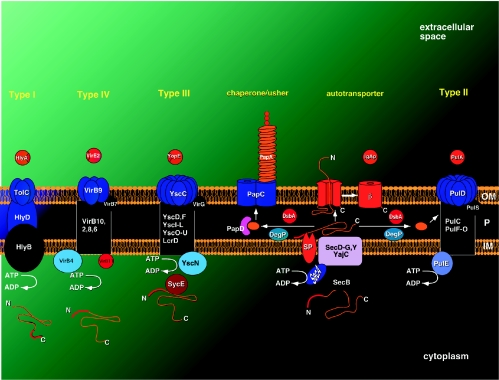
Gram-negative bacteria secrete a wide range of proteins whose functions include biogenesis of organelles, such as pili and flagella; nutrient acquisition; virulence; and efflux of drugs and other toxins. Export of these proteins to the bacterial surface involves transport across the inner membrane (IM), periplasm, and outer membrane (OM) of the cell envelope. Several pathways have evolved to fulfill the task of secretion (Table 1). These pathways can be divided into two main groups: (i) Sec dependent and (ii) Sec independent (Fig. 1) (64).
TABLE 1.
Representative members of the eight secretion systems
| Secretion system | Substrate | Organism | Function |
|---|---|---|---|
| Sec dependent | |||
| AT | IgA1 protease | Neisseria gonorrhoeae | Protease of IgA1 |
| Tsh | Avian pathogenic Escherichia coli | Protease, adhesin | |
| Hap | Haemophilus influenzae | Adhesin, protease | |
| TPS | FHA | Bordetella pertussis | Hemagglutinin |
| ShlA | Serratia marcescens | Cytolysin | |
| HMW1A | Haemophilus influenzae | Adhesin | |
| CU | Components of P pili | Uropathogenic Escherichia coli | Adhesin |
| Components of type 1 pili | Uropathogenic Escherichia coli | Adhesin | |
| T2S | PulA | Klebsiella oxytoca | Starch-hydrolyzing lipoprotein |
| CT | Vibrio cholerae | Toxin | |
| LipA | Pseudomonas aeruginosa | Lipase | |
| ENP | CsgA | Escherichia coli | Curli component |
| Sec independent | |||
| T1S | HlyA | Enteropathogenic Escherichia coli | Hemolysin |
| PtrA | Erwinia chrysanthemi | Protease | |
| AprA | Pseudomonas aeruginosa | Alkaline protease | |
| T3S | Yops | Yersinia pestis | Multiple functions |
| InvA | Salmonella typhimurium | Invasin | |
| T4S | VirD2/transfer DNA | Agrobacterium tumefaciens | Tumorigenesis |
| PT | Bordetella pertussis | Toxin | |
| CagA | Helicobacter pylori | Activator of HGF/SF signaling pathway |
FIG. 1.
Schematic presentation of Sec-dependent and Sec-independent secretion pathways in gram-negative bacteria. The type I pathway is exemplified by the hemolysin A (HlyA) secretion in pathogenic E. coli, the type II pathway by YopE secretion in Yersinia pestis, the type IV pathway by the VirE2 protein of Agrobacterium tumefaciens, the chaperone/usher pathway by P pilus subunit (PapA) secretion in uropathogenic E. coli, the autotransporter pathway by IgA1 protease (IgAp) secretion in Neisseria gonorrhoeae, and the type II pathway by pullulanase (PulA) secretion in Klebsiella oxytoca. Secreted polypeptides are shown in orange, with their amino- or carboxyl-terminal signal peptides drawn in red. Only proteins secreted by Sec-dependent pathways contain cleavable signal peptides. Also, proteins secreted by Sec-dependent pathways may fold, fully or partially, in the periplasm with the assistance of periplasmic accessory factors (e.g., DsbA), whereas misfolded secreted proteins in the periplasm are degraded by periplasmic proteases (e.g., DegP). P, periplasm; SP, signal peptide; β, β-domain of autotransporter IgA1 protease; N, amino terminus; C, carboxy terminus.
Proteins secreted via the Sec-dependent pathways utilize a common machinery, the Sec translocase, for transport across the IM and are mainly differentiated based on their mechanisms of secretion across the OM (64). Sec-dependent pathways include the type II secretion (T2S); the autotransporter (AT), or type V secretion; the two-partner secretion (TPS); and the chaperone/usher (CU) secretion systems (34, 46, 58, 70, 73). The type IV secretion (T4S), or adapted-conjugation, pathway can be Sec dependent but is mostly considered Sec independent (21). Sec-independent pathways tend to allow direct export from the cytoplasm to the extracellular environment in one step and do not involve periplasmic intermediates. These pathways also include the type I secretion (T1S), or ABC (ATP-binding cassette), exporters (5) and the type III secretion (T3S) systems (31). An alternative Sec-independent pathway known as twin-arginine translocation (Tat) is employed for transport of already-folded proteins across the IM. Generally, Tat substrates are targeted to the periplasm of the cell (4) but can also be transported across the OM via the type II pathway, such as the phospholipases of Pseudomonas aeruginosa (77). The Sec and Tat machineries for transport across the IM will not be discussed further.
In this review, we summarize the current knowledge of OM protein secretion in terms of the structure of the exporters and the mechanism of secretion and examine similarities and differences among the different export pathways (Table 2). Since many virulence factors are secreted proteins, a more complete understanding of the mechanisms underlying bacterial protein secretion is crucial to the development of new vaccines and treatments for a wide range of human pathogens.
TABLE 2.
Structural features of the OM components of protein secretion systems
| Secretion system | OM component | Structural features
|
References | ||
|---|---|---|---|---|---|
| Size of precursor protein (aa)a | Structural organization | Size of export channel (Å) | |||
| Sec dependent | |||||
| AT | NalP | 1,083 | Monomer, 12-stranded β-barrel with N-terminal α-helix | 10 by 12.5 | 52 |
| TPS | HMW1B | 545 | Homotetramer, predicted 22-stranded β-barrel | 27 | 67 |
| CU | PapC | 836 | Dimer, β-barrel pore structure | 20-30 | 29, 41 |
| T2S | PulD | 660 | Homomultimer (12 monomers), extensive β-barrel structure predicted | 76 | 49 |
| ENP | CsgG? | 277 | ? | ? | 43, 64 |
| Sec independent | |||||
| T1S | TolC | 495 | Homotrimer, each monomer contributing 4 β-strands to form 12-stranded β-barrel | 35 | 38 |
| T3S | YscC | 607 | Multimer, ring-like structure | 40 | 9, 40 |
| T4S | VirB9 | 293 | Heterodimer, complexes with VirB7 via disulfide bonds | ? | 2 |
aa, amino acids.
AT SECRETION
The AT pathway, also known as the type V secretion system, is one of the most widely distributed secretion systems among the gram-negative bacteria (80). Its main characteristic is the simplicity that marks protein translocation across the OM, since its substrates can mediate their own transport across the outer lipid bilayer (33, 46). Proteins secreted by the AT pathway are typically virulence factors with diverse roles in pathogenesis (28, 39).
ATs are synthesized as large multidomain precursors consisting of an N-terminal Sec-dependent signal sequence; an internal passenger domain, or α-domain; and a C-terminal β-domain. Following transport across the IM and cleavage of the signal sequence, the C-terminal domain inserts into the OM, driving export of the passenger domain to the cell surface. Thereafter, the passenger domain may either remain attached to the bacterial cell or be released to the external milieu (33, 46).
Most of the currently proposed models for AT secretion suggest that upon insertion into the OM, the C-terminal translocator domain folds into a β-barrel with a central hydrophilic channel (33, 44, 46). An α-helical region located at the N terminus of the translocator domain, called the “linker” region, is believed to adopt a hairpin structure that inserts into the pore, pulling the passenger domain to the cell surface (45). Recently, the crystal structure of the translocator domain of the NalP AT from Neisseria meningitidis was solved, revealing a monomeric 12-stranded β-barrel that encloses a pore of 10 by 12.5 Å. The β-barrel exhibits all the features of other OM porin-type structures, and most remarkably, it has both termini located on the periplasmic side of the membrane (52).
A different structural configuration was predicted for the translocator domain of the Hia AT from Haemophilus influenzae, YadA from Yersinia enterocolitica, and NhhA from N. meningitidis. Studies with these ATs supported the formation of a trimeric 12-stranded β-barrel in which each monomer contributes four transmembrane β-strands (54, 56, 66). All these proteins are characterized by a very short translocator domain ∼70 residues in length (33, 56, 66).
A third structural motif for the translocator domain has been proposed for intimin and invasin, which are believed to be putative ATs (46, 75). In this case, the β-barrel is formed by the N-terminal part of the protein, which inserts into the OM, possibly driving export of the C-terminal domain to the cell surface (46). Studies with intimin suggested formation of a dimer (75).
Based on the current knowledge of the AT translocator domain, various models have been predicted for transport of the passenger domain across the OM. The monomeric nature of the NalP translocator domain supports secretion of the mature AT through a monomeric β-barrel pore. According to this model, the linker region inserts into the channel, pulling the passenger domain to the bacterial surface and plugging the pore after secretion. This is in accordance with the pore activity of NalP (52). Similarly, blockage of the pore was reported for the recombinant β-domain of the immunoglobulin A (IgA) protease (76) and the AIDA β-domain (45). However, the pore dimensions measured for NalP reveal a rather narrow channel that could host only two fully extended polypeptides at a time. This suggests that the linker region can adopt its α-helical conformation only after the passenger domain has exited the pore completely, and thus translocation and folding may be coupled processes (52).
An alternative model of AT secretion has been proposed by Veiga et al. (76), whose studies with a recombinant C-terminal domain of the IgA protease from N. gonorrhoeae revealed the formation of an oligomeric β-barrel of at least six monomers, with a central pore of ∼2 nm, that can accommodate transport of folded or partially folded proteins (76).
Following export to the cell surface, the passenger domain adopts its native conformation (33, 46), either with the help of an intramolecular chaperone (51) or by an unknown mechanism. Thereafter, the folded protein may remain attached to the bacterial surface (33, 46) or be cleaved and released to the external environment (30, 33, 46, 65).
TPS
The TPS system shares many similarities with the AT pathway (33, 46). Like the AT system, the TPS pathway is widespread among gram-negative bacteria (80) and is dedicated to the secretion of large proteins with virulence functions. However, the main distinguishing feature of this system is that it involves one accessory protein for translocation of the exoprotein across the OM (33, 34, 46).
Proteins secreted by the TPS pathway, called TpsA proteins, transverse the IM with the help of a Sec-dependent N-terminal signal peptide (33, 34). A 110-residue conserved “TPS domain” located in their N termini is essential for secretion across the OM (33), a process that involves an integral OM channel-forming protein, TpsB (34). During OM transport, TpsA adopts its native conformation (33, 34), and in the case of cytolysins, this step is coupled to protein activation (62). Thereafter, TpsA may either remain attached to the bacterial cell or be released to the external medium (33).
No TpsB transporter has been crystallized to date, and thus information on their structure comes from sequence analyses and experimental data, derived mainly from studies of three members of the family, Serratia marcescens ShlB, Bordetella pertussis FhaC, and H. influenzae HMW1/2B. TpsB transporters share only a few conserved residues, and their homology is generally limited to structural motifs (34, 80). They are rich in beta structure and are predicted to fold into a β-barrel (33, 46, 80), which appears to be restricted to their C-terminal portion (27). A highly conserved region with the consensus sequence D/EXAHyXAHyGGXBS/THyRGY/F, where X can be any residue and Hy can be any hydrophobic residue, was found in most TpsB transporters. This motif localizes in a predicted surface loop (80), and studies with FhaC revealed that it might possess an important role in secretion (27). Recent studies with HMW1B demonstrated formation of a tetrameric pore with an estimated size of ∼2.7 nm (67). Similar pore dimensions were estimated for FhaC as well (32).
In the process of secretion, the TpsA proteins interact physically with the corresponding TpsB transporter on the periplasmic side of the OM, possibly through secretion signals present in the TPS domain (34). Such a direct interaction was recently demonstrated between HMW1B and the N terminus of the HMW1 adhesin (67). The fact that each transporter is responsible for secretion of a specific exoprotein (34) has led to the suggestion that interactions between an exoprotein and its transporter may occur via variant rather than invariant residues of the TPS domain (14). The exoproteins are believed to cross the membrane in an extended conformation from the N to the C terminus and to fold progressively as they reach the cell surface. It is speculated that the TPS domain is involved in nucleating folding (14). Once on the surface, the TpsA protein usually remains noncovalently attached to the membrane, but in some cases it may be released to the external environment (33). FhaC appears to undergo a conformational change during FHA secretion, suggesting that TpsB transporters may be gated by their substrates (27). Thus, interaction with the exoprotein may lead to opening of the transporter's channel, which closes after TpsA has reached the bacterial surface and remains in a closed state until it interacts with another exoprotein. This is consistent with the hypothesis that each transporter can secrete several of its substrate exoproteins and is in agreement with the substoichiometric amounts of FhaC produced and FHA secreted (33).
CU SECRETION
The CU pathway is dedicated to the assembly and secretion of a superfamily of virulence-associated surface structures. This pathway typically assembles rod-like fibers called pili or fimbriae but also may assemble amorphous, capsule-like structures. The biogenesis of both structures occurs by similar mechanisms and requires the actions of two secretion components working in concert: a periplasmic chaperone and an outer membrane protein called an usher.
The molecular basis for many aspects of fiber assembly has been revealed by crystal structures, and the CU pathway is one of the best-understood protein secretion pathways. Following translocation across the IM as unfolded polypeptides via the Sec system, subunits must interact with the periplasmic chaperone. The chaperone acts by a mechanism called donor strand complementation to allow proper folding of the subunits while simultaneously preventing premature subunit-subunit interactions (12, 59). Chaperone-subunit complexes next target the OM usher, which serves as a platform for fiber assembly and secretion to the cell surface. Fiber assembly is thought to occur at the periplasmic face of the usher, concomitant with secretion of the fiber through the usher to the cell surface (70). Pili are assembled in a top-down fashion, with the adhesin incorporated first. The usher facilitates this by differentially recognizing chaperone-subunit complexes according to their final positions in the pilus (22, 61). At the usher, chaperone-subunit interactions are exchanged for subunit-subunit interactions in a process called donor strand exchange, which couples chaperone dissociation with fiber assembly (60, 81). A significant finding from recent crystal structures is that the chaperone maintains subunits in a high-energy conformation (60, 81). Subunit-subunit interactions and donor strand exchange then allow subunits to undergo a topological transition to a more compact, lower-energy state. This topological transition presumably provides the driving force for fiber formation and secretion at the usher.
The molecular details of how the usher coordinates fiber assembly and secretion across the OM are less well understood, but structural and functional analyses are beginning to elucidate this process for pilus biogenesis. Chaperone-subunit complexes initially target an N-terminal region of the usher consisting of the first 124 to 139 residues of the mature N terminus (47, 48). Each usher specifically interacts with its own chaperone-subunit complexes, and the N-terminal targeting domain may confer specificity on the usher (47, 72). Binding of a chaperone-adhesin complex may induce a conformational change in the usher, priming the usher for subsequent assembly events (61). Subsequent events include stable interaction of chaperone-subunit complexes with the usher C terminus, which is required for subunit assembly into pili and secretion to the cell surface (61, 72). The P pilus usher PapC forms channels 2 to 3 nm in diameter, which are large enough to allow secretion of folded pilus subunits (71). A recent high-resolution electron microscopy study revealed that PapC assembles as a dimeric complex, with apparent channels located at the center of each monomer (41). Thus, the usher assembles in the OM as a twin-pore secretion complex. Interestingly, a similar twin-pore structure has been identified for the unrelated TOM protein translocase of the mitochondrial OM (1). Exactly how the twin-pore usher complex functions in the biogenesis of surface structures remains to be determined. The dimeric usher presumably would allow the simultaneous binding of two chaperone-subunit complexes, potentially positioning the complexes to favor donor strand exchange between the subunits. The energy generated from donor strand exchange would then be used to drive fiber assembly and secretion through the usher to the cell surface.
T2S
The T2S system is responsible for the extracellular transport of a wide variety of hydrolytic enzymes and toxins (58, 64). Production of the type IV pili, responsible for adhesion and twitching motility of several bacteria, is also believed to occur through T2S (53, 64). Its secretion apparatus is much more complex in terms of protein composition than the previously described Sec-dependent machineries.
Substrates of the T2S system cross the IM via the Sec translocase and fold in the periplasm with the help of the enzymes present there (58, 64). Transport across the OM involves the products of 12 to 15 genes, depending on the species (58). One of the most intriguing features of the T2S pathway is that it contains only one integral OM protein, protein D, which is believed to form the translocation channel (49, 53, 69). The rest of the proteins that comprise the secretion apparatus seem to be associated with the IM and periplasm, with the exception of the lipoprotein S, which when present, is located on the periplasmic side of the OM (49, 57, 58, 64).
Protein D is a member of the secretin family (58, 64, 69, 80). Secretins range from 50 to 70 kDa in mass (69). In addition to T2S systems, secretins are also essential components of the T3S system and the system for filamentous phage export and assembly (7, 50, 69, 80). As a typical secretin, protein D forms very stable multimeric complexes in the OM, with a highly conserved C-terminal half and a more diverse N-terminal domain (58, 69, 80). The N-terminal protein domain extends into the periplasm and seems to be important for substrate recognition, interaction with the cytoplasmic components of the secretion apparatus, protein multimerization, and possibly gating of the formed channel (49). The C-terminal domain of protein D is thought to be embedded in the membrane, forming the pore through which the substrate is exported to the cell surface (7, 49, 50, 69). This secretin domain is predicted to fold into a β-barrel (69). In addition, some D proteins contain a small C-terminal domain which binds a secretin-specific lipoprotein S, responsible for proper targeting and insertion of the secretin into the OM (58, 64).
Studies with the D-S complex from Klebsiella oxytoca (PulDS) have provided insights into the structure of the channel. Scanning transmission electron microscopy analyses of the PulDS complexes revealed a ring-like structure of 12 PulD subunits surrounded by 12 radial spokes of PulS (49). The enclosed central cavity has an estimated internal diameter of 7.6 nm, sufficient for passage of folded proteins (49). Side views of the PulDS complex indicate two ring-shaped structures stacked one on top of the other. The broader ring is probably integrated in the OM, whereas the smaller ring seems to protrude into the periplasm as far as the IM, folding back inside the channel, and thus this structure most likely corresponds to the N-terminal domain of PulD (50). This is in agreement with the high-density region observed within the ring structures of both PulD and the Pseudomonas secretin XcpQ (7, 49). Recently, the structure of the secretin PilQ was determined by cryoelectron microscopy to 12-Å resolution (15), revealing a continuous channel, sealed at both the top and bottom, with a diameter of 87 Å at its widest part. These structural features suggested more than a passive role for the secretins and have important implications for their function in OM secretion (15).
Several models have been proposed for secretion across the OM through the T2S pathway. One of them suggests secretion through a central channel formed by the outer surfaces of the β-barrels of 12 protein D monomers. Alternatively, each D monomer may contribute several β-strands, leading to formation of a single β-barrel with a large central pore (69). Formation of macromolecular superstructures has also been proposed, in which several D oligomers of 12 subunits each form higher-ordered pore structures. In this model, secretion may occur either through the central pore formed by the outer surfaces of the oligomers or through the individual pores formed by each D dodecamer of the superstructure (58). This model is further supported by the structural analysis of the secretin PilQ, which suggests formation of a tetramer of PilQ trimers (15). It is speculated that during secretion, the pseudopilin components of the T2S apparatus may act as a piston, pushing the exported protein through the secretin pore (49).
T1S
The T1S system exports proteins directly from the cytoplasm to the extracellular environment (5, 37). Its substrates are usually high-molecular-weight exoenzymes or toxins. Type I secretion is best exemplified by the secretion system of α-hemolysin (HlyA) of pathogenic Escherichia coli (37).
Proteins secreted by the T1S systems do not contain a cleavable N-terminal signal peptide; instead, they possess a noncleavable C-terminal signal sequence that targets them to the secretion apparatus (5, 6). The latter consists of three polypeptides: the two components of the IM translocase, which are a member of the ABC superfamily (e.g., HlyB) (5) and a member of the membrane fusion protein family (e.g., HlyD), and the integral OM protein that forms a β-barrel with a central hydrophilic pore (e.g., TolC) (36). In the case of the E. coli HlyA secretion system, upon interaction with the substrate, the IM translocase recruits TolC via the adaptor HlyD, forming a continuous channel that spans the cell envelope (36). A 38-residue charged N-terminal cytosolic domain of HlyD is essential for TolC recruitment and T1S (3, 36).
The crystal structure of TolC, obtained at 2.1-Å resolution, revealed a homotrimer of three 471-residue monomers that form a single channel (38). The most striking characteristic of TolC is its 100-Å-long periplasmic domain. The TolC channel can be divided into three units: (i) a 12-stranded right-handed β-barrel OM domain (the channel domain), (ii) a left-handed 12-stranded α-helical barrel domain that protrudes deep into the periplasm (the tunnel domain), and (iii) a mixed α/β equatorial domain that forms a strap around the midsection of the α-helical barrel. In the TolC trimer, each monomer contributes four antiparallel β-strands, four antiparallel α-helices, and shorter helices and strands that form one-third of the equatorial domain (36, 38). On the extracellular side of the OM, the TolC channel is fully open and does not have any inward-folding loop to constrict the formed β-barrel or any plug domain to control channel permeability. At the periplasmic end, on the other hand, the helices of the TolC tunnel are arranged into pairs of coiled coils, which taper to close the pore (36). As a result, the periplasmic entrance of TolC is nearly closed, with an effective diameter of ∼3.9 Å, and thus must undergo conformational change to allow substrate passage. The periplasmic entrance of TolC is the only constriction of the TolC pore (36).
Transition of TolC from a closed resting state to an open state is a crucial step in protein export (36). Opening of the TolC periplasmic entrance occurs in a substrate-dependent manner and is thought to involve an iris-like rearrangement of the periplasmic tunnel entrance helices (36). This model was further supported by in vivo experiments with E. coli, where the TolC entrance was fixed by introduced disulfide bonds. When the TolC coiled coils were locked into the narrowest entrance configuration, HlyA was no longer exported. The locked TolC, however, was found to be recruited properly by the IM translocase, indicating that the untwisting of the TolC entrance helices is essential for proper TolC-dependent export, as it opens the entrance pore to allow passage of the substrate (23). Once the substrate is exported, TolC reverts to its resting closed state (36).
T3S
The T3S pathway can mediate delivery of bacterial virulence factors directly into host cells, involving protein export not only across the bacterial cell envelope, but the plasma membrane of the eukaryotic cell as well (16). It was first characterized for the Yop proteins of Yersinia and has much in common with the flagellar export system (55).
T3S is often associated with virulence plasmids and pathogenicity islands and consists of three types of proteins: (i) structural components of the injection apparatus, (ii) effector (secreted) proteins, and (iii) regulatory factors controlling expression of structural and effector proteins (25). The injection apparatus consists of at least 20 different proteins (74) that assemble into a needle complex, which allows protein secretion upon contact with the eukaryotic target cell; hence the term “contact-dependent pathway” that is often used to describe T3S (25). Targeting of the substrates to the T3S apparatus is believed to occur via N-terminal signal peptides, although it has also been proposed that secretion signals might reside in the 5′ ends of the corresponding mRNAs (16).
As many as 11 proteins are conserved in the T3S system (31). The OM component of the needle apparatus is one of these proteins, and it is exemplified by YscC from Y. enterocolitica. YscC belongs to the secretin family, and it is the only component of the Yop system that is clearly found in the OM (40). Homologues of YscC include the secretins InvG of Salmonella enterica serovar Typhimurium, EscC of enteropathogenic E. coli, MxiD of Shigella flexneri, and HrcC of the plant pathogen Pseudomonas syringae (20, 26, 40, 63). Being a secretin, YscC multimerizes and forms very stable ring-shaped structures (40) with a molecular mass of ∼1 MDa (9). Electron microscopy demonstrated that YscC exhibits a 13-fold angular order, which is indicative of an oligomer of 13 subunits (9). Analyses of the formed channels demonstrated a stain-accessible central pore (9, 40) with an external diameter of ∼14 nm and an inner diameter of ∼4 nm (9). YscC has an estimated total length of 11 to 12 nm and is composed of two stacked rings and a conical domain. These findings suggest that YscC probably corresponds to the upper two rings of the basal body of the needle complex (9).
Both YscC and InvG require an OM lipoprotein (YscW and InvH, respectively) for proper OM insertion (17, 40). Absence of YscW from Y. enterocolitica results in a lower yield of the YscC secretin and reduced substrate secretion via the T3S pathway (40). Interestingly, YscW is not a stable component of the oligomeric structure (9), as is the case for the PulDS proteins of the T2S pathway. The lipoprotein binding site has been identified in the C termini of various secretins. However, studies with a C terminally truncated YscC revealed that the C terminus is not essential for OM localization and function of YscC in the secretion process or for interaction with the YscW lipoprotein (8). These findings suggest that the C terminus of YscC may render the secretin lipoprotein dependent for OM insertion, which may in turn increase the efficiency of the secretion pathway (8). In addition, YscW promotes YscC oligomerization, which indicates that interaction with YscC occurs prior to the formation of the secretin multimer (8). Studies with the EscC and MxiD secretins have indicated that correct insertion and function of these two proteins in the OM depends on the presence of other T3S apparatus components (26, 63).
Secretion through the T3S pathway is generally believed to occur in one step without the involvement of periplasmic intermediates. In this model, the secreted proteins are exported directly across the two membranes via a continuous channel that spans the cell envelope. The formation of such a continuous conduit by the T3S apparatus has been supported by both biochemical studies and electron microscopic images (16, 26).
T4S
T4S systems are characterized by a remarkable functional versatility, as they can transfer both proteins and single-stranded-DNA-protein complexes in a cell-contact-dependent or cell-contact-independent mechanism (11, 42). They are closely related to bacterial conjugation and are thought to have evolved from the conjugation machinery (11, 21). T4S is generally Sec independent, with the remarkable exceptions of the Sec-dependent transport of the B. pertussis PT toxin (and possibly the coupling protein [CP]-independent systems [see below] of Brucella spp. and Bartonella tribocorum) (11).
The T-DNA transfer system of A. tumefaciens has provided the basis of what is known about T4S (11, 13). Conjugative T4S systems are composed of three distinct substructures: the CP hexamer, which targets the substrate to the transenvelope apparatus; the transenvelope protein complex for transport of the substrate across the IM and OM; and the conjugative T pilus, for substrate delivery to the target cell. The last two substructures consist of mating-pair formation (Mpf) proteins, which in the case of A. tumefaciens are the VirB1-VirB11 proteins (11, 42). Recent studies have proposed that the proteins VirB2, VirB6, VirB8, VirB9, and VirB11 correspond to channel subunits of the T4S apparatus, whereas VirB4, VirB5, VirB7, and VirB10 interact indirectly with the secreted substrate through their direct association with the channel subunits (10). The rest of the Mpf proteins do not interact with the transfer intermediate (10).
VirB7 and VirB9 are the OM constituents of the translocation pore. Both proteins are predicted to possess large periplasmic domains (19, 24). VirB9 is a 292-residue secretin-like protein that interacts through its C-terminal third with the 55-amino-acid residue OM lipoprotein VirB7 to form a disulfide-linked heterodimeric protein complex (2). This complex is anchored to the OM by a covalent interaction of VirB7 with this lipid bilayer (18, 24) and requires the IM protein VirB6 for proper formation (35). In addition, VirB9 can form homodimers and has also been shown to form intermolecular complexes with the IM proteins VirB8 and VirB10. Formation of VirB9-VirB8 and VirB9-VirB10 complexes involves the N-terminal third of VirB9, whereas self-interacting domains lie in both the N and C termini of VirB9 (18). VirB7 has also been shown to form homodimers that interact with subunits of the T pilus (35). VirB7 and VirB9 homologues have been detected in Helicobacter pylori as components of its surface filamentous structures, indicating that the H. pylori T4S apparatus is a filamentous macromolecular complex that protrudes from the cell envelope (68). The VirB2 pilin has been proposed to act in concert with VirB9 for translocation across the OM. Deletion of either of these two proteins arrests OM transport (10).
Two working models have been proposed for secretion via the T4S system. The “channel model” supports the idea that substrates are transported across the OM through the lumen of a pilus-like structure encased by the OM components VirB7 and VirB9. The “piston model,” on the other hand, involves transport through a retractile pilus that extends and pushes the substrate through the OM pore (11). The piston model has been shown to be correct in the case of the F-plasmid T4S machinery and possibly in the case of the B. pertussis Ptl system (11, 42). Future studies are expected to provide a better understanding of the molecular mechanisms that govern substrate export through T4S systems.
ENP
A distinct secretion mechanism, called the extracellular nucleation-precipitation (ENP) pathway, is envisioned for the production of curli, the adhesive fibers on the surface of several pathogenic bacteria, responsible for specific interactions with eukaryotic hosts and biofilm formation (43, 64). In E. coli, the two curli components, CsgA and CsgB, and the OM lipoprotein CsgG cross the IM via the Sec system. Although knowledge of the secretion mechanism is very limited, experimental data indicate that CsgG is required for secretion of both CsgA and CsgB across the OM. Two models have been proposed for OM export; one supports the idea that CsgG acts as a chaperone stabilizing the two curli components, whereas the other favors formation of a CsgG channel through which CsgA and CsgB are secreted (43, 64). Further work is needed to determine the exact mechanism of curli secretion.
CONCLUDING REMARKS
At least eight different pathways are currently characterized in gram-negative bacteria for secretion of proteins across the cell envelope (Table 1). Their OM transporters can be grouped into six distinct families: the AT β-domain, the TpsB protein (of the TPS), the pilus usher protein, the secretin (of T2S and T3S systems), the outer membrane component of T1S (exemplified by E. coli TolC), and the outer membrane component of T4S (exemplified by the Agrobacterium VirB9 protein). The OM component (a lipoprotein) of the curli secretion system may eventually be added to this growing list of protein transporters, provided that more information becomes available regarding its structure and role in curli biogenesis (43) (Table 2).
Two high-resolution crystal structures of OM transporters have been determined to date, the structure of an AT β-domain (NalP) and that of the OM component of a T1S system (TolC). Information regarding the structures and functions of the other OM transporters (Table 1) has been based primarily on molecular, biochemical, and bioinformatics studies. Comparison of these six protein families indicates that (i) most protein transporters appear to be oligomeric or multimeric, perhaps with the exception of the AT β-domain; (ii) internal channel dimensions range between 10 and 100 Å, which is significantly larger than those of the general OM porins; and (iii) at least three different protein families, the ushers, the secretins, and the TpsB proteins, fold into two distinct protein domains. The N-terminal domain of the secretins and TpsB proteins appears to be localized in the periplasm and is required for substrate recognition, whereas the C-terminal domain folds into a β-barrel, providing the pore channel for substrate secretion across the membrane. In the case of the usher, recent analyses indicate that the N terminus, which contains the substrate recognition site, forms part of the β-barrel, whereas the C terminus folds as a separate periplasmic domain tightly associated with the N-terminal β-barrel (29).
Despite the vast knowledge acquired over the past years regarding protein transport across the bacterial OM, how these transporters insert and assemble in the membrane remains vague. For many years, it was thought that insertion of proteins into the OM occurs spontaneously without the requirement for a proteinaceous machinery. However, a recent study challenges this model. Voulhoux and colleagues have reported that a highly conserved OM protein, named Omp85 in N. meningitidis, plays an essential role in OM biogenesis (78). Omp85 was found to be directly involved in the biogenesis of general porins, a secretin of a T2S system, OM enzymes, and an AT protein. Although the details remain to be revealed, it is now proposed that biogenesis of OM proteins follows a “periplasmic chaperone (Skp)/usher (Omp85)-like pathway” (79). Future studies will no doubt be directed toward the molecular characterization of this mechanism and the unraveling of the architecture of the Omp85 secretion machinery.
Many more questions still remain to be addressed regarding protein export across the bacterial OM. What is the folding state of the secreted proteins before and during their transport across the OM? How are unfolded or partially folded proteins protected against proteolytic degradation? How do secreted proteins and macromolecular secretion machines get through the peptidoglycan layer? If energy sources are not present in the periplasm and the OM, how do molecules as large as the secreted proteins overcome the unfavorable entropy constraints and pass through the hydrophobic membrane environment? Are any chaperones universally required? What is the fate of an OM transporter after secretion? How do the OM pores prevent leakage of small periplasmic molecules? What mechanisms are involved in channel gating? How is specificity of secreted proteins assured in general? What is the architecture of the T4S and T3S machineries? What are the structures of the components of the different secretion apparatuses? Answers to these questions will provide a more complete picture of the secretion process and will lead to more efficient means of protection against bacterial infections.
Acknowledgments
We are grateful to Vassilis Koronakis, James Nataro, Joseph St Geme III, Rachel Fernandez, and Hye-Jeong Yeo for critical reading of the manuscript.
Our research is supported by grants from the USDA (94-37204-1091) and the Welch Foundation (E-1548) to C.S, and from the NIH (GM62987) to D.G.T.
REFERENCES
- 1.Ahting, U., C. Thun, R. Hegerl, D. Typke, F. E. Nargang, W. Neupert, and S. Nussberger. 1999. The TOM core complex: the general protein import pore of the outer membrane of mitochondria. J. Cell Biol. 147:959-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, L. B., A. V. Hertzel, and A. Das. 1996. Agrobacterium tumefaciens VirB7 and VirB9 form a disulfide-linked protein complex. Proc. Natl. Acad. Sci. USA 93:8889-8894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balakrishnan, L., C. Hughes, and V. Koronakis. 2001. Substrate-triggered recruitment of the TolC channel-tunnel during type I export of hemolysin by Escherichia coli. J. Mol. Biol. 313:501-510. [DOI] [PubMed] [Google Scholar]
- 4.Berks, B. C., T. Palmer, and F. Sargent. 2003. The Tat protein translocation pathway and its role in microbial physiology. Adv. Microb. Physiol. 47:187-254. [DOI] [PubMed] [Google Scholar]
- 5.Binet, R., S. Letoffe, J. M. Ghigo, P. Delepelaire, and C. Wandersman. 1997. Protein secretion by gram-negative bacterial ABC exporters. Folia Microbiol. (Prague) 42:179-183. [DOI] [PubMed] [Google Scholar]
- 6.Blight, M. A., and I. B. Holland. 1994. Heterologous protein secretion and the versatile Escherichia coli haemolysin translocator. Trends Biotechnol. 12:450-455. [DOI] [PubMed] [Google Scholar]
- 7.Brok, R., P. Van Gelder, M. Winterhalter, U. Ziese, A. J. Koster, H. de Cock, M. Koster, J. Tommassen, and W. Bitter. 1999. The C-terminal domain of the Pseudomonas secretin XcpQ forms oligomeric rings with pore activity. J. Mol. Biol. 294:1169-1179. [DOI] [PubMed] [Google Scholar]
- 8.Burghout, P., F. Beckers, E. de Wit, R. van Boxtel, G. R. Cornelis, J. Tommassen, and M. Koster. 2004. Role of the pilot protein YscW in the biogenesis of the YscC secretin in Yersinia enterocolitica. J. Bacteriol. 186:5366-5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burghout, P., R. van Boxtel, P. Van Gelder, P. Ringler, S. A. Muller, J. Tommassen, and M. Koster. 2004. Structure and electrophysiological properties of the YscC secretin from the type III secretion system of Yersinia enterocolitica. J. Bacteriol. 186:4645-4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cascales, E., and P. J. Christie. 2004. Definition of a bacterial type IV secretion pathway for a DNA substrate. Science 304:1170-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cascales, E., and P. J. Christie. 2003. The versatile bacterial type IV secretion systems. Nat. Rev. Microbiol. 1:137-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choudhury, D., A. Thompson, V. Stojanoff, S. Langermann, J. Pinkner, S. J. Hultgren, and S. D. Knight. 1999. X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli. Science 285:1061-1066. [DOI] [PubMed] [Google Scholar]
- 13.Christie, P. J. 1997. Agrobacterium tumefaciens T-complex transport apparatus: a paradigm for a new family of multifunctional transporters in eubacteria. J. Bacteriol. 179:3085-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clantin, B., H. Hodak, E. Willery, C. Locht, F. Jacob-Dubuisson, and V. Villeret. 2004. The crystal structure of filamentous hemagglutinin secretion domain and its implications for the two-partner secretion pathway. Proc. Natl. Acad. Sci. USA 101:6194-6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collins, R. F., S. A. Frye, A. Kitmitto, R. C. Ford, T. Tonjum, and J. P. Derrick. 2004. Structure of the Neisseria meningitidis outer membrane PilQ secretin complex at 12 Å resolution. J. Biol. Chem. 279:39750-39756 [DOI] [PubMed] [Google Scholar]
- 16.Cornelis, G. R. 2002. The Yersinia Ysc-Yop ‘type III’ weaponry. Nat. Rev. Mol. Cell. Biol. 3:742-752. [DOI] [PubMed] [Google Scholar]
- 17.Daefler, S., and M. Russel. 1998. The Salmonella typhimurium InvH protein is an outer membrane lipoprotein required for the proper localization of InvG. Mol. Microbiol. 28:1367-1380. [DOI] [PubMed] [Google Scholar]
- 18.Das, A., and Y. H. Xie. 2000. The Agrobacterium T-DNA transport pore proteins VirB8, VirB9, and VirB10 interact with one another. J. Bacteriol. 182:758-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Das, A., and Y. H. Xie. 1998. Construction of transposon Tn3phoA: its application in defining the membrane topology of the Agrobacterium tumefaciens DNA transfer proteins. Mol. Microbiol. 27:405-414. [DOI] [PubMed] [Google Scholar]
- 20.Deng, W. L., and H. C. Huang. 1999. Cellular locations of Pseudomonas syringae pv. syringae HrcC and HrcJ proteins, required for harpin secretion via the type III pathway. J. Bacteriol. 181:2298-2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding, Z., K. Atmakuri, and P. J. Christie. 2003. The outs and ins of bacterial type IV secretion substrates. Trends Microbiol. 11:527-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dodson, K. W., F. Jacob-Dubuisson, R. T. Striker, and S. J. Hultgren. 1993. Outer-membrane PapC molecular usher discriminately recognizes periplasmic chaperone-pilus subunit complexes. Proc. Natl. Acad. Sci. USA 90:3670-3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eswaran, J., C. Hughes, and V. Koronakis. 2003. Locking TolC entrance helices to prevent protein translocation by the bacterial type I export apparatus. J. Mol. Biol. 327:309-315. [DOI] [PubMed] [Google Scholar]
- 24.Fernandez, D., T. A. Dang, G. M. Spudich, X. R. Zhou, B. R. Berger, and P. J. Christie. 1996. The Agrobacterium tumefaciens virB7 gene product, a proposed component of the T-complex transport apparatus, is a membrane-associated lipoprotein exposed at the periplasmic surface. J. Bacteriol. 178:3156-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galan, J. E., and A. Collmer. 1999. Type III secretion machines: bacterial devices for protein delivery into host cells. Science 284:1322-1328. [DOI] [PubMed] [Google Scholar]
- 26.Gauthier, A., J. L. Puente, and B. B. Finlay. 2003. Secretin of the enteropathogenic Escherichia coli type III secretion system requires components of the type III apparatus for assembly and localization. Infect. Immun. 71:3310-3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guedin, S., E. Willery, J. Tommassen, E. Fort, H. Drobecq, C. Locht, and F. Jacob-Dubuisson. 2000. Novel topological features of FhaC, the outer membrane transporter involved in the secretion of the Bordetella pertussis filamentous hemagglutinin. J. Biol. Chem. 275:30202-30210. [DOI] [PubMed] [Google Scholar]
- 28.Henderson, I. R., and J. P. Nataro. 2001. Virulence functions of autotransporter proteins. Infect. Immun. 69:1231-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henderson, N. S., S. Shu Kin So, C. Martin, R. Kulkarni, and D. G. Thanassi. 2004. Topology of the outer membrane usher PapC determined by site-fluorescence labeling. J. Biol. Chem. 279:53747-53754. [DOI] [PubMed] [Google Scholar]
- 30.Hendrixson, D. R., M. L. de la Morena, C. Stathopoulos, and J. W. St Geme III. 1997. Structural determinants of processing and secretion of the Haemophilus influenzae Hap protein. Mol. Microbiol 26:505-518. [DOI] [PubMed] [Google Scholar]
- 31.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jacob-Dubuisson, F., C. El-Hamel, N. Saint, S. Guedin, E. Willery, G. Molle, and C. Locht. 1999. Channel formation by FhaC, the outer membrane protein involved in the secretion of the Bordetella pertussis filamentous hemagglutinin. J. Biol. Chem. 274:37731-37735. [DOI] [PubMed] [Google Scholar]
- 33.Jacob-Dubuisson, F., R. Fernandez, and L. Coutte. 2004. Protein secretion through autotransporter and two-partner pathways. Biochem. Biophys. Acta 1694:235-257. [DOI] [PubMed] [Google Scholar]
- 34.Jacob-Dubuisson, F., C. Locht, and R. Antoine. 2001. Two-partner secretion in Gram-negative bacteria: a thrifty, specific pathway for large virulence proteins. Mol. Microbiol. 40:306-313. [DOI] [PubMed] [Google Scholar]
- 35.Jakubowski, S. J., V. Krishnamoorthy, and P. J. Christie. 2003. Agrobacterium tumefaciens VirB6 protein participates in formation of VirB7 and VirB9 complexes required for type IV secretion. J. Bacteriol. 185:2867-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koronakis, V., J. Eswaran, and C. Hughes. 2004. Structure and function of TolC: the bacterial exit duct for proteins and drugs. Annu. Rev. Biochem. 73:467-489. [DOI] [PubMed] [Google Scholar]
- 37.Koronakis, V., E. Koronakis, and C. Hughes. 1989. Isolation and analysis of the C-terminal signal directing export of Escherichia coli hemolysin protein across both bacterial membranes. EMBO J. 8:595-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koronakis, V., A. Sharff, E. Koronakis, B. Luisi, and C. Hughes. 2000. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 405:914-919. [DOI] [PubMed] [Google Scholar]
- 39.Kostakioti, M., and C. Stathopoulos. 2004. Functional analysis of the Tsh autotransporter of an avian pathogenic Escherichia coli strain. Infect. Immun. 72:5548-5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koster, M., W. Bitter, H. de Cock, A. Allaoui, G. R. Cornelis, and J. Tommassen. 1997. The outer membrane component, YscC, of the Yop secretion machinery of Yersinia enterocolitica forms a ring-shaped multimeric complex. Mol. Microbiol. 26:789-797. [DOI] [PubMed] [Google Scholar]
- 41.Li, H., L. Qian, Z. Chen, D. Thahbot, G. Liu, T. Liu, and D. G. Thanassi. 2004. The outer membrane usher forms a twin-pore secretion complex. J. Mol. Biol. 344:1397-1407. [DOI] [PubMed] [Google Scholar]
- 42.Llosa, M., and D. O'Callaghan. 2004. Euroconference on the Biology of Type IV Secretion Processes: bacterial gates into the outer world. Mol. Microbiol 53:1-8. [DOI] [PubMed] [Google Scholar]
- 43.Loferer, H., M. Hammar, and S. Normark. 1997. Availability of the fibre subunit CsgA and the nucleator protein CsgB during assembly of fibronectin-binding curli is limited by the intracellular concentration of the novel lipoprotein CsgG. Mol. Microbiol. 26:11-23. [DOI] [PubMed] [Google Scholar]
- 44.Loveless, B. J., and M. H. Saier, Jr. 1997. A novel family of channel-forming, autotransporting, bacterial virulence factors. Mol. Membr. Biol. 14:113-123. [DOI] [PubMed] [Google Scholar]
- 45.Maurer, J., J. Jose, and T. F. Meyer. 1999. Characterization of the essential transport function of the AIDA-I autotransporter and evidence supporting structural predictions. J. Bacteriol. 181:7014-7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Newman, L. C., and C. Stathopoulos. 2004. Autotransporter and two-partner secretion: delivery of large-size virulence factors by gram-negative bacterial pathogens. Crit. Rev. Microbiol. 30:275-286. [DOI] [PubMed] [Google Scholar]
- 47.Ng, T. W., L. Akman, M. Osisami, and D. G. Thanassi. 2004. The usher N terminus is the initial targeting site for chaperone-subunit complexes and participates in subsequent pilus biogenesis events. J. Bacteriol. 186:5321-5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nishiyama, M., M. Vetsch, C. Puorger, I. Jelesarov, and R. Glockshuber. 2003. Identification and characterization of the chaperone-subunit complex-binding domain from the type 1 pilus assembly platform FimD. J. Mol. Biol. 330:513-525. [DOI] [PubMed] [Google Scholar]
- 49.Nouwen, N., N. Ranson, H. Saibil, B. Wolpensinger, A. Engel, A. Ghazi, and A. P. Pugsley. 1999. Secretin PulD: association with pilot PulS, structure, and ion-conducting channel formation. Proc. Natl. Acad. Sci. USA 96:8173-8177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nouwen, N., H. Stahlberg, A. P. Pugsley, and A. Engel. 2000. Domain structure of secretin PulD revealed by limited proteolysis and electron microscopy. EMBO J. 19:2229-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oliver, D. C., G. Huang, E. Nodel, S. Pleasance, and R. C. Fernandez. 2003. A conserved region within the Bordetella pertussis autotransporter BrkA is necessary for folding of its passenger domain. Mol. Microbiol. 47:1367-1383. [DOI] [PubMed] [Google Scholar]
- 52.Oomen, C. J., P. Van Ulsen, P. Van Gelder, M. Feijen, J. Tommassen, and P. Gros. 2004. Structure of the translocator domain of a bacterial autotransporter. EMBO J. 23:1257-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peabody, C. R., Y. J. Chung, M. R. Yen, D. Vidal-Ingigliardi, A. P. Pugsley, and M. H. Saier, Jr. 2003. Type II protein secretion and its relationship to bacterial type IV pili and archaeal flagella. Microbiology 149:3051-3072. [DOI] [PubMed] [Google Scholar]
- 54.Peak, I. R., Y. Srikhanta, M. Dieckelmann, E. R. Moxon, and M. P. Jennings. 2000. Identification and characterisation of a novel conserved outer membrane protein from Neisseria meningitidis. FEMS Immunol. Med. Microbiol. 28:329-334. [DOI] [PubMed] [Google Scholar]
- 55.Plano, G. V., K. Schesser, and M. L. Nilles. 2003. Type III secretion systems, p. 95-114. In D. L. Burns et al. (ed.), Bacterial protein toxins. ASM Press, Washington, D.C.
- 56.Roggenkamp, A., N. Ackermann, C. A. Jacobi, K. Truelzsch, H. Hoffmann, and J. Heesemann. 2003. Molecular analysis of transport and oligomerization of the Yersinia enterocolitica adhesin YadA. J. Bacteriol. 185:3735-3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Russel, M. 1998. Macromolecular assembly and secretion across the bacterial cell envelope: type II protein secretion systems. J. Mol. Biol. 279:485-499. [DOI] [PubMed] [Google Scholar]
- 58.Sandkvist, M. 2001. Biology of type II secretion. Mol. Microbiol. 40:271-283. [DOI] [PubMed] [Google Scholar]
- 59.Sauer, F. G., K. Futterer, J. S. Pinkner, K. W. Dodson, S. J. Hultgren, and G. Waksman. 1999. Structural basis of chaperone function and pilus biogenesis. Science 285:1058-1061. [DOI] [PubMed] [Google Scholar]
- 60.Sauer, F. G., J. S. Pinkner, G. Waksman, and S. J. Hultgren. 2002. Chaperone priming of pilus subunits facilitates a topological transition that drives fiber formation. Cell 111:543-551. [DOI] [PubMed] [Google Scholar]
- 61.Saulino, E. T., D. G. Thanassi, J. S. Pinkner, and S. J. Hultgren. 1998. Ramifications of kinetic partitioning on usher-mediated pilus biogenesis. EMBO J. 17:2177-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schonherr, R., R. Tsolis, T. Focareta, and V. Braun. 1993. Amino acid replacements in the Serratia marcescens haemolysin ShIA define sites involved in activation and secretion. Mol. Microbiol. 9:1229-1237. [DOI] [PubMed] [Google Scholar]
- 63.Schuch, R., and A. T. Maurelli. 2001. MxiM and MxiJ, base elements of the Mxi-Spa type III secretion system of Shigella, interact with and stabilize the MxiD secretin in the cell envelope. J. Bacteriol. 183:6991-6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stathopoulos, C., D. R. Hendrixson, D. G. Thanassi, S. J. Hultgren, J. W. St Geme III, and R. Curtiss III. 2000. Secretion of virulence determinants by the general secretory pathway in gram-negative pathogens: an evolving story. Microbes Infect. 2:1061-1072. [DOI] [PubMed] [Google Scholar]
- 65.Stathopoulos, C., D. L. Provence, and R. Curtiss III. 1999. Characterization of the avian pathogenic Escherichia coli hemagglutinin Tsh, a member of the immunoglobulin A protease-type family of autotransporters. Infect. Immun. 67:772-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Surana, N. K., D. Cutter, S. J. Barenkamp, and J. W. St Geme III. 2004. The Haemophilus influenzae Hia autotransporter contains an unusually short trimeric translocator domain. J. Biol. Chem. 279:14679-14685. [DOI] [PubMed] [Google Scholar]
- 67.Surana, N. K., S. Grass, G. G. Hardy, H. Li, D. G. Thanassi, and J. W. Geme III. 2004. Evidence for conservation of architecture and physical properties of Omp85-like proteins throughout evolution. Proc. Natl. Acad. Sci. USA 101:14497-14502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tanaka, J., T. Suzuki, H. Mimuro, and C. Sasakawa. 2003. Structural definition on the surface of Helicobacter pylori type IV secretion apparatus. Cell Microbiol. 5:395-404. [DOI] [PubMed] [Google Scholar]
- 69.Thanassi, D. G. 2002. Ushers and secretins: channels for the secretion of folded proteins across the bacterial outer membrane. J. Mol. Microbiol. Biotechnol. 4:11-20. [PubMed] [Google Scholar]
- 70.Thanassi, D. G., E. T. Saulino, and S. J. Hultgren. 1998. The chaperone/usher pathway: a major terminal branch of the general secretory pathway. Curr. Opin. Microbiol. 1:223-231. [DOI] [PubMed] [Google Scholar]
- 71.Thanassi, D. G., E. T. Saulino, M. J. Lombardo, R. Roth, J. Heuser, and S. J. Hultgren. 1998. The PapC usher forms an oligomeric channel: implications for pilus biogenesis across the outer membrane. Proc. Natl. Acad. Sci. USA 95:3146-3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thanassi, D. G., C. Stathopoulos, K. Dodson, D. Geiger, and S. J. Hultgren. 2002. Bacterial outer membrane ushers contain distinct targeting and assembly domains for pilus biogenesis. J. Bacteriol. 184:6260-6269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thanassi, D. G., C. Stathopoulos, A. Karkal, and H. Li. 2005.. Protein secretion in the absence of ATP: the autotransporter, two-partner secretion and chaperone/usher pathways of Gram negative bacteria. Mol. Membr. Biol. 22:63-72. [DOI] [PubMed]
- 74.Thomas, N. A., and B. Brett Finlay. 2003. Establishing order for type III secretion substrates—a hierarchical process. Trends Microbiol. 11:398-403. [DOI] [PubMed] [Google Scholar]
- 75.Touze, T., R. D. Hayward, J. Eswaran, J. M. Leong, and V. Koronakis. 2004. Self-association of EPEC intimin mediated by the beta-barrel-containing anchor domain: a role in clustering of the Tir receptor. Mol. Microbiol. 51:73-87. [DOI] [PubMed] [Google Scholar]
- 76.Veiga, E., E. Sugawara, H. Nikaido, V. de Lorenzo, and L. A. Fernandez. 2002. Export of autotransported proteins proceeds through an oligomeric ring shaped by C-terminal domains. EMBO J. 21:2122-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Voulhoux, R., G. Ball, B. Ize, M. L. Vasil, A. Lazdunski, L. F. Wu, and A. Filloux. 2001. Involvement of the twin-arginine translocation system in protein secretion via the type II pathway. EMBO J. 20:6735-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Voulhoux, R., M. P. Bos, J. Geurtsen, M. Mols, and J. Tommassen. 2003. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science 299:262-265. [DOI] [PubMed] [Google Scholar]
- 79.Voulhoux, R., and J. Tommassen. 2004. Omp85, an evolutionarily conserved bacterial protein involved in outer-membrane-protein assembly. Res. Microbiol. 155:129-135. [DOI] [PubMed] [Google Scholar]
- 80.Yen, M. R., C. R. Peabody, S. M. Partovi, Y. Zhai, Y. H. Tseng, and M. H. Saier. 2002. Protein-translocating outer membrane porins of Gram-negative bacteria. Biochim. Biophys. Acta 1562:6-31. [DOI] [PubMed] [Google Scholar]
- 81.Zavialov, A. V., J. Berglund, A. F. Pudney, L. J. Fooks, T. M. Ibrahim, S. MacIntyre, and S. D. Knight. 2003. Structure and biogenesis of the capsular F1 antigen from Yersinia pestis: preserved folding energy drives fiber formation. Cell 113:587-596. [DOI] [PubMed] [Google Scholar]