Abstract
Cardiovascular disease (CVD) remains a global economic burden even in the 21st century with 85% of deaths resulting from heart attacks. Despite efforts in reducing the risk factors, and enhancing pharmacotherapeutic strategies, challenges persist in early identification of disease progression and functional recovery of damaged hearts. Targeting mitochondrial dysfunction, a key player in the pathogenesis of CVD has been less successful due to its role in other coexisting diseases. Additionally, it is the only organelle with an agathokakological function that is a remedy and a poison for the cell. In this review, we describe the origins of cardiac mitochondria and the role of heteroplasmy and mitochondrial subpopulations namely the interfibrillar, subsarcolemmal, perinuclear, and intranuclear mitochondria in maintaining cardiac function and in disease-associated remodeling. The cumulative evidence of mitochondrial retrograde communication with the nucleus is addressed, highlighting the need to study the genotype-phenotype relationships of specific organelle functions with CVD by using approaches like genome-wide association study (GWAS). Finally, we discuss the practicality of computational methods combined with single-cell sequencing technologies to address the challenges of genetic screening in the identification of heteroplasmy and contributory genes towards CVD.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12964-024-01899-x.
Keywords: Cardiovascular disease, Organellogenesis, Mitochondrial subpopulations, Cell-organelle communication, Computational biology, Mitochondrial genetics
Background
The recently concluded Wellcome Conference on Mitochondrial Medicine (2024) held in the UK brought together world leaders to discuss the advances and voids in mitochondrial disease research [1]. Keynote speakers highlighted the need to depart from traditional therapies addressing biochemical changes and adopt strategies to replace mutant mitochondria, specific to the disease and organ affected. However, such targeted approaches need well-characterized models as the same mitochondrial genetic variations may express different phenotypes across individuals, making identification of the best therapeutic course of action a challenge in many studies. Advanced technologies in sequencing identifying the causal variants, gene therapies, and large human trial data need to be utilized for this purpose. Likewise, the National Heart, Lung, and Blood Institute’s (MD, USA) expert group on Mitochondrial research in the Cardiovascular system has highlighted the lack of rigorous and quantitative information in the field to explore mitochondrial therapies that can treat heart failure (HF) [2]. The primary recommendation of the group was to foster a cross-disciplinary approach by bringing together Biologists, Bioinformaticians, Engineers, and Clinicians in designing team science proposals to understand mitochondrial functions. To do so, there needs to be a comprehensive review covering the current knowledge and systems-based approaches that can help researchers in multidisciplinary teams identify promising future directions. Our review covers fundamental aspects of mitochondria in cardiac development, its role in pathology, repair, and remodeling along with the various signaling pathways involved in the quality control of this energetic organelle. However, it is incomplete without providing solutions that answer many of the key questions that remain elusive in the community such as the role of distinct sub-populations of mitochondria in cardiomyocytes, the contribution of mitochondrial DNA (mtDNA) variants, and the effects of mitochondria-nucleus crosstalk to cardiovascular health. Therefore, we discuss some of the existing state-of-the-art tools available to the community to provide computational rigor in unraveling some of these mysteries, benefit from the data available in some of the repositories, and generate new approaches to study the role of mitochondria in cardiovascular disease. In recent times, several genomic technologies such as long-range sequencing, single-cell RNAseq, deep-sequencing etc., have made it possible to study the organelle in extreme detail and the data generated should not remain underutilized by multidisciplinary groups. Our aim of the review is to bring these groups together to study this organelle and design therapeutic approaches that would treat cardiovascular disease.
Mitochondria in cardiac development
Respiratory transitions in cardiogenesis
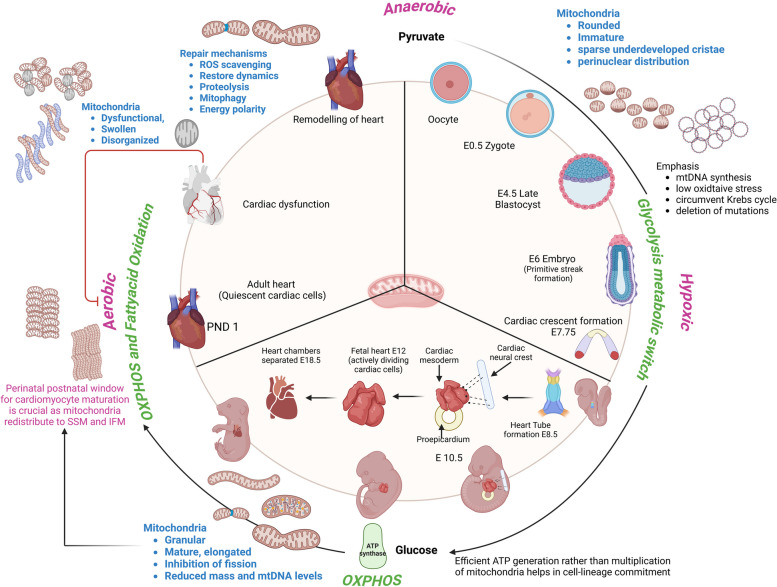
Cardiogenesis is a well-orchestrated event beginning from the heart tube formation to the first beat of the embryo at 3 weeks into gestation in humans. The fascinating arrangement of complex cell types in the heart arises from the maturation of progenitor cells in the fertilized egg from the cardiac mesoderm, proepicardium, and cardiac neural crest [3]. Given the hypoxic environment in which embryogenesis occurs, meeting the energy requirements of the cell is critical in determining the growth and fate of the embryo. Hence it is not surprising to note that 100,000 mitochondria are harbored in the oocytes compared to 5000–8000 in an adult cardiomyocyte [4, 5]. Rather, the metabolic transition in embryogenesis is intricate and involves signaling molecules that drive the differentiation and transition processes as outlined in Table 1. Disturbances in the transition process as observed during preterm birth predispose individuals to cardiovascular-related mortality in adulthood [6].
Table 1.
Metabolic transitions in cardiogenesis and re-distribution of mitochondria
| Cell type | Respiration | Energy substrate | Mitochondrial distribution and features | Ref. |
|---|---|---|---|---|
| Oocyte | anaerobic | Pyruvate > Lactate > fatty acid > Glucose |
Distribution: Structured subpopulations of mitochondria exist with 90% of the ooplasm occupied by mitochondria mostly distributed in the perinuclear region. They do not exhibit usual morphology and have arc-like cristae Features: Even in presence of oxygen and functional mitochondria, oocyte utilizes lactate/pyruvate for energy by PPP rather than glucose by glycolysis because the energy is instant. Use of the adenosine salvage pathway was found prominent in case of bovine oocyte |
[7–11] |
| Blastocyst | aerobic (TE cells) and anaerobic/quiescent (ICM) | Glucose > glutamate > Pyruvate/Lactate > Fatty acids |
Distribution: During late morula, mitochondria appear vesicular with sparse cristae due to anaerobic glycolysis and gradually transform to a lamellated form with the maturation of embryo to accommodate ETC and start with the aerobic oxidation but have immature cristae. In TE mitochondria are long and slender while the ICM mitochondria are spherical due to greater oxygen consumption by TE compared to ICM Features: Accumulation of mitochondria around ER is a feature observed in this stage. Rapid increase in oxygen consumption compared to the single cell zygote stage. The vesicular state also prevents accumulation of toxic ROS products. At implantation, first the trophectoderm and then the entire embryo gain capacity to replicate mtDNA. The mtDNA replication does not take place up to and including the morula stage and the number of mtDNA copies/blastomere is progressively decreased after each embryonic cell division. This results in these blastomeres progressively losing their capacity to generate ATP through OXPHOS as they become more reliant on anaerobic respiration |
[11–14] |
|
Mesoderm (Lateral plate mesoderm > Cardiogenic mesoderm > heart fields) |
High dependence on OXPHOS (Key point of metabolic switch with highest basal RCR) | Pyruvate/Glutamate |
Distribution: Mitochondria transform from a granular and fragmented structure to a filamentous reticular elongated network with mature cristae that can produce energy more effectively Features: Mitochondrial fission is inhibited at this stage to conserve and produce more energy and less ROS. Despite reduction in mitochondrial mass and mtDNA levels, cells show high mitochondrial efficiency The Drp1 gene silencing induces metabolic switch with reliance on anaerobic glycolysis |
[15, 16] |
| Endoderm | Dependent on OXPHOS | Glucose |
Distribution: Mitochondria are elongated and matured as they differentiate from mesoendoderm to endoderm and show high respiratory capacity Most studies are reported in stem cells and results in vivo systems is not known |
[17, 18] |
| Cardiac neural crest | aerobic glycolysis and low OXPHOS | Glucose > cholesterol (Shh pathway) |
Features: Mitochondria are responsible for differentiation of neural crest cells [19]. metabolic abnormalities leading to neurocristopathy is the reason behind Leigh syndrome |
[20] |
| Proepicardium/epicardium | Aerobic | Glucose | Features: Undergo mesenchymal to epithelial transformation to form the epicardium protecting the heart | [21] |
PPP pentose phosphate pathway, ETC electron transport chain, RCR respiratory control ratio, TE trophoectoderm, ICM Inner cell mass, ER Endoplasmic reticulum, Shh Sonic hedgehog pathway
The transition from an oocyte to fetal, neonatal, and adult is immensely complex and involves a metabolic phase transition from hypoxic to anoxic to aerobic environments linked to a shift from glycolysis to fatty acid oxidation [7, 8, 22]. Figure 1 summarizes the key aspects of mitochondrial participation in the course of this cardiac development phase. During this phase transition, the mitochondria actively provide energy to the cell while simultaneously undergoing a dramatic transition from an immature, rounded organelle with sparsely developed cristae to a mature, organized, and elongated structure with dense cristae architecture [12, 17, 22]. Recently Beutner et al. (2024) compared mitochondria from myocytes of mouse embryonic hearts at different intracellular locations and confirmed that location dictates mitochondrial structure and function [23]. While the known spatially distinct populations in an adult heart are interfibrillar, subsarcolemmal, and perinuclear, the study defined a new “cytoplasmic” category to differentiate the predominant mitochondria found during the embryonic stages (E9.5) that are not associated with any particular structure, and that disappear by post-natal day1. The perinatal window is the critical stage during which changes to mitochondria structure and distribution increase the mitochondrial volume in myocytes. Interfibrillar and subsarcolemmal subtypes are localized predominantly in the postnatal myocytes while perinuclear mitochondria support the embryo predominantly at E16.5. To accommodate the structural maturation, more copies of mtDNA are made to promote the electron transport chain (ETC) gene expression, supporting the increasing demands of the developing heart. The hypoxic conditions help in mtDNA synthesis and avoid oxidative stress in the immature mitochondria by circumventing the Krebs cycle. Oocytes have an evolutionarily conserved strategy of suppressing the complex-I function to maintain a ROS-free environment for healthy embryogenesis [24]. Next, the shift in energy substrate from pyruvate to glucose (10–85%) during the development of blastocyst leads to a metabolic switch to oxidative phosphorylation (OXPHOS) to generate adenosine triphosphate (ATP) more efficiently as the number of mitochondria per cell is diminished. A plausible explanation for such adaptations is that these processes reduce stress-induced mutations in mtDNA, which is important as they lack protective histones or repair mechanisms. Thus, mtDNA is highly stable, and pathological mutations are selectively eliminated before embryogenesis in a process called purifying selection [25]. Abnormalities in glucose metabolism/utilization at this stage have resulted in physiological complications due to abnormal mitochondrial energy production and division [26]. On the contrary, supplementation of oocytes with autologous mitochondria resulted in compromised cardiac architecture (mucoid degeneration of the valves) and weight gain in the offspring despite enhancing fertility and thereby raises questions about the use of the technique for assisted reproductive technologies [27]. Arribat et al., (2019) demonstrated in mice that the epigenetic influence of the additional copies of mtDNA on the nuclear DNA (nDNA) resulted in the cardiac abnormalities highlighting the tight regulation of mitochondrial quality control and the organelle-organ communication [28]. Further post-natal stages show a rapid conversion to fatty acid oxidation (80%) from glucose owing to its increased uptake by the mitochondria, activated by several key regulators of the tricarboxylic acid (TCA) cycle and the availability of oxygen [29, 30]. Although metabolic switching seems beneficial, it is identified as the key perpetrator of regeneration as the process was found reversible by inhibition of Pdk4, which enhances glucose oxidation [31, 32]. Therefore, for grasping the mitochondria’s involvement in cardiomyocyte maturation and regeneration under pathological conditions, it is imperative to understand the transcriptional signals and inter-organelle communications that drive the process.
Fig. 1.
A visualized summary of mitochondrial participation in cardiac development
Mitochondrial adaptations and their impact on cardiogenesis
Mitochondria play a critical role in eukaryotic cell differentiation. Typically, the immature mitochondria in stem cells, influenced by metabolic transitions, develop complex structures, propelling the organelle to undergo fission, a prerequisite for differentiation [33]. The process is triggered by the Wnt and Notch signaling pathways through the Forkhead box O (FoxO) transcription factors that control the mitochondrial fission-1 (Fis1) gene. Contrary to this hypothesis, Lee (2020) found that the morphological changes in mitochondria that could trigger differentiation were discrete from the expression of Fis1 and depended on a balance between fusion and fission as explained by the ratio of genes mitofusion2(Mfn2) and dynamin-1-like (Dnm1L) [34]. An in-depth understanding of the process of organogenesis and organellogenesis during development demonstrated that mtDNA quality control is regulated by 11 proteins (TFAM, POLG, TWINKLE, TOM complex, TIM complex, HSP60, CLPP, LONP1, OPA1, MFN1/2, and DRP1) that regulate the OXPHOS pathway [27, 28]. When mtDNA replication is promoted by lineage-specific markers (vimentin, nestin, β-tubulin), upregulation of PolgA, PolgB, Tfam, ATPase5b, COXI is observed, and the organelle differentiates to an elongated form with swollen cristae to consume more oxygen and generate higher membrane potential and ATP. This adaptation of organelle with organ goes hand-in-hand with distinct patterns of mitochondrial distribution observed in the myogenesis of zebrafish (promoted via the Sonic hedgehog pathway) and the mtDNA mosaicism (Heteroplasmy: existence of both mutant and wild-type mtDNA genomes within a cell) in human development reported recently [35].
Cardiac mesoderm is considered as the starting point of mammalian cardiogenesis. The differentiation of mesodermal cells to the heart-forming area is controlled by signaling molecules involved in the Wnt/β-catenin pathway, TGFβ and BMP2. Several detailed reports of the process and commitment to various cell types and their contribution to portions of the heart have been presented in previous reviews [36, 37]. Here we emphasize the role of mitochondria to the development process as there are recent fascinating reports about the ways in which mitochondrial dynamics are involved in cardiogenesis [15, 38]. Mostafavi (2021), observed that despite higher mitochondrial activity, coupling efficiency and higher ATP production during mesodermal differentiation, there was a decrease in organelle content and its DNA compared to undifferentiated human pluripotent stem cells (PSCs) [38]. This could be caused by a process of morphological adaptation to drive cell-lineage commitment rather than multiplication as demonstrated earlier in induced PSCs. Involvement of mitochondrial fission gene Drp1 was found to limit the mesodermal differentiation as in this case, the use of Mdivi-1 a selective Drp1 inhibitor elevated the mesodermal expression of cardiac genes in the stem cells accompanied by increased respiratory activity in the mitochondria and decreased glycolysis. In a recent similar study, depletion of Tfam also reduced mesoderm differentiation efficiency and the commitment of human PSCs to cell-lineages highlighting the regulatory role of mitochondria in cell-fate determination and demonstrating that any defects in the process could lead to fetal cardiac abnormalities [39].
Evidence suggests that the maturation of cardiomyocytes is driven in part by changes in the distribution of mitochondria among its subcellular locations, notably the subsarcolemmal and interfibrillar populations [40]. Cardiac cells transition from an actively dividing state in the fetus to a quiescent state postnatally. Their independent respiratory ability is supported by well-developed mitochondria which support the heart with the energy required for its uninterrupted contractility. The perinatal-postnatal window is a crucial phase of cardiomyocyte maturation and mitochondria contribute to the maturation process as they move from perinuclear spaces to interfibrillar and subsarcolemmal spaces with designated functional and biochemical independency [22]. The interfibrillar mitochondria (IFM) has higher ETC activity compared to the subsarcolemmal mitochondria (SSM) and supports energy for contractile activity while the SSM provides energy to support the biochemical reactions in the muscle cells [41]. Although maturation of cardiomyocytes (CMs) and mitochondria are intertwined, there is evidence to suggest that mitochondria respond to metabolic stimuli first and drive subsequent cell maturation [30]. First, stimulation by the hypoxic environment drives the perinuclear distribution of mitochondria and fission-fusion dynamics through AMPK, HIF-1α and mTOR signaling pathways. Second, the availability of several metabolic drivers to generate ATP such as glucose, fatty acids, branched-chain amino acids (BCAA), hormones, glucocorticoids, estrogen, retinoic acid, angiotensin-II, Noncoding RNA and O-GlcNAcylation, drive the crosstalk between genetic and metabolic signaling [40]. The fetal dependence on glycolysis shifts to OXPHOS post-natally from breast milk which contains higher amounts of fatty acids [42]. This is associated with the maturation of CMs. Recent empirical evidence by Li (2023) confirmed that activation of AMPK is a driving factor for the maturation of CMs derived from human-induced PSC’s preceded by mitochondrial maturation [43]. Activating AMPK increased cellular ATP, promoted biogenesis, increased the membrane potential and fatty acid uptake, clearly indicating metabolically mature organelle drive cardiomyocyte development. Thus, the maturation of mitochondria may not be a consequence of cardiomyocyte maturation but may actually trigger the maturation due to their crucial function in maintaining the metabolic phenotype [44]. The process of postnatal heart development needs to be systematically studied because nutrient changes during the first few days after birth may have significant effects on mitochondrial structure, physiology, biochemical processes and regulate the cell cycle.
Inter-organelle communication and the role of mitochondria
During development, particularly in the perinatal-postnatal window, changing metabolic environments prompt dynamic interactions among cell organelles [45]. Contact sites between organelles are extremely close (~ 10 nm) and allow for efficient signaling with loss of contact known to cause autophagic responses. Among these communications, mitochondria play an essential role as they occupy 35% of the cell volume of an adult cardiomyocyte and are responsible for handling the high energy demands of the heart and calcium dynamics required for rhythmic electrical activity. Hence it is not surprising that imbalances in such inter-organelle communication in the heart frequently result in pathological conditions. More importantly, these could be divided into two aspects (a) mitochondria-nuclear communication and (b) mitochondria-cell organelle communication (mainly endoplasmic reticulum (ER), Golgi, and lysosome). Here, we focus on the role of mitochondria-organelle crosstalk in the context of cardiac development and the healthy heart.
Mitochondria communication with the nucleus
Unknown until 1991, mitochondrial communication with the nucleus has been recognized as a key process to maintain organelle homeostasis in yeast [46, 47]. Butow and Avadhani [45] were the earliest to investigate the mito-nuclear signaling mechanism in yeast models & mammalian cells, identifying the key regulatory factors converging at TOR signaling in the former while it was Ca2+ dynamics in the latter. In order to control its own metabolism, physiological functions, and respond to stress stimuli, mitochondria have to signal the nucleus to regulate relevant genes by communicating through retrograde signaling [48]. Physically mitochondria are known to interact with the other organelle via MAM (mitochondria associate membranes) [49, 50]. They help to regulate calcium, lipid and intracellular trafficking and include multiple protein complexes & ion channels. Many such ‘Mitonuclear’ communications have been reviewed by Quirós et al. and are essential for orchestrating cellular events [51]. The complex interplay however needs additional evaluation through in vivo models at different genetic scales, to understand this complex regulation during heart development. Despite the fascinating ways by which the mitochondria communicate with the nucleus, direct nucleus-mitochondrial contact has not been reported yet [52].
Berg and Kurland hypothesized that the retrograde translocation of genes to the nucleus is purely based on selection and high mutation in mitochondrial genes under stress [53]. The emerging reports on carrier-mediated translocation of mitochondrial proteins under stressed environment has proven the hypothesis with identification of GPS2 protein as an intermediary for mitochondrial retrograde signaling that can activate nuclear-encoded mitochondrial genes and mediate translocation of AIFm2 (primarily located in the mitochondrial intermembrane space) to nucleus by forming HNE adducts (transport mechanisms specific for translocation) only under pathophysiological states [54–56]. A mouse model of cardiomyocyte-specific knockout of MnSOD was the first evidence of embryonic lethality caused by the translocation of AIFm2 to the nucleus resulting in apoptosis [57]. As the nuclear effectors of mitochondrial signaling are well-known to control the quality of the organelle, mitochondrial signaling is known to help the cell eliminate dysfunctional organelles and is distinctly bidirectional based on stress responses. To date, some of the canonical signals include calcium, ROS, Adenine nucleotides (ADP, AMP, NAD, NADH) and TCA intermediates (Acetyl-CoA, αKG, succinate, fumarate) which have been studied extensively in yeast systems [58]. Though several studies have evaluated signaling proteins that function retrogradely [59], there are very few studies that specifically identified the role of retrograde signaling in the mammalian heart and have been listed in Table 2. Novel techniques such as in-situ mapping of RNA-genome interactions lately provided evidence of a previously unidentified phenomenon of transcriptional regulation of nuclear signaling by the mitochondria in endothelial cells [57]. The study revealed an increasing association of mitochondrial RNA with nuclear chromatin in diabetic stress, resulting in the activation of inflammatory responses. A compelling model in the mammalian system has been developed recently with mice engineered to have different mtDNA backgrounds that showed differences in cardiac metabolism and aging [60]. The increased non-pathogenic heteroplasmy in this model caused progressive metabolic stress during adulthood possibly due to nuclear-mitochondrial cross-talk. Such models hold promise for future studies to explore inter-organelle communication. Computational advancements from Hodgkinson [51], Ryten [61], and Chiu [62] labs hold promising approaches for the future to decipher the role of retrograde signaling in cardiac development. Several molecular mechanisms in this area have been studied only in yeast systems, whose role needs evaluation in the development of the developing heart.
Table 2.
Identified roles of retrograde (bi-directional) signals that control mitochondrial-nuclear signaling established in heart
| Retrograde signal | Role in cardiac physiology | Ref. |
|---|---|---|
| p53 | Cause stress induced apoptosis. Absence of p53 reduced mitochondrial injury and improved cardiac function | [63] |
| AIFm2 | Translocation induces apoptosis | [55, 56] |
| SP1 | Calcium regulation | [64] |
| SIRT3 | Cardioprotective in stress induced cell death | [65] |
| miR-378 | Cardioprotective antiapoptotic miRNA | [66] |
| HIG2A | Resistance to hypoxia | [67, 68] |
| UPRmt | Stress response to recover defective mitochondria (Critical pathways involved: a) mtHSP70 and HSF1, b) ERα-NRF1-HTRA2, c) ATF4/ATF5-CHOP, Sirt3-FOXO3a-SOD2) | [69, 70] |
| Atfs-1 | Not known | - |
| CLK-1 | Not known | - |
| Rtg1P/Rtg3P | Not known | - |
| Fumarase | Elevated fumarate is Cardioprotective via activation of the Nrf2 Antioxidant Pathway | [71] |
| TIN2 | Not known | - |
| RECQL4 | Not known | - |
Contrary to the belief that the mammalian mtDNA is a naked, circular, unshielded structure with multiple individual copies in an organelle, it is actually packaged into heritable units of distinct ellipsoid mtDNA-protein complexes consisting of multiple mtDNA copies termed as the ‘mitochondrial nucleoid’ [72]. Because of its compact shape, lack of introns, and frequent overlapping of reading frames between nearby genes, mtDNA has a higher gene density than nDNA [46]. Despite its small size, mtDNA-transcribed mRNA has a remarkable presence across the total cardiac mRNA (30%) [47]. The mitochondrial nucleoid is composed of more than 50 proteins that provide stability to the mtDNA much like histones with nuclear DNA. Of critical importance among these proteins is Tfam, which maintains the packaging, replication, copy number and transmission of mtDNA. Several studies have reported that inactivation of Tfam caused lethality in embryonic and adult hearts characterized by myocardial wall thickening, dilated cardiomyopathy, and mitochondrial dysfunction [73, 74]. Recently, the presence of nuclear-encoded transcription factors (MOF, AP1, CEBPB, MEF2D), and the contribution of nuclear spliceosome complex to mtDNA-splicing provided strong evidence of control of mitochondrial replication/transcription demonstrating evolutionary adaptation of the mammalian mtDNA in the nucleoid [75]. Apart from the proteins forming the mitochondrial nucleoid, the mitochondria are regulated by the non-coding RNA that contributes to cardiac homeostasis. Most widely studied among them are the microRNAs termed as ‘mito-miRs’ which have a preferential mitochondrial localization. Although their origin has been debated considering the presence of pre-miRNA in mitochondria which suggests their origin from mtDNA, their expression has been critical in regulation of OXPHOS in infarcted hearts (miR-1 and miR181c) [76]. The import-export mechanism for these small non-coding RNA are topics of ongoing research to reveal the nuclear-mitochondrial crosstalk. Likewise, mitochondria lack import for long non-coding RNAs and their presence within the organelle could well direct their origins to mtDNA. The LIPCAR (long noncoding RNA uc022bqs.1) has been the only mitochondrial long noncoding RNA upregulated in patients with LV remodelling following MI [77]. In conjunction, evidence of circular RNAs in the mitochondria could be the early evidence for nuclear-mitochondrial crosstalk [78] but their cardiac-specific roles are yet to be uncovered. However, these could be of potential diagnostic or therapeutic use as suppressing the mitochondrial fission and apoptosis-related circular RNA (MFACR), the first circular RNA to be discovered in the heart, was found to be cardioprotective in mice. MFACR suppression resulted in reduced mitochondrial fission mediated by the interaction of miR-652-3p and MTP18 [79].
Mitochondria communication with other organelles
The positioning of organelles in heart muscle cells is governed by tightly regulated processes such as Ca2+ regulation, autophagy, and lipid metabolism. Through early electron-microscopy studies on embryonic heart development, we know that the myocardium is not a uniform compact tissue and that myocytes contain scattered free ribosomes, Golgi, mitochondria, granular endoplasmic reticulum, and phagocytes. Progressive development brings order and differentiation to heart tissue. The proteins required for the recruitment of additional cell types to the developing heart are processed by these organelles to create extra-cellular matrix and collagen and the development of additional layers of fibroblasts [48]. It is this reorganization phase that necessitates contact among organelles to meet the requirements of cardiomyocyte function [52]. While it is known that embryonic development is accompanied by changes in the volumes of organelles, we lack information on how these organelles communicate to maintain their volumes according to cellular requirements [45].
Recently, Kim (2022) mapped physical interactions at high resolution between mitochondria, sarcoplasmic reticulum, and lipid droplets in mice hearts using focused ion beam scanning electron microscopy. The images showed the highest surface area interaction by postnatal day 14. This is supported by the fact that postnatal cardiomyocyte contraction is accompanied by the maturation of Sarcoendoplasmic Reticulum Calcium ATPase (SERCA) pumps by day 21 [80]. Any altercation of these interactions can severely affect the heart function. In 2023, Jan Boren’s group demonstrated that PCSK9-deficient mice had impaired contractile function leading to cardiomyopathy and premature death due to altered mitochondria-ER contact [81]. This is also the case in Mfn2 deficient mice (Mfn2 tethers mitochondria to ER) [82]. A more direct contact with ER was established by FUNDC1, an outer mitochondrial membrane protein, by binding to the Inositol triphosphate (IP3) receptor which was significantly suppressed in patients with HF [83]. Thus, cardiac remodeling during postnatal development is accompanied by organelle interactions which is critical for calcium handling and energy distribution. A comparison of interactions across mitochondrial subtypes has not been made yet that would reveal developmental profiles that vary across regions where these organelles are distributed.
The mitochondria and Golgi positioning in cardiomyocyte development is a topic of interest and needs more research as they complement each other by acting as Ca2+ sinks and regulating protein trafficking. Given the highly dynamic environment of the embryo, Yuri Morozov recently found that stress impacts the Golgi apparatus even before structural changes can manifest in the mitochondria [84]. The impact of an early malfunction in the Golgi on the mitochondria has not been investigated particularly in the heart although upcoming reports hint at the control of mitochondrial plasticity through Golgi network vesicles, which is interesting to study in heart development [85, 86].
Lysosomes traditionally facilitate the degradation and recycling of cellular waste through autophagy. Canonically, this process also maintains mitochondrial quality in the cell by selective removal of damaged mitochondria, called mitophagy. A recent review has sufficiently summarized the pathways leading to mitophagy and its role in cardioprotection. However, most of these reported signals are transmitted during stress or from damaged mitochondria [87]. We thereby sought to identify driving factors in cardiac developmental stress that promote mitochondria-lysosome communication. Gong et al., provided strong evidence of mitochondrial maturation in perinatal stages driven by Parkin-dependent mitophagy [88]. The increased oxygenation drives Mfn2-Parkin interaction to remove fetal mitochondria causing a metabolic shift from glycolytic to fatty acid metabolism in the mouse heart. The process is likely driven by the loss of mitochondria-ER contact sites and the recruitment of optineurin that drives the binding of autophagosome protein LC3 forming a mitophagy complex around the mitochondria [89, 90]. Studies on such mitochondria-lysosome communication are rudimentary and require an in-depth understanding of how they regulate cardiac maturation. Besides mitophagy, new roles for mitochondria-lysosomes interactions have been identified such as controlling calcium dynamics, forming hybrid organelle, and compartment-selective microautophagy [91–93]. However, their activation or role in embryonic heart development is yet to be explored.
Mitochondrial dysfunction and cardiac remodeling
Organelle phenotype and cardiac physiology
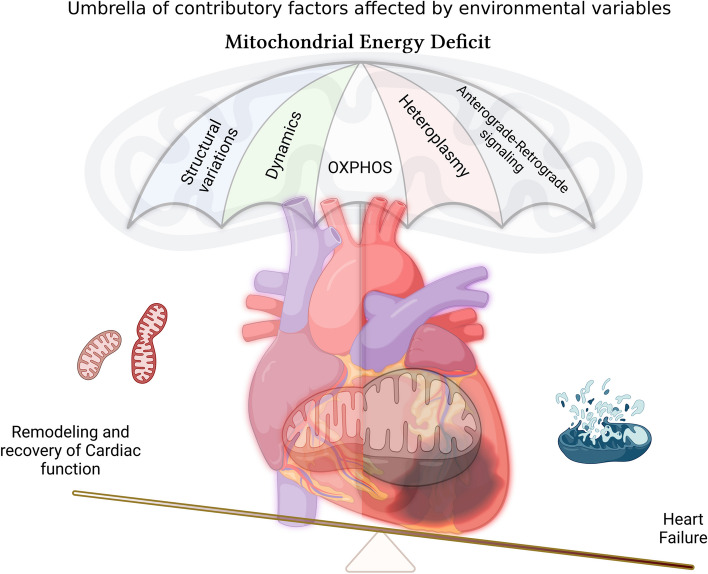
Early death in children with inherited mitochondrial diseases often involves cardiovascular complications which in part is due to the dysfunctional organelle contributing to the disease phenotype [94–96]. Abnormalities in mitochondria can significantly contribute to impaired OXPHOS deficiency leading to lower oxygen utilization and thereby contractile dysfunction, apoptosis, and cell death. To understand the holistic impact, studies must integrate cardiac physiology and structural alterations with mitochondrial attributes such as structure, respiratory capacity, and dynamics. When combined with integrating the consequence with genomic changes such as expression or mutation, it provides an enormous understanding of the mitochondrial basis for cardiovascular disease. Unfortunately, few studies (mostly pre-clinical) have recognized such integration to provide a comprehensive understanding of how an organelle dysfunction leads to phenotypic changes and/or remodeling (Additional Table 1 and Fig. 2). The etiology of mitochondrial dysfunction in the development of disease is complicated and is spatio-temporally associated with elevation of sympathetic signaling, oxidative stress, inflammatory response, and metabolic syndromes such as diabetes [97]. Additionally, the same mutation can lead to cardiomyopathy symptoms in some patients while others may remain asymptomatic. Current therapies targeting the reactive oxygen species (ROS), and oxidative stress were unsuccessful, exposing the dearth of knowledge we have in this field. We hereby discuss a brief outcome of studies that have integrated the heart’s function and its mitochondrial characteristics under pathological milieu (Additional Table 1). The table also provides information on attempted interventions and their targets in mitochondria.
Fig. 2.
The image portrays key factors contributing to the mitochondrial health which in-turn affect the cardiac function
Structural
Diseased hearts show distinct morphological changes in mitochondria which are in sync with functional alterations. The organelle swells due to edema, lacks cohesive cristae structure, and appears vacuolar under conditions of stress due to ischemia-reperfusion injury (IRI) [98–104], hypertrophy [105], cardiomyopathy [106–108] and acute myocardial infarction (AMI) [109, 110]. Some of these HF conditions are also encountered in comorbidities such as obesity and diabetes, which additionally contribute to the organelle’s appearance [98, 99, 111]. Notably, such hearts showed reduced ejection fraction (EF) and fractional shortening (FS) with prominent diastolic dysfunction due to stiffening without any changes in the chamber dimensions. While tachycardia and fibrillation were prominent in IRI with limited recovery of ventricular function, hypertrophy caused by diabetes resulted in reduction in E/A ratio, and left ventricular (LV) mass. The above changes were observed along with disorganized mitochondrial (irregular patterns) instead of the ‘pearl string appearance’ (linear arrangement) [99]. Interestingly, these changes did not alter the heart rate, which could have led to heart failure with preserved ejection fraction (HFpEF).
The severity of disease condition can sometimes be observed by a loss of mitochondrial number in conditions of hypoxia, and hypertrophy especially in hemodialysis and diabetes [111–113]. Quantification of the dimensions of the diseased mitochondria reveals a reduction in cross-sectional area, perimeter, and Feret diameter with increased roundedness due to IRI. These structural changes were in addition to the loss of membrane potential and lower oxygen consumption that led to premature beats.
Dynamics in remodeling
Cardiac stress is well-known to promote fission of mitochondria as observed in animal models of pressure overload and cardiomyopathy. Traditionally, these hearts show higher levels of fission protein DRP1 [2, 107, 114, 115]. However, this might not hold true in all cases as a recent report on neonatal cardiomyopathy revealed downregulation of fusion (Mfn1 and Mfn2) and fission (Drp1 and Fis1) [106]. Furthermore, diabetic hearts did not show any changes in fission/fusion protein despite the increase in area/size [113]. In addition, the study showed significant downregulation of mitofilin in the interfibrillar mitochondria from diabetic mouse hearts. These dynamic changes in cardiac mitochondria were associated with diastolic dysfunction, increased posterior wall, septum thickness along with drop in EF and FS. Recent studies indicate that mitochondrial fission is critical in promoting cardiac injury [116] and inhibition of fission using drugs (Midvi1, RTA408 and Dapagliflozin) alone was found to be cardioprotective [117–119] even in cases of comorbidities such as hypertension and diabetes [120].
OXPHOS
OXPHOS is critical for the supply of ATP required for myocardial contractility. Dysfunction of the apparatus leads to promotion of oxidative stress by elevating the release of free radicals. In the case of the OXPHOS, rather than changes in protein abundance, it is frequently the presence of functional mutations that disrupt the machinery. Dysfunctional / mutated OXPHOS proteins have been implicated in several mitochondrial myopathies involving mutations in genes encoding for OXPHOS proteins [96]. Although a decline in any of the complex activities means reduced respiratory capacity, reports integrating ETC-complex activity with physiology have shown innate differences across the mitochondrial subpopulations with respect to susceptibility to the pathology [115]. During pressure overload, rat hearts showed reduced complex-IV activity and uncoupling with TCA cycle but studies with an underlying condition of diabetes showed that complex I-V activities declined in IFM alone but not in SSM [113]. More recently, researchers showed that reduction in respiratory activity of IFM as a key factor for remodeling in hypertrophic cardiomyopathy in human heart tissue [121]. Given that comorbidities are a crucial aspect of CVD cases, studies in preclinical model should emphasize further studies in models with comorbidities rather than normal ones [60, 98].
Mitophagy
Imbalances in clearing of dysfunctional mitochondria can trigger apoptosis due to release of mitochondrial contents into the cytosol. Insufficient mitophagy is a characteristic of hearts affected by CVD and has been associated with impaired EF, FS, and high energy consumption. Autophagosomes with damaged mitochondria were a prominent observation in hypertrophic hearts which had reduced EF and increased end-diastolic left ventricular posterior wall thickness (LVPWd) without changes in LV diameter [105]. While an elevated expression of autophagy genes (Lc3-I and II, Lamp2, Pink1 and Parkin) and proteins are featured in hypertrophy [105] and pressure-overload [115] animal models, it is more a complex phenomenon in the human heart especially in patients with diabetic cardiomyopathy (DCM) [122]. The phenotypic differences in diabetic cardiomyopathy vary across the two major types (Type1 and type2 diabetes mellitus) partially owing to opposing effects on the expression of autophagy-related pathways [123, 124]. Compelling evidence in human hearts before and after bypass surgery showed mitophagy after successful bypass which was simultaneously associated with increased biogenesis although the study lacked physiological data on cardiac function [125]. Thus, increasing turnover of mitochondria is beneficial but the differences in lab and clinical observations have posed a challenge in the translation of animal studies to patient care and should not be overlooked. Despite the differences, the characteristic physiological changes in hearts from some of these studies did not differ from those observed in hypertrophy. Rather, stabilizing functional mitochondria looks to be a promising tool in improving cardiac function associated with mitochondrial impairment [121].
Copy-number
Mitochondrial DNA Copy number obtained from the leukocyte is recognized as an independent predictor of CVD [126] and is inversely related to the risk of HF [127]. Recently, Genome wide association studies (GWAS) identified the influence of mtDNA on nDNA methylation through the use of multi-center datasets drawn from several major CVD trials [128]. In its simplest form, copy number is interpreted as the ratio of mtDNA/nDNA. While most clinical studies have reported this number from the blood, preclinical approaches have measured the tissue levels and found similar results. Andres et al., observed increased copy number in heart tissue perfused with blood post-cardiopulmonary bypass as being associated with better functional recovery and improved biogenesis of the organelle, missing the link with cardiac physiology [125].
Overall, in the backdrop of robust mechanisms of mitochondrial genesis and clearance, abnormalities in the mitochondria seemed to be disease and comorbidity-related. The lack of comprehensive understanding of mitochondrial physiology with cardiac physiology has made it difficult to understand CVD pathology which is why drug discovery in certain areas is lacking such as cardiac IRI and AMI. Also, differences across species and limitations of studies reasserting the results in comorbidities have made it difficult for translation to bedside. Studies aimed at co-morbidities and their influence on mitochondria in cardiac IRI were broadly reported in pre-clinical studies by the Kurian group, showing loss of cardioprotection in DCM was due to mitochondrial dysfunction [129–132]. Recently, a mouse model of non-pathologic mtDNA heteroplasmy has been developed which exhibits multiple comorbidities such as HF, pulmonary hypertension, sarcopenia, frailty and premature death in adulthood [60]. Exploring such models will redefine our understanding of the canonical signaling pathways in cardioprotection and help design and identify new drug targets.
Mitochondria in cardiac repair
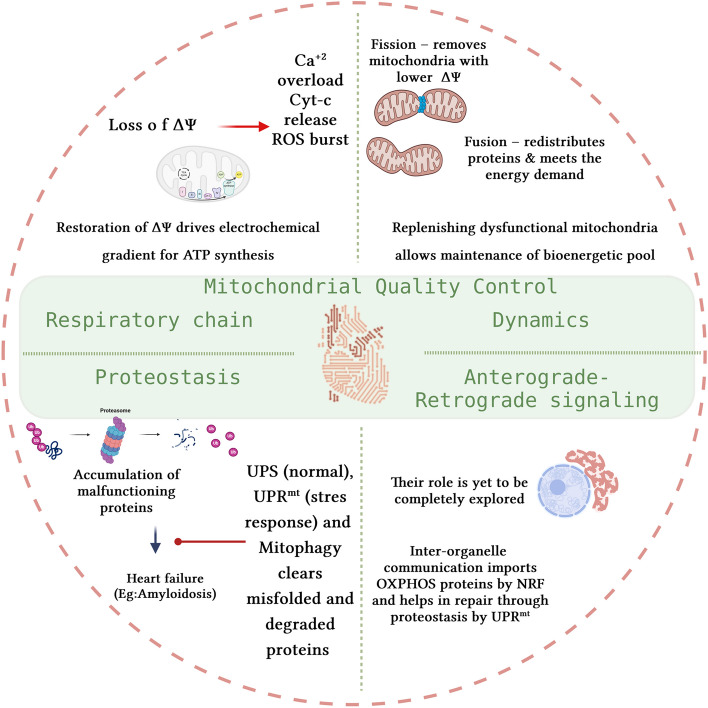
While we have seen how mitochondrial dysfunction contributes to CVD risk, it is equally important to investigate the other side of the coin. Here we discuss how cardiac repair progresses following mitochondrial recovery and helps in remodeling to improve the heart’s function. A recent study indicated the responsiveness of dysfunctional mitochondria to drug treatment in diseased heart tissues that could treat cardiomyopathy in the future [121]. Several mechanisms of quality control are involved in cardiac repair and have been extensively reviewed by others [133]. We discuss here the perturbations that mitochondria undergo during the cardiac repair process and how they influence cardiac function as presented in Fig. 3. Mitochondria have been implicated as a metabolic driver of cardiovascular complications in induced and acquired CVD [134]. Hence the heart must maintain a fine balance of this organelle’s function to generate energy and limit ROS. Mitochondria have developed innate quality control mechanisms to sustain their function in times of imbalance to maintain the cardiac function through repair mechanism and homeostasis is imperative for mitochondrial function and cardiac health. Discussed further in the review is the role of the major contributors to quality control for mitochondria including ROS, proteostasis, dynamics, and antero-retrograde signaling.
Fig. 3.
Mitochondrial contribution to cardiac homeostasis through quality control. ΔΨ-mitochondrial membrane potential, Cyt-c-Cytochrome-c, UPS-ubiquitin proteasome system, and UPRmt-mitochondrial unfolded protein response
ROS
The DNA repair mechanisms in mitochondria are immature, which makes the mtDNA susceptible to damage by the oxidative stress caused by ROS. The ROS is a generated both as a consequence of normal ETC (Complex-I and III) activity or due to mitochondrial dysfunction, for example, monoamine oxidase in the heart [135]. Seminal studies by Hoppel and Lesnefsky group have significantly advanced our comprehension of ROS damage to mitochondria in the heart. These works demonstrate that inhibiting the ETC confers mitochondrial protection in ischemic hearts by preventing ROS-induced damage to cardiolipin and cytochrome-c [136, 137]. On the contrary, a balanced ROS level is necessary for signaling that promotes growth & proliferation as in the case of angiogenesis, contraction by stimulation of calcium channels, and regulates autophagy [138]. The antioxidant system (SOD, catalase, GPx, thioredoxin, and peroxiredoxin) in the mitochondria helps to balance this ROS and maintain physiological signaling and avoid undesirable effects. ROS scavenging has been a primary target to orchestrate cardiac repair followed by ROS-activated pathways such as the AMPK, and RAS and inhibition of ROS-producing enzymes. On the other hand, ROS promotes the transient opening of the mitochondrial permeability transition pore (mPTP) which is beneficial for heart function [139]. Multiple animal studies support the fact that reducing ROS generation through the modulation of the antioxidant system can enhance cardiac health and delay premature aging [140].
ROS is a double-edged sword, requiring tight regulation of its levels and posing a challenge for the proper administration of therapies. For instance, the antioxidant properties of H2S and its donors have proven cardioprotective benefits but the endogenous levels of H2S and functional mitochondria in heart determine the dose and a positive outcome for heart experiencing ischemia-reperfusion injury [141]. Hence, many mitochondria-targeted ROS scavengers such as MitoQ, SkQ, and SS31 more effectively protect the heart than the general ROS scavengers partly due to their effectiveness in preserving functional mitochondria and eliminating pathological ROS. Another important cardioprotective effect of ROS involving the mitochondria is provided by PKCε activation and translocation to mitochondria, induced by ROS. But a rather opposite effect is observed by PKCδ translocation, also triggered by ROS owing to the species involved which in this case was OH* radical [142, 143]. Given that the ROS triggers several pathways, its cardioprotective effect depends on the functional mitochondria status.
The broad effects of ROS-mediated damage are obvious in endothelial dysfunction, hypertension, atherosclerosis, and HF. In particular, the involvement of mitochondria was noted by Lin et al. in patients with atrial fibrillation due to mtDNA damage leading to enhanced ROS [144]. Overall, we have a more complete understanding of the damage of ROS rather than its benefits. Studies of the benefits of ROS observed that ROS modulation regulated adipogenesis [145] in equine adipocytes, which is of potential use to prevent post-infarction remodeling of the heart where lipid deposition and fibrosis are the key reasons for HF. Gangwar et al., demonstrated the benefits of reactive oxygen and nitrogen species in the promotion of cell survival mediated by nitrosylation in murine cardiomyocytes subjected to hypoxia [146]. In a study by Long et al. (2020), muscle regeneration properties of flavonoid silibinin were attributed to increased ROS levels [147]. More recently, Li (2022) reviewed the impact of exercise on muscle health and proposed that exercise increases ROS and improves skeletal muscle activity by inducing epigenetic changes [148]. Despite these findings, the molecular mechanism behind protective ROS signaling remains elusive. It is also not clear if this signaling helps in the regeneration and recovery of damaged hearts and if so what is the role of mitochondria in the process.
Dynamics in cardiac repair
The fusion/fission dynamics of mitochondria provide the organelle with much-required structural integrity in environments of stress [149]. It helps to maintain a healthy pool of mitochondria and therefore altered dynamics of the process are a cause of HF. Recent reviews by Lin et al. (2021) [150], Poznyak et al. (2022) [151], Quiles et al. (2022) [152], and Hausenloy group (2023) [153], extensively discuss how these opposing processes of fusion and fission of mitochondria are regulated through fusion proteins (MFN1,MFN2, OPA1) and fission proteins (DRP1, FIS1, MFF, MiD49/51). At present, we have a reasonable amount of evidence supporting that striking a balance between the two processes is key as low fusion or an excess of fission can trigger apoptosis. At a steady state, metabolic and energetic changes in the cell act as stimuli to promote the maintenance of energy production and are usually associated with morphological changes to mitochondria. The OPA1 protein has been recognized as a key regulator of this process by playing the dual role of being pro or anti-fusion [154, 155]. OPA1 is spliced into 2 forms by OMA1, the long unprocessed form (l-OPA1) and the short-processed form (s-OPA1). The ratio of these forms decides the fusion and pore opening of the inner membrane in the pro-fusion state. Enrichment of s-OPA1 was found to suppress fusion. In heart failure, reduced OPA1 levels have been reported from both human and animal heart tissues although the contribution of these forms in the heart is still unclear [156]. Supporting evidence shows that stress-induced depolarization of mitochondrial membrane due to ROS increases the activity of OMA1, which in turn converts l-OPA1 to s-OPA1, shifting the equilibrium to a pro-fission state [157, 158]. Besides fusion, OPA1 has pleiotropic effects of maintaining cristae morphology and respiratory efficiency, making it a sought-after drug target [159]. In 2018, ablation of OMA1 provided evidence of cardioprotection in three different mouse models of heart failure [158]. Developing OMA1 modulating drugs is a challenge given their off-target effects of cardiotoxicity which needs to be addressed [160]. The approach to promoting fusion or inhibiting fission for cardioprotection has taken center stage in recent times, especially with the development of a new class of drugs. These include small molecules such as M1 fusion promoter, and Mdivi1, a fission inhibitor that proved effective in several in vivo studies [161–164]. Additionally, the repurposing of known drug molecules that target mitochondrial dynamics is also being extensively explored for cardioprotection [165]. In particular, these approaches hold promise in diabetic cases where the underlying cause of heart failure is disengaged mitochondrial dynamics [164, 166]. Translation of these therapies to the clinic requires extensive evaluation of safety and efficacy in humans which is currently lacking for this new class of compounds. More details on such protective strategies have been recently reviewed by Hausenloy’s group (2023) [145]. However, as we found that mitochondrial subtypes are specifically prone to damage in various heart failure conditions such as IRI, finding the subtype affected and targeting them could help in faster recovery of the heart [167].
The present consensus is that improving mitochondrial fusion and/or reducing mitochondrial fission preserves the myocardium and several reviews have consolidated those findings [168, 169]. But the significance of this target strategy lies in the fact that this is well executed even in underlying pathological states such as diabetes [170]. Empirically, pathological remodeling is different from regeneration and stands as a barrier for stem cell applications to promote differentiation and proliferation in post-infarcted hearts. Mitochondrial dynamics holds as a promising target to promote regeneration as noted in vascular smooth muscle cells by antagonizing the GLP1 receptor [171]. On the other hand, GLP1 agonists have shown promising benefits in cardiovascular trials in diabetic patients [172] and hypertension [173], but whether they are effective in remodeling the same heart if challenged with infarction remains to be tested. A recent meta-analysis to date also supported GLP1Ra therapy to have only a moderate benefit in treating coronary lesions (MACE-Trial) [174].
Proteostasis
Proteostasis is a process of regulated homeostasis of protein translation, folding, and degradation [175]. It involves numerous chaperones and regulatory proteins that are indispensable for the renewal of cellular proteins to maintain normal cardiac contractility. Normally, mitochondrial protein homeostasis is maintained by the folding and maturation of imported proteins, the guided refolding of misfolded proteins, and the degradation of non-assembled, damaged and misfolded proteins [176]. Interestingly, more than 40 independent proteases for mitochondria have been recognized that not only maintain proteostasis but also act as central regulators of organelle plasticity [177]. Chen et al., (2021) collectively reviewed the role of these mitoproteases in diseased hearts and emphasized that perturbations in this system precede mitochondrial dysfunction [178]. Dysfunctional proteostasis could be caused by oxidative modification of proteolytic proteins (LONP1) or mutation in controlling genes (MIPEP, CLPP, HTRA2) [176, 179, 180]. Rescuing these malfunctioning systems provides mitochondrial stability thereby protecting the heart from mitochondriopathies [179, 181, 182]. Overall, ‘mitochondrial proteostasis’ is controlled by the Ubiquitin proteasome system (UPS) via mitochondria-associated degradation (MAD), the unfolded protein response (UPRmt), and mitophagy. These systems have been extensively studied as therapeutic targets for diseases linked to mitochondrial quality control as recently reviewed by Hong et al. (2024) [183]. There is an extensive understanding of each of these systems as has been recently reviewed by others [184–189]. Although these systems work in tandem to remove unwanted proteins, the UPS primarily tags targeted proteins with ubiquitin and transports them to the cytosol for degradation while the UPRmt is a stress response activated by the accumulation of unfolded/misfolded proteins within the mitochondria and degrades them via transcriptional activation of proteases through retrograde signaling [190].
The UPS has been implicated in several pathologic cardiac states such as hypertrophy, ischemia-reperfusion injury, and mitochondriopathies which have increased ubiquitin-positive deposits. Given its frequent role in HF, targeting specific proteases instead of upregulation of the entire system has shown a good therapeutic response [191]. Similarly, the enhancement of the UPRmt response protects the heart from pathologic stress, making it a potentially valuable drug target [192]. On the contrary, while it is found that mitophagy markers are elevated in failing hearts, recent studies suggest that increasing mitophagy can alleviate microvascular endothelial dysfunction to prevent heart failure from ischemia/reperfusion injury [193, 194]. Miranda-Silva et al., studied this ambiguous role of mitophagy in remodeling but could not explain the elevation in mitophagy found both during the presence (overload) and absence (unload) of stress [195]. It is postulated to be an important quality control action to maintain mitochondrial bioenergetics. Hence it is not clear if mitophagy is beneficial or harmful for cardiac remodeling and is a subject of research focus. Given current knowledge of the role of proteostasis in heart failure, future studies may wish to focus on how these systems are interconnected with an emphasis on the time course of drug intervention and monitoring mitochondrial quality.
Nuclear-mitochondrial crosstalk
The topic of nuclear-mitochondrial crosstalk merits an entire review to itself and therefore this section only briefly highlights studies relevant to cardiac function. The OXPHOS machinery has only 13 proteins encoded by the mtDNA while the rest of the proteins (1,140 mouse) are imported from the cytosol [196]. The translocation is necessary to produce functional mitochondria to meet the energy demands of the cell and suggests the need for a well-orchestrated signaling mechanism from both ends to meet the working demands of the myocardium. This bidirectional signaling; anterograde (nucleus to mitochondria) and retrograde (mitochondria to nucleus) is critical for cell-plasticity but not comprehensively explored as to how they maintain the cardiac physiology and contribute to pathological states [2]. Evolving studies in the field of mitochondrial research such as the evidence of mitochondrial splicing variants generated by nuclei-encoded spliceosome complexes suggest that there is an ocean of information hidden in the signaling process yet to be discovered [75]. One such emerging case study reported nuclear control of mtDNA replication by topoisomerase (TOP3A) and its involvement in dilated cardiomyopathy [197]. The study observed multiple mtDNA deletions with predominant myalgia and weakness suggesting energy deficiency disorder, but further studies on TOP3A’s function and pathological role are yet to be established.
Analogous to the above case study, a well-studied nuclear-driven signal that controls mitochondria is the energy shift from glucose/lactate in the prenatal hypoxic environment to fatty acids during postnatal conditions, driven by the metabolic sensor Hypoxia-inducible factor (HIF), whose discovery was awarded the Nobel Prize in 2019 [198, 199]. Regarded as the master regulator for oxygen homeostasis, HIF helps muscles generate ATP from lactate during stress, reducing its dependence on OXPHOS. Additionally, HIF upregulates SOD2 to suppress ROS in mitochondria. Semenza (2014) has comprehensively reviewed HIF’s role in cardioprotection, part of which is through metabolic reprogramming that helps recovery of heart following ischemic injury [200]. Following prolonged hypoxia/ischemia, the perinuclear mitochondrial distributes and activates the HIF leading to cardiomyopathy in the long term due to chronic expression impairing heart function. Thus, HIF-stabilization has been a key strategy for promoting contraction and reduction of infarction [201, 202]. Factually, HIF induction is controlled by mitochondria as depletion of mitochondria DNA in ρ0cells or inhibition of ETC function prevents degradation of HIF [203]. Recent evidence also suggests that mitochondria-targeted HIF protects the liver from oxidative stress by downregulating mtDNA-encoded mRNA without affecting the organelle content emphasizing its non-transcriptional role under hypoxia [204]. However such roles with relation to HIF or its mitolocalizing isoforms (HIF3α) or such signaling mechanisms in general are not fully understood in cardiovascular disease [204].
The endosymbiotic origins of mitochondria in eukaryotic cells have repositioned its genome to integrate with the host’s nucleus [205]. Most of the nuclear-mitochondrial crosstalk is ‘anterograde’(95%), that is, nuclear transcripts influencing mitochondrial gene expression, which is evident from the presence N-terminal mitochondrial targeting sequences (NUMTs) present in protein-precursors imported to the mitochondria; garnering significant research attention [206]. Thus, it would be an understatement to say that mitochondria depend on the nucleus for their existence. Prominently, the anterograde signaling pathways promote mitochondrial health by activation of genes encoding mitochondrial proteins through transcriptions factors such as NRF1&2. In response to energy status (level of ATP/ADP, NAD/NADH, TCA cycle enzymes and membrane potential), the activation of nuclear receptors (PPAR delta and EER) driven by PGC1α promotes the genes required for regulation of TCA cycle and ETC-proteins. Such regulation is mediated by signaling involving the AMPK, AKT and SIRTUIN pathways. For instance, AMPK activation promotes biogenesis, and increases membrane potential and fatty acid oxidation, thereby providing energy for cardiac contractile function, and differentiation as demonstrated in a recent work on human induced PSC derived cardiomyocytes [43].
Unlike the role of retrograde signaling in the developing heart, the role of it in pathologically challenged hearts is poorly described. Two major pathways identified in retrograde response are the RTG (retrograde) and TOR pathways studied extensively in yeast systems and the lack of mammalian homologs of the proteins in the pathway has left us with unvalidated results [207]. It is known that mitochondrial stress initiates the translocation of transcription factors such as SIRT, RTG and FOXO that drive the transcription of genes that promote antioxidant response or apoptosis dependent on the signal. Even the molecules released from the mitochondria (mainly the Ca2+, ROS and protein breakdown products of UPRmt response) that affect nuclear gene transcription are accepted as canonical retrograde signals [208]. One such gatekeeping retrograde signal (ATFS-1) protected C.elegans from anoxic stress [209]. Under proteolytic stress conditions, ATFS-1 import to the mitochondria is blocked and it accumulates in the nucleus to activate the target genes. Emerging evidence suggests that its mammalian ortholog family of genes; Activating transcription factors (ATF) play an important role in cardioprotection as well [210–213]. The stimulus for activation is thought to come from protein misfolding response (UPRmt), which has been identified as an emerging drug target for cardioprotection [213, 214]. The immunostimulatory role of mtDNA is also one of the factors in eliciting retrograde response. Especially, a supercoiled confirmation of mtDNA called the Z-DNA is reported to activate the ZBP1 expression and sustain an inflammatory response that contributes to Doxorubicin-induced cardiotoxicity [215]. ZBP1 is a key innate immune regulator, whose role in CVD pathologies is yet to be explored. More recently, mitochondria-derived peptides (humanin and MOTS-c) were discovered to be involved in CVD pathologies [216]. However, their targets and signaling mechanisms must be studied before they can be harnessed for treatment [217].
Despite knowing these signals, identifying the networks and responses in the cardiac physiology is still a challenge that needs to be worked on [2]. New methods of single-cell sequencing and GWAS studies can help to understand these pathways in in-silico systems and can be validated in working models. Likewise, the somatic mutations and the post-translational modifications that affect the signaling and its interaction are poorly understood (Ex: OPA1). There is now a demand for animal studies focusing on nuclear-mitochondria crosstalk to identify individual populations at risk, drugs that interact with signaling (toxicogenomics) and identify new targets (pharmacogenomic).
The energy polarity in maintaining cardiac function
The heart is dynamically able to derive energy from numerous metabolic sources (e.g., fatty acids, carbohydrates, BCAA, and ketone bodies). These substrates compete for the OXPHOS at the mitochondria by providing acetyl-CoA [218]. Despite availability of all the substrates, ATP production is contributed to by the oxidation of fatty acids and carbohydrates (90%). Under normal physiological conditions, BCAA and ketone bodies are known to have less (10%) contribution to the overall ATP production. Since most CVD complications arise in patients with metabolic alterations as a temporal phase transition, it is important to analyze the influence of increased levels of specific intermediary fuels on cardiac function and progression of disease. Also, groups have reported that targeting mitochondrial dynamics and proteolytic pathways seem to improve cardiac function by metabolic remodeling even in disease states but without a clear consensus of the process to derive energy for remodeling [170, 219]. This emphasizes the need for basic research to explore the energy polarity for various substrates and their relation to cardiac function, signaling, genetic influences, and recovery from pathological states.
Cardiac energy substrates clearly have a metabolic relationship to one another and also function as signaling molecules influencing the mitochondria. Muoio and Neufer (2012) proposed that the nutrient induced mitochondrial stress caused by accumulation of acetyl CoA fosters hyperacetylation leading to ROS production, decrease in OXPHOS capacity, impairment of glucose uptake and insulin sensitivity [220]. Similarly, ketone bodies inhibit HDAC protein to control gene expression, and in pathological states, the heart relies on ketone metabolism to derive energy [220–222]. The mechanistic link between the energy substrates, their metabolism and contractile function remains obscure and establishing them could be of potential benefit for metabolic therapy (recovery) of failing hearts [223].
Aubert et.al’s (2016) quantitative proteomics of heart failure in a mouse model showed downregulated fatty acid metabolism and elevated ketone oxidation in mouse hearts [222]. Recently several studies have recognized ketone therapy as a clinical opportunity to treat heart failure giving a new direction to the heart as a ‘metabolic omnivore’ [224]. Opposing views have been proposed by Brahma (2022) stating that ketones are not crucial for the maintenance of normal homeostasis and are utilized only in the presence of heart failure [225]. Likewise, catabolic defect in BCAA was noted as a feature of heart failure [226] and the clearance of accumulated intermediates recovered the cardiac function. Benefits could be derived by simultaneously targeting the mitochondria as well since the elevated BCAA suppressed OXPHOS and increased ROS in the isolated mitochondria from the same study.
Genetic regulation of energy utilization in heart
Under pathological conditions, short-term energy substrate regulation is based on demand/supply, controlled by signaling mechanisms induced by biomolecules such as insulin and glucose. But in long-term conditions like hypertension, atherosclerosis, and diabetes, the induced changes in metabolism are regulated transcriptionally. The major transcriptional regulator in such cases is the HIF family of proteins, the activation of which promotes glycolysis and suppresses fatty acid (FA) oxidation. Liu (2020) found that suppressed myocardial HIF-1 in cyanotic congenital heart disease resulted in higher levels of glycolytic intermediates, and enhanced substrate utilization by the mitochondria for glucose instead of FA [227]. HIF also regulates cardiac lipid metabolism as elevated HIF family genes contribute to atherosclerosis [228], hypertrophy and remodeling [229, 230]. In the case of type-2 diabetes mellitus (T2DM), abnormal metabolic phenotype of HIF-1α destabilization is noted as less tolerance to hypoxia and rapid HF progression and its stabilization by Molidustat was effective in the treatment of diabetic heart from post-ischemic recovery [231]. Several such specific nuclear receptor transcription factors and co-activators regulate the genes and control the metabolic substrate preference which is yet to be explored. The significance of the shift in substrates for ATP synthesis in failing heart can also trigger cardioprotection as observed in hormetic strategies adopted to reduce myocardial IRI such as ischemic preconditioning, postconditioning, remote ischemic conditioning either by transient activation of signals or genetic changes. Such cardioprotective strategies are known to offer a second window of protection, which is far from being completely understood [232, 233].
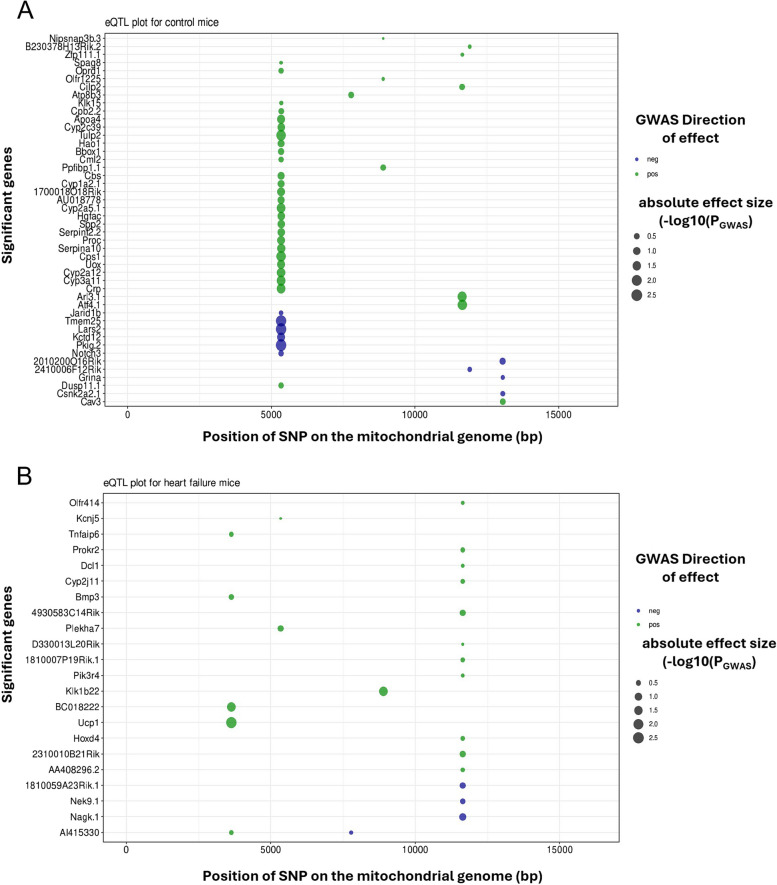
Studying the crosstalk between metabolic substrates and cardiac phenotype in heart failure is limited by the fact that large number of factors and signaling influences play a role in controlling the cardiac metabolism and function. Such situations could be managed by the use of computational tools (GWAS, and Network analysis) to find the role played by the energy substrates in maintaining the cardiac function. For instance, using systems genetics approaches, we previously published GWAS for heart failure in the hybrid mouse diversity panel (HMDP), a genetic reference population consisting of a large number of inbred mouse strains [234]. The study identified previously unrecognized genetic variants on the nuclear genome that contributed to heart failure-associated phenotype in a large panel of genetically diverse inbred mice. When we examined the mitochondrial single nucleotide variants (mtSNV) associated with the mouse strains from the HMDP for similar phenotypic links (Figs. 4 and 5), we found a significant association of several physiological parameters with isoproterenol-induced HF. Interestingly, heart mtDNA copy number appears to be negatively associated with the diversity in these mice [235] which supports studies in the field that associate reduction in mtDNA copy-number as a risk factor for HF [125, 127, 235, 236]. Further studies to characterize various mitochondria-related phenotypes as discussed in Sect. 2 and establishing a clear role for the mtDNA SNPs across the subtypes in CVD will unfold new signaling mechanisms.
Fig. 4.
QTL map associating mtSNPs at significant (LOD > 3) loci with cardiac traits in HMDP mice from (A) control and (B) isoproterenol challenged groups. Lung-Lung weight, mtCN- mitochondrial copy number, A-Late mitral inflow velocity-, MSNER-Mean normalized systolic ejection rate, Vcf- velocity of circumferential fiber shortening, PWs- posterior wall thickness at end systole, IVSs- interventricular septal wall thickness at systole, IVSd- interventricular septal wall thickness at diastole, and wLung- normalized lung weight
Fig. 5.
eQTL Plot of significant (LOD-threshold > 10) gene interactions with mtSNPs of HMDP mice strains from (A) control and (B) isoproterenol-challenged groups
Metabolic shift as a compensatory mechanism
ATP is an essential energy source for heart function. The imbalance in ATP generation/utilization due to heart failure is of great concern. In the heart, the majority of ATP is generated by fatty acid metabolism rather than glucose metabolism but at the expenditure of high oxygen consumption [237]. This preferential dependence of heart, liver and muscles towards fatty acids can undergo a shift in limited oxygen conditions, switching to glucose oxidation. How efficient these two processes are in sustaining the contractile function of the heart remains a topic of debate. Additionally, ketones and BCAAs also can be consumed to produce energy for the heart in processes that are often seen in HF [218]. Despite several substrates present, most of the processes end up in the mitochondria for ATP synthesis and calcium uptake, which is why it is essential to connect mitochondrial (organelle) function to the mechanical functions of the heart. At this point, it is unclear whether ATP depletion is the reason for HF because it limits the hemodynamic function of the heart, often termed the energy starvation hypothesis [238] or whether disease progression leads to metabolic dysregulation of energy generating processes. In a recent study by Abdalla (2023), evidence indicates that complex epigenetic regulation drives ATP depletion at a very early stage even before the loss of cardiac function, and is associated with the downregulation of genes encoding the proteins for mitochondrial NAD+ synthesis pathway [239]. Translational advantage of NAD+ supplementation to restore mitochondrial respiration in hypertrophic hearts was demonstrated in human hypertrophic cardiomyopathy by Nollet (2023), clearly indicating the cause and the solution [121]. Nollet’s group observed that in patients with obstructive hypertrophic cardiomyopathy, improper arrangement of mitochondria was strongly associated with dysfunctional mitochondrial respiration. Restoration of NADH levels by supplementation with NAD+ restored mitochondrial respiration and could be a valuable treatment option [240].
ATP production and utilization are highly regulated processes and its yield from substrates is in the order of FA(106) > BCAA(40) > glucose(32) > ketones(22). Though BCAA generates a substantial amount of ATP, its contribution is < 5% in the heart under normal circumstances as FA oxidation meets the ATP demand. In conditions of high demand, ATP production is supplemented by glycolysis and the phosphotransferase reactions [241]. However, chronic high demand, as observed in CVD, activates the sensors controlling metabolic remodelling leading to reduced utilization of ATP along with impairment in its production by any of these routes (fatty acid, glycolysis, BCAA, ketones, creatine kinase, and adenylate kinase). Thus, rather than simply regulating ATP synthesis/degradation, the pathways that control its production/utilization play a key role in cardiac pathology as explained by Ingwall [238, 241]. In fact, impaired activation of mitochondrial Potassium channels appears to be the reason for impaired responsiveness to conditioning strategies/drugs, especially in aged hearts [242]. These reports force us to rethink the actions of various drugs to study their role in ATP generation or their effects on the processes/pathways/channels that require ATP. As demonstrated by Molidustat and NAD+ administration, modulation of metabolic demand can act as a powerful treatment against HF progression by restoring mitochondrial function [121, 231]. In conclusion, it is essential to assess the metabolic demand of the heart, and its function based on substrates and effectively design strategies for recovery from HF.
Canonical pathways of cardioprotection via mitochondrial signaling
Pathways
From our earlier discussions, we have sufficient evidence indicating that targeting the mitochondria can rescue the failing heart as most of the disease-implicated signaling pathways converge at mitochondrial organelle function. Table 3 summarizes the list of cardiac-involved pathways and the changes that disruption of these pathways brings about in the organelle and its function. The outcome of disrupted signaling is an altered cell metabolism and/or an inhibition of apoptosis. Additional emphasis should be placed on the presence of functional mitochondria for effective cardioprotection, the absence of which could significantly derail the efforts of targeted therapies [141].
Table 3.
Mitochondrial convergence of cardioprotective pathways
| S.No. | Major Pathway | Key downstream molecule affecting mitochondria | Interaction with mitochondrial component | Phenotype changes in mitochondria due to activation of cardioprotective pathways | Ref |
|---|---|---|---|---|---|
| 1 | RISK (PI3K/Akt, MEK/ERK) | GSK3β, PIM1, cGMP/PKG | mPTP | Closing of mPTP prevents swelling and rupture of mitochondria | [243] |
| 2 | SAFE (JAK-STAT3) | STAT3, GSK3β | mPTP and complex-I | Opening of mitoKATP channels causes depolarization and reduces Ca2+ uptake to maintain the matrix volume | [244] |
| 3 |
NO/PKG-PKC pathway |
GSK3β, NO, PKG and PKC |
PKG and PKC with mPTP and mitoKATP channels, NO is inhibitor of cyt-c oxidase activity and respiration competitively to O2, S-nitrosation of proteins |
NO causes increased mitochondrial membrane potential and activation of cytoprotective signaling | [245–247] |
| 4 | PI3K/Akt/mTORC1 (Insulin mediated) | mTORC1/MCL-1 | ATP citrate lyase, MCL-1 | - | [248, 249] |
| 5 | Adenosinergic (Activation of A1 and A2A AR) | Multiple effectors such as Protein kinases, HSPs and HIF1α | mPTP and mitoKATP Channels | - | [250] |
| 6 | Mitophagy and Ubiquitin-Proteasome |
PINK1 (membrane depolarization sensor), PARKIN, MFN2 and Ubiquitination of adaptor proteins (p62/SQSTM1 and Optineurin) Short lived mis-folded mutant proteins |
PINK1 functions beyond mitophagy to control mitochondrial functions by regulating complex-I activity in response to stress while Deubiquitinases play key role as recruitment, activation, import and export of proteins to the mitochondria | Activation maintains mitochondrial integrity and stabilizes membrane potential apart from promoting mitophagy | [251–253] |
| 7 | PKCε | PKCε phosphorylates Cx43 | PKCε interacts with VDAC, ANT, and HKII to control the permeability pore transition. It Also activates mKATP channels | Increased mitochondrial swelling is prevented by inhibition of PKCε | [254] |
| 8 | Mito Connexin 43 | Cx43 is a component of IMM only in SSM | Regulates K + fluxes via mitoKATP channels | The sites of phosphorylation determine the phenotype and is yet to be explored | [255, 256] |
| 9 | Autocoid stimulation-GPCR (Adenosine, Bradykinin, Opioids) | PKCε,NO, STAT3 and GSK3β. | Cx43, mPTP and mitoKATP | Connexin reduction is associated with reduced K+ influx and swelling. It lowers ROS and reduces cardiac infarct size | [257, 258] |
| 10 | MOSPD contact site with ER- novel area of search | NA | NA | NA | [259] |
| 11 | Uncoupling proteins | ROS | UCP3, 2 and 1 | Transporters that regulate discharge of proton gradient and acts as calcium regulator to maintain thermogenesis and structure | [260, 261] |
RISK reperfusion induced salvage kinase pathway, SAFE survivor activating factor enhancement pathway, mPTP mitochondrial permeability transition pore, mitoKATP ATP activated mitochondrial potassium channels, PKC protein kinase C, PKG protein kinase G, VDAC voltage-dependent anion channel, ANT adenine nucleotide translocator, UCP uncoupling protein, CX43 connexin 43, GSK glycogen synthase kinase, mTOR mammalian target of rapamycin, NA not available and yet to be explored
Differential impact
A larger puzzle that remains to be solved is the differential impact of signaling on the interfibrillar (IFM), subsarcolemmal (SSM), perinuclear (PNM) and intranuclear (INM) mitochondrial subpopulations present in the heart. The seminal investigations led by Palmer, Hoppel, and Tandler in the 1970s–80s demonstrated that IFM had superior substrate oxidation capacity and better calcium handling compared to SSM [262, 263]. These adaptations of mitochondrial subtypes are critical in pathological conditions as they shape the recovery and remodeling of the heart. However, such observations agree to differ with the animal model and pathology as best described in a recent work by Shekar (2022), demonstrating that IFM and SSM in porcine hearts subjected to AMI underwent the same significant dysfunction without any difference in calcium handling or ATP production [264]. According to the group, oxygen consumption was the biggest difference among the subtypes. At this juncture, we need to rethink if the sub-type differences help in remodeling especially using more clinically relevant large animal models. Another key aspect of mitochondrial subtype differences is their response to drugs which has been demonstrated to impact the recovery from ischemia-reperfusion injury [167, 265, 266]. Studying mitochondrial toxicity in drug discovery is a different aspect dealt with elsewhere [267]. Besides the obvious spatial and functional differences among these mitochondrial sub-populations, exceptionally little information is available as to how these subtypes behave differently in the presence of acute and chronic stress [268, 269]. For instance, the pioneering study by Hoppel (1982) provided evidence of selective involvement of IFM in cardiomyopathy [270]. Subsequent studies in diabetic hearts also showed declined respiratory activity in IFM but not in SSM [113]. Such differences also impact the protective strategies and signaling that otherwise work in normal conditions [129] and additionally negate the cardioprotective effect of previously established drugs [271]. Interestingly, a recent study by Rajab (2022) found structural alterations in mitochondria induced by early diabetes in mice [272]. The recent data from the Antentor Hinton lab suggests that the mitochondria undergo various morphological changes due to aging and can potentially link to HF phenotypes observed in humans [273]. A major drawback in the field is that most of this information is available for only the most widely studied subpopulations: IFM and SSM due to their relative abundance and established isolation protocols. Even these relatively more studied subpopulations suffer from a lack of research interest: the last paper to address exclusive differences between IFM and SSM subtypes in terms of markers expressed was published in 2009, in which it reported that the presence of connexin43 is localized in SSM but not IFM [274]. In a recent thematic issue in the Philosophical Transactions of the Royal Society, Voglhuber (2022) compared for the first time IFM and PNM in heart failure [275]. Their study reported the role of PNM in shaping nucleoplasmic calcium levels thereby controlling the cell microenvironment and hypertrophy progression. Advancements in single-cell omics platforms have provided more insights into subpopulations of cardiomyocytes in hypertrophy, providing more challenges for researchers to explore, leaving behind clues for investigating the mitochondrial subpopulations in a similar manner [276]. Together they may form the deciding factors in shaping the cardiac phenotypes.
Genetic regulation
Somatic mutations in mtDNA are increasingly being recognized as contributors of cardiovascular disease but our understanding of its role is restricted by the lack of mechanisms that explain the outcomes due to heteroplasmy and the threshold effect [134, 277, 278]. Age and haplogroups remain a major contributor of these mutations apart from underlying conditions such as diabetes, hypercholesteremia, and hypertension which are typically associated with CVD cases [279]. Though it has been recognized that most mtDNA mutations are present in the D-loop region and the 12 S rRNA, a recent report by Calabrese (2022) has recognized no specific mutational hotspots exist for these widely distributed mtSNV across stroke, hypertension, or ischemic heart disease [279]. But these mutations are, however, strongly associated with age and could possibly be a result of genomic mutations caused in the mitochondrial regulatory genes such as TFAM and POLG [280]. In 2019, McManus demonstrated for the first time the interaction between mtDNA and nDNA augmenting the progression of cardiomyopathy in C57BL/6JEiJ mice [281] and was later elaborated by Lim (2021) in humans [282]. Lim identified 11 different genotype-specific cardiac involvements in patients with mitochondrial mutations suggesting a diagnostic use for the mtDNA mutations. This rapidly evolving research in the field could be translated to new therapies in development by harvesting the benefits of single cell/nucleus/mitochondrial profiling and gene editing for treating CVD. For instance, using our data of nuclear gene expression in HMDP, we constructed an eQTL (Expression quantitative trait loci (eQTL) – statistical method linking phenotypic data (traits/gene expression) and genotypic data (molecular markers/SNPs) [283]) plot explaining the interaction of mtSNPs (mitochondrial Single nucleotide polymorphisms) with differential gene expression in HF induced by isoproterenol (Figs. 4 and 5). The eQTL suggested a significant number of genes involved to be associated with loci in the mtDNA, whose expressions were affected by the mtSNPs in the HMDP from both control and heart failure groups. In the coming sections, we provide insights into how newer computational approaches provide useful information on the regulation and function of mitochondria and their relationship with disease progression.
Computational approaches provide insights into mitochondrial function and disease
Over the past two decades, computational and ‘omics-scale approaches have revolutionized our ability to understand the heart [284]. For example, gene expression analysis has progressed from traditional qPCR to microarrays before transitioning to the era of next-generation sequencing with bulk and now single-cell and spatial transcriptomics sequencing approaches. Additionally, advances in genotyping, DNA sequencing, and GWAS approaches have revealed new insights into the relationships between polymorphisms and disease. Taken together, these new approaches provide new insights into CVDs, mitochondrial anomalies, and overall physiological and pathological states in the heart [285, 286]. Concomitantly, mutations in mtDNA are associated with CVDs and can originate from maternal inheritance, epigenetic modification, environmental and lifestyle factors as discussed in detail elsewhere [287]. In this section, we describe some recent advancements in the study of mitochondria in the heart using systems genetics.
Bulk RNAseq and mitochondria
In mitochondria-rich cardiomyocytes, up to 80% of all sequencing reads are soaked up by mitochondrial genes, yet an early filtering step in most RNAseq protocols is the removal of all mtDNA-associated gene expression [288]. This results in a loss of significant amounts of useful data and leaves the researcher with only ~ 20% of their original reads for downstream analysis especially in tissue like heart which comprises of highest percentage of mtDNA [289]. It is likely that a closer examination of this ‘junk’ RNA would reveal powerful indicators, mediators, and drivers of CVD. For example, mitochondrial dysfunction predisposed murine hearts to RV failure in a study by Hwang (2021) as it turned out to that the most significantly affected pathway has genes related to ETC [290]. Similarly, increased non-pathologic heteroplasmy in a mouse model predisposed them to metabolic stress in adulthood leading to not only heart failure but also affected other organs, causing premature death [60].Although, the hearts were sequenced on a Illumina HiSeq platform, the authors missed out on the mtDNA sequencing which could have given more critical information on mRNA abundances, long non-coding genes, mRNA information on mitochondria encoded peptides, as elucidated by Dunin (2019) in fish hearts [291]. Another advantage offered by mtRNA sequencing is that the heteroplasmy is transcribed with < 5% difference from mtDNA [292] which could be used to explain the cardiac phenotypes. Bulk RNAseq, however, has several limitations that other methods, such as sc/sn-RNAseq or long-read sequencing are able to address more clearly. For one, bulk RNAseq struggles to differentiate Nuclear-Mitochondrial DNA segments (NUMT) from genuine mtDNA transcripts which can significantly reduce the efficacy of the method. Furthermore, bulk RNAseq reports results on a heterogenous population of cell types, locations, and temporal profiles, whereas other approaches such as snSeq/scSeq or spatial transcriptomics combined with computational approaches avoids these issues by focusing on mitochondrial dynamics at the individual cell level.
Single Cell/Nucleus-RNAseq and mitochondria
Single Cell approaches have exploded in popularity over the past few years as a means of understanding the underlying role of individual cells in physiology and disease. Limitations in the most popular single cell sequencing (scSeq) platforms (e.g. Fluidigm or 10x Chromium) prevent researchers from studying cardiomyocytes specifically due to their size and asymmetrical shape [293], however there is robust data generated at the nuclear level and exciting new developments [294, 295] that suggest that cell-level analyses will soon be possible in the heart. Despite these challenges, sc/snSeq approaches have proven specifically useful in understanding the heterogenous cellular populations of the heart, providing researchers with enormous amounts of data compiled in Tabula Muris [296], human heart [297] and the ENCODE project [298] on transcriptomic heterogeneity not only in normal but also in diseased hearts across multiple distinct cell populations. When combined with new advances in spatial transcriptomic mapping [299], reports suggest that we may be at the start of a new generation of precision medicine in which drug responses are understood at an individual level by analyzing patient-specific cellular heterogeneity in disease states. Although such an approach has many hurdles to pass before it is able to be implemented in the clinic [300], the big-data generated in these approaches helps to pave the way for integration of studies to identify disease transcriptional patterns [301] and establishing comprehensive cell-atlases for the heart [297, 302]. With rapid advancements in the field of scRNAseq, causal links of cellular lineages towards experimental myocardial infarction have been identified [301]. Despite these advances, most of the above referenced studies totally ignore the data captured from the mitochondria due to faulty assumptions regarding data cleaning owing to maintaining the quality of analysis, heteroplasmic nature of the mtDNA and experimental necessities of using nuclei rather than whole cell. Given the importance of transcriptomic anterograde and retrograde signaling from nuclear and mtDNA respectively, it is necessary the organelle be not neglected.
In 2018, Nomura et al. found transcriptional increases in mitochondrial genes associated with oxidative phosphorylation and biogenesis due to pressure overload in mice. They mapped single cardiomyocyte transcriptomes by reconstructing the trajectory of signatures that lead to hypertrophy and showed an association of increased mitochondrial biogenesis with cardiac hypertrophy as a compensatory mechanism for high energy demand [303]. A direct evidence of heteroplasmy affecting metabolic functions of cells and causing multiple comorbidities including heart failure has been reported by the José Antonio Enríquez’s group in a well-characterized mouse model [60]. Cell-type specific gene regulatory networks were also noted in snRNA-seq studies on postnatal hearts which served as a rich source of data sets for analysis of mitochondrial cardiomyopathies [304]. Additional studies using these previously overlooked sources of mitochondrial information are crucial to further advance our understanding of cardiac signaling and the relationship between this crucial organelle and organismal homeostasis and survival.
Long Read Sequencing and the mitochondria
Despite the successes of many sequencing approaches to studying the role of polymorphisms and structural variations that underly the phenotypic diversity of the heart in both stressed and unstressed conditions, there are key downsides to the commonly used short-read paradigm that are being addressed by the advent of new long-read sequencing approaches such as the nanopore by Oxford Nanopore Technologies, single-molecule real-time sequencing by PacBio, and the synthetic technologies adopted by Element Biosciences, MGI, Illumina and 10XGenomics [305, 306]. One key flaw of short-read sequences is the inability to distinguish between large-scale duplications of DNA sequences. This is of key importance for mitochondria as NUMTs are broadly misidentified as mitochondrial sequences when they are, in fact, nuclear in origin.
Long read sequencing relies on microfluidics and light or electric current-based technology to generate significantly longer reads than are possible using the standard synthesis and ligation-based approaches. Long Read sequences are easily able to reach 16Kb size of mitochondrial genome in less than 2 h and offer a unique means by which researchers can fully understand the entire structure and sequence of the mtDNA across its sub-cellular populations. For example: Pollet (2020) provided a benchmark mtDNA variant identification in equines which could be applied to other animal species [307] and Vandiver’s data establishes the relation between mtDNA deletion and aging using this rapidly rising technology [308]. This is an exciting field of active research that should be more fully developed for cardiovascular research in the coming years for the detection of rare mitochondrial genetic variants across the mitochondrial subtypes.
Resources and repositories for mtDNA studies
In contrast to the large number of repositories for other forms of genetic/transcriptomic/phenotypic data, there exist limited working online repositories for mammalian mtDNA-associated research. Currently, human mitochondrial genome research benefits from MITOMAP (https://mitomap.org/foswiki/bin/view/MITOMAP/WebHome), MSeqDR (https://mseqdr.org/mitobox.php), and HmtDB (https://www.hmtdb.uniba.it/about) which host the database and tools for analyses of human mtDNA variations as reviewed by Cappa [309]. Murine data is nonexistent on these platforms and is difficult to acquire through other means. For example, the two major resources MitoCarta and MioBreak are limited to an inventory of mouse genes encoding proteins that localize to the mitochondria and mtDNA break points. Researchers exploring the NIH’s sequence read archive (SRA) and gene expression omnibus (GEO) find it difficult to specifically obtain data on the mitochondrial genome as it is not a prioritized feature in these datasets. Hence, a well-curated database of studies exploring the mitochondrial genome in non-human species under various pathological conditions is required. There are, however, some growing resources for analyzing mtDNA sequences once they are obtained such as the MitoMiner 4.0 [310], MitoZ 3.5 [311], MitoSeek [312], MitoFinder [313], BamSignals [314], MseqDR [315], and Mitohelper [316]. These tools help in implementing wide range of functions such as data mining, annotation, heteroplasmy and mutation detection, and visualization either as a standalone function or integrating into an environment of graphical step by step usage such as Galaxy [317]or mitoXplorer [318]. A major caveat in these resources is the lack of curation, species specificity, inter-platform integration and user-restricted access. With the increasing advent of sequencing technologies, the above-discussed data will become common and require resolute, streamlined storage and analysis platforms.
Aggregation of data points into predictive models in the mitochondria
Due to the low effect sizes of many individual GWAS loci, there has been an evolving trend in the field to instead develop polygenic risk scores (PRS),;measures that integrate large amounts of information about individual polymorphisms into a single predictive measurement. This score estimates the genetic liability for an individual to acquire a specific disease or phenotype. Although PRS loses the ability to pinpoint a single gene or mutation as causal for disease, studies have used PRS approaches to reach predictive powers similar to those seen in much more penetrant single-gene-driven diseases, including in coronary artery disease [319]. This risk stratification strategy is helpful for disease prediction and precision medicine and helps to better evaluate the effectiveness of treatment strategies by stratifying patients into risk groups. A major limitation of PRS is its reliance on the presence of known polymorphisms that are associated with disease and numerous groups have highlighted the difficulty of applying PRS appropriately to understudied populations outside of European ancestries [320]. Some research groups have begun to apply the principles of PRS to study individuals based on variations in mitochondrial haplotype. In these scenarios, the mitochondrial haplotype proved to be a synergistic factor that influences phenotypic diversity and disease progression as seen in Alzheimer’s Disease [321] and obesity [322]. Efforts to apply mitochondrial PRS to the cardiovascular system have been largely lacking and infrequently studied by other groups such as inclusion of mitochondrial genes as a driver in PRS estimation of Torsade de Pointes in the context of patients taking antipsychotic medication for treatment of schizophrenia [323]. Truby (2021) specifically used mitochondrial PRS to predict HF in European population, based on nuclear genes affecting synthesis of long chain acyl carnitine [324]. However, studies based on mitochondrial encoded genes, wider populations (non-European) and inclusive of several cardiometabolic disease traits are necessary. Of late, mtDNA copy-number (mtDNAcn) has become de facto biomarker of healthy heart and computational methods have utilized this to generate ‘mitoscore’ based PRS for the ESTHER study [325]. This has clearly associated novel risk loci with variation in mtDNAcn, but more research is needed to thoroughly link these scores to downstream cardiovascular events in a wider population and mtDNA variants.
Detecting heteroplasmy with new techniques
Cardiovascular disease is the most common aging-associated condition, with mitochondrial dysfunction widely recognized as its root cause [326]. Within each cell, multiple copies of mtDNA exist. As we age, the accumulation of ROS partially mutates the mtDNA, leading to a mix of unaltered and mutated forms within the same cell termed ‘heteroplasmy.’ Over time, with increased heteroplasmy, the heart cell’s ability to produce energy diminishes, failing to meet the organ’s demands as mutated mtDNA becomes predominant. This process is accompanied by disrupted mitochondrial dynamics and reduced mitophagy, resulting in the buildup of ROS-generating mitochondria. This creates a vicious cycle, contributing to heart failure. Mitochondrial heteroplasmy has been linked to pathogenic phenotypes in disease based on their frequency and function (synonymous vs. non-synonymous) [327]. However, the majority of research done on heteroplasmy has relied on modest estimation of total heteroplasmic mitochondrial content, rather than the actual effects of different variants seen within the heteroplasmic heart. This is because classic means of studying mtDNA heteroplasmy has been limited to PCR or bulk sequencing techniques that have not offered insights into the variability present between individuals. One approach to this issue is to saturate the space through massively parallel sequencing approaches such as performed by Zhang (2012), in which researchers combined long-range PCR and parallel sequencing methods to detect and quantify heteroplasmy with 100% sensitivity from human samples with known mutations [328]. However, the heteroplasmy in terms of sub-population, and distinguishing micro (within single mitochondria) and macro (within a cell) heteroplasmy remains unresolved. Recently, steps have been taken to creatively overcome this issue through sophisticated analysis approaches. For instance, researchers developed methods combining enzyme histochemistry and single-cell laser microdissection to study mtDNA within individual skeletal muscle fibers by sequencing [329]. Nevertheless, deciphering results in most of the sequencing approaches can prove difficult as they are hindered by NUMT from the nDNA. More recently, single-cell mitochondrial sequencing has helped to establish cell lineages in hematopoiesis and could be potentially useful in the future for establishing protocols to study the spatial distribution of somatic mutations and heteroplasmy in the heart [330, 331]. More research is needed on this topic, due to its outsized impact on cardiovascular phenotypes.
Conclusions and call for research
In this review, we have set out to describe the current state of the field in terms of understanding the role of mitochondria in cardiovascular health. The vast research in the last decade has proved beyond the fact that the mitochondria no longer are confined to the powerhouse analogy and have become signal processors of the cell [332]. We have described how mitochondria are vital to cardiac development, homeostasis of a physiologically healthy heart, the ways in which the mitochondria are affected by pathological remodeling of the heart and how they can, in their own way, contribute to disease progression. Finally, we have highlighted the advent of new computational approaches to better understand the role of mitochondria in phenotype variability. A common theme of our review is the repeated observation that mitochondria are often overlooked as a driver of phenotype, either ignored or, in some cases, deliberately filtered out from the data in order to ‘clean’ it. Recent advances in the field have shown that mitochondria are associated with many phenotypes (spatially distinct, genetically heterogenous, and functionally specialized) and are a key player in CVD. It is vitally important that these ‘discarded’ castoffs of the filtering process for, for example, RNAseq analyses are recovered and studied by interested groups as these existing datasets represent large amounts of untapped but crucially relevant data.
Supplementary Information
Authors’ contributions
S.R. designed this review, performed the selection of literature, and prepared the figures. S.R. and C.D.R. collected the related references, drafted the manuscript, participated in the discussion, and revised the manuscript. Both authors have read and approved the final manuscript.
Funding
This review was supported by the National Institutes of Health (NIH) National Heart, Lung, and Blood Institute (NHLBI) under award numbers: R01HL162636 and R00HL138301. We gratefully acknowledge the funding provided by the NIH, which made this work possible.
Data availability
No datasets were generated or analysed during the current study.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mitochondrial medicine - Therapeutic Development Wellcome Genome Campus, UK. https://coursesandconferences.wellcomeconnectingscience.org/event/mitochondrial-medicine-therapeutic-development-20240318/. Accessed 03 Mar 2024.
- 2.Tian R, Colucci WS, Arany Z, Bachschmid MM, Ballinger SW, Boudina S, Bruce JE, Busija DW, Dikalov S, Dorn GW, et al. Unlocking the secrets of mitochondria in the cardiovascular system. Circulation. 2019;140:1205–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brade T, Pane LS, Moretti A, Chien KR, Laugwitz KL. Embryonic heart progenitors and cardiogenesis. Cold Spring Harb Perspect Med. 2013;3:a013847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ankel-Simons F, Cummins JM. Misconceptions about mitochondria and mammalian fertilization: implications for theories on human evolution. Proc Natl Acad Sci. 1996;93:13859–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang X, Zhang X, Wu D, Huang Z, Hou T, Jian C, Yu P, Lu F, Zhang R, Sun T, et al. Mitochondrial flashes regulate ATP homeostasis in the heart. eLife. 2017;6:e23908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen ED, Roethlin K, Yee M, Woeller CF, Brookes PS, Porter GA, O’Reilly MA. PPARγ drives mitochondrial stress signaling and the loss of atrial cardiomyocytes in newborn mice exposed to hyperoxia. Redox Biol. 2024;76:103351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Blerkom J. Mitochondria in early mammalian development. Semin Cell Dev Biol. 2009;20:354–64. [DOI] [PubMed] [Google Scholar]
- 8.May-Panloup P, Boguenet M, Hady El H, Pierre-Emmanuel B, Reynier P. Embryo and its mitochondria. Antioxidants. 2021;10:139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trebichalská Z, Kyjovská D, Kloudová S, Otevřel P, Hampl A, Holubcová Z. Cytoplasmic maturation in human oocytes: an ultrastructural study †. Biol Reprod. 2021;104:106–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirillova A, Smitz JEJ, Sukhikh GT, Mazunin I. The role of mitochondria in oocyte maturation. Cells. 2021;10:2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Houghton FD, Thompson JG, Kennedy CJ, Leese HJ. Oxygen consumption and energy metabolism of the early mouse embryo. Mol Reprod Dev. 1996;44:476–85. [DOI] [PubMed] [Google Scholar]
- 12.Shepard TH, Muffley LA, Smith LT. Mitochondrial ultrastructure in embryos after implantation. Hum Reprod. 2000;15(Suppl 2):218–28. [DOI] [PubMed] [Google Scholar]
- 13.Houghton FD. Energy metabolism of the inner cell mass and trophectoderm of the mouse blastocyst. Differentiation. 2006;74:11–8. [DOI] [PubMed] [Google Scholar]
- 14.Dumollard R, Carroll J, Duchen MR, Campbell K, Swann K. Mitochondrial function and redox state in mammalian embryos. Semin Cell Dev Biol. 2009;20:346–53. [DOI] [PubMed] [Google Scholar]
- 15.Hoque A, Sivakumaran P, Bond ST, Ling NXY, Kong AM, Scott JW, Bandara N, Hernández D, Liu GS, Wong RCB, et al. Mitochondrial fission protein Drp1 inhibition promotes cardiac mesodermal differentiation of human pluripotent stem cells. Cell Death Discov. 2018;4:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu V, Dahan P, Ahsan FM, Patananan AN, Roy IJ, Torres A Jr, Nguyen RMT, Huang D, Braas D, Teitell MA. Mitochondrial metabolism and glutamine are essential for mesoderm differentiation of human pluripotent stem cells. Cell Res. 2019;29:596–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi J, Seo BJ, La H, Yoon SH, Hong YJ, Lee JH, Chung HM, Hong K, Do JT. Comparative analysis of the mitochondrial morphology, energy metabolism, and gene expression signatures in three types of blastocyst-derived stem cells. Redox Biol. 2020;30:10143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lv J, Yi Y, Qi Y, Yan C, Jin W, Meng L, Zhang D, Jiang W. Mitochondrial homeostasis regulates definitive endoderm differentiation of human pluripotent stem cells. Cell Death Discovery. 2022;8:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Costa R, Muccioli S, Brillo V, Bachmann M, Szabò I, Leanza L. Mitochondrial dysfunction interferes with neural crest specification through the FoxD3 transcription factor. Pharmacol Res. 2021;164:105385. [DOI] [PubMed] [Google Scholar]
- 20.Bhattacharya D, Khan B, Simoes-Costa M. Neural crest metabolism: at the crossroads of development and disease. Dev Biol. 2021;475:245–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kocabas F, Mahmoud AI, Sosic D, Porrello ER, Chen R, Garcia JA, DeBerardinis RJ, Sadek HA. The hypoxic epicardial and subepicardial microenvironment. J Cardiovasc Transl Res. 2012;5:654–65. [DOI] [PubMed] [Google Scholar]
- 22.Porter GA Jr, Hom J, Hoffman D, Quintanilla R, de Mesy Bentley K, Sheu SS. Bioenergetics, mitochondria, and cardiac myocyte differentiation. Prog Pediatr Cardiol. 2011;31:75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beutner G, Burris JR, Collins MP, Kulkarni CA, Nadtochiy SM, de Mesy Bentley KL, Cohen ED, Brookes PS, Porter GA. Jr.: coordinated metabolic responses to cyclophilin D deletion in the developing heart. iScience. 2024;27:109157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodríguez-Nuevo A, Torres-Sanchez A, Duran JM, De Guirior C, Martínez-Zamora MA, Böke E. Oocytes maintain ROS-free mitochondrial metabolism by suppressing complex I. Nature. 2022;607:756–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fan W, Waymire KG, Narula N, Li P, Rocher C, Coskun PE, Vannan MA, Narula J, Macgregor GR, Wallace DC. A mouse model of mitochondrial disease reveals germline selection against severe mtDNA mutations. Science. 2008;319:958–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arhin SK, Lu J, Xi H, Jin X. Energy requirements in mammalian oogenesis. Cell Mol Biol (Noisy-le-grand). 2018;64:12–9. [PubMed] [Google Scholar]
- 27.Makanji SJJC, Johnson Y, Tsai JL, Lagondar T-S, Rodda S, Sun F, Pangestu X, Chen M, Temple-Smith P. The transgenerational effects of oocyte mitochondrial supplementation. Sci Rep. 2019;9:6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arribat Y, Grepper D, Lagarrigue S, Richard J, Gachet M, Gut P, Amati F. Mitochondria in embryogenesis: an organellogenesis perspective. Front cell Dev Biology. 2019;7:282–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lopaschuk GD, Jaswal JS. Energy metabolic phenotype of the cardiomyocyte during development, differentiation, and postnatal maturation. J Cardiovasc Pharmacol. 2010;56:130–40. [DOI] [PubMed] [Google Scholar]
- 30.Persad KL, Lopaschuk GD. Energy metabolism on mitochondrial maturation and its effects on cardiomyocyte cell fate. Front Cell Dev Biology. 2022;10:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cardoso AC, Lam NT, Savla JJ, Nakada Y, Pereira AHM, Elnwasany A, Menendez-Montes I, Ensley EL, Bezan Petric U, Sharma G, et al. Mitochondrial substrate utilization regulates cardiomyocyte cell-cycle progression. Nat Metabolism. 2020;2:167–78. [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao Y, Gao C, Pan X, Lei K. Emerging roles of mitochondria in animal regeneration. Cell Regen. 2023;12:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ludikhuize MC, Meerlo M, Gallego MP, Xanthakis D, Burgaya Julià M, Nguyen NTB, Brombacher EC, Liv N, Maurice MM, Paik JH, et al. Mitochondria define intestinal stem cell differentiation downstream of a FOXO/Notch Axis. Cell Metabol. 2020;32:889-e900887. [DOI] [PubMed] [Google Scholar]
- 34.Lee JE, Seo BJ, Han MJ, Hong YJ, Hong K, Song H, Lee JW, Do JT. Changes in the expression of mitochondrial morphology-related genes during the differentiation of murine embryonic stem cells. Stem Cells Int. 2020;2020:9369268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mertens J, Regin M, De Munck N, Couvreu de Deckersberg E, Belva F, Sermon K, Tournaye H, Blockeel C, Van de Velde H, Spits C. Mitochondrial DNA variants segregate during human preimplantation development into genetically different cell lineages that are maintained postnatally. Hum Mol Genet. 2022;31:3629–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu SM, Chien KR, Mummery C. Origins and fates of cardiovascular progenitor cells. Cell. 2008;132:537–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Vliet P, Wu SM, Zaffran S, Pucéat M. Early cardiac development: a view from stem cells to embryos. Cardiovasc Res. 2012;96:352–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mostafavi S, Balafkan N, Pettersen IKN, Nido GS, Siller R, Tzoulis C, Sullivan GJ, Bindoff LA. Distinct mitochondrial remodeling during mesoderm differentiation in a human-based stem cell model. Front Cell Dev Biol. 2021;9:744777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qi Y, Ye Y, Wang R, Yu S, Zhang Y, Lv J, Jin W, Xia S, Jiang W, Li Y, Zhang D. Mitochondrial dysfunction by TFAM depletion disrupts self-renewal and lineage differentiation of human PSCs by affecting cell proliferation and YAP response. Redox Biol. 2022;50:102248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garbern JC, Lee RT. Mitochondria and metabolic transitions in cardiomyocytes: lessons from development for stem cell-derived cardiomyocytes. Stem Cell Res Ther. 2021;12:177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hollander JM, Thapa D, Shepherd DL. Physiological and structural differences in spatially distinct subpopulations of cardiac mitochondria: influence of cardiac pathologies. Am J Physiol Heart Circ Physiol. 2014;307:H1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paredes A, Justo-Méndez R, Jiménez-Blasco D, Núñez V, Calero I, Villalba-Orero M, Alegre-Martí A, Fischer T, Gradillas A, Sant’Anna VAR, et al. γ-Linolenic acid in maternal milk drives cardiac metabolic maturation. Nature. 2023;618:365–73. [DOI] [PubMed] [Google Scholar]
- 43.Li D, Armand LC, Sun F, Hwang H, Wolfson D, Rampoldi A, Liu R, Forghani P, Hu X, Yu W-M, et al. AMPK activator-treated human cardiac spheres enhance maturation and enable pathological modeling. Stem Cell Res Ther. 2023;14:322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao Q, Sun Q, Zhou L, Liu K, Jiao K. Complex regulation of mitochondrial function during cardiac development. J Am Heart Assoc. 2019;8:e012731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Knaapen MW, Vrolijk BC, Wenink AC. Ultrastructural changes of the myocardium in the embryonic rat heart. Anat Rec. 1997;248:233–41. [DOI] [PubMed] [Google Scholar]
- 46.Yasukawa T, Kang D. An overview of mammalian mitochondrial DNA replication mechanisms. J Biochem. 2018;164:183–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mercer TR, Neph S, Dinger ME, Crawford J, Smith MA, Shearwood AM, Haugen E, Bracken CP, Rackham O, Stamatoyannopoulos JA, et al. The human mitochondrial transcriptome. Cell. 2011;146:645–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Manasek FJ. Embryonic development of the heart: II. Formation of the epicardium. Development. 1969;22:333–48. [PubMed] [Google Scholar]
- 49.Bréhat J, Leick S, Musman J, Su JB, Eychenne N, Giton F, Rivard M, Barel LA, Tropeano C, Vitarelli F, et al. Identification of a mechanism promoting mitochondrial sterol accumulation during myocardial ischemia-reperfusion: role of TSPO and STAR. Basic Res Cardiol. 2024;119:481–503. [DOI] [PubMed] [Google Scholar]
- 50.Ovciarikova J, Shikha S, Lacombe A, Courjol F, McCrone R, Hussain W, Maclean A, Lemgruber L, Martins-Duarte ES, Gissot M, Sheiner L. Two ancient membrane pores mediate mitochondrial-nucleus membrane contact sites. J Cell Biol. 2024;223:e202304075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quirós PM, Mottis A, Auwerx J. Mitonuclear communication in homeostasis and stress. Nat Rev Mol Cell Biol. 2016;17:213–26. [DOI] [PubMed] [Google Scholar]
- 52.Kim Y, Ajayi PT, Bleck CKE, Glancy B. Three-dimensional remodelling of the cellular energy distribution system during postnatal heart development. Philos Trans R Soc Lond B Biol Sci. 2022;377:20210322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Berg OG, Kurland CG. Why mitochondrial genes are most often found in nuclei. Mol Biol Evol. 2000;17:951–61. [DOI] [PubMed] [Google Scholar]
- 54.Cardamone MD, Tanasa B, Cederquist CT, Huang J, Mahdaviani K, Li W, Rosenfeld MG, Liesa M, Perissi V. Mitochondrial retrograde signaling in mammals is mediated by the transcriptional cofactor gps2 via direct mitochondria-to-nucleus translocation. Mol Cell. 2018;69:757-e772757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miriyala S, Chandra M, Orr W, Kevil C, Bhuiyan MS, Panchatcharam M. Abstract 20021: mitochondrial mediated retrograde signaling in MnSOD cardiomyocyte-specific knockout: role in heart failure development and progression. Circulation. 2017;136:A20021-20021. [Google Scholar]
- 56.Miriyala S, Thippakorn C, Chaiswing L, Xu Y, Noel T, Tovmasyan A, Batinic-Haberle I, Vander Kooi CW, Chi W, Latif AA, et al. Novel role of 4-hydroxy-2-nonenal in AIFm2-mediated mitochondrial stress signaling. Free Radic Biol Med. 2016;91:68–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sriram K, Qi Z, Yuan D, Malhi NK, Liu X, Calandrelli R, Luo Y, Tapia A, Jin S, Shi J, et al. Regulation of nuclear transcription by mitochondrial RNA in endothelial cells. eLife. 2024;13:e86204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Butow RA, Avadhani NG. Mitochondrial signaling: the retrograde response. Mol Cell. 2004;14:1–15. [DOI] [PubMed] [Google Scholar]
- 59.Monaghan RM, Whitmarsh AJ. Mitochondrial proteins moonlighting in the Nucleus. Trends Biochem Sci. 2015;40:728–35. [DOI] [PubMed] [Google Scholar]
- 60.Lechuga-Vieco AV, Latorre-Pellicer A, Calvo E, Torroja C, Pellico J, Acín-Pérez R, García-Gil ML, Santos A, Bagwan N, Bonzon-Kulichenko E, et al. Heteroplasmy of wild-type mitochondrial DNA variants in mice causes metabolic heart disease with pulmonary hypertension and frailty. Circulation. 2022;145:1084–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fairbrother-Browne A, Ali AT, Reynolds RH, Garcia-Ruiz S, Zhang D, Chen Z, Ryten M, Hodgkinson A. Mitochondrial-nuclear cross-talk in the human brain is modulated by cell type and perturbed in neurodegenerative disease. Commun Biol. 2021;4:1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chiu ST, Tseng WW, Wei AC. Mathematical modeling and analysis of mitochondrial retrograde signaling dynamics. iScience. 2022;25:105502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Velez JM, Miriyala S, Nithipongvanitch R, Noel T, Plabplueng CD, Oberley T, Jungsuwadee P, Van Remmen H, Vore M, St Clair DK. p53 regulates oxidative stress-mediated retrograde signaling: a novel mechanism for chemotherapy-induced cardiac injury. PLoS ONE. 2011;6:e18005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ji L, Liu F, Jing Z, Huang Q, Zhao Y, Cao H, Li J, Yin C, Xing J, Li F. MICU1 alleviates diabetic cardiomyopathy through mitochondrial Ca2+–dependent antioxidant response. Diabetes. 2017;66:1586–600. [DOI] [PubMed] [Google Scholar]
- 65.Sack MN. Emerging characterization of the role of SIRT3-mediated mitochondrial protein deacetylation in the heart. Am J Physiol Heart Circ Physiol. 2011;301:H2191-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fang J, Song XW, Tian J, Chen HY, Li DF, Wang JF, Ren AJ, Yuan WJ, Lin L. Overexpression of microRNA-378 attenuates ischemia-induced apoptosis by inhibiting caspase-3 expression in cardiac myocytes. Apoptosis. 2012;17:410–23. [DOI] [PubMed] [Google Scholar]
- 67.Salazar C, Elorza AA, Cofre G, Ruiz-Hincapie P, Shirihai O, Ruiz LM. The OXPHOS supercomplex assembly factor HIG2A responds to changes in energetic metabolism and cell cycle. J Cell Physiol. 2019;234:17405–19. [DOI] [PubMed] [Google Scholar]
- 68.Salazar C, Barros M, Elorza AA, Ruiz LM. Dynamic distribution of HIG2A between the mitochondria and the nucleus in response to hypoxia and oxidative stress. Int J Mol Sci. 2021;23:389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nandi SS, Maresh GM, Patel KP. Decreased mitochondrial unfolded protein response (UPRmt) in HFpEF. Faseb J. 2022;36.
- 70.Liu J, He X, Zheng S, Zhu A, Wang J. The mitochondrial unfolded protein response: a novel protective pathway targeting cardiomyocytes. Oxid Med Cell Longev. 2022;2022:6430342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ashrafian H, Czibik G, Bellahcene M, Aksentijević D, Smith Anthony C, Mitchell Sarah J, Dodd Michael S, Kirwan J, Byrne Jonathan J, Ludwig C, et al. Fumarate is cardioprotective via activation of the Nrf2 antioxidant pathway. Cell Metabol. 2012;15:361–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee SR, Han J. Mitochondrial nucleoid: Shield and switch of the mitochondrial genome. Oxid Med Cell Longev. 2017;2017:8060949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ghazal N, Peoples JN, Mohiuddin TA, Kwong JQ. Mitochondrial functional resilience after TFAM ablation in the adult heart. Am J Physiology-Cell Physiol. 2021;320:C929-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Peralta S, Wang X, Moraes CT. Mitochondrial transcription: lessons from mouse models. Biochim Biophys Acta. 2012;1819:961–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Herai RH, Negraes PD, Muotri AR. Evidence of nuclei-encoded spliceosome mediating splicing of mitochondrial RNA. Hum Mol Genet. 2017;26:2472–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Feng Y, Huang W, Paul C, Liu X, Sadayappan S, Wang Y, Pauklin S. Mitochondrial nucleoid in cardiac homeostasis: bidirectional signaling of mitochondria and nucleus in cardiac diseases. Basic Res Cardiol. 2021;116:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang Z, Gao W, Long QQ, et al. Increased plasma levels of lncRNA H19 and LIPCAR are associated with increased risk of coronary artery disease in a Chinese population. Sci Rep. 2017;7:7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gao Y, Wu M, Fan Y, Li S, Lai Z, Huang Y, Lan X, Lei C, Chen H, Dang R. Identification and characterization of circular RNAs in Qinchuan cattle testis. Royal Soc Open Sci. 2018;5:180413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang K, Gan T-Y, Li N, Liu C-Y, Zhou L-Y, Gao J-N, Chen C, Yan K-W, Ponnusamy M, Zhang Y-H, Li P-F. Circular RNA mediates cardiomyocyte death via miRNA-dependent upregulation of MTP18 expression. Cell Death Differ. 2017;24:1111–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Piquereau J, Novotova M, Fortin D, Garnier A, Ventura-Clapier R, Veksler V, Joubert F. Postnatal development of mouse heart: formation of energetic microdomains. J Physiol. 2010;588:2443–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Laudette M, Lindbom M, Arif M, Cinato M, Ruiz M, Doran S, Miljanovic A, Rutberg M, Andersson L, Klevstig M, et al. Cardiomyocyte-specific PCSK9 deficiency compromises mitochondrial bioenergetics and heart function. Cardiovasc Res. 2023;119:1537–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen Y, Liu Y, Dorn GW 2. Mitochondrial fusion is essential for organelle function and cardiac homeostasis. Circ Res. 2011;109:1327–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wu S, Lu Q, Wang Q, Ding Y, Ma Z, Mao X, Huang K, Xie Z, Zou MH. Binding of FUN14 domain containing 1 with Inositol 1,4,5-Trisphosphate receptor in Mitochondria-Associated endoplasmic reticulum membranes maintains mitochondrial dynamics and function in hearts in vivo. Circulation. 2017;136:2248–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Morozov YM, Rakic P. Disorder of golgi apparatus precedes anoxia-induced pathology of mitochondria. Int J Mol Sci. 2023;24:4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tábara LC, Morris JL, Prudent J. The complex dance of organelles during mitochondrial division. Trends Cell Biol. 2021;31:241–53. [DOI] [PubMed] [Google Scholar]
- 86.Nagashima S, Tábara LC, Tilokani L, Paupe V, Anand H, Pogson JH, Zunino R, McBride HM, Prudent J. Golgi-derived PI(4)P-containing vesicles drive late steps of mitochondrial division. Science. 2020;367:1366–71. [DOI] [PubMed] [Google Scholar]
- 87.Moyzis A, Gustafsson ÅB. Multiple recycling routes: canonical vs. non-canonical mitophagy in the heart. Biochim Biophys Acta Mol Basis Dis. 2019;1865:797–809. [DOI] [PubMed] [Google Scholar]
- 88.Gong G, Song M, Csordas G, Kelly DP, Matkovich SJ, Dorn GW. 2nd: parkin-mediated mitophagy directs perinatal cardiac metabolic maturation in mice. Science. 2015;350:aad2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.McLelland GL, Goiran T, Yi W, Dorval G, Chen CX, Lauinger ND, Krahn AI, Valimehr S, Rakovic A, Rouiller I, et al. Mfn2 ubiquitination by PINK1/parkin gates the p97-dependent release of ER from mitochondria to drive mitophagy. Elife. 2018;7:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, Sideris DP, Fogel AI, Youle RJ. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524:309–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Peng W, Wong YC, Krainc D. Mitochondria-lysosome contacts regulate mitochondrial Ca < sup > 2+ dynamics via lysosomal TRPML1. Proc Natl Acad Sci 2020;117:19266–19275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ma X, Manley S, Qian H, Li Y, Zhang C, Li K, Ding B, Guo F, Chen A, Zhang X, et al. Mitochondria-lysosome-related organelles mediate mitochondrial clearance during cellular dedifferentiation. Cell Rep. 2023;42:113291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Prashar A, Bussi C, Fearns A, Capurro MI, Gao X, Sesaki H, Gutierrez MG, Jones NL. Lysosomes drive the piecemeal removal of mitochondrial inner membrane. Nature. 2024;632:1110–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Behjati M, Sabri MR, Etemadi Far M, Nejati M. Cardiac complications in inherited mitochondrial diseases. Heart Fail Rev. 2021;26:391–403. [DOI] [PubMed] [Google Scholar]
- 95.Meyers DE, Basha HI, Koenig MK. Mitochondrial cardiomyopathy: pathophysiology, diagnosis, and management. Tex Heart Inst J. 2013;40:385–94. [PMC free article] [PubMed] [Google Scholar]
- 96.Holmgren D, Wåhlander H, Eriksson BO, Oldfors A, Holme E, Tulinius M. Cardiomyopathy in children with mitochondrial disease; clinical course and cardiological findings. Eur Heart J. 2003;24:280–8. [DOI] [PubMed] [Google Scholar]
- 97.Kumar AA, Kelly DP, Chirinos JA. Mitochondrial dysfunction in heart failure with preserved ejection fraction. Circulation. 2019;139:1435–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang J, Cai X, Zhang Q, Li X, Li S, Ma J, Zhu W, Liu X, Wei M, Tu W, et al. Hydrogen sulfide restores sevoflurane postconditioning mediated cardioprotection in diabetic rats: role of SIRT1/Nrf2 signaling-modulated mitochondrial dysfunction and oxidative stress. J Cell Physiol. 2021;236:5052–68. [DOI] [PubMed] [Google Scholar]
- 99.Zemljic-Harpf AE, See Hoe LE, Schilling JM, Zuniga-Hertz JP, Nguyen A, Vaishnav YJ, Belza GJ, Budiono BP, Patel PM, Head BP, et al. Morphine induces physiological, structural, and molecular benefits in the diabetic myocardium. Faseb j. 2021;35:e21407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Subramani J, Kundumani-Sridharan V, Das KC. Thioredoxin protects mitochondrial structure, function and biogenesis in myocardial ischemia-reperfusion via redox-dependent activation of AKT-CREB- PGC1α pathway in aged mice. Aging. 2020;12:19809–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Demeter-Haludka V, Kovács M, Petrus A, Patai R, Muntean DM, Siklós L, Végh Á. Examination of the role of mitochondrial morphology and function in the cardioprotective effect of Sodium Nitrite Administered 24 h before Ischemia/Reperfusion Injury. Front Pharmacol. 2018;9:286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li Q, Lian C, Zhou R, Li T, Xiang X, Liu B. Pretreatment with Xenon protected immature rabbit Heart from Ischaemia/Reperfusion Injury by opening of the mitoKATP Channel. Heart Lung Circulation. 2013;22:276–83. [DOI] [PubMed] [Google Scholar]
- 103.Elrod JW, Calvert JW, Morrison J, Doeller JE, Kraus DW, Tao L, Jiao X, Scalia R, Kiss L, Szabo C, et al. Hydrogen sulfide attenuates myocardial ischemia-reperfusion injury by preservation of mitochondrial function. Proc Natl Acad Sci U S A. 2007;104:15560–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shinohara T, Takahashi N, Kohno H, Yamanaka K, Ooie T, Wakisaka O, Murozono Y, Taniguchi Y, Torigoe Y, Hara M, et al. Mitochondria are targets for geranylgeranylacetone-induced cardioprotection against ischemia-reperfusion in the rat heart. Am J Physiol Heart Circ Physiol. 2007;293:H1892-1899. [DOI] [PubMed] [Google Scholar]
- 105.Fan G, Chen MJ, Wei J. Involvement of phosphatase and tensin homolog-induced putative kinase 1/Parkin-mediated autophagy in angiotensin II-induced cardiac hypertrophy in C57BL/6 mice. J Int Med Res. 2020;48:300060519896143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ravizzoni Dartora D, Flahault A, Pontes CNR, He Y, Deprez A, Cloutier A, Cagnone G, Gaub P, Altit G, Bigras JL, et al. Cardiac left ventricle mitochondrial dysfunction after neonatal exposure to Hyperoxia: relevance for Cardiomyopathy after Preterm Birth. Hypertension. 2022;79:575–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Elezaby A, Sverdlov AL, Tu VH, Soni K, Luptak I, Qin F, Liesa M, Shirihai OS, Rimer J, Schaffer JE, et al. Mitochondrial remodeling in mice with cardiomyocyte-specific lipid overload. J Mol Cell Cardiol. 2015;79:275–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shirasaka T, Miyagawa S, Fukushima S, Saito A, Shiozaki M, Kawaguchi N, Matsuura N, Nakatani S, Sakai Y, Daimon T, et al. A slow-releasing form of prostacyclin agonist (ONO1301SR) enhances endogenous secretion of multiple cardiotherapeutic cytokines and improves cardiac function in a rapid-pacing–induced model of canine heart failure. J Thorac Cardiovasc Surg. 2013;146:413–21. [DOI] [PubMed] [Google Scholar]
- 109.Chen H, Dong Y, He X, Li J, Wang J. Paeoniflorin improves cardiac function and decreases adverse postinfarction left ventricular remodeling in a rat model of acute myocardial infarction. Drug Des Devel Ther. 2018;12:823–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Katz MG, Gubara SM, Hadas Y, Weber T, Kumar A, Eliyahu E, Bridges CR, Fargnoli AS. Effects of genetic transfection on calcium cycling pathways mediated by double-stranded adeno-associated virus in postinfarction remodeling. J Thorac Cardiovasc Surg. 2020;159:1809-e18191803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tao L, Huang X, Xu M, Yang L, Hua F. MiR-144 protects the heart from hyperglycemia-induced injury by regulating mitochondrial biogenesis and cardiomyocyte apoptosis. Faseb J. 2020;34:2173–97. [DOI] [PubMed] [Google Scholar]
- 112.Wu QS, He Q, He JQ, Chao J, Wang WY, Zhou Y, Lou JZ, Kong W, Chen JF. The role of mitofilin in left ventricular hypertrophy in hemodialysis patients. Ren Fail. 2018;40:252–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Thapa D, Nichols CE, Lewis SE, Shepherd DL, Jagannathan R, Croston TL, Tveter KJ, Holden AA, Baseler WA, Hollander JM. Transgenic overexpression of mitofilin attenuates diabetes mellitus-associated cardiac and mitochondria dysfunction. J Mol Cell Cardiol. 2015;79:212–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tian L, Neuber-Hess M, Mewburn J, Dasgupta A, Dunham-Snary K, Wu D, Chen KH, Hong Z, Sharp WW, Kutty S, Archer SL. Ischemia-induced Drp1 and Fis1-mediated mitochondrial fission and right ventricular dysfunction in pulmonary hypertension. J Mol Med (Berl). 2017;95:381–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chaanine AH, Kohlbrenner E, Gamb SI, Guenzel AJ, Klaus K, Fayyaz AU, Nair KS, Hajjar RJ, Redfield MM. FOXO3a regulates BNIP3 and modulates mitochondrial calcium, dynamics, and function in cardiac stress. Am J Physiol Heart Circ Physiol. 2016;311:H1540-1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ma L, Chang X, Gao J, Zhang Y, Chen Y, Zhou H, Zhou N, Du N, Li J, Bi J, et al. METTL3 boosts mitochondrial fission and induces cardiac fibrosis after ischemia/reperfusion injury. Int J Biol Sci. 2024;20:433–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yang ZJ, Guo CL, Gong YX, Li L, Wang LL, Liu HM, Cao JM, Lu ZY. Dapagliflozin suppresses isoprenaline-induced cardiac hypertrophy through inhibition of mitochondrial fission. J Cardiovasc Pharmacol. 2024;83:193–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Preston KJ, Kawai T, Torimoto K, et al. Mitochondrial fission inhibition protects against hypertension induced by angiotensin II. Hypertens Res. 2024;47:1338–49. [DOI] [PubMed]
- 119.Hao J, Zhou J, Hu S, Zhang P, Wu H, Yang J, Zhao B, Liu H, Lin H, Chi J, Lou D. RTA 408 ameliorates diabetic cardiomyopathy by activating Nrf2 to regulate mitochondrial fission and fusion and inhibiting NF-κB-mediated inflammation. Am J Physiol Cell Physiol. 2024;326:C331-47. [DOI] [PubMed] [Google Scholar]
- 120.Maneechote C, Palee S, Kerdphoo S, Jaiwongkam T, Chattipakorn SC, Chattipakorn N. Pharmacological inhibition of mitochondrial fission attenuates cardiac ischemia-reperfusion injury in pre-diabetic rats. Biochem Pharmacol. 2020;182:114295. [DOI] [PubMed] [Google Scholar]
- 121.Nollet EE, Duursma I, Rozenbaum A, Eggelbusch M, Wüst RCI, Schoonvelde SAC, Michels M, Jansen M, van der Wel NN, Bedi KC, et al. Mitochondrial dysfunction in human hypertrophic cardiomyopathy is linked to cardiomyocyte architecture disruption and corrected by improving NADH-driven mitochondrial respiration. Eur Heart J. 2023;44:1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Prandi FR, Evangelista I, Sergi D, et al. Mechanisms of cardiac dysfunction in diabetic cardiomyopathy: molecular abnormalities and phenotypical variants. Heart Fail Rev. 2023;28:597–606. [DOI] [PubMed]
- 123.Xu X, Kobayashi S, Chen K, Timm D, Volden P, Huang Y, Gulick J, Yue Z, Robbins J, Epstein PN, Liang Q. Diminished autophagy limits cardiac injury in mouse models of type 1 diabetes. J Biol Chem. 2013;288:18077–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Munasinghe PE, Riu F, Dixit P, Edamatsu M, Saxena P, Hamer NS, Galvin IF, Bunton RW, Lequeux S, Jones G, et al. Type-2 diabetes increases autophagy in the human heart through promotion of Beclin-1 mediated pathway. Int J Cardiol. 2016;202:13–20. [DOI] [PubMed] [Google Scholar]
- 125.Andres AM, Tucker KC, Thomas A, Taylor DJ, Sengstock D, Jahania SM, Dabir R, Pourpirali S, Brown JA, Westbrook DG, et al. Mitophagy and mitochondrial biogenesis in atrial tissue of patients undergoing heart surgery with cardiopulmonary bypass. JCI Insight. 2017;2:e89303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ashar FN, Zhang Y, Longchamps RJ, Lane J, Moes A, Grove ML, Mychaleckyj JC, Taylor KD, Coresh J, Rotter JI, et al. Association of mitochondrial DNA copy number with cardiovascular disease. JAMA Cardiol. 2017;2:1247–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hong YS, Longchamps RJ, Zhao D, Castellani CA, Loehr LR, Chang PP, Matsushita K, Grove ML, Boerwinkle E, Arking DE, Guallar E. Mitochondrial DNA Copy Number and Incident Heart failure. Circulation. 2020;141:1823–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Castellani CA, Longchamps RJ, Sumpter JA, Newcomb CE, Lane JA, Grove ML, Bressler J, Brody JA, Floyd JS, Bartz TM, et al. Mitochondrial DNA copy number can influence mortality and cardiovascular disease via methylation of nuclear DNA CpGs. Genome Med. 2020;12:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kurian GA, Ansari M, Prem PN. Diabetic cardiomyopathy attenuated the protective effect of ischaemic post-conditioning against ischaemia-reperfusion injury in the isolated rat heart model. Arch Physiol Biochem. 2020;129(3):711–22. [DOI] [PubMed]
- 130.Ansari M, Kurian GA. Diabetic animal fed with high-fat diet prevents the protective effect of myocardial ischemic preconditioning effect in isolated rat heart perfusion model. J Biochem Mol Toxicol. 2020;34:e22457. [DOI] [PubMed] [Google Scholar]
- 131.Kurian AM. Mitochondrial dysfunction plays a key role in the abrogation of cardioprotection by sodium hydrosulfide post-conditioning in diabetic cardiomyopathy rat heart. Naunyn Schmiedebergs Arch Pharmacol. 2020;393:339–48. [DOI] [PubMed] [Google Scholar]
- 132.Ansari M, Kurian GA. Mechanism of hydrogen sulfide preconditioning-associated protection against ischemia–reperfusion injury differs in diabetic heart that develops myopathy. Cardiovasc Toxicol. 2020;20:155–67. [DOI] [PubMed] [Google Scholar]
- 133.Broughton KM, Wang BJ, Firouzi F, Khalafalla F, Dimmeler S, Fernandez-Aviles F, Sussman MA. Mechanisms of cardiac repair and regeneration. Circul Res. 2018;122:1151–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Poznyak AV, Ivanova EA, Sobenin IA, Yet S-F, Orekhov AN. The role of mitochondria in cardiovascular diseases. Biology. 2020;9:137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mongirdienė A, Skrodenis L, Varoneckaitė L, Mierkytė G, Gerulis J. Reactive oxygen species Induced pathways in Heart failure pathogenesis and potential therapeutic strategies. Biomedicines. 2022;10:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chen Q, Moghaddas S, Hoppel CL, Lesnefsky EJ. Ischemic defects in the electron transport chain increase the production of reactive oxygen species from isolated rat heart mitochondria. Am J Physiol Cell Physiol. 2008;294:C460-466. [DOI] [PubMed] [Google Scholar]
- 137.Lesnefsky EJ, Chen Q, Moghaddas S, Hassan MO, Tandler B, Hoppel CL. Blockade of electron transport during ischemia protects cardiac mitochondria. J Biol Chem. 2004;279:47961–7. [DOI] [PubMed] [Google Scholar]
- 138.Brown DI, Griendling KK. Regulation of signal transduction by reactive oxygen species in the cardiovascular system. Circul Res. 2015;116:531–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hausenloy D, Wynne A, Duchen M, Yellon D. Transient mitochondrial permeability transition pore opening mediates preconditioning-induced protection. Circulation. 2004;109:1714–7. [DOI] [PubMed] [Google Scholar]
- 140.Peoples JN, Saraf A, Ghazal N, Pham TT, Kwong JQ. Mitochondrial dysfunction and oxidative stress in heart disease. Exp Mol Med. 2019;51:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Nandi S, Ravindran S, Kurian GA. Role of endogenous hydrogen sulfide in cardiac mitochondrial preservation during ischemia reperfusion injury. Biomed Pharmacother. 2018;97:271–9. [DOI] [PubMed] [Google Scholar]
- 142.Kornfeld OS, Hwang S, Disatnik MH, Chen CH, Qvit N, Mochly-Rosen D. Mitochondrial reactive oxygen species at the heart of the Matter. Circul Res. 2015;116:1783–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Garlid AO, Jaburek M, Jacobs JP, Garlid KD. Mitochondrial reactive oxygen species: which ROS signals cardioprotection? Am J Physiol Heart Circ Physiol. 2013;305:H960-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lin PH, Lee SH, Su CP, Wei YH. Oxidative damage to mitochondrial DNA in atrial muscle of patients with atrial fibrillation. Free Radic Biol Med. 2003;35:1310–8. [DOI] [PubMed] [Google Scholar]
- 145.Han J, Choi HY, Dayem AA, Kim K, Yang G, Won J, Do SH, Kim J-H, Jeong K-S, Cho S-G. Regulation of adipogenesis through differential modulation of ROS and kinase signaling pathways by 3,4′-Dihydroxyflavone treatment. J Cell Biochem. 2017;118:1065–77. [DOI] [PubMed] [Google Scholar]
- 146.Gangwar A, Paul S, Ahmad Y, Bhargava K. Competing trends of ROS and RNS-mediated protein modifications during hypoxia as an alternate mechanism of NO benefits. Biochimie. 2018;148:127–38. [DOI] [PubMed] [Google Scholar]
- 147.Long X, Gao Y, Liu W, Liu X, Hayashi T, Mizuno K, Hattori S, Ikejima T. Natural flavonoid silibinin promotes the migration and myogenic differentiation of murine C2C12 myoblasts via modulation of ROS generation and down-regulation of estrogen receptor α expression. Mol Cell Biochem. 2020;474:243–61. [DOI] [PubMed] [Google Scholar]
- 148.Li J, Wang Z, Li C, Song Y, Wang Y, Bo H, Zhang Y. Impact of exercise and aging on mitochondrial homeostasis in skeletal muscle: roles of ROS and Epigenetics. Cells. 2022;11:2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Marín-García J, Akhmedov AT. Mitochondrial dynamics and cell death in heart failure. Heart Fail Rev. 2016;21:123–36. [DOI] [PubMed] [Google Scholar]
- 150.Lin KL, Chen SD, Lin KJ, Liou CW, Chuang YC, Wang PW, Chuang JH, Lin TK. Quality Matters? The Involvement of Mitochondrial Quality Control in Cardiovascular Disease. Front Cell Dev Biol. 2021;9:636295. [DOI] [PMC free article] [PubMed]
- 151.Poznyak AV, Kirichenko TV, Borisov EE, Shakhpazyan NK, Kartuesov AG, Orekhov AN. Mitochondrial implications in cardiovascular aging and diseases: the specific role of mitochondrial dynamics and shifts. Int J Mol Sci. 2022;23:2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Quiles JM, Gustafsson ÅB. The role of mitochondrial fission in cardiovascular health and disease. Nat Rev Cardiol. 2022;19:723–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hernandez-Resendiz S, Prakash A, Loo SJ, Semenzato M, Chinda K, Crespo-Avilan GE, Dam LC, Lu S, Scorrano L, Hausenloy DJ. Targeting mitochondrial shape: at the heart of cardioprotection. Basic Res Cardiol. 2023;118:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Lee H, Smith SB, Yoon Y. The short variant of the mitochondrial dynamin OPA1 maintains mitochondrial energetics and cristae structure. J Biol Chem. 2017;292:7115–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ge Y, Shi X, Boopathy S, McDonald J, Smith AW, Chao LH. Two forms of Opa1 cooperate to complete fusion of the mitochondrial inner-membrane. Elife 2020, 9. [DOI] [PMC free article] [PubMed]
- 156.Chen L, Gong Q, Stice JP, Knowlton AA. Mitochondrial OPA1, apoptosis, and heart failure. Cardiovasc Res. 2009;84:91–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.MacVicar TD, Lane JD. Impaired OMA1-dependent cleavage of OPA1 and reduced DRP1 fission activity combine to prevent mitophagy in cells that are dependent on oxidative phosphorylation. J Cell Sci. 2014;127:2313–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Acin-Perez R, Lechuga-Vieco AV, Del Mar Muñoz M, Nieto-Arellano R, Torroja C, Sánchez-Cabo F, Jiménez C, González-Guerra A, Carrascoso I, Benincá C, et al. Ablation of the stress protease OMA1 protects against heart failure in mice. Sci Transl Med. 2018;10:eaan4935. [DOI] [PubMed] [Google Scholar]
- 159.Burke N, Hall AR, Hausenloy DJ. OPA1 in cardiovascular health and disease. Curr Drug Targets. 2015;16:912–20. [DOI] [PubMed] [Google Scholar]
- 160.Alavi MV. Recent advances in, and challenges of, designing OMA1 drug screens. Pharmacol Res. 2022;176:106063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Maneechote C, Palee S, Kerdphoo S, Jaiwongkam T, Chattipakorn SC, Chattipakorn N. Balancing mitochondrial dynamics via increasing mitochondrial fusion attenuates infarct size and left ventricular dysfunction in rats with cardiac ischemia/reperfusion injury. Clin Sci (Lond). 2019;133:497–513. [DOI] [PubMed] [Google Scholar]
- 162.Maneechote C, Khuanjing T, Ongnok B, Arinno A, Prathumsap N, Chunchai T, Arunsak B, Nawara W, Chattipakorn SC, Chattipakorn N. Promoting mitochondrial fusion in doxorubicin-induced cardiotoxicity: a novel therapeutic target for cardioprotection. Clin Sci (Lond). 2022;136:841–60. [DOI] [PubMed] [Google Scholar]
- 163.Tran DT, Tu Z, Alawieh A, Mulligan J, Esckilsen S, Quinn K, Sundararaj K, Wallace C, Finnegan R, Allen P, et al. Modulating donor mitochondrial fusion/fission delivers immunoprotective effects in cardiac transplantation. Am J Transpl. 2022;22:386–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ding M, Zeng K, Yu M, Fu F. Abstract 10792: mitochondrial Fusion promoter effectively alleviates mitochondrial dysfunction and improves cardiac function in Diabetic rats. Circulation. 2019;140:A10792-10792. [Google Scholar]
- 165.Wang T, Xing G, Fu T, Ma Y, Wang Q, Zhang S, Chang X, Tong Y. Role of mitochondria in doxorubicin-mediated cardiotoxicity: from molecular mechanisms to therapeutic strategies. Int J Med Sci. 2024;21:809–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Franco A, Li J, Kelly DP, Hershberger RE, Marian AJ, Lewis RM, Song M, Dang X, Schmidt AD, Mathyer ME, et al. A human mitofusin 2 mutation can cause mitophagic cardiomyopathy. Elife. 2023;12:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ravindran S, Ansari Banu S, Kurian GA. Hydrogen sulfide preconditioning shows differential protection towards interfibrillar and subsarcolemmal mitochondria from isolated rat heart subjected to revascularization injury. Cardiovasc Pathol. 2016;25:306–15. [DOI] [PubMed] [Google Scholar]
- 168.Dai SH, Wu QC, Zhu RR, Wan XM, Zhou XL. Notch1 protects against myocardial ischaemia-reperfusion injury via regulating mitochondrial fusion and function. J Cell Mol Med. 2020;24:3183–91. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 169.Maneechote C, Palee S, Kerdphoo S, Jaiwongkam T, Chattipakorn Siriporn C, Chattipakorn N. Differential temporal inhibition of mitochondrial fission by Mdivi-1 exerts effective cardioprotection in cardiac ischemia/reperfusion injury. Clin Sci. 2018;132:1669–83. [DOI] [PubMed] [Google Scholar]
- 170.Feng X, Wang S, Yang X, Lin J, Man W, Dong Y, Zhang Y, Zhao Z, Wang H, Sun D. Mst1 knockout alleviates mitochondrial fission and mitigates left ventricular remodeling in the development of diabetic cardiomyopathy. Front Cell Dev Biol. 2020;8:628842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Torres G, Morales PE, García-Miguel M, Norambuena-Soto I, Cartes-Saavedra B, Vidal-Peña G, Moncada-Ruff D, Sanhueza-Olivares F, San Martín A, Chiong M. Glucagon-like peptide-1 inhibits vascular smooth muscle cell dedifferentiation through mitochondrial dynamics regulation. Biochem Pharmacol. 2016;104:52–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Ma X, Liu Z, Ilyas I, Little PJ, Kamato D, Sahebka A, Chen Z, Luo S, Zheng X, Weng J, Xu S. GLP-1 receptor agonists (GLP-1RAs): cardiovascular actions and therapeutic potential. Int J Biol Sci. 2021;17:2050–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Deng Y, Li S, Chen Z, Wang W, Geng B, Cai J. Mdivi-1, a mitochondrial fission inhibitor, reduces angiotensin-II- induced hypertension by mediating VSMC phenotypic switch. Biomed Pharmacother. 2021;140:111689. [DOI] [PubMed] [Google Scholar]
- 174.Giugliano D, Scappaticcio L, Longo M, Caruso P, Maiorino MI, Bellastella G, Ceriello A, Chiodini P, Esposito K. GLP-1 receptor agonists and cardiorenal outcomes in type 2 diabetes: an updated meta-analysis of eight CVOTs. Cardiovasc Diabetol. 2021;20:189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Moehle EA, Shen K, Dillin A. Mitochondrial proteostasis in the context of cellular and organismal health and aging. J Biol Chem. 2019;294:5396–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Eldomery MK, Akdemir ZC, Vögtle FN, Charng WL, Mulica P, Rosenfeld JA, Gambin T, Gu S, Burrage LC, Al Shamsi A, et al. MIPEP recessive variants cause a syndrome of left ventricular non-compaction, hypotonia, and infantile death. Genome Med. 2016;8:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Deshwal S, Fiedler KU, Langer T. Mitochondrial proteases: multifaceted regulators of mitochondrial plasticity. Annu Rev Biochem. 2020;89:501–28. [DOI] [PubMed] [Google Scholar]
- 178.Chen Z, Huang L, Tso A, Wang S, Fang X, Ouyang K, Han Z. Mitochondrial chaperones and proteases in cardiomyocytes and heart failure. Front Mol Biosci. 2021;8: 630332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Hoshino A, Okawa Y, Ariyoshi M, Kaimoto S, Uchihashi M, Fukai K, Iwai-Kanai E, Matoba S. Oxidative post-translational modifications develop LONP1 dysfunction in pressure overload heart failure. Circ Heart Fail. 2014;7:500–9. [DOI] [PubMed] [Google Scholar]
- 180.Kang S, Louboutin JP, Datta P, Landel CP, Martinez D, Zervos AS, Strayer DS, Fernandes-Alnemri T, Alnemri ES. Loss of HtrA2/Omi activity in non-neuronal tissues of adult mice causes premature aging. Cell Death Differ. 2013;20:259–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Seiferling D, Szczepanowska K, Becker C, Senft K, Hermans S, Maiti P, König T, Kukat A, Trifunovic A. Loss of CLPP alleviates mitochondrial cardiomyopathy without affecting the mammalian UPRmt. EMBO Rep. 2016;17:953–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Bhuiyan MS, Fukunaga K. Inhibition of HtrA2/Omi ameliorates heart dysfunction following ischemia/reperfusion injury in rat heart in vivo. Eur J Pharmacol. 2007;557:168–77. [DOI] [PubMed] [Google Scholar]
- 183.Hong WL, Huang H, Zeng X, Duan CY. Targeting mitochondrial quality control: new therapeutic strategies for major diseases. Mil Med Res. 2024;11:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Deng J, Wang D, Shi Y, Lin L, Gao W, Sun Y, Song X, Li Y, Li J. Mitochondrial unfolded protein response mechanism and its cardiovascular protective effects. Biomed Pharmacother. 2024;177:116989. [DOI] [PubMed] [Google Scholar]
- 185.Titus AS, Sung EA, Zablocki D, Sadoshima J. Mitophagy for cardioprotection. Basic Res Cardiol. 2023;118:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Alam S, Abdullah CS, Aishwarya R, Morshed M, Bhuiyan MS. Molecular perspectives of mitochondrial adaptations and their role in cardiac proteostasis. Front Physiol. 2020;11:1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Guerra J, Matta L, Bartelt A. Cardiac proteostasis in obesity and cardiovascular disease. Herz. 2024;49:118–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ghosh R, Vinod V, Symons JD, Boudina S. Protein and Mitochondria Quality Control Mechanisms and Cardiac Aging. Cells. 2020;9:933. [DOI] [PMC free article] [PubMed]
- 189.Tang Y, Xu W, Liu Y, Zhou J, Cui K, Chen Y. Autophagy protects mitochondrial health in heart failure. Heart Fail Rev. 2024;29:113–23. [DOI] [PubMed] [Google Scholar]
- 190.Torres AK, Fleischhart V, Inestrosa NC. Mitochondrial unfolded protein response (UPRmt): what we know thus far. Front Cell Dev Biol. 2024;12:1405393. [DOI] [PMC free article] [PubMed]
- 191.Barac YD, Emrich F, Krutzwakd-Josefson E, Schrepfer S, Sampaio LC, Willerson JT, Robbins RC, Ciechanover A, Mohr FW, Aravot D, Taylor DA. The ubiquitin-proteasome system: a potential therapeutic target for heart failure. J Heart Lung Transpl. 2017;36:708–14. [DOI] [PubMed] [Google Scholar]
- 192.Smyrnias I, Gray SP, Okonko DO, Sawyer G, Zoccarato A, Catibog N, López B, González A, Ravassa S, Díez J, Shah AM. Cardioprotective effect of the mitochondrial unfolded protein response during chronic pressure overload. J Am Coll Cardiol. 2019;73:1795–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Qipshidze N, Tyagi N, Metreveli N, Lominadze D, Tyagi SC. Autophagy mechanism of right ventricular remodeling in murine model of pulmonary artery constriction. Am J Physiol Heart Circ Physiol. 2012;302:H688-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Wu D, Ji H, Du W, Ren L, Qian G. Mitophagy alleviates ischemia/reperfusion-induced microvascular damage through improving mitochondrial quality control. Bioengineered. 2022;13:3596–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Miranda-Silva D, Alves PGR, Rizo E, Fonseca D, Lima A, Baganha T, Conceição F, Sousa G, Gonçalves C. Mitochondrial reversible changes determine diastolic function adaptations during myocardial (reverse) remodeling. Circ Heart Fail. 2020;13:e006170. [DOI] [PubMed] [Google Scholar]
- 196.Rath S, Sharma R, Gupta R, Ast T, Chan C, Durham TJ, Goodman RP, Grabarek Z, Haas ME, Hung WHW, et al. MitoCarta3.0: an updated mitochondrial proteome now with sub-organelle localization and pathway annotations. Nucleic Acids Res. 2021;49:D1541-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Primiano G, Torraco A, Verrigni D, Sabino A, Bertini E, Carrozzo R, Silvestri G, Servidei S. Novel TOP3A variant Associated with mitochondrial disease: expanding the clinical spectrum of Topoisomerase III Alpha-Related diseases. Neurol Genet. 2022;8:e200007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Semenza GL, Wang GL. A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol Cell Biol. 1992;12:5447–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Cadenas S, Aragonés J, Landázuri MO. Mitochondrial reprogramming through cardiac oxygen sensors in ischaemic heart disease. Cardiovascular Res. 2010;88:219–28. [DOI] [PubMed] [Google Scholar]
- 200.Semenza GL. Hypoxia-inducible factor 1 and cardiovascular disease. Annu Rev Physiol. 2014;76:39–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Hölscher M, Schäfer K, Krull S, Farhat K, Hesse A, Silter M, Lin Y, Pichler BJ, Thistlethwaite P, El-Armouche A, et al. Unfavourable consequences of chronic cardiac HIF-1α stabilization. Cardiovascular Res. 2012;94:77–86. [DOI] [PubMed] [Google Scholar]
- 202.Thomas LW, Ashcroft M. Exploring the molecular interface between hypoxia-inducible factor signalling and mitochondria. Cell Mol Life Sci. 2019;76:1759–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Huang X, Zhao L, Peng R. Hypoxia-inducible factor 1 and mitochondria: an intimate connection. Biomolecules. 2022;13:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Li HS, Zhou YN, Li L, Li SF, Long D, Chen XL, Zhang JB, Feng L, Li YP. HIF-1α protects against oxidative stress by directly targeting mitochondria. Redox Biol. 2019;25:101109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Hoffman H, Grigg GW. An electron microscopic study of mitochondria formation. Exp Cell Res. 1958;15:118–31. [DOI] [PubMed] [Google Scholar]
- 206.Schmidt O, Pfanner N, Meisinger C. Mitochondrial protein import: from proteomics to functional mechanisms. Nat Rev Mol Cell Biol. 2010;11:655–67. [DOI] [PubMed] [Google Scholar]
- 207.Vögtle F-N. Open questions on the mitochondrial unfolded protein response. FEBS J. 2021;288:2856–69. [DOI] [PubMed] [Google Scholar]
- 208.Soledad RB, Charles S, Samarjit D. The secret messages between mitochondria and nucleus in muscle cell biology. Arch Biochem Biophys. 2019;666:52–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Nargund AM, Pellegrino MW, Fiorese CJ, Baker BM, Haynes CM. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science. 2012;337:587–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Wang YT, Lim Y, McCall MN, Huang KT, Haynes CM, Nehrke K, Brookes PS. Cardioprotection by the mitochondrial unfolded protein response requires ATF5. Am J Physiol Heart Circ Physiol. 2019;317:H472-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Shao Y, Li Y, Liu Y, Zhu S, Wu J, Ma K, Li G, Huang S, Wen H, Zhang C, et al. ATF3 coordinates the survival and proliferation of cardiac macrophages and protects against ischemia-reperfusion injury. Nat Cardiovasc Res. 2024;3:28–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Hofmann C, Aghajani M, Alcock CD, Blackwood EA, Sandmann C, Herzog N, Groß J, Plate L, Wiseman RL, Kaufman RJ, et al. ATF6 protects against protein misfolding during cardiac hypertrophy. J Mol Cell Cardiol. 2024;189:12–24. [DOI] [PubMed] [Google Scholar]
- 213.Yan G, Han Z, Kwon Y, Jousma J, Nukala SB, Prosser BL, Du X, Pinho S, Ong SB, Lee WH, Ong SG. Integrated stress response potentiates Ponatinib-Induced Cardiotoxicity. Circ Res. 2024;134:482–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Li HX, Wang TH, Wu LX, Xue FS, Zhang GH, Yan T. Role of Keap1-Nrf2/ARE signal transduction pathway in protection of dexmedetomidine preconditioning against myocardial ischemia/reperfusion injury. Biosci Rep. 2022;42(9):BSR20221306. [DOI] [PMC free article] [PubMed]
- 215.Lei Y, VanPortfliet JJ, Chen YF, Bryant JD, Li Y, Fails D, Torres-Odio S, Ragan KB, Deng J, Mohan A, et al. Cooperative sensing of mitochondrial DNA by ZBP1 and cGAS promotes cardiotoxicity. Cell. 2023;186:3013-e30323022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Merry TL, Chan A, Woodhead JST, Reynolds JC, Kumagai H, Kim SJ, Lee C. Mitochondrial-derived peptides in energy metabolism. Am J Physiol Endocrinol Metab. 2020;319:E659-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Dabravolski SA, Nikiforov NG, Starodubova AV, Popkova TV, Orekhov AN. The role of mitochondria-derived peptides in cardiovascular diseases and their potential as therapeutic targets. Int J Mol Sci. 2021;22:8770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Lopaschuk GD, Ussher JR. Evolving concepts of myocardial energy metabolism. Circul Res. 2016;119:1173–6. [DOI] [PubMed] [Google Scholar]
- 219.Xu M, Xue R-Q, Lu Y, Yong S-Y, Wu Q, Cui Y-L, Zuo X-T, Yu X-J, Zhao M, Zang W-J. Choline ameliorates cardiac hypertrophy by regulating metabolic remodelling and UPRmt through SIRT3-AMPK pathway. Cardiovascular Res. 2018;115:530–45. [DOI] [PubMed] [Google Scholar]
- 220.Muoio Deborah M, Neufer PD. Lipid-Induced mitochondrial stress and insulin action in muscle. Cell Metabol. 2012;15:595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Newman JC, Verdin E. Ketone bodies as signaling metabolites. Trends Endocrinol Metabolism. 2014;25:42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Aubert G, Martin OJ, Horton JL, Lai L, Vega RB, Leone TC, Koves T, Gardell SJ, Krüger M, Hoppel CL, et al. The failing heart relies on Ketone bodies as a fuel. Circulation. 2016;133:698–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Pascual F, Coleman RA. Fuel availability and fate in cardiac metabolism: a tale of two substrates. Biochim Biophys Acta. 2016;1861:1425–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Takahara S, Soni S, Maayah ZH, Ferdaoussi M, Dyck JRB. Ketone therapy for heart failure: current evidence for clinical use. Cardiovasc Res. 2022;118:977–87. [DOI] [PubMed] [Google Scholar]
- 225.Brahma MK, Wende AR, McCommis KS. CrossTalk opposing view: ketone bodies are not an important metabolic fuel for the heart. J Physiol. 2022;600:1005–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Sun H, Olson KC, Gao C, Prosdocimo DA, Zhou M, Wang Z, Jeyaraj D, Youn J-Y, Ren S, Liu Y, et al. Catabolic defect of branched-chain amino acids promotes heart failure. Circulation. 2016;133:2038–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Liu Y, Luo Q, Su Z, Xing J, Wu J, Xiang L, Huang Y, Pan H, Wu X, Zhang X, et al. Suppression of myocardial HIF-1 by pubertal insulin resistance compromises metabolic adaptation and impairs cardiac function in patients with cyanotic congenital heart disease. Eur Heart J. 2020;41:ehaa946-2168. [DOI] [PubMed] [Google Scholar]
- 228.Maier A, Wu H, Cordasic N, Oefner P, Dietel B, Thiele C, Weidemann A, Eckardt KU, Warnecke C. Hypoxia-inducible protein 2 Hig2/Hilpda mediates neutral lipid accumulation in macrophages and contributes to atherosclerosis in apolipoprotein E-deficient mice. Faseb j. 2017;31:4971–84. [DOI] [PubMed] [Google Scholar]
- 229.Mylonis I, Simos G, Paraskeva E. Hypoxia-inducible factors and the regulation of lipid metabolism. Cells. 2019;8:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Krishnan J, Suter M, Windak R, Krebs T, Felley A, Montessuit C, Tokarska-Schlattner M, Aasum E, Bogdanova A, Perriard E, et al. Activation of a HIF1alpha-PPARgamma axis underlies the integration of glycolytic and lipid anabolic pathways in pathologic cardiac hypertrophy. Cell Metab. 2009;9:512–24. [DOI] [PubMed] [Google Scholar]
- 231.Sousa Fialho ML, Purnama U, Dennis KMJH, Montes Aparicio CN, Castro-Guarda M, Massourides E, Tyler DJ, Carr CA, Heather LC. Activation of HIF1α rescues the hypoxic response and reverses metabolic dysfunction in the diabetic heart. Diabetes. 2021;70:2518–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Kovács M, Gönczi M, Kovács E, Végh A. Time course analysis of cardiac pacing-induced gene expression changes in the canine heart. Mol Cell Biochem. 2013;372:257–66. [DOI] [PubMed] [Google Scholar]
- 233.Pavo N, Lukovic D, Zlabinger K, Zimba A, Lorant D, Goliasch G, Winkler J, Pils D, Auer K, Jan Ankersmit H, et al. Sequential activation of different pathway networks in ischemia-affected and non-affected myocardium, inducing intrinsic remote conditioning to prevent left ventricular remodeling. Sci Rep. 2017;7:43958–43958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Rau CD, Wang J, Avetisyan R, Romay MC, Martin L, Ren S, Wang Y, Lusis AJ. Mapping genetic contributions to cardiac pathology induced by beta-adrenergic stimulation in mice. Circulation: Cardiovasc Genet. 2015;8:40–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Cao Y, Vergnes L, Wang Y-C, Pan C, Chella Krishnan K, Moore TM, Rosa-Garrido M, Kimball TH, Zhou Z, Charugundla S, et al. Sex differences in heart mitochondria regulate diastolic dysfunction. Nat Commun. 2022;13:3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Huang J, Tan L, Shen R, Zhang L, Zuo H, Wang DW. Decreased peripheral mitochondrial DNA Copy Number is Associated with the risk of heart failure and long-term outcomes. Medicine. 2016;95:e3323-3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Leverve X, Batandier C, Fontaine E. Choosing the right substrate. Novartis Found Symp. 2007;280:108–21 discussion 121 – 107, 160 – 104. [PubMed] [Google Scholar]
- 238.Ingwall JS, Weiss RG. Is the failing heart energy starved? Circul Res. 2004;95:135–45. [DOI] [PubMed] [Google Scholar]
- 239.Ahmed A, Syed JN, Chi L, Wang Y, Perez-Romero C, Lee D, Kocaqi E, Caballero A, Yang J, Escalante-Covarrubias Q, et al. KDM8 epigenetically controls cardiac metabolism to prevent initiation of dilated cardiomyopathy. Nat Cardiovasc Res. 2023;2:174–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Sequeira V, Batzner A, Maack C. Targeting mitochondria in hypertrophic cardiomyopathy. Eur Heart J. 2023;44:1186–8. [DOI] [PubMed] [Google Scholar]
- 241.Ingwall JS. Energy metabolism in heart failure and remodelling. Cardiovasc Res. 2009;81:412–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Lee T-M, Su S-F, Chou T-F, Lee Y-T, Tsai C-H. Loss of preconditioning by attenuated activation of myocardial ATP-Sensitive Potassium channels in Elderly patients undergoing coronary angioplasty. Circulation. 2002;105:334–40. [DOI] [PubMed] [Google Scholar]
- 243.Rossello X, Yellon DM. The RISK pathway and beyond. Basic Res Cardiol. 2018;113:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Hadebe N, Cour M, Lecour S. The SAFE pathway for cardioprotection: is this a promising target? Basic Res Cardiol. 2018;113:9. [DOI] [PubMed] [Google Scholar]
- 245.Shvedova M, Anfinogenova Y, Popov SV, Atochin DN. Connexins and nitric oxide inside and outside mitochondria: significance for cardiac protection and adaptation. Front Physiol. 2018;9:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Boveris A, Costa LE, Cadenas E, Poderoso JJ. [20] regulation of mitochondrial respiration by adenosine diphosphate, oxygen, and nitric oxide. Methods Enzymol. 1999;301:188–98 Academic Press. [DOI] [PubMed] [Google Scholar]
- 247.Erusalimsky JD, Moncada S. Nitric oxide and mitochondrial signaling. Arterioscler Thromb Vasc Biol. 2007;27:2524–31. [DOI] [PubMed] [Google Scholar]
- 248.Fulda S. Shifting the balance of mitochondrial apoptosis: therapeutic perspectives. Front Oncol. 2012;2:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Linke M, Fritsch SD, Sukhbaatar N, Hengstschläger M, Weichhart T. mTORC1 and mTORC2 as regulators of cell metabolism in immunity. FEBS Lett. 2017;591:3089–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Escobales N, Nuñez RE, Javadov S. Mitochondrial angiotensin receptors and cardioprotective pathways. Am J Physiol Heart Circ Physiol. 2019;316:H1426-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Liang WJ, Gustafsson ÅB. The aging heart: mitophagy at the center of rejuvenation. Front Cardiovasc Med. 2020;7:294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Poole AC, Thomas RE, Andrews LA, McBride HM, Whitworth AJ, Pallanck LJ. The PINK1/Parkin pathway regulates mitochondrial morphology. Proceedings of the National Academy of Sciences. 2008;105:1638–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Vranas M, Lu Y, Rasool S, Croteau N, Krett JD, Sauvé V, Gehring K, Fon EA, Durcan TM, Trempe J-F. Selective localization of Mfn2 near PINK1 enables its preferential ubiquitination by Parkin on mitochondria. Open Biology. 2022;12:210255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Baines CP, Song CX, Zheng YT, Wang GW, Zhang J, Wang OL, Guo Y, Bolli R, Cardwell EM, Ping P. Protein kinase cepsilon interacts with and inhibits the permeability transition pore in cardiac mitochondria. Circ Res. 2003;92:873–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Penna C, Perrelli M-G, Pagliaro P. Mitochondrial pathways, permeability transition pore, and Redox Signaling in Cardioprotection: therapeutic implications. Antioxid Redox Signal. 2012;18:556–99. [DOI] [PubMed] [Google Scholar]
- 256.Boengler K, Leybaert L, Ruiz-Meana M, Schulz R. Connexin 43 in Mitochondria: what do we really know about its function? Front Physiol. 2022;13:928934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Rodríguez-Sinovas A, Ruiz-Meana M, Denuc A, García-Dorado D. Mitochondrial Cx43, an important component of cardiac preconditioning. Biochim et Biophys Acta (BBA) - Biomembr. 2018;1860:174–81. [DOI] [PubMed] [Google Scholar]
- 258.Heusch G, Boengler K, Schulz R. Cardioprotection. Circulation. 2008;118:1915–9. [DOI] [PubMed] [Google Scholar]
- 259.Cabukusta B, Berlin I, van Elsland DM, Forkink I, Spits M, de Jong AWM, Akkermans JJLL, Wijdeven RHM, Janssen GMC, van Veelen PA, Neefjes J. Human VAPome analysis reveals MOSPD1 and MOSPD3 as membrane contact site proteins interacting with FFAT-Related FFNT motifs. Cell Rep. 2020;33:108475. [DOI] [PubMed] [Google Scholar]
- 260.Cadenas S. Mitochondrial uncoupling, ROS generation and cardioprotection. Biochim et Biophys Acta (BBA) - Bioenergetics. 2018;1859:940–50. [DOI] [PubMed] [Google Scholar]
- 261.Graier WF, Trenker M, Malli R. Mitochondrial Ca2+, the secret behind the function of uncoupling proteins 2 and 3? Cell Calcium. 2008;44:36–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Palmer JW, Tandler B, Hoppel CL. Heterogeneous response of subsarcolemmal heart mitochondria to calcium. Am J Physiol. 1986;250:H741-748. [DOI] [PubMed] [Google Scholar]
- 263.Palmer JW, Tandler B, Hoppel CL. Biochemical properties of subsarcolemmal and interfibrillar mitochondria isolated from rat cardiac muscle. J Biol Chem. 1977;252:8731–9. [PubMed] [Google Scholar]
- 264.Chandra Shekar K, Yannopoulos D, Kosmopoulos M, Riess ML. Differential effects of reperfusion on cardiac mitochondrial subpopulations in a preclinical porcine model of acute myocardial infarction. Front Cell Dev Biol. 2022;10:843733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Banu SA, Ravindran S, Kurian GA. Hydrogen sulfide post-conditioning preserves interfibrillar mitochondria of rat heart during ischemia reperfusion injury. Cell Stress Chaperones. 2016;21:571–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Shanmugam K, Prem PN, Boovarahan SR, Sivakumar B, Kurian GA. FIsetin Preserves Interfibrillar Mitochondria to protect against myocardial ischemia-reperfusion Injury. Cell Biochem Biophys. 2022;80:123–37. [DOI] [PubMed] [Google Scholar]
- 267.Stoker ML, Newport E, Hulit JC, West AP, Morten KJ. Impact of pharmacological agents on mitochondrial function: a growing opportunity? Biochem Soc Trans. 2019;47:1757–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Kurian GA, Berenshtein E, Saada A, Chevion M. Rat cardiac mitochondrial sub-populations show distinct features of oxidative phosphorylation during ischemia, reperfusion and ischemic preconditioning. Cell Physiol Biochem. 2012;30:83–94. [DOI] [PubMed] [Google Scholar]
- 269.Kurian GA, Berenshtein E, Kakhlon O, Chevion M. Energy status determines the distinct biochemical and physiological behavior of interfibrillar and sub-sarcolemmal mitochondria. Biochem Biophys Res Commun. 2012;428:376–82. [DOI] [PubMed] [Google Scholar]
- 270.Hoppel CL, Tandler B, Parland W, Turkaly JS, Albers LD. Hamster cardiomyopathy. A defect in oxidative phosphorylation in the cardiac interfibrillar mitochondria. J Biol Chem. 1982;257:1540–8. [PubMed] [Google Scholar]
- 271.Ansari M, Prem PN, Kurian GA. Hydrogen sulfide postconditioning rendered cardioprotection against myocardial ischemia-reperfusion injury is compromised in rats with diabetic cardiomyopathy. Microvasc Res. 2022;141:104322. [DOI] [PubMed] [Google Scholar]
- 272.Rajab BS, Kassab S, Stonall CD, Daghistani H, Gibbons S, Mamas M, Smith D, Mironov A, AlBalawi Z, Zhang YH, et al. Differential remodelling of mitochondrial subpopulations and mitochondrial dysfunction are a feature of early stage diabetes. Sci Rep. 2022;12:978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Vue Z, Neikirk K, Vang L, Garza-Lopez E, Christensen TA, Shao J, Lam J, Beasley HK, Marshall AG, Crabtree A, et al. Three-dimensional mitochondria reconstructions of murine cardiac muscle changes in size across aging. Am J Physiol Heart Circ Physiol. 2023;325:H965-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Boengler K, Stahlhofen S, van de Sand A, Gres P, Ruiz-Meana M, Garcia-Dorado D, Heusch G, Schulz R. Presence of connexin 43 in subsarcolemmal, but not in interfibrillar cardiomyocyte mitochondria. Basic Res Cardiol. 2009;104:141–7. [DOI] [PubMed] [Google Scholar]
- 275.Voglhuber J, Holzer M, Radulović S, Thai PN, Djalinac N, Matzer I, Wallner M, Bugger H, Zirlik A, Leitinger G, et al. Functional remodelling of perinuclear mitochondria alters nucleoplasmic ca < sup > 2+ signalling in heart failure. Philosophical Trans Royal Soc B: Biol Sci. 2022;377:20210320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Wehrens M, de Leeuw AE, Wright-Clark M, Eding JEC, Boogerd CJ, Molenaar B, van der Kraak PH, Kuster DWD, van der Velden J, Michels M, et al. Single-cell transcriptomics provides insights into hypertrophic cardiomyopathy. Cell Rep. 2022;39:110809. [DOI] [PubMed] [Google Scholar]
- 277.Wang L, Zhang Q, Yuan K, Yuan J. mtDNA in the Pathogenesis of Cardiovascular Diseases. Disease Markers 2021, 2021:7157109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Heimlich JB, Bick AG. Somatic mutations in cardiovascular disease. Circul Res. 2022;130:149–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Calabrese C, Pyle A, Griffin H, Coxhead J, Hussain R, Braund PS, Li L, Burgess A, Munroe PB, Little L, et al. Heteroplasmic mitochondrial DNA variants in cardiovascular diseases. PLoS Genet. 2022;18:e1010068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Choudhury S, Huang AY, Kim J, Zhou Z, Morillo K, Maury EA, Tsai JW, Miller MB, Lodato MA, Araten S, et al. Somatic mutations in single human cardiomyocytes reveal age-associated DNA damage and widespread oxidative genotoxicity. Nat Aging. 2022;2:714–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.McManus MJ, Picard M, Chen H-W, De Haas HJ, Potluri P, Leipzig J, Towheed A, Angelin A, Sengupta P, Morrow RM, et al. Mitochondrial DNA variation dictates expressivity and progression of nuclear DNA mutations causing cardiomyopathy. Cell Metabol. 2019;29:78-e9075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Lim AZ, Jones DM, Bates MGD, Schaefer AM, O’Sullivan J, Feeney C, Farrugia ME, Bourke JP, Turnbull DM, Gorman GS, et al. Risk of cardiac manifestations in adult mitochondrial disease caused by nuclear genetic defects. Open Heart. 2021;8:e001510. [Google Scholar]
- 283.Miles C, Wayne M. Quantitative trait locus (QTL) analysis. Nat Educ. 2008;1(1):208.
- 284.Rau CD, Lusis AJ, Wang Y. Systems genetics for mechanistic discovery in heart diseases. Circul Res. 2020;126:1795–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Tcheandjieu C, Zhu X, Hilliard AT, Clarke SL, Napolioni V, Ma S, Lee KM, Fang H, Chen F, Lu Y, et al. Large-scale genome-wide association study of coronary artery disease in genetically diverse populations. Nat Med. 2022;28:1679–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Shah S, Henry A, Roselli C, Lin H, Sveinbjörnsson G, Fatemifar G, Hedman ÅK, Wilk JB, Morley MP, Chaffin MD, et al. Genome-wide association and mendelian randomisation analysis provide insights into the pathogenesis of heart failure. Nat Commun. 2020;11:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Dabravolski SA, Khotina VA, Sukhorukov VN, Kalmykov VA, Mikhaleva LM, Orekhov AN. The role of mitochondrial DNA mutations in cardiovascular diseases. Int J Mol Sci. 2022;23:952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Molenaar B, Rooij EV. single-cell sequencing of the mammalian heart. Circul Res. 2018;123:1033–5. [DOI] [PubMed] [Google Scholar]
- 289.Osorio D, Cai JJ. Systematic determination of the mitochondrial proportion in human and mice tissues for single-cell RNA-sequencing data quality control. Bioinf (Oxford England). 2021;37:963–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Hwang HV, Sandeep N, Nair RV, Hu D-Q, Zhao M, Lan IS, Fajardo G, Matkovich SJ, Bernstein D, Reddy S. Transcriptomic and functional analyses of mitochondrial dysfunction in pressure overload-induced right ventricular failure. J Am Heart Association. 2021;10:e017835-017835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Dubin A, Jørgensen TE, Jakt LM, Johansen SD. The mitochondrial transcriptome of the anglerfish Lophius piscatorius. BMC Res Notes. 2019;12:800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Zhang R, Nakahira K, Choi AMK, Gu Z. Heteroplasmy concordance between mitochondrial DNA and RNA. Sci Rep. 2019;9:12942–12942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Chen Z, Wei L, Duru F, Chen L. Single-cell RNA sequencing: in-depth decoding of heart biology and cardiovascular diseases. Curr Genom. 2020;21:585–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Yekelchyk M, Guenther S, Preussner J, Braun T. Mono- and multi-nucleated ventricular cardiomyocytes constitute a transcriptionally homogenous cell population. Basic Res Cardiol. 2019;114:36–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Wang M, Gu M, Liu L, Liu Y, Tian L. Single-cell RNA sequencing (scRNA-seq) in cardiac tissue: applications and limitations. Vasc Health Risk Manag. 2021;17:641–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.The Tabula Muris Consortium, Overall coordination, Logistical coordination, et al. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature. 2018;562:367–72. [DOI] [PMC free article] [PubMed]
- 297.Litviňuková M, Talavera-López C, Maatz H, Reichart D, Worth CL, Lindberg EL, Kanda M, Polanski K, Heinig M, Lee M, et al. Cells of the adult human heart. Nature. 2020;588:466–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Luo Y, Hitz BC, Gabdank I, Hilton JA, Kagda MS, Lam B, Myers Z, Sud P, Jou J, Lin K, et al. New developments on the Encyclopedia of DNA elements (ENCODE) data portal. Nucleic Acids Res. 2020;48:D882-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Kuppe C, Ramirez Flores RO, Li Z, Hayat S, Levinson RT, Liao X, Hannani MT, Tanevski J, Wünnemann F, Nagai JS, et al. Spatial multi-omic map of human myocardial infarction. Nature. 2022;608:766–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Paik DT, Cho S, Tian L, Chang HY, Wu JC. Single-cell RNA sequencing in cardiovascular development, disease and medicine. Nat Reviews Cardiol. 2020;17:457–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Zhuang L, Lu L, Zhang R, Chen K, Yan X. Comprehensive Integration of single-cell transcriptional profiling reveals the heterogeneities of non-cardiomyocytes in healthy and ischemic hearts. Front Cardiovasc Med. 2020;7:615161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Feng W, Bais A, He H, Rios C, Jiang S, Xu J, Chang C, Kostka D, Li G. Single-cell transcriptomic analysis identifies murine heart molecular features at embryonic and neonatal stages. Nat Commun. 2022;13:7960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Nomura S, Satoh M, Fujita T, Higo T, Sumida T, Ko T, Yamaguchi T, Tobita T, Naito AT, Ito M, et al. Cardiomyocyte gene programs encoding morphological and functional signatures in cardiac hypertrophy and failure. Nat Commun. 2018;9:4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Hu P, Liu J, Zhao J, Wilkins BJ, Lupino K, Wu H, Pei L. Single-nucleus transcriptomic survey of cell diversity and functional maturation in postnatal mammalian hearts. Genes Dev. 2018;32:1344–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Pollard MO, Gurdasani D, Mentzer AJ, Porter T, Sandhu MS. Long reads: their purpose and place. Hum Mol Genet. 2018;27:R234-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Marx V. Method of the year: long-read sequencing. Nat Methods. 2023;20:6–11. [DOI] [PubMed] [Google Scholar]
- 307.Dhorne-Pollet S, Barrey E, Pollet N. A new method for long-read sequencing of animal mitochondrial genomes: application to the identification of equine mitochondrial DNA variants. BMC Genomics. 2020;21:785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Vandiver AR, Hoang AN, Herbst A, Lee CC, Aiken JM, McKenzie D, Teitell MA, Timp W, Wanagat J. Nanopore sequencing identifies a higher frequency and expanded spectrum of mitochondrial DNA deletion mutations in human aging. Aging Cell. 2023;22:e13842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Cappa R, de Campos C, Maxwell AP, McKnight AJ. Mitochondrial toolbox – a review of online resources to explore mitochondrial genomics. Front Genet. 2020;11:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Smith AC, Robinson AJ. MitoMiner v4.0: an updated database of mitochondrial localization evidence, phenotypes and diseases. Nucleic Acids Res. 2018;47:D1225-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Meng G, Li Y, Yang C, Liu S. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 2019;47:e63-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Guo Y, Li J, Li CI, Shyr Y, Samuels DC. MitoSeek: extracting mitochondria information and performing high-throughput mitochondria sequencing analysis. Bioinformatics. 2013;29:1210–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Allio R, Schomaker-Bastos A, Romiguier J, Prosdocimi F, Nabholz B, Delsuc F. MitoFinder: efficient automated large-scale extraction of mitogenomic data in target enrichment phylogenomics. Mol Ecol Resour. 2020;20:892–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Mammana A, Helmuth J. bamsignals: Extract read count signals from bam files. R package version 1.36.0. 2024. https://github.com/lamortenera/bamsignals.
- 315.Falk MJ, Shen L, Gonzalez M, Leipzig J, Lott MT, Stassen APM, Diroma MA, Navarro-Gomez D, Yeske P, Bai R, et al. Mitochondrial disease sequence Data Resource (MSeqDR): a global grass-roots consortium to facilitate deposition, curation, annotation, and integrated analysis of genomic data for the mitochondrial disease clinical and research communities. Mol Genet Metab. 2015;114:388–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Lim SJ, Thompson LR. Mitohelper: a mitochondrial reference sequence analysis tool for fish eDNA studies. Environ DNA. 2021;3:706–15. [Google Scholar]
- 317.Community TG. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2022 update. Nucleic Acids Res. 2022;50:W345–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Marchiano F, Haering M, Habermann BH. The mitoXplorer 2.0 update: integrating and interpreting mitochondrial expression dynamics within a cellular context. Nucleic Acids Res. 2022;50:W490-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, Natarajan P, Lander ES, Lubitz SA, Ellinor PT, Kathiresan S. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet. 2018;50:1219–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Duncan L, Shen H, Gelaye B, Meijsen J, Ressler K, Feldman M, Peterson R, Domingue B. Analysis of polygenic risk score usage and performance in diverse human populations. Nat Commun. 2019;10:3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Andrews SJ, Fulton-Howard B, Patterson C, McFall GP, Gross A, Michaelis EK, Goate A, Swerdlow RH, Pa J. Mitonuclear interactions influence Alzheimer’s disease risk. Neurobiol Aging. 2020;87:e138137-138114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Aw WC, Towarnicki SG, Melvin RG, Youngson NA, Garvin MR, Hu Y, Nielsen S, Thomas T, Pickford R, Bustamante S, et al. Genotype to phenotype: Diet-by-mitochondrial DNA haplotype interactions drive metabolic flexibility and organismal fitness. PLoS Genet. 2018;14:e1007735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Lu Z, Zhang Y, Yan H, Su Y, Guo L, Liao Y, Lu T, Yu H, Wang L, Li J, et al. ATAD3B and SKIL polymorphisms associated with antipsychotic-induced QTc interval change in patients with schizophrenia: a genome-wide association study. Transl Psychiatry. 2022;12:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Truby LK, Giamberardino S, Kwee L, Kraus WE, Abdulrahim J, Regan JA, Shah SH. Abstract 13874: a mitochondrial polygenic risk score is Associated with metabolite levels implicated in heart failure. Circulation. 2021;144:A13874-13874. [Google Scholar]
- 325.Gentiluomo M, Giaccherini M, Gào X, Guo F, Stocker H, Schöttker B, Brenner H, Canzian F, Campa D. Genome-wide association study of mitochondrial copy number. Hum Mol Genet. 2021;31:1346–55. [DOI] [PubMed] [Google Scholar]
- 326.Elorza AA, Soffia JP. mtDNA heteroplasmy at the core of aging-Associated Heart failure. An integrative view of OXPHOS and mitochondrial life cycle in Cardiac mitochondrial physiology. Front Cell Dev Biol. 2021;9:625020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Ye K, Lu J, Ma F, Keinan A, Gu Z. Extensive pathogenicity of mitochondrial heteroplasmy in healthy human individuals. Proc Natl Acad Sci. 2014;111:10654–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Zhang W, Cui H, Wong L-JC. Comprehensive one-step molecular analyses of mitochondrial genome by massively parallel sequencing. Clin Chem. 2012;58:1322–31. [DOI] [PubMed] [Google Scholar]
- 329.Payne BAI, Cree L, Chinnery PF: Single-cell analysis of mitochondrial DNA. In Mitochondrial medicine: volume I, Probing mitochondrial function. Edited by Weissig V, Edeas M. New York, NY: Springer New York; 2015: 67–76. [DOI] [PubMed]
- 330.Lareau CA, Ludwig LS, Muus C, Gohil SH, Zhao T, Chiang Z, Pelka K, Verboon JM, Luo W, Christian E, et al. Massively parallel single-cell mitochondrial DNA genotyping and chromatin profiling. Nat Biotechnol. 2021;39:451–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Miller TE, Lareau CA, Verga JA, DePasquale EAK, Liu V, Ssozi D, Sandor K, Yin Y, Ludwig LS, El Farran CA, et al. Mitochondrial variant enrichment from high-throughput single-cell RNA sequencing resolves clonal populations. Nat Biotechnol. 2022;40:1030–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Picard M, Shirihai OS. Mitochondrial signal transduction. Cell Metab. 2022;34:1620–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed during the current study.