Abstract
Background
Although the representative treatment for colorectal cancer (CRC) is radiotherapy, cancer cells survive due to inherent radioresistance or resistance acquired after radiation treatment, accelerating tumor malignancy and causing local recurrence and metastasis. However, the detailed mechanisms of malignancy induced after radiotherapy are not well understood. To develop more effective and improved radiotherapy and diagnostic methods, it is necessary to clearly identify the mechanisms of radioresistance and discover related biomarkers.
Methods
To analyze the expression pattern of miRNAs in radioresistant CRC, sequence analysis was performed in radioresistant HCT116 cells using Gene Expression Omnibus, and then miR-1226-5p, which had the highest expression in resistant cells compared to parental cells, was selected. To confirm the effect of miR-1226-5 on tumorigenicity, Western blot, qRT-PCR, transwell migration, and invasion assays were performed to confirm the expression of EMT factors, cell mobility and invasiveness. Additionally, the tumorigenic ability of miR-1226-5p was confirmed in organoids derived from colorectal cancer patients. In CRC cells, IRF1, a target gene of miR-1226-5p, and circSLC43A1, which acts as a sponge for miR-1226-5p, were discovered and the mechanism was analyzed by confirming the tumorigenic phenotype. To analyze the effect of tumor-derived miR-1226-5p on macrophages, the expression of M2 marker in co-cultured cells and CRC patient tissues were confirmed by qRT-PCR and immunohistochemical (IHC) staining analyses.
Results
This study found that overexpressed miR-1226-5p in radioresistant CRC dramatically promoted epithelial-mesenchymal transition (EMT), migration, invasion, and tumor growth by suppressing the expression of its target gene, IRF1. Additionally, we discovered circSLC43A1, a factor that acts as a sponge for miR-1226-5p and suppresses its expression, and verified that EMT, migration, invasion, and tumor growth are suppressed by circSLC43A1 in radioresistant CRC cells. Resistant CRC cells-derived miR-1226-5p was transferred to macrophages and contributed to tumorigenicity by inducing M2 polarization and secretion of TGF-β.
Conclusions
This study showed that the circSLC43A1/miR-1226-5p/IRF1 axis is involved in radioresistance and cancer aggressiveness in CRC. It was suggested that the discovered signaling factors could be used as potential biomarkers for diagnosis and treatment of radioresistant CRC.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12967-024-05797-1.
Keywords: Radioresistance of colorectal cancer, miR-1226-5p, circSLC43A1, IRF1, M2 macrophage, Tumor malignancy, TGF-β
Background
Colorectal cancer (CRC) is a malignant tumor that occurs in the colon and rectum and is the third most common type of cancer [1]. Treatment approaches for colorectal cancer typically include a combination of surgery, radiotherapy, chemotherapy, and targeted therapy [2, 3]. Intrinsic resistance of cancer cells or resistance acquired after radiotherapy can lead to survival of cancer cells, accelerate malignant progression, and potentially lead to local recurrence and metastasis [4, 5]. Recent technological advances in radiotherapy (RT) have enabled more accurate irradiation and reduced side effects on normal tissues [6], but acquisition of radioresistance and tumor recurrence still remain major challenges. Recent research trends on the acquisition mechanism of resistance focus on the relationship between tumors and the microenvironment, especially immune cells.
Solid tumors are influenced by the tumor microenvironment, including infiltration of immune cells, angiogenesis, and fibroblast proliferation [7–9]. After radiotherapy, continuous interaction between tumor cells and the microenvironment induces an inflammatory response that promotes the progression of tumor malignancy and resistance [7, 8, 10]. Macrophages are one of the most important immune cells within the tumor microenvironment and are characterized by high plasticity [11]. Macrophages have the ability to differentiate into two types: pro-inflammatory (M1) macrophages and anti-inflammatory (M2) macrophages, which perform distinct functions in cancer progression [12]. M1 macrophages mediate pathogen resistance and release inflammatory cytokines [13]. M2 macrophages display a pro- tumor phenotype and are involved in EMT, angiogenesis and metastasis [14]. Vascular endothelial growth factor (VEGF) and platelet-derived growth factor (PDGF) are secreted from M2 macrophages to promote the creation of an environment for tumor development, and CCL22 is secreted to induce the infiltration of regulatory T cells and suppress anti-tumor immunity. These processes ultimately increase tumor malignancy and radioresistance [7, 15]. Previous studies have reported that macrophages interact with tumor cells and other immune factors to influence CRC progression [11]. However, research on the mechanism of tumor malignancy according to M2 polarization of macrophages in radioresistant CRC is not fully understood. Therefore, the discovery of related signaling factors by analyzing the tumor malignancy mechanism of radioresistant CRC and macrophages will be useful as a target for diagnosis and treatment to overcome radioresistance.
MicroRNAs (miRNAs) are non-coding RNAs that are 23 nucleotides in length and function to suppress post-transcriptional gene expression by binding to the RNA 3'UTR of target factors [16]. In CRC, miRNAs are known to be involved in the regulation of proliferation, metastasis, angiogenesis, autophagy, apoptosis, and radioresistance by regulating the expression of various targets [16]. Oncogenic miRNAs are involved in regulating interactions between cancer cells and other cells, leading to immune evasion and therapeutic resistance [17]. Since miRNAs are easily transferred between cancer cells and surrounding cells, it has been reported that tumor-derived miRNAs translocate to macrophages and induces macrophage polarization, thereby inducing tumor malignancy mechanisms [18, 19]. For example, it has been reported that miR-143-3p derived from esophageal squamous cell carcinoma induces M2 polarization of macrophages and induces radioresistance [20]. In addition, miR-106b-5p in colorectal cancer activates PI3K/Akt/mTOR signaling to induce M2 polarization, thereby increasing tumor cell radioresistance, migration, invasion, and metastasis [21]. These studies suggest that tumor-derived miRNAs may be involved in radioresistance by altering macrophage polarization.
This study revealed that miR-1226-5p overexpressed in radioresistant CRC cells induces the mechanism of tumor malignancy by suppressing the expression of the target factor, IRF1, and promoting the activation of M2 macrophages and the secretion of TGF-β, thereby inducing radioresistance. These results improved our understanding of the mechanisms of radioresistance-induced cancer malignancy. In addition, the factors discovered through the mechanism analysis of miR-1226-5p in radioresistant CRC were verified at the cellular level, animal models, patient-derived organoids, and specimens showing their potential as biomarkers for the diagnosis and treatment of patients with radioresistant CRC.
Methods
Patient specimens
Tissue and plasma from patients with colorectal cancer were Institutional Review Board (IRB) approved in Korea Institute of Radiological and Medical Sciences (KIRAMS). The plasma specimens used for this study were distributed by the Korea Institute of Radiological and Medical Sciences (KIRAMS) Radiation Biobank (KRB) in Republic of Korea (KRB-2021-I002, KRB-2023-I001). The bio-specimens and data used in this study were provided by the Radiation Tissue Resources Bank of Korea Cancer Center Hospital (TB-2021-02-B/P50, C/P50, L/P40). For human cancer tissues, samples were obtained from patients with colorectal cancer whose primary tumor site was the colon or rectum. In the miR-1226-5p expression analysis, no classification was made according to patient information, and experiments were performed on adjacent normal and tumor tissues. Human tissue specimens used for tissue staining were classified according to the patient's radiotherapy status. Human plasma samples were classified according to the patient's radiotherapy status and analyzed for miR-1226-5p expression.
Cell culture
HCT116 (MSI, CIMP-positive), SW480 (MSS, CIMP-negative), and THP-1 cells were purchased from Korean Cell Line Bank (KCLB, Korea). The identity of the cell lines was confirmed by short tandem repeat (STR) profiling, and the analysis was provided by KCLB (Korea). In addition, the cells used in this study were confirmed to be free of mycoplasma contamination. HCT116, SW480, and THP-1 cells were cultured in RPMI 1640 media (Corning, NY, USA). RPMI 1640 media used for cell culture were supplemented with 10% Fetal Bovine Serum (FBS, Corning, NY, USA) and 1% penicillin streptomycin (Corning, NY, USA). Cells were cultured in an incubator at 37 °C and 5% CO2. miR-1226-5p mimic and miR-1226-5p inhibitor were synthesized by Genolution Inc. (Korea). All siRNAs against TGF-β and STAT6 purchased from Santa Cruz Biotechnology (TX, USA). The IRF1 gene was inserted into the pClneoFH vector. The circSLC43A1 was inserted into the pcDNA3.1( +) CircRNA Mini Vector. The primers used for cloning are listed in Table 1. Plasmid, miRNA, and siRNA were transfected into cells using Lipofectamine 2000 reagent (Invitrogen, MA, USA) following the manufacturer’s instructions.
Table 1.
Primer sequences used in experiments
| Primer for qRT-PCR | Sequences (5'–3') |
|---|---|
| miR-1226-5p | GTGAGGGCATGCAGGCCTG |
| miR-3529-3p | AACAACAAAATCACTAGTCTTCCA |
| miR-522-3p | AAAATGGTTCCCTTTAGAGTGT |
| miR-4706 | GGAGGAAGTGGGCGCTGCTT |
| miR-3184-3p | AAAGTCTCGCTCTCTGCCCCTCA |
| miR-1237-3p | TCCTTCTGCTCCGTCCCC |
| RASSF1 | F: TGTACTTGCGGAAGCTGTTG |
| R: CCTTCAGGACAAAGCTCAGG | |
| PTEN | F: AGACCATAACCCACCACAGC |
| R: TTCGTCCCTTTCCAGCTTTA | |
| GADD45a | F: GGAGGAAGTGCTCAGCAAAG |
| R: CAGGCACAACACCACGTTAT | |
| IRF1 | F: AGGGGAAAAGGAGCCAGATC |
| R: CCTCCTCGATATCTGGCAGG | |
| circSLC43A1 | F: CCTCATCAGTGCTGTGTTCG |
| R: AGGTTGAGCATCTCGTCCTG | |
| circPDK3 | F: CATTACAAGACCACGCCTGA |
| R: GTGTTAGCCAGCCGCACAG | |
| circPPEF1 | F: TGGTCCTCGGCTACAATTTC |
| R: ATTTGGCCTTGTTCATCAGC | |
| circPHF8 | F: TGATGATGATGACCCTGCTT |
| R: TGCTCCAGCACTCAAAGAGA | |
| SLC43A1 | F: ATCAGTGCCTTCACCCTGAC |
| R: ACAATGGTGTGCAGGACAAA | |
| CD163 | F: CCAGTCCCAAACACTGTCCT |
| R: CACTCTCTATGCAGGCCACA | |
| CD206 | F: ACGGACTGGGTTGCTATCAC |
| R: TGATCCCCAAAAGTGTGTCA | |
| CD11b | F: ACGTAAATGGGGACAAGCTG |
| R: GATCCTGAGGTTCCGTGAAA | |
| ARG1 | F: GGAGACCACAGTTTGGCAAT |
| R: CCACTTGTGGTTGTCAGTGG | |
| IL-8 | F: TTCTGCAGCTCTGTGTGAAG |
| R: TTGGGGTGGAAAGGTTTGGA | |
| IL-6 | F: TACATCCTCGACGGCATCTC |
| R: AGTGCCTCTTTGCTGCTTTC | |
| IL-1β | F: GGACAAGCTGAG GAAGATGC |
| R: TCGTTATCCCATGTGTCGAA | |
| VEGF | F: GACAGACAGACAGACACCGCC |
| R: GAACAGCCC AGAAGTTGGACG | |
| CCL17 | F: AGCCATTCCCCTTAGAAAGC |
| R: CTGCCCTGCACAGTTACAAA | |
| CCL22 | F: CGCGTGGTGAAACACTTCTA |
| R: ATCTTCACCCAGGGCACTCT | |
| CCL24 | F: GCCTTCTGTTCCTTGGTGTC |
| R: GACCACTCGGTTCTCAGGAA | |
| TGF-β | F: GTGGAAACCCACAACGAAAT |
| R: CGGAGCTCTGATGTGTTGAA | |
| GAPDH | F: CATCTCTGCCCCCTCTGCTGA |
| R: GGATGACCTTGCCCACAGCCT | |
| Primer for cloning | Sequences (5'–3') |
| IRF1 | F: ATGCCCATCACTCGGATGCG |
| R: CCTGCTACGGTGCACAGGGA | |
| circSLC43A1 | F: CTGAGAGCAGCACCAACACC |
| R: CCAGAAGGGCTCTCCTTTCAGG |
Organoid culture
The patients had a clinical stage of T3N + prior to treatment. These patients received neoadjuvant chemoradiation (NCRT) and were evaluated using MRI and PET scans before and after treatment. The patients were classified into radiosensitive responses (referred to as RS) and radioresistant responses (referred to as RR) based on the tumor regression grade (TRG) after NCRT [22]. Methods for isolation and culture of patient-derived organoids were previously described [22]. The patient-derived organoid was cultured in DMEM/F12 media contained B27 (Gibco, NY, USA), N-acetyl cysteine (United States Pharmacopeia, MD, USA), human epidermal growth factor (BioVision, MA, USA), human Noggin (Peprotech, NJ, USA), gastrin (Sigma-Aldrich, MO, USA), and A83-01 (BioVision, MA, USA).
Colony formation assay
Cells were seeded into 60 mm dish at 100, 300, 1000, and 5000 cells/dish. After 16 h, cells were exposed to 3, 5, and 7 Gy radiation. It is stained with crystal violet 10–14 days after radiation exposure. Count the number of colonies and plot a survival curve.
WST-8 assay
The indicated cells were seeded into 96-well plates at 4000 cells/well. Cells were exposed to γ-rays with 137Cs γ-rays source with a dose rate of 3.81 Gy/min. After irradiation, the WST-8 cell viability assay kit (Dyne, Korea) was used to measure cell viability according to the manufacturer’s instructions. Absorbance is measured at 450 nm using a microplate reader.
Transwell-migration and invasion assays
For the transwell migration assay, the bottom of the transwell was coated with 0.2% gelatin. Cells were seeded on the transwell and filled the lower chamber with complete media. After 16 h, the transwell was fixed with methanol and stained with crystal violet. For the invasion assay, a transwell with 8 μm pores (Corning, NY, USA) was coated with matrigel (Corning, NY, USA). The cells were seeded on the matrigel-coated transwell and filled the lower chamber with complete media. After 16 h, the transwell was fixed with methanol and stained with crystal violet.
Western blot analysis
Proteins were obtained from cells by using a lysis buffer (10 mM Tris–HCl with pH 7.4, 150 mM NaCl, 1% NP-40, 1 mM EDTA, 0.1% SDS) that included protease inhibitors and phosphatase inhibitors (Roche, Switzerland). Samples were separated by electrophoresis on SDS-PAGE gel and then transferred to PVDF membranes. The membrane was incubated overnight at 4 °C using the following antibodies; E-cadherin (610182) was purchased from BD bioscience (NJ, USA). SNAIL (#3879), ZEB1 (#3396), p-STAT6 (#56554), and IRF1 (#8478) were obtained from Cell signaling (MA, USA). TWIST (ab50887), N-cadherin (ab18203), and γ-H2AX (ab11174) were purchased from Abcam (UK). STAT6 (sc-374021) and β-actin (sc-47778) were obtained from Santa Cruz Biotechnology (TX, USA). Horseradish peroxidase (HRP) conjugated secondary antibodies (Bio-rad, CA, USA) were incubated for 1 h in room temperature.
Total RNA isolation and quantitative real-time PCR
RNA was extracted from cells, tissue and plasma using Trizol reagent (Qiagen, Germany). Complementary DNA (cDNA) was synthesized according to the manufacturer’s instructions of the SensiFAST™ cDNA Synthesis Kit (Bioline, OH, USA) and Mir-X™ miRNA First-Strand Synthesis Kit (Takara, JAPAN). Real-time PCR was performed with Power SYBR™ Green PCR Master Mix (Applied Biosystems, MA, USA). Primers used for real-time PCR are listed in Table 1.
Luciferase reporter assay
The binding site of miR-1226-5p in IRF1 3’UTR or circSLC43A1 was inserted into the vector, pmirGLO dual-luciferase miRNA target expression vector (Promega, WI, USA). Luciferase vectors, miR-1226-5p mimics and renilla were co-transfected into HCT116 cells. After 24 h, luciferase activity was measured using the dual-luciferase reporter assay system (Promega, WI, USA).
AGO2-RNA immunoprecipitation
This experiment was performed using RIP-assay kit for microRNA (MBL International Corporation, MA, USA) according to the manufacturer’s instructions. Cell lysates were obtained from HCT116 cells transfected with miR-1226-5p. The cell lysate was mixed immobilized Agarose beads (Santa Cruz Biotechnology, TX, USA) with AGO2 antibody (Sigma-Aldrich, MO, USA) for immunoprecipitation. RNA is isolated from the formed AGO2 antibody-agarose bead-ribonucleoprotein (RNP) complex, and RNA level of IRF1 was measured using real-time PCR.
In vivo assay
These studies were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of Korea Institute of Radiological & Medical Science. HCT116 parental, and radioresistant cells (HCT116-RR) were subcutaneously injected into the right flank of 6-week-old Balb/c nude female mice. Cells were injected at 1 × 107 cells per mice. Based on the day of irradiation, they were sacrificed 2 weeks later. The tumor volume was measured width and length and calculated as (width × width × length)/2. HCT116 radioresistant cells (HCT116-RR) overexpressing NC and miR-1226-5p inhibitor (anti-miR-1226-5p) were injected into right flank of 6-week-old Balb/c nude female mice. Cells were injected at 1 × 107 cells per mice. When the tumor volume of the mouse was 150–200 mm3, 5 Gy was irradiated to the tumor using X-RAD 320 (PXi, CT, USA). Based on the day of irradiation, they were sacrificed 3 weeks later.
Immunohistochemistry (IHC) staining
Tumor tissue from patients with colorectal cancer is fixed in 4% formaldehyde was embedded in paraffin. Paraffin sections are deparaffinized and hydrated. For IHC, paraffin sections were subjected to Target retrieval solution (Dako, CA, USA), and endogenous peroxidase was blocked using H2O2. Antibodies used in IHC were CD206 (ab64693, Abcam, UK). After reacting with biotinylated secondary antibody, DAB staining was performed. The signal was detected using cellSens (Olympus, Japan).
Statistical analysis
GraphPad software was used for all data analysis. All data are expressed as mean ± SD. Statistical calculations were performed with Student’s t test or One-way ANOVA followed by bonferroni comparison test.
Results
Characterization of established radioresistant CRC cells
To elucidate the main mechanism of colorectal cancer (CRC) patients’ resistance to radiotherapy, radioresistant cell lines (HCT116-RR and SW480-RR) were established by exposing CRC cells HCT116 and SW480 to 5 Gy radiation 15 times (total 75 Gy). HCT116-RR and SW480-RR were irradiated with various radiation doses, respectively, and several radioresistance characteristics were confirmed. As a result, cell viability of parental cells was reduced in a radiation dose-dependent manner, whereas the viability of HCT116-RR and SW480-RR cells that acquired radioresistance was dramatically increased compared to that in HCT116 and SW480 cells (Supplemental Fig. 1A). The colony formation assay also demonstrated that HCT116-RR and SW480-RR cells had higher survival rates when exposed to radiation than the HCT116 and SW480 cells (Supplemental Fig. 1B). In addition, the expression of γ-H2AX protein, a DNA damage marker, was significantly increased in HCT116 and SW480 cells than in HCT116-RR and SW480-RR cells after irradiation (Supplementary Fig. 1C). Additionally, the tumorigenic ability of radioresistant cells was confirmed in vivo. When mice injected with parental HCT116 cells were irradiated, the tumorigenic ability was significantly reduced compared to the control group; however, in mice administered the resistant cell line HCT116-RR, there was little difference in tumor formation ability between the irradiated group and the control group (Supplemental Fig. 1D).
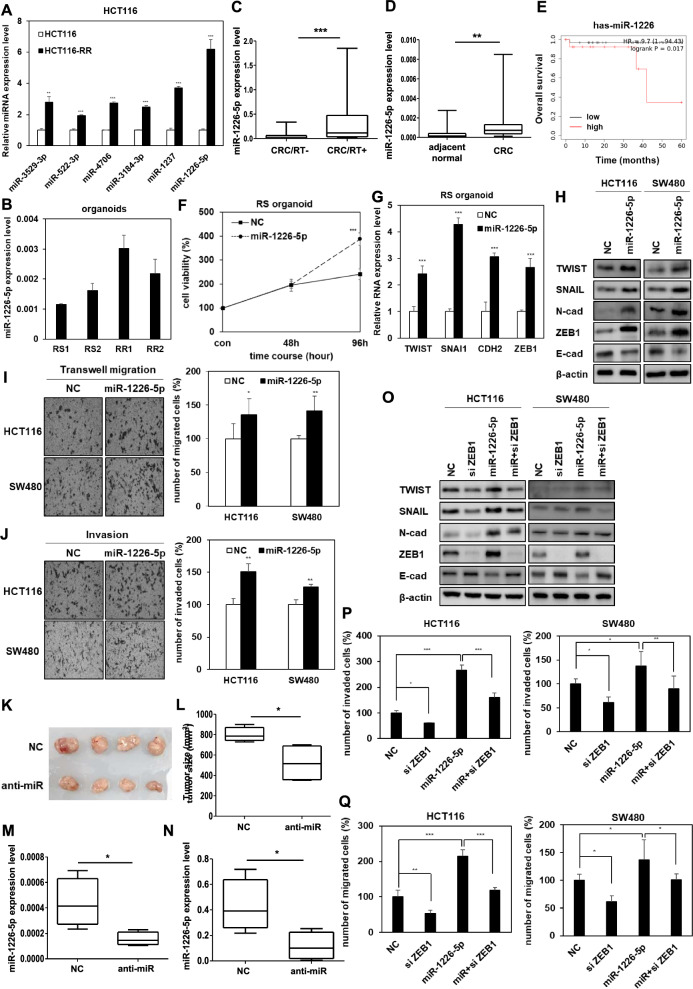
miR-1226-5p is involved in the EMT, mobility, invasiveness, and tumor growth capacity of radioresistant CRC cells
To discover miRNAs involved in the malignancy of radioresistant CRC, six miRNAs (miR-3529-3p, miR-522-3p, miR-4706, miR-3184-3p, miR-1237, and miR-1226-5p) were first selected through the GEO (Gene Expression Omnibus) series GSE159528, which is miRNA sequencing data analyzed from parental and radioresistant HCT116. We then compared the expression patterns of six candidate miRNAs in parental and HCT116-RR cells, and selected miR-1226-5p as the most dramatically higher expressed miRNA in radioresistant cells than in parental cells (Fig. 1A). We additionally measured the expression of six candidate miRNAs in SW480 and SW480-RR cells. As a result, among the candidates, the expression of miR-1226-5p was the highest in SW480-RR cells compared to parental cells, showing a pattern similar to that of HCT116-RR cells (Supplemental Fig. 2). Organoids derived from radiosensitive (RS) and radioresistant (RR) CRC patients classified by radiotherapy efficacy were utilized to clinically validate the selected miR-1226-5p [22]. As a result, the expression of miR-1226-5p was higher in RR organoids than in RS organoids (Fig. 1B). The selected miR-1226-5p was expressed more highly in the plasma of CRC patients who received radiotherapy than those of non-radiotherapy patients (Fig. 1C). The expression of miR-1226-5p in radioresistant HCT116 and SW480 cells increased in a radiation dose-dependent manner compared to the parental cells (Supplemental Fig. 3). These results not only support the selection of miR-1226-5p as a target for CRC radioresistance research, but also suggest the possibility that miR-1226-5p may be involved in cancer malignancy and radioresistance. The expression of miR-1226-5p in CRC patient tissues was also higher than that in adjacent normal tissues (Fig. 1D). As a result of analysis using Kaplan–Meier plot, high expression of miR-1226-5p lowered the overall survival rate (Fig. 1E). These results suggest that miR-1226-5p is closely related to the radioresistance mechanism in CRC.
Fig. 1.
miR-1226-5p promotes tumor progression by upregulating ZEB1 in radioresistant CRC cells HCT116 and SW480. A Six miRNAs (miR-3529-3p, miR-522-3p, miR-4706, miR-3184-3p, miR-1237, and miR-1226-5p) were selected based on miRNA sequencing data from the GEO series GSE159528. The expression of six miRNAs was measured by qRT-PCR in HCT116 and HCT116-RR cells. B The levels of miR-1226-5p were determined in radioresistant or radiosensitive patient-derived organoids. (RS; radiosensitivity; RR; radioresistance). C Levels of miR-1226-5p were measured in plasma of CRC patients with or without radiotherapy (CRC/RT-, n = 37; CRC/RT + , n = 37). qRT-PCR was normalized to U6. D The levels of miR-1226-5p were measured in adjacent normal and tumor tissues of CRC patients (adjacent normal, n = 39; tumor, n = 40). E Kaplan–Meier overall survival curve of stage II rectal adenocarcinoma patients according to the expression level of miR-1226-5p. F, G Radiosensitive CRC patient-derived organoids were transfected with negative control (NC) or miR-1226-5p. F Cell viability was measured overexpression of miR-1226-5p in radiosensitive organoids derived from CRC patients by MTT assay. G TWIST, SNAIL, CDH2, and ZEB1 mRNA expression levels were measured by qRT-PCR. qRT-PCR was normalized to GAPDH. H-J HCT116 and SW480 cells were transfected with negative control (NC) or miR-1226-5p mimic. H Expression of TWIST, SNAIL, N-cadherin, ZEB1, and E-cadherin in the indicated cells was confirmed through Western blot analysis. β-Actin was used for normalization in Western blot analysis. Transwell migration (I), and matrigel coated invasion assays (J) were subjected on the designated cells. K-N HCT116 RR cells overexpressing the negative control (NC) or the miR-1226-5p inhibitor (anti-miR-1226-5p) were subcutaneously injected into the flank of Balb/c nude mice (n = 4; 1 × 107 cells/mouse). Whole tumor images (K) and tumor size (L) of negative control (NC) and miR-1226-5p inhibitor (anti-miR-1226-5p) overexpressed mice are shown. Scale bar is 1 cm. The expression of miR-1226-5p in tumor tissue (M) and plasma (N) of mice was analyzed by qRT-PCR. qRT-PCR was normalized to U6. O–Q To elucidate the tumorigenicity mechanism of miR-1226-5p, HCT116 and SW480 cells were transfected with siRNA against ZEB1 or miR-1226-5p alone or together. O Expression of TWIST, SNAIL, N-cadherin, ZEB1, and E-cadherin was confirmed by Western blot analysis in the indicated cells. β-Actin was used for normalization in Western blot analysis. Transwell migration (P) and matrigel-coated invasion assays (Q) were applied to HCT116 and SW480 cells. Scale bar 200um. The experiment was repeated with triplicates and representative Western blotting images are shown. The data are presented as the mean ± S.D. after triplicate. *P < 0.05; **P < 0.01; ***P < 0.001. Student’s t test, and One-way ANOVA followed by bonferroni comparison test
The tumorigenic properties of miR-1226-5p were analyzed in CRC patient-derived organoids, HCT116, and SW480 cells. First, overexpression of miR-1226-5p promoted cell proliferation in organoids and increased mRNA expression of EMT factors TWIST, SNAI1 (encoding SNAIL), CDH2 (encoding N-cadherin), and ZEB1 (Fig. 1F, G). In addition, overexpression of miR-1226-5p in HCT116 and SW480 increased the expression of mesenchymal marker proteins (TWIST, SNAIL, N-cadherin, and ZEB1), but decreased the expression of E-cadherin, an epithelial cell marker (Fig. 1H). And, miR-1226-5p also enhanced cell migratory ability and invasiveness (Fig. 1I, J). To confirm the efficacy of miR-1226-5p in vivo, HCT116-RR cells, whose expression was suppressed with miR-1226-5p inhibitor (anti-miR-1226-5p), were injected into mice, and tumor growth ability was confirmed. Compared with the control group, the tumor size was reduced in the miR-1226-5p inhibitor, and the expression of miR-1226-5p in the tissues and plasma of mice was reduced (Fig. 1K–N). These results suggest that miR-1226-5p promotes EMT, migration, invasiveness, and tumorigenic capacity in CRC.
For an in-depth understanding of the signaling mechanisms of EMT, migration, and invasion induced by miR-1226-5p, we focused on the relationship between the key factor ZEB1, and miR-1226-5p. It has been reported that ZEB1, a representative EMT factor, promotes CRC invasion and metastasis [23] and is upregulated by miR-1226-5p. HCT116 and SW480 cells were transfected with miR-1226-5p and siRNA for ZEB1. miR-1226-5p-induced the mesenchymal markers (TWIST, SNAIL, and N-cadherin) were reduced by ZEB1 siRNA, but miR-1226-5p-reduced the expression of E-cadherin was increased by ZEB1 siRNA (Fig. 1O). In addition, cell migratory and invasive abilities induced by miR-1226-5p were reduced upon ZEB1 inhibition (Fig. 1P, Q, Supplementary Fig. 4A, B). These results suggest that the oncogenic effects of miR-1226-5p are induced through the ZEB1 signaling pathway.
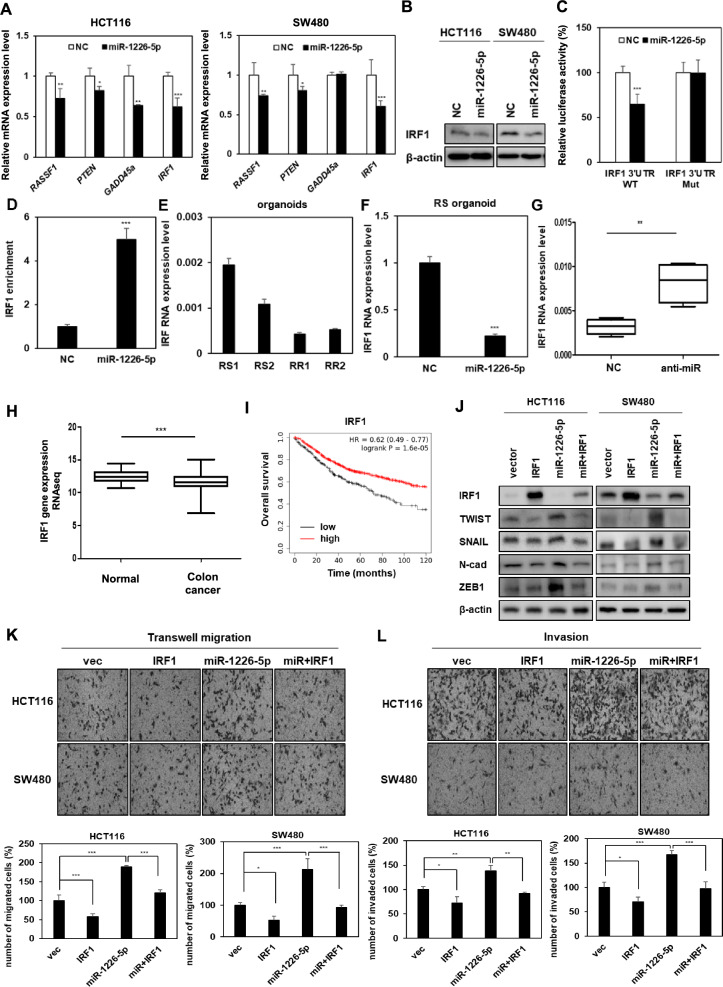
miR-1226-5p increases tumorigenicity by suppressing the expression of IRF1 in CRC cells
Using TargetScan, a microRNA target prediction tool, candidate genes regulated by miR-1226-5p were discovered. Among the four candidate factors, RASSF1, PTEN, GADD45a, and IRF1 (interferon regulatory factor 1), the IRF1 mRNA was most dramatically repressed upon overexpression of miR-1226-5p (Fig. 2A). Additionally, it was confirmed that protein expression of IRF1 was suppressed in HCT116 and SW480 cells overexpressing miR-1226-5p (Fig. 2B). To determine whether miR-1226-5p downregulates IRF1 expression by directly binding to the 3'UTR, wild- or mutant-type 3'UTR constructs of IRF1 were generated and luciferase activity was evaluated. As a result, luciferase activity decreased in the IRF1 wild-type group, while there was no significant difference in the mutant group (Fig. 2C). The complex of miRNA and AGO2 protein binds to target gene mRNA and induces translational repression or mRNA degradation [24]. Therefore, AGO2-RNA immunoprecipitation analysis was performed to confirm direct binding between miR-1226-5p and the target factor, IRF1. It was confirmed that the mRNA of IRF1 bound to AGO2 protein was enriched when overexpressing miR-1226-5p (Fig. 2D). These results support that the complex of miR-1226-5p and AGO2 protein binds to the binding site of IRF1 3’UTR and directly reduces its expression.
Fig. 2.
miR-1226-5p increases the tumorigenic phenotype by directly suppressing IRF1 expression. A The mRNA levels of candidates selected from TargetScan, a target prediction site for miRNAs, were measured in HCT116 and SW480, respectively. B Protein expression of IRF1 was confirmed in both CRC cells overexpressing miR-1226-5p. C HCT116 cells were co-transfected with wild-type (WT) or mutant (Mut) vectors of IRF1 3'UTR in the presence or absence of miR-1226-5p mimic, and then luciferase activity was examined. D IRF1 enrichment level was confirmed using AGO2-RNA-immunoprecipitation (AGO2-IP) and qRT-PCR analysis in SW480 cells overexpressing NC or miR-1226-5p. E Basal levels of IRF1 were identified in RS and RR CRC patient-derived organoids. F The mRNA level of IRF1 was measured by qRT-PCR when miR-1226-5p was overexpressed in RS organoids. G HCT116-RR cells overexpressing the negative control (NC) or the miR-1226-5p inhibitor (anti-miR-1226-5p) were injected subcutaneously into the flanks of Balb/c nude mice (n = 4; 1 × 107 cells/mouse). The mRNA expression of IRF1 in formed tumor tissues was performed by qRT-PCR. H Expression levels of IRF1 in normal patients (n = 41) and colon cancer patients (n = 469) of TCGA colon cancer (COAD) in Genomic Data Common (GDC) are shown as box and whisker plots. I Kaplan–Meier overall survival curves of colon adenocarcinoma patients according to the expression level of IRF1 were shown. J–L HCT116 and SW480 cells were co-transfected with miR-1226-5p mimic and IRF1 overexpression vector. J Expression of TWIST, SNAIL, N-cadherin, and ZEB1 was confirmed by Western blot analysis in the indicated cells. Transwell migration (K) and matrigel-coated invasion assays (L) were subjected on indicated cells. qRT-PCR was normalized to GAPDH. β-Actin was used for normalization in Western blot analysis. The experiment was repeated three times and representative Western blot images are shown. The data are presented as the mean ± S.D. *P < 0.05; **P < 0.01; ***P < 0.001. Student’s t test and One-way ANOVA followed by bonferroni comparison test
Additionally, a comparison of the mRNA expression pattern of IRF1 in radioresponsive CRC-derived organoids showed that its expression was lower in RR organoids compared to RS organoids (Fig. 2E). It was confirmed again that the mRNA expression of IRF1 decreased when miR-1226-5p was overexpressed in RS organoids (Fig. 2F). For in vivo validation, negative control and miR-1226-5p inhibitor (anti-miR-1226-5p) were transfected into HCT116-RR cells and then injected into mice, respectively. As a result, IRF1 expression was increased in tumor tissues of mice overexpressing miR-1226-5p inhibitor compared to the control group (Fig. 2G). These results suggest that miR-1226-5p downregulates the expression of IRF1 at the cellular and in vivo levels.
Using the public dataset Cancer Genome Atlas (TCGA) database, IRF1 expression was lower in colon cancer (COAD) compared to the normal group (Fig. 2H). Kaplan–Meier plot analysis showed that low IRF1 expression was associated with poor prognosis in patients with colon adenocarcinoma (Fig. 2I). Additionally, we verified the relationship between the two factors by the effect on tumorigenic phenotype depending on the expression of IRF1 and miR-1226-5p. The expression of TWIST, SNAIL, N-cadherin, and ZEB1 (Fig. 2J), cancer cell migration ability (Fig. 2K), and invasiveness (Fig. 2L) induced by miR-1226-5p were reduced by IRF1 overexpression. Overall, these results suggest that miR-1226-5p directly binds to its target IRF1 to regulate its expression, rather than through the influence of ZEB1, a downstream signaling pathway of miR-1226-5p.
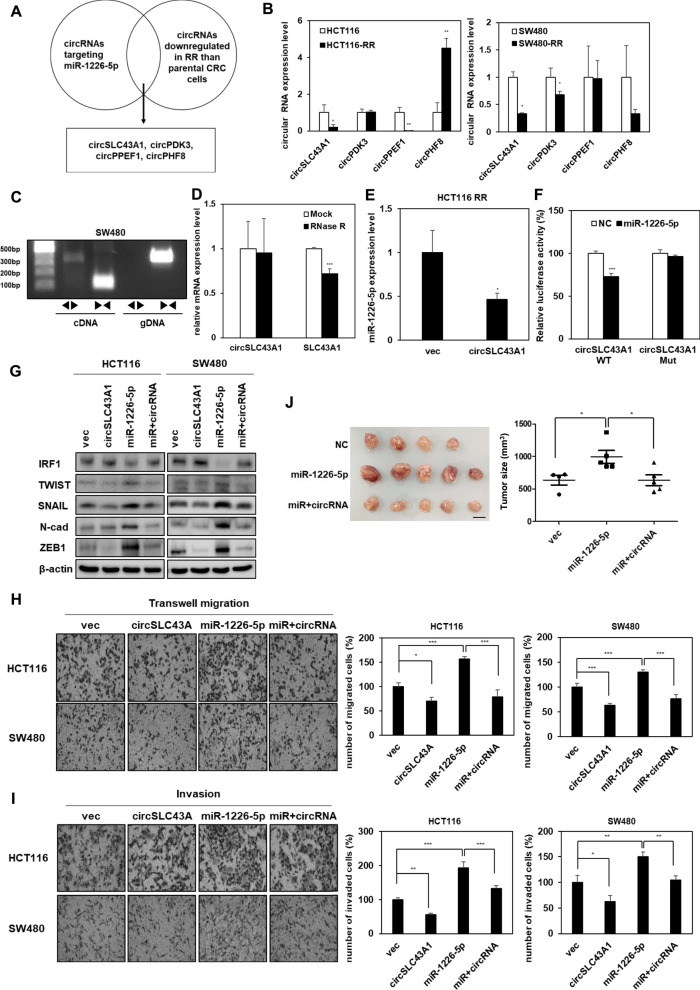
circSLC43A1 suppresses tumorigenicity by suppressing the expression of miR-1226-5p
Circular RNAs (circRNAs) are known to act as sponges that regulate the expression of miRNAs [25]. Therefore, we sought to determine whether inhibition of miR-1226-5p, which is highly expressed in radioresistant CRC cells, is possible as a therapeutic target for CRC. Using circBank, a circRNA prediction site [26], we selected four candidate circRNAs (circSLC43A1, circPDK3, circPPEF1, and circPHF8) that suppress the expression of miR-1226-5p (Fig. 3A). As a result of confirming the expression of selected candidate factors in parental, HCT116-RR and SW480-RR cells, circSLC43A1 (hsa_circ_0022141), which showed the lowest expression level in radioresistant CRC cells, was selected (Fig. 3B). According to circBank, circSLC43A1 is derived from exons 3–14 of SLC43A1 and is located at chr11: 57254567–5726880 (http://www.circbank.cn/). To determine the characteristics of circSLC43A1, circSLC43A1 was amplified using divergent primers derived from complementary DNA (cDNA) rather than genomic DNA (gDNA) obtained from SW480 cells (Fig. 3C). Additional validation experiments using ribonuclease R (RNase R) showed that SLC43A1 mRNA was degraded by RNase R, whereas circSLC43A1 was not affected (Fig. 3D). This was supported by reports that circRNA has stable cytoplasmic expression against degradation by RNase compared to linear RNA [27]. Upon circSLC43A1 overexpression in HCT116-RR, the expression of miR-1226-5p was decreased, demonstrating the potential of circSLC43A1 as a sponge for miR-1226-5p (Fig. 3E). To determine whether circSLC43A1 directly binds to and regulates the expression of miR-1226-5p, we performed a luciferase assay. As a result, binding of the wild-type (WT) vector of circSLC43A1 and miR-1226-5p reduced luciferase activity compared to the control group. Meanwhile, in the case of the mutant-type (Mut) vector of circSLC43A1, there was no change in its activity compared to the control group. These results showed that circSLC43A1 inhibited the expression of miR-1226-5p by directly binding to it (Fig. 3F).
Fig. 3.
circSLC43A1 reduces EMT, cell migratory ability, invasiveness, and tumor growth by directly downregulating miR-1226-5p expression. A Schematic diagram of the selection of candidate circRNAs that directly inhibit the expression of miR-1226-5p. B The expression levels of four candidate circRNAs, selected using the circbank database, were compared in radiosensitive or resistant HCT116 and SW480 cells. C circSLC43A1 was quantified by amplification of cDNA of circSLC43A1 in SW480 cells using divergent and convergent primers. D After RNase R treatment in HCT116, the mRNA levels of circSLC43A1 and SLC43A1 were determined using qRT-PCR. E The expression level of miR-1226-6p was measured when circSLC43A1 was overexpressed in HCT116 RR cells. F After co-transfecting cells with miR-1226-5p mimic and the wild-type (WT) or mutant-type (Mut) vector of circSLC43A1, the binding of the two factors was confirmed in HCT116 cells using dual luciferase assay. G-I circSLC43A1 was overexpressed alone or together with miR-1226-5p in HCT116 and SW480 cells, respectively. G TWIST, SNAIL, N-cadherin, and ZEB1 protein expression was detected by Western blot analysis. Transwell migration (H) and matrigel-coated invasion assays (I) were applied to the indicated cells. J HCT116 RR overexpressing miR-1226-5p alone or together with circSLC43A1 were injected subcutaneously into the right flank of Balb/c nude mice (n = 4; 1 × 107 cells/mouse). At 2 weeks after injection, tumor tissues were harvested from sacrificed mice. A whole image of the tumor (left) and the volume of the tumor (right) were quantified and displayed in a graph. Scale bar 1 cm. qRT-PCR were normalized to GAPDH and U6. β-Actin was used for normalization in Western blot analysis. The experiment was repeated with triplicates and representative Western blotting images are shown. The data are presented as the mean ± S.D. *P < 0.05; **P < 0.01; ***P < 0.001. Student’s t test, and One-way ANOVA followed by bonferroni comparison test
Overexpression of circSLC43A1 in HCT116-RR and SW480-RR cells significantly reduced the protein expression of TWIST, SNAIL, N-cadherin, and ZEB1 (Supplemental Fig. 5A), as well as cell motility and invasion (Supplemental Fig. 5B, C). The effect of downregulation of miR-1226-5p expression by circSLC43A1 on the mechanism of tumorigenicity in CRC cells was confirmed. Next, we investigated the effects on the cancer process depending on the expression of the circSLC43A1 and miR-1226-5p. The expression of IRF1, which was decreased by miR-1226-5p, was restored by circSLC43A1 (Fig. 3G). As confirmed at the cellular level and in vivo, the expression of EMT markers (TWIST, SNAIL, N-cadherin, and ZEB1), cell motility, invasion, and tumor growth induced by miR-1226-5p were reduced by overexpression of circSLC43A1 (Fig. 3G–J). Taken together, these results suggest that overexpression of circSLC43A1 may reduce tumor malignancy induced by miR-1226-5p/ZEB1 in radioresistant CRC.
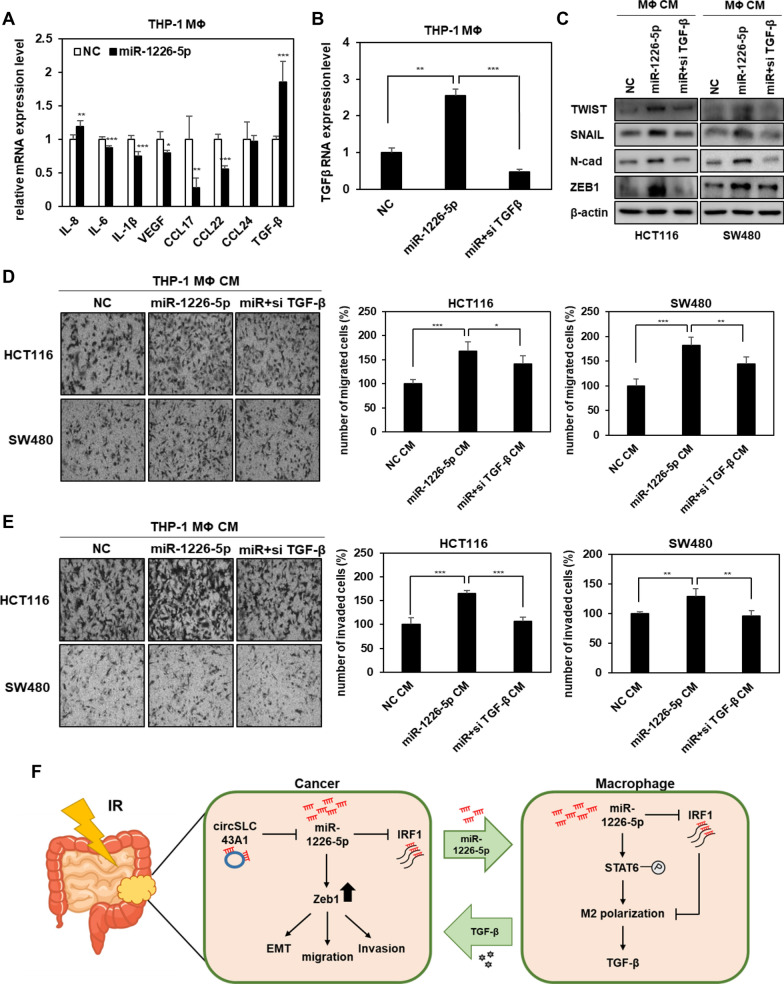
Fig. 5.
miR-1226-5p-mediated TGF-β secretion in M2 macrophages enhances the EMT, migratory and invasive capacity of CRC cells. A After overexpressing miR-1226-5p in THP-1-derived macrophages, the mRNA expression levels of cytokines and growth factors were screened by qRT-PCR. B THP-1-derived macrophages were overexpressed with miR-1226-5p and siRNA targeting TGF-β, and the mRNA expression of TGF-β was measured. C-E Conditioned media harvested from THP-1-derived macrophages overexpressing miR-1226-5p and siRNA targeting TGF-β were treated with HCT-116 and SW480 cells, respectively. C Expression of TWIST, SNAIL, N-cadherin, and ZEB1 was confirmed by Western blot analysis. Transwell migration (D) and matrigel-coated invasion assays (E) were applied to the indicated cells. Scale bar 200um. F Schematic diagram showing the mechanism by which radioresistant CRC cell-derived miR-1226-5p induces cancer malignancy by enhancing M2 polarization of macrophages. The experiment was repeated three times and representative Western blot images are shown. The data are presented as the mean ± S.D. *P < 0.05; **P < 0.01; ***P < 0.001. One-way ANOVA followed by bonferroni comparison test
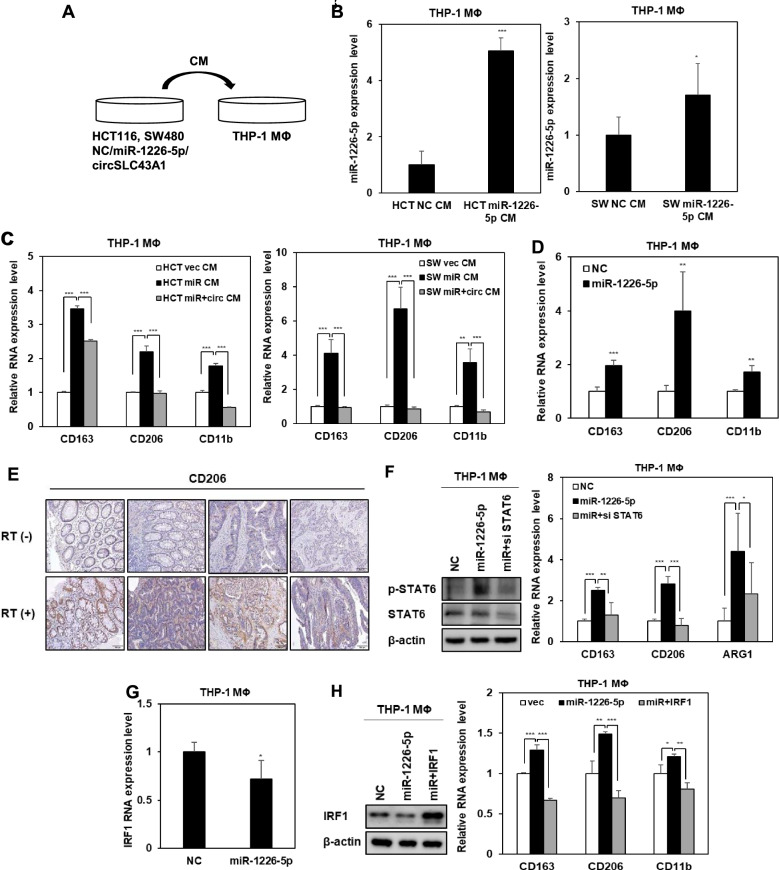
Radioresistant CRC cell-derived miR-1226-5p promotes polarization of M2 macrophages
The tumor microenvironment (TME) and related signaling pathways have a significant impact on the development and progression of CRC [28]. In particular, macrophages play a significant role in the changes of the tumor microenvironment by regulating various immune responses and increasing angiogenesis and tumor cell invasion, and migration [29]. Among the two types of macrophages, M1 macrophages induce apoptosis of tumor cells, whereas M2 macrophages contribute to angiogenesis and EMT of tumor cells and induce an immunosuppressive microenvironment [11, 13]. To determine whether CRC cells-derived miR-1226-5p promotes M2 polarization in macrophages, THP-1-derived macrophages were treated with conditioned media from HCT116 and SW480 cells overexpressing miR-1226-5p (Fig. 4A). When THP-1-derived macrophages were treated with conditioned media derived from CRC cells overexpressing miR-1226-5p, the expression of miR-1226-5p was confirmed to increase compared to the control group (Fig. 4B). When THP-1 derived macrophages were treated with conditioned media derived from CRC cells overexpressing miR-1226-5p, the expression of M2 macrophage markers CD163, CD206, and CD11b was increased. Meanwhile, the expression of M2 macrophage markers was reduced upon treatment with conditioned media from HCT116 and SW480 cells that simultaneously overexpressed miR-1226-5p and circSLC43A1 (Fig. 4C). Additionally, the same results as above were seen when THP-1-derived macrophages were directly transfected with the miR-1226-5p mimic (Fig. 4D). These results suggest that tumor-derived miR-1226-5p plays an important role in inducing M2 polarization and that inhibition of miR-1226-5p secretion by circSLC43A1 can inhibit M2 polarization of macrophages. CRC patients were classified into two groups according to the presence or absence of radiation treatment, and the expression of CD206, an M2 macrophage marker, was confirmed. As a result, the expression was increased in the radiation treatment group (Fig. 4E). These results suggest that increased miR-1226-5p in CRC cells by radiation induces M2 polarization of macrophages. To analyze the potential mechanism promoting M2 polarization of macrophages, we confirmed the expression of STAT6, a known M2 polarization-involved factor [30]. As a result, the expression of p-STAT6 was increased by overexpression of miR-1226-5p in macrophages (Fig. 4F). We investigated whether the induction of M2 polarization by miR-1226-5p was mediated through the STAT6 pathway. The miR-1226-5p-induced upregulation of CD163, CD206, and ARG1 mRNA expression was reduced by STAT6 inhibition (Fig. 4F). In macrophages, STAT6 reduces IRF1 expression, thereby potentiating M2 polarization.
Fig. 4.
miR-1226-5p induces M2 polarization via IRF1 inhibition in THP-1-derived macrophages. A–D Human monocytes, THP-1, were differentiated into macrophages by treating them with PMA (100 ng/ml) for 24 h. A Conditioned media harvested from HCT116 and SW480 overexpressing either miR-1226-5p and circSLC43A1 were treated with THP-1-derived macrophages. B The level of miR-1226-5p in the indicated THP-1-derived macrophages was confirmed by qRT-PCR. C The mRNA levels of CD163, CD206, and CD11b were measured in the indicated THP-1-derived macrophages. D After overexpression of the miR-1226-5p mimic in THP-1-derived macrophages, the expression of CD163, CD206, and CD11b was confirmed by qRT-PCR. E The expression of CD206 in tissues of CRC patients with and without radiotherapy was compared by IHC. Scale bar 100 um. F After co-transfection of siRNA against STAT6 and miR-1226-5p in THP-1-derived macrophages, protein expression of p-STAT6, and STAT6 was compared through Western blot analysis (left). The mRNA levels of CD163, CD206, and ARG1 were measured in the indicated cells (right). G The mRNA expression of IRF1 upon overexpression of miR-1226-5p in THP-1-derived macrophages was analyzed by qRT-PCR. H After co-transfecting THP-1-derived macrophages with miR-1226-5p and IRF1 overexpression vector, the protein expression of IRF1 was compared through Western blot analysis (left). The mRNA levels of CD163, CD206, and CD11b were measured in the indicated cells (right). qRT-PCR was normalized to GAPDH and U6. β-Actin was used for normalization in Western blot analysis. The experiment was repeated three times and representative Western blot images are shown. The data are presented as the mean ± S.D. *P < 0.05; **P < 0.01; ***P < 0.001. Student’s t test, and One-way ANOVA followed by bonferroni comparison test
There is a report that inhibition of IRF1 induces M2 polarization of macrophages [31]. Therefore, we analyzed the effects of miR-1226-5p and its target IRF1 on M2 macrophage polarization. When overexpressing miR-1226-5p in THP-1-derived macrophages, the mRNA and protein expressions of IRF1 were decreased (Fig. 4G, H). At this time, miR-1226-5p-induced expression of M2 markers, including CD163, CD206, and CD11b, was downregulated by the overexpression of IRF1 (Fig. 4H). Taken together, in THP-1-derived macrophages, miR-1226-5p suppressed the expression of IRF1 and stimulated the activation of STAT6, thereby enhancing M2 polarization.
TGF-β induced by miR-1226-5p in M2 macrophages increases tumorigenic ability in CRC cells
M2 macrophages secrete various growth factors, matrix metalloproteinases (MMPs), and other cytokines that not only enhance EMT and metastasis of CRC but also attenuate immune responses [11]. Among various candidate cytokines and growth factors, the mRNA expression of TGF-β was highest in THP-1-derived macrophages overexpressing miR-1226-5p (Fig. 5A). To investigate the relationship between selected TGF-β and the circSLC43A1/miR-1226-5p axis, conditioned media from HCT116 and SW480 cells overexpressing circSLC43A1 and miR-1226-5p was obtained and treated with THP-1-derived macrophages. TGF-β expression, which was increased by the conditioned media for miR-1226-5p overexpression, was reduced when treated with the conditioned media for circSLC43A1 overexpression (Supplemental Fig. 6A, B). These results suggested that THP-1-derived macrophages secreted TGF-β expression was regulated by the circSLC43A1/miR-1226-5p axis in CRC cells. In addition, it was confirmed that the expression of TGF-β was regulated through STAT6, a major pathway of miR-1226-5p-induced M2 polarization (Supplemental Fig. 7). To confirm that TGF-β secreted from M2 macrophages induced by miR-1226-5p is a factor influencing cancer malignancy, THP-1-derived macrophages co-transfected with miR-1226-5p and siRNA against TGF-β. Conditioned media was obtained from THP-1-derived macrophages (Fig. 5B). In HCT116 and SW480 cells treated with conditioned media from macrophages overexpressing miR-1226-5p, increased expression of TWIST, SNAIL, N-cadherin, and ZEB1 (Fig. 5C), cell motility (Fig. 5D), and invasiveness (Fig. 5E) were decreased due to TGF-β inhibition. These findings suggest that miR-1226-5p, which is highly expressed in radioresistant CRC cells, promotes tumorigenic ability by suppressing IRF1 expression. In addition, tumor-derived miR-1226-5p is delivered to macrophages within the tumor microenvironment and increases STAT6, thereby inducing M2 polarization and increasing TGF-β production, ultimately contributing to improving cancer aggressiveness (Fig. 5F).
Discussion
This study analyzed the tumor malignancy mechanism of miR-1226-5p derived from radioresistant CRC cells on tumors and microenvironment, especially macrophages. As a result, it was revealed that miR-1226-5p not only increases the expression of ZEB1 and suppresses the expression of IRF1 to induce tumorigenicity, but also induces M2 polarization and secretion of TGF-β by being transferred to macrophages, ultimately increasing tumor malignancy. miR-1226-5p, which is upregulated in radioresistant CRC cells and patient-derived organoids, promoted the expression of EMT markers (TWIST, SNAIL, and N-cadherin), cell mobility, and invasiveness by increasing ZEB1, and enhanced tumor growth in animal models (Fig. 1). ZEB1, which is upregulated by miR-1226-5p in this signaling mechanism, is known as an oncogenic transcription factor that plays a critical role in all stages of tumor progression [32, 33]. It has been reported that increased ZEB1 expression in prostate or pancreatic cancer leads to treatment resistance [34]. ATM stabilizes CHK1 through ZEB1 to promote DNA repair and radioresistance [35]. ATM induces radioresistance in lung adenocarcinoma through miR-183 [36]. These reports accumulate the possibility that ZEB1 is a major factor contributing to radioresistance.
Since miRNAs function by suppressing target genes expression [37], the tumor suppressor IRF1 was selected as the target gene of miR-1226-5p. It was verified that miR-1226-5p induces cancer malignancy by directly reducing the expression of IRF1 in CRC cells and organoids (Fig. 2A, B, F). Additionally, IRF1 showed higher expression in radiosensitive than radioresistant CRC organoids and lower expression in CRC patients than normal (Fig. 2E, F). According to previous reports, IRF1 is known to induce apoptosis and inhibit cell proliferation by regulating the expression of cell cycle regulatory genes in various cancers (e.g., colon [38], breast [39], and gastric [40]). These reports support our findings that IRF1 functions as a tumor suppressor. In addition, the expression of IRF1, a target factor of miR-1226-5p, was low in radioresistant head and neck squamous cell carcinoma cells [41]. It has been reported that overexpression of IRF1 in colon cancer inhibits proliferation and tumor growth and increases radiosensitivity [42]. IRF1 has been proposed as an important radiospecific biomarker for malignant tumors based on microarray and bioinformatics analysis [43]. In summary, these reports suggest that IRF1 is negatively correlated with radioresistance. Meanwhile, the transcription of IRF1 is promoted by STAT1, which is activated by the IFN-γ-mediated receptor on macrophages [44]. IRF1 plays an important role in M1 polarization by inducing the production of iNOS, IL-6, IL-12, and TNF-α [45, 46]. However, when the expression of IRF1 is suppressed by miR-1226-5p, leading to the M2 polarization of macrophages (Fig. 4).
To analyze the regulatory mechanism of miR-1226-5p, which is upregulated in radioresistant CRC, circRNAs were selected using to circBank predicted site. circRNAs are a type of noncoding RNA characterized by a closed loop structure resulting from back-splicing and are distinguished from linear RNAs by the absence of a 5' cap and a 3' polyadenylated tail [27]. Recent reports have shown that dysregulation of circRNAs is associated with cancer development [47], and circRNAs have potential as valuable therapeutic targets due to their stable and characteristic expression in various tissues and cell types [47, 48]. In CRC cells, circLONP2 was reported to be upregulated by directly interacting with DDX1, enhancing cell migration and invasion, and inhibition of circLONP2 by antisense oligonucleotides blocked CRC metastasis [49]. In our study, circSLC43A1 effectively suppressed the expression of miR-1226-5p, thereby inhibiting the EMT, cell migration, and invasiveness of radioresistant CRC cells and reducing the tumor size in vivo (Fig. 3). In addition, the suppression of miR-1226-5p expression by circSLC43A1 decreased the M2 polarization of macrophages and the mRNA expression level of TGF-β (Fig. 4G, Supplementary Fig. 5). These results suggest the potential of circSLC43A1 as an effective therapeutic target for radioresistant CRC with high expression of miR-1226-5p.
Macrophages play an important role in mediating phagocytosis of cancer cells and cytotoxic tumor killing, and mediate processes such as angiogenesis, extracellular matrix remodeling, cancer cell proliferation, metastasis, immunosuppression, and resistance to chemotherapy. It is an important component of the microenvironment [50]. Macrophages have phenotypic plasticity to polarize into M1 or M2 macrophages in response to a variety of stimuli, where miRNAs play an important role in controlling polarization and contributing to the immune response [19]. Studies of the tumor microenvironment and CRC based on these conflicting abilities of macrophages are needed. Many studies have shown that miR-9, miR-127, miR-155, and miR-125b induce M1 polarization of macrophages, while miR-34a, miR-132, miR-146a, and miR-223 induce M2 polarization [51]. After confirming that miR-1226-5p, which was highly expressed in radioresistant CRC, was delivered to macrophages, it was revealed that M2 polarization of macrophages was induced by activation of STAT6 and inhibition of IRF1 induced by miR-1226-5p. This result verified that miR-1226-5p is a factor that plays an important role in macrophage M2 polarization (Fig. 4).
Several previous studies have reported that M2 macrophage polarization is associated with radioresistance. For example, CD163-positive M2 macrophages infiltrating the tissue of HPV-negative head and neck squamous cell carcinoma induce radioresistance by releasing HB-EGF [52]. CCL2 secreted from irradiated CAFs polarizes M2 macrophages with CD163 and CD206-positive phenotypes and promotes radioresistance in cervical cancer [53]. In a mouse model of irradiated pancreatic cancer, a significant proportion of macrophages in pancreatic tumors had an M2-like phenotype, including the CD206 marker. In addition, radiation promotes MCSF production in Pancreatic ductal adenocarcinoma cells, ultimately inducing radioresistance [54]. These studies suggest that M2 macrophage polarization and radioresistance are closely related. Therefore, in our study, we also showed that miR-1226-5p promoted M2 polarization by increasing the expression of M2 markers CD163, CD206, and CD11b. Figure 4E showed that the tissues of patients who received radiotherapy had higher expression of CD206 than those of patients who did not receive treatment, further supporting the positive correlation between radiation and M2 polarization.
miR-1226-5p induced polarization of M2 macrophages and subsequent release of TGF-β, creating an immunosuppressive tumor microenvironment and promoting tumor progression. In addition, inhibition of TGF-β secreted by M2 macrophages suppressed EMT and cell migratory and invasive abilities of CRC cells (Fig. 5, Supplemental Figs. 6, 7). TGF-β is a strong stimulator of EMT and a factor that increases the invasiveness, metastasis, and radioresistance of cancer cells [55, 56]. TGF-β signaling regulates the DNA damage response (DDR) pathway, affecting the repair mechanism of radiation-induced DNA double-strand breaks and potentially contributing to radioresistance [57]. These results suggest that TGF-β may be an important indicator of radiation-induced cytotoxicity and DNA damage [57]. According to these reports, it can be suggested that TGF-β secreted from miR-1226-5p-induced M2 macrophages is positively correlated with radioresistance. In summary, our study suggested that tumor-derived miR-1226-5p was involved in radioresistance by activating M2 polarization of macrophages and promoting the secretion of TGF-β. TGF-β plays an important role in immune evasion within the tumor microenvironment by suppressing the activity of cytotoxic T cells and natural killer cells [57–59]. Therefore, targeting TGF-β signaling has become an attractive cancer treatment strategy. Treatment combining TGF-β inhibitors with radiotherapy offers the potential to overcome radioresistance by inhibiting pro-tumorigenic effects on cancer cells and the tumor microenvironment [60].
Recent studies have shown that miRNAs can be reliably detected in plasma and serum. Therefore, the levels of specific miRNAs in body fluids are valuable as diagnostic and prognostic biomarkers of disease [61]. The discovery of biomarkers associated with radioresistance can help predict radiotherapy response, allowing for a personalized medical approach for each patient based on the unique characteristics and biological profile of the tumor [62, 63]. The development of biomarkers for radioresistance can provide insight into the underlying resistance mechanisms, guiding the development of strategies to overcome treatment resistance as well as reduce side effects, thereby improving patient outcomes [62, 63]. In this way, the potential of using miRNAs as biomarkers for radiation resistance in colorectal cancer (CRC) has been established [64]. For instance, increased levels of miR-100 and miR-630 enhance radiosensitivity [65, 66], but, the upregulation of miR-622 and miR-106b leads to radioresistance [67, 68] in CRC cells. Therefore, miR-1226-5p is highly expressed in radioresistant cells and is associated with tumor malignancy in CRC, suggesting its potential as a biomarker for diagnosing malignancy in CRC.
This study revealed the interaction and pathway of miR-1226-5p and TGF-β signaling between tumors and macrophages for improved treatment efficacy, ultimately suggesting the possibility of use as targets for diagnosis and treatment.
Conclusions
In summary, miR-1226-5p, whose expression is increased in radioresistant CRC, promotes tumorigenicity by suppressing the expression of its target, IRF1, and activating ZEB1. Additionally, circSLC43A1 was identified as a sponge that suppresses the expression of miR-1226-5p, and it was found to suppress tumor aggressiveness in radioresistant CRC. Tumor-derived miR-1226-5p created an immunosuppressive environment by promoting M2 polarization of macrophages and increasing STAT6 activation and TGF-β secretion. Tumor treatment strategies targeting radioresistance-inducing miR-1226-5p and related factors in CRC would be useful for improving treatment outcomes and the prognosis of patients with radioresistant CRC. Additionally, the factors discovered in this study suggest potential as targets for diagnosing and treating radioresistant CRC.
Supplementary Information
Acknowledgements
Not applicable.
Abbreviations
- CRC
Colorectal cancer
- EMT
Epithelial–mesenchymal transition
- RR
Radioresistant
- RS
Radiosensitive
- THP-1 MΦ
THP-1-derived macrophages
- circRNA
Circular RNA
- IHC
Immunohistochemistry
- qRT-PCR
Quantitative Real-Time PCR
- Km plot
Kaplan–Meier plot
- miRNA
MicroRNA
- IRF1
Interferon regulatory factor
Author contributions
IHB supervised all research work. IHB and JYC designed all the experiments, analyzed data, and drafted the manuscript. IHB, JYC, HJS, and DHL participated in animal experiments. All authors discussed the results and commented on the manuscript. JHK and USS provided organoid and analyzed the data. ICS discussed the results commented on the manuscript. All authors have read and approved the article.
Funding
This work was supported by grants of the National Research Foundation of Korea and KIRAMS, funded by Ministry of Science and ICT (MSIT) in Republic of Korea. [No. NRF-2021R1A2C2005966(50698-2023), NRF-2017M2A2A7A01018542(50035-2019), and 50531-2024].
Availability of data and materials
The TCGA data used in the analysis was provided by UCSC Xena [https://xenabrowser.net/datapages/]. The Km plot used in the analysis was provided by Kaplan–Meier plotter [https://www.kmplot.com/]. The miRNA sequencing data used in the analysis was provided by Gene Expression Omnibus (GEO). The series number is GSE159528.
Declarations
Ethics approval and consent to participate
Human lung tissue and plasma from lung cancer patients were Institutional Review Board (IRB) approved in Korea Institute of Radiological and Medical Sciences (KIRAMS).
Consent for publication
All authors involved in this study agreed to this manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hossain MS, Karuniawati H, Jairoun AA, Urbi Z, Ooi J, John A, et al. Colorectal cancer: a review of carcinogenesis, global epidemiology, current challenges, risk factors, preventive and treatment strategies. Cancers (Basel). 2022. 10.3390/cancers14071732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xie YH, Chen YX, Fang JY. Comprehensive review of targeted therapy for colorectal cancer. Signal Transduct Target Ther. 2020;5(1):22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kumar A, Gautam V, Sandhu A, Rawat K, Sharma A, Saha L. Current and emerging therapeutic approaches for colorectal cancer: a comprehensive review. World J Gastrointest Surg. 2023;15(4):495–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buckley AM, Lynam-Lennon N, O’Neill H, O’Sullivan J. Targeting hallmarks of cancer to enhance radiosensitivity in gastrointestinal cancers. Nat Rev Gastroenterol Hepatol. 2020;17(5):298–313. [DOI] [PubMed] [Google Scholar]
- 5.Jin Y, Chen Z, Dong J, Wang B, Fan S, Yang X, et al. SREBP1/FASN/cholesterol axis facilitates radioresistance in colorectal cancer. FEBS Open Bio. 2021;11(5):1343–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olivares-Urbano MA, Grinan-Lison C, Marchal JA, Nunez MI. CSC radioresistance: a therapeutic challenge to improve radiotherapy effectiveness in cancer. Cells. 2020. 10.3390/cells9071651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suwa T, Kobayashi M, Nam JM, Harada H. Tumor microenvironment and radioresistance. Exp Mol Med. 2021;53(6):1029–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orimo A, Gupta PB, Sgroi DC, Arenzana-Seisdedos F, Delaunay T, Naeem R, et al. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell. 2005;121(3):335–48. [DOI] [PubMed] [Google Scholar]
- 9.Belli C, Trapani D, Viale G, D’Amico P, Duso BA, Della Vigna P, et al. Targeting the microenvironment in solid tumors. Cancer Treat Rev. 2018;65:22–32. [DOI] [PubMed] [Google Scholar]
- 10.Dehne N, Mora J, Namgaladze D, Weigert A, Brune B. Cancer cell and macrophage cross-talk in the tumor microenvironment. Curr Opin Pharmacol. 2017;35:12–9. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, Zhao Y, Li Q, Wang Y. Macrophages, as a promising strategy to targeted treatment for colorectal cancer metastasis in tumor immune microenvironment. Front Immunol. 2021;12: 685978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao L, Che X, Qiu X, Li Z, Yang B, Wang S, et al. M2 macrophage infiltration into tumor islets leads to poor prognosis in non-small-cell lung cancer. Cancer Manag Res. 2019;11:6125–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu J, Geng X, Hou J, Wu G. New insights into M1/M2 macrophages: key modulators in cancer progression. Cancer Cell Int. 2021;21(1):389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boutilier AJ, Elsawa SF. Macrophage polarization states in the tumor microenvironment. Int J Mol Sci. 2021. 10.3390/ijms22136995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sica A, Larghi P, Mancino A, Rubino L, Porta C, Totaro MG, et al. Macrophage polarization in tumour progression. Semin Cancer Biol. 2008;18(5):349–55. [DOI] [PubMed] [Google Scholar]
- 16.Zhang N, Hu X, Du Y, Du J. The role of miRNAs in colorectal cancer progression and chemoradiotherapy. Biomed Pharmacother. 2021;134: 111099. [DOI] [PubMed] [Google Scholar]
- 17.Pedroza-Torres A, Romero-Cordoba SL, Montano S, Peralta-Zaragoza O, Velez-Uriza DE, Arriaga-Canon C, et al. Radio-miRs: a comprehensive view of radioresistance-related microRNAs. Genetics. 2024. 10.1093/genetics/iyae097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cui M, Wang H, Yao X, Zhang D, Xie Y, Cui R, et al. Circulating microRNAs in cancer: potential and challenge. Front Genet. 2019;10:626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Curtale G, Rubino M, Locati M. MicroRNAs as molecular switches in macrophage activation. Front Immunol. 2019;10:799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao LR, Zhang J, Huang N, Deng W, Ni W, Xiao Z, et al. Tumor-derived exosomal miR-143-3p induces macrophage M2 polarization to cause radiation resistance in locally advanced esophageal squamous cell carcinoma. Int J Mol Sci. 2024. 10.3390/ijms25116082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang C, Dou R, Wei C, Liu K, Shi D, Zhang C, et al. Tumor-derived exosomal microRNA-106b-5p activates EMT-cancer cell and M2-subtype TAM interaction to facilitate CRC metastasis. Mol Ther. 2021;29(6):2088–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee J, Kwon J, Kim D, Park M, Kim K, Bae I, et al. Gene expression profiles associated with radio-responsiveness in locally advanced rectal cancer. Biology (Basel). 2021. 10.3390/biology10060500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang F, Sun G, Peng C, Chen J, Quan J, Wu C, et al. ZEB1 promotes colorectal cancer cell invasion and disease progression by enhanced LOXL2 transcription. Int J Clin Exp Pathol. 2021;14(1):9–23. [PMC free article] [PubMed] [Google Scholar]
- 24.Jungers CF, Djuranovic S. Modulation of miRISC-mediated gene silencing in eukaryotes. Front Mol Biosci. 2022;9: 832916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Panda AC. Circular RNAs act as miRNA sponges. Adv Exp Med Biol. 2018;1087:67–79. [DOI] [PubMed] [Google Scholar]
- 26.Liu M, Wang Q, Shen J, Yang BB, Ding X. Circbank: a comprehensive database for circRNA with standard nomenclature. RNA Biol. 2019;16(7):899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang C, Zhang M, Liu Y, Cui D, Gao L, Jiang Y. CircRNF10 triggers a positive feedback loop to facilitate progression of glioblastoma via redeploying the ferroptosis defense in GSCs. J Exp Clin Cancer Res. 2023;42(1):242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Novoa Diaz MB, Martin MJ, Gentili C. Tumor microenvironment involvement in colorectal cancer progression via Wnt/beta-catenin pathway: providing understanding of the complex mechanisms of chemoresistance. World J Gastroenterol. 2022;28(26):3027–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin Y, Xu J, Lan H. Tumor-associated macrophages in tumor metastasis: biological roles and clinical therapeutic applications. J Hematol Oncol. 2019;12(1):76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang N, Liang H, Zen K. Molecular mechanisms that influence the macrophage m1–m2 polarization balance. Front Immunol. 2014;5:614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin J, Yu G. Hypoxic lung cancer cell-derived exosomal miR-21 mediates macrophage M2 polarization and promotes cancer cell proliferation through targeting IRF1. World J Surg Oncol. 2022;20(1):241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goossens S, Vandamme N, Van Vlierberghe P, Berx G. EMT transcription factors in cancer development re-evaluated: beyond EMT and MET. Biochim Biophys Acta Rev Cancer. 2017;1868(2):584–91. [DOI] [PubMed] [Google Scholar]
- 33.Krebs AM, Mitschke J, Lasierra Losada M, Schmalhofer O, Boerries M, Busch H, et al. The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat Cell Biol. 2017;19(5):518–29. [DOI] [PubMed] [Google Scholar]
- 34.Wang Z, Chen Y, Lin Y, Wang X, Cui X, Zhang Z, et al. Novel crosstalk between KLF4 and ZEB1 regulates gemcitabine resistance in pancreatic ductal adenocarcinoma. Int J Oncol. 2017;51(4):1239–48. [DOI] [PubMed] [Google Scholar]
- 35.Zhang P, Wei Y, Wang L, Debeb BG, Yuan Y, Zhang J, et al. ATM-mediated stabilization of ZEB1 promotes DNA damage response and radioresistance through CHK1. Nat Cell Biol. 2014;16(9):864–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang Y, Zhang M, Li Y, Luo J, Wang Y, Geng W, et al. miR-183 promotes radioresistance of lung adenocarcinoma H1299 cells via epithelial-mesenchymal transition. Braz J Med Biol Res. 2021;54(5): e9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Min H, Yoon S. Got target? Computational methods for microRNA target prediction and their extension. Exp Mol Med. 2010;42(4):233–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Y, Lin B, Yang S, Huang J. IRF1 suppresses colon cancer proliferation by reducing SPI1-mediated transcriptional activation of GPX4 and promoting ferroptosis. Exp Cell Res. 2023;431(1): 113733. [DOI] [PubMed] [Google Scholar]
- 39.Bouker KB, Skaar TC, Riggins RB, Harburger DS, Fernandez DR, Zwart A, et al. Interferon regulatory factor-1 (IRF-1) exhibits tumor suppressor activities in breast cancer associated with caspase activation and induction of apoptosis. Carcinogenesis. 2005;26(9):1527–35. [DOI] [PubMed] [Google Scholar]
- 40.Gao J, Senthil M, Ren B, Yan J, Xing Q, Yu J, et al. IRF-1 transcriptionally upregulates PUMA, which mediates the mitochondrial apoptotic pathway in IRF-1-induced apoptosis in cancer cells. Cell Death Differ. 2010;17(4):699–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen X, Liu L, Mims J, Punska EC, Williams KE, Zhao W, et al. Analysis of DNA methylation and gene expression in radiation-resistant head and neck tumors. Epigenetics. 2015;10(6):545–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu X, Wu Y, Yi K, Hu Y, Ding W, Xing C. IRF1 regulates the progression of colorectal cancer via interferon-induced proteins. Int J Mol Med. 2021. 10.3892/ijmm.2021.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eschrich S, Zhang H, Zhao H, Boulware D, Lee JH, Bloom G, et al. Systems biology modeling of the radiation sensitivity network: a biomarker discovery platform. Int J Radiat Oncol Biol Phys. 2009;75(2):497–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao C, Mirando AC, Sove RJ, Medeiros TX, Annex BH, Popel AS. A mechanistic integrative computational model of macrophage polarization: implications in human pathophysiology. PLoS Comput Biol. 2019;15(11): e1007468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kashfi K, Kannikal J, Nath N. Macrophage reprogramming and cancer therapeutics: role of iNOS-derived NO. Cells. 2021. 10.3390/cells10113194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Recchia Luciani G, Barilli A, Visigalli R, Sala R, Dall’Asta V, Rotoli BM. IRF1 mediates growth arrest and the induction of a secretory phenotype in alveolar epithelial cells in response to inflammatory cytokines IFNgamma/TNFalpha. Int J Mol Sci. 2024. 10.3390/ijms25063463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He AT, Liu J, Li F, Yang BB. Targeting circular RNAs as a therapeutic approach: current strategies and challenges. Signal Transduct Target Ther. 2021;6(1):185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9(9): e1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Han K, Wang F-W, Cao C-H, Ling H, Chen J-W, Chen R-X, et al. CircLONP2 enhances colorectal carcinoma invasion and metastasis through modulating the maturation and exosomal dissemination of microRNA-17. Mol Cancer. 2020;19:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mantovani A, Allavena P, Marchesi F, Garlanda C. Macrophages as tools and targets in cancer therapy. Nat Rev Drug Discov. 2022;21(11):799–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Essandoh K, Li Y, Huo J, Fan GC. MiRNA-mediated macrophage polarization and its potential role in the regulation of inflammatory response. Shock. 2016;46(2):122–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fu E, Liu T, Yu S, Chen X, Song L, Lou H, et al. M2 macrophages reduce the radiosensitivity of head and neck cancer by releasing HB-EGF. Oncol Rep. 2020;44(2):698–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sheng Y, Zhang B, Xing B, Liu Z, Chang Y, Wu G, et al. Cancer-associated fibroblasts exposed to high-dose ionizing radiation promote M2 polarization of macrophages, which induce radiosensitivity in cervical cancer. Cancers (Basel). 2023. 10.3390/cancers15051620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seifert L, Werba G, Tiwari S, Giao Ly NN, Nguy S, Alothman S, et al. Radiation therapy induces macrophages to suppress T-cell responses against pancreatic tumors in mice. Gastroenterology. 2016;150(7):1659-1672.e1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hao Y, Baker D, Ten Dijke P. TGF-beta-mediated epithelial-mesenchymal transition and cancer metastasis. Int J Mol Sci. 2019. 10.3390/ijms20112767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang J, Xu Z, Wang Z, Du G, Lun L. TGF-beta signaling in cancer radiotherapy. Cytokine. 2021;148: 155709. [DOI] [PubMed] [Google Scholar]
- 57.Farhood B, Hoseini-Ghahfarokhi M, Motevaseli E, Mirtavoos-Mahyari H, Musa AE, Najafi M. TGF-β in radiotherapy: mechanisms of tumor resistance and normal tissues injury. Pharmacol Res. 2020;155: 104745. [DOI] [PubMed] [Google Scholar]
- 58.Chen J, Liu X, Zeng Z, Li J, Luo Y, Sun W, et al. Immunomodulation of NK cells by ionizing radiation. Front Oncol. 2020;10:874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Flavell RA, Sanjabi S, Wrzesinski SH, Licona-Limon P. The polarization of immune cells in the tumour environment by TGFbeta. Nat Rev Immunol. 2010;10(8):554–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vanpouille-Box C, Diamond JM, Pilones KA, Zavadil J, Babb JS, Formenti SC, et al. TGFbeta Is a Master Regulator of Radiation Therapy-Induced Antitumor Immunity. Cancer Res. 2015;75(11):2232–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chakrabortty A, Patton DJ, Smith BF, Agarwal P. miRNAs: potential as biomarkers and therapeutic targets for cancer. Genes (Basel). 2023. 10.3390/genes14071375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meng L, Xu J, Ye Y, Wang Y, Luo S, Gong X. The combination of radiotherapy with immunotherapy and potential predictive biomarkers for treatment of non-small cell lung cancer patients. Front Immunol. 2021;12: 723609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chang L, Graham P, Hao J, Bucci J, Malouf D, Gillatt D, et al. Proteomics discovery of radioresistant cancer biomarkers for radiotherapy. Cancer Lett. 2015;369(2):289–97. [DOI] [PubMed] [Google Scholar]
- 64.Yang IP, Yip KL, Chang YT, Chen YC, Huang CW, Tsai HL, et al. MicroRNAs as predictive biomarkers in patients with colorectal cancer receiving chemotherapy or chemoradiotherapy: a narrative literature review. Cancers (Basel). 2023. 10.3390/cancers15051358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang XD, Xu XH, Zhang SY, Wu Y, Xing CG, Ru G, et al. Role of miR-100 in the radioresistance of colorectal cancer cells. Am J Cancer Res. 2015;5(2):545–59. [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang Y, Yu J, Liu H, Ma W, Yan L, Wang J, et al. Novel epigenetic CREB-miR-630 signaling axis regulates radiosensitivity in colorectal cancer. PLoS One. 2015;10(8): e0133870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zheng L, Zhang Y, Liu Y, Zhou M, Lu Y, Yuan L, et al. MiR-106b induces cell radioresistance via the PTEN/PI3K/AKT pathways and p21 in colorectal cancer. J Transl Med. 2015;13:252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ma W, Yu J, Qi X, Liang L, Zhang Y, Ding Y, et al. Radiation-induced microRNA-622 causes radioresistance in colorectal cancer cells by down-regulating Rb. Oncotarget. 2015;6(18):15984–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The TCGA data used in the analysis was provided by UCSC Xena [https://xenabrowser.net/datapages/]. The Km plot used in the analysis was provided by Kaplan–Meier plotter [https://www.kmplot.com/]. The miRNA sequencing data used in the analysis was provided by Gene Expression Omnibus (GEO). The series number is GSE159528.