Abstract
Deep learning-based image analysis offers great potential in clinical practice. However, it faces mainly two challenges: scarcity of large-scale annotated clinical data for training and susceptibility to adversarial data in inference. As an example, an artificial intelligence (AI) system could check patient positioning, by segmenting and evaluating relative positions of anatomical structures in medical images. Nevertheless, data to train such AI system might be highly imbalanced with mostly well-positioned images being available. Thus, we propose the use of synthetic X-ray images and annotation masks forward projected from 3D photon-counting CT volumes to create realistic non-optimally positioned X-ray images for training. An open-source model (TotalSegmentator) was used to annotate the clavicles in 3D CT volumes. We evaluated model robustness with respect to the internal (simulated) patient rotation on real-data-trained models and real&synthetic-data-trained models. Our results showed that real&synthetic- data-trained models have Dice score percentage improvements of 3% to 15% across different groups compared to the real-data-trained model. Therefore, we demonstrated that synthetic data could be supplementary used to train and enrich heavily underrepresented conditions to increase model robustness.
Keywords: Robustness, Adversarial Training, Synthetic X-ray, Computed Tomography, Segmentation
Subject terms: Radiography, Biomedical engineering
Introduction
In clinical imaging examinations, proper patient positioning is crucial for accurately capturing anatomical structures. This can aid in medical image analysis1, longitudinal disease monitoring, radiotherapy planning2, thereby enhancing diagnostic confidence. To ensure correct patient positioning, an automatic positioning analysis system could be developed to support clinicians and technical assistants, so to optimize patient management by reducing the time or need for retakes. There are several quality criteria for patient positioning in chest X-rays (CXRs), such as the clavicle-spine distance and the visibility of certain thoracic anatomical landmarks3. A segmentation approach could identify the key anatomical structures in the images, allowing for the computation of quality metrics like distances and overlaps between anatomies.
Deep learning has emerged as a powerful tool for medical image analysis, ranging from segmentation, disease detection to report writing. It relies on large quantities of annotated images and could be the basis for the aforementioned segmentation task. However, obtaining large-scale annotated clinical CXRs, especially those with non-optimal positioning, is challenging. In addition, the robustness of current segmentation models in adversarial-positioned CXR is unknown. In realm of this chicken-and-egg dilemma, we first explore the robustness current segmentation models, and then supplementary train the models with adversarial CXR to improve robustness.
In order to reduce vulnerability to adversarial attacks when deploying deep learning models in real-world applications, robustness certification and adversarial training have been studied over the years to enhance model resilience. Robustness is the degree that a model’s performance changes in the presence of perturbations or uncertainties. Some studies certified robustness by using an abstract domain such as Zonotope to capture the effect of affine transformations inside neural networks4,5. On the other hand, various methods have been suggested to generate adversarial perturbations with respect to the input and learned features6–9, including fast gradient sign method10, DeepFool11, saliency map attacks12, expectation over transformation13 and curriculum adversarial training14. Studies have been conducted that apply the adversarial pertubations in natural red-blue-green images15 and skin lesion classification in medical imaging16,17. Besides classification, robustness benchmarking was also demonstrated by crafting adversarial examples using fast gradient sign method, DeepFool and saliency map attacks on whole brain segmentation18.
There is an emerging usage of realistic synthetic data for machine learning in medicine19–22, as curation of large-scale annotated clinical data is challenging due to scarcity or ethical issues, especially adversarial data. Synthetic image generation was studied in a range of imaging modalities including pathological images on skin lesions23,24, retinal images25, and in generation of synthetic CT images from MR images26–29. Particularly in X-ray imaging, synthetic X-rays (also known as digitally reconstructed radiographs, DRR)30 can be also generated from 3D CT volumes by analytic forward projection or GANs. Only a few studies have been carried out for using synthetic X-rays as training, for example to detect lung lesions31 or to quantify patient rotation32. A CNN trained with synthetic X-rays from CT volumes to quantify airspaces achieved an accuracy on the level of radiologists for a COVID lesion segmentation task33. Gao et al.34 used synthetic X-rays for lesion segmentation, landmark detection and surgical tool detection tasks, and their ground truth annotations were obtained by automatic segmentation or forward kinematics.
We propose to generate synthetic X-ray images from 3D CT volumes also for the use case to generate large amount of normal and adversarial x-ray images and in-image ground truth annotations systematically at the same time. We used a state-of-the-art CT segmentation tool TotalSegmentator35 to obtain ground truth for left and right clavicles in 3D CT domain. Both 3D image and annotations were forward projected to 2D X-ray domain and are characterized by non-optimal patient positioning. We trained clavicle segmentation models using real data and additionally with synthetic data for robustness evaluation. A model is rated robust if the Dice scores are consistent to slight changes in adversarial features, and in this study, we evaluated the Dice scores across different projection angles. We further evaluated the performance of open available CXR segmentation model TorchXRayVision36 as baseline comparison.
Our contribution in this paper is threefold: We first demonstrated the generation of synthetic CXR and their corresponding segmentations from CT volumes. Subsequently, we explored two distinct applications of synthetic images: as a mean to test and interpret model performance on adversarial data, and to augment an existing training dataset with synthetic adversarial cases, thereby enhancing model performance and robustness. Moreover, we used the open source models TotalSegmentator and TorchXRayVision to enable a more reproducible research.
Methods
Overview
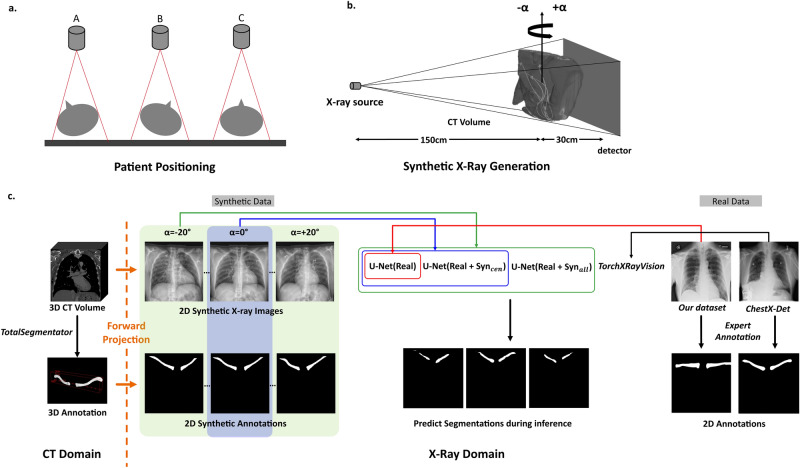
Figure 1 shows our concept and X-ray simulation. It shows the forward projection setup for generating synthetic X-ray images (Figure 1a). Figure 1b illustrates how rotated (adversarial) and non-rotated (normal) data are position by rotating patient volume along y-axis. Figure 1c shows the pipeline from synthetic X-ray images and annotations to adversarial robustness measurement.
Fig. 1.
(a) Patient positioning possibilities in clinical X-Ray examination, A and B represent adversarial positioning and C represents correct positioning; (b) Simulation setup for generating rotated and non-rotated synthetic X-rays with an angle in the range of [, ]; (c) Our proposed workflow: In the CT domain, we generated ground truth segmentation masks using TotalSegmentator. Paired CT image and segmentation volumes were forward projected to X-ray domain as normal and adversarial data for training and testing, and to quantify robustness in segmentation models TorchXRayVision, U-Net(Real), U-Net() and U-Net().
Generation of synthetic X-ray images and ground truth masks from computed tomography scans
A total of 116 photon-counting CT (NAEOTOM Alpha, Siemens Healthineers AG, Forchheim, Germany) datasets from individual patients were used to generate synthetic X-ray images. Each CT volume has a voxel size of 0.50.50.7, and 1000 slices. Each CT volume underwent forward projection by ray tracing based on a cone-beam geometry. Virtual X-ray beams traverse the CT volume, and interacts with internal anatomies as attenuation and scattering. By accumulating the attenuation values along these paths, a simulated X-ray image is generated, representing the intensity of X-ray transmission through the volume from various angles37. With parameters similar to clinical chest X-ray examinations38, the X-ray source-to-patient distance is 150 cm, patient-to-detector distance is 30 cm, and the simulated detector has a matrix of pixels. To simulate the adversarial patient positioning, projection parameters were set such that X-ray source is rotated along the y-axis with angle in range of [, ], with a step size of and the central projection at (Figure 1b). Furthermore, standard radiographic image post-processing was applied to images.
To generate the synthetic ground truth annotations, each photon-counting CT dataset was segmented by the open-source deep learning segmentation toolbox TotalSegmentator35 (version 2.0.5). TotalSegmentator could segment 104 anatomic structures in CT images and was trained on nnUNet39 using 1204 patient datasets. Python-API was used to call TotalSegmentator and ’roi-subset’ option was used to segment left and right clavicles separately. The resulting left and right volumes were combined such that each photon-counting CT has a corresponding 3D segmentation volume of clavicles (Fig. 1c). The annotations were then forward projected using the same setup and parameters as in their image domain, and binarized to obtain the segmentation mask.
Datasets and training
TorchXRayVision36 is a Python library that contains chest X-ray datasets and models including disease classification and segmentation. The segmentation model in TorchXRayVision was trained by the ChestX-Det40, which is a subset of the NIH ChestX-14 dataset. ChestX-Det contains 3575 images with segmentation annotations for chest anatomies including clavicles and lung, which were annotated by three board certified radiologists. The chest segmentation model in TorchXRayVision used a pretrained PSPNet41 and was trained with 3575 images. Instead of only using only an internal model, we included TorchXRayVision model as a baseline and to enable reproducible research.
For our own segmentation model, the real X-ray dataset consists of 3434 CXR images, with clavicle masks annotated manually by experts. 350 real CXR images were randomly separated for testing. A total of 3434 images were used for training (train: 3134, valid: 387). From the 116 photon-counting CT examinations, we generated 21 synthetic X-ray images for each dataset, yielding a total of 2436 synthetic X-ray images and the corresponding clavicle annotations. Subject-specific splitting were performed to randomly select 96 datasets (2016 images) for training and 20 datasets (420 images) for testing. With an addition of 288 well-positioned synthetic X-ray images, U-Net() has a total of 3731 training images. U-Net() uses 2016 synthetic well- and adversarial- positioned X-ray images, resulting in a total of 5450 training images. Table 1 provides detailed information about the dataset and the models. Same image preprocessing steps were applied to real and synthetic X-ray images before training, which include resize and normalization. Both real and synthetic X-ray and respective clavicle masks were resized to an image dimension of by bilinear interpolation and zero padding. Subsequently, the pixel values in real and synthetic images were normalized into the range of [0, 1]. U-Net42 was used to train our models, Dice loss and Adam optimizer with a learning rate of 0.01 were used and early stopping was applied when the model did not improve in the last 30 epochs.
Table 1.
Dataset distribution for public and self-trained models.
| Training | Testing | ||||
|---|---|---|---|---|---|
| Real | Synthetic | Total | Real | Synthetic | |
| TorchXRayVision | 3575 | 0 | 3575 | 350 | 420 |
| U-Net(Real) | 3434 | 0 | 3434 | ||
| U-Net(Real+Syn_cen) | 3434 | 288 | 3731 | ||
| U-Net(Real+Syn_all) | 3434 | 2016 | 5450 | ||
Robustness evaluation
An ablation study was used to select the optimal network hyperparameters. By varying the batch sizes, depth of U-Net and loss functions, the Dice scores were evaluated. For the loss function in the network, Dice similarity coefficient43 resulted as Dice loss44 calculation:
| 1 |
where y indicates the ground truth, indicates the predicted segmentation, and is used to avoid division by 0 so to ensure loss function stability. Multiplication of y and indicate the intersected region of ground truth and predicted segmentation. Whereas the Dice with binary cross entropy (BCE) loss is defined as:
| 2 |
| 3 |
both Dice and DiceBCE loss functions were evaluated in the ablation study. The real and synthetic X-ray images were evaluated on TorchXRayVision, images were resized to and normalized to [-1024, 1024] as in their pipeline. The first two classes in the output predictions among 14 classes are left and right clavicles respectively, and combined to form the clavicle prediction. Both real and synthetic X-ray images were evaluated on our self-trained models U-Net(Real), U-Net() and U-Net(). For each model’s output, the prediction was defined as the true class when the predicted probability is >0.5.
Robustness was shown as Dice score in a boxplot for 20 randomly selected patients across 21 projections, resulting 420 images. For simpler representation, internal patient rotation values are divided into five groups. Well-positioned X-ray images denoted as group “Center” ( to ). Adversarial images are categorized into “Moderate Negative” ( to ), “Low Negative” ( to ), “Low Positive” ( to ), and “Moderate Positive” ( to ). Interquartile range (IQR) is represented by the upper and lower box edges which indicates 75th percentile (or third quartile, Q3) and 25th percentile (or first quartile, Q1) respectively. The whiskers extend to the farthest data point lying within 1.5 IQR from the box, with upper whiskers , while lower whiskers . Statistical analysis on the mean Dice score and standard deviation for four models were evaluated on synthetic test images, and real test images as a baseline. To further demonstrate the Dice changes for patients at different angles, we performed a box plot comparing U-Net(Real) with U-Net() or U-Net() on angles , , , and .
Furthermore, the distance metrics Hausdorff Distance (HD) and the 95th percentile of Hausdorff Distance (HD95) are also evaluated. The ground truth Y and predicted segmentation could be represented by respective point sets and . It is a measure of the distance between two subsets of a metric space. With is the Euclidean distance between y and , the Hausdorff Distance of Y to is defined as:
| 4 |
Instead of the maximum of the nearest point between Y and , HD95 is defined as 95th percentile of the distances. This can reduce the sensitivity to outliers and provide a more robust comparison.
Results
Performance of real data-trained models
Figure 2 shows the robustness analysis of models trained with real X-ray images as a boxplot, robustness for TorchXRayVision is black and U-Net(Real) is red in color. Internal patient rotation values are divided into five groups. Well-positioned X-ray images are grouped as center. The boxplot shows a clear, symmetrically distributed trend across the five groups of values, delineated by a curve-shaped progression in terms of central tendency measures and quartile ranges. Highest median values are observed in central values for both TorchXRayVision and U-Net(Real) model, as depicted by the line within the box. Mild negative and positive demonstrate successively lower median, while moderate negative and positive exhibit the lowest median values. A similar pattern is also observed in Table 2 which shows the Dice score, HD and HD95 in mean and standard deviation. Interquartile range (IQR) is represented by the upper and lower box edges which indicates 75th and 25th percentile respectively. IQR are large in moderate negative and positive values, while narrowing towards the central values. This pattern suggests a decrease in variability towards X-rays positioned closer to the center. This pattern is further supported by the whiskers, which extend to the farthest data point lying within 1.5 IQR from the box. The upper and lower whiskers for both models exhibit a consistent decrease from the central towards the outer values. Notably, the moderate negative and positive values exhibit large distance in their lower whiskers. This highlighted the increase in model performance variability at higher values. Overall, the observed curve-shaped pattern in median and quartile range, coupled with the increasing distances of the whiskers, underscores the decrease in robustness of models in adversarial data.
Fig. 2.

Robustness as Dice score for real data-trained segmentation models in boxplot on well- and adversarial- positioned data. TorchXRayVision is black and U-Net(Real) is red in color. Internal patient rotation values are divided into five groups. Well-positioned X-ray images are grouped as center. While adversarial data indicated by Moderate Negative (-), Low Negative (-), Low Positive (+), and Moderate Positive (+). The ranges includes values with a step size of . Ie. center represent values of , , and .
Table 2.
Robustness of four models. Dice score, Hausdorff Distance (HD) and 95th percentile of HD (HD95) are measured between the ground truth and predicted segmentation by four models on synthetic data.
| Moderate - | Low - | No | Low + | Moderate + | ||
|---|---|---|---|---|---|---|
| TorchXRayVision | Dice | 48.67 ± 21.86 | 58.06 ± 21.53 | 62.26 ± 20.32 | 60.21 ± 22.25 | 53.27 ± 26.12 |
| HD | 2077.14 ± 115.53 | 2219.49 ± 32.33 | 2287.61 ± 32.06 | 2285.14 ± 41.69 | 2176.00 ± 82.62 | |
| HD95 | 1539.31 ± 100.88 | 1796.58 ± 90.51 | 1913.68 ± 49.05 | 1870.52 ± 84.12 | 1630.44 ± 45.73 | |
| U-Net(Real) | Dice | 79.12 ± 8.90 | 82.94 ± 10.46 | 83.79 ± 13.24 | 86.37 ± 6.61 | 76.71 ± 17.69 |
| HD | 6.91 ± 0.30 | 6.38 ± 0.11 | 6.31 ± 0.12 | 6.14 ± 0.17 | 6.69 ± 0.35 | |
| HD95 | 5.07 ± 0.23 | 4.38 ± 0.10 | 4.15 ± 0.19 | 4.01 ± 0.03 | 4.84 ± 0.39 | |
| U-Net() | Dice |
87.14 ± 4.58 (10.14%) |
88.84 ± 4.52 (7.11%) |
89.61 ± 2.78 (6.95%) |
89.14 ± 3.30 (3.21%) |
86.02 ± 7.57 (12.14%) |
| HD | 5.57 ± 0.27 | 5.43 ± 0.10 | 5.56 ± 0.08 | 5.64 ± 0.10 | 5.76 ± 0.13 | |
| HD95 | 4.08 ± 0.24 | 3.77 ± 0.03 | 3.63 ± 0.10 | 3.78 ± 0.06 | 4.07 ± 0.39 | |
| U-Net() | Dice |
88.50 ± 3.45 (11.86%) |
89.03 ± 3.47 (7.34%) |
89.21 ± 3.49 (6.47%) |
89.32 ± 2.19 (3.42%) |
88.95 ± 3.39 (15.96%) |
| HD | 5.36 ± 0.10 | 5.37 ± 0.09 | 5.36 ± 0.07 | 5.50 ± 0.06 | 5.40 ±0.04 | |
| HD95 | 3.71 ± 0.10 | 3.67 ± 0.04 | 3.63 ± 0.05 | 3.64 ± 0.06 | 3.62 ± 0.07 | |
Values are shown in groups of normal and adversarial data using the internal rotation feature . Values are shown as mean ± standard deviation. Values in bracket show the percentage increase of the real and synthetic data-trained models compared to real data-trained model U-Net(Real). Bold values indicate the best results for each internal rotation category.
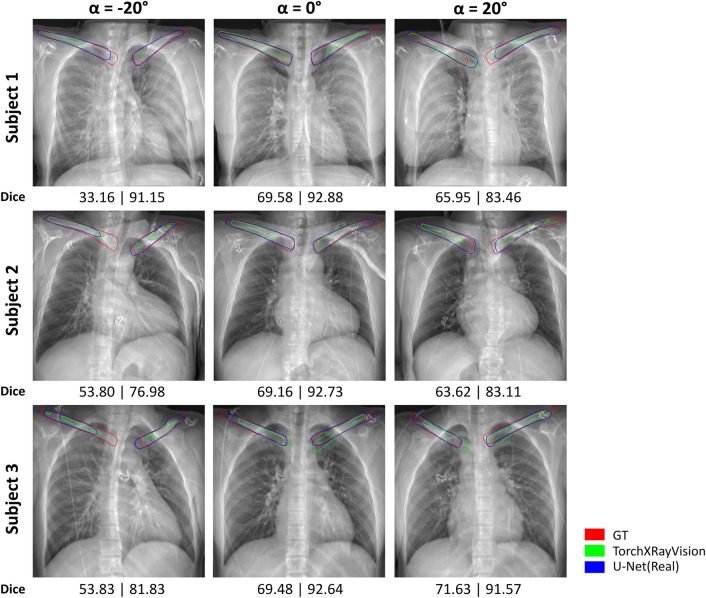
Figure 3 shows synthetic X-ray of three test subjects with segmentation contours of ground truth (red color) and predictions from TorchXRayVision (green color) and U-Net(Real) (blue color). In , both TorchXrayVision and U-Net(Real) segmentation contour are most similar to the ground truth masks. In most cases of and , the clavicle contours for both models are either longer or shorter than the ground truth.
Fig. 3.
Clavicle segmentation results of real-data-trained segmentation models. Three subjects with adversarial-positioned ( and ) and well-positioned () images are shown.The ground truth (GT) segmentation contours from TotalSegmentator are red, TorchXRayVision are green, and U-Net(Real) are blue in color. The Dice score analysis with respect to GT for each patient and angle are shown below each image, left is TorchXRayVision and right is U-Net(Real).
Improvement when trained with real and synthetic adversarial data
To assess the influence of the synthetic data, we further trained the U-Net model with real and synthetic X-ray images, as U-Net() and U-Net(). Figure 4 shows the robustness analysis of models trained with real X-ray images as a boxplot, robustness for U-Net() is blue and U-Net() is green in color. There is a consistent increase in the median in both U-Net() and U-Net() models compared to U-Net(Real). The IQR and the distance between the upper and lower whiskers exhibit a consistent decrease across all five groups of values when synthetic data is incorporated into training for both U-Net() and U-Net(). And the IQR and whiskers range in U-Net() even reduced more than U-Net(). This indicates a positive impact of adversarial synthetic data incorporation on model performance.
Fig. 4.
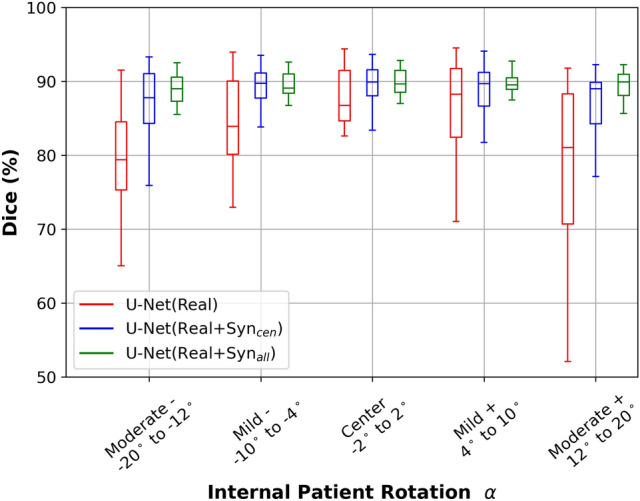
Robustness as Dice scores for real and synthetic data-trained segmentation models in boxplot on well- and adversarial- positioned data. Values in mean and standard deviation are shown in Table 2. U-Net(Real) is red, U-Net() is blue and U-Net( is green in color. Description for the boxplot is similar as Fig. 2.
Furthermore, when considering the trend across various values, the median, IQR and whiskers ranges of U-Net() are equally consistent across all groups. While the symmetric curve trend still exists in both U-Net() and U-Net(Real). The two models trained with only well-positioned data have lower median in moderate negative and positive than center value. Also, the IQR has a larger decrease in moderate than in mild values, with most pronounced decrease in mild positive. While in center values, the IQR of U-Net() and U-Net() are similar. The above results collectively underscore the efficacy of synthetic adversarial data-incorporated training, which enhanced the resilience and robustness of deep learning segmentation models. In addition, similar pattern are observed quantitatively in Table 2. Percentage increase in Dice score are larger in moderate than center or low values.
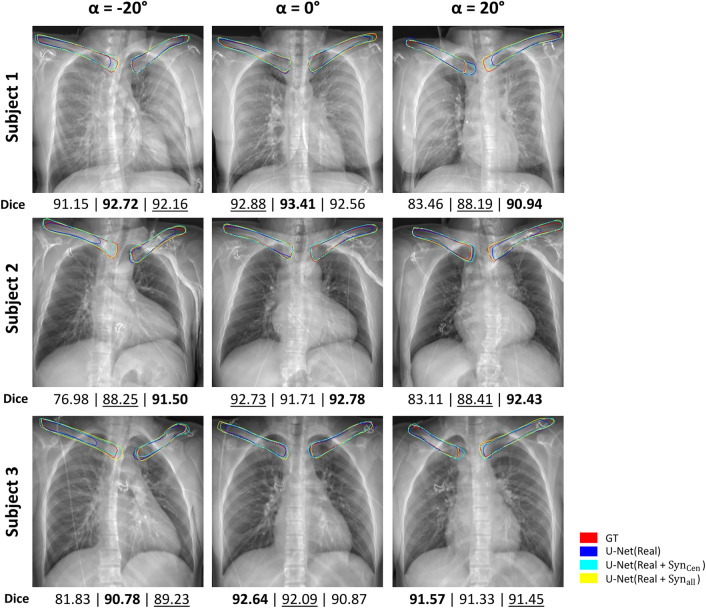
Figure 5 shows synthetic X-ray of three test subjects with segmentation contours of ground truth in red, predictions from U-Net(Real) is blue, U-Net() is cyan, and U-Net() is yellow in color. In , the segmentation contours from all three models have a high similarity to the ground truth, with U-Net(Real) and U-Net() slightly under or over predict on Subject 1, and 3. In and , the contour U-Net(Real) fall short in at least one clavicle in each subject. Moreover, U-Net() has generally a higher similarity to ground truth than U-Net(). This is supported by the Dice score analysis shown below each image. U-Net() generally shows the highest or second-highest Dice score.
Fig. 5.
Clavicle segmentation results of real and synthetic data-trained segmentation models. Three subject images where the patient is adversarial-positioned ( and ) and well-positioned (). The segmentation contours color for ground truth (GT) are red, U-Net(Real) are blue and U-Net() are cyan, and U-Net() are yellow. The Dice score analysis with respect to GT for each patient and angle are shown below each image, left is U-Net(Real), middle is U-Net(), and right is U-Net(). Bold indicates the highest value among the triplets, and underline indicates the second-highest value.
Network and ablation study
The training of the three models was performed on a NVIDIA A40 48GB GPU. Each epoch takes 300 seconds, early stopping was applied when model did not improve within the last 30 epochs. An ablation study was performed on the U-Net(Real) where different components or loss terms are tested during training to evaluate the efficacy of each component. By varying the batch sizes, depth of U-Net and loss functions, the Dice score were evaluated. Table 3 shows the Dice score in mean and standard deviation of the ablation study, bold values indicates the highest Dice. The hyperparameters with highest Dice score is when using Dice loss, batch size of 32 and 5 layer in U-Net. Hence, these hyperparameters were also used in U-Net() and U-Net() training. As a baseline comparison, we also test the other three models TorchXRayVision, U-Net() and U-Net() on 350 real X-ray images. Respective Dice scores for four models are shown in Table 4.
Table 3.
Ablation study on U-Net(Real). 18 combinations resulted when varying depth of U-Net, loss function and batch size. Dice score were shown as mean and standard deviation (SD). Bold indicates highest Dice score.
| Exp | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
|---|---|---|---|---|---|---|---|---|---|
| Depth | 3 | 4 | |||||||
| Loss | Dice | DiceBCE | Dice | ||||||
| Batch | 8 | 16 | 32 | 8 | 16 | 32 | 8 | 16 | 32 |
| Mean | 89.79 | 89.88 | 90.00 | 89.42 | 89.28 | 88.60 | 91.63 | 91.37 | 91.33 |
| SD | 10.84 | 11.08 | 9.24 | 10.26 | 11.45 | 9.78 | 8.68 | 8.44 | 8.82 |
| Exp | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 |
|---|---|---|---|---|---|---|---|---|---|
| Depth | 4 | 5 | |||||||
| Loss | DiceBCE | Dice | DiceBCE | ||||||
| Batch | 8 | 16 | 32 | 8 | 16 | 32 | 8 | 16 | 32 |
| Mean | 91.17 | 91.45 | 91.23 | 92.04 | 92.14 | 92.58 | 92.26 | 91.53 | 90.96 |
| SD | 8.12 | 8.48 | 9.00 | 7.07 | 7.05 | 7.14 | 7.55 | 7.58 | 7.38 |
Table 4.
Evaluation of four models on real X-ray images. Dice score were shown as mean and standard deviation (SD). Bold indicates highest Dice score.
| TorchXRayVision | U-Net(Real) | U-Net() | U-Net() | |
|---|---|---|---|---|
| Mean | 42.03 | 92.58 | 91.96 | 92.30 |
| SD | 32.62 | 7.14 | 9.47 | 8.29 |
Discussion
To separate the effect of domain learning when measuring adversarial robustness, we define the model U-Net() for which only well-positioned synthetic X-ray were added to training. This model did not seen any adversarial data. The increase in median, and reduction of IQR and whiskers range in U-Net() than U-Net(Real) across all values demonstrated the transfer learning between real and synthetic X-ray. However, U-Net() still exhibit a curve shape trend, ie. higher Dice in the center while lower Dice on the large values. In addition, the curve shape trend still exists when using the open-source TorchXRayVision as a baseline comparison. Yet, the primary focus of this analysis is not on the overall and absolute performance of different network, but rather on observing the trend of the Dice score across various adversarial data, ie. angles. We can see there is an agreement of the Dice score trend across different angles which agrees to our model trained with internal data.
Compared to U-Net(), U-Net() has a reduced whisker range in all groups and reduction in IQR in moderate negative, mild and moderate positive angles. With only 96 images added per adversarial features, angle , to the training, the spread of Dice score reduced and the model performances improved. Furthermore, U-Net() has similar median Dice, IQR and whisker range for all angles. Robustness refers to the ability of a model to maintain its performance and generalization capabilities when faced with perturbations. Hence, similar variability shows the resilience and reduced misclassification in clavicle segmentations. On the other hand in the center group, both U-Net() and U-Net() have similar IQR and median, but slightly more reduced whiskers range. Both models contain the same well-positioned synthetic X-ray images, while U-Net() contains additional adversarial-positioned synthetic images. With addition of adversarial training data, the peripheral Dice score is reduced, thus indicating the learning of adversarial features. As a rule of thumb, the increase in training data might contribute to the combined effect of the performance improvement. Yet the performance improvement in the center category is not substantial, for instance in Figure 4, the blue box and green box in the center category even with an increase of total training data from 3731 to 5450. The increase of performance only appears in adversarial angles. This suggests that the increase in adversarial data can provide a larger feature distribution in the data, thus allowing the network to learn the adversarial features. Moreover, this is also the insight of our study with more data, which is to showcase the generation of synthetic data. These findings collectively support the theory that adversarial patient positioning would contribute to segmentation model robustness. Furthermore, the addition of synthetic adversarial training data enhanced the consistency and performance of the deep learning-based bone segmentation model. Future works will include collection and testing on real CXRs which is shown to have the projection angle variation.
In radiomics, it is common to use image perturbations to determine features’ robustness and stability, with the goal of enhancing the reliability of radiomic analysis by using the robust features45–48. The perturbations includes rotation which also want to simulate the patient position variation during imaging. However, these studies mostly focus on CT images with rotation along z-axis. Our study focus on rotation along y-axis, as this patient rotation might happen during examination, and unlike z-axis rotation, it cannot be adjusted retrospectively. Also studies in deep learning models robustness have only been focusing on modifying the pixel intensity or translation, random cropping10,16–18, and have not bring forward to a clinical use case such as patient rotation. Therefore we propose the novel use of synthetic X-rays from CT for generating the adversarial patient positioned images for testing and training. Forward projected images could moreover reflect the actual attenuation effects of photon in X-ray beam on the anatomical changes when patient is rotated. Even though other generative view synthesis approaches might be applied, forward projection is a systematical reconstruction and a comparison of them is not our main focus. Extending beyond prior research32–34, our study further investigate in which aspect could the mixture of real and synthetic data-trained models outperform real data only trained models.
Conclusion
This study has demonstrated the potential and effectiveness of applying adversarial synthetic X-rays generated from 3D photon-counting CT and TotalSegmentator annotations to quantify and increase robustness of bone segmentation models in X-ray. In real data-trained models, we found the models are less robust. As Dice scores increase in absolute value and spread with increasing internal patient rotation. Adding adversarial synthetic X-rays to training data reduces the variations and thus enhances model robustness. The focus of this study was on CXR and clavicle segmentation, but the underlying principle has great potential for applications in other domains including segmentation for hip replacement. Conclusively, we presented a systematic way of generating synthetic X-rays which can be used as an option to improve the robustness of deep learning models supplementary to standard approaches.
Disclaimer
The presented methods in this paper are not commercially available and their future availability cannot be guaranteed.
Author contributions
WYRF and AF conceived the experiments. WYRF implemented and conducted the experiments. WYRF, CH, MB implemented the code. AF, CH, RB collected data. WYRF, AF and SS analysed the results. WYRF, AF and SS drafted the manuscript. AF, BG and SK designed and supervised the study. All authors reviewed and approved the final version of the manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Data availability
The datasets generated during and/or analysed during the current study are not publicly available due to company data privacy, but might be available on reasonable request to the corresponding author.
Declarations
Competing interests
WYRF, AF, CH, RB, MB, BG and SK are employees of Siemens Healthineers AG. SS declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Berlin, L. The importance of proper radiographic positioning and technique. AJR. American journal of roentgenology 166, 769–771 (1996). [DOI] [PubMed] [Google Scholar]
- 2.Kubota, Y. et al. Development of an automatic evaluation method for patient positioning error. Journal of Applied Clinical Medical Physics 16, 100–111 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hamer, O., Zorger, N., Feuerbach, S. & Müller-Wille, R. Grundkurs Thoraxröntgen: Tipps und Tricks für die systematische Bildanalyse (Springer-Verlag, 2013).
- 4.Singh, G., Gehr, T., Mirman, M., Püschel, M. & Vechev, M. Fast and effective robustness certification. In Bengio, S. et al. (eds.) Advances in Neural Information Processing Systems, vol. 31 (Curran Associates, Inc., 2018).
- 5.Gehr, T. et al. Ai2: Safety and robustness certification of neural networks with abstract interpretation. In 2018 IEEE Symposium on Security and Privacy (S &P), 3–18 (IEEE, 2018).
- 6.Biggio, B. et al. Evasion attacks against machine learning at test time. In Machine Learning and Knowledge Discovery in Databases: European Conference, ECML PKDD 2013, Prague, Czech Republic, September 23-27, 2013, Proceedings, Part III 13, 387–402 (Springer, 2013).
- 7.Szegedy, C. et al. Intriguing properties of neural networks. arXiv preprint arXiv:1312.6199 (2013).
- 8.Madry, A., Makelov, A., Schmidt, L., Tsipras, D. & Vladu, A. Towards deep learning models resistant to adversarial attacks. arXiv preprint arXiv:1706.06083 (2017).
- 9.Gowal, S. et al. Improving robustness using generated data. Advances in Neural Information Processing Systems 34, 4218–4233 (2021). [Google Scholar]
- 10.Goodfellow, I. J., Shlens, J. & Szegedy, C. Explaining and harnessing adversarial examples. arXiv preprint arXiv:1412.6572 (2014).
- 11.Moosavi-Dezfooli, S., Fawzi, A. & Frossard, P. Deepfool: A simple and accurate method to fool deep neural networks. In 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2574–2582 (IEEE Computer Society, Los Alamitos, CA, USA, 2016).
- 12.Papernot, N. et al. The limitations of deep learning in adversarial settings. In 2016 IEEE European Symposium on Security and Privacy (EuroS &P), 372–387 (IEEE Computer Society, Los Alamitos, CA, USA, 2016).
- 13.Athalye, A., Engstrom, L., Ilyas, A. & Kwok, K. Synthesizing robust adversarial examples. In 2018 International Conference on Machine Learning, 284–293 (PMLR, 2018).
- 14.Cai, Q.-Z., Du, M., Liu, C. & Song, D. Curriculum adversarial training. arXiv preprint arXiv:1805.04807 (2018).
- 15.Kamann, C. & Rother, C. Benchmarking the robustness of semantic segmentation models. In 2020 Conference on Computer Vision and Pattern Recognition, 8828–8838 (2020).
- 16.Finlayson, S. G. et al. Adversarial attacks on medical machine learning. Science 363, 1287–1289 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghaffari Laleh, N. et al. Adversarial attacks and adversarial robustness in computational pathology. Nature Commun. 13, 5711 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paschali, M., Conjeti, S., Navarro, F. & Navab, N. Generalizability vs. robustness: Investigating medical imaging networks using adversarial examples. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2018: 21st International Conference, Granada, Spain, September 16-20, 2018, Proceedings, Part I, 493–501 (Springer, 2018).
- 19.Frangi, A. F., Tsaftaris, S. A. & Prince, J. L. Simulation and synthesis in medical imaging. IEEE Trans. Med. Imaging 37, 673–679 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen, R. J., Lu, M. Y., Chen, T. Y., Williamson, D. F. & Mahmood, F. Synthetic data in machine learning for medicine and healthcare. Nature Biomedical Engineering 5, 493–497 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goncalves, A. et al. Generation and evaluation of synthetic patient data. BMC Med. Res. Methodol. 20, 1–40 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu, Y. et al. Machine learning for synthetic data generation: A review. arXiv preprint arXiv:2302.04062 (2023).
- 23.Ghorbani, A., Natarajan, V., Coz, D. & Liu, Y. DermGAN: Synthetic generation of clinical skin images with pathology. In Machine learning for Health Workshop, 155–170 (PMLR, 2020).
- 24.Mahmood, F. et al. Deep adversarial training for multi-organ nuclei segmentation in histopathology images. IEEE Trans. Med. Imaging 39, 3257–3267 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Costa, P. et al. End-to-end adversarial retinal image synthesis. IEEE Trans. Med. Imaging 37, 781–791 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Su, K.-H. et al. Generation of brain pseudo-CTs using an undersampled, single-acquisition UTE-mDixon pulse sequence and unsupervised clustering. Med. Phys. 42, 4974–4986 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnstone, E. et al. Systematic review of synthetic computed tomography generation methodologies for use in magnetic resonance imaging-only radiation therapy. International Journal of Radiation Oncology* Biology* Physics 100, 199–217 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Owrangi, A. M., Greer, P. B. & Glide-Hurst, C. K. MRI-only treatment planning: Benefits and challenges. Phys. Med. Biol. 63, 05TR01 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim, H. et al. Clinical feasibility of deep learning-based synthetic ct images from t2-weighted mr images for cervical cancer patients compared to mrcat. Sci. Rep. 14, 8504 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Unberath, M. et al. DeepDRR–a catalyst for machine learning in fluoroscopy-guided procedures. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2018: 21st International Conference, Granada, Spain, September 16-20, 2018, Proceedings, Part IV 11, 98–106 (Springer, 2018).
- 31.Moturu, A. & Chang, A. Creation of synthetic X-rays to train a neural network to detect lung cancer. Journal Beyond Sciences Initiative, University of Toronto, in Toronto (2018).
- 32.Fok, W. Y. R. et al. Learning patient rotation using synthetic X-ray images from 3D CT volumes. In Medical Imaging with Deep Learning, short paper track (2023).
- 33.Barbosa, E. J. M. Jr. et al. Automated detection and quantification of COVID-19 airspace disease on chest radiographs: A novel approach achieving expert radiologist-level performance using a deep convolutional neural network trained on digital reconstructed radiographs from computed tomography-derived ground truth. Invest. Radiol. 56, 471–479 (2021). [DOI] [PubMed] [Google Scholar]
- 34.Gao, C. et al. Synthetic data accelerates the development of generalizable learning-based algorithms for X-ray image analysis. Nature Machine Intelligence 5, 294–308 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wasserthal, J. et al. Totalsegmentator: Robust segmentation of 104 anatomic structures in CT images. Radiology: Artificial Intelligence 5 (2023). [DOI] [PMC free article] [PubMed]
- 36.Cohen, J. P. et al. TorchXRayVision: A library of chest X-ray datasets and models. In Medical Imaging with Deep Learning (2022).
- 37.Siddon, R. L. Fast calculation of the exact radiological path for a three-dimensional ct array. Medical Physics 12, 252–255 (1985). [DOI] [PubMed] [Google Scholar]
- 38.Puddy, E. & Hill, C. Interpretation of the chest radiograph. Continuing Education in Anaesthesia, Critical Care & Pain 7, 71–75 (2007). [Google Scholar]
- 39.Isensee, F., Jaeger, P. F., Kohl, S. A., Petersen, J. & Maier-Hein, K. H. nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. Nature Methods 18, 203–211 (2021). [DOI] [PubMed] [Google Scholar]
- 40.Lian, J. et al. A structure-aware relation network for thoracic diseases detection and segmentation (IEEE Trans. Med, Imaging, 2021). [DOI] [PubMed] [Google Scholar]
- 41.Zhao, H., Shi, J., Qi, X., Wang, X. & Jia, J. Pyramid scene parsing network. In 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 6230–6239 (2017).
- 42.Ronneberger, O., Fischer, P. & Brox, T. U-Net: Convolutional networks for biomedical image segmentation. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2015: 18th international conference, Munich, Germany, October 5-9, 2015, proceedings, part III 18, 234–241 (Springer, 2015).
- 43.Zou, K. H. et al. Statistical validation of image segmentation quality based on a spatial overlap index. Acad. Radiol. 11, 178–189 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sudre, C. H., Li, W., Vercauteren, T., Ourselin, S. & Jorge Cardoso, M. Generalised dice overlap as a deep learning loss function for highly unbalanced segmentations. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support: Third International Workshop, DLMIA 2017, and 7th International Workshop, ML-CDS 2017, Held in Conjunction with MICCAI 2017, Québec City, QC, Canada, September 14, Proceedings 3, 240–248 (Springer, 2017). [DOI] [PMC free article] [PubMed]
- 45.van Timmeren, J. E. et al. Test-retest data for radiomics feature stability analysis: Generalizable or study-specific?. Tomography 2, 361–365 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zwanenburg, A. et al. Assessing robustness of radiomic features by image perturbation. Sci. Rep. 9, 614 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Teng, X. et al. Improving radiomic model reliability using robust features from perturbations for head-and-neck carcinoma. Frontiers in Oncology 12, 974467 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mukherjee, S. et al. Assessing the robustness of a machine-learning model for early detection of pancreatic adenocarcinoma (PDA): Evaluating resilience to variations in image acquisition and radiomics workflow using image perturbation methods. Abdominal Radiology 1–11 (2024). [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and/or analysed during the current study are not publicly available due to company data privacy, but might be available on reasonable request to the corresponding author.