Abstract
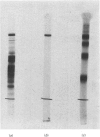
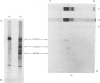
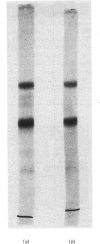
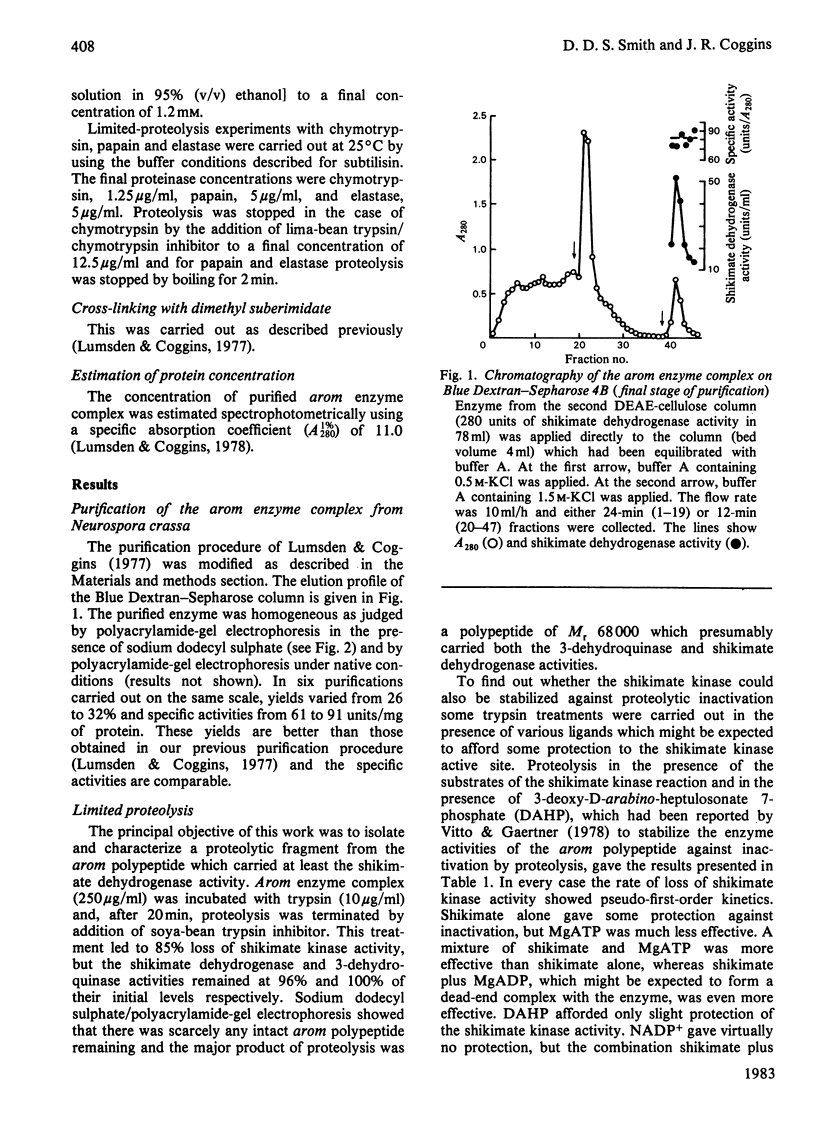
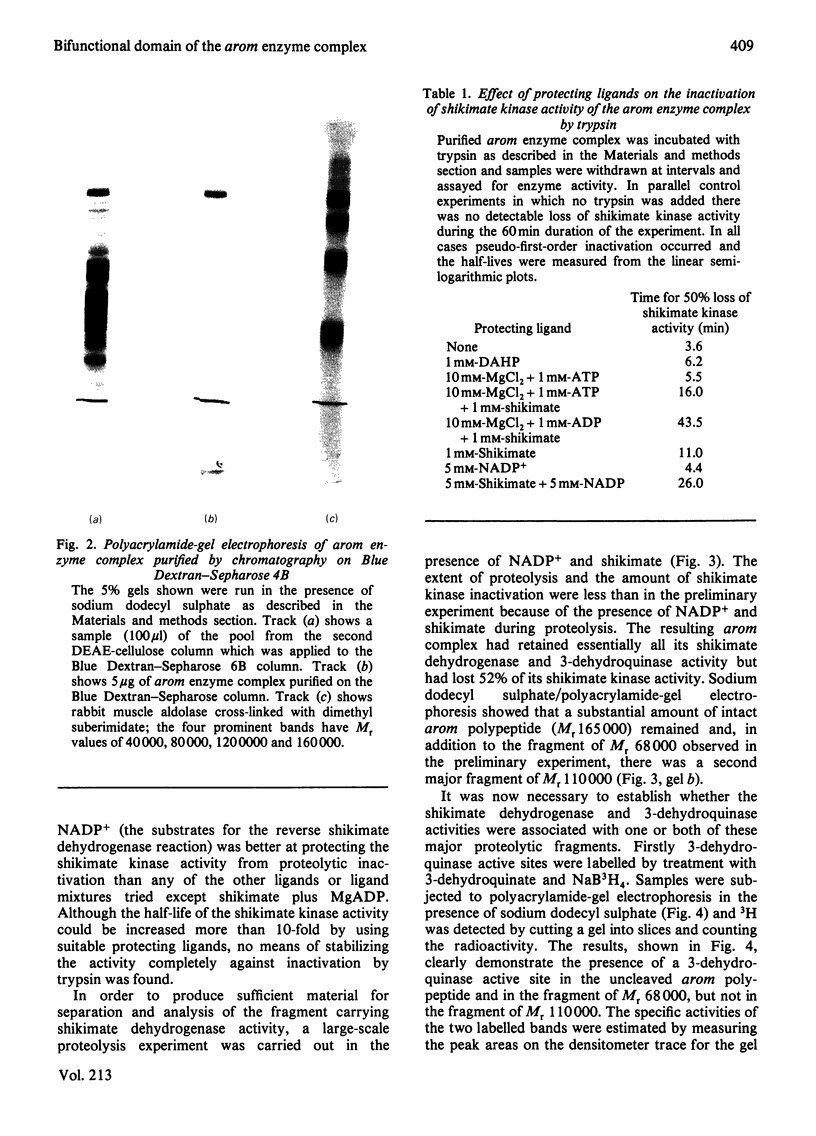
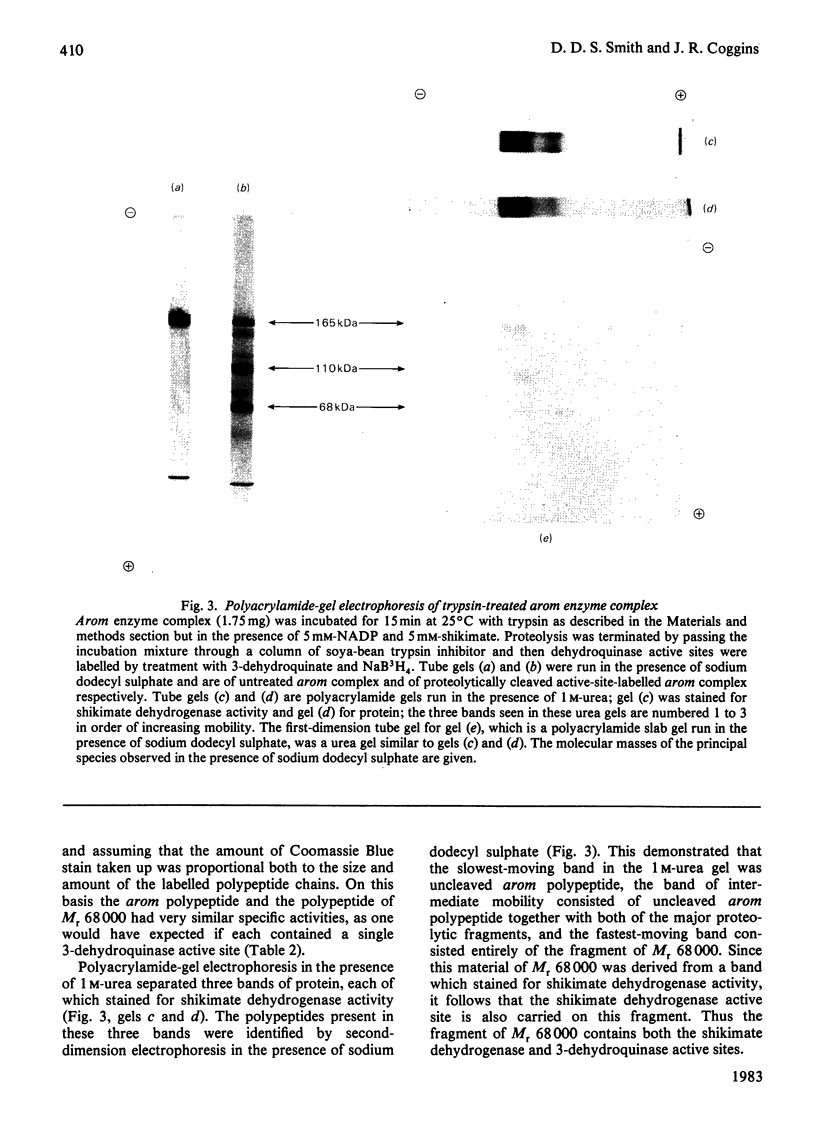
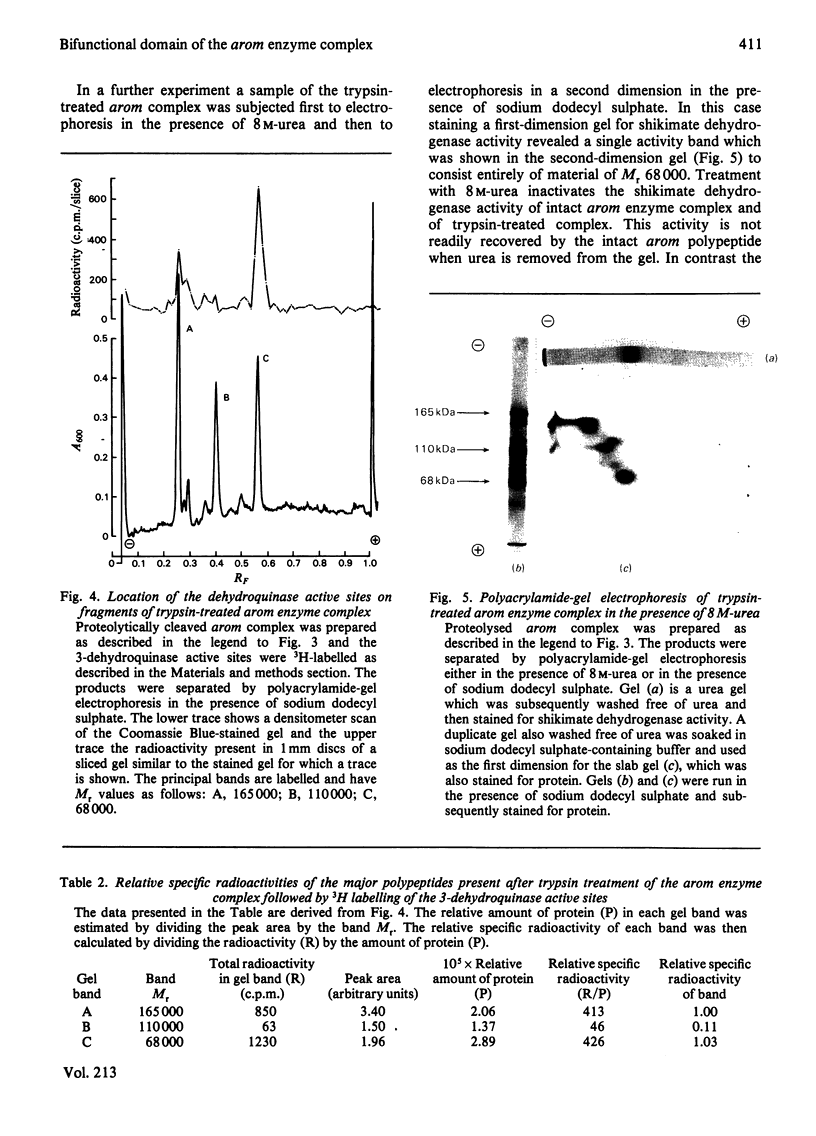
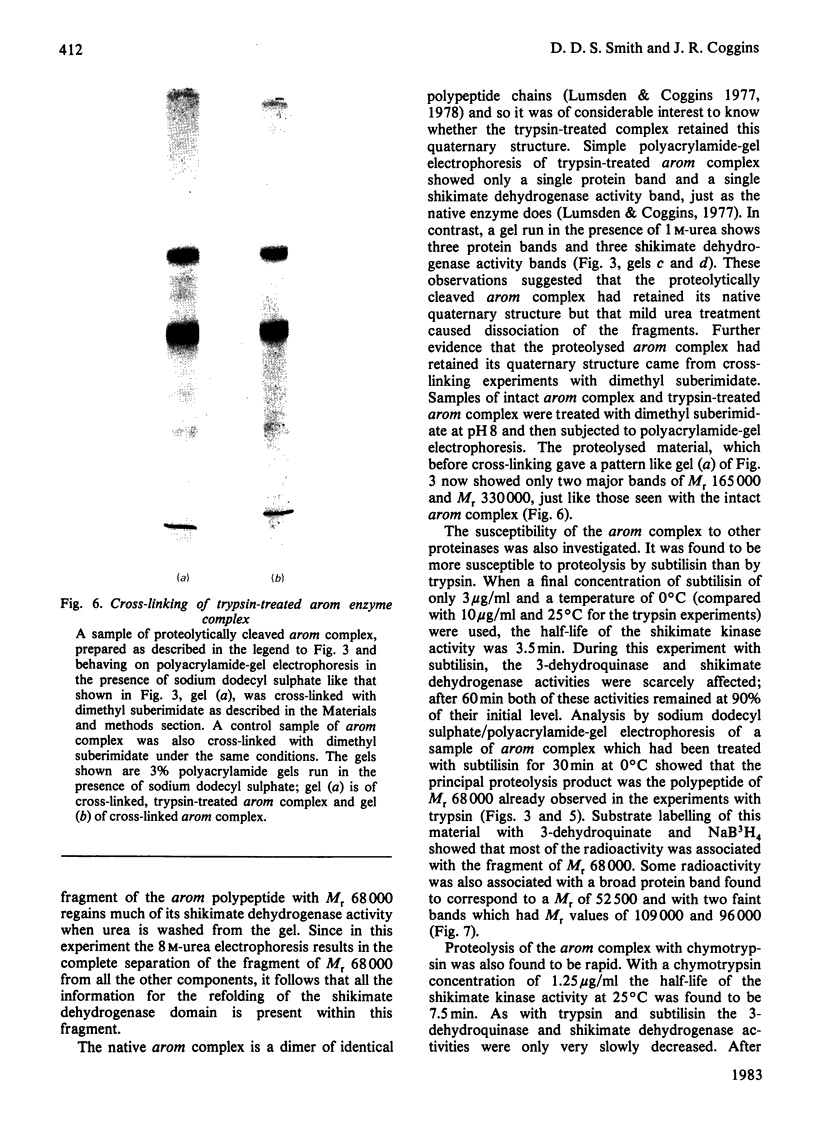
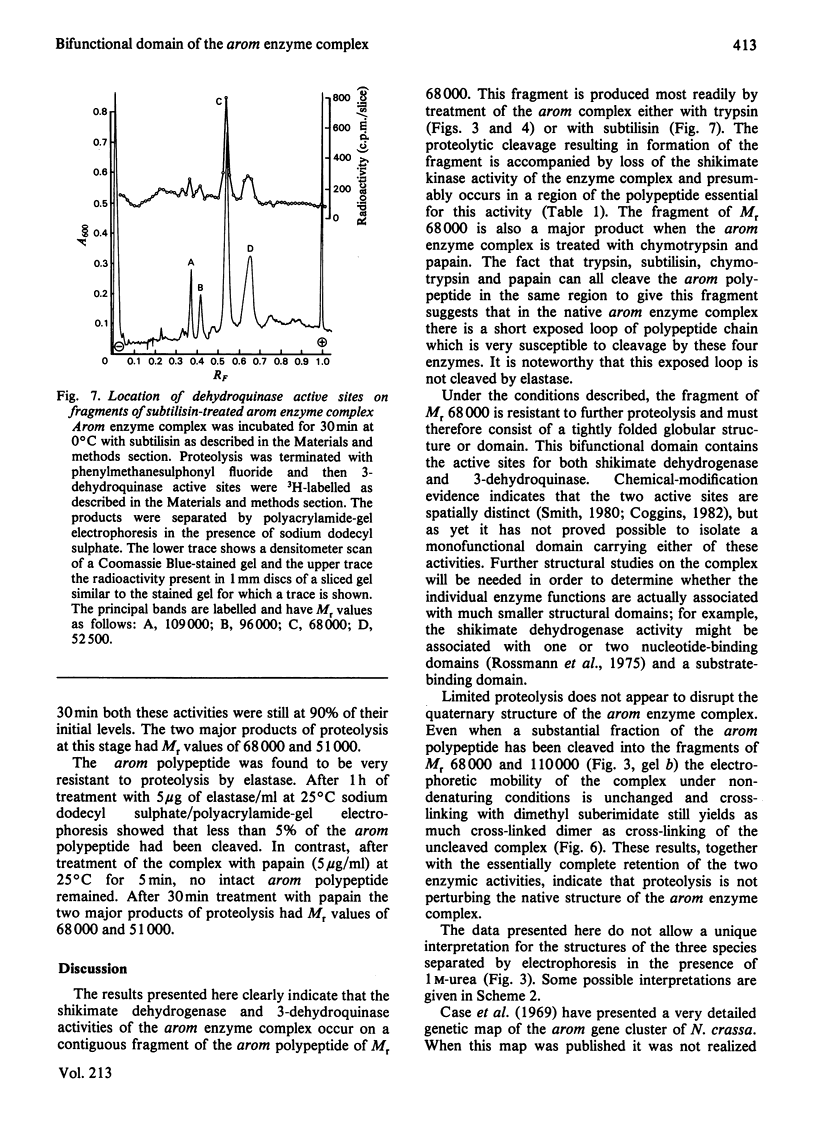
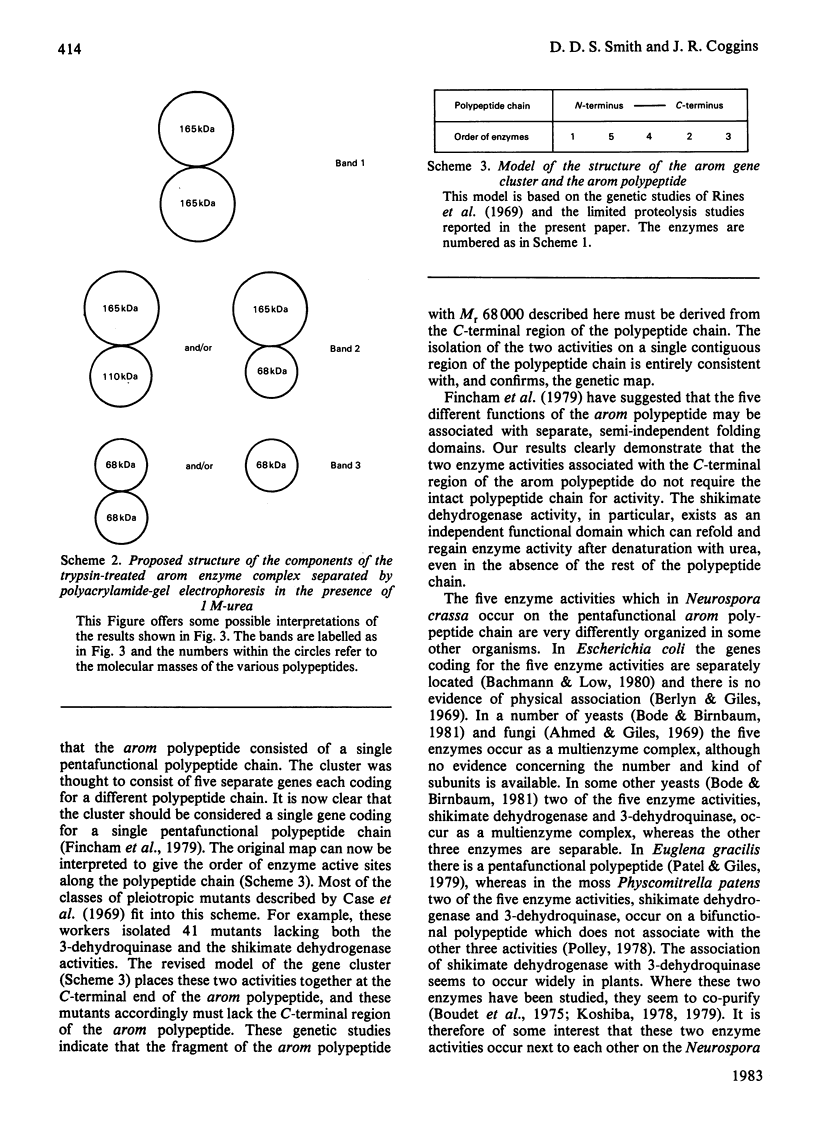
Limited proteolysis of the arom enzyme complex of Neurospora crassa by trypsin or subtilisin yielded a stable fragment of Mr 68000. This fragment, which was purified by two-dimensional polyacrylamide-gel electrophoresis, was shown by activity staining to contain the shikimate dehydrogenase active site, and by substrate labelling with 3-dehydroquinate and NaB3H4 to contain the 3-dehydroquinase active site. The fragment thus constitutes a bifunctional domain containing the two enzymic activities that are known, from genetic evidence, to be located adjacently at the C-terminal end of the pentafunctional arom polypeptide.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahmed S. I., Giles N. H. Organization of enzymes in the common aromatic synthetic pathway: evidence for aggregation in fungi. J Bacteriol. 1969 Jul;99(1):231–237. doi: 10.1128/jb.99.1.231-237.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J., Low K. B. Linkage map of Escherichia coli K-12, edition 6. Microbiol Rev. 1980 Mar;44(1):1–56. doi: 10.1128/mr.44.1.1-56.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D. L., Harrison R. A., Perham R. N. The stoichiometry of polypeptide chains in the pyruvate dehydrogenase multienzyme complex of E. coli determined by a simple novel method. FEBS Lett. 1975 Dec 15;60(2):427–430. doi: 10.1016/0014-5793(75)80764-2. [DOI] [PubMed] [Google Scholar]
- Berlyn M. B., Giles N. H. Organization of enzymes in the polyaromatic synthetic pathway: separability in bacteria. J Bacteriol. 1969 Jul;99(1):222–230. doi: 10.1128/jb.99.1.222-230.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode R., Birnbaum D. Aggregation und Trennbarkeit der Enzyme des Shikimat-Pathway bei Hefen. Z Allg Mikrobiol. 1981;21(6):417–422. doi: 10.1002/jobm.3630210602. [DOI] [PubMed] [Google Scholar]
- Case M. E., Burgoyne L., Giles N. H. In vivo and in vitro complementation between DHQ synthetase mutants in the arom gene cluster of Neurospora crassa. Genetics. 1969 Nov;63(3):581–588. doi: 10.1093/genetics/63.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coggins J. R., Lumsden J., Malcolm A. D. A study of the quaternary structure of Escherichia coli RNA polymerase using bis(imido esters). Biochemistry. 1977 Mar 22;16(6):1111–1116. doi: 10.1021/bi00625a013. [DOI] [PubMed] [Google Scholar]
- Danson M. J., Hale G., Johnson P., Perham R. N., Smith J., Spragg P. Molecular weight and symmetry of the pyruvate dehydrogenase multienzyme complex of Escherichia coli. J Mol Biol. 1979 Apr 25;129(4):603–617. doi: 10.1016/0022-2836(79)90471-6. [DOI] [PubMed] [Google Scholar]
- Dautry-Varsat A., Garel J. R. Refolding of a bifunctional enzyme and its monofunctional fragment. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5979–5982. doi: 10.1073/pnas.75.12.5979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dileepan K. N., Lin C. Y., Smith S. Release of two thioesterase domains from fatty acid synthetase by limited digestion with trypsin. Biochem J. 1978 Oct 1;175(1):199–206. doi: 10.1042/bj1750199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ERLANGER B. F., KOKOWSKY N., COHEN W. The preparation and properties of two new chromogenic substrates of trypsin. Arch Biochem Biophys. 1961 Nov;95:271–278. doi: 10.1016/0003-9861(61)90145-x. [DOI] [PubMed] [Google Scholar]
- Gaertner F. H., Cole K. W. A cluster-gene: evidence for one gene, one polypeptide, five enzymes. Biochem Biophys Res Commun. 1977 Mar 21;75(2):259–264. doi: 10.1016/0006-291x(77)91037-3. [DOI] [PubMed] [Google Scholar]
- Giles N. H., Case M. E., Partridge C. W., Ahmed S. I. A gene cluster in Nuerospora crassa coding for an aggregate of five aromatic synthetic enzymes. Proc Natl Acad Sci U S A. 1967 Oct;58(4):1453–1460. doi: 10.1073/pnas.58.4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner K., Bisswanger H. Multifunctional proteins. Annu Rev Biochem. 1976;45:143–166. doi: 10.1146/annurev.bi.45.070176.001043. [DOI] [PubMed] [Google Scholar]
- Koshiba T. Purification of two forms of the associated 3-dehydroquinate hydro-lyase and shikimate:NADP+ oxidoreductase in Phaseolus mungo seedlings. Biochim Biophys Acta. 1978 Jan 12;522(1):10–18. doi: 10.1016/0005-2744(78)90317-0. [DOI] [PubMed] [Google Scholar]
- Lumsden J., Coggins J. R. The subunit structure of the arom multienzyme complex of Neurospora crassa. A possible pentafunctional polypeptide chain. Biochem J. 1977 Mar 1;161(3):599–607. doi: 10.1042/bj1610599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumsden J., Coggins J. R. The subunit structure of the arom multienzyme complex of Neurospora crassa. Evidence from peptide 'maps' for the identity of the subunits. Biochem J. 1978 Feb 1;169(2):441–444. doi: 10.1042/bj1690441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy A. D., Hardie D. G. The multifunctional polypeptide chains of rabbit-mammary fatty-acid synthase. Stoichiometry of active sites and active-site mapping using limited proteolysis. Eur J Biochem. 1983 Jan 17;130(1):185–193. doi: 10.1111/j.1432-1033.1983.tb07135.x. [DOI] [PubMed] [Google Scholar]
- Patel V. B., Giles N. H. Purification of the arom multienzyme aggregate from Euglena gracilis. Biochim Biophys Acta. 1979 Mar 16;567(1):24–34. doi: 10.1016/0005-2744(79)90168-2. [DOI] [PubMed] [Google Scholar]
- Polley L. D. Purification and characterization of 3-dehydroquinate hydrolase and shikmate oxidoreductase. Evidence for a bifunctional enzyme. Biochim Biophys Acta. 1978 Sep 11;526(1):259–266. doi: 10.1016/0005-2744(78)90310-8. [DOI] [PubMed] [Google Scholar]
- Rines H. W., Case M. E., Giles N. H. Mutants in the arom gene cluster of Neurospora crassa specific for biosynthetic dehydroquinase. Genetics. 1969 Apr;61(4):789–800. doi: 10.1093/genetics/61.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan L. D., Vestling C. S. Rapid purification of lactate dehydrogenase from rat liver and hepatoma: a new approach. Arch Biochem Biophys. 1974 Jan;160(1):279–284. doi: 10.1016/s0003-9861(74)80035-4. [DOI] [PubMed] [Google Scholar]
- Tan L. U., MacKenzie R. E. Methylenetetrahydrofolate dehydrogenase-methenyltetrahydrofolate cyclohydrolase-formyltetrahydrofolate synthetase from porcine liver. Location of the activities in two domains of the multifunctional polypeptide. Can J Biochem. 1979 Jun;57(6):806–812. doi: 10.1139/o79-100. [DOI] [PubMed] [Google Scholar]
- Vitto A., Gaertner F. H. Proteolytic inactivation of a pentafunctional enzyme conjugate: coordinate protection by the first substrate. Biochem Biophys Res Commun. 1978 Jun 14;82(3):977–981. doi: 10.1016/0006-291x(78)90879-3. [DOI] [PubMed] [Google Scholar]