Abstract
The mechanisms involved in the normal developmental regulation of globin gene expression, and the response to pharmacological agents that elevate fetal hemoglobin, may be expected to involve either changes in each cell or a selection process affecting subsets of differentiating erythroid cells. To study these mechanisms we have developed assays to measure mRNA levels in single erythroid cells. The assay involved the use of globin-specific probes, with no detectable cross-reactivity, in real-time, fluorescence-based quantitative PCR (Q-PCR). We had previously used this Q-PCR method to measure globin mRNA levels in cultures of primary erythroid cells demonstrating that drugs like hydroxyurea, 5-azacytidine and butyric acid each yielded increases in γ/( γ + β) mRNA ratios, with differential effects on β-globin levels. We have now extended this approach to measure globin mRNA levels in single K562 cells, a human erythroleukemic cell line, with and without 30 µM hemin treatment. Hemin exposure increases total hemoglobin levels by ~9-fold and total α-, ɛ- and γ-globin mRNA levels by 1.5–2.3-fold. Single cell analyses showed initial wide distributions of each of the three individual globin mRNA levels with most cells having detectable but very low levels of each globin transcript. Hemin induction shifted the distributions to higher levels, with a tendency to residual left skewing as some cells remained with very low expression levels despite the effect of hemin in increasing expression in most of these low expressing cells. Thus transcriptional heterogeneity remains a crucial variable, even in this extensively used model of human erythroid biology, and clearly influences strongly the response to inducing agents. These methods may enable us to define better possible molecular and/or cellular models of globin gene modulation.
INTRODUCTION
The developmental (1), as well as the pharmacological (2), regulation of globin gene expression have been the subject of considerable investigation. The ontogenetic changes in the expression of globin genes during development, from embryonic to fetal to adult types, have long been studied as a model of temporally-controlled gene regulation. Pharmacologic modulation of globin gene expression is the basis of therapeutic agents for the treatment of certain genetic diseases of hemoglobin such as thalassemias and sickle cell disease (3,4) by increasing fetal hemoglobin (HbF, α2γ2), which can replace the impaired adult hemoglobins. The precise molecular and cellular mechanism(s) involved in the pharmacologic modulation of γ-globin gene expression by clinically useful drugs, such as 5-azacytidine, hydroxyurea and butyric acid derivatives, are as yet unclear.
The mechanisms of these pharmacologic agents are expected to involve either a general molecular process in which all erythroid cells yield a response (transcriptional and/or translational) or a selective one in which only subsets of differentiating cells are selected by the agents as responders. The latter mechanism can either be stochastic or reflect pre-existing heterogeneity in the target erythroid cell populations. Distinguishing between these possible cellular mechanisms would be best addressed by an assay system capable of determining the level of globin gene(s) expressed in single erythroid cells. The limited quantities of nucleic acid material present in a single cell, however, places constraints on method(s) suitable for such analyses. The power of nucleic acid amplification methods (e.g. PCR) has previously been harnessed for methods with similarly limiting quantities of template material such as pre-implantation diagnosis from single cells for in vitro fertilization (5), cDNA library construction (6), gene expression analysis in single live neurons (7) as well as for profiling gene expression in single precursor cells prior to hematopoietic lineage commitment (8,9). However, these previous investigations have been qualitative rather than quantitative in nature and were designed for the most part to demonstrate the presence or absence of specific mutations or markers.
Recently, we reported the use of a fluorescence-based, real-time quantitative PCR (Q-PCR) method in the analysis of globin gene expression in primary erythroid cells in culture (10). The assay involves the inclusion of a dual fluorescently-labeled oligonucleotide probe within the PCR with the 5′ nuclease activity of Taq polymerase producing one active fluorophore for each new strand of cDNA synthesized during PCR (11). With this method, quantitation is possible over a wide range of starting template concentrations since the fluorescence intensity is directly proportional to the quantity of amplified copies. Assay of very low copy number templates has been demonstrated with this procedure (12) and may be suitable for the purpose of assaying the levels of transcript present in single cells. The ability to accurately determine levels of messenger RNA expression of various genes in single cells should clarify the mechanism by which an enhancer increases the level of expression of a cloned transgene. Two models of enhancer action may be considered, one in which the rate of transcription for each cell in a population is increased in response to enhancer action versus one in which the transgene increases the number of expressing cells within that population (13,14). The end result in either case is an increase in gene expression for the cell population as a whole. In addition, similar ‘all versus some’ situations in which gene expression is affected by environmental conditions such as drug-induced changes in gene expression or commitment to differentiation should also be amenable to investigation using such techniques.
The K562 erythroleukemia cell line has previously been shown to be Philadelphia-chromosome-positive (15) and the expression of both embryonic [Hb Gower (α2ɛ2) and Hb Portland (ζ2ɛ2)] and fetal (HbF) hemoglobins is markedly increased after exposure to hemin (16,17). This cell line, and its response to hemin and other globin gene inducers, has been used extensively as a model system for the investigation of human globin gene regulation for its relatively homogeneous characteristics and the reversibility of induction.
We report here the assay of α-, γ- and ɛ-globin levels in single K562 cells in response to 30 µM hemin induction. Single K562 cells were isolated by limiting dilution after 48 h exposure to hemin and transcript levels assayed; the results and their logarithmic transformation are presented, as are statistical analyses of these data. We find that the response to hemin is a variegated one and does not involve each and every cell; rather, cells at the lower end of the expression spectrum for each gene seem disproportionately affected by the induction process. This methodology is now being applied to the assay of changes in globin message expression in single primary human erythroid cells in response to clinically relevant drugs such as hydroxyurea. It is anticipated that results from such studies will enable us to define possible molecular and/or cellular models of pharmacologically-induced globin gene modulation.
MATERIALS AND METHODS
Cell culture
Erythroleukemic K562 cells were obtained from ATCC (Manassas, VA) and grown in RPMI 1640 culture medium supplemented with 10% fetal bovine serum, 100 U/ml penicillin and 100 µg/ml streptomycin (Biofluids, Rockville, MD). Cultures were incubated at 37°C in an atmosphere of 5% CO2 in air with extra humidity. Hemin was obtained from Sigma (St Louis, MO) and a 5 mM stock solution prepared as previously described (18). Briefly, 32.5 mg bovine hemin was dissolved in 0.5 ml 1 M NaOH for 30 min, mixed with 0.5 ml of 0.5 M Tris base followed by 10 ml of 10% bovine serum albumin. The mixture was neutralized by addition of 0.5 ml of 1 M HCl, the pH verified and then filtered through a 0.45 µm filter. The solution was stored at 4°C for no more than 1 week before use. Hemin was added to K562 cultures at a final concentration of 30 µM after which they were returned to the incubator. Total RNA was extracted from ~106 cells using the RNeasy mini kit (Qiagen, Santa Clarita, CA) and quantified by spectrophotometry. Single K562 cells were isolated by limiting dilution in serum-free medium to ~1 cell/µl and dispensing 1 µl aliquots into individual wells of a 72-well Terasaki plate. Wells containing single cells were identified by visual inspection under an inverted microscope and processed for Q-PCR analysis.
TaqMan assay
Single cells suspended in 1 µl serum-free medium were lysed by addition of 4 µl lysis cocktail containing 1% Nonidet P-40 (Sigma), 6 mM dithiothreitol (DTT) and 2 U/µl RNasin (Promega, Madison, WI). The lysate was mixed by pipetting and transferred to a 0.2 ml reaction tube and 15 µl enzyme mix added. The enzyme mix consisted of 4 µl of 5× RT buffer, 2 µl 0.1 M DTT, 1 µl 10 mM dNTP mix, 0.5 µg oligo(dT)24 and 200 U Superscript II (Life Technologies, Gaithersburg, MD) in DEPC H2O. The tubes were incubated at 42°C for 60 min followed by 10 min at 80°C to inactivate the enzyme.
Quantitative real-time PCR assay of transcripts was carried out using gene-specific double fluorescently-labeled probes in a 7700 Sequence Detector (PE Biosystems, Foster City, CA). All probes are designed to span exon junctions in the fully processed message in order to prevent reporting of amplification of any possible contaminating genomic DNA. All primers and probes were made using reagents from Glen Research (Chantilly, VA) on an ABI 394 synthesizer (PE Biosystems). 6-Carboxy fluorescein (FAM) was used as the 5′ fluorescent reporter while tetramethylrhodamine (TAMRA) was added to the 3′ end as quencher. Probe and primers used for quantitation of γ-globin have been previously reported (10). In addition, the following primers and probes for α- and ɛ-globins were synthesized and used to quantify their respective message levels. α-globin forward primer: 5′-CTCTTCTGGTCCCCACAGACT-3′, α-globin reverse primer: 5′-GGCCTTGACGTTGGTCTTG-3′, α-globin probe: 5′-FAM-ACCATG-GTGCTGTCTCCTGCCGTAMRA-3′, ɛ-globin forward primer: 5′-CAAG-CCCGCCTTTGCTAA-3′, ɛ-globin reverse primer: 5′-TTGCCAAAGTGAGTAGCCAGAA-3′, ɛ-globin probe: 5′-FAMACTTCAACTCCTGGGTAACGTGATGGTGATTATTAMRA-3′. Double fluorescently-labeled probes were HPLC purified on a 250 × 10 mm Biovantage C8 reverse phase column (Thomson, Chantilly, VA) using a 10–35% acetonitrile gradient in 0.1 M triethylamine acetate pH 7 with the detector set to monitor column eluate at 260, 494 and 565 nm. Products were lyophilized and characterized by absorption spectroscopy as well as DNase I treatment to confirm a ≥2-fold increase in fluorescence. All oligonucleotide primers and probes were quantitated by absorbance at 260 nm. Specific transcripts within cDNA reaction products were quantitated in a reaction mix consisting of 10 mM Tris pH 8.3, 50 mM KCl, 3 mM MgCl2, 200 µM dNTP (dTTP replaced with dUTP at 400 µM), 0.01 U/µl uracil DNA glycosylase (Roche Molecular) and 0.025 U/µl Platinum Taq polymerase (Life Technologies Inc). Standard curves were constructed using dilutions of an accurately determined plasmid containing the cDNA of interest as template. A dynamic range of five log-orders of concentration or greater was routinely achieved for each transcript of interest.
Hemoglobin spectroscopy
Cells were pelleted from the culture medium by centrifugation, washed in DPBS then suspended and lysed in sterile distilled water at 0°C for 15 min. After pelleting the debris for 10 min at 4°C (10 000 g), the supernatant was collected and absorption spectra from 300 to 600 nm acquired using a Hewlett Packard 8452 diode array spectrophotometer. Protein concentration was determined using bicinchoninic acid reagent (absorbance read at 562 nm) with a bovine serum albumin standard (Pierce, Rockford, IL). The change in absorbance at 413 nm (wavelength of maximum absorption) for each lysate was normalized to the protein concentration yielding a specific measure of hemoglobin content.
Statistical analyses
All statistical analyses were performed using GraphPad Prism version 3 (GraphPad Software, San Diego, CA) except for skew values, which were calculated with StatView version 4 and the significance levels for skewness with Mathematica version 4. Log transformed absolute expression levels for single cells were tested for deviation from normality using the Dallal and Wilkinson implementation of Lilliefors’ test (19).
RESULTS AND DISCUSSION
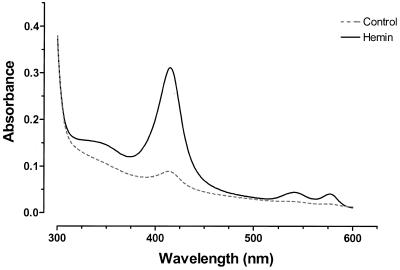
The change in the level of hemoglobin in K562 cells cultured with or without 30 µM hemin for 48 h is shown in Figure 1. After normalizing for total protein content there was an 8.7-fold increase in hemoglobin content of treated K562 cells as reflected in the change in absorbance at 413 nm/mg protein content (ΔA413/mg total protein). Increased levels of mRNA for all three (α-, γ- and ɛ-) globins assayed were also found in total RNA from hemin-treated cells (Table 1). In comparison with the increase observed for hemoglobin (protein) levels, more modest increases were observed for the mRNA levels (1.5–2.3-fold). These findings are in agreement with earlier findings regarding induction of globin gene expression by hemin in K562 cells (16,20). It is evident however that such assays yield very little information about the breadth of individual cellular responses to hemin induction. Indeed, whether the observed increases are the result of a uniform cellular response or that of a subset of cells to hemin induction is not known.
Figure 1.
Absorption spectroscopy of K562 cell lysates. Spectra of lysates from control (dashed line) and cells treated with 30 µM hemin for 48 h (solid line) are shown from 300 to 625 nm. The change in absorbance at 413 nm normalized to the protein concentration (ΔA413/mg total protein) was determined and used as an indicator of hemoglobin expression level.
Table 1. Levels of globin mRNAs in control and hemin treated K562 cells: pooled and single cell determinations.
| amol globin mRNA/µg total RNA | mean amol globin mRNA/cell | |||
|---|---|---|---|---|
| Globin message | Control | Hemin treated | Control | Hemin treated |
| α-globin |
1100 ± 110 |
2400 ± 230 |
0.017 ± 0.021 (1 × 104)a |
0.052 ± 0.058 (3.1 × 104) |
| γ-globin |
6300 ± 660 |
9300 ± 480 |
1.4 ± 1.6 (8.4 × 105) |
3.7 ± 2.4 (2.2 × 106) |
| ɛ-globin | 120 ± 6 | 270 ± 16 | 0.0015 ± 0.0025 (9 × 102) | 0.0062 ± 0.0046 (3.7 × 103) |
aCopy number.
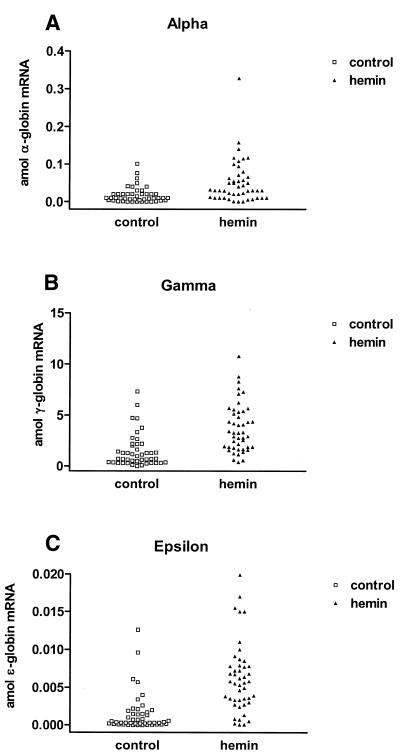
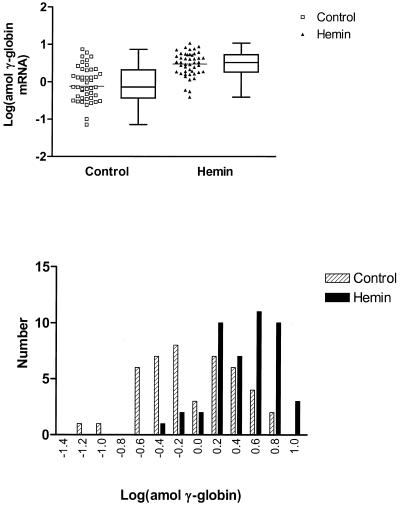
To test these alternatives, we measured mRNA levels in each of 46 K562 cells with and without hemin induction. Figures 2–5 present these data showing responses in single K562 cells for levels of α-, γ- and ɛ-globin genes, respectively. Figure 2 shows the untransformed data while in Figures 3–5 the data are presented after logarithmic transformation in order to minimize deviations in the distributions of results from normality. The results of t-tests as well as F-tests are reported at 95% confidence intervals for the transformed data to compare differences in means and variances, respectively.
Figure 2.
Plots of transcript levels for control (squares) and 30 µM hemin treated (triangles) K562 cells. Data for α-globin (A), γ-globin (B) and ɛ-globin (C) are shown. Mean values for globin transcript levels were as follows: 1.4 ± 1.6 amol/cell γ-globin, 0.017 ± 0.021 amol/cell α-globin and 0.0015 ± 0.0025 amol/cell ɛ-globin for control cells increasing to 3.7 ± 2.4 amol/cell γ-globin, 0.052 ± 0.058 amol/cell α-globin and 0.0062 ± 0.0046 amol/cell ɛ-globin respectively for hemin-treated cells (mean ± standard deviation).
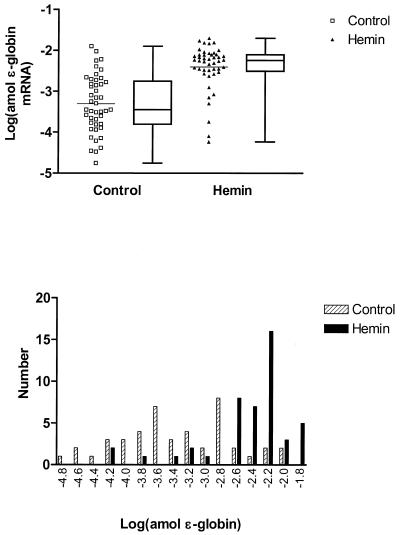
Figure 5.
Logarithmic transformation of ɛ-globin transcript levels in single K562 cells. The upper panel presents plots of transformed data for single-cell expression in control (squares) and hemin-treated (triangles) K562 cells in which the mean (bars) was increased from –3.30 to –2.40 (P < 0.0001, t-test), respectively, while the variance was decreased from 0.49 to 0.31 (P = 0.06, F-test). Box and whisker plots are presented alongside the scatter plots showing the 25th and 75th percentile and median values for transcript level. Frequency histograms of the transformed data are shown in the lower panel with controls shown as hatched columns and hemin-treated cells as solid columns. The skew in the distributions for the transformed data for control and treated cells is 0.005 (P = 0.988) and –1.781 (P < 0.001) respectively.
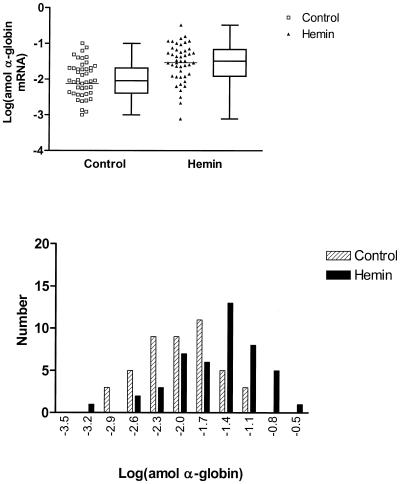
Figure 3.
Logarithmic transformation of α-globin transcript levels in single K562 cells. The upper panel presents plots of transformed data for single-cell expression in control (squares) and hemin-treated (triangles) K562 cells. The bars represent the mean values for logarithm of the globin level which increased from –2.13 to –1.54 (P = 0.0004, t-test), respectively. Concomitantly, the variance declined from 0.90 to 0.28 (P < 0.0001, F-test). Box and whisker plots are presented alongside the scatter plots showing the 25th and 75th percentile and median values for transcript level. Frequency histograms of the transformed data are shown in the lower panel with controls shown as hatched columns and hemin-treated cells as solid columns. The skew in the distributions for the transformed data for control and treated cells is 0.001 (P = 1.00) and –0.690 (P = 0.041), respectively.
The untransformed data presented in Figure 2 summarize changes at the single-cell level for α-, γ- and ɛ-globin transcripts in single K562 cells treated with hemin. The mean transcript levels for both the control and hemin-treated cells were in the order γ > α > ɛ, an analogous order of abundance is apparent in the cDNAs from total RNA isolated from ~106 cells (Table 1). The mean message levels for all three globin transcripts in single cells were increased after treatment and are summarized in Table 1. Control cells had mean transcript levels (expressed in attomoles, 10–18 mol) of 1.4 ± 1.6 amol/cell γ-globin, 0.017 ± 0.021 amol/cell α-globin and 0.0015 ± 0.0025 amol/cell ɛ-globin which increased to 3.7 ± 2.4 amol/cell γ-globin, 0.052 ± 0.058 amol/cell α-globin and 0.0062 ± 0.0046 amol/cell ɛ-globin, respectively (mean ± standard deviation). Translated to copy number, these levels correspond to 900 (ɛ-globin), 104 (α-globin) and 8.4 × 105 (γ-globin) copies per cell for controls versus 3.7 × 103 (ɛ-globin), 3.1 × 104 (α-globin) and 2.2 × 106 (γ-globin) copies per cell after hemin treatment. Interestingly, the data reveal considerable diversity in the values of each transcript both in the control and treated cell populations.
It will be noted that hemin induction increases the percentage of cells expressing larger amounts of each globin message but that a significant number of cells always remain at very low values; that is, a sub-population of K562 cells exhibit considerable resistance to hemin-mediated induction of globin gene expression. These very low values are higher than zero, as shown by the zero values obtained when no cells are added to the reaction mixture (data not shown). Also, as with values for globin message/µg of total RNA (Table 1) γ-globin is by far the predominant globin mRNA species present, with an appreciable α/non-α transcript imbalance (although we do not have data for ζ and δ). Previous reports based on isoelectric focusing and cellulose acetate electrophoresis have also indicated considerable α/non-α hemoglobin chain imbalance in K562 cells, analogous to that observed for the mRNA levels in the present study (21).
The Dallal and Wilkinson implementation of Lilliefors’ test (19) for deviation from (Gaussian) normality shows that for each message, the control population is probably not representative of a normal distribution, emphasizing the necessity for transformations which could normalize or render the data symmetrical for statistical analyses. The upper panel in Figure 3 shows the result of a log transformation of individual α-globin levels in both control and hemin-treated cells, while the resulting frequency distribution histogram is shown in the lower panel. The initial symmetric distribution becomes skewed to the left (lower values) after induction, suggesting that the effect of hemin is not uniform on each cell, i.e. some cells appear to be non-responders. The P values from t- and F-tests reveal significant differences both in the means and variances between control and hemin-treated cells (Table 2). Similar results are obtained for the means and variances of the log-transformed γ-globin levels (Fig. 4). Again, the initial symmetry is replaced by a left-skewing after induction. In the case of similarly transformed ɛ-globin levels (Fig. 5), the same changes in skewing are apparent but the residual left skewing is greatest and most apparent; the means but not the variances were found to be significantly different. The values for skewing are reported in Table 2.
Table 2. Summary of statistical parameters for globin mRNA levels in single cells.
| Globin message | Mean | S.D.a | S.E.M.b | Variance | Skew |
|---|---|---|---|---|---|
| α-globin (control) | –2.006 | 0.488 | 0.072 | 0.238 | 0.001 (P = 1.000) |
| α-globin (hemin) | –1.538 | 0.532 | 0.078 | 0.283 | –0.690 (P = 0.041) |
| γ-globin (control) | –0.058 | 0.461 | 0.068 | 0.213 | –0.025 (P = 0.940) |
| γ-globin (hemin) | 0.472 | 0.322 | 0.047 | 0.104 | –0.658 (P = 0.052) |
| ɛ-globin (control) | –0.058 | 0.461 | 0.352 | 0.213 | 0.005 (P = 0.988) |
| ɛ-globin (hemin) | 0.472 | 0.322 | 0.048 | 0.104 | –1.781 (P < 0.001) |
aStandard deviation.
bStandard error of the mean.
Figure 4.
Logarithmic transformation of γ-globin transcript levels in single K562 cells. The upper panel presents plots of transformed data for single cell expression in control (squares) and hemin-treated (triangles) K562 cells in which the mean (bars) was increased from –0.12 to 0.47 (P < 0.0001, t-test) respectively while the variance was decreased from 0.40 to 0.10 (P < 0.0001, F-test). Box and whisker plots are presented alongside the scatter plots showing the 25th and 75th percentile and median values for transcript level. Frequency histograms of the transformed data are shown in the lower panel with controls shown as hatched columns and hemin-treated cells as solid columns. The skew in the distributions for the transformed data for control and treated cells is –0.025 (P = 0.94) and –0.658 (P = 0.052) respectively.
Thus in all three instances the frequency distribution histograms of the transformed data indicate a significant right shift to higher levels of expression in the case of hemin-treated cells, but with continued heterogeneity and strong residual skewing indicating the existence of poorly responding cells. This method of presenting the data shows more strikingly than in Figure 2 that the hemin effect to increase globin mRNA expression leaves significant numbers of cells with very low levels of mRNA. In general, the data presented above on log distributions of single genes appear similar to distributions of logs of calculated copies per cell for very large numbers of genes (6000–40 000) reported in previous studies using bulk cultures, tissues or organisms (e.g. yeast) (22). In addition, in these studies it has been shown that relatively few (0.1–2%) of the monitored genes show changes in expression levels in response to biochemical or genetic perturbations greater than ~2-fold (22,23). These changes are comparable to those that we find in even the highly expressed, differentiated globin gene program.
With respect to molecular mechanisms, the very low globin transcript levels in individual K562 cells may be an analog of the general phenomenon of ‘illegitimate’ transcription (24,25) or, more specifically, may represent the recently recognized process of intergenic transcription within the globin cluster of erythroid cells (26,27). In either case, however, what is striking is the heterogeneity of the cells for all three globin genes and that this heterogeneity remains after induction, suggesting either that cell selection is a paramount phenomenon in this variation or that effects of hemin on transcription mechanisms cannot overcome variations due to cell heterogeneity. One possible cause of the diversity of expression levels is that although K562 cells represent a remarkably uniform population, they may be heterogeneous from individual cells being assayed at different stages of the cell cycle. Further, there have been suggestions that subclones of K562 cells with somewhat different hemoglobin phenotypes could be isolated (21,28).
The present results do not distinguish between cell selection and molecular genetic mechanisms for hemin induction. In either case, however, it is clear that the induction process is superimposed on pre-existing heterogeneity of globin gene transcription. Multiple assays on single cells during the course of induction might help to resolve this conundrum.
The similarity in the changes in the distribution of mRNA expression levels of all three globin genes, despite the fact that they are found on two different chromosomes, suggests a similarity in the mechanism of hemin action for all three genes. Previous reports using Friend erythroleukemia cells have established that hemin-induced modulation of globin gene expression occurs both at the pretranslational and translational levels (29–31). Similar results have been reported with K562 cells (32,33).
The clinical treatment of disorders such as sickle cell anemia with drugs that increase HbF expression, such as hydroxyurea, is effective only in a subset of patients and the cellular mechanism(s) involved in this increase are as yet unclear. The development of in vitro models of hydroxyurea-mediated increases in HbF in human erythroid cells (10) makes it possible to apply quantitative assays of globin gene expression to single primary erythroid cells in culture, although clearly these will be more complex as there is already clear heterogeneity in these cells, even when fully-purified, and the induction process may well be irreversible. Efforts are currently underway to characterize the hydroxyurea-induced increase in γ-globin transcript levels in single primary erythroid cells in culture.
Previous use of PCR in characterizing nucleic acid templates from single cells has been limited for the most part to genotyping single cells prior to zygote implantation for in vitro fertilization (5) or amplification of total transcript content for detection of specific sequences by labeled probes after blotting (8,9). In such applications, precise quantitation of template from single cells is either unnecessary or not feasible because ensuring uniform, unbiased amplification of all sequences is difficult due to the inherent variability in transcript length and complexity. The implementation of precise single-cell quantitation presents an opportunity for the investigation of the behavior of an ensemble of cells in response to pharmacologic or environmental manipulation. In so doing it is possible to determine whether all cells respond to the applied stimulus or only a subset of cells is involved.
This approach may also be expected to increase our understanding of mechanisms of control of gene expression by cis-acting enhancer/control elements which are now widely used in transgenic and transfection experiments. Two models of the mode of action of such elements currently predominate in the literature: one in which the enhancer acts to increase the rate of transcription of a linked gene in all cells (34,35) and the other which postulates that enhancers act to increase the probability that a linked gene will establish and maintain transcriptional activity (13,14). In order to distinguish between these models of possible modes of action, quantitative analysis of gene expression in single cells will be required.
REFERENCES
- 1.Baron M.H. (1996) Developmental regulation of the vertebrate globin multigene family. Gene Expr., 6, 129–137. [PMC free article] [PubMed] [Google Scholar]
- 2.Rodgers G.P. and Rachmilewitz,E.A. (1995) Novel treatment options in the severe β-globin disorders. Br. J. Haematol., 91, 263–268. [DOI] [PubMed] [Google Scholar]
- 3.Olivieri N.F. and Weatherall,D.J. (1998) The therapeutic reactivation of fetal haemoglobin. Hum. Mol. Genet., 7, 1655–1658. [DOI] [PubMed] [Google Scholar]
- 4.Atweh G.F., Sutton,M., Nassif,I., Boosalis,V., Dover,G.J., Wallenstein,S., Wright,E., McMahon,L., Stamatoyannopoulos,G., Faller,D.V. and Perrine,S.P. (1999) Sustained induction of fetal hemoglobin by pulse butyrate therapy in sickle cell disease. Blood, 93, 1790–1797. [PMC free article] [PubMed] [Google Scholar]
- 5.Snabes M.C., Chong,S.S., Subramanian,S.B., Kristjansson,K., DiSepio,D. and Hughes,M.R. (1994) Preimplantation single-cell analysis of multiple genetic loci by whole-genome amplification. Proc. Natl Acad. Sci. USA, 91, 6181–6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brady G. and Iscove,N.N. (1993) Construction of c(DNA) libraries from single cells. Methods Enzymol., 225, 611–623. [DOI] [PubMed] [Google Scholar]
- 7.Eberwine J., Yeh,H., Miyashiro,K., Cao,Y., Nair,S., Finnell,R., Zettel,M. and Coleman,P. (1992) Analysis of gene expression in single live neurons. Proc. Natl Acad. Sci. USA, 89, 3010–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng T., Shen,H., Giokas,D., Gere,J., Tenen,D.G. and Scadden,D.T. (1996) Temporal mapping of gene expression levels during the differentiation of individual primary hematopoietic cells. Proc. Natl Acad. Sci. USA, 93, 13158–13163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu M., Krause,D., Greaves,M., Sharkis,S., Dexter,M., Heyworth,C. and Enver,T. (1997) Multilineage gene expression precedes commitment in the hemopoietic system. Genes Dev., 11, 774–785 [DOI] [PubMed] [Google Scholar]
- 10.Smith R.D., Li,J., Noguchi,C.T. and Schechter,A.N. (2000) Quantitative (PCR) analysis of (HbF) inducers in primary human adult erythroid cells. Blood, 95, 863–869. [PubMed] [Google Scholar]
- 11.Livak K.J., Flood,S.J., Marmaro,J., Giusti,W. and Deetz,K. (1995) Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting (PCR) product and nucleic acid hybridization. PCR Methods Appl., 4, 357–362. [DOI] [PubMed] [Google Scholar]
- 12.Morrison T.B., Weis,J.J. and Wittwer,C.T. (1998) Quantification of low-copy transcripts by continuous (SYBR) Green (I) monitoring during amplification. Biotechniques, 24, 954–962. [PubMed] [Google Scholar]
- 13.Sutherland H.G., Martin,D.I. and Whitelaw,E. (1997) A globin enhancer acts by increasing the proportion of erythrocytes expressing a linked transgene. Mol. Cell. Biol., 17, 1607–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hume D.A. (2000) Probability in transcriptional regulation and its implications for leukocyte differentiation and inducible gene expression. Blood, 96, 2323–2328. [PubMed] [Google Scholar]
- 15.Lozzio C.B. and Lozzio,B.B. (1975) Human Chronic myelogenous leukemia cell-line with positive Philadelphia chromosome. Blood, 45, 321–334. [PubMed] [Google Scholar]
- 16.Rutherford T.R., Clegg,J.B. and Weatherall,D.J. (1979) K562 human leukaemic cells synthesise embryonic haemoglobin in response to haemin. Nature, 280, 164–165. [DOI] [PubMed] [Google Scholar]
- 17.Benz E.J. Jr, Murnane,M.J., Tonkonow,B.L., Berman,B.W., Mazur,E.M., Cavallesco,C., Jenko,T., Snyder,E.L., Forget,B.G. and Hoffman,R. (1980) Embryonic-fetal erythroid characteristics of a human leukemic cell line. Proc. Natl Acad. Sci. USA, 77, 3509–3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fibach E., Kollia,P., Schechter,A.N., Noguchi,C.T. and Rodgers,G.P. (1995) Hemin-induced acceleration of hemoglobin production in immature cultured erythroid cells: preferential enhancement of fetal hemoglobin. Blood, 85, 2967–2974. [PubMed] [Google Scholar]
- 19.Dallal G.E. and Wilkinson,L. (1986) An analytic approximation to the distribution of Lilliefors’s test statistic for normality. Am. Statistician, 40, 294–296 [Google Scholar]
- 20.Dean A., Ley,T.J., Humphries,R.K., Fordis,M. and Schechter,A.N. (1983) Inducible transcription of five globin genes in (K)562 human leukemia cells. Proc. Natl Acad. Sci. USA, 80, 5515–5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Testa U., Vainchenker,W., Beuzard,Y., Rouyer-Fessard,P., Guerrasio,A., Titeux,M., Lapotre,P., Bouguet,J., Breton-Gorius,J. and Rosa,J. (1982) Hemoglobin expression in clones of (K)562 cell line. Eur. J. Biochem., 121, 649–655. [DOI] [PubMed] [Google Scholar]
- 22.Lockhart D.J. and Winzeler,E.A. (2000) Genomics, gene expression and DNA arrays. Nature, 405, 827–836. [DOI] [PubMed] [Google Scholar]
- 23.Ly D.H., Lockhart,D.J., Lerner,R.A. and Schultz,P.G.), (2000) Mitotic misregulation and human aging. Science, 287, 2486–2492. [DOI] [PubMed] [Google Scholar]
- 24.Kimoto Y. (1998) A single human cell expresses all messenger ribonucleic acids: the arrow of time in a cell. Mol. Gen. Genet., 258, 233–239. [DOI] [PubMed] [Google Scholar]
- 25.Negrier C., Vinciguerra,C., Attali,O., Grenier,C., Larcher,M.E. and Dechavanne,M. (1998) Illegitimate transcription: its use for studying genetic abnormalities in lymphoblastoid cells from patients with glanzmann’s thrombasthenia. Br. J. Haematol., 100, 33–39. [DOI] [PubMed] [Google Scholar]
- 26.Collis P., Antoniou,M. and Grosveld,F. (1990) Definition of the minimal requirements within the human β-globin gene and the dominant control region for high level expression. EMBO J., 9, 233–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ashe H.L., Monks,J., Wijgerde,M., Fraser,P. and Proudfoot,N.J. (1997) Intergenic transcription and transinduction of the human β-globin locus. Genes Dev., 11, 2494–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mookerjee B., Arcasoy,M.O. and Atweh,G.F. (1992) Spontaneous delta- to β-globin switching in (K)562 human leukemia cells. Blood, 79, 820–825. [PubMed] [Google Scholar]
- 29.Ross J. and Sautner,D. (1976) Induction of globin m(RNA) accumulation by hemin in cultured erythroleukemic cells. Cell, 8, 513–520. [DOI] [PubMed] [Google Scholar]
- 30.Rutherford T.R. and Weatherall,D.J. (1979) Deficient heme synthesis as the cause of noninducibility of hemoglobin synthesis in a Friend erythroleukemia cell line. Cell, 16, 415–423. [DOI] [PubMed] [Google Scholar]
- 31.Lowenhaupt K. and Lingrel,J.B. (1979) Synthesis and turnover of globin m(RNA) in murine erythroleukemia cells induced with hemin. Proc. Natl Acad. Sci. USA, 76, 5173–5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kollia P., Fibach,E., Najjar,S.M., Schechter,A.N. and Noguchi,C.T. (1996) Modifications of (RNA) processing modulate the expression of hemoglobin genes. Proc. Natl Acad. Sci. USA, 93, 5693–5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kollia P., Fibach,E., Politou,M., Noguchi,C.T., Schechter,A.N. and Loukopoulos,D. (1998) Hydroxyurea and hemin affect both the transcriptional and post-transcriptional mechanisms of some globin genes in human adult erythroid cells. Ann. N. Y. Acad. Sci., 850, 449–451 [DOI] [PubMed] [Google Scholar]
- 34.Banerji J., Rusconi,S. and Schaffner,W. (1981) Expression of a beta-globin gene is enhanced by remote (SV)40 DNA sequences. Cell, 27, 299–308. [DOI] [PubMed] [Google Scholar]
- 35.Treisman R. and Maniatis,T. (1985) Simian virus 40 enhancer increases number of (RNA) polymerase (II) molecules on linked DNA. Nature, 315, 73–75. [DOI] [PubMed] [Google Scholar]