Abstract
Background
The metabolomic profiles of individuals with different clinical manifestations of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection have not been clearly characterized.
Methods
We performed metabolomics analysis of 166 individuals, including 62 healthy controls, 16 individuals with asymptomatic SARS-CoV-2 infection, and 88 patients with moderate (n = 42) and severe (n = 46) symptomatic 2019 coronavirus disease (COVID-19; 17 with short-term and 34 with long-term nucleic-acid test positivity). By examining differential expression, we identified candidate metabolites associated with different SARS-CoV-2 infection presentations. Functional and machine learning analyses were performed to explore the metabolites’ functions and verify their candidacy as biomarkers.
Results
A total of 417 metabolites were detected. We discovered 70 differentially expressed metabolites that may help differentiate asymptomatic infections from healthy controls and COVID-19 patients with different disease severity. Cyclamic acid and N-Acetylneuraminic Acid were identified to distinguish symptomatic infected patients and asymptomatic infected patients. Shikimic Acid, Glycyrrhetinic acid and 3-Hydroxybutyrate can supply significant insights for distinguishing short-term and long-term nucleic-acid test positivity.
Conclusion
Metabolomic profiling may highlight novel biomarkers for the identification of individuals with asymptomatic SARS-CoV-2 infection and further our understanding of the molecular pathogenesis of COVID-19.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12985-024-02523-7.
Keywords: SARS-CoV-2, COVID-19, Asymptomatic infection, Long-term nucleic acid test positive, Metabolomics
Introduction
The 2019 coronavirus disease (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has created a global public health crisis. A proportion of SARS-CoV-2–infected individuals are asymptomatic subjects (ASs) [1]; these individuals are difficult to identify and can rapidly spread the disease to unprotected and otherwise susceptible people. In addition, nucleic-acid test results remain positive for long periods for some symptomatic patients (SMs) under treatment [i.e., long-term nucleic-acid test positivity (LTNP)], whereas they become negative after shorter periods for others [i.e., short-term nucleic-acid test positivity (STNP)]. The asymptomatic harboring of SARS-CoV-2 and LTNP contribute greatly to economic burdens and reduced quality of life and further threaten global public health. However, biological metabolites and mechanisms distinguishing ASs from healthy controls (HCs) and individuals with LTNP from those with STNP had not been previously identified.
Viruses, including SARS-CoV-2, hijack host cells to obtain nucleotides, lipids, and amino acids for replication [2]. In addition, viral infections can cause metabolic changes in host cells via various mechanisms [3, 4]. For example, lipidome reprogramming by Zika virus contributes to placental damage [5], and serum plasmalogen phosphatidylethanolamine levels are increased in patients infected with this virus [6]. Aberrant neutrophil metabolism, which can contribute to cellular dysfunction, has been observed in patients with severe COVID-19, and glyceraldehyde-3-phosphate dehydrogenase has been found to suppress neutrophil extracellular trap formation via a cell-intrinsic mechanism [7].
Metabolomics – the study of metabolite species in biofluids, cells, and tissues – was introduced by Nicholson and colleagues and is used widely for biomarker discovery [8]. Such discovery, based on changes in metabolite profiles, continues to contribute substantially to medical diagnostics. Various metabolic biomarkers have been proposed to reflect the future risk of contracting severe infectious disease in patients with cardiovascular disease and diabetes, respectively [9, 10], and non-alcoholic fatty liver disease has been associated with certain metabolic biomarkers and mechanisms [11]. Serum metabolite alterations have been observed in hospitalized SARS-CoV-2–infected patients [12, 13], and the severity of COVID-19 has been associated with lipid, amino-acid, and energy metabolism [13]. Some metabolites in the kynurenine pathway (kynurenine, tryptophan, and 3-hydroxykynurenine) have been proposed to be COVID-19 prognostic markers [14]. Therefore, understanding host cell metabolism and metabolites alters could help to find potential biomarkers to distinguish different severity of COVID-19 for further medical treatments, especially for asymptomatic patients and LTNP.
In this study, to explore the potential biomarkers of asymptomatic SARS-CoV-2 infection and LTNP, we characterized the metabolomes of plasma samples from patients with COVID-19 and HCs. We sought differentially expressed metabolites associated with SARS-CoV-2 infection, COVID-19 severity, and viral persistence. In addition, the associations of potential biomarkers with disease severity were verified using machine learning.
Methods
Ethics statement
The ethics committees of the First Affiliated Hospital of Xi’an Jiaotong University (XJTU1AF2020LSK-015) and Renmin Hospital of Wuhan University (WDRY2020-K130) approved this study. All participants or their surrogates provided written informed consent. High-throughput sequencing was performed on plasma samples collected during standard diagnostic testing, avoiding the need for further sample collection for the purposes of this study.
Samples and disease classification
This study was conducted with plasma samples from HCs, ASs, and SMs. The classification standards of SD and MD were identified and agreed by three professional doctors according to the guidelines of the World Health Organization and China’s National Health Commission [15–17]. SM cases were divided into cases of moderate disease (MD) and severe disease (SD) and, when nucleic-acid test results were available, cases of LTNP (≥ 60 days) and STNP (≤ 45 days) based on clinical observations of the predominant timing of test negativity (≤ 45 days) among patients with COVID-19 at Renmin Hospital.
Blood sample collection and processing
At various timepoints during hospital visitation, peripheral blood samples were collected from study participants into EDTA-K2 Vacuette tubes (cat. no. Y09012282; Jiangsu Yuli Medical Equipment Co., Ltd., China) and were stored at room temperature for no more than 0.5 h, then stored at 4℃ and processed within 4–6 h. To obtain plasma samples, the whole blood samples were centrifuged at 2500 rpm for 20 min and stored at − 80℃. All procedures were performed in the biosafety level–2 laboratory of Renmin Hospital’s Department of Clinical Diagnostics.
For metabolite analysis by liquid chromatography electrospray ionization tandem mass spectrometry (LC-ESI-MS/MS), samples were thawed on ice and vortexed for 10s. Pure methanol (150µL/50µL plasma) was added, followed by vortexing for 3 min and centrifugation at 12,000 rpm and 4℃ for 10 min. The supernatants were collected and centrifuged at the same speed and temperature for 5 min, followed by final supernatant collection.
Sample analysis
The samples were analyzed using an LC-ESI-MS/MS system [Shim-pack ultrafast LC CBM 30 A, Shimadzu; triple-quadrupole linear ion trap (QTRAP®) with ESI turbo ion-spray interface, Sciex; Analyst 1.6.3 software, Sciex; ACQeITY ultra-performance liquid chromatography (UPLC) HSS T3 C18 column (1.8 μm, 2.1 × 100 mm), Waters]. The UPLC conditions were: column temperature, 40℃; injection volume, 2 µL; flow rate, 0.4 mL/min; solvent system, water (0.1% acetic acid): acetonitrile (0.1% acetic acid); and gradient sequence, 95:5 V/V at 0 min, 10:90 V/V at 11.0 min, 10:90 V/V at 12.0 min, 95:5 V/V at 12.1 min, and 95:5 V/V at 14.0 min. Linear ion trap and triple quadrupole (QQQ) sequences were acquired, with 10- and 100-µmol/L polypropylene glycol solutions, respectively, used for instrument tuning and mass calibration. The ESI parameters for metabolite analysis were: source temperature, 500℃; ion spray voltages, 5500 and − 4500 V; and ion source gas I, gas II, and curtain gas pressures, 55, 60, and 25 psi, respectively. Declustering potential and collision energy optimization was performed for the multiple-reaction monitoring of individual transitions. For each period, a specific set of transitions was monitored according to the hydrophilic and hydrophobic metabolites eluted during that period (Table S1).
Data analysis
The MS data were processed using Analyst 1.6.3 (Sciex). Quality control (QC) samples are mixed, with repetitive techniques and interspersed in the front, middle, and back positions of the detected samples. The TIC stacking curves of mix-QC samples have high overlap, that is, the retention time and peak intensity are consistent, indicating that the signal stability of mass spectrometry is good when detecting the same sample at different times. The high stability of the instrument provides important guarantees for the repeatability and reliability of data. Standard samples were used to establish theoretical Q1 (MS1), Q3 (MS2), and retention time (RT) databases. Subsequently, biological samples were directly collected and analyzed qualitatively and quantitatively based on the information stored in the databases, ensuring an RT deviation of less than 0.2 min. Apart from the standard substances, a subset of compounds in the extensive target library were based on high quality MS2 spectrum matched from public databases, with standard substances constituting 70% of the total compounds. The QQQ scans were screened to determine the characteristic ions of each substance detected. Using MultiQuant software, the chromatographic peaks for each metabolite detected in different samples were integrated and corrected according to the peak type and retention time.
Differences in metabolite expression between groups (SARS-CoV-2 infection vs. HC, AS vs. HC, SM vs. AS, SD vs. MD, and LTNP vs. STNP) were assessed using orthogonal partial-least-squares discriminant analysis (OPLS-DA) based on the MetaboAnalyst package (version 1.0.1). During the OPLS-DA analysis, the data underwent log2 transformation and “meanCenter” processing. These differences were used to perform Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. Metabolites with fold change (FC) values ≥ 2 or ≤ 0.5 (≥ 1.5 or ≤ 0.67 for LTNP vs. STNP analyses) and variable importance in projection (VIP) values ≥ 1 were selected as differentially expressed metabolites. Machine learning was performed using the “xgboost” (extreme gradient boosting) package (version 1.4.1.1) [18] to select relevant metabolites from the training set according to the intensity of differential expression. For each pair of groups compared, normalized metabolite intensities were used as modeling data (75% assigned to the training and 25% assigned to the testing sets). All of analyses were performed in R (version 4.1.2).
Results
Sample characteristics
The study comprised 220 plasma samples from 166 individuals (62 HCs, 16 ASs, and 88 SMs). Of the SMs, 42 had MD and 46 had SD; 17 cases of LTNP and 34 cases of STNP were identified. Two or more samples were included from each of 5 HCs, 12 SMs with MD, and 23 SMs with SD. The characteristics of the patients from which the samples were obtained are provided in Table S2. In total, 417 metabolites were identified in this study (Table S3). The total ion chromatogram (TIC), extracted ion chromatogram (XIC), and superposition diagram of TIC detected by mix-QC sample mass spectrometry for healthy controls and COVID-19 patients were shown in Figure S1.
Metabolites associated with SARS-CoV-2 infection and COVID-19 characteristics
SARS-CoV-2 infection
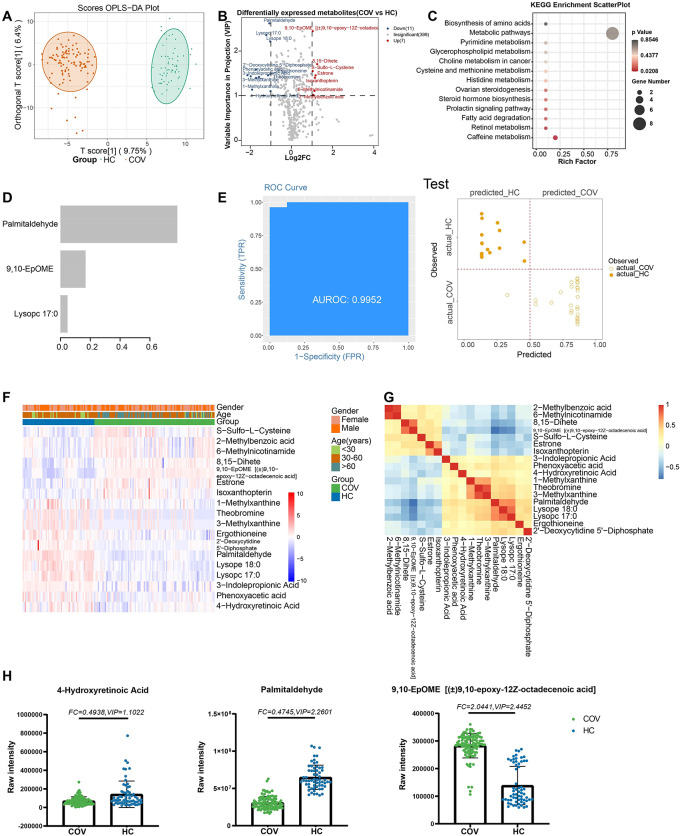
Differential metabolite expression between groups is summarized in Table S4, and OPLS-DA model results are shown in Figure S2. Eighteen metabolites were differentially expressed between patients with SARS-CoV-2 infection and HCs (Fig. 1A, B). Their KEGG pathways were related mainly to caffeine metabolism (ko00232) and retinol metabolism (ko00830; Fig. 1C, Table S5). Three metabolites in this set were prioritized in the machine learning model: lysophosphatidylcholine, palmitadehyde, and 9,10-epoxy-12Z-octadecenoic acid (Fig. 1D, F, G, H). The areas under the curve (AUCs) for the training and testing sets were 0.9822 and 0.9952, respectively, with 94.35% and 97.62% accuracy (Fig. 1E; Figure S3).
Fig. 1.
Metabolites associated with SARS-COV-2 infections. (A) The OPLS-DA scores plot of COV vs. HC. The Y axis represents orthogonal T score, and the X axis represents T score; (B) The volcano plot of COV vs. HC. Red dots represent the up-regulated metabolites (log2FC ≥ 1, VIP ≥ 1); blue dots represent the down-regulated metabolites (log2FC≤-1, VIP ≥ 1); gray dots represent metabolites without significant changes (-1 < log2FC < 1, VIP ≤ 1). (C) The KEGG analysis of the DEMs in COV vs. HC. (D) The important metabolites prioritized by xgboost analysis. (E) Receiver operating characteristic (ROC) and performance of the xgboost model in the test sets. (F) The heatmap of 18 metabolites in COV vs. HC. (G) The correlation heatmap of DEMs. (H) the raw intensity of 4- hydroxyretinoic acid, palmitadehyde and 9,10-EpOME
Asymptomatic SARS-CoV-2 infection
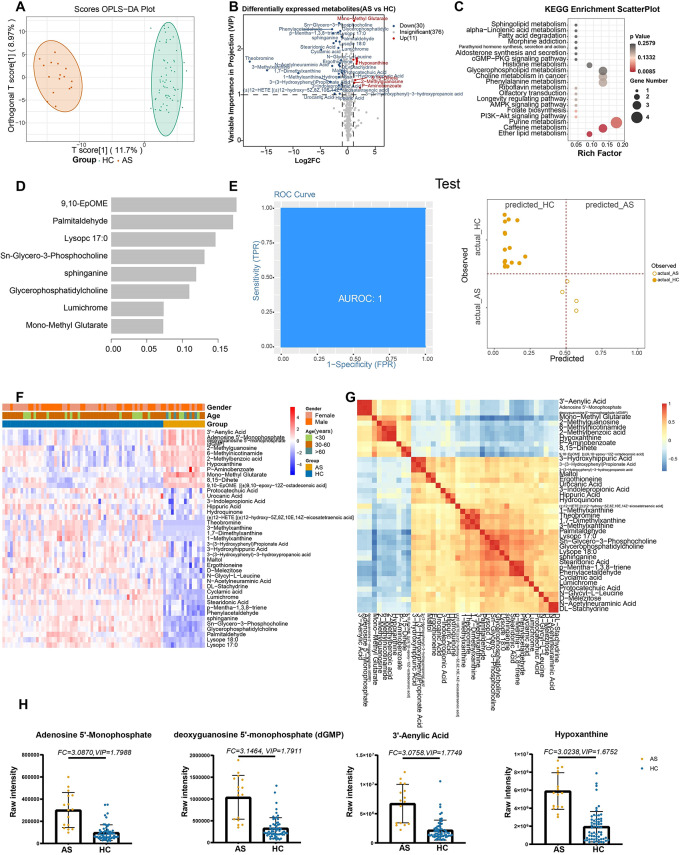
Forty-one metabolites were differentially expressed between ASs and HCs (Fig. 2A, B). Their KEGG pathways were related mainly to ether lipid metabolism (ko00565), caffeine metabolism (ko00232), and synthesis and degradation of ketone bodies (ko00072; Fig. 2C). In addition, deoxyguanosine 5’-monophosphate (FC = 3.146), 3’-aenylic acid (FC = 3.076), hypoxanthine (FC = 3.024), and adenosine 5’-monophosphate (AMP; FC = 3.087), involved in purine metabolism, were upregulated in ASs relative to HCs, whereas lumichrome (FC = 0.304) and Sn-glycero-3-phosphocholine (FC = 0.329) were down-regulated (Table S5). The machine learning analysis prioritized eight metabolites, the top three of which were those identified in the SARS-CoV-2 infection vs. HC analysis (Fig. 2D, F, G). The AUC was 1 for the training and test sets, with 100% and 95% accuracy, respectively (Fig. 2E; Figure S3.
Fig. 2.
Metabolites associated with the asymptomatic infection of SARS-COV-2. (A) The OPLS-DA scores plot of AS vs. HC. The Y axis represents orthogonal T score, and the X axis represents T score; (B) The volcano plot of AS vs. HC. Red dots represent the up-regulated metabolites(log2FC ≥ 1, VIP ≥ 1); blue dots represent the down-regulated metabolites (log2FC≤-1, VIP ≥ 1); gray dots represent metabolites without significant changes (-1 < log2FC < 1, VIP ≤ 1). (C) The KEGG analysis of the DEMs and in AS vs. HC. (D) The important metabolites prioritized by xgboost analysis. (E) Receiver operating characteristic (ROC) and performance of the xgboost model in the test sets. (F) The heatmap of 41 metabolites in AS vs. HC. (G) The correlation heatmap of DEMs. (H) the raw intensity of Adenosine 5’-Monophosphate (AMP), deoxyguanosine 5’-monophosphate (dGMP), 3’-Aenylic Acid and Hypoxanthine
Symptomatic COVID-19
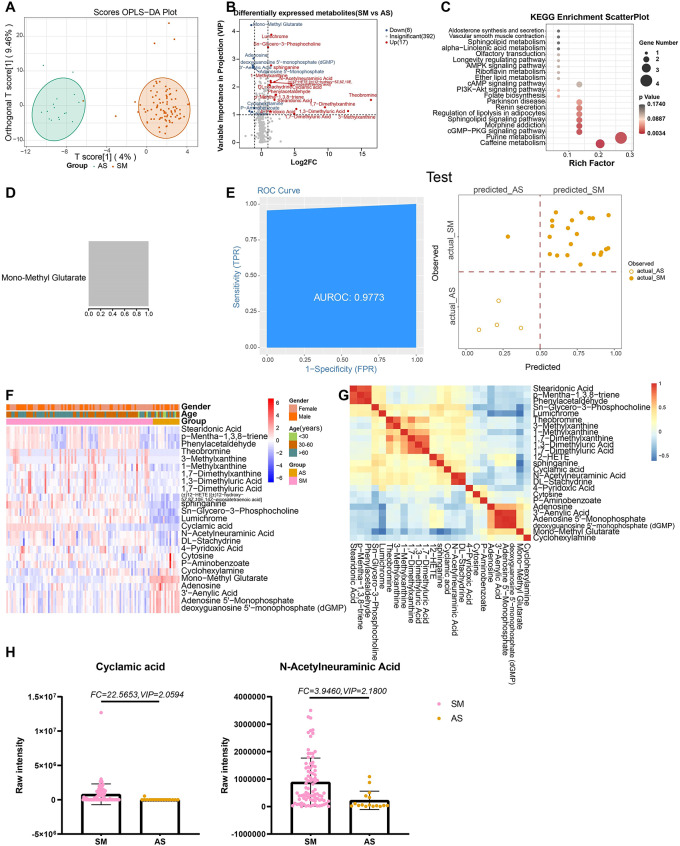
Twenty-five metabolites, including theobromine (FC = 87,787.345, VIP = 1.534) and lumichrome (FC = 3.883, VIP = 2.848), were differentially expressed between ASs and SMs (Fig. 3A, B). KEGG pathways associated with symptomatic COVID-19 were related mainly to cyclic guanosine monophosphate-protein kinase G signaling pathway and purine metabolism (Fig. 3C). For the machine learning model, only one metabolite (mono-methyl glutarate) had a high importance score (Fig. 3D, F, G). The AUCs for the training and testing sets were 0.9924 and 0.9773, respectively, with 98.72% and 96.15% accuracy (Fig. 3E; S3).
Fig. 3.
Metabolites associated with the disease severity of COVID-19. (A) The OPLS-DA scores plot of SD vs. MD. The Y axis represents orthogonal T score, and the X axis represents T score; (B) The volcano plot of SD vs. MD. Red dots represent the up-regulated metabolites (log2FC ≥ 1, VIP ≥ 1); blue dots represent the down-regulated metabolites (log2FC≤-1, VIP ≥ 1); gray dots represent metabolites without significant changes (-1 < log2FC < 1, VIP ≤ 1). (C) The KEGG analysis of the DEMs in SD vs. MD. (D) The important metabolites prioritized by xgboost analysis. (E) Receiver operating characteristic (ROC) and performance of the xgboost model in the test sets. (F) The heatmap of 13 metabolites in SD vs. MD. (G) The correlation heatmap of DEMs. (H) the raw intensity of cyclamic acid and N-Acetylneuraminic acid
COVID-19 severity
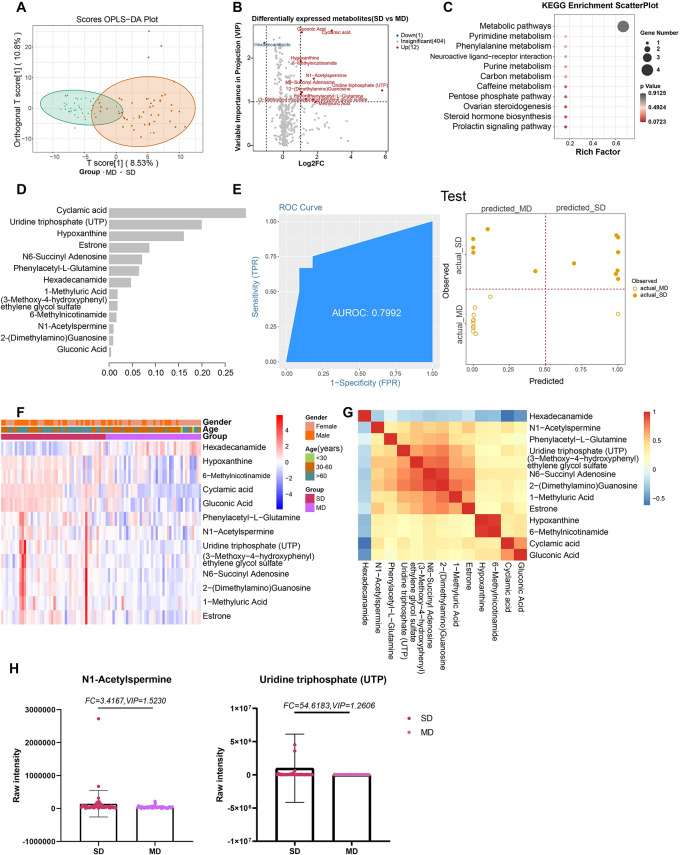
Thirteen metabolites, including cyclamic acid (FC = 7.088, VIP = 2.630) and uridine triphosphate (UTP; FC = 54.618, VIP = 1.261), were differentially expressed between MD and SD cases (Fig. 4A, B). KEGG analysis of these metabolites yielded no meaningful result (Fig. 4C). Eleven metabolites with high importance scores were included in the machine learning model (Fig. 4D, F, G). The AUCs for the training and testing sets were 1 and 0.7992, respectively, with 100% and 73.91% accuracy (Fig. 4E; Figure S3).
Fig. 4.
Metabolites associated with the asymptomatic and symptomatic of COVID-19. (A) The OPLS-DA scores plot of SM vs. AS. The Y axis represents orthogonal T score, and the X axis represents T score; (B) The volcano plot of SM vs. AS. Red dots represent the up-regulated metabolites (log2FC ≥ 1, VIP ≥ 1); blue dots represent the down-regulated metabolites (log2FC≤-1, VIP ≥ 1); gray dots represent metabolites without significant changes (-1 < log2FC < 1, VIP ≤ 1). (C) The KEGG analysis of the DEMs in SM vs. AS. (D) The important metabolites prioritized by xgboost analysis. (E) Receiver operating characteristic (ROC) and performance of the xgboost model in the test sets. (F) The heatmap of 25 metabolites in SM vs. AS. (G) The correlation heatmap of DEMs. (H) the raw intensity of uridine triphosphate (UTP) and N1-Acetylspermine
Viral persistence
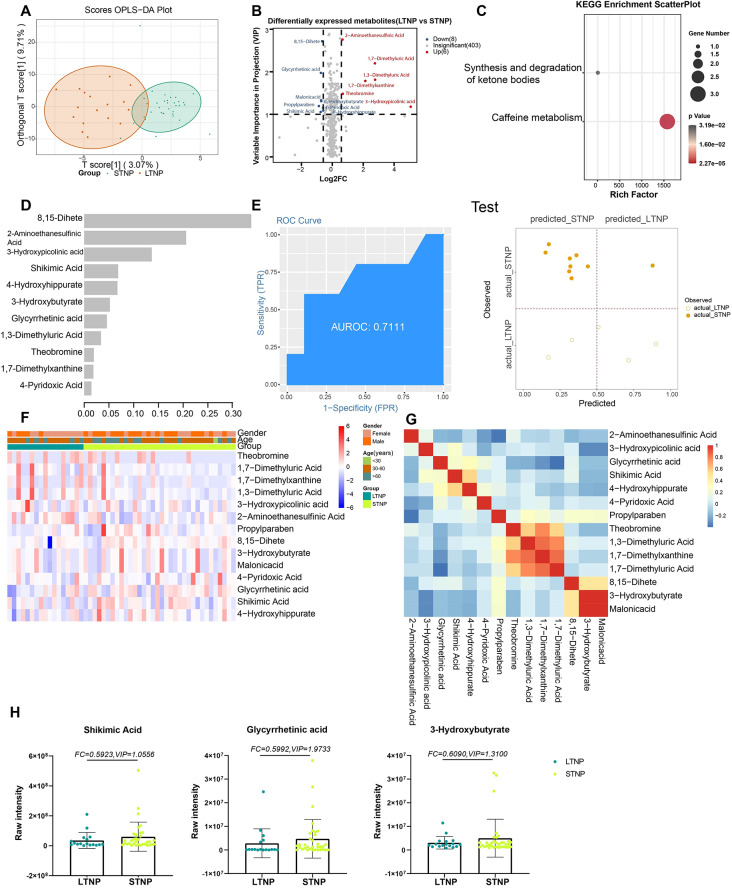
Fourteen metabolites were differentially expressed between STNP and LTNP cases (Fig. 5A, B). Caffeine metabolism (ko00232) was identified as a significant KEGG pathway for these metabolites (Fig. 5C). The machine learning model was based on 11 metabolites with high importance scores (Fig. 5D, F, G). The AUCs for the training and testing sets were 0.9808 and 0.7111, respectively, with 88.89% and 78.57% accuracy (Fig. 5E; Figure S3).
Fig. 5.
Metabolites associated with the viral persistence of SARS-CoV-2 infection. (A) The OPLS-DA scores plot of LTNP vs. STNP. The Y axis represents orthogonal T score, and the X axis represents T score; (B) The volcano plot of LTNP vs. STNP. Red dots represent the up-regulated metabolites (log2FC ≥ 0.58, VIP ≥ 1); blue dots represent the down-regulated metabolites (log2FC≤-0.58, VIP ≥ 1); gray dots represent metabolites without significant changes (-0.58 < log2FC < 0.58, VIP ≤ 1). (C) The KEGG analysis of the DEMs in LTNP vs. STNP. (D) The important metabolites prioritized by xgboost analysis. (E) Receiver operating characteristic (ROC) and performance of the xgboost model in the test sets. (F) The heatmap of 14 metabolites in LTNP vs. STNP. (G) The correlation heatmap of DEMs. (H) the raw intensity of Shikimic Acid, Glycyrrhetinic acid and 3-Hydroxybutyrate
Discussion
In this study, we performed metabolomic profiling of 220 plasma samples and identified candidate metabolites associated with SARS-CoV-2 infection and COVID-19 symptomatic status and severity. We identified 417 metabolites, 70 of which were differentially expressed in at least one of the five group comparisons performed. Machine learning led to the identification of multiple biomarkers distinguishing COVID-19 cases of different severities from HCs.
4-Hydroxyretinoic acid, which participates in retinal metabolism, was down-regulated in patients with SARS-CoV-2 infection relative to HCs in this study. The identification of the associated mechanism could provide a new perspective on COVID-19 treatment [19]. Palmitaldehyde and phosphoethanolamine are produced by the cleavage of sphingosine-1-phosphate (S1P) by S1P lyase [20]. Compared to HCs, down-regulated palmitaldehyde levels in SARS-CoV-2 infection indicated by our data suggest that S1P levels are also decreased in individuals with COVID-19, consistent with the finding that the significant reduction of the serum S1P level is a potential indicator of COVID-19 severity [21]. The inflammatory cell function involve S1P signaling via S1P receptors, and B and T cells have distinctive profiles of these receptors, which were found to provide significant beneficial effects towards counteracting SARS-CoV-2 infection [22, 23].
Metabolites involved in purine metabolism, including AMP [generated from adenosine triphosphate (ATP)], were upregulated in ASs relative to HCs in this study. Extracellular ATP signaling alerts the immune system to tissue damage [24]. ATP is primarily pro-inflammatory and is released by necrotic, endothelial, and activated immune cells and under hypoxia [25]. In addition, lumichrome, a riboflavin derivative, was down-regulated in AS relative to HCs. The functions of riboflavin include protection against oxidative inflammatory lung damage, neutrophil infiltration, and peripheral activated granulocyte accumulation (which reduces inflammatory responses), and riboflavin deficiency exacerbates oxidative stress [26–28]. Thus, the low lumichrome levels indicated by our data suggest that ASs also have low riboflavin levels, with important immune consequences [29]. Besides, ketogenic Diet and ketone bodies are suggested to be potential clinical nutritional interventions to assist in the treatment of COVID-19, which is consistent with our functions of the identified metabolites associated with Synthesis and degradation of ketone bodies [30].
Metabolites found to be associated with symptomatic infected vs. asymptomatic infection included N-acetylneuraminic acid. N-acetylneuraminic acid, which had high VIP and FC values, has been found to be associated with a key SARS-CoV-2 spike protein domain and thus the virus’s cell entry [31]. In addition, the performance of its derivatives luciferyl N-acetylneuraminic acid and luciferyl 4,7-di-O-methyl-N-acetylneuraminic acid in detecting neuraminidases from influenza virus has been evaluated [32]. Theobromine also had high FC values for SMs relative to ASs. Theobromine was not detected in ASs; it was found only in SMs in this study. It may regulate systemic and intestinal antibody concentrations, and was found to influence immune-system regulation by determining the concentrations of key immunoglobulins in rats [33]. In addition, purine metabolism is relevant to viral replication and immune response, which is associated with COVID-19 [34].
UTP and N1-acetylspermine distinguished MD from SD in this study, with the highest FC and high VIP values. UTP, an extracellular nucleotide, activates P2 purinergic receptors on phagocyte surfaces and induces immune and physiological responses in several other cell types [35, 36]. In mice, its use as a vaccine adjuvant enhanced the production of anti-rabies antibodies [37]. The level of N1-acetylspermine, which participates in polyamine catabolism, was found to be elevated in human breast cancer relative to normal breast tissue [38, 39]. Polyamines play critical roles in transcription, translation, cell cycling, and the duration of viral infection [40]; they are required for many steps of virus replication [41]. Nevertheless, given that COVID-19 is an acute infectious disease, it is imperative to focus on the disease progression in future research endeavors.
1,7-Dimethylxanthine was upregulated and shikimic acid (SA), 3-hydroxybutyrate, and glycyrrhetinic acid (GA) were down-regulated in LTNP cases relative to STNP cases in this study. 1,7-Dimethylxanthine is associated with cholestatic liver fibrosis and acute pulmonary and systemic inflammation [42, 43]. SA has been found to have antioxidant properties and to interfere with retinal inflammatory mediator release [44]. 3-Hydroxybutyrate exerts beneficial hemodynamic effects without reducing myocardial external energy efficiency in patients with chronic heart failure with reduced ejection fraction [45]. GA was found to be a potential inhibitory effect on the cytokine storm (CS) triggered by coronavirus disease 2019 (COVID-19) [46].Thus, SA and GA down-regulation may protect against inflammation and apoptosis in patients with COVID-19 characterized by LTNP.
The AUC values for the five comparison groups in the training data all exceeded 0.9, indicating high performance. In the test data, the AUC values for COV vs. HC, AS vs. HC, and SM vs. AS also surpassed 0.9, while the AUC values for SD vs. MD and LTNP vs. STNP were 0.7992 and 0.7111, respectively, falling below 0.8. This highlights a potential limitation stemming from the insufficient sample size for robust machine learning analysis. Therefore, additional sample collection and further validation of the differentially expressed metabolites are warranted.
Conclusions
In this study, we identified multiple metabolites and related pathways associated with SARS-CoV-2 infection and persistence and COVID-19 severity. Our findings contribute to the understanding of metabolite contributions to the pathogenesis of COVID-19 and highlight novel biomarkers for the disease’s diagnosis and treatment.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Material 1: Supplementary Fig. 1| (A) Superposition diagram of total ion chromatogram (TIC) detected by mix-quality control (QC) samples mass spectrometry for healthy controls. Different color means different mix-QC samples; (B) Superposition diagram of TIC detected by mix-QC samples mass spectrometry for COVID-19. Different color means different mix-QC samples; (C) extracted ion chromatogram (XIC) of healthy controls through multiple reaction monitoring (MRM). Chromatographic peaks of different colors represent different metabolites detected; (D) XIC of COVID-19 samples through MRM. Chromatographic peaks of different colors represent different metabolites detected; (E) TIC of mix-QC samples for healthy controls; (F) TIC of mix-QC samples for COVID-19. Supplementary Fig. 2| (A) the PCA plot for mix samples and our samples. (B) OPLS-DA permutation Test of 5 compared groups; R2X and R2Y represent the interpretation degree of the model to x and Y matrix respectively; Q2 represents predictive ability of the model. The x-axis represents the accuracy of the model. The y-axis represents frequency of accuracy in 200 models. Supplementary Fig. 2| Receiver operating characteristic (ROC) and performance of the xgboost model in the training sets of 5 compared groups
Supplementary Material 2: Table S1| List of HPLC conditions and data acquisition used in this analysis
Supplementary Material 3: Table S2| the detailed characteristic information of samples used in this study
Supplementary Material 4: Table S3| the raw intensity for all samples
Supplementary Material 5: Table S4| the differentially expressed metabolites in 5 compared groups
Supplementary Material 6: Table S5| the KEGG results for differentially expressed metabolites in 5 compared groups
Author contributions
CSZ, BYS, QX and XL designed and conceptualized this study. JY, XRW, FH, HYQ and MJ acquired the data. XQ, HB, YJL, JG, LJL, YWP, JX, YXC, YXL, and JPM performed data generation, processing and statistical analyses. All of the authors wrote and revised the manuscript.
Funding
This study is supported in part by the Department of Science and Technology of Shaanxi Province (Grant No. 2020ZDXM2-SF-02) and the operational funds from the First Affiliated Hospital of Xi’an Jiaotong University.
Data availability
The metabolomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository with the dataset identifier IPXD0003289001.
Declarations
Ethics approval and consent to participate
The ethics committees of the First Affiliated Hospital of Xi’an Jiaotong University (XJTU1AF2020LSK-015) and Renmin Hospital of Wuhan University (WDRY2020-K130) approved this study. All participants or their surrogates provided written informed consent.
Consent for publication
Prior to sample collection, all participants provided written informed consent for the study publication.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Qi Xin and Xiao Liang contributed equally to this work.
Contributor Information
Chengsheng Zhang, Email: cszhang99@126.com.
Bingyin Shi, Email: shibingy@126.com.
References
- 1.Han D, Li R, Han Y, et al. COVID-19: insight into the asymptomatic SARS-COV-2 infection and transmission. Int J Biol Sci. 2020;16(15):2803–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thaker SK, Ch’ng J, Christofk HR. Viral hijacking of cellular metabolism. BMC Biol. 2019;17(1):59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeVito SR, Ortiz-Riaño E, Martínez-Sobrido L, et al. Cytomegalovirus-mediated activation of pyrimidine biosynthesis drives UDP-sugar synthesis to support viral protein glycosylation. Proc Natl Acad Sci U S A. 2014;111(50):18019–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vastag L, Koyuncu E, Grady SL, et al. Divergent effects of human cytomegalovirus and herpes simplex virus-1 on cellular metabolism. PLoS Pathog. 2011;7(7):e1002124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Q, Gouilly J. Metabolic reprogramming by Zika virus provokes inflammation in human placenta. Nat Commun. 2020;11(1):2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Queiroz A, Pinto IFD, Lima M, et al. Lipidomic Analysis Reveals Serum Alteration of Plasmalogens in patients infected with ZIKA Virus. Front Microbiol. 2019;10:753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y, Hook JS. Neutrophil metabolomics in severe COVID-19 reveal GAPDH as a suppressor of neutrophil extracellular trap formation. Nat Commun. 2023;14(1):2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nicholson JK, Lindon JC, Holmes E. Metabonomics’: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica. 1999;29(11):1181–9. [DOI] [PubMed] [Google Scholar]
- 9.Deelen J, Kettunen J. A metabolic profile of all-cause mortality risk identified in an observational study of 44,168 individuals. Nat Commun. 2019;10(1):3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ritchie SC, Würtz P, Nath AP, et al. The Biomarker GlycA is Associated with chronic inflammation and predicts long-term risk of severe infection. Cell Syst. 2015;1(4):293–301. [DOI] [PubMed] [Google Scholar]
- 11.Svegliati-Baroni G, Pierantonelli I, Torquato P, et al. Lipidomic biomarkers and mechanisms of lipotoxicity in non-alcoholic fatty liver disease. Free Radic Biol Med. 2019;144:293–309. [DOI] [PubMed] [Google Scholar]
- 12.Shen B, Yi X, Sun Y, et al. Proteomic and metabolomic characterization of COVID-19 patient sera. Cell. 2020;182(1):59–e7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bruzzone C, Conde R, Embade N, et al. Metabolomics as a powerful tool for diagnostic, pronostic and drug intervention analysis in COVID-19. Front Mol Biosci. 2023;10:1111482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hasan MR, Suleiman M, Pérez-López A. Metabolomics in the diagnosis and prognosis of COVID-19. Front Genet. 2021;12:721556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.COVID-19: Surveillance, case investigation and epidemiological protocols(World Health Organization, 22 October 2020). https://www.who.int/internal-publications-detail/considerations-in-the-investigation-of-cases-and-clusters-of-covid-19
- 16.Laboratory testing for 2019 novel coronavirus (2019-nCoV) in suspected human cases. (World Health Organization,19 March 2020). https://www.who.int/health-topics/coronavirus/laboratory-diagnostics-for-novel-coronavirus
- 17.New coronavirus pneumonia prevention and control program (8nd ed.) (Government of 640 China, 14 May 2021);. http://www.nhc.gov.cn/jkj/s3577/202105/6f1e8ec6c4a540d99fafef52fc86d0f8/files/4a860a7e85d14d55a22fbab0bbe77cd9.pdf
- 18.Chen T, Guestrin C. XGBoost: A Scalable Tree Boosting System, in Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. 2016, Association for Computing Machinery: San Francisco, California, USA. pp. 785–794.
- 19.Sarohan AR. COVID-19: endogenous retinoic acid theory and retinoic acid depletion syndrome. Med Hypotheses. 2020;144:110250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pyne NJ, Pyne S. Sphingosine 1-phosphate and cancer. Nat Rev Cancer. 2010;10(7):489–503. [DOI] [PubMed] [Google Scholar]
- 21.Marfia G, Navone S, Guarnaccia L, et al. Decreased serum level of sphingosine-1-phosphate: a novel predictor of clinical severity in COVID-19. EMBO Mol Med. 2021;13(1):e13424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goetzl EJ, Rosen H. Regulation of immunity by lysosphingolipids and their G protein-coupled receptors. J Clin Invest. 2004;114(11):1531–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGowan EM, Haddadi N, Nassif NT et al. Targeting the SphK-S1P-SIPR pathway as a potential Therapeutic Approach for COVID-19. Int J Mol Sci, 2020. 21(19). [DOI] [PMC free article] [PubMed]
- 24.Mortaz E, Braber S, Nazary M, et al. ATP in the pathogenesis of lung emphysema. Eur J Pharmacol. 2009;619(1–3):92–6. [DOI] [PubMed] [Google Scholar]
- 25.Faas MM, Sáez T, de Vos P. Extracellular ATP and adenosine: the Yin and Yang in immune responses? Mol Aspects Med. 2017;55:9–19. [DOI] [PubMed] [Google Scholar]
- 26.Powers HJ. Current knowledge concerning optimum nutritional status of riboflavin, niacin and pyridoxine. Proc Nutr Soc. 1999;58(2):435–40. [DOI] [PubMed] [Google Scholar]
- 27.Seekamp A, Hultquist DE, Till GO. Protection by vitamin B2 against oxidant-mediated acute lung injury. Inflammation. 1999;23(5):449–60. [DOI] [PubMed] [Google Scholar]
- 28.Verdrengh M, Tarkowski A. Riboflavin in innate and acquired immune responses. Inflamm Res. 2005;54(9):390–3. [DOI] [PubMed] [Google Scholar]
- 29.Peterson CT, Rodionov DA. B vitamins and their role in Immune Regulation and Cancer. Nutrients, 2020. 12(11). [DOI] [PMC free article] [PubMed]
- 30.Bolesławska I, Kowalówka M. Ketogenic Diet and Ketone bodies as clinical support for the treatment of SARS-CoV-2-Review of the evidence. Viruses, 2023. 15(6). [DOI] [PMC free article] [PubMed]
- 31.Zhu X, Yi Y, Fan Z, et al. Novel mono- and multivalent N-acetylneuraminic acid glycoclusters as potential broad-spectrum entry inhibitors for influenza and coronavirus infection. Eur J Med Chem. 2023;260:115723. [DOI] [PubMed] [Google Scholar]
- 32.Yang W, Liu X, Peng X, et al. Synthesis of novel N-acetylneuraminic acid derivatives as substrates for rapid detection of influenza virus neuraminidase. Carbohydr Res. 2012;359:92–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Camps-Bossacoma M, Pérez-Cano FJ, Franch À, et al. Theobromine is responsible for the effects of Cocoa on the antibody Immune Status of rats. J Nutr. 2018;148(3):464–71. [DOI] [PubMed] [Google Scholar]
- 34.Alexandre LS, Carrilho E. Post-COVID-19 metabolomics: pursuing the sequels of a pandemic. Covid-19 metabolomics and diagnosis: volume 2. Nature Switzerland: Cham: Springer; 2023. pp. 95–111. F.N. Crespilho, Editor. [Google Scholar]
- 35.Bours MJ, Swennen EL, Di Virgilio F, et al. Adenosine 5’-triphosphate and adenosine as endogenous signaling molecules in immunity and inflammation. Pharmacol Ther. 2006;112(2):358–404. [DOI] [PubMed] [Google Scholar]
- 36.Burnstock G. The past, present and future of purine nucleotides as signalling molecules. Neuropharmacology. 1997;36(9):1127–39. [DOI] [PubMed] [Google Scholar]
- 37.Iwaki Y, Sakai Y, Ochiai K, et al. Enhancement of antibody production against Rabies virus by uridine 5′-triphosphate in mice. Microbes Infect. 2014;16(3):196–202. [DOI] [PubMed] [Google Scholar]
- 38.Karasawa K, Qiu X, Lee T. Purification and characterization from rat kidney membranes of a novel platelet-activating factor (PAF)-dependent transacetylase that catalyzes the hydrolysis of PAF, formation of PAF analogs, and C2-ceramide. J Biol Chem. 1999;274(13):8655–61. [DOI] [PubMed] [Google Scholar]
- 39.Lee T. Acetylation of sphingosine by PAF-dependent transacetylase. Adv Exp Med Biol. 1996;416:113–9. [DOI] [PubMed] [Google Scholar]
- 40.Lee TC, Ou MC, Shinozaki K, et al. Biosynthesis of N-acetylsphingosine by platelet-activating factor: sphingosine CoA-independent transacetylase in HL-60 cels. J Biol Chem. 1996;271(1):209–17. [DOI] [PubMed] [Google Scholar]
- 41.Li B, Yang J, Zhao F, et al. Prevalence and impact of cardiovascular metabolic diseases on COVID-19 in China. Clin Res Cardiol. 2020;109(5):531–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kühn H, Barnett J, Grunberger D, et al. Overexpression, purification and characterization of human recombinant 15-lipoxygenase. Biochim Biophys Acta. 1993;1169(1):80–9. [DOI] [PubMed] [Google Scholar]
- 43.Geraets L, Haegens A, Weseler AR, et al. Inhibition of acute pulmonary and systemic inflammation by 1,7-dimethylxanthine. Eur J Pharmacol. 2010;629(1–3):132–9. [DOI] [PubMed] [Google Scholar]
- 44.Al-Malki AL. Shikimic acid from Artemisia absinthium inhibits protein glycation in diabetic rats. Int J Biol Macromol. 2019;122:1212–6. [DOI] [PubMed] [Google Scholar]
- 45.Nielsen R, Møller N, Gormsen LC, et al. Cardiovascular effects of Treatment with the Ketone body 3-Hydroxybutyrate in Chronic Heart failure patients. Circulation. 2019;139(18):2129–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li H, You J, Yang X, et al. Glycyrrhetinic acid: a potential drug for the treatment of COVID-19 cytokine storm. Phytomedicine. 2022;102:154153. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 1: Supplementary Fig. 1| (A) Superposition diagram of total ion chromatogram (TIC) detected by mix-quality control (QC) samples mass spectrometry for healthy controls. Different color means different mix-QC samples; (B) Superposition diagram of TIC detected by mix-QC samples mass spectrometry for COVID-19. Different color means different mix-QC samples; (C) extracted ion chromatogram (XIC) of healthy controls through multiple reaction monitoring (MRM). Chromatographic peaks of different colors represent different metabolites detected; (D) XIC of COVID-19 samples through MRM. Chromatographic peaks of different colors represent different metabolites detected; (E) TIC of mix-QC samples for healthy controls; (F) TIC of mix-QC samples for COVID-19. Supplementary Fig. 2| (A) the PCA plot for mix samples and our samples. (B) OPLS-DA permutation Test of 5 compared groups; R2X and R2Y represent the interpretation degree of the model to x and Y matrix respectively; Q2 represents predictive ability of the model. The x-axis represents the accuracy of the model. The y-axis represents frequency of accuracy in 200 models. Supplementary Fig. 2| Receiver operating characteristic (ROC) and performance of the xgboost model in the training sets of 5 compared groups
Supplementary Material 2: Table S1| List of HPLC conditions and data acquisition used in this analysis
Supplementary Material 3: Table S2| the detailed characteristic information of samples used in this study
Supplementary Material 4: Table S3| the raw intensity for all samples
Supplementary Material 5: Table S4| the differentially expressed metabolites in 5 compared groups
Supplementary Material 6: Table S5| the KEGG results for differentially expressed metabolites in 5 compared groups
Data Availability Statement
The metabolomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository with the dataset identifier IPXD0003289001.