Abstract
Immunoscreening of a Candida albicans expression library resulted in the isolation of a novel gene encoding a 32.9-kDa polypeptide (288 amino acids), with 27.7% homology to the product of Saccharomyces cerevisiae YGR106c, a putative vacuolar protein. Heterozygous mutants in this gene displayed an altered budding growth pattern, characterized by the formation of chains of buds, decreasingly in size towards the apex, without separation of the daughter buds. Consequently, this gene was designated ABG1. A conditional mutant for ABG1 with the remaining allele under the control of the MET3 promoter did not grow in the presence of methionine and cysteine, demonstrating that ABG1 was essential for viability. Western analysis revealed the presence of a major 32.9-kDa band, mainly in a particulate fraction (P40) enriched in vacuoles, and tagging with green fluorescent protein confirmed that Abg1p localized to the vacuole. Vacuole inheritance has been linked to the regulation of branching frequency in C. albicans. Under repressing conditions, the conditional mutant had an increased frequency of branching under hyphal inducing conditions and an altered sensitivity to substances that interfered with cell wall assembly. Repression of ABG1 in the conditional mutant strain caused disturbance of normal size and number of vacuoles both in yeast and mycelial cells and also in the asymmetric vacuole inheritance associated with the characteristic pattern of germ tubes and branching in C. albicans. These observations indicate that ABG1 plays a key role in vacuole biogenesis, cytokinesis, and hyphal branching.
Candida albicans is the major fungal pathogen of humans (10) and now represents the fourth most common agent of microbial lethal septicemia in immunocompromised patients (20). Yeast-hypha morphogenesis has often been considered to be a component of the repertoire of factors influencing virulence in this fungus (12, 26, 37, 44). Hyphal forms are invasive, and this property could promote tissue penetration during the early stages of infection, whereas the yeast form might be more suited for dissemination in the bloodstream (57). The cell cycle of C. albicans is intimately involved with the regulation of this morphogenetic process correlated with growth in vivo, since the cell cycle must be regulated to enable changes in cell shape (5). Recently, correlation between vacuolar inheritance and modulation of the cell cycle of C. albicans during true hyphal growth has been described (4). While vacuolar volume of mother and daughter cells of C. albicans at cytokinesis in yeast and pseudohyphal forms is similar (67), hyphal cell division is asymmetric and cytoplasm is partitioned predominantly to the proximal apical cell and most vacuole is inherited by the subapical mother yeast cell or hyphal intercalary compartment (4). Accordingly, the concept of size-regulated cell cycle control has been refined to propose that it is the cytoplasmic volume minus vacuolar volume (and that of other organelles) rather than total cell volume that is relevant in cell size-regulated control of the eukaryotic cell cycle (4). Consequently, mutations in genes that influence normal vacuolar inheritance will in turn influence the branching frequency of C. albicans.
During mycelial growth, vegetative growth is achieved by elongation and branching of hyphal filaments. Germ tube emergence is accompanied by a rapid increase in the vacuolar volume in the mother yeast cell (4, 27), but the cellular cytoplasmic volume remains relatively constant while the overall cell volume increases linearly. The remaining cell space in these expanding cells is occupied by an expanding vacuole or vacuoles. The vacuolation process has been reported to be a general feature of the growth of the C. albicans mycelial form (27, 28) and of a number of other fungi that also exhibit extensive vacuolation during hyphal growth (50, 56). Formation of secondary germ tubes from mother cells or of branches from subapical compartments occurs only after regeneration of the cytoplasm at the expense of the vacuole.
The mechanism by which vacuole expansion occurs during germ tube formation and its inheritance remains mostly unknown, although it has been reported that a C. albicans vac8Δ mutant, defective in vacuolar inheritance, had an increased branching frequency (C. Barelle, R. Mathias, C. Gaillardin, N. Gow, and A. Brown, Abstr. Yeast Gen. Mol. Biol. Meet., abstr. 103, 2000). A C. albicans vps11Δ mutant, homologue of the S. cerevisiae class C VPS genes, has also been described recently (45). This mutant presented phenotypes that closely resembled those of class C vps mutants of S. cerevisiae, including the absence of a vacuolar compartment, and was defective in yeast-to-hypha morphogenesis; it was concluded that C. albicans VPS11 is required for vacuole biogenesis and also for germ tube emergence. Results reported by Palmer et al. (45) suggest that vacuole protein-sorting pathways, as well as vacuolar inheritance, are important during the yeast-to-hypha transition. In addition to the evidence that the vacuole plays a role in the morphological transition and in branching, it has also been suggested that vacuolar ABC transporters directly influence fungal virulence (60).
Here, we report the isolation, sequencing, and expression analysis of ABG1, a novel and essential C. albicans gene that codes for a vacuolar protein. The cellular localization of the protein was determined by tagging with the green fluorescent protein (GFP). Functional characterization of Abg1p included the construction of a conditional null strain. ABG1 heterozygous and conditional mutants were analyzed and shown to be defective in vacuole biogenesis and cytokinesis and also exhibited an increased rate of hyphal branching.
MATERIALS AND METHODS
Microorganisms, media, and growth conditions.
C. albicans strains used in this study are listed in Table 1. Cells were routinely grown in YPD (2% glucose, 1% yeast extract, 2% Bacto peptone [Difco]) or YNB (0.67% yeast nitrogen base without amino acids, 2% glucose) media at 28°C with shaking. Media were supplemented with uridine (25 μg/ml) when appropriate. For repressing conditions for the MET3 promoter, C. albicans strains were grown in YNB with 5 mM methionine and 2 mM cysteine. For sensitivity assays, plates containing solid (1.5% agar) YNB plus 2 mM methionine and 0.5 mM cysteine were supplemented with Congo red (200 μg/ml), sodium dodecyl sulfate (SDS) (0.025%), Calcofluor white (125 μg/ml), NaCl (1 M), glycerol (2.5 M), and H2O2 (6 mM).
TABLE 1.
Strains of C. albicans
| Strain | Parental strain | Genotype | Source or reference |
|---|---|---|---|
| 26555 | Wild type | ATCCa | |
| SC5314 | Wild type | 24 | |
| CAI4 | SC5314 | ura3Δ::λimm434/ura3Δ::λimm434 | 21 |
| CAI4-URA3 | CAI4 | ura3Δ::λimm434/ura3Δ::λimm434, RPS10::URA3 | This work |
| WO-1 | 52 | ||
| CJ1 | CAI4 | ura3Δ::λimm434/ura3Δ::λimm434; abg1Δ::URA3-dp1200/ABG1 | This work |
| CJ2 | CJ1 | ura3Δ::λimm434/ura3Δ::λimm434; abg1Δ::dp1200/ABG1 | This work |
| CV1 | CAI4 | ura3Δ::λimm434/ura3Δ::λimm434; abg1Δ::hisG-URA3-hisG/ABG1 | This work |
| CV2 | CV1 | ura3Δ::λimm434/ura3Δ::λimm434; abg1Δ::hisG/ABG1 | This work |
| CV3 | CV2 | ura3Δ::λimm434/ura3Δ::λimm434; abg1Δ::hisG/abg1Δ:URA3-MET3p-ABG1 | This work |
| CGA4 | CAI4 | CAI4 derivative, pACT-ABG1-GFP | This work |
American Type Culture Collection.
Different conditions were used to induce germ tube formation: (i) 10% (vol/vol) fetal calf serum (Sigma) in water (27), (ii) changes in pH and temperature (pH 4 to 7.5 and 25 to 37°C) in modified Lee's medium (47), or (iii) starvation-stimulated dimorphism (15). In this latter case, cells were grown in Lee's medium at 28°C overnight. Cells were next collected by centrifugation, washed twice in sterile distilled water, and resuspended in sterile distilled water at a final concentration of 2 × 107 cells/ml. This cellular suspension was incubated at 28°C with shaking (200 rpm) for 3 h and then subjected to a starvation period at 4°C for 48 h. After this period, cells were collected by centrifugation and resuspended in Lee's medium, prewarmed at 37°C to give the same final cell density mentioned above. To obtain yeast cells under similar conditions, the same protocol was followed but final incubation was performed at 28°C. For further phenotypic analysis, the CAI4 strain was transformed with CIp10 integrating vector using the RPS10 locus for the URA3 gene integration (42) to obtain the URA+ CAI4-URA3 strain. Reintegration of URA3 at an appropriate expression locus such as RPS10 offsets most problems related to the phenotypic changes associated with ectopic expression of URA3 during Ura-Blaster-mediated gene disruptions (6).
Transformation of C. albicans.
C. albicans was transformed by the lithium acetate procedure, described by Gietz et al. (24). Ura+ segregants were selected in minimal medium containing uridine and 5-fluoroorotic acid (1 μg/ml).
Isolation and cloning of the ABG1 gene.
Immunoscreening of a cDNA library of C. albicans ATCC 26555 in the expression vector λgt11 with a specific germ tube polyclonal antibody, previously obtained by our group (15), was performed by standard methods (1). Eight immunoreactive clones were detected, and cDNAs from these clones were amplified by PCR and sequenced by using λgt11 universal primers. Sequence analysis was carried out in the Stanford Genome Technology Center (http://sequence.standford.edu/group/candida/search.html) and Minnesota University (http://alces.med.umn.edu/gbsearch/ybc.html) databases. The isolated clone described in this work contained an incomplete open reading frame (ORF) which encodes a putative polypeptide of 288 amino acids showing 99% identity with contig 5-3205. Primers FC4 and RC4 (Table 2) amplify a 2,200-bp product from CAI4 genomic DNA that includes the entire ORF plus 700 bp of upstream sequence and 500 bp of downstream sequence. This product was digested with BamHI and HindIII and cloned into the vector pUC19 (66), also digested with BamHI and HindIII.
TABLE 2.
Primers
| Primer | Sequence (5′-3′) |
|---|---|
| λgt11F | GGTGGCGGACGACTCCTGGAGCCCG |
| λgt11R | TTGACACCAGACCAACTGGTAATG |
| FC4 | GAAAGGATCCAATTTTGGCTCG |
| RC4 | TGCTAAGCTTTGGTGACTTAGCGTGG |
| FP4 | GCGCGGATCCATGGTTTCACTTTCTAATTT |
| RP4 | GCGCGAATTCTTATTTTTATCAAAATCAATTTG |
| PF1F | GCGCAAGCTTCTGGATAAATTGGAAATGAAGGCC |
| PR1F | GCGCCTGCAGTATCTCTAGTGCTCCGTATA |
| PF2F | GCGCAGATCTCTGGACTTGAATTGTTATTC |
| PR2F | GCGCGGTACCTCTTCCGCTCAGCAAATAAACAA |
| N4F | AGTAGCAAAAGAGATAAACAAAGTGACAATGAAAATGATGCCGAAATCGAACAAGAAATTGCATTTTTTAACCAATAGGCCG |
| M4R | AGTATCGAAAATGCTGACAGGGTCTGACTCCTCGGAAGCTAAGGATTCAGCACCTTCAAAATTGTGTGGAATTGTGAGCGGATA |
| PORF | GCGCCTGCAGCTTTCCTTAATAATTTCATCT |
| RGFP | CCCGGGTTCATTTTTTTTATC |
Genomic library screening.
Hybridization screening of a C. albicans genomic library (strain WO-1) constructed in the plasmid vector pEMBLYe23 (2) was carried out as described elsewhere (52), using as a probe a 500-bp fragment from the ORF downstream sequence amplified with primers PF2F and PF2R (Table 2).
DNA and RNA manipulation methods.
Plasmid and phage DNA purification, digestion with restriction enzymes, subcloning, and Southern and Northern blotting were carried out by standard protocols (1). The digoxigenin detection kit was used for nonradioactive probe labeling and hybridization. All reagents were purchased from Roche Molecular Biochemicals. Genomic DNA was extracted from C. albicans cells as reported by Rose et al. (51), and total RNA was isolated using a RNeasy kit (QIAGEN) according to the manufacturer's instructions and extracted as described by Ausubel et al. (1).
ABG1 gene disruption.
The PCR-based gene disruption method (65) was used to attempt to generate an ABG1 knockout strain. Primers N4F and M4R (Table 2) were designed to contain 20 bp homologous to PCR disruption plasmid (pDDB57) flanked with 80 bp of sequence to direct homologous integration into the ABG1 ORF. Ura-Blaster protocol (22) was also used to attempt to generate an ABG1 null mutant. One 700-bp HindIII-PstI fragment from the 5′ ABG1 upstream noncoding region was amplified by PCR by using PF1F and PR1F primers (Table 2), digested, and cloned into the HindIII-PstI site from vector pBB510 (7, 46) to generate p510V. A 500-bp fragment from the 3′ downstream noncoding region was amplified by PCR by using PF2F and PR2F primers (Table 2), digested with BglII and KpnI, and cloned into p510V also digested with these restriction enzymes, to form plasmid p510VV. The ABG1 disruption cassette was excised from plasmid p510VV with HindIII and KpnI before transformation into the C. albicans CAI4 strain. For the construction of the ABG1 conditional mutant, a 200-bp PCR product was amplified from the ABG1 locus of the CAI4 strain using primers FP4 and PORF (Table 2). The amplified product was cut with BamHI and PstI and ligated in pCaDis plasmid (14) to generate pMD plasmid. Before transformation into ABG1 heterozygous strain (abg1Δ::hisG/ABG1), pMD was linearized with BsgI (New England Biolabs).
Abg1p localization by fusion with GFP.
Plasmid pAG-ADH-ABG1-GFP for expression of GFP-tagged Abg1p from the constitutive promoter ADH1 was created by amplifying ABG1 ORF from CAI4 genomic DNA with primers FP4 and RGFP (Table 2) and ligated into the ADH-GFP plasmid (pAG1) at BamHI and SmaI sites. The pAG-ADH-ABG1-GFP construction was used to transform the CAI4 strain, generating strain CGA4.
Microscopy.
Wet mounts of yeast cells grown in liquid culture medium and hyphal cells grown on poly-l-lysine-coated microscope slides (18), placed in petri dishes containing 10% (vol/vol) newborn calf serum (Gibco) in water plus amino acids, were examined with a Nikon Eclipse E800 microscope. Chitin localization was assessed with Calcofluor white staining as described previously (48). Nuclei were stained with a DAPI (4′,6-diamidino-2-phenylindole) (1 mg/ml), and vacuoles were stained with the vital stain FM4-64 (Molecular Probes) as described by Vida and Emr (62). GFP, DAPI, Calcofluor, and FM4-64 preparations were examined with a Leica DMR fluorescence microscope.
Antibodies.
The germ tube-specific polyclonal antibody (PAb anti-gt) against purified walls from mycelial cells of C. albicans was obtained as described elsewhere (15). An antibody recognizing Abg1p protein was prepared by Sigma-Genosys, by using a synthetic peptide selected from the deduced amino acid sequence from the ABG1 gene. PAb anti-Abg1p was raised against a 16-mer residue (ISSKRDKQSDNENDAC) derived from the N-terminal domain of the protein.
Subcellular fractionation.
Cells grown in YPD liquid medium at 28°C overnight were collected by centrifugation (4,000 × g, 10 min) and washed twice with chilled 1 mM phenylmethylsulfonyl fluoride in 10 mM Tris hydrochloride buffer (pH 7.2) (buffer A) and broken by shaking with glass beads (425 to 600 μm; Sigma). The procedure resulted in complete cell breakage as assessed by examination of the preparation in a phase-contrast microscope. The cell walls were pelleted (4,000 × g for 10 min) from the cell-free homogenate, washed four times with chilled buffer A, and then boiled for 5 min with 2% SDS in glass-distilled water to remove noncovalently bound proteins; they were finally washed four more times with buffer A. After sedimentation, the purified cell walls were digested in buffer A containing 0.5 mg/ml of Zymolyase 20T (ICN Biomedicals Inc.) for 3 h at 28°C. After treatment, the wall residue was removed by centrifugation (3,000 × g, 30 min) and discarded. The solubilized material was concentrated by freeze-drying. The supernatant fluid obtained subsequent to cell breakage (after cell wall sedimentation) was centrifuged at 40,000 × g for 40 min to obtain a mixed membrane fraction (P40), and the resulting supernatant was then centrifuged at 100,000 × g for 1 h to obtain a microsomal fraction (P100). The supernatant obtained (cytosolic fraction) was kept. The total sugar content in the cell wall digests was determined by the method of Dubois et al. (19), whereas the protein contents in the other samples (P40, P100, and the cytosolic fraction) were determined following the method described by Lowry et al. (39).
SDS-PAGE and Western blotting.
SDS-polyacrylamide gel electrophoresis (PAGE) under denaturing conditions was performed basically as described by Laemmli (36) with 12% linear gels. Electrophoretic transfer to nitrocellulose paper was carried out as described previously (15). Blotted proteins were immunodetected by using the primary specific antibodies (PAb anti-gt and PAb anti-Abg1p; see above) diluted (1:1,000 and 1:500, respectively) in 0.01 M Tris-HCl buffer (pH 7.4), containing 0.9% NaCl, 0.05% Tween 20, and 3% bovine serum albumin as a blocking agent. Peroxidase-conjugated secondary antibodies (Bio-Rad Laboratories) were used at 1:2,000 dilution. Immunoreactive species were detected by the enhanced chemiluminescent method from Amersham Biosciences, following the manufacturer's instructions.
Nucleotide sequence accession number.
The newly identified sequence homologous to assembly 5 was submitted to DBBJ/EMB/GenBank (accession number AY193774).
RESULTS
Isolation and cloning of the ABG1 gene.
An expression library of C. albicans strain ATCC 26555 was immunoscreened with PAb anti-gt (15). Several positive clones were isolated, and the cDNAs of these clones were amplified by PCR using the universal λ primers; the resulting sequences were analyzed by BLAST against the genome database of Candida albicans (http://Candida.stanford.edu/btComb.html). The nucleotide sequence of the clone described in this work was shown to include a novel incomplete ORF contained in contig 5-3205. This ORF encodes a putative polypeptide of 288 amino acids with a calculated molecular mass of 32,921 Da and a theoretical pI of 4.79. Heterozygous and conditional strains for this ORF displayed an altered budding growth pattern (see below). For this reason this gene was designated ABG1. A genomic clone of the gene was obtained by PCR using two primers deduced from the contig sequence (Table 2; see Materials and Methods). The 2,200-bp BamHI-HindIII fragment amplified contained the complete ORF and upstream and downstream sequences and was cloned in pUC19. The nucleotide sequence of the fragment was shown to be identical to contig 5-3205.
Revision and analysis of the ABG1 sequence.
In the last Stanford's Assembly of Candida albicans Sequence Project (assembly 19), contig 5-3205 containing the ABG1 sequence was restructured. Thus, the ABG1 upstream noncoding region is now contained in contig 19-1523. In this assembly, a 366-nucleotide sequence, including part of the ORF, is missing and the last fragment of the gene was contained in the contigs 19-10122 and 19-20122, encoding a different putative polypeptide of 189 amino acids (Fig. 1A). A similar arrangement for ABG1 exists in the European Candida albicans Database (http://genolist.pasteur.fr/CandidaDB/). To address this, a hybridization screening of a C. albicans WO-1 genomic library was performed using as a probe the region of ABG1 that remained unchanged between assemblies 5 and 19 and that corresponded to the downstream noncoding region. Two positive clones were obtained and subsequently sequenced. The nucleotide sequence of both clones was shown to be identical and exhibited 100% identity with our preliminary results and with assembly 5. These results were also confirmed by Southern analysis (Fig. 1B). Chromosomal DNA of two different C. albicans strains, SC5314 and CAI4, was digested with different combinations of restriction enzymes and hybridized using the same probe employed for the hybridization screening (the downstream noncoding region of the gene). In all cases, one hybridizing band that matched the restriction map provided in assembly 5 was found, confirming our previous data and suggesting that there is only one homologous sequence in the genomic DNA. Database searches revealed that ABG1 displayed no homology with any mammalian gene. The best match (27.7% overall identity) was found with a hypothetical ORF from Saccharomyces cerevisiae (YGR106c; accession number NC001139) that encoded a putative polypeptide of 293 amino acids. Analysis of the two sequences performed with the Dense Alignment Surface Program (16) revealed that both predicted polypeptides share a potential transmembrane domain and a signal peptide sequence. Thus, Abg1p has a putative signal peptide from positions 7 to 22, with a possible cleavage site between amino acids 22 and 23 and one transmembrane domain between amino acids 241 and 260, while the YGR106c gene product had a putative signal peptide at positions 8 to 16 and a transmembrane domain between amino acids 222 to 244.
FIG. 1.
Revision of the C. albicans ABG1 sequence. (A) Diagram of contigs 5-3205, 19-10122, 19-20122, and 19-1523 showing the location of the ABG1 gene (striped arrow). (B) Southern analysis of the ABG1 alleles. Genomic DNA from SC5314 (lanes 1, 2, and 3) and CAI4 (lanes 4, 5, and 6) strains was digested with BamHI/HindIII (lanes 1 and 4), BamHI/SmaI (lanes 2 and 5), and with XbaI/HindIII (lanes 3 and 6). A 1,400-bp DNA fragment from the coding sequence of ABG1 in contig 5-3205 was used as a probe. In all cases fragment sizes observed were consistent with the restriction map of contig 5-3205 from Candida albicans Sequence Project (Stanford University).
ABG1 is transcribed in both yeast and hyphal C. albicans cells.
The expression profile of the ABG1 transcript was determined by Northern analysis during C. albicans starvation-stimulated morphological transition. Yeast and hyphal cells of strain CAI4 were obtained (see Materials and Methods) and total RNA was isolated from starved cells at 30, 60, and 120 min after inoculation. The ABG1 ORF was used as a probe, and ABG1 mRNA expression was detected 30 min after the induction of the morphological yeast-to-mycelium transition; mRNA levels remained constant for at least 2 h in all samples. ABG1 was also expressed in cells grown under other germ tube induction conditions tested (see Materials and Methods). This pattern of mRNA expression was found to be consistent with the results from the gene disruption experiments performed, since a constitutive expression could be expected for an essential gene (see below).
Abg1p resides in the vacuolar membrane of C. albicans yeast and hyphal cells.
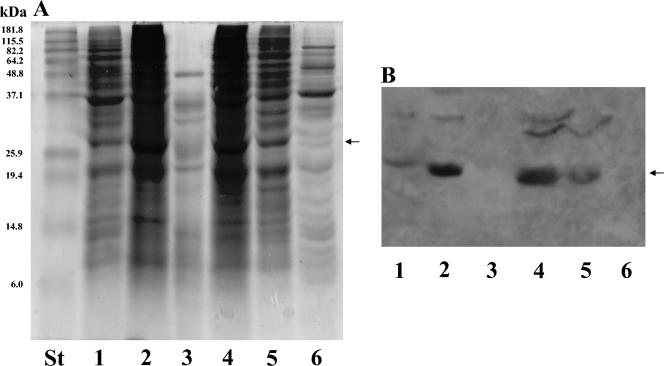
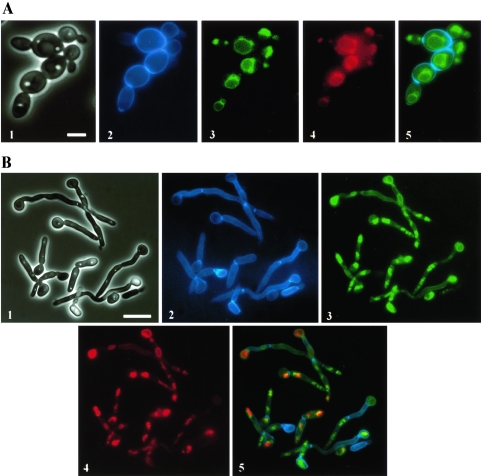
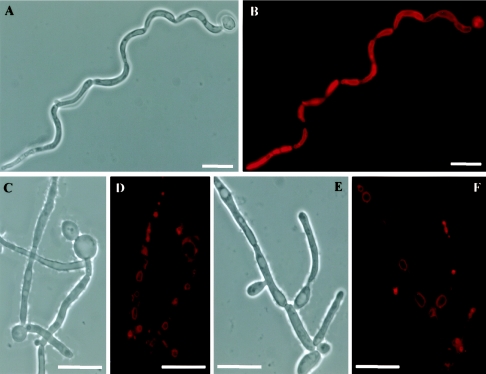
To identify the gene product encoded by ABG1, the protein species present in several subcellular fractions (different cell wall extracts, total whole-cell homogenate, P40, P100, and cytosol; see Materials and Methods) obtained from the CAI4 strain were analyzed by SDS-PAGE and Western analysis using PAb anti-Abg1p as a probe. An immunoreactive 32.9-kDa major species was detected in all samples assayed except SDS cell wall extract and cytosol fraction (Fig. 2). These results were consistent with the suggested cytoplasmic membrane localization for the Abg1p protein. Although PAb anti-Abg1p appears to be specific, reactivity of the antibody was also detected in the same samples towards species of 35 and 37 kDa which may correspond to polypeptides sharing common epitopes with Abg1p species or, alternatively, may represent different states (e.g., phosphorylation levels, since bioinformatic analysis revealed several possible phosphorylation sites in the Abg1p sequence) of the Abg1p polypeptide that could affect its electrophoretic mobility. To examine Abg1p localization in live Candida cells, a fragment encoding the codon-optimized yEGFP (17) was fused in frame to the 3′ end of ABG1. The fusion construct was placed under control of the ADH1 promoter in an ARS/URA3-containing plasmid. Tagged cells containing this construction and growing under yeast and hyphal induction conditions showed ring-like GFP fluorescence within the cytoplasm (Fig. 3). Costaining with FM4-64, a membrane-specific styryl dye that labels the compartments of the endocytotic pathway and ultimately the vacuolar membrane (62), revealed that the Abg1p-GFP fusion proptein was associated mainly with the vacuole in C. albicans (Fig. 3). However, the Abg1p species could also be associated with other subcellular membranes (i.e., plasma membrane), as revealed by the reactivity towards PAb anti-Abg1p observed in the SDS extract (Fig. 2, lane 2) and in the P100 sample (Fig. 2, lane 5).
FIG. 2.
SDS-PAGE and Western analysis of subcellular fractions from the C. albicans CAI4 strain. Linear 12% polyacrylamide gels were loaded (50 μg of each sample expressed as total protein content per well) with whole-cell lysate (lane 1), SDS cell wall extract (lane 2), Zymolyase cell wall digest (lane 3), P40 fraction (lane 4), P100 (lane 5), and soluble (cytosol) fraction (lane 6). After electrophoresis, gels were stained with Coomassie blue (panel A) and polypeptides were subsequently transferred to nitrocellulose sheets and immunodetected with PAb anti-Abg1p (panel B). Molecular weight standards were run in parallel with the different samples (lane labeled St in panel A). The arrows point to the band corresponding to Abg1p.
FIG. 3.
Abg1p localizes in the vacuole membrane of C. albicans yeast cells (A) and mycelial filaments (B). Phase-contrast image (1), Calcofluor-stained cells (2), Abg1p-GFP (3) and FM4-64 staining (4) fluorescence micrographs, and a merge image (5) of CAI4 cells expressing GFP-tagged Abg1p. Bars in panels 1 of part A (5 μm) and part B (10 μm) are representative of the corresponding series of panels (1 to 5) for each part of the figure.
ABG1 is an essential gene for C. albicans.
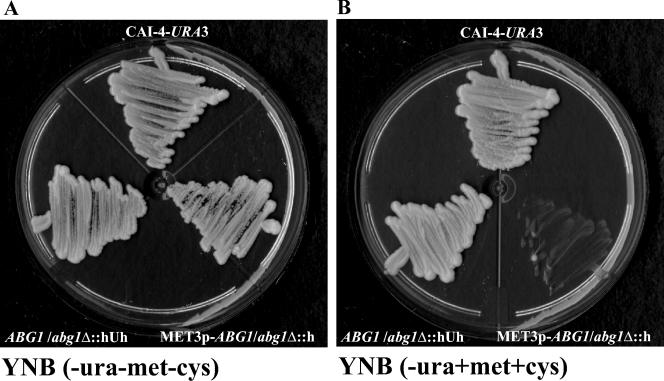
Initial attempts to construct an abg1Δ/abg1Δ null mutant using a PCR-based gene disruption method (65) were unsuccessful. The first copy of ABG1 was disrupted by integration of the URA3 cassette (see Materials and Methods). More than 100 transformants recovered after the second round of transformation were analyzed, but all of them still contained a wild-type ABG1 allele. Attempts to disrupt the second allele using the Ura-Blaster method also failed. These results suggested that ABG1 was essential. In order to test this possibility, the second ABG1 allele in the heterozygous strain (CV2 strain; Table 2) was placed under the control of the MET3 promoter (14). pMD cassette (see Materials and Methods) was used to transform the CV2 strain, generating a conditional mutant (CV3). Southern analysis was used to confirm the genotypes of strains CV1 and CV2 and the disruption and replacement of the second ABG1 allele with a new Met3p-driven ABG1 allele in the conditional strain (data not shown). To check the regulation of ABG1 expression by the MET3 promoter in the conditional strain, Northern analysis was performed. CV3 strain was grown in YNB for 24 h and then inoculated at a low cell density (optical density at 600 nm of 0.1) in YNB supplemented with methionine and cysteine. After 4 h under repressing conditions, no ABG1 expression was detected, although normal ABG1 expression was found when the CV3 strain was inoculated in YNB without amino acids. This loss of expression was not found to be influenced by methionine or cysteine supplementation of growth media since in the presence of both amino acids normal ABG1 expression was found in the parental CAI4-URA3 strain (data not shown). When CV3 was plated on solid YNB supplemented with amino acids, no growth was detected, although CAI4-URA3 and the heterozygous Ura+ strain grew normally. When plated in YNB without methionine and cysteine, CV3 grew as well as CAI4-URA3 and the Ura+ heterozygous strain, thus confirming that ABG1 is an essential gene in C. albicans (Fig. 4).
FIG. 4.
ABG1 is an essential gene. CAI4-URA3 and ABG1 heterozygous (CV1) and hemizygous (CV3) strains were plated on YNB medium in the absence (A) and in the presence (B) of methionine and cysteine.
Repression of ABG1 results in morphological alterations in yeast and hyphal cells and in vacuolar morphology defects.
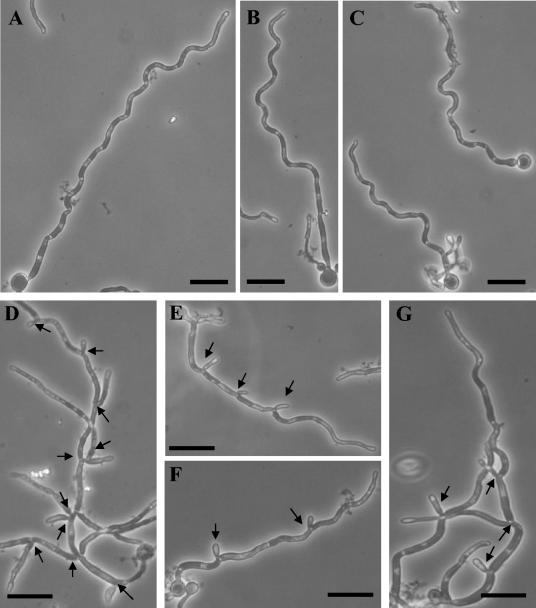
Heterozygous strains obtained by both disruption techniques displayed an altered budding growth pattern, characterized by the formation of chains of buds that decreased in size towards the chain end (Fig. 5B). Southern blot analysis revealed no polymorphisms in the ABG1 sequence (see above; Fig. 1B); consequently, the phenotype displayed by the heterozygous mutant appears to not be allele specific. Since the heterozygous mutant displayed a morphological phenotype, overexpression of ABG1 was tested by introduction of an extra copy of ABG1 under the control of ACT promoter, but no apparent effect on phenotype was observed. The conditional strain grown in the absence of amino acids behaves exactly the same way as the heterozygous strain; therefore, all phenotypic studies were done in the presence of both methionine and cysteine, since parental and heterozygous strains were not affected by the presence of these two amino acids in the culture medium (data not shown). When the conditional mutant was incubated under conditions that normally support budding growth (YPD medium supplemented with methionine and cysteine, incubated at 28°C), it displayed defects in cytokinesis (Fig. 5C and D) similar to those observed in the heterozygous strain (see above). When the conditional CV3 mutant strain was incubated under hypha-inducing conditions, the branching frequency was higher than that observed in the parental strain. This is predicted from previous models inferring that asymmetric vacuole inheritance is associated with nonbranching hyphae (4). Since previous reports (4) also indicated that the delay between cytokinesis and the formation of a germ tube or branch in the newly formed subapical compartment was dependent on the serum concentration, we examined the branching frequency of the conditional ABG1 mutant cells growing overnight in 10% (vol/vol) serum on poly-l-lysine-coated microscope slides. Since the lengths of cell compartments were found to be similar in the parental and conditional mutant strains, parameters employed to quantify branching frequency were (i) the number of branches per hypha and (ii) the number of cell compartments that exhibited a branch. Using these criteria, about 95% ± 5% of hyphae in the CV3 strain had one or more branches, compared to 20% ± 5% in the case of the CAI4-URA3 strain. Moreover, 37% ± 11.4% of cell compartments in hyphal filaments in the CV3 strain had one branch (Fig. 6D to G) compared to 6% ± 4% in the control strain.
FIG. 5.
Phase-contrast microscopy observations of CAI4-URA3 (A), CV2 (B), and CV3 (C and D) strains grown under conditions to induce budding growth (see Materials and Methods). Cells in panels A, C, and D were grown in YPD medium supplemented with methionine and cysteine, whereas cells in panel B were grown in the absence of both amino acids. Bar = 10 μm in all panels.
FIG. 6.
Phase-contrast microscopy observations of CAI4-URA3 (A, B, and C) and CV3 (D, E, F, and G) incubated under conditions to induce hyphal growth (see Materials and Methods). Bar = 10 μm in all panels. Arrows in panels D, E, F, and G point to branches in hyphal filaments.
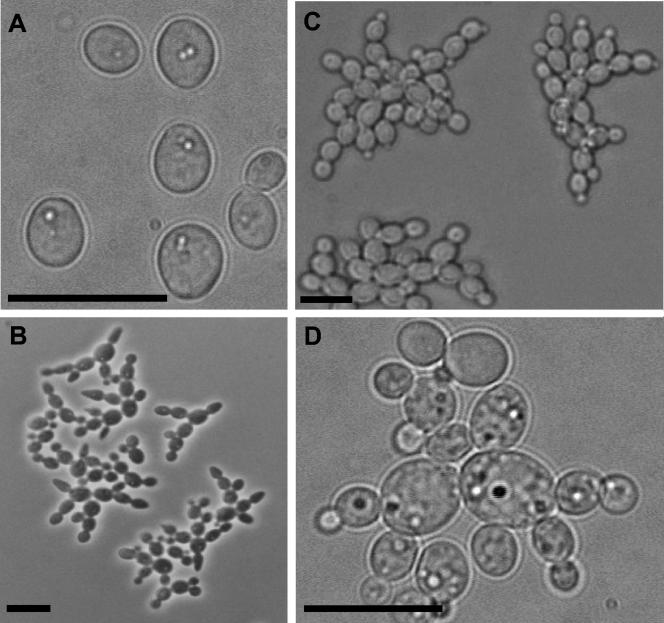
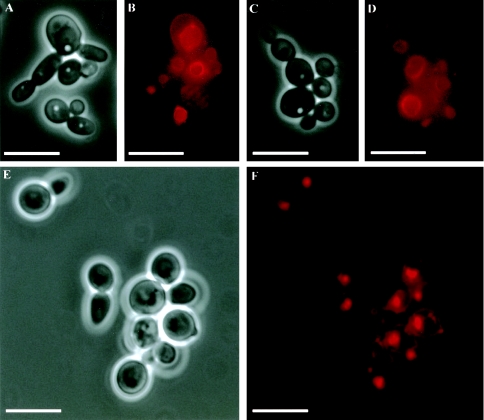
Because the ABG1 gene product was located in the vacuolar membrane (see above), vacuolar morphology in the conditional mutant was examined with the vital dye FM4-64 (see Materials and Methods). Vacuolar compartments in yeast (Fig. 7A to D) and hyphal (Fig. 8A to B) cells of the wild-type strain occupied a significant portion of the cell, whereas in the conditional mutant strain vacuoles were smaller (Fig. 7E to F; Fig. 8C to F). Moreover, in CV3 yeast cells, single, apparently fragmented vacuolar compartments were observed (Fig. 7E to F), whereas in hyphal cells, several nonfragmented, smaller vacuoles were observed in each cellular compartment (Fig. 8C to F). Despite these vacuolar defects, distribution and morphology of other subcellular organelles such as nuclei were unaffected in the ABG1 conditional strain. In all cases, there were no apparent defects in the uptake and/or the transport to the vacuole of FM4-64, as concluded from the homogeneous vacuolar membrane staining observed.
FIG. 7.
Vacuole morphology in yeast cells. Phase-contrast (A, C, and E) and FM4-64-staining fluorescence observations (B, D, and F) of CAI4-URA3 (A-D) and CV3 (E-F) strains. Cells of the wild-type strain exhibited single, large vacuolar compartments (B and D), while in the conditional mutant strain cells a single vacuole was also observed, although in this case the vacuoles were smaller and showed a collapsed appearance (F). Bar = 10 μm in all panels.
FIG. 8.
Vacuole morphology in hyphal cells. Phase-contrast (A, C, and E) and FM4-64-staining fluorescence observations (B, D, and F) of CAI4-URA3 (A and B) and CV3 (C-F) strains. ABG1 conditional mutant cells had several small vacuoles in each cellular compartment (D, F), while in the wild-type cells a single, large vacuole that occupied almost all the cytoplasmic volume was observed (B). Bar = 10 μm in all panels.
ABG1 heterozygous and conditional mutants have defects in the cell wall.
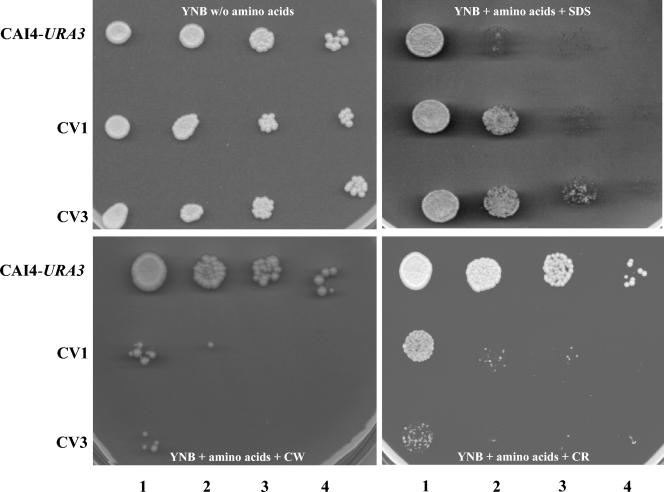
Phenotypes indicative of defective cell wall structure and/or composition in ABG1 heterozygous and conditional mutants were assessed by determining the sensitivity of CAI4-URA3, CV1, and CV3 strains to Calcofluor white, SDS, and Congo red. These tests were carried out in YNB agar supplemented with amino acids, since total repression of ABG1 expression in the CV3 strain was required and the presence of methionine and cysteine in the culture medium does not affect the response of CAI4-URA3 and CV1 strains to the agents assayed (data not shown). Heterozygous (CV1) and ABG1 conditional (CV3) strains were unable to grow on YNB agar supplemented with 125 μg/ml Calcofluor white, an agent which binds to nascent chitin chains and prevents cross-linking of the chitin chains to form microfibrils. In this context, mutants with elevated chitin levels or with other additional defects in cell wall are hypersensitive to Calcofluor white (49). The CV3 strain also exhibited an increased sensitivity to Congo red and a decreased sensitivity to SDS, thus suggesting an additional defect in the β-1-3 d-glucan assembly (34) and an alteration of the chemical composition of the cell wall (Fig. 9).
FIG. 9.
Sensitivity to cell wall-disturbing agents. Drops (5 μl) of serial cell suspensions containing from 106 to 103 cells per ml (from left to right in each panel) of parental (CAI4-URA3), heterozygous (CV1), and conditional (CV3) strains were spotted onto YNB medium supplemented with SDS (0.025%), Calcofluor white (CW; 125 μg/ml), and Congo red (CR; 200 μg/ml).
DISCUSSION
Immunoscreening of a cDNA library with a germ tube-specific polyclonal antibody (15) led to the isolation of a novel gene encoding a putative polypeptide of 288 amino acids, which on the basis of the altered budding growth pattern exhibited by the heterozygous and conditional strains for this gene, was designated as ABG1. A search for sequence similarities in the current protein databases revealed that there was no obvious homologue of the ABG1 gene product (Abg1p) in mammals, although the best match (27.7% overall identity) was found with a hypothetical ORF from the most-related organism S. cerevisiae that encodes a putative polypeptide of 293 amino acids. In this work we report isolation and characterization of ABG1, a novel C. albicans gene that encodes a vacuolar protein. Evidence supporting the vacuolar localization for the ABG1 gene product included analysis of the deduced amino acid sequence from Abg1p, which showed the presence of a potential transmembranal domain and a signal peptide, and the use of specific PAb anti-Abg1p antibody, which allowed the identification in Western immunoblots of a 32.9-kDa immunoreactive polypeptide mostly in the mixed membrane fraction (P40) (but also in SDS cell wall extract and in the P100 fraction); in this context, it should be stressed that Western blotting was the only suitable procedure for immunological detection of ABG1 product, since the presence of a thick cell wall in C. albicans represents an unavoidable obstacle (along with the plasma membrane) for immunoglobulins to access cytosolic space. Besides, the GFP-tagged gene product of the possible S. cerevisiae homolog (YGR106c) for the ABG1 C. albicans gene was found to be located in yeast vacuolar membrane (32); and finally, Abg1p tagged with the GFP localized in the vacuolar membrane of living C. albicans cells, in both yeast and hyphal forms. Although vacuolar mislocalization of GFP-tagged proteins has been reported (35), these species were NH2-terminal GFP fusion products in contrast with the COOH-terminal fusion protein used in this work, and vacuolar targeting signals are usually within the NH2-terminal sequences of proteins (33, 40). Besides, reverse transcription-PCR analysis demonstrated that ABG1 endogenous promoter has a similar expression level as ACT promoter (results not shown), so an overproduction of Abg1p-GFP product which may be responsible for its vacuolar mislocalization is not likely; on the other hand, the location of Abg1p is consistent with its involvement in vacuole morphology and inheritance, since mutant strain cells exhibited an altered vacuolar morphological appearance, as revealed by fluorescence microscopy observation of FM4-64-stained cells.
Despite the low homology between ABG1 and YGR106c, both gene products share the characteristic of being located at the vacuolar membrane level, so complementation experiments to confirm that these two genes actually are homologs could be an interesting possibility, although this experimental approach has severe limitations since (i) the S. cerevisiae YGR106cΔ mutant has no detectable phenotype and (ii) the YGR106c ORF contains at least one CUG codon which in C. albicans is translated as serine instead of the universal leucine (53).
The fact that antibodies raised against purified cell wall preparations may also cross-react with nonwall cell components released during the isolation and purification processes is not unusual since similar results have also been reported by different authors (4, 21, 23). Since the vacuole is a large compartment in the cytoplasm of hyphal cells (27), it is possible that contamination with vacuolar components may occur during the cell wall purification process, and consequently polyclonal antiserum raised towards purified cell walls (PAb anti-gt) may also contain antibodies to vacuolar components. Construction of a conditional strain for ABG1 under MET3 promoter control enabled us to demonstrate that this novel gene coding for a vacuolar protein is essential for C. albicans, since the growth of the conditional strain was completely inhibited in the presence of methionine and cysteine at levels that allowed the normal growth of the CAI4-URA3 strain. Methionine- and cysteine-mediated repression of ABG1 in the conditional strain was used to study the effects of Abg1p depletion on the morphogenesis of C. albicans. Under repressing conditions, yeast cells of the conditional strain exhibited a morphological phenotype characterized by a failure in the cytokinesis that lead to the formation of bud chains. When hyphal growth was induced in the presence of amino acids, normal hyphal induction was observed in the conditional strain but an altered hyphal branching vacuolation pattern was observed. Overall, these observations suggest that Abg1p may be involved in cytokinesis and vacuole biogenesis. The observed alterations in the number and in the morphology and size of vacuoles may be correlated with the increased rate of hyphal branching, which is in agreement with the relationship between vacuolar volume and cell cycle progression as reported by Barelle et al. (4). Loss of ABG1 function also resulted in pleiotropic effects on structure and/or composition of the cell wall.
Such changes have been the focus of the research of numerous groups, since many of the biological functions related to the biology, pathogenicity, and virulence in this microorganism reside in the cell wall (9, 11, 13, 31, 38, 41, 43, 54). The conditional mutant exhibited more sensitivity to Calcofluor white and Congo red, although the cytokinesis defects observed could also be related to alterations in wall structure, which in turn may account for the increased sensitivity to cell wall-perturbing agents. Chitin deposition in the mutant strain did not appear to be affected; however, the sensitivity of CV1 and CV3 strains to SDS was decreased. The secretory pathway could also be affected by ABG1 disruption, which in turn may affect, directly or indirectly, cell wall biogenesis.
However, little is known about the role that intracellular organelles, such as the vacuole, play in growth and pathogenicity of C. albicans. The vacuole is a multifunctional compartment involved in hydrolysis like the mammalian lysosome and in storage, calcium ion homeostasis, and osmoregulation in the case of the plant vacuole; in fungal cells, vacuoles are required for compartmentalization of metabolites as well as toxic substances (33, 59).
Known roles suggested for vacuoles in yeast are described as degradation of cellular proteins, cytoplasmic homeostasis, and survival during periods of stress (3, 58). In this context, several genes encoding proteins related to vacuolar morphology and/or functions have been characterized. None of the phenotypes displayed by C. albicans mutants for VAM, VMA, VAC, and VPS genes (i.e., sensitivity to calcium chloride and 37°C temperature, sensitivity to osmotic stress [e.g., glycerol or sodium chloride], vacuolar morphology, or inheritance) (8, 45, 61, 63, 64) resemble the characteristic phenotype exhibited by ABG1 mutants, which appear to be affected in cytokinesis and branching processes and were not sensitive to osmotic, oxidative, and temperature stress (data not shown).
How organelle biogenesis and inheritance is linked to cell division is poorly understood in fungi. Loss of Abg1p leads to a defect in yeast cell cytokinesis, causing an altered budding growth and also an increase in branching frequency during hyphal development. Interestingly, Cln3p, a G1 cyclin described in S. cerevisiae, controls initiation of cell division and also vacuolar biogenesis and segregation (30). Organelle contribution to overall cell size is not usually taken into consideration in studies of cell size control, but the results from increased branching in hyphae in the CV3 conditional strain suggest that vacuoles are playing an essential role in the cell size control. Thus, the CV3 cells have an increased frequency of division, indicated by a higher number of branches in hyphae, with respect to the parental strain which correlated with the presence of more but smaller vacuoles in each cellular hyphal compartment when compared with the wild-type strain. On the basis of the observed correlation between vacuolation and cell cycle progression (4), the concept of size-regulated cell cycle control must be refined; that is to say, the cell size is related to its cytoplasmic volume minus the vacuole (and other organelle) space rather than total cell volume. The gene product of ABG1 links vacuole biology with cellular morphogenesis, cell wall biosynthesis, and cell cycle progression. All of these processes are intimately associated with virulence traits in C. albicans. In addition, Abg1p is the first essential vacuolar protein described in C. albicans and has no homology with mammalian genes, suggesting that this gene product could be regarded as a potential target for antifungal chemotherapy.
Acknowledgments
This work has been supported by grants BMC2001-2975 from the Programa Nacional de Promoción General del Conocimiento, Ministerio de Ciencia y Tecnología, and GRUPOS/04/04 from the Subsecretaria de la Oficina de Ciencia y Tecnología de la Generalitat Valenciana, Valencia, Spain (to J.P.M). N.A.R.G. acknowledges grants from the BBSRC, MRC, and Wellcome Trust. A.D. acknowledges grants MCRTN-CT-2003-504148 from the European Commission and BIO-2002-02124 from DGCyT, Ministerio de Ciencia y Tecnología. V.V. was the recipient of a predoctoral grant from Ministerio de Educación, Cultura y Deporte, Spain.
We acknowledge W. L. Chaffin and B. B. Magee for the kind gift of expression and genomic libraries. We thank A. Mitchell, B. Braun, P. Sudbery, and E. López for plasmid cession. Collaboration from N. Martín, E. López, and R. Dégano in this work is acknowledged.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1992. Current protocols in molecular biology. Wiley Interscience, New York, N.Y.
- 2.Baldari, C., and G. Cesareni. 1985. Plasmids pEMBLY: a new single stranded shuttle vectors for the recovery and analysis of yeast DNA sequences. Gene 35:27-32. [DOI] [PubMed] [Google Scholar]
- 3.Banta, L. M., J. S. Robinson, D. J. Klionsky, and S. D. Emr. 1988. Organelle assembly in yeast: characterization of yeast mutants defective in vacuolar biogenesis and protein sorting. J. Cell Biol. 107:1369-1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barelle, C. J., E. A. Bohula, S. J. Kron, D. Wessels, D. R. Soll, A. Schafer, A. J. P. Brown, and N. A. R. Gow. 2003. Asynchronous cell cycle and asymmetric vacuolar inheritance in true hyphae of Candida albicans. Eukaryot. Cell 2:398-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berman, J., and N. A. R. Gow. 2004. Cell cycle of fungal pathogens, p. 101-125. In G. San-Blas and R. A. Calderone (ed.), Pathogenic fungi: structural biology and taxonomy. Caister Academic Press, Norfolk, United Kingdom.
- 6.Brand, A., D. M. MacCallum, A. J. Brown, N. A. Gow, and F. C. Odds. 2004. Ectopic expression of URA3 can influence the virulence phenotypes and proteome of Candida albicans but can be overcome by targeted reintegration of URA3 at the RPS10 locus. Eukaryot. Cell. 3:900-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braun, R. B., and A. D. Johnson. 2000. TUP1, CPH1 and EFG1 make independent contributions to filamentation in Candida albicans. Genetics 155:57-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruckmann, A., W. Künkel, K. Augsten, R. Wetzker, and R. Eck. 2001. The deletion of CaVPS34 in the human pathogenic yeast Candida albicans causes defects in vesicle-mediated protein sorting and nuclear segregation. Yeast 18:343-353. [DOI] [PubMed] [Google Scholar]
- 9.Calderone, R. A. 1993. Recognition between Candida albicans and host cells. Trends Microbiol. 1:55-58. [DOI] [PubMed] [Google Scholar]
- 10.Calderone, R. A. (ed.). 2002. Candida and candidiasis. American Society for Microbiology, Washington, D.C.
- 11.Calderone, R. A., and P. C. Braun. 1991. Adherence and receptor relationships of Candida albicans. Microbiol. Rev. 55:1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calderone, R. A., and W. A. Fonzi. 2001. Virulence factors of Candida albicans. Trends Microbiol. 9:327-335. [DOI] [PubMed] [Google Scholar]
- 13.Chaffin, W. L., J. L. López-Ribot, M. Casanova, D. Gozalbo, and J. P. Martínez. 1998. Cell wall and secreted proteins of Candida albicans: identification, function, and expression. Microbiol. Mol. Biol. Rev. 62:130-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Care, R. S., J. Trevethick, K. M. Binley, and P. E. Sudbery. 1999. The MET3 promoter: a new tool for Candida albicans molecular genetics. Mol. Microbiol. 34:792-798. [DOI] [PubMed] [Google Scholar]
- 15.Casanova, M., M. L. Gil, L. Cardeñoso, J. P. Martínez, and R. Sentandreu. 1989. Identification of wall-specific antigens synthesized during germ tube formation by Candida albicans. Infect. Immun. 57:262-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cserzo, M., E. Wallin, I. Simon, G. Von Heijne, and A. Elofsson. 1997. Prediction of transmembrane α-helices in prokaryotic membrane proteins: the dense alignment surface methods. Protein Eng. 10:673-676. [DOI] [PubMed] [Google Scholar]
- 17.Cormack, C. P., G. Bertram, M. Egerton, N. A. R. Gow, S. Falkow, and A. J. P. Brown. 1997. Yeast-enhanced green fluorescent protein (yEGFP): a reporter of gene expression in Candida albicans. Microbiology 143:303-311. [DOI] [PubMed] [Google Scholar]
- 18.Crombie, T., N. A. R. Gow, and G. W. Gooday. 1990. Influence of applied electrical fields on yeast and hyphal growth of Candida albicans. J. Gen. Microbiol. 136:311-317. [DOI] [PubMed] [Google Scholar]
- 19.Dubois, M., K. A. Gilles, J. K. Hamilton, P. A. Rebers, and F. Smith. 1956. Colorimetric method for determination of sugars and related substances. Anal. Chem. 28:350-356. [Google Scholar]
- 20.Edmond, M. B., S. E. Wallace, D. K. McClish, M. A. Pfaller, R. N. Jones, and R. P. Wenzel. 1999. Nosocomial bloodstream infections in United States hospitals: a three-year analysis. Clin. Infect. Dis. 29:239-244. [DOI] [PubMed] [Google Scholar]
- 21.Eroles, P., M. Sentandreu, M. V. Elorza, and R. Sentandreu. 1997. The highly immunogenic enolase and Hsp70p are adventitious Candida albicans cell wall proteins. Microbiology 143:313-320. [DOI] [PubMed] [Google Scholar]
- 22.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galán, A., M. Casanova, A. Murgui, D. M. MacCallum, F. C. Odds, N. A. R. Gow, and J. P. Martínez. 2004. The Candida albicans pH-regulated KER1 gene encodes a lysine/glutamic-acid-rich plasma-membrane protein that is involved in cell aggregation. Microbiology 150:2641-2651. [DOI] [PubMed] [Google Scholar]
- 24.Gietz, R. D., R. H. Schiestl, A. R. Willems, and R. A. Woods. 1995. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11:355-360. [DOI] [PubMed] [Google Scholar]
- 25.Gillum, A. M., E. Y. H. Tsay, and D. R. Kirsch. 1984. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of Saccharomyces cerevisiae URA3 and E. coli pyr mutations. Mol. Gen. Genet. 198:179-182. [DOI] [PubMed] [Google Scholar]
- 26.Gow, N. A. R. 2002. Cell biology and cell cycle of Candida albicans, p. 145-158. In R. A. Calderone (ed.), Candida and candidiasis. American Society for Microbiology, Washington, D.C.
- 27.Gow, N. A. R., and G. W. Gooday. 1982. Vacuolation, branch production and linear growth of germ tubes of Candida albicans. J. Gen. Microbiol. 128:2195-2198. [DOI] [PubMed] [Google Scholar]
- 28.Gow, N. A. R., and G. W. Gooday. 1987. Cytological aspects of dimorphism in Candida albicans. Crit. Rev. Microbiol. 15:73-78. [DOI] [PubMed] [Google Scholar]
- 29.Gow, N. A. R., G. Henderson, and G. W. Gooday. 1986. Cytological interrelationships between the cell cycle and duplication cycle of Candida albicans. Microbios 47:97-105. [PubMed] [Google Scholar]
- 30.Han, B., R. Amarayo, and M. Polymenis. 2003. The G1 cyclin Cln3p controls vacuolar biogenesis in Saccharomyces cerevisiae. Genetics 165:467-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hostetter, M. K. 1994. Adhesins and ligands involved in the interaction of Candida spp. with epithelial and endothelial surfaces. Clin. Microbiol. Rev. 7:29-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huh, W., J. V. Falvo, L. C. Gerke, A. S. Carrol, R. W. Howson, J. S. Weissman, and E. K. O'Shea. 2003. Global analysis of protein localization in budding yeast. Nature 425:686-691. [DOI] [PubMed] [Google Scholar]
- 33.Klionsky, D. J., P. K. Herman, and S. D. Emr. 1990. The fungal vacuole: composition, function, and biogenesis. Microbiol. Rev. 54:266-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kopecka, M., and M. Gabriel. 1992. The influence of congo red on the cell wall and (1-3)-β-d-glucan microfibril biogenesis in Saccharomyces cerevisiae. Arch. Microbiol. 158:115-126. [DOI] [PubMed] [Google Scholar]
- 35.Kunze, I. I., G. Hensel, K. Adler, J. Bernard, B. Neubohn, C. Nilsson, R. Stoltenburg, S. D. Kohlwein, and G. Kunze. 1999. The green fluorescent protein targets secretory proteins to the yeast vacuole. Biochim. Biophys. Acta 1410:287-298. [DOI] [PubMed] [Google Scholar]
- 36.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 37.Lo, H. J., J. R. Kohler, B. DiDomenico, D. Loebenberg, A. Cacciapuoti, and G. R. Fink. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90:939-949. [DOI] [PubMed] [Google Scholar]
- 38.López-Ribot, J. L., M. Casanova, A. Murgui, and J. P. Martínez. 2004. Antibody response to Candida albicans cell wall antigens. FEMS Immunol. Med. Microbiol. 41:87-196. [DOI] [PubMed] [Google Scholar]
- 39.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 40.Martínez, E., B. Segui-Real, E. Silles, M. J. Mazón, and I. V. Sandoval. 1999. The prepropeptide of vacuolar aminopeptidase I is necessary and sufficient to target the fluorescent reporter protein GFP to the vacuole of yeast by the Ccvt pathway. Mol. Microbiol. 33:52-62. [DOI] [PubMed] [Google Scholar]
- 41.Martínez, J. P., J. L. López-Ribot, M. L. Gil, and W. L. Chaffin. 1998. Serologic response to cell wall mannoproteins and proteins of Candida albicans. Clin. Microbiol. Rev. 11:121-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murad, A. M. A., P. R. Lee, I. D. Broadbent, C. J. Barelle, and A. J. P. Brown. 2000. CIp10, an efficient and convenient integrating vector for Candida albicans. Yeast 16:325-327. [DOI] [PubMed] [Google Scholar]
- 43.Naglik, J. R., S. J. Challacombe, and B. Hube. 2003. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol. Mol. Biol. Rev. 67:400-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Odds, F. C., N. A. R. Gow, and A. J. P. Brown. 2001. Fungal virulence studies come of age. Genome Biol. 2:1009.1-1009.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Palmer, G. E., A. Cashmore, and J. Sturtevant. 2003. Candida albicans VPS11 is required for vacuole biogenesis and germ tube formation. Eukaryot. Cell 2:411-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pérez-Martin, J., J. A. Uría, and A. D. Johnson. 1999. Phenotypic switching in Candida albicans is controlled by a SIR2 gene. EMBO J. 18:2580-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Porta, A., A. M. Ramón, and W. A. Fonzi. 1999. PRR1, a homolog of Aspergillus nidulans palF, controls pH-dependent gene expression and filamentation in Candida albicans. J. Bacteriol. 181:7516-7523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pringle, J. R., A. E. M. Adams, D. G. Drubin, and B. K. Haarer. 1991. Immunofluorescence methods for yeast, vol. 194. Academic Press Ltd., London, England. [DOI] [PubMed]
- 49.Ram, A. F. J., A. Wolters, R. Ten Hoopen, and F. M. Klis. 1994. A new approach for isolating cell wall mutants in Saccharomyces cerevisiae by screening for hypersensitive to Calcofluor white. Yeast 10:1019-1030. [DOI] [PubMed] [Google Scholar]
- 50.Robinow, C. F. 1963. Observations on cell growth, mitosis, and division in the fungus Basidiobolus ranarum. J. Cell Biol. 17:123-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rose, M. D., F. Winston, and D. Hieter. 1990. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 52.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning. A laboratory manual, 2nd edition. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 53.Santos, M. A., G. Keith, and M. F. Tuite. 1993. Non-standard translational events in Candida albicans mediated by an unusual seryl-tRNA with a 5′-CAG-3′ (leucine) anticodon. EMBO J. 12:607-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sentandreu, R., J. P. Martínez, M. V. Elorza, and S. Mormeneo. 1991. Relationships between dimorphism, cell wall structure, and surface activities in Candida albicans, p. 72-88. In R. Prasad (ed.), Candida albicans: cellular and molecular biology. Springer-Verlag, Berlin, Heidelberg, Germany.
- 55.Soll, D. R., C. J. Langtimm, J. P. McDowell, J. Hicks, and R. Galask. 1987. High-frequency switching in Candida albicans. J. Clin. Microbiol. 25:1611-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Steinberg, G., M. Schliwa, C. Lehmler, M. Bölker, R. Kahmann, and J. R. McIntosh. 1998. Kinesin from the plant pathogenic fungus Ustilago maydis is involved in vacuole formation and cytoplasmic migration. J. Cell Sci. 111:2235-2246. [DOI] [PubMed] [Google Scholar]
- 57.Sudbery, P., N. Gow, and J. Berman. 2004. The distinct morphogenic states of Candida albicans. Trends Microbiol. 12:317-324. [DOI] [PubMed] [Google Scholar]
- 58.Takeshige, K., M. Baba, S. Tsuboi, T. Noda, and Y. Ohsumi. 1992. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J. Cell Biol. 119:301-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Teter, S. A., and D. J. Klionsky. 2000. Transport of proteins to the yeast vacuole: autophagy, cytoplasm-to-vacuole targeting, and role of the vacuole in degradation. Semin. Cell Dev. Biol. 11:173-179. [DOI] [PubMed] [Google Scholar]
- 60.Theiss, S., M. Kretschmar, T. Nichterlein, H. Hof, N. Agabian, J. Hacker, and G. A. Köhler. 2002. Functional analysis of a vacuolar ABC transporter in wild-type Candida albicans reveals its involvement in virulence. Mol. Microbiol. 43:571-584. [DOI] [PubMed] [Google Scholar]
- 61.Thumm, M. 2000. Structure and function of the yeast vacuole and its role in autophagy. Microsc. Res. Tech. 51:563-572. [DOI] [PubMed] [Google Scholar]
- 62.Vida, T. A., and S. D. Emr. 1995. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 128:779-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wada, Y., Y. Ohsumi, and Y. Anraku. 1992. Genes for directing vacuolar morphogenesis in Saccharomyces cerevisiae. I. Isolation and characterization of two classes of vam mutants. J. Biol. Chem. 267:18665-18670. [PubMed] [Google Scholar]
- 64.Weisman, L. S. 2003. Yeast vacuole inheritance and dynamics. Annu. Rev. Genet. 37:435-460. [DOI] [PubMed] [Google Scholar]
- 65.Wilson, R. B., D. Davis, B. M. Enloe, and A. P. Mitchell. 2000. A recyclable cassette for PCR product-directed gene disruptions. Yeast 16:65-70. [DOI] [PubMed] [Google Scholar]
- 66.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide references of the M13 mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 67.Yokoyama, K., and K. Takeo. 1983. Differences of asymmetrical division between the pseudomycelial and yeast forms of Candida albicans and their effect on multiplication. Arch. Microbiol. 134:251-253. [DOI] [PubMed] [Google Scholar]