Abstract
Over 200 genetic mutations in copper-zinc superoxide dismutase (SOD1) have been linked to amyotrophic lateral sclerosis (ALS). Among these, two ALS-causing mutants, histidine-46→arginine (H46R) and glycine-85→arginine (G85R), exhibit a decreased capacity to bind metal ions. Here, we report two cryo–electron microscopy structures of amyloid fibrils formed by H46R and G85R. These mutations lead to the formation of amyloid fibrils with unique structures distinct from those of the native fibril. The core of these fibrils features a serpentine arrangement with seven or eight β strands, secured by a hydrophobic cavity and a salt bridge between arginine-85 and aspartic acid–101 in the G85R fibril. We demonstrate that these mutant fibrils are notably more toxic and capable of promoting the aggregation of wild-type SOD1 more effectively, causing mitochondrial impairment and activating ferroptosis in cell cultures, compared to wild-type SOD1 fibrils. Our study provides insights into the structural mechanisms by which SOD1 mutants aggregate and induce cytotoxicity in ALS.
SOD1 mutants H46R and G85R form amyloid fibril structures, induce templated misfolding, and activate ferroptosis in ALS.
INTRODUCTION
Amyotrophic lateral sclerosis (ALS), also known as motor neuron disease, is a progressive, devastating neurodegenerative disease characterized by the selective death of motor neurons in the brain and spinal cord (1–6). The fundamental unanswered question remains as to how motor neurons die in ALS. Ferroptosis, an iron-dependent form of nonapoptotic cell death (7–10), is regulated by glutathione peroxidase 4 (GPX4) (11, 12), and GPX4 has been found to be down-regulated in the spinal cord of patients with ALS (7, 13). However, the molecular mechanism underlying the mediation of ferroptosis in ALS remains unclear (14, 15). Approximately 90% of ALS cases are sporadic, while about 10% are familial and are typically passed down with inheritance (1–4, 16–18). There are now more than 50 genes that have been implicated in ALS pathogenesis (1). Among them, the sod1 gene, the first gene to be associated with a familial form of ALS (19), is the second most common cause of the disease (1, 17, 18). About 2 to 6% of cases of ALS are caused by mutations in the antioxidant enzyme copper-zinc superoxide dismutase (SOD1) (1, 2, 4–6, 16–18). Notably, more than 200 genetic mutations of SOD1 have been identified in the familial form of ALS (https://alsod.ac.uk/) (1–5, 16, 19–39). These mutations have remarkably diverse effects on the structure, activity, and stability of the native state of SOD1 (1–3, 16, 22, 29, 31, 32, 35, 38, 39). The cytoplasmic aggregation of these mutants and wild-type SOD1 in motor neurons is a pathological feature of ALS (1–4, 16–18, 22–26, 29, 30, 32, 34, 35). It is, however, currently unknown whether different ALS-causing SOD1 mutations produce distinct SOD1 strains that influence the evolution of the disease (1, 40, 41) and whether they promote SOD1 aggregation by fundamentally distinct mechanisms (1–4, 24–26, 29–32, 36, 40–42).
Since SOD1 was found to be associated with familial ALS in 1993 (19), great efforts have been dedicated to unravel the mysteries of the atomic structure of SOD1 aggregates (1, 43–47) and SOD1 strains (1, 40, 41). Recently, we reported a cryo–electron microscopy (cryo-EM) structure of the amyloid fibril formed by the disulfide-reduced, apo form of full-length wild-type human SOD1 featuring an in-register intramolecular β strand architecture (43), which is stabilized by salt bridges, hydrophobic cavities, and hydrogen-bonding networks and provides structural insights into the conversion of SOD1 from its immature form into an aggregated form during ALS pathogenesis. Cytoplasmic aggregation of transactive response DNA binding protein-43 (TDP-43) in neurons is another pathological feature of ALS (48–50). Very recently, Sharma and coworkers (48) reported cryo-EM structures of in vitro–generated amyloid fibrils from full-length TDP-43, which are different from those of TDP-43 amyloid fibrils purified from the brains of patients with ALS and frontotemporal lobar degeneration (49, 50). However, despite three decades of investigation (1–5, 16–50), atomic structural information of in vivo–derived SOD1 amyloid fibrils is not available, and the molecular mechanisms by which mutations in SOD1 cause the familial form of ALS remain a mystery.
There are two subsets of mutations in SOD1 linked to the familial form of ALS. Metal-binding region mutants, such as H46R, H46D, G85R, D125H, and S134N, have mutations localized in and around the metal-binding sites in SOD1, which have substantially altered biophysical properties relative to the wild-type protein (1–5, 16, 19–25, 27, 29, 31–43). By contrast, wild-type–like mutants, such as A4V, D90A, G93A, D101G, and D101N, retain most of their properties similar to wild-type SOD1 (1–5, 16, 19–26, 28–32, 38–41, 43). In this study, we focus specifically on two ALS-causing SOD1 mutant proteins, H46R and G85R, because of the following reasons. First, H46R and G85R are metal-binding region mutants with reduced affinity for metal ions (zinc in the case of G85R and copper and zinc for H46R) (2, 16, 20, 21, 31, 38); the aggregates or inclusions formed by H46R and G85R exhibit prion-like properties (36, 37). Second, transgenic mice expressing H46R and G85R develop a similar ALS-like phenotype comprising paralysis and muscle loss at several months of age (1, 3, 23, 24, 33–35, 37). However, the mechanism behind the phenomenon remains unclear. Third, injection of in vitro–generated amyloid fibrils from H46R and G85R into the spinal cords of newborn transgenic mice expressing G85R accelerates the progression of ALS (51, 52), showing the relevance of the in vitro–prepared fibrils to human disease. Fourth, the atomic structure of wild-type SOD1 fibrils has shown that several familial mutations including H46R, H46D, G85R, D101G, and D101N may alter crucial interactions (salt bridges) in the cytotoxic SOD1 fibril structure (43), but whether ALS-causing SOD1 mutants H46R and G85R form similar amyloid fibril structures is unknown.
Here, we generated homogeneous amyloid fibrils from the apo forms of two ALS-causing SOD1 mutant proteins, H46R and G85R, under reducing conditions and determine the atomic structure using cryo-EM. We demonstrate that H46R and G85R form amyloid fibril structures, induce templated misfolding of native SOD1, and significantly promote ferroptosis in neuronal cells. Our findings provide structural insights into the molecular mechanisms by which mutations in SOD1 exhibit cytotoxicity, promote ferroptosis regulated by GPX4, and cause the familial form of ALS.
RESULTS
The cryo-EM structures of the H46R fibril and the G85R fibril are compared with each other
We first treated the apo forms of H46R and G85R with 5 mM tris(2-carboxyethyl)phosphine (TCEP). TCEP, a highly stable disulfide-reducing agent, can be used to generate a reduced state that is able to mimic physiological reducing environments (43). We produced amyloid fibrils from recombinant, full-length apo human SOD1 (residues 1 to 153) with H46R mutation or G85R mutation overexpressed in Escherichia coli, by incubating the purified apoproteins in 20 mM tris-HCl buffer (pH 7.4) containing 5 mM TCEP and shaking at 37°C for 40 to 48 hours (see Materials and Methods). Amyloid fibrils formed by H46R and G85R under these reducing conditions were concentrated to ~30 μM in a centrifugal filter (Millipore) and examined by electron microscopy without further treatment.
Negative-staining transmission electron microscopy (TEM) images showed that the apo forms of ALS-causing SOD1 mutants H46R and G85R formed homogeneous and unbranched fibrils under reducing conditions (fig. S1, A and B). We then compared the images of amyloid fibrils assembled from H46R and G85R by atomic force microscopy (AFM) (fig. S1, C and D) and determined the atomic structures of the H46R fibril and the G85R fibril by cryo-EM (Figs. 1 to 3 and Table 1). The AFM images, cryo-EM micrographs, and two-dimensional (2D) class average images using RELION3.1 (53) showed that both the H46R fibril and the G85R fibril were composed of a single protofilament with a left-handed helical twist (fig. S1, C to F) and were arranged in a staggered manner (fig. S2, A and B). The helical pitch was 170 ± 9 nm for the H46R fibril (fig. S1C) and 168 ± 7 nm for the G85R fibril (fig. S1D). The fibrils were morphologically homogeneous (fig. S1, C to F), showing a fibril full width of 9.8 ± 1.0 nm for the H46R fibril (fig. S1C) and 10.0 ± 1.4 nm for the G85R fibril (fig. S1D).
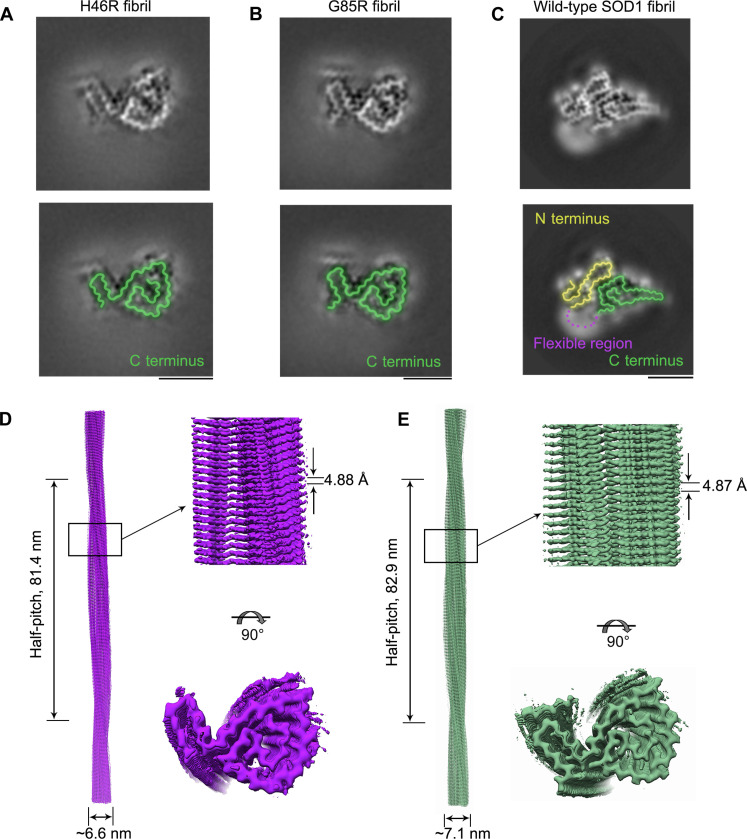
Fig. 1. Comparison of the cryo-EM structures of the H46R fibril and the G85R fibril.
(A and B) Cross-sectional view of the 3D map of the H46R fibril (A) or the G85R fibril (B) showing a protofilament comprising a C-terminal segment (green). (C) Cross-sectional view of the 3D map of the wild-type SOD1 fibril, however, showing a protofilament comprising not only a C-terminal segment (green) but also an N-terminal segment (yellow) with an unstructured flexible fragment (magenta dashed line) (43). Scale bars, 5 nm. For full clarity, we false color the equivalent regions in (A) to (C). (D and E) 3D map of the H46R fibril (D) or the G85R fibril (E) showing a single protofilament [in purple for (D) and green for (E)] intertwined into a left-handed helix, with a fibril core width of ~6.6 nm (D) or ~7.1 nm (E) and a half-helical pitch of 81.4 nm (D) or 82.9 nm (E) (left). Enlarged section of the H46R fibril (D) or the G85R fibril (E) showing a side view of the density map (top right). Close-up view of the density map on the left showing that the subunit in a protofilament stacks along the fibril axis with a helical rise of 4.88 Å (D) or 4.87 Å (E) (top right). Top view of the density map of the H46R fibril (D) or the G85R fibril (E) (bottom right).
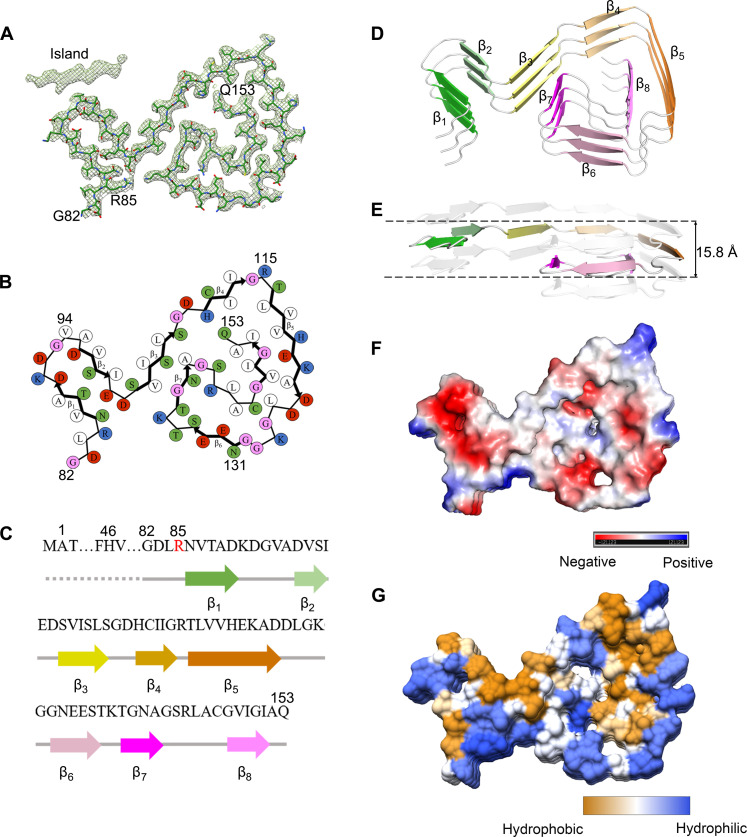
Fig. 3. The ALS-causing SOD1 mutant G85R also forms an amyloid fibril structure.
(A) Cryo-EM map of the G85R fibril with the atomic model overlaid. The G85R fibril core comprises a C-terminal segment (residues 82 to 153) colored purple. (B) Schematic view of the G85R fibril core. The residues are colored as follows: white, hydrophobic; green, polar; red and blue, negatively charged and positively charged, respectively; and magenta, glycine. β Strands are indicated with bold lines. (C) Sequence of the G85R fibril core comprising residues 82 to 153 from the full-length human G85R SOD1 (1 to 153) with the observed eight β strands colored green (β1), light green (β2), yellow (β3), gold (β4), orange (β5), pink (β6), magenta (β7), and light magenta (β8) in the C-terminal segment. The dotted line corresponds to residues 1 to 81 not modeled in the cryo-EM density. The ALS-causing mutation site R85 is highlighted in red. (D) Ribbon representation of the structure of a G85R fibril core containing three molecular layers and a C-terminal segment. We show the secondary structure panel in the same orientation as the other panels. (E) As in (D), but viewed perpendicular to the helical axis, revealing that the height of one layer along the helical axis is 15.8 Å. (F) Electrostatic surface representation of the structure of a G85R fibril core containing three molecular layers and a C-terminal segment. (G) Hydrophobic surface representation of the structure of a G85R fibril core as in (D). The surface of the G85R fibril core is shown according to the electrostatic properties (red, negatively charged; blue, positively charged) (F) or the hydrophobicity (yellow, hydrophobic; blue, hydrophilic) (G) of the residues.
Table 1. Cryo-EM data collection, refinement, and validation statistics.
| H46R fibril (EMD-35460, PDB 8IHV) | G85R fibril (EMD-35459, PDB 8IHU) | |
|---|---|---|
| Data collection and processing | ||
| Magnification | 105,000 | 105,000 |
| Voltage (kV) | 300 | 300 |
| Camera | Gatan K3 (Krios G4) | Gatan K3 (Krios G4) |
| Frame exposure time (s) | 0.08 | 0.08 |
| Movie frames (n) | 40 | 40 |
| Electron exposure (e−/Å2) | 60 | 60 |
| Defocus range (μm) | −2.0 to −1.2 | −2.0 to −1.2 |
| Pixel size (Å) | 0.84 | 0.84 |
| Symmetry imposed | C 1 | C 1 |
| Box size (pixel) | 400 | 400 |
| Interbox distance (Å) | 33.6 | 33.6 |
| Micrographs collected (n) | 8,680 | 6,366 |
| Segments extracted (n) | 1,062,069 | 626,757 |
| Segments after Class2D (n) | 235,369 | 305,672 |
| Segments after Class3D (n) | 59,857 | 53,707 |
| Map resolution (Å) | 3.11 | 2.97 |
| FSC threshold | 0.143 | 0.143 |
| Map resolution range (Å) | 3.01–4.83 | 2.87–4.79 |
| Refinement | ||
| Initial model used | De novo | De novo |
| Model resolution (Å) | 3.11 | 2.97 |
| FSC threshold | 0.143 | 0.143 |
| Model resolution range (Å) | 3.11 | 2.97 |
| Map sharpening B factor (Å2) | −78.38 | −82.56 |
| Model composition | ||
| Nonhydrogen atoms | 1,446 | 1,527 |
| Protein residues | 207 | 216 |
| Ligands | 0 | 0 |
| B factors (Å2) | ||
| Protein | 81.95 | 52.90 |
| RMSDs | ||
| Bond lengths (Å) | 0.005 | 0.005 |
| Bond angles (°) | 0.722 | 0.713 |
| Validation | ||
| MolProbity score | 2.39 | 2.25 |
| Clashscore | 12.57 | 9.90 |
| Poor rotamers (%) | 0 | 0 |
| Ramachandran plot | ||
| Favored (%) | 76.12 | 80.00 |
| Allowed (%) | 23.88 | 20.00 |
| Disallowed (%) | 0 | 0 |
| Model versus data (CC) | 0.84 | 0.82 |
Using helical reconstruction in RELION3.1 (53), we determined density maps of the ordered cores of H46R fibril and G85R fibril, with overall resolutions of 3.11 and 2.97 Å, respectively, which featured well-resolved side-chain densities and clearly separated β strands along the fibril axis (Fig. 1, A and B, and fig. S3, A and B). Cross-sectional views of the 3D maps of the H46R fibril and the G85R fibril showed a protofilament comprising a C-terminal segment (Fig. 1, A and B). Cross-sectional view of the 3D map of the wild-type SOD1 fibril, however, showed a protofilament comprising not only an N-terminal segment but also a C-terminal segment, with an unstructured flexible region in between (Fig. 1C) (43). 3D maps of the H46R fibril and the G85R fibril showed a single protofilament intertwined into a left-handed helix, with fibril core widths of ~6.6 and ~7.1 nm and half-helical pitches of 81.4 and 82.9 nm, respectively (Fig. 1, D and E). The subunits within the protofilaments of H46R and G85R stacked along the fibril axis with helical rises of 4.88 and 4.87 Å and twists of −1.079° and −1.058°, respectively (Fig. 1, D and E). Together, the data showed that under reducing conditions, H46R fibril and G85R fibril displayed similar structures.
ALS-causing SOD1 mutant proteins H46R and G85R form amyloid fibril structures
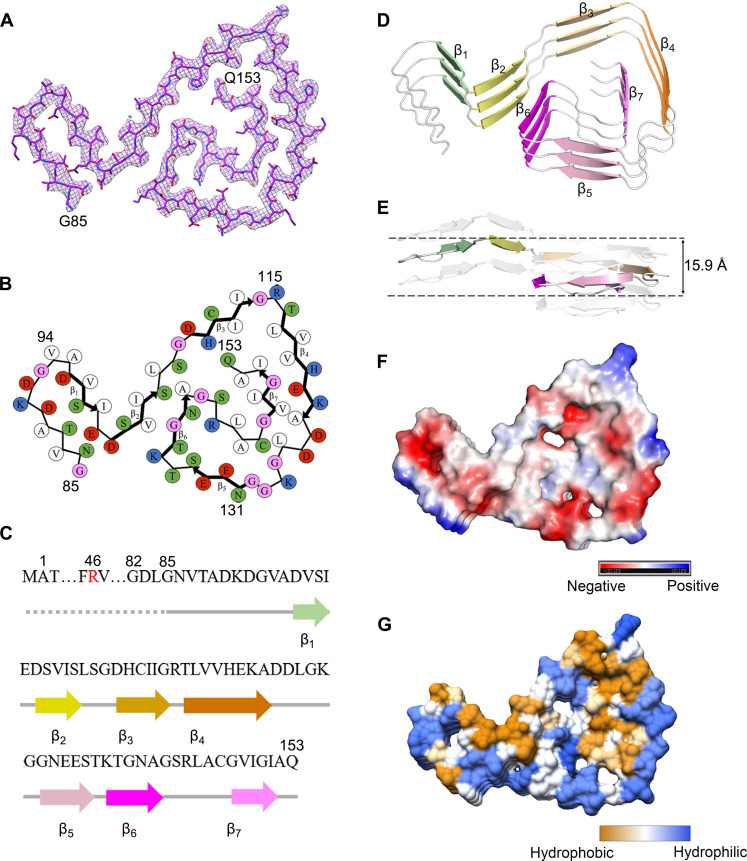
We unambiguously built a structure model of H46R fibril comprising a C-terminal segment (residues 85 to 153) at 3.11 Å (Fig. 2) and that of G85R fibril comprising a C-terminal segment (residues 82 to 153) at 2.97 Å (Fig. 3). Side-chain densities for many residues in the H46R fibril and most residues in the G85R fibril had high local resolution (3.00 to 3.125 Å) (fig. S3, C and D). Side chains for the residues in the fibril cores of H46R and G85R can be well accommodated into the density maps (Figs. 2A and 3A). We observed one unidentified density flanking the protofilament in the G85R fibril, termed an island (Fig. 3A). This island is located on the opposing side of hydrophobic side chains of V94 and A95 in the G85R fibril (Fig. 3A), and it would seem likely that this island results from part of the N-terminal SOD1 segment. A different island is observed in the structure of an amyloid fibril formed by full-length human prion protein with E196K mutation, a genetic Creutzfeldt-Jakob disease–related mutation (54). The exteriors of the fibril cores of H46R and G85R are partly hydrophilic, carrying many negatively charged or positively charged residues, whereas side chains of most hydrophobic residues are mainly located in the interiors of the H46R/G85R fibril fold (Figs. 2 and 3, B to G). A hydrophobic core (Figs. 2 and 3, B and G), three hydrogen bonds (figs. S4, A and B, and S5, A to F), a salt bridge (fig. S4, C and D), and a very compact fold (Figs. 2 and 3, B and D) help stabilize the fibril cores, as described in detail below.
Fig. 2. The ALS-causing SOD1 mutant H46R forms an amyloid fibril structure.
(A) Cryo-EM map of the H46R fibril with the atomic model overlaid. The H46R fibril core comprises a C-terminal segment (residues 85 to 153) colored purple. (B) Schematic view of the H46R fibril core. The residues are colored as follows: white, hydrophobic; green, polar; red and blue, negatively charged and positively charged, respectively; and magenta, glycine. β Strands are indicated with bold lines. N86 in (B) is depicted as facing inward to the cavity. (C) Sequence of the H46R fibril core comprising residues 85 to 153 from the full-length human H46R SOD1 (1 to 153) with the observed seven β strands colored light green (β1), yellow (β2), gold (β3), orange (β4), pink (β5), magenta (β6), and light magenta (β7) in the C-terminal segment. The dotted line corresponds to residues 1 to 84 not modeled in the cryo-EM density. The ALS-causing mutation site R46 is highlighted in red. (D) Ribbon representation of the structure of an H46R fibril core containing three molecular layers and a C-terminal segment. We show the secondary structure (D) in the same orientation as the other panels. (E) As in (D), but viewed perpendicular to the helical axis, revealing that the height of one layer along the helical axis is 15.9 Å. (F) Electrostatic surface representation of the structure of an H46R fibril core containing three molecular layers and a C-terminal segment. (G) Hydrophobic surface representation of the structure of an H46R fibril core as in (D). The surface of the H46R fibril core is shown according to the electrostatic properties (red, negatively charged; blue, positively charged) (F) or the hydrophobicity (yellow, hydrophobic; blue, hydrophilic) (G) of the residues.
Hydrophobic side chains of I112, L117, V119, A123, L126, L144, V148, I149, I151, and A152 are found to locate in the interior of H46R fibrils (Fig. 2B) to form a stable hydrophobic core. In sharp contrast, hydrophilic side chains of N86, T88, D90, D96, S98, E100, and D101 are found to locate in the interior of H46R fibrils to form a hydrophilic cavity (Fig. 2B). Hydrophilic side chains of S105, S107, H110, S142, and Q153 and those of E132, S134, T137, N139, and R143 are found to locate in the interior of H46R fibrils (Fig. 2B) to form the second and the third hydrophilic cavities, respectively. Such a stable hydrophobic core and three hydrophilic cavities are common in the structures of H46R fibrils (the common fold; Fig. 2B) and G85R fibrils (Fig. 3B), except that hydrophilic side chains of R85, N86, T88, D90, D96, S98, E100, and D101 are found to locate in the interior of G85R fibrils to form a hydrophilic cavity (Fig. 3B).
R85 and D101 form a salt bridge with a distance of 2.5 Å to stabilize the G85R fibril core (fig. S4, C and D), whereas N86 and D101 form a hydrogen bond with a distance of 2.6 Å to stabilize the H46R fibril core (fig. S4, A and B). Two pairs of amino acids (H110 and Q153 and S134 and R143) form two or three hydrogen bonds to stabilize the fibril cores (fig. S5, A to F). The fibril core structures of H46R and G85R only comprise a C-terminal segment containing residues 85 to 153 and 82 to 153, respectively (Figs. 2 and 3, A to D). The wild-type SOD1 fibril core structure, however, comprises not only an N-terminal segment (residues 3 to 55) but also a C-terminal segment (residues 86 to 153), with an unstructured flexible region in between (43). Thus, the H46R mutation and the G85R mutation alter crucial salt bridges in the wild-type SOD1 fibril (figs. S4 and S5), resulting in amyloid fibrils with distinct structures compared to the wild-type fibril (Figs. 1 to 3).
The fibril cores feature a very compact fold and exhibit a serpentine fold containing seven or eight β strands stabilized by a hydrophobic cavity (Figs. 2 and 3, B, D, and G). Seven β strands (β1 to β7) and eight β strands (β1 to β8) are present in the fibril core structures of H46R and G85R, respectively (Figs. 2 and 3, B to D). The height of one layer of the H46R fibril core (or the G85R fibril core) along the helical axis is 15.9 Å (or 15.8 Å), which is the distance between the highest point in the loop between β6 and β7 (or between β7 and β8) and the lowest point in the loop between β1 and β2 (Figs. 2E and 3E). Together, these results demonstrate that H46R and G85R form similar amyloid fibril structures.
Fibril seeds from H46R and G85R exhibit cytotoxicity and activate ferroptosis in neuronal cells
SOD1 fibrils produced under reducing conditions have functions to induce mitochondrial damage (43, 55). Ferroptosis has functional importance in mediating motor neuron death in ALS (7, 8, 13). Specifically, mitochondria in ferroptotic cells appear smaller than normal and show increased mitochondrial membrane density, decreased mitochondrial cristae, and rupture of the outer membrane (9, 56, 57). Given that two ALS-causing SOD1 mutants, H46R and G85R, do form similar amyloid fibril structures (Figs. 2 and 3), we predicted that these two mutants might perform similar functions implicated in ALS, exhibiting similar toxicity and ability to induce mitochondrial damage and activate ferroptosis in neuronal cells.
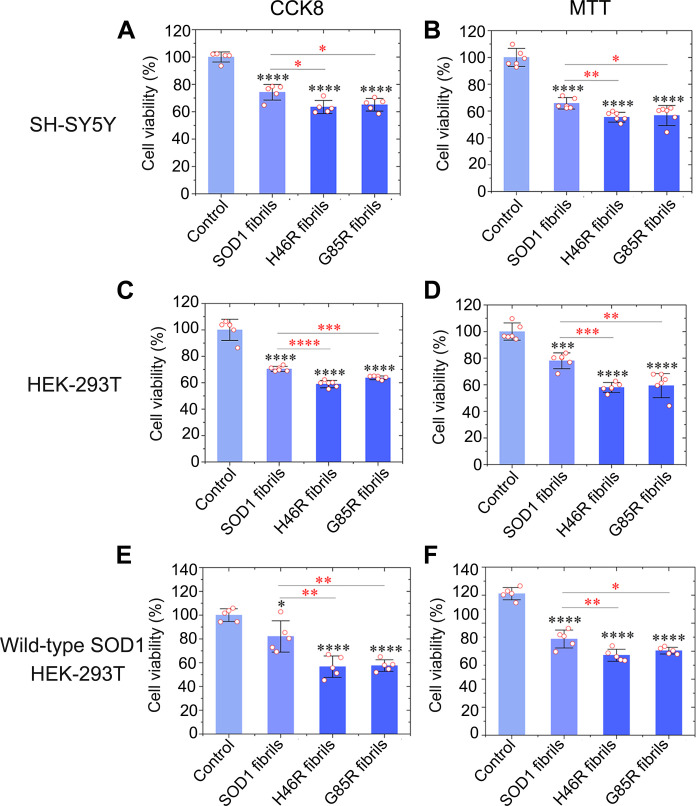
We focus on cytotoxicity to lend biological relevance to these fibril structures. SH-SY5Y neuroblastoma cells, human embryonic kidney (HEK) 293T cells, and HEK-293T cells stably expressing FLAG-tagged wild-type SOD1 were cultured for 1 day; then incubated with 0 μM SOD1 fibril seeds (control), 10 μM wild-type SOD1 fibril seeds, 10 μM H46R fibril seeds, and 10 μM G85R fibril seeds, respectively, for 1.5 days; and further investigated by Cell Counting Kit-8 (CCK8) reduction assay and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) reduction assay (Fig. 4, A to F). Notably, fibril seeds from H46R and G85R exhibited significantly higher cytotoxicity to SH-SY5Y cells (Fig. 4, A and B), HEK-293T cells (Fig. 4, C and D), and HEK-293T cells stably expressing FLAG-tagged wild-type SOD1 (Fig. 4, E and F) than wild-type SOD1 fibril seeds (P = 0.012, 0.024, 0.0033, 0.045, 0.000063, 0.00026, 0.00022, 0.0048, 0.0074, 0.0046, 0.00996, and 0.026, respectively). Together, the data showed that fibril seeds from H46R and G85R are more cytotoxic to cultured cells than wild-type SOD1 fibril seeds generated under the same conditions.
Fig. 4. Fibril seeds from H46R and G85R are more cytotoxic to cultured cells than wild-type SOD1 fibril seeds generated under the same conditions.
(A to F) Cytotoxicity of fibril seeds from H46R and G85R to SH-SY5Y neuroblastoma cells (A and B), HEK-293T cells (C and D), or HEK-293T cells stably expressing FLAG-tagged wild-type SOD1 (E and F) assessed by the CCK8 assay (A, C, and E) and the MTT assay (B, D, and F), compared with that of fibril seeds from wild-type SOD1. Cells were cultured for 1 day and then incubated with 0 μM SOD1 fibril seeds, 10 μM wild-type SOD1 fibril seeds, 10 μM H46R fibril seeds, and 10 μM G85R fibril seeds, respectively, for 1.5 days. The cell viability (%) (open red circles shown in scatter plots) was expressed as the means ± SD (with error bars) of values obtained in n = 5 (A, E, and F) or 6 (B to D) biologically independent experiments. SOD1 fibrils, P = 0.000030, 0.000011, 0.000041, 0.00051, 0.0227, and 0.000014 (A to F); H46R fibrils, P = 0.00000089, 0.0000011, 0.0000045, 0.0000015, 0.000015, and 0.00000023 and 0.012, 0.0033, 0.000063, 0.00022, 0.0074, and 0.00996 (A to F); and G85R fibrils, P = 0.0000010, 0.000011, 0.0000084, 0.000037, 0.0000011, and 0.000000084 and 0.024, 0.045, 0.00026, 0.0048, 0.0046, and 0.026 (A to F). Statistical analyses were performed using two-sided Student’s t test. Values of P < 0.05 indicate statistically significant differences. The following notation is used throughout: *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 relative to control. Cells treated with 20 mM tris-HCl buffer (pH 7.4) containing 5 mM TCEP for 1.5 days were used as a control.
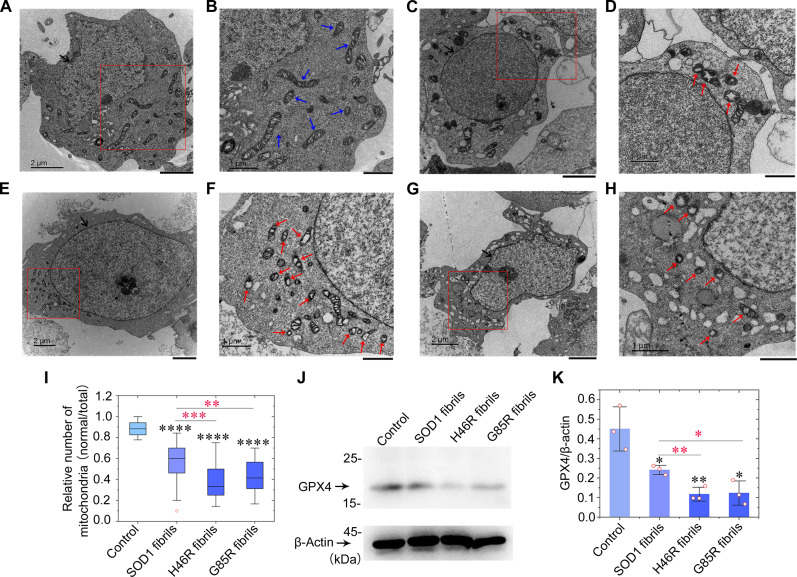
We next used ultrathin section TEM and Western blotting to further test this hypothesis. The morphology of normal mitochondria in SH-SY5Y cells incubated with 0 μM SOD1 fibril seeds (control), which are highlighted by blue arrows, was tubular or round (Fig. 5, A and B). Wild-type SOD1 fibril seed treatment (10 μM) caused serious mitochondrial impairment and induced ferroptosis in SH-SY5Y cells (Fig. 5, C and D). H46R fibril seed treatment (10 μM) (Fig. 5, E and F) and G85R fibril seed treatment (10 μM) (Fig. 5, G and H) caused severe mitochondrial impairment and promoted ferroptosis in SH-SY5Y cells. About 40% mitochondria in wild-type SOD1 fibril seed–treated cells (Fig. 5, D and I) and about 70% mitochondria in H46R fibril seed– and G85R fibril seed–treated cells (Fig. 5, F, H, and I) became smaller than normal mitochondria with increased mitochondrial membrane density and reduction of mitochondrial cristae (shrunken mitochondria) or became swollen and vacuolized. These abnormal mitochondria with morphological features of ferroptosis or mitochondrial vacuolization are highlighted by red arrows (Fig. 5, D, F, and H). A significantly lower number of normal mitochondria was observed in SH-SY5Y cells treated by wild-type SOD1 fibril seeds, H46R fibril seeds, and G85R fibril seeds than did in control cells treated by tris-HCl buffer containing TCEP (P = 3.3 × 10−12, 5.6 × 10−21, and 5.9 × 10−22, respectively) (Fig. 5I). A significantly lower number of normal mitochondria was observed in SH-SY5Y cells treated by H46R fibril seeds and G85R fibril seeds than did in control cells treated by wild-type SOD1 fibril seeds (P = 0.00020 and 0.0016, respectively) (Fig. 5I). To gain a quantitative understanding of how SOD1 mutations regulate neuronal cell ferroptosis, we detected a master regulator of ferroptosis, GPX4 (7, 11, 12), in the above cells using the anti-GPX4 antibody (Fig. 5J). Upon incubation with 10 μM fibril seeds for 3 days, GPX4 was down-regulated in SH-SY5Y cells treated by wild-type SOD1 fibril seeds where GPX4 protein levels were reduced by ~46% (P = 0.0346) (Fig. 5, J and K). We observed that GPX4 was strongly down-regulated in SH-SY5Y cells treated by H46R fibril seeds and G85R fibril seeds for 3 days, and GPX4 protein levels were reduced by ~74% in H46R fibril seed–treated cells (P = 0.0081) and ~73% in G85R fibril seed–treated cells (P = 0.0116), compared to control cells treated by tris-HCl buffer containing TCEP when normalized to β-actin (Fig. 5, J and K). Notably, GPX4 protein levels in H46R fibril seed– and G85R fibril seed–treated cells were significantly decreased compared with control cells treated by wild-type SOD1 fibril seeds (P = 0.0072 and 0.0367, respectively), indicating that treatment of cells with 10 μM H46R fibril seeds or 10 μM G85R fibril seeds significantly promoted ferroptosis in SH-SY5Y cells (Fig. 5K). These results demonstrate that the fibril seeds from H46R and G85R cause more severe mitochondrial impairment and activate ferroptosis in neuronal cells in a similar way, compared with those from wild-type SOD1. It is possible that adding SOD1 fibrils to the outside of cells causes mitochondrial dysfunction through the accumulation of misfolded SOD1 on mitochondrial membranes. Together, these data demonstrate that the ALS-causing SOD1 mutant proteins H46R and G85R form similar amyloid fibril structures and strongly suggest that these different SOD1 mutants exhibit similar toxicity and the ability to induce mitochondrial damage and activate ferroptosis in neuronal cells, contributing to ALS pathology.
Fig. 5. Fibril seeds from H46R and G85R cause severe mitochondrial impairment and induce ferroptosis in neuronal cells.
(A to H) SH-SY5Y cells were cultured for 1 day and then incubated with 0 μM SOD1 fibril seeds (A and B), 10 μM wild-type SOD1 fibril seeds (C and D), 10 μM H46R fibril seeds (E and F), and 10 μM G85R fibril seeds (G and H), respectively, for 3 days. Nuclei and normal mitochondria in SH-SY5Y cells are highlighted using black arrows (A, C, E, and G) and blue arrows (B), respectively. Abnormal mitochondria with morphological features of ferroptosis or mitochondrial vacuolization are highlighted by red arrows (D, F, and H). Samples were negatively stained using 2% uranyl acetate and lead citrate. Scale bars, 2 μm (A, C, E, and G) and 1 μm (B, D, F, and H). (I) Box plot analyzing the relative number of mitochondria (normal/total) in SH-SY5Y cells treated with SOD1 fibril seeds and showing the quantification of TEM images in n = 30 SH-SY5Y cells examined over three independent experiments. (J) Western blot for GPX4 in the cell lysates from SH-SY5Y cells incubated with fibril seeds from H46R and G85R, compared with those incubated with wild-type SOD1 fibril seeds. β-Actin served as the protein loading control. (K) The relative amount of GPX4 in the above cell lines (open red circles shown in scatter plots) was determined as a ratio of the density of GPX4 band over the density of β-actin band in cell lysates and expressed as the means ± SD (with error bars) of values obtained in three independent experiments.
Fibril seeds from H46R and G85R significantly promote templated misfolding of native SOD1 in cells
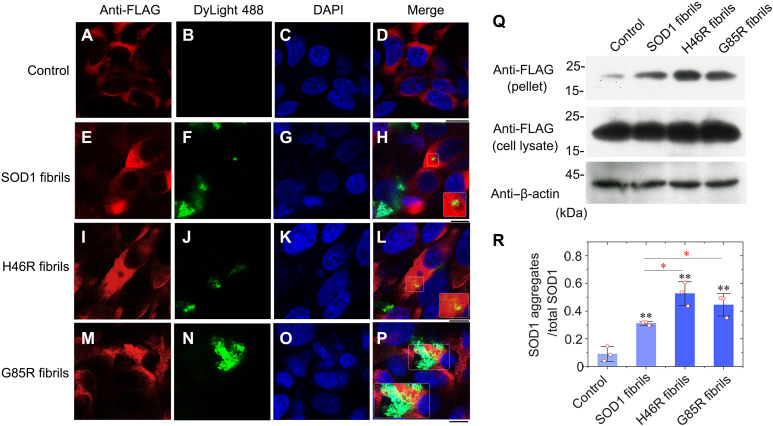
Given that two ALS-causing SOD1 mutant proteins, H46R and G85R, do exhibit high toxicity and the ability to induce mitochondrial damage and activate ferroptosis in neuronal cells (Figs. 4 and 5), we predicted that fibril seeds from these two mutants might induce templated misfolding of native SOD1 in previous healthy cells, an important pathway for prion-like propagation of mutant SOD1 misfolding (36, 37, 58). We next used confocal microscopy and Western blotting to test this hypothesis. HEK-293T cells stably expressing FLAG-tagged wild-type SOD1 were cultured for 1 day; then incubated with 0 μM DyLight 488–labeled SOD1 fibril seeds (control), 10 μM DyLight 488–labeled wild-type SOD1 fibril seeds, 10 μM DyLight 488–labeled H46R fibril seeds, and 10 μM DyLight 488–labeled G85R fibril seeds, respectively, for 1 day; and further investigated by immunofluorescence (Fig. 6, A to P). FLAG-tagged wild-type SOD1 (red) was mainly located in the cytoplasm of HEK-293T cells after incubation without or with SOD1 fibril seeds for 1 day (Fig. 6, A to P). DyLight 488–labeled SOD1 fibril seeds (green), internalized possibly through endocytosis, were also located in the cytoplasm of the cells (Fig. 6, E to P). Notably, abundant orange puncta indicate colocalization of abundant cytoplasmic aggregates of wild-type SOD1 and fibril seeds from H46R and G85R in granules (Fig. 6, L and P). In sharp contrast, only a few yellow puncta were observed, indicating colocalization of only a few cytoplasmic aggregates of wild-type SOD1 and wild-type SOD1 fibril seeds in granules (Fig. 6H). Thus, fibril seeds from H46R and G85R induce templated misfolding and aggregation of wild-type SOD1 in previous healthy cells, much stronger than those from wild-type SOD1. To gain a quantitative understanding of how SOD1 mutations promote templated misfolding of wild-type SOD1 in cells, we used HEK-293T cells stably expressing FLAG-tagged wild-type SOD1, which were cultured for 1 day and then incubated with 0 μM SOD1 fibril seeds (control), 10 μM wild-type SOD1 fibril seeds, 10 μM H46R fibril seeds, and 10 μM G85R fibril seeds, respectively, for 2 days. The sarkosyl-insoluble ultracentrifugation pellets from the above cells were probed with anti-FLAG antibody, and the corresponding cell lysates were probed using anti-FLAG antibody and anti–β-actin antibody (Fig. 6Q). We found that ~31, ~53, and ~45% of total SOD1 ended up in the pellet when the cells were treated with fibril seeds from wild-type SOD1 and its variants H46R and G85R, respectively (Fig. 6R). Notably, the amount of insoluble SOD1 aggregates in H46R fibril seed– and G85R fibril seed–treated cells was significantly increased compared with control cells treated by wild-type SOD1 fibril seeds (P = 0.012 and 0.049, respectively), indicating that treatment of cells with 10 μM H46R fibril seeds or 10 μM G85R fibril seeds significantly promoted templated misfolding of wild-type SOD1 in HEK-293T cells (Fig. 6R). These results demonstrate that the fibril seeds from H46R and G85R significantly promote templated misfolding and aggregation of native SOD1 in cells in a similar way, compared with those from wild-type SOD1. Together, these data strongly suggest that the ALS-causing SOD1 mutant proteins H46R and G85R exhibit similar abilities to induce templated misfolding of native SOD1 in previous healthy cells, contributing to ALS pathology.
Fig. 6. Fibril seeds from H46R and G85R are more efficient at seeding the aggregation of FLAG-tagged wild-type SOD1 in cultured cells, compared to wild-type SOD1 fibril seeds generated under the same conditions.
(A to P) Immunofluorescence imaging of HEK-293T cells stably expressing FLAG-tagged wild-type SOD1 incubated with 0 μM DyLight 488–labeled SOD1 fibril seeds (A to D; control), 10 μM DyLight 488–labeled wild-type SOD1 fibril seeds (E to H), 10 μM DyLight 488–labeled H46R fibril seeds (I to L), and 10 μM DyLight 488–labeled G85R fibril seeds (M to P), respectively, using an anti-FLAG antibody against FLAG-tagged SOD1 (red) and staining with DyLight 488 (green) and 4′,6-diamidino-2-phenylindole (DAPI) (blue). Orange puncta indicate colocalization of cytoplasmic SOD1 aggregates and fibril seeds from H46R and G85R in granules (L and P). Scale bars, 10 μm. (Q) Western blot for SOD1 in the sarkosyl-insoluble pellets and the corresponding cell lysates from HEK-293T cells stably expressing FLAG-tagged wild-type SOD1 incubated with fibril seeds from H46R and G85R, compared with those incubated with wild-type SOD1 fibril seeds. β-Actin served as the protein loading control. Cells were cultured for 1 day and then incubated with 0 μM SOD1 fibril seeds, 10 μM wild-type SOD1 fibril seeds, 10 μM H46R fibril seeds, and 10 μM G85R fibril seeds, respectively, for 2 days. (R) The relative amount of insoluble SOD1 aggregates in the above cell lines (open red circles shown in scatter plots) was determined as a ratio of the density of insoluble SOD1 aggregate bands over the density of the total SOD1 bands in cell lysates and expressed as the means ± SD (with error bars) of values obtained in three independent experiments. SOD1 fibrils, P = 0.0022; H46R fibrils, P = 0.0017 and 0.012; G85R fibrils, P = 0.0032 and 0.049. Statistical analyses were performed using two-sided Student’s t test. Values of P < 0.05 indicate statistically significant differences. *P < 0.05, **P < 0.01, and ***P < 0.001 relative to control. Cells treated with 20 mM tris-HCl buffer (pH 7.4) containing 5 mM TCEP and cells incubated with 10 μM wild-type SOD1 fibril seeds were used as controls, respectively.
The H46R mutation and the G85R mutation significantly decrease the conformational stability of SOD1 fibrils
We propose that H46R and G85R SOD1 fibrils are more cytotoxic because of their higher capacity to template misfolding of native SOD1 in cells as compared to the fibrils formed from the wild-type protein (Figs. 4 to 6). However, what is lacking is some mechanistic explanation why the mutant SOD1 fibrils would have such a higher templating capacity in cells. We first examined whether H46R and G85R SOD1 fibrils exhibit distinct conformational stability from wild-type SOD1 fibrils. Chemical denaturation was used to evaluate the conformational stability of the SOD1 fibrils. A strong chaotropic salt, guanidine thiocyanate (GdnSCN), was used in our denaturation assay (fig. S6A). Notably, the C1/2 values of the H46R fibril and the G85R fibril are 0.74 ± 0.33 and 0.70 ± 0.36 M (fig. S6B), respectively, which are significantly lower than the wild-type fibril (1.90 ± 0.13 M; fig. S6B), suggesting that the H46R fibril and the G85R fibril are less stable than the wild type. Together, the data showed that the H46R fibril and the G85R fibril have a significantly lower conformational stability compared to the wild-type fibril, which is one of the reasons why the mutant SOD1 fibrils have such a higher templating capacity in cells.
We next performed seeding experiments with purified proteins in the test tube. We unexpectedly found that wild-type SOD1 fibril seeds, H46R fibril seeds, and G85R fibril seeds significantly nucleated aggregation of wild-type SOD1 in a similar way (fig. S7, A and B). The fibril formation lag time in wild-type SOD1 in the presence of 2% preformed seed fibrils of wild-type SOD1, H46R, and G85R is 6.95 ± 2.30, 9.85 ± 3.70, and 7.97 ± 0.73 hours, respectively (fig. S7B), which is significantly lower than that in the absence of preformed seed fibrils (32.71 ± 0.90 hours; P = 0.000055, 0.00048, and 0.0000032, respectively; fig. S7B), but the lag time of fibril formation of wild-type SOD1 in the presence of H46R fibril seeds and G85R fibril seeds is not significantly lower than that in the presence of the wild-type fibril seeds (P = 0.313 and 0.504, respectively; fig. S7B). Thus, H46R and G85R SOD1 fibrils are able to template misfolding of wild-type SOD1 in vitro, similar to fibrils formed from the wild-type protein.
Wild-type SOD1 is templated into fibrils with H46R/G85R morphology rather than the wild-type SOD1 fibril morphology
We used anti-FLAG immunofluorescence microscopy and Western blotting to follow the level of coaggregation of FLAG-tagged wild-type SOD1 and H46R or G85R (Fig. 6). These data suggest that the mutant fibrils induce a higher level of aggregation of the wild-type protein and that wild-type SOD1 is templated into fibrils with H46R/G85R morphology rather than the wild-type SOD1 fibril morphology. To ascertain the nature of H46R fibril– and G85R fibril–seeded aggregates of wild-type SOD1 in vitro and in cells, we first conducted immunogold electron microscopy. Amyloid fibrils were produced from the apo form of recombinant FLAG-tagged wild-type SOD1 incubated in 20 mM tris-HCl buffer (pH 7.4) containing 5 mM TCEP and shaking at 37°C for 28 hours, in the presence of 2% preformed seed fibrils of H46R (fig. S8A) or G85R (fig. S8B), and labeled by gold particles conjugated with anti-FLAG antibody. Wild-type SOD1 aggregates purified from HEK-293T cells stably expressing FLAG-tagged wild-type SOD1 incubated with preformed seed fibrils of H46R (fig. S8C) or G85R (fig. S8D) for 2 days and also labeled by gold particles conjugated with anti-FLAG antibody. Notably, the amyloid fibrils in the above samples were recognized by anti-FLAG antibody and decorated with 10-nm gold labels, and anti-FLAG immunogold staining demonstrated the incorporation of wild-type SOD1 into the amyloid fibrils seeded by H46R and G85R (fig. S8, A to D).
We next seeded in vitro amyloid fibril formation of wild-type SOD1 with H46R and G85R, respectively, and document the morphology of the fibrils by TEM, AFM, and cryo-EM (fig. S9, A to F). Negative-staining TEM images showed that the apo form of recombinant wild-type SOD1 incubated in 20 mM tris-HCl buffer (pH 7.4) containing 5 mM TCEP and shaking at 37°C for 28 hours, in the presence of 2% preformed seed fibrils of H46R and G85R, respectively, also formed homogeneous and unbranched fibrils under reducing conditions (fig. S9, A and B), with morphology similar to that of H46R fibrils and G85R fibrils (fig. S1, A and B). We called these fibrils H46R fibril– and G85R fibril–seeded fibrils of wild-type SOD1, respectively (fig. S9, A to F). The AFM images, cryo-EM micrographs, and 2D class average images using RELION3.1 (53) showed that both H46R fibril– and G85R fibril–seeded fibrils of wild-type SOD1 were composed of a single protofilament with a left-handed helical twist (fig. S9, C to F) and were arranged in a staggered manner (fig. S10, A and B). The helical pitch was 173 ± 9 nm for H46R fibril–seeded fibrils of wild-type SOD1 (fig. S9C) and 170 ± 10 nm for G85R fibril–seeded fibrils of wild-type SOD1 (fig. S9D), which is similar to the helical pitch for H46R fibrils and G85R fibrils (fig. S1, C and D) but longer than the wild-type fibril (43). The 2D class average images showed that at least in vitro, the wild-type SOD1 fibril adopts the H46R/G85R fibril morphology (figs. S2, A and B, and S10, A and B) but does not adopt the wild-type fibril morphology (fig. S10C).
Together, these data strongly suggest that wild-type SOD1 is templated into fibrils with H46R/G85R morphology rather than the wild-type SOD1 fibril morphology. The seeding experiments indicate that the wild-type SOD1 fibril adopts the more cytotoxic H46R/G85R fibril morphology, which is another reason why the mutant SOD1 fibrils have such a higher templating capacity in cells. We plan to isolate FLAG-tagged aggregates from their H46R/G85R fibril–seeded cells and document morphology by cryo-EM 2D classes or AFM in the near future.
DISCUSSION
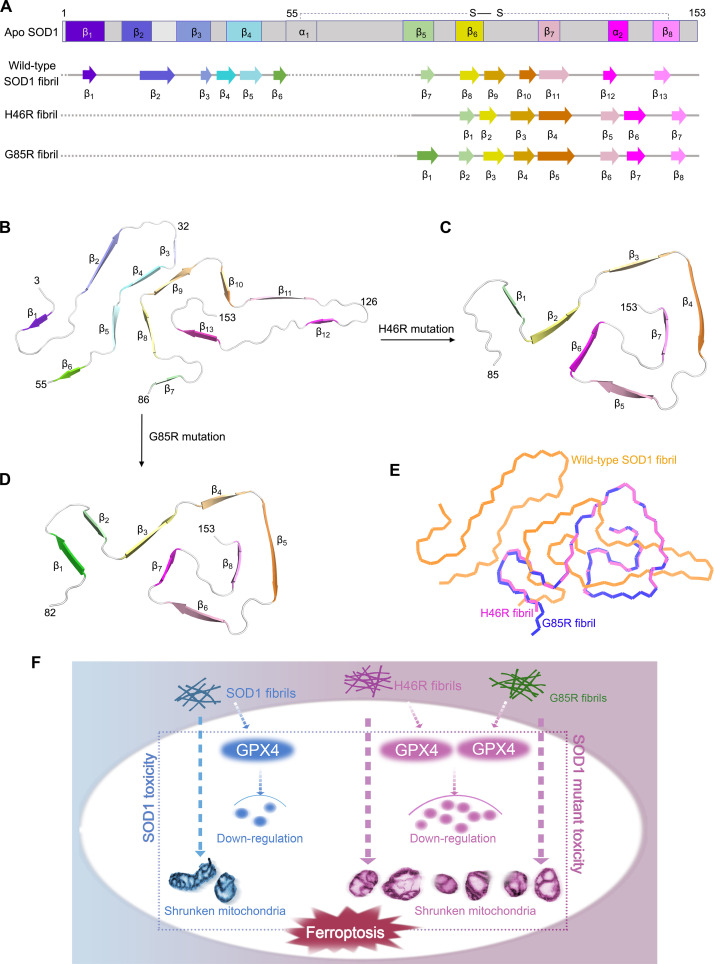
Mutations in SOD1 account for about 2 to 6% of all ALS (1, 2, 16–18, 59). Because familial mutations in SOD1, such as H46R and G85R, are involved in the pathogenesis of the motor neuron disease ALS where it is observed to form intracellular fibrillar inclusions (3, 16, 22, 34, 35, 37, 60), it has generally been thought that these proteinaceous inclusions could be responsible for neuronal cell death in patients with ALS (3, 16, 37). Here, we compared the structures of apo SOD1, the wild-type SOD1 fibril, the H46R fibril, and the G85R fibril (Fig. 7, A to E). Notably, the SOD1 molecule adopts largely distinctive secondary and tertiary structures in three different SOD1 structures (apo SOD1, the wild-type SOD1 fibril, and the H46R/G85R fibril) (Fig. 7, A to D), highlighting the phenotypic diversity of SOD1 in physiological and fibrillar states. The full-length apo human SOD1 monomer contains eight β strands (β1 to β8), two α helices (α1 and α2), and a single disulfide bond between C57 in α1 and C146 in β8 (Fig. 7A) (61). Once it folds into its fibrillar form under reducing conditions, the SOD1 subunit undergoes a totally conformational rearrangement. The wild-type human SOD1 fibril core contains six β strands (β1 to β6) by its N-terminal segment (residues 3 to 55) and seven β strands (β7 to β13) by its C-terminal segment (residues 86 to 153), exhibiting an in-register intramolecular β strand architecture (Fig. 7, A and B) (43). In sharp contrast, the fibril cores of H46R and G85R only comprise a C-terminal segment with residues 85 to 153 and 82 to 153 containing seven β strands (β1 to β7) and eight β strands (β1 to β8), respectively (Fig. 7, A, C, and D). In the wild-type SOD1 fibril structure [Protein Data Bank (PDB) 7VZF] (43), where G85 is not present in the core, H46 is involved in a weak salt bridge with E100 that should be strengthened in the presence of the H46R mutation; in the G85R fibril structure (PDB 8IHU), R85 is involved in a new salt bridge with D101 (fig. S4, C and D). Thus, the H46R mutation and the G85R mutation result in amyloid fibrils with distinct structures compared to the wild-type fibril (Fig. 7, B to E). H46R and G85R form almost isomorphous fibrils and similar amyloid fibril structures with an root mean square deviation (RMSD) of 0.273 Å (55 to 55 Cα atoms), while the wild-type SOD1 fibril could hardly align with the H46R fibril and the G85R fibril with RMSD of 16.775 Å (68 to 68 Cα atoms) and 16.776 Å (68 to 68 Cα atoms), respectively (Fig. 7E).
Fig. 7. Comparison of the structures of the apo form of SOD1, the wild-type SOD1 fibril, the H46R fibril, and the G85R fibril.
(A) Sequence alignment of the full-length apo human SOD1 monomer [Protein Data Bank (PDB) 1HL4] (61) with eight β strands (β1 to β8), two α helices, and a single disulfide bond. Sequence alignment of the wild-type SOD1 fibril core comprising residues 3 to 55 and 86 to 153 from the full-length wild-type human SOD1 (PDB 7VZF) (43) with the observed 13 β strands (β1 to β13). Sequence alignment of the H46R fibril core comprising residues 85 to 153 from the full-length human H46R SOD1 with the observed seven β strands colored light green (β1), yellow (β2), gold (β3), orange (β4), pink (β5), magenta (β6), and light magenta (β7) in the C-terminal segment. The dotted line corresponds to residues 1 to 84 not modeled in the cryo-EM density. Sequence alignment of the G85R fibril core comprising residues 82 to 153 from the full-length human G85R SOD1 with the observed eight β strands colored green (β1), light green (β2), yellow (β3), gold (β4), orange (β5), pink (β6), magenta (β7), and light magenta (β8) in the C-terminal segment (bottom). The dotted line corresponds to residues 1 to 81 not modeled in the cryo-EM density. (B to D) Ribbon representation of the structures of a wild-type SOD1 fibril core (B), an H46R fibril core (C), and a G85R fibril core (D), all of which contain one molecular layer and a monomer. (E) Overlay of the structures of a wild-type SOD1 fibril core (orange), an H46R fibril core (magenta), and a G85R fibril core (blue). (F) A hypothetical model shows how fibril seeds from H46R and G85R exhibit higher cytotoxicity and have a significantly higher ability to cause mitochondrial impairment and activate ferroptosis in neuronal cells.
This work builds on our previous study (43) and provides high-resolution cryo-EM structures of two types of in vitro–generated amyloid fibrils from familial ALS-linked SOD1 mutants. The recent surge in structural studies of amyloid fibrils derived from patient tissue has pointed to an association between specific diseases and amyloid polymorphs (49, 50). In the current absence of structures of in vivo–derived SOD1 amyloid fibrils, our work provides important structural foundations for thinking about SOD1-mediated neuropathogenesis in familial ALS. We address the relevance of the in vitro–prepared amyloid fibrils to human disease below. We show that in vitro–generated amyloid fibrils from H46R and G85R are more cytotoxic and efficient at seeding the aggregation of native SOD1 in cultured cells than wild-type SOD1 fibril seeds generated under the same conditions. That is, these amyloid fibrils show heightened virulence in cells and show an increased ability to nucleate the aggregation of native SOD1. It has been reported that these amyloid fibrils induce ferroptosis in cultured cells (this work) and injection of these amyloid fibrils into the spinal cords of newborn transgenic mice expressing G85R accelerates the progression of ALS (51, 52). These amyloid fibrils may be biochemically a forme fruste of those accumulating in patients with ALS and the same SOD1 mutations. Although the SOD1 fibril structures determined here may not to be identical to those accumulating in patients with ALS and the same SOD1 mutations, the work provides important initial insights into the potential structural underpinnings of how these SOD1 mutants might aggregate and cause cytotoxicity in ALS. We plan to collect structural data on the H46R fibril and the G85R fibril purified from the brains of patients with ALS or ALS transgenic mice in the near future.
Previous studies proposed two alternative models of the core of G85R fibrils extracted from spinal cords of transgenic mice carrying this mutation and developing an ALS-like phenotype, based on protease digestion assay, mass spectrometric analysis, and binary epitope mapping assay (25, 40). One “three key region model” predicts that the G85R fibril core contains one N-terminal segment comprising residues 1 to 30, one segment comprising residues ~70 to 90, and one C-terminal segment comprising residues ~103 to 153 (25). This is partly compatible with our model, wherein β1 and β3 to β8 would correspond to the two C-terminal segments in the model of G85R fibril core proposed by Furukawa and coworkers (25). They also showed that amino acids 1 to 30 are found in the core from purified H46R fibrils under test-tube conditions and predicted that the H46R fibril core contains one N-terminal segment comprising residues 1 to 30 and two C-terminal segments comprising residues ~85 to 120 and ~130 to 153 (25). This is partly compatible with our model, wherein β1 to β4 and β5 to β7 would correspond to the two C-terminal segments in the model of H46R fibril core (25). Another three key region model predicts that the G85R fibril core contains two N-terminal segments comprising residues 43 to 57 and 57 to 72 and one C-terminal segment comprising residues 131 to 153 (40). This is compatible with our model to a lesser extent, wherein β6 to β8 would correspond to the C-terminal segment in the model of G85R fibril core proposed by Bergh and coworkers (40). The Hart laboratory determined the crystal structure of the ALS-causing SOD1 mutant H46R and proposed that a conformational change in H46R permits a gain of interaction between dimers that aggregate into zigzag filaments, highlighting the role of gain of interaction in pathogenic SOD1 (62). This is partly compatible with our model, wherein the H46R fibril core exhibits a serpentine fold containing seven β strands. It should be mentioned that in our model, the fibril cores of H46R and G85R contain the C-terminal segment comprising residues 85 to 153 and 82 to 153, respectively. The N-terminal segment in the fibril cores was not observed, except an island in the G85R fibril. This differentiates from Furukawa et al. (25) and Bergh et al. (40) who found that one or two N-terminal segments are present in the fibril cores of H46R and G85R. We then consider the impact of cofactors that might be present in patients and mouse models of ALS, but not in our cell-free fibrillization. Notably, these cofactors might promote the inclusion of more N-terminal residues in the fibril core as suggested for these mutants by Furukawa et al. (25) and Bergh et al. (40) and as are present in the wild-type SOD1 fibril core (43).
It should be mentioned that the two cryo-EM structures have loops formed by residues 90 to 96 in which the anionic side chains of two D residues, D90 and D96, are stuffed, without apparent charge compensation, into a tightly packed internal cavity (Figs. 2 and 3, A to D). Meanwhile, the cationic side chain of K91, adjacent to D90, projects outward into the solvent (Figs. 2 and 3, A and B). Thus, it would seem to make more sense if the side chain of K91 were pointing into the cavity to compensate for the charge of D96 (at least). We are confident in the relative positioning of the hydrophilic side chains of D90, K91, and D96 in our model because, first, our model was refined by PHENIX 1.15.2 and the side chains of D90 and D96 toward the hydrophilic cavity are correct. The side-chain map of D90 in the G85R fibril was toward the hydrophilic cavity (Fig. 3, A and B). Although the density map is equivocal for these side chains in the H46R fibril, it is highly similar to that of the G85R fibril, and we can also see the trend of the density map when the map contour value has been enlarged (Fig. 2, A and B). Second, the two carboxylic acids in D90 and D96 of the H46R fibril and the G85R fibril form two hydrogen bonds with distances of 3.7 and 4.3 Å, respectively, to stabilize the hydrophilic cavity (Figs. 2 and 3, A and B).
The H46R fibril and the G85R fibril in this study were both formed under reducing conditions, which lack copper and zinc ions and the native disulfide bond in the structures. These immature forms have been proposed to be the origin for cytotoxic misfolded conformations of SOD1 (20–22, 63, 64). Previous studies have shown that the amyloid-like aggregates isolated from ALS transgenic mice or cells expressing ALS-causing SOD1 mutants contain metal-deficient and disulfide-reduced SOD1, suggesting their pathogenic potential (23, 24, 65). More than 10 ALS-causing SOD1 mutant proteins, including H46R and G85R, have been found to be susceptible to disulfide reduction (21), four of which are enriched in the spinal cord and brain throughout the lifetime of transgenic mice carrying these mutations, lacking the native disulfide bond (23, 66). Thus, structural determination of SOD1 mutant fibrils is of importance for understanding the pathogenic mechanism of SOD1 mutants in ALS.
In summary, on the one hand, two SOD1 metal-binding mutants, H46R and G85R, which have prion-like properties (36, 37) and have previously found to induce ALS-like disease in a mouse model (23, 24, 33–35, 37), form amyloid fibril structures revealed by cryo-EM; the H46R fibril and the G85R fibril consist of a single protofilament with a fibril core comprising residues 85 to 153 and 82 to 153, respectively. The reported high-resolution cryo-EM structures of H46R fibril and G85R fibril reveal unusual overall structures when compared to the wild-type fibril, characterized by an alteration of crucial salt bridges, a C-terminal fibril core, an island flanking the protofilament in the G85R fibril, 3 hydrophilic cavities, and 7 or 8 instead of 13 β strands in the core. On the other hand, fibril seeds from H46R and G85R exhibit higher cytotoxicity and have a significantly higher ability to cause mitochondrial impairment and activate ferroptosis in neuronal cells via templated misfolding compared to the wild-type fibril seeds (Fig. 7F). We find a direct link between amyloid fibrils formed by genetic mutations of SOD1 and GPX4-regulated ferroptosis related to ALS (Fig. 7F). The fibril structures will be valuable to understanding the structural basis underlying the functions of familial mutations in an amyloid state and inspiring future research on the molecular mechanisms by which mutations in SOD1 exhibit cytotoxicity, promote ferroptosis, induce templated misfolding of native SOD1 in cells, and cause the familial form of ALS.
MATERIALS AND METHODS
Protein expression and purification
A plasmid-encoding, full-length human SOD1 (1 to 153) was a gift from T. O’Halloran (Chemistry of Life Processed Institute, Northwestern University). The sequence for SOD1 (1 to 153) was expressed from the vector pET-3d, and two SOD1 mutants H46R and G85R were constructed by site-directed mutagenesis using a wild-type SOD1 template; the primers are shown in table S1. All SOD1 plasmids were transformed into E. coli BL21 (DE3) cells (Novagen, Merck, Darmstadt, Germany). Recombinant full-length wild-type human SOD1 and its variants H46R and G85R as well as recombinant FLAG-tagged wild-type human SOD1 were expressed from the vector pET-30a(+) in E. coli BL21 (DE3) cells. SOD1 proteins were purified to homogeneity by Q-Sepharose chromatography as described by Chattopadhyay et al. (67) and Xu et al. (68). After purification, recombinant wild-type SOD1 and its variants H46R and G85R were demetallated by dialysis in 10 mM EDTA and 10 mM NaAc buffer (pH 3.8) five times, as described by Chattopadhyay et al. (67) and Xu et al. (68). In all, 10 mM NaAc buffer (pH 3.8) and 20 mM tris-HCl buffer (pH 7.4) were used for further dialysis. The apo forms of wild-type SOD1, H46R, and G85R were then concentrated, filtered, and stored at −80°C. AAnalyst-800 atomic absorption spectrometer (PerkinElmer) was used to quantify metal content of SOD1 samples. Samples of wild-type SOD1, H46R, and G85R contained <5% of residual metal ions, indicating that the samples were indeed in the apo state. SDS–polyacrylamide gel electrophoresis (PAGE) and mass spectrometry were used to confirm that the purified apo SOD1 proteins were single species with an intact disulfide bond. A NanoDrop OneC Microvolume UV-Vis Spectrophotometer (Thermo Fisher Scientific) was used to determine the concentration of apo SOD1 proteins according to their absorbances at 214 nm with a standard calibration curve drawn from bovine serum albumin (BSA).
SOD1 fibril formation and seeding experiments
The apo forms of recombinant full-length wild-type human SOD1 and its variants H46R and G85R were incubated in 20 mM tris-HCl buffer (pH 7.4) containing 5 mM TCEP and shaking at 37°C for 40 to 48 hours, and the SOD1 fibrils were collected. Large amorphous aggregates in SOD1 fibril samples were removed by centrifugation for 5000g at 4°C for 10 min. The supernatants (purified amyloid fibrils of SOD1) were then concentrated to ~30 μM in a centrifugal filter (Millipore). SDS-PAGE and mass spectrometry were used to confirm that full-length SOD1 had been incorporated into the SOD1 fibrils. To prepare SOD1 fibril seeds for the following functional experiments, the above concentrated amyloid fibrils of SOD1 were sonicated for 5 min (5-s on and 5-s off) on ice. A NanoDrop OneC Microvolume UV-Vis Spectrophotometer (Thermo Fisher Scientific) was used to determine the concentrations of the wild-type SOD1 fibril, the H46R fibril, and the G85R fibril according to their absorbances at 214 nm with a standard calibration curve drawn from BSA. For seeding experiments, amyloid fibrils were produced from the apo form of recombinant wild-type SOD1 (30 μM) incubated in 20 mM tris-HCl buffer (pH 7.4) containing 5 mM TCEP and shaking at 37°C for 28 hours, in the presence of 2% (v/v) H46R fibril seeds and 2% (v/v) G85R fibril seeds, respectively.
TEM of H46R fibrils, G85R fibrils, and wild-type SOD1 fibrils seeded by H46R fibrils and G85R fibrils
H46R fibrils and G85R fibrils as well as H46R fibril– and G85R fibril–seeded fibrils of wild-type SOD1 were examined by TEM of negatively stained samples. Ten microliters of SOD1 mutation fibril samples (~30 μM) were loaded on copper grids for 30 s and washed with H2O for 10 s. Samples on grids were then stained with 2% (w/v) uranyl acetate for 30 s and dried in air at 25°C. The stained samples were examined using a JEM-1400 Plus transmission electron microscope (JEOL) operating at 100 kV for H46R fibrils and G85R fibrils, and a Talos L120C TEM (Thermo Fisher Scientific) operating at 120 kV for wild-type SOD1 fibrils seeded by H46R fibrils and G85R fibrils.
AFM of H46R fibrils, G85R fibrils, and wild-type SOD1 fibrils seeded by H46R fibrils and G85R fibrils
H46R fibrils and G85R fibrils as well as H46R fibril– and G85R fibril–seeded fibrils of wild-type SOD1 were produced as described above. Ten microliters of SOD1 mutation fibril samples (~30 μM) were incubated on a freshly cleaved mica surface for 2 min, followed by rinsing three times with 10 μl of pure water to remove the unbound fibrils and drying at room temperature. The fibrils on the mica surface were probed in air by the Dimension Icon scanning probe microscope (Bruker) with ScanAsyst mode. The measurements were realized using a SCANASYST-AIR probe (Bruker) with a spring constant of 0.4 N/m and a resonance frequency of 70 kHz. A fixed resolution (256 × 256 data points) of the AFM images was acquired with a scan rate at 1 Hz and analyzed using NanoScope Analysis 2.0 software (Bruker).
Cryo-EM of H46R fibrils, G85R fibrils, and wild-type SOD1 fibrils seeded by H46R fibrils and G85R fibrils
H46R fibrils and G85R fibrils as well as H46R fibril– and G85R fibril–seeded fibrils of wild-type SOD1 were produced as described above. An aliquot of 3.5 μl of ~30 μM SOD1 mutation fibril solution was applied to glow-discharged holey carbon grids (Quantifoil Cu R1.2/1.3, 300 mesh), blotted for 3.5 s, and plunge-frozen in liquid ethane using a Vitrobot Mark IV. The grids were examined using a Glacios transmission electron microscope, operated at 200 kV, and equipped with a field emission gun and a Ceta-D complementary metal-oxide semiconductor camera (Thermo Fisher Scientific). The cryo-EM micrographs were acquired on a Krios G4 transmission electron microscope operated at 300 kV (Thermo Fisher Scientific) and equipped with a Bio-Quantum K3 direct electron detector (Gatan). A total of 8680 movies for H46R fibrils, 6366 movies for G85R fibrils, 4851 movies for wild-type SOD1 fibrils seeded by H46R fibrils, and 5888 movies for wild-type SOD1 fibrils seeded by G85R fibrils were collected in superresolution mode at a nominal magnification of ×105,000 (physical pixel size, 0.84 Å) and a dose of 18.75 e− Å−2 s−1 (see Table 1). An exposure time of 3.2 s was used, and the resulting videos were dose fractionated into 40 frames. A defocus range of −1.2 to −2.0 μm was used.
Helical reconstruction
All image-processing steps, which include manual picking, particle extraction, 2D and 3D classifications, 3D refinement, and postprocessing, were performed by RELION-3.1 (53). For the H46R fibril, 63,230 fibrils were picked manually from 8680 micrographs, and 1024- and 686-pixel boxes were used to extract particles by 90% overlap scheme. 2D classification of 1024–box size particles was used to calculate the initial twist angle. In regard to helical rise, 4.8 Å was used as the initial value. Particles were extracted into 400 box sizes for further processing. After several iterations of 2D and 3D classifications, particles with the same morphology were picked out. Local searches of symmetry in 3D classification were used to determine the final twist angle and rise value. The 3D initial model was a cylinder that was built by RELION helix toolbox; 3D classification was performed several times to generate a proper reference map for 3D refinement. 3D refinement of the selected 3D classes with appropriate reference was performed to obtain final reconstruction. The final map of the H46R fibril was convergent with a rise of 4.88 Å and a twist angle of −1.079°. Postprocessing was performed to sharpen the map with a B factor of −78.38 Å2. On the basis of the gold standard Fourier shell correlation (FSC) = 0.143 criteria, the overall resolution was reported as 3.11 Å. The statistics of cryo-EM data collection and refinement is shown in Table 1. For the G85R fibril, 37,434 fibrils were picked manually from 6366 micrographs, and 1024- and 686-pixel boxes were used to extract particles by 90% overlap scheme. 2D classification of 1024–box size particles was used to calculate the initial twist angle. In regard to helical rise, 4.8 Å was used as the initial value. Particles were extracted into 400 box sizes for further processing. After several iterations of 2D and 3D classifications, particles with the same morphology were picked out. Local searches of symmetry in 3D classification were used to determine the final twist angle and rise value. The 3D initial model was a cylinder that was built by RELION helix toolbox, and because of the similarity of the density maps between the H46R fibril and the G85R fibril, we next used the H46R fibril density map with an initial low-pass filter of 30 Å as a reference map; 3D classification was performed several times to generate a proper reference map for 3D refinement. 3D refinement of the selected 3D classes with appropriate reference was performed to obtain final reconstruction. The final map of the G85R fibril was convergent with a rise of 4.87 Å and a twist angle of −1.058°. Postprocessing was performed to sharpen the map with a B factor of −82.56 Å2. On the basis of the gold standard FSC = 0.143 criteria, the overall resolution was reported as 2.97 Å. The statistics of cryo-EM data collection and refinement is shown in Table 1.
For wild-type SOD1 fibrils seeded by H46R fibrils and G85R fibrils, 14,256 and 19,899 fibrils were picked manually from 4851 and 5888 micrographs, respectively, and 1024- and 686-pixel boxes were used to extract particles by 90% overlap scheme for 2D classification. After several iterations of 2D classification, particles with the same morphology were picked out.
Atomic model building and refinement
Coot 0.8.9.2 (69) was used to build de novo and modify the atomic models of the H46R fibril and the G85R fibril. The models with three adjacent layers were generated for structure refinement. The models were refined using the real-space refinement program in PHENIX 1.15.2 (70). All density map–related figures were prepared in Chimera1.15. Ribbon representation of the structure of SOD1 fibril was prepared in PyMOL 2.3.
Cell culture and transfection
SH-SY5Y neuroblastoma cells (catalog number GDC0210) and HEK-293T cells (catalog number GDC0187) were obtained from China Center for Type Culture Collection (Wuhan, China) and cultured in minimum essential medium and in Dulbecco’s modified Eagle’s medium (Gibco, Invitrogen), respectively, supplemented with 10% (v/v) fetal bovine serum (Gibco), streptomycin (100 U/ml), and penicillin (100 U/ml) in 5% CO2 at 37°C. HEK-293T cell lines stably expressing wild-type human SOD1 or FLAG-tagged wild-type human SOD1 were constructed with a lentiviral vector construction system (phage-puro). The target DNA fragments were inserted into the lentiviral vector, and the plasmids containing target DNA, a plasmid encoding the VSVG gene, and p976 were packaged in HEK-293T cells at a ratio of 2:1:1 by Lipofectamine 2000 (Invitrogen). The ratio of liposome to DNA was 2:1. After 48 hours of transfection, the viruses were harvested and filtered, and then HEK-293T cells were infected with the packaged lentivirus twice for 12 hours each with a 12-hour interval. To establish the stable cell lines, puromycin was used to screen overexpressed cells. The expression of each protein was detected by Western blot.
Cell viability assays
SH-SY5Y cells, HEK-293T cells, or HEK-293T cells stably expressing FLAG-tagged wild-type SOD1 were plated in 96-well plates in minimum essential medium. After incubation for 24 hours, wild-type SOD1 fibril seeds, H46R fibril seeds, or 10 μM G85R fibril seeds at a final concentration of 10 μM were added into the medium for 36 hours. The MTT stock solution (5 mg/ml) was diluted with phosphate-buffered saline (PBS) and added into the well for 4 hours until formazan was formed in the cells. The final concentration of MTT was 0.5 mg/ml. Last, the dark blue formazan crystal was dissolved with dimethyl sulfoxide, followed by measuring its absorbance at 492 nm using a Thermo Multiskan MK3 microplate reader (Thermo Fisher Scientific). Cells were incubated in a medium containing 10% CCK8 for 2 to 4 hours, and the absorbance of the orange formazan was also measured with a microplate reader at 450 nm. Cell viability was expressed as the percentage ratio of the absorbance of wells containing the treated samples to that of wells containing cells treated with 20 mM tris-HCl buffer (pH 7.4) containing 5 mM TCEP. The cell viability data, analyzed using OriginPro software version 8.0724 (Origin Laboratory), are expressed as the means ± SD (with error bars) of values obtained from five or six independent experiments. All experiments were further confirmed by biological repeats.
Ultrathin section TEM
SH-SY5Y cells were cultured in six-well plates in minimum essential medium for 1 day and then cultured with 0 μM SOD1 fibril seeds, 10 μM wild-type SOD1 fibril seeds, 10 μM H46R fibril seeds, and 10 μM G85R fibril seeds, respectively, for 3 days, with cells cultured in 20 mM tris-HCl buffer (pH 7.4) containing 5 mM TCEP as a negative control. After prefixation with 3% paraformaldehyde and 1.5% glutaraldehyde in 1× PBS (pH 7.4), the cells were harvested and postfixed in 1% osmium tetroxide for 1 hour using an ice bath; the samples were then dehydrated in graded acetone and embedded in 812 resins. Ultrathin sections of the cells were prepared using a Leica Ultracut S Microtome and negatively stained using 2% uranyl acetate and lead citrate. The doubly stained ultrathin sections of cells were examined using a JEM-1400 Plus transmission electron microscope (JEOL) operating at 100 kV. The TEM images were analyzed using OriginPro software version 8.0724 (Origin Laboratory). All experiments were further confirmed by biological repeats. The enlarged regions display the detailed structures of mitochondria in cells. Box plot analyzes the relative number of mitochondria (normal/total) in SH-SY5Y cells treated with SOD1 fibril seeds and shows the quantification of TEM images in n = 30 SH-SY5Y cells examined over three independent experiments. The experiments were done blind and average 12 to 16 mitochondria were present in each cell. The boxes (blue) extend from the 25th to 75th percentile (quantiles 1 and 3). Minima, maxima, center, and bounds of box represent quantile 1 − 1.5 × interquartile range, quantile 3 + 1.5 × interquartile range, median (black line), and quantiles 1 and 3, respectively. Bounds of whiskers (minima and maxima) and outliers (open red circles).
Western blotting
For analysis by Western blotting, SH-SY5Y cells were cultured in six-well plates in minimum essential medium for 1 day and then cultured with 0 μM SOD1 fibril seeds, 10 μM wild-type SOD1 fibril seeds, 10 μM H46R fibril seeds, and 10 μM G85R fibril seeds, respectively, for 3 days, with cells cultured in 20 mM tris-HCl buffer (pH 7.4) containing 5 mM TCEP as a negative control. The cells were harvested and resuspended in lysis buffer (pH 7.6) containing 1% Triton X-100, 50 mM tris, 150 mM NaCl, 1 mM phenylmethanesulfonyl fluoride, and protease inhibitors (Beyotime) on ice for half an hour. The cell lysates were centrifuged at 10,000g for 10 min. The supernatants were boiled in SDS-PAGE loading buffer for 15 min, then subjected to 12.5% SDS-PAGE, and probed with the following specific antibodies: rabbit anti-GPX4 monoclonal antibody (Abcam, ab125066; 1:2000) and mouse anti–β-actin (Beyotime, AA128; 1:1000). The amount of loaded protein was normalized using a BCA Protein Quantification kit (Beyotime). For calculating the amounts of GPX4 and β-actin, ImageJ software (National Institutes of Health, Bethesda, MD) was used to assess the densitometry of the corresponding protein bands. The normalized amount of GPX4 in SH-SY5Y cells was calculated as the ratio of the density of GPX4 band over the density of β-actin band in cell lysates and expressed as the means ± SD (with error bars) of values obtained in three independent experiments. Statistical analyses were performed using two-sided Student’s t test. Values of P < 0.05 indicate statistically significant differences. The following notation is used throughout: *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 relative to control. Cells treated with 20 mM tris-HCl buffer (pH 7.4) containing 5 mM TCEP and cells incubated with 10 μM wild-type SOD1 fibril seeds were used as controls, respectively.
Laser scanning confocal analysis
Fibril seeds from wild-type human SOD1 and its variants H46R and G85R were labeled by DyLight 488 using the DyLight 488 Antibody Labeling Kit (Thermo Fisher Scientific, 53024). HEK-293T cells stably expressing FLAG-tagged full-length wild-type human SOD1 were cultured in minimum essential medium for 1 day and then cultured with 0 μM DyLight 488–labeled SOD1 fibril seeds, 10 μM DyLight 488–labeled wild-type SOD1 fibril seeds, 10 μM DyLight 488–labeled H46R fibril seeds, and 10 μM DyLight 488–labeled G85R fibril seeds, respectively, for 1 day at 37°C, with cells cultured in 20 mM tris-HCl buffer (pH 7.4) containing 5 mM TCEP as a negative control. Cells were fixed, permeabilized, stained with DyLight 488 (green), immunostained with the mouse anti-FLAG monoclonal antibody (Sigma-Aldrich, F1804; 1:2000) and a secondary antibody conjugated with Alexa Fluor 647 (red) and stained with 4tained with secondary antib (DAPI) (blue). Images of DyLight 488–labeled SOD1 fibril seed (green) and FLAG-tagged wild-type SOD1 (red) were captured using a Leica TCS SP8 laser scanning confocal microscope (Wetzlar, Germany). The enlarged regions in the bottom right corner of the merged images show enlarged images from the same images.
Sarkosyl-insoluble Western blotting
Sarkosyl-insoluble Western blotting was used to investigate templated misfolding of wild-type SOD1 in cells induced by fibril seeds from H46R and G85R. HEK-293T cells stably expressing FLAG-tagged full-length wild-type human SOD1 were cultured in six-well plates in minimum essential medium for 1 day and then cultured with 0 μM SOD1 fibril seeds, 10 μM wild-type SOD1 fibril seeds, 10 μM H46R fibril seeds, and 10 μM G85R fibril seeds, respectively, for 2 days at 37°C, with cells cultured in 20 mM tris-HCl buffer (pH 7.4) containing 5 mM TCEP as a negative control. The cells were harvested and resuspended in lysis buffer (pH 7.6) containing 1% Triton X-100, 50 mM tris, 150 mM NaCl, 1 mM phenylmethanesulfonyl fluoride, and protease inhibitors (Beyotime) on ice for half an hour. The cell lysates were centrifuged at 10,000g for 10 min. Cell lysates from the above HEK-293T stable cells were centrifuged at 10,000g for 10 min at 4°C to remove the cell debris. Half of the supernatant was incubated with 1% sarkosyl for 30 min at 37°C. The mixture was then ultracentrifuged at 150,000g for 30 min, and the supernatant was carefully removed. The sarkosyl-insoluble pellets were boiled in the SDS-PAGE loading buffer for 15 min. The other half of the supernatant, which served as the total protein sample, was also boiled in the SDS-PAGE loading buffer for 15 min. The samples were separated by 12.5% SDS-PAGE and then Western blotted as follows. The samples were transferred to polyvinylidene difluoride membranes (Millipore). The membranes were blocked with 5% fat-free milk in 25 mM tris-buffered saline buffer containing 0.047% Tween 20. Then, the sarkosyl-insoluble pellets from the above cells were probed with the anti-FLAG antibody, and the corresponding cell lysates were probed using anti-FLAG and anti–β-actin antibodies. The amount of loaded protein was normalized using a BCA Protein Quantification kit (Beyotime). For calculating the amounts of sarkosyl-insoluble SOD1, the ImageJ software (National Institutes of Health, Bethesda, MD) was used to assess the densitometry of SOD1 bands. The relative amount of insoluble SOD1 aggregates in HEK-293T cells stably expressing wild-type SOD1 was determined as a ratio of the density of insoluble SOD1 aggregate bands over that of the total SOD1 bands in cell lysates and expressed as the means ± SD (with error bars) of values obtained in three independent experiments. Statistical analyses were performed using two-sided Student’s t test. Values of P < 0.05 indicate statistically significant differences. The following notation is used throughout: *P < 0.05, **P < 0.01, and ***P < 0.001 relative to control. Cells treated with 20 mM tris-HCl buffer (pH 7.4) containing 5 mM TCEP and cells incubated with 10 μM wild-type SOD1 fibril seeds were used as controls, respectively.
Global denaturation of H46R, G85R, and wild-type fibrils analyzed by thioflavin T fluorescence
Amyloid fibrils were produced from the apo forms of recombinant wild-type SOD1 and its variants H46R and G85R incubated in 20 mM tris-HCl buffer (pH 7.4) containing 5 mM TCEP and shaking at 37°C for 40 to 48 hours. Samples (10 μM) of the SOD1 fibrils were incubated for 1 hour at 25°C in the presence of different concentrations of GdnSCN. The concentration of GdnSCN was then adjusted to 0.35 M, followed by a thioflavin T (ThT) binding assay. A Cytation 3 Cell Imaging Multi-Mode Reader (BioTek) was used to the ThT fluorescence produced, with excitation at 450 nm and emission at 480 nm. The half concentration at which the ThT fluorescence intensity of SOD1 fibrils is decreased by 50% (C1/2) of the H46R fibril, the G85R fibril, and the wild-type fibril was determined using a sigmoidal equation (67, 71) using the above ThT fluorescence data. Statistical analyses were performed using two-sided Student’s t test. Values of P < 0.05 indicate statistically significant differences. The following notation is used throughout: *P < 0.05, **P < 0.01, and ***P < 0.001 relative to the wild-type SOD1 fibril (a control).
Amyloid fibril formation of H46R, G85R, and wild-type SOD1 analyzed by a ThT binding assay
Amyloid fibrils were formed from the apo form of recombinant wild-type SOD1 (30 μM) incubated in 20 mM tris-HCl buffer (pH 7.4) containing 5 mM TCEP and shaking at 37°C for 56 hours, in the absence and presence of 2% (v/v) wild-type SOD1 fibril seeds, 2% (v/v) H46R fibril seeds, or 2% (v/v) G85R fibril seeds, and then analyzed by a ThT binding assay. The final concentrations of wild-type SOD1 and ThT were 1 and 125 μM, respectively. A Cytation 3 Cell Imaging Multi-Mode Reader (BioTek) was used to the ThT fluorescence produced, with excitation at 450 nm and emission at 480 nm. The fibril formation lag time in wild-type SOD1 in the absence and presence of wild-type SOD1 fibril seeds, H46R fibril seeds, or G85R fibril seeds was determined using a sigmoidal equation (67, 71) using the ThT fluorescence data. Statistical analyses were performed using two-sided Student’s t test. Values of P < 0.05 indicate statistically significant differences. The following notation is used throughout: *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 relative to the lag time of wild-type SOD1 alone (a control).
Immunogold electron microscopy of wild-type SOD1 fibrils seeded by H46R fibrils and G85R fibrils
Amyloid fibrils were produced from the apo form of recombinant FLAG-tagged wild-type SOD1 incubated in 20 mM tris-HCl buffer (pH 7.4) containing 5 mM TCEP and shaking at 37°C for 28 hours, in the presence of 2% (v/v) H46R fibril seeds and 2% (v/v) G85R fibril seeds, respectively. Sample aliquots of 10 μl wild-type SOD1 fibrils seeded by H46R fibrils and G85R fibrils were absorbed onto copper grids for 30 s, and then washed two times with water for 30 s. HEK-293T cells stably expressing FLAG-tagged wild-type SOD1 were cultured in six-well plates in minimum essential medium for 1 day and then incubated with 10 μM H46R fibril seeds and 10 μM G85R fibril seeds, respectively, for 2 days at 37°C. The above HEK-293T cells were washed twice with cold PBS and harvested with an extraction buffer containing 10 mM tris-HCl (pH 7.4), 10% sucrose, 0.8 M NaCl, and 1 mM EGTA. Cell extracts were sonicated on ice for 5 min using 30 cycles of 5-s on and 5-s off and centrifuged at 17,000g for 30 min at 4°C to remove the cell debris. The supernatant was incubated with 2% sarkosyl for 1 hour at 37°C. The mixture was then centrifuged at 10,000g for 10 min, the supernatant was ultracentrifuged at 150,000g for 1 hour at 4°C, and the supernatant was carefully removed. The sarkosyl-insoluble pellets were resuspended in the extraction buffer (50 μl) and centrifuged at 3000g for 10 min. The supernatant was then diluted threefold in 50 mM tris-HCl buffer (pH 7.4) containing 0.15 M NaCl, 10% sucrose, and 0.2% sarkosyl, followed by ultracentrifugation at 150,000g for 1 hour at 4°C. The final pellets were resuspended in 20 mM tris-HCl buffer (pH 7.4). Sample aliquots of 10 μl of wild-type SOD1 aggregates purified from HEK-293T cells stably expressing FLAG-tagged wild-type SOD1 seeded by H46R fibrils and G85R fibrils were absorbed onto copper grids for 30 s and then washed two times with water for 30 s. Samples on grids were incubated with rabbit anti-FLAG polyclonal antibody (1:100) for 30 min at room temperature and then blocked with 0.1% BSA in 20 mM tris-HCl buffer (pH 7.4) for 15 min. Ten-nanometer gold-labeled homologous secondary antibody (goat anti-rabbit immunoglobulin G H&L, Abcam, ab27234; 1:20) was used to incubate the grids for 20 min at 37°C. Unbound gold-labeled homologous secondary antibodies were removed by washing with 200 μl of water drop by drop. Samples on grids were then stained with 2% (w/v) uranyl acetate for 1 min. The stained samples were examined using a Talos L120C transmission electron microscope (Thermo Fisher Scientific) operating at 120 kV.
Statistical analysis
The data shown for each experiment were based on at least three technical replicates, as indicated in the individual figure legends. The data are presented as the means ± SD, and P values were determined using a two-sided Student’s t test. Differences were considered statistically significant when P < 0.05. All experiments were further confirmed by biological repeats.
Acknowledgments
Cryo-EM data were collected at the Core Facility of Wuhan University, China. All Cryo-EM data were processed at High Performance Computing Center in Interdisciplinary Research Center on Biology and Chemistry, Chinese Academy of Sciences, and in Wuhan University. We thank T. O’Halloran (Northwestern University) for the gift of the human SOD1 plasmid; D. Li (Cryo-EM Unit, Core Facility of Wuhan University) and X.-N. Li (Cryo-EM Unit, Core Facility of Wuhan University) for excellent assistance with cryo-EM; X.-D. Zhou (AFM Unit, Core Facility of Wuhan University) for technical assistance with AFM; Z. Song and W. Zou (College of Life Sciences, Wuhan University) for technical assistances with the TEM of ultrathin sections of cells; L.-T. Hu (College of Life Sciences, Wuhan University) for technical assistance with TEM; and R. He (College of Pharmacy, Jinan University) and Y. Wang (Institute of Biophysics, Chinese Academy of Sciences) for suggestions.
Funding: Y.L. was supported by the National Natural Science Foundation of China (32271326, 32071212, and 31770833) and the Key Project of Basic Research, Science and Technology R&D Fund of Shenzhen (JCYJ20200109144418639). C.L. was supported by the National Natural Science Foundation of China (82188101 and 32171236), the Science and Technology Commission of Shanghai Municipality (22JC1410400), Shanghai Basic Research Pioneer Project, Shanghai Pilot Program for Basic Research–Chinese Academy of Sciences, Shanghai Branch (CYJ-SHFY-2022-005), and the CAS Project for Young Scientists in Basic Research (YSBR-095). L.-Q.W. was supported by the National Natural Science Foundation of China (32201040) and China Postdoctoral Science Foundation (2021TQ0252 and 2021 M700103). D.L. was supported by the National Natural Science Foundation of China (92353302 and 32170683). W.L. was supported by the National Natural Science Foundation of China (82271524). L.Z. was supported by the Key Project of Basic Research, Science and Technology R&D Fund of Shenzhen (JCYJ20200109144418639).
Author contributions: Conceptualization: Y.L., C.L., L.-Q.W., H.-Y.Y., D.L., L.Z., and W.L. Methodology: Y.L., C.L., L.-Q.W., Y.M., H.-Y.Y., W.X., and L.Z. Investigation: L.-Q.W., Y.M., M.-Y.Z., H.-Y.Y., X.-N.L., K.Z., W.X., J.C., and Y.L. Visualization: L.-Q.W., Y.M., M.-Y.Z., H.-Y.Y., K.Z., L.Z., W.L., C.L., and Y.L. Resources: Y.M., X.H., J.C., L.Z., and Z.W. Validation: L.-Q.W., Y.M., M.-Y.Z., H.-Y.Y., X.-N.L., K.Z., W.X., and Y.L. Formal analysis: L.-Q.W., Y.M., M.-Y.Z., H.-Y.Y., W.X., X.H., L.Z., Z.W., and Y.L. Funding acquisition: Y.L., C.L., L.Z., and W.L. Supervision: Y.L., C.L., L.Z., and W.L. Writing—original draft: Y.L., C.L., L.-Q.W., Y.M., H.-Y.Y., and D.L. Writing—review and editing: Y.L., C.L., Y.M., H.-Y.Y., K.Z., D.L., and Z.W.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Cryo-EM density maps and the atomic models of human H46R SOD1 fibrils and human G85R SOD1 fibrils are available through the Electron Microscopy Data Bank and Protein Data Bank with accession codes EMD-35460, EMD-35459, PDB 8IHV, and PDB 8IHU, respectively.
Supplementary Materials
The PDF file includes:
Table S1
Figs. S1 to S10
Legends for data S1 to S10
Uncropped Western blots
Other Supplementary Material for this manuscript includes the following:
Data S1 to S10
REFERENCES AND NOTES
- 1.Ayers J. I., Borchelt D. R., Phenotypic diversity in ALS and the role of poly-conformational protein misfolding. Acta Neuropathol. 142, 41–55 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shaw B. F., Valentine J. S., How do ALS-associated mutations in superoxide dismutase 1 promote aggregation of the protein? Trends Biochem. Sci. 32, 78–85 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Chattopadhyay M., Valentine J. S., Aggregation of copper-zinc superoxide dismutase in familial and sporadic ALS. Antioxid. Redox Signal. 11, 1603–1614 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cleveland D. W., Rothstein J. D., From Charcot to Lou Gehrig: Deciphering selective motor neuron death in ALS. Nat. Rev. Neurosci. 2, 806–819 (2001). [DOI] [PubMed] [Google Scholar]
- 5.Miller T. M., Cudkowicz M. E., Genge A., Shaw P. J., Sobue G., Bucelli R. C., Chiò A., Damme P. V., Ludolph A. C., Glass J. D., Andrews J. A., Babu S., Benatar M., McDermott C. J., Cochrane T., Chary S., Chew S., Zhu H., Wu F., Nestorov I., Graham D., Sun P., McNeill M., Fanning L., Ferguson T. A., Fradette S., VALOR and OLE Working Group , Trial of antisense oligonucleotide tofersen for SOD1 ALS. N. Engl. J. Med. 387, 1099–1110 (2022). [DOI] [PubMed] [Google Scholar]
- 6.Mead R. J., Shan N., Reiser H. J., Marshall F., Shaw P. J., Amyotrophic lateral sclerosis: A neurodegenerative disorder poised for successful therapeutic translation. Nat. Rev. Drug Discov. 22, 185–212 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang T., Tomas D., Perera N. D., Cuic B., Luikinga S., Viden A., Barton S. K., McLean C. A., Samson A. L., Southon A., Bush A. I., Murphy J. M., Turner B. J., Ferroptosis mediates selective motor neuron death in amyotrophic lateral sclerosis. Cell Death Differ. 29, 1187–1198 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang D., Liang W., Huo D., Wang H., Wang Y., Cong C., Zhang C., Yan S., Gao M., Su X., Tan X., Zhang W., Han L., Zhang D., Feng H., SPY1 inhibits neuronal ferroptosis in amyotrophic lateral sclerosis by reducing lipid peroxidation through regulation of GCH1 and TFR1. Cell Death Differ. 30, 369–382 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dixon S. J., Lemberg K. M., Lamprecht M. R., Skouta R., Zaitsev E. M., Gleason C. E., Patel D. N., Bauer A. J., Cantley A. M., Yang W. S., Morrison B. III, Stockwell B. R., Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 149, 1060–1072 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stockwell B. R., Ferroptosis turns 10: Emerging mechanisms, physiological functions, and therapeutic applications. Cell 185, 2401–2421 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang W. S., SriRamaratnam R., Welsch M. E., Shimada K., Skouta R., Viswanathan V. S., Cheah J. H., Clemons P. A., Shamji A. F., Clish C. B., Brown L. M., Girotti A. W., Cornish V. W., Schreiber S. L., Stockwell B. R., Regulation of ferroptotic cancer cell death by GPX4. Cell 156, 317–331 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Angeli J. P. F., Schneider M., Proneth B., Tyurina Y. Y., Tyurin V. A., Hammond V. J., Herbach N., Aichler M., Walch A., Eggenhofer E., Basavarajappa D., Rådmark O., Kobayashi S., Seibt T., Beck H., Neff F., Esposito I., Wanke R., Förster H., Yefremova O., Heinrichmeyer M., Bornkamm G. W., Geissler E. K., Thomas S. B., Stockwell B. R., O’Donnell V. B., Kagan V. E., Schick J. A., Conrad M., Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat. Cell. Biol. 16, 1180–1191 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tu L.-F., Zhang T.-Z., Zhou Y.-F., Zhou Q.-Q., Gong H.-B., Liang L., Hai L.-N., You N.-X., Su Y., Chen Y.-J., Mo X.-K., Shi C.-Z., Luo L.-P., Sun W.-Y., Duan W.-J., Kurihara H., Li Y.-F., He R.-R., GPX4 deficiency-dependent phospholipid peroxidation drives motor deficits of ALS. J. Adv. Res. 43, 205–218 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y., Lv M. N., Zhao W. J., Research on ferroptosis as a therapeutic target for the treatment of neurodegenerative diseases. Ageing Res. Rev. 91, 102035 (2023). [DOI] [PubMed] [Google Scholar]
- 15.Wang Y., Wu S., Li Q., Sun H., Wang H., Pharmacological inhibition of ferroptosis as a therapeutic target for neurodegenerative diseases and strokes. Adv. Sci. 10, e2300325 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valentine J. S., Doucette P. A., Zittin Potter S., Copper-zinc superoxide dismutase and amyotrophic lateral sclerosis. Annu. Rev. Biochem. 74, 563–593 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Polymenidou M., Cleveland D. W., The seeds of neurodegeneration: Prion-like spreading in ALS. Cell 147, 498–508 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ajroud-Driss S., Siddique T., Sporadic and hereditary amyotrophic lateral sclerosis (ALS). Biochim. Biophys. Acta 1852, 679–684 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Rosen D. R., Siddique T., Patterson D., Figlewicz D. A., Sapp P., Hentati A., Donaldson D., Goto J., O'Regan J. P., Deng H. X., Rahmani Z., Krizus A., McKenna-Yasek D., Cayabyab A., Gaston S. M., Berger R., Tanzi R. E., Halperin J. J., Herzfeldt B., den Bergh R. V., Hung W. Y., Bird T., Deng G., Mulder D. W., Smyth C., Laing N. G., Soriano E., Pericak–Vance M. A., Haines J., Rouleau G. A., Gusella J. S., Horvitz H. R., Brown R. H. Jr., Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 362, 59–62 (1993). [DOI] [PubMed] [Google Scholar]
- 20.Tiwari A., Xu Z., Hayward L. J., Aberrantly increased hydrophobicity shared by mutants of Cu,Zn-superoxide dismutase in familial amyotrophic lateral sclerosis. J. Biol. Chem. 280, 29771–29779 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Tiwari A., Hayward L. J., Familial amyotrophic lateral sclerosis mutants of copper/zinc superoxide dismutase are susceptible to disulfide reduction. J. Biol. Chem. 278, 5984–5992 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Furukawa Y., Kaneko K., Yamanaka K., O’Halloran T. V., Nukina N., Complete loss of post-translational modifications triggers fibrillar aggregation of SOD1 in the familial form of amyotrophic lateral sclerosis. J. Biol. Chem. 283, 24167–24176 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jonsson P. A., Graffmo K. S., Andersen P. M., Brännström T., Lindberg M., Oliveberg M., Marklund S. L., Disulphide-reduced superoxide dismutase-1 in CNS of transgenic amyotrophic lateral sclerosis models. Brain 129, 451–464 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Shaw B. F., Lelie H. L., Durazo A., Nersissian A. M., Xu G., Chan P. K., Gralla E. B., Tiwari A., Hayward L. J., Borchelt D. R., Valentine J. S., Whitelegge J. P., Detergent-insoluble aggregates associated with amyotrophic lateral sclerosis in transgenic mice contain primarily full-length, unmodified superoxide dismutase-1. J. Biol. Chem. 283, 8340–8350 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Furukawa Y., Kaneko K., Yamanaka K., Nukina N., Mutation-dependent polymorphism of Cu,Zn-superoxide dismutase aggregates in the familial form of amyotrophic lateral sclerosis. J. Biol. Chem. 285, 22221–22231 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan P. K., Chattopadhyay M., Sharma S., Souda P., Gralla E. B., Borchelt D. R., Whitelegge J. P., Valentine J. S., Structural similarity of wild-type and ALS-mutant superoxide dismutase-1 fibrils using limited proteolysis and atomic force microscopy. Proc. Natl. Acad. Sci. U.S.A. 110, 10934–10939 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arisato T., Okubo R., Arata H., Abe K., Fukada K., Sakoda S., Shimizu A., Qin X. H., Izumo S., Osame M., Nakagawa M., Clinical and pathological studies of familial amyotrophic lateral sclerosis (FALS) with SOD1 H46R mutation in large Japanese families. Acta Neuropathol. 106, 561–568 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Cudkowicz M. E., McKenna-Yasek D., Sapp P. E., Chin W., Geller B., Hayden D. L., Schoenfeld D. A., Hosler B. A., Horvitz H. R., Brown R. H., Epidemiology of mutations in superoxide dismutase in amyotrophic lateral sclerosis. Ann. Neurol. 41, 210–221 (1997). [DOI] [PubMed] [Google Scholar]
- 29.Prudencio M., Hart P. J., Borchelt D. R., Andersen P. M., Variation in aggregation propensities among ALS-associated variants of SOD1: Correlation to human disease. Hum. Mol. Genet. 18, 3217–3226 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ayers J., Lelie H., Workman A., Prudencio M., Brown H., Fromholt S., Valentine J., Whitelegge J., Borchelt D., Distinctive features of the D101N and D101G variants of superoxide dismutase 1; two mutations that produce rapidly progressing motor neuron disease. J. Neurochem. 128, 305–314 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodriguez J. A., Valentine J. S., Eggers D. K., Roe J. A., Tiwari A., Brown R. H. Jr., Hayward L. J., Familial amyotrophic lateral sclerosis-associated mutations decrease the thermal stability of distinctly metallated species of human copper/zinc superoxide dismutase. J. Biol. Chem. 277, 15932–15937 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Wang Q., Johnson J. L., Agar N. Y. R., Agar J. N., Protein aggregation and protein instability govern familial amyotrophic lateral sclerosis patient survival. PLOS Biol. 6, e170 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ripps M. E., Huntley G. W., Hof P. R., Morrison J. H., Gordon J. W., Transgenic mice expressing an altered murine superoxide dismutase gene provide an animal model of amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. U.S.A. 92, 689–693 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bruijn L. I., Becher M. W., Lee M. K., Anderson K. L., Jenkins N. A., Copeland N. G., Sisodia S. S., Rothstein J. D., Borchelt D. R., Price D. L., Cleveland D. W., ALS-linked SOD1 mutant G85R mediates damage to astrocytes and promotes rapidly progressive disease with SOD1-containing inclusions. Neuron 18, 327–338 (1997). [DOI] [PubMed] [Google Scholar]
- 35.Nagano S., Takahashi Y., Yamamoto K., Masutani H., Fujiwara N., Urushitani M., Araki T., A cysteine residue affects the conformational state and neuronal toxicity of mutant SOD1 in mice: Relevance to the pathogenesis of ALS. Hum. Mol. Genet. 24, 3427–3439 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Münch C., O’Brien J., Bertolotti A., Prion-like propagation of mutant superoxide dismutase-1 misfolding in neuronal cells. Proc. Natl. Acad. Sci. U.S.A. 108, 3548–3553 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ayers J. I., Fromholt S. E., O'Neal V. M., Diamond J. H., Borchelt D. R., Prion-like propagation of mutant SOD1 misfolding and motor neuron disease spread along neuroanatomical pathways. Acta Neuropathol. 131, 103–114 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sekhar A., Rumfeldt J. A. O., Broom H. R., Doyle C. M., Sobering R. E., Meiering E. M., Kay L. E., Probing the free energy landscapes of ALS disease mutants of SOD1 by NMR spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 113, E6939–E6945 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sekhar A., Kay L. E., An NMR view of protein dynamics in health and disease. Annu. Rev. Biophys. 48, 297–319 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Bergh J., Bergh J., Zetterström P., Andersen P. M., Brännström T., Graffmo K. S., Jonsson P. A., Lang L., Danielsson J., Oliveberg M., Marklund S. L., Structural and kinetic analysis of protein-aggregate strains in vivo using binary epitope mapping. Proc. Natl. Acad. Sci. U.S.A. 112, 4489–4494 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bidhendi E. E., Bergh J., Zetterström P., Andersen P. M., Marklund S. L., Brännström T., Two superoxide dismutase prion strains transmit amyotrophic lateral sclerosis-like disease. J. Clin. Invest. 126, 2249–2253 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bruijn L. I., Houseweart M. K., Kato S., Anderson K. L., Anderson S. D., Ohama E., Reaume A. G., Scott R. W., Cleveland D. W., Aggregation and motor neuron toxicity of an ALS-linked SOD1 mutant independent from wild-type SOD1. Science 281, 1851–1854 (1998). [DOI] [PubMed] [Google Scholar]
- 43.Wang L. Q., Ma Y., Yuan H. Y., Zhao K., Zhang M. Y., Wang Q., Huang X., Xu W. C., Dai B., Chen J., Li D., Zhang D., Wang Z., Zou L., Yin P., Liu C., Liang Y., Cryo-EM structure of an amyloid fibril formed by full-length human SOD1 reveals its conformational conversion. Nat. Commun. 13, 3491 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iwakawa N., Morimoto D., Walinda E., Shirakawa M., Sugase K., Multiple-state monitoring of SOD1 amyloid formation at single-residue resolution by Rheo-NMR spectroscopy. J. Am. Chem. Soc. 143, 10604–10613 (2021). [DOI] [PubMed] [Google Scholar]
- 45.Sangwan S., Sawaya M. R., Murray K. A., Hughes M. P., Eisenberg D. S., Atomic structures of corkscrew-forming segments of SOD1 reveal varied oligomer conformations. Protein Sci. 27, 1231–1242 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sangwan S., Zhao A., Adams K. L., Jayson C. K., Sawaya M. R., Guenther E. L., Pan A. C., Ngo J., Moore D. M., Soriaga A. B., Do T. D., Goldschmidt L., Nelson R., Bowers M. T., Koehler C. M., Shaw D. E., Novitch B. G., Eisenberg D. S., Atomic structure of a toxic, oligomeric segment of SOD1 linked to amyotrophic lateral sclerosis (ALS). Proc. Natl. Acad. Sci. U.S.A. 114, 8770–8775 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ivanova M. I., Sievers S. A., Guenther E. L., Johnson L. M., Winkler D. D., Galaleldeen A., Sawaya M. R., Hart P. J., Eisenberg D. S., Aggregation-triggering segments of SOD1 fibril formation support a common pathway for familial and sporadic ALS. Proc. Natl. Acad. Sci. U.S.A. 111, 197–201 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sharma K., Stockert F., Shenoy J., Berbon M., Abdul-Shukkoor M. B., Habenstein B., Loquet A., Schmidt M., Fändrich M., Cryo-EM observation of the amyloid key structure of polymorphic TDP-43 amyloid fibrils. Nat. Commun. 15, 486 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arseni D., Hasegawa M., Murzin A. G., Kametani F., Arai M., Yoshida M., Ryskeldi-Falcon B., Structure of pathological TDP-43 filaments from ALS with FTLD. Nature 601, 139–143 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arseni D., Chen R., Murzin A. G., Peak-Chew S. Y., Garringer H. J., Newell K. L., Kametani F., Robinson A. C., Vidal R., Ghetti B., Hasegawa M., Ryskeldi-Falcon B., TDP-43 forms amyloid filaments with a distinct fold in type A FTLD-TDP. Nature 620, 898–903 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ayers J. I., Diamond J., Sari A., Fromholt S., Galaleldeen A., Ostrow L. W., Glass J. D., Hart P. J., Borchelt D. R., Distinct conformers of transmissible misfolded SOD1 distinguish human SOD1-FALS from other forms of familial and sporadic ALS. Acta Neuropathol. 132, 827–840 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ayers J. I., Xu G., Dillon K., Lu Q., Chen Z., Beckman J., Moreno-Romero A. K., Zamora D. L., Galaleldeen A., Borchelt D. R., Variation in the vulnerability of mice expressing human superoxide dismutase 1 to prion-like seeding: A study of the influence of primary amino acid sequence. Acta Neuropathol. Commun. 9, 92 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scheres S. H. W., Amyloid structure determination in RELION-3.1. Acta Crystallogr. D Struct. Biol. 76 ( Pt 2), 94–101 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang L.-Q., Zhao K., Yuan H.-Y., Li X.-N., Dang H.-B., Ma Y., Wang Q., Wang C., Sun Y., Chen J., Li D., Zhang D., Yin P., Liu C., Liang Y., Genetic prion disease-related mutation E196K displays a novel amyloid fibril structure revealed by cryo-EM. Sci. Adv. 7, eabg9676 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Salehi M., Nikkhah M., Ghasemi A., Arab S. S., Mitochondrial membrane disruption by aggregation products of ALS-causing superoxide dismutase-1 mutants. Int. J. Biol. Macromol. 75, 290–297 (2015). [DOI] [PubMed] [Google Scholar]
- 56.Xie Y., Hou W., Song X., Yu Y., Huang J., Sun X., Kang R., Tang D., Ferroptosis: Process and function. Cell Death Differ. 23, 369–379 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin Z.-H., Liu Y., Xue N.-J., Zheng R., Yan Y.-Q., Wang Z.-X., Li Y.-L., Ying C.-Z., Song Z., Tian J., Pu J.-L., Zhang B.-R., Quercetin protects against MPP+/MPTP-induced dopaminergic neuron death in Parkinson’s disease by inhibiting ferroptosis. Oxid. Med. Cell. Longev. 2022, 7769355 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Arnold F. J., Nguyen A. D., Bedlack R. S., Bennett C. L., La Spada A. R., Intercellular transmission of pathogenic proteins in ALS: Exploring the pathogenic wave. Neurobiol. Dis. 184, 106218 (2023). [DOI] [PubMed] [Google Scholar]
- 59.Forsberg K. M., Graffmo K. S., Stenvall E., Tabikh N., Marklund S. L., Brännström T., Andersen P. M., Widespread CNS pathology in amyotrophic lateral sclerosis homozygous for the D90A SOD1 mutation. Acta Neuropathol. 145, 13–28 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang J., Xu G., Gonzales V., Coonfield M., Fromholt D., Copeland N. G., Jenkins N. A., Borchelt D. R., Fibrillar inclusions and motor neuron degeneration in transgenic mice expressing superoxide dismutase 1 with a disrupted copper-binding site. Neurobiol. Dis. 10, 128–138 (2002). [DOI] [PubMed] [Google Scholar]
- 61.Strange R. W., Antonyuk S., Hough M. A., Doucette P. A., Rodriguez J. A., Hart P. J., Hayward L. J., Valentine J. S., Hasnain S. S., The structure of holo and metal-deficient wild-type human Cu,Zn superoxide dismutase and its relevance to familial amyotrophic lateral sclerosis. J. Mol. Biol. 328, 877–891 (2003). [DOI] [PubMed] [Google Scholar]
- 62.Elam J. S., Taylor A. B., Strange R., Antonyuk S., Doucette P. A., Rodriguez J. A., Hasnain S. S., Hayward L. J., Valentine J. S., Yeates T. O., Hart P. J., Amyloid-like filaments and water-filled nanotubes formed by SOD1 mutant proteins linked to familial ALS. Nat. Struct. Biol. 10, 461–467 (2003). [DOI] [PubMed] [Google Scholar]
- 63.Sala F. A., Wright G. S. A., Antonyuk S. V., Garratt R. C., Hasnain S. S., Molecular recognition and maturation of SOD1 by its evolutionarily destabilised cognate chaperone hCCS. PLoS Biol. 17, e3000141 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Luchinat E., Banci L., In-cell NMR in human cells: Direct protein expression allows structural studies of protein folding and maturation. Acc. Chem. Res. 51, 1550–1557 (2018). [DOI] [PubMed] [Google Scholar]
- 65.Bourassa M. W., Brown H. H., Borchelt D. R., Vogt S., Miller L. M., Metal-deficient aggregates and diminished copper found in cells expressing SOD1 mutations that cause ALS. Front. Aging Neurosci. 6, 110 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zetterström P., Stewart H. G., Bergemalm D., Jonsson P. A., Graffmo K. S., Andersen P. M., Brännström T., Oliveberg M., Marklund S. L., Soluble misfolded subfractions of mutant superoxide dismutase-1s are enriched in spinal cords throughout life in murine ALS models. Proc. Natl. Acad. Sci. U.S.A. 104, 14157–14162 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chattopadhyay M., Durazo A., Sohn S. H., Strong C. D., Gralla E. B., Whitelegge J. P., Valentine J. S., Initiation and elongation in fibrillation of ALS-linked superoxide dismutase. Proc. Natl. Acad. Sci. U.S.A. 105, 18663–18668 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu W.-C., Liang J.-Z., Li C., He Z.-X., Yuan H.-Y., Huang B.-Y., Liu X.-L., Tang B., Pang D.-W., Du H.-N., Yang Y., Chen J., Wang L., Zhang M., Liang Y., Pathological hydrogen peroxide triggers the fibrillization of wild-type SOD1 via sulfenic acid modification of Cys-111. Cell Death Dis. 9, 67 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66 ( Pt 4), 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mo Z. Y., Zhu Y. Z., Zhu H. L., Fan J. B., Chen J., Liang Y., Low micromolar zinc accelerates the fibrillization of human Tau via bridging of Cys-291 and Cys-322. J. Biol. Chem. 284, 34648–34657 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1
Figs. S1 to S10
Legends for data S1 to S10
Uncropped Western blots
Data S1 to S10