Abstract
The SR proteins constitute a family of nuclear phosphoproteins which are required for constitutive splicing and also influence alternative splicing regulation. They have a modular structure consisting of one or two RNA recognition motifs (RRMs) and a C-terminal domain, rich in arginine and serine residues. The functional role of the different domains of SR proteins in constitutive splicing activity has been extensively studied in vitro; however, their contribution to alternative splicing specificity in vivo has not been clearly established. We sought to address how the modular domains of SR proteins contribute to alternative splicing specificity. The activity of a series of chimeric proteins consisting of domain swaps between different SR proteins showed that splice site selection is determined by the nature of the RRMs and that RRM2 of SF2/ASF has a dominant role and can confer specificity to a heterologous protein. In contrast, the identity of the RS domain is not important, as the RS domains are functionally interchangeable. The contribution of the RRMs to alternative splicing specificity in vivo suggests that sequence-specific RNA binding by SR proteins is required for this activity.
INTRODUCTION
The SR proteins are a family of structurally and functionally related polypeptides that play an important role in constitutive and regulated pre-mRNA splicing (reviewed in 1–4). They are involved in multiple steps of the constitutive splicing reaction: among other functions they promote the assembly of the earliest pre-spliceosomal complex E (5), bridge the 5′ and 3′ splice sites, recruit the U1snRNP particle to the 5′ splice site and participate in recruitment of the U4/U6·U5 tri-snRNP (for a review see 6). In addition, the discovery that some SR and SR-like proteins remain associated with the mRNA products after the splicing reaction and that a subset of SR proteins shuttle from the nucleus to the cytoplasm suggested that SR proteins may have roles not only in nuclear pre-mRNA splicing, but may also have additional functions such as mRNA transport or be involved in cytoplasmic events (7–9).
The role of SR proteins in alternative splicing regulation is antagonised by members of the hnRNP A/B family of proteins, in such a way that increased levels of SR proteins lead to the selection of proximal 5′ splice sites, whereas an excess of hnRNP A/B proteins promotes the selection of distal 5′ splice sites (10–13). Thus, the relative level and activity of members of the SR and hnRNP A/B families of proteins may represent an important determinant of alternative splicing regulation (9). However, activities that antagonise SR protein function are not restricted to the hnRNP A/B family of proteins, as illustrated by the recent identification in Drosophila of a new antagonist of SR protein function, termed RSF1 (14). In addition, individual SR proteins can sometimes antagonise each other in alternative splice site selection, as in the case of the antagonistic effects of SF2/ASF and SC35 on the regulation of β-tropomyosin (15) and of SF2/ASF and SRp20 on the regulation of SRp20 pre-mRNA alternative splicing (16). In addition to the 10 identified members of the SR family of proteins in mammals, a class of related RS domain-containing proteins, termed SR protein-related polypeptides (SRrp) or SR-like proteins, are also involved in splicing regulation (for review see 1,17)
The SR proteins seem to be functionally redundant in constitutive splicing, as illustrated by the ability of any individual SR protein to complement an otherwise inactive cytosolic HeLa S100 extract (reviewed in 4,18). However, several differences in the ability of these proteins to regulate alternative splicing, as well as the ability of individual SR proteins to commit different pre-mRNAs to the splicing pathway, suggested that individual SR proteins may have unique functions in splicing regulation (19–23). Genetic approaches used to address this question showed that SF2/ASF was essential for cell viability in the DT40 chicken cell line (24). In addition, genetic disruption of SRp55/B52 in Drosophila (25) and of SRp20 in the mouse (26) caused early embryonic lethality, strongly suggesting essential and unique functions for these genes. However, more recent experiments in which the whole complement of Caenorhabditis elegans SR proteins were individually depleted using RNA interference have demonstrated that certain SR proteins are functionally redundant (27).
The SR proteins have a modular structure that consists of one or two RNA recognition motifs (RRMs) in the N-terminus and a C-terminal domain rich in arginine and serine residues (termed the RS domain). An extensive structure–function analysis of SF2/ASF, which is the prototype member of the SR family of splicing regulators (28,29), indicated that both RRMs in SF2/ASF are required for efficient binding to RNA (30,31). The use of conventional and functional SELEX protocols allowed the identification of high affinity binding sites for individual SR proteins (32–37) and led to the conclusion that SR proteins are sequence-specific RNA-binding proteins with distinct RNA-binding specificities (for a review see 18). Individual SR proteins have been found to associate with exonic splicing enhancers in many different genes, leading to activation of otherwise inefficient upstream 3′ splice sites (38–42; for reviews see 4,43). In addition, SR proteins can also negatively regulate splicing of an intron, as shown in the adenovirus late transcript, where binding of SF2/ASF to an intronic purine-rich sequence inhibits splicing of a 3′ splice site (44).
The RS domains have been proposed to mediate protein–protein interactions (45) and to be important determinants of subcellular localisation and nucleo-cytoplasmic shuttling (8,46–48). They are able to activate splicing even when fused to a heterologous RRM, which led to the proposal that they function as activators of splicing (49).
An important issue is how the modular domains of SR proteins contribute to splicing specificity, both in constitutive and regulated splicing, and also how critical is sequence-specific high affinity RNA binding for the function of SR proteins in both splicing processes. It has been shown that both the RRMs and the RS domain are essential for constitutive splicing in vitro (30,31). The role of the modular domains of SR proteins in constitutive splicing has recently been studied using domain swaps between SC35 and SF2/ASF or deletion mutants of SF2/ASF. Using a functional splicing commitment assay and in vitro splicing it was shown that substrate specificity in constitutive splicing is determined by the nature of the RRMs and that the identity of the RS domain did not affect splicing specificity, suggesting that the RS domains may have redundant functions in constitutive splicing (23,50). These experiments clearly demonstrated that individual domains in SR proteins function as modules in constitutive splicing.
In contrast, the role of the modular domains of SR proteins in regulated splicing has been less extensively studied. Analysis of SF2/ASF deletion mutant proteins in a nuclear switch assay in vitro established that both RRMs were necessary and sufficient to influence the selection of proximal 5′ splice sites and, surprisingly, that the RS domain was dispensable for this activity. These experiments suggested that RNA binding is required for alternative splicing (30). The alternative splicing activity of SF2/ASF has also been shown to correlate with its ability to promote multiple occupancy of alternative 5′ splice sites by U1 snRNP (51). It is not clear, however, whether sequence-specific binding to target RNAs play a role in splice site selection.
We sought to investigate the role of the modular domains of different SR proteins in alternative splicing regulation in living cells. To this end, we transiently overexpressed chimeric proteins consisting of individual domains of SF2/ASF and SRp40, SF2/ASF and SC35 and SF2/ASF and SRp20 and assayed changes in the pattern of splicing of two different reporters, the adenovirus E1A pre-mRNA and a fibronectin minigene. We demonstrate that the RRMs determine the alternative splicing specificity in vivo and that the RS domains are functionally interchangeable. These results demonstrate the modularity of SR proteins in alternative splicing regulation.
MATERIALS AND METHODS
Epitope-tagged expression plasmids
The epitope-tagged SR protein expression plasmids were previously described. Briefly, they were constructed by PCR amplification of cDNA clones coding for the respective SR proteins with specific primers and the resulting PCR products were subcloned into the pCGTHCFFLT7 expression vector (52), to generate the pCGT7-SR constructs. Transcription is driven by the CMV enhancer/promoter and the coding sequence begins with an N-terminal epitope tag, MASMTGGQQMG. This epitope tag corresponds to the first 11 residues of the bacteriophage T7 gene 10 capsid protein and is recognised by the T7.tag monoclonal antibody (Novagen). For the SF2/ASF and the SC35 constructs PCR products were amplified with specific primers and subcloned into the pCGT7 expression vector as XbaI–BamHI fragments. In the case of the SRp20 constructs, due to the presence of an internal XbaI site in the SRp20 cDNA, the amplified fragments were designed with SpeI and BamHI sites and were subcloned into the XbaI and BamHI sites of the pCGT7 expression vector. In the case of the SRp40 constructs, due to the presence of an internal BamHI site in the SRp40 cDNA, the amplified fragments were designed with XbaI and BclI sites and were subcloned into the XbaI–BamHI sites of the pCGT7 expression vector. Construction of the SF2/SRp40 and SF2/SC35 chimeras was described previously (8). Briefly, by swapping domains between the corresponding SR proteins we took advantage of natural SacI and ApaI sites in the SF2/ASF cDNA, between RRM1 and RRM2 and between RRM2 and the RS domain, respectively. PCR products comprising the indicted domains were amplified from cDNA clones using specific primer pairs. The resulting PCR products were assembled using the SacI and ApaI sites and subcloned into the pCGT7 expression vector. The resulting chimeric proteins comprise the following residues: F1F240RS (previously referred to as ISF2IISF2RS40; 8) consists of residues 3–197 (RRM1 and RRM2) of SF2/ASF followed by residues 184–271 (RS domain) of SRp40; F1402FRS (previously termed ISF2II40RSSF2) consists of residues 3–107 (RRM1) and 196–248 (RS domain) of SF2/ASF separated by residues 86–181 (RRM2) of SRp40; 401F240RS (previously termed I40IISF2RS40) comprises residues 2–83 (RRM1) and 184–271 (RS domain) of SRp40 separated by residues 107–197 (RRM2) of SF2/ASF; C1FRS (previously termed I35RSSF2) comprises residues 2–114 (RRM) of SC35 followed by residues 197–248 (RS domain) of SF2/ASF.
Two new chimeric proteins, F120RS and F220RS, which comprise the first or second RRM of SF2/ASF followed by the RS domain of SRp20, were first generated and subsequently used as templates to generate the 201F120RS and the 201F220RS chimeric proteins.
In the case of 201F220RS, SF2/ASF and SRp20 were independently amplified with oligo pairs 1/2 and 3/4, respectively. The two PCR products, which have a 20 bp overlap, were used in equimolar amounts for a second PCR amplification with oligos 1 and 4. The resulting PCR product was purified, digested with SpeI and BamHI and subcloned into the corresponding sites of the pCG-T7 vector to generate the F220RS chimera, which comprises RRM2 of SF2/ASF and the RS domain of SRp20. Subsequently, the SRp20 and F220RS chimeras were independently amplified with oligo pairs 5/6 and 7/4, respectively. The two PCR products, which have a 20 bp overlap, were used in equimolar amounts for a second PCR amplification with oligos 5 and 4. The resulting PCR product was purified, digested with SpeI and BamHI and subcloned into the corresponding sites of the pCG-T7 vector to generate the 201F220RS chimera, which consists of residues 2–85 (RRM) and 105–164 (RS domain) of SRp20 separated by residues 107–202 (RRM2) of SF2/ASF.
In the case of 201F120RS, SF2/ASF and SRp20 were independently amplified with oligo pairs 8/9 and 10/4, respectively. The two PCR products, which have a 20 bp overlap, were used in equimolar amounts for a second PCR amplification with oligos 8 and 4. The resulting PCR product was purified, digested with SpeI and BamHI and subcloned into the corresponding sites of the pCG-T7 vector to generate the F120RS chimera, which comprises RRM1 of SF2/ASF and the RS domain of SRp20. Subsequently, the SRp20 and the F120RS chimeras were independently amplified with oligo pairs 5/11 and 12/4, respectively. The two PCR products, which have a 20 bp overlap, were used in equimolar amounts for a second PCR amplification with oligos 5 and 4. The resulting PCR product was purified, digested with SpeI and BamHI and subcloned into the corresponding sites of the pCG-T7 vector to generate the 201F120RS chimera, which consists of residues 2–85 (RRM) and 105–164 (RS domain) of SRp20 separated by residues 2–97 (RRM1) of SF2/ASF.
Two steps of PCR amplification were employed to generate the 201FRS chimeric protein expression plasmid. First, SRp20 and SF2 were independently amplified with oligo pairs 5/13 and 14/15, respectively. The two PCR products, which have a 20 bp overlap, were used in equimolar amounts for a second PCR amplification with oligo pairs 5/15. The resulting PCR product was purified, digested with SpeI and BamHI and subcloned into the corresponding sites of the pCG-T7 vector to generate the 201FRS chimera, which comprises residues 2–85 (RRM) of SRp20 followed by residues 204–248 (RS domain) of SF2/ASF.
Oligonucleotides
#1, TCGACTAGTGCTCCCCGAGGTCGCTATGGC; #2, TCCTCCTACGATAGCTCGGGCTACG; #3, CCCGAGCTATCGTAGGAGGAGTCCT; #4, TCGGGATCCCTATTTCCTTTCATTTGA; #5, TCGACTAGTCATCGTGATTCCTGTCCA; #6, CTCGGGGAGCTTTTTCACCATTCGA; #7, TGGTGAAAAAGCTCCCCGAGGTCGC; #8, TCGACTAGTTCGGGAGGTGGTGTGATT; #9, TCCTCCTACGTCGGCCTGTTCCACG; #10, AACAGGCCGACGTAG GAGGAGTCCT; #11, CACCTCCCGATTTTTCACCATTCGA; #12, TGGTGAAAAATCGGGAGGTGGTGTG; #13, TGCGGCTACGTTTTTCACCATTCGA; #14, TGGTGAAAAACGTAGCCGCAGCCGT; #15, CTTGGATCCTTAGGTACGAGA.
Cell culture and transfections
HeLa cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum (FCS). Transfections of HeLa cells and purification of total RNA were as previously described (12,53). Briefly, 1 µg of expression plasmid was co-transfected into HeLa cells with 6 µg of the adenovirus E1A reporter plasmid pMTE1A (54) in the presence of 20 µg of Lipofectin (Gibco BRL). The E1A gene plasmid pMTE1A used in the alternative splicing assays was described previously (12,54) and was kindly provided by B. Moran. The cells were grown to 60–75% confluence in 60 mm dishes, plasmid DNA was removed 12–16 h later and DMEM containing 10% FCS was added for an additional 24 h. RNA was extracted using the Total RNA Isolation Reagent (Advanced Biotechnologies). Total RNA (5 µg) was analysed by RT–PCR with Superscript II reverse transcriptase (Life Technologies) and AmpliTaq DNA polymerase (Perkin Elmer), as described previously (53). E1A mRNA detection was carried out with the exon 1 forward primer 5′-GTTTTCTCCTCCGAGCCGCTCCGA-3′ and the 5′-end-labelled exon 2 reverse primer 5′-CTCAGGCTCAGGTTCAGACACAGG-3′. Amplified products separated by urea–PAGE were detected by autoradiography and quantitated by PhosphorImager analysis (BAS2000; Fujix).
For analyisis of fibronectin EDI splicing in vivo, 60–70% confluent Hep3B cells in 60 mm dishes were transfected with 2 µg of the fibronectin minigene driven by a mutant fibronectin promoter and 400 ng of SR expression plasmids using the Fugene transfection reagent (Roche Molecular Biochemicals) according to the manufacturer’s conditions. Analysis of the in vivo splicing reaction was carried out as previously described (55).
Western analysis
For western blot analysis cell lysates were made with RIPA buffer as described previously (53). The electrophoretically separated proteins were transferred onto Hybond P membranes (Amersham Pharmacia Biotech). Non-specific binding was blocked by incubating the blot with 5% non-fat dry milk in TBST (20 mM Tris, pH 7.6, 137 mM NaCl and 0.1% Tween 20). Proteins were detected by subsequent incubation with the primary monoclonal antibody anti-T7.tag (Novagen) in TBST, which recognises the epitope tag at the N-terminus of the proteins. After extensive rinsing with TBST the blots were incubated with secondary antibodies conjugated to horseradish peroxidase at a 1:7000 dilution. After further rinsing in TBST the blots were developed using ECL.
RESULTS
Domain requirements for alternative splicing specificity
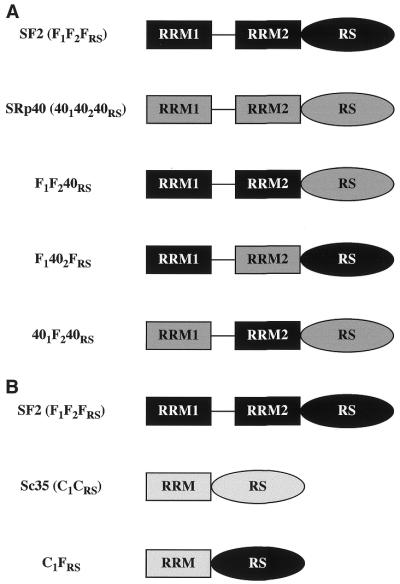
In order to investigate the role of individual domains of SR proteins in alternative splicing, we analysed the activity of a series of chimeric SR proteins in regulated splicing of two reporters, adenovirus E1A pre-mRNA and a fibronectin minigene. The first series of SR chimeric proteins were created by substituting individual domains of SF2/ASF with those of SRp40 and by replacing the RS domain of SC35 with that of SF2/ASF. These chimeras are depicted in Figure 1 and have been previously described (8).
Figure 1.
The domain structures of SF2/ASF, SRp40, SC35 and chimeric proteins consisting of domain swaps are shown schematically. SF2/ASF (first row, A and B) and SRp40 (second row, A) are composed of two RRMs and a C-terminal RS domain, whereas SC35 (second row, B) has only one N-terminal RRM. The nomenclature is based on the origin of each domain: F indicates a domain from SF2/ASF; 40 indicates a domain from SRp40; C, indicates a domain from SC35. The subscript 1 or 2 indicates whether the domain derives from RRM1 or RRM2, whereas the subscript RS indicates an RS domain.
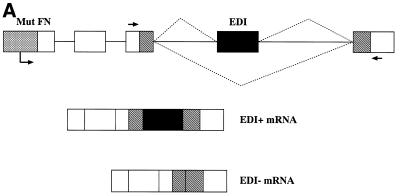
We first evaluated the effects of these chimeric SR proteins on alternative splicing of the EDI fibronectin exon in vivo. We used a well-characterised system, the α-globin/fibronectin minigene, in which the human fibronectin EDI exon, its flanking introns and part of its neighbouring exons are inserted in the third exon of the human α-globin gene. The expression of this minigene is driven by a mutant version of the human fibronectin promoter (Fig. 2A) (56–58). This alternative splicing event involves inclusion or skipping of the alternative EDI exon, which encodes a facultative type III repeat of fibronectin (59). Inclusion of the EDI exon, also known as EDA or EIIIA, depends on the presence of an exonic enhancer located within the central region of the alternative exon (38,57,60) and is highly regulated during embryo development and in proliferative processes such as healing or liver regeneration (reviewed in 61). Binding of SR proteins to this splicing enhancer promotes inclusion of this alternative exon and, in particular, it has been shown that SF2/ASF and 9G8 promote EDI inclusion in vivo and that this effect requires the presence of an intact EDI exonic enhancer (55).
Figure 2.
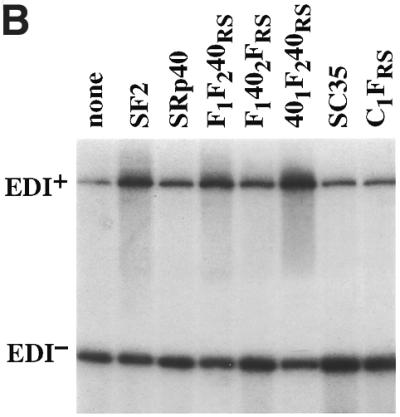
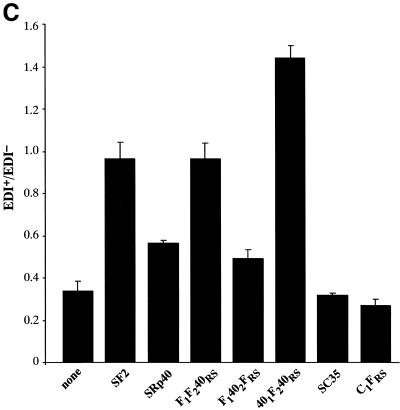
Role of the structural domains of SF2, SRp40 and SC35 proteins in the regulation of fibronectin (FN)–EDI alternative splicing. (A) Structure of the α-globin/fibronectin construct. All exons are included constitutively, except for EDI, which is alternatively spliced. Empty boxes represent human globin sequences; dashed boxes, human FN sequences; mut FN indicates a 220 bp fragment of the mutant (CRE–/CCAAT–) human FN promoter. The transcription start site is indicated by an arrow underneath the promoter. Arrows indicate the locations of primers that overlap globin/FN exon boundaries that were used for RT–PCR analysis. (B) Alternative splicing activity of the chimeric SR proteins. Wild-type SF2/ASF, SRp40 and SC35 proteins and each of the indicated chimeric proteins was co-transfected with the FN–EDI reporter gene. RNA was harvested at 36 h post-transfection and analysed by RT–PCR with a labelled reverse primer, denaturing PAGE and autoradiography, as described in Materials and Methods. (C) The histogram display the ratios between the level of radioactivity in EDI+ and EDI– products. Each experiment was repeated at least three times and data represent averages, with bars indicating standard errors.
Hep3B cells were co-transfected with a version of this fibronectin minigene driven by the mutant fibronectin promoter (Fig. 2A) (55,58) and plasmids expressing cDNAs for wild-type SF2/ASF, SRp40, SC35 or the chimeric proteins described in Figure 1. This resulted in different degrees of EDI inclusion, compared to the extent of EDI splicing in the absence of any co-transfected SR protein (Fig. 2). In agreement with previous results, the highest inclusion of EDI was caused by co-transfection of SF2/ASF, SRp40 was less stimulatory whereas SC35 did not significantly affect the extent of EDI inclusion (55). Two main conclusions could be extracted from the activity of the chimeric SR proteins used in this assay. First, the identity of RRM2 is of crucial importance, as a chimera in which the second RRM of SF2/ASF was replaced by that of SRp40 showed a reduction in EDI inclusion, comparable to the effect observed with wild-type SRp40 (F1402FRS, Fig. 2B and C). Likewise, the reciprocal chimera in which RRM2 of SRp40 was replaced with that of SF2/ASF was even more active than wild-type SF2/ASF in promoting EDI inclusion (401F240RS, Fig. 2B and C). In contrast, replacing the RS domain of SF2/ASF with that of SRp40 did not affect the activity of the resultant chimeric protein (F1F240RS), which behaved as wild-type SF2/ASF, suggesting that RS domains are interchangeable and do not affect alternative splicing activity and/or specificity. Likewise, the C1FRS mutant, in which the RS domain of SC35 was replaced with that of SF2/ASF, displays similar activity to wild-type SC35, again demonstrating that the RS domains are interchangeable for alternative splicing. In every case we observed that the specificity of splice site selection was determined by the RRMs and that the RS domains were functionally interchangeable. The SF2/SRp40 and SF2/SC35 chimeric proteins localise to the nuclear speckles, despite differences in their ability to shuttle from the nucleus to the cytoplasm, which was dependent on the presence of the RS domain of SF2/ASF (8). When the activities of the same chimeric proteins were analysed with the E1A splicing reporter similar conclusions were obtained. As previously reported, SF2/ASF strongly promotes selection of the most proximal 5′ splice site, giving rise to the 13S isoform, and SRp40 has a similar activity, although the switch towards the most proximal site is less pronounced (20). We found that the switch to the most proximal 5′ splice site in the E1A adenovirus reporter was more or less pronounced, depending on whether RRM2 was from SF2/ASF or from SRp40, respectively (data not shown).
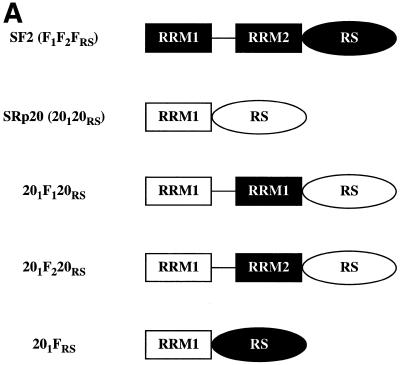
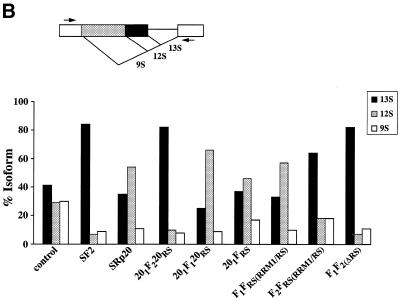
To further evaluate the role of the RRMs in splice selection, and in particular of RRM2 of SF2/ASF, we made two additional chimeric constructs in which either the first (RRM1) or the second (RRM2) RRM of SF2/ASF was inserted between the natural RRM and RS domain of SRp20, converting this one RRM- into a two RRM-containing SR protein (Fig. 3A). In addition, we constructed a chimeric protein in which the RS domain of SRp20 was replaced with that of SF2/ASF. We overexpressed wild-type SF2/ASF, deletion mutants of this protein, wild-type SRp20 and the chimeric proteins described above and assayed changes in the patterns of alternative splicing of an adenovirus E1A splicing reporter. A purine-rich bidirectional splicing enhancer that is located upstream of the 12S 5′ splice site and bound by a subset of SR proteins, among them SF2/ASF, has recently been identified in the E1A pre-mRNA, but it is not clear whether this cis-acting sequence mediates the SF2/ASF effect on the most proximal 5′ splice site (62). All expressed proteins accumulated to similar levels in transfected HeLa cells, as verified by western blot analysis of whole cell lysates (Fig. 3B). Insertion of either RRM1 or RRM2 of SF2/ASF into SRp20 did not alter the subcellular localisation of the resulting chimeric proteins, which localise to the nuclear speckles (not shown).
Figure 3.
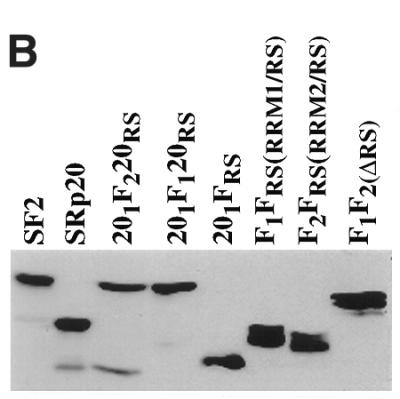
(A) The domain structures of SF2/ASF, SRp20 and chimeric proteins consisting of domain swaps are shown schematically. SF2/ASF (first row) is composed of two RRMs and a C-terminal RS domain, whereas SRp20 has only one N-terminal RRM. The nomenclature is based on the origin of each domain: F indicates a domain from SF2/ASF; 20 indicates a domain from SRp20. The subscript 1 or 2 indicates whether the domain derives from RRM1 or RRM2, whereas the subscript RS denotes an RS domain. (B) Western blotting analysis of wild-type and chimeric SR proteins. All expressed proteins accumulated to similar levels in transfected Hep3B cells (this figure) and HeLa cells (data not shown)
In agreement with previous results, wild-type SF2/ASF and the mutant lacking RRM1 (F2FRS) strongly activated the 13S 5′ splice site, whereas SRp20 and the SF2/ASF mutant that lacks RRM2 (F1FRS) strongly and reproducibly stimulated the 12S 5′ splice site (Fig. 4) (20,48). The rationale behind the design of the SF2/SRp20 chimeras was to test whether the presence of RRM2 of SF2/ASF in the context of the SRp20 protein could act in a dominant manner and confer SF2/ASF specificity to the chimeric protein. This was indeed the case, whereas insertion of RRM1 of SF2/ASF between the RRM and RS domain of SRp20 does not change the splicing specificity of the resulting chimera (201F120RS), insertion of RRM2 produced a drastic shift in the splice site selection since the mutant protein, 201F220RS, was as active as SF2/ASF wild-type and strongly activated the most proximal 5′ splice site that gives rise to the 13S isoform. This experiment confirmed that the nature of the RRM determines the selection of a particular 5′ splice site. The RS domain of SF2/ASF is not required for alternative splicing in vivo, as shown by the activity of the SF2/ASF mutant, which lacks the RS domain (F1F2 in Fig. 4; 48). The 201FRS mutant in which the RS domain of SRp20 was replaced with the corresponding domain of SF2/ASF has similar activity to wild-type SRp20, strongly suggesting that the RS domains are interchangeable. Quantitation of the relative use of the E1A 5′ splice sites upon overexpression of the different proteins is shown in Figure 4B.
Figure 4.
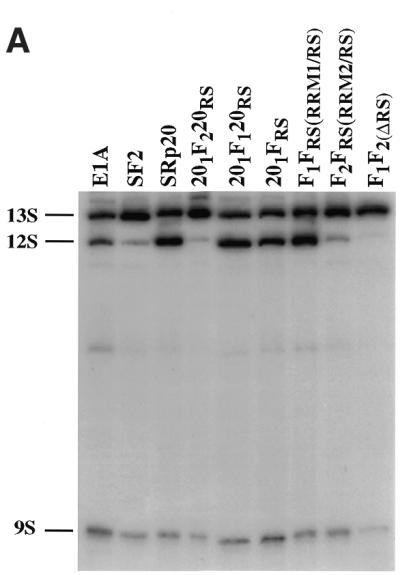
Role of SF2/SRp20 structural domains in regulating alternative splicing of adenovirus E1A pre-mRNA. (A) Alternative splicing activity of the SF2/SRp20 chimeric proteins. Each of the indicated chimeric proteins and wild-type control proteins was overexpressed from plasmids co-transfected with the E1A reporter gene. RNA was harvested at 36 h post-transfection and analysed by RT–PCR with a labelled reverse primer, denaturing PAGE and autoradiography, as described in Materials and Methods. The positions of 13S, 12S and 9S spliced mRNAs are indicated on the left. (B) Quantitation of E1A mRNA isoforms in transfected cells. A diagram of the E1A reporter gene indicates the alternative 5′ splice sites and splicing events that generate 13S, 12S and 9S mRNAs. The locations of the exon primers used for RT–PCR analysis are shown. The relative amounts of 13S, 12S and 9S E1A mRNAs were calculated from the data in (A), using a PhosphorImager, and the percentage of each isoform is shown. Nearly identical results were obtained in three independent experiments.
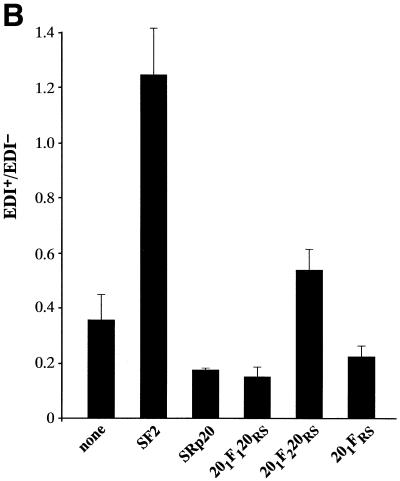
Next, we analysed the effects of the SF2/SRp20 chimeric proteins described above on alternative splicing of the fibronectin splicing reporter. Hep3B cells were co-transfected with a version of the fibronectin minigene driven by the mutant fibronectin promoter and plasmids expressing cDNAs for the SR proteins SF2/ASF and SRp20 in their wild-type versions or the SF2/SRp20 chimeric proteins. Overexpression of wild-type SF2/ASF greatly enhanced EDI inclusion, whereas wild-type SRp20 had a slightly inhibitory effect. Replacing the RS domain of SRp20 with that of SF2/ASF (Fig. 5, 201FRS mutant) did not confer SF2/ASF activity to the resulting chimeric protein, confirming that the RS domains do not determine specificity in alternative splicing and are functionally interchangeable. The construct that inserts RRM1 of SF2/ASF into SRp20 did not significantly affect inclusion of EDI (Fig. 5, 201F1120RS). In contrast, insertion of RRM2 of SF2/ASF into SRp20 resulted in a shift to inclusion of EDI, but the effects were modest (Fig. 5, 201F1220RS).
Figure 5.
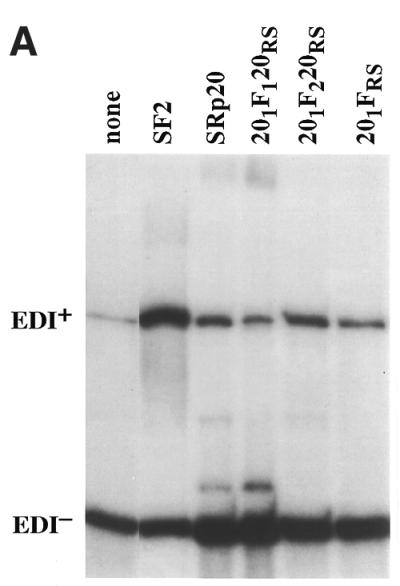
Role of the structural domains of SF2/SRp20 chimeric proteins in regulating alternative splicing of FN–EDI mRNA. (A) Alternative splicing activity of the SF2/SRp20 chimeric proteins. Each of the indicated chimeric proteins and wild-type control proteins was overexpressed from plasmids co-transfected with the FN–EDI reporter gene. RNA was harvested at 36 h post-transfection and analysed by RT–PCR with a labelled reverse primer, denaturing PAGE and autoradiography, as described in Materials and Methods. (B) The histogram displays the ratios between the levels of radioactivity in EDI+ and EDI– products. Each experiment was repeated at least three times and data represent averages, with bars indicating standard errors.
In summary, we have shown that the specificity of splice site selection in living cells is determined by the nature of the RRMs, which function as modules, and that the RS domains do not contribute to alternative splicing specificity and are functionally interchangeable. We also found that SF2/ASF specificity in alternative splicing resides in RRM2, as was demonstrated for constitutive splicing in vitro (50), which suggests that the basic mechanisms of binding to targets are common in both processes. More importantly, the functions of the two RRM modules can be separated, as shown by the dominant role of RRM2 of SF2/ASF in different contexts.
DISCUSSION
The SR proteins function in both constitutive and alternative splicing, but the role of individual domains of SR proteins has only been analysed for the constitutive splicing reaction. Although both the RRMs and the RS domain of SR proteins are required for constitutive splicing, the substrate specificity of SR proteins in constitutive splicing is determined by the RRMs and the RS domains are functionally interchangeable (23,50).
It has not been clearly established whether general and/or sequence-specific RNA binding by SR proteins is required for their activity in alternative splicing regulation. In fact, contradictory results have been obtained using different experimental systems. Using in vitro assays it was shown that mutants of SF2/ASF that lack either RRM1 or RRM2 were inactive in alternative splicing, suggesting a correlation between RNA binding and alternative splicing activity (30,31). It is possible, although unlikely, that the protein tags used in these various in vitro and in vivo systems are affecting protein function. Our previous observations with wild-type untagged SR expression vectors show that these constructs give the same effects as the T7 epitope-tagged versions (20). In contrast, the same SF2/ASF mutant proteins were shown to be active in E1A pre-mRNA alternative splicing in vivo, although with an altered splice site specificity, suggesting that sequence-specific interactions may play a role in selecting particular splice sites for activation (48). In addition, similar SF2/ASF mutant proteins lacking either RRM1 or RRM2 were still able to facilitate U1 snRNP binding to a pre-mRNA containing a functional 5′ splice site (63). It was proposed that SF2/ASF mediates alternative splicing by promoting the indiscriminate binding of U1 snRNP to alternative 5′ splice sites and it has recently been shown that hnRNP A1 antagonises SF2/ASF action by causing reduced U1 snRNP binding to the same sites (51,64). More importantly, mutations in hnRNP A1 and SF2/ASF showed that the opposite effects of the proteins on 5′ splice site choice are correlated with their effects on U1 snRNP binding (64).
In this paper, we have sought to address how the modular domains of SR proteins contribute to alternative splicing specificity and whether sequence-specific binding to high affinity sites in the RNA is required for the function of SR proteins in regulated splicing.
Adenovirus E1A pre-mRNA generates three major mRNA isoforms that represent alternative 5′ splice site utilisation and it has been previouly shown that SF2/ASF and hnRNP A1 have antagonistic functions in vivo, with SF2/ASF promoting the selection of the most proximal 13S 5′splice site and hnRNP A1 selecting the most distal 9S 5′splice site (12,13). It was also shown that a mutant SF2/ASF protein that lacks RRM2, as well as wild-type SRp20, which naturally lacks a second RRM, selected the 12S 5′ splice site in transient transfection assays (48).
Here we show that insertion of RRM2 of SF2/ASF between the RRM and the RS domain of SRp20 rendered a chimeric protein that was as active as wild-type SF2/ASF in promoting selection of the most proximal 5′ splice site, whereas insertion of RRM1 of SF2/ASF did not alter the activity of SRp20. This experiment clearly demonstrates that the nature of the RRM determines splice site selection and, in particular, that RRM2 of SF2/ASF has a dominant role. These conclusions were further confirmed by the activity of the SF2/SRp40 chimeric proteins on alternative splicing of the fibronectin EDI exon, which is dependent on binding of SR proteins to an exonic splicing enhancer. Thus, the findings in this paper show that splicing specificity is determined by the RRMs and, in particular, the identity of RRM2 is important.
These experiments demonstrate that the presence of RRM2 of SF2/ASF determines specificity of splice site selection. One likely explanation for these results is that the RNA binding specificity of the chimeric proteins was altered in such a way that SF2/ASF specificity of binding was conferred to those chimeric proteins that contain RRM2 of SF2/ASF. However, it is not clear how the RRMs in the SR proteins bind to RNA. It has been shown that both RRMs of SF2/ASF together bind with strong synergy to RNA. In addition, the high affinity binding sites selected by SELEX with an SF2/ASF protein comprising both RRMs are distinct from the sites selected with RRM1 alone, suggesting that both RRMs of SF2/ASF act together in binding RNA (32). In contrast, in other RNA-binding proteins, such as the U1snRNP-associated U1A protein, individual RRMs have their own RNA-binding specificity and interact independently with distinct RNA elements in pre-mRNA (65)
In the case of the hnRNP A1 protein, structural and functional analysis of variant proteins comprising duplications, deletions or swaps of the RRMs showed that the two RRMS have naturally evolved as part of a double RRM structure (66,67). It is highly likely that each RRM in the two RRM-containing SR proteins function as a bipartite RNA-binding domain while retaining significant modular character, similar to what has been shown for hnRNP A1 (67).
An alternative explanation for the dominant role of RRM2 of SF2/ASF in alternative splicing may lie in the establishment of new protein–protein rather than protein–RNA contacts. In such a scenario RRM2 of SF2/ASF may help recruit additional splicing factors to the splicesosome and modulate splicing specificity.
It has been shown that individual SR proteins interact with each other, with the small subunit of U2AF (U2AF35) and with the U1snRNP protein U170K (45,68). This has led to the suggestion that the SR proteins may act as bridging factors to facilitate functional interactions between the 5′ and 3′ splice sites. In addition, a subset of SR proteins interacts with a protein that binds to S/MAR regions, known as scaffold attachment factor B or SAF-B (69), and SRp20 has been shown to bind RBMp, a mammalian germ cell-specific RNA-binding protein (70).
Most of the SR protein–protein interactions have been shown to be dependent on the presence of the RS domain in the interacting proteins (45,68,71). However, the RBMp–SRp20 interaction is mediated via the SRp20 RRM (70), which suggests that protein–protein interaction domains are present in the RRMs. It is possible that RRM2 could help mediate homodimeric interactions recruiting wild-type SF2/ASF protein or, alternatively, sequestering endogenous factors such as SRp20. This activity would explain why the sole presence of RRM2 of SF2/ASF results in EDI inclusion in the fibronectin gene and selection of the 13S 5′ splice site in the E1A adenovirus pre-mRNA, both activities that are strongly favoured by wild-type SF2/ASF protein. It should be taken into account that the alternative splicing assays, both in vivo and in vitro, are carried out in the presence of multiple endogenous SR proteins which could be recruited to the spliceosome via protein–protein interactions with the exogenously expressed proteins. Thus, it is not possible at this stage to exclude the possibility that, in addition to directly binding the substrate pre-mRNA, the RRM2 domain can also function as a protein–protein interaction domain to recruit other SR proteins involved in alternative splicing of the substrate pre-mRNA.
The RS domain is a distinctive feature of the SR family of proteins and is also present in a family of related splicing factors, the so-called SR-like proteins. It has been proposed as being a domain that directs protein–protein interactions and has also been shown to be an important determinant of subcellular localisation, but it is not clear whether individual RS domains have redundant or unique functions. In fact, contradictory results have been obtained using different experimental systems. Specific roles for the RS domains of individual SR proteins have been suggested by the high phylogenetic conservation of specific sequences within the RS domains (72). The activities of chimeric proteins containing different RS domains fused to the RRM of the bacteriophage MS2 protein in the splicing of substrates containing a single MS2-binding site correlated directly with the number of RS dipeptides present in the RS domain, suggesting that different RS domains have unique activities (73). Individual RS domains have also been shown to have unique properties in directing subcellular localisation and influencing the ability of SR proteins to shuttle from the nucleus to the cytoplasm (8,46–48). However, the RS domains are functionally interchangeable in vivo, as shown by the ability of chimeric proteins consisting of the RRMs of SF2/ASF fused to RS domains of different SR proteins to rescue cell viability in a chicken B cell line, DT40 (74). In contrast, it was recently shown that RS domains are not all functionally equivalent in vivo in Drosophila, as different RS domains varied considerably in their ability to restore Tra2 function (75).
The RS domains are essential for constitutive splicing activity, but dispensable for regulated splicing, both in vitro and in vivo (30,31,48). When its function was assayed in constitutive splicing it was shown that RS domains are functionally interchangeable and do not contribute to splicing specificity (23,50). We show here that replacing the RS domain of SF2/ASF by that of SRp40 or that of SC35 by that of SF2/ASF did not affect the activity of the resultant chimeric proteins (F1F240RS and C1FRS, respectively) in alternative splicing of the fibronectin reporter, suggesting that these RS domains are interchangeable and do not affect alternative splicing activity and/or specificity (Fig. 2). Similarly, replacing the RS domain of SRp20 with that of SF2/ASF (201FRS mutant) did not alter the activity of the chimeric protein with either the E1A or the fibronectin splicing reporters, confirming that the RS domains are interchangeable (Figs 4 and 5).
In summary, this present study has shown that substrate specificity in alternative splicing is determined by the nature of the RRMs and that the RS domains are interchangeable, as has been demonstrated for the constitutive splicing reaction. These results demonstrate the modularity of SR proteins for their alternative splicing function in vivo. Our previous results showing different splice site activation specificities of the SF2/ASF mutants depending on which of the two RRMs was deleted, together with the results shown here, strongly suggest that sequence-specific interactions may play a role in selecting particular splice sites for activation. The dominant role of RRM2 of SF2/ASF could be explained by two non-mutually exclusive models. It is possible that the presence of this domain confers SF2/ASF RNA binding specificity to the chimeric proteins. Alternatively, RRM2 of SF2/ASF could mediate protein–protein interactions, bringing wild-type SF2/ASF or other SR proteins into play.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Alberto Kornblihtt (Buenos Aires) for the generous gift of the FN/EDI constructs. We are grateful to Ian Eperon (Leicester) and Andres Muro (Trieste) for critical reading of the manuscript. We are indebted to Douglas Stuart (MRC HGU Photography Department) for help in the preparation of figures. J.F.C. was supported by the Medical Research Council and W.v.d.H.v.O by the Wellcome Trust.
REFERENCES
- 1.Fu X.D. (1995) The superfamily of arginine/serine-rich splicing factors. RNA, 1, 663–680. [PMC free article] [PubMed] [Google Scholar]
- 2.Krämer A. (1996) The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu. Rev. Biochem., 65, 367–409. [DOI] [PubMed] [Google Scholar]
- 3.Valcárcel J. and Green,M.R. (1996) The SR protein family: pleiotropic functions in pre-mRNA splicing. Trends Biochem. Sci., 21, 296–301. [PubMed] [Google Scholar]
- 4.Cáceres J.F. and Krainer,A.R. (1997) Mammalian pre-mRNA splicing factors. In Krainer,A.R. (ed.), Eukaryotic mRNA Processing. IRL Press, Oxford, UK, pp. 174–212
- 5.Staknis D. and Reed,R. (1994) SR proteins promote the first specific recognition of pre-mRNA and are present together with the U1 small nuclear ribonucleoprotein particle in a general splicing enhancer complex. Mol. Cell. Biol., 14, 7670–7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graveley B.R. (2000) Sorting out the complexity of SR protein functions. RNA, 6, 1197–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blencowe B.J., Issner,R., Nickerson,J.A. and Sharp,P.A. (1998) A coactivator of pre-mRNA splicing. Genes Dev., 12, 996–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cáceres J.F., Screaton,G.R. and Krainer,A.R. (1998) A specific subset of SR proteins shuttles continuously betweeen the nucleus and the cytoplasm. Genes Dev., 12, 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanamura A., Cáceres,J.F., Mayeda,A., Franza,B.R.Jr and Krainer,A.R. (1998) Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. RNA, 4, 430–444. [PMC free article] [PubMed] [Google Scholar]
- 10.Mayeda A. and Krainer,A.R. (1992) Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell, 68, 365–375. [DOI] [PubMed] [Google Scholar]
- 11.Mayeda A., Munroe,S.H., Cáceres,J.F. and Krainer,A.R. (1994) Function of conserved domains of hnRNP A1 and other hnRNP A/B proteins. EMBO J., 13, 5483–5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cáceres J.F., Stamm,S., Helfman,D.M. and Krainer,A.R. (1994) Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science, 265, 1706–1709. [DOI] [PubMed] [Google Scholar]
- 13.Yang X., Bani,M.R., Lu,S.-J., Rowan,S., Ben-David,Y. and Chabot,B. (1994) The A1 and A1B proteins of heterogenous nuclear ribonucleoparticles modulate 5′ splice site selection in vivo. Proc. Natl Acad. Sci. USA, 91, 6924–6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Labourier E., Bourbon,H.M., Gallouzi,I.E., Fostier,M., Allemand,E. and Tazi,J. (1999) Antagonism between RSF1 and SR proteins for both splice-site recognition in vitro and Drosophila development. Genes Dev., 13, 740–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gallego M.E., Gattoni,R., Stevenin,J., Marie,J. and Expert-Bezancon,A. (1997) The SR splicing factors ASF/SF2 and SC35 have antagonistic effects on intronic enhancer-dependent splicing of the beta-tropomyosin alternative exon 6A. EMBO J., 16, 1772–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jumaa H. and Nielsen,P.J. (1997) The splicing factor SRp20 modifies splicing of its own mRNA and ASF/SF2 antagonizes this regulation. EMBO J., 16, 5077–5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blencowe B.J., Bowman,J.A., McCracken,S. and Rosonina,E. (1999) SR-related proteins and the processing of messenger RNA precursors. Biochem. Cell Biol., 77, 277–291. [PubMed] [Google Scholar]
- 18.Tacke R. and Manley,J.L. (1999) Determinants of SR protein specificity. Curr. Opin. Cell Biol., 11, 358–362. [DOI] [PubMed] [Google Scholar]
- 19.Zahler A.M., Neugebauer,K.M., Lane,W.S. and Roth,M.B. (1993) Distinct functions of SR proteins in alternative pre-mRNA splicing. Science, 260, 219–222. [DOI] [PubMed] [Google Scholar]
- 20.Screaton G.R., Cáceres,J.F., Mayeda,A., Bell,M.V., Plebanski,M., Jackson,D.G., Bell,J.I. and Krainer,A.R. (1995) Identification and characterization of three members of the human SR family of pre-mRNA splicing factors. EMBO J., 14, 4336–4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J. and Manley,J.L. (1995) Overexpression of the SR proteins ASF/SF2 and SC35 influences alternative splicing in vivo in diverse ways. RNA, 1, 335–346. [PMC free article] [PubMed] [Google Scholar]
- 22.Fu X.D. (1993) Specific commitment of different pre-mRNAs to splicing by single SR proteins. Nature, 365, 82–85. [DOI] [PubMed] [Google Scholar]
- 23.Chandler S.D., Mayeda,A., Yeakley,J.M., Krainer,A.R. and Fu,X.D. (1997) RNA splicing specificity determined by the coordinated action of RNA recognition motifs in SR proteins. Proc. Natl Acad. Sci. USA, 94, 3596–3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J., Takagaki,Y. and Manley,J.L. (1996) Targeted disruption of an essential vertebrate gene: ASF/SF2 is required for cell viability. Genes Dev., 10, 2588–2599. [DOI] [PubMed] [Google Scholar]
- 25.Ring H.Z. and Lis,J.T. (1994) The SR protein B52/SRp55 is essential for Drosophila development. Mol. Cell. Biol., 14, 7499–7506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jumaa H., Wei,G. and Nielsen,P.J. (1999) Blastocyst formation is blocked in mouse embryos lacking the splicing factor SRp20. Curr. Biol., 9, 899–902. [DOI] [PubMed] [Google Scholar]
- 27.Longman D., Johnstone,I.L. and Cáceres,J.F. (2000) Functional characterization of SR and SR related genes in Caenorhabditis elegans. EMBO J., 19, 1625–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ge H., Zuo,P. and Manley,J.L. (1991) Primary structure of the human splicing factor ASF reveals similarities with Drosophila regulators. Cell, 66, 373–382. [DOI] [PubMed] [Google Scholar]
- 29.Krainer A.R., Mayeda,A., Kozak,D. and Binns,G. (1991) Functional expression of cloned human splicing factor SF2: homology to RNA-binding proteins, U1 70K, and Drosophila splicing regulators. Cell, 66, 383–394. [DOI] [PubMed] [Google Scholar]
- 30.Cáceres J.F. and Krainer,A.R. (1993) Functional analysis of pre-mRNA splicing factor SF2/ASF structural domains EMBO J., 12, 4715–4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zuo P. and Manley,J.L. (1993) Functional domains of the human splicing factor ASF/SF2. EMBO J., 12, 4727–4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tacke R. and Manley,J.L. (1995) The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J., 14, 3540–3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heinrichs V. and Baker,B.S. (1995) The Drosophila SR protein RBP1 contributes to the regulation of doublesex alternative splicing by recognizing RBP1 RNA target sequences. EMBO J., 14, 3987–4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tacke R., Chen,Y. and Manley,J.L. (1997) Sequence-specific RNA binding by an SR protein requires RS domain phosphorylation: creation of an SRp40-specific splicing enhancer. Proc. Natl Acad. Sci. USA, 94, 1148–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi H., Hoffman,B.E. and Lis,J.T. (1997) A specific RNA hairpin loop structure binds the RNA recognition motifs of the Drosophila SR protein B52. Mol. Cell. Biol., 17, 2649–2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu H.X., Zhang,M. and Krainer,A.R. (1998) Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev., 12, 1998–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cavaloc Y., Bourgeois,C.F., Kister,L. and Stevenin,J. (1999) The splicing factors 9G8 and SRp20 transactivate splicing through different and specific enhancers. RNA, 5, 468–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lavigueur A., La Branche,H., Kornblihtt,A.R. and Chabot,B. (1993) A splicing enhancer in the human fibronectin alternate ED1 exon interacts with SR proteins and stimulates U2 snRNP binding. Genes Dev., 7, 2405–2417. [DOI] [PubMed] [Google Scholar]
- 39.Sun Q., Mayeda,A., Hampson,R.K., Krainer,A.R. and Rottman,F.M. (1993) General splicing factor SF2/ASF promotes alternative splicing by binding to an exonic splicing enhancer. Genes Dev., 7, 2598–2608. [DOI] [PubMed] [Google Scholar]
- 40.Ramchatesingh J., Zahler,A.M., Neugebauer,K.M., Roth,M.B. and Cooper,T.A. (1995) A subset of SR proteins activates splicing of the cardiac troponin T alternative exon by direct interactions with an exonic enhancer. Mol. Cell. Biol., 15, 4898–4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lynch K.W. and Maniatis,T. (1996) Assembly of specific SR protein complexes on distinct regulatory elements of the Drosophila doublesex splicing enhancer. Genes Dev., 10, 2089–2101. [DOI] [PubMed] [Google Scholar]
- 42.Selvakumar M. and Helfman,D.M. (1999) Exonic splicing enhancers contribute to the use of both 3′ and 5′ splice site usage of rat beta-tropomyosin pre-mRNA. RNA, 5, 378–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blencowe B.J. (2000) Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem. Sci., 25, 106–110. [DOI] [PubMed] [Google Scholar]
- 44.Kanopka A., Muhlemann,O. and Akusjarvi,G. (1996) Inhibition by SR proteins of splicing of a regulated adenovirus pre-mRNA. Nature, 381, 535–538. [DOI] [PubMed] [Google Scholar]
- 45.Wu J.Y. and Maniatis,T. (1993) Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell, 75, 1061–1070. [DOI] [PubMed] [Google Scholar]
- 46.Li H. and Bingham,P.M. (1991) Arginine/serine rich domains of the su(wa) and tra RNA processing regulators target proteins to a subnuclear compartment implicated in splicing. Cell, 67, 335–342. [DOI] [PubMed] [Google Scholar]
- 47.Hedley M.L., Amrein,H. and Maniatis,T. (1995) An aminoacid sequence motif sufficient for subnuclear localization of an arginine/serine rich splicing factor. Proc. Natl Acad. Sci. USA, 92, 11524–11528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cáceres J.F., Misteli,T., Screaton,G.R., Spector,D.L. and Krainer,A.R. (1997) Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J. Cell Biol., 138, 225–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Graveley B.R. and Maniatis,T. (1998) Arginine/serine-rich domains of SR proteins can function as activators of pre-mRNA splicing. Mol. Cell, 1, 765–771. [DOI] [PubMed] [Google Scholar]
- 50.Mayeda A., Screaton,G.R., Chandler,S.D., Fu,X.D. and Krainer,A.R. (1999) Substrate specificities of SR proteins in constitutive splicing are determined by their RNA recognition motifs and composite pre-mRNA exonic elements. Mol. Cell. Biol., 19, 1853–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eperon I.C., Ireland,D.C., Smith,R.A., Mayeda,A. and Krainer,A.R. (1993) Pathways for selection of 5′ splice sites by U1 snRNPs and SF2/ASF. EMBO J., 12, 3607–3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilson A.C., Peterson,M.G. and Herr,W. (1995) The HCF repeat is an unusual proteolytic cleavage signal. Genes Dev., 9, 2445–2458. [DOI] [PubMed] [Google Scholar]
- 53.van der Houven van Oordt W., Diaz-Meco,M.T., Lozano,J., Krainer,A.R., Moscat,J. and Cáceres,J.F. (2000) The MKK3/6-p38 signaling cascade alters the subcellular distribution of hnRNP A1 and modulates alternative splicing regulation. J. Cell Biol., 149, 307–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zerler B., Moran,B., Maruyama,K., Moomaw,J., Grodzicker,T. and Ruley,H.E. (1986) Adenovirus E1A coding sequences that enable ras and pmt oncogenes to transform cultured primary cells. Mol. Cell. Biol., 6, 887–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cramer P., Cáceres,J.F., Cazalla,D., Kadener,S., Muro,A.F., Baralle,F.E. and Kornblihtt,A.R. (1999) Coupling of transcription with alternative splicing: RNA pol II promoters modulate SF2/ASF and 9G8 effects on an exonic splicing enhancer. Mol. Cell, 4, 251–258. [DOI] [PubMed] [Google Scholar]
- 56.Vibe-Pedersen K., Kornblihtt,A.R. and Baralle,F.E. (1984) Expression of a human alpha-globin/fibronectin gene hybrid generates two mRNAs by alternative splicing. EMBO J., 3, 2511–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Caputi M., Casari,G., Guenzi,S., Tagliabue,R., Sidoli,A., Melo,C.A. and Baralle,F.E. (1994) A novel bipartite splicing enhancer modulates the differential processing of the human fibronectin EDA exon. Nucleic Acids Res., 22, 1018–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cramer P., Pesce,C.G., Baralle,F.E. and Kornblihtt,A.R. (1997) Functional association between promoter structure and transcript alternative splicing. Proc. Natl Acad. Sci. USA, 94, 11456–11460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kornblihtt A.R, Vibe-Pedersen,K. and Baralle,F.E. (1984) Human fibronectin: molecular cloning evidence for two mRNA species differing by an internal segment coding for a structural domain. EMBO J., 3, 221–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mardon H.J., Sebastio,G. and Baralle,F.E. (1987) A role for exon sequences in alternative splicing of the human fibronectin gene. Nucleic Acids Res., 15, 7725–7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kornblihtt A.R., Pesce,C.G., Alonso,C.R., Cramer,P., Srebrow,A., Werbajh,S. and Muro,A.F. (1996) The fibronectin gene as a model for splicing and transcription studies. FASEB J., 10, 248–257. [DOI] [PubMed] [Google Scholar]
- 62.Bourgeois C.F., Popielarz,M., Hildwein,G. and Stevenin,J. (1999) Identification of a bidirectional splicing enhancer: differential involvement of SR proteins in 5′ or 3′ splice site activation. Mol. Cell. Biol., 19, 7347–7356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jamison S.F., Pasman,Z., Wang,J., Will,C., Luhrmann,R., Manley,J.L. and Garcia-Blanco,M.A. (1995) U1 snRNP-ASF/SF2 interaction and 5′ splice site recognition: characterization of required elements. Nucleic Acids Res., 23, 3260–3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Eperon I.C., Makarova,O., Mayeda,A., Munroe,S.H., Cáceres,J.F. and Krainer,A.R. (2000) Selection of alternative 5′ splice sites: the role of U1 snRNP and models for the antagonistic effects of SF2/ASF and hnRNP A1. Mol. Cell. Biol., 20, 8303–8318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lutz C.S. and Alwine,J.C. (1994) Direct interaction of the U1 snRNP-A protein with the upstream efficiency element of the SV40 late polyadenylation signal. Genes Dev., 8, 576–586. [DOI] [PubMed] [Google Scholar]
- 66.Xu R.M., Jokhan,L., Cheng,X., Mayeda,A. and Krainer,A.R. (1997) Crystal structure of human UP1, the domain of hnRNP A1 that contains two RNA-recognition motifs. Structure, 5, 559–570. [DOI] [PubMed] [Google Scholar]
- 67.Mayeda A., Munroe,S.H., Xu,R.M. and Krainer,A.R. (1998) Distinct functions of the closely related tandem RNA-recognition motifs of hnRNP A1. RNA, 4, 1111–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kohtz J.D., Jamison,S.F., Will,C.L., Zuo,P., Lührmann,R., Garcia-Blanco,M.A. and Manley,J.L. (1994) Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature, 368, 119–124. [DOI] [PubMed] [Google Scholar]
- 69.Nayler O., Stratling,W., Bourquin,J.P., Stagljar,I., Lindemann,L., Jasper,H., Hartmann,A.M., Fackelmayer,F.O., Ullrich,A. and Stamm,S. (1998) SAF-B protein couples transcription and pre-mRNA splicing to SAR/MAR elements. Nucleic Acids Res., 26, 3542–3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Elliott D.J., Bourgeois,C.F., Klink,A., Stevenin,J. and Cooke,H.J. (2000) A mammalian germ cell-specific RNA-binding protein interacts with ubiquitously expressed proteins involved in splice site selection. Proc. Natl Acad. Sci. USA, 97, 5717–5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Amrein H., Hedley,M.L. and Maniatis,T. (1994) The role of specific protein-RNA and protein-protein interactions in positive and negative control of pre-mRNA splicing by Transformer 2. Cell, 76, 735–746. [DOI] [PubMed] [Google Scholar]
- 72.Birney E., Kumar,S. and Krainer,A.R. (1993) Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res., 21, 5803–5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Graveley B.R., Hertel,K.J. and Maniatis,T. (1998) A systematic analysis of the factors that determine the strength of pre-mRNA splicing enhancers. EMBO J., 17, 6747–6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang J., Xiao,S.H. and Manley,J.L. (1998) Genetic analysis of the SR protein ASF/SF2: interchangeability of RS domains and negative control of splicing. Genes Dev., 12, 2222–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dauwalder B. and Mattox,W. (1998) Analysis of the functional specificity of RS domains in vivo. EMBO J., 17, 6049–6060. [DOI] [PMC free article] [PubMed] [Google Scholar]