Abstract
We have developed microtiter assays for detecting catalysis by type IB topoisomerases and retroviral integrases. Each assay employs model DNA substrates containing biotin in one strand and digoxigenin in another. In each case action of the enzyme results in the formation of a single DNA strand containing both groups. This allows the reaction product to be quantified by capturing biotinylated product DNA on avidin-coated plates followed by detection using an anti-digoxigenin ELISA. The order of addition of reactants and inhibitors can be varied to distinguish effects of test compounds on different steps in the reaction. These assays were used to screen compound libraries for inhibitors active against mammalian topoisomerase or HIV integrase. We identified (–)-epigallocatechin 3-O-gallate, as a potent inhibitor of religation by mammalian topoisomerase (IC50 of 26 nM), potentially explaining the anti-cancer properties previously attributed to this compound. New integrase inhibitors were also identified. A similar strategy may be used to develop microtiter assays for many further DNA modifying enzymes.
INTRODUCTION
We present a suite of assays for analysis of DNA modifying enzymes. These methods are designed for use in microtiter plates, allowing high throughput and potential automation. We present model applications to studies of poxvirus topoisomerase, mammalian type IB topoisomerase and HIV integrase, though we note that the approach can be employed for many more enzyme types than those described. Model applications to inhibitor development are also described.
Type IB topoisomerases relax supercoiled DNA by introducing transient nicks into one DNA strand. The topoisomerase becomes covalently bound to the DNA 3′ end at the nicked site by a phosphotyrosine bridge, thereby conserving phosphate bond energy for rejoining. After DNA relaxation, the 5′ OH group at the nick attacks the phosphotyrosine linkage and a transesterification reaction reseals the nicked DNA (1). Mammalian topoisomerase IB (henceforth EU-TOPO) displays relatively little discrimination between primary DNA sequences, though preferred DNA sites have been identified in a few cases (2,3). Poxviruses also encode type IB topoisomerases, which differ from the mammalian cellular enzyme in being much smaller (35 versus 91 kDa) and highly sequence specific (4–6). Here we describe studies of the topoisomerase of the poxvirus molluscum contagiosum virus (MCV), which causes opportunistic infections in immunocompromised patients (7–10).
Topoisomerases are targets for several important anti-tumor and anti-infective drugs (11,12). Many of the most effective inhibitors trap the covalent topoisomerase–DNA intermediate, leading to the persistence of DNA breaks that disrupt cell growth. For example, camptothecin traps covalent intermediates made by human topoisomerase IB and thereby kills rapidly dividing cancer cells. Emphasizing the toxicity of trapped covalent intermediates is the recent discovery of a DNA repair enzyme dedicated to removing stalled intermediates from cellular DNA (13).
Like topoisomerases, retroviral integrase proteins also carry out DNA breaking and joining reactions, but via a different catalytic mechanism (14,15). Retroviral integrase enzymes insert retroviral cDNA into host DNA during the early stages of retroviral infection. The integration substrate is a linear cDNA copy of the viral RNA genome. Prior to integration, 2 nt are removed from each 3′ end in the linear cDNA precursor (16–18). This reaction may be important for preparing a defined substrate for subsequent reaction steps, since the reverse transcriptase often adds heterogeneous non-templated bases to the 3′ ends of non-integrated cDNAs (19,20) The recessed 3′ ends are then joined to 5′ protruding ends of breaks made in the target DNA. The remaining DNA strands are then filled in and ligated, probably by the action of host DNA repair enzymes, to complete formation of an integrated provirus (21). The terminal cleavage and strand transfer reactions can be modeled in reactions containing purified integrase (22–27) and the gap filling and ligation reactions can be modeled with purified DNA repair enzymes (21).
Integration is a required step in retroviral replication, as indicated for example by the finding that HIV mutants defective in integration cannot replicate (14). The HIV integrase enzyme is today the only one of the three HIV enzymes for which clinically useful inhibitors are not available. The strand transfer step is particularly attractive as a target for inhibitors, since preintegration complexes with cleaved ends are a prominent intermediate in infected cells (17,18,20), and the complex of integrase and cleaved DNA ends has been validated as an inhibitor target in vivo (28).
Here we present improved assays for identifying inhibitors of type IB topoisomerase and retroviral integrase enzymes. Existing gel-based assays for topoisomerase function are not convenient for monitoring large numbers of tests. It is also desirable to have efficient assays for religation of covalent topoisomerase–DNA intermediates, since clinically useful inhibitors often block this step. We describe microtiter plate assays that monitor religation by two type IB topoisomerases, EU-TOPO and that of MCV (MCV-TOP). For integrase enzymes, two microtiter assays for the strand transfer reaction have been presented previously (29,30). In this study, we present an improved version of the assay described by Craigie et al. (29). The assays were validated with previously reported inhibitors, as well as used to identify new ones. The methods described can potentially be applied to studies of many DNA modifying enzymes.
MATERIALS AND METHODS
Chemicals
Camptothecin, coumermycin A1, (–)-epigallocatechin 3-O-gallate (EGCG), and psychosine (PSYCH) were purchased from Sigma. 4-(4-Bromophenyl)-2,4-dioxobutanoic acid (BDBA) and 2-hydroxy-4-oxo-4-phenylbut-2-enoic acid (HOPE) were from Ryan Scientific, Inc. L-731 988, L-708 906 and L-731 942 were a kind gift from D. Hazuda at Merck Research Laboratories. Chemicals screened were obtained from Sigma/Aldrich, Salor or collaborators J. Siegel at UCSD or W. Fenical and J. Faulkner at the Scripps Institute of Oceanography.
Topoisomerase activity assays
MCV-TOP was purified using nickel-chelating Sepharose as described (5). Standard reaction mixtures for assaying MCV-TOP contained 10 nM MCV-TOP, 2 nM substrate oligodeoxyribonucleotide, 20 mM Tris–Cl (pH 8.0), 200 mM potassium glutamate, 1 mM DTT, 0.1% Triton X-100 and 10% DMSO in a volume of 50 µl. The MCV-TOP substrate used for this assay was derived from a previously characterized optimal sequence (sub a) (5). In the version of the substrate used here (MCV sub a′, see Fig. 1B), the 5-base duplex extension 3′ of the 5′-CCCTT-3′ sequence can dissociate upon covalent complex formation. This traps the covalent complex but permits religation to an added complementary DNA strand. Following covalent complex formation, religation was initiated by addition of 2-fold excess of a labeled 15mer single strand DNA (HW378, see Fig. 1B). To test inhibition of covalent complex formation, chemicals were added to the enzyme and preincubated for 5 min and then the reaction was started with addition of oligonucleotide substrate. To test inhibition of the religation step, topoisomerase was mixed with substrate and incubated to allow covalent complex formation. Test chemicals were then added to the reaction followed by the religation substrate. Reactions were incubated for 1 h. All steps were carried out at 37°C.
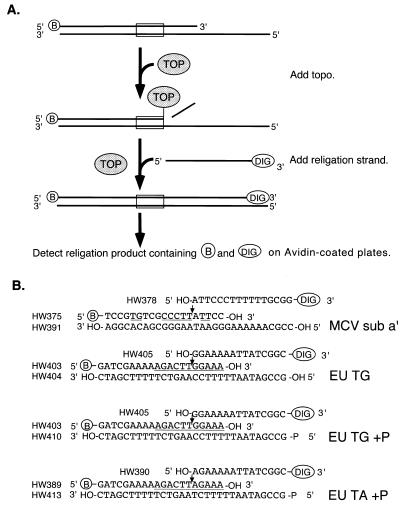
Figure 1.
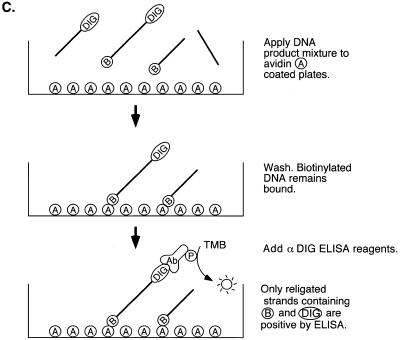
Rapid microtiter assays for type IB topoisomerases. (A) Strategy for using the topoisomerase religation reaction for rapid product detection. The suicide substrate containing biotin on the 5′ end of the top strand (indicated by ‘B’) was incubated with a type IB topoisomerase (top). Formation of the covalent complex cleaves the DNA and releases the short duplex extension 3′ of the cleavage site (middle), thereby trapping the covalent complex. This allows the religation reaction to be carried out by addition of a religation strand containing digoxigenin (bottom). The extent of product formation is then monitored by capturing the strand containing biotin and digoxigenin. (B) DNA sequence of substrates for MCV-TOP and EU-TOPO. The arrows indicate the points of cleavage in the covalent intermediate. Topoisomerase recognition sites are underlined. ‘B’ indicates biotin; ‘DIG’ indicates digoxigenin modifications. ‘HW’ numbers indicate the names of oligonucleotide strands. MCV sub a′, MCV-TOP substrate set. EU TG, mammalian-type IB topoisomerase substrate, containing TG nucleotides flanking the point of cleavage. EU TG+P, same as EU TG substrate but containing a 5′ phosphate on strand HW410 to block self-religation hairpin formation. EU TA+P: same as EU TG+P except an A/T base pair is substituted for G/C 3′ of the point of cleavage. (C) Diagram of the method for detecting religation product. ‘A’ indicates avidin molecules bound to a microtiter plate. ‘Ab’ and ‘P’ indicate the antibody peroxidase conjugate. ‘TMB’ indicates the 3,3′-5,5′-tetramethylbenzidine peroxidase substrate. After the topoisomerase reaction, the product mixture was added to avidin-coupled microtiter wells. Unbound DNA was removed by washes. Bound DNA was then quantitated by detecting digoxigenin using an anti-DIG ELISA.
Standard reaction mixtures for assaying EU-TOPO (from calf thymus) contained 2.5 U EU-TOPO, 2 nM substrate oligonucleotide, 20 mM Tris–Cl (pH 8.0), 72 mM KCl, 5 mM MgCl2, 5 mM DTT and 10% DMSO. Reactions were incubated for 1 h. All steps were carried out at 37°C. EU-TOPO was obtained from Gibco BRL. The EU-TOPO substrate was based on an optimal sequence found in a Tetrahymena rRNA gene (2,3) (Fig. 1B).
HIV-1 integrase assays
HIV-1 integrase was modified to contain an N-terminal His Tag and purified using nickel-chelating Sepharose as described (31). Standard reaction mixtures for assaying HIV-1 integrase contained 312 nM (400 ng) integrase, 7 nM (6 ng) substrate oligonucleotide, 20 mM HEPES (pH 7.5), 5% PEG, 10 mM MgCl2, 10 mM DTT, 0.1 mg/ml BSA and 10% DMSO in a final reaction volume of 40 µl. Integrase protein was diluted in 0.1 mg/ml BSA, 0.5 M NaCl, 5 mM EDTA and 20 mM HEPES (pH 7.5) prior to use. To test for inhibition of strand transfer, the standard reaction minus the DMSO was preincubated for 5 min at room temperature. Test chemicals, dissolved in 100% DMSO, were then added to the reaction. The reaction was carried out for 1 h at 37°C. The detection of strand transfer products was carried out as described for products of topoisomerase assays.
Detection of reaction products in avidin-coupled microtiter wells
After the reactions were completed, mixtures were adjusted to 20 mM Tris–Cl (pH 8.0), 400 mM NaCl, 10 mM EDTA and 0.1 mg/ml sonicated salmon sperm DNA in a final volume of 100 µl. NaCl (400 nM) was omitted in the EU-TOPO assay. The samples were added to avidin-coupled microtiter wells (Boehringer Mannheim), which were then gently agitated at room temperature for 1 h. DNA was denatured and unbound DNA was removed by three washes (200 µl, 5 min each) with 30 mM NaOH, 200 mM NaCl, 1 mM EDTA. Bound DNA was then adjusted to 10 mM Tris–Cl (pH 8.0) and 1 mM EDTA. The relative activity was determined using an anti-digoxigenin ELISA. Anti-digoxigenin-peroxidase Fab fragments (0.01 U, Pierce or Boehringer Mannheim) were added into wells and incubated for 1 h at 37°C. Unbound anti-digoxigenin peroxidase Fab fragments were removed by five washes (300 µl) with PBS containing 0.1% Tween-20. Bound DNA was then incubated with 200 µl of 3,3′-5,5′-tetramethylbenzidine (TMB) solution (Boehringer Mannheim) until sufficient blue product accumulated. The reaction was terminated by adding 100 µl of 1 M sulfuric acid. The resulting yellow color was quantitated at 450 nm using a Molecular Dynamics plate reader with SOFTmax PRO software. Positive control reactions typically had values from 0.7 to 1.8; negative control reactions typically had values from 0.04 to 0.07.
Gel assay for activity of EU TOPO
Substrate HW406 of sequence d(GATCGAAAAAGACTTGGAAA) was labeled on the 5′ end with [γ-32P]ATP and T4 polynucleotide kinase. Labeled HW406 was annealed with HW404 for EU TG or HW410 for EU TG+P, and 50 nM DNA was incubated with 12 U of protein in the buffer described for assays above. The formation of covalent complexes was carried out for 12 h, and then the religation reaction was started with addition of 100 nM religation strand HW407 of sequence d(GGAAAAATTATCGGC) and incubated for 2 h at room temperature. The reaction products were analyzed by electrophoresis on denaturing polyacrylamide gels and visualized by autoradiography.
RESULTS
A microtiter plate assay for MCV topoisomerase
The sequence-specific binding of MCV-TOP (5,6) was exploited to position a topoisomerase binding site near the end of a short duplex oligonucleotide (MCV sub a′, Fig. 1A and B). Formation of the covalent complex cleaves the top strand of MCV sub a′ (strand HW375; Fig. 1A, top). The 5-base strand 3′ of the resulting DNA nick is too short to remain annealed, and so dissociates and is lost by dilution which traps the covalent complex (Fig. 1A, middle). Such substrates have been called ‘suicide substrates’ in the literature (32–35). Addition of a new DNA strand (HW378) complementary to the single stranded region in the suicide substrate allows attack of the 5′ OH of the added strand at the topoisomerase–DNA phosphotyrosine linkage. This reaction joins the newly added ‘religation strand’ (HW378) to the starting top strand (HW375; Fig. 1A, bottom).
To permit detection of the religation product, a biotin group is attached to the 5′ end of the cleavable strand (HW375), and a digoxigenin group is attached to the 3′ end of the religation strand (HW378). Thus, the religation reaction generates a DNA strand with biotin on one end and digoxigenin on the other. This DNA strand is then captured by applying the reaction products to an avidin-coated plate (Fig. 1C). After washing, the amount of product is detected by addition of an anti-digoxigenin Fab linked to peroxidase, followed by washing and addition of a colorimetric substrate. The colored product formed by the peroxidase reaction is then quantified by spectrophotometry. Each step in the process is carried out in 96-well microtiter plates, thereby speeding throughput compared to earlier gel-based assays.
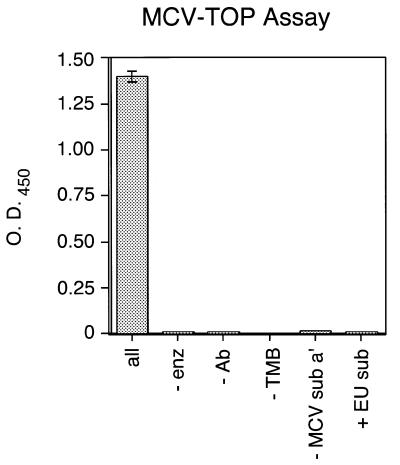
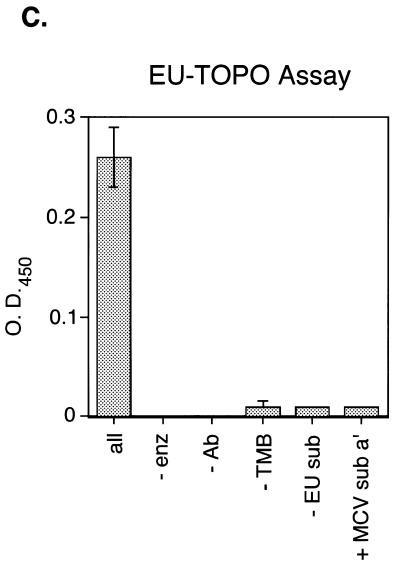
Test reactions carried out under a variety of conditions are presented in Figure 2. No product formation was detected in reactions lacking MCV-TOP, anti-digoxigenin peroxidase-Fab, DNA substrate MCV sub a′ or TMB peroxidase substrate. In addition, no activity was detected when a DNA substrate for EU-TOPO was substituted for MCV sub a′, confirming the sequence specificity of catalysis by MCV-TOP.
Figure 2.
Requirements for the microtiter assay of MCV-TOP activity. Reactions contained the complete set of reagents (‘all’) or lacked the indicated ingredient (–). In the right-most lane (+EU sub), the EU TG+P substrate was substituted for MCV sub a′. All reactions were repeated at least three times.
A microtiter plate assay for EU-TOPO
We also developed a similar assay for function of EU-TOPO, a known target for anticancer drugs. The DNA substrate was designed to contain a preferred site for topoisomerase IB action identified in Tetrahymena DNA (2,3).
Initial tests using substrate EU TG (Fig. 1B) revealed relatively low activity in the microtiter plate assay (data not shown). To investigate this, assays were carried out with oligonucleotides matching substrate EU TG except lacking the biotin and digoxigenin modifications (strands HW406 and HW407 respectively; see Materials and Methods for sequences). Oligonucleotide HW406 was 5′ labeled with 32P, annealed with HW404 (sequence in Fig. 1B), then incubated with EU-TOPO. Labeled DNA products were visualized after gel electrophoresis by autoradiography (Fig. 3A). Oligodeoxynucleotides matching expected reaction products were synthesized, labeled and subjected to electrophoresis in adjacent lanes. These assays revealed much less formation of the religated DNA strand than in comparable assays with MCV-TOP (Fig. 3A, lanes 4 and 5, ‘religated products’; for MCV-TOP see 36,37).
Figure 3.
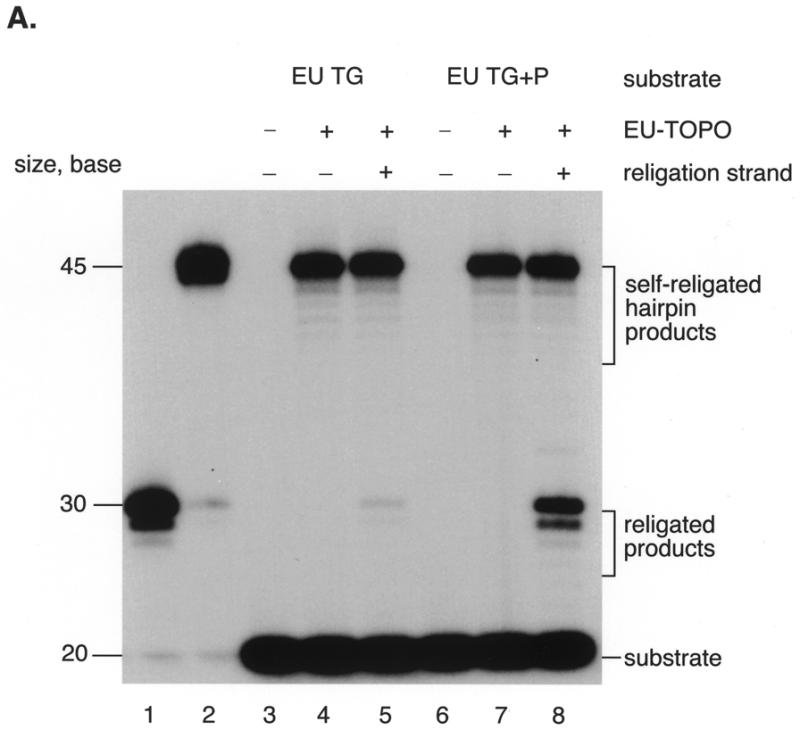
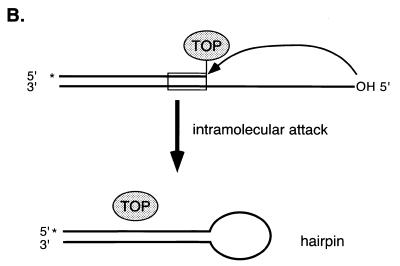
Assays of EU-TOPO activity. (A) Comparison of religation activities of substrates EU TG and EU TG+P using 32P-labeled substrates and gel electrophoresis. Substrate DNAs were labeled with 32P on the 5′ end of strand HW406. Covalent complexes were formed by incubation with EU-TOPO, then further incubated after addition of the religation strand (HW407). Lane 1, 30 base marker for the expected religation product; lane 2, 45 base marker matching the self-religated hairpin product; lane 3, substrate EU TG; lane 4, substrate EU TG + EU-TOPO; lane 5, substrate EU TG + EU-TOPO + religation strand; lane 6, substrate EU TG+P only (see sequence HW410, Fig. 2B); lane 7, substrate EU TG+P + EU-TOPO; lane 8, substrate EU TG+P + EU-TOPO + religation strand. The sequences of the markers were d(GATCGAAAAAGACTTGGAAAAATTATCGGC) (lane 1, HW408, the expected religation product) and d(GATCGAAAAAGACTTGCCGATAATTTTTCCAAGTCTTTTTCGATC) (lane 2, HW412, the self-religated hairpin product). (B) Diagram of formation of the self-religated hairpin products. DNA strands are indicated by the thick lines, the ball marked ‘TOP’ indicates EU-TOPO, and the asterisk indicates the point of 5′ end labeling. (C) A. Requirements for the EU-TOPO microtiter assay. Reactions contained the complete set of reagents (‘all’) or lacked the indicated ingredient (–). In the right-most lane (+MCV sub a′), the MCV sub a′ substrate was substituted for EU TG+P. All reactions were repeated at least three times.
However, an unexpected DNA form was produced in abundance (Fig. 3A, ‘self religated hairpin products’). These products migrated with a DNA marker (HW412) matching that expected from joining the 5′ end of strand HW404 at the covalent phosphotyrosine bond. This product was probably formed by an intramolecular reaction in which the single stranded 5′ end region of oligonucleotide HW404 attacks a EU-TOPO covalent complex, forming a hairpin, as reported previously (35) (Fig. 3B). Alternatively, this product could have been formed by a second copy of strand HW404 on another DNA duplex attacking the covalent intermediate. To reduce formation of this side product, the reactive 5′ OH end of oligonucleotide HW404 was blocked by phosphorylation (Fig. 1B, EU TG+P, strands HW410 and HW413). Blockage resulted in reduced formation of the ‘self-religated’ product and much greater production of the desired religation strand (Fig. 3A, lanes 7 and 8, ‘religated products’). The substrates used in the plate assay were then also modified by phosphorylation, resulting in a 4- to 6-fold increase in product formation (data not shown). Phosphorylation did not block hairpin formation completely, however, due to incomplete phosphorylation of the strand HW410 during chemical synthesis (data not shown). Why EU-TOPO but not MCV-TOP favored hairpin formation is unclear but of interest for possible insight into the reaction mechanism.
The requirements for product formation for the EU-TOPO reaction are shown in Figure 3C. No activity was detected when EU-TOPO, anti-digoxigenin-peroxidase Fab fragment, TMB or EU TG+P substrate DNA was omitted from the reaction. In addition, no activity was detected when oligonucleotide MCV sub a′ was substituted for EU TG+P, documenting the sequence specificity of EU-TOPO in this assay.
Response of the microtiter assay to known topoisomerase inhibitors
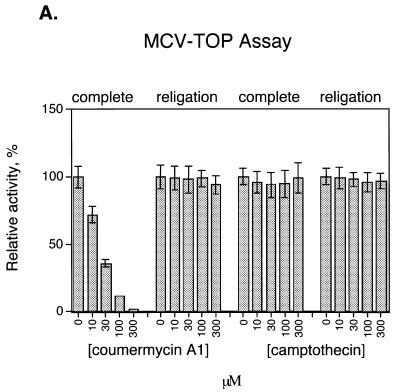
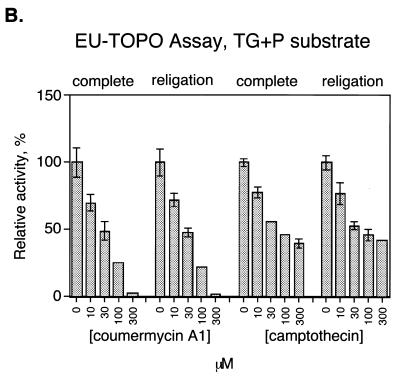
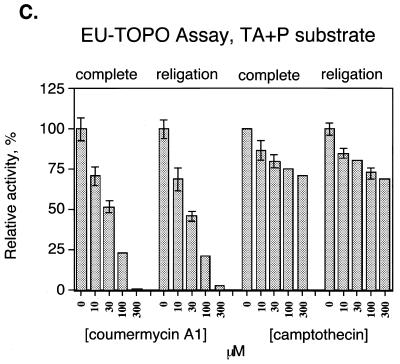
As a first step in applying this assay to inhibitor identification, the effect of known topoisomerase inhibitors was tested (Fig. 4A). Two orders of addition were compared. Reactions were carried out in which enzyme was preincubated with inhibitor before addition of DNA substrate (Fig. 4A, ‘complete’), which would reveal inhibition at any step in the reaction pathway. Reactions were also carried out with a different order of addition, in which covalent complexes were first formed, then inhibitor added, and the religation strand added last (Fig. 4A, ‘religation’). This order of addition tested inhibition of the religation step only.
Figure 4.
Detecting inhibition of MCV-TOP and EU-TOPO using the microtiter assays. (A) Assays of MCV-TOP. The concentrations of inhibitor used are listed beneath the bar graph. ‘Complete’ indicates that the inhibitor was added to the topoisomerase before addition of substrate, thereby assaying for inhibition at any step in the reaction. ‘Religation’ indicates that the inhibitor was added after covalent complex formation, thus assaying inhibition of the religation step. Concentrations in µM are shown at the bottom. Assays were normalized to the uninhibited control (100% activity). Measurements were repeated at least 10 times. (B) Assays of EU-TOPO using EU TG+P substrate. (C) Assays of EU-TOPO using EU TA+P substrate.
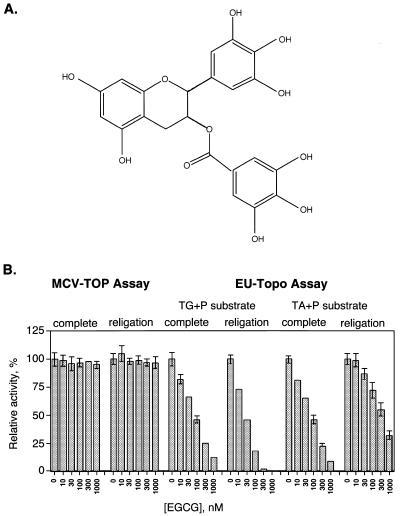
Coumermycin A1, a known MCV-TOP inhibitor, was tested and found to inhibit the complete reaction, but not religation by MCV-TOP (Fig. 4A). Camptothecin, an inhibitor of EU-TOPO, was not active against MCV-TOP.
For EU-TOPO, we tested two different substrate DNAs, containing TA or TG as the bases flanking the scissile phosphate. It has been suggested that some EU-TOPO religation inhibitors form a ternary complex involving contacts to the base 3′ of the point of incision (38–40), so we sought to discriminate such effects using the microtiter assay.
Coumermycin A1 also inhibited EU-TOPO (Fig. 4B and C). In this case inhibition of the complete reaction and religation were comparable. Evidently for EU-TOPO coumermycin is capable of acting on the covalent intermediate. Inhibition by coumermycin was not sensitive to the identity of the base 3′ of the scissile phosphate (compare Fig. 4B and C). Inhibition of EU-TOPO by camptothecin, in contrast, was more efficient when the 3′ base was G, as expected from previous reports (40,41).
Screening for inhibitors of topoisomerases
We next used the microtiter assays for MCV-TOP and EU-TOPO to screen libraries of pure compounds and natural product extracts for inhibitory activities. These compounds were mainly collected from commercial sources based on their resemblance to known inhibitors of DNA modifying enzymes. Other compounds were natural products or natural product extracts isolated in the laboratories of W. Fenical and J. Faulkner at the Scripps Institute of Oceanography. Altogether about 10 000 compounds were screened in at least one of the assays and most in at least two of the three (including integrase, described below).
The most potent topoisomerase inhibitor identified was EGCG (Fig. 5A). EGCG inhibited religation by EU-TOPO with an IC50 of 26 nM for the EU TG+P substrate. On the EU TA+P substrate, however, the IC50 was 500 nM, indicating possible selectivity for the base 3′ of the scissile phosphate (Fig. 5B). Addition of EGCG to reactions prior to formation of the covalent complex also resulted in inhibition of both substrates with an IC50 of 60–70 nM. EGCG had no effect on MCV-TOP in this concentration range. EGCG has been reported to have anticancer properties based on previous studies in vivo (42–44). Our data suggest that EGCG may exert its anticancer effects at least in part by inhibiting religation by EU-TOPO.
Figure 5.
Potent inhibition of EU-TOPO by EGCG. (A) Structure of EGCG. (B) Assays of MCV-TOP and EU-TOPO in the presence of different concentrations of EGCG. Figure labels are as in Figure 4 except the inhibitor concentrations are in nM.
A microtiter plate assay for HIV-1 integrase
Two previous reports have presented microtiter assays for strand transfer by HIV integrase (29,30). Craigie et al. previously described a method in which the action of integrase covalently connects a biotin group to a reporter molecule (Fig. 6A). The sequence of the oligonucleotide used matches one end of the non-integrated HIV-1 cDNA (Fig. 6B). The duplex oligonucleotide contains biotin on one strand at the 5′ end distal to the point of joining, and a label on the 3′ end of the other strand. Covalent integration forms a single strand containing the biotin group and the label. Note that different duplexes of the same sequence serve as models for the viral cDNA end and the target DNA.
Figure 6.
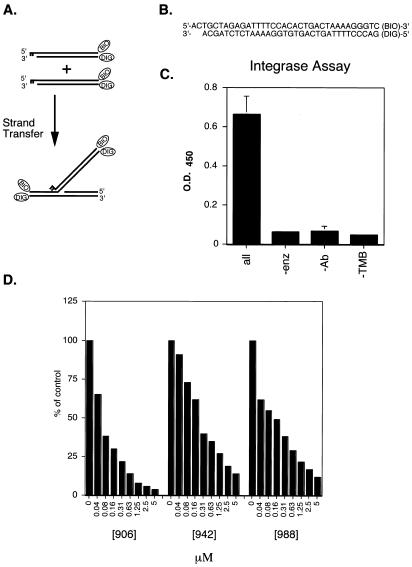
An improved microtiter assay for HIV-1 integrase. (A) Principle of the assay method. The oligonucleotide shown mimics the terminus of the HIV cDNA after cleavage by integrase protein. Integration results in the covalent integration of one oligonucleotide into another. (B) Sequence of the duplex oligonucleotide substrate used, illustrating the location of the biotin and digoxigenin groups. (C) Requirements for the integrase microtiter assay. The assay components indicated below the bar graph were left out of the indicated assays. (D) IC50 measurements of the di-keto acid inhibitors. Concentrations of inhibitor added are indicated in µM.
Here we describe three improvements of the method of Craigie et al. (29). (i) The biotin–digoxigenin combination described above is used instead of the radioactive detection used previously, which speeds throughput and simplifies the procedure. (ii) The DNA substrates have been lengthened (Fig. 6B) to allow the reaction to be carried out in the presence of Mg2+ instead of Mn2+, which was used previously (45–47). Magnesium is widely thought to be the more likely cofactor in vivo. (iii) The integrase and DNA substrates are preassembled by a preincubation step prior to addition of the inhibitor. Steps (ii) and (iii) yield complexes that better mimic integration complexes in vivo and greatly alter the response of the assay to inhibitors (for discussion see 28,48,49). Some of the requirements for the integrase assay are documented in Figure 6C.
Response of the integrase assay to known inhibitors
The response of the plate assay was tested using the di-keto acid family of integrase inhibitors. These compounds have been shown to inhibit integrase in preassembled reactions in vitro and to inhibit HIV replication by selectively blocking integration in cell culture (28). As can be seen in Figure 6D, the three di-keto acid inhibitors tested inhibited integrase efficiently in the microtiter assay, with IC50 values ranging from 0.1 to 0.3 µM.
Screening for new inhibitors of HIV integrase
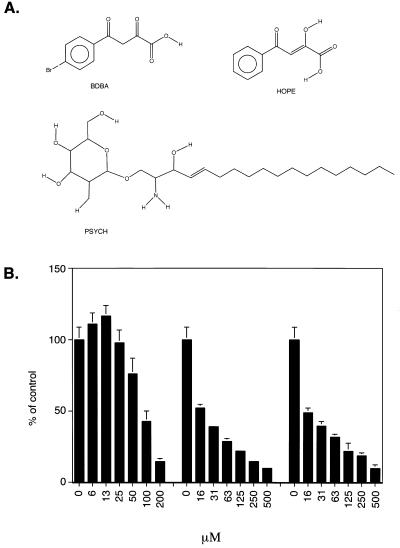
The microtiter plate assay was then used to screen our library of small molecules and natural product extracts. Three of the compounds displaying inhibitory activity are shown in Figure 7A. Assay results are shown in Figure 7B. Inhibitors included two relatives of the di-keto acids, BDBA and HOPE, and the hydroxylated lipid psychosine. IC50 values were 15, 17 and 84 µM respectively. Although the IC50 values were all higher than for the reported di-keto acid inhibitors (28), these tests document the utility of the microtiter assay in identifying new inhibitors.
Figure 7.
New integrase inhibitors identified using the microtiter assay. (A) Structures of the integrase inhibitors. (B) Inhibition of integrase assays by PSYCH, HOPE and BDBA. Concentrations are in µM.
DISCUSSION
Here we describe rapid microtiter assays for function of eukaryotic type IB topoisomerases and retroviral integrases. In each case the method exploits the ability of the enzyme to connect previously separate DNA strands. A biotin group is attached to one DNA strand and digoxigenin to the other. Action of the enzyme connects the two, allowing simple detection by capturing product on an avidin-coated plate followed by anti-digoxigenin ELISA. These methods are in daily use in our laboratory and have greatly increased throughput in characterization of enzyme fractions and inhibitors.
Identification of inhibitors
The EU-TOPO microtiter assay screen was used to identify a compound, EGCG, which inhibited EU-TOPO religation with an IC50 of 26 nM. This documents the utility of this assay in identifying potent inhibitors. EGCG has been previously reported to have anti-cancer properties. Asian folklore and modern epidemiological studies hold that people who drink large quantities of green tea are less prone to gastric and colorectal cancers (42,43). Fractionation of green tea concentrates yielded EGCG, which was found to obstruct tumor formation in several models. For example, induction of carcinomas by N-ethyl-N′-nitro-N-nitrosoguanidine was reduced by cotreatment with EGCG (50), and EGCG was reported to reduce metastasis in mice (44). Our data suggest that EGCG may act at least in part by trapping covalent protein–DNA complexes made by EU-TOPO, leading to DNA break formation and death of rapidly dividing cells, the mechanism previously reported for camptothecin.
For HIV integrase, the assay identified new di-keto acid inhibitors, members of a chemical family known to be active against integrase in vivo (28). Previous work has established that the di-keto acids bind only to integrase in complex with DNA, and not to free integrase. We find that our assays are inhibited by the di-keto acids, documenting that our reaction conditions do result in assembly of integrase with substrates. To date the only integrase inhibitors that clearly act against integrase in vivo are the di-keto acids, so we favored addressing this step in our screens. It will be of interest to see whether the leads reported here such as psychosine can be developed into compounds active against integrase in vivo.
Application of this assay method to other enzymes
Variations of the described method could be used to detect the activity of many other viral or cellular DNA modifying enzymes. In each case the principle would be the same, using the enzyme to covalently attach a digoxigenin-labeled DNA to a biotin-containing DNA. In all cases the product is captured on an avidin-coated plate and assayed by anti-digoxigenin ELISA.
For example, DNA ligases could readily assayed. One double-stranded DNA would contain the biotin group, the second the digoxigenin group. The DNA 5′ ends on the opposite ends of the DNA chain from the tags would contain 5′ phosphates to permit ligation. Action of ligase would then connect the two covalently. The products would be captured on the avidin plate and detected by ELISA. Ligase cofactors (e.g. XRCC1 and XRCC4) could be assayed by arranging appropriate ligation reactions to depend on the added cofactors. Assays of DNA kinase could be carried out as for assays of DNA ligation, but the reactant DNAs would not be phosphorylated on the DNA 5′ ends. Lack of the 5′ phosphate would prevent ligase from connecting the DNA molecules. Addition of an appropriate DNA kinase and ATP allows the DNA ends to be phosphorylated, which permits subsequent ligation. The DNA products are then captured and assayed as above. Endoucleases could be assayed using substrates where the biotin and digoxigenin start out linked and the enzyme activity is detected by separating the two. Polymerases could be detected by assaying incorporation of digoxigenin-modified nucleotides into substrates containing biotin. Other variations could be devised to detect most types of DNA modifying enzymes.
Acknowledgments
ACKNOWLEDGEMENTS
We thank members of the Bushman laboratory and Heather Carlson for helpful discussions, Allison Bocksruker for artwork and Lynn Artale for help preparing the manuscript. This work was supported by NIH grant AI46222-01 and GM56553, the James B. Pendleton Charitable Trust, the Berger Foundation and Cornelia Mackey. F.D.B. is a Scholar of the Leukemia and Lymphoma Society of America.
REFERENCES
- 1.Wang J.C. (1996) DNA topoisomerases. Annu. Rev. Biochem., 65, 635–692. [DOI] [PubMed] [Google Scholar]
- 2.Andersen A.H., Gocke,E., Bonven,B.J., Nielsen,O.F. and Westergaard,O. (1985) Topoisomerase I has a strong binding preference for a conserved hexadecameric sequence in the promoter region of the rRNA gene from Tetrahymena pyriformis. Nucleic Acids Res., 13, 1543–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonven B.J., Bocke,E. and Westergaard,O. (1985) A high affinity topoisomerase I binding sequence is clustered at DNase I hypersensitive sites in tetrahymena R-chromatin. Cell, 41, 541–551. [DOI] [PubMed] [Google Scholar]
- 4.Shuman S. (1998) Vaccinia virus DNA topoisomerase: a model eukaryotic type IB enzyme. Biochim. Biophys. Acta, 1400, 321–339. [DOI] [PubMed] [Google Scholar]
- 5.Hwang Y., Wang,B. and Bushman,F.D. (1998) Molluscum contagiosum virus topoisomerase: purification, activities and response to inhibitors. J. Virol., 72, 3401–3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hwang Y., Burgin,A. and Bushman,F.D. (1999) DNA contacts stimulate catalysis by a poxvirus topoisomerase. J. Biol. Chem., 274, 9160–9168. [DOI] [PubMed] [Google Scholar]
- 7.Gottlieb S.L. and Myskowski,P.L. (1994) Molluscum contagiosum. Int. J. Dermatol., 33, 453–461. [DOI] [PubMed] [Google Scholar]
- 8.Porter C.D., Blake,N.W., Cream,J.J. and Archard,L.C. (1992) In Wright,D. and Archer,A.L. (eds), Molecular and Cell Biology of Sexually Transmitted Diseases. Chapman and Hall, London.
- 9.Petersen C.S. and Gerstoft,J. (1992) Molluscum contagiosum in HIV-infected patients. Dermatology, 184, 19–21. [DOI] [PubMed] [Google Scholar]
- 10.Schwartz J.J. and Myskowski,P.L. (1992) Molluscum contagiosum in patients with human immunodeficiency virus infection. J. Am. Acad. Dermatol., 27, 583–588. [DOI] [PubMed] [Google Scholar]
- 11.Kornberg A. and Baker,T. (1991) DNA Replication. W.H. Freeman and Company, New York, NY.
- 12.Pommier Y., Pourquier,P., Fan,Y. and Strumberg,D. (1998) Mechanism of action of eukaryotic DNA topoisomerase I and drugs targeted to the enzyme. Biochim. Biophys. Acta, 1400, 83–106. [DOI] [PubMed] [Google Scholar]
- 13.Pouliot J.J., Yao,K.C., Robertson,C.A. and Nash,H.A. (1999) Yeast gene for a Tyr-DNA phosphodiesterase that repairs topoisomerase I complexes. Science, 286, 552–555. [DOI] [PubMed] [Google Scholar]
- 14.Coffin J.M., Hughes,S.H. and Varmus,H.E. (1997) Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 15.Hansen M.S.T., Carteau,S., Hoffmann,C., Li,L. and Bushman,F. (1998) In Setlow,J.K. (ed.), Genetic Engineering. Principles and Methods. Plenum Press, New York and London, Vol. 20, pp. 41–62. [DOI] [PubMed]
- 16.Brown P.O., Bowerman,B., Varmus,H.E. and Bishop,J.M. (1989) Retroviral integration: structure of the initial covalent complex and its precursor and a role for the viral IN protein. Proc. Natl Acad. Sci. USA, 86, 2525–2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujiwara T. and Mizuuchi,K. (1988) Retroviral DNA integration: structure of an integration intermediate. Cell, 54, 497–504. [DOI] [PubMed] [Google Scholar]
- 18.Roth M.J., Schwartzberg,P.L. and Goff,S.P. (1989) Structure of the termini of DNA intermediates in the integration of retroviral DNA: dependence of IN function and terminal DNA sequence. Cell, 58, 47–54. [DOI] [PubMed] [Google Scholar]
- 19.Patel P.H. and Preston,B.D. (1994) Marked infidelity of human immunodeficiency virus type 1 reverse transcriptase at RNA and DNA template ends. Proc. Natl Acad. Sci. USA, 91, 549–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller M.D., Farnet,C.M. and Bushman,F.D. (1997) Human immunodeficiency virus type 1 preintegration complexes: studies of organization and Composition. J. Virol., 71, 5382–5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoder K.E. and Bushman,F.D. (2000) Repair of gaps in retroviral DNA integration intermediates. J. Virol., 74, 11191–11200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katzman M., Katz,R.A., Skalka,A.M. and Leis,J. (1989) The avian retroviral integration protein cleaves the terminal sequences of linear viral DNA at the in vivo sites of integration. J. Virol., 63, 5319–5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sherman P.A. and Fyfe,J.A. (1990) Human immunodeficiency virus integration protein expressed in Escherichia coli possesses selective DNA cleaving activity. Proc. Natl Acad. Sci. USA, 87, 5119–5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Craigie R., Fujiwara,T. and Bushman,F. (1990) The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell, 62, 829–837. [DOI] [PubMed] [Google Scholar]
- 25.Bushman F.D., Fujiwara,T. and Craigie,R. (1990) Retroviral DNA integration directed by HIV integration protein in vitro. Science, 249, 1555–1558. [DOI] [PubMed] [Google Scholar]
- 26.Katz R.A., Merkel,G., Kulkosky,J., Leis,J. and Skalka,A.M. (1990) The avian retroviral IN protein is both necessary and sufficient for integrative recombination in vitro. Cell, 63, 87–95. [DOI] [PubMed] [Google Scholar]
- 27.Bushman F.D. and Craigie,R. (1991) Activities of human immunodeficiency virus (HIV) integration protein in vitro: specific cleavage and integration of HIV DNA Proc. Natl Acad. Sci. USA, 88, 1339–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hazuda D.J., Felock,P., Witmer,M., Wolfe,A., Stillmock,K., Grobler,J.A., Espeseth,A., Gabryelski,L., Schleif,W., Blau,C. and Miller,M.D. (2000) Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science, 287, 646–650. [DOI] [PubMed] [Google Scholar]
- 29.Craigie R., Mizuuchi,K., Bushman,F.D. and Engelman,A. (1991) A rapid in vitro assay for HIV DNA integration. Nucleic Acids Res., 19, 2729–2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hazuda D.J., Hastings,J.C., Wolfe,A.L. and Emini,E.A. (1994) A novel assay for the DNA strand-transfer reaction of HIV-1 integrase Nucleic Acids Res., 22, 1121–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bushman F.D., Engelman,A., Palmer,I., Wingfield,P. and Craigie,R. (1993) Domains of the integrase protein of human immunodeficiency virus type 1 responsible for polynucleotidyl transfer and zinc binding. Proc. Natl Acad. Sci. USA, 90, 3428–3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Richet E., Abcarian,P. and Nash,H.A. (1988) Synapsis of attachment sites during lambda integrative recombination involves capture of a naked DNA by a Protein-DNA Complex. Cell, 52, 9–17. [DOI] [PubMed] [Google Scholar]
- 33.Burgin A.B.J., Huizenga,B.N. and Nash,H.A. (1995) A novel suicide substrate for DNA topoisomerases and site-specific recombinases. Nucleic Acids Res., 23, 2973–2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shuman S. (1992) DNA strand transfer reactions catalyzed by Vaccinia topoisomerase I. J. Biol. Chem., 267, 8620–8627. [PubMed] [Google Scholar]
- 35.Pommier Y., Jenkins,J., Kohlhagen,G. and Leteurtre,F. (1995) DNA recombinase activity of eukaryotic DNA topoisomerase I; effects of camptothecin and other inhibitors. Mutat. Res., 337, 135–145. [DOI] [PubMed] [Google Scholar]
- 36.Hwang Y., Rowley,D., Rhodes,D., Gertsch,J., Fenical,W. and Bushman,F.D. (1999) Mechanism of inhibition of a poxvirus topoisomerase by the marine natural product sansalvamide A. Mol. Pharmacol., 55, 1049–1053. [DOI] [PubMed] [Google Scholar]
- 37.Hwang Y., Park,M., Fischer,W.H. and Bushman,F. (1999) Domain structure of the type-1B topoisomerase encoded by molluscum contagiosum virus. Virology, 262, 479–491. [DOI] [PubMed] [Google Scholar]
- 38.Jaxel C., Capranico,G., Kerrigan,D., Kohn,K.W. and Pommier,Y. (1991) Effect of local DNA sequence on topoisomerase I cleavage in the presence or absence of camptothecin. J. Biol. Chem., 266, 20418–20423. [PubMed] [Google Scholar]
- 39.Tanizawa A., Kohn,K.W. and Pommier,Y. (1993) Induction of cleavage in topoisomerase I c-DNA by topoisomerase I enzymes from calf thymus and wheat germ in the presence and absence of camptothecin Nucleic Acids Res., 21, 5157–5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fan Y., Weinstein,J.N., Kohn,K.W., Shi,L.M. and Pommier,Y. (1998) molecular modeling studies of the DNA-topoisomerase I ternary cleavable complex with camptothecin. J. Med. Chem., 41, 2216–2226. [DOI] [PubMed] [Google Scholar]
- 41.Redinbo M.R., Stewart,L., Kuhn,P., Champoux,J.J. and Hol,W.G.J. (1998) Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA. Science, 279, 1504–1513. [DOI] [PubMed] [Google Scholar]
- 42.Kono S., Ikeda,M., Tokudome,S. and Kuratsune,M. (1988) A case-control study of gastric cancer and diet in northern Kyushu, Japan. Jpn J.Cancer Res., 79, 1067–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tajima K. and Tominaga,S. (1985) Dietary habits and gastro-intestinal cancers: a comparative case-control study of stomach and large intestinal cancers in Nagoya, Japan. Jpn J.Cancer Res., 76, 705–716. [PubMed] [Google Scholar]
- 44.Taniguchi S., Fujiki,H., Kobayashi,H., Go,H., Miyado,K., Sadano,H. and Shimokawa,R. (1992) Effect of (-)-epigallocatechin gallate, the main constituent of green tea, on lung metastasis with mouse B16 melanoma cell lines. Cancer Lett., 65, 51–54. [DOI] [PubMed] [Google Scholar]
- 45.Lee S.P., Kim,H.G., Censullo,M.L. and Han,M.K. (1995) Characterization of magnesium-dependent 3′-processing activity for human immunodeficiency virus type 1 integrase in vitro: Real-time kinetic studies using fluorescence resonance energy transfer. Biochemistry, 34, 10205–10214. [DOI] [PubMed] [Google Scholar]
- 46.Miller M., Bor,Y.-C. and Bushman,F.D. (1995) Target DNA capture by HIV-1 integration complexes. Curr. Biol., 5, 1047–1056. [DOI] [PubMed] [Google Scholar]
- 47.Engelman A. and Craigie,R. (1995) Efficient magnesium-dependent human immunodeficiency virus type 1 integrase activity. J. Virol., 69, 5908–5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hazuda D.J., Felock,P.J., Hastings,J.C., Pramanik,B. and Wolfe,A.L. (1997) Differential divalent cation requirements uncouple the assembly and catalytic reactions of human immunodeficiency virus type 1 integrase. J. Virol., 71, 7005–7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hansen M.S.T., Smith,G.J.I., Kafri,T., Molteni,V., Siegel,J.S. and Bushman,F.D. (1999) Integration complexes derived from HIV vectors for rapid assays in vitro. Nature Biotech., 17, 578–582. [DOI] [PubMed] [Google Scholar]
- 50.Yamane T., Takahashi,T., Kuwata,K., Oya,K., Inagake,M., Kitao,Y., Suganuma,M. and Fujiki,H. (1995) Inhibition of N-methyl-N′-nitro-N-nitrosoguanidine-induced carcinogenesis by (-)-epigallocatechin gallate in the rat glandular stomach. Cancer Res., 55, 2081–2084. [PubMed] [Google Scholar]