve searching for DNA patterns that are overrepresented in a set of related sequences relative to a set of unrelated sequences. The putative promoter regions of the genes in the cluster that had lower RNA levels upon SCS exposure and that contain multiple Salmonella invasion genes was subjected to motif detection using Gibbs sampling (67, 99, 100).
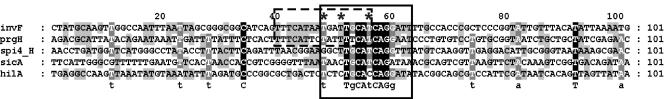
We identified a motif that partially overlaps the previously described HilA box (59) (Fig. 1). The HilA box was initially suggested to consist of two nearly perfect 6-nucleotide direct repeats centered around a T in the prgH and invF promoters, TTTCATNNTNNTTkCAT (59). The overrepresented motif in the cluster revealed by our in silico analysis (tN3TgCAtCAGga) overlaps the HilA box and includes the three nucleotides shown to be essential for HilA binding (59) (Fig. 1). We detected this motif in the promoter regions of prgH and invF, known HilA targets (7). In addition, the tN3TgCAtCAGga motif is present in the promoters of three additional genes, sicA, hilA, and spi4_H (Fig. 1).
FIG. 1.
Alignment of the putative HilA-box. Motif detection revealed the presence of a HilA-box (59). However, the consensus sequence retrieved by motif detection is shifted by 9 nucleotides and is indicated with a black box; consensus sequences as described in the literature are boxed with a dotted line for comparison. Sequences upstream of the translational start site of the indicated genes were taken from the complete genome sequence of S. enterica serovar Typhimurium (69) (NC_003197). Intergenic sequences were aligned using the motif positions as seeds and edited in GeneDoc (76). Color coding: black indicates conserved in all aligned sequences, dark grey indicates conserved in at least 80% of the aligned sequences, and light grey indicates conserved in at least 60% of the aligned sequences. The three nucleotides critical for binding and activation (59) are indicated with an asterisk.
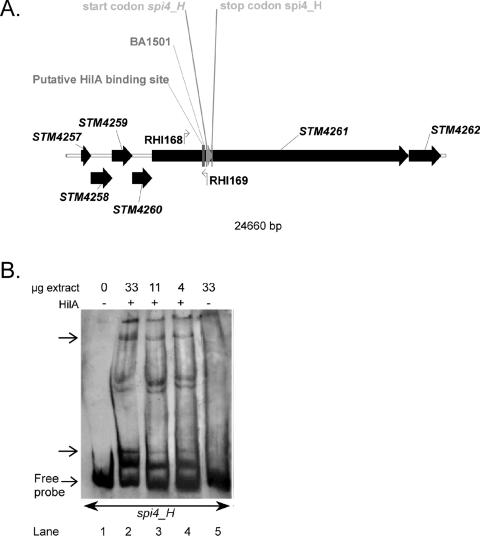
HilA box found in promoter region of spi4_H.
SPI4 has a major role in influencing intestinal colonization of mammalian species (74). The SPI4 gene spi4_H (GenBank entry AF060869) was originally described by Wong et al. (103). However, the current annotation of the S. enterica serovar Typhimurium LT2 genome (69) lacks spi4_H and the original spi4_H sequence is now located within a strikingly large (16,679 bp) gene, STM4261, recently named both icgA (invasion-coregulated gene A) (35) and siiE (Salmonella intestinal infection gene E) (74). icgA/siiE is a putative homologue of HlyA (α-hemolysin) (29) and is predicted to encode a type 1 exported RTX (repeat in toxin) pore-forming toxin or adhesin (35). Based on MudJ fusion experiments, icgA/siiE was suggested to be directly or indirectly regulated by HilA (35).
We found that the upstream region of the originally described spi4_H gene contains a putative HilA box (Fig. 1 and 2A). In addition, gel mobility shift assays demonstrated the binding of HilA to the spi4_H promoter (Fig. 2B). These results are consistent with the original description of spi4_H as a gene that is regulated by SirA in a HilA-dependent manner (BA1501 MudJ fusion, depicted in Fig. 2B) (1). Ellermeier and Slauch (35) also described HilA regulation of icgA::MudJ, however, they did not describe where their MudJ insertion occurred in STM4261 (icgA). Therefore, the lacZ expression they observed could correspond to that of spi4_H.
FIG. 2.
A. Genetic organization of S. enterica serovar Typhimurium SPI4. The systematic number designation (STM) of open reading frames annotated in the S. enterica serovar Typhimurium genome (69) is given. The start and stop codons of the spi4_H genes, as annotated in GenBank entry AF060869, have been indicated. Additional features on the diagram of SPI4 are as follows: the detected putative HilA box, the binding sites of the primers used for amplification of the spi4_H promoter region (RHI-168 and RHI-169), and the insertion position of MudJ in BA1501, a SirA and HilA-regulated fusion (1). B. HilA+ extract alters the gel mobility of the spi4_H promoter DNA fragment. Gel mobility shift assays were performed with the spi4_H probe as outlined in Materials and Methods. Lane 1 is a control showing the migration of the probe in the absence of any added protein. Lanes 2 to 4 contain decreasing amount of HilA+ extract (i.e., 33, 11, and 4 μg). Lane 5 contains 33 μg total proteins of the HilA− extract. The arrows indicate the suggested DNA-protein interaction.
However, under the conditions we used, LB medium and low oxygen, the spi4_H promoter we amplified could not drive the expression of the lacZY gene (data not shown). It is possible that the promoter region we amplified was incomplete or that other environmental cues are required to induce spi4_H expression. In sum, these data support the idea that the newly annotated STM4261 locus likely either contains at least two genes, icgA/siiE and spi4_H, or icgA/siiE and spi4_H correspond to the same gene. However, this should be interpreted with caution. Although gel mobility shift assays indicated that HilA interacts with the amplified spi4_H upstream region, as long as we cannot determine the right conditions to switch on spi4_H expression and prove that it encodes a functional gene, the role of HilA and its interaction with the motif found upstream of this possible gene is premature.
HilA binds to and regulates sicA via the HilA box.
The presence of an HilA consensus sequence in the sicA promoter region has not been previously reported, and HilA is not generally believed to directly activate the sicA promoter. To determine whether sicA is regulated by HilA, expression studies using an episomal sicA reporter gene fusion were conducted. The sicA reporter contained either a wild-type (pHD11 (22) or a mutant (pCMPG5402) HilA box. The third T (italic) in the HilA box consensus sequence, tN3TgCAtCAGg, was previously shown to be critical for HilA DNA binding to the prgH and invF promoters (Fig. 1) (59, 60). Therefore, in the sicA promoter, we substituted this T with a C (Fig. 1). Reporter gene fusion assays were performed under multiple environmental conditions and in wild-type and hilA deletion backgrounds (Fig. 3A).
In a hilA deletion background, sicA reporter expression is significantly reduced, as reported previously (24). The single T-to-C substitution in the putative HilA box of the sicA reporter construct (pCMPG5402, i.e., sicA*-lacZY) completely abolished expression in both the wild-type and hilA deletion backgrounds (Fig. 3A). A similar observation was made by Lostroh et al. (59) regarding the invF and prgH promoters. Thus, the putative HilA box is important for HilA-dependent sicA induction.
To determine whether HilA binds to the putative HilA box within the sicA promoter region, we performed gel mobility shift assays (Fig. 3B). The mobility of the sicA promoter fragment decreased in the presence of HilA+ extracts (Fig. 3B, lanes 2 to 4). The addition of unlabeled promoter DNA as a specific competitor diminished the amount of labeled sicA fragment that shifted (Fig. 3B, lanes 6 to 7), confirming the specificity of the protein-DNA interaction. Less of the mutant sicA* (Fig. 3B, lane 11) than the wild-type promoter fragment (Fig. 3B, lane 9) seemed to have altered mobility upon the addition of the HilA+ extract. These results suggest that HilA specifically binds the putative HilA box in the sicA promoter region. Since the putative HilA box coincides with the transcription start site (25), it is possible that the abrogated expression of the mutated sicA reporter is due to ineffective DNA polymerase binding at the promoter. While this cannot be ruled out, it is clear that HilA binds to this site and likely that this binding plays an important role in sicA activation.
It has been suggested that expression of sicA occurs via read-through transcription of invFGEABCIJspaOPQRSsicAsipBCDA from a HilA-dependent promoter upstream of invF (22). In this model, basal levels of SicA, along with InvF, activate sicA expression from an InvF-dependent promoter located between spaS and sicA (22). Our experimental and in silico results indicate that sicA is directly regulated by HilA via the putative HilA box immediately upstream of sicA. However, HilA is not sufficient for heterologous sicA transcription in E. coli (22). Thus, it seems likely that SicA, InvF, and HilA act in concert to activate sicA.
HilA acts as an autorepressor under repressing conditions.
The identification of a putative HilA box in the hilA promoter suggests that HilA may be autoregulated. Previous reports have suggested that HilA self-regulates, but there are conflicting data as to whether the regulation is positive or negative. Specifically, a chromosomal hilA β-galactosidase reporter strain in a hilA mutant background produced 50% less β-galactosidase when the hilA lesion was complemented with a plasmid-encoded hilA gene, in comparison to the noncomplemented strain (8). This suggested that the autoregulation is negative. In contrast, an episomal hilA reporter gene produced 30% more β-galactosidase in a hilA+ than in a hilA deletion background, suggesting that the autoregulation is positive (8).
While activators usually bind upstream of open reading frames, repressors can bind both upstream and downstream (6). Particularly in the case of autoregulation, downstream repressor binding sites predominate (20). Examples of Salmonella genes with repressor binding sites that are 3′ of transcription start sites are metF, regulated by MetR (21), and cysB (autoregulation) (80). Since the putative HilA box at +80 to >+92, i.e., downstream of the transcription start of hilA (90), it seemed likely that HilA represses its own expression.
In Fig. 4, we investigated the role of HilA in hilA expression using an episomal hilA reporter gene fusion, pLS31 (90). Our results support a role of HilA as an autorepressor. Under derepressing conditions (high osmolarity, low oxygen) (8), no clear difference in hilA expression is observed between a wild-type and hilA deletion background (Fig. 4A). However, under repressing conditions (low osmolarity and high oxygen) (8, 60) hilA-lacZY expression was significantly higher in a hilA deletion background relative to a wild-type background (Fig. 4B). These data indicate that under derepressing conditions, HilA has no effect on the hilA promoter, but under repressing conditions, HilA significantly reduces its own expression level.
Identified HilA box in the hilA promoter is important for hilA regulation.
As mentioned above, the third T (italic) in the HilA box consensus sequence, tN3TgCAtCAGg, was previously shown to be critical for HilA DNA binding to the prgH and invF promoters (Fig. 1) (59, 60). In the hilA promoter the motif contains a C at the same position (Fig. 1). We tested the effect of the single C→ T base pair substitution in the HilA box of the hilA-lacZYA (pCMPG5401) reporter construct, i.e., hilA*-lacZY, on HilA-mediated expression. Compared to the hilA-lacZY reporter, expression from the hilA* promoter was reduced in both wild-type and hilA deletion backgrounds, in both derepressing and repressing conditions (Fig. 4A and B). This implies that the mutation not only interferes with the putative HilA binding at the HilA box but also influences hilA transcription level through a second mechanism, e.g., through interference with the action of either DNA polymerase or of an unidentified regulatory protein. In contrast to the hilA-lacZY fusion, under derepressing conditions, the hilA* promoter was repressed in the wild-type background (Fig. 4A).
These data suggest that the identity of the nucleotide at the critical position in the HilA box may determine how tightly HilA binds. To test this notion, in vitro DNA-binding assays using Myc- and His-tagged HilA protein were performed. Figure 4C confirms the binding of HilA to the hilA promoter. Indeed, incubation of labeled hilA promoter fragments with extracts made from E. coli cells expressing the tagged HilA protein from the arabinose-induced PBAD promoter (HilA+) impeded the migration of the probe into a native gel (Fig. 4C, lanes 2 and 6). In contrast, a retarded band was not observed when the hilA probe was incubated with extracts lacking HilA (HilA−, Fig. 4C, lane 3), demonstrating that the retardation of the probe requires HilA. The addition of unlabeled competitor DNA seemed to diminish the sequestration of the HilA-DNA complex (Fig. 4C, lane 4), indicating that HilA binds specifically to the hilA promoter. Labeled hilA* promoter fragments seemed to be more strongly bound by HilA than the hilA promoter fragment (Fig. 4C, lane 8). These data suggest that the C→ T substitution in the hilA promoter putative HilA box allows for increased binding of HilA.
A T→ C substitution in the HilA boxes of sicA, invF, and prgH (i.e., T→ C in caTcaggaw Fig. 1) appeared to result in reduced HilA binding and severely reduced HilA-dependent activation (Fig. 3A, 4A, and 4B) (59, 60). This is consistent with the known importance of the T in the invF and prgH promoters for HilA binding (59, 60). In contrast, a C→ T substitution in the hilA promoter putative HilA box seemed to result in increased HilA binding, which could be explained by the fact that, in this promoter, HilA acts as a repressor and thereby could reduce hilA expression (Fig. 4A and B) irrespective of the conditions.
Concluding remarks.
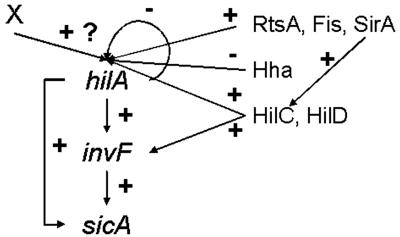
Gene expression profiling experiments followed by in silico motif detection on a cluster of coexpressed S. enterica serovar Typhimurium genes revealed a motif, previously described as the HilA box, in the promoter regions of spi4_H, sicA, and hilA. Site-directed mutagenesis, reporter gene expression, and gel mobility shift assays indicated that sicA expression requires HilA and that hilA is negatively autoregulated. Thus, HilA appears to act as an activator for the sicA gene and as a repressor for the hilA gene. These results allow some reflection on the design of the HilA transcriptional network. Combining all knowledge of the HilA regulator, the hilA-invF-sicA regulatory network could be categorized as a feedforward loop network motif (93) (Fig. 5).
FIG. 5.
Postulated HilA transcriptional regulation network. HilA-InvF-SicA constitute a feedforward loop: the transcription factor HilA regulates a second transcription factor, InvF, and both jointly regulate sicA. hilA expression is proposed to be negatively autoregulated. An additional transcriptional factor(s) X could be required for hilA expression. Other transcriptional regulators, HilC (33, 78, 79, 83, 90), HilD (78, 90), Hha (38, 79), SirA/BarA (1, 3, 49, 83, 97), Fis (9, 91, 102) and RtsA (35), known to regulate hilA expression by binding to hilA promoter region, are included. HilD and HilC also activate expression of a subset of SPI1 genes independently of HilA through activation of invF transcription (2). SirA can bypass the hilA gene to regulate invasion determinants (83) through directly binding to the hilC promoter (97). +, induction; −, repression. For clarity, regulation of hilA expression by the following genes is not depicted: phoPQ (10, 81), envZ/ompR (64), phoBR (65), hilE (9, 37), H-NS and HU (hupB) (91, 102), csrA (3), lon (12, 96), ams (RNase E) (37), pag (37), orgC (26, 37), integration host factor (37), fadD (65), cpxA (75), and fliZ (33, 48, 65).
A feedforward loop rejects transient activation signals from general transcription factors and responds only to persistent signals. In addition, a feedforward loop allows for rapid system shutdown. Together, this results in increased specificity and tight temporal regulation. In this model, HilA and InvF act in an AND-gate-like manner to control sicA expression. When hilA is activated, the signal is transmitted to the output sicA by two pathways, a direct one from HilA and a delayed one through InvF (Fig. 5). If hilA activation is transient, InvF cannot reach the level needed to significantly activate sicA, and the input signal is not transduced through the circuit. Only when HilA signals long enough to allow InvF to accumulate is sicA activated. Once hilA is deactivated, sicA shuts down rapidly. Tight temporal regulation of SicA through this feedforward loop should avoid useless energy investment in production of effector proteins when the environmental conditions are not optimal for invasion. Especially in light of the SicA role as a chaperone to InvF (24, 25), this feedforward loop could hold biological relevance.
Experimental evidence also suggests a negative autoregulation feedback (hilA) superimposed on the feedforward loop. Negative autoregulation feedback appears in over 40% of known transcription factors in E. coli (85). Negative autoregulation feedback reduces the rise time (i.e., the delay from the initiation of production until half-maximal product concentration is reached) (85), favoring the dynamic behavior of the transcription network (68). A shorter rise time is possible because the unrepressed promoter can be activated rapidly. Later, a freshly produced repressor can shut off its own production and the required steady-state concentration can be quickly reached. A strong nonautoregulated promoter will reach any given concentration faster but will stabilize at a much higher steady state, which is undesirable due to metabolic cost, possible toxic effects, and the long time required for its subsequent dilution when production is ceased (66, 77, 85). It would be interesting to characterize the kinetic behavior of the different regulatory circuit elements controlling gene expression during invasion of salmonellae once all regulatory mechanisms for hilA expression and HilA activity and all targets of HilA are identified.
Acknowledgments
S. De Keersmaecker and K. Marchal were Research Associates of the Belgian Fund for Scientific Research (FWO-Vlaanderen) when this study was conducted. K. Engelen is a research assistant of IWT. The work was initiated in the laboratory of S. Falkow, Stanford University, under support from the National Institutes of Health (AI-26195). C. Detweiler was additionally supported by an American Cancer Society Fellowship (PF-99-146-01-MBC) and the University of Colorado at Boulder. This work is also partially supported by STWW-00162 and GBOU-SQUAD-20160 of the IWT.
We gratefully acknowledge C. Lee, V. Miller, S. Busby, and W. de Vos for kindly providing strains and plasmids used in this study.
REFERENCES
- 1.Ahmer, B. M., J. van Reeuwijk, P. R. Watson, T. S. Wallis, and F. Heffron. 1999. Salmonella SirA is a global regulator of genes mediating enteropathogenesis. Mol. Microbiol. 31:971-982. [DOI] [PubMed] [Google Scholar]
- 2.Akbar, S., L. M. Schechter, C. P. Lostroh, and C. A. Lee. 2003. AraC/XylS family members, HilD and HilC, directly activate virulence gene expression independently of HilA in Salmonella typhimurium. Mol. Microbiol. 47:715-728. [DOI] [PubMed] [Google Scholar]
- 3.Altier, C., M. Suyemoto, A. I. Ruiz, K. D. Burnham, and R. Maurer. 2000. Characterization of two novel regulatory genes affecting Salmonella invasion gene expression. Mol. Microbiol. 35:635-646. [DOI] [PubMed] [Google Scholar]
- 4.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 5.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. H. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Babu, M. M., and S. A. Teichmann. 2003. Functional determinants of transcription factors in Escherichia coli: protein families and binding sites. Trends Genet. 19:75-79. [DOI] [PubMed] [Google Scholar]
- 7.Bajaj, V., C. Hwang, and C. A. Lee. 1995. HilA is a novel OmpR/ToxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol. Microbiol. 18:715-727. [DOI] [PubMed] [Google Scholar]
- 8.Bajaj, V., R. L. Lucas, C. Hwang, and C. A. Lee. 1996. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol. Microbiol. 22:703-714. [DOI] [PubMed] [Google Scholar]
- 9.Baxter, M. A., T. F. Fahlen, R. L. Wilson, and B. D. Jones. 2003. HilE interacts with HilD and negatively regulates hilA transcription and expression of the Salmonella enterica serovar Typhimurium invasive phenotype. Infect. Immun. 71:1295-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Behlau, I., and S. I. Miller. 1993. A PhoP-repressed gene promotes Salmonella typhimurium invasion of epithelial cells. J. Bacteriol. 175:4475-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benson, D. A., I. Karsch-Mizrachi, D. J. Lipman, J. Ostell, B. A. Rapp, and D. L. Wheeler. 2002. GenBank. Nucleic Acids Res. 30:17-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boddicker, J. D., and B. D. Jones. 2004. Lon protease activity causes down-regulation of Salmonella pathogenicity island 1 invasion gene expression after infection of epithelial cells. Infect. Immun. 72:2002-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 14.Bullas, L. R., and J. I. Ryu. 1983. Salmonella typhimurium LT2 strains which are r- m+ for all three chromosomally located systems of DNA restriction and modification. J. Bacteriol. 156:471-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bumann, D. 2002. Examination of Salmonella gene expression in an infected mammalian host using the green fluorescent protein and two-colour flow cytometry. Mol. Microbiol. 43:1269-1283. [DOI] [PubMed] [Google Scholar]
- 16.Carter, P. B., and F. M. Collins. 1974. The route of enteric infection in normal mice. J. Exp. Med. 139:1189-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan, K., S. Baker, C. C. Kim, C. S. Detweiler, G. Dougan, and S. Falkow. 2003. Genomic comparison of Salmonella enterica serovars and Salmonella bongori by use of an S. enterica serovar Typhimurium DNA microarray. J. Bacteriol. 185:553-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coconnier, M. H., V. Lievin, M. F. Bernet-Camard, S. Hudault, and A. L. Servin. 1997. Antibacterial effect of the adhering human Lactobacillus acidophilus strain LB. Antimicrob. Agents Chemother. 41:1046-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coessens, B., G. Thijs, S. Aerts, K. Marchal, F. De Smet, K. Engelen, P. Glenisson, Y. Moreau, J. Mathys, and B. De Moor. 2003. INCLUSive: A web portal and service registry for microarray and regulatory sequence analysis. Nucleic Acids Res. 31:3468-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collado-Vides, J., B. Magasanik, and J. D. Gralla. 1991. Control site location and transcriptional regulation in Escherichia coli. Microbiol. Rev. 55:371-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cowan, J. M., M. L. Urbanowski, M. Talmi, and G. V. Stauffer. 1993. Regulation of the Salmonella typhimurium metF gene by the MetR protein. J. Bacteriol. 175:5862-5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Darwin, K. H., and V. L. Miller. 1999. InvF is required for expression of genes encoding proteins secreted by the SPI1 type III secretion apparatus in Salmonella typhimurium. J. Bacteriol. 181:4949-4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Darwin, K. H., and V. L. Miller. 1999. Molecular basis of the interaction of Salmonella with the intestinal mucosa. Clin. Microbiol. Rev. 12:405-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Darwin, K. H., and V. L. Miller. 2000. The putative invasion protein chaperone SicA acts together with InvF to activate the expression of Salmonella typhimurium virulence genes. Mol. Microbiol. 35:949-959. [DOI] [PubMed] [Google Scholar]
- 25.Darwin, K. H., and V. L. Miller. 2001. Type III secretion chaperone-dependent regulation: activation of virulence genes by SicA and InvF in Salmonella typhimurium. EMBO J. 20:1850-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Day, J. B., and C. A. Lee. 2003. Secretion of the orgC gene product by Salmonella enterica serovar Typhimurium. Infect. Immun. 71:6680-6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Man, J. C., M. Rogosa, and M. E. Sharpe. 1960. A medium for the cultivation of lactobacilli. J. Appl. Bacteriol. 23:130-135. [Google Scholar]
- 28.De Smet, F., J. Mathys, K. Marchal, G. Thijs, B. De Moor, and Y. Moreau. 2002. Adaptive quality-based clustering of gene expression profiles. Bioinformatics 18:735-746. [DOI] [PubMed] [Google Scholar]
- 29.Detweiler, C. S., D. M. Monack, I. E. Brodsky, H. Mathew, and S. Falkow. 2003. virK, somA and rcsC are important for systemic Salmonella enterica serovar Typhimurium infection and cationic peptide resistance. Mol. Microbiol. 48:385-400. [DOI] [PubMed] [Google Scholar]
- 30.Durant, J. A., D. E. Corrier, L. H. Stanker, and S. C. Ricke. 2000. Expression of the hilA Salmonella typhimurium gene in a poultry Salmonella enteritidis isolate in response to lactate and nutrients. J. Appl. Microbiol. 89:63-69. [DOI] [PubMed] [Google Scholar]
- 31.Durant, J. A., D. E. Corrier, L. H. Stanker, and S. C. Ricke. 2000. Salmonella enteritidis hilA gene fusion response after incubation in spent media from either S. enteritidis or a poultry Lactobacillus strain. J. Environ. Sci. Health B. 35:599-610. [DOI] [PubMed] [Google Scholar]
- 32.Eichelberg, K., and J. E. Galan. 1999. Differential regulation of Salmonella typhimurium type III secreted proteins by pathogenicity island 1 (SPI-1)-encoded transcriptional activators InvF and HilA. Infect. Immun. 67:4099-4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eichelberg, K., W. D. Hardt, and J. E. Galan. 1999. Characterization of SprA, an AraC-like transcriptional regulator encoded within the Salmonella typhimurium pathogenicity island 1. Mol. Microbiol. 33:139-152. [DOI] [PubMed] [Google Scholar]
- 34.Eisen, M. B., and P. O. Brown. 1999. DNA arrays for analysis of gene expression. Methods Enzymol. 303:179-205. [DOI] [PubMed] [Google Scholar]
- 35.Ellermeier, C. D., and J. M. Slauch. 2003. RtsA and RtsB coordinately regulate expression of the invasion and flagellar genes in Salmonella enterica serovar Typhimurium. J. Bacteriol. 185:5096-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Engelen, K., B. Coessens, K. Marchal, and B. De Moor. 2003. MARAN: normalizing micro-array data. Bioinformatics 19:893-894. [DOI] [PubMed] [Google Scholar]
- 37.Fahlen, T. F., N. Mathur, and B. D. Jones. 2000. Identification and characterization of mutants with increased expression of hilA, the invasion gene transcriptional activator of Salmonella typhimurium. FEMS Immunol. Med. Microbiol. 28:25-35. [DOI] [PubMed] [Google Scholar]
- 38.Fahlen, T. F., R. L. Wilson, J. D. Boddicker, and B. D. Jones. 2001. Hha is a negative modulator of transcription of hilA, the Salmonella enterica serovar Typhimurium invasion gene transcriptional activator. J. Bacteriol. 183:6620-6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Food and Agriculture Organization/World Health Organization. 2001. Evaluation of health and nutritional properties of powder milk and live lactic acid bacteria. Food and Agriculture Organization, Rome, Italy.
- 40.Fuller, R. 1989. Probiotics in man and animals. J. Appl. Bacteriol. 66:365-378. [PubMed] [Google Scholar]
- 41.Galan, J. E. 1996. Molecular genetic bases of Salmonella entry into host cells. Mol. Microbiol. 20:263-271. [DOI] [PubMed] [Google Scholar]
- 42.Garcia-Vescovi, E., F. C. Soncini, and E. A. Groisman. 1996. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell 84:165-174. [DOI] [PubMed] [Google Scholar]
- 43.Groisman, E. A. 2001. The pleiotropic two-component regulatory system PhoP-PhoQ. J. Bacteriol. 183:1835-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hentschel, U., U. Dobrindt, and M. Steinert. 2003. Commensal bacteria make a difference. Trends Microbiol. 11:148-150. [DOI] [PubMed] [Google Scholar]
- 45.Hoiseth, S. K., and B. A. Stocker. 1981. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291:238-239. [DOI] [PubMed] [Google Scholar]
- 46.Hooper, L. V., L. Bry, P. G. Falk, and J. I. Gordon. 1998. Host-microbial symbiosis in the mammalian intestine: exploring an internal ecosystem. Bioessays 20:336-343. [DOI] [PubMed] [Google Scholar]
- 47.Hudault, S., V. Lievin, M. F. Bernet-Camard, and A. L. Servin. 1997. Antagonistic activity exerted in vitro and in vivo by Lactobacillus casei (strain GG) against Salmonella typhimurium C5 infection. Appl. Environ. Microbiol. 63:513-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iyoda, S., T. Kamidoi, K. Hirose, K. Kutsukake, and H. Watanabe. 2001. A flagellar gene fliZ regulates the expression of invasion genes and virulence phenotype in Salmonella enterica serovar Typhimurium. Microb. Pathog. 30:81-90. [DOI] [PubMed] [Google Scholar]
- 49.Johnston, C., D. A. Pegues, C. J. Hueck, A. Lee, and S. I. Miller. 1996. Transcriptional activation of Salmonella typhimurium invasion genes by a member of the phosphorylated response-regulator superfamily. Mol. Microbiol. 22:715-727. [DOI] [PubMed] [Google Scholar]
- 50.Jones, B. D., N. Ghori, and S. Falkow. 1994. Salmonella typhimurium initiates murine infection by penetrating and destroying the specialized epithelial M cells of the Peyer's patches. J. Exp. Med. 180:15-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kaniga, K., S. Tucker, D. Trollinger, and J. E. Galan. 1995. Homologs of the Shigella IpaB and IpaC invasins are required for Salmonella typhimurium entry into cultured epithelial cells. J. Bacteriol. 177:3965-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kerr, M. K., and G. A. Churchill. 2001. Bootstrapping cluster analysis: assessing the reliability of conclusions from microarray experiments. Proc. Natl. Acad. Sci. USA 98:8961-8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kerr, M. K., and G. A. Churchill. 2001. Statistical design and the analysis of gene expression microarray data. Genet. Res. 77:123-128. [DOI] [PubMed] [Google Scholar]
- 54.Kerr, M. K., M. Martin, and G. A. Churchill. 2000. Analysis of variance for gene expression microarray data. J. Comput. Biol. 7:819-837. [DOI] [PubMed] [Google Scholar]
- 55.Lawrence, C. E., S. F. Altschul, M. S. Boguski, J. S. Liu, A. F. Neuwald, and J. C. Wootton. 1993. Detecting subtle sequence signals: a Gibbs sampling strategy for multiple alignment. Science 262:208-214. [DOI] [PubMed] [Google Scholar]
- 56.Lee, C. A., and S. Falkow. 1990. The ability of Salmonella to enter mammalian cells is affected by bacterial growth state. Proc. Natl. Acad. Sci. USA 87:4304-4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lehto, E. M., and S. J. Salminen. 1997. Inhibition of Salmonella typhimurium adhesion to Caco-2 cell cultures by Lactobacillus strain GG spent culture supernate: only a pH effect? FEMS Immunol. Med. Microbiol. 18:125-132. [DOI] [PubMed] [Google Scholar]
- 58.Lodge, J., J. Fear, S. Busby, P. Gunasekaran, and N. R. Kamini. 1992. Broad host range plasmids carrying the Escherichia coli lactose and galactose operons. FEMS Microbiol. Lett. 74:271-276. [DOI] [PubMed] [Google Scholar]
- 59.Lostroh, C. P., V. Bajaj, and C. A. Lee. 2000. The cis requirements for transcriptional activation by HilA, a virulence determinant encoded on SPI-1. Mol. Microbiol. 37:300-315. [DOI] [PubMed] [Google Scholar]
- 60.Lostroh, C. P., and C. A. Lee. 2001. The HilA box and sequences outside it determine the magnitude of HilA-dependent activation of P-prgH from Salmonella pathogenicity island 1. J. Bacteriol. 183:4876-4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lostroh, C. P., and C. A. Lee. 2001. The Salmonella pathogenicity island-1 type III secretion system. Microbes. Infect. 3:1281-1291. [DOI] [PubMed] [Google Scholar]
- 62.Lu, L., and W. A. 2001. Walker. Pathologic and physiologic interactions of bacteria with the gastrointestinal epithelium. Am. J. Clin. Nutr. 73:1124S-1130S. [DOI] [PubMed] [Google Scholar]
- 63.Lucas, R. L., and C. A. Lee. 2000. Unravelling the mysteries of virulence gene regulation in Salmonella typhimurium. Mol. Microbiol. 36:1024-1033. [DOI] [PubMed] [Google Scholar]
- 64.Lucas, R. L., and C. A. Lee. 2001. Roles of hilC and hilD in regulation of hilA expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 183:2733-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lucas, R. L., C. P. Lostroh, C. C. DiRusso, M. P. Spector, B. L. Wanner, and C. A. Lee. 2000. Multiple factors independently regulate hilA and invasion gene expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 182:1872-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lutz, R., and H. Bujard. 1997. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 25:1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Marchal, K., G. Thijs, S. De Keersmaecker, P. Monsieurs, B. De Moor, and J. Vanderleyden. 2003. Genome-specific higher-order background models to improve motif detection. Trends Microbiol. 11:61-66. [DOI] [PubMed] [Google Scholar]
- 68.McAdams, H. H., and A. Arkin. 1997. Stochastic mechanisms in gene expression. Proc. Natl. Acad. Sci. USA 94:814-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Y. Du, S. F. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 70.Michael, B., J. N. Smith, S. Swift, F. Heffron, and B. M. M. Ahmer. 2001. SdiA of Salmonella enterica is a LuxR homolog that detects mixed microbial communities. J. Bacteriol. 183:5733-5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 72.Miller, S. I., and D. A. Pegues. 2000. Salmonella species, including Salmonella typhi, p. 2344-2363. In G. L. Mandell, J. E. Bennett, and R. Dolin (ed.), Principles and practice in infectious diseases. Churchill Livingstone, Philadelphia, Pa.
- 73.Moreau, Y., F. De Smet, G. Thijs, K. Marchal, and B. De Moor. 2002. Functional bioinformatics of microarray data: from expression to regulation. Proc. IEEE 90:1722-1743. [Google Scholar]
- 74.Morgan, E., J. D. Campbell, S. C. Rowe, J. Bispham, M. P. Stevens, A. J. Bowen, P. A. Barrow, D. J. Maskell, and T. S. Wallis. 2004. Identification of host-specific colonization factors of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 54:994-1010. [DOI] [PubMed] [Google Scholar]
- 75.Nakayama, S., A. Kushiro, T. Asahara, R. Tanaka, L. Hu, D. J. Kopecko, and H. Watanabe. 2003. Activation of hilA expression at low pH requires the signal sensor CpxA, but not the cognate response regulator CpxR, in Salmonella enterica serovar Typhimurium. Microbiology 149:2809-2817. [DOI] [PubMed] [Google Scholar]
- 76.Nicholas, K. B., H. B. J. Nicholas, and D. W. I. Deerfield. 1997. GeneDoc: analysis and visualization of genetic variation. EMBNEW News 4:14. [Google Scholar]
- 77.Oehmichen, R., G. Klock, L. Altschmied, and W. Hillen. 1984. Construction of an E. coli strain overproducing the Tn10-encoded TET repressor and its use for large scale purification. EMBO J. 3:539-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Olekhnovich, I. N., and R. J. Kadner. 2002. DNA-binding activities of the HilC and HilD virulence regulatory proteins of Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:4148-4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Olekhnovich, I. N., and R. J. Kadner. 2004. Contribution of the RpoA C-terminal domain to stimulation of the Salmonella enterica hilA promoter by HilC and HilD. J. Bacteriol. 186:3249-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ostrowski, J., and N. M. Kredich. 1991. Negative autoregulation of cysB in Salmonella typhimurium: in vitro interactions of CysB protein with the cysB promoter. J. Bacteriol. 173:2212-2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pegues, D. A., M. J. Hantman, I. Behlau, and S. I. Miller. 1995. PhoP/PhoQ transcriptional repression of Salmonella typhimurium invasion genes: evidence for a role in protein secretion. Mol. Microbiol. 17:169-181. [DOI] [PubMed] [Google Scholar]
- 82.Provence, D. L., and R. Curtiss. 1994. Gene transfer in Gram-negative bacteria, p. 317-347. In P. Gerhardt, R. G. E. Murray, W. A. Wood, and N. R. Krieg (ed.), Methods for general and molecular bacteriology. American Society for Microbiology, Washington, D.C.
- 83.Rakeman, J. L., H. R. Bonifield, and S. I. Miller. 1999. A HilA-independent pathway to Salmonella typhimurium invasion gene transcription. J. Bacteriol. 181:3096-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rodriguez, C. R., L. M. Schechter, and C. A. Lee. 2002. Detection and characterization of the S. typhimurium HilA protein. BMC Microbiol. 2:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rosenfeld, N., M. B. Elowitz, and U. Alon. 2002. Negative autoregulation speeds the response times of transcription networks. J. Mol. Biol. 323:785-793. [DOI] [PubMed] [Google Scholar]
- 86.Salama, N., K. Guillemin, T. K. McDaniel, G. Sherlock, L. Tompkins, and S. Falkow. 2000. A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc. Natl. Acad. Sci. USA 97:14668-14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning. a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 88.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Santos, R. L., R. M. Tsolis, A. J. Baumler, and L. G. Adams. 2003. Pathogenesis of Salmonella-induced enteritis. Braz. J. Med. Biol. Res. 36:3-12. [DOI] [PubMed] [Google Scholar]
- 90.Schechter, L. M., S. M. Damrauer, and C. A. Lee. 1999. Two AraC/XylS family members can independently counteract the effect of repressing sequences upstream of the hilA promoter. Mol. Microbiol. 32:629-642. [DOI] [PubMed] [Google Scholar]
- 91.Schechter, L. M., S. Jain, S. Akbar, and C. A. Lee. 2003. The small nucleoid-binding proteins H-NS, HU, and Fis affect hilA expression in Salmonella enterica serovar Typhimurium. Infect. Immun. 71:5432-5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schechter, L. M., and C. A. Lee. 2001. AraC/XylS family members, HilC and HilD, directly bind and derepress the Salmonella typhimurium hilA promoter. Mol. Microbiol. 40:1289-1299. [DOI] [PubMed] [Google Scholar]
- 93.Shen-Orr, S. S., R. Milo, S. Mangan, and U. Alon. 2002. Network motifs in the transcriptional regulation network of Escherichia coli. Nat. Genet. 31:64-68. [DOI] [PubMed] [Google Scholar]
- 94.Sherwood, L., and M. D. Gorbach. 1996. The discovery of Lactobacillus GG. Nutr. Today 31:2S-4S. [Google Scholar]
- 95.Silva, M., N. V. Jacobus, C. Deneke, and S. L. Gorbach. 1987. Antimicrobial substance from a human Lactobacillus strain. Antimicrob. Agents Chemother. 31:1231-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Takaya, A., T. Tomoyasu, A. Tokumitsu, M. Morioka, and T. Yamamoto. 2002. The ATP-dependent Lon protease of Salmonella enterica serovar Typhimurium regulates invasion and expression of genes carried on Salmonella pathogenicity island 1. J. Bacteriol. 184:224-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Teplitski, M., R. I. Goodier, and B. M. Ahmer. 2003. Pathways leading from BarA/SirA to motility and virulence gene expression in Salmonella. J. Bacteriol. 185:7257-7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Thijs, G., M. Lescot, K. Marchal, S. Rombauts, B. De Moor, P. Rouze, and Y. Moreau. 2001. A higher-order background model improves the detection of promoter regulatory elements by Gibbs sampling. Bioinformatics 17:1113-1122. [DOI] [PubMed] [Google Scholar]
- 99.Thijs, G., K. Marchal, M. Lescot, S. Rombauts, B. De Moor, P. Rouze, and Y. Moreau. 2002. A Gibbs sampling method to detect overrepresented motifs in the upstream regions of coexpressed genes. J. Comput. Biol. 9:447-464. [DOI] [PubMed] [Google Scholar]
- 100.Thijs, G., Y. Moreau, F. De Smet, J. Mathys, M. Lescot, S. Rombauts, P. Rouze, B. De Moor, and K. Marchal. 2002. INCLUSive: INtegrated clustering, upstream of sequence retrieval and motif sampling. Bioinformatics. 18:331-332. [DOI] [PubMed] [Google Scholar]
- 101.Wallis, T. S., and E. E. Galyov. 2000. Molecular basis of Salmonella-induced enteritis. Mol. Microbiol. 36:997-1005. [DOI] [PubMed] [Google Scholar]
- 102.Wilson, R. L., S. J. Libby, A. M. Freet, J. D. Boddicker, T. F. Fahlen, and B. D. Jones. 2001. Fis, a DNA nucleoid-associated protein, is involved in Salmonella typhimurium SPI-1 invasion gene expression. Mol. Microbiol. 39:79-88. [DOI] [PubMed] [Google Scholar]
- 103.Wong, K. K., M. McClelland, L. C. Stillwell, E. C. Sisk, S. J. Thurston, and J. D. Saffer. 1998. Identification and sequence analysis of a 27-kilobase chromosomal fragment containing a Salmonella pathogenicity island located at 92 minutes on the chromosome map of Salmonella enterica serovar Typhimurium LT2. Infect. Immun. 66:3365-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 105.Zhang, S., R. A. Kingsley, R. L. Santos, H. Andrews-Polymenis, M. Raffatellu, J. Figueiredo, J. Nunes, R. M. Tsolis, L. G. Adams, and A. J. Baumler. 2003. Molecular pathogenesis of Salmonella enterica serotype Typhimurium-induced diarrhea. Infect. Immun. 71:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhou, D. G., and J. Galan. 2001. Salmonella entry into host cells: the work in concert of type III secreted effector proteins. Microbes Infect. 3:1293-1298. [DOI] [PubMed] [Google Scholar]