Abstract
Yeast Saccharomyces cerevisiae Cdc13p is the telomere-binding protein that protects telomeres and regulates telomere length. It is documented that Cdc13p binds specifically to single-stranded TG1–3 telomeric DNA sequences and interacts with Stn1p. To localize the region for single-stranded TG1–3 DNA binding, Cdc13p mutants were constructed by deletion mutagenesis and assayed for their binding activity. Based on in vitro electrophoretic mobility shift assay, a 243-amino-acid fragment of Cdc13p (amino acids 451–693) was sufficient to bind single-stranded TG1–3 with specificity similar to that of the native protein. Consistent with the in vitro observation, in vivo one-hybrid analysis also indicated that this region of Cdc13p was sufficient to localize itself to telomeres. However, the telomere-binding region of Cdc13p (amino acids 451–693) was not capable of complementing the growth defects of cdc13 mutants. Instead, a region comprising the Stn1p-interacting and telomere-binding region of Cdc13p (amino acids 252–924) complemented the growth defects of cdc13 mutants. These results suggest that binding to telomeres by Cdc13p is not sufficient to account for the cell viability, interaction with Stn1p is also required. Taken together, we have defined the telomere-binding domain of Cdc13p and showed that both binding to telomeres and Stn1p by Cdc13p are required to maintain cell growth.
INTRODUCTION
Telomeres are the specialized structure at the very ends of eukaryotic chromosomes. Telomeres are essential for the maintenance of chromosome integrity (1,2). They prevent end-to-end fusion of chromosomes, protect chromosome from degradation by nucleases and facilitate the complete replication of chromosomes. They also suppress the expression of nearby genes, a phenomenon known as telomere position effect (TPE) (3). Telomeres might also position chromosomes within the nucleus (4). In most of the cloned telomeric sequences, telomeres are composed of short tandem repeated sequences with the strand running in a 5′ to 3′ direction toward the guanine (G)-rich ends (1). The sequence and the number of repeats vary considerably in different species. For example, the telomere sequences of yeast Saccharomyces cerevisiae are ∼250–300 bp of TG1–3/C1–3A, whereas those of human are ∼10 kb TTAGGG/CCCTAA repeats (1,2). Besides the double-stranded telomeric DNA repeats, telomeres in all of the organisms that have been analyzed, also contain a G-rich single-stranded tail (5–7). In S.cerevisiae, a >30 base single-stranded TG1–3 tail was detected in late S phase of the cell cycle (8). This single-stranded tail was postulated as an intermediate during telomere replication (8,9).
Protein factors that interact with telomeres participate in telomere functions (10). Factors that bind to the double-stranded telomeric DNA sequences have been identified from many organisms (11–16). These proteins are essential for the maintenance of telomere functions and cell viability (11,12,16). For example, in S.cerevisiae, Rap1p has been shown to bind double-stranded telomeric DNA (11,17,18). Mutations on Rap1p affect the length of telomere, TPE, localization of the telomere within the nucleus, telomere recombination and cell viability (19–21). Moreover, protein factors bound to the single-stranded telomeric DNA were identified in several organisms (22–30). Among these protein factors, Oxytricha telomere-binding protein is well characterized. The protein is heterodimeric and is composed of an α subunit and a β subunit (31–33). The α subunit is a single-stranded DNA-binding protein that binds to the G4T4 single-stranded end of the telomere. Although the β subunit is not directly involved in binding, it is required for terminus-specific binding. In the yeast genome, there is no homologue of the Oxytricha α- and β-like binding proteins. Instead, Cdc13p binds to single-stranded TG1–3 sequences in vitro and interacts with telomeres in vivo (24,25,34). It is believed that, in yeast, Cdc13p is the functional equivalent of Oxytricha α and β binding proteins despite there being no sequence similarity between Oxytricha telomere-binding proteins and yeast Cdc13p (J.-J.Lin, unpublished observation). Furthermore, Cdc13p dissociates from single-stranded TG1–3 DNA at 300 mM NaCl (J.J.Lin and V.A.Zakian, unpublished result) whereas the binding of Oxytricha proteins to G4T4 DNA remains associated even at 2 M NaCl (33). Thus, Cdc13p might represent a class of single-stranded telomere-binding protein that is different from the Oxytricha protein.
CDC13 is an essential gene (35). It is involved in cell cycle control, because a temperature-sensitive allele of CDC13, cdc13-1, causes the cell cycle to arrest in G2/M phase at the non-permissive temperature (35). This cell cycle control is dependent on the gene product of RAD9, which was postulated to be involved in surveying the integrity of chromosomes (36). Cdc13p might enable Rad9p to differentiate whether the ends of a linear DNA are telomeres or broken ends by binding to telomeres. It is also demonstrated that cdc13-1 cells at the non-permissive temperature accumulate single-stranded G-rich DNA near telomeres (35). Nevertheless, it is unclear whether this single-stranded G-rich DNA is recognized as a damage to trigger the checkpoint mechanism. Since a mutation allele of CDC13, cdc13est, causes gradual loss of telomere, Cdc13p also appears to regulate the length of telomeres (24). At least a portion of the telomere function of Cdc13p is contributed by its interaction with Stn1p. STN1 was identified from a high copy suppressor screening to rescue the temperature-sensitive lethality of cdc13-1 (37) and direct interaction of Cdc13p with Stn1p was demonstrated by two-hybrid assays (37). Moreover, telomere defects caused by STN1 mutation are similar, if not identical, to CDC13 mutation, suggesting that these two genes act together in maintaining telomere function (37).
CDC13 encodes a 924-amino-acid protein with molecular mass of 104 895 kDa (35). Sequence comparison between Cdc13p and known DNA- or RNA-binding proteins did not provide any information on the region within Cdc13p responsible for interacting with telomeres (J.-J.Lin, unpublished observation). Thus, Cdc13p might contain a novel DNA-interacting motif for binding to telomeres. In order to identify the functional domains of Cdc13p, we have generated deletion mutants and examined the single-stranded TG1–3-binding activity and their ability to interact with Stn1p. Here we showed that Cdc13(451–693)p, a Cdc13p fragment ranging from amino acids 451 to 693, retain the single-stranded TG1–3-binding activity. We also showed that binding to telomeres by Cdc13p is not sufficient to maintain cell viability. Moreover, amino acids 252–924, which covered the telomere-binding and Stn1p-interacting activities of Cdc13p, are sufficient to account for the essential function of Cdc13p.
MATERIALS AND METHODS
Strains
Escherichia coli strain DH5α was used as host for plasmid construction and propagation, and strain BL21(DE3)pLysS for Cdc13(451–693)p purification. Yeast strain YPH499 [MATa ura3-52 lys2-801amber ade2-101ochre trp1-Δ63 his3-Δ200 leu2-Δ1 (38)] with URA3 and ADE2 placed near the telomere of chromosomes VII-L and V-R (YPH499UTAT), respectively, was used as the host for analyzing TPE. Yeast strains, HIS-Tel and HIS-Int-CA, used for one-hybrid analysis were kindly provided by V. A. Zakian [Princeton University (34)]. Strain 2758-8-4b (MATa cdc13-1 his7 leu2-3, 112 ura3-52 trp1-289, provided by L. Hartwell, University of Washington) was used as the host for complementation tests.
Plasmid construction
Deletion mutants of Cdc13p (Fig. 1A) were generated by fusion of various regions of CDC13 with the glutathione S-transferase (GST) gene of pGEX plasmids (Pharmacia). The 4.4-kb SmaI–HpaI fragment containing full length CDC13 from pTHA-CDC13 (25) was ligated to the SmaI-digested pGEX-3X to generate pGST-CDC13. Plasmid pGST-CDC13 was digested with BglII and self-ligated to generate pGST-CDC13(1–600), which deleted the BglII–BglII fragment within CDC13. Plasmid pGST-CDC13(451–924) was constructed by ligating the 2.9-kb BamHI–HpaI fragment containing half of CDC13 to BamHI- and SmaI-digested pGEX-3X. Plasmid pGST-CDC13(451–924) was digested with either PvuII or BglII, and self-ligated to generate pGST-CDC13(451–871) and pGST-CDC13(451–600), respectively. Plasmid pGST-CDC13(451–693) was constructed by digesting pGST-CDC13(451–924) with SalI and NruI, treating with T4 DNA polymerase and self-ligation. A 1.4-kb EcoRI–SalI fragment of pTHA-CDC13 was ligated to EcoRI- and SalI-digested pGEX-4T-1 to generate pGST-CDC13(510–924). To construct pGST-CDC13(601–856), a 0.8-kb BglII–BglII fragment from pTHA-CDC13 was inserted into BamHI-digested pGEX-1. The NruI–SmaI fragment of pGST-CDC13(601–856) was deleted to generate pGST-CDC13(601–693). Expression of these fusion proteins was induced by the addition of isopropyl β-d-thiogalactoside (IPTG) and confirmed by western blotting analysis using antibody against GST (data not shown).
Figure 1.
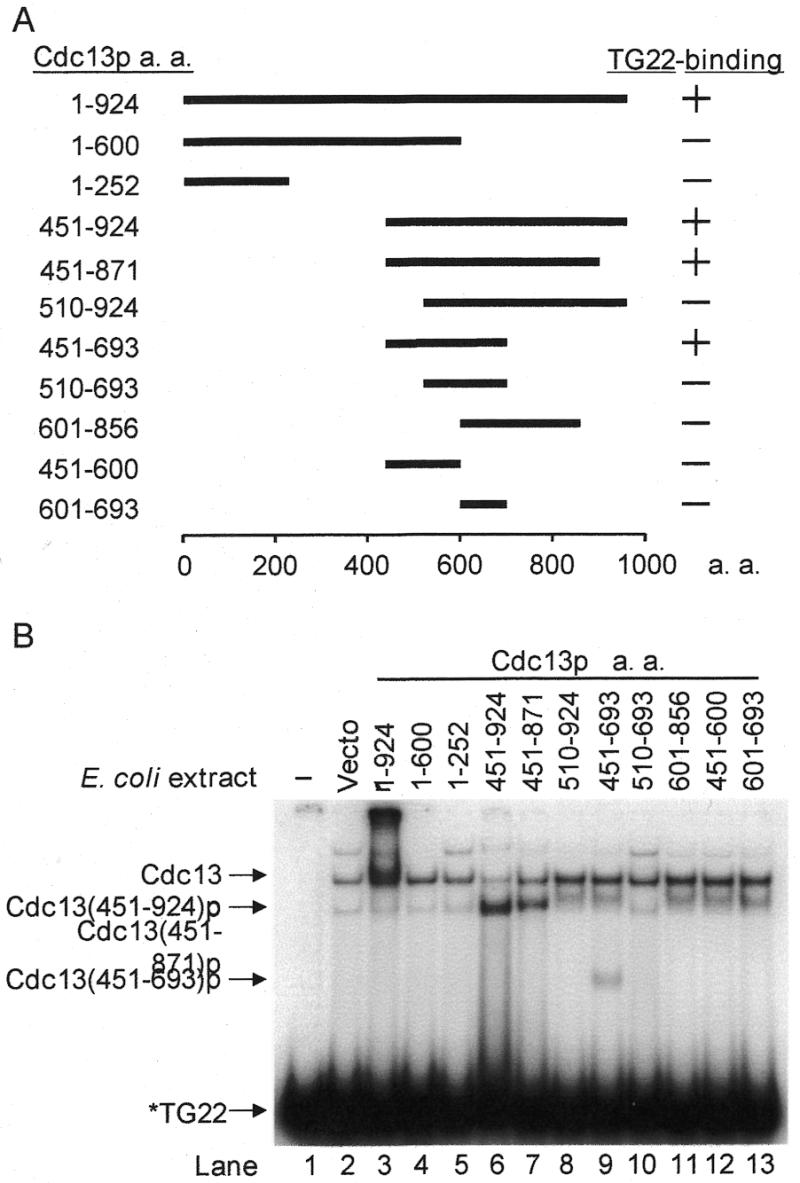
Single-stranded TG1–3-binding domain of Cdc13p. (A) Schematic representation of Cdc13p deletion mutants. The wild-type protein is 924 amino acids in length. The relative locations of the fragments (thick line) and the first as well as the last amino acids are indicated. All these mutants were fused in-frame to a GST protein on their N terminus. On the right are the single-stranded TG22-binding activities of mutants that were analyzed by EMSA. (B) Cdc13(451–693)p contains the single-stranded TG1–3-binding domain of Cdc13p. Escherichia coli extracts (15 µg) were mixed with 32P-labeled TG22 in 15-µl reaction mixtures. The reactions were incubated at room temperature for 10 min. A 3-µl volume of 80% glycerol was added to each reaction before analysis of the reaction products on an 8% polyacrylamide gel. Extracts carrying the GST fusion of Cdc13p deletion mutants are indicated. The first lane has no extracts. Migration of free DNA (TG22), Cdc13(451–693)p, Cdc13(451–871)p, Cdc13(451–924)p and Cdc13p are indicated.
Plasmid pET6H-CDC13(451–693), which was used to purify Cdc13(451–693)p, was constructed by inserting the NcoI–NruI fragment of pTHA-NLS-CDC(451–693) into NcoI/SmaI-digested pET6H (a gift from C.-H. Hu., National Marine University, Taiwan). The resulting plasmid was used to express 6× His-tagged Cdc13(451–693)p under the control of the T7 promoter.
To construct a plasmid for one-hybrid analysis, the DNA fragment encoding Cdc13(451–693)p was amplified by PCR using pTHA-CDC13 as the template and with primers ML11 (5′-CCGCTCGAGATCCTGTGGGACAATGAC-3′) and ML12 (5′-CCGCTCGAGATGAGAACCGTTTCT-3′). The 0.7-kb PCR product was subcloned into SmaI-digested pUC18 to generate pUC-CDC13(451–693). The 0.7-kb DNA fragment from XhoI-digested pUC-CDC13(451–693) was then ligated with the XhoI-digested pJG4-5 (39) or pRF4-6NL (34) to generate pJG-CDC13(451–693) or pRF-CDC13(451–693), respectively.
Since Cdc13p is a telomere-binding protein, the cellular localization of Cdc13p should be in the nucleus. To make certain that the truncated Cdc13p would be delivered into the nucleus, two oligonucleotides NLSw (5′-CTAGCCCCAAGAAGAAGCGGAAGGTCGC-3′) and NLSc (5′-CATGGCGACCTTCCGCTTCTTCTTTGGGG-3′) encoding the nuclear localization sequence (NLS) of SV40 large T antigen (39,40) were annealed and ligated with SpeI- and NcoI-digested pTHA-CDC13. The resulting plasmid, pTHA-NLS-CDC13, encoded a fusion protein with three HA-tag and NLS at the N-terminal of Cdc13p. Similarly, plasmid vector pTHA-NLS was constructed by inserting NLS sequences into pTHA (25). To construct plasmid pTHA-NLS-CDC13(451–924), a 1.6-kb DNA fragment encoding amino acids 451–924 of Cdc13p was PCR amplified with primers ML01 (5′-TGCCATGGGGATCCTGTGGGACAAT-3′) and CDC133 (5′-AACTGCAGACTAGTCGACTCTTGCTTCTTACC-3′) using Vent DNA polymerase (New England Biolabs). The PCR product was digested with NcoI and SalI, and was inserted into NcoI- and SalI-digested pTHA-NLS. To generate pTHA-NLS-CDC13(451–693), plasmid pTHA-NLS-CDC13(451–924) was digested with NruI and SalI, blunted by T4 DNA polymerase and self-ligated. Plasmid pTHA-NLS-CDC13(1–252) was constructed by ligating the NcoI–EcoRI fragment (0.8 kb) encoding the N-terminal 252-amino-acid polypeptide, of pAS-CDC13(1–252) to NcoI- and EcoRI-digested pTHA-NLS. A 2-kb NcoI–SalI fragment of pAS-CDC13(252–924) was ligated with NcoI- and SalI-digested pTHA-NLS to generate pTHA-NLS-CDC13(252–924). Expression of full-length and truncated CDC13 was confirmed by western blotting analysis on the extracts prepared from yeast cells with anti-HA antibody 12CA5 (data not shown).
Two-hybrid plasmids were constructed by subcloning STN1 into pACT2 and several fragments of CDC13 into pAS2 (37,41). Plasmid pACT-STN1 was constructed by ligating a NcoI–BamHI 1.6-kb fragment containing full-length STN1 from pJG-STN1 (unpublished construct) into pACT2 with the same sites. To construct full-length CDC13 into pAS2, the NcoI–SalI fragment (3 kb) of pTHA-CDC13 (25) was inserted into the NcoI- and SalI-digested pAS2. Plasmid carrying the temperature-sensitive allele of CDC13, cdc13-1, was constructed by replacing the 1.4-kb fragment of pAS-CDC13 with the equivalent of pTHA-CDC13-1 (25). Plasmid pAS-CDC13(252–924) was constructed by first ligating the 1.4-kb EcoRI–SalI CDC13 fragment of pTHA-CDC13 into EcoRI- and SalI-digested pAS2-1 followed by inserting a 0.8-kb EcoRI–EcoRI fragment of CDC13 isolated from pTHA-CDC13 with EcoRI digestion.
Preparation of E.coli extracts
To prepare protein extracts from E.coli, a 10-ml culture of E.coli containing a Cdc13p-deletion construct was grown in LB medium with 50 µg/ml ampicillin at 30°C to OD600 = 0.6. At this point, IPTG was added to a final concentration of 1 mM and the cells were allowed to grow at 30°C for an additional 16 h before harvesting by centrifugation. The pellets were resuspended in 0.5 ml buffer A [50 mM Tris–HCl pH 7.5, 1 mM EDTA, 1× protease inhibitor cocktail (Calbiochem) and 50 mM glucose] and were sonicated. Cell debris was removed by centrifugation in an Eppendorf microfuge for 10 min at 4°C, the supernatants were aliquoted and frozen in a dry ice-ethanol bath, and stored at –70°C. The concentration of protein in the E.coli extract was ∼2–6 mg/ml as determined using a Bio-Rad protein assay kit with bovine serum albumin as a standard.
Purification of 6× His-tagged Cdc13(451–693)p
To purify 6× His-tagged Cdc13(451–693)p, a 500-ml culture of IPTG-induced E.coli harboring pET6H-CDC13(451–693) was collected by centrifugation. Cells were resuspended in 30 ml of GdHCl buffer (6 M guanidine–HCl, 0.1 M NaH2PO4, 0.01 M Tris pH 8.0), followed by gentle shaking for 1 h at 4°C. The suspension was then centrifuged at 13 000 g for 15 min at 4°C. The clear supernatant was collected and applied to a 5 ml Ni-NTA–agarose column (Qiagen) previously equilibrated with GdHCl buffer. The column was washed stepwise with 20 ml of GdHCl buffer containing 1 mM imidazole followed by 20 ml of GdHCl buffer containing 20 mM imidazole. The bound protein was eluted by 12 ml of GdHCl buffer containing 200 mM imidazole. To renature the purified Cdc13(451–693)p, protein eluted from the Ni-NTA–agarose column was diluted to ∼50 µg/ml using GdHCl buffer and dialyzed against renaturation buffer (100 mM Tris pH 8.0, 2 mM EDTA, 2 mM DTT, 0.4 M l-arginine, 20% glycerol) at 4°C for 12 h.
Electrophoretic mobility shift assay (EMSA)
Oligonucleotide TG22 (5′-GTGGTGGGTGGGTGTGTGTGGG-3′), TG10 (5′-GGGTGTGGTG-3′), TG15 (5′-TGTGTGGGTGTGGTG-3′), TG20 (5′-TGGTGTGTGTGGGTGTGGTG-3′), TG25 (5′-GGGTGTGGTGTGTGTGGGTGTGGTG-3′), TG30 (5′-TGTGTGGGTGTGGTGTGTGTGGGTGTGGTG-3′) or TG35 (5′-TGTGGTGTGTGGGTGTGGTGTGTGTGGGTGTGGTG-3′) was first 5′ end-labeled with [γ-32P]ATP (3000 mCi/mM, NEN) using T4 polynucleotide kinase (New England Biolabs) and subsequently purified from a 10% sequencing gel after electrophoresis. To perform the assays, cell extracts were mixed with 2.0 ng of 32P-labeled TG22 or TG15 DNA with a total volume of 15 µl containing 50 mM Tris–HCl pH 7.5, 1 mM EDTA, 50 mM NaCl, 1 mM DTT and 1 µg of single stranded poly(dI–dC). The mixtures were allowed to incubate at room temperature for 10 min. Then, 3 µl of 80% glycerol was added and the mixtures were loaded on an 8% non-denaturing polyacrylamide gel, which was pre-run at 125 V for 10 min. Electrophoresis was carried out in TBE (89 mM Tris–borate/2 mM EDTA) at 125 V for 105 min. The gels were dried and autoradiographed. For competition analysis, 2.0 ng 32P-labeled TG15 was mixed with varying amounts of non-radioactive competitors before addition of the cell extracts. Binding activity was quantified with a PhosphorImager (Molecular Dynamics).
One-hybrid analysis
One-hybrid analysis was performed using the methods described by Bourns et al. (34). Plasmid pJG4-5, pJG-CDC13(451–693) or pRF-CDC13(451–693) was first transformed into yeast strains HIS-Tel and HIS-Int-CA, and selected on plates lacking tryptophan. To test for telomere-binding activity in vivo, cells were grown in liquid medium lacking tryptophan for ∼16 h. Then, cells were spotted in 10-fold serial dilutions on yeast synthetic complete medium (YC) plates lacking tryptophan, or lacking histidine, or with 10, 20 or 40 mM of 3-amino-1,2,4-triazole (3-AT, Sigma, St Louis, MO). Plates were incubated at 30°C until the colonies could be observed. Because HIS-Tel cells carrying the B42 transcription-activation domain fused with the DNA-binding region of Cdc13p did not grow well on plates with galactose, all the experiments were performed in plates containing 1.5% galactose and 1% glucose. The HIS-Int-CA strain grew better in this condition than in plates containing 3% galactose.
Complementation of cdc13Δ by cdc13 fragments using plasmid loss experiments
Plasmid loss experiments were carried out to test if binding of single-stranded TG1–3 and/or Stn1p-interacting activity of Cdc13p is sufficient to complement the lack of viability caused by the cdc13Δ mutation. Briefly, plasmid YEP24-CDC13 [abbreviation of plasmid YEP24-CDC13-161-4 (35)] was transformed into a diploid strain YJL401-UTAT [CDC13/cdc13Δ::HIS3 (25)] carrying one allele of the cdc13 null mutation. The transformants were sporulated and subjected to tetrad analysis. Haploid strain YJL501 was selected from the spores that were Ura+ (YEP24-CDC13), His+ (cdc13Δ::HIS3) and Ade+ (ADE2 near the telomere of chromosome V-R). Such a strain requires a plasmid carrying CDC13 (YEP24-CDC13) for its viability. Subsequently, plasmid pTHA-NLS, pTHA-NLS-CDC13, pTHA-NLS-CDC13(1–252), pTHA-NLS-CDC13(252–924), pTHA-NLS-CDC13(451–924) or pTHA-NLS-CDC13(451–693) was separately transformed into YJL501. The resulting transformants were spotted onto plates containing 0.5 mg/ml 5-fluoroorotic acid (5-FOA) and incubated at 30°C until colonies formed.
Two-hybrid analysis
Plasmids pACT2 and pACT-STN1 were transformed separately into yeast strain Y190. The resulting strains were then transformed with plasmids pAS2 or pAS2 containing CDC13 fragments. The HIS3 reporter system was also used to evaluate the interaction between Cdc13p and Stn1p. In the assays, 5–10 fresh transformed colonies from each transformation were mixed and spotted in 10-fold serial dilutions onto YC plates lacking histidine without or with 25 mM 3-AT. Plates were kept at 25 or 30°C until colonies formed.
RESULTS
Cdc13(451–693)p bound to single-stranded TG1–3 DNA in vitro
To identify the telomeric DNA-interacting region in Cdc13p, plasmids suitable to express GST fusions with various fragments of Cdc13p were constructed (Fig. 1A). Using 32P-labeled 22-base TG1–3 oligonucleotides (TG22) as substrate, the DNA-binding activity of these truncated Cdc13p was determined by EMSA. By comparing the gel-shift pattern with that of the vector alone, extra single-stranded TG1–3-binding activity was observed in E.coli extracts expressing Cdc13p fragments containing amino acids 451–924, 451–871 or 451–693 (Fig. 1B). Among these fusion proteins, Cdc13(451–693)p was the shortest single-stranded TG1–3 DNA-binding fragment of Cdc13p. Western blotting analysis using anti-GST antibody confirmed that these truncated Cdc13p polypeptides were indeed expressed, albeit degradation of the full-length and some of these truncated forms of Cdc13p was also observed (data not shown). Therefore, while it is not clear if the fragment can be shortened further, it was evident that the single-stranded TG1–3-binding activity of Cdc13p is located within the fragment comprising residues 451–693.
We also have expressed a 6× His-tagged-Cdc13(451–693)p in E.coli and purified this tagged protein using Ni-NTA–agarose (Fig. 2A). Using 32P-labeled single-stranded TG1–3 oligonucleotides as substrate, the DNA-binding ability of this recombinant polypeptide was determined by EMSA. The result shown in Figure 2B further demonstrated that this 6× His-tagged Cdc13(451–693)p is capable of forming complexes with the single-stranded TG1–3. Evidently, the single-stranded TG1–3 binding activity of Cdc13p was located within amino acids 451–693. Results shown in Figure 2B also indicate that the length of DNA substrate affected the electrophoretic mobility of the Cdc13(451–693)p–DNA complex. Interestingly, a second migration band was apparent on TG30 or TG35 but not on TG25; the identity of this second migration band is uncertain (Fig. 2B, lanes 19–24 and 25–30). Under our assay condition, Cdc13(451–693)p bound TG15 with an apparent binding constant of 120 nM.
Figure 2.
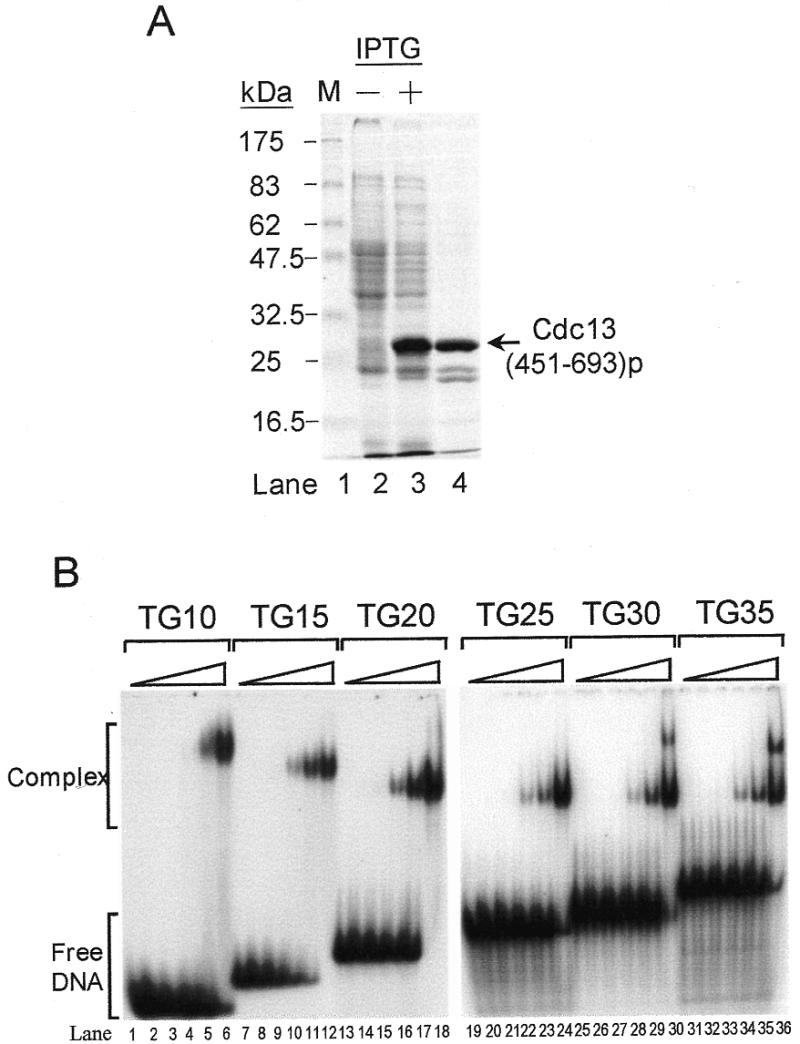
Purified Cdc13(451–693)p binds single-stranded TG1–3 telomeric DNA. (A) Purification of Cdc13(451–693)p. A 6× His-tagged Cdc13(451–693)p was purified from E.coli using a Ni-NTA–agarose column (see Materials and Methods). A Coomassie blue-stained 10% SDS–polyacrylamide gel is given. Lane 1 shows the molecular weight marker; lanes 2 and 3 were 50 µl of E.coli cultures harboring the pET6H-CDC13(451–693) plasmid grown without and with IPTG induction, respectively; lane 4 was 5 µg of purified Cdc13(451–693)p. (B) Cdc13(451–693)p contains the single-stranded TG1–3-binding domain of Cdc13p. Approximately 27 nM each of 32P-labeled TG10 (lanes 1–6), TG15 (lanes 7–12), TG20 (lanes 13–18), TG25 (lanes 19–24), TG30 (lanes 25–30) and TG35 (lanes 31–36) were mixed with several concentrations of the purified Cdc13(451–693)p and then gel shift assay was carried out. Cdc13(451–693)p used in each set of experiments were 0, 7, 22, 66, 200 and 600 nM. Autoradiograms are shown.
Cdc13(451–693)p specifically bound to single-stranded TG1–3 DNA
To evaluate the selectivity of the Cdc13(451–693)p binding activity, purified protein was mixed with 32P-labeled single-stranded TG1–3 and various amounts of unlabeled nucleic acid competitors before being subjected to EMSA analysis. As shown in Figure 3, unlabeled TG15 competed efficiently with 32P-labeled TG15. The binding was reduced by ∼50% when the competitor was presented at equal concentrations (Fig. 3A, lanes 3–5 and B). On the other hand, vertebrate (T2AG3), Oxytricha (T4G4) and Tetrahymena (T2G4) telomeric DNA did not compete for the binding activity of Cdc13(451–693)p to TG15 (Fig. 3A, lanes 6–14). Total yeast RNA, single-stranded C1–3A DNA or duplex TG1–3/ C1–3A DNA did not compete for Cdc13(451–693)p binding either (Fig. 3A, lanes 15–17, and data not shown). This result indicated that Cdc13(451–693)p bound specifically to single-strand TG1–3 telomeric DNA. With the exception of vertebrate telomeric DNA, which partially competed away the binding of Cdc13p to TG22 (25), Cdc13(451–693)p bound specifically to single-stranded TG1–3 telomeric DNA similarly to Cdc13p.
Figure 3.
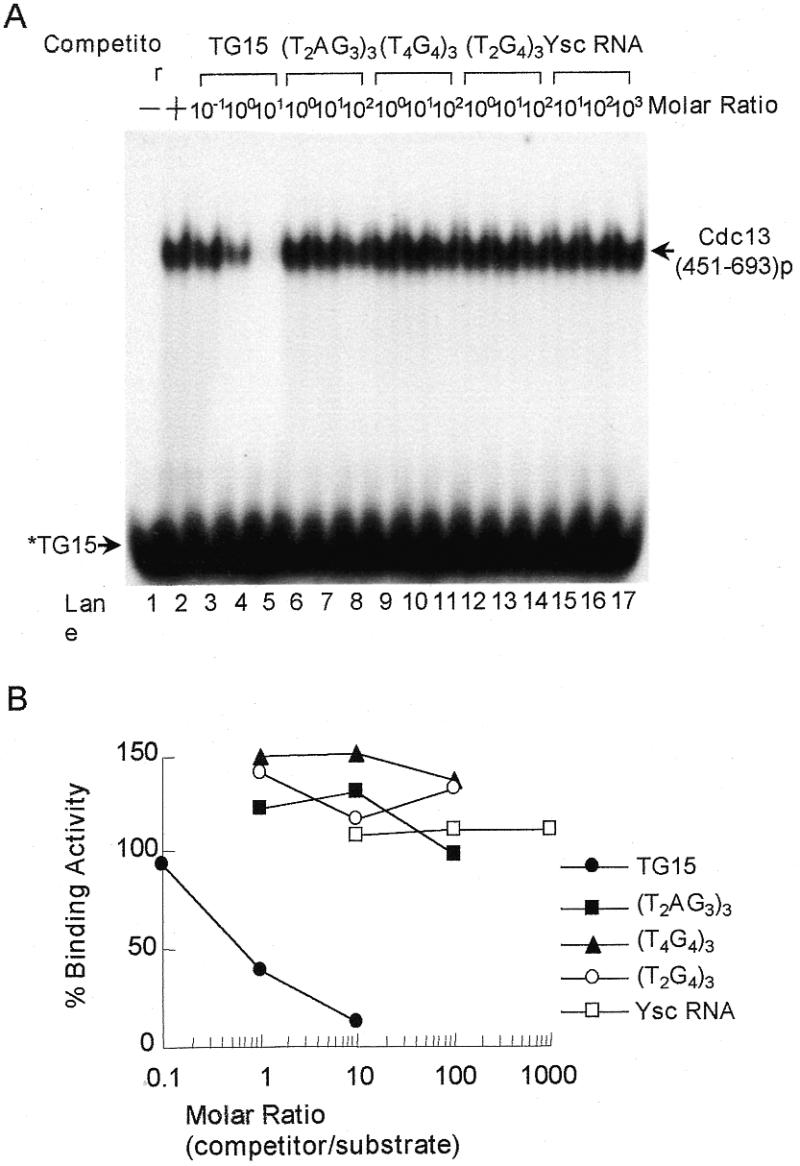
Competition analysis of Cdc13(451–693)p telomeric DNA-binding activity. (A) 32P-labeled TG15 (2 ng) was mixed with several concentrations of different competitors and then 0.2 µg of the purified Cdc13(451–693)p was added to the mixtures. Competitors were TG22 (yeast), (T2AG3)3 (vertebrate), (T4G4)3 (Oxytricha), (T2G4)3 (Tetrahymena) and total yeast RNA. Gel shift assay was then carried out. An autoradiogram is shown. (B) Quantification of the Cdc13(451–693)p-binding activity. The relative level of the binding activity was quantified by a PhosphorImager and the binding activity in the absence of competitor was taken as 100 [(A) lane 2]. The data show the average from three experiments. Symbols used are: TG15 (yeast, closed circles), (T2AG3)3 (vertebrate, closed squares), (T4G4)3 (Oxytricha, closed triangles), (T2G4)3 (Tetrahymena, open circles) and total yeast RNA (open squares), respectively.
Cdc13(451–693)p bound telomere in vivo
Previously, a one-hybrid system was developed to examine whether a protein interacts with telomeres in vivo (34). In that system, a promoter-defective allele of HIS3 is placed near the telomere of chromosome VII-L, HIS-Tel. The protein to be tested is fused to the E.coli B42 transcription-activation domain. When this fusion protein interacts with telomeres, it activates the expression of HIS3. Thus, expression of His3p can be used as a means to identify telomere-interacting protein. To verify whether Cdc13(451–693)p binds telomere in vivo, this fragment was fused with the B42 transcription-activation domain (Act-Cdc13-DB) and expressed in yeast HIS-Tel or HIS-Int-CA. The HIS-Int-CA strain carried internal HIS3 with C1–3A sequences at the 5′ region (34). Dilutions of cells were spotted onto plates without histidine to evaluate the expression of HIS3. HIS-Tel cells carrying plasmid vector alone (Act) or Cdc13(451–693)p without the B42 transcription-activation domain (Cdc13-DB) cannot grow on plates lacking histidine (Fig. 4, left panel). However, cells carrying the B42 transcription-activation domain fused with the DNA-binding region of Cdc13p (Act-Cdc13-DB) grew on plates lacking histidine (Fig. 4, left panel). The levels of cell growth with 3-AT for the HIS-Int-CA strain carrying plasmid vector alone (Act), Cdc13-DB or Act-Cdc13-DB were similar, indicating that Cdc13(451–693)p would not bind internal TG1–3/C1–3A duplex DNA (Fig. 4, right panel). Thus, Cdc13(451–693)p is sufficient to position itself to telomeres. Taken together, both in vitro and in vivo evidence indicate that the DNA-binding domain of Cdc13p was located in amino acids 451–693.
Figure 4.
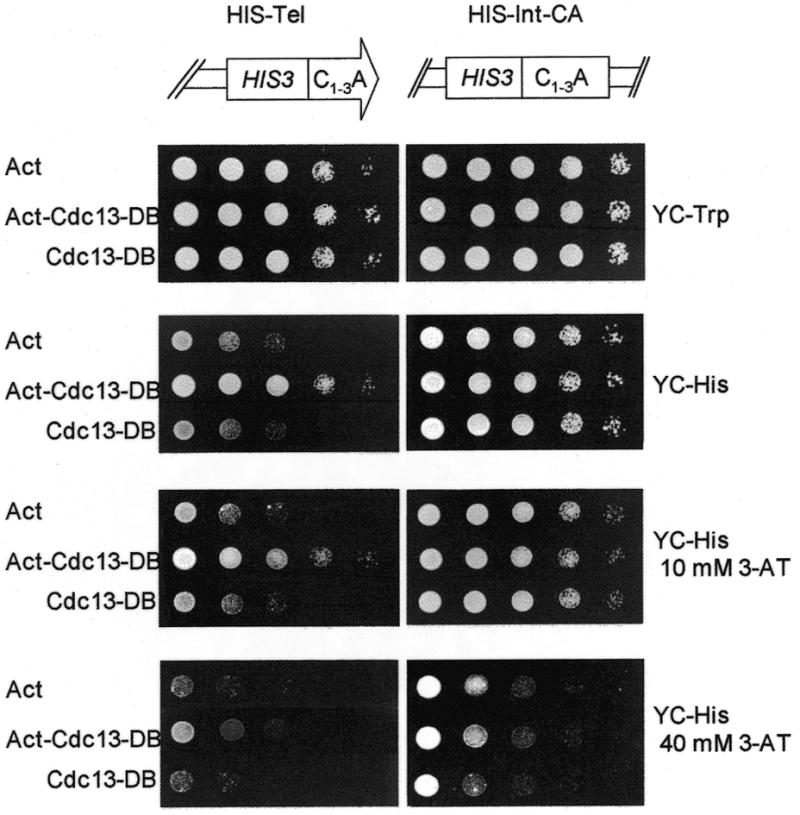
Cdc13(451–693)p binds telomere in vivo. HIS-Tel (left panel) or HIS-Int-CA (right panel) cells harboring pJG4-5 (Act), pJG-CDC13(451–693) (Act-CDC13-DB) or pRF-CDC13(451–693) (CDC13-DB) were spotted in 10-fold serial dilutions onto plates lacking tryptophan (YC-Trp), lacking histidine (YC-His), YC-His plates with 10 mM 3-AT and YC-His plates with 40 mM 3-AT. Photographs were taken after the plates were incubated at 30°C for 3 days.
Cdc13(451–693)p was not sufficient to complement cdc13 mutations
It has been known that Cdc13p is essential for cell viability. The next question that was considered in this study was whether binding of Cdc13p to telomere is sufficient to account for its essentiality. Here, plasmids expressing Cdc13p, Cdc13(1–252)p, Cdc13(252–924)p, Cdc13(451–924)p or Cdc13(451–693)p were constructed and transformed into yeast strain 2758-8-4b, a temperature-sensitive mutant of CDC13 (cdc13-1). Yeast strain 2758-8-4b (cdc13-1) grows normally at 25°C and arrests at G2/M phase of the cell cycle at 30°C. Full-length CDC13 complemented the temperature-sensitive phenotype of cdc13-1 at 30 or 37°C (Fig. 5A). Cells expressing Cdc13(252–924)p grew at all three temperatures tested. However, cells expressing other fragments of Cdc13p did not complement the temperature-sensitive phenotype of cdc13-1 at 30 or 37°C (Fig. 5A). Since Cdc13(451–693)p could not complement the growth arrest caused by the cdc13-1 mutation, telomere-binding activity alone was therefore not sufficient to account for the essentiality of Cdc13p.
Figure 5.
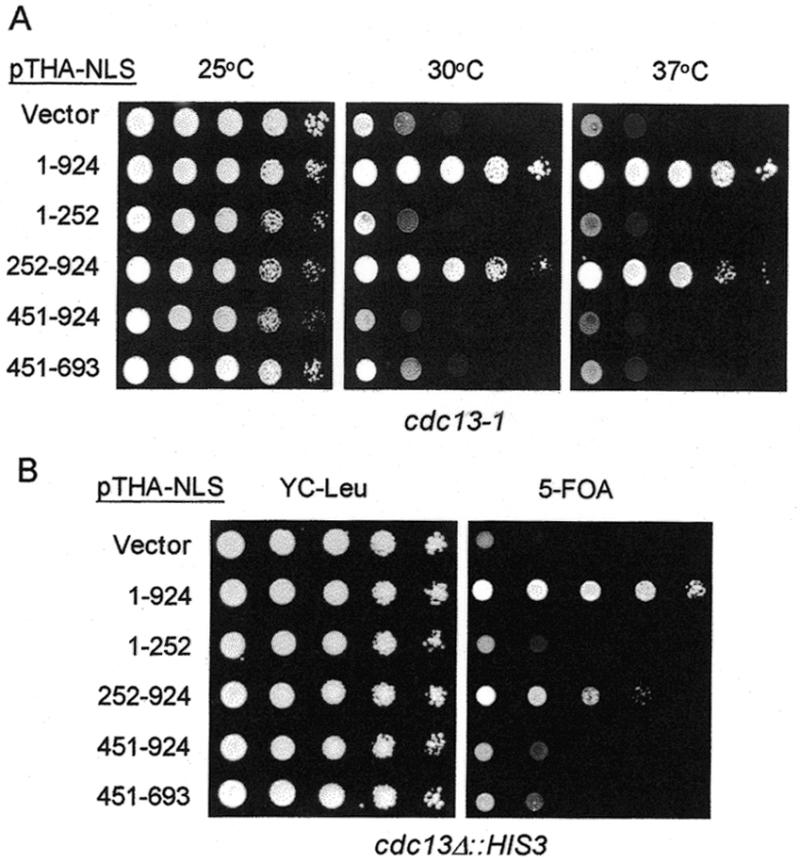
The telomere-binding domain of Cdc13p cannot rescue the growth defect phenotype of cdc13 mutants. (A) Yeast 2758-8-4b (cdc13-1) carrying plasmid pTHA-NLS (vector), pTHA-NLS-CDC13 (1–924), pTHA-NLS-CDC13(1–252), pTHA-NLS-CDC13(252–924), pTHA-NLS-CDC13(451–924) or pTHA-NLS-CDC13(451–693) were spotted in 10-fold serial dilutions on YC-Leu plates and grown at 25°C (left), 30°C (middle) or 37°C (right) until colonies formed. (B) Yeast strain YJL501 (cdc13Δ::HIS3/YEP24-CDC13) carrying plasmids pTHA-NLS (vector), pTHA-NLS-CDC13 (1–924), pTHA-NLS-CDC13(1–252), pTHA-NLS-CDC13(252–924), pTHA-NLS-CDC13(451–924) or pTHA-NLS-CDC13(451–693) were spotted in 10-fold serial dilutions on YC-Leu plates or plates containing 5-FOA and incubated at 30°C until colonies formed. Photographs of the plates are shown.
To test whether Cdc13(252–924)p complements the null allele of cdc13, a plasmid loss experiment was conducted. Here, the cdc13Δ::HIS3 strain YJL501 requires the CDC13-bearing plasmid (YEP24-CDC13, with URA3 marker) to grow. If a second plasmid introduced into the yeast expresses functional CDC13, YJL501 then no longer requires plasmid YEP24-CDC13 for viability. Growth on 5-FOA is used to monitor the loss of YEP24-CDC13. As shown in Figure 5B, 5-FOA-resistant cells were observed in YJL501 transformed with plasmid expressing Cdc13p. Similarly, 5-FOA-resistant cells were observed in YJL501 expressing Cdc13(252–924), although this rescue was ∼10- to 100-fold less efficient than that of Cdc13p. However, transformation of YJL501 with plasmid vector, pTHA-NLS, or plasmids expressing other fragments of CDC13 did not yield any 5-FOA-resistant cells (Fig. 5B). We have also analyzed the meiotic products from CDC13/cdc13Δ::HIS3 diploid cells harboring plasmid pTHA-NLS-CDC13 or pTHA-NLS-CDC13(252–924). Having analyzed ∼2000 spore products each from CDC13/cdc13Δ::HIS3 cells carrying plasmid pTHA-NLS-CDC13 (Leu2 marker) or pTHA-NLS-CDC13(252–924) (Leu2 marker), we obtained 465 and 69 His+ Leu+ haploid cells, respectively (data not shown). Thus, even though it remains to be tested whether this complementation was caused by overexpression of Cdc13(252–924)p, Cdc13(252–924)p was sufficient to complement cdc13-1 and cdc13Δ mutations.
Cdc13(252–924)p was capable of interacting with Stn1p
Cdc13p was shown to interact with Stn1p. To test if this interaction is required for the essential function of Cdc13p, the interaction between Cdc13(252–924)p and Stn1p was evaluated. Two-hybrid analysis was used previously to establish the interaction between Cdc13p and Stn1p (37). Here, we used the same approach to dissect the region within Cdc13p that interacts with Stn1p. Plasmids were constructed in which CDC13 or its fragments were fused to the DNA-binding domain of GAL4. These plasmids were transformed into yeast strain Y190 carrying a plasmid with STN1 fused to the activation domain of GAL4 (pACT-STN1) to analyze for their interaction. The ability to grow on medium lacking histidine was used as the criterion to evaluate the interaction between Stn1p and various truncated forms of Cdc13p. As shown in Figure 6, under our assay conditions, His+ colonies were apparent at 25 or 30°C in Y190 harboring plasmids pAS-CDC13 and pACT-STN1. This result was consistent with the previous report that Cdc13p interacts with Stn1p (37). His+ colonies were also apparent at 25 or 30°C in Y190 harboring plasmids pAS-CDC13(252–924) and pACT-STN1 indicating that Cdc13(252–924)p was capable of interacting with Stn1p. We also examined if Cdc13-1p, with Pro371 being replaced by Ser (24,25), might interact with Stn1p. Interestingly, the HIS3 expression level in Y190/pACT-STN1 carrying pAS-CDC13-1 cells was comparable to those expressing either Cdc13p or Cdc13(252–924)p at 25°C (Fig. 6, top panel). However, the HIS3 expression level of cells carrying Cdc13-1p was relatively low at 30°C (Fig. 6, bottom panel). Similarly, using β-galactosidase as the reporter system, the color-forming ability of Cdc13-1p was reproducibly reduced, as compared with that of Cdc13p at 30 or 37°C (data not shown). These results indicated that Cdc13(252–924)p was capable of interacting with Stn1p. Moreover, the interaction of Stn1p with Cdc13-1p appeared reduced at higher temperature, suggesting that the temperature-sensitive lethality phenotype of cdc13-1 might be due to a decrease in interaction with Stn1p.
Figure 6.
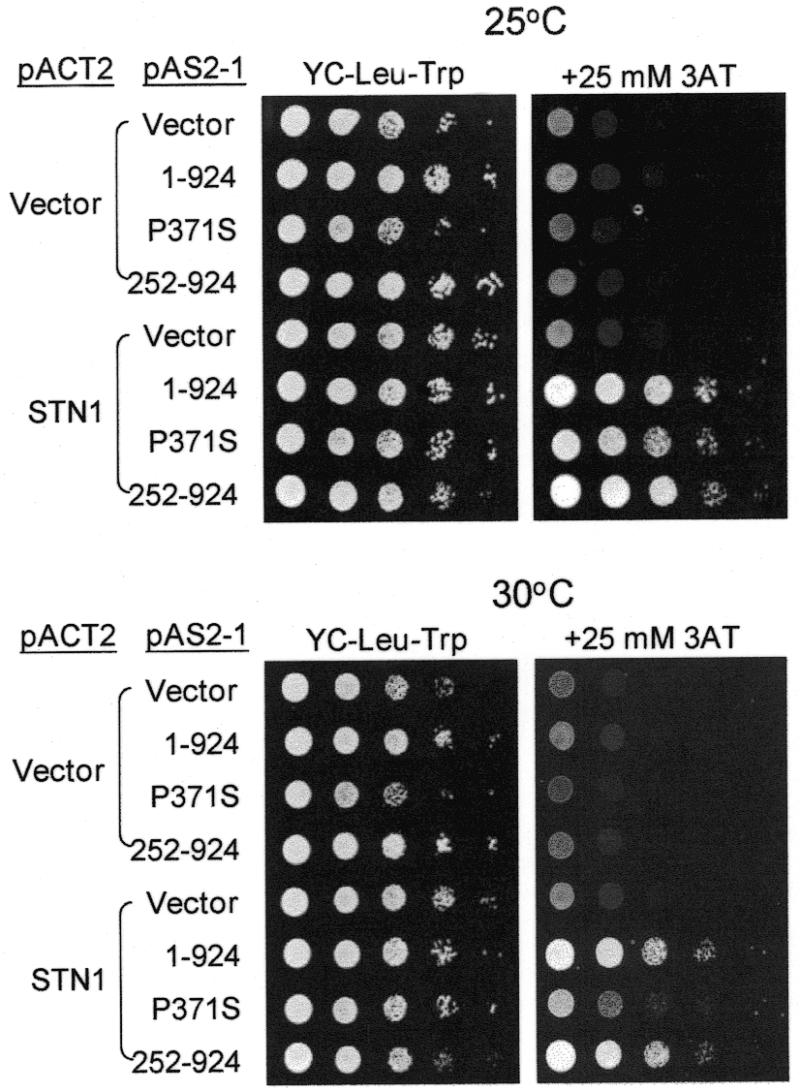
Cdc13(251–924)p interacts with Stn1p. Yeast cells Y190/pACT2 or Y190/pACT-STN1 carrying plasmid pAS2 (vector), pAS-CDC13 (1–924), pAS-CDC13-1 (P371S) or pAS-CDC13(252–924) (252–924) were grown on YC medium without leucine and tryptophan for 16 h at 30°C. Ten-fold serial dilutions of yeast cells were spotted on plates without leucine and tryptophan (YC-Leu-Trp), or without leucine, tryptophan and histidine with the addition of 25 mM of 3-AT (YC-Leu-Trp-His+3-AT), and incubated at 25°C (top panel) or 30°C (bottom panel) until colonies formed. The photographs of the plates are shown.
To address the possibility that reduced interaction between Cdc13-1p and Stn1p was due to the reduced stability of Cdc13-1p, the Cdc13-1p level at non-permissive temperature was evaluated. Here, we applied an immunoblotting assay using polyclonal antibodies raised against Cdc13(1–252)p (T.-L.Pang and J.-J.Lin, unpublished data) to evaluate the cellular level of Cdc13p. As shown in Figure 7, under the condition that >90% of the cells had arrested at G2/M phase, the Cdc13-1p level at the non-permissive temperature (30°C) was similar to the level of Cdc13-1p at the permissive temperature (25°C). Our results suggested that reduced interaction between Cdc13-1p and Stn1p was not due to the reduced stability of Cdc13-1p.
Figure 7.
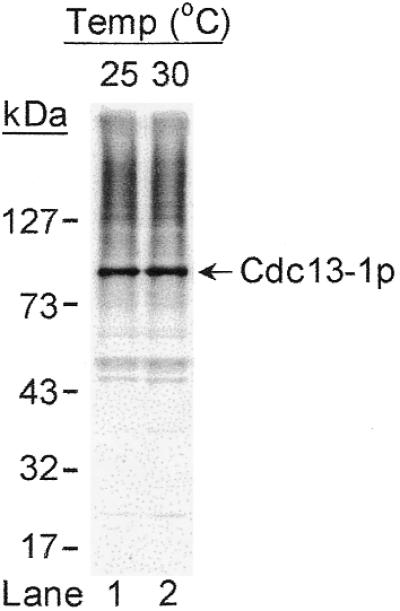
Western blotting analysis of Cdc13-1p. Strain 2758-8-4b (cdc13-1) was grown at permissive temperature (25°C) and shifted to non-permissive temperature (30°C) for 2 h. Total cell extracts prepared from these cells were separated by 10% SDS–PAGE and subjected to immunoblotting analysis using polyclonal antibodies raised against Cdc13(1–252)p. Bound antibodies were visualized by chemiluminescence using an ECL kit (Amersham-Pharmacia). Molecular markers are indicated on the left.
DISCUSSION
Cdc13p binds specifically to the single-stranded TG1–3 tail of yeast telomere. Here, we have delineated the regions of Cdc13p responsible for this interaction. The single-stranded TG1–3-binding domain of Cdc13p is within amino acids 451–693, and binding of the single-stranded TG1–3 is specific. However, binding to telomeres by Cdc13p is not sufficient to account for the essential function of Cdc13p. Judging from the results the C-terminal 673-amino-acid polypeptide was sufficient to complement the growth defect phenotype of several cdc13 mutants and interaction with Stn1p. Our results indicated that the Stn1p-interaction function and the telomere-binding activity of Cdc13p were essential for cell growth.
Upon proteolytic degradation of the Cdc13p–DNA complex, a fragment of Cdc13p that covers amino acids 557 to ∼690 was shown to associate with single-stranded TG1–3 DNA (42). In this report, our strategy to identify the DNA-binding region of Cdc13p was the subcloning of restriction enzyme-digested CDC13 fragments followed by evaluation of the single-stranded TG1–3-binding activities of these expressed CDC13 fragments. Using this approach, the smallest fragment that still contained the single-stranded TG1–3-binding activities of Cdc13p was within amino acids 451–693 (Fig. 1). Smaller fragments such as Cdc13(510–693), Cdc13(451–600) or Cdc13(601–693) did not show detectable single-stranded TG1–3 binding activity. It is unclear whether Cdc13(510–693)p, which covers amino acids 557 to ∼690, did not interact with single-stranded TG1–3 DNA. Nevertheless, our identification of Cdc13(451–693)p as a stably expressed telomeric-binding domain of Cdc13p provides useful information for future understanding of how Cdc13p binds and modulates telomere function.
The identity of the second migration bands using TG30 or TG35 as DNA substrate is unclear (Fig. 2). The simplest explanation for the appearance of this second migration band would be that TG30 or TG35 provides enough space for two molecules of Cdc13(451–693)p to bind. This result would also imply that the optimal binding site of Cdc13(451–693)p on DNA was ∼13–15 bases, an estimation that is in reasonable agreement with the results of using a 34-kDa Cdc13 DBD by Huges et al. (42). However, this explanation is complicated by observations from several reports that telomeric DNA-binding proteins can also promote the formation of a G-quartet structure (43,44). It will be interesting to know whether Cdc13p promotes the formation of a G-quartet structure upon binding.
Contrary to the ciliate telomere-binding proteins, both Cdc13p and Cdc13(451–693)p preferred low salt for binding (data not shown). Sequence analysis of this 243-amino-acid region did not provide information on which residues within this region are responsible for interacting with telomeres. Evidence was presented that single-stranded DNA-binding proteins interact with DNA via hydrophobic interactions between aromatic side chains of the protein and the DNA bases (45–47). For example, the single-stranded DNA-binding protein T4 gp32 utilizes residues Tyr84, 99, 106, 115, 137, 186 and Phe138 to form a hydrophobic pocket to interact with the bases of single-stranded DNA (46). In addition, the Oxytricha α and β complex uses a series of aromatic amino acids including Tyr130α, Tyr293α, Tyr134β, His292α, Phe109β and Tyr239α to interact with the extended bases of single-stranded T4G4 DNA (47). It is likely that aromatic residues within the 243 amino acids of Cdc13p play a role in the interaction with telomeres. Interestingly, this region indeed has a high content of aromatic residues. However, it remains to be determined which amino acid residues are responsible for this protein–DNA interaction.
Using the one-hybrid system, Bourns et al. (34) showed previously that the N-terminal 251 amino acids of Cdc13p interacted with telomere, but that amino acids 252–508 or 509–924 of Cdc13p failed to do so. Since the N-terminal 251 amino acids were sufficient to target Cdc13p to telomere, it was concluded that the telomere-binding domain was within this region of Cdc13p. In contrast to their study, we showed that the telomere-binding domain was mapped to within the 451–693 amino acids of Cdc13p both in vitro and in vivo (Figs 1 and 4). Given that the N-terminal 1–251 amino acids of Cdc13p did not bind to single-stranded TG1–3 DNA in vitro (Fig. 1), it would be interesting to know how this region of Cdc13p interacts with telomeres in vivo. Clearly, in vivo one-hybrid system detected not only proteins that interact directly with telomeric DNA but also those that interact indirectly (34). For example, Sir and Rif proteins interact with telomeres by their ability to bind Rap1p (48–50). One possible explanation is that this N-terminal 251-amino-acid fragment interacts indirectly with the telomere. Conceivably, protein factors that associated with telomeres including Rap1p, Rif proteins and Sir proteins, are potential candidates for recruiting the N-terminal 251-amino-acid polypeptide of Cdc13p to telomeres.
A Pro371 to Ser substitution caused the phenotype of Cdc13-1p (24,25). On the basis of gel mobility-shift assay, the cdc13-1 mutation did not affect the telomere-binding activity of Cdc13p [J.-J.Lin and V.A.Zakian, unpublished result (24,42)]. In our two-hybrid system, the interaction of Cdc13-1p with Stn1p was temperature dependent and the interaction was reduced at the non-permissive temperature (Fig. 6). This result indicated that interaction between Cdc13p and Stn1p is essential for cell survival. While we cannot rule out that another uncharacterized change in Cdc13-1p is responsible for the cell cycle arrest at the non-permissive temperature, relatively weak interaction between Cdc13-1p and Stn1p might indeed cause the accumulation of single-stranded G-rich DNA near telomeres (35) leading to this phenotype.
Acknowledgments
ACKNOWLEDGEMENTS
We thank members of J.-J.L.’s laboratory for their help. We also thank Drs W. J. Lin and C. Wang for critical reading of the manuscript. This work was supported by NSC grants 87-2314-B-010-004, 88-2314-010-070 and in part by grant 89-B-FA22-2-4 (Program for Promoting Academic Excellence of Universities).
REFERENCES
- 1.Zakian V.A. (1995) Science, 270, 1601–1607. [DOI] [PubMed] [Google Scholar]
- 2.Blackburn E.H. (1990) J. Biol. Chem., 265, 5919–5921. [PubMed] [Google Scholar]
- 3.Gottschling D.E., Aparicio,O.M., Billington,B.L. and Zakian,V.A. (1990) Cell, 63, 751–762. [DOI] [PubMed] [Google Scholar]
- 4.Klein F., Laroche,T., Cardenas,M.E., Hofmann,J.F.X., Schweizer,D. and Gasser,S.M. (1992) J. Cell Biol., 117, 935–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henderson E.R. and Blackburn,E.H. (1989) Mol. Cell. Biol., 9, 345–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klobutcher L.A., Swanton,M.T., Donini,P. and Prescott,D.M. (1981) Proc. Natl Acad. Sci. USA, 78, 3015–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pluta A.F., Kaine,B.P. and Spear,B.B. (1982) Nucleic Acids Res., 10, 8145–8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wellinger R.J., Wolf,A.J. and Zakian,V.A. (1993) Cell, 72, 51–60. [DOI] [PubMed] [Google Scholar]
- 9.Wellinger R.J., Ethier,K., Labrecque,P. and Zakian,V.A. (1996) Cell, 85, 423–433. [DOI] [PubMed] [Google Scholar]
- 10.Shore D. (1997) Trends Biochem. Sci., 22, 233–235. [DOI] [PubMed] [Google Scholar]
- 11.Conrad M.N., Wright,J.H., Wolf,A.J. and Zakian,V.A. (1990) Cell, 63, 739–750. [DOI] [PubMed] [Google Scholar]
- 12.Cooper J.P., Nimmo,E.R., Allshire,R.C. and Cech,T.R. (1997) Nature, 385, 744–747. [DOI] [PubMed] [Google Scholar]
- 13.Bilaud T., Brun,C., Ancelin,K., Koering,C.E., Laroche,T. and Gilson,E. (1997) Nature Genet., 17, 236–239. [DOI] [PubMed] [Google Scholar]
- 14.Broccoli D., Smogorzewska,A., Chong,L. and de Lange,T. (1997) Nature Genet., 17, 231–235. [DOI] [PubMed] [Google Scholar]
- 15.Chong L., van Steensel,B., Broccoli,D., Erdjument-Bromage,H., Hanish,J., Tempst,P. and de Lange,T. (1995) Science, 270, 1663–1667. [DOI] [PubMed] [Google Scholar]
- 16.van Steensel B., Smogorzewska,A. and de Lange,T. (1998) Cell, 92, 401–413. [DOI] [PubMed] [Google Scholar]
- 17.Wright J.H. and Zakian,V.A. (1995) Nucleic Acids Res., 23, 1454–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wright J.H., Gottschling,D.E. and Zakian,V.A. (1992) Genes Dev., 6, 197–210. [DOI] [PubMed] [Google Scholar]
- 19.Kyrion G., Boakye,K.A. and Lustig,A.J. (1992) Mol. Cell. Biol., 12, 5159–5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lustig A.J., Kurtz,S. and Shore,D. (1990) Science, 250, 549–553. [DOI] [PubMed] [Google Scholar]
- 21.Stavenhagen J.B. and Zakian,V.A. (1998) Genes Dev., 12, 3044–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin J.-J. (1993) Bioessays, 15, 555–557. [DOI] [PubMed] [Google Scholar]
- 23.Lin J.-J. and Zakian,V.A. (1994) Nucleic Acids Res., 22, 4906–4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nugent C.I., Hughes,T.R., Lue,N.F. and Lundblad,V. (1996) Science, 274, 249–252. [DOI] [PubMed] [Google Scholar]
- 25.Lin J.-J. and Zakian,V.A. (1996) Proc. Natl Acad. Sci. USA, 93, 13760–13765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petracek M.E., Konkel,L.M.C., Kable,M.L. and Berman,J. (1998) EMBO J., 13, 3648–3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McKay S.J. and Cooke,H. (1992) Nucleic Acids Res., 20, 6461–6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Konkel L.M.C., Enomoto,S., Chamberlain,E.M., McCune-Zierath,P., Iyadurai,S.J.P. and Berman,J. (1995) Proc. Natl Acad. Sci. USA, 92, 5558–5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishikawa F., Matunis,M.J., Dreyfuss,G. and Cech,T.R. (1993) Mol. Cell. Biol., 13, 4301–4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Virta-Pearlman V., Morris,D.K. and Lundblad,V. (1996) Genes Dev., 10, 3094–3104. [DOI] [PubMed] [Google Scholar]
- 31.Fang G., Gray,J.T. and Cech,T.R. (1993) Genes Dev., 7, 870–882. [DOI] [PubMed] [Google Scholar]
- 32.Fang G. and Cech,T.R. (1993) Proc. Natl Acad. Sci. USA, 90, 6056–6060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gottschling D.E. and Zakian,V.A. (1986) Cell, 47, 195–205. [DOI] [PubMed] [Google Scholar]
- 34.Bourns B.D., Alexander,M.K., Smith,A.M. and Zakian,V.A. (1998) Mol. Cell. Biol., 18, 5600–5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garvik B., Carson,M. and Hartwell,L. (1995) Mol. Cell. Biol., 15, 6128–6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weinert T.A. and Hartwell,L.H. (1988) Science, 241, 317–322. [DOI] [PubMed] [Google Scholar]
- 37.Grandin N., Reed,S.I. and Charbonneau,M. (1997) Genes Dev., 11, 512–527. [DOI] [PubMed] [Google Scholar]
- 38.Sikorski R.S. and Hieter,P. (1989) Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gyuris J., Golemis,E., Chertkov,H. and Brent,R. (1993) Cell, 75, 791–803. [DOI] [PubMed] [Google Scholar]
- 40.Chevray P.M. and Nathans,D. (1992) Proc. Natl Acad. Sci. USA, 89, 5789–5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harper J.W., Adami,G.R., Wei,N., Keyomarsi,K. and Elledge,S.J. (1993) Cell, 75, 805–816. [DOI] [PubMed] [Google Scholar]
- 42.Hughes T.R., Weilbaecher,R.G., Walterscheid,M. and Lundblad,V. (2000) Proc. Natl Acad. Sci. USA, 97, 6457–6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fang G. and Cech,T.R. (1993) Cell, 74, 875–885. [DOI] [PubMed] [Google Scholar]
- 44.Giraldo R. and Rhodes,D. (1998) EMBO J., 13, 2411–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Venkataram Prasad B.V. and Chiu,W. (1987) J. Mol. Biol., 193, 579–584. [DOI] [PubMed] [Google Scholar]
- 46.Shamoo Y., Friedman,A.M., Parsons,M.R., Konigsberg,W.H. and Steitz,T.A. (1995) Nature, 376, 362–366. [DOI] [PubMed] [Google Scholar]
- 47.Horvath M.P., Schweiker,V.L., Bevilacqua,J.M., Ruggles,J.A. and Schultz,S.C. (1998) Cell, 95, 963–974. [DOI] [PubMed] [Google Scholar]
- 48.Moretti P., Freeman,K., Coodly,L. and Shore,D. (1994) Genes Dev., 8, 2257–2269. [DOI] [PubMed] [Google Scholar]
- 49.Hardy C.F., Sussel,L. and Shore,D. (1992) Genes Dev., 6, 801–814. [DOI] [PubMed] [Google Scholar]
- 50.Wotton D. and Shore,D. (1997) Genes Dev., 11, 748–760. [DOI] [PubMed] [Google Scholar]


