Abstract
Background
Hemoplasma infections in cattle are caused by Mycoplasma wenyonii and Candidatus Mycoplasma haemobos and induce asymptomatic or chronic infections but occasionally lead to life-threatening hemolytic anemia. Despite the global distribution of bovine hemoplasmas, information regarding their transmission vectors and prevalence is still lacking in the Republic of Korea. Therefore, this study aims to investigate the infection rate of bovine hemoplasma in cattle and houseflies and to assess the risk factors associated with hemoplasma infection in cattle.
Methods
Overall, 376 blood samples were collected from Korean indigenous cattle (male, 10−13 months old), along with 2,690 houseflies (Musca domestica) from the same farm where the cattle were raised. PCR assays targeting the 16S rRNA gene were performed to detect hemoplasmas, and positive samples were sequenced.
Results
The infection rate of bovine hemoplasmas was 50.8% (191/376) in cattle and 7.4% in pooled houseflies. Among cattle, 18.6% (70/376) and 20.0% (75/376) tested positive for M. wenyonii and Candidatus M. haemobos, respectively. Conversely, in houseflies, Candidatus M. haemobos was more frequently detected (5.9%) than M. wenyonii (0.7%). Co-infection was 12.2% (46/376) in cattle and 0.7% in flies. Furthermore, hemoplasma infection was significantly associated with the grazing experience of their dams. Cattle born to cows with grazing experience exhibited a higher risk for M. wenyonii infection (odds ratio [OR] = 1.62; 95% confidence interval [CI]: 1.03−2.55; P = 0.045), whereas these cattle had a lower risk for Candidatus M. haemobos infection (OR = 0.32, 95% CI: 0.19−0.74; P = 0.000) than animals born to cows without grazing experience. The sequences obtained from houseflies were confirmed as Candidatus M. haemobos, which displayed high similarity (98.2−100%) to those from cattle obtained in this study.
Conclusions
To our knowledge, this study represents the first report of bovine hemoplasmas identified in houseflies. This molecular evidence suggests that houseflies may be possible vectors for Candidatus M. haemobos.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12917-024-04343-x.
Keywords: Bovine hemoplasma, Mycoplasma wenyonii, Candidatus mycoplasma haemobos, Houseflies, Transmission route
Background
Hemotropic mycoplasma (hemoplasmas) are small, pleomorphic, and wall-less bacteria that parasitize the surface of red blood cells, causing infections in various animals, including humans [1, 2]. Hemoplasmas are emerging pathogens in a wide variety of animals [2]. To date, two distinct hemoplasmas have been identified in cattle: Mycoplasma wenyonii and Candidatus Mycoplasma haemobos [3, 4]. The clinical signs of these bovine hemoplasmas include anemia, pyrexia, lymphadenopathy, anorexia, decreased milk production, and reproductive problems; however, most infections in animals remain subclinical [1, 4–8]. Acute infections are rare in cattle, but occasionally they can lead to overt life-threatening hemolytic anemia [9, 10].
Bovine hemoplasmas are widely distributed worldwide [4, 8, 11–16], but their transmission routes are poorly understood. Several studies have reported that transmission can occur through direct contact with infected blood, contaminated surgical instruments, blood-sucking arthropods (such as ticks, flies, lice, and mosquitoes), or via transplacental infection [3, 14, 17–20]. Although M. wenyonii and Candidatus M. haemobos DNA has been detected in several vectors [17, 20–23], the evidence that arthropod vectors were involved has not been sufficiently proven in the Republic of Korea (ROK).
The housefly (Musca domestica) is the most abundant and widely distributed fly species [24] and can carry more than 100 pathogens responsible for diseases in animals and humans [25] and consequently poses a substantial health risk [26]. However, despite their role in disease transmission, little is known about the specific pathogens that houseflies carry, and farmers often overlook their influence. To date, houseflies were not considered to be potential candidate vectors for transmitting bovine hemoplasmas. However, previous results have detected bovine hemoplasmas in blood-sucking flies [17]. Moreover, a recent study showed that houseflies could be potential carriers of Mycoplasma spp. [27]. Flies are predominantly found at cattle farms during the summer and autumn and readily contaminate food and eating utensils. Considering their adaptability, mobility, and affinity for various environments, houseflies may be involved in disseminating bovine mycoplasmas with other pathogens between animals or on farms. Since the exact vector of this disease has not yet been identified, the role of houseflies in transmitting bovine mycoplasmas needs to be determined.
Compared to other countries, a minimal number of studies on hemoplasmas have been conducted in the ROK. So far, only one study on bovine hemoplasmas is available [12]. A limitation of previous study was focused on elucidating the presence of hemoplasmas and the association between hemoplasma infection and anemia [12]. Therefore, considering that houseflies are found abundantly on farms, we aimed to investigate the occurrence of bovine hemoplasmas in houseflies, evaluate the possible transmission routes of bovine hemoplasma using molecular analysis, and determine the risk factors associated with hemoplasma infection in cattle.
Materials and methods
Ethical statement
All animal-related experimental procedures were conducted in strict adherence to the relevant guidelines and regulations for the use of animal samples as approved by the Institutional Animal Care and Use Committee (IACUC) at Kyungpook National University (No. KNU-2023-0280). Written informed consent was obtained from the cattle owners for the collection of blood samples.
Sample collection
During the summer of 2023, 376 blood samples were collected from the jugular vein of each bull reared on a single farm in the ROK. The cattle used for blood collection were 10–13 months old, male, clinically healthy, and had never grazed on pasture. To assess potential variation in infection rates based on the grazing experience of their dams, the animals were classified into two groups: those with grazing experience (n = 127) and those without (n = 249). Because some of the dams were pregnant, the owner did not want to collect blood from them. Moreover, some had already been sold to other farms and blood collection from cows cannot be conducted. For these reasons, this study focused on how the grazing experience of their dams affects bovine hemoplasma infection in their offspring. The difference in infection rate was also evaluated according to age: <1 year old (n = 346) and over 1 year old (n = 30). Heparin−anticoagulated blood was kept on ice and transferred to the laboratory at Kyungpook National University and then used for DNA extraction. A total of 2,690 houseflies were trapped on the same farm where the cattle were raised during the summer. This sample size was deemed sufficient balancing logistical feasibility and process. The collections were performed near the bulls using an entomological net. All fly specimens were promptly frozen (−80 °C) upon collection and transferred to the laboratory for species identification and hemoplasma detection. Other fly species were excluded.
DNA extraction and polymerase chain reaction
DNA was extracted from blood and fly samples using the AccuPrep® Genomic DNA Extraction Kit (Bioneer, Daejeon, ROK) according to the manufacturer’s instructions. For the blood samples, 200 µL was used. Regarding fly samples, 12 specimens were pooled to obtain approximately 25 mg, resulting in the extraction of 269 DNA. Subsequently, all DNA were stored at − 80 °C until further analysis. Extracted DNA were screened for M. wenyonii and Candidatus M. haemobos detection using a PCR assay targeting the 16S rRNA gene. The PCR methods for detecting hemoplasmas and identifying fly species were performed as previously described [8, 28]. Positive and negative controls were included in each PCR run. Two samples confirmed as positive in our previous study were used as positive controls [12] and distilled water was used for negative control. All PCR products were separated via electrophoresis on 1−1.5% agarose gels and then visualized by staining with ethidium bromide.
Sequencing and phylogenetic analyses
All PCR products were purified using the AccuPrep® PCR Purification Kit (Bioneer) according to the manufacturer’s instructions and then directly sequenced (Bioneer). The nucleotide sequences obtained from this study were aligned using the BioEdit software (version 7.2) and compared among themselves and then with other sequences from the GenBank database using the BLAST in National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST). The percentage of nucleotide sequences similarities between our sample sequences and the reference sequences was assessed in Geneious Prime software. Phylogenetic analysis of hemoplasmas identified in cattle and houseflies was conducted using the maximum-likelihood method implemented in MEGA11. The best substitution model used in this study was Hasegawa-Kishino-Yano (HKY) + G. The reliabilities of clusters were assessed using bootstrap values with 1,000 replicates. The nucleotide sequences obtained in this study were assigned the following accession numbers: PP544180−PP544184 and PP544185−PP544194 for M. wenyonii and Candidatus M. haemobos, respectively.
Statistical analysis
Statistical analysis was performed using the SPSS Statistics 29 software package for Windows (SPSS Inc, Chicago, IL, USA). Associations between bovine hemoplasma-positive samples and the grazing experience of dams were analyzed using the Chi-square test. Binary logistic regression analysis was used to determine the relationship between the prevalence of hemoplasma infections and the grazing experience of the dams. Odds ratio (OR) with 95% confidence intervals (CIs) were calculated to assess the likelihood of association. A P-value of < 0.05 (two-tailed) was considered statistically significant.
Results
The overall infection rate of hemoplasmas in cattle was 50.8% (191/376; 95% CI: 45.7−55.9). Among them, 70 (18.6%; 95% CI: 14.7−22.5) and 75 cattle (19.7%; 95% CI: 15.9−23.9) tested positive for M. wenyonii and Candidatus M. haemobos, respectively. Furthermore, 46 cattle (12.1%; 95% CI:8.9−15.5) were co−infected with both species (Table 1). The flies used in this study were identified as houseflies (Musca domestica) using species identification primers (Supplementary Fig. 1). The positive infection rate of bovine hemoplasmas in pooled houseflies was 7.4% (95% CI: 4.3−10.5). Candidatus M. haemobos DNA was most frequently detected (5.9%; 95% CI: 3.1−8.7), while the positivity rate of M. wenyonii was 0.7% (95% CI: -0.3−1.7). Co−infection with both species was 0.7% (95% CI: -0.3−1.7). Overall, Candidatus M. haemobos was more frequently identified in houseflies than M. wenyonii (Table 1).
Table 1.
The infection rate of hemoplasmas detected in cattle (n = 376) and pooled houseflies (n = 269)
| Pathogens | No. of positive (%) in cattle | No. of positive (%) in flies* |
|---|---|---|
| M. wenyonii | 70 (18.6%) | 2 (0.7%) |
| Candidatus M. haemobos | 75 (20.0%) | 16 (5.9%) |
| Co-infection | 46 (12.2%) | 2 (0.7%) |
| Total | 191 (50.8%) | 20 (7.4%) |
*To extract DNA from housefly samples, 12 pooled specimens were used
The associations between hemoplasma-positive samples and the age of cattle and the grazing experience of their dams were evaluated. As shown in Table 2, no significant difference in hemoplasma infections was noted according to the age of cattle. However, the grazing experience of the dams was statistically significant for both types of hemoplasma infections. Compared with cattle born from housed cows, the infection rates of M. wenyonii were significantly greater in cattle born to cows with grazing experience (χ2 = 3.86, P = 0.0495), whereas Candidatus M. haemobos infection rates were significantly lower in these cattle (χ2 = 18.38, P = 0.000). Additionally, the risk of being positive for M. wenyonii was 1.62-fold higher in cattle born to cows with grazing experience (95% CI: 1.03−2.55; P = 0.045), while the likelihood of being positive to Candidatus M. haemobos was 0.32-fold lower in these cattle (95% CI: 0.19−0.74; P = 0.000; Table 3).
Table 2.
Risk factors associated with hemoplasma infection
| Variables | M. wenyonii | Candidatus M. haemobos | ||
|---|---|---|---|---|
| No (%). of positive | χ2 (P-value) | No (%). of positive | χ2 (P-value) | |
| Age | ||||
| <1 year (n = 346) | 30.6% (106/346) | 0.094 (0.759) | 32.4% (112/346) | 0.071 (0.790) |
| ≥1 year (n = 30) | 33.3% (10/30) | 30.3% (9/30) | ||
| Grazing | ||||
| No (n = 249) | 27.3% (68/249) | 3.86 (0.049)* | 39.8% (99/249) | 18.38 (0.000)*** |
| Yes (n = 127) | 37.8% (48/127) | 17.3% (22/127) | ||
Grazing refers to the mother cows that were grazing before delivery
*P < 0.05, **P < 0.005, *** P < 0.001
Table 3.
Univariate logistic regression analysis for each hemoplasma infection according to grazing experience of the mother cows
| Grazing | M. wenyonii | Candidatus M. haemobos | ||||
|---|---|---|---|---|---|---|
| P-value | OR (95% CI) | P-value | OR (95% CI) | |||
| No (Ref.) | − | 1.00 | − | 1.00 | ||
| Yes | 0.045* | 1.62 (1.03−2.55) | 0.000** | 0.32 (0.19−0.74) | ||
*P < 0.05, **P < 0.005 vs. reference (Ref.)
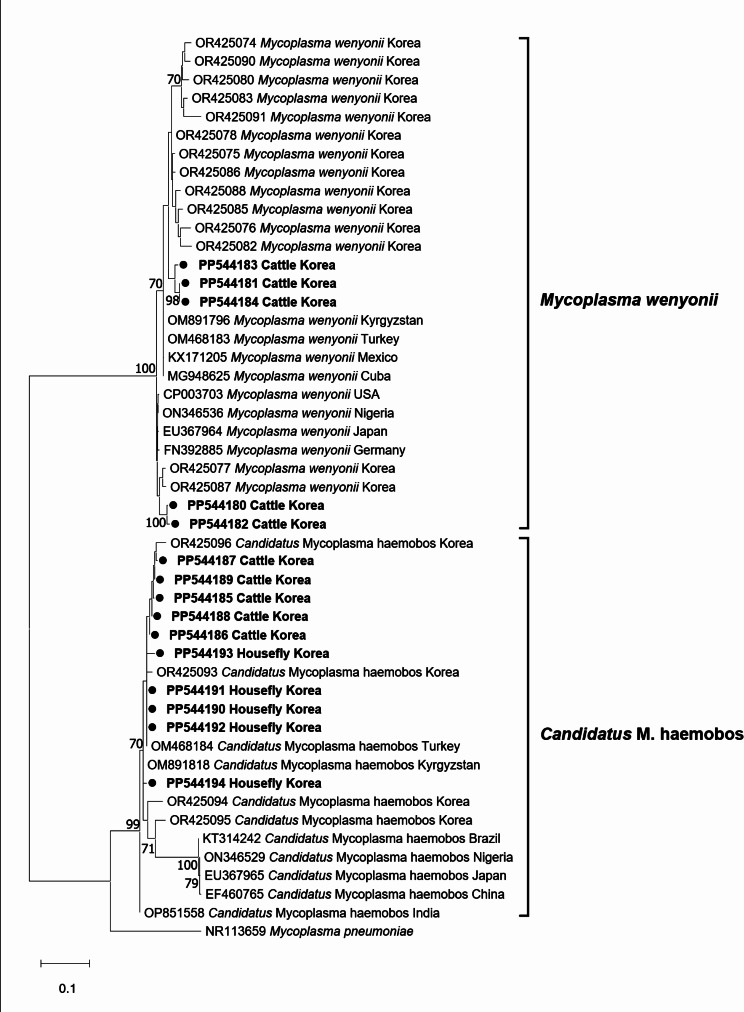
To investigate the genetic relationships of bovine hemoplasmas detected in cattle and houseflies, ten sequences (five for M. wenyonii and five for Candidatus M. haemobos) from cattle blood samples were selected and successfully obtained, whereas only five sequences for Candidatus M. haemobos were obtained from houseflies. Therefore, five M. wenyonii and ten Candidatus M. haemobos sequences were included in our analysis. According to the phylogenetic analysis based on the 16S rRNA gene, our sequences were classified into M. wenyonii and Candidatus M. haemobos (Fig. 1). The five M. wenyonii sequences identified from cattle exhibited 98.0−100% identity to each other and demonstrated 93.9−99.7% identity to those previously reported in the ROK [12], whereas these sequences showed 97.5−100% identity to cattle sequences reported from Cuba, Germany, Japan, Kyrgyzstan, Mexico, Nigeria, Turkey, and USA. Five sequences of Candidatus M. haemobos from houseflies showed 98.0−100% identity, whereas five from cattle exhibited 98.7−99.8% identity. Five sequences obtained from houseflies displayed 98.2−100% identity to cattle detected in this study, 99.5−100% identity to cattle in Brazil, 98.2−99.3% identity to cattle in China and Japan, 98.4−99.6% identity to cattle in India, and 98.8−100% identity to cattle in Kyrgyzstan, Nigeria, and Turkey.
Fig. 1.
Phylogenetic tree inferred using maximum-likelihood analysis, employing the HKY + G model. This tree represents the16S rRNA gene sequence of Mycoplasma wenyonii and Candidatus M. haemobos. Bootstrap values, expressed as a percentage of 1,000 replicates, are presented at the nodes. Values less than 70% are not shown. The scale bar represents the number of nucleotide substitutions per site. Sequences analyzed in this study are indicated by bold circle symbol
Discussion
In this study, M. wenyonii and Candidatus M. haemobos were detected in both cattle and houseflies. While bovine hemoplasmas are not typically considered pathogenic in the ROK, they may have persisted for a long time under various environmental conditions. Our findings revealed that the sequences of Candidatus M. haemobos from houseflies closely resembled those found in cattle. To our knowledge, this is the first study to identify the presence of bovine hemoplasmas in houseflies (Musca domestica).
Hemoplasmas infection in cattle has been frequently reported in several countries [4, 11–16]. However, there is little information on hemoplasmas in the ROK. Nevertheless, the occurrence of hemoplasmas in this study was relatively high compared to other countries [8, 10, 15, 21, 29]. Several studies have shown that hemoplasma infections are associated with agroclimatic zones, sex, age, and rearing system [10, 15, 30]. However, the cattle used in this study were all of one sex (male) with a single indoor raising system and differed only in age. Nevertheless, our findings revealed that bovine hemoplasma infections were not associated with the age of cattle, contrary to previous studies that indicated adult cattle (≥ 1 year) have a higher risk of hemoplasma infection than young animals [13, 31–33]. It is believed that other factors, such as farm management, hygiene, and environment, may influence hemoplasma infection. One limitation of this study is that all animals were of the same sex (male), and the number of samples was inconsistent between the two groups, preventing a comparison based on sex and age from being performed. The clinical importance and economic impact of hemoplasmas on the livestock industry have not been fully explored in the ROK; however, given their high infection rate, it should not be overlooked.
According to a recent study performed by our group, M. wenyonii was found to be more prevalent in grazing cattle than Candidatus M. haemobos [12]. However, this study showed a slightly higher prevalence of Candidatus M. haemobos (20%) than M. wenyonii (18.6%), which differs from previous findings [12]. The most significant difference between the two studies is that the cattle in this study had not experienced grazing and were raised solely in the barn. The grazing experience of their dams may have influenced M. wenyonii and Candidatus M. haemobos infections. Our results showed that in cattle born from cows with grazing experience, M. wenyonii infection was high, whereas Candidatus M. haemobos infection was low. Given that the dams had grazed prior to delivery, the possibility that they were infected by ticks cannot be excluded. Thus, M. wenyonii could have been transmitted to the dams by ticks and passed the infection to their offspring via the placenta. In contrast, cattle born from cows with no grazing experience demonstrated a high positive rate of Candidatus M. haemobos infection, suggesting that vectors other than ticks may be involved in its transmission. Previous studies suggest that vertical transmission could be a possible route for bovine hemoplasmas [14, 18, 19]. However, another limitation of the current study is that we could not examine hemoplasma infection from their dams; hence, we cannot confirm whether M. wenyonii and Candidatus M. haemobos are transmitted by transplacental infection. Therefore, further studies are warranted to examine offspring and their dams to elucidate vertical transmission.
Bovine hemoplasma infection is a vector-borne disease; however, the primary vectors responsible for transmitting bovine hemoplasmas remain unclear in the ROK. Several studies have reported the identification of M. wenyonii DNA in ticks, lice, flies, and mosquitoes [9, 11, 17, 34–36], whereas the presence of Candidatus M. haemobos was confirmed in flies and Rhipicephalus microplus and Haemaphysalis longicornis ticks [20, 23]. However, experimental proof of bovine hemoplasma transmission by arthropods is rare. Compared to findings from previous literature, our findings demonstrated that Candidatus M. haemobos DNA was predominantly detected in houseflies, with identical sequence homology between cattle and fly DNA. Additionally, M. wenyonii DNA was also identified in houseflies, although its sequence could not be obtained, possibly owing to the small amount of M. wenyonii DNA present in houseflies. To our knowledge, this is the first time Candidatus M. haemobos DNA has been identified in houseflies. The infection rate of Candidatus M. haemobos was significantly decreased in cattle born to cows with grazing experience, whereas its infection rate was 2.3-fold higher in cattle born to cows with no grazing experience. Furthermore, considering that Candidatus M. haemobos DNA was detected in houseflies and that the bulls used in this experiment were raised in the barn, it is possible that Candidatus M. haemobos infection may be transmitted to cattle by flies rather than ticks. Collectively, these findings indicate that the vectors responsible for transmitting each hemoplasma may differ.
Phylogenetic analysis revealed that the sequences of Candidatus M. haemobos from cattle and houseflies shared high similarity with those reported in other countries. The infection rate of Candidatus M. haemobos was low in houseflies; however, the genomic relatedness of Candidatus M. haemobos DNA sequences from cattle detected at the same farm suggests that houseflies can serve as a vector for Candidatus M. haemobos. According to a previous study, M. arginini was identified in houseflies, and their role as carriers of Mycoplasma spp. was also demonstrated [27]. Although M. arginini and Candidatus M. haemobos cause different clinical symptoms in cattle, our results support that houseflies may act as possible vectors for Mycoplasma spp. At this point, how houseflies transmit disease to cattle remains unclear, as they are not blood-sucking vectors. However, houseflies may use mucous membranes, such as the nasal area, to transmit the bacteria [37]. Given that flies can travel 7–32 km daily, keeping them away from cattle is crucial [38, 39]. Most importantly, houseflies comprised 100% of all Diptera collected and identified on this farm. In the cattle farm environment, feces and manure attract houseflies, making them carriers of many pathogens [40]. These findings suggest that houseflies may be potential vectors of Candidatus M. haemobos and may influence livestock health. Therefore, our findings underscore the importance of managing and controlling houseflies through environmental improvement practices to protect animal health.
Conclusion
The present study has established the first molecular evidence of Candidatus M. haemobos in houseflies. Our findings suggest that bovine hemoplasma infection is high in the ROK. The sequences of Candidatus M. haemobos identified from houseflies showed high genomic relatedness to cattle detected at the same farm. Additionally, this study provides new insights into the vector transmission of bovine hemoplasmas in the ROK. Further studies are warranted to identify the transmission routes of hemoplasma infection through ocular or nasal discharges in cattle to evaluate the role of houseflies as vectors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to thank all the veterinarians who participated in the study as well as all the colleagues who contributed to sample collection and sample preparation.
Abbreviations
- 16S rRNA
16S ribosomal RNA
- Candidatus M. haemobos
Candidatus Mycoplasma haemobos
- CO1
cytochrome c oxidase subunit 1
- M. wenyonii
Mycoplasma wenyonii
- PCR
polymerase chain reaction
- ROK
Republic of Korea
Author contributions
KSC designed the study. MHP and SJC performed laboratory analysis. YK performed sampling. HCC, YJP, MJJ, and JS analyzed data. MHP, SJC, and KSC wrote the manuscript. All authors read and approved the final manuscript.
Funding
This research was supported by the National Research Foundation of Korea (NRF), which is funded by the Mid-Career Research Program (grant No. NRF-2021R1A2C1011579).
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request. The nucleotide sequences obtained in this study were assigned the following accession numbers: PP544180-PP544184 for M. wenyonii and PP544185-PP544194 for Candidatus M. haemobos, and are available in the repository, WEB LINK (https://www.ncbi.nlm.nih.gov/nuccore/PP544180.1−PP544184.1).
Declarations
Ethical approval and consent to participate
This study was reviewed and approved by the ethics committee of Institutional Animal Care and Use Committee (IACUC) at Kyungpook National University (approval number: KNU-2023-0280). Written informed consent was obtained from the cattle owners for the collection of blood samples.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Min-Ho Park, Seok-Jin Cho and Youngjun Kim contributed equally to this work.
References
- 1.Messick JB. Hemotrophic mycoplasmas (hemoplasmas): a review and new insights into pathogenic potential. Vet Clin Pathol. 2004;33:2–13. 10.1111/j.1939-165x.2004.tb00342.x. [DOI] [PubMed] [Google Scholar]
- 2.Maggi RG, Mascarelli PE, Balakrishnan N, Rohde CM, Kelly CM, Ramaiah L, et al. Candidatus Mycoplasma haemomacaque and Bartonella quintana bacteremia in cynomolgus monkeys. J Clin Microbiol. 2013;51:1408–11. 10.1128/JCM.03019-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hofmann-Lehmann R, Meli ML, Dreher UM, Gonczi E, Deplazes P, Braun U, et al. Concurrent infections with vector-borne pathogens associated with fatal hemolytic anemia in a cattle herd in Switzerland. J Clin Microbiol. 2004;42:3775–80. 10.1128/JCM.42.8.3775-3780.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tagawa M, Matsumoto K, Inokuma H. Molecular detection of Mycoplasma wenyonii and ‘Candidatus Mycoplasma haemobos’ in cattle in Hokkaido. Japan Vet Microbiol. 2008;132:177–80. 10.1016/j.vetmic.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 5.Smith JA, Thrall MA, Smith JL, Salman MD, Ching SV, Collins JK. Eperythrozoon wenyonii infection in dairy cattle. J Am Vet Med Assoc. 1990;196:1244–50. 10.2460/javma.1990.196.08.1244. [PubMed] [Google Scholar]
- 6.Hoelzle K, Winkler M, Kramer MM, Wittenbrink MM, Dieckmann SM, Hoelzle LE. Detection of Candidatus Mycoplasma haemobos in cattle with anaemia. Vet J. 2011;187:408–10. 10.1016/j.tvjl.2010.01.016. [DOI] [PubMed] [Google Scholar]
- 7.Baggenstos R, Wenzinger B, Meli ML, Hofmann-Lehmann R, Knubben-Schweizer G. Haemoplasma infection in a dairy cow. Tierarztl Prax Ausg G Grosstiere Nutztiere. 2012;40:397–400. 10.5167/uzh-69252. [PubMed] [Google Scholar]
- 8.Erol U, Sahin OF, Altay K. Molecular prevalence of bovine hemoplasmosis in Turkey with first detection of Mycoplasma wenyonii and Candidatus Mycoplasma haemobos in cattle and water buffalo. Vet Res Commun. 2023;47:207–15. 10.1007/s11259-022-09943-2. [DOI] [PubMed] [Google Scholar]
- 9.Neimark H, Johansson KE, Rikihisa Y, Tully JG. Proposal to transfer some members of the genera Haemobartonella and Eperythrozoon to the genus Mycoplasma with descriptions of ‘Candidatus Mycoplasma haemofelis’, ‘Candidatus Mycoplasma haemomuris’, ‘Candidatus Mycoplasma haemosuis’ and ‘Candidatus Mycoplasma wenyonii’. Int J Syst Evol Microbiol. 2001;51:891–9. 10.1099/00207713-51-3-891. [DOI] [PubMed] [Google Scholar]
- 10.Murugesan AC, Kumaragurubaran K, Gunasekaran K, Murugasamy SA, Arunachalam S, Annamalai R, et al. Molecular detection of hemoplasma in animals in Tamil Nadu, India and hemoplasma genome analysis. Vet Res Commun. 2024;48:955–68. 10.1007/s11259-023-10263-2. [DOI] [PubMed] [Google Scholar]
- 11.Thongmeesee K, Chonglomkrod B, Srisakdi C, Saributr M, Suksai P, Kamkong P, et al. Molecular detection of Mycoplasma wenyonii and its closely related hemotropic Mycoplasma sp. in blood-sucking flies from a buffalo farm in Chachoengsao province, Thailand. Acta Trop. 2022;235:106647. 10.1016/j.actatropica.2022.106647. [DOI] [PubMed] [Google Scholar]
- 12.Kim Y, Kim H, Choi JH, Cho HC, Ji MJ, Park YJ, et al. Preliminary report of Mycoplasma Wenoynii and Candidatus Mycoplasma haemobos infection in Korean native cattle. BMC Vet Res. 2024;20:121. 10.1186/s12917-024-03976-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diaz-Sanchez AA, Corona-Gonzalez B, Meli ML, Alvarez DO, Canizares EV, Rodriguez OF, et al. First molecular evidence of bovine hemoplasma species (Mycoplasma spp.) in water buffalo and dairy cattle herds in Cuba. Parasit Vectors. 2019;12:78. 10.1186/s13071-019-3325-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niethammer FM, Ade J, Hoelzle LE, Schade B. Hemotrophic mycoplasma in Simmental cattle in Bavaria: prevalence, blood parameters, and transplacental transmission of ‘Candidatus Mycoplasma haemobos’ and Mycoplasma wenyonii. Acta Vet Scand. 2018;60:74. 10.1186/s13028-018-0428-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamani J, Shand M, Shekaro A, Laminu B, Toyin O, Abasiama MS, et al. Mycoplasma wenyonii and Candidatus Mycoplasma Haemobos in pastoralists cattle in Nigeria. Acta Parasitol. 2023;68:430–8. 10.1007/s11686-023-00683-0. [DOI] [PubMed] [Google Scholar]
- 16.Schambow RA, Poulsen K, Bolin S, Krahn D, Norby B, Sockett D, et al. Apparent prevalence of Mycoplasma wenyonii, Candidatus Mycoplasma haemobos, and bovine leukemia virus in Wisconsin and Michigan dairy cattle herds. JDS Commun. 2021;2:261–6. 10.3168/jdsc.2020-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hornok S, Micsutka A, Meli ML, Lutz H, Hofmann-Lehmann R. Molecular investigation of transplacental and vector-borne transmission of bovine haemoplasmas. Vet Microbiol. 2011;152:411–4. 10.1016/j.vetmic.2011.04.031. [DOI] [PubMed] [Google Scholar]
- 18.Sasaoka F, Suzuki J, Hirata T, Ichijo T, Furuhama K, Harasawa R, et al. Vertical transmission of Mycoplasma wenyonii in cattle, supported by analysis of the ribonuclease P RNA gene - short communication. Acta Vet Hung. 2015;63:271–4. 10.1556/004.2015.025. [DOI] [PubMed] [Google Scholar]
- 19.Girotto-Soares A, Soares JF, Bogado ALG, de Macedo CAB, Sandeski LM, Garcia JL, et al. Candidatus Mycoplasma haemobos’: transplacental transmission in dairy cows (Bos taurus). Vet Microbiol. 2016;195:22–4. 10.1016/j.vetmic.2016.08.020. [DOI] [PubMed] [Google Scholar]
- 20.Shi H, Duan L, Liu F, Hu Y, Shi Z, Chen X, et al. Rhipicephalus (Boophilus) microplus ticks as reservoir and vector of ‘Candidatus Mycoplasma haemobos’ in China. Vet Parasitol. 2019;274:108929. 10.1016/j.vetpar.2019.108929. [DOI] [PubMed] [Google Scholar]
- 21.Ferrari LDR, Hassan-Kadle AA, Collere FCM, Coradi VS, Ibrahim AM, Osman AM, et al. Hemoplasmas and ticks in cattle from Somalia. Acta Trop. 2022;236:106696. 10.1016/j.actatropica.2022.106696. [DOI] [PubMed] [Google Scholar]
- 22.Ybanez AP, Ybanez RHD, Armonia RKM, Chico JKE, Ferraren KJV, Tapdasan EP, et al. First molecular detection of Mycoplasma wenyonii and the ectoparasite biodiversity in dairy water buffalo and cattle in Bohol, Philippines. Parasitol Int. 2019;70:77–81. 10.1016/j.parint.2019.02.004. [DOI] [PubMed] [Google Scholar]
- 23.Arendt M, Stadler J, Ritzmann M, Ade J, Hoelzle K, Hoelzle LE. Hemotrophic mycoplasmas-vector transmission in livestock. Microorganisms. 2024;12:7. 10.3390/microorganisms12071278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bertolini AB, Thyssen PJ, Brandao PE, Prado AM, Silva SOD, Mioni MDR, et al. Flies (Insecta, Diptera) collected in the environment of dairy farms as carriers of rotavirus A and betacoronavirus. J Appl Microbiol. 2023;134:lxad020. 10.1093/jambio/lxad020. [DOI] [PubMed] [Google Scholar]
- 25.Issa R. Musca domestica acts as transport vector hosts. Bull Natl Res Cent. 2019;43:73. 10.1186/s42269-019-0111-0. [Google Scholar]
- 26.Hamdan M, Kamalanathan T, Iqbal A, Gnanaprakasam AR, Shajahan S, Alsadeq MH, et al. Kdr kdr mutations and deltamethrin resistance in houseflies in Abu Dhabi, UAE. Parasit Vectors. 2024;17:47. 10.1186/s13071-024-06128-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gioia G, Severgnini M, Cremonesi P, Castiglioni B, Freeman J, Sipka A, et al. Genomic characterization of Mycoplasma arginini isolated from a housefly on a dairy farm and comparison with isolates from bovine milk and lung tissue. Microbiol Spectr. 2023;11:e0301022. 10.1128/spectrum.03010-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park SH, Zhang Y, Piao H, Yu DH, Jeong HJ, Yoo GY, et al. Use of cytochrome c oxidase subunit i (COI) nucleotide sequences for identification of the Korean Luciliinae fly species (Diptera: Calliphoridae) in forensic investigations. J Korean Med Sci. 2009;24:1058–63. 10.3346/jkms.2009.24.6.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santos NJR, Brito DRB, Abate HL, Paixao SF, Soares EDS, Vieira T, et al. Hemotropic mycoplasmas infection in water buffaloes (Bubalus bubalis) from Northeastern Brazil. Comp Immunol Microbiol Infect Dis. 2018;56:27–9. 10.1016/j.cimid.2017.12.003. [DOI] [PubMed] [Google Scholar]
- 30.Byamukama B, Tumwebaze MA, Tayebwa DS, Byaruhanga J, Angwe MK, Li J, et al. First molecular detection and characterization of hemotropic Mycoplasma species in cattle and goats from Uganda. Anim (Basel). 2020;10:1624. 10.3390/ani10091624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Congli Y, Hong Y, Zhonghai Z, Zhibiao Y, Li C, Jianguo Z, et al. Prevalence of Mycoplasma wenyonii infection on seven dairy farms in Shanghai, China. Thai J Vet Med. 2011;41:179–84. 10.56808/2985-1130.2294. [Google Scholar]
- 32.Girotto A, Zangirolamo AF, Bogado AL, Souza AS, da Silva GC, Garcia JL, et al. Molecular detection and occurrence of ‘Candidatus Mycoplasma haemobos’ in dairy cattle of Southern Brazil. Rev Bras Parasitol Vet. 2012;21:342–4. 10.1590/S1984-29612012000300034. [DOI] [PubMed] [Google Scholar]
- 33.Tagawa M, Ybanez AP, Matsumoto K, Yokoyama N, Inokuma H. Prevalence and risk factor analysis of bovine hemoplasma infection by direct PCR in Eastern Hokkaido, Japan. J Vet Med Sci. 2012;74:1171–6. 10.1292/jvms.12-0118. [DOI] [PubMed] [Google Scholar]
- 34.Song Q, Wang L, Fang R, Khan MK, Zhou Y, Zhao J. Detection of Mycoplasma wenyonii in cattle and transmission vectors by the loop-mediated isothermal amplification (LAMP) assay. Trop Anim Health Prod. 2013;45:247–50. 10.1007/s11250-012-0197-y. [DOI] [PubMed] [Google Scholar]
- 35.Mohd Hasan LI, Kho KL, Koh FX, Hassan Nizam QN, Tay ST. Molecular evidence of hemoplasmas in Malaysian cattle and ticks. Trop Biomed. 2017;34:668–74. [PubMed] [Google Scholar]
- 36.Stevanovic O, Jurkovic D, Polkinghorne A, Celes A, Ilic T, Dimitrijevic S, et al. Molecular detection of Babesia divergens and Mycoplasma wenyonii infection in cattle from Bosnia and Herzegovina. Parasitol Res. 2020;119:1423–7. 10.1007/s00436-020-06630-6. [DOI] [PubMed] [Google Scholar]
- 37.Espinosa-Gongora C, Dahl J, Elvstrom A, van Wamel WJ, Guardabassi L. Individual predisposition to Staphylococcus aureus colonization in pigs on the basis of quantification, carriage dynamics, and serological profiles. Appl Environ Microbiol. 2015;81:1251–6. 10.1128/AEM.03392-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nazni WA, Luke H, Wan Rozita WM, Abdullah AG, Sa’diyah I, Azahari AH, et al. Determination of the flight range and dispersal of the housefly, Musca domestica (L.) using mark release recapture technique. Trop Biomed. 2005;22:53–61. [PubMed] [Google Scholar]
- 39.Nayduch D, Neupane S, Pickens V, Purvis T, Olds C. Houseflies are underappreciated yet important reservoirs and vectors of microbial threats to animal and human health. Microorganisms. 2023;11:583. 10.3390/microorganisms11030583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alegbeleye OO, Singleton I, Sant’Ana AS. Sources and contamination routes of microbial pathogens to fresh produce during field cultivation: a review. Food Microbiol. 2018;73:177–208. 10.1016/j.fm.2018.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request. The nucleotide sequences obtained in this study were assigned the following accession numbers: PP544180-PP544184 for M. wenyonii and PP544185-PP544194 for Candidatus M. haemobos, and are available in the repository, WEB LINK (https://www.ncbi.nlm.nih.gov/nuccore/PP544180.1−PP544184.1).