Abstract
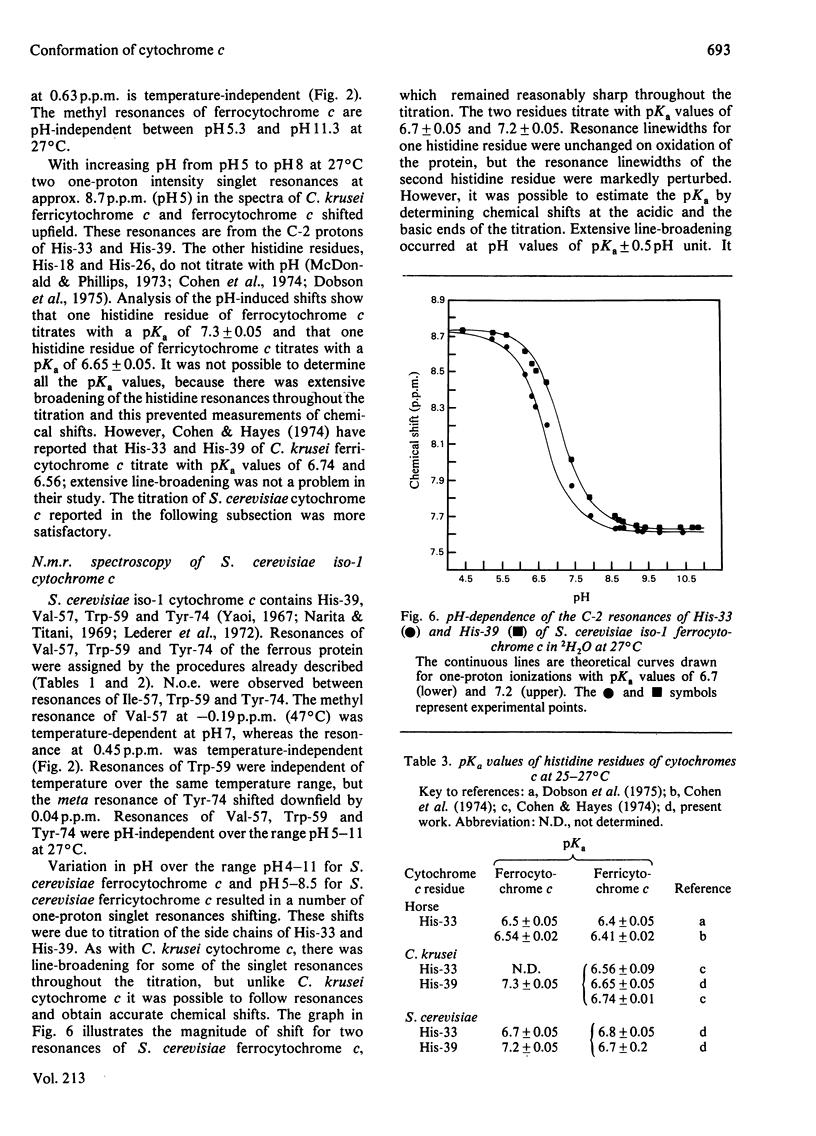
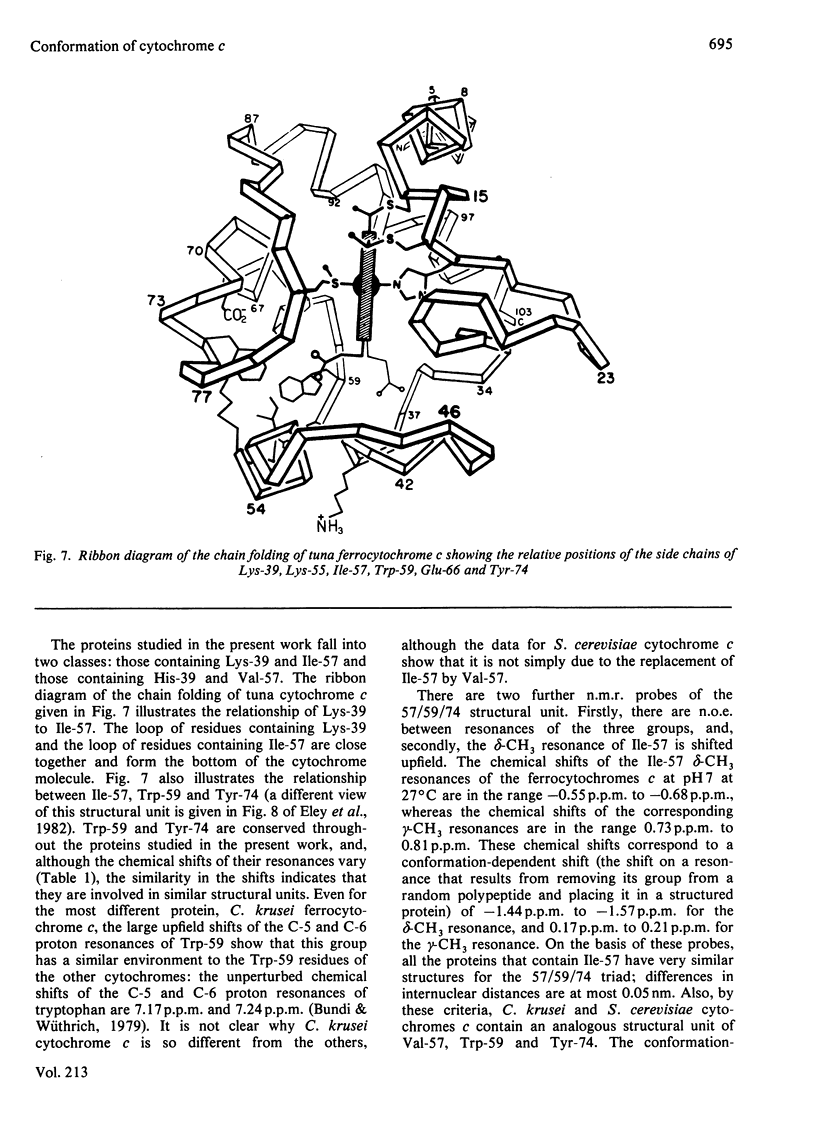
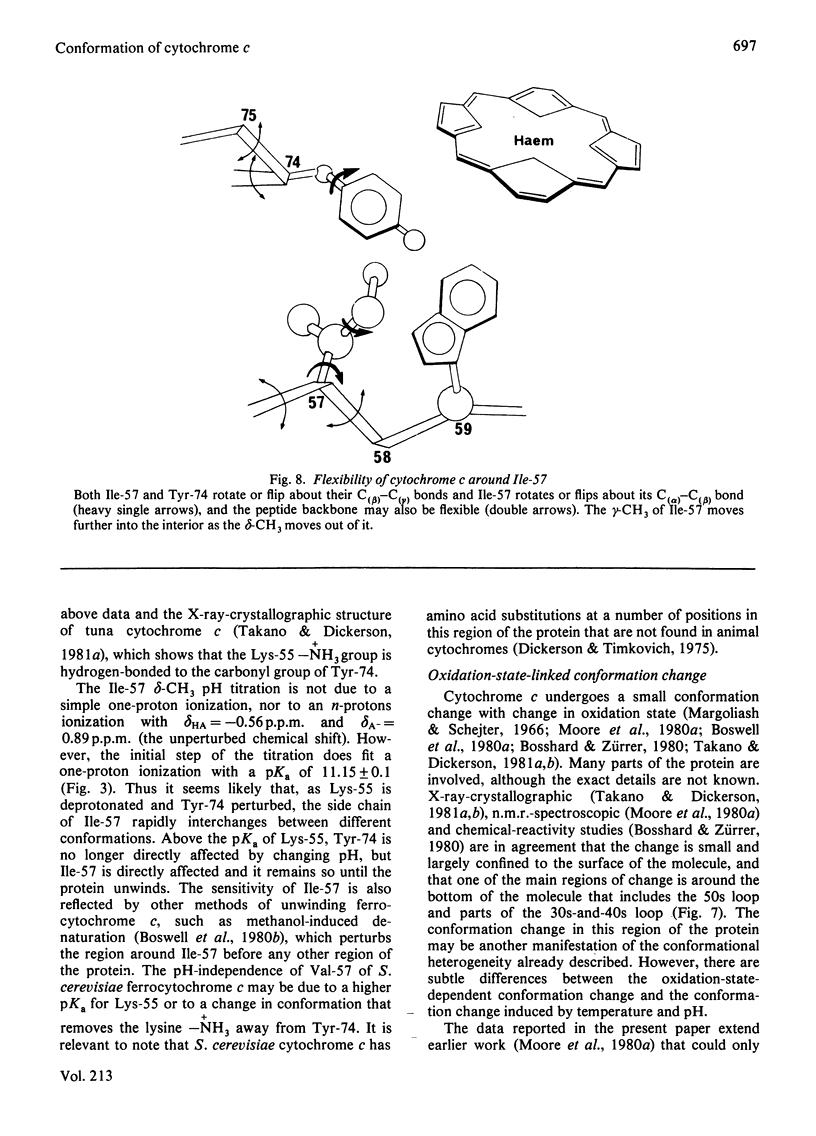
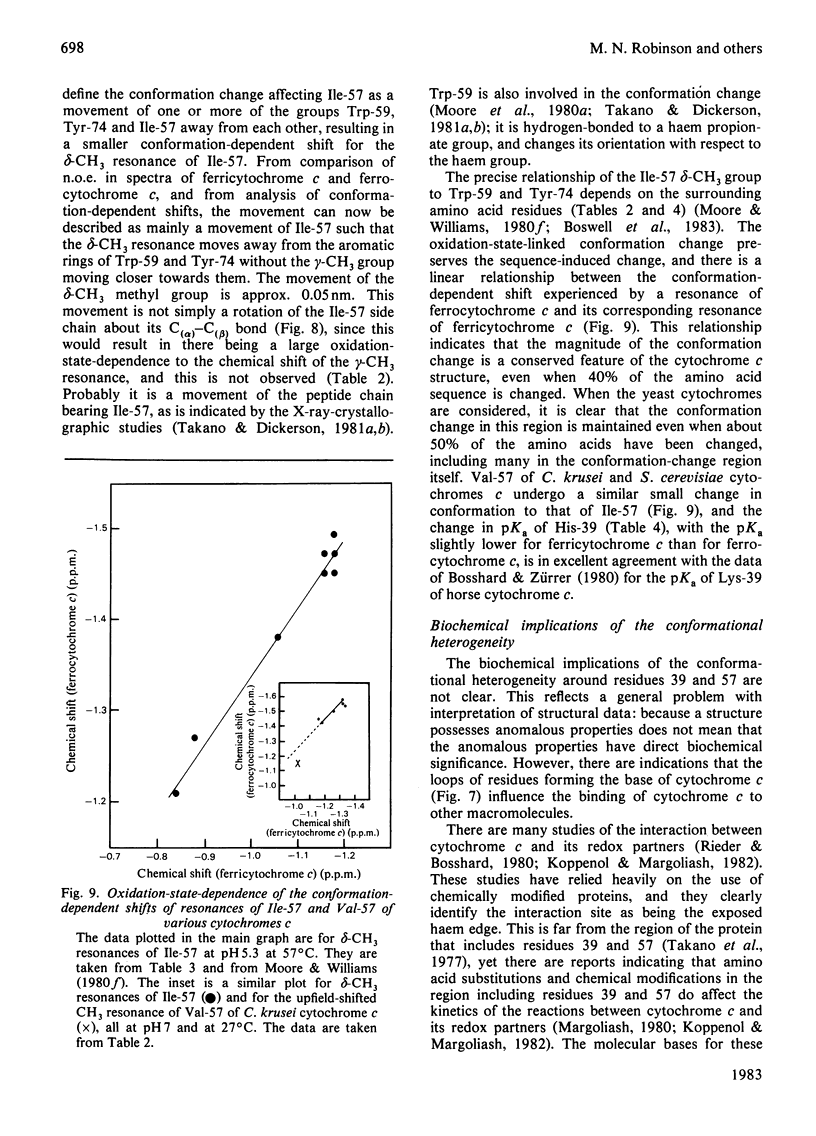
1H-n.m.r. studies of horse, tuna, Candida krusei and Saccharomyces cerevisiae cytochromes c showed that each of the proteins contains a similar cluster of residues at the bottom of the protein that assists in shielding the haem from the solvent. The relative positions of the residues forming these clusters vary continuously with temperature, and they change with the change in protein redox state. This conformational heterogeneity is discussed with reference to the conformational flexibility of cytochrome c around residues 57, 59 and 74. Spectroscopic measurements of pKa values for Lys-55 (horse and tuna cytochromes c) and His-33 and His-39 (C. krusei and S. cerevisiae cytochromes c) are in excellent agreement with expectations based on chemical-modification studies of horse cytochrome c. [Bosshard & Zürrer (1980) J. Biol. Chem. 255, 6694-6699] and on the X-ray-crystallographic structure of tuna cytochrome c [Takano & Dickerson (1981) J. Mol. Biol. 153, 79-94, 95-115].
Full text
PDF

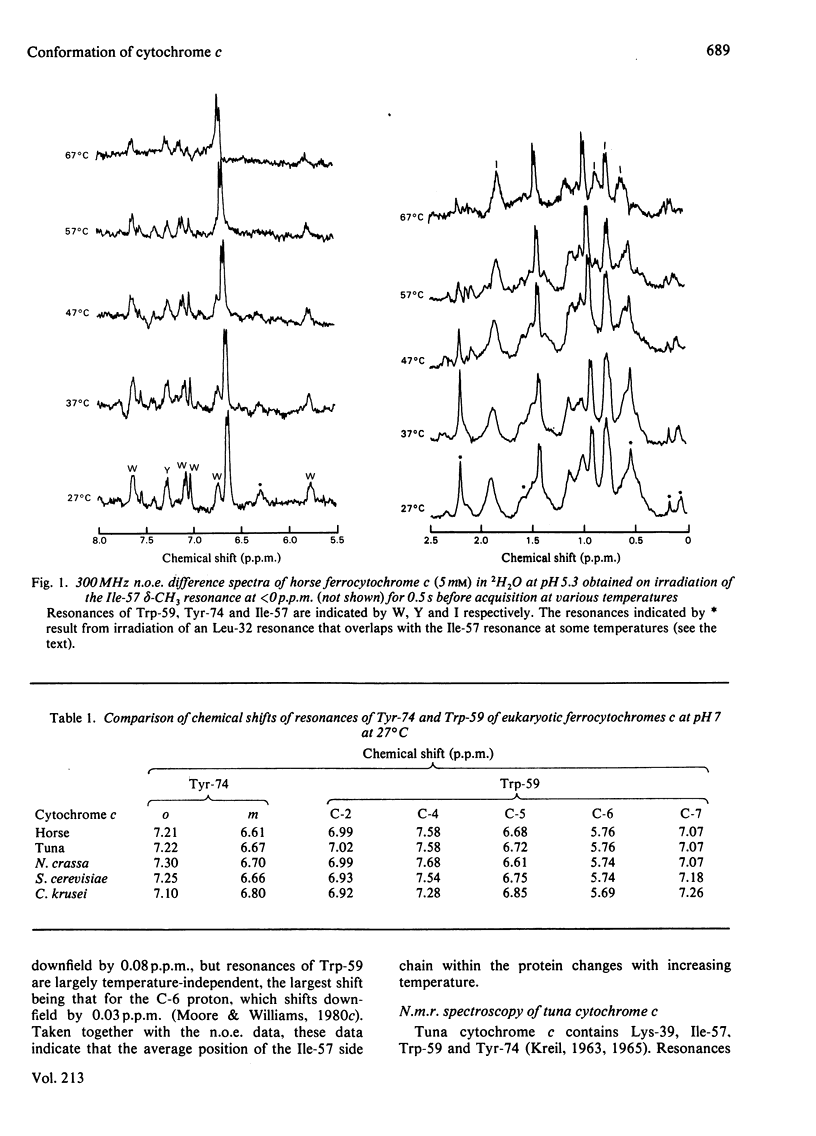
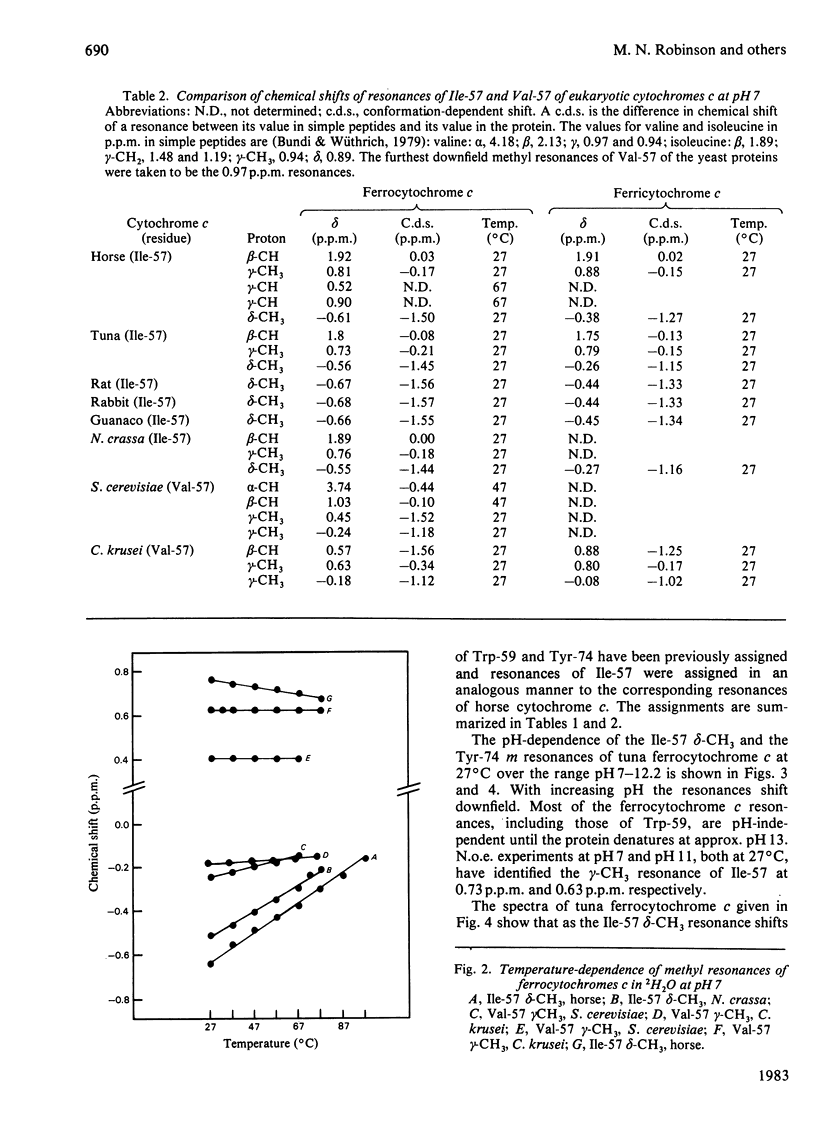
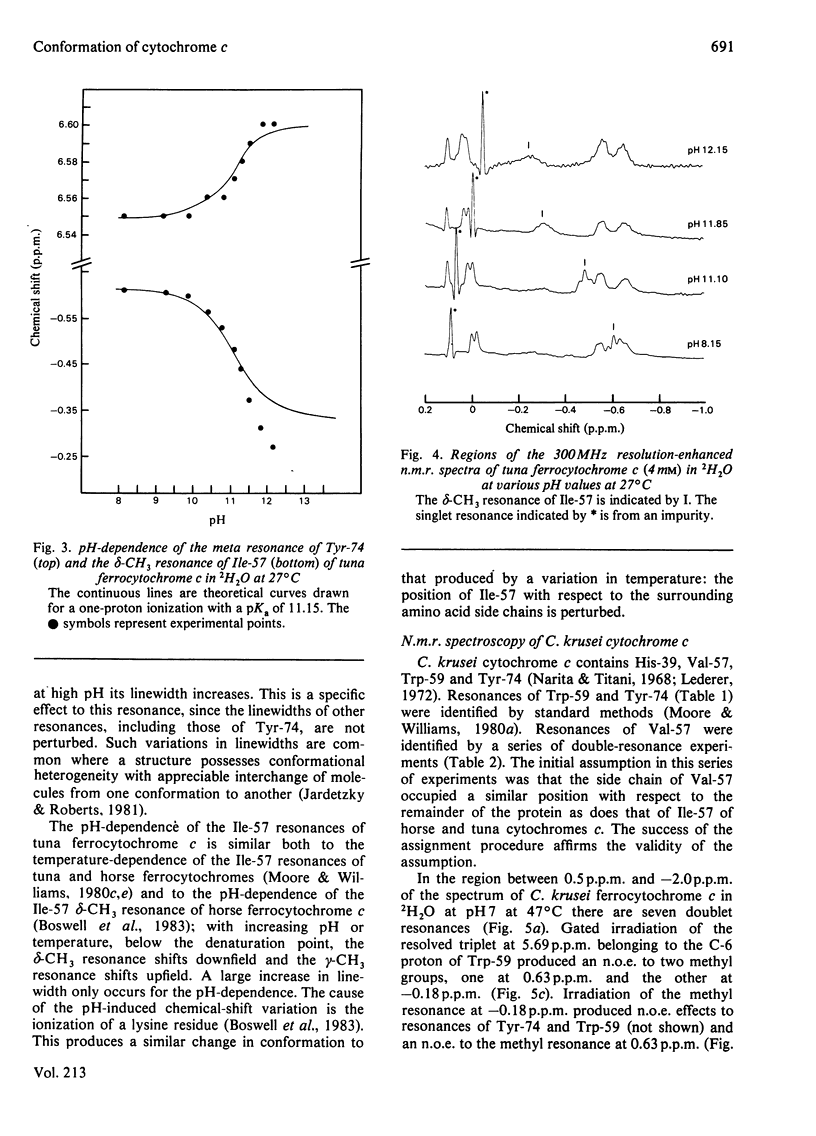
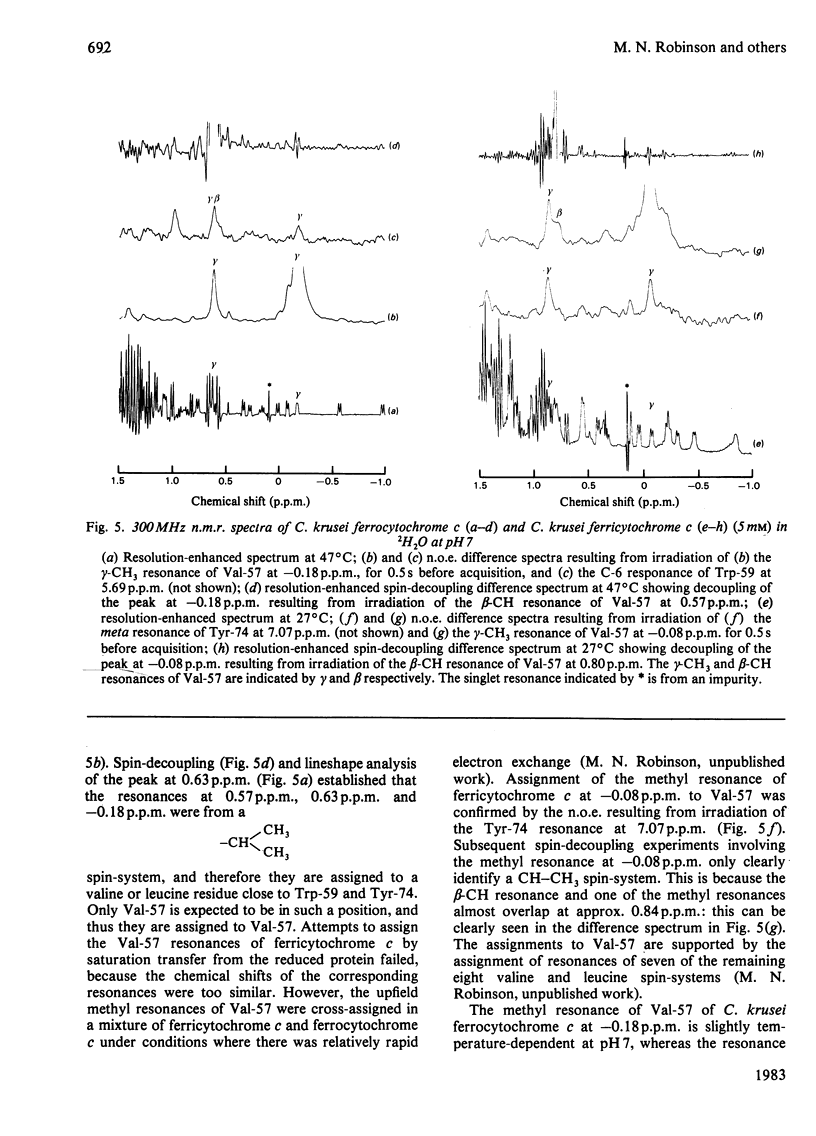




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bosshard H. R., Zürrer M. The conformation of cytochrome c in solution. Localization of a conformational difference between ferri- and ferrocytochrome c on the surface of the molecule. J Biol Chem. 1980 Jul 25;255(14):6694–6699. [PubMed] [Google Scholar]
- Boswell A. P., Moore G. R., Williams R. J., Harris D. E., Wallace C. J., Bocieck S., Welti D. Ionization of tyrosine and lysine residues in native and modified horse cytochrome c. Biochem J. 1983 Sep 1;213(3):679–686. doi: 10.1042/bj2130679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boswell A. P., Moore G. R., Williams R. J. Nuclear-magnetic-resonance studies of the methanol-induced denaturation of ferricytochrome c and ferrocytochrome c. Biochem Soc Trans. 1980 Oct;8(5):638–639. doi: 10.1042/bst0080638. [DOI] [PubMed] [Google Scholar]
- Carlson S. S., Mross G. A., Wilson A. C., Mead R. T., Wolin L. D., Bowers S. F., Foley N. T., Muijsers A. O., Margoliash E. Primary structure of mouse, rat, and guinea pig cytochrome c. Biochemistry. 1977 Apr 5;16(7):1437–1442. doi: 10.1021/bi00626a031. [DOI] [PubMed] [Google Scholar]
- Cohen J. S., Fisher W. R., Schechter A. N. Spectroscopic studies on the conformation of cytochrome c and apocytochrome c. J Biol Chem. 1974 Feb 25;249(4):1113–1118. [PubMed] [Google Scholar]
- Cohen J. S., Hayes M. B. Nuclear magnetic resonance titration curves of histidine ring protons. V. Comparative study of cytochrome c from three species and the assignment of individual proton resonances. J Biol Chem. 1974 Sep 10;249(17):5472–5477. [PubMed] [Google Scholar]
- Dobson C. M., Moore G. R., Williams R. J. Assignment of aromatic amino acid PMR resonances of horse ferricytochrome c. FEBS Lett. 1975 Mar 1;51(1):60–65. doi: 10.1016/0014-5793(75)80854-4. [DOI] [PubMed] [Google Scholar]
- Eley C. G., Moore G. R., Williams R. J., Neupert W., Boon P. J., Brinkhof H. H., Nivard R. J., Tesser G. I. Structural role of the tyrosine residues of cytochrome c. Biochem J. 1982 Jul 1;205(1):153–165. doi: 10.1042/bj2050153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris D. E., Offord R. E. A functioning complex between tryptic fragments of cytochrome c. A route to the production of semisynthetic analogues. Biochem J. 1977 Jan 1;161(1):21–25. doi: 10.1042/bj1610021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller J., Smith E. L. Neurospora crassa cytochrome c. II. Chymotryptic peptides, tryptic peptides, cyanogen bromide peptides, and the complete amino acid sequence. J Biol Chem. 1966 Jul 10;241(13):3165–3180. [PubMed] [Google Scholar]
- KREIL G. DIE C-TERMINALE AMINOSAEURESEQUENZ DES THUNFISCH-CYTOCHROMS C. Hoppe Seylers Z Physiol Chem. 1965;340:86–87. doi: 10.1515/bchm2.1965.340.1-2.86. [DOI] [PubMed] [Google Scholar]
- KREIL G. UBER DIE ARTSPEZIFITAET VON CYTOCHROM C: VERGLEICH DER AMINOSAEURESEQUENZ DES THUNFISCH-CYTOCHROMS C MIT DER DES PFERDE-CYTOCHROMS C. Hoppe Seylers Z Physiol Chem. 1963;334:154–166. doi: 10.1515/bchm2.1963.334.1.154. [DOI] [PubMed] [Google Scholar]
- Keller R. M., Wüthrich K. 1H-NMR studies of structural homologies between the heme environments in horse cytochrome c and in cytochrome c-552 from Euglena gracilis. Biochim Biophys Acta. 1981 Apr 28;668(2):307–320. doi: 10.1016/0005-2795(81)90038-6. [DOI] [PubMed] [Google Scholar]
- Keller R. M., Wüthrich K. Assignment of the heme c resonances in the 360 MHz H NMR spectra of cytochrome c. Biochim Biophys Acta. 1978 Mar 28;533(1):195–208. doi: 10.1016/0005-2795(78)90564-0. [DOI] [PubMed] [Google Scholar]
- Koppenol W. H., Margoliash E. The asymmetric distribution of charges on the surface of horse cytochrome c. Functional implications. J Biol Chem. 1982 Apr 25;257(8):4426–4437. [PubMed] [Google Scholar]
- Lederer F. Candida krusei chtochrome c: a correction to the sequence. Glutamine-16, an invariant residue in mitochondrial cytochrome c? Eur J Biochem. 1972 Nov 21;31(1):144–147. doi: 10.1111/j.1432-1033.1972.tb02512.x. [DOI] [PubMed] [Google Scholar]
- Lederer F., Simon A. M. Neurospora crassa and Humicola lanuginosa cytochromes c: more homology in the heme region. Biochem Biophys Res Commun. 1974 Jan 23;56(2):317–323. doi: 10.1016/0006-291x(74)90844-4. [DOI] [PubMed] [Google Scholar]
- Lederer F., Simon A. M., Verdière J. Saccharomyces cereviaiae iso-cytochromes c: revision of the amino acid sequence between the cysteine residues. Biochem Biophys Res Commun. 1972 Apr 14;47(1):55–58. doi: 10.1016/s0006-291x(72)80009-3. [DOI] [PubMed] [Google Scholar]
- MARGOLIASH E., SMITH E. L., KREIL G., TUPPY H. Amino-acid sequence of horse heart cytochrome c. Nature. 1961 Dec 23;192:1125–1127. doi: 10.1038/1921125a0. [DOI] [PubMed] [Google Scholar]
- Mandel N., Mandel G., Trus B. L., Rosenberg J., Carlson G., Dickerson R. E. Tuna cytochrome c at 2.0 A resolution. III. Coordinate optimization and comparison of structures. J Biol Chem. 1977 Jul 10;252(13):4619–4636. [PubMed] [Google Scholar]
- Margoliash E., Schejter A. Cytochrome c. Adv Protein Chem. 1966;21:113–286. doi: 10.1016/s0065-3233(08)60128-x. [DOI] [PubMed] [Google Scholar]
- McDonald C. C., Phillips W. D. Proton magnetic resonance studies of horse cytochrome c. Biochemistry. 1973 Aug 14;12(17):3170–3186. doi: 10.1021/bi00741a006. [DOI] [PubMed] [Google Scholar]
- Moore G. R., Williams R. J. Nuclear-magnetic-resonance studies of eukaryotic cytochrome c. Assignment of resonances of aliphatic amino acids. Eur J Biochem. 1980 Feb;103(3):503–512. doi: 10.1111/j.1432-1033.1980.tb05974.x. [DOI] [PubMed] [Google Scholar]
- Moore G. R., Williams R. J. Nuclear-magnetic-resonance studies of eukaryotic cytochrome c. Assignment of resonances of aromatic amino acids. Eur J Biochem. 1980 Feb;103(3):493–502. doi: 10.1111/j.1432-1033.1980.tb05973.x. [DOI] [PubMed] [Google Scholar]
- Moore G. R., Williams R. J. Nuclear-magnetic-resonance studies of ferrocytochrome c. pH and temperature dependence. Eur J Biochem. 1980 Feb;103(3):513–521. doi: 10.1111/j.1432-1033.1980.tb05975.x. [DOI] [PubMed] [Google Scholar]
- Narita K., Chitani K. The amino acid sequence of cytochrome C from Candida krusei. J Biochem. 1968 Feb;63(2):226–241. doi: 10.1093/oxfordjournals.jbchem.a128765. [DOI] [PubMed] [Google Scholar]
- Narita K., Chitani K. The complete amino acid sequence in baker's yeast cytochrome c. J Biochem. 1969 Feb;65(2):259–267. [PubMed] [Google Scholar]
- Redfield A. G., Gupta R. K. Pulsed NMR study of the structure of cytochrome c. Cold Spring Harb Symp Quant Biol. 1972;36:405–411. doi: 10.1101/sqb.1972.036.01.052. [DOI] [PubMed] [Google Scholar]
- Rieder R., Bosshard H. R. Comparison of the binding sites on cytochrome c for cytochrome c oxidase, cytochrome bc1, and cytochrome c1. Differential acetylation of lysyl residues in free and complexed cytochrome c. J Biol Chem. 1980 May 25;255(10):4732–4739. [PubMed] [Google Scholar]
- Shelnutt J. A., Rousseau D. L., Dethmers J. K., Margoliash E. Protein influences on porphyrin structure in cytochrome c: evidence from Raman difference spectroscopy. Biochemistry. 1981 Oct 27;20(22):6485–6497. doi: 10.1021/bi00525a030. [DOI] [PubMed] [Google Scholar]
- Takano T., Dickerson R. E. Conformation change of cytochrome c. II. Ferricytochrome c refinement at 1.8 A and comparison with the ferrocytochrome structure. J Mol Biol. 1981 Nov 25;153(1):95–115. doi: 10.1016/0022-2836(81)90529-5. [DOI] [PubMed] [Google Scholar]
- Takano T., Dickerson R. E. Redox conformation changes in refined tuna cytochrome c. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6371–6375. doi: 10.1073/pnas.77.11.6371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano T., Kallai O. B., Swanson R., Dickerson R. E. The structure of ferrocytochrome c at 2.45 A resolution. J Biol Chem. 1973 Aug 10;248(15):5234–5255. [PubMed] [Google Scholar]
- Takano T., Swanson R., Kallai O. B., Dickerson R. E. Conformational changes upon reduction of cytochrome c. Cold Spring Harb Symp Quant Biol. 1972;36:397–404. doi: 10.1101/sqb.1972.036.01.051. [DOI] [PubMed] [Google Scholar]
- Takano T., Trus B. L., Mandel N., Mandel G., Kallai O. B., Swanson R., Dickerson R. E. Tuna cytochrome c at 2.0 A resolution. II. Ferrocytochrome structure analysis. J Biol Chem. 1977 Jan 25;252(2):776–785. [PubMed] [Google Scholar]
- Urbanski G. J., Margoliash E. Topographic determinants on cytochrome c. I. The complete antigenic structures of rabbit, mouse, and guanaco cytochromes c in rabbits and mice1. J Immunol. 1977 Apr;118(4):1170–1180. [PubMed] [Google Scholar]
- Vanderkooi J., Erecińska M., Chance B. Cytochrome c interaction with membranes. II. Comparative study of the interaction of c cytochromes with the mitochondrial membrane. Arch Biochem Biophys. 1973 Aug;157(2):531–540. doi: 10.1016/0003-9861(73)90672-3. [DOI] [PubMed] [Google Scholar]
- Yaoi Y. Comparison of the primary structures of cytochromes c from wild and respiration-deficient mutant yeasts. J Biochem. 1967 Jan;61(1):54–58. doi: 10.1093/oxfordjournals.jbchem.a128520. [DOI] [PubMed] [Google Scholar]


