Abstract
Pseudomonas aeruginosa colonizes the pulmonary tissue of patients with cystic fibrosis (CF), leading to biofilm-associated infections. The pulmonary fluid of CF patients usually contains elevated concentrations of cations and may contain the P. aeruginosa redox-active pigment pyocyanin, which is known to disrupt calcium homeostasis of host cells. Since divalent cations are important bridging ions for bacterial polysaccharides and since they may play regulatory roles in bacterial gene expression, we investigated the effect of calcium ions on the extracellular matrix constituents of P. aeruginosa biofilms. For mucoid strain P. aeruginosa FRD1, calcium addition (1.0 and 10 mM as CaCl2) resulted in biofilms that were at least 10-fold thicker than biofilms without added calcium. Scanning confocal laser microscopy showed increased spacing between cells for the thick biofilms, and Fourier transform infrared spectroscopy revealed that the material between cells is primarily alginate. An algD transcriptional reporter demonstrated that calcium addition caused an eightfold increase in alg gene expression in FRD1 biofilms. Calcium addition also resulted in increased amounts of three extracellular proteases (AprA, LasB, and PrpL). Immunoblots of the biofilm extracellular material established that AprA was harbored within the biofilm extracellular matrix. An aprA deletion mutation and a mutation in gene for a putative P. aeruginosa calmodulin-like protein did not significantly affect calcium-induced biofilm structure. Two-dimensional gel electrophoresis showed increased amounts of phenazine biosynthetic proteins in FRD1 biofilms and in calcium-amended planktonic cultures. Spectrochemical analyses showed that the calcium addition causes a three- to fivefold increase in pyocyanin production. These results demonstrate that calcium addition affects the structure and extracellular matrix composition of mucoid P. aeruginosa biofilms, through increased expression and stability of bacterial extracellular products. The calcium-induced extracellular matrix of mucoid P. aeruginosa consists primarily of the virulence factor alginate and also harbors extracellular proteases and perhaps pyocyanin, a biomolecule that may further disrupt cellular calcium levels.
Patients with cystic fibrosis (CF) are highly susceptible to bacterial pulmonary infections, particularly chronic infections with Pseudomonas aeruginosa (reviewed in reference 38). P. aeruginosa pulmonary infections are considered biofilm-associated diseases, where the bacteria colonize the surface of pulmonary tissue (36), are often encapsulated in extracellular polysaccharide matrix material (25, 36, 48), secrete quorum-sensing signaling compounds (58), and have increased resistance to antibiotics (reviewed in reference 59) and to host defenses (2, 30, 45). Mutations in the cystic fibrosis transmembrane regulator of CF patients lead to physiological alteration of pulmonary fluid, with the most characteristic manifestation being the increased concentrations of thick pulmonary mucus that includes DNA and actin from lysed neutrophils (38). Disruption of the CF transmembrane regulator also results in secretions with elevated concentrations of sodium and other cations. In addition to these cellular manifestations, extracellular products from infectious bacteria may contribute to the pulmonary abnormalities. The extracellular redox cycling compound produced by P. aeruginosa, pyocyanin, has been shown to disrupt cytosolic calcium concentrations and calcium homeostasis of epithelial cells (8). Pyocyanin has been detected in the pulmonary fluid of patients infected with P. aeruginosa (66). Therefore, this disruption may occur in vivo.
Acute P. aeruginosa infections are usually caused by nonmucoid strains, which do not produce the capsular polysaccharide alginate. Mucoid P. aeruginosa strains typically produce alginate, a high-molecular-weight polymer of O-acetylated d-mannuronate linked by β 1-4 glycosidic bonds to its C5 epimer, l-guluronate (14, 20). Once P. aeruginosa strains begin to produce this polymer, pulmonary infections may become chronic and patient prognosis declines. The switch from the nonmucoid to the mucoid phenotype is complex, involving a hierarchy of regulatory proteins (71), including the alternative sigma factor σe (11, 39, 74), whose activity is repressed by the antisigma factors MucA and MucB (21, 56). Mutations in mucA are often responsible for the switch to the mucoid phenotype in clinical isolates (5). Antibiotics as well as components of host defensive processes may contribute to mucA mutations and to the accompanying phenotypic switch from non-alginate-producing strains and acute infections to alginate-producing strains and chronic infections (42, 53).
Extracellular polymeric substances form the structural material that holds the bacteria within the three-dimensional matrices of biofilms. There are seemingly conflicting reports over the composition of the bacterial extracellular matrix material for P. aeruginosa biofilms. These reports may reflect the differences in strains used in different studies. With mucoid P. aeruginosa FRD1 strains, alginate appears to be the primary constituent of the biofilm extracellular matrix material (48). Studies with this CF isolate and its mutant derivatives showed that alginate affects the three-dimensional architecture of P. aeruginosa FRD1 biofilms. Fourier transform infrared spectroscopy (FTIR) demonstrated that alginate was associated with biofilms of this mucoid strain. Similar results were obtained using a mutant derivative of the nonmucoid strain P. aeruginosa PAO1 engineered to overproduce alginate (25, 60). On the other hand, results using wild-type P. aeruginosa PAO1 and PA14 showed that alginate was not associated with biofilms (72) and that a second polysaccharide likely encoded by a separate polysaccharide biosynthetic gene cluster may provide the extracellular matrix material for those cells (18, 19, 29). In another report, using P. aeruginosa PAO1, DNA was suggested to be the extracellular matrix material, since the addition of DNase to biofilms dispersed the bacteria (64). Many bacterial extracellular polysaccharides, including alginate, contain negatively charged uronic acid residues. These sugar subunits bind calcium ions by ionic interactions, allowing cross-bridging and gelling of the extracellular polysaccharides (28). In the case of alginate produced by algae and the bacterium Azotobacter vinelandii, blocks of l-guluronate residues bind calcium to alginate (12). Although P. aeruginosa does not produce l-guluronate blocks, calcium apparently binds the uronic acid residues and affects the polymer's physical properties (59).
A recent study characterizing biofilms of the marine bacterium Vibrio cholerae implicated calcium ions in O-antigen polysaccharide binding and biofilm maintenance (32). When this bacterium undergoes a switch from higher calcium ion concentrations, such as those found in seawater, to a lower concentration, the biofilms disperse, suggesting that the loss of a calcium ion from an O-antigen polysaccharide may result in disruption of the biofilms and release of individual cells. This destabilizing effect may be specific for calcium, since it was not observed with the addition of magnesium. In addition to their role in biofilm matrix stability, calcium ions may affect bacterial gene expression. In eukaryotic cells, calcium is an important signaling molecule, and it may play a regulatory role in bacteria (27, 49). Many bacteria contain genes for calmodulin-like proteins with characteristic EF-hand motifs (46), including P. aeruginosa PA4107 and possibly plcR (6), which has been shown to bind calcium (61). In P. aeruginosa, secretion and/or stability of certain extracellular proteins is affected by calcium ions. Toxins secreted by the type III secretion system are repressed by calcium (75). The amounts of extracellular elastase (LasB) and LasA, secreted by the type II secretory pathway, increase in the presence of added calcium (52).
Since the calcium ion is known to affect biofilm architecture, we investigated its role in the structure and extracellular matrix constituents of P. aeruginosa biofilms. Surprisingly, we found that added calcium resulted in biofilms of mucoid P. aeruginosa FRD1 that were 10- to 20-fold thicker than biofilms without added calcium. We used this phenomenon to characterize alginate amounts and expression of alg biosynthetic genes within these biofilms and to characterize proteins within the extracellular matrix material. Amounts of extracellular proteases increased in Ca2+-amended biofilms, and the proteases were found to be harbored within the Ca2+-augmented alginate matrix. Amounts of biosynthetic proteins for the extracellular virulence factor pyocyanin also increased with added calcium. The results indicate that, in mucoid strains of P. aeruginosa, alginate is the primary extracellular matrix material of the biofilms and that this matrix material can act as a reservoir for other extracellular virulence factors.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Pseudomonas aeruginosa FRD1 and PAO1 were used in this study. P. aeruginosa FRD1 is an alginate-overproducing (mucoid) CF pulmonary isolate (51), and P. aeruginosa PAO1 is a nonmucoid non-CF isolate. The PA4107 and PA1249 genes were deleted from the chromosomes of both FRD1 and PAO1, by using allelic exchange, resulting in replacement of each gene with a gentamicin omega fragment. PA4107 encodes a putative calmodulin-like protein containing predicted Ca2+ binding EF-hand motifs, and the strains produced by this deletion were labeled P. aeruginosa PAO1043 ΔcalA::aacCIΩ and FRD1043 ΔcalA::aacCIW. PA1249 (aprA) encodes alkaline metalloprotease, and its deletions were labeled PAO1252 ΔaprA::aacCIΩ and FRD1252 ΔaprA::aacCIΩ. P. aeruginosa FRD875 algD>xylE (69) was generously provided by Daniel Wozniak (Wake Forest University). For scanning confocal laser microscopy (SCLM) the strains expressed the gfpmut2 from a constitutive promoter from plasmid pMF230 (48).
Escherichia coli and P. aeruginosa were routinely cultured in L broth (10 g tryptone, 5 g yeast extract, 5 g NaCl, per liter). Pseudomonas isolation agar (Difco) was used to select for P. aeruginosa following matings with E. coli. Antibiotics when used were at the indicated concentrations (per milliliter): ampicillin at 100 μg, carbenicillin at 300 μg, and gentamicin at 15 μg for E. coli and 100 μg for P. aeruginosa. General DNA manipulations were performed as described previously (1). Restriction endonucleases were purchased from New England Biolabs. Oligonucleotide primers were synthesized by IDTDNA Corp.
Biofilm minimal medium (BMM) contained (per liter) 9.0 mM sodium glutamate, 50 mM glycerol, 0.02 mM MgSO4, 0.15 mM NaH2PO4, 0.34 mM K2HPO4, 145 mM NaCl, 20 μl trace metal solution, and 1 ml vitamin solution. Trace metal solution (per liter of 0.83 M HCl) contained 5.0 g CuSO4 · 5H2O, 5.0 g ZnSO4 · 7H2O, 5.0 g FeSO4 · 7H2O, and 2.0 g MnCl2 · 4H2O. Vitamin solution (per liter) contained 0.5 g thiamine and 1 mg biotin. The pH of the medium was adjusted to 7.0. MgSO4, CaCl2 · 2H2O, trace minerals, and vitamins were added to the medium after autoclaving. When used, CaCl2 · 2H2O was added to final concentrations from 0.5 to 10.0 mM. Since BMM is at neutral pH and the free phosphate concentration is less than 50 mg/liter, addition of CaCl2 · 2H2O does not cause the removal of phosphate as a calcium phosphate precipitate (7, 63). However, to ensure that the observed results were due to Ca2+ addition, rather than to phosphate starvation from insoluble calcium phosphate precipitation, experiments were performed with various concentrations of CaCl2 · 2H2O (0 to 10 mM) and phosphate (0.05, 0.49, 4.9, and 49 mM). The free phosphate in the culture supernatant was measured by using the vanadomolybdophosphoric acid method (23) following cell growth and centrifugation to remove cells and any possible precipitate. Calcium phosphate precipitates were observed when the phosphate concentration was raised to 49 mM, and when phosphate was 4.9 mM phosphate and calcium was 10 mM. No precipitate was observed in other media (including in BMM), and free phosphate was available after cell growth. Biofilms were cultivated in the absence of carbenicillin, since plasmid pMF230 is stable in these cells over the time course of these experiments without antibiotic selection and since antibiotics may influence biofilm structure.
Biofilm growth.
The flow cell system for the SCLM experiments was similar to that described previously (48). Briefly, the system consisted of a once-flowthrough flow cell with a medium reservoir, pump, silicone tubing, and a waste container. The flow cell was composed of a polycarbonate support with inlet and outlet ports and a glass coverslip, for SCLM observation of the biofilms, sealed onto the polycarbonate support with a Viton gasket. The SCLM flow cells were maintained in an incubator at 37 ± 1°C during biofilm formation. Prior to inoculation into the flow cells, each strain was incubated in BMM for 16 to 20 h at 37°C. When the cell density reached approximately 1 × 107 cells/ml, as determined by absorption/scattering (optical density at 600 nm [OD600]), the cultures were diluted with an appropriate volume of sterile 0.85% NaCl solution to make a 1 × 106-cell/ml inoculum. Approximately 2 ml of this culture was used to inoculate the flow cells under quiescent conditions for 20 min. Flow of sterile BMM was then initiated at a rate of 1.2 ml/min.
For FTIR, algD expression, and proteomic studies, P. aeruginosa biofilms were cultivated on the walls of silicone tubing (0.6-m length, size 18 tubing with interior volume of 40 ml), similarly to the experiments described by Sauer et al. (55). The flow system and inoculation procedure were similar to those described for the SCLM experiments. Following inoculation, sterile BMM was pumped through the tubing at a rate of 2 ml/min for 72 h. Attached cells were removed from the interior surface of the tubing by using the plunger of a 3-ml syringe. Biofilm cells were resuspended in saline solution during harvesting (approximately 150 ml).
FTIR of biofilms.
Biofilms were removed from the silicone tubing and spotted onto cards with infrared transparent films (Spectrotech). Prior to spotting, the biofilms were diluted from 2- to 10-fold in saline, to optimize absorbance intensity for each biofilm. The biofilms were dried onto the cards in a laminar flow hood and stored under desiccation prior to analysis. The spectra were collected and processed as described previously (48).
Spectrometric analyses of biofilms.
The concentrations of alginate were determined by using the modified carbazole method of Knutson and Jeanes (17, 33). Total protein concentration was determined by a modified version of the method of Lowry et al. (37) using reagents from Sigma Chemical Corp. (catalog no. 690-A).
SDS-PAGE analysis of extracellular proteins.
For analysis of extracellular proteins from planktonic cultures, P. aeruginosa PAO1 and FRD1 were incubated overnight in BMM and then diluted 1/100 in fresh BMM and incubated with shaking at 37°C until mid-exponential phase (OD600 of 0.4). Cells were removed by two centrifugations (12,000 × g, 30 min each). Cell pellets were used for two-dimensional gel electrophoresis analysis (described below). Trichloroacetic acid (TCA) (final concentration, 20%) was added to the supernatants, and the mixture was incubated on ice for 60 min to precipitate extracellular proteins. The mixture was centrifuged (15,000 × g, 40 min), and the protein pellets were washed five or six times with ice-cold acetone to remove residual TCA. Proteins were solubilized in denaturing protein sample buffer using a water bath sonicator. Proteins (10 μg) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using 4% stacking gels and 12% resolving gels (35) and analyzed by Coomassie blue staining. For analysis of extracellular proteins in biofilms, the biofilms were cultivated on silicone tubing as described above. The biofilms were harvested by adding saline to the tubing and removing biofilms with the plunger of a 3-μl syringe. The final cell solution (approximately 150 ml) was centrifuged twice to remove cells, and the cell pellets were analyzed by two-dimensional gel electrophoresis. Extracellular proteins were precipitated from the supernatants using TCA as described for the planktonic cells. N-terminal sequence analysis of proteins was performed by David McCourt (Midwest Scientific) using the Edman degradation method. Matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry of proteins was performed by Anthony Haag (Mass Spectrometry Lab, University of Texas Medical Branch, Galveston, TX).
Immunoblot analysis.
Extracellular extracts (5 μg protein per sample) from planktonic culture and from biofilms were separated by SDS-PAGE and then electroblotted onto nitrocellulose membranes (1). The membranes were probed with AprA antibodies (generously provided by Jeffrey Hobden, Wayne State University). Goat anti-rabbit immunoglobulin G, conjugated to horseradish peroxidase, was used as the secondary antibody. Antibody binding was detected by chemiluminescent analysis (1).
Two-dimensional gel electrophoresis of cellular proteins.
Cells from planktonic and biofilm cultures were obtained as described above. Cell pellets were washed twice with saline solution and resuspended in TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) containing 0.3 mg/ml of phenylmethylsulfonyl fluoride. Cells were disrupted by sonication (12 times for 10 s, 4 W, 4°C), and the cell debris and unbroken cells were removed by centrifugation (12,000 × g, 60 min, 4°C). The protein concentrations of the supernatants were determined by using the modified Lowry assay, and 200 μg of protein was loaded per gel. Protein samples were stored as aliquots at −80°C. Two-dimensional gel electrophoresis was performed similarly to the protocol described previously (55). Proteins were loaded by in gel rehydration using 18-cm IPG strips with a pH range of 4 to 7 and rehydrated for 20 to 24 h. Solubilization buffer consisted of 9 M urea, 2.14 M thiourea, 4% CHAPS (3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate), 2% (wt/vol) carrier ampholytes, 0.037 M dithiothreitol, and a trace amount of bromphenol blue. Isoelectric focusing was conducted using a Maltiphor II (Pharmacia) at 20°C. Proteins were focused for a total 28 kV · h. IPG strips were reduced and alkylated for 15 min by 11.3 mM dithiothreitol and 12 mM iodacetamide, in buffer containing 6 M urea, 4% (wt/vol) SDS, 30% (vol/vol) glycerol, 50 mM Tris-HCl, and a trace amount of bromphenol blue, pH 6.8. Strips were embedded on top of SDS-polyacrylamide gels using 0.75% (wt/vol) agarose in electrophoresis buffer. Second-dimension electrophoresis was carried out on 11% polyacrylamide gels (230 by 200 by 1 mm) using a vertical Hoefer Dalt system (Pharmacia) at 10°C using 10 mA/gel for 4 h and then 40 mA/gel for 14 to 16 h. Gels were stained for 72 h with 0.03% (wt/vol) Coomassie brilliant blue G-250 in 16% (wt/vol) ammonium sulfate, 24% (vol/vol) methanol, and 4.8% (vol/vol) orthophosphoric acid.
Catechol-2,3-dioxygenase assays for algD gene expression.
The assay of XylE activity was performed as described previously (57, 68). For analysis of planktonic cultures, P. aeruginosa FRD875 algD-xylE was incubated in BMM overnight and then diluted to an OD540 of 0.1 in fresh BMM. Cultures were incubated with shaking at 37°C and 250 rpm and sampled hourly. One-ml samples were pelleted at 13,000 × g for 15 min, and the pellets were resuspended in 1 ml of assay buffer (50 mM potassium phosphate buffer, pH 7.5, 10% acetone). Samples were kept on ice until assayed. An aliquot (20 to 50 μl) of each sample was combined with 940 to 970 μl of assay buffer containing 1.0 mM catechol, and the OD375 was monitored over time for 10 min, during incubation at 25°C. Catechol-2,3-dioxygenase activity was measured by monitoring the formation of the reaction product 2-hydroxymuconic semialdehyde at 375 nm (ɛ = 4.4 M−1 cm−1). Increase in OD375 was measured against a 1.0-ml blank containing only assay buffer-catechol solution. The enzyme activity was recorded as the amount of enzyme required to produce 1 nmol of product per minute per μg of protein at 25°C. For analysis of gene expression activity in biofilms, P. aeruginosa FRD858 algD-xylE was cultivated on silicone tubing as described above. Attached cells were removed from the interior surface of tubing using a 3-ml syringe plunger at 24, 48, and 72 h of incubation. The cells were resuspended in BMM to give an optical density of 0.40 to 0.43 (OD540). The catechol-2,3-dioxygenase assays were performed as described for the planktonic cells.
Pyocyanin analysis.
Pyocyanin production was determined as described previously (13). P. aeruginosa was incubated for 18 h at 37°C in peptone broth (1% peptone, 1% NaCl, 1% glycerol) with 0 to 10 mM CaCl2 and with 0 to 49 mM added phosphate. Pyocyanin was extracted from 5 ml of culture with 1 ml of chloroform. The chloroform extraction was repeated four times, and the organic fractions were pooled and evaporated. The residue was dissolved in 1 ml of 0.2 N HCl, and the amount of extracted pyocyanin was determined spectrophotometrically at 520 nm, with conversion to μg of pyocyanin per ml of culture by using the coefficient 17.1 (13, 34). Pyocyanin production is reported as μg of pyocyanin per mg of cellular protein.
SCLM analysis of biofilms.
The bacteria were observed using a Leica TCS-SP confocal microscope (Leica Microsystems, Heidelberg, Germany) using a 10× or 40× objective. The green fluorescent protein was excited with the 488-nm line of an argon laser, and emission was collected at 500 to 550 nm. Confocal stacks were collected, and images were processed using Imaris 3.0 (Bitplane AG) and Adobe Photoshop. Image analyses were performed using the COMSTAT program, as described by Heydorn et al. (26).
RESULTS
Scanning confocal laser microscopy reveals increased thickness of mucoid P. aeruginosa biofilms with added Ca2+.
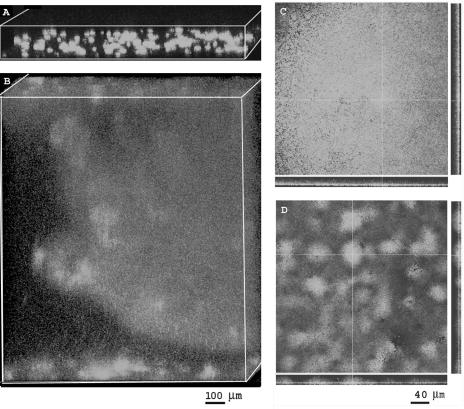
Our previous results using alginate-producing and alginate-impaired mutant strains of P. aeruginosa demonstrated that alginate contributes to the thickness of P. aeruginosa biofilms, allowing them to form microcolonies that extend from the substratum (48). Here, we observed similar results for biofilms of mucoid strain P. aeruginosa FRD1 (Fig. 1A). Using COMSTAT analysis of SCLM images, the mean and standard deviation for the thickness of FRD1 biofilms was 35 ± 10 μm at 24 h and 95 ± 21 μm at 48 h, similar to our previous findings. When biofilms were grown in the presence of added Ca2+ (1.0 and 10.0 mM CaCl2), we observed as much as a 20-fold increase in biofilm thickness at 24 h (Fig. 1B), with the FRD1 biofilms averaging 670 ± 80 μm thick. The maximum thickness of these biofilms extended to both sides of the flow cell (approximately 1 mm). By 48 h the Ca2+-amended biofilm completely filled the flow chamber, with only small channels where the back pressure apparently caused disruption of the biofilm and allowed flow to proceed. The COMSTAT results demonstrated an approximately twofold-greater cellular biomass per area in the FRD1 biofilms without added Ca2+ than in the Ca2+-amended biofilms, suggesting that the Ca2+-amended biofilms were thicker but had less cellular material and greater spacing between cells. Viable cell counts, performed at 18 h of biofilm growth, also showed fewer cells in the Ca2+-amended biofilms. The average cell density of Ca2+-amended biofilms was 2.5 × 107 cells/cm2 compared to 1.2 × 108 cells/cm2 in the biofilms cultivated without added Ca2+.
FIG. 1.
SCLM images of P. aeruginosa biofilms at 24 h of growth. (A) P. aeruginosa FRD1 biofilm cultivated in BMM without added CaCl2, viewed at an angle; magnification, ×100. (B) Angle view of FRD1 biofilm cultivated in BMM with 10 mM CaCl2; magnification, ×100. (C) Top view and confocal reconstructed transverse sections of P. aeruginosa PAO1 biofilm, cultivated in BMM without added CaCl2; magnification, ×400. (D) Top view and transverse reconstructions of PAO1 biofilm, cultivated in BMM with 10 mM CaCl2; magnification, ×400. All cells expressed the gfp-mut2 from plasmid pMF230 for visualization with SCLM.
Biofilms of the nonmucoid strain P. aeruginosa PAO1 showed a small increase in thickness with added Ca2+ (Fig. 1C), compared to biofilms without added Ca2+. At 48 h, the PAO1 biofilms averaged 120 ± 25 μm with Ca2+ and 95 ± 20 μm without Ca2+. Cell counts showed a slight increase in the PAO1 biofilms with added Ca2+. The Ca2+-amended biofilms had 9.5 × 108 cells/cm2 at 18 h, compared to 6.1 × 108 cells/cm2 in biofilms without added Ca2+. A difference in the surface roughness of the biofilms was observed by SCLM and by COMSTAT analysis for the PAO1 biofilms with added Ca2+, compared to biofilms without added Ca2+ (Fig. 1C and D).
The increased thickness of FRD1 biofilms in the presence of Ca2+ is due to an increased amount of alginate in the biofilm extracellular matrix.
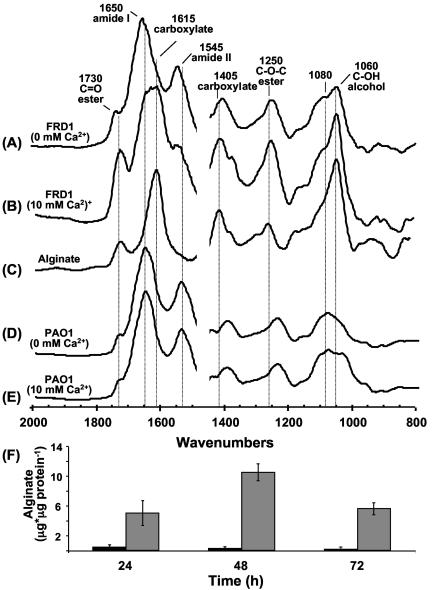
We characterized the biofilms by using FTIR to provide information on the chemical constituents of the biofilm. For these experiments, biofilms were cultivated on silicone tubing and then harvested for FTIR analysis as described in Materials and Methods. Upon visual inspection, the biofilms associated with the silicone tubing showed a noticeable increase in thickness with added Ca2+, similar to observations of biofilms on glass substrata. FTIR revealed differences in the relative amounts of biofilm constituents in the presence or absence of added Ca2+. In FRD1 biofilms with no added Ca2+, the spectra showed dominant absorbance bands at the following (cm−1): 1,650 (amide I), 1,545 (amide II), 1,405 (due in part to stretching of the carboxylate ion), 1,250 (due to P=O stretching, C—O—C stretching, and/or amide III), and 1,080 (P—O and C—OH stretching). These absorbance bands are primarily the result of cellular components including protein and DNA and are found in both alginate- and non-alginate-producing strains (48). In addition to these absorbance bands, the spectra of the FRD1 biofilms had absorbances at 1,730 (C=O stretching of esters), 1,615 (asymmetric stretching of the carboxylate ion—seen as a shoulder on the amide I band), 1,250 (C—O—C of the esters), and 1,060 (C—OH stretching of alcohols). These additional absorbance bands are due to alginate within the biofilm matrix (48) and are identical to the bands observed for purified alginate (Fig. 2C). For FRD1 biofilms cultivated in the presence of added Ca2+ in the medium, the relative abundances of the absorbance bands changed, with the alginate-associated bands becoming dominant. In particular, the band at 1,615 (carboxylate ion) increased and partially obscured the amide I (1,650) and amide II (1,545) bands. The bands at 1,060 (C—OH) and 1,410 (carboxylate) also showed increases relative to the amide I and amide II bands, indicating an increase in the ratio of carbohydrate to cell protein. The bands associated with the ester linkage to the alginate O-acetyl group (1,730 and 1,250) also demonstrated significant increases in biofilms with Ca2+ addition. The increased ratio of alginate to protein in FRD1 biofilms cultivated with added Ca2+ was verified by using chemical analysis of these components (Fig. 2F). When biofilms were incubated with no added Ca2+, they produced from 0.5 to 1.0 μg of alginate per μg of cellular protein. However, when Ca2+ was added, the biofilms contained approximately a 10-fold increase in the amount of alginate per cellular protein, with the FRD1 biofilms having 6 μg alginate per μg of protein at 24 h and 10 μg alginate per μg of protein at 48 h.
FIG. 2.
Infrared spectra of P. aeruginosa biofilms originally cultivated on silicone tubing for 72 h. (A) P. aeruginosa FRD1 cultured in BMM with no added CaCl2. Absorbance bands associated with cell biomass include (cm−1) 1,650 (amide I), 1,545 (amide II), 1,405 (stretching of the carboxylate ion), 1,250 (P=O stretching, C—O—C stretching, and/or amide III), and 1,080 (P—O and C—OH stretching). Absorbance bands due to alginate include (cm−1) 1,735 (C=O stretching of esters), 1,615 (asymmetric stretching of the carboxylate ion), 1,250 (C—O—C of the esters), and 1,060 (C—OH stretching of alcohols). (B) FTIR spectrum of FRD1 biofilm cultivated in BMM with 10 mM CaCl2, showing shift in ratios of alginate-associated bands relative to bands associated with cellular material. (C) FTIR spectrum of alginate purified from FRD1. (D) FTIR spectrum of P. aeruginosa PAO1 biofilm, cultivated in BMM with no added CaCl2, showing no bands associated with alginate. (E) FTIR spectrum of PAO1 biofilm, cultivated in BMM with 10 mM CaCl2, also showing no alginate-associated bands. (F) Amounts of alginate and protein in FRD1 biofilms, determined by colorimetric assays. Black bars, biofilms in BMM with no added CaCl2. Grey bars, biofilms in BMM with 10 mM added CaCl2.
Previous results from Wozniak et al. indicated that alginate was not a component of P. aeruginosa PAO1 biofilms (72). The FTIR spectra here verify those results. In the PAO1 biofilms without added Ca2+, the absorbance bands associated with cell protein were present, but not the alginate-associated bands (Fig. 2D). A similar spectrum of PAO1 biofilms was observed in the presence of added Ca2+ (Fig. 2E), indicating that the additional thickness and surface roughness associated with the Ca2+-amended PAO1 biofilms, as seen by SCLM, were not due to alginate production by these bacteria. An absorbance band at 1,060 cm−1 was observed in PAO1 biofilms with added Ca2+. However, subtraction spectra of biofilms with and without added Ca2+ did not give the spectrum of pure alginate (data not shown), as was the case for the FRD1 biofilms (48), indicating that alginate was not associated with either of these PAO1 biofilms.
Transcriptional reporter demonstrates increased algD gene expression in FRD1 planktonic culture and in biofilms with Ca2+ addition.
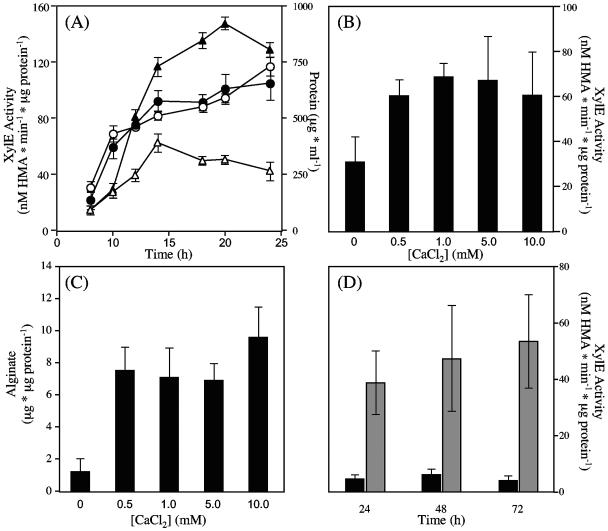
The increase in biofilm thickness and the increased amount of alginate in the P. aeruginosa FRD1 biofilms in the presence of added Ca2+ may have been due in part to the cross-bridging of Ca2+ to the alginate uronic acid residues, thereby retaining the alginate as a gel within these biofilms. However, it is also possible that Ca2+ addition resulted in the increased expression of the alg gene cluster and increased biosynthesis of alginate. To test this hypothesis, we used an FRD1-based algD-xylE transcriptional reporter to assay the relative amounts of algD transcription in the presence and absence of added Ca2+. The algD-xylE reporter is a single-copy chromosomally encoded reporter that disrupts the alg operon and blocks alginate production. As a result, cell growth of the reporter strain was similar both with and without Ca2+ (Fig. 3A), since carbon was not shuttled from cellular material to alginate. In planktonic culture, algD transcription was approximately threefold greater with the addition of 10 mM CaCl2 to the medium (Fig. 3A). The increase in algD transcription with added Ca2+ appeared to plateau at 0.5 mM CaCl2, having approximately the same level of transcriptional reporter activity at all added [CaCl2] (Fig. 3B). In addition, an increased amount of accumulated alginate was detected in the culture supernatants of cells at all of the added [CaCl2] (Fig. 3C). When the reporter strain was cultivated in biofilms, algD transcription increased in the presence of added Ca2+ (Fig. 3D). At 24 and 48 h of growth algD transcription was approximately eightfold greater than without added Ca2+. The increase in transcription with added Ca2+ continued at 72 h. The results indicate that the alginate increase observed by FTIR was due at least in part to increased transcription of the alg gene cluster with added Ca2+ and not solely due to increased cross-bridging of the alginate polymer.
FIG. 3.
Effect of Ca2+ on alg expression measured by using an algD>xylE transcriptional fusion. (A) XylE activity of P. aeruginosa FRD875 algD>xylE cells in planktonic culture showing increased alg expression with added Ca2+. Open triangles, XylE activity of cells in BMM. Closed triangles, XylE activity data in BMM with 10 mM CaCl2. Open circles, cell growth of P. aeruginosa FRD875 algD>xylE in BMM measured as total cellular protein. Closed circles, total protein of cells grown in BMM with 10 mM CaCl2. Error bars represent the standard deviations of four samples using duplicate experiments. (B) Effect of [CaCl2] on alg expression at 18 h of planktonic growth, measured as XylE activity. (C) Effect of [CaCl2] on alginate production of FRD1 in planktonic culture at 18 h. (D) Effect of Ca2+ on alg expression by P. aeruginosa FRD875 algD>xylE, cultivated in biofilms. Black bars, XylE activity of cells cultured in BMM. Grey bars, XylE activity of cells in BMM with 10 mM CaCl2. Error bars represent the standard deviations of three to four replicates. Cells exposed to 0.5 to 10.0 mM CaCl2 exhibited significantly different XylE activity (P < 0.05) and alginate production (P < 0.01) compared to cells cultured in 0 mM CaCl2 HMA, 2-hydroxymuconic semialdehyde.
The alginate matrix in Ca2+-induced biofilms harbors other extracellular virulence factors.
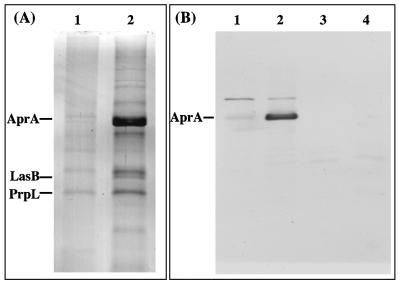
An inverse regulatory relationship was found to exist between alginate production and the extracellular protease LasA (47). However, Ca2+ is known to affect the production and/or stability of certain extracellular proteins of P. aeruginosa (52, 75). To determine if Ca2+ addition affects expression and/or stability of extracellular proteins from an alginate-producing strain, we extracted cell supernatants of P. aeruginosa FRD1 cultured in BMM with and without added Ca2+. With no added Ca2+, strain FRD1 had little extracellular protein, as predicted from the previous studies (47). However, with increased [CaCl2], strain FRD1 had three dominant bands in culture supernatants. We excised the bands and identified the proteins by N-terminal sequence analysis. The 49-kDa band was identified as alkaline metalloprotease AprA (24), the 33-kDa band was identified as elastase LasB (4, 31), and the 27-kDa band is the PrpL protease (65). To ensure that the increased amounts of proteases were due to Ca2+ addition and not due to PO43− limitation (by possible PO43− precipitation as calcium phosphate), experiments were performed with various concentrations of Ca2+ and PO43− as described in Materials and Methods. At each PO43− concentration there was an increase in the extracellular proteases with increasing Ca2+, including medium with greater concentrations of PO43− to Ca2+ (1 mM Ca and 4.9 mM PO43−), indicating that the increased amounts of proteases were due primarily to Ca2+ addition, and not to PO43− starvation.
Since Ca2+ addition caused an increase in the amounts of both alginate and extracellular proteases, we tested the possibility that AprA (the most dominant band in the culture supernatants) was harbored within the alginate biofilm matrix. The extracellular matrix of biofilms from Ca2+-amended cultures and cultures lacking added Ca2+ was extracted, and the alginates were degraded by using an alginate lyase to release the extracellular proteins from the biofilm matrix. Immunoblot assays were performed on the extracted matrix material using an AprA antibody. Only a faint band was observed in the matrix of P. aeruginosa FRD biofilms cultivated without added Ca2+, and no band was observed in biofilms of the aprA deletion mutant strain, P. aeruginosa FRD1252 ΔaprA::aacCIΩ (Fig. 4B). However, a pronounced AprA band was observed in biofilm extracts of the Ca2+-amended biofilms, indicating that this protein was maintained in the matrix of these biofilms.
FIG. 4.
(A) Coomassie blue-stained gel of extracellular proteins produced by planktonic cultures of P. aeruginosa FRD1. Lane 1, proteins from cells cultured in BMM. Lane 2, proteins from cells cultured in BMM with 10 mM CaCl2, showing increased amounts of AprA, LasB, and PrpL. Proteins were identified by N-terminal sequence analysis. (B) Immunoblot of extracellular matrix material from 72-h P. aeruginosa FRD1 biofilms using AprA antibodies. Lane 1, FRD1 in BMM. Lane 2, FRD1 in BMM with 10 mM CaCl2. Lane 3, FRD1252 ΔaprA::aacCIΩ in BMM. Lane 4, FRD1252 ΔaprA::aacCIΩ in BMM with 10 mM CaCl2.
We tested whether the AprA protein contributes to biofilm three-dimensional structure, by comparing SCLM images of P. aeruginosa FRD1 and P. aeruginosa FRD1252 ΔaprA::aacCIΩ. No significant difference was observed between biofilms of these strains (data not shown), indicating that AprA does not have a substantial effect on the structural integrity of the FRD1 biofilms. We also found no difference between the structure of the FRD1 biofilm and those of biofilms of P. aeruginosa FRD1043 ΔcalA::aacCIΩ (data not shown), indicating that the gene PA4107, which encodes a putative calmodulin-like protein, also does not play a significant role in calcium-induced biofilm structure.
Phenazine biosynthetic proteins are affected by calcium and by biofilm growth.
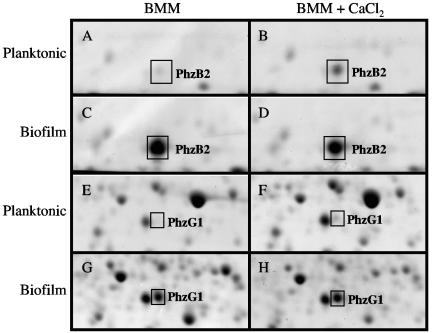
In addition to the extracellular proteins, we analyzed cellular proteins by two-dimensional gel electrophoresis to determine if Ca2+ addition and biofilm growth affect the presence or amounts of certain cellular proteins. Of the protein differences observed, several of the proteins were gel extracted and analyzed by MALDI-TOF mass spectroscopy. Two of the proteins identified as being induced by Ca2+ in planktonic culture were the phenazine biosynthetic proteins PhzB2 and PhzG (Fig. 5). These proteins are involved in the synthesis of the phenazine-based redox cycling compound pyocyanin and are encoded on two separate operons (44, 62). To compare protein amounts of PhzB2 and PhzG, we compared protein spot signal intensities for duplicate gels under the four conditions (planktonic growth versus biofilms at 0 mM versus 10 mM added CaCl2). The results showed that Ca2+ addition resulted in a threefold increase in PhzB2 amount during bacterial growth in planktonic culture (Fig. 5A and B). PhzB2 showed increased amounts in biofilms compared to planktonic culture (Fig. 5C and D), with an approximately eightfold increase in biofilms versus planktonic culture at 0 mM CaCl2 and a threefold increase in biofilms at 10 mM CaCl2. In biofilms [CaCl2] had little effect on PhzB2 amounts, with the protein spot signal intensities being similar at both 0 and 10 mM CaCl2. The amount of PhzG increased approximately fourfold in planktonic culture with the addition of CaCl2 to the medium (Fig. 5E and F). In biofilms, this protein was 25- to 30-fold greater than its amount in planktonic cultures (Fig. 5G and H) whether or not Ca2+ was added.
FIG. 5.
(A to D) Sections of two-dimensional gels showing the effect of Ca2+ and biofilm growth on pyocyanin biosynthetic protein PhzB2 of P. aeruginosa FRD1 under the following conditions: (A) planktonic culture in BMM, (B) planktonic culture in BMM plus 10 mM CaCl2, (C) biofilm culture in BMM, and (D) biofilm culture in BMM plus 10 mM CaCl2. (E to H) Sections of two-dimensional gels showing effect of Ca2+ and biofilm growth on PhzG of P. aeruginosa FRD1 under the following conditions: (E) planktonic culture in BMM, (F) planktonic culture in BMM plus 10 mM CaCl2, (G) biofilm culture in BMM, and (H) biofilm cultured in BMM plus 10 mM CaCl2.
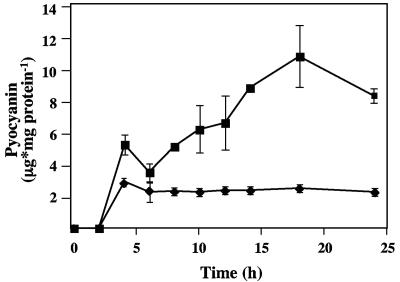
To determine if Ca2+ affects production of the P. aeruginosa phenazine product pyocyanin, we assayed pyocyanin production in planktonic culture with and without added CaCl2. Over time, pyocyanin production was approximately two- to threefold greater when cultures were incubated with 10 mM CaCl2 than when no CaCl2 was added to the medium (Fig. 6). When the [PO43−] was varied over a range of 0.05 to 4.9 mM, an increase in pyocyanin production was observed for cultures with 1 mM added Ca2+ compared to cultures with no added Ca2+, indicating that the observed increase was due to Ca2+ addition and not to PO43− starvation.
FIG. 6.
Effect of Ca2+ on pyocyanin biosynthesis by P. aeruginosa FRD1. Diamonds, pyocyanin production in peptone broth planktonic culture. Squares, pyocyanin production in peptone broth plus 10 mM CaCl2.
DISCUSSION
The results presented here show that the biofilm extracellular matrix of a mucoid strain of P. aeruginosa in Ca2+-amended cultures is composed primarily of alginate. Extracellular proteases are present in the extracellular material with the alginate matrix acting as a reservoir to maintain these proteases in the biofilms. Although present, the protease AprA does not play a significant role in the structural integrity of these biofilms. The biofilm matrix also likely contains pyocyanin, since proteins for pyocyanin biosynthesis show increased amounts in biofilm cultures and in cultures amended with Ca2+. The extracellular matrix of a mucoid P. aeruginosa biofilm where little Ca2+ is present is composed primarily of alginate, with lesser amounts of extracellular proteins.
These results confirm previous findings of both Nivens et al. and Wozniak et al. that alginate is not required for biofilm formation by nonmucoid strains of P. aeruginosa (48, 72). Nivens et al. demonstrated that nonmucoid variants of P. aeruginosa FRD1 were capable of biofilm formation and that no alginate was associated with those biofilms. Using P. aeruginosa PAO1, which does not produce alginate under known conditions, Wozniak et al. demonstrated that this strain was capable of biofilm formation in the absence of alg gene expression. The FTIR results shown here (Fig. 2) confirm that no alginate is associated with PAO1 biofilms. However, in biofilms of the mucoid CF pulmonary isolate FRD1, alginate constitutes the major component of the extracellular matrix material.
The surprising finding here is that Ca2+ addition has a major effect on the extracellular matrix of the mucoid P. aeruginosa biofilms, including the amount of alginate produced. Alginate biosynthesis is regulated by a complex hierarchy of proteins (71), including the stress response sigma factor σe (11, 15, 41). In nonmucoid strains, isolated from environmental sources other than chronic CF pulmonary infections, this sigma factor is sequestered in the membrane by the antisigma factor MucAB (40, 43). Nonmucoid strains show no evidence of alginate production under any environmental conditions described, including in biofilms (72). Alginate-producing strains isolated from the pulmonary fluid of CF patients often have a mutation in mucA (40), causing release of σe from membrane sequestration and subsequent expression of the σe-controlled regulon, including the alginate biosynthetic gene cluster (43). Dehydration and high osmolarity have been shown to play roles in alg gene expression in a mucoid strain (3, 10). The results here show that another environmental parameter, Ca2+, also affects the amount of alginate produced by a mucoid strain (Fig. 2 and 3). The addition of Ca2+ causes at least a threefold increase in alg gene expression during P. aeruginosa FRD1 growth in planktonic culture and as much as an eightfold increase in alg gene expression during biofilm growth. This increase in alg expression results in biofilms that contained 5 to 10 mg of alginate per mg of protein. Biofilms without added Ca2+ had approximately 0.5 to 1.0 mg alginate per mg of protein. The increased amount of alginate in Ca2+-amended cultures results in biofilms that are as much as 20-fold thicker than those without added Ca2+ (Fig. 1). The mechanism for enhanced alg expression caused by Ca2+ addition is not known. However, the results suggest that a Ca2+-controlled factor(s) may be involved in alg gene regulation, either through a previously identified regulator, such as one of the two-component sensory systems known to be involved in alg regulation (9, 22, 70), or through another regulatory factor that has not yet been identified. In biofilms, Ca2+ may play an additional role in the observed increase in biofilm thickness by ionic cross-linking and gelling of the uronic acid residues, thereby modifying the biofilm architecture.
Another surprising finding is the Ca2+-induced production of proteinaceous extracellular material in mucoid P. aeruginosa. Previously, an inverse regulatory relationship between expression of alginate biosynthetic genes and expression of LasA was described (47). When P. aeruginosa establishes chronic infections on the pulmonary tissue of CF patients, it synthesizes the immunoprotective compound alginate and represses other immune stimulatory factors, such as extracellular proteins and flagellin (67). In the absence of added Ca2+, this phenomenon is corroborated here, since little to no extracellular proteins were observed in cultures where no Ca2+ was added to the medium (Fig. 4). However, when Ca2+ was added, intense protein bands of AprA, LasB, and PrpL were observed. These proteins were expressed concomitantly with the alg genes, indicating that, although an inverse relationship between alg expression and protease expression may exist, this relationship is dependent on environmental factors, and particularly on the presence of Ca2+. The proteases are maintained in the alginate extracellular material, making them components of the P. aeruginosa biofilm extracellular matrix. Since these data are based on protein amounts, Ca2+ may affect gene expression, protein stability, or a combination of those two factors.
Also present in the extracellular matrix of Ca2+-induced mucoid P. aeruginosa biofilms is the redox cycling compound pyocyanin. The results show that at least two of the pyocyanin biosynthetic proteins, one on each phenazine biosynthetic operon, are induced by Ca2+ addition and are also induced by biofilm-associated growth (Fig. 5). The chemical analysis showed increased production of pyocyanin with Ca2+ addition in P. aeruginosa FRD1 (Fig. 6). Pyocyanin has been detected in the pulmonary fluid of CF patients (66), suggesting that these operons may be induced in vivo. In addition, using DNA microarray analyses, Wolfgang et al. have demonstrated that the phenazine biosynthetic genes, including both phzB2 and phzG, are induced by mucopurulent respiratory liquids from CF patients (67). Therefore, it is likely that the in vivo Ca2+ concentration in pulmonary fluid of CF patients is sufficient for induction of the phenazine biosynthetic genes. Pyocyanin is a redox cycling compound that has a number of effects on host cells, including an effect on the vacuolar ATPase (54). Among other functions, the vacuolar ATPase is important for maintaining cytosolic Ca2+ levels in Saccharomyces cerevisiae (16). Pyocyanin has been shown to disrupt calcium homeostasis of human epithelial cells (8). Therefore, pyocyanin production, induced in biofilms and by Ca2+, may contribute to altered calcium levels in pulmonary fluid of CF patients and to further induction of the other Ca2+-regulated factors.
The mechanism for calcium regulation in bacteria is not known. Calcium plays a regulatory role for channels and transporters (50). Calmodulin-like proteins have been proposed to have originated in high-G+C gram-positive bacteria and ultimately to have been acquired by eukaryotes (76). A calmodulin-like protein and regulator have been described in Sinorhizobium etli and shown to play a role in development of bacteroids during symbiosis of this bacterium with its leguminous host (73). P. aeruginosa encodes a protein with EF-hand motifs (PA4107) (62) characteristic of calmodulin-like proteins. However, deletions of the gene for this protein did not have a regulatory effect on any of the Ca2+-induced factors described here, indicating that this protein does not play a regulatory role in the production of these compounds.
Acknowledgments
We thank David McCourt for providing the N-terminal sequence analyses and Anthony Haag for providing the MALDI-TOF analysis. We thank Daniel Wozniak for providing P. aeruginosa FRD875 algD>xylE and Jeffrey Hobden for providing the AprA antiserum.
This work was supported by Public Health Service grant AI-46588 (M.J.F.) from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1993. Current protocols in molecular biology, vol. 2. John Wiley & Sons, Inc., New York, N.Y.
- 2.Baltimore, R. S., and M. Mitchell. 1982. Immunologic investigations of mucoid strains of Pseudomonas aeruginosa: comparison of susceptibility to opsonic antibody in mucoid and nonmucoid strains. J. Infect. Dis. 141:238-247. [DOI] [PubMed] [Google Scholar]
- 3.Berry, A., J. D. DeVault, and A. M. Chakrabarty. 1989. High osmolarity is a signal for enhanced algD transcription in mucoid and nonmucoid Pseudomonas aeruginosa strains. J. Bacteriol. 171:2312-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bever, R. A., and B. H. Iglewski. 1988. Molecular characterization and nucleotide sequence of the Pseudomonas aeruginosa elastase structural gene. J. Bacteriol. 170:4309-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boucher, J. C., H. Yu, M. H. Mudd, and V. Deretic. 1997. Mucoid Pseudomonas aeruginosa in cystic fibrosis: characterization of muc mutations in clinical isolates and analysis of clearance in a mouse model of respiratory infection. Infect. Immun. 65:3838-3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cota-Gomez, A., A. I. Vasil, J. Kadurugamuwa, T. J. Beveridge, H. P. Schweizer, and M. L. Vasil. 1997. PlcR1 and PlcR2 are putative calcium-binding proteins required for secretion of the hemolytic phospholipase C of Pseudomonas aeruginosa. Infect. Immun. 65:2904-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de-Bashan, L. E., and Y. Bashan. 2004. Recent advances in removing phosphorus from wastewater and its future use as fertilizer (1997-2003). Water Res. 38:4222-4246. [DOI] [PubMed] [Google Scholar]
- 8.Denning, G. M., M. A. Railsback, G. T. Rasmussen, C. D. Cox, and B. E. Britigan. 1998. Pseudomonas pyocyanine alters calcium signaling in human airway epithelial cells. Am. J. Physiol. 274:L893-L900. [DOI] [PubMed] [Google Scholar]
- 9.Deretic, V., R. Dikshit, W. M. Konyecsni, A. M. Chakrabarty, and T. K. Misra. 1989. The algR gene, which regulates mucoidy in Pseudomonas aeruginosa, belongs to a class of environmentally responsive genes. J. Bacteriol. 171:1278-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeVault, J. D., K. Kimbara, and A. M. Chakrabarty. 1990. Pulmonary dehydration and infection in cystic fibrosis: evidence that ethanol activates gene expression and induction of mucoidy in Pseudomonas aeruginosa. Mol. Microbiol. 4:737-745. [DOI] [PubMed] [Google Scholar]
- 11.DeVries, C. A., and D. E. Ohman. 1994. Mucoid-to-nonmucoid conversion in alginate-producing Pseudomonas aeruginosa often results from spontaneous mutations in algT, encoding a putative alternate sigma factor, and shows evidence for autoregulation. J. Bacteriol. 176:6677-6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Draget, K. I., B. Strand, M. Hartmann, S. Valla, O. Smidsrod, and G. Skjak-Braek. 2000. Ionic and acid gel formation of epimerised alginates; the effect of AlgE4. Int. J. Biol. Macromol. 27:117-122. [DOI] [PubMed] [Google Scholar]
- 13.Essar, D. W., L. Eberly, A. Hadero, and I. P. Crawford. 1990. Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: interchangeability of the two anthranilate synthases and evolutionary implications. J. Bacteriol. 172:884-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans, L. R., and A. Linker. 1973. Production and characterization of the slime polysaccharide of Pseudomonas aeruginosa. J. Bacteriol. 116:915-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flynn, J. L., and D. E. Ohman. 1988. Cloning of genes from mucoid Pseudomonas aeruginosa which control spontaneous conversion to the alginate production phenotype. J. Bacteriol. 170:1452-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Förster, C., and P. M. Kane. 2000. Cytosolic Ca2+ homeostasis is a constitutive function of the V-ATPase in Saccharomyces cerevisiae. J. Biol. Chem. 275:38245-38253. [DOI] [PubMed] [Google Scholar]
- 17.Franklin, M. J., and D. E. Ohman. 1996. Identification of algI and algJ in the Pseudomonas aeruginosa alginate biosynthetic gene cluster which are required for alginate O acetylation. J. Bacteriol. 178:2186-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friedman, L., and R. Kolter. 2004. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51:675-690. [DOI] [PubMed] [Google Scholar]
- 19.Friedman, L., and R. Kolter. 2004. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J. Bacteriol. 186:4457-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gacesa, P. 1988. Alginates. Carbohydr. Polym. 8:161-182. [Google Scholar]
- 21.Goldberg, J. B., W. L. Gorman, J. L. Flynn, and D. E. Ohman. 1993. A mutation in algN permits trans activation of alginate production by algT in Pseudomonas species. J. Bacteriol. 175:1303-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldberg, J. B., and D. E. Ohman. 1987. Construction and characterization of Pseudomonas aeruginosa algB mutants: role of algB in high-level production of alginate. J. Bacteriol. 169:1593-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greenberg, A. E., L. S. Clesceri, and A. D. Eaton (ed.). 1992. Standard methods for the examination of water and wastewater. American Public Health Association, Washington, D.C.
- 24.Guzzo, J., M. Murgier, A. Filloux, and A. Lazdunski. 1990. Cloning of the Pseudomonas aeruginosa alkaline protease gene and secretion of the protease into the medium by Escherichia coli. J. Bacteriol. 172:942-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hentzer, M., G. M. Teitzel, G. J. Balzer, A. Heydorn, S. Molin, M. Givskov, and M. R. Parsek. 2001. Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J. Bacteriol. 183:5395-5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heydorn, A., A. T. Nielsen, M. Hentzer, C. Sternberg, M. Givskov, B. K. Ersboll, and S. Molin. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395-2407. [DOI] [PubMed] [Google Scholar]
- 27.Holland, I. B., H. E. Jones, A. K. Campbell, and A. Jacq. 1999. An assessment of the role of intracellular free Ca2+ in E. coli. Biochimie 81:901-907. [DOI] [PubMed] [Google Scholar]
- 28.Hoyle, B. D., and J. W. Costerton. 1989. Transient exposure to a physiologically-relevant concentration of calcium confers tobramycin resistance upon sessile cells of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 51:339-341. [DOI] [PubMed] [Google Scholar]
- 29.Jackson, K. D., M. Starkey, S. Kremer, M. R. Parsek, and D. J. Wozniak. 2004. Identification of psl, a locus encoding a potential exopolysaccharide that is essential for Pseudomonas aeruginosa PAO1 biofilm formation. J. Bacteriol. 186:4466-4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jesaitis, A. J., M. J. Franklin, D. Berglund, M. Sasaki, C. M. Lord, J. B. Bleazard, J. E. Duffy, H. Beyenal, and Z. Lewandowski. 2003. Compromised host defense on Pseudomonas aeruginosa biofilms: characterization of neutrophil and biofilm interactions. J. Immunol. 171:4329-4339. [DOI] [PubMed] [Google Scholar]
- 31.Kessler, E., and M. Safrin. 1988. Synthesis, processing, and transport of Pseudomonas aeruginosa elastase. J. Bacteriol. 170:5241-5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kierek, K., and P. I. Watnick. 2003. The Vibrio cholerae O139 O-antigen polysaccharide is essential for Ca2+-dependent biofilm development in sea water. Proc. Natl. Acad. Sci. USA 100:14357-14362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knutson, C. A., and A. Jeanes. 1968. A new modification of the carbazole analysis: application to heteropolysaccharides. Anal. Biochem. 24:470-481. [DOI] [PubMed] [Google Scholar]
- 34.Kurachi, M. 1958. Studies on the biosynthesis of pyocyanine. Isolation and determination of pyocyanine. Bull. Inst. Chem. Res. Kyoto Univ. 36:163-173. [Google Scholar]
- 35.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 36.Lam, J., R. Chan, K. Lam, and J. W. Costerton. 1980. Production of mucoid microcolonies by Pseudomonas aeruginosa within infected lungs in cystic fibrosis. Infect. Immun. 28:546-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 38.Lyczak, J. B., C. L. Cannon, and G. B. Pier. 2002. Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 15:194-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin, D. W., B. W. Holloway, and V. Deretic. 1993. Characterization of a locus determining the mucoid status of Pseudomonas aeruginosa: AlgU shows sequence similarities with a Bacillus sigma factor. J. Bacteriol. 175:1153-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin, D. W., M. J. Schurr, M. H. Mudd, J. R. W. Govan, B. W. Holloway, and V. Deretic. 1993. Mechanism of conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. Proc. Natl. Acad. Sci. USA 90:8377-8381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin, D. W., M. J. Schurr, H. Yu, and V. Deretic. 1994. Analysis of promoters controlled by the putative sigma factor AlgU regulating conversion to mucoidy in Pseudomonas aeruginosa: relationship to σE and stress response. J. Bacteriol. 176:6688-6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mathee, K., O. Ciofu, C. Sternberg, P. W. Lindum, J. I. Campbell, P. Jensen, A. H. Johnsen, M. Givskov, D. E. Ohman, S. Molin, N. Hoiby, and A. Kharazmi. 1999. Mucoid conversion of Pseudomonas aeruginosa by hydrogen peroxide: a mechanism for virulence activation in the cystic fibrosis lung. Microbiology 145:1349-1357. [DOI] [PubMed] [Google Scholar]
- 43.Mathee, K., C. J. McPherson, and D. E. Ohman. 1997. Posttranslational control of the algT (algU)-encoded σ22 for expression of the alginate regulon in Pseudomonas aeruginosa and localization of its antagonist proteins MucA and MucB (AlgN). J. Bacteriol. 179:3711-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mavrodi, D. V., R. F. Bonsall, S. M. Delaney, M. J. Soule, G. Phillips, and L. S. Thomashow. 2001. Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J. Bacteriol. 183:6454-6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meluleni, G. J., M. Grout, D. J. Evans, and G. B. Pier. 1995. Mucoid Pseudomonas aeruginosa growing in a biofilm in vitro are killed by opsonic antibodies to the mucoid exopolysaccharide capsule but not by antibodies produced during chronic lung infection in cystic fibrosis patients. J. Immunol. 155:2029-2038. [PubMed] [Google Scholar]
- 46.Michiels, J., C. Xi, J. Verhaert, and J. Vanderleyden. 2002. The functions of Ca2+ in bacteria: a role for EF-hand proteins? Trends Microbiol. 10:87-93. [DOI] [PubMed] [Google Scholar]
- 47.Mohr, C. D., L. Rust, A. M. Albus, B. H. Iglewski, and V. Deretic. 1990. Expression patterns of genes encoding elastase and controlling mucoidy: co-ordinate regulation of two virulence factors in Pseudomonas aeruginosa isolates from cystic fibrosis. Mol. Microbiol. 4:2103-2110. [DOI] [PubMed] [Google Scholar]
- 48.Nivens, D. E., D. E. Ohman, J. Williams, and M. J. Franklin. 2001. Role of alginate and its O acetylation in formation of Pseudomonas aeruginosa microcolonies and biofilms. J. Bacteriol. 183:1047-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Norris, V., M. Chen, M. Goldberg, J. Voskuil, G. McGurk, and I. B. Holland. 1991. Calcium in bacteria: a solution to which problem? Mol. Microbiol. 5:775-778. [DOI] [PubMed] [Google Scholar]
- 50.Norris, V., S. Grant, P. Freestone, J. Canvin, F. N. Sheikh, I. Toth, M. Trinei, K. Modha, and R. I. Norman. 1996. Calcium signalling in bacteria. J. Bacteriol. 178:3677-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ohman, D. E., and A. M. Chakrabarty. 1981. Genetic mapping of chromosomal determinants for the production of the exopolysaccharide alginate in a Pseudomonas aeruginosa cystic fibrosis isolate. Infect. Immun. 33:142-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Olson, J. C., and D. E. Ohman. 1992. Efficient production and processing of elastase and LasA by Pseudomonas aeruginosa require zinc and calcium ions. J. Bacteriol. 174:4140-4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pina, S. E., and S. J. Mattingly. 1997. The role of fluoroquinolones in the promotion of alginate synthesis and antibiotic resistance in Pseudomonas aeruginosa. Curr. Microbiol. 35:103-108. [DOI] [PubMed] [Google Scholar]
- 54.Ran, H., D. J. Hassett, and G. W. Lau. 2003. Human targets of Pseudomonas aeruginosa pyocyanin. Proc. Natl. Acad. Sci. USA 100:14315-14320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sauer, K., A. K. Camper, G. D. Ehrlich, J. W. Costerton, and D. G. Davies. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184:1140-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schurr, M. J., H. Yu, J. M. Martinez-Salazar, J. C. Boucher, and V. Deretic. 1996. Control of AlgU, a member of the σE-like family of stress sigma factors, by the negative regulators MucA and MucB and Pseudomonas aeruginosa conversion to mucoidy in cystic fibrosis. J. Bacteriol. 178:4997-5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schweizer, H. P., and T. T. Hoang. 1995. An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene 158:15-22. [DOI] [PubMed] [Google Scholar]
- 58.Singh, P. K., A. L. Schaefer, M. R. Parsek, T. O. Moninger, M. J. Welsh, and E. P. Greenberg. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762-764. [DOI] [PubMed] [Google Scholar]
- 59.Skjåk-Bræk, G., F. Zanetti, and S. Paoletti. 1989. Effect of acetylation on some solution and gelling properties of alginates. Carbohydr. Res. 185:131-138. [Google Scholar]
- 60.Stapper, A. P., G. Narasimhan, D. E. Ohman, J. Barakat, M. Hentzer, S. Molin, A. Kharazmi, N. Hoiby, and K. Mathee. 2004. Alginate production affects Pseudomonas aeruginosa biofilm development and architecture, but is not essential for biofilm formation. J. Med. Microbiol. 53:679-690. [DOI] [PubMed] [Google Scholar]
- 61.Stonehouse, M. J., A. Cota-Gomez, S. K. Parker, W. E. Martin, J. A. Hankin, R. C. Murphy, W. Chen, K. B. Lim, M. Hackett, A. I. Vasil, and M. L. Vasil. 2002. A novel class of microbial phosphocholine-specific phospholipases C. Mol. Microbiol. 46:661-676. [DOI] [PubMed] [Google Scholar]
- 62.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 63.van der Houwen, J. A. M., G. Cressey, B. A. Cressey, and E. Valsami-Jones. 2003. The effect of organic ligands on the crystallinity of calcium phosphate. J. Cryst. Growth 249:572-583. [Google Scholar]
- 64.Whitchurch, C. B., T. Tolker-Nielsen, P. C. Ragas, and J. S. Mattick. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. [DOI] [PubMed] [Google Scholar]
- 65.Wilderman, P. J., A. I. Vasil, Z. Johnson, M. J. Wilson, H. E. Cunliffe, I. L. Lamont, and M. L. Vasil. 2001. Characterization of an endoprotease (PrpL) encoded by a PvdS-regulated gene in Pseudomonas aeruginosa. Infect. Immun. 69:5385-5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wilson, R., D. A. Sykes, D. Watson, A. Rutman, G. W. Taylor, and P. J. Cole. 1988. Measurement of Pseudomonas aeruginosa phenazine pigments in sputum and assessment of their contribution to sputum sol toxicity for respiratory epithelium. Infect. Immun. 56:2515-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wolfgang, M. C., J. Jyot, A. L. Goodman, R. Ramphal, and S. Lory. 2004. Pseudomonas aeruginosa regulates flagellin expression as part of a global response to airway fluid from cystic fibrosis patients. Proc. Natl. Acad. Sci. USA 101:6664-6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Woolwine, S. C., A. B. Sprinkle, and D. J. Wozniak. 2001. Loss of Pseudomonas aeruginosa PhpA aminopeptidase activity results in increased algD transcription. J. Bacteriol. 183:4674-4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Woolwine, S. C., and D. J. Wozniak. 1999. Identification of an Escherichia coli pepA homolog and its involvement in suppression of the algB phenotype in mucoid Pseudomonas aeruginosa. J. Bacteriol. 181:107-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wozniak, D. J., and D. E. Ohman. 1991. Pseudomonas aeruginosa AlgB, a two-component response regulator of the NtrC family, is required for algD transcription. J. Bacteriol. 173:1406-1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wozniak, D. J., and D. E. Ohman. 1994. Transcriptional analysis of the Pseudomonas aeruginosa genes algR, algB, and algD reveals a hierarchy of alginate gene expression which is modulated by algT. J. Bacteriol. 176:6007-6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wozniak, D. J., T. J. Wyckoff, M. Starkey, R. Keyser, P. Azadi, G. A. O'Toole, and M. R. Parsek. 2003. Alginate is not a significant component of the extracellular polysaccharide matrix of PA14 and PAO1 Pseudomonas aeruginosa biofilms. Proc. Natl. Acad. Sci. USA 100:7907-7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xi, C., E. Schoeters, J. Vanderleyden, and J. Michiels. 2000. Symbiosis-specific expression of Rhizobium etli casA encoding a secreted calmodulin-related protein. Proc. Natl. Acad. Sci. USA 97:11114-11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xie, Z.-D., C. D. Hershberger, S. Shankar, R. W. Ye, and A. M. Chakrabarty. 1996. Sigma factor-anti-sigma factor interaction in alginate synthesis: inhibition of AlgT by MucA. J. Bacteriol. 178:4990-4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yahr, T. L., L. M. Mende-Mueller, M. B. Friese, and D. W. Frank. 1997. Identification of type III secreted products of the Pseudomonas aeruginosa exoenzyme S regulon. J. Bacteriol. 179:7165-7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang, K. 2001. Prokaryotic calmodulins: recent developments and evolutionary implications. J. Mol. Microbiol. Biotechnol. 3:457-459. [PubMed] [Google Scholar]