Abstract
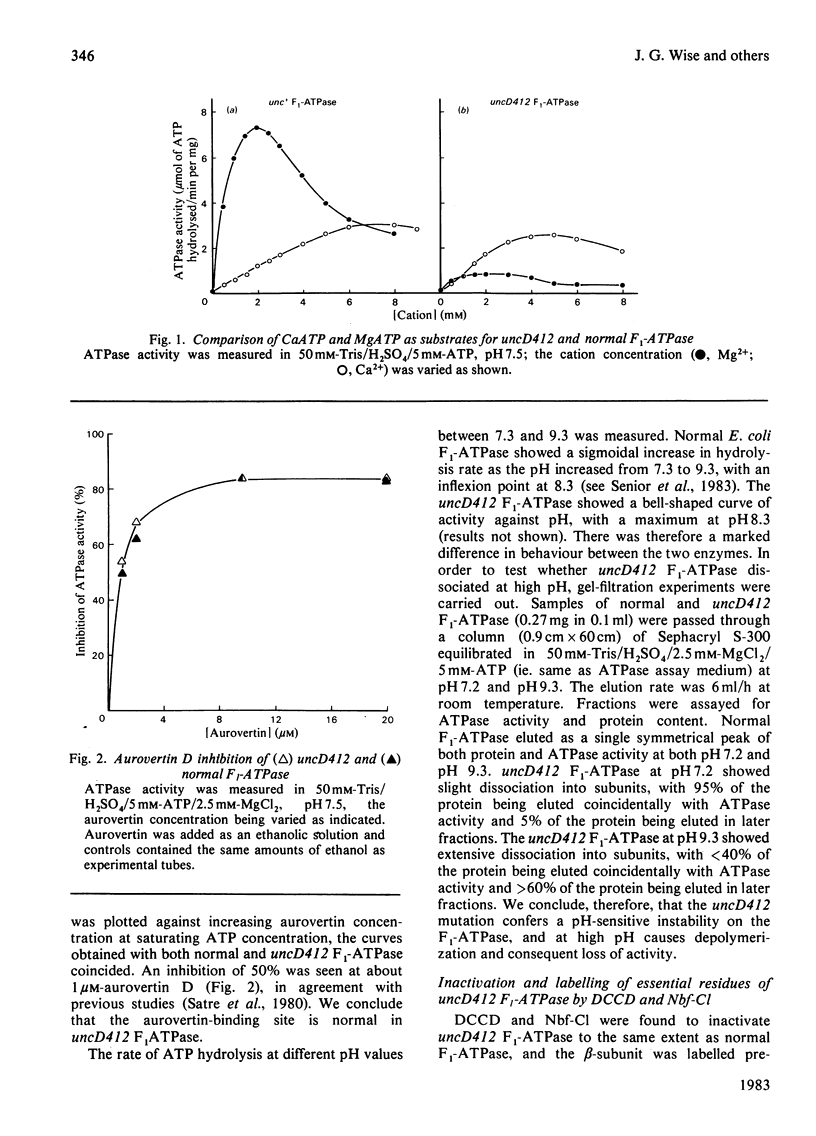
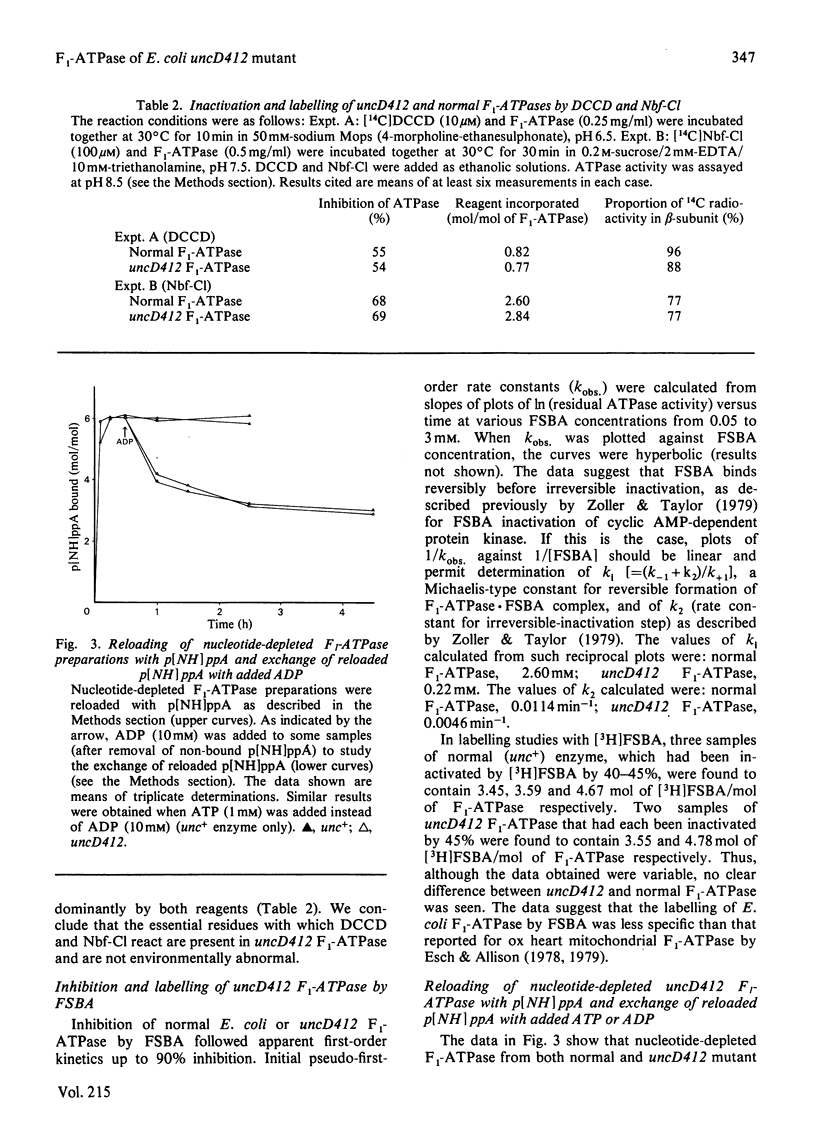
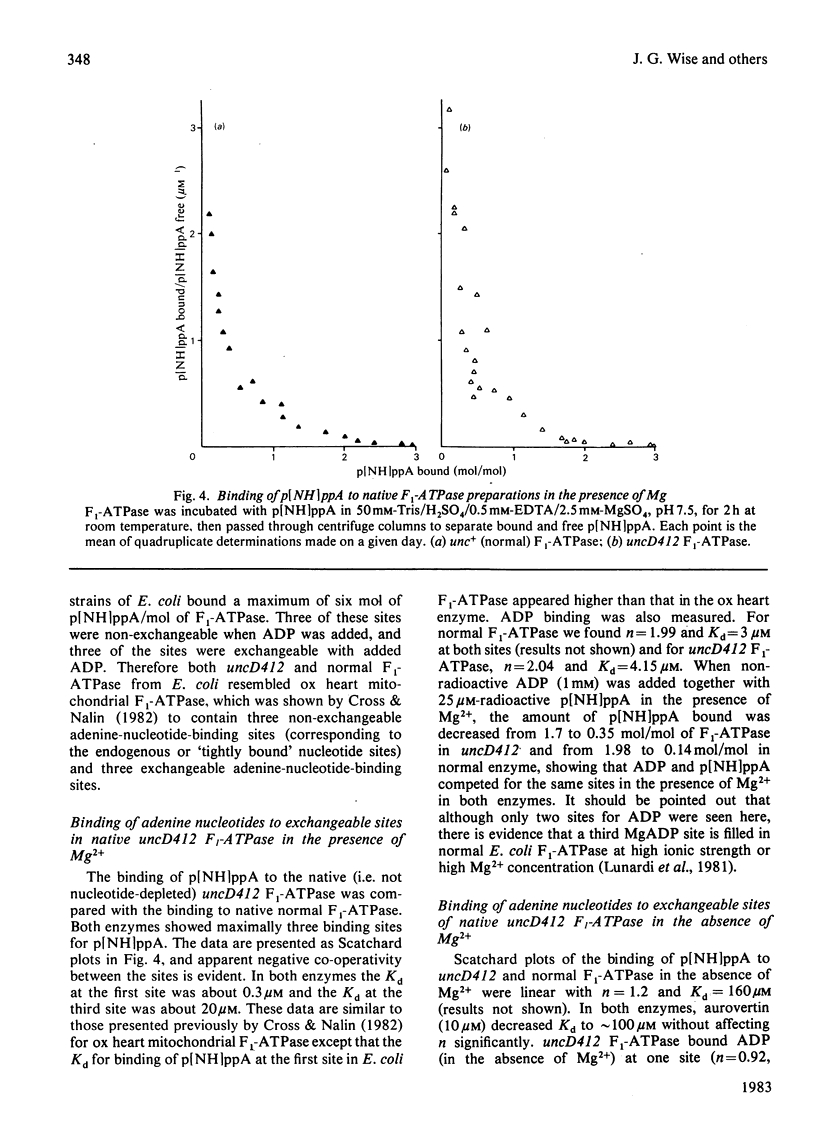
Properties of purified F1-ATPase from Escherichia coli mutant strain AN484 (uncD412) have been studied in an attempt to understand why the amino acid substitution in the beta-subunit of this enzyme causes a tenfold reduction from normal MgATP hydrolysis rate. In most properties that were studied, uncD412 F1-ATPase resembled normal E. coli F1-ATPase. Both enzymes were found to contain a total of six adenine-nucleotide-binding sites, of which three were found to be non-exchangeable and three were exchangeable (catalytic) sites. Binding of the non-hydrolysable substrate analogue adenosine 5'-[beta gamma-imido]triphosphate (p[NH]ppA) to the three exchangeable sites showed apparent negative co-operativity. The binding affinities for p[NH]ppA, and also ADP, at the exchangeable sites were similar in the two enzymes. Both enzymes were inhibited by efrapeptin, aurovertin and p[NH]ppA, and were inactivated by dicyclohexylcarbodi-imide, 4-chloro-7-nitrobenzofurazan and p-fluorosulphonyl-benzoyl-5'-adenosine. Km values for CaATP and MgATP were similar in the two enzymes. uncD412 F1-ATPase was abnormally unstable at high pH, and dissociated into subunits readily with consequent loss of activity. The reason for the impairment of catalysis in uncD412 F1-ATPase cannot be stated with certainty from these studies. However we discuss the possibility that the mutation interrupts subunit interaction, thereby causing a partial impairment in the site-site co-operativity which is required for 'promotion' of catalysis in this enzyme.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cox G. B., Downie J. A., Langman L., Senior A. E., Ash G., Fayle D. R., Gibson F. Assembly of the adenosine triphosphatase complex in Escherichia coli: assembly of F0 is dependent on the formation of specific F1 subunits. J Bacteriol. 1981 Oct;148(1):30–42. doi: 10.1128/jb.148.1.30-42.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross R. L., Grubmeyer C., Penefsky H. S. Mechanism of ATP hydrolysis by beef heart mitochondrial ATPase. Rate enhancements resulting from cooperative interactions between multiple catalytic sites. J Biol Chem. 1982 Oct 25;257(20):12101–12105. [PubMed] [Google Scholar]
- Cross R. L., Kohlbrenner W. E. The mode of inhibition of oxidative phosphorylation by efrapeptin (A23871). Evidence for an alternating site mechanism for ATP synthesis. J Biol Chem. 1978 Jul 25;253(14):4865–4873. [PubMed] [Google Scholar]
- Cross R. L., Nalin C. M. Adenine nucleotide binding sites on beef heart F1-ATPase. Evidence for three exchangeable sites that are distinct from three noncatalytic sites. J Biol Chem. 1982 Mar 25;257(6):2874–2881. [PubMed] [Google Scholar]
- Esch F. S., Allison W. S. Identification of a tyrosine residue at a nucleotide binding site in the beta subunit of the mitochondrial ATPase with p-fluorosulfonyl[14C]-benzoyl-5'-adenosine. J Biol Chem. 1978 Sep 10;253(17):6100–6106. [PubMed] [Google Scholar]
- Esch F. S., Allison W. S. On the subunit stoichiometry of the F1-ATPase and the sites in it that react specifically with p-fluorosulfonylbenzoyl-5'-adenosine. J Biol Chem. 1979 Nov 10;254(21):10740–10746. [PubMed] [Google Scholar]
- Garrett N. E., Penefsky H. S. Interaction of adenine nucleotides with multiple binding sites on beef heart mitochondrial adenosine triphosphatase. J Biol Chem. 1975 Sep 10;250(17):6640–6647. [PubMed] [Google Scholar]
- Gibson F. Biochemical and genetic studies on the assembly and function of the F1-F0 adenosine triphosphatase of Escherichia coli. Biochem Soc Trans. 1983 Jun;11(3):229–240. doi: 10.1042/bst0110229. [DOI] [PubMed] [Google Scholar]
- Grubmeyer C., Cross R. L., Penefsky H. S. Mechanism of ATP hydrolysis by beef heart mitochondrial ATPase. Rate constants for elementary steps in catalysis at a single site. J Biol Chem. 1982 Oct 25;257(20):12092–12100. [PubMed] [Google Scholar]
- Kanazawa H., Horiuchi Y., Takagi M., Ishino Y., Futai M. Coupling factor F1 ATPase with defective beta subunit from a mutant of Escherichia coli. J Biochem. 1980 Sep;88(3):695–703. doi: 10.1093/oxfordjournals.jbchem.a133022. [DOI] [PubMed] [Google Scholar]
- Lunardi J., Satre M., Vignais P. V. Exploration of adenosine 5'-diphosphate-adenosine 5'-triphosphate binding sites of Escherichia coli adenosine 5'-triphosphatase with arylazido adenine nucleotides. Biochemistry. 1981 Feb 3;20(3):473–480. doi: 10.1021/bi00506a005. [DOI] [PubMed] [Google Scholar]
- Penefsky H. S. Reversible binding of Pi by beef heart mitochondrial adenosine triphosphatase. J Biol Chem. 1977 May 10;252(9):2891–2899. [PubMed] [Google Scholar]
- Satre M., Bof M., Vignais P. V. Interaction of Escherichia coli adenosine triphosphatase with aurovertin and citreoviridin: inhibition and fluorescence studies. J Bacteriol. 1980 Jun;142(3):768–776. doi: 10.1128/jb.142.3.768-776.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senior A. E., Downie J. A., Cox G. B., Gibson F., Langman L., Fayle D. R. The uncA gene codes for the alpha-subunit of the adenosine triphosphatase of Escherichia coli. Electrophoretic analysis of uncA mutant strains. Biochem J. 1979 Apr 15;180(1):103–109. doi: 10.1042/bj1800103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senior A. E., Fayle D. R., Downie J. A., Gibson F., Cox G. B. Properties of membranes from mutant strains of Escherichia coli in which the beta-subunit of the adenosine triphosphatase is abnormal. Biochem J. 1979 Apr 15;180(1):111–118. doi: 10.1042/bj1800111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senior A. E., Langman L., Cox G. B., Gibson F. Oxidative phosphorylation in Escherichia coli. Characterization of mutant strains in which F1-ATPase contains abnormal beta-subunits. Biochem J. 1983 Feb 15;210(2):395–403. doi: 10.1042/bj2100395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senior A. E., Wise J. G. The proton-ATPase of bacteria and mitochondria. J Membr Biol. 1983;73(2):105–124. doi: 10.1007/BF01870434. [DOI] [PubMed] [Google Scholar]
- TAUSSKY H. H., SHORR E. A microcolorimetric method for the determination of inorganic phosphorus. J Biol Chem. 1953 Jun;202(2):675–685. [PubMed] [Google Scholar]
- Wise J. G., Latchney L. R., Senior A. E. The defective proton-ATPase of uncA mutants of Escherichia coli. Studies of nucleotide binding sites, bound aurovertin fluorescence, and labeling of essential residues of the purified F1-ATPase. J Biol Chem. 1981 Oct 25;256(20):10383–10389. [PubMed] [Google Scholar]
- Wyatt J. L., Colman R. F. Affinity labeling of rabbit muscle pyruvate kinase by 5'-p-fluorosulfonylbenzoyladenosine. Biochemistry. 1977 Apr 5;16(7):1333–1342. doi: 10.1021/bi00626a015. [DOI] [PubMed] [Google Scholar]
- Zoller M. J., Taylor S. S. Affinity labeling of the nucleotide binding site of the catalytic subunit of cAMP-dependent protein kinase using p-fluorosulfonyl-[14C]benzoyl 5'-adenosine. Identification of a modified lysine residue. J Biol Chem. 1979 Sep 10;254(17):8363–8368. [PubMed] [Google Scholar]


