Abstract
The heat shock transcription factor Hsf1 of the yeast Saccharomyces cerevisiae regulates expression of genes encoding heat shock proteins and a variety of other proteins as well. To better understand the cellular roles of Hsf1, we screened multicopy suppressor genes of a temperature-sensitive hsf1 mutation. The RIM15 gene, encoding a protein kinase that is negatively regulated by the cyclic AMP-dependent protein kinase, was identified as a suppressor, but Rim15-regulated stress-responsive transcription factors, such as Msn2, Msn4, and Gis1, were unable to rescue the temperature-sensitive growth phenotype of the hsf1 mutant. Another class of suppressors encoded cell wall stress sensors, Wsc1, Wsc2, and Mid2, and the GDP/GTP exchange factor Rom2 that interacts with these cell wall sensors. Activation of a protein kinase, Pkc1, which is induced by these cell wall sensor proteins upon heat shock, but not activation of the Pkc1-regulated mitogen-activated protein kinase cascade, was necessary for the hsf1 suppression. Like Wsc-Pkc1 pathway mutants, hsf1 cells exhibited an osmotic remedial cell lysis phenotype at elevated temperatures. Several of the other suppressors were found to encode proteins functioning in cell wall organization. These results suggest that Hsf1 in concert with Pkc1 regulates cell wall remodeling in response to heat shock.
All organisms respond to thermal stress by activating a gene expression program governed by stress-responsive transcription factors. The heat shock transcription factor (HSF), a protein evolutionarily conserved from yeasts to humans, regulates expression of a set of proteins called heat shock proteins (HSPs), many of which function as molecular chaperones (28, 37). In the yeast Saccharomyces cerevisiae, other transcriptional networks are also induced by heat shock. A pair of partially redundant transcription factors, Msn2 and Msn4, whose activity is controlled by the cyclic AMP (cAMP)-dependent protein kinase (PKA), activates expression of genes encoding several HSPs, enzymes for carbohydrate metabolism, and proteins involved in protection against oxidative stress (48). The transcription factors Rlm1 and Swi4, which are targets of a stress-inducible mitogen-activated protein kinase (MAPK), stimulate transcription of cell wall protein genes and cell cycle-regulated genes, respectively (14, 17).
Among these transcription factors, only HSF encoded by the HSF1 locus is essential for the growth of S. cerevisiae at normal, as well as elevated temperatures. Like HSFs of other eukaryotes, yeast Hsf1 forms a homotrimer and binds to a regulatory sequence, the heat shock element (HSE), of target genes. The HSE consists of multiple inverted repeats of the 5-bp sequence nGAAn (where n is any nucleotide). Both continuous (nTTCnnGAAnnTTCn) and discontinuous [e.g., nTTCnnGAAn(5 bp)nGAAn] arrays of repeats can function as HSEs (1, 28, 37). A genome-wide Hsf1-binding analysis revealed that Hsf1 binds to the 5′ upstream region of approximately 165 of 6,200 loci in the yeast genome (15). An expression analysis with an hsf1 mutant showed that Hsf1 activates transcription of at least 59 genes upon heat shock (53). The products of these genes are implicated in a broad range of biological functions, including protein folding and maturation, energy generation, carbohydrate metabolism, maintenance of cell integrity, cell signaling, and transcription (15, 53).
The Hsf1 protein consists of discrete domains necessary for DNA binding, for trimer formation, for activation of transcription (named AR1 and AR2), for repression of the activation ability (CE2 [for conserved element 2]), and for regulation of the CE2 function (CTM [for C-terminal modulator]) (19, 32, 44, 47). Notably, the CTM domain is required for the growth of yeast at elevated temperatures for heat-induced hyperphosphorylation of Hsf1 and for transcriptional activation of genes containing the discontinuous HSE but not the continuous HSE. All of the defects associated with loss of CTM function are bypassed when CE2 has simultaneously been deleted, suggesting that CE2 inhibits hyperphosphorylation and HSE architecture-specific transcriptional activation and that in response to heat shock CTM restrains the inhibitory functions of CE2 (16).
Whereas the DNA-binding domain of Hsf1 is essential for viability of yeast, the other domains exhibit differential requirements for growth (16, 19, 32, 47). Here, we isolated multicopy suppressor genes that rescue the temperature sensitivity of an hsf1 mutant lacking the CTM function. Analyses of these isolates revealed the involvement of Hsf1 in cell wall remodeling and, additionally, showed functional interactions between Hsf1 and two protein kinases, Pkc1, an upstream regulator of the MAPK cascade, and Rim15, a downstream target of PKA.
MATERIALS AND METHODS
Yeast strains and media.
The yeast strains used in the present study are listed in Table 1. All strains were derived from HS126 (16). Cells containing various hsf1 derivatives were created by plasmid shuffling, and null mutations were introduced by using a one-step gene disruption method (16, 44, 53). Rich medium containing glucose (YPD) and enriched synthetic glucose medium (ESD) were prepared as described previously (44).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype (plasmid) |
|---|---|
| HS126 | MATα ade2 ura3 leu2 his3 trp1 can1 hsf1::HIS3 YCp-URA3-HSF1 (pSK906) |
| HS131 | YCp-TRP1-hsf1-AR1Δ-ba1 (pK137) of HS134 |
| HS133 | YCp-URA3-hsf1-AR1Δ-ba1 (pK136) of HS134 |
| HS134 | LEU2::SSA4-lacZ of HS126 |
| HS144 | YCp-LEU2-hsf1-AR1Δ-N583-HSF1(316-529) (pK144 shown as Hsf1-Hs) of HS126 |
| HS154 | YCp-LEU2-hsf1-AR1Δ-N583-hsf+(269-594) (pK154 shown as Hsf1-Sp-CTMΔ) of HS126 |
| HS170T | YCp-TRP1-HSF1 (pK157) of HS126 |
| HS171T | YCp-TRP1-hsf1-ba1 (pK159) of HS126 |
| HS174 | gis1::LEU2 YCp-TRP1-HSF1 (pK157) of HS126 |
| HS175 | gis1::LEU2 YCp-TRP1-hsf1-ba1 (pK159) of HS126 |
| HS176 | msn2::LEU2 msn4::ADE2 YCp-TRP1-HSF1 (pK157) of HS126 |
| HS177 | msn2::LEU2 msn4::ADE2 YCp-TRP1-hsf1-ba1 (pK159) of HS126 |
| HS194 | wsc1::ADE2 YCp-TRP1-HSF1 (pK157) of HS134 |
| HS195 | wsc1::ADE2 YCp-TRP1-hsf1-ba1 (pK159) of HS134 |
| YAY9 | YCp-TRP1-hsf1-F256S (pAY9) of HS134 |
| YN49 | YCp-TRP1-hsf1-N583 (pN49) of HS134 |
Multicopy suppressor screening.
Strain HS131 (YCp-TRP1-hsf1-AR1Δ-ba1) was transformed with a genomic library cloned into the YEp24 multicopy vector (YEp-URA3). Transformants were selected for uracil prototrophy on ESD medium lacking uracil and were incubated at 38°C to identify plasmids, allowing growth of hsf1-AR1Δ-ba1 cells at the restrictive temperature. Plasmids were recovered from the cells, and the nucleotide sequences of the regions flanking the inserts were determined. To identify genes responsible for suppression, candidate genes were subcloned into YEp24 or YEplac195 (YEp-URA3) and were retested for the ability to suppress the temperature sensitivity. The original isolates bearing ROM2 did not contain the coding sequence of the N-terminal 326 amino acids. The full-length ROM2 gene amplified by PCR was used for further analysis. The HSP82, MSN2, MSN4, GIS1, PKC1, MPK1, RLM1, and SWI4 genes were amplified by PCR from yeast genomic DNA and were cloned into YEp24 or YEplac195. Plasmids bearing constitutively active alleles PKC1R398P, BCK1-20, and MKK1S386P were kindly provided by Kunio Matsumoto.
RNA analysis.
Cells were grown to an optical density of 1.0 at 600 nm (OD600) under the conditions described in the figure legends. Total RNA was prepared from the cells, quantified by determining the absorbance at 260 nm, and subjected to reverse transcription-PCR (RT-PCR) analysis as described previously (16). The amounts of PCR products were compared after normalizing RNA samples to the levels of control ACT1 mRNA (encoding actin) (16, 44).
Immunoblot analysis.
Wild-type HSF1 and hsf1-ba1 cells were grown in YPD medium at 28 or 39°C to an OD600 of 2.0. Cells were harvested from 2 ml of culture, and protein extracts were prepared as described previously (22). Identical protein samples were separated on two sodium dodecyl sulfate-polyacrylamide gels; one gel was stained with Coomassie brilliant blue to confirm equivalent loading of protein samples, and the other was subjected to immunoblotting with an antibody recognizing phosphorylated Mpk1 (Phospho-p44/42 MAPK antibody; Cell Signaling Technology, Inc.).
Cell lysis assay.
Cell suspensions were spotted on YPD medium or YPD containing 1 M sorbitol, incubated at 28°C for 1 day, and then incubated at 38°C overnight. Subsequently, the plates were overlaid with an alkaline phosphatase assay solution as described previously (34).
RESULTS
Isolation of multicopy suppressor genes of the temperature-sensitive growth defect associated with CTM mutations.
CTM function is inactivated by the “ba1” mutation in which two arginine residues in the CTM are replaced by glutamic acid residues (Fig. 1A) (44). Cells expressing the Hsf1-ba1 protein exhibit slow-growth at elevated temperatures and, when combined with deletion of the nonessential activation domain AR1 (Fig. 1A, hsf1-AR1Δ-ba1 mutation), show a severe growth defect at 38°C (16). Unlike other hsf1 mutants, whose temperature sensitivity is suppressed by elevated expression of Hsp90 (27, 55), introduction of a multicopy plasmid bearing the Hsp90 gene HSP82 into hsf1-AR1Δ-ba1 cells failed to recover normal growth at 38°C (Fig. 1B).
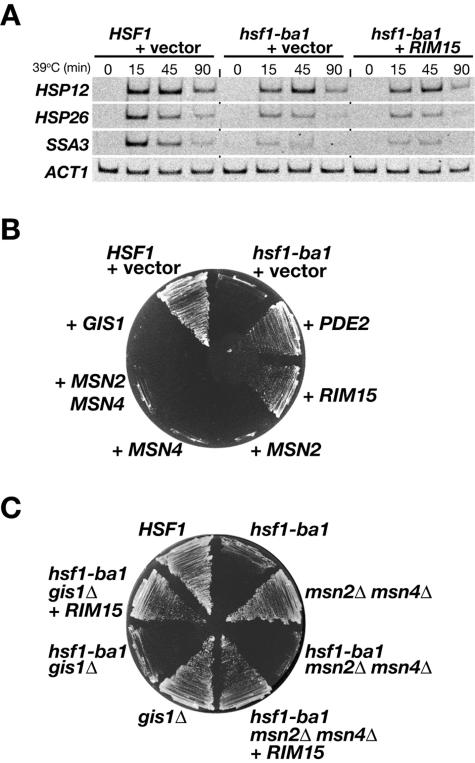
FIG. 1.
Characterization of multicopy suppressor genes. (A) Schematic diagram of structural motifs of Hsf1 and Hsf1 mutant constructs. The motifs indicated above Hsf1 are as follows: AR1 and AR2, activation domains; DBD, DNA-binding domain; oligomer, oligomerization domain; CE2, conserved element 2; CTM, C-terminal modulator. Numbers represent amino acid positions. Hsf1-Sp-CTMΔ contains amino acids 269 to 594 of S. pombe HSF but lacks the C-terminal 15 amino acids that function as a CTM domain. Hsf1-Hs contains amino acids 316 to 529 of human HSF1. (B) Multicopy suppression of growth defects at elevated temperature of hsf1-AR1Δ-ba1 cells. Wild-type HSF1 cells harboring empty vector (+ vector) and hsf1-AR1Δ-ba1 cells harboring empty vector or the vector bearing the suppressor genes (indicated by “+” and the gene name) were streaked on YPD medium and were incubated at 38°C for 2 days. The growth of hsf1-AR1Δ-ba1 cells harboring YEp-HSP82 is also shown. (C) RT-PCR analysis of multicopy suppressor gene effects on heat shock response target gene transcription. Wild-type HSF1 cells harboring empty vector (+ vector)and hsf1-ba1 cells harboring empty vector, YEp-RIM15, YEp-ROM2, YEp-RTS1, or YEp-ZDS1 were grown in ESD medium lacking uracil at 28°C, and then the temperature was shifted to 39°C. At the indicated times, aliquots of cells were removed and stored at −80°C. Total RNA prepared from each sample was subjected to RT-PCR analysis with primers for several heat-inducible genes (CUP1, CPR6, HSP42, and HSP78) and a control gene, ACT1.
To explore cellular functions of Hsf1 further, we screened multicopy suppressor genes that rescue the temperature-sensitive growth of hsf1-AR1Δ-ba1 cells (Fig. 1B). In addition to the expected wild-type HSF1 gene, various genes were identified as suppressors (Table 2). The PDE2 gene encodes a cAMP phosphodiesterase that downregulates PKA-dependent responses (48). The RIM15 gene product is a protein kinase containing a PAS domain that is known to act as a sensor for a variety of stimuli, and its kinase activity is negatively regulated by PKA (5, 39, 52). The plasma membrane proteins encoded by WSC1, WSC2, and MID2 play the role of stress sensors and bind to and activate Rom2, a GDP/GTP exchange protein for the small GTP-binding protein Rho1 (12, 18, 22, 33, 36, 38, 51). Rho1 is an upstream regulator of the protein kinase Pkc1, which affects actin filament organization and cell wall biogenesis (14, 17). EXG1 and KRE6 encode the major exo-1,3-β-glucanase and a protein required for β-1,6-glucan synthesis, respectively (24, 41). The B′ regulatory subunit of protein phosphatase 2A encoded by RTS1 was also involved in suppression of the hsf1-AR1Δ-ba1 phenotype (9). The ZDS1 gene, the most frequently isolated gene in this screen, and its paralog ZDS2 have been identified in numerous other screens designed to isolate genes that act as negative regulators of CDC42 (3), positive effectors of replication origin function (54), or stabilizers of linear centromeric plasmids (43). Other studies show that Zds1 has properties reminiscent of the PKA anchoring proteins (13). However, the exact functions of Zds1 and Zds2 have not been established. The YGR146C gene has been recognized as an Hsf1-bound gene, but the molecular function of its product is not known (15). All of the suppressors were also able to improve the slow-growth phenotype of hsf1-ba1 cells at 38°C (data not shown), and we used hsf1-ba1 cells in the analyses presented below.
TABLE 2.
List of multicopy suppressor genes
| Gene | Description and producta | No. of isolates |
|---|---|---|
| HSF1 | Heat shock transcription factor | 2 |
| PDE2 | High-affinity cAMP phosphodiesterase | 3 |
| RIM15 | Trehalose-associated protein kinase related to S. pombe cek1+ | 1 |
| WSC1 | Cell wall integrity and stress response component 1 | 16 |
| WSC2 | Cell wall integrity and stress response component 2 | 13 |
| MID2 | Protein required for mating | 1 |
| ROM2 | GDP/GTP exchange protein for Rho1 and Rho2 | 3 |
| EXG1 | Exo-1,3-β-glucanase | 1 |
| KRE6 | Protein required for β-1,6-glucan biosynthesis | 6 |
| RTS1 | B′-type regulatory subunit of protein phosphatase 2A | 2 |
| ZDS1 | Zillion different screens 1 | 33 |
| ZDS2 | Zillion different screens 2 | 2 |
| YGR146C | Unknown | 6 |
Derived from the Saccharomyces Genome Database and/or the MIPS Comprehensive Yeast Genome Database.
Rts1 affects transcription activation by Hsf1.
We first examined whether Hsf1 regulates transcription of the suppressors identified above. Hsf1 did not bind to or activate any of the suppressor genes, with the exception of YGR146C, as judged from previous genome-wide analyses (15, 53). Although the 5′ upstream region of YGR146C contains an HSE and binds Hsf1 (15), the mRNA levels of YGR146C were not affected by the hsf1-ba1 mutation (data not shown). We thus concluded that none of the suppressors are the direct targets of Hsf1.
We then tested the effects of suppressors on transcription activation by Hsf1-ba1. We analyzed the mRNA levels of Hsf1 target genes by using quantitative RT-PCR (16). As shown in Fig. 1C, heat-induced accumulation of the mRNAs from CUP1 and CPR6, which contain the discontinuous HSE, was severely compromised in hsf1-ba1 cells relative to wild type. In contrast, the hsf1-ba1 mutation did not significantly affect transcriptional activation through the continuous HSEs of HSP42 and HSP78. Among the suppressor genes we tested, the heat shock response of CUP1 and CPR6 in the hsf1-ba1 cells was only restored by introduction of RTS1 (Fig. 1C and data not shown). The RTS1 gene has been previously isolated as a multicopy suppressor of hsp60-ts, a temperature-sensitive allele of HSP60 that encodes a mitochondrial GroEL homologue (46). Null mutations of RTS1 result in a low-level of heat-induced transcription of Hsf1 target genes, such as HSP60 and HSP10 (a mitochondrial GroES homologue) (46). Our finding that overexpression of RTS1 in hsf1-ba1 cells restores the heat shock response of CUP1 and CPR6 supports the hypothesis that Rts1 affects the ability of Hsf1 to activate transcription. How Rts1 regulates the Hsf1 activity will be the focus of a future study. The products of the other suppressor genes may collaborate with unknown protein(s), whose expression or function is affected by the hsf1-ba1 mutation, so as to enable cells to grow at elevated temperatures.
Suppression of the temperature sensitivity of hsf1-ba1 cells by RIM15.
Cells with deficient PKA activity exhibit increased resistance toward heat stress (48). Accordingly, downregulation of PKA by Pde2 phosphodiesterase and overexpression of Rim15, a kinase acting immediately downstream of and negatively regulated by PKA, were responsible for suppression of the hsf1-ba1 mutation. The Rim15 kinase is required for proper establishment of the G0 program and for extension of life span (10, 35, 39). In response to nutrient limitation, the transcription factors Msn2, Msn4, and Gis1 cooperatively mediate the entire Rim15-dependent transcription response and induce expression of various genes, including HSP12, HSP26, and SSA3 (5, 35, 39).
We analyzed effect of RIM15 overexpression in the hsf1-ba1 cells on the heat shock response of HSP12, HSP26, and SSA3 (Fig. 2A). In logarithmically growing hsf1-ba1 cells, heat-induced accumulation of the HSP12, HSP26, and SSA3 mRNAs was reduced by ca. 60, 40, and 15% compared to HSF1 wild-type controls, respectively, and the levels were not affected by multiple copies of RIM15. We then examined the growth of hsf1-ba1 cells harboring multiple copies of MSN2, MSN4, and GIS1 and found that they are unable to rescue the growth defect (Fig. 2B). Wild-type HSF1 cells containing either msn2Δ msn4Δ double null mutations or a gis1Δ null mutation were able to grow at 38°C (Fig. 2C). When the hsf1-ba1 mutation was combined with the null mutations of these genes, the combinations did not exacerbate the heat sensitivity of hsf1-ba1 cells. Furthermore, RIM15 rescued the temperature sensitivity of hsf1-ba1 msn2Δ msn4Δ and hsf1-ba1 gis1Δ cells. Taken together, we conclude that Msn2, Msn4, and Gis1 are dispensable for suppression by Rim15 and suggest that Rim15 regulates the functions of different sets of proteins in response to distinct stressors, heat and nutrient limitation.
FIG. 2.
Effect of components of the PKA pathway on the growth of hsf1-ba1 cells. (A) RT-PCR analysis of RIM15 overexpression effects on HSP12, HSP26, and SSA3 transcription. Wild-type HSF1 cells harboring empty vector (+ vector) and hsf1-ba1 cells harboring empty vector or YEp-RIM15 were grown in ESD medium lacking uracil, and total RNA prepared from each sample was subjected to RT-PCR analysis, as described for Fig. 1C. (B) Growth of hsf1-ba1 cells harboring multiple copies of the PKA pathway genes at elevated temperature. Wild-type HSF1 cells harboring empty vector (+ vector) and hsf1-ba1 cells harboring empty vector or the vector bearing various genes (indicated by “+” and the gene name) were streaked onto YPD medium and were incubated at 38°C for 2 days. (C) Growth of hsf1-ba1 cells containing mutations in the PKA pathway genes at elevated temperature. Wild-type HSF1 cells, their derivatives containing the indicated mutations, and mutant cells harboring YEp-RIM15 were streaked on YPD medium and were incubated at 38°C for 2 days.
Suppression of the temperature sensitivity of hsf1-ba1 cells by activation of the Wsc-Pkc1 pathway.
Upon heat shock, plasma membrane sensor proteins encoded by WSC1, WSC2, and MID2 activate Rom2 to promote GTP loading of Rho1, which in turn activates Pkc1 (36). Pkc1 then activates the downstream MAPK cascade consisting of Bck1, a pair of redundant MAPK kinases Mkk1 and Mkk2, and a MAPK Mpk1/Slt2 (14, 17). Mpk1 activates the transcription factors Rlm1 and Swi4, which regulate expression of cell wall genes and cell cycle-regulated genes, respectively (2, 20).
We examined whether the temperature sensitivity of hsf1-ba1 cells could be suppressed by overexpressing components of the Pkc1-MAPK pathway. As shown in Fig. 3A, a multicopy plasmid bearing PKC1 enabled hsf1-ba1 cells to grow at 38°C. A constitutively active allele of PKC1 (PKC1R398P) also rescued the temperature sensitivity, indicating that activation of Pkc1 is correlated with suppression of the hsf1-ba1 phenotype. However, introduction of the downstream MAPK cascade components as constitutively active alleles (BCK1-20 and MKK1S386P) or a multicopy gene (MPK1) was not sufficient to support the growth of hsf1-ba1 cells at 38°C. Overexpression of RLM1 or SWI4 did not rescue the growth defect. These data suggest that an alternative Pkc1 pathway mediates suppression of the temperature sensitivity of hsf1-ba1 cells (see Discussion).
FIG. 3.
Effect of components of the Wsc-Pkc1-Mpk1 pathway on the growth of hsf1-ba1 cells. (A) Suppression of temperature-sensitive growth defects of hsf1-ba1 cells by activation of the Wsc-Pkc1 pathway. Wild-type HSF1 cells harboring empty vector (+ vector) and hsf1-ba1 cells harboring empty vector, YEp-ROM2, YEp-PKC1, YCp-PKC1R398P, YCp-BCK1-20, YCp-MKK1S386P, YEp-MPK1, YEp-RLM1, or YEp-SWI4 were streaked on YPD medium and were incubated at 38°C for 2 days. (B) Heat-induced phosphorylation of Mpk1 in hsf1-ba1 cells. Wild-type HSF1 and hsf1-ba1 cells were grown in YPD medium at 28°C, and then the temperature was shifted to 39°C. At the indicated times, aliquots of cells were removed and protein extracts were prepared. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and subjected to immunoblot analysis with an antibody recognizing phosphorylated Mpk1.
Because the components of the Wsc-Pkc1 pathway are necessary for the growth of cells at normal or elevated temperatures (14, 17), it is possible that the temperature sensitivity of hsf1-ba1 cells is due to inefficient activation of this pathway. Heat-responsive activation of the Wsc-Pkc1 pathway causes phosphorylation of Mpk1 and increases its catalytic activity (21). We analyzed the activated form of Mpk1 by using an antibody that recognizes only phosphorylated Mpk1 (51). When the temperature of control HSF1 cells was shifted from 28 to 39°C, the amount of phosphorylated Mpk1 increased significantly, as shown by immunoblot analysis (Fig. 3B). Similar levels of the activated Mpk1 were detected in the extracts of hsf1-ba1 cells grown either at 28 or at 39°C. Therefore, the hsf1-ba1 mutation does not affect the heat-regulated activation of the Wsc-Pkc1-Mpk1 pathway.
Cell wall defect of hsf1-ba1 cells.
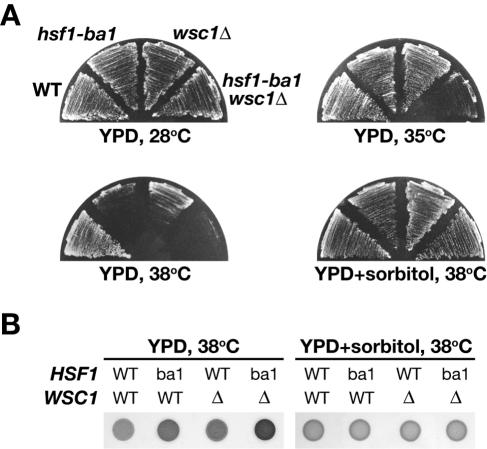
Inactivation of the Wsc-Pkc1 pathway leads to cell wall defects (12, 18, 22, 25, 33, 34, 38, 51). As reported previously (12, 18, 51), wsc1Δ cells exhibit a weak heat sensitivity, and the wild-type phenotype was restored by addition of an osmotic stabilizer, such as sorbitol, to the medium (Fig. 4A). We found that hsf1-ba1 cells are able to grow on medium containing sorbitol at 38°C. To test the cell lysis phenotype of the hsf1-ba1 mutant, we conducted a cell lysis assay, in which leakage of alkaline phosphatase from lysed cells is detected by a nonpermeable alkaline phosphatase substrate added to the culture plates (34). Cells expressing wild-type Hsf1 were negative controls, since the wild-type cells do not lyse even at elevated temperatures. The hsf1-ba1 cells, as well as wsc1Δ cells, underwent cell lysis on the standard plate but not on the osmotically stabilized plate (Fig. 4B). We then combined the hsf1-ba1 mutation with wsc1Δ. The growth of hsf1-ba1 wsc1Δ cells was inhibited at 35°C, but the addition of sorbitol to the medium allowed this mutant to grow even at 38°C (Fig. 4A). The osmotic remedial cell lysis phenotype of hsf1-ba1 cells was significantly exacerbated by combination with the wsc1Δ mutation, but sorbitol protected the cells from lysis, as judged by the alkaline phosphatase leakage assay (Fig. 4B). These results show that a cell wall defect is responsible for the temperature-sensitive growth inhibition of hsf1-ba1 cells and that Hsf1 is necessary for proper cell wall remodeling upon heat shock.
FIG. 4.
Suppression of the lysis phenotype of hsf1-ba1 cells by osmotic stabilization. (A) Growth of cells containing hsf1-ba1 and wsc1Δ mutations under various conditions. Wild-type HSF1 cells (WT) and their derivatives containing hsf1-ba1 and wsc1Δ mutations were streaked onto YPD medium or YPD medium containing 1 M sorbitol and were incubated at 28, 35, or 38°C for 2 days. (B) Cell lysis assay of hsf1-ba1 and wsc1Δ cells grown at elevated temperature. Suspensions of the indicated cells were spotted on YPD medium or YPD medium containing 1 M sorbitol. Plates were incubated at 28°C for 1 day and then switched to 38°C and incubated overnight. The plate was overlaid with an alkaline phosphatase assay solution and incubated at 38°C for 1 h.
Cell wall integrity of various hsf1 mutants.
To confirm the involvement of Hsf1 in cell wall remodeling, we analyzed the growth phenotype of cells containing various temperature-sensitive mutations in HSF1 (see Fig. 1A). The Hsf1-Sp-CTMΔ construct is the fusion protein of the central region (DNA-binding, oligomerization, and CE2 domains) of S. cerevisiae Hsf1 and the C-terminal region (without CTM domain) of Schizosaccharomyces pombe HSF. The Hsf1-Hs fusion contains the central region of S. cerevisiae Hsf1 and the C-terminal activation domain of human HSF1 (16). Hsf1-N583 is the C-terminally truncated form of Hsf1 lacking the AR2 and CTM domains (27, 44, 47). Hsf1-F256S contains a substitution of phenylalanine to serine at the 256 position in the DNA-binding domain (53). Among these hsf1 mutants, the temperature sensitivity of cells expressing hsf1-Sp-CTMΔ or hsf1-Hs was rescued when ROM2 was overexpressed or when the medium contained sorbitol (Fig. 5). Thus, several hsf1 mutations cause defects in cell wall organization at elevated temperatures.
FIG. 5.
Growth of various hsf1 mutants under cell wall-stabilizing conditions. Cells expressing Hsf1 (WT), Hsf1-Sp-CTMΔ (Sp-CTMΔ), Hsf1-Hs (Hs), Hsf1-F256S (F256S), or Hsf1-N583 (N583) were streaked onto YPD medium or YPD containing 1 M sorbitol. Plates were incubated at 28 or at 37°C for 2 days. The lower left panel shows growth of the indicated hsf1 mutant cells harboring YEp-ROM2 on YPD medium at 37°C.
DISCUSSION
The temperature-sensitive growth phenotype of hsf1-ba1 cells was suppressed by activation of the Wsc-Pkc1 pathway, which mediates maintenance of cell wall integrity. The hsf1-ba1 mutation consistently led to an osmotic remedial cell lysis phenotype at elevated temperatures. The activation of the Wsc-Pkc1 pathway or inclusion of an osmotic stabilizer in the medium rescued the growth defects of several hsf1 mutants at the restrictive temperature. Thus, the present study disclosed a novel cellular role of Hsf1: regulation of cell wall remodeling for adaptation to a high-temperature environment.
Although activation of the Wsc-Pkc1 pathway, which is known to activate a MAPK cascade, causes suppression of the temperature sensitivity of hsf1-ba1 cells, expression of individual components of the MAPK cascade or Mpk1-regulated transcription factors was unable to do so. Deletion of PKC1, but not the downstream components, results in the osmotic remedial cell lysis phenotype even at normal growth temperatures, implying that Pkc1 affects cell wall organization through a pathway other than the MAPK cascade (25, 34, 42). In addition, Pkc1, but not the MAPK cascade, regulates cellular functions such as heat-induced depolarization of the actin cytoskeleton (7), attenuation of ribosome biogenesis upon interruption of the secretory pathway (26, 31), and nuclear perturbation caused by high osmolarity (30). We suggest that Pkc1 regulates cell wall organization through an alternative signaling pathway in collaboration with Hsf1 to prevent cell lysis at elevated temperatures.
In addition to the components of the Wsc-Pkc1 pathway, the following observations implicate additional suppressor genes in cell wall maintenance. The S. cerevisiae cell wall consists of β-1,3-glucan, β-1,6-glucan, chitin, and mannoproteins (23). The EXG1 gene encoding the major exo-1,3-β-glucanase has been identified as a multicopy suppressor of the osmotic remedial cell lysis phenotype of ypk1-1ts ypk2Δ mutant cells. The Ypk1 and Ypk2 protein kinases function in parallel with the Pkc1-dependent pathway for maintenance of cell wall integrity (40). We also found that multiple copies of EXG1 improved the slow-growth phenotype of wsc1Δ cells at 38°C (data not shown). The KRE6 product, which may function as a glycoside hydrolase or transglycosidase in the Golgi complex, is required for the synthesis of β-1,6-glucan (29, 41). Mild overexpression of KRE6 rescues the cell lysis phenotype of pkc1Δ, indicating a functional interaction between Kre6 and Pkc1 (42). The ZDS1 gene, in addition to WSC1 and ROM2, has been identified as a multicopy suppressor of fks1-1154 fks2Δ, a temperature-sensitive mutant of cell wall 1,3-β-glucan synthase (45). A large-scale two-hybrid experiment showed Zds1 and Zds2 interactions with diverse gene products, including Rho1 and Pkc1 (8). Although the function of the YGR146C product is unknown, its transcription, as well as transcription of EXG1 and KRE6, is induced upon transient cell wall damage (11). In addition, null mutations of PDE2 cause a loss of cell wall strength and overexpression of PDE2 suppresses the sorbitol dependence of a mutant strain with fragile cell walls, implicating the PKA pathway in the maintenance of cell wall integrity (49).
Although Msn2 and Msn4 share various target genes with Hsf1 (4, 50), and Rlm1 activates transcription of cell wall genes in response to heat shock (20), multiple copies of MSN2, MSN4, and RLM1 failed to rescue the cell wall defect associated with the hsf1-ba1 mutation. Rather, the kinases controlling these transcription activators are functioning with Hsf1. Hsf1 regulates heat-induced transcription of several cell wall genes, including CWP1, SPI1, HOR7, YGP1, and ZEO1 (53). Our preliminary observations showed that the mRNA levels of CWP1, SPI1, and ZEO1 were slightly reduced in hsf1-ba1 cells relative to wild-type but that multiple copies of these genes were not sufficient to rescue the temperature sensitivity of hsf1-ba1 cells (data not shown). It has been estimated that more than 1,200 S. cerevisiae genes are in some way related to cell wall biosynthesis (6). Our data suggest that Hsf1 regulates expression of not only HSPs but also an additional set of unknown proteins that are involved in cell wall formation and remodeling.
Acknowledgments
We thank Naoyuki Hayashi for providing the yeast gene library, Kunio Matsumoto for plasmids, Ayaka Saka and Yoshikazu Ohya for advice on immunoblotting, and Toshio Fukasawa for critically reading the manuscript and for helpful discussions.
This study was supported in part by grants-in-aid for Scientific Research from the Ministry of Education, Sciences, Sports, and Culture to H.S.
REFERENCES
- 1.Amin, J., J. Ananthan, and R. Voellmy. 1988. Key features of heat shock regulatory elements. Mol. Cell. Biol. 8:3761-3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baetz, K., J. Moffat, J. Haynes, M. Chang, and B. Andrews. 2001. Transcriptional coregulation by the cell integrity mitogen-activated protein kinase Slt2 and the cell cycle regulator Swi4. Mol. Cell. Biol. 21:6515-6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bi, E., and J. R. Pringle. 1996. ZDS1 and ZDS2, genes whose products may regulate Cdc42p in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:5264-5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boy-Marcotte, E., G. Lagniel, M. Perrot, F. Bussereau, A. Boudsocq, M. Jacquet, and J. Labarre. 1999. The heat shock response in yeast: differential regulations and contributions of the Msn2p/Msn4p and Hsf1p regulons. Mol. Microbiol. 33:274-283. [DOI] [PubMed] [Google Scholar]
- 5.Cameroni, E., N. Hulo, J. Roosen, J. Winderickx, and C. De Virgilio. 2004. The novel yeast PAS kinase Rim15 orchestrates G0-associated antioxidant defense mechanisms. Cell Cycle 3:462-468. [PubMed] [Google Scholar]
- 6.De Groot, P. W. J., C. Cristina Ruiz, C. R. Vazquez de Aldana, E. Dueas, V. J. Cid, F. Del Rey, J. M. Rodriquez-Pena, P. Perez, A. Andel, J. Caubin, J. Arroyo, J. C. Garcia, C. Gil, M. Molina, L. J. Garcia, C. Nombela, and F. M. Klis. 2001. A genomic approach for the identification and classification of genes involved in cell wall formation and its regulation in Saccharomyces cerevisiae. Comp. Funct. Genomics 2:124-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delley, P. A., and M. N. Hall. 1999. Cell wall stress depolarizes cell growth via hyperactivation of RHO1. J. Cell Biol. 147:163-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drees, B. L., B. Sundin, E. Brazeau, J. P. Caviston, G. C. Chen, W. Guo, K. G. Kozminski, M. W. Lau, J. J. Moskow, A. Tong, L. R. Schenkman, A. McKenzie, P. Brennwald, M. Longtine, E. Bi, C. Chan, P. Novick, C. Boone, J. R. Pringle, T. N. Davis, S. Fields, and D. G. Drubin. 2001. A protein interaction map for cell polarity development. J. Cell Biol. 154:549-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evangelista, C. C., Jr., A. M. Rodriguez Torres, M. P. Limbach, and R. S. Zitomer. 1996. Rox3 and Rts1 function in the global stress response pathway in baker's yeast. Genetics 142:1083-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fabrizio, P., F. Pozza, S. D. Pletcher, C. M. Gendron, and V. D. Longo. 2001. Regulation of longevity and stress resistance by Sch9 in yeast. Science 292:288-290. [DOI] [PubMed] [Google Scholar]
- 11.Garcia, R., C. Bermejo, C. Grau, C. R. Perez, J. M. Rodriguez-Pena, J. Francois, C. Nombela, and J. Arroyo. 2004. The global transcriptional response to transient cell wall damage in Saccharomyces cerevisiae and its regulation by the cell integrity signaling pathway. J. Biol. Chem. 279:15183-15195. [DOI] [PubMed] [Google Scholar]
- 12.Gray, J. V., J. P. Ogas, Y. Kamada, M. Stone, D. E. Levin, and I. Herskowitz. 1997. A role for the Pkc1 MAP kinase pathway of Saccharomyces cerevisiae in bud emergence and identification of a putative upstream regulator. EMBO J. 16:4924-4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griffioen, G., S. Swinnen, and J. M. Thevelein. 2003. Feedback inhibition on cell wall integrity signaling by Zds1 involves Gsk3 phosphorylation of a cAMP-dependent protein kinase regulatory subunit. J. Biol. Chem. 278:23460-23471. [DOI] [PubMed] [Google Scholar]
- 14.Gustin, M. C., J. Albertyn, M. Alexander, and K. Davenport. 1998. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62:1264-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hahn, J. S., Z. Hu, D. J. Thiele, and V. R. Iyer. 2004. Genome-wide analysis of the biology of stress responses through heat shock transcription factor. Mol. Cell. Biol. 24:5249-5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hashikawa, N., and H. Sakurai. 2004. Phosphorylation of the yeast heat sock transcription factor is implicated in gene-specific activation dependent on the architecture of the heat shock element. Mol. Cell. Biol. 24:3648-3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heinisch, J. J., A. Lorberg, H. P. Schmitz, and J. J. Jacoby. 1999. The protein kinase C-mediated MAP kinase pathway involved in the maintenance of cellular integrity in Saccharomyces cerevisiae. Mol. Microbiol. 32:671-680. [DOI] [PubMed] [Google Scholar]
- 18.Jacoby, J. J., S. M. Nilius, and J. J. Heinisch. 1998. A screen for upstream components of the yeast protein kinase C signal transduction pathway identifies the product of the SLG1 gene. Mol. Gen. Genet. 258:148-155. [DOI] [PubMed] [Google Scholar]
- 19.Jakobsen, B. K., and H. R. B. Pelham. 1991. A conserved heptapeptide restrains the activity of the yeast heat shock transcription factor. EMBO J. 10:369-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jung, U. S., and D. E. Levin. 1999. Genome-wide analysis of gene expression regulated by the yeast cell wall integrity signalling pathway. Mol. Microbiol. 34:1049-1057. [DOI] [PubMed] [Google Scholar]
- 21.Kamada, Y., U. S. Jung, J. Piotrowski, and D. E. Levin. 1995. The protein kinase C-activated MAP kinase pathway of Saccharomyces cerevisiae mediates a novel aspect of the heat shock response. Genes Dev. 9:1559-1571. [DOI] [PubMed] [Google Scholar]
- 22.Ketela, T., R. Green, and H. Bussey. 1999. Saccharomyces cerevisiae Mid2p is a potential cell wall stress sensor and upstream activator of the PKC1-MPK1 cell integrity pathway. J. Bacteriol. 181:3330-3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klis, F. M., P. Mol, K. Hellingwerf, and S. Brul. 2002. Dynamics of cell wall structure in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 26:239-256. [DOI] [PubMed] [Google Scholar]
- 24.Larriba, G., E. Andaluz, R. Cueva, and R. D. Basco. 1995. Molecular biology of yeast exoglucanases. FEMS Microbiol. Lett. 125:121-126. [DOI] [PubMed] [Google Scholar]
- 25.Levin, D. E., and E. Bartlett-Heubusch. 1992. Mutants in the S. cerevisiae PKC1 gene display a cell cycle-specific osmotic stability defect. J. Cell Biol. 116:1221-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, Y., R. D. Moir, I. K. Sethy-Coraci, J. R. Warner, and I. M. Willis. 2000. Repression of ribosome and tRNA synthesis in secretion-defective cells is signaled by a novel branch of the cell integrity pathway. Mol. Cell. Biol. 20:3843-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morano, K. A., N. Santoro, K. A. Koch, and D. J. Thiele. 1999. A trans-activation domain in yeast heat shock transcription factor is essential for cell cycle progression during stress. Mol. Cell. Biol. 19:402-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morimoto, R. I. 1998. Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 12:3788-3796. [DOI] [PubMed] [Google Scholar]
- 29.Montijn, R. C., E. Vink, W. H. Muller, A. J. Verkleij, H. Van Den Ende, B. Henrissat, and F. M. Klis. 1999. Localization of synthesis of β1,6-glucan in Saccharomyces cerevisiae. J. Bacteriol. 181:7414-7420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nanduri, J., and A. M. Tartakoff. 2001. Perturbation of the nucleus: a novel Hog1p-independent, Pkc1p-dependent consequence of hypertonic shock in yeast. Mol. Biol. Cell 12:1835-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nierras, C. R., and J. R. Warner. 1999. Protein kinase C enables the regulatory circuit that connects membrane synthesis to ribosome synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 274:13235-13241. [DOI] [PubMed] [Google Scholar]
- 32.Nieto-Sotelo, J., G. Wiederrecht, A. Okuda, and C. S. Parker. 1990. The yeast heat shock transcription factor contains a transcriptional activation domain whose activity is repressed under nonshock conditions. Cell 62:807-817. [DOI] [PubMed] [Google Scholar]
- 33.Ozaki, K., K. Tanaka, H. Imamura, T. Hihara, T. Kameyama, H. Nonaka, H. Hirano, Y. Matsuura, and Y. Takai. 1996. Rom1p and Rom2p are GDP/GTP exchange proteins (GEPs) for the Rho1p small GTP binding protein in Saccharomyces cerevisiae. EMBO J. 15:2196-2207. [PMC free article] [PubMed] [Google Scholar]
- 34.Paravicini, G., M. Cooper, L. Friedli, D. J. Smith, J. L. Carpentier, L. S. Klig, and M. A. Payton. 1992. The osmotic integrity of the yeast cell requires a functional PKC1 gene product. Mol. Cell. Biol. 12:4896-4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pedruzzi, I., N. Burckert, P. Egger, and C. De Virgilio. 2000. Saccharomyces cerevisiae Ras/cAMP pathway controls post-diauxic shift element-dependent transcription through the zinc finger protein Gis1. EMBO J. 19:2569-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Philip, B., and D. E. Levin. 2001. Wsc1 and Mid2 are cell surface sensors for cell wall integrity signaling that act through Rom2, a guanine nucleotide exchange factor for Rho1. Mol. Cell. Biol. 21:271-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pirkkala, L., P. Nykanen, and L. Sistonen. 2001. Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J. 15:1118-1131. [DOI] [PubMed] [Google Scholar]
- 38.Rajavel, M., B. Philip, B. M. Buehrer, B. Errede, and D. E. Levin. 1999. Mid2 is a putative sensor for cell integrity signaling in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:3969-3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reinders, A., N. Burckert, T. Boller, A. Wiemken, and C. De Virgilio. 1998. Saccharomyces cerevisiae cAMP-dependent protein kinase controls entry into stationary phase through the Rim15p protein kinase. Genes Dev. 12:2943-2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roelants, F. M., P. D. Torrance, N. Bezman, and J. Thorner. 2002. Pkh1 and Pkh2 differentially phosphorylate and activate Ypk1 and Ykr2 and define protein kinase modules required for maintenance of cell wall integrity. Mol. Biol. Cell 13:3005-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roemer, T., and H. Bussey. 1991. Yeast β-glucan synthesis: KRE6 encodes a predicted type II membrane protein required for glucan synthesis in vivo and for glucan synthase activity in vitro. Proc. Natl. Acad. Sci. USA 88:11295-11299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roemer, T., G. Paravicini, M. A. Payton, and H. Bussey. 1994. Characterization of the yeast (1->6)-β-glucan biosynthetic components, Kre6p and Skn1p, and genetic interactions between the PKC1 pathway and extracellular matrix assembly. J. Cell Biol. 127:567-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roy, N., and K. W. Runge. 1999. The ZDS1 and ZDS2 proteins require the Sir3p component of yeast silent chromatin to enhance the stability of short linear centromeric plasmids. Chromosoma 108:146-161. [DOI] [PubMed] [Google Scholar]
- 44.Sakurai, H., and T. Fukasawa. 2001. A novel domain of the yeast heat shock factor that regulates its activation function. Biochem. Biophys. Res. Commun. 285:696-701. [DOI] [PubMed] [Google Scholar]
- 45.Sekiya-Kawasaki, M., M. Abe, A. Saka, D. Watanabe, K. Kono, M. Minemura-Asakawa, S. Ishihara, T. Watanabe, and Y. Ohya. 2002. Dissection of upstream regulatory components of the Rho1p effector, 1,3-β-glucan synthase, in Saccharomyces cerevisiae. Genetics 162:663-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shu, Y., and R. L. Hallberg. 1995. SCS1, a multicopy suppressor of hsp60-ts mutant alleles, does not encode a mitochondrially targeted protein. Mol. Cell. Biol. 15:5618-5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sorger, P. K. 1990. Yeast heat shock factor contains separable transient and sustained response transcriptional activators. Cell 62:793-805. [DOI] [PubMed] [Google Scholar]
- 48.Thevelein, J. M., and J. H. de Winde. 1999. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 33:904-918. [DOI] [PubMed] [Google Scholar]
- 49.Tomlin, G. C., G. E. Hamilton, D. C. Gardner, R. M. Walmsley, L. I. Stateva, and S. G. Oliver. 2000. Suppression of sorbitol dependence in a strain bearing a mutation in the SRB1/PSA1/VIG9 gene encoding GDP-mannose pyrophosphorylase by PDE2 overexpression suggests a role for the Ras/cAMP signal-transduction pathway in the control of yeast cell-wall biogenesis. Microbiology 146:2133-2146. [DOI] [PubMed] [Google Scholar]
- 50.Treger, J. M., A. P. Schmitt, J. R. Simon, and K. McEntee. 1998. Transcriptional factor mutations reveal regulatory complexities of heat shock and newly identified stress genes in Saccharomyces cerevisiae. J. Biol. Chem. 273:26875-26879. [DOI] [PubMed] [Google Scholar]
- 51.Verna, J., A. Lodder, K. Lee, A. Vagts, and R. Ballester. 1997. A family of genes required for maintenance of cell wall integrity and for the stress response in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 94:13804-13809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vidan, S., and A. P. Mitchell. 1997. Stimulation of yeast meiotic gene expression by the glucose-repressible protein kinase Rim15p. Mol. Cell. Biol. 17:2688-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamamoto, A., Y. Mizukami, and H. Sakurai. 2005. Identification of a novel class of target genes and a novel type of binding sequence of heat shock transcription factor in Saccharomyces cerevisiae. J. Biol. Chem. 280:11911-11919. [DOI] [PubMed] [Google Scholar]
- 54.Yu, Y., Y. W. Jiang, R. J. Wellinger, K. Carlson, J. M. Roberts, and D. J. Stillman. 1996. Mutations in the homologous ZDS1 and ZDS2 genes affect cell cycle progression. Mol. Cell. Biol. 16:5254-5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zarzov, P., H. Boucherie, and C. Mann. 1997. A yeast heat shock transcription factor (Hsf1) mutant is defective in both Hsc82/Hsp82 synthesis and spindle pole body duplication. J. Cell Sci. 110:1879-1891. [DOI] [PubMed] [Google Scholar]