Abstract
Aerobic, glucose-limited chemostat cultures of Saccharomyces cerevisiae CEN.PK113-7D were grown with different nitrogen sources. Cultures grown with phenylalanine, leucine, or methionine as a nitrogen source contained high levels of the corresponding fusel alcohols and organic acids, indicating activity of the Ehrlich pathway. Also, fusel alcohols derived from the other two amino acids were detected in the supernatant, suggesting the involvement of a common enzyme activity. Transcript level analysis revealed that among the five thiamine-pyrophospate-dependent decarboxylases (PDC1, PDC5, PDC6, ARO10, and THI3), only ARO10 was transcriptionally up-regulated when phenylalanine, leucine, or methionine was used as a nitrogen source compared to growth on ammonia, proline, and asparagine. Moreover, 2-oxo acid decarboxylase activity measured in cell extract from CEN.PK113-7D grown with phenylalanine, methionine, or leucine displayed similar broad-substrate 2-oxo acid decarboxylase activity. Constitutive expression of ARO10 in ethanol-limited chemostat cultures in a strain lacking the five thiamine-pyrophosphate-dependent decarboxylases, grown with ammonia as a nitrogen source, led to a measurable decarboxylase activity with phenylalanine-, leucine-, and methionine-derived 2-oxo acids. Moreover, even with ammonia as the nitrogen source, these cultures produced significant amounts of the corresponding fusel alcohols. Nonetheless, the constitutive expression of ARO10 in an isogenic wild-type strain grown in a glucose-limited chemostat with ammonia did not lead to any 2-oxo acid decarboxylase activity. Furthermore, even when ARO10 was constitutively expressed, growth with phenylalanine as the nitrogen source led to increased decarboxylase activities in cell extracts. The results reported here indicate the involvement of posttranscriptional regulation and/or a second protein in the ARO10-dependent, broad-substrate-specificity decarboxylase activity.
Saccharomyces cerevisiae has a narrow range of carbon sources that support growth (1) but is considerably more flexible with respect to the utilization of nitrogen sources (2). Most amino acids can be utilized as sole nitrogen sources but not as sole carbon sources for growth (28). The most common mechanism for utilizing amino acids as nitrogen sources is transamination, using 2-oxoglutarate or other 2-oxo acids as amino acceptors. This process leaves the carbon skeleton of the amino acid intact, in the form of a 2-oxo acid. For some amino acids (e.g., alanine), the resulting 2-oxo acid, pyruvate, can be readily cometabolized in central metabolism. In other cases, such as for the aromatic and branched-chain amino acids, the 2-oxo acids resulting from transamination are not intermediates of central metabolism. Even though they cannot be used as auxiliary carbon sources, these compounds are often transformed by the yeast cells before they are excreted into the growth medium.
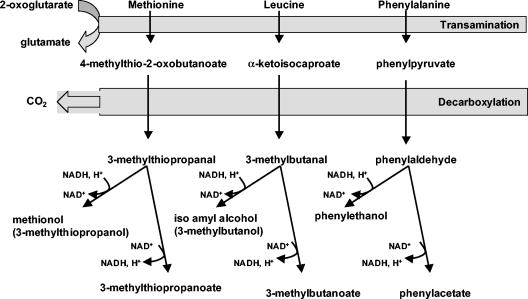
An important and common pathway for catabolism of amino acids by yeasts is called the Ehrlich pathway (7-12, 37). This pathway is initiated by transamination of the amino acid to the corresponding 2-oxo acid. This 2-oxo acid is then decarboxylated to the corresponding aldehyde. Depending on the redox status of the cells (44), the aldehydes can then be reduced by alcohol dehydrogenases (yielding a group of compounds commonly referred to as fusel alcohols) or be oxidized to the corresponding organic acid (“fusel acids”) by aldehyde dehydrogenases (Fig. 1). The fusel alcohols and their esters are especially important contributors to the flavor and aroma of fermented beverages (6, 16, 45). Phenylethanol, which has a typical rose-like flavor, can be produced by biotransformation of phenylalanine with S. cerevisiae cell suspensions (38, 39).
FIG. 1.
Formation of fusel alcohols and fusel organic acids during the catabolism of the amino acids leucine, phenylalanine, and methionine.
The identity of the decarboxylase(s) that catalyzes the initial step of the Ehrlich pathway has recently been investigated in our laboratories (7, 9-11, 44). The S. cerevisiae genome contains five genes that share sequence similarities with genes encoding thiamine pyrophosphate (TPP)-dependent decarboxylases (19, 20, 27) (for a review, see reference 21). Three of these genes (PDC1, PDC5, and PDC6) encode pyruvate decarboxylases. PDC1 and PDC5 encode the major pyruvate decarboxylases under most cultivation conditions (15, 20); PDC6 is specifically expressed under low-sulfur conditions and encodes a pyruvate decarboxylase that has a low content of sulfur-containing amino acids (4, 14). Mutants in which all three PDC genes have been inactivated, and which completely lack pyruvate decarboxylase activity, still express branched-chain and aromatic 2-oxo acid decarboxylase activities (7, 40, 44). The other two members of this gene family are ARO10 and THI3. Based on studies with deletion mutants, both have been implicated in the decarboxylation of branched-chain and aromatic 2-oxo acids (7, 10, 44). In addition, Thi3p has been assumed to be a positive regulator of the thiamine biosynthetic pathway. Upon its deletion, the transcription of all the genes of thiamine biosynthesis was negatively affected (13, 21). An aro10 thi3 double-deletion mutant completely lacks phenylpyruvate decarboxylase activity, whereas the single-deletion mutants in these genes retain this enzyme activity (44). This might lead to the simple conclusion that both genes encode active phenylpyruvate decarboxylases. However, the situation is more complicated, as pdc1 pdc5 pdc6 thi3 quadruple-deletion mutants, but not pdc1 pdc5 pdc6 aro10 mutants, express phenylpyruvate decarboxylase activity (7, 10, 44). These and other observations have led to the proposal that THI3 may not by itself encode an active phenylpyruvate decarboxylase but requires the simultaneous expression of one of the PDC genes to contribute to phenylpyruvate decarboxylase activity (44). This provided a first indication that the regulation and substrate specificities of the TPP-dependent decarboxylases in S. cerevisiae may be more complicated than a simple situation in which substrate specificity is determined by a mixture of five decarboxylases with defined—if overlapping—substrate specificities and kinetics.
With the exception of the transcriptional regulation of ARO10 by aromatic amino acids modulated by the positive transcription factor ARO80 (24), comparatively little is known about the regulation of fusel alcohol production in S. cerevisiae and the impact of the expression levels of the decarboxylase genes on the rates of production of the different decarboxylases.
The aim of the present study was to analyze the substrate specificity of the ARO10-dependent decarboxylase activity in S. cerevisiae, its impact on the production of fusel alcohols and acids, and the importance of transcriptional regulation in controlling its in vivo activity. To this end, we correlated the expression of ARO10 (as well as that of the other decarboxylase genes) with the levels of fusel alcohols and acids in chemostat cultures of S. cerevisiae grown with different nitrogen sources. Subsequently, we investigated the substrate specificity of the ARO10-dependent decarboxylase activity and the impact of transcriptional regulation of ARO10 on this activity by constitutively expressing ARO10 in a wild-type S. cerevisiae strain, as well as in a pdc1 pdc5 pdc6 aro10 thi3 quintuple-null mutant.
MATERIALS AND METHODS
Strains.
The Saccharomyces cerevisiae strains used in this study are listed in Table 1.
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| CEN-PK 113-7D | MATaMAL2-8c SUC2 isogenic prototrophic strain | P. Köttera |
| CEN-PK 113-5D | MATaMAL2-8c SUC2 ura3 | P. Kötter |
| CEN-PK 555-4D | MATaMAL2-8c SUC2 aro10Δ | 44 |
| CEN-PK 711-7C | MATaMAL2-8c SUC2 ura3 pdc1Δ pdc5Δ pcd6Δ aro10Δ thi3Δ | This study |
| IMZ001 | MATaMAL2-8c SUC2 ura3 pdc1Δ pdc5Δ pcd6Δ aro10Δ thi3Δ p426GPD (URA3) | This study |
| IMZ002 | MATaMAL2-8c SUC2 ura3 pdc1Δ pdc5Δ pcd6Δ aro10Δ thi3Δ pUDe001 (URA3 TDH3p-ARO10) | This study |
| IME003 | MATaMAL2-8c SUC2 ura3 pUDe001 (URA3 TDH3p-ARO10) | This study |
| IME004 | MATaMAL2-8c SUC2 ura3 p426GPD (URA3) | This study |
Institut fur Mikrobiologie der J. W. Goethe Universitat, Frankfurt, Germany.
Recombinant-DNA techniques.
Standard protocols were followed for plasmid isolation, restriction, ligation, transformation, and gel electrophoresis (30). Yeast chromosomal DNA was isolated by a method described previously (22). S. cerevisiae strains were transformed using the lithium acetate-single-stranded carrier DNA-polyethylene glycol method (17).
Overexpression of ARO10.
The ARO10 (YDR380W) open reading frame was PCR amplified from CEN.PK113-7D genomic DNA using primers ARO10-fwd (GGTCTAGAATGGCACCTGTTACAATTGAAAAG) and ARO10-rev (GGCTCGAGCTATTTTTTATTTCTTTTAAGTGCCGC), designed to introduce restriction sites (underlined) for endonuclease XbaI upstream of the ATG and XhoI downstream of the stop codon, respectively. The PCR product and the vector p426GPD (31) were digested by XbaI and XhoI. The XbaI-XhoI PCR fragment was directionally cloned behind the glyceraldehyde-3-phosphate dehydrogenase promoter (TDH3p) into p426GPD, resulting in plasmid pUDe001. The ARO10 open reading frame sequence was confirmed by sequencing. The plasmid pUDe001 was transformed by the lithium acetate-single-stranded carrier DNA-polyethylene glycol method (17) into the S. cerevisiae CEN.PK 113-5D strain, resulting in strain IME003, and into strain CEN.PK 711-7C, resulting in strain IMZ002. Similarly, CEN.PK 113-5D and CEN.PK 711-7C were transformed with p426GPD (31), resulting in IME002 and IMZ001, respectively.
Chemostat cultivation.
Aerobic chemostat cultivation was performed at 30°C in 1-liter working volume laboratory fermentors (Applikon, Schiedam, The Netherlands) at a stirrer speed of 800 rpm, pH 5.0, with a dilution rate (D) of 0.10 h−1 as described previously (42), with the exception of the strains IMZ001 and IMZ002, which were grown at a dilution rate of 0.05 h−1. The pH was kept constant, using an ADI 1030 biocontroller (Applikon, Schiedam, The Netherlands), via the automatic addition of 2 M KOH. The fermentor was flushed with air at a flow rate of 0.5 liter min−1 using a Brooks 5876 mass-flow controller (Brooks Instruments, Veenendaal, The Netherlands). The dissolved-oxygen concentration was continuously monitored with an Ingold model 34 100 3002 probe (Mettler-Toledo, Greifensee, Switzerland) and was above 50% of air saturation.
Carbon-limited steady-state chemostat cultures of S. cerevisiae strains were grown as described previously (43) on synthetic medium containing 7.5 g of glucose liter−1 or 5.7 g liter−1 of ethanol, keeping molar carbon equivalence constant at 0.25 M, and either 5.0 g liter−1 (NH4)2SO4, 5.0 g liter−1 of l-phenylalanine (44), 10 g liter−1 l-leucine, 11.3 g liter−1 l-methionine, 5 g liter−1 l-asparagine, or 8.8 g liter−1 l-proline as the sole nitrogen source. The absence of (NH4)2SO4 was compensated for by the addition of equimolar amounts of K2SO4 when phenylalanine, leucine, methionine, proline, or asparagine was used as the only nitrogen source.
Culture dry weight.
Culture dry weights were determined via filtration as described previously (35).
Extracellular-metabolite analysis.
For the determination of phenylalanine, leucine, and methionine catabolism products and carbon recovery, culture supernatants and media were analyzed by high-performance liquid chromatography (HPLC), fitted with an AMINEX HPX-87H ion-exchange column (300 by 7.8 mm; Bio-Rad) mounted in a Waters Alliance 2690 HPLC apparatus, at 60°C using H2SO4 as the mobile phase with a flow rate of 0.6 ml · min−1. Metabolites were detected by a dual-wavelength absorbance detector (Waters 2487) and a refractive-index detector (Waters 2410) and integrated with Chrompack Maitre 2.5 software.
Identification of metabolites by NMR spectroscopy.
After lyophilization, samples of culture supernatants were dissolved in D2O. 1H, 1H-1H TOCSY, and 1H-13C correlation spectra (direct and long range) were measured at 300 K on a Bruker Avance 600 nuclear magnetic resonance (NMR) spectrometer equipped with an inverse triple-resonance probe and a pulse field gradient system. Quantitative 1H-NMR experiments were also performed at 600 MHz. To 0.5 ml of supernatant, an equal amount of a standard solution containing maleic acid and EDTA was added. After lyophilization, the residue was dissolved in D2O and the 1H-NMR spectrum was measured using a relaxation delay of 30 seconds, ensuring full relaxation of all the hydrogen atoms between pulses. The integrals of the characteristic resonances for each component and the internal standard (singlet at 6.1 ppm) were measured, and the contents of the individual components were calculated.
Preparation of cell extracts.
For the preparation of cell extracts, culture samples were harvested by centrifugation; washed twice with 10 mM potassium-phosphate buffer, pH 7.5, containing 2 mM EDTA; concentrated fourfold; and stored at −20°C. Before cell breakage, the samples were thawed at room temperature, washed, and resuspended in 100 mM potassium phosphate buffer, pH 7.5, containing 2 mM MgCl2 and 2 mM dithiothreitol. Extracts were prepared by sonication with 0.7 mm glass beads at 0°C for 2 min at 0.5-min intervals with an MSE sonicator (150-W output; 8-μm peak-to-peak amplitude). Unbroken cells and debris were removed by centrifugation at 4°C (20 min; 36,000 × g). The purified cell extract was used for enzyme assays.
2-Oxo acid decarboxylase assays.
2-Oxo acid decarboxylase activity was measured at 30°C immediately after preparation of cell extracts using a coupled reaction. Activity was measured by following the reduction of NAD+ at 340 nm in the presence of excess aldehyde dehydrogenase from yeast. The reaction mixtures contained, in a total volume of 1 ml, 100 mM KH2PO4/K2HPO4 buffer, pH 7.0; 2 mM NAD+; 5 mM MgCl2; 15 mM pyrazole; 0.2 mM thiamine diphosphate; 1.75 U of yeast aldehyde dehydrogenase (Sigma-Aldrich, Zwijndrecht, The Netherlands) (dissolved in 1 mM dithiothreitol); and 2 mM phenylpyruvic acid, α-ketoisocaproate, α-ketoisovalerate, α-ketomethylvalerate, 3-methylthio-α-ketobutyrate, or pyruvate to initiate the reaction. Reaction rates were linearly proportional to the amount of cell extract added.
Activity data normalization.
The per-strain normalization accounts for the difference in detection efficiency between 2-oxo acid decarboxylase activities. It also allows comparison of the relative change in activity levels, as well as displaying these levels in similar scales on the same graph. GeneSpring (Silicon Genetics, Redwood City, CA) uses the following formula to normalize to the median for each strain: (activity of strain X on substrate Y)/(median of every measurement of strain X).
Protein determination.
Protein concentrations in cell extracts were determined by the Lowry method (29). Bovine serum albumin (fatty acid free; Sigma, St. Louis, Mo.) was used as a standard.
Microarray analysis.
DNA microarray analyses were performed with the S98 Yeast GeneChip arrays from Affymetrix as previously described (34). Cells were transferred directly from chemostats into liquid nitrogen and processed according to the manufacturer's instructions (Affymetrix technical manual; Affymetrix, Santa Clara, CA.). Data analyses were performed with the Affymetrix software packages Microarray Suite v5.0, MicroDB v3.0, and Data Mining Tool v3.0. The Significance Analysis of Microarrays (SAM version 1.12) (41) add-in to Microsoft Excel was used for comparisons of replicate array experiments.
RESULTS
Measurement of phenylalanine and phenylethanol in chemostat culture of S. cerevisiae grown on various nitrogen sources.
The S. cerevisiae reference strain CEN.PK113-7D was grown on synthetic medium in aerobic, glucose-limited chemostat cultures with different nitrogen sources: ammonium sulfate, phenylalanine, leucine, methionine, proline, or asparagine. During growth on phenylalanine as the nitrogen source, HPLC analysis of culture supernatants revealed the presence of high concentrations of phenylethanol and phenylacetate, consistent with the operation of the Ehrlich pathway. Surprisingly, low but significant concentrations of these metabolites were also observed when leucine or methionine was the sole nitrogen source (Table 2). The concentrations of phenylethanol and phenylacetate in leucine- and methionine-grown cultures were 20- to 50-fold higher than in cultures grown with ammonium sulfate as the nitrogen source (Table 2). Similarly, 2-methylpropanoate and p-hydroxyphenylacetate, which are Ehrlich pathway-derived catabolites of valine and tyrosine, respectively, were also detectable when phenylalanine, leucine, or methionine was used as the sole nitrogen source. Although 3-methylbutanol, an expected product of leucine catabolism, could not be detected in our HPLC setup, the compound was detected by 1H-NMR in leucine, phenylalanine, and methionine cultures (data not shown). Conversely, none of these metabolites were detected in cultures grown with ammonium sulfate, proline, or asparagine as the nitrogen source (Table 2).
TABLE 2.
Concentrations of fusel alcohols and corresponding organic acidsa
| N source | Phenylethanol (mM) | Phenylacetate (mM) | 3-Methylbutanoate (mM) | 2-Methylpropanoate (mM) | 3-Methylthiopropanol (mM) | 3-Methylthiopropanoate (mM) | p-Hydroxyphenylacetate (mM) | p-Hydroxyphenylethanol (mM) |
|---|---|---|---|---|---|---|---|---|
| Ammonia | 0.003 ± 0.000 | 0.003 ± 0.000 | ND b | ND | ND | ND | ND | ND |
| Leucine | 0.059 ± 0.013 | 0.137 ± 0.036 | 4.538 ± 0.351 | 0.310 ± 0.000 | ND | ND | 0.135 ± 0.031 | ND |
| Methionine | 0.225 ± 0.071 | 0.180 ± 0.045 | ND | 0.204 ± 0.000 | 0.757 ± 0.309 | ND | 0.190 ± 0.017 | 0.054 ± 0.025 |
| Phenylalanine | .261 ± 0.141 | 9.915 ± 0.681 | ND | 0.161 ± 0.021 | ND | ND | 0.076 ± 0.020 | ND |
| Proline | ND | ND | ND | ND | 0.037 ± 0.007 | ND | ND | ND |
| Asparagine | ND | ND | ND | ND | 0.026 ± 0.004 | ND | ND | ND |
In aerobic, glucose-limited chemostat cultures (D = 0.10 h−1) of S. cerevisiae CEN.PK 113-7D grown with different nitrogen sources. Data are presented as average ± mean deviation of metabolite quantification from two independent chemostat cultures. 3-Methylbutanol (derived from leucine), 2-methylpropanol (derived from valine), 2-methylbutanol, and 2-methylbutanoate (derived from isoleucine) were not detected by the HPLC setup used in the present study.
ND, not detected.
These results can be explained in two different ways. First, growth on amino acids whose catabolism involves the Ehrlich pathway may lead to coordinate induction of Ehrlich pathway enzymes with different substrate specificities. Alternatively, these amino acids may induce Ehrlich pathway enzymes with broad substrate specificities. To further investigate this phenomenon, we focused on the irreversible decarboxylase reaction.
Decarboxylation of 2-oxo acids by cell extracts of wild-type S. cerevisiae grown on various nitrogen sources.
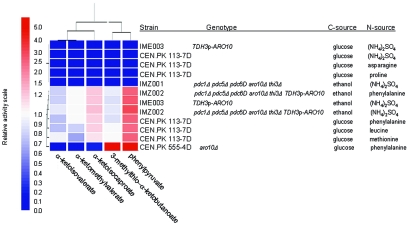
2-Oxo-acid-decarboxylase activities involved in the Ehrlich pathway were analyzed in cell extracts of S. cerevisiae CEN.PK113-7D grown in aerobic carbon-limited chemostat cultures with different amino acids as the sole nitrogen source (Table 3). Phenylpyruvate, α-ketoisovalerate, α-ketoisocaproate, α-ketomethylvalerate, and 3-methylthio-α-ketobutyrate were selected as substrates based on the observed metabolite profiles (Table 2). Significant activities with all five substrates were detected in cultures grown with leucine, methionine, or phenylalanine as the nitrogen source (Table 3). Conversely, no activity was measured in cell extracts from cultures grown on ammonium, asparagine, or proline, in good agreement with the absence of alcohols and acids in the corresponding culture supernatants (Table 2). When activities were expressed relative to the activity with phenylpyruvate, the substrate specificity did not differ markedly as a function of the nitrogen source for growth. This suggested involvement of a single common decarboxylase activity in the catabolism of leucine, methionine, and phenylalanine (Fig. 2).
TABLE 3.
Specific activities of 2-oxo acid decarboxylation by cell extractsa
| Nitrogen source | Specific decarboxylase activity [nmol · min−1 · (mg protein)−1]
|
||||
|---|---|---|---|---|---|
| Phenylpyruvate (phenylalanine) | α-Ketoisovalerate (valine) | α-Ketoisocaproate (leucine) | α-Ketomethylvalerate (isoleucine) | 3-Methylthio α-ketobutyrate (methionine) | |
| Ammonia | BDb | BD | BD | BD | BD |
| Leucine | 13.5 ± 0.7 (100) | 4 ± 0.01 (29) | 6.5 ± 0.7 (48) | 4.5 ± 0.7 (33) | 5.5 ± 0.6 (41) |
| Methionine | 22.25 ± 1.8 (100) | 8.5 ± 0.5 (38) | 9.25 ± 0.5 (42) | 5.5 ± 0.9 (25) | 9 ± 0.01 (40) |
| Phenylalanine | 67.5 ± 0.7 (100) | 19 ± 0 (28) | 29.5 ± 0.7 (43) | 25.7 ± 3.8 (38) | 22 ± 0 (32) |
| Proline | BD | BD | BD | BD | BD |
| Asparagine | BD | BD | BD | BD | BD |
Prepared from aerobic, glucose-limited chemostat cultures of S. cerevisiae CEN.PK 113-7D grown with different amino acids as the sole nitrogen source. Data are the average ± mean deviation of assays from two independent chemostat cultures. The relative 2-oxo acid activities, expressed as a percentage of phenylpyruvate activity, are in parentheses. The column headings include in parentheses the amino acid which the 2-oxo acid used as a substrate is derived from.
BD, below detection limit.
FIG. 2.
Eisen representation of relative 2-oxo acid decarboxylase activity. Cell extracts of CEN.PK113-7D, CEN.PK 555-4D, IME003, IMZ001, and IMZ002 grown in aerobic carbon-limited (glucose or ethanol) chemostat cultures with different nitrogen sources were measured for 2-oxo-acid decarboxylase activity. Each cell extract was tested for conversion of phenylpyruvate, α-ketoisovalerate, α-ketoisocaproate, α-ketomethylvalerate, and 3-methylthio-α-ketobutyrate. The activity data were normalized to the mean and clustered by hierarchical clustering using Genespring (Silicon Genetics, Redwood City, CA). The so-called normalized data were displayed on a scale from 0 to 5 (see Materials and Methods).
Transcript levels of TPP-dependent decarboxylase genes in wild-type S. cerevisiae grown on various nitrogen sources.
The pyruvate-decarboxylase genes PDC1, PDC5, and PDC6 and the related genes THI3 and ARO10 have all been implicated in the production of fusel alcohols and fusel acids by S. cerevisiae in the literature (7, 9-11, 44), but their substrate specificities and catalytic contributions remain unknown. To check whether the induction of a “broad-substrate-specificity decarboxylase activity” observed in cell extracts could be correlated with the transcriptional induction of a single gene, expression of the five decarboxylase genes was analyzed.
The levels of the ACT1 transcript, a commonly used “loading standard” for mRNA analysis (32), were the same for all six nitrogen sources (Table 4). PDC5, PDC6, and THI3 were transcribed at a constant, very low level. PDC1 showed much higher transcript levels, but they did not significantly differ for the six nitrogen sources (t test analysis at P < 0.01). Only ARO10 was differentially transcribed for the different nitrogen sources (Table 4). In cultures grown with leucine, phenylalanine, or methionine as the nitrogen source, the transcript level was at least 15-fold higher than in cultures grown with ammonium sulfate as the nitrogen source. Moreover, cultures grown with proline or asparagine as the nitrogen source yielded the same very low ARO10 transcript levels as ammonium sulfate-grown cultures (Table 4).
TABLE 4.
Transcript levels of genes with sequence similarity to thiamin-pyrophosphate-dependent decarboxylases in aerobic, glucose-limited chemostat cultures of S. cerevisiae CEN.PK113-7D grown with different amino acids as the sole nitrogen sourcea
| Nitrogen source | ACT1 | ARO10 | PDC1 | PDC5 | PDC6 | THI3 |
|---|---|---|---|---|---|---|
| Ammonia | 2,488 ± 81 | 67 ± 3 | 1,123 ± 147 | 95 ± 4 | 66 ± 4 | 92 ± 9 |
| Leucine | 2,149 ± 204 | 1,045 ± 167 | 1,311 ± 90 | 73 ± 7 | 31 ± 8 | 128 ± 11 |
| Methionine | 2,831 ± 624 | 1,335 ± 130 | 1,459 ± 226 | 87 ± 14 | 17 ± 3 | 126 ± 22 |
| Phenylalanine | 2,917 ± 575 | 1,996 ± 201 | 894 ± 319 | 87 ± 32 | 25 ± 5 | 109 ± 17 |
| Proline | 2,294 ± 127 | 37 ± 6 | 1,505 ± 173 | 81 ± 8 | 76 ± 8 | 115 ± 12 |
| Asparagine | 2,416 ± 122 | 61 ± 11 | 1,170 ± 109 | 64 ± 6 | 46 ± 8 | 105 ± 47 |
Transcript levels were determined with Affymetrix Gene Chips. Data are the average ± standard deviation of three independent chemostat cultures. The ACT1 transcript is included as a reference.
ARO10 encodes a broad-substrate-specificity 2-oxo-acid-decarboxylase in S. cerevisiae.
The transcriptional regulation of ARO10; the similar substrate specificities of decarboxylase activities in cell extracts of leucine-, methionine-, and phenylalanine-grown cultures; and the metabolite profiles in these cultures all suggested that Aro10p is responsible for a broad-substrate-specificity decarboxylase activity involved in the production of fusel alcohols and acids. To test this hypothesis, an S. cerevisiae strain lacking all five TPP-dependent decarboxylase genes (CEN-PK711-7C pdc1Δ pdc5Δ pdc6Δ thi3Δ aro10Δ ura3Δ) was constructed. The ura3 genotype was complemented by transformation either with the empty expression vector p426GPD (strain IMZ001) or with the same vector carrying ARO10 under the control of the constitutive TDH3 promoter (strain IMZ002). Strains IMZ001 and IMZ002 could not grow on glucose synthetic media as a result of the pdc1Δ pdc5Δ pdc6Δ genotype (15). Therefore, ethanol was used as a carbon source.
Cell extracts of the quintuple-deletion strain IMZ001, grown in aerobic, ethanol-limited chemostat cultures at a dilution rate of 0.05 h−1 and with ammonium sulfate as the nitrogen source, did not exhibit any decarboxylase activity (Table 5). Constitutive expression of ARO10 in this genetic background (strain IMZ002) restored decarboxylase activity with the 2-oxo acids derived from leucine, phenylalanine, and methionine. Interestingly, no activity could be measured with pyruvate as a decarboxylase substrate (Table 5). The relative specific activities with the nonpyruvate substrates were similar to those observed in cell extracts of the reference strain CEN.PK 113-7D grown with phenylalanine, leucine, or methionine as the nitrogen source (Fig. 2 and Tables 3 and 5).
TABLE 5.
Substrate specificity of the ARO10-dependent 2-oxo-acid-decarboxylase activity in S. cerevisiaea
| Substrate | Enzyme activity nmol min−1 (mg protein)−1
|
|||
|---|---|---|---|---|
| IMZ0001 [(NH4)2SO4c] | IMZ002 [(NH4)2SO4c] | IMZ002 [phenylalaninec] | Ratiob | |
| Phenylpyruvate | BDd | 61.75 ± 1.71 (100) | 270 ± 6.98 (100) | 4.37 |
| α-Ketoisovalerate | BD | 16.75 ± 2.21 (27) | 78.25 ± 4.1 (29) | 4.67 |
| α-Ketoisocaproate | BD | 25 ± 0.82 (40) | 118 ± 7.53 (44) | 4.72 |
| α-Ketomethylvalerate | BD | 21 ± 1.63 (34) | 97 ± 6.68 (36) | 4.62 |
| 3-Methylthio-α-ketobutyrate | BD | 18.5 ± 1.29 (30) | 87.7 ± 7.69 (32) | 4.74 |
| Pyruvate | BD | BD | BD | |
Strain IMZ001 is pdc1Δ pdc5Δ pdc6Δ aro10Δ thi3Δ carrying the empty expression vector p426GPD (2μ URA3 TDH3p). Strain IMZ002 is the same strain carrying the plasmid pUDe001 (2μ URA3 TDH3p-ARO10). Both strains were grown in aerobic, ethanol-limited chemostat cultures with ammonia as the nitrogen source. Enzyme activities were assayed in cell extracts. Data are the average±average deviation of the mean from assays of two independent chemostat cultures. The relative 2-oxo acid activities, expressed as a percentage of phenylpyruvate activity, are in parentheses.
Ratio of phenylalanine versus (NH4)2SO4.
N source.
BD, below detection limit.
The quintuple-deletion strain IMZ001 was unable to grow in ethanol-limited chemostat cultures (D = 0.05 h−1) when phenylalanine was the sole nitrogen source. This ability was recovered in strain IMZ002, which constitutively expresses ARO10 from the TDH3 promoter. Unexpectedly, decarboxylase activities in cell extracts of strain IMZ002 grown with phenylalanine as the nitrogen source were 4.5-fold higher than in cultures grown with ammonium sulfate as the nitrogen source (Table 5). Reintroduction of ARO10 in the quintuple-deletion strain also restored the production of fusel alcohols and acids. HPLC analysis of ethanol-limited, ammonium-grown chemostat cultures of IMZ002 revealed low but significant concentrations of phenylethanol (0.11 ± 0.01 mM) and phenylacetate (0.20 ± 0.03 mM). The concentrations of these compounds were below the HPLC detection limit (0.003 mM) in chemostat cultures of the quintuple-deletion strain IMZ001. When the ARO10-expressing strain was grown with phenylalanine as the nitrogen source, high concentrations of phenylethanol (2.69 ± 0.06 mM) and phenylacetate (7.29 ± 0.34 mM) were observed in culture supernatants. Furthermore, low concentrations of 3-methylthiopropanol (0.67 mM) and p-hydroxyphenylacetate (0.16 mM) were identified, confirming the involvement of Aro10p in the synthesis of a broad range of fusel alcohols and acids in vivo.
Overexpression of ARO10 in the isogenic reference strain CEN.PK 113-5D.
To investigate whether overexpression of ARO10 can be used to modify fusel alcohol production by wild-type S. cerevisiae strains, the expression vector carrying ARO10 under the control of the TDH3 promoter was introduced into the reference strain CEN.PK113-5D (resulting in strain IME003 [Table 1]). Surprisingly, except for pyruvate decarboxylase, no 2-oxo acid decarboxylase activity was detectable in cell extracts of this strain when it was grown in glucose-limited chemostat cultures with ammonium sulfate as the nitrogen source (Table 6). Monitoring of the ARO10 transcript level by quantitative PCR in strain IM003 grown in a glucose-limited chemostat with ammonium sulfate as the nitrogen source revealed expression of the TDH3-driven construct. The level of expression was equivalent to half of the ACT1 reference transcript signal. In the meantime, no ARO10 transcript was detected in strain IME004 grown under similar conditions. Furthermore, decarboxylase activities in glucose-limited chemostat cultures grown with phenylalanine as the nitrogen source were the same as those of the empty-vector reference strain IME004 (Table 6). When ethanol instead of glucose was used as the carbon source, the presence of the ARO10 expression vector did result in increased decarboxylase activities relative to an empty-vector reference strain (Table 6). These results contradict the simple view that ARO10 encodes a fully functional decarboxylase whose expression is primarily regulated at the level of transcription.
TABLE 6.
Regulation of decarboxylase activities in the reference S. cerevisiae strain IME004 (CEN.PK113-5D, p426GPD) and in an isogenic strain expressing a multicopy plasmid-borne ARO10 gene from a constitutive TDH3 promoter, strain IME003a
| Substrate for decarboxylase assay | IME004 [ura3 p426GPD (URA3)]
|
IME003 [ura3 pUDe001 (URA3 TDH3p-ARO10)]
|
||||||
|---|---|---|---|---|---|---|---|---|
| Ethanol
|
Glucose
|
Ethanol
|
Glucose
|
|||||
| (NH4)2SO4 | Phenylalanine | (NH4)2SO4 | Phenylalanine | (NH4)2SO4 | Phenylalanine | (NH4)2SO4 | Phenylalanine | |
| Phenylpyruvate | BDb | 64 ± 1.2 | BD | 67.5 ± 0.7 | 35 ± 5.7 | 76.5 ± 1.5 | BD | 82.75 ± 26.73 |
| α-Ketoisovalerate | BD | 17 ± 2.2 | BD | 19 ± 0 | 19 ± 0 | 21.75 ± 1.7 | BD | 22.4 ± 6.4 |
| α-Ketoisocaproate | BD | 28 ± 3.1 | BD | 29.5 ± 0.7 | 15 ± 1.4 | 30.5 ± 3 | BD | 31.5 ± 9.8 |
| α-Ketomethylvalerate | BD | 23 ± 0.7 | BD | 25.7 ± 3.8 | 12.5 ± 0.7 | 28 ± 4.0 | BD | 28.3 ± 13.6 |
| 3-Methylthio-α-ketobutyrate | BD | 20.5 ± 1.2 | BD | 22 ± 0 | 11 ± 0 | 23.5 ± 1.2 | BD | 24.25 ± 9.1 |
Both strains were grown in aerobic, carbon-limited chemostat cultures with glucose or ethanol as a carbon source and ammonia or phenylalanine as a carbon source and ammonia or phenylalanine as a nitrogen source. Enzyme activities were assayed in cell extracts of independent duplicate cultures and are expressed as nmol·min−1·(mg protein)−1. Data are presented as average ± average deviation of the mean of two chemostat cultures.
BD, below detection limit.
DISCUSSION
Formation of fusel alcohols by S. cerevisiae.
In brewery and wine fermentations, S. cerevisiae is responsible for the production of a variety of metabolites that contribute to flavor and aroma. Among the volatile flavor compounds, an important class consists of higher alcohols that are less volatile than ethanol (18). These higher alcohols are derived from the carbon skeletons of amino acids, which can in theory be synthesized de novo but in brewery and wine fermentations are generally taken up from the wort or grape must. It is commonly accepted that branched-chain (9-11) and aromatic (7, 44) amino acid-derived alcohols originate from the Ehrlich pathway (12) (Fig. 1). Our results support the notion that this pathway is also involved in the production of 3-methylthiopropanol (methionol) and 3-methylthiopropanoate from methionine. These sulfur-containing compounds are relevant to the production of alcoholic beverages. Methionol, which has a raw-potato odor, is commonly measured in wine and is known to negatively affect white wine and red wine aroma above 0.6 mg/liter and 2 to 3 mg/liter, respectively (3) (Table 1).
Our results indicated that induction of an Ehrlich pathway for catabolism of one amino acid leads to the formation of significant amounts of fusel alcohols and acids from other amino acids. This suggested that conversion of branched-chain, aromatic, and sulfur-containing amino acids to the corresponding fusel alcohols and acids, via an Ehrlich pathway, involves common broad-substrate-specificity enzyme activities. Furthermore, as our experiments were performed with synthetic media to which only single amino acids were added, these results indicated that the decarboxylase activity involved in the Ehrlich pathway could compete for 2-oxo acids with the transaminases involved in de novo amino acid biosynthesis. The chemostat conditions used in this study were designed to reveal the molecular nature of the decarboxylase step of the Ehrlich pathway. Although these conditions are different from typical alcoholic fermentation processes, the conclusion drawn about the role of ARO10 is relevant for interpreting the patterns of flavor production in wine and beer fermentation.
Aro10p is involved in a broad-substrate-specificity Ehrlich pathway decarboxylase activity.
Transcript analysis demonstrated that the induction of Ehrlich pathway activity by the amino acids leucine, phenylalanine, and methionine coincided with the transcriptional up-regulation of ARO10, but not with that of the other four genes encoding (putative) thiamine pyrophosphate-dependent decarboxylases. Indeed, overexpression of ARO10 in a strain in which the five chromosomal decarboxylase genes had been deleted was sufficient to restore a broad-substrate-specificity decarboxylase activity. The substrate specificity profile of this strictly ARO10-dependent activity was the same as those of the activities induced by leucine, phenylalanine, and methionine in wild-type cells.
Previous research in S. cerevisiae with aro10 null mutants has indicated the presence of an ARO10-independent decarboxylase activity (44). This alternative activity has been reported to require the simultaneous expression of at least one of the three pyruvate decarboxylase genes (PDC 1, -5, and -6) and the putative decarboxylase gene THI3. Based on previous work by Dickinson and coworkers, the last gene has also been implicated in the decarboxylation of the 2-oxo acids derived mainly from leucine (11) and to a lesser extent isoleucine (10). This ARO10-independent 2-oxo acid decarboxylase activity exhibited a completely different substrate specificity profile. In particular, the decarboxylase activity observed in cultures of an aro10 null mutant grown with phenylalanine as the nitrogen source showed no activity with α-ketoisovalerate and α-ketomethylvalerate as the substrate (Fig. 2, strain CEN.PK 555-4D).
The results described here support the notion that the ARO10-dependent, broad-substrate-specificity decarboxylase is primarily responsible for the Ehrlich pathway decarboxylation reaction in wild-type S. cerevisiae. The molecular basis and substrate specificity of the ARO10-independent activity that is detected in aro10 null mutants (44) (Table 5), as well as its possible involvement in (off-)flavor production by wild-type strains, require further research.
Transcriptional regulation of ARO10.
Previous work has shown that transcription of ARO10 is induced by tryptophan (24) and phenylalanine (44) and is dependent on the transcriptional regulator Aro80p (24). Other studies (7, 9-11) suggested that expression of ARO10 might also be up-regulated by valine and isoleucine, based on metabolite profiling; however, this conclusion was not backed up by expression analysis. Our results clearly show that ARO10 expression was strongly up-regulated in the presence of leucine and methionine (Table 3), consistent with its proposed role as a broad-substrate-specificity decarboxylase.
Further analysis of the transcriptome data revealed that ARO9 (aromatic amino transferase II) (23) was coexpressed with ARO10 in cultures grown with different nitrogen sources (data not shown). This suggested that the transaminase activity of Aro9p might not be restricted to aromatic amino acids (24) but, similar to the Aro10p-dependent decarboxylase activity, might have a broad substrate specificity. It remains to be investigated whether and to what extent Aro80p is involved in the transcriptional up-regulation of ARO9 and ARO10 by the nonaromatic amino acids leucine and methionine. This question could not be resolved by the microarray analyses, since ARO80 transcript levels were extremely low and did not differ significantly for the nitrogen sources studied (data not shown). Further research with aro80 null strains is required to investigate whether the regulatory role of Aro80p extends beyond aromatic amino acid metabolism expression control or, alternatively, another regulatory protein or proteins control the upregulation of ARO9 and ARO10 in leucine- and methionine-grown cultures. A comprehensive discussion of the genomewide transcriptional responses of S. cerevisiae to the six nitrogen sources used in this study will be published elsewhere (V. M. Boer, S. L. Tai, Z. Vuralhan, Y. Afrifin, M. C. Walsh, M. D. W. Piper, J. H. de Winde, J.-M. Daran, and J. T. Pronk, unpublished data).
Involvement of other factors in the activity and regulation of Aro10p.
Earlier works (7, 9-11, 15, 40, 44) on the decarboxylation of branched-chain and aromatic 2-oxo acid decarboxylation were based on the implicit assumption that single proteins (e.g., Aro10p and/or Thi3p) would act as thiamine pyrophosphate-dependent decarboxylase enzymes. While the present study proves that Aro10p plays a key role in broad-substrate-specificity decarboxylase activity, it also provides several clear indications that additional factors are involved in this activity and its regulation.
Our attempt to overexpress ARO10 under the control of the TDH3 promoter in order to uncouple its expression from environmental parameters, such as the presence of phenylalanine, yielded unexpected results. In cultures grown with ammonium sulfate as the nitrogen source, the TDH3p-ARO10 construct yielded activity in ethanol-grown cultures but, surprisingly, not when glucose was the carbon source. This unexpected dependency on the carbon source was independent of the expression of the other four decarboxylase genes. As the TDH3 promoter is known to give very high transcript levels in glucose- as well as ethanol-grown cultures, this observation suggests that transcription of the ARO10 gene is not sufficient to yield an active broad-substrate-specificity decarboxylase activity. Furthermore, in ethanol-grown cultures of the “ARO10 constitutive” strains, addition of phenylalanine to culture media caused a strong increase in the broad-substrate-specificity decarboxylase activities in cell extracts.
These observations may indicate that the functional expression of the ARO10 gene is regulated at a posttranscriptional level in a carbon and nitrogen source-dependent manner. Alternatively, the catalytic activity and/or stability of Aro10p may require the presence of one or more additional proteins whose expression is carbon and nitrogen source dependent.
Recent protein interactome studies based on the two-hybrid approach (25) identified two potential Aro10p interaction partners. Fit2p is possibly involved in iron uptake (33, 36), and Ena5p is an ATP-driven sodium transporter, a member of the Na+-transporting ATPase family in the superfamily of P-type ATPases (5, 26). Taking into account the subcellular localization of Fit2p and Ena5p (the cell wall and plasma membrane, respectively), it is difficult to envision them as key factors in controlling the activity or stability of Aro10p. Of these two putative partners, only FIT2 would show an expression profile that would corroborate our assumption (data not shown).
The present study has clearly established the importance of Aro10p in the key decarboxylation step of the Ehrlich pathway. At the same time, it has raised new and important questions about the additional factors involved in the molecular composition, posttranscriptional regulation, and/or stability of the Aro10p-dependent decarboxylase activity. These questions need to be resolved before strategies can be devised to rationally modify the production of volatile flavor compounds by S. cerevisiae in beverages and fine-chemical production, e.g., via genetic modification of in vivo decarboxylase activity. Purification and characterization of the broad-substrate-specificity decarboxylase from cell extracts is likely to be essential to resolve the outstanding issues.
Acknowledgments
This work was financially supported by the Board of the Delft University of Technology (“Beloning Excellent Onderzoek” Program), the Dutch government (CW-NOW program “Transition towards Sustainable Technology”), and the Kluyver Center for Genomics of Industrial Fermentation. Z.V. and V.M.B. were financially supported by DSM.
We thank Pascale Daran-Lapujade and the DSM discussion group for helpful comments on the manuscript and Matthew Piper for his contribution in the initial phase of the project.
REFERENCES
- 1.Barnett, J. A. 1992. Some controls on oligosaccharide utilization by yeasts: The physiological basis of the Kluyver effect. FEMS Microbiol. Lett. 100:371-378. [DOI] [PubMed] [Google Scholar]
- 2.Barnett, J. A., R. W. Payne, and D. Yarrow. 1983. Yeasts: characteristics and identification, 3rd ed. Cambridge University Press, Cambridge, United Kingdom.
- 3.Baumes, R., R. Cordonnier, S. Nitz, and F. Drawert. 1986. Identification and determination of volatile constituents in wine from different vine cultivars. J. Sci. Food Agric. 37:927-943. [Google Scholar]
- 4.Boer, V. M., J. H. de Winde, J. T. Pronk, and M. D. Piper. 2003. The genome-wide transcriptional responses of Saccharomyces cerevisiae grown on glucose in aerobic chemostat cultures limited for carbon, nitrogen, phosphorus, or sulfur. J. Biol. Chem. 278:3265-3274. [DOI] [PubMed] [Google Scholar]
- 5.Catty, P., D. A. de Kerchove, and A. Goffeau. 1997. The complete inventory of the yeast Saccharomyces cerevisiae P-type transport ATPases. FEBS Lett. 409:325-332. [DOI] [PubMed] [Google Scholar]
- 6.Clark, G. S. 1990. Phenylethyl alcohol. Perfumer Flavorist 15:37-44. [Google Scholar]
- 7.Dickinson, J. R., L. E. J. Salgado, and J. E. Hewlins. 2003. The catabolism of amino acids to long chain and complex alcohols in Saccharomyces cerevisiae. J. Biol. Chem. 278:8028-8034. [DOI] [PubMed] [Google Scholar]
- 8.Dickinson, J. R., and I. W. Dawes. 1992. The catabolism of branched-chain amino acids occurs via 2-oxoacid dehydrogenase in Saccharomyces cerevisiae. J. Gen. Microbiol. 138:2029-2033. [DOI] [PubMed] [Google Scholar]
- 9.Dickinson, J. R., S. J. Harrison, and M. J. E. Hewlins. 1998. An investigation of the metabolism of valine to isobutyl alcohol in Saccharomyces cerevisiae. J. Biol. Chem. 273:25751-25756. [DOI] [PubMed] [Google Scholar]
- 10.Dickinson, J. R., S. J. Harrison, J. A. Dickinson, and J. E. Hewlins. 2000. An investigation of the metabolism of isoleucine to active amyl alcohol in Saccharomyces cerevisiae. J. Biol. Chem. 275:10937-10942. [DOI] [PubMed] [Google Scholar]
- 11.Dickinson, J. R., M. Lanterman, D. Danner, B. Pearson, P. Sanz, S. Harrison, and M. Hewlins. 1997. A 13C nuclear magnetic resonance investigation of the metabolism of leucine to isoamyl alcohol in Saccharomyces cerevisiae. J. Biol. Chem. 272:26871-26878. [DOI] [PubMed] [Google Scholar]
- 12.Ehrlich, F. 1907. Uber die Bedingungen der Fuselolbildung and uber ihren Zusammenhang mit dem Eiweissaufbau der Hefe. Ber. Dtsch. Chem. Ges. 40:1027-1047. [Google Scholar]
- 13.Enjo, F., K. Nosaka, M. Ogata, A. Iwashima, and H. Nishimura. 1997. Isolation and characterization of a thiamin transport gene, THI10, from Saccharomyces cerevisiae. J. Biol. Chem. 272:19165-19170. [DOI] [PubMed] [Google Scholar]
- 14.Fauchon, M., G. Lagniel, J. C. Aude, L. Lombardia, P. Soularue, C. Petat, G. Marguerie, A. Sentenac, M. Werner, and J. Labarre. 2002. Sulfur sparing in the yeast proteome in response to sulfur demand. Mol. Cell 9:713-723. [DOI] [PubMed] [Google Scholar]
- 15.Flikweert, M. T., L. van der Zanden, W. M. Janssen, H. Y. Steensma, J. P. Van Dijken, and J. T. Pronk. 1996. Pyruvate decarboxylase: an indispensable enzyme for growth of Saccharomyces cerevisiae on glucose. Yeast 12:247-257. [DOI] [PubMed] [Google Scholar]
- 16.Furia, T. E., and N. Bellanca. 1971. Fenaroli's handbook of flavor ingredients, synthetic flavors. CRC Press, Cleveland, Ohio.
- 17.Gietz, R. D., and R. A. Woods. 2002. Transformation of yeast by the Liac/SS carrier DNA/PEG method. Methods Enzymol. 350:87-96. [DOI] [PubMed] [Google Scholar]
- 18.Harrison, G. A. F. 1970. The flavor of beer: a review. J. Inst. Brew. 76:486-495. [Google Scholar]
- 19.Hohmann, S. 1991. PDC6, a weakly expressed pyruvate decarboxylase gene from yeast, is activated when fused spontaneously under the control of the PDC1 promotor. Curr. Genet. 20:373-378. [DOI] [PubMed] [Google Scholar]
- 20.Hohmann, S., and H. Cederberg. 1990. Autoregulation may control the expression of yeast pyruvate decarboxylase structural genes PDC1 and PDC5. Eur. J. Biochem. 188:615-621. [DOI] [PubMed] [Google Scholar]
- 21.Hohmann, S., and P. A. Meacock. 1998. Thiamin metabolism and thiamin diphosphate-dependent enzymes in the yeast Saccharomyces cerevisiae: genetic regulation. Biochim. Biophys. Acta 1385:201-219. [DOI] [PubMed] [Google Scholar]
- 22.Holm, C., D. W. Meeks-Wagner, W. L. Fangman, and D. Botstein. 1986. A rapid, efficient method for isolating DNA from yeast. Gene 42:169-173. [DOI] [PubMed] [Google Scholar]
- 23.Iraqui, I., S. Vissers, M. Cartiaux, and A. Urrestarazu. 1998. Characterisation of Saccharomyces cerevisiae ARO8 and ARO9 genes encoding aromatic aminotransferases I and II reveals a new aminotransferase subfamily. Mol. Gen. Genet. 257:238-248. [DOI] [PubMed] [Google Scholar]
- 24.Iraqui, I., S. Vissers, B. Andre, and A. Urrestarazu. 1999. Transcriptional induction by aromatic amino acids in S. cerevisiae. Mol. Cell. Biol. 19:3360-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ito, T., T. Chiba, R. Ozawa, M. Yoshida, M. Hattori, and Y. Sakaki. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 98:4569-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jesus Ferreira, M. C., X. Bao, V. Laize, and S. Hohmann. 2001. Transposon mutagenesis reveals novel loci affecting tolerance to salt stress and growth at low temperature. Curr. Genet. 40:27-39. [DOI] [PubMed] [Google Scholar]
- 27.Kellermann, E., P. G. Seeboth, and C. P. Hollenberg. 1986. Analysis of the primary structure and promoter function of a pyruvate decarboxylase gene (PDC1) from Saccharomyces cerevisiae. Nucleic Acids Res. 14:8963-8977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Large, P. J. 1986. Degradation of organic nitrogen compounds by yeasts. Yeast 2:1-34. [Google Scholar]
- 29.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randell. 1951. Protein measurement with the folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 30.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 31.Mumberg, D., R. Muller, and M. Funk. 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156:119-122. [DOI] [PubMed] [Google Scholar]
- 32.Ng, R., and J. Abelson. 1980. Isolation and sequence of the gene for actin in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 77:3912-3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Philpott, C. C., O. Protchenko, Y. W. Kim, Y. Boretsky, and M. Shakoury-Elizeh. 2002. The response to iron deprivation in Saccharomyces cerevisiae: expression of siderophore-based systems of iron uptake. Biochem. Soc. Trans. 30:698-702. [DOI] [PubMed] [Google Scholar]
- 34.Piper, M. D., P. Daran-Lapujade, C. Bro, B. Regenberg, S. Knudsen, J. Nielsen, and J. T. Pronk. 2002. Reproducibility of oligonucleotide microarray transcriptome analyses. An interlaboratory comparison using chemostat cultures of Saccharomyces cerevisiae. J. Biol. Chem. 277:37001-37008. [DOI] [PubMed] [Google Scholar]
- 35.Postma, E., C. Verduyn, W. A. Scheffers, and J. P. Van Dijken. 1989. Enzymic analysis of the crabtree effect in glucose-limited chemostat cultures of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 55:468-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Protchenko, O., T. Ferea, J. Rashford, J. Tiedeman, P. O. Brown, D. Botstein, and C. C. Philpott. 2001. Three cell wall mannoproteins facilitate the uptake of iron in Saccharomyces cerevisiae. J. Biol. Chem. 276:49244-49250. [DOI] [PubMed] [Google Scholar]
- 37.Sentheshanmuganathan, S. 1960. The mechanism of the formation of higher alcohols from amino acids by Saccharomyces cerevisiae. Biochem. J. 74:568-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stark, D., D. Zala, T. Munch, B. Sonnleitner, I. W. Marison, and U. Von Stockar. 2003. Inhibition aspects of the bioconversion of phenylalanine to 2-phenylethanol by Saccharomyces cerevisiae. Enzyme Microbiol. Technol. 32:212-223. [Google Scholar]
- 39.Stark, D., T. Munch, B. Sonnleitner, I. W. Marison, and U. Von Stockar. 2002. Extractive bioconversion of 2-phenylethanol from l-phenylalanine by Saccharomyces cerevisiae. Biotechnol. Prog. 18:514-523. [DOI] [PubMed] [Google Scholar]
- 40.Ter Schure, E. G., M. Flikweert, J. P. van Dijken, J. T. Pronk, and C. T. Verrips. 1998. Pyruvate decarboxylase catalyzes decarboxylation of branched-chain 2-oxo acids but is not essential for fusel alcohol production by Saccharomyces cerevisiae. Appl. Environ. Microbiol. 64:1303-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van den Berg, M. A., P. de Jong-Gubbels, C. J. Kortland, J. P. Van Dijken, J. T. Pronk, and H. Y. Steensma. 1996. The two acetyl-coenzyme A synthetases of Saccharomyces cerevisiae differ with respect to kinetic properties and transcriptional regulation. J. Biol. Chem. 271:28953-28959. [DOI] [PubMed] [Google Scholar]
- 43.Verduyn, C., E. Postma, W. A. Scheffers, and J. P. Van Dijken. 1990. Physiology of Saccharomyces cerevisiae in anaerobic glucose-limited chemostat cultures. J. Gen. Microbiol. 136:395-403. [DOI] [PubMed] [Google Scholar]
- 44.Vuralhan, Z., M. A. Morais, S. L. Tai, M. D. Piper, and J. T. Pronk. 2003. Identification and characterization of phenylpyruvate decarboxylase genes in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 69:4534-4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Welsh, F. W., W. D. Murray, and R. E. Williams. 1989. Microbiological and enzymatic production of flavor and fragrance chemicals. Crit. Rev. Biotechnol. 8:105-169. [Google Scholar]