Abstract
The evolution of morphinan alkaloid biosynthesis in plants of the genus Papaver includes permutation of several processes including gene duplication, fusion, neofunctionalization, and deletion resulting in the present chemotaxonomy. A critical gene fusion event resulting in the key bifunctional enzyme reticuline epimerase (REPI), which catalyzes the stereochemical inversion of (S)-reticuline, was suggested to precede neofunctionalization of downstream enzymes leading to morphine biosynthesis in opium poppy (Papaver somniferum). The ancestrally related aldo-keto reductases 1,2-dehydroreticuline reductase (DRR), which occurs in some species as a component of REPI, and codeinone reductase (COR) catalyze the second and penultimate steps, respectively, in the pathway converting (S)-reticuline to morphine. Orthologs for each enzyme isolated from the transcriptomes of 12 Papaver species were shown to catalyze their respective reactions in species that capture states of the metabolic pathway prior to key evolutionary events, including the gene fusion event leading to REPI, thus suggesting a patchwork model for pathway evolution. Analysis of the structure and substrate preferences of DRR orthologs in comparison with COR orthologs revealed structure-function relationships underpinning the functional latency of DRR and COR orthologs in the genus Papaver, thus providing insights into the molecular events leading to the evolution of the pathway.
Subject terms: Secondary metabolism, Plant evolution
Conservation across the Papaver genus of two aldo-keto reductases catalyzing the second and penultimate steps in morphine biosynthesis reveals the latent activity of several enzymes and suggests a patchwork model of pathway evolution in opium poppy.
Introduction
Benzylisoquinoline alkaloids (BIAs) comprise a large family of structurally and functionally diverse plant specialized metabolites found primarily in the order Ranunculales1. BIAs exhibit potent pharmacological properties and include the vasodilator papaverine, the tumor suppressant and antitussive agent noscapine, the analgesics morphine and codeine, and semisynthetic derivatives of thebaine, such as oxycodone and the opioid antagonist naloxone2. Opium poppy (Papaver somniferum) as a cultivated crop remains the sole commercial source of all pharmaceutical opiates3, and is the only plant known to produce morphine and codeine, although other Papaver species accumulate the morphinan pathway intermediate thebaine.
The first committed step in morphinan alkaloid biosynthesis is the stereochemical inversion of the central branchpoint intermediate, (S)-reticuline1, from which most BIAs are derived. The inversion is a result of carbon-carbon and carbon-oxygen mediated ring formation and rearrangement. In opium poppy, (S)-reticuline is converted to (R)-reticuline by a unique fusion protein, reticuline epimerase (REPI), consisting of a translational coupling between a cytochrome P450 (CYP) and an aldo-keto reductase (AKR)4,5. Eight additional enzymes convert (R)-reticuline to morphine (Fig. 1A), including another CYP4–6, AKR4,5,7, one short chain dehydrogenase (SDR)8, two pathogenesis-related 10 proteins (PR10s)9,10, two Fe2+/2-oxoglutarate-dependent dioxygenases (DIOXs)11, and one acetyl-CoA-dependent acyltransferase (AT)12. The repeated recruitment of functionally distinct enzymes belonging to a limited number of protein families is a common theme in BIA metabolism. Recently, whole genome sequencing and comparative transcriptome analyzes, coupled with targeted alkaloid profiling, have begun to reveal key evolutionary events that have shaped BIA biosynthesis across members of the genus Papaver. Current models predict a dynamic evolutionary process whereby gene duplication, clustering, fusion, and deletion events have contributed to the recruitment and neofunctionalization of new biosynthetic enzymes, which ultimately facilitated the production of morphinan alkaloids in some Papaver species and, more specifically, the unique occurrence of morphine in opium poppy13–16.
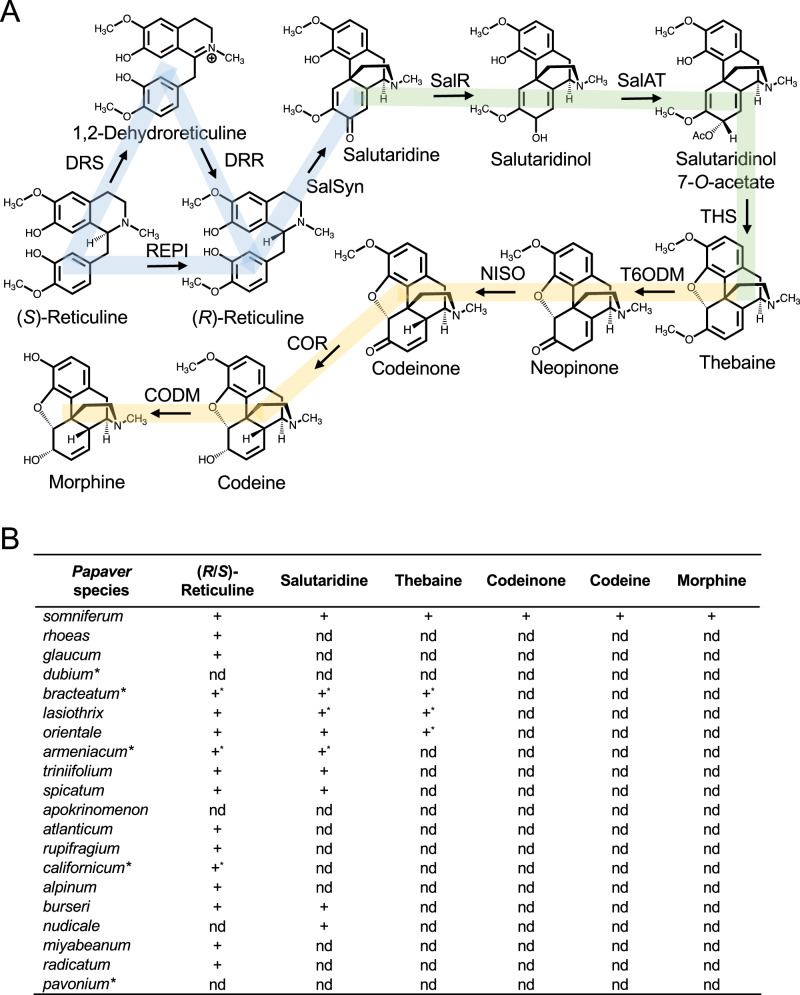
Fig. 1. Alkaloid profiling of various Papaver species.
A Morphinan alkaloid biosynthetic pathway in opium poppy. Steps from (S)-reticuline to salutaridine are highlighted in blue, the pathway from salutaridine to thebaine is highlighted in green, and the conversion of thebaine to morphine is highlighted in yellow. B Alkaloids in the pathway from (S)-reticuline to morphine detected in various Papaver species. ‘+’ indicates that the corresponding alkaloid was identified and the abbreviation ‘nd’ indicates that no alkaloid was detected. ‘+*’ indicate detected alkaloids reported in other published work15,30.
A patchwork model of metabolic pathway evolution17,18, which has been proposed for morphine14 and caffeine19 biosynthesis among other plant specialized metabolites, predicts that the latent catalytic promiscuity of some metabolic enzymes can be co-opted into new biosynthetic pathways20,21. A background network of latent enzyme promiscuity, dubbed the ‘underground metabolism’22, provides a platform for the recruitment of novel biosynthetic pathways whereby blocks of consecutive latent enzymatic activities may be linked together by key neofunctionalization events. The patchwork mechanism provides an attractive alternative to recruitment models suggesting the involvement of a specific selective pressure for the emergence of each new biosynthetic step by reducing the actual number of selection events. Gene duplication, via whole genome or local repetition processes, provides new genetic material for neofunctionalization and selection without compromising the function of the original gene.
REPI is an example of gene sequential duplication, clustering and fusion, and its role as the gateway into morphinan biosynthesis has been the focus of several recent studies14–16. REPI is a fusion of the CYP, 1,2-dehydroreticuline synthase (DRS), and the AKR, 1,2-dehydroreticuline reductase (DRR). The two-domain enzyme converts (S)-reticuline to (R)-reticuline via a two-step process, starting with the oxidation of (S)-reticuline by DRS yielding the iminium ion, 1,2-dehydroreticuline, from which the carbon-nitrogen double bond is subsequently reduced by DRR to form (R)-reticuline4,5. Phenol coupling of (R)-reticuline by the stereoselective CYP, salutaridine synthase (SalSyn), yields the pro-morphinan alkaloid salutaridine6. The fusion of DRS and DRR into REPI has been proposed as a pivotal event in the evolution of morphinan alkaloid biosynthesis15,16. Of particular interest is the relationship between DRR and codeinone reductase (COR), which catalyzes the penultimate step in the formation of morphine by reducing the ketone in codeinone to an alcohol in codeine and is the only other known AKR involved in BIA metabolism7. The AKR superfamily occurs in all phyla and plays diverse biological roles in plants, including detoxification of reactive molecules, carbon assimilation, iron acquisition, and the biosynthesis of specialized metabolites23, such as isoflavonoids24 and tropane alkaloids25. AKRs reduce a variety of functional groups, typically carbonyls such as aldehydes, ketones and quinones, atypically steroid double bonds, and in the case of REPI/DRR iminium ion carbon-nitrogen double bonds23,26. AKRs canonically employ a ‘push and pull’ mechanism involving a central catalytic tyrosine, which protonates or deprotonates the substrate with the aid of a histidine or lysine and an aspartate, while NADP(H) donates or accepts a hydride23. Atypical AKR mechanisms have been demonstrated primarily in the AKR1D family of steroid 5B-reductases27,28. Sequence, structural, and mutagenesis studies have shown COR to possess the canonical AKR tetrad, whereas homology modeling of DRR has revealed novel substitutions indicative of an atypical AKR mechanism29. The occurrence of different catalytic mechanisms poses intriguing questions about the evolutionary divergence of DRR and COR genes, and the fusion of DRR and DRS to form REPI.
In this work, we explore the evolution of morphinan alkaloid biosynthesis with a focus on the AKR enzymes, DRR, COR and REPI, and a consideration of early steps in the pro-morphinan alkaloid pathway. Using transcriptomics, kinetic characterization, structural reasoning based on homology models and molecular docking, and heterologous expression in yeast, we show the ubiquitous occurrence of functional orthologs for both DRR and COR in members of the genus Papaver.
Results
Alkaloid profiling of Papaver species
Alkaloid profiling was performed on 12 Papaver species (Fig. S1) and combined with previously published data on the distribution of morphinan alkaloids (Fig. 1B; Table S1). Phylogenetic relationships assembled from previous work8,15,30 are shown in Fig. 2B. Previously reported morphinan alkaloid profiles that mostly agree with data presented herein are available for 6 of the 12 analyzed Papaver species. The only notable discrepancies were (i) the empirical detection of salutaridine in P. nudicaule, which was previously reported not to accumulate pro-morphinan or morphinan alkaloids15, and (ii) the lack of salutaridine and thebaine in P. lasiothrix, which have been previously reported in this species30. In the 20 Papaver species shown in Fig. 1B, (i) P. armeniacum, P. triniifolium, P. spicatum, P. burseri, and P. nudicaule all accumulated salutaridine, (ii) P. bracteatum, P. lasiothrix, and P. orientale accumulated thebaine, and (iii) only P. somniferum accumulated codeinone, codeine, and morphine. P. orientale has also been reported to accumulate oripavine15, which is formed by the 3-O-demethylation of thebaine.
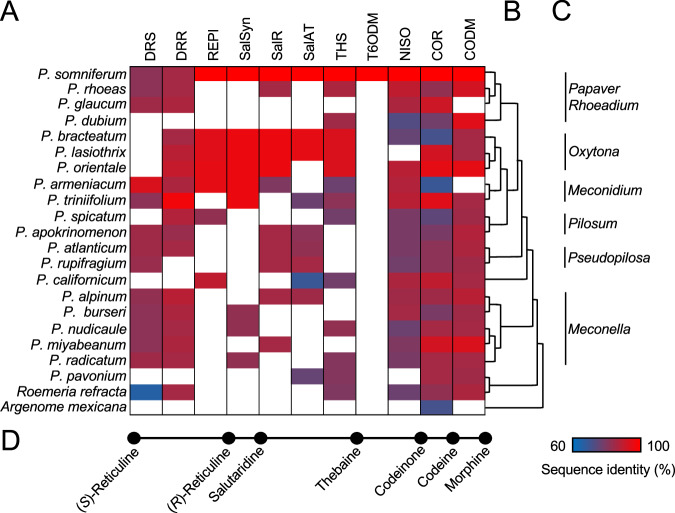
Fig. 2. Transcript profiling of various Papaver species.
A Occurrence of transcripts encoding proteins exhibiting greater than 60% amino acid sequence identity relative to enzymes involved in the conversion of (S)-reticuline to morphine in opium poppy. Nucleotide sequencing and contig assembly statistics are provided for each transcriptome in Table S8. Transcripts showing relatively higher predicted amino acid sequence identity with a different enzyme were omitted. Numerical percent identities and indication of partial and truncated transcript with the corresponding percent coverage are provided in Fig S2. Relative amino acid sequence identity with the corresponding opium poppy biosynthetic enzyme is shown as a gradient from red (100%) to blue (60%). White indicates that no transcript was annotated. Transcriptomes from previously published work were used for Papaver rhoeas31,32, P. dubium P. bracteatum P. orientale P. armeniacum P. californicum and P. pavonium15, R. refracta34, and A. mexicana33. B Consensus phylogenetic relationship among Papaver species based on previous published work15,80. C Selected section denominations of Papaver species. D Simplified diagram of the morphine biosynthetic pathway showing key metabolites from Fig. 1B and their corresponding position in panel A.
Transcriptome analysis of morphinan biosynthetic genes in Papaver species
Gene orthologs corresponding to putative pro-morphinan and morphinan alkaloid biosynthetic genes were detected (Figs. 2 and S2) by nucleotide BLAST analysis of the 12 transcriptomes sequenced and assembled herein, and previously reported libraries15,31–34 using P. somniferum biosynthetic genes as queries (Table S2). BLAST hits were sorted based on nucleotide sequence identity compared with known opium poppy biosynthetic enzymes (Fig. S3) and omitted if a higher sequence identity to a different biosynthetic enzyme was observed. For species with available genomes (i.e., P. bracteatum, P. armeniacum, P. atlanticum, P. californicum, and P. nudicaule), transcripts were cross-referenced to genomic data for further validation (Fig. S4 and table S3). Orthologous gene families (orthogroups) were determined35 to supplement BLAST analysis and investigate evolutionary relationships (Table S4).
The difficulty of accurately annotating transcripts based solely on sequence similarity is highlighted by several sets of enzyme families and subfamilies that share considerable amino acid sequence identity between pairs or within a group: AKR (i.e., DRR and COR), PR10 (i.e., THS and NISO), CYP719 [i.e., SalSyn and cheilanthifoline synthase (CheSyn)] and, in particular, DIOX (i.e., T6ODM, CODM, PODA, and DIOX6) family enzymes. A gradient of similarity across related enzymes precluded a trivial classification without functional characterization. AKR homologs of DRR and COR were initially classified based on catalytic tetrads whereby COR-like transcripts encoded proteins possessing the canonical AKR tetrad motif D51Y56K86H119 (COR numbering, abbreviated as DYKH), whereas DRR-like transcripts encoded proteins possessing the previously reported DYMP29 and additional variants DYML and DYNP (Figs. 3 and S3E). No similar and clearly differentiating architectural features were found in other putative pro-morphinan or morphinan biosynthetic enzymes belonging to the PR10, CYP719, SDR, AT, or DIOX families; thus, transcripts corresponding to these enzymes were annotated solely based on sequence identity. The CYP719s, SalSyn and CheSyn more clearly separated from each other (Fig. S3C). SalSyn was restricted to salutaridine-producing species; most distantly in the Meconella P. burseri, P. radicatum, and P. miyabeanum. DIOX-like transcripts were more ambiguous with all sequences sharing substantial identity with each other, which agrees with the reported substrate promiscuity of these enzymes36. DIOX-like transcripts were subcategorized as CODM-like, PODA-like, or DIOX6-like (Fig. S3A, B). Based on the criteria used, no DIOX-like transcripts were annotated as T6ODM-like. Although PODA-like transcripts from P. rhoeas, P. triniifolium and P. miyabeanum all shared >90% sequence identity with T6ODM, all three transcripts showed greater sequence identity with PODA (Fig. 3A, B). Interestingly, trace T6ODM activity has been observed in P. somniferum PODA36. Putative enzymes corresponding to PR10 proteins showed a gradient between THS- and NISO-like sequences (Fig. S3D). No closely related biosynthetic enzymes for other morphinan biosynthetic enzymes (i.e., DRS (CYP82Y2), SalR, and SalAT) were identified in these Papaver species.
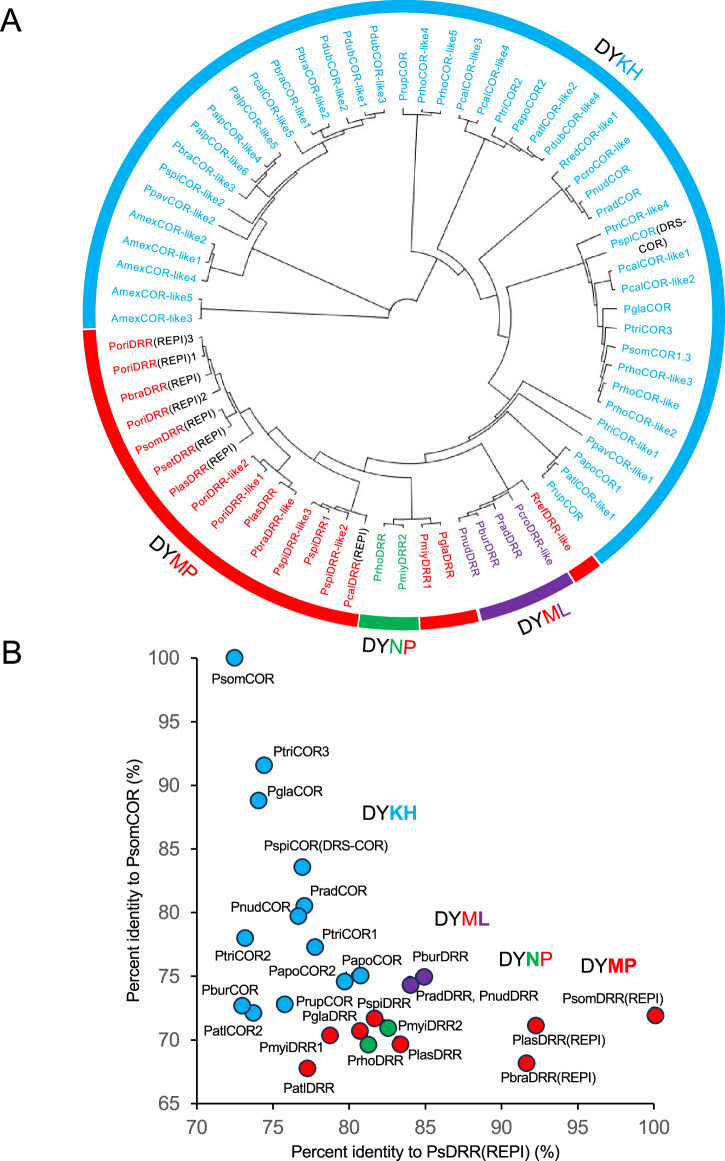
Fig. 3. Phylogenetic and sequence similarity analysis of AKR homologs from various Papaver species.
A Phylogenetic tree showing the relationships among full-length AKR transcripts found in transcriptomes from various Papaver species. Annotations of ‘COR-like’ or ‘DRR-like’ was based on the catalytic tetrad found in each predicted polypeptide. COR-like proteins containing the DYKH catalytic tetrad are shown in cyan, whereas DRR-like proteins are indicated in red (DYMP), purple (DYML), or green (DYNP). The function of transcripts lacking a ‘like’ suffix have been empirically confirmed. AKRs occurring as a fusion with a cytochrome P450 (i.e., a REPI homolog) are shown in black. The neighbor-joining tree was constructed using the Geneious tree-building Tamura-Nei model with a cost matrix of 65% similarity (5.0/−4.0), a gap open-penalty of 12, and a gap extension-penalty of 3 (nodes: 136; tips: 69). B Sequence similarity analysis of selected AKRs found in transcriptome from various Papaver species. Predicted proteins were plotted based on amino acid sequence identity compared with COR and DRR (from REPI), calculated using Clustal Omega81. COR-like proteins containing the DYKH catalytic tetrad are shown in cyan, whereas DRR-like proteins are indicated in red (DYMP), purple (DYML), or green (DYNP).
The observed orthologous transcripts were consistent with the metabolite profiling for most Papaver species, with some key exceptions. Despite the occurrence of salutaridine, no transcripts corresponding to REPI were identified in P. triniifolium, P. burseri, P. nudicaule, and to a similar extant P. spicatum. However, transcripts with moderate to high sequence identity compared with the DRS and DRR moieties of P. somniferum REPI were detected, unfused (Fig. 2 and S2). In these species, unfused DRS and DRR may catalyze the conversion of (S)- to (R)-reticuline instead of REPI. An unusual DRS-COR fusion found in P. spicatum cannot be ruled out as a transcriptome assembly error, although this is unlikely. SalSyn transcripts were also missing in P. spicatum and SalAT in P. orientale; the latter having been previously reported12. Similar DRS and DRR pairs are found throughout the genus Papaver. Notably, unfused DRS-DRR pairs predicted to convert (S)-reticuline to (R)-reticuline in place of REPI are found in the more distantly related Meconella section before the earliest known instance of REPI in P. californicum (Figs. 2 and S2).
Pertinent to the concept of ‘underground’ metabolism, was the observation of orthologous transcripts in Papaver species lacking the associated pro-morphinan or morphinan alkaloids. Most striking, COR-like transcripts were found in all Papaver species, and detected in other members of the Papaveraceae including R. refracta, Argemone mexicana, Chelidonium majus37, and Corydalis yanhusuo38,39. Transcripts from P. glaucum, P. lasiothrix, P. orientale, P. triniifolium, and most distantly P. miyabeanum shared over 90% sequence identity to P. somniferum COR. The roles of such COR-like transcripts are unknown since only P. somniferum accumulates the known COR substrates and products. DIOX-like transcripts with sequence identities closest to P. somniferum CODM were nearly ubiquitous across the Papaver genus and occurred even more broadly in other members of the Papaveraceae including Roemeria refracta. As mentioned above, the closely related DIOXs T6ODM, CODM, PODA, and DIOX6 preclude simple sequence-based assignment. Similar with COR-like transcripts, CODM, PODA, and DIOX6-like transcripts sharing over 90% sequence identity to their P. somniferum counterparts were found sporadically across the genus Papaver, most distantly in the Meconella section.
Papaver AKR orthologs show either DRR or COR activity
The role of DRR in REPI as a gateway step into morphinan alkaloid biosynthesis together with a shared ancestry with COR, and their apparent ubiquity in the Papaver genus, suggest that DRR and COR are important components in the evolutionary history of the pathway. DRR and COR transcripts were categorized based on overall sequence identity compared with P. somniferum DRR(REPI) and COR, although a comparison of their catalytic tetrad motifs was a more robust feature to predict COR or DRR activity. COR-like transcripts possessed the canonical AKR tetrad, DYKH, whereas DRR-like transcripts contained the variations DYMP, DYML or DYNP. All enzymes containing the DYKH tetrad showed only COR activity, whereas all enzymes with either DYMP, DYML or DYNP motifs displayed only DRR activity. Six of the 12 CORs and 2 of the 11 DRRs showed weak activity detected only in long (overnight) assays (Figs. S5 and S6). Interestingly, the putative DRS-AKR fusion from P. spicatum contained the DYKH catalytic tetrad and showed weak COR activity, but not DRR activity (Fig. S6), suggesting a DRS-COR fusion. The REPI-like DRS-AKR transcript in P. spicatum contained the COR-like DYKH catalytic tetrad in its AKR moiety and exhibited COR activity (Fig. S6). Specific activities (Figs. 4 and 5) were measured for 9 DRRs and 6 CORs, and 4 representatives of each were further characterized (Tables 1, 2).
Fig. 4. Functional characterization of DRR homologs from various Papaver species.
Specific activities of select DRR homologs. Corresponding enzyme Papaver sections and catalytic tetrad motifs are shown underneath the X-axis and the reaction coordinate above the plots. A,B Black bars indicate specific activity in ‘forward’ reductive enzyme assays measuring the formation of (R)-reticuline from 20 μM 1,2-dehydroreticuline and 500 μM NADPH. Reactions were carried out in 100 mM Bis-tris propane buffer, pH 7.0. A Data for all characterized DRRs. B Data for a subset of DRRs with relatively low reductive activity. C,D Gray bars indicate specific activity in ‘reverse’ oxidative enzyme assays measuring the formation of 1,2-dehydroreticuline from 20 μM (R)-reticuline and 500 μM NADP+. Reactions were carried out in 100 mM Bis-tris propane buffer, pH 8.8. C Data for all characterized DRRs. D Data for a subset of DRRs with relatively low oxidative activity. The protein concentrations and reaction times required to achieve less than 10% substrate conversion for each enzyme are provided in Table S10. Bars represent the mean ± standard deviation of three independent replicates.
Fig. 5. Functional characterization of COR homologs from various Papaver species.
Specific activities of select COR homologs. Corresponding enzyme Papaver sections and catalytic tetrad motifs are shown underneath the X-axis and the reaction coordinate above the plots. A,B Black bars indicate specific activity in ‘forward’ reductive enzyme assays measuring the formation of codeinone from 50 μM codeine and 1 mM NADPH. Reactions were carried out in 100 mM Bis-tris propane buffer, pH 8.0. A Data for all characterized CORs. B Data for a subset of CORs with relatively low reductive activity. C,D Gray bars indicate specific activity in ‘reverse’ oxidative enzyme assays measuring the formation of 1,2-dehydroreticuline from 50 μM codeine and 1 mM NADP+. Reactions were carried out in 100 mM Bis-tris propane buffer, pH 8.8. C Data for all characterized CORs. D Data for a subset of CORs with relatively low oxidative activity. The protein concentrations and reaction times required to achieve less than 10% substrate conversion for each enzyme are provided in Table S10. Bars represent the mean ± standard deviation of three independent replicates.
Table 1.
Kinetic parameters of selected DRRs from various Papaver species
| Enzyme | Substrate | Product | kcat (s−1) | Km (μM) | Ki (μM) | kcat/Km (M−1 s−1) |
|---|---|---|---|---|---|---|
| PburDRR | 1,2-DR | (R)-R | 5.40 ± 0.47 | 5.5 ± 1.0 | 79.5 ± 18.7 | 977,706 |
| PsomDRR(REPI) | 1,2-DR | (R)-R | 5.36 ± 1.71 | 7.3 ± 2.2 | 43.9 ± 15.4 | 735,245 |
| PglaDRR | 1,2-DR | (R)-R | 0.57 ± 0.05 | 16.4 ± 2.3 | 108.8 ± 23.4 | 35,078 |
| PlasDRR(REPI) | 1,2-DR | (R)-R | 0.22 ± 0.02 | 12.3 ± 2.2 | 177.9 ± 63.2 | 17,544 |
| PburDRR | (R)-R | 1,2-DR | 1.03 + 0.03 | 14.5 ± 1.8 | NA | 71,776 |
| PsomDRR(REPI) | (R)-R | 1,2-DR | 0.43 + 0.01 | 37.3 ± 4.0 | NA | 11,559 |
| PgalDRR | (R)-R | 1,2-DR | 0.31 + 0.04 | 906.6 ± 176.6 | NA | 340 |
| PlasDRR(REPI) | (R)-R | 1,2-DR | 0.150 + 0.003 | 47.2 ± 2.9 | NA | 3097 |
Kinetic curves are shown in Fig. S17 and reaction conditions are provided in Table S12. Ki was determined from substrate inhibition curve fit. ‘NA’, not applicable, represents substrate inhibition was not observed. 1,2-dehydroreticuline is abbreviated as 1,2-DR and (R)-reticuline as (R)-R. Errors represent the mean ± standard deviation of three independent replicates.
Table 2.
Kinetic parameters of selected CORs from various Papaver species
| Enzyme | Substrate | Product | kcat (s−1) | Km (μM) | kcat/Km (M−1 s−1) |
|---|---|---|---|---|---|
| PsomCOR | Codeinone | Codeine | 3.33 ± 0.13 | 17.4 ± 1.6 | 191,331 |
| PtriCOR3 | Codeinone | Codeine | 0.99 ± 0.03 | 10.0 ± 0.9 | 98,801 |
| PapoCOR1 | Codeinone | Codeine | 0.0261 ± 0.002 | 116.8 ± 17.8 | 224 |
| PglaCOR | Codeinone | Codeine | 0.0236 ± 0.0007 | 78.8 ± 5.6 | 300 |
| PsomCOR | Codeine | Codeinone | 0.81 ± 0.03 | 49.9 ± 4.1 | 16,285 |
| PtriCOR3 | Codeine | Codeinone | 0.39 ± 0.01 | 62.7 ± 5.2 | 6223 |
| PapoCOR1 | Codeine | Codeinone | 0.0060 ± 0.0004 | 72.1 ± 9.4 | 89 |
Unexpectedly, P. somniferum DRR(REPI) did not display the highest specific activity among DRR homologs in the Papaver genus. P. bracteatum DRR(REPI) displayed 2.7-fold higher reductive specific activity, whereas P. radicatum, P. nudicaule and P. burseri showed 7.2-, 6.2- and 5.6-fold higher reductive, and 10.1-, 4.8- and 5.0-fold higher oxidative activity, respectively, compared with P. somniferum DRR(REPI) (Fig. 4; Table S5). Kinetic characterization of P. burseri DRR showed a 1.3- and 6.2-fold increase in reductive and oxidative kcat/Km, respectively, compared with P. somniferum DRR(REPI) (Table 1, S5). P. radicatum, P. nudicaule and P. burseri DRRs are independent enzymes (i.e., not fused with DRS) that all share the DYML motif and are closely related in the Meconella section. The other tested DRRs belonging to the Meconella section, P. miyabeanum DRR1 and DRR2, contain the DYMP and DYNP motifs, respectively, showed 7.8- and 42.3-fold lower specific activity in the reductive assay, and 40.8- and 100.0-fold lower specific activity in the oxidative assay, respectively, compared with P. somniferum DRR(REPI) (Fig. 4; Table S5). Kinetic characterization of P. burseri DRR, P. somniferum DRR(REPI), P. glaucum DRR, and P. lasiothrix DRR(REPI) supported the observed trends in specific activity. Although less difference was apparent when comparing the reductive kcat/Km of P. burseri and P. somniferum DRR(REPI), and the oxidative kcat/Km of P. lasiothrix DRR(REPI) and P. somniferum DRR(REPI) (Table 1, S5)).
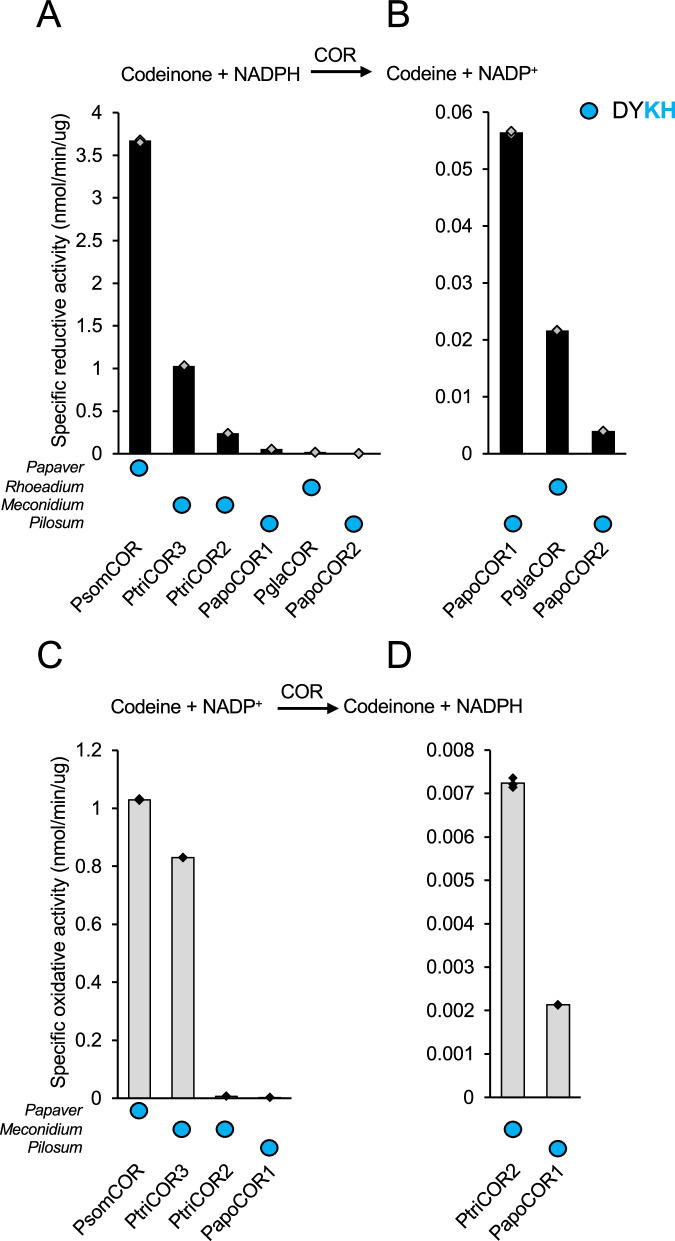
In contrast to the pattern of DRR activity, P. somniferum COR displayed the highest specific activity among COR homologs in the Papaver genus. Among the six enzymes that displayed the highest specific activity, P. triniifolium COR3 showed the most similar specific activity compared with P. somniferum COR (i.e., 3.6- and 1.2-fold lower with respect to reductive and oxidative reactions) (Fig. 5; Table S5). P. triniifolium COR2, P. apokrinomenon COR1, P. glaucum COR, and P. apokrinomenon COR2 showed 15.3-, 65.5-, 166.8-, and 917.5-fold lower specific activities in the reductive direction, whereas P. triniifolium COR2, P. apokrinomenon COR1 displayed 143.1- and 490.5-fold lower specific activity in the oxidative direction. kcat/Km for P. somniferum COR, P. triniifolium COR3, P. apokrinomenon COR1, and P. glaucum COR (Table 2, S5) supported trends in specific activity although P. apokrinomenon COR1 and P. glaucum COR showed a more dramatic decrease in reductive kcat/Km.
Among the Papaver AKRs, higher sequence identity to the P. somniferum enzyme (COR or DRR(REPI)) is correlated to increased activity among CORs but not DRRs (Table S6). The correlation suggests that Papaver CORs followed a more linear evolution compared with Papaver DRRs, which potentially followed a more branched evolutionary path. Unfused Meconella DRRs likely faced different selection pressures compared with fused DRR(REPI)s after divergence of the resulting lineages. The detection of highly active CORs in P. triniifolium, P. apokrinomenon, and P. glaucum was unexpected since none of these species accumulate COR substrates (i.e., codeinone or morphinone) or products (i.e., codeine or morphine). Moreover, only P. triniifolium accumulates even the pro-morphinan salutaridine (Fig. 1B).
AKR evolution and catalysis as revealed by P. somniferum DRR(REPI) mutagenesis
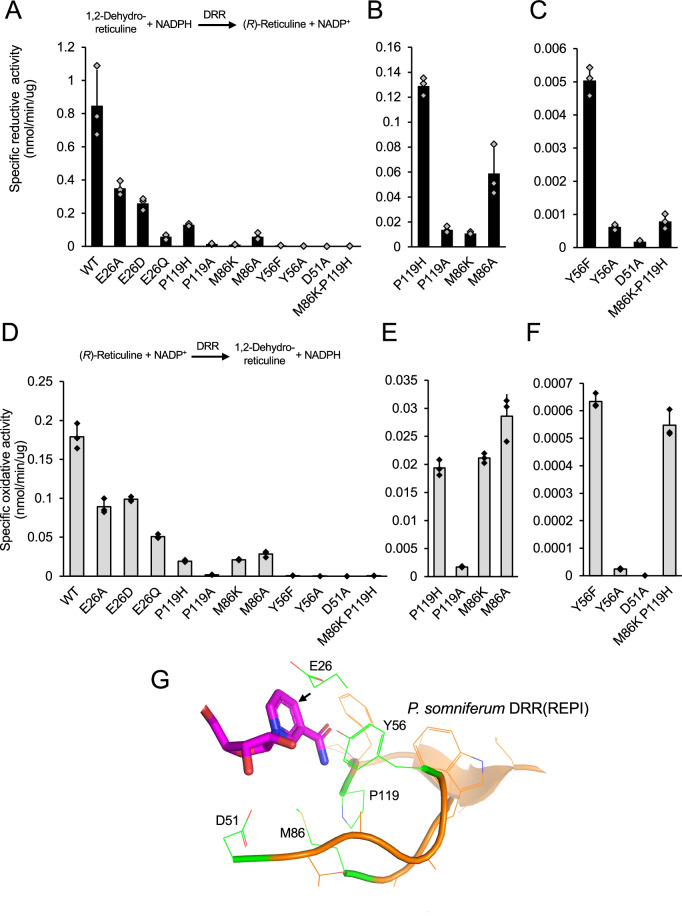
To investigate the atypical substitutions to the canonical AKR catalytic tetrad in DRRs, site-directed mutagenesis targeting the DYMP motif was applied to P. somniferum DRR(REPI) (Fig. 6). Two strategies were followed in mutant selection. First, substitutions were designed to negate the role of specific residues (E26A, E26D, E26Q, D51A, Y56A, Y56F, M86A, and P119A) and second, other substitutions were intended to reconstitute the canonical AKR catalytic tetrad of DYKH (M86K, P119H, and M86K-P119H). The E26 mutants were designed to investigate a hypothesized catalytic role of E2629. The mutants E26A, E26D, and E26Q showed 2.6-, 3.7-, and 19.5-fold decrease in reductive activity and 2.0-, 1.8-, and 3.5-fold decrease in oxidative activity, respectively (Fig. 6; Table S5). The retainment of modest to low DRR activity in the E26 mutants, particularly E26A, suggests that E26 plays a non-critical role in DRR catalysis. All mutations affecting the DYMP motif retained at least a low level of activity except for D51A in the oxidative direction, for which no activity was detected even in overnight assays. In the reductive direction, D51A showed a 4667-fold decrease in activity compared with wild type DRR(REPI), which represented the largest decrease in activity among the mutants (Fig. 6; Table S5). Y56A and Y56F substitutions showed the second largest decreases in activity (i.e., 1355- and 168-fold lower for the reductive reaction, and 7500- and 286-fold lower for the oxidative direction, respectively) compared with wild type DRR(REPI) (Fig. 6; Table S5). The higher activity of Y56F versus Y56A was expected, owing to the similarity of phenylalanine with tyrosine compared to alanine. It was nevertheless surprising that the Y56A and Y56F substitutions retain even a low level of activity based on the canonical importance of the titratable group contributed by Y56. Both D51 and Y56 are ubiquitously conserved across all AKR transcripts in the Papaver genus, which coincides with substitutions resulting in the most detrimental effect on P. somniferum DRR(REPI) activity and highlighting their importance. M86 and P119 are less conserved across Papaver DRRs (i.e., natural M86N and P119L substitutions occur in active DRRs), although these positions are conserved as K86 and H119 across Papaver CORs. P. somniferum DRR(REPI) P119A and P119H substitutions resulted in a 60.0- and 6.5-fold decrease in activity for the reductive reaction, and a 105.9- and 9.5-fold decrease in the oxidative direction, respectively, compared with wild type DRR(REPI) (Fig. 6; Table S5). M86A and M86K substitutions resulted in 19.1- and 76.4-fold decreases in activity for the reductive reaction, and 6.2- and 8.6-fold decreases in the oxidative direction, respectively (Fig. 6; Table S5). The modest decrease in activity of M86 and P119 mutants and the observed natural amino acid substitutions at these positions among Papaver DRRs suggest that they are not required for catalysis. This is contrary to CORs where K86 and H119, along with D51, form a proton relay promoting Y56 as general acid or base.
Fig. 6. Mutagenesis of the DRR component of REPI from opium poppy reveals a novel catalytic tetrad.
A–C Black bars indicate specific activity of DRR mutants in ‘forward’ reductive enzyme assays measuring the formation of (R)-reticuline from 20 μM 1,2-dehydroreticuline and 500 μM NADPH. Reactions were carried out in 100 mM Bis-tris propane buffer, pH 7.0. A Data for all characterized DRRs. B Data for a subset of DRRs with relatively moderate reductive activity. C Data for a subset of DRRs with relatively low reductive activity. D–F Gray bars indicate specific activity of DRR mutants in ‘reverse’ oxidative enzyme assays measuring the formation of 1,2-dehydroreticuline from 20 μM (R)-reticuline and 500 μM NADP+. Reactions were carried out in 100 mM Bis-tris propane buffer, pH 8.8. D Data for all characterized DRRs. E Data for a subset of DRRs with relatively moderate oxidative activity. F Data for a subset of DRRs with relatively low oxidative activity. The protein concentrations and reaction times required to achieve less than 10% substrate conversion for each enzyme are provided in Table S11. Bars represent the mean ± standard deviation of three independent replicates. Reaction coordinates are shown above the plots. G Depiction of the active site residues selected for mutagenesis from the homology model of P. somniferum DRR from REPI. Mutagenesis candidate residues are shown in green, NADPH in magenta, and the pro R face of NADPH is designated with an arrow.
To investigate the apparent proton relay disruption, the canonical AKR catalytic tetrad of DYKH was reconstituted in P. somniferum DRR(REPI) by substituting M86K and P119H. The single substitutions retained a low level of DRR activity as described above, whereas the double substitution, M86K-P119H showed a 1063- and 327-fold decrease in DRR activity in the reductive and oxidative directions, respectively, compared with wild type DRR(REPI) (Fig. 6; Table S5). The M86K and M86K-P119H substitutions also exhibited a low level of COR activity (Fig. S7), which is attributed to the M86K substitution since no COR activity was detected with a P119H substitution. The low level of COR activity enabled by the M86K substitution is presumably caused by the reconstitution of a proton relay between D51, K86, and Y56.
Homology modeling and substrate docking of Papaver AKRs
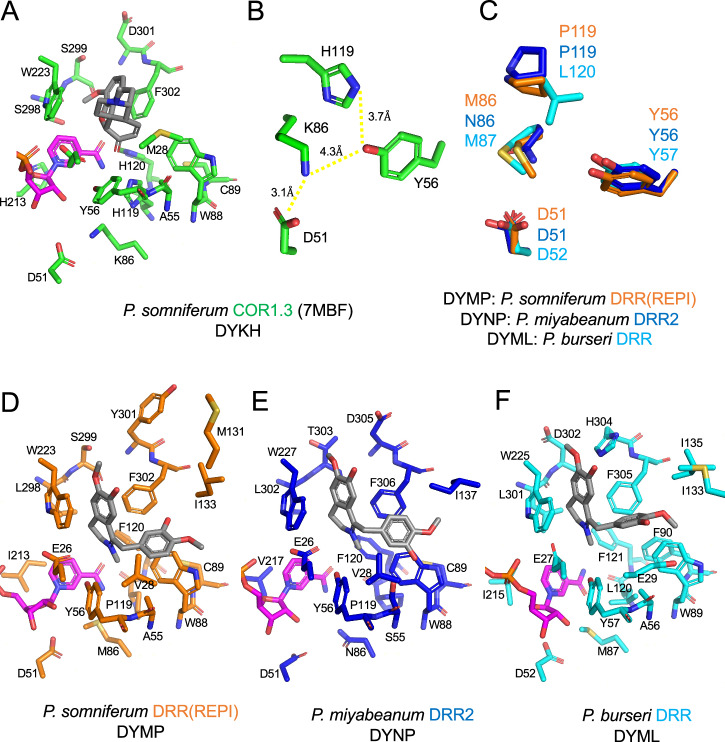
The atypical DYMP motif in P. somniferum DRR(REPI) was further investigated by preparing a homology model of the holoenzyme bound to substrate and cofactor. Additionally, models of representative homologs possessing either the canonical AKR catalytic tetrad (DYKH) or one of the other observed substituted motifs (DYML or DYNP) were prepared. The DYKH catalytic tetrad was represented by the P. somniferum COR crystal structure (PDB: 7MBF), whereas homologs bearing the DYML and DYNP motifs were represented by homology models of P. burseri DRR and P. miyabeanum DRR2, respectively (Fig. 7, S8,S9). Homology models were prepared using P. somniferum COR crystal structure (PDB: 7MBF) as a template, and NADPH and alkaloid substrates were modeled by molecular docking.
Fig. 7. Characterization of the binding pocket in AKRs from various Papaver species.
A Crystal structure of COR (7MBF) from opium poppy (P. somniferum)29 shown in green. B Arrangement of the catalytic tetrad in COR from P. somniferum. C Superposition of the respective motifs of substitutions to the canonical AKR catalytic tetrad in P. somniferum DRR(REPI) (orange), P. miyabeanum DRR2 (blue), and P. burseri DRR (cyan). D the DRR component of REPI from P. somniferum shown in orange, (E) DRR2 from P. miyabeanum shown in blue, and F DRR from P. burseri shown in cyan. In all cases, a docked molecule of NADPH is shown in magenta and docked ligand molecules (i.e., codeinone for COR and 1,2-dehydroreticuline for DRR) are shown in gray. Carbon atoms are represented by the base color of each enzyme, with red corresponding to oxygen, blue to nitrogen, and yellow to sulfur. Hydrogen bonding and electrostatic interactions are shown in yellow (predicted) and red (disrupted) dashed lines. Red arrows depict predicted movements of sidechains and substrates based on binding pocket residue substitutions. PyMOL was used to generate images of the models82.
The following structural analysis employs P. somniferum DRR(REPI) numbering unless stated otherwise. The modeled DRRs adopt the canonical AKR TIM-barrel fold from which the binding pocket is formed by five large loops (Fig. S8A), previously described29. The NADP(H) binding pocket is formed by loops B, C, β1α1, and β2α2 (Fig S8A). Inspection of the NADP(H) binding pocket reveal residues that are both positionally and sequentially conserved (G23, T24, E26, D51, Y56, N166, Q187, S214, L216, W223, A246, V261, V262, K263, S264, F265, E272, N273) while others are positionally conserved with substitutions (F/A/F/F25, A/A/A/G53, M/K/M/N86, P/H/L/P119, F/H/F/F120, S/C/S/S165, I/H/I/V213, S/A/S/S218, N/I/N/N219, G/C/G/G220, T/A/T/T221, L/M/M/M229, R/R/N/R269, L/S/L/L298) (Fig. S8B), P. somniferum DRR(REPI)/P. somniferum COR/P. burseri DRR/P. miyabeanum DRR2. Papaver AKR multiple sequence alignment shows further conservative substitutions to Q187, V261, and F265 (Fig. S10). Notably, the ring stacking interaction between residue 213 and the nicotinamide ring of NADP(H) is not conserved in REPI/DRRs, where the typically aromatic 213 is substituted with a short aliphatic residue (Fig. 7, S8B)), even though this distinctive structural feature is highly conserved with the AKR superfamily40. Other residues and key interactions between protein side chains and NADP(H) are well conserved, as observed across the entire AKR superfamily: carboxamide side chain (S/C165, N166, Q190), nicotinamide ribose (D51, T24), pyrophosphate backbone (L216, S214), and adenosine 2’-monophosphate (S264, F265, R269*, E272, N273)40. R269 is substituted with an asparagine in several DRRs, but the carboxamide side chain likely still mediates hydrogen-bond interactions with the adenosine 2’-monophosphate of NADP(H) that are functionally similar with the guanidino functional group of arginine (Fig. S8B).
The substrate binding pocket is formed by contributions from all five loops (Fig. S8A). Closer inspection reveals the conservation of key residues (E26, Y56, W88, I133, W223, F302) and substitutions to residues (V/M/E/V28, A/A/A/S55, C/C/F/C89, P/H/L/P119, F/H/F/F120, M/N/M/M131, L/S/L/L298, S/S/D/T299, Y/N/H/N301), P. somniferum DRR(REPI)/P. somniferum COR/P. burseri DRR/P. miyabeanum DRR2 (Fig. 7). Papaver AKR multiple sequence alignments show additional substitutions to I133, L198, and minimally F302 (Fig. S10). Exclusive to the Meconella DYML DRRs is the conservation of E28, F89, and H301. F89 is otherwise substituted by cysteine, serine, or glycine in other homologs, whereas E28 is substituted with methionine in P. somniferum COR and its two most similar homologs, and otherwise primarily substituted by a short aliphatic residue. Position 301 is primarily conserved as an aspartic or glutamic acid in CORs and weak REPI/DRRs (P. glaucum DRR being to most active of these (Fig. 4)) and is a tyrosine in REPI homologs (excluding the weak P. lasiothrix REPI). Position 131 is more generally conserved as a methionine in REPI/DRRs and displays more variability in CORs. Coordination between all four catalytic residues is evident in the COR catalytic tetrad (DYKH) (Fig. 7B)29, where H119, K86, and D51 act to alter the apparent pKa of Y5623. Substitutions to the canonical AKR catalytic tetrad observed in DRRs (i.e., DYMP, DYML, and DYNP) preclude the formation of the canonical hydrogen-bonding network with Y56 (Fig. 7C). This, alongside the loss of H213 stacking with the nicotinamide ring of NADP(H) and mutagenesis results, suggests that some homologs may utilize an alternative catalytic mechanism lacking some of the features assisting with the general acid/base functions of Y56 and allowing for slightly altered orientations of NADP(H). Additionally, E2629 and K2741 have been proposed as alternative catalytic residues to act in place of the substituted lysine and histidine in P. somniferum DRR(REPI). Retention of modest to low DRR activity in the site-directed mutants generated for E26 suggests roles for the side chain of this residue in substrate binding or coordination, but the side chain does not appear to play a critical role in catalysis (Fig. 6). K27 was not included in our mutagenesis and is conserved in REPIs and several CORs, but it is commonly substituted with threonine in DRRs (Fig. S10). The lack of K27 conservation between REPI and DRRs raises questions about its proposed roles in catalysis. However, both substitutions possess titratable sidechains and thus cannot be discredited without proper biochemical analysis.
This work is complementary to our previous modeling of P. somniferum DRR(REPI)29 and more recent AlphaFold modeling and molecular dynamics analyzes of P. somniferum REPI41. Investigation of the DRR(REPI) model generated by molecular dynamics (as shown in Fig. 6 from41) alongside a superposition of the AlphaFold REPI model (https://alphafold.ebi.ac.uk) with our DRR(REPI) homology model reveal several similarities and differences. All three models suggest that substitutions to the canonical AKR catalytic tetrad residues (K86 and H119) are unlikely to alter the architecture of the active site, but the loss of functional groups created by these substitutions suggest a more simple catalytic mechanism or that alternate functional groups for catalysis may be supplied by the side chains at nearby positions: E2629 and K2741. The molecular dynamics study41 proposed an alternate mode of 1,2-dehydroreticline binding characterized by a more closed conformation of loop A, pushing 1,2-dehydroreticuline deeper into the TIM-barrel (Fig. S9). In the absence of experimentally determined REPI or DRR structures, the validity of these models should be cautiously assessed.
Functional characterization of Papaver DRS, DRR, REPI and SalSyn in yeast
To further investigate the gateway into morphinan alkaloid biosynthesis across the Papaver genus, available DRS, DRR, REPI, and SalSyn orthologs were characterized in engineered yeast. The anchor domains of Papaver CYPs were modified to promote membrane insertion empirically more conducive to catalytic activity42,43 (Table S7). Engineered DRS orthologs from P. rhoeas, P. triniifolium, P. nudicaule, and P. burseri transiently expressed in yeast with genome-integrated benzylisoquinoline uptake permease 1 (BUP1) and cytochrome P450 reductase 2 (CPR2) were able to convert (S)-reticuline to 1,2-dehydroreticuline (Figs. 8A and S11). Congruent to in vitro characterization, DRR orthologs from P. nudicaule, P. burseri, and P. spicatum converted 1,2-dehydroreticuline to (R)-reticuline when transiently expressed in yeast with genome-integrated BUP1 and CRP2 (Figs. 8D and S12). Papaver REPI orthologs from P. somniferum, P. lasiothrix, and the putative DRS-COR fusion from P. spicatum were transiently expressed in yeast with genome-integrated P. somniferum SalSyn, BUP1, and CPR2, in which the (R)-reticuline produced from (S)-reticuline by REPI was further converted to salutaridine by SalSyn, allowing detection of REPI activity without chiral separation of (S/R)-reticuline. P. somniferum and P. lasiothrix REPI orthologs converted (S)-reticuline as shown by the detection of salutaridine (Figs. 8E and S13A). Unlike yeast expressing P. somniferum REPI, in which only salutaridine was detected, yeast expressing P. lasiothrix REPI also accumulated 1,2-dehydroreticuline suggesting that the DRR domain is less efficient than the DRS component of the fusion protein (Figs. 8E and S13A). This result is consistent with the kinetic characterization of P. lasiothrix DRR(REPI), which showed a 39-fold decrease in catalytic efficiency compared with P. somniferum DRR(REPI) (Table 1). The putative P. spicatum DRS-COR fusion showed DRS activity by converting (S)-reticuline to 1,2-dehydroreticuline, although (R)-reticuline was not detected. This result is also consistent with the in vitro characterization of P. spicatum COR(DRS-COR), which showed COR, but not DRR, activity (Fig. S6). The COR activity associated with the P. spicatum DRS-COR fusion was also confirmed in transformed yeast (Fig. S13B). However, transient expression of the P. spicatum DRS-COR fusion, together with unfused P. spicatum DRR in yeast containing integrated SalSyn, BUP1, and CPR2 resulted in the conversion of (S)-reticuline to (R)-reticuline, evident by the accumulation of salutaridine (Figs. 8E and S14A). In this case, (S)-reticuline was apparently converted to 1,2-dehydroreticuline by P. spicatum DRS(DRS-COR) and subsequently converted to (R)-reticuline by the unfused P. spicatum DRR. Similarly, unfused DRS and DRR orthologous pairs from P. burseri and P. nudicaule also converted (S)-reticuline to (R)-reticuline, which was shown by the detection of salutaridine (Figs. 8E and S14A), suggesting that unfused DRS and DRR enzymes can catalyze the conversion of (S)-reticuline to (R)-reticuline in these species. The DRS-COR fusion converted (S)-reticuline to (R)-reticuline when expressed in yeast (Figs. 8E and S13A).
Fig. 8. Functional characterization of DRR, DRS, REPI, and SalSyn homologs from various Papaver species in engineered yeast strains.
A DRS, (B,C) SalSyn, (D) DRR, (E) REPI and unfused DRS-DRR pairs were transiently expressed in one of two yeast strains: (i) a BUP1- and CPR2-chromosomally integrated strain (used for DRS, SalSyn and DRR) and (ii) a BUP1-, CPR2- and SalSyn-chromosomally integrated strain (used for REPI, and unfused DRS and DRR pairs). Yeast strains were feed 50 μM of the indicated substrate (i.e., (S)-reticuline for DRS, (R)-reticuline for SalSyn, 1,2-dehydroreticuline for DRR, and (S)-reticuline for REPI, and unfused DRS and DRR pairs. Yeast cultures were incubated for the indicated time (i.e., 12, 24 or 48 h) and conversion products (i.e., 1,2-dehydroreticuline for DRS, salutaridine for SalSyn, (R)-reticuline for DRR, and salutaridine, (S)-reticuline and 1,2-dehydroreticuline for REPI, and unfused DRS and DRR pairs) were detected and quantified using a five-point standard curve. Error bars correspond to the mean ± standard deviation of three biological replicates (i.e., three independent colonies). The catalyzed reaction and sequences are shown above each plot.
Finally, P. orientale, P. somniferum, P. triniifolium, P. lasiothrix, and P. burseri SalSyn orthologs were transiently expressed in yeast with genome integrated BUP1 and CPR2, and all showed SalSyn activity, evident by the conversion of (R)-reticuline to salutaridine (Figs. 8B,C and S15). None of the enzymes encoded by these SalSyn orthologs accepted (S)-reticuline. Importantly, identity of the SalSyn ortholog from P. burseri was confirmed.
Discussion
Herein, we demonstrate the widespread occurrence across the genus Papaver of orthologous genes putatively encoding morphine biosynthetic enzymes, notably in several species that do not accumulate pro-morphinan or morphinan alkaloids. In vitro characterization of enzymes encoded by DRR and COR orthologs revealed the latency and ubiquity of DRR and COR activities in the genus Papaver and, more unexpectedly, the occurrence of enzymes with specific activities higher than those found in P. somniferum. Whereas highly active DRRs could potentially participate in the stereochemical inversion of (S)-reticuline in salutaridine biosynthesis, highly active CORs in species that do not accumulate morphinan alkaloids (i.e., P. triniifolium P. glaucum, and P. apokrinomenon) cannot be explained according to the canonical function of COR. Although not functionally characterized in this study, similar trends are apparent for the ubiquitous CODM and NISO orthologs, and the more taxonomically restricted SalR, SalAT, and THS orthologs. We present two hypotheses to explain these phenomena, which could at least partially explain the presence of morphinan alkaloid biosynthetic enzymes in Papaver species that do not accumulate pro-morphinan or morphinan alkaloids.
In one hypothesis, DRR and COR genes in the genus Papaver could be remnants of ancestral pro-morphinan or morphinan biosynthetic pathways that have been partially lost, leaving behind a latent biosynthetic fingerprint. This has been proposed in the broader context of BIA biosynthesis in angiosperms not known to accumulate BIAs based on phylogenetic analysis of norcoclaurine synthase (NCS), berberine bridge enzyme, and selected O-methyltransferases44. Recently, characterization of REPI from the non-morphinan producing P. californicum was also suggested to be a remnant of ancestral pro-morphinan alkaloid biosynthesis15. Assuming monophyletic origin, the lack of pro-morphinan biosynthetic enzymes in species that diverged after the earliest known occurrence of pro-morphinan biosynthesis can be attributed to gene deletion15.
In a second hypothesis, several morphinan alkaloid biosynthetic enzymes could be multifunctional with roles in other metabolic pathways. This is especially attractive for the DIOX enzymes, which display broad BIA substrate ranges36. CODM was shown to be potentially involved in the biosynthesis of protopines and protoberberines, in addition to its role in morphinan alkaloid biosynthesis. Based on the purported emergence of protopine and protoberberine alkaloid biosynthesis before the divergence of the Papaver genus44, the ubiquitous distribution of CODM-like transcript in Papaver species could be the result of orthologous roles in other BIA biosynthetic pathways. CORs in the Papaver genus might also be multifunctional, although substrates other than morphinan alkaloids have not been tested. Latent CORs may represent ancestral AKRs, with separate physiological roles, later recruited into morphinan alkaloid biosynthesis based on their non-canonical ability to convert codeinone to codeine. Similar reasoning can be applied to other morphinan biosynthetic enzymes including SalR, which is involved in the conversion of salutaridine to thebaine, having also been shown to occur widely in the genus Papaver beyond a correlation with thebaine distribution45. This hypothesis, however, does not explain the occurrence, in non-morphinan producing Papaver species, of high amino acid sequence identity and highly active COR enzymes. These features are most likely remnants of an ancient morphinan alkaloid biosynthetic pathway.
These hypotheses align with the patchwork model of pathway evolution, which has been recently suggested for morphinan alkaloid biosynthesis14, whereby a limited number of key evolutionary events bridge gaps between clusters of pre-existing latent biosynthetic enzymes. The background of latent enzymes, dubbed ‘underground metabolism’, provides a foundation for the emergence of novel biosynthetic pathways22. Gene duplication, either through whole genome or local replication processes, provides new genetic material for the recruitment of novel enzymes without disrupting existing biological functions. The focus of this work was the emergence of the AKRs DRR and COR, however intriguing patterns in transcript distribution particularly in the DIOX family merit discussion. Additionally, the taxonomical restriction of SalSyn- and T6ODM-like transcripts to salutaridine and thebaine-producing Papaver species suggests their emergence be important evolutionary events. SalSyn catalyzes the formation of the pro-morphinan scaffold, which represents a potentially selective trait. Recruitment of T6ODM, possibly from its closest and potentially more widespread paralog PODA, which exhibits latent T6ODM activity36, could facilitate the formation of codeine and morphine. In pyrrilizidine alkaloid biosynthesis, homospermidine synthase was recruited from the primary metabolic enzyme deoxyhypusine synthase, which demonstrates latent homospermidine synthase activity46. Additionally, caffeine biosynthesis arose several times independently from benzoate and salicylate O-methyltransferases harboring latent xanthine N-methyltransferase activity, which has been maintained for over 100 million years19. Moreover, recent work on the characterization of PR10 proteins from P. somniferum suggests that the enzymes NCS, THS and NISO evolved from BIA binding proteins47. Binding affinity may be considered a form of pre-enzymatic latency, from which neofunctionalization can yield novel enzyme function48,49.
A growing body of research has highlighted the importance of transcriptional regulation in the evolution of specialized metabolism50, including BIA biosynthesis14,51,52. The evolution of regulatory elements, such as transcription factors, has been proposed in plant specialized metabolism, specifically nicotine biosynthesis in the genus Nicotiana53,54. Whole genome triplication in the Solanaceae family is thought to have led to an enrichment of transposable elements responsible for the derivation of new transcription factor binding sites and subsequent root-specific expression of nicotine biosynthetic genes54. In this case, whole genome triplication not only contributes to the neofunctionalization of duplicated primary metabolic genes, but also provides a mechanism for the evolution of gene regulation. Based on the localized expression of pro-morphinan and morphinan alkaloid biosynthetic enzymes in either sieve elements and/or laticifers of P. somniferum, similar processes could be relevant in BIA metabolism.
Given that DRR and COR are the only AKRs known to be involved in BIA biosynthesis, the inferred evolutionary history of these two enzymes help to shed light on how genes and enzyme activities can be recruited and modified to catalyze distinct steps during the evolution of complex metabolic pathways. The AKR superfamily is a diverse family of enzymes found in all phyla, possessing a well conserved canonical catalytic tetrad DYKH and most commonly catalyzing the relatively simple reduction of carbonyl oxygens23. However, variations in catalytic tetrads and chemical reactivity have also been observed23,27,28. We show that CORs possess the canonical AKR catalytic tetrad of DYKH, whereas DRRs possess atypical DYMP (as previously documented in P. somniferum DRR(REPI)29) and the newly documented DYML or DYNP tetrads. The K86M/N and H119P/L substitutions in DRR motifs clearly disrupt their canonical role in altering the apparent pKa of Y56, as demonstrated in structural analysis and supported by activity assays of mutants at positions 86 and 119 in P. somniferum DRR(REPI). Congruently, reconstitution of the D-K-Y proton relay in P. somniferum DRR(REPI) by the gain-of-function M86K mutant allowed for the reduction of codeinone and oxidation of codeine. Parallel to the catalytic tetrad substitutions, DRRs possess the atypical loss of aromaticity at position 213 which canonically interacts with the nicotinamide ring of NADP(H), from which the hydride is donated or accepted. These patterns suggest that the catalysis of reduction of 1,2-dehydroreticuline does not strictly require alterations of pKa in Y56 by K86 and H119, and perhaps is more tolerant of suboptimal orientations of the hydride donor and the substrate. This is supported by the higher reactivity of 1,2-iminium ion substrates55. Similar alterations to canonical catalytic residues have also been observed in alcohol dehydrogenases (ADHs) with biosynthetic roles in monoterpene indole alkaloid (MIA) biosynthesis catalyzing 1,4- and/or 1,2-iminium ion reductions56–59. Substitutions to typically conserved residues involved in both proton relay and zinc ion coordination have led to the suggestion that the proper positioning of the reactive iminium ion substrate and NADPH is sufficient for reduction to occur in the absence of general acid/base catalysis56. DRR, and MIA ADHs are structurally different than other imine reductases (IREDs) which typically possess the conserved IRED fold60,61. IRED imine reductions typically proceed through enzyme catalyzed protonation of the imine substrate to form an iminium ion intermediate, which is readily reduced by NADPH60,62–64. 1,2-Dehydroreticuline, like most substrates of the atypical MIA ADHs, is present as an iminium ion; thus, the initial imine protonation observed in IREDs is not required. DRR and the atypical MIA ADHs demonstrate an intriguing case of convergent evolution, in which iminium ion reduction was recruited in independent alkaloid biosynthetic pathways, BIA and MIA, from different classes of reductive enzymes, AKRs and ADHs, respectively. As proposed for atypical MIA ADHs, reduction of the iminium ion form of 1,2-dehyroreticuline by DRR might only require proper coordination of the substrate and cofactor allowing for hydride attack at the C1 carbon to yield (R)-reticuline. Alternative catalytic residues, E2629 and K2741, have been proposed for P. somniferum DRR(REPI). Our mutagenesis studies of P. somniferum DRR(REPI) do not support the catalytic involvement of E26, while lack of conservation within DRRs questions the contribution of K27. Additionally, the apparent reactivity of 1,2-dehydroreticuline requiring only hydride attack without protonation/deprotonation, suggests that the catalytic mechanism of DRR may not require additional titratable residues to assist with the hydride transfer reaction. 1,2-Dehydroreticuline has been shown to exist as an iminium ion and in basic conditions, an enamine65. However, feeding experiments in P. somniferum suggest that iminium ion to enamine conversion does not occur66. As illustrated by molecular dynamics simulations of REPI41, the fusion of DRS and DRR in REPI is expected to promote the channeling of the 1,2-dehydroreticuline produced by DRS to DRR, precluding enamine formation. However, substrate channeling between separate DRS and DRR enzyme molecules has not been demonstrated, and it is more likely that 1,2-dehydroreticuline occurred in its enamine form in earlier Papaver species, prior to the REPI fusion. This provides an ancestral role for Y56 and other accessory catalytic residues, in which general acid/base catalysis could assist with the conversion of the enamine to the 1,2-dehydroretciuline iminium ion to assist with its reduction to (R)-reticuline. This scenario also suggests that metabolic channeling may have provided more selective pressure for the REPI fusion event than catalytic enhancement.
Interestingly, a single M86K substitution in the P. somniferum DYMP catalytic tetrad, which partially reconstituted the canonical DYKH tetrad, resulted in a bifunctional DRR/COR enzyme. Structural analysis by homology modeling and molecular docking suggests that the partial reconstitution of the canonical DYKH tetrad as DYKP allows K86 and D51 to promote Y56 protonation/deprotonation of the substrate. This demonstrates that a single amino acid substitution is sufficient to toggle latent enzymatic activity, yielding a different enzyme with the potential to participate in specialized metabolism. The recruitment of xanthine methyltransferase occurred repeatedly during the convergent evolution of caffeine biosynthesis from latent enzyme activity facilitated by single proline to serine and histidine to asparagine substitutions in an ancestral methyltransferase19. Other examples of perturbations to enzyme specificity are reviewed in ref. 67, including cases in glucosinolate biosynthesis68 and acyl-sugar biosynthesis69. These examples demonstrate the relative ease with which latent activities can be neofunctionalized and, subsequently, undergo selection.
Owing to its broad conservation across plant AKRs70 and placement in the same orthogroup, DRR and COR likely evolved from a common AKR ancestor. Presumably, the DRR/COR AKR ancestor possessed the canonical DYKH catalytic tetrad, and single or double amino acid substitutions forming the DYMP motif led to neofunctionalization and subsequent selection of DRR. The DYMP motif is most widespread, being associated with P. somniferum DRR(REPI) and DRRs in the more distantly related Meconella section and, thus, assumed to be the ancestral DRR catalytic tetrad. Interestingly, the DYML DRR motif, and several other substrate binding pocket residues are only conserved in the Meconella section, in P. radicatum, P. burseri, and P. nudicaule, corresponding to DRRs with higher activity compared with P. somniferum DRR(REPI). P. burseri, and P. nudicaule were also shown to accumulate salutaridine, suggesting that their DRRs are involved in the stereochemical inversion of (S)-reticuline. Neofunctionalization leading to COR is less straightforward in the absence of a clear structural marker such as the DRR motifs. COR catalyzes the canonical reduction of a carbonyl oxygen and might have emerged from the general promiscuity of the AKR family. Our study failed to identify native Papaver AKRs lacking both COR and DRR activity. Ancestral enzyme reconstruction or analysis of AKRs beyond the Papaver genus could better define the structural underpinnings of COR evolution.
The Meconella species, P. burseri and P. nudicaule, both accumulate salutaridine and possess transcripts for unfused DRS and DRR orthologs in place of REPI, consistent with the recently published draft genome of P. nudicaule15. The conversion of (S)-reticuline to (R)-reticuline in these species is apparently catalyzed by unfused DRS and DRR enzymes. Importantly, these findings suggest that the stereochemical inversion of (S)-reticuline emerged prior to the REPI fusion from previously functionalized DRS and DRR orthologs and before the divergence of the Meconella section. Following this hypothesis, the fusion event yielding REPI likely occurred after the Meconella divergence and before the emergence of P. californicum, potentially under the selective pressure of enhancing pro-morphinan alkaloid accumulation, rather than being the pivotal step in its evolution as previously suggested14–16. In addition to precluding enamine formation and taking into consideration the prevalence of gene clusters in BIA metabolism71, the REPI fusion can be considered the ultimate in ‘gene clustering’, ensuring spatial/temporal co-expression of DRS and DRR as a means of optimizing the formation of (R)-reticuline and downstream pathway intermediates. A summary of the proposed, revised evolutionary history of pro-morphinan and morphinan biosynthesis in the genus Papaver is illustrated in Fig. S19.
Mapping the pro-morphinan and morphinan alkaloid genetic landscape in the genus Papaver is revealing a dynamic evolutionary history involving the existence of promiscuous catalysts with latent functions, multifunctional enzymes, remnants of ancestral biosynthetic pathways, and only marginally understood transcriptional regulation. Our work contributes to the narrative that evolution of the intriguing REPI fusion protein represents a significant step in the process15,16. Our systematic and mechanistic analyzes of AKRs from the Papaver genus highlight the timing and structural features relevant to the evolution of pro-morphinan and morphinan alkaloids, serve as a model to understand the evolution of plant specialized metabolism in general.
Methods
Chemicals
Codeinone, 1,2-dehydroreticuline, and salutaridine were purchased from Toronto Research Chemicals (https://trc-canada.com/). (S)-Reticuline and (R)-reticuline were purchased from Cayman Chemicals (https://caymanchem.com/). Codeine was a gift from Sanofi-Aventis (https://www.sanofi.com/).
Plant material
Papaver seeds were obtained from Plant World Seeds (https://plant-world-seeds.com) and were cultivated in growth chambers at 20/18 °C (light/dark) with a 16-h photoperiod with cool white fluorescent and incandescent lights. Rosette stems with leaves removed at their base were harvested from three independent plants grown to anthesis. Rosette stems were flash frozen using liquid nitrogen and stored at −80 °C until used for RNA and or alkaloid extraction. Tissues were extracted with 1.2 mL acetonitrile and centrifuged (10,000 g) to remove insoluble debris, which was extracted twice with 500 μL acetonitrile, centrifuged after each extraction, and all supernatants were combined in a fresh tube. The remaining insoluble debris was used to determine the dry weight of each sample for normalization purposes. The insoluble debris was heated to 60 °C under vacuum for 3 days for complete desiccation prior to weighing. To ensure a narrow dynamic range of metabolites in the high-resolution liquid chromatography-mass spectrometry (LC-MS) analysis, dry weights were also used to adjust supernatant concentrations between replicates so that all extracts corresponded to an equivalent amount of dry matter. Following this adjustment, 500 μL from each acetonitrile extract was diluted 1:100 or 1:1000 with acetonitrile. P. somniferum was grown with appropriate license and government approval.
Transcriptomes
RNA from Papaver species was extracted using the trimethyl ammonium bromide (CTAB) method72 from frozen rosette stems, which were ground to a fine powder using a TissueLyser (Qiagen; Hilden, Germany). Libraries were prepared using the NEBNext Ultra II Directional RNA Library Prep Kit (New England Biolabs). Illumina sequencing and transcriptome assembly were performed at the Genome Quebec Innovation Center (http://gqinnovationcenter.com/index.aspx/) based on HiSeq PE100 (Illumina; San Diego, CA) (Table S8). Previously published transcriptomes were included in our analyzes. Short read assemblies (SRAs) were assembled using trinity for Roemeria refracta (SRR19536717)34. NCBI transcriptome shotgun assemblies were used for P. dubium (GJOS00000000), P. bracteatum (GJOQ00000000), P. orientale (GJOT00000000), P. armeniacum (GJOO00000000), P. californicum (GJOY00000000), and P. pavonium (GJOU00000000)15; and Argemone mexicana (GJVJ00000000)33. The One Thousand Plant Transcriptomes database was used for P. rhoeas31,32. Transcripts were annotated based on sequence identity to P. somniferum morphinan biosynthetic enzymes, with a 60% sequence identity cut-off (Table S2). Transcripts were omitted if higher sequence identity to a different biosynthetic enzyme was observed. For example, many SalSyn-like transcripts above 70% identity to somniferum SalSyn showed over 90% identity to cheilanthifoline or stylopine synthase. In these obvious cases, transcripts were not assigned ‘SalSyn-like’ annotation. Partial and truncated transcripts were included, and their percent coverage reported (Fig. S2). Annotated transcripts with reported percent identities and coverage, when applicable, are shown in Fig. S2. A simplified version of the results is shown in Fig. 2. Annotated transcripts were cross-reference to available genomic data for further validation for P. bracteatum (PUWZ01), P. armeniacum (JAJJWX01), P. atlanticum (JAJJMB01), P. californicum (JAJJMC01), P. nudicaule (JAJJMA01) (Fig. S4 and Table S3). Orthogroups were identified using OrthoFinder V2.5.535 (Table S4).
Morphinan alkaloid profiling
High-resolution ESI-LTQ-Orbitrap-XL MS was performed as described previously73 either (i) by direct infusion via a syringe pump or (ii) by LC performed using an Accela HPLC (Thermo Fisher; Waltham, MA) equipped with a Poroshell 120 SB-C18 column, 2.1×50 mm, 2.7 μm (Agilent Technologies; Santa Clara, CA). Direct infusion was performed in conjunction with isocratic solvent flow (50:50 Solvent A:Solvent B, 0.5 mL/min) using a T-bar union. Briefly, for each standard, a steady flow (5 μL/min) of a methanolic (100%) or partially aqueous (50:50 water:methanol) solution (1 μg/mL) was achieved using a 500-μL Hamilton syringe pump. This flow was streamed into HPLC solvent flow, followed by ESI under the following conditions: source voltage 3.00 kV, current ~100 μA, capillary temperature 380 °C, voltage 6.00 V, vaporizer temperature 400 °C, tube lens 45 V. Owing to the thermal stability of alkaloids, and their ease of ionization, the same source conditions were used in all cases. Continuous injection of alkaloid permitted opportunity to optimize MS conditions for each standard; for example, different collision energies were tried for every standard to determine best settings. To observe the retention time for each standard with our chosen HPLC-based analysis method, fractionation was performed on five microliters of alkaloid at 5 μM, injected into solvent flow (0.5 mL/min) with a gradient of Solvent A [10 mM ammonium acetate, pH 5.5, 5% (v/v) acetonitrile] and Solvent B (100% acetonitrile). Specifically, the gradient was as follows: 100–80% Solvent A over 5 min, 80–50% over 3 min, 50–0% over 3 min, isocratic at 0% for 2 min, 0–100% over 0.1 min, and isocratic at 100% for 1.9 min. The total run time was 15 min with data collected for 10 min. The ESI conditions were identical to those used for direct infusion analyzes of standards (via T-bar union and coupled HPLC flow, described above). Identical source conditions were used for the analysis of poppy extract. Following ionization, LTQ-Orbitrap-XL analysis was conducted using a parent-ion list ( > 100 m/z) of known alkaloid masses targeted for collision-induced dissociation (CID) spectra acquisition.
AKR expression and purification
Synthetic constructs of AKRs were ordered from TWIST biosciences. Sequences from P. apokrinomenon, P. burseri, P. glaucum, P. lasiothrix, P. miyabeanum, P. nudicaule, P. radicatum, P. rupifragium, P. spicatum, and P. triniifolium were obtained from transcriptome analysis. AKR sequences from REPI were ordered without the fused DRS from P. somniferum and P. bracteatum from NCBI accession numbers KP985721 and KP985719 respectively and P. lasiothrix, and P. spicatum from transcriptomes analysis. P. somniferum COR (isoform 1.3) sequence was obtained from NCBI accession number AAF13738. Synthetic constructs were cloned into pET47b expression vector, recombinantly expressed using Arctic Express E. coli cells, and purified as previously described29. Starter cultures were grown overnight in 50 mL Luria–Bertani (Miller) broth supplemented with 30 mg/L kanamycin and 35 mg/L gentamycin (LBKG) at 25 °C with shaking at 170 rpm to an OD595 value of ~0.4, and subsequently used to inoculate 1-liter cultures using LBKG broth. Cultures were grown at 30 °C to an OD595 value of 0.5–0.6 and cooled to 16 °C for 30 min. Isopropyl β-D-1-thiogalactopyranoside was added to a final concentration of 1 mM to induce recombinant protein expression and cultures were incubated at 16 °C for 24 h. Cells were harvested by centrifugation and cell pellets were resuspended in lysis buffer [50 mM sodium phosphate, pH 8.0, 300 mM NaCl, 15% (v/v) glycerol]. Resuspended pellets stored at −80 °C were thawed and lysed by sonication in the presence of lysozyme and DNase, cell debris was subsequently removed by centrifugation at 4 °C. Supernatant was incubated with 1 mL of lysis buffer equilibrated TALON resin (Clontech; Kusatsu, Japan) with shaking at 65 rpm for 45 min on ice. The resin was washed with 10 mL of lysis buffer followed by a 40 mL lysis buffer wash incubated for 45 min with shaking at 65 rpm on ice. The resin was then washed with 20 mL lysis buffer supplemented with 5 mM imidazole, followed by 2 mL lysis buffer supplemented with 10 mM imidazole, and finally recombinant protein was eluted with 4 mL of lysis buffer supplemented with 200 mM imidazole. Imidazole concentration was reduced to less than 1 mM by ultrafiltration (30k). Protein purity was determined by SDS-PAGE (Figs. S16 and S20). Protein concentration was measured by absorbance at 280 nm based on a theoretical extinction coefficient74.
Mutagenesis
Site-directed mutagenesis was performed in the pET47b plasmid using a previously described method75 used in our previous work29. Briefly, nucleotide substitutions were introduced into target codons by PCR site directed mutagenesis using Q5 High-Fidelity DNA polymerase (New England Biolabs; Ipswich, MA) and oligonucleotide primers (Integrated DNA Technologies; Coralville, IA) containing targeted substitutions76 (Table S9). Constructs were verified by dideoxynucleotide chain-termination sequencing.
DRR enzyme assays
Reductive and oxidation enzyme assays were performed as previously described4 with the following modifications. Reductive assays were run at 37 °C with 20 μM 1,2-dihydroreticuline, 500 μM NADPH, 100 mM Bis-tris propane, pH 7.0 in 50 μL reactions. Oxidative assays were run at 37 °C with 20 μM (R)-reticuline, 500 μM NADP+, 100 mM Bis-tris propane, pH 8.8, in 50 μL reactions. Enzyme concentration and assay time were run within the linear range, less than 10% substrate conversion, for Papaver DRRs (Table S10) and P. somniferum DRR(REPI) mutants (Table S11). Enzymes with relatively weak DRR activity precluded proper determination of specific activity and were assayed overnight using 2 μg of recombinant protein under otherwise standard reduction or oxidation conditions. Reductive kinetic assays were performed using 500 μM NADPH and 1 μM to 80 μM 1,2-dehydroreticuline with optimized pH (100 mM Bis-tris), temperature, and assay times (Table S12). Oxidative kinetic assays were run using 500 μM NADP+ and 2.5 μM to 500 μM (R)-reticuline with optimized pH (100 mM Bis-tris), temperature, and assay times (Table S12). Reactions were quenched with 300 μL of methanol and diluted to 1 μM total alkaloid with 650 μL of LC-MS solvent A [0.08% (v/v) acetic acid: acetonitrile (95:5)] before analysis by LC-MS as previously described4. Enzyme assays were performed in triplicate and analytes quantified using a five-point standard curve. For enzyme kinetics, quenching methanol was evaporated and resuspended in LC-MS solvent A. Michaelis–Menton and substrate inhibition kinetics were determined using Graphpad Prism 5 (Insight Partners, New York, NY) software.
COR enzyme assays
Reductive and oxidation in vitro enzyme assays were conducted as previously described74, employed in our previous work29, with the following modifications. Reductive assays were run at 25 °C with 50 μM codeinone, 1 mM NADPH, 100 mM Bis-tris propane pH 8.0 in 50 μL reactions. Oxidative assays were run at 25 °C with 50 μM codeine, 1 μM NADP+, 100 mM Bis-tris propane pH 8.8 in 50 μL reactions. Enzyme concentration and assay time were run within the linear range, less than 10% substrate conversion (Table S10). Steady state enzyme kinetics were carried out for representative CORs ranging from higher to lower specific activities. Reductive kinetic assays were run with 1 mM NADPH and 1 μM to 400 μM codeinone with optimized pH (100 mM Bis-tris), temperature, and assay times (Table S12). Oxidative kinetic assays were run with 1 mM NADP+ and 1 μM to 400 μM codeine with optimized pH (100 mM Bis-tris), temperature, and assay times (Table S12). Reactions were quenched with 300 μL of methanol before analysis by LC-MS as previously described75. Enzyme assays were performed in triplicates and analytes quantified using a five-point standard curve. For enzyme kinetics, quenching methanol was evaporated and resuspended in LC-MS solvent A (10 mM ammonium acetate, pH 5.5). Michaelis–Menton kinetic analysis were preformed using Graphpad Prism 5 software.
Transient expression of DRS, DRR, REPI, and SalSyn in yeast
Synthetic constructs of DRS, DRR, REPI, and SalSyn were ordered from Twist Biosciences (San Francisco, CA). Sequences from P. burseri, P. lasiothrix, P. nudicaule, P. orientale, P. triniifolium, and P. spicatum were obtained from transcriptome analysis. P. rhoeas DRS and P. somniferum SalSyn and REPI were obtained from NCBI accession numbers KP985722, EF451150 and KP985721 respectively. To avoid incorrect membrane insertion of cytochrome P450 domains the N-terminal of DRS and REPI were substituted with that of germacrene A oxidase (GAO) as previously reported43 and SalSyn N-terminal substituted with that of cheilanthifoline synthase (CFS) as previously reported42. All genes were codon optimized (GenScript; Piscataway, NJ) and are shown in Table S7. For transient expression, Papaver DRS, DRR, REPI, and SalSyn synthetic genes were cloned into pESC-His vector under PGK1 promoter (DRS, REPI, SalSyn) and TDH3 (DRR). DRS, DRR, and SalSyn genes were transformed into the engineered yeast strain BC with chromosome integrated P. somniferum BIA uptake permease 1 (BUP1) and CYP reductase 2 (CPR2). REPI and unfused DRS-DRR genes were transformed into the strain BC with an integrated P. somniferum SalSyn, strain BCSS, used to distinguish between (S)- and (R)-reticuline by conversion of (R)-reticuline to salutaridine. All transformations were preformed using the LiAc/PEG/single-stranded carrier DNA (ssDNA) method77. Chromosomal integration of BUP1, CRP2, and SalSyn were engineered from the CEN.PK parent strain using CRISPR-Cas9 technology78.
Yeast substrate feeding
Three colonies from transformed strain BC or BCSS were used to inoculate 500 μL SD-his medium starter cultures grown overnight at 30 °C and 900 rpm using a gyratory microplate shaker. Feeding experiments were carried out by inoculating 200 μL of fresh SD-His medium containing 50 μM of substrate with 10 μL of starter culture and subsequently grown for 12, 24, or 48 h. Reactions were quenched by adding two volumes of acetonitrile and centrifuged for 30 min at 2000g. Supernatants were then analyzed by LCMS using the same protocol as for COR assays and analytes were quantified using five-point standard curves.
Modeling of DRR and COR structures
Homology models of P. somniferum DRR(REPI), P. miyabeanum DRR2, and P. burseri DRR were prepared with Modeller79 using P. somniferum COR as a template (PDB: 7MBF). Induced-fit substrate and cofactor docking were preformed using Molecular Operating Environment (MOE) (https://www.chemcomp.com/products.htm). Standard default docking settings were used. P. somniferum COR coordinates (PDB: 7MBF) were used in docking studies. PyMOL was used to visualize AKR models (https://pymol.org/2/).
Statistics and Reproducibility
Error bars in all figures correspond to the mean ± standard deviation of three independent biological replicates.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
This work was funded by an Alberta Innovates Strategic Research Project grant, and by Natural Sciences and Engineering Research Council of Canada Discovery grants to P.J.F. and K.K.S.N.
Author contributions
S.C.C. designed the experiments, acquired and analyzed data, and wrote the manuscript. F.R. acquired and analyzed data. X.C. and J.M.H. performed the transcriptomics and metabolomics, respectively. K.K.S.N. reviewed the manuscript. P.J.F. conceived the study, supervised the research, revised the manuscript, and approved it for submission.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Huijuan Guo and David Favero. A peer review file is available.
Data availability
Data used to plot all graphs is available as Supplementary Data 1. All other data are available from the corresponding author on reasonable request. Other supplementary data is available in the Supplementary Information file. Raw nucleotide sequence data have been deposited in the National Center for Biotechnology Information (NCBI) Short Read Archive (SRA) database under the following accession codes: SRR27145420, SRR27235872, SRR27235873, SRR27235874, SRR27235875, SRR27212614, SRR27212567, SRR27212568, SRR27212569, SRR27283754, SRR27283755, and SRR27283756 under the bioproject PRJNA1050196. Assembled transcriptomes have been deposited in the NCBI Transcriptome Shotgun Assembly (TSA) database under the following accession codes: GKSI00000000, GKSF00000000, GKSE00000000, GKSG00000000, GKSD00000000, GKSB00000000, GKSA00000000, GKRY00000000, GKRX00000000, GKRW00000000, GKRV00000000, GKRS00000000 under the bioproject PRJNA1050196. Nucleotide sequence data have been deposited in the NCBI Genbank database under the following accession codes: AKRs (OR829681, OR829682, OR829683, OR829684, OR829685, OR829686, OR829687, OR829688, OR829689, OR829690, OR829691, OR829692, OR829693, OR829694, OR829695, OR829696, OR829697, OR829698, OR29699); DRS-AKR fusions (OR829700, OR829701); DRSs (OR838695, OR838696, OR838697); and SalSyns (OR829703, OR829702, OR838698).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-024-07100-w.
References
- 1.Dastmalchi, M., Park, M. R., Morris, J. S. & Facchini, P. Family portraits: the enzymes behind benzylisoquinoline alkaloid diversity. Phytochem. Rev.17, 249–277 (2018). [Google Scholar]
- 2.World Health Organization. World health organization model list of essential medicines. Ment. Holist. Health.: Some Int. Perspect.21, 119–134 (2019). [Google Scholar]
- 3.Board, I. N. C. Narcotic Drugs - Technical Report. (2019).
- 4.Farrow, S. C., Hagel, J. M., Beaudoin, G. A. W., Burns, D. C. & Facchini, P. J. Stereochemical inversion of (S)-reticuline by a cytochrome P450 fusion in opium poppy. Nat. Chem. Biol.11, 728–732 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Winzer, T. et al. Morphinan biosynthesis in opium poppy requires a P450-oxidoreductase fusion protein. Science349, 309–312 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Gesell, A. et al. CYP719B1 is salutaridine synthase, the C-C phenol-coupling enzyme of morphine biosynthesis in opium poppy. J. Biol. Chem.284, 24432–24442 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lenz, R. & Zenk, M. H. Purification and properties of codeinone reductase (NADPH) from Papaver somniferum cell cultures and differentiated plants. Eur. J. Biochem.233, 132–139 (1995). [DOI] [PubMed] [Google Scholar]
- 8.Ziegler, J. et al. Comparative transcript and alkaloid profiling in Papaver species identifies a short chain dehydrogenase/reductase involved in morphine biosynthesis. Plant J.48, 177–192 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Dastmalchi, M. et al. Neopinone isomerase is involved in codeine and morphine biosynthesis in opium poppy. Nat. Chem. Biol.15, 384–390 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Chen, X. et al. A pathogenesis-related 10 protein catalyzes the final step in thebaine biosynthesis. Nat. Chem. Biol.14, 738–743 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Hagel, J. M. & Facchini, P. J. Dioxygenases catalyze the O-demethylation steps of morphine biosynthesis in opium poppy. Nat. Chem. Biol.6, 273–275 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Grothe, T., Lenz, R. & Kutchan, T. M. Molecular characterization of the salutaridinol 7-O-acetyltransferase involved in ormphine biosynthesis in opium poppy Papaver somniferum. J. Biol. Chem.276, 30717–30723 (2001). [DOI] [PubMed] [Google Scholar]
- 13.Li, Y., Winzer, T., He, Z. & Graham, I. A. Over 100 million years of enzyme evolution underpinning the production of morphine in the Papaveraceae family of flowering plants. Plant Commun.1, 100029 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang, X. et al. Three chromosome-scale Papaver genomes reveal punctuated patchwork evolution of the morphinan and noscapine biosynthesis pathway. Nat. Commun.12, 6030 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Catania, T. et al. A functionally conserved STORR gene fusion in Papaver species that diverged 16.8 million years ago. Nat. Commun.13, 3150 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang, R.-G. et al. Subgenome-aware analyses suggest a reticulate allopolyploidization origin in three Papaver genomes. Nat. Commun.14, 2204 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jensen, R. A. Enzyme recruitment in evolution of new function. Annu. Rev. Microbiol.30, 409–425 (1976). [DOI] [PubMed] [Google Scholar]
- 18.Yčas, M. On earlier states of biochemical systems. J. Theor. Biol.44, 145–160 (1974). [DOI] [PubMed] [Google Scholar]
- 19.Huang, R., O’Donnell, A. J., Barboline, J. J. & Barkman, T. J. Convergent evolution of caffeine in plants by co-option of exapted ancestral enzymes. Proc. Natl Acad. Sci. USA113, 10613–10618 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rieseberg, T. P. et al. Crossroads in the evolution of plant specialized metabolism. Sem. Cell Devel. Biol.134, 37–58 (2023). [DOI] [PubMed] [Google Scholar]
- 21.Khersonsky, O., Roodveldt, C. & Tawfik, D. Enzyme promiscuity: evolutionary and mechanistic aspects. Curr. Opin. Chem. Biol.10, 498–508 (2006). [DOI] [PubMed] [Google Scholar]
- 22.D’Ari, R. & Casadesús, J. Underground metabolsim. Bioessays20, 181–186 (1998). [DOI] [PubMed] [Google Scholar]
- 23.Penning, T. M. The aldo-keto reductases (AKRs): Overview. Chem. -Biol. Interact.234, 236–246 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Welle, R. & Grisebach, H. Isolation of a novel NADPH-dependent reductase which coacts with chalcone synthase in the biosynthesis of 6′-deoxychalcone. FEBS Lett.236, 221–225 (1988). [Google Scholar]
- 25.Jirschitzka, J. et al. Plant tropane alkaloid biosynthesis evolved independently in the Solanaceae and Erythroxylaceae. Proc. Natl Acad. Sci. USA109, 10304–10309 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De-Eknamkul, W. & Zenk, M. H. Purification of properties of 1,2-dehydroreticuline reductase from Papaver somniferum seedlings. Phytochemisty31, 813–821 (1992). [Google Scholar]
- 27.Di Costanzo, L., Drury, J. E., Christianson, D. W. & Penning, T. M. Structure and catalytic mechanism of human steroid 5β-reductase (AKR1D1). Mol. Cell. Endocrinol.301, 191–198 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schlegel, B. P., Ratnam, K. & Penning, T. M. Retention of NADPH-linked quinone reductase activity in an aldo-keto reductase following mutation of the catalytic tyrosine. Biochemistry37, 11003–11011 (1998). [DOI] [PubMed] [Google Scholar]
- 29.Carr, S. C., Torres, M. A., Morris, J. S., Facchini, P. J. & Ng, K. K. S. Structural studies of codeinone reductase reveal novel insights into aldo-keto reductase function in benzylisoquinoline alkaloid biosynthesis. J. Biol. Chem.297, 101211 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sariyar, G. Biodiversity in the alkaloids of Turkish Papaver species. Pure Appl. Chem.74, 557–574 (2002). [Google Scholar]
- 31.One Thousand Plant Transcriptomes Initiative. One thousand plant transcriptomes and the phylogenomics of green plants. Nature574, 679–685 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carpenter, E. J. et al. Access to RNA-sequencing data from 1,173 plant species: The 1000 Plant transcriptomes initiative (1KP). GigaScience8, giz126 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loza-Muller, L., Shitan, N., Yamada, Y. & Vázquez-Flota, F. AmABCB1, an alkaloid transporter from seeds of Argemone mexicana L (Papaveraceae). Planta254, 122 (2021). [DOI] [PubMed] [Google Scholar]
- 34.Mazuecos-Aguilera, I. & Suárez-Santiago, V. N. Identification of candidate genes involved in the determinism of pollen grain aperture morphology by comparative transcriptome analysis in Papaveraceae. Plants12, 1570 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emms, D. M. & Kelly, S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol.20, 238 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farrow, S. C. & Facchini, P. J. Dioxygenases catalyze O-demethylation and O,O-demethylenation with widespread roles in benzylisoquinoline alkaloid metabolism in opium poppy. J. Biol. Chem.288, 28997–29012 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pourmazaheri, H. et al. Comparative analysis of the root and leaf transcriptomes in Chelidonium majus L. PLoS ONE14, e0215165 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liao, D. et al. Identification and developmental expression profiling of putative alkaloid biosynthetic genes in Corydalis yanhusuo bulbs. Sci. Rep.6, 19460 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu, D. et al. Integration of full-length transcriptomics and targeted metabolomics to identify benzylisoquinoline alkaloid biosynthetic genes in Corydalis yanhusuo. Hort. Res.8, 16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jez, J. M., Bennett, M. J., Schlegel, B. P., Lewis, M. & Penning, T. M. Comparative anatomy of the aldo–keto reductase superfamily. Biochem. J.326, 625–636 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Diaz-Bárcena, A., Fernandez-Pacios, L. & Giraldo, P. Structural characterization and molecular dynamics study of the REPI fusion protein from Papaver somniferum L. Biomolecules14, 2 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Galanie, S., Thodey, K., Trenchard, I. J., Filsinger Interrante, M. & Smolke, C. D. Complete biosynthesis of opioids in yeast. Science349, 1095–1100 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fossati, E. et al. Reconstitution of a 10-gene pathway for synthesis of the plant alkaloid dihydrosanguinarine in Saccharomyces cerevisiae. Nat. Commun.5, 3283 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Liscombe, D. K., MacLeod, B. P., Loukanina, N., Nandi, O. I. & Facchini, P. J. Evidence for the monophyletic evolution of benzylisoquinoline alkaloid biosynthesis in angiosperms. Phytochemistry66, 1374–1393 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Ziegler, J. et al. Evolution of morphine biosynthesis in opium poppy. Phytochemistry70, 1696–1707 (2009). [DOI] [PubMed] [Google Scholar]
- 46.Ober, D. & Hartmann, T. Homospermidine synthase, the first pathway-specific enzyme of pyrrolizidine alkaloid biosynthesis, evolved from deoxyhypusine synthase. Proc. Natl Acad. Sci. USA96, 14777–14782 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ozber, N. et al. Alkaloid binding to opium poppy major latex proteins triggers structural modification and functional aggregation. Nat. Commun.13, 6768 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clifton, B. E. et al. Evolution of cyclohexadienyl dehydratase from an ancestral solute-binding protein. Nat. Chem. Biol.14, 542–547 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Kaltenbach, M. et al. Evolution of chalcone isomerase from a noncatalytic ancestor. Nat. Chem. Biol.14, 548–555 (2018). [DOI] [PubMed] [Google Scholar]
- 50.Méteignier, L.-V., Nützmann, H.-W., Papon, N., Osbourn, A. & Courdavault, V. Emerging mechanistic insights into the regulation of specialized metabolism in plants. Nat. Plants9, 22–30 (2022). [DOI] [PubMed] [Google Scholar]
- 51.Jia, Y. et al. The tissue-specific chromatin accessibility landscape of Papaver somniferum. Front. Genet.14, 1136736 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu, Y. et al. Evolutionary analysis of conserved non‐coding elements subsequent to whole‐genome duplication in opium poppy. Plant J.116, 1804–1824 (2023). [DOI] [PubMed] [Google Scholar]
- 53.Kajikawa, M. et al. Genomic insights into the evolution of the nicotine biosynthesis pathway in tobacco. Plant Physiol.174, 999–1011 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu, S. et al. Wild tobacco genomes reveal the evolution of nicotine biosynthesis. Proc. Natl Acad. Sci. USA114, 6133–6138 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schrittwieser, J. H., Velikogne, S. & Kroutil, W. Biocatalytic imine reduction and reductive amination of ketones. Adv. Synth. Catal.357, 1655–1685 (2015). [Google Scholar]
- 56.Langley, C. et al. Expansion of the catalytic repertoire of alcohol dehydrogenases in plant metabolism. Angew. Chem.61, e202210934 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hong, B. et al. Biosynthesis of strychnine. Nature607, 617–622 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schotte, C. et al. Directed biosynthesis of mitragynine stereoisomers. J. Am. Chem. Soc.145, 4957–4963 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stavrinides, A. et al. Structural investigation of heteroyohimbine alkaloid synthesis reveals active site elements that control stereoselectivity. Nat. Commun.7, 12116 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mangas-Sanchez, J. et al. Imine reductases (IREDs). Curr. Opin. Chem. Biol.37, 19–25 (2017). [DOI] [PubMed] [Google Scholar]
- 61.Wu, K., Huang, J. & Shao, L. Imine reductases: Multifunctional biocatalysts with varying active sites and catalytic mechanisms. ChemCatChem14, e202200921 (2022).
- 62.Czekster, C. M., Vandemeulebroucke, A. & Blanchard, J. S. Kinetic and chemical mechanism of the dihydrofolate reductase from Mycobacterium tuberculosis. Biochemistry50, 367–375 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Meneely, K. M. & Lamb, A. L. Two structures of a thiazolinyl imine reductase from Yersinia enterocolitica provide insight into catalysis and binding to the nonribosomal peptide synthetase module of HMWP1. Biochemistry51, 9002–9013 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rodríguez-Mata, M. et al. Structure and activity of NADPH-dependent reductase Q1EQE0 from Streptomyces kanamyceticus, which catalyses the (R)-selective reduction of an imine substrate. ChemBioChem14, 1372–1379 (2013). [DOI] [PubMed] [Google Scholar]
- 65.He, X. & Brossi, A. 1,2-dehydroreticuline: Conversion of iminium salts into enamines. J. Nat. Prod.56, 973–975 (1993). [Google Scholar]
- 66.Battersby, A. R., Foulkes, D. M., Hirst, M., Parry, G. V. & Staunton, J. Alkaloid Biosynthesis. Part X1. Studies related to the formation and oxidation of reticuline in morphine biosynthesis. L. Chem Soc. C 210–216 (1968).
- 67.Leong, B. J. & Last, R. L. Promiscuity, impersonation and accommodation: evolution of plant specialized metabolism. Curr. Opin. Struct. Biol.47, 105–112 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.He, Y. et al. Structural and functional evolution of isopropylmalate dehydrogenases in the leucine and glucosinolate pathways of Arabidopsis thaliana. J. Biol. Chem.286, 28794–28801 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fan, P. et al. In vitro reconstruction and analysis of evolutionary variation of the tomato acylsucrose metabolic network. Proc. Natl. Acad. Sci. USA113, E239–48(2016). [DOI] [PMC free article] [PubMed]
- 70.Krishnamurthy, P., Pothiraj, R., Suthanthiram, B., Somasundaram, S. M. & Subbaraya, U. Phylogenomic classification and synteny network analyses deciphered the evolutionary landscape of aldo–keto reductase (AKR) gene superfamily in the plant kingdom. Gene816, 146169 (2022). [DOI] [PubMed] [Google Scholar]
- 71.Guo, L. et al. The opium poppy genome and morphinan production. Science362, 343–347 (2018). [DOI] [PubMed] [Google Scholar]
- 72.Gambino, G., Perrone, I. & Gribaudo, I. A Rapid and affective method for RNA extraction from different tissues of grapevine and other woody plants. Phytochem. Anal.19, 520–525 (2008). [DOI] [PubMed] [Google Scholar]
- 73.Perdomo-Menéndez, I. M., Hagel, J. M. & Facchini, P. J. Benzylisoquinoline alkaloid analysis using high-resolution Orbitrap LC-MSn. J. Mass Spectrom.56, e4683 (2020). [DOI] [PubMed] [Google Scholar]
- 74.Gill, S. C. & von Hippel, P. H. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem.182, 319–326 (1989). [DOI] [PubMed] [Google Scholar]
- 75.Dastmalchi, M., Chang, L., Torres, M. A., Ng, K. K. S. & Facchini, P. J. Codeinone reductase isoforms with differential stability, efficiency and product selectivity in opium poppy. Plant J.95, 631–647 (2018). [DOI] [PubMed] [Google Scholar]
- 76.Zheng, L. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res.32, e115 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gietz, R. D. & Schiestl, R. H. Frozen competent yeast cells that can be transformed with high efficiency using the LiAc/SS carrier DNA/PEG method. Nat. Protoc.2, 1–4 (2007). [DOI] [PubMed] [Google Scholar]
- 78.Jensen, N. B. et al. EasyClone: method for iterative chromosomal integration of multiple genes Saccharomyces cerevisiae. FEMS Yeast Res.14, 238–248 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Webb, B. & Sali, A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinform.54, 5.6.1–5.6.37 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Carolan, J. C., Hook, I. L. I., Chase, M. W., Kadereit, J. W. & Hodkinson, T. R. Phylogenetics of Papaver and related genera based on DNA sequences from ITS nuclear ribosomal DNA and plastid trnL intron and trnL–F intergenic spacers. Ann. Bot.98, 141–155 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Madeira, F. et al. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res.50, W276–W279 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.The PyMOL Molecular Graphics System, Version 2.2.2 Schrödinger, LLC.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
Data used to plot all graphs is available as Supplementary Data 1. All other data are available from the corresponding author on reasonable request. Other supplementary data is available in the Supplementary Information file. Raw nucleotide sequence data have been deposited in the National Center for Biotechnology Information (NCBI) Short Read Archive (SRA) database under the following accession codes: SRR27145420, SRR27235872, SRR27235873, SRR27235874, SRR27235875, SRR27212614, SRR27212567, SRR27212568, SRR27212569, SRR27283754, SRR27283755, and SRR27283756 under the bioproject PRJNA1050196. Assembled transcriptomes have been deposited in the NCBI Transcriptome Shotgun Assembly (TSA) database under the following accession codes: GKSI00000000, GKSF00000000, GKSE00000000, GKSG00000000, GKSD00000000, GKSB00000000, GKSA00000000, GKRY00000000, GKRX00000000, GKRW00000000, GKRV00000000, GKRS00000000 under the bioproject PRJNA1050196. Nucleotide sequence data have been deposited in the NCBI Genbank database under the following accession codes: AKRs (OR829681, OR829682, OR829683, OR829684, OR829685, OR829686, OR829687, OR829688, OR829689, OR829690, OR829691, OR829692, OR829693, OR829694, OR829695, OR829696, OR829697, OR829698, OR29699); DRS-AKR fusions (OR829700, OR829701); DRSs (OR838695, OR838696, OR838697); and SalSyns (OR829703, OR829702, OR838698).