Abstract
Tumor progression is driven by dynamic interactions between cancer cells and their surrounding microenvironment. Investigating the spatiotemporal evolution of tumors can provide crucial insights into how intrinsic changes within cancer cells and extrinsic alterations in the microenvironment cooperate to drive different stages of tumor progression. Here, we integrate high-resolution spatial transcriptomics and evolving lineage tracing technologies to elucidate how tumor expansion, plasticity, and metastasis co-evolve with microenvironmental remodeling in a Kras;p53-driven mouse model of lung adenocarcinoma. We find that rapid tumor expansion contributes to a hypoxic, immunosuppressive, and fibrotic microenvironment that is associated with the emergence of pro-metastatic cancer cell states. Furthermore, metastases arise from spatially-confined subclones of primary tumors and remodel the distant metastatic niche into a fibrotic, collagen-rich microenvironment. Together, we present a comprehensive dataset integrating spatial assays and lineage tracing to elucidate how sequential changes in cancer cell state and microenvironmental structures cooperate to promote tumor progression.
INTRODUCTION
Tumor progression is driven by the dynamic interactions between cancer cells1,2 and the their surrounding microenvironment3,4. In this process, as cancer cells accumulate genetic and epigenetic alterations, the microenvironment exerts selective pressures through factors such as spatial constraints5,6, signaling molecules7, nutrient and oxygen availability8,9, and immune infiltration3,10 among other phenomena. In turn, tumor growth remodels the surrounding microenvironment, for example, by restructuring the extracellular matrix and altering the composition and state of infiltrating stromal cells11. Systematically characterizing the cell intrinsic and extrinsic effects that drive tumor subclonal selection, cellular plasticity, and metastasis will not only provide insights into the principles of tumor evolution but also carry clinical implications. To accomplish this, one must study a tumor’s evolutionary dynamics alongside its microenvironmental composition in the native spatial context.
Integrating tumor phylogenetic analysis, the study of lineage relationships of cancer cells within a tumor12–17, with spatial information provides a comprehensive framework for understanding the interplay between tumor microenvironment and progression. Specifically, spatially resolved phylogenetic studies enable one to approach key questions in cancer evolution such as, what are the major spatial communities that exist in tumors, and how do these relate to tumor stage? From which spatial niches do subclonal expansions arise during tumor progression, and how does this relate to tumor plasticity and the capacity to seed metastases? And, how does the spatial growth pattern of tumor progression shape the surrounding microenvironment? Early studies reconstructing tumor phylogenies from multi-region sampling of patient tumors uncovered the spatial heterogeneity of genetic changes within tumors and have demonstrated the dynamics of tumor growth and spatially-constrained origins of metastatic dissemination18–24. More recently, spatial genomics approaches have further elucidated how the spatial distribution of genome alterations leads to clonal outgrowth, dispersion of subclones with distinct driver mutations, interactions with the immune system, and metastasis25–29. While these studies have greatly enhanced our understanding of how tumors grow in space and time, they can be limited in their ability to either resolve high-resolution spatial organization, infer deeper phylogenetic relationships of cancer cells, or simultaneously measure the microenvironmental composition and gene expression.
The development of molecular recording technologies that install evolving lineage-tracing barcodes30–40 and associated computational tools41–46 enable the reconstruction of high-resolution phylogenies for studying tumor evolution13. Typically, these lineage-tracing technologies employ genome-editing tools, such as CRISPR/Cas9, to introduce heritable and irreversible mutations progressively at defined genomic loci, which can be transcribed and thus profiled with single-cell RNA-seq. In cancer, initial studies applied this technology to track the metastatic dynamics of cancer cell lines transplanted into mice47–49. Previously, we described a lineage-tracing enabled genetically-engineered mouse model of KrasLSL-G12D/+;Trp53fl/fl-driven lung adenocarcinoma (KP-Tracer) to continuously track tumor evolution from nascent transformation of single cells to aggressive metastasis50. In this system, intratracheal delivery of Cre recombinase using viral vectors simultaneously induces Cas9-based lineage tracing and tumor initiation. This model recapitulates the major steps of the evolution of human lung adenocarcinoma, both molecularly and histopathologically51–55. Using this system, we recently identified subclonal expansions, quantified tumor plasticity, traced metastatic origins and routes, and disentangled the effect of genetic drivers on tumor evolution. However, as our previous applications have relied on studying dissociated single cells, it has remained unclear how key tumor evolutionary properties are associated with microenvironmental changes.
Here, we present an integrated lineage and spatial platform for tracking tumor evolution in situ by applying high-resolution spatial transcriptomics to our lineage tracing-enabled KP-Tracer model. Using two complementary spatial transcriptomics assays – Slide-seq56,57 with spot-based coverage at 10μm near-cell resolution of large tissue fields-of-view, and Slide-tags58 with higher molecular sensitivity and spatial profiling of individual nuclei – we produce a comprehensive spatial transcriptomics dataset of Kras;p53-driven lung adenocarcinoma evolution. Integrating these spatial transcriptomics data with inferred cancer cell lineages uncovered robust spatial communities associated with tumor progression, including the formation of a hypoxic tumor interior during rapid tumor subclonal expansion. Our analysis additionally reveals that this hypoxic environment is associated with pervasive tissue remodeling characterized by fibrosis, priming of immune cells, and the emergence of a pro-metastatic epithelial-to-mesenchymal transition (EMT). Together, this study provides a scalable platform for studying the relationship between tissue architecture and tumor progression, revealing key insights into the ecological and evolutionary dynamics underpinning tumor evolution at unprecedented resolution.
RESULTS
An integrated lineage and spatial platform for studying tumor evolution
To study tumor evolution while preserving the native spatial context of cancerous and stromal tissue, we integrated spatial transcriptomics methods with Cas9-based lineage-tracing technology in our previously described KP-Tracer model of lung adenocarcinoma50. This model is built upon the well-characterized model of Kras;Trp53-driven lung adenocarcinoma51,52,54,55 and is equipped with a Cre-inducible Cas9-based evolving lineage tracer that is able to continuously record high-resolution cell lineages over months-long timescales32,41. Introduction of Cre into individual lung cells in the adult animal both induces the oncogene mutations (i.e., expression of KrasG12D and homozygous loss of p53) and initiates Cas9 expression. Cas9 then introduces irreversible and heritable insertions and deletions (“indels”) at defined genomic “target sites”, each discernable by a random 14bp integration barcode (“intBC”) and expressed as a polyadenylated transcript. As most sequencing-based spatial transcriptomics assays capture polyadenylated transcripts from tissue sections56–60, applying these assays to the KP-Tracer model yields simultaneous measurement of spatially-resolved cell transcriptional states and lineage relationships.
We initiated lung tumors and lineage-tracing in alveolar type II (AT2) cells (a major cell of origin for lung adenocarcinoma) by intratracheally delivering adenovirus expressing Cre recombinase under the control of an AT2 cell-specific, surfactant Protein C (SPC) gene promoter61. Twelve to sixteen weeks post tumor initiation, tumor bearing lungs were harvested for cryopreservation, and then sectioned and applied to spatial transcriptomics arrays (Figure 1A; Methods). To comprehensively profile the spatiotemporal evolution of tumor progression, we utilized two complementary spatial transcriptomics technologies: Slide-seq56,57 that captures transcriptomic states of “spots” at near-cellular 10μm resolution in continuous, large fields-of-view (up to 1cm × 1cm); and Slide-tags58 that sparsely samples individual nuclei for transcriptomic profiling and provides accurate spatial localization for a subset of these nuclei (typically ~50–70%). Together, this combination marries the scale of Slide-seq and true single-nucleus resolution of Slide-tags to jointly measure spatially resolved cell lineage and unbiased transcriptomic states in the native tumor microenvironment.
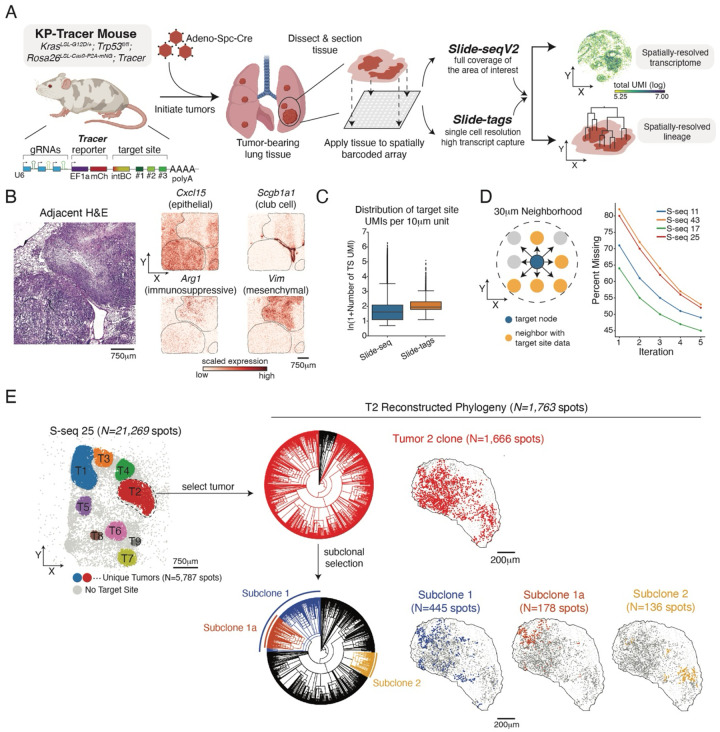
Figure 1. An integrated lineage and spatial platform enables high-resolution analysis of tumor evolution in vivo.
(A) Schematic of experimental workflow for integrated, spatially resolved lineage and cell state analysis. In KP-tracer mice, oncogenic KrasG12D/+;Trp53−/− mutations and Cas9-based lineage tracing were simultaneously activated upon administration of adenovirus carrying SPC promoter-driven Cre recombinase. After 12–16 weeks, mice were sacrificed, and cryopreserved tumor-bearing lungs were sectioned for spatial profiling with Slide-seq and Slide-tags technologies. Libraries were prepared and sequenced to study spatially resolved lineages and transcriptional patterns. S-seq 30 is used as a representative example for total UMI capture in a spatial array. Biorender was used to create parts of this schematic.
(B) Representative H&E staining and spatially resolved gene expression data for a lung section carrying three tumors (black line). Log-normalized, scaled counts for epithelial-like (Cxcl15 and Scgb1a1), immunosuppressive myeloid (Arg1), and mesenchymal cells (Vim) are shown.
(C) Distribution of the number of target-site UMIs for Slide-seq and Slide-tags data. Ln(1+x) counts are shown.
(D) Schematic of spatial imputation of lineage-tracing data in 30μm neighborhoods (left) and representative examples of missingness left after each of 5 iterations of spatial imputation. (E) Representative spatially resolved lineages in spatial array S-seq 25 profiling a lung section carrying 9 distinct tumors. Reconstructed lineages are displayed for a representative tumor, T2. Successive nested subclones displaying both shared and distinct lineage states in unique colors are indicated on the phylogenetic tree and mapped spatially. Lineages marked in black spots not included in the designated subclone. Overall, spots that are more related in lineage tend to be spatially coherent.
With these two technologies, we comprehensively profiled tumor-bearing lungs across various stages of progression with 44 Slide-seq arrays and 5 Slide-tags arrays (Figure S1A–C; Methods; Supplementary Table 1). The resulting datasets provided spatial profiling of distinct domains in tumor-bearing tissues characterized by the expression of canonical marker genes and corroborated by paired H&E: for example, in the tumor-bearing lung we found that Cxcl15 and Scgb1a1 marked epithelial-like domains, representing alveolar and club cells, respectively. Moreover, histologically aggressive regions were marked by Vim (characteristic of mesenchymal-like cancer cells) and Arg1 (characteristic of immunosuppressive myeloid cells62) (Figure 1B). Altogether, these datasets provide high-resolution views into the microenvironmental context and organization of tumors.
Computational tools enable the inference of spatially resolved cancer cell phylogenies
As the KP-Tracer system expresses lineage tracing target-sites as poly-adenylated transcripts, we next turned to evaluating the recovery of these target sites from the complementary spatial transcriptomics platforms. Reassuringly, we detected target-site transcripts robustly across tens-of-thousands of spots or nuclei in these spatial datasets, with Slide-tags data having more consistent detection of target-sites as expected (Figure 1C; Figure S1D–E).
While Slide-tags provided true single-cell measurements and thus were amenable to previously-described lineage reconstruction approaches41,44, there were two predominant analytical challenges in reconstructing tumor phylogenies of tens-of-thousands of spots observed in Slide-seq data. First, Slide-seq captures RNA molecules with near-cellular resolution, meaning that each spot may contain RNAs originating from multiple cells57; similarly, cells with distinct lineage states can be captured in a single spot, which we term “conflicting states”. As prior phylogenetic reconstruction algorithms for Cas9-lineage tracing data presume mapping of cells to single states, we first implemented new Cassiopeia-Greedy41 and Neighbor-Joining63 variants that could use many conflicting states during reconstruction (Methods). We also tested the effects of three strategies for preprocessing conflicting states via simulation: (1) a strategy that used all conflicting states observed in a spot along with the abundance of each state in that spot (“all states”); (2) all conflicting states observed in a spot, but without considering their abundance (“collapse duplicates”); or (3) a strategy that used only the most abundant state (“most abundant”). We found that the second strategy (“collapse duplicates”) performed most robustly (Figure S1F; Methods).
A second challenge is that Slide-seq assays (and to a lesser extent Slide-tags) have an increased missing data rate relative to droplet-based single-cell assays64. As expected, we observed overall lower target-site transcript capture (and thus higher missing data) in Slide-seq datasets (Figure S1D,G). We hypothesized that spatial relationships could be used to overcome this sparsity, which was supported by our observations that indel states were coherent within small spatial neighborhoods (Figure S1H–I). We therefore developed an inferential approach that predicted missing lineage-tracing states from spatial neighbors (within 30μm of a target node) with sufficient recovery (at least 3 UMI supporting a target site intBC-indel combination; Figure 1D). We first tested the feasibility of this approach using simulations of lineage tracing data on spatial arrays using Cassiopeia (Methods). We found that missing lineage-tracing barcodes could consistently be recovered at high accuracy (Figure S1J), and that spatial imputation followed by tree inference by a hybrid algorithm consisting of the Cassiopeia-Greedy and Neighbor-Joining algorithms resulted in the best reconstructions, especially in high-dropout regimes (Figure S1K–L; Methods). Next, we tested our ability to recover held-out target site data from real Slide-seq data and similarly found that missing data could be robustly recovered by spatial predictions, resulting in a median accuracy of 90% on imputing held-out data across all experiments, matching our simulation results (random predictions had a median accuracy of 67% and yielded 29% fewer imputations; Figure S1M). As expected, more frequent alleles had higher imputation accuracy (Figure S1N; Methods). Over multiple iterations of this imputation algorithm, we found that we could recover up to 58% of missing data (4–58%, on average 31% across datasets), resulting in comparable missing data rates to previous reports using single-cell approaches that have enabled robust tree reconstruction and biological insights (Figure 1D, Figure S1O). Though we only retain high-confidence imputations, and our benchmarks point to the promise of this spatial imputation in this context, there are notable caveats especially in the case of cell migration (see Limitations of this Study). Combining Slide-seq data and validation from orthogonal trees provided by Slide-tags establish a foundation for studying the spatial lineages of cancer cells.
Together, these computational improvements enabled us to build lineages of cancer cells in the native context of a tumor’s microenvironment at unprecedented resolution (Figure 1E). Our lineages revealed phylogenetic relationships in structured spatial environments and enabled us to explore the spatial localization of increasingly related subclones within the same tumor (Figure 1E, Figure S1P). With these data and approaches, we turned to investigating the relationship between changes to the microenvironmental architecture and tumor progression.
Spatial transcriptomics reveal the ecosystems of lung adenocarcinoma
While recent efforts have studied the composition of tumors in this model using single-cell approaches50,54,55, it has remained challenging to profile the spatial organization of these cell types. To address this, we leveraged the complementary insights gained from the high sensitivity, true single-nucleus measurements of Slide-tags and the broad field-of-view of Slide-seq to perform a systematic analysis of tumor spatial organization across stages of progression observed in our 49 spatial transcriptomics arrays representing more than 100 tumors.
Focusing first on the true single nuclei profiled with Slide-tags, we performed fine-grained annotation of clusters consisting of normal epithelial, stromal, immune, and tumor cells (determined by canonical marker genes and the presence of active lineage-tracing edits) (Figure 2A–B; Figure S2A; Methods). In addition to annotating previously described tumor and normal epithelial cells in this model50,55, we identified a previously undescribed tumor cell state characterized by the expression of neuronal genes such Piezo2 and Robo1, the endothelial marker Pecam1, maintenance of the lung-lineage transcription factor Nkx2-1, and absence of Vim (Figure S2B–C). Although this cell type expressed active lineage tracing marks in our system, it is likely that this cell type was excluded in previous studies50,55,65 by purifying cancer cells against CD31 expression (also known as Pecam1, expressed in this population) prior to transcriptomic profiling; this highlights the advantage of spatial transcriptomics in profiling all cells and communities, eliminating potential biases arising from tissue dissociation and preparation. In the immune and stromal compartment, we observed large macrophage, fibroblast, and endothelial populations with lower representation of B cells and dendritic cells (Figure 2A; Figure S2A). Among macrophages, we detected SiglecF+ tissue-resident alveolar macrophages and three distinct tumor-associated macrophage (TAM) populations: Vegfa+ TAMs, immunosuppressive Arg1+ TAMs, and proangiogenic Pecam1+ TAMs (Figure 2A). We additionally detected a diverse set of cancer-associated fibroblasts (CAFs): a mesothelial-like Wt1+ population, an inflammatory-like CAF (“iCAF”) population expressing the complement gene C7 and Abca8a, and a myofibroblast-like CAF (“myCAF”) population expressing Postn (Figure 2A, Figure S2A).
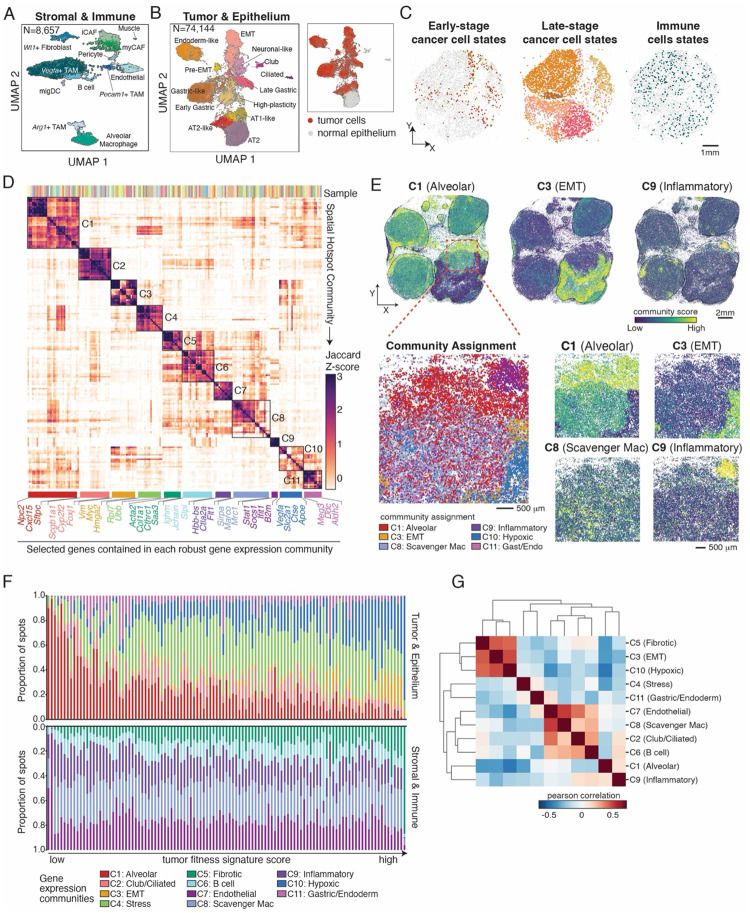
Figure 2. Diverse spatial gene expression communities emerge during KP-tracer tumor progression.
(A-B) UMAP projections of Slide-tags data on tumor bearing lungs from KP-Tracer mice, annotated by cell type. (A) Slide-tags data corresponding to all stromal and immune cell types: Cd45+ immune cells and other non-epithelial stromal cells. (B) Slide-tags data corresponding to all cancer and normal epithelial cells. Inset indicates where cancer cells are found in this projection.
(C) Representative spatial projections of early-stage and late-stage cancer cell states, and immune cell types from Slide-tags analysis of KP-Tracer tumor bearing lung (shown on S-tags 3). Colors correspond to those in UMAP projections in (A-B).
(D) Heatmap of Z-scored Jaccard overlap between genes contained in spatial gene expression communities. Each row or column is a community, defined as a set of spatially autocorrelated genes identified with Hotspot, and robust spatial gene expression communities are determined by hierarchical clustering and indicated by annotated blocks. The Slide-seq sample from which a community is identified is indicated by unique colors on the top of the heatmap. Representative genes specific to each spatial community are highlighted at the bottom of the heatmap.
(E) Community scores of selected spatial communities projected onto a representative Slide-seq dataset of a tumor bearing lung with 4 major tumors (S-seq 43). Tumor boundaries are indicated with black lines (top). Zoom in of region showing community assignments and scores for a selection of communities (bottom).
(F) Proportion of gene expression community assignments across all KP lung tumors in the Slide-seq dataset, ordered by increasing fitness signature scores. Each bar indicates a single segmented tumor in the Slide-seq dataset. Top: communities that are more related to tumor or epithelial programs. Bottom: communities that are related to stromal and immune programs. (G) Heatmap reporting Pearson correlation of community abundances across all tumors in the Slide-seq data.
To explore the spatial localization of these diverse cell states, we assigned spatial locations to Slide-tags nuclei and spatially projected cell identities. Consistent with previous characterizations of Slide-tags spatial mapping rates14, we found that approximately 50% of nuclei could be confidently assigned to a spatial location (Figure S2D). Across the five Slide-tags arrays, we observed a distinct pattern where less aggressive, “early-stage” tumor cell states (i.e., AT2- and AT1-like cancer cells, indicated by expression of active lineage marks and distinct gene expression from normal AT2 and AT1 cells) co-localized on the periphery of tumor sections consisting of more aggressive “late stage” tumor cells (Figure 2C, Figure S2E). Similar to previous work in this model66, we also found that distinct immune and stromal cell types exhibited differential infiltration – for example, Alveolar Macrophages and iCAFs were typically found outside tumors, whereas Arg1+ TAMs and myCAFs were more likely to be found within tumors (Figure 2C, Figure S2E).
The spatially-localized transcriptional signatures observed with Slide-tags motivated us to pair this approach with Slide-seq assays to survey the spatial gene expression communities across large tissue areas in tumors. We thus turned to the 44 tissue sections assayed with Slide-seq that collectively represent more than 100 tumors at various tumor stages. To identify modules of genes that were recurrently spatially co-expressed across multiple samples, we employed the Hotspot33 algorithm (Methods). Our analysis revealed 11 recurrent spatial gene modules, hereafter referred to as “communities” (Figure 2D–E), that we annotated by inspecting the genes contained within communities and evaluating the expression level of community genes (captured in a “community score”) in cell types identified by Slide-tags data (Figure S2F–G).
The genes contained within these transcriptional communities represent a variety of co-localized gene expression states: for example, an early-stage alveolar-like community contained genes marking epithelial cells such as Sftpc and Cxcl15 (“C1: Alveolar”), a hypoxic community contained canonical marker genes of hypoxia such as Slc2a1 (also known as Glut1) (“C10: Hypoxia”), and an epithelial-to-mesenchymal (EMT) community contained genes such as Vim, up-regulation of Myc signaling, and metastasis-related genes such as Hmga2 (“C3: EMT”; Figure 2D–E, Figure S2G). In addition to fibroblast (C5), B cell (C6), and endothelial (C7) communities, we identified two distinct immunoregulatory-related communities. The first community contained genes associated with scavenger-like macrophages like Marco and Mrc1 (“C8: Scavenger Mac”); a second community contained genes characteristic of inflammation such as B2m, Stat1, and Ifit1 (“C9: Inflammatory”). As these communities describe genes co-expressed in spatial proximity, they provide insights into possible intercellular interactions. For example, the EMT and hypoxic communities (C3 and C10) contained genes associated with macrophage recruitment (e.g. Csf1) and polarization to immunosuppressive states that have been previously reported to promote aggressive cancer phenotypes (e.g., Arg162 and Spp167), while the Inflammatory community (C9) contained Cxcl9 that has been previously reported in anti-tumor macrophage polarization67 (Figure S2G).
To inspect the distribution of these communities across large tissue sections profiled with Slide-seq, we quantified community scores for each spot and assigned spots to the community with the highest score (Figure 2E–F, Figure S2H–I). In comparing histology from an adjacent layer to the community scores, we found co-localization between areas indicating high tumor grade (as indicated by histology) and high scores for EMT, hypoxic, and fibrotic communities (C3, C10, C5; Figure S2H). We next asked how the distribution of community assignments varied over tumor stages using a gene set signature we previously identified to robustly associate with tumor progression (termed a “fitness signature”)50 (Figure 2F; Figure S2I; Methods). Specifically, this fitness signature contains genes that are associated with subclonal expansions in this model, and their collective activity (i.e., “score”) reflects tumor progression towards an aggressive, pro-metastatic state. Consistent with the definition of this signature, after ranking tumors by their fitness signature score and inspecting the proportion of community assignments, we observed that early-stage tumors were dominated by epithelial, endothelial, and inflammatory communities (C1, C7, and C8, respectively) but that late-stage tumors had larger fractions of EMT, hypoxic, and fibroblast communities (C3, C10, and C5, respectively; Figure 2F, Figure S2I). Moreover, we found that overall abundances of EMT, hypoxic, and fibroblast community assignments (C3, C10, and C5, respectively) were correlated across all tumors; conversely, they were anticorrelated with the abundances of alveolar and inflammatory communities (C1 and C8, respectively) (Figure 2G).
Together, these analyses unite the unique advantages of Slide-tags and Slide-seq assays to provide a consensus set of spatial communities that highlight differential immune and stromal activation and localization patterns across tumor progression in KP tumors. These observations motivated us to next integrate our phylogenies to understand how the spatiotemporal dynamics of these communities are associated with tumor plasticity and subclonal expansion.
Rapid tumor subclonal expansion contributes to a hypoxic niche with decreased cancer cell plasticity
Integrating cell state information with high-resolution phylogenies can offer new insights into various aspects of tumor evolution, such as the historical record of subclonal growth rates (i.e, “phylogenetic fitness”) or the kinetics of tumor cell state transitions (which can be quantified as a “clonal plasticity” score for each cell). In our previous work, we described a model whereby KP-Tracer tumor progression is driven by the loss of an initial AT2-like cell state and accompanying increases in single-cell clonal plasticity and transcriptional heterogeneity; in turn, these high-plasticity cells provide a diverse pool of transcriptional states from which high-fitness, low-plasticity subclones with increased metastatic ability and expression for EMT markers like Vim and Hmga2 are selected50. Consistent with this previous work, the tumors studied with this spatial-lineage platform showed an overall distribution where transient increases in plasticity are followed by the selection of low-plasticity, high fitness subclones (Figure S3A). Using this platform, we sought to understand how our previously described model unfolds spatially and associates with changes to the surrounding microenvironment.
As the measurement of phylogenetic fitness reports on the history of subclonal growth, spatially-resolved phylogenies are well suited to understanding the growth patterns in tumors and their molecular consequences22,68. In one representative Slide-seq example (S-seq 40), we found an expanding subclone with high phylogenetic fitness localized to a tumor interior characterized by late-stage Hypoxic and EMT communities (C10 & C3) while the tumor periphery had lower phylogenetic fitness and was marked by the Alveolar community (C1) (Figure 3A). This co-localization of high phylogenetic fitness with hypoxic regions was supported by three lines of evidence: first, we found that phylogenetic fitness was correlated with the orthogonal, previously-described fitness signature50 (Pearson’s r = 0.4; Figure S3B). Second, in a systematic analysis of all Slide-seq tumors, we found that the EMT and Hypoxic communities were most strongly correlated with phylogenetic fitness (Figure S3C). Finally, across all high-resolution Slide-tags arrays, we similarly found that the late-stage states (e.g., EMT and Endoderm-like) were most likely to be found in regions that had previously undergone subclonal expansion (Figure S3D). These orthogonal data collectively support the observation that the co-localization of expansion and hypoxia is consistent across tumors and is not an artifact of tree reconstruction or the near-cell resolution of Slide-seq.
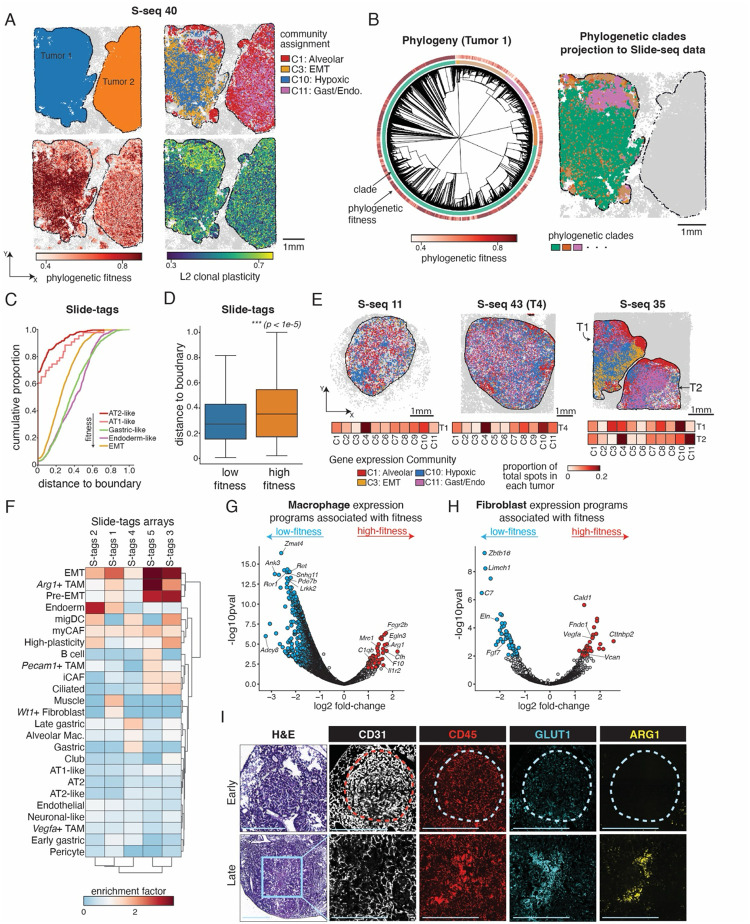
Figure 3. Subclonal expansions associate with microenvironmental remodeling towards a hypoxic, fibrotic, and immunosuppressive state.
(A) A representative Slide-seq array containing two tumors (S-seq 40) is shown with spatial projections of tumor annotations, selected gene expression community assignments, phylogenetic fitness, and L2 clonal plasticity.
(B) Reconstructed phylogeny and spatial localization of phylogenetic subclades for Tumor 1 from the representative Slide-seq dataset (S-seq 40) example shown in (A). The phylogeny is annotated by subclonal clade assignment (inner color track) and phylogenetic fitness (outer color track).
(C) Cumulative density distributions for normalized Euclidean distance to nearest non-tumor cell (i.e., tumor boundary) for five selected major cancer cell states across all Slide-tags arrays. Cancer cells in high-fitness-associated cell states (e.g. EMT, Endoderm-like, Gastric-like) locate further away from the tumor boundary than those in low-fitness-associated states (AT2-like, AT1-like). Distance is normalized to unit scale (0–1).
(D) Distribution of normalized Euclidean distances to nearest non-tumor cell (i.e., tumor boundary) for high-fitness and low-fitness cells (defined here as having phylogenetic fitness greater than the 90th or less than the 10th percentiles, respectively). High-fitness cells are significantly further away from the tumor boundary (p<1e-5, wilcoxon rank-sums test).
(E) Representative Slide-seq examples showing the evolution of the spatial gene expression communities following tumor progression (left to right). Selected community assignments are displayed, and full proportion of assignments are reported in 1D heatmaps under each spatial dataset.
(F) Clustered heatmap of enrichments of cell type abundances in spatial neighborhoods of high- and low-fitness cells in 5 Slide-tags arrays. Values > 1 indicate that a cell type is more abundant (i.e., enriched) in neighborhoods of cells with high fitness. Cell type names are identical to those reported in Figure 2A–B.
(G-H) Differential expression analysis of (G) macrophage and (H) fibroblast polarization states in neighborhoods of high- and low-fitness cells from Slide-tags arrays. Each dot is a gene, and significant hits (log2|FC| >= 1 and false-discovery-rate adjusted p-value < 0.05) are reported in red and blue. Red genes are up-regulated in neighborhoods of high-fitness cells, and blue genes are down-regulated. Significant GO terms are reported in Supplementary Table 1.
(I) H&E and paired immunofluorescence staining of endothelial-cell marker CD31, immune cell marker CD45, hypoxia-reporter GLUT1, and immunosuppressive myeloid marker ARG1 in representative KP tumors. The interior of large, late-stage tumors is marked with a decrease of endothelial cells (CD31) and increases of hypoxia (GLUT1) and immunosuppressive myeloid cells (ARG1, CD45). Scale bars = 1mm.
The localization of expanding subclones characterized by aggressive gene expression states in a representative Slide-seq example (S-seq 40) prompted us to hypothesize that rapid subclonal expansions may create a layered environment whereby expanding subclones dominate a core surrounded by non-expanding cells (Figure 3A–B). Focusing first on this representative Slide-seq example, we observed that multiple low-fitness areas of Tumor 1 could be grouped together in a phylogenetic subclade despite being geographically distant (though many indels were shared across the tree, these low-fitness, distant cells were marked by the shared absence of indels marking the expanding region) (Figure 3A–B; Figure S3E). Though this pattern could be generated many ways (e.g., independent migration of several subclones), the most parsimonious interpretation suggests that these scattered low-fitness cells were in close spatial proximity during the early stage of tumor growth but were later pushed to the tumor periphery because of a subclonal expansion event. To investigate the consistency of this phenomenon, we next quantified the phylogenetic fitness of individual cancer cells derived from high-resolution Slide-tags arrays on multiple tumors and inspected the spatial distribution of subclonal expansion. In this analysis, we also found that the tumor core in Slide-tags data was more likely to contain cells with more aggressive gene expression states (e.g., Endoderm-like and EMT states) and higher phylogenetic fitness as inferred from reconstructed trees (Figure 3C–D p < 1e-5, wilcoxon rank-sums test; Figure 2C; Figure S2E).
The observed data supporting a model in which subclonal expansion creates an aggressive, hypoxic interior led us to next explore whether the transitions between gene expression states also occur in a spatially coherent manner. As demonstrated in our previous work, integrating high-resolution lineage tracing offers a unique opportunity to quantitatively measure the frequency of cell state transitions, or “single-cell clonal plasticity”50,69. Starting in the representative Slide-seq example (S-seq 40), we observed that low-plasticity clones in Tumor 1 co-localized with high-fitness regions in the tumor interior whereas the high-plasticity regions of Tumor 2 (which lacked a subclonal expansion) appeared to lack spatial organization (Figure 3A). Consistent with this, we found that the high-fitness Hypoxic and EMT communities, and related states, were associated with lower plasticity across all Slide-seq and Slide-tags datasets (Figure S3F–G). To better understand how transient increases in plasticity contribute to the subclonal expansions observed across Slide-seq datasets (Figure S3A), we further examined the transition to subclonal expansion in arrays profiled with Slide-tags (Figure S4H–J). Across our Slide-tags data, we found there was little spatial organization of high-plasticity cells in tumors without detectable subclonal expansion (as measured by Moran’s I autocorrelation statistic70), whereas low-plasticity cells were spatially localized to the tumor center in tumors after expansion (Figure S3I–J; Methods). This suggests that subclonal expansion, and its associated molecular changes, are important for coherent spatial organization during tumor progression.
Collectively, these data support a model whereby the tumor microenvironment is sequentially remodeled by subclonal expansion, culminating in a hypoxic core and eventually the emergence of a late-stage, pro-metastatic EMT state. As evidenced by examples of tumors across various stages, this model is characterized by the exclusion of early-stage communities (e.g., C1: Alveolar) to the tumor periphery while subclonal expansions contribute to the acquisition of a low-plasticity, high-fitness Hypoxic community (C10) and eventual transition to an EMT community (C3) (Figure 3E; Figure 2F; Figure S2I).
Subclonal expansion is accompanied by immunosuppressive and fibrotic microenvironmental remodeling
As our Slide-seq data suggest that the microenvironment is remodeled during subclonal expansion, we next exploited Slide-tags data to dissect the expansion-associated cell state transitions at single-nucleus resolution. After quantifying phylogenetic fitness on trees inferred from Slide-tags data, we stratified nuclei into high- and low-fitness groups and inspected the cell type abundances in their spatial neighborhoods (Figure 3F; Figure S3K; Methods). As expected, we found that the EMT cancer cell state was most consistently enriched in neighborhoods surrounding high-fitness nuclei (Figure 3F). With respect to differential enrichment of specific immune and stromal populations, we found that Arg1+ TAMs and myCAF populations were consistently enriched in spatial neighborhoods of high-fitness cells whereas iCAFs and other TAMs were not (Figure 3F). To more systematically probe the polarization states of macrophages and fibroblasts associated with subclonal expansions, we performed differential expression within these cell types in spatial neighborhoods of high- and low-fitness cells (Figure 3G–H). In addition to high Arg1 expression, macrophages in spatial neighborhoods of high-fitness cells were characterized by the presence of the hypoxia-induced factor Egnl3, the Fcg-receptor Fcgr2b, the macrophage scavenger receptor Mrc1, and enriched for programs indicating increased endocytosis and complement activity (Figure 3G; Table S1). Fibroblasts associated with spatial neighborhoods of high-fitness cells were characterized by higher expression of genes implicated in hypoxia, collagen synthesis, and fibrosis such as Vcan, Fndc1, Cald1 and Vegfa (Figure 3H; Table S1).
To inspect the generalizability of these patterns, we returned to the comprehensive dataset of 44 Slide-seq arrays. Indeed, a systematic analysis of our Slide-seq arrays revealed that spatial neighborhood surrounding high-fitness, low-plasticity spots were most enriched for EMT, Hypoxic, and Fibrotic communities (C3, C10, and C5, respectively) and depleted for Alveolar, Endothelial, and Inflammatory communities (C1, C7, and C9, respectively) (Figure S3L–M; Methods). Moreover, consistent with our finding in this mouse model, reanalysis of published spatial transcriptomics data of human lung adenocarcinoma40 demonstrated that expression of the hypoxia-reporter SLC2A1 (also known as GLUT1) in tumors was associated with cell proliferation (as measured by MKI67), TGFβ signaling, EMT (SNAI2), and immunosuppressive macrophage polarization (FCGR2B) (Figure S3N–O).
Together, these differential gene expression programs suggest a model whereby subclonal expansion promotes a hypoxic tumor interior that polarizes immune and stromal cells into pro-tumor immunosuppressive and fibrotic states and facilitates the emergence of a pro-metastatic cancer cell state. Indeed, in returning to our previous Slide-seq analysis of community program assignments across tumor progression, we observed that the Hypoxic community (C10) appears prior to EMT (C3) when ranked by the transcriptional fitness signature (Figure 2F; Figure S2I). In further support of this, immunofluorescence staining of KP-Tracer tumors revealed that hypoxia (as evidenced by the canonical hypoxia marker GLUT1 [Slc2a1] protein levels71,72) preceded the emergence of immunosuppressive ARG1+ immune cells (Figure 3I).
Spatially resolved lineages reveal the evolution of metastasis-initiating niches in the primary tumor
Metastasis, the ultimate stage of tumor progression, accounts for approximately 90% of cancer-related mortality and is associated with pervasive microenvironmental remodeling73–77. However, it has remained challenging to delineate the specific microenvironmental features associated with tumor evolutionary dynamics during metastasis progression. Outstanding questions include: do the niches surrounding subclones giving rise to metastases differ from those surrounding other subclones? How do these gene expression programs change during metastatic spread? Our spatial-lineage platform is well-suited to identify the spatial localization of metastasis-initiating subclones and characterize the microenvironmental remodeling associated with each step of the metastatic cascade.
We began by performing spatial transcriptomics on a KP-Tracer mouse with multiple primary lung tumors and widespread metastases in the mediastinal lymph node, rib cage, and diaphragm (Figure 4A, Figure S4A). To maximize the probability of detecting metastasis-initiating subclones in primary tumors, we sampled multiple representative layers of the tumor-bearing lung at approximately 200–500um intervals, enabling us to study multiple large primary tumors from top-to-bottom. Tumor segmentation of Slide-seq data from these sections and coarse-grained spatial alignment determined by shared lineage states revealed four major tumors that could be tracked across layers (Figure 4B).
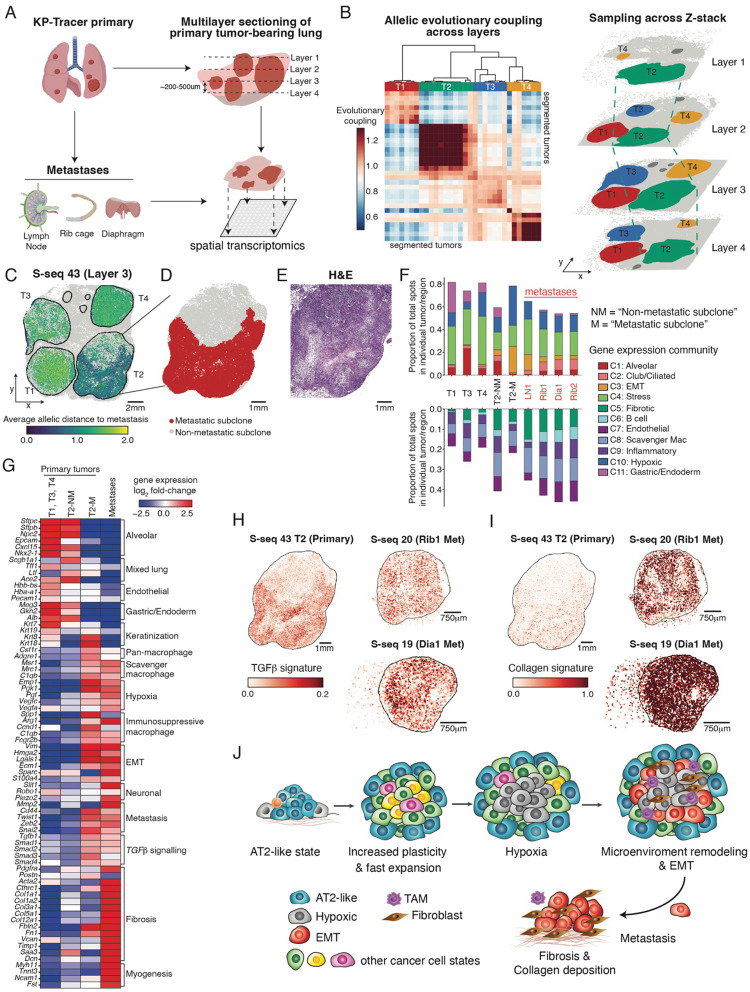
Figure 4. Tracing the evolution of subclonal niches across the metastatic cascade.
(A) Schematic of spatial transcriptomics workflow from a KP-Tracer mouse with large primary lung tumors and paired metastases from the lymph node, rib cage, and diaphragm. Multiple lung sections with four large primary tumors were harvested and subjected to both Slide-seq and Slide-tags assays. Biorender was used to create parts of this schematic.
(B) Coarse-grained alignment of Slide-seq spatial transcriptomics data (based on lineage-tracing edits) from four representative layers (Layer 1 – Layer 4) of a KP tumor bearing lung at approximately 200–500μm intervals from different z position. (Left) A clustered heatmap of allelic evolutionary coupling scores across all Slide-seq datasets from the tumor-bearing lung identifies the four major tumors. Each row or column is a single tumor from one Slide-seq dataset. (Right) 3D reconstruction of aligned datasets, annotated by one of four major tumors. Individual tumors are labeled in different colors.
(C) Representative spatial projection (S-seq 43) of allelic distances – summarizing how different lineage-tracing edits are between cells – for each spot with lineage-tracing data. Distance was computed to a consensus metastatic parental allele and normalized between 0 and 2.
(D-E) The metastasis-initiating subclone in T2 was segmented from cells with high relatedness to metastatic tumors and labeled in red. (E) H&E staining of T2.
(F) Proportion of gene expression community across representative stages of the metastatic cascade, including primary lung tumors (T1,3,4) without relatedness to metastases, the metastasis-initiating (M) and non-metastatic-initiating (NM) subclones in the primary tumor (T2) that gave rise to metastases, and four metastases. Top: communities that are more related to tumor or epithelial programs. Bottom: communities that are related to stromal/immune programs.
(G) Heatmap of gene expression log2-fold-changes between environmental niche (primary tumors without metastatic relationship, non-metastasis-initiating (NM) and metastasis-initiating (M) subclones within T2, and metastases). Genes are manually organized into ontologies.
(H-I) Spatial projection of gene expression scores of the Hallmark TGFβ and Collagen gene signatures on the metastasis-initiating primary tumor and selected metastases. Tumor 2 on S-seq 43 is used as the representative layer.
(J) A schematic model of KP tumor evolution and microenvironmental remodeling.
Our spatial-lineages in the large Slide-seq assays provide an opportunity to both compare the trajectory of multiple tumors and understand the transcriptional evolution of the niche surrounding the metastasis-initiating subclone in a single primary tumor. To do so, we first identified the spatial localization of subclones giving rise to metastasis by inspecting the allelic similarities between primary tumors and metastases (Figure 4C). This analysis revealed that metastases from all 3 locations were phylogenetically related to a spatially coherent subclone in primary Tumor 2 (“T2”). T2 could be identified in each layer independently and could be thus tracked across all sampled layers of this primary tumor (Figure S4B–C). This pattern was consistent in matched Slide-tags data, overlapped with subclonal expansions identified from our phylogenies, and was associated with regions exhibiting poorly differentiated histological features (Figure 4C–E; Figure S4C–F). Because all metastases shared indels with an expanding subclone that could be found across layers, it is most likely that all metastases arose after subclonal expansion.
To understand the phylogenetic and gene expression programs underlying metastatic potential in this region of T2, we segmented this tumor into a niche surrounding the cells giving rise to metastases (“T2-Met”) or otherwise (“T2-NonMet”) and compared their gene expression patterns (Figure 4D–E; Figure S4D–F; Methods). The T2-Met niche had higher proportions of the EMT and Hypoxic communities (C3 & C10, respectively) and lower proportions of the Gastric/Endoderm and Alveolar communities (C11 & C1, respectively) (Figure 4F). The T2-Met niche additionally down-regulated genes associated with Gastric and Endoderm states (e.g., Gkn2 and Meg3), and had higher expression of genes marking cancer cell EMT (e.g., Vim), scavenger macrophages (e.g., Mrc1 and Msr1), immunosuppressive macrophages (e.g., Arg1 and Fcgr2b), TGFβ signaling (e.g., Tgfb1 and Smad4), and fibrosis (e.g., Cthrc1 and Postn) (Figure 4G). Orthogonal analysis with Slide-tags data corroborated these findings, as Arg1+ TAMs and myCAFs were most enriched in spatial neighborhoods of cells in the primary tumor related to metastases (Figure S4G). Moreover, immunofluorescence staining confirmed that ARG1+ cells co-localized with the metastasis-initiating VIM+ region of the T2 primary tumor (Figure S4H). Together, these results nominate several key molecular processes as potential drivers of the pro-metastatic niche, including fibrosis, TGFβ signaling, and intercellular interactions between cancer cells, activated fibroblasts, and Arg1+ immunosuppressive macrophages.
Metastatic colonization is accompanied by increased collagen deposition and fibrosis
Beyond the evolution within the primary tumors, we next investigated whether the microenvironments at distant metastatic sites are remodeled to resemble, or diverge from, the metastasis-initiating niche within the primary tumor. Comparing the niches surrounding metastases and the T2-Met subclone in the primary tumor, we found that metastases contained proportionally more regions annotated by stromal or immune communities and showed specifically higher representation of the Fibrotic community (C5) (Figure 4F). As these communities represent several gene programs and may mask fine-scaled cell type changes, we further characterized the differential gene expression changes distinguishing niches of the primary tumor and metastases (Figure 4G). While metastases up-regulated genes also found to distinguish the T2-Met niche – such as the EMT markers Vim and Hmga2 and TGFβ-related genes – metastases displayed large up-regulation of genes associated with collagen deposition (e.g., Col1a1 and Col12a1) and myogenesis (Tnnt3 and Ncam1) (Figure 4G). After quantifying the activity of these gene expression programs in Slide-seq spots, we confirmed that these aggregated gene expression signals were spatially localized to tumor regions: metastatic tumors generally resembled the metastasis-initiating subclone in the primary tumor (for example with respect to TGFβ signaling: log2FC = −0.14, t-test p=1.0; Figure 4H) but substantially up-regulated collagen-related genes as compared to the primary tumor (log2FC = 3.81, t-test p<1e-5) (Figure 4I). Consistent with this finding in Slide-seq data, immunofluorescence staining showed a marked increase in COL3A1 protein in metastases as compared to primary tumors (Figure S4I). Collectively, these results complement recent findings that TGFβ signaling is critical for EMT and metastatic seeding in this model74, and highlight that while certain expression programs – such as TGFβ signaling – precede metastasis in the metastasis-initiating subclone, the resulting metastatic tumor is remodeled to have increased fibrosis and collagen-related gene program activity.
DISCUSSION
In this study, we integrated high-resolution spatial transcriptomics with Cas9-based lineage tracing in a genetically engineered mouse model of lung adenocarcinoma to dissect the dynamic interplay between tumor evolution and microenvironmental remodeling in a spatially resolved fashion. Our analysis uncovered spatial communities associated with different stages of tumor progression; revealed relationships between tumor growth, plasticity and microenvironmental remodeling; and identified metastasis-initiating subclones that informed on the spatiotemporal evolution of gene expression along the metastatic cascade. These results present an unprecedented spatial map of lung adenocarcinoma evolution, showcasing the power of integrating spatially resolved transcriptomics and lineages to dissect the complex tumor dynamics underlying cancer progression.
The insights into spatiotemporal dynamics offered by this spatial-lineage platform contributes new dimensions to our previous model of KP tumor evolution (Figure 4J). Our previous results provided several lines of evidence that tumors, following the initial loss of an AT2-like state, are characterized by a cancer-cell-intrinsic increase in clonal plasticity, leading to gains in transcriptional heterogeneity and subsequent subclonal expansion50. In the present study, we find that rapid subclonal expansion pushes early-stage cells to the tumor periphery and contributes to the formation of a hypoxic microenvironment in the tumor core. This hypoxic niche promotes additional microenvironmental remodeling characterized by Arg1+ immunosuppressive myeloid subsets and myCAF-like fibroblasts; for example, by recruiting myeloid cells through hypoxia-induced chemokine secretion (e.g., Ccl2, Ccl6, and Csf1) and polarizing immune and stromal cells through hypoxia-induced signaling cascades (e.g., Hif1a and Vegfa) as previously suggested78–82 (Figure S2A,G; Figure 3G–H). In turn, this hypoxic, immunosuppressive, and fibrotic niche may contribute to another wave of cancer cell state transitions and the emergence of a pro-metastatic EMT state, for example through TGFβ signaling as shown in our analysis (Figure 4G–H) and detailed in a recent study74. As these cells metastasize, the metastatic environment is further remodeled to an enhanced fibrotic niche marked by increased collagen deposition.
Epigenetic remodeling is a hallmark of cancer and has been shown to play a critical role in cancer progression and drug resistance83–85. Our proposed model of tumor progression provides key insights into how cancer-intrinsic alterations and external signals integrate to regulate tumor cell states. Building on previous work in this model which has shown that tumor progression is driven by epigenetic rather than somatic changes50,54, our analysis adds more granularity into this process and suggests an appealing hypothesis that epigenetic remodeling can be disentangled into two distinct phases. First, following the loss of the AT2-like state, cancer cells enter a permissive epigenetic phase characterized by increased plasticity and transcriptional heterogeneity. As high-plasticity regions of these tumors do not appear to be spatially coherent (Figure 3A, Figure S3H–I), this suggests that this phase of epigenetic remodeling is mostly driven by cell-intrinsic changes accompanying the loss of the AT2-like state.
In contrast, the second phase of epigenetic changes follows subclonal expansions that drive microenvironmental remodeling towards a hypoxic state characterized by immunosuppressive and fibrotic communities. As several lines of evidence suggest that hypoxia precedes the formation of the EMT state (Figure 2F, Figure S2I, Figure 3E), we postulate that these environmental changes contribute to the induction and selection of an epigenetically-stable, pro-metastatic EMT state. This hypothesis aligns with prior reports associating hypoxia with genomic instability and EMT22,86–88, including in human lung adenocarcinoma89, and here our spatial-lineage data provide new evidence linking subclonal expansion as a mechanism driving hypoxia and tumor progression. In addition to our observation that human lung adenocarcinoma tumors contain spatially-defined hypoxic regions90 (Figure S3N–O), hypoxia has also been shown to play critical roles in lung adenocarcinoma91 and other cancers (e.g., glioma92 and clear cell renal cell carcinoma22); thus, further dissecting the relationships between subclonal expansions and hypoxia in these cancers may reveal opportunities for therapies spanning multiple cancer types. Together, these findings provide fundamental insights into how cancer cell states are regulated by both intrinsic and extrinsic changes and highlight the possible therapeutic ramifications of this regulation.
While our study elucidates new aspects of how tumor evolution unfolds spatially, it also sets the foundation for further studies. First, mechanistic studies will be needed to dissect how the hypoxic niche polarizes immune and stromal subsets, and how this might lead to an aggressive, mesenchymal tumor state. As we have previously reported that plasticity plays an important role in tumor progression28,50,55,83, one area of research will be how hypoxia affects the high-plasticity cell states in lung cancer. Second, the platform we developed here can be adapted to study the spatiotemporal dynamics of tumor evolution in other models or under different perturbations. Notably, our platform is also amenable to modeling the effect of additional genetic perturbations as Cas9 is continuously expressed for tracing50. Third, while we introduced new computational approaches for phylogenetic reconstruction approaches that address the sparsity, resolution, and scale of these data, there remain opportunities to build new algorithms specifically tailored to the spatial aspect of data and statistically infer how spatial organization affects phylogenetic patterns.
In summary, our study unites the insights provided by spatially resolved lineages and transcriptomics to investigate the fundamental patterns of tumor growth and its interactions with the microenvironment. Our analyses lead to a comprehensive model of how a tumor grows from a single, transformed cell into a large and complex ecosystem and provided new evidence for how tumor expansion-associated microenvironmental remodeling may contribute to a distinct wave of cell state reprogramming towards pro-metastatic states. As one of the most comprehensive datasets of spatial tumor evolution to date, we anticipate that this resource will help pioneer new computational methods and quantitative and predictive models of tumor evolution.
Limitations of the study
While our study reveals new aspects of tumor progression, there are limitations in the interpretation and extensibility of the approaches applied here. First, a single slide section may not represent the entirety of clonal dynamics in a tumor. To minimize this potential bias, we corroborated phylogenetic patterns with histology, orthogonal gene expression signatures derived from our previous single-cell lineage-tracing data (derived from unbiased sampling of whole tumors) and analyzing representative sections at different depths of tumors from a tumor-bearing lung in Figure 4. As scaling spatial transcriptomics experiments becomes more affordable, future studies can more densely sample three-dimensional structure to entirely account for this bias. Second, as a consequence of profiling tumor sections, we observe less indel diversity in spatial lineage tracing data than in previous applications with unbiased sampling, leading to lower resolution phylogenetic relationships. This may be ameliorated by optimizing the lineage-tracing kinetics and adapting tools for recording past molecular signaling events93,94. Third, the molecular sparsity and resolution of Slide-seq data pose a challenge in reconstructing phylogenies and detecting smaller spatial neighborhoods. While we provide a spatial imputation algorithm to account for these technical issues, and benchmark its effectiveness in a variety of simulated and held-out experiments, we anticipate that this imputation approach may have limitations in cases where lineage data is not spatially coherent, for example in systems with higher degrees of cell migration. In these scenarios, either alternative technologies with improved capture and resolution or new algorithms for performing spatial imputation and detecting robust spatial communities will be necessary. Finally, the trees presented in this study are only estimates of true phylogenetic relationships, and may not truly reflect cell division histories; when possible, our study uses orthogonal data and approaches to substantiate all claims.
METHODS
EXPERIMENTAL MODELS AND SUBJECT DETAILS
KP-Tracer mouse was generated by generating chimeric mice from blastocyst injection of engineered, lineage tracer enabled mouse embryonic stem cells harboring conditional alleles KrasLSL-G12D/+;Trp53fl/fl; Rosa26LSL-Cas9-P2A-mNeonGreen as previously described50. Eight-to-twelve-week-old KP-Tracer mice were infected intratracheally with ad5-SPC-Cre virus (1×10^8 Pfu) purchased from University of Iowa viral vector core for tumor initiation. This enables specific tumor initiation and lineage-tracing in Alveolar Type II (AT2) cells, the major cell-type of origin of lung adenocarcinoma. All studies were performed under an animal protocol approved by the Massachusetts Institute of Technology (MIT) Committee on Animal Care. Mice were assessed for morbidity according to MIT Division of Comparative Medicine guidelines and humanely sacrificed prior to natural expiration.
METHODS DETAILS
Sample processing
Tumor-bearing lungs were harvested and re-inflated with ~2ml of 50% OCT (1:1 mix with PBS) and 1:100 of RNase inhibitor (NEB M0314L). After cleaning up excess blood and liquid, the whole tissue was embedded in 100% OCT and frozen using dry ice-methanol bath. Frozen samples were kept at −80C until sectioning for further analysis.
Spatial transcriptomics with Slide-seqV2
For 3 mm and 5.5 mm arrays.
Fresh frozen tissues were cryo-sectioned at a thickness of 10 μm using a Cryostat (CM1950, Leica) set at −17 to −18 °C. The tissue sections were carefully transferred onto precooled arrays, which were placed on top of a glass slide inside the cryostat. A finger was briefly placed underneath the slide to melt the tissue and adhere it to the array. Immediately after, the tissue and array were transferred together into a 1.5 ml or 2 ml Eppendorf tube containing 200 μl (for 3 mm arrays) or 500 μl (for 5.5 mm arrays) of hybridization buffer (6x SSC with 2 U μl−1 Lucigen NxGen RNase inhibitor, Lucigen, 30281). The samples were incubated in the hybridization buffer for 15 minutes to 1 hour at room temperature, allowing the RNA to bind to the oligonucleotides on the beads. After incubation, the tissue and array were briefly dipped into 1x Maxima RT buffer to wash off the hybridization buffer and then transferred to the reverse transcription (RT) reaction mixture (1x Maxima RT buffer, 1 mM dNTPs (NEB, N0477L), 2 U μl−1 Lucigen NxGen RNase inhibitor, 2.5 μM template switch oligonucleotide, 10 U/μL Maxima H Minus reverse transcriptase (Thermofisher Scientific, EP0753)). The tissue and array were incubated in 200 μl (for 3 mm arrays) and 500 μl (for 5.5 mm arrays) of the RT reaction mixture for 30 minutes at room temperature, followed by 1.5 hours at 52 °C. To digest the tissue, 200 μl (for 3 mm arrays) or 500 μl (for 5.5 mm arrays) of tissue digestion buffer (200 mM Tris-Cl pH 8, 400 mM NaCl, 4% SDS, 10 mM EDTA and 32 U ml−1 proteinase K (NEB, P8107S)) was added to the reaction mixture and incubated at 37 °C for 30 minutes. Following digestion, 200 μl (for 3 mm arrays) or 500 μl (for 5.5 mm arrays) of wash buffer (10 mM Tris pH 8.0, 1 mM EDTA and 0.01% Tween-20) was added, and a P200 pipette was used to carefully triturate the beads off the array. The beads were centrifuged at 3000g for 2 minutes, followed by three washes with wash buffer. To remove RNA strands, the beads were incubated in 0.1N NaOH for 5 minutes, followed by a wash with wash buffer and 1x TE buffer, and centrifuged again at 3000g for 2 minutes. Second-strand synthesis was performed by mixing the beads with 200 μl (for 3 mm arrays) or 500 μl (for 5.5 mm arrays) of second-strand synthesis mixture (1x Maxima RT buffer, 1 mM dNTPs, 10 μM dN-SMRT oligonucleotide and 12.5U μl−1 Klenow enzyme (NEB, M0210)) and incubating at 37 °C for 1 hour. The beads were then washed three times with wash buffer and once with water. cDNA amplification was carried out by resuspending the beads in 200 μl (for 3mm arrays) or 1.2 ml (for 5.5 mm arrays) of cDNA amplification mixture (1x Terra Direct PCR mix buffer (Takara Biosciences, 639270), 1.25 U μl−1 of Terra polymerase (Takara Biosciences, 639270), 2.5 μM TruSeq PCR handle primer and 2.5 μM SMART PCR primer). The reaction was divided into 50 μl aliquots and amplified using the following PCR program: 95 °C for 3 min; four cycles of 98 °C for 20 s, 65 °C for 45 s and 72 °C for 3 min; nine cycles of 98 °C for 20 s, 67 °C for 20 s and 72 °C for 3 min; 72 °C for 5 min; hold at 4 °C. The cDNA product was purified twice using SPRI beads (Beckman Coulter, B23318) at a 0.8x bead-to-sample ratio, eluting in a final volume of 20 μl (for 3mm arrays) and 60 μl (for 5.5 mm arrays). A total of 1 ng (for 3 mm arrays) or 3× 1ng (for 5.5 mm arrays) of cDNA was used for Illumina sequencing library construction. The Nextera XT kit (Illumina, FC-131–1096) was used for tagmentation, followed by amplification with TruSeq5 and N700 series barcoded index primers. Libraries were cleaned with SPRI beads according to the manufacturer’s instructions at a 0.6x bead-to-sample ratio and resuspended in 10 μl of water per reaction. Lineage tracing target site libraries were amplified from cDNA and prepared fpr Illumina sequencing using previously described protocols50.
For Curio 1 cm arrays.
The buffers and enzymes used were the same as those described for the 3 mm and 5.5 mm arrays but adjusted for scale. In brief, hybridization, dipping, washing, RT reaction and tissue digestion were performed using the reservoirs provided by Curio with 500 μl volume for each step. After tissue digestion the beads were divided into 2 tubes for wash buffer washes and combined for cDNA amplification. A total of 4.8 ml of cDNA amplification mixture was prepared, and the reaction was divided into 50 μl aliquots for cDNA amplification in 96-well PCR plates, following the same PCR program as outlined previously. cDNA was purified twice using 0.8x SPRI beads and eluted in a final volume of 80 μl. 8× 1ng cDNA products were used for Illumina sequencing library preparation through tagmentation with a Nextera XT kit, followed by amplification and cleanup as stated above. Lineage tracing target site libraries were amplified from cDNA and prepared fpr Illumina sequencing using previously described protocols50.
Spatial transcriptomics with Slide-tags
Fresh frozen tissues were cryo-sectioned at 20 μm thickness using a Cryostat set at −17 to −18 °C. Precooled 6 mm square custom-made biopsy punches were used to punch and isolate regions of interest from the tissue sections. The isolated tissue regions were carefully transferred onto a precooled array, which was placed on top of a glass slide. A finger was briefly placed underneath the slide to melt the tissue onto the array. Immediately after, the tissue, array, and slide were placed on ice, and approximately 10 μl of dissociation buffer (82 mM Na2SO4, 30 mM K2SO4, 10 mM glucose, 10 mM HEPES, 5 mM MgCl2) was gently pipetted onto the tissue to ensure it was fully covered. The array was then exposed to an ultraviolet (UV) light source (0.42 mW mm−2, Thorlabs, M365LP1-C5, Thorlabs, LEDD1B) for 1 minute to cleave spatial barcode oligonucleotides off the beads. After photo-cleavage, the array was incubated on ice for 7.5 minutes before being transferred to a well of a 12-well plate. To release the tissue from the array, 1 ml of extraction buffer (dissociation buffer with 1% Kollidon VA64, 0.2% Triton X-100, 1% BSA, 666 U ml−1 RNase-inhibitor) was gently dispensed onto the array, and the buffer was carefully triturated up and down over the tissue 10–15 times. This process was repeated until the tissue was completely released from the array. The array was then discarded, and mechanical dissociation of the tissue was performed by triturating the supernatant 100–150 times using a 1 ml pipette to fully release the nuclei from the tissue. The extraction buffer containing the nuclei was transferred to a 15 ml tube. The well was washed three times with 1 ml of wash buffer (dissociation buffer with 1%BSA and 1: 100 RNase-inhibitor) and the washes were pooled into the same 15 ml tube. The final volume of the wash buffer was adjusted to 10 ml. The nuclei were centrifuged at 600g for 10 minutes at 4 °C. After centrifugation, 9.5 ml of the supernatant was carefully removed. The pellet was resuspended and passed through a precooled 40 μm cell strainer (Corning, 431750) into a 1.5 eppendorf tube. The 15 ml tube and cell strainer were washed with 1 ml of wash buffer, and the nuclei were pelleted again by centrifuging at 600g for 10 minutes at 4 °C. After centrifugation, the supernatant was carefully removed, leaving approximately 50 μl of wash buffer for nuclei resuspension. To determine cell count, 2 μl of resuspended nuclei was mixed with 18 μl of 1: 100 diluted DAPI, and the nuclei were manually counted using a C-Chip Fuchs-Rosenthal disposable hemocytometer (INCYTO, DHC-F01–5). Based on the cell count, up to 25,000 nuclei were processed using the Chromium Next GEM Single Cell 3’ Reagent Kits v3.1 (with Feature Barcode technology for Cell Surface Protein, 10x Genomics, PN-1000268). Lineage tracing target site libraries were amplified from cDNA and prepared fpr Illumina sequencing using previously described protocols50.
H&E staining
H&E was performed with a Leica ST5010 Autostainer XL and Leica CV5030 Fully Automated Glass Coverslipper. Bright-field images were taken using the Leica Aperio VERSA Brightfield, Fluorescence & FISH Digital Pathology Scanner under a ×10 objective. Tumor grade was analyzed in H&E-stained sections using an automated deep neural network developed by Aiforia.
Sequencing
Sequencing was performed at using NovaSeq S4. For Slide-seq gene expression libraries: read1: 50bp, read2: 50bp, index1: 8bp was used. For Slide-seq Target Site libraries: read1: 44bp, read2: 260bp, index1: 8bp was used. For Slide-tags gene expression libraries: read1: 28bp, read2: 90bp, index1: 10bp, index2: 10bp was used. For Slide-tags gene expression libraries: read1: 28bp, read2: 260bp, index1: 8bp setting was used.
Immunofluorescence staining & imaging
15 μm-20 μm tissue sections were fixed in 4% PFA at room temperature for 10–15 min. The sections were washed twice in 1x PBS. Antigen retrieval was performed by boiling 1X IHC Antigen Retrieval Solution (ThermoFisher Scientific, 00-4955-58) and incubating tissue sections inside for 30 min until the solution cooled down, followed by washing tissue sections with 1x PBS and incubated in 0.3% PBST (0.3% Triton X-100 in PBS) at room temperature for 10 min. Three times of 1x PBS wash was then performed. Blocking (0.5% BSA and 0.1% Triton X-100 in 1x PBS) was performed at room temperature for 1 hour. Tissue sections were incubated with primary antibodies: VIM (1: 200, Biotechne, AF2105), CD31 (1: 200, Biotechne, AF3628), ARG1 (1: 200; Cell Signaling Technology, 93668), GLUT1 (1: 100; AbCam, ab195020), CD45 (1: 200, Cell Signaling Technology, 70257), and COL3A1 (1: 200, Proteintech, 22734–1-AP) at 4 °C overnight. Tissue sections were washed three times with 1x PBS and further incubated with secondary antibodies (donkey anti-goat 405, 1: 1000, ThermoFisher Scientific, A-48259; donkey anti-mouse 647, 1: 1000, ThermoFisher Scientific, A-31571; donkey anti-rabbit 647, 1: 1000, ThermoFisher Scientific) at room temperature for 2–3 hours. Tissue sections were then washed three times with 1x PBS, mounted and imaged using Dragonfly 201–40 High Speed Confocal Imaging Platform.
QUANTIFICATION AND STATISTICAL ANALYSIS
Slide-seqV2 gene expression quantification and quality-control
A python implementation of Kallisto-bustools95 (kb_python, version 0.27.3 available at https://github.com/pachterlab/kb_python) was used for transcript quantification and processing from raw FASTQs produced with Slide-seq. Specifically, we utilized the count procedure implemented in Kallisto that quantifies the number of UMIs in a Slide-seq library that map to each transcript sequence in the provided reference (here, mm10). To account for the unique read structure of the Slide-seq library, we invoked the count procedure with the flag -x “0,0,8,0,26,32:0,32,41:1,0,0”. To determine a whitelist of barcodes to use during quantification, we matched barcodes identified with kallisto to the spatial barcodes and their coordinates observed during in situ sequencing of the Slide-seq array during fabrication56,57. We then used a custom script to assign spatial coordinates, identified during in situ sequencing of the Slide-seq array prior to running the assay, to quantifications from the kallisto pipeline and returned an AnnData structure containing the spatially-resolved transcript abundances for each spot. To supplement the barcode filtering during the kallisto pipeline, we applied an extra filter requiring at least 150 UMIs observed in a spot. For most analyses, we utilize log-normalized counts where each cell’s UMI total is scaled to the median library size and a log1p transformation is applied. When scaled counts are used, we additionally use Scanpy’s scale function with a max value of 10.
Slide-tags gene expression quantification and quality-control
Similar to Slide-seq processing, we utilized the python implementation of Kallisto-bustools95 (kb_python, version 0.27.3 available at https://github.com/pachterlab/kb_python) to quantify transcript abundance from FASTQ data. As this data represents reads from sequencing single-nuclei with the 10X V3 kit, we utilized the --umi-gene, --workflow nucleus, and -x 10XV3 flags. Similar to the Slide-seq analysis, we utilized the mm10 transcriptome reference.
After transcript quantification, we applied several quality-control procedures. First, we removed background gene expression signal from ambient RNA by applying Cellbender96 (version 0.3.0, available at https://github.com/broadinstitute/CellBender) to the unfiltered gene expression counts. We used default settings for all libraries, except for 10X Library 9 where we used the following flags: --empty-drop-training-fraction 0.15, --total-droplets-included 20000, --learning-rate 0.0001, and --epochs 300. After running Cellbender, we applied further cell-filters to remove outliers with high mitochondrial or ribosomal content (between 5–15% for libraries). We further inspected the count distribution in each library and removed nuclei with excessively high UMI content (approximately 20,000 UMIs). All quality-control was performed with Scanpy97 (version 1.10.0, downloaded via pip). For most analyses, we utilize log-normalized counts where each cell’s UMI total is scaled to the median library size and a log1p transformation is applied. When scaled counts are used, we additionally use Scanpy’s scale function with a max value of 10.
Slide-seq lineage tracing target-site data processing
To begin processing target-site data, we trimmed reads from Slide-seq libraries using cutadapt98 (version 4.1) with the following flags: -m :250 --max-n 0.2 --discard-untrimmed -O 10 --no-indels --match-read-wildcards -e 2 -j 16 --action retain -G AATCCAGCTAGCTGTGCAGC. We then applied Cassiopeia41 (version 2.0.0, available at https://github.com/YosefLab/Cassiopeia) to trimmed FASTQs using the “slideseq2” chemistry and specific parameters for Slide-seq libraries. First, to account for the possibility of multiple cells observed in a given spot, we allowed allele conflicts (allow_allele_conflicts = True) and did not enable doublet filtering. While we performed intBC whitelist correction, we did not perform additional error correction to remove intBCs with conflicting alleles (this is similarly motived by the fact more than one cell can be observed in a given spot). We additionally relaxed the UMI/cell threshold to account for reduced capture of Slide-seq assays (min_umi_per_cell = 2). Finally, we utilized the “likelihood” method for UMI collapsing, with max_hq_mismatches = 3 and max_indels = 2. Other settings remained default. This pipeline produced a cleaned allele table, reporting the set of intBCs and alleles for each observed spot, that was used for tree reconstruction.
Slide-tags lineage tracing target-site data processing
Cassiopeia41 (version 2.0.0, available at https://github.com/YosefLab/Cassiopeia) was used to process FASTQs containing target-site data. As Slide-tags represents single-nucleus data, we utilized default settings except for a more relaxed UMI/cell cutoff (min_umi_per_cell = 5) to reflect the reduced sensitivity of single-nucleus sequencing. As a part of default settings, we corrected cell barcodes to those observed after quality-control filtering, corrected intBCs to a whitelist for the corresponding mESC (E1) with a distance threshold of 1, and performed UMI (with a maximum distance of 2) and intBC error correction (minimum UMI support of 5) to correct for conflicting target sites observed in the same nuclei. Doublets were filtered out using the default conflicting threshold of 35%. This pipeline produced a cleaned allele table, reporting the set of intBCs and alleles for each observed spot, that was used for tree reconstruction.
Slide-tags spatial barcode processing
Spatial mapping of Slide-tags nuclei was achieved as previously described58. Briefly, reads from spatial barcode FASTQ files were filtered for those containing the spatial barcode universal primer constant sequence and cell barcode sequences from a called cell barcode whitelist generated by the gene expression pipeline (see above section entitled “Slide-tags gene expression quantification and quality-control”). Spatial barcode sequences were matched with a whitelist of in situ sequenced spatial barcodes, assigning spatial coordinates to each true spatial barcode. The set of spatial barcodes and the corresponding x,y coordinates for each cell barcode were clustered with DBSCAN99 (implemented in the R package dbscan, version 1.1−11). For cell barcodes with a single cluster of spatial barcodes, spatial barcodes not contained in the cluster were filtered out and a UMI-weighted centroid of the remaining spatial barcodes represented the x,y coordinates of the cell barcode. DBSCAN parameters were determined from a sweep of minPts values (3 to 15) under a constant eps = 50. The chosen minPts positioned the highest proportion of cell barcodes.
Spatial imputation of lineage-tracing data
To recover lineage-tracing data for reconstruction on spatial assays, we performed spatially-informed imputation of target site data. To begin, we first created a character matrix from the allele tables constructed from target-site lineage tracing processing. In this character matrix, denoted as X, each row corresponds to a cell (or spot) and each column corresponds to a particular cut site in an integration barcode (intBC). For clarity of notation, we refer to each cut-site/intBC pair as a character, and thus in our system a character matrix will have (|intBCs| × 3) columns. The entry X[i, j] denotes the edit (which we refer to as a “state”) observed at the ith cell/spot in the jth character. The missing data rate refers to the proportion of entries in this character matrix that do not have data that pass our quality-control filters.
To perform spatial imputation, we first constructed a spatial nearest-neighbor graph (N) such that each spot was connected to all other spots within 30μm of the spot. For each missing entry in character matrix, i, j we queried the frequency of states at character j in all neighbors of spot i in N. If the concordance of a particular state was higher than 80% in these neighbors, then we replaced the entry X[i, j] with this state. To minimize the effect of nearby stromal cells in a neighborhood – which should not have active lineage-tracing – we did not allow this state to be 0, the uncut state. To maximize the alleles were used during spatial imputation, we required each state to be supported by at least 3 UMIs. We reported this procedure for each missing entry in the character matrix for a total of 5 iterations which continued to remove missing data from the character matrices with no apparent reduction in accuracy in simulations or held-out real data (Figure S1J–N).
Benchmarks of imputation and reconstruction accuracy
To benchmark the accuracy of spatial imputation and downstream effects on tree reconstruction, we utilized two different strategies:
Synthetic data: First, we utilized the Cas9-based lineage-tracing simulation framework in Cassiopeia41 (version 2.0.0, available at https://github.com/YosefLab/Cassiopeia). Specifically, we simulated trees using Cassiopeia’s BirthDeathSimulator with the following parameters: 5000 extant cells, and utilized a LogNormal birth-waiting distribution parameterized by log (f) where f is a fitness coefficient that accumulates with each cell division (in each cell division, a new coefficient f ~ N(0, 0.25) is drawn and added to the base fitness) and a standard deviation of 0.5. Then, we simulated lineage tracing data onto the tree with Cassiopeia’s Cas9LineageTracingDataSimulator with desired mutation proportion of 0.7, 100 states, 39 cut sites (representing our system with approximately 13 intBCs, each with 3 cut-sites), and no missing data rates at this point. Then, we simulated spatial coordinates on each tree using the ClonalSpatialDataSimulator over a shape of (1,1,1) and sampled a 2D slice from this 3D simulation at random. Finally, we subsampled from this spatial array using the UniformLeafSubsampler in Cassiopeia with a rate of 0.4 (resulting in lineages with 2,000 observations) and induced random dropout at various rates: [0.1, 0.25, 0.5, 0.6, 0.7, 0.9]. We simulated 10 trees for each parameter combination. As the spatial array simulated does not exactly match that from Slide-seq, we applied a modified k-nearest-neighbor graph construction approach, linking together spots to their closest 10 neighbors and performed spatial imputation (see section titled “Spatial imputation of lineage-tracing data”). We required concordance of 0.8 for the selected state and at least 5 votes. Since this simulated data does not include any normal cells, we do allow the imputation of the state 0. We reported the accuracy of this imputation strategy in Figure S1J). Then, we compared the tree reconstructing accuracies using the triplets_correct function in Cassiopeia for reconstructions with or without imputation and for different reconstruction strategies: modified Neighbor-Joining, Cassiopeia-Greedy, or a hybrid of these two approaches (see section “Phylogenetic reconstruction”).
Simulated held-out Slide-seq data: In the next experiment, we assessed the accuracy of recovering target-site data that was held-out from real Slide-seq data. To do this, for a given Slide-seq array, we masked out 10% of the observed data (supported by at least 3 UMIs) and performed spatial imputation in neighborhoods of 30μm using the strategy described previously (see section titled “Spatial imputation of lineage-tracing data). Similarly, we required a concordance of 0.8 and at least 5 votes in support of the imputed allele. We only considered samples where at least 10 states were imputed. Random predictions were obtained by shuffling the node labels in the neighborhood graph. We reported the average accuracy and total number of imputed values over five replicates in Figure S1M.
Simulation benchmarks of lineage-tracing pre-processing
As a feature of the Slide-seq is that multiple cells may be observed in one spot57, multiple conflicting alleles can be observed for a given target site in a single spot. Typically, this would break the assumption of the Cassiopeia reconstruction pipeline (in single-cell approaches, we assume that only one allele can be tied to a given intBC and perform error correction or filtering otherwise). However, we implemented new reconstruction algorithms that can handle multiple conflicting states in each spot (see section entitled “Phylogenetic reconstruction”) and simulated the effects of various pre-processing techniques.
First, we simulated trees on two-dimensional surfaces where various proportions of cells would be grouped together based on their spatial location. To do so, we simulated simple binary trees of 2000 cells and overlaid lineage-tracing data with Cassiopeia’s Cas9LineageTracingDataSimulator function using the following parameters: 39 characters, a mutation proportion of 0.5, and no missing data. We then merged together cells using Cassiopeia’s SupercellularSampler method with rates of [0.1, 0.2, 0.3, 0.4, 0.5, 0.6]. We simulated 32 replicates.
For each replicate, we pre-processed character matrices according to three strategies. Here, the entry of the ith cell and jth character (denoted as X[i, j]) would contain a set of states X[i, j] = {s1, s2, …, sk}, each state occurring at some frequency f(si) = fi. In the first strategy (“collapse duplicates”) we take the unique set of states so that X[i, j] = {s1, s2, …, sk′} s. t. fi = 1 ∀ i ∈ k′; in the second strategy (“most common”) we take the most common state, such that ; and the third strategy (“all states”) we do not perform any filtering. In Figure S1F we report the tree reconstruction error (measured with normalized Robinson-Foulds distance) for trees reconstructed with Neighbor-Joining63.
Phylogenetic reconstruction on Slide-seq data
To enable phylogenetic reconstruction on Slide-seq data in which multiple cells can be contained in a single spot and thus conflicting alleles are present, we implemented a Hybrid Cassiopeia-Greedy & Neighbor-Joining algorithm that could utilize conflicting allele states.
For Cassiopeia-Greedy, we modified the splitting decision rule to account for all states observed in a spot. Cassiopeia-Greedy is a simple, heuristic-based algorithm for reconstructing phylogenies that iteratively finds the most common state in a given population and splits samples into groups based on the presence or absence of the state. It is based on a perfect-phylogeny reconstruction algorithm100 and has an efficient runtime of O(mn) for a population of n samples and m characters. Here, we changed the procedure to find the state with the highest frequency by allowing each sample to carry multiple states in a character. The runtime of this algorithm is still polynomial in the size of the sample population – O(n(ms)) where in the worst case scenario every single state is observed in every single character; given the size of the spatial array, this is exceedingly uncommon and typically 1–3 cells are captured per spot57.
For Neighbor-Joining, we utilized the standard algorithm63 but with a modified distance map that accounts for multiple states per spot. Specifically, we implemented a new dissimilarity metric that takes in two sets of states S1 and S2 and computes all the pairwise allelic dissimilarities and reports a linkage similar to hierarchical clustering. Here, we use the modified allelic dissimilarity for two states si, sj to compute distances between pairs of states, previously described41,47,50:
In the case where weights are passed in, then the dissimilarity function is computed as follows:
Then, we utilized a single linkage function such that only the smallest modified allelic dissimilarity across all pairs of states in S1 and S2 was used. This is to maintain such that if the same state is observed in two spots, the dissimilarity returned is 0.
For the hybrid reconstruction, we utilized the modified Cassiopeia-Greedy algorithm described above until subpopulations of size 1000 cells were found, at which point Neighbor-Joining with the modified dissimilarity metric was used to resolve phylogenetic relationships. We utilized state probabilities inferred from all Slide-seq and Slide-tags datasets and used the weight − log(pi) for state si during tree reconstruction.
Phylogenetic reconstruction on Slide-tags data
We utilized the standard Cassiopeia-Hybrid41 algorithm for reconstructing Slide-tags phylogenies. Briefly, this approach applies the heuristic-based Cassiopeia-Greedy algorithm to reconstruct relationships between the major subclones and then applies the maximum-parsimony-based Cassiopeia-ILP algorithm to solve fine-grained phylogenetic structure in smaller populations. As previously described in detail41, Cassiopeia-ILP proceeds by building a potential graph of all possible ancestral states (constrained in size by a user-defined parameter) and solves for the maximum-parsimony phylogeny by reconstructing a Steiner Tree on this data structure. The Steiner Tree problem is solved via an Integer Linear Program (ILP) allowed a certain time to converge. Here, the transition between Cassiopeia-Greedy and -ILP algorithms is determined by the distance to the latest common ancestor (LCA) of a subpopulation.
We applied the Cassiopeia-Hybrid algorithm with state priors inferred from all samples41,47,50, determined the switch between Greedy and ILP algorithms using an LCA cutoff of 20, devised a potential graph of 10000 nodes with a maximum distance of 15 across nodes (maximum_potential_graph_lca_distance=15), and allowed the ILP 12600s to converge.
Slide-tags cell type annotation
After performing quality-control on Slide-tags gene expression data, we assigned cell types first by integrating Slide-tags data with an annotated single-cell gene expression reference dataset of KP-Tracer tumors50 with scANVI101. To do so, first identified 4,750 variable genes using Scanpy’s97 highly_variable_genes function using the flavor=“seurat_v3” and raw counts. We then trained an scVI model102,103 on the joint dataset and these variable genes using 3 layers and 70 latent dimensions over 1000 epochs. Then, we transferred labels from the single-cell reference dataset to the Slide-tags nuclei with scANVI utilizing 200 samples per label and 100 epochs. Through this, we used the gene_likelihood=“nb” setting in training models and used the technology – Slide-tags or single-cell – variable to signify batch.
After training this model, subset to the scANVI embeddings to the Slide-tags data only and re-clustered the data with Scanpy97 using the Leiden algorithm104 and resolution 1.2. We then split clusters into those that appeared to derive from tumor/epithelial cells or those that derived from the stroma. To call tumor or epithelial clusters, we evaluated if a cluster had an abundance of tumor nuclei (defined as nuclei with target site data and at least 20% of their sites containing indels) or expressed the epithelial-lineage marker Nxk2-1. Immune cell clusters were identified based on the marker Cd45 (Ptprc) and other stromal cells were identified by expression of Pdgfra, Col1a1, or Col5a1 (fibroblasts) or Pecam1 (endothelial cells). For each subsetted dataset (tumor/epithelia or stromal), we reclustered the data and annotated cell types based on annotations predicted with scANVI and differentially expressed genes identified with Scanpy’s rank_genes_group function (using the Wilcoxon test).
Assessment of Slide-tags tumor cell type signatures in previous KP-Tracer data
To test the portability and accuracy of the tumor clusters identified in Slide-tags, we assessed the activity of gene signatures in the previous KP-Tracer data50. Specifically, we for each cell-type identified in Slide-tags, we computed the top 100 differentially-expressed genes using the Wilcoxon test in Scanpy97 and further filtered genes to have a log-fold change > 1 and an FDR-corrected p-value <= 0.01, and an AUROC of at least 0.6. We then used these genes to define a transcriptional signature for each Slide-tags cell type. each of these signatures, we scored the activity in cell types identified in Slide-tags data and the previous KP-Tracer dataset using the score_genes function using n_bins=30 and ctrl_size equal to the number of genes in the gene set. Signatures were computed on scaled, log-normalized counts. The result of this analysis is presented in Figure S2B.
Slide-seq spatial community detection and scoring
To identify spatial communities in Slide-seq data, first applied the Hotspot105 algorithm for detecting spatially autocorrelated gene sets on each sample. In the spatial mode, this algorithm constructs a nearest neighbor graph based on spatial coordinates, computes an autocorrelation statistic for each gene, and then identifies modules of genes that have significant pairwise autocorrelation values. Here, we applied Hotspot with 20 neighbors, and FDR threshold of 0.01 to identify spatially autocorrelated genes, and a minimum module size of 50 genes.
Then, to identify robust modules of genes that appear across tumors, we assessed the Jaccard overlap between all pairs of modules across all tumors and filtered out modules that did not have a Jaccard overlap of at least 0.2 with at most one other module. We then performed Z-normalization on these Jaccard statistics and clustered these using hierarchical clustering (using the “ward” method on Euclidean distances) and identified 11 clusters, representing robust spatial modules.
As these robust modules are collections of modules across all samples we analyzed, we distilled these down to a set of genes – representing what we call a “spatial community” in this study – by taking genes that appear in at least 25% of the modules in the robust module. Using these genes in the spatial community, we compute the activity of these communities for each spot (termed “community scores”) using the score_genes function in Scanpy97 with ctrl_size=100 and n_bins=30. We computed these scores on scaled, log-normalized gene expression counts. To obtain community assignments for each spot, we took the community with the highest score.
Tumor segmentation
To segment tumors, we utilized the SpatialData106 package and the napari-spatialdata viewer for interactive annotation. To identify tumor areas on a sample, we overlaid phylogenetic subclones and the number of target-site UMIs detected and manually segmented areas that appeared to be (a) phylogenetically related and (b) had elevated target-site UMIs indicative of tumor regions. We saved these annotations and used the segmentations to perform downstream analysis on a tumor-by-tumor basis.
Fitness signature calculation
To quantify fitness signature scores, we utilized a gene set that was found to be associated with changes in fitness from our previous single-cell KP-Tracer study50. Using this gene set, we quantified the transcriptional activity for each spot in Slide-seq data by applying the score_genes function in Scanpy97 with ctrl_size=100 and n_bins=30. We computed these scores on scaled, log-normalized gene expression counts.
Phylogenetic fitness inference
We quantified fitness on Slide-seq and Slide-tags phylogenies by utilizing the LBIFitness fitness estimator in Cassiopeia41. This function wraps a fitness estimator based on the “local branching index” as previously described107. This procedure has been previously used in our system50. Primed by the true single-cell resolution of Slide-tags trees, we estimated branch lengths using the IIDExponentialMLE branch length estimator in Cassiopeia. This function implements a function that provides maximum-likelihood branch lengths on a tree topology given the pattern of edits observed in the leaves and an assumptions about the irreversibility of Cas9 editing108. Using the branch lengths determined by this maximum-likelihood procedure, we estimated single-cell fitness on Slide-tags trees.
Due to the increased missingness on Slide-seq trees and the fact that MLE-based branch length approaches have not been benchmarked on Slide-seq data, we performed a more conservative branch length estimation, as done previously50. Here, branches had a length of 1 if they had any mutations along them, otherwise they had a branch length of 0. Using these branch lengths, we estimated single-cell fitness on Slide-seq trees.
Single-cell clonal plasticity quantification
To estimate single-cell clonal plasticity on phylogenies, we applied approaches described in our previous studies50,69. Specifically, on Slide-tags data where we have true single-cell data and associated cell type identities, we applied the score_small_parsimony procedure to all nodes in a tree using meta_item=“cell_type” and normalized by the number of leaves in the subtree induced by the node. Then, we computed plasticity for each cell by averaging together all the normalized parsimonies.
Since we do not have true single-cell resolution for Slide-seq data, we employed the L2 plasticity score described in our previous study50, using community scores. Specifically, let Ci be the vector of community scores associated with spot i. For this spot i we found its closest phylogenetic neighbors (denoted by set N) and then computed the L2-Plasticity (L20(i)) for this spot by the average Euclidean distance to the vector of community scores for these neighbors: All scores were unit scaled.
Differential expression and abundance in neighborhoods of high-fitness cells
To identify changes in gene expression and spatial communities associated with fitness, we first stratified cells into high- and low-fitness groups. In Slide-seq data, we computed single-cell fitness scores (see section above entitled “Phylogenetic fitness inference”) and identified a threshold separating two modes using scipy.signal.argrelmin in the merged fitness distributions and split spots into high-fitness groups and low-fitness groups based on this threshold. Only tumors with at least 200 observations with lineage-tracing data were used. As each fitness distribution is normalized within individual tumors to be unit-scaled, this approach finds a global pattern in high- and low-fitness cells. Then, we constructed a neighborhood graph connecting each spot to all other spots within 30μm. The community scores for all communities were computed in these neighborhoods and the distributions in neighborhoods of high- and low-fitness cell were reported in Figure S3L.
In Slide-tags data, high and low-fitness cells were similarly determined from the distribution of all fitnesses using scipy.signal.argmin. As Slide-tags is sparser than Slide-seq, we constructed neighborhoods using the closest 20 cells (an example is shown in Figure S3K). We then identified the differentially-expressed genes in neighborhoods of high- and low-fitness cells of all Macrophage and Fibroblast subsets using the t-test as implemented in Scanpy’s97 rank_genes_groups function. For the Macrophage analysis, we evaluated the Alveolar Macrophages, Arg1+ TAMs, Pecam1+ TAMs, and Vegfa+ TAMs; for the Fibroblast analysis we evaluated the Wt1+ fibroblast, iCAF-like and myCAF-like populations. Genes expressed in fewer than 50 cells were filtered out, and the differential expression statistics for the top 10,000 genes were computed. Genes with an absolute log2-fold-change > 1 and an FDR-corrected p-value < 0.01 were marked as significantly differentially expressed. To compute enrichments in these neighborhoods, we computed the frequency of cell types in neighborhoods of high- and low-fitness cells and divided by the expected fraction of these cell types given the overall distribution and size of the Slide-tags array.
GO Term analysis of differentially-expressed genes was performed using gseapy109 (version 1.1.3) with the following gene sets: “WikiPathways_2019_Mouse”, “Reactome_2022”, “GO_Biological_Process_2023”, “GO_Molecular_Function_2023”, and “KEGG_2019_Mouse”. Significant terms are reported in Supplementary Table 2.
Differential expression in neighborhoods of high-plasticity cells in Slide-seq
Similar to the fitness-based analysis (see section entitled “Differential expression in neighborhoods of high-fitness cells”), we stratified cells into high- and low-plasticity groups. After quantifying the L2-clonal plasticity score in Slide-seq data, we determined a threshold separating high- and low-plasticity regions if a cell had greater plasticity than the 60th percentile or less than the 40th percentile, respectively. Then, we constructed a neighborhood graph connecting each spot to all other spots within 30μm. The community scores for all communities were computed in these neighborhoods and the distributions in neighborhoods of high- and low-plasticity cells were reported in Figure S3M.
Coarse-grained alignment of Slide-seq data
To track the three-dimensional structure of clones across sampled layers in Figure 4, we utilized the non-imputed processed target-site data (see section entitled “Slide-seq lineage tracing target-site data processing”). To maximize fidelity of slide registration, we enforced hard quality-control cutoffs, requiring each spot be supported by at least 7 UMIs and then subsequently each intBC-allele to be supported by at least 5 UMIs. We filtered out spots that had less than 20% of their sites reporting indels, or more than 70% missing data. We then computed modified allelic distances (see section above entitled “Phylogenetic reconstruction on Slide-seq data”) between all pairs of spots across layers. Modified allelic distances here are normalized by the number characters shared between two spots (thus are normalized to values between 0–2). For computational reasons, we did not allow ambiguous alleles (taking only the most frequent allele per intBC in a spot) as the distance calculation is memory- and time-intensive. Using this distance matrix, we computed allelic evolutionary couplings using compute_evolutionary_coupling function in Cassiopeia with the following parameters: minimum_proportion = 0.0002, number_of_shuffles = 100. We then normalized the evolutionary coupling as previously described50, as so:
Where E(i,j) denotes the allelic evolutionary coupling between spot i and j and max(E[i’, j’]) indicates the maximum value across all evolutionary couplings. Clusters identified via hierarchical clustering of the normalized allelic evolutionary coupling matrix were used as registered Tumor IDs in Figure 4B.
Detection of metastasis-initiating subclones
To detect metastasis-initiating subclones in primary tumors, we created a shared character matrix between all lung sections profiled with 1cm × 1cm Curio arrays and Slide-seq samples of metastases. We filtered out spots that did not have at least 2 UMIs intBC-alleles that were not supported by at least 2 UMIs. We further filtered out spots that had fewer than 20% of their target-sites cut and more than 70% missingness. For computational reasons, we did not allow ambiguous alleles (taking only the most frequent allele per intBC in a spot) as the distance calculation is memory- and time-intensive. We then computed a shared metastatic parental allele state by taking states that were shared amongst 60% of spots in metastases profiled with Slide-seq. From this parental state, we computed the modified allelic distance (normalized by the number of shared characters) to all spots in the lung sample. We performed a similar analysis in paired Slide-tags data, computing the normalized modified allelic distances from all nuclei to the metastatic parental allele state.
Differential expression across metastatic cascade
We identified gene expression changes across niches associated with the metastatic cascade by employing the distances computed in the section above entitled “Detection of metastasis-initiating subclones”. We identified the metastasis-originating subclone as localizing to T2, so T1, T3 and T4 were determined to be Primary tumors without any relationship to the metastases. Focusing on T2, we further segmented it into a metastasis-initiating subclone (T2-Met) and other subclones (T2-NonMet). Specifically, we assigned cells to a metastatic subclone if their normalized modified allelic distance was less than 0.8. Then, using these assignments, we performed watershed segmentation with a custom procedure. Specifically, we binned signal into bins of 100 adjacent spots, applied a Gaussian filter with a sigma of 1.5 (with the Python package skimage) and then applied an Otsu threshold and dilation. We then applied an exact distance transform with scipy.ndimage.distance_transform_edt and computed a Watershed mask over peaks identified with skimage.feature.peak_local_max with a goal of identifying one tumor. This segmented subclone was labeled as T2-Met, and the remainder of the tumor was called T2-NonMet. We then performed differential expression across the library-size-normalized, logged counts of four groups (Primary tumors without metastatic relationship; T2-Met; T2-NonMet; and metastases) using a t-test implemented in Scanpy’s97 rank_genes_groups and reported the log2-fold-change in Figure 4G.
Signature scores for TGFβ signaling were computed using MSigDB’s “HALLMARK_TGF_BETA_SIGNALING” signature. Signature scores for collagen were computed for a custom gene set consisting of Acta2, Col1a1, Col2a1, Col3a1, Col5a1, and Col12a1. Significance was computed using a one-sided t-test assessing if signature scores were higher in the metastatic tumor as compared to the primary tumor.
Differential cell type abundance in metastatic neighborhoods
Similar to analyses stratifying Slide-tags cells into neighborhoods of high- and low-fitness cells, we stratified cells into neighborhoods of cells closely related to metastases. As with determining cells related to metastases in Slide-seq data, we computed the distance to the parental metastatic allele and assigned cells with distances smaller than 0.8 as related to metastases. Then, we reconstructed spatial neighborhoods of the closest 20 cells and quantified cell type enrichments based on the frequencies of cell types in these neighborhoods and the overall frequency in a Slide-tags array.
Supplementary Material
Table S1: Fitness-neighborhood differential expression and GO Term analyses.
ACKNOWLEDGEMENTS
We thank Jack Rose, Can Ergen, Chen Weng, Pu Zheng, Sean-Luc Shanahan, Yun Zhang, Anjali Saqi, Meaghan McGery, Santiago Naranjo, Michelle Chan, Romain Lopez, Adam Gayoso, and all members of the Weissman, Yang, Yosef, Chen, and Chang labs for helpful discussions. We thank Cristen Muresan, Anne Odera, Maria Gould, Daniel Braslavsky, Maxim Litvinov, and Nicole Dow for administrative support. We thank the Whitehead Institute and Broad Institute Sequencing Facility for sequencing support.
M.G.J. is supported by an NCI Pathway to Independence Award (NIH K99CA286968). N.M.A. was supported by a NIH F30 fellowship (1F30CA278495). K.E.Y. was supported by the National Cancer Institute of the National Institutes of Health under Award Number K00CA253729. L.W.K. is supported by a Helen Hay Whitney Postdoctoral Fellowship. T.J. laboratory currently also receives funding from The Lustgarten Foundation for Pancreatic Cancer Research, but this funding did not support the research described in this manuscript. This work was supported in part by the Cancer Center Support (core) grant P30-CA14051 from the National Cancer Institute and by the NIH grant R35CA274464. T.J. is the David H. Koch Professor of Biology and a Daniel K. Ludwig Scholar. J.S.W. is supported by the Howard Hughes Medical Institute, NCI Cancer Target Discovery and Development (CTD^2) and NIH Centers of Excellence in Genomic Science (CEGS). Both J.S.W. and T.J. received fundings from Ludwig Center at MIT. D.Y. is supported by a Damon Runyon Dale Frey Award, an NCI Transition Career Development Award 1K22CA289207 and an NIH Director’s New Innovator Award 1DP2OD037078. N.Y. is supported in part by an NIH grant R56-HG013117 and by the European Union Council (ERC, Tx-phylogeography, 101089213). Views and opinions expressed are however those of the authors only and do not necessarily reflect those of the European Union or the European Research Council Executive Agency. Neither the European Union nor the granting authority can be held responsible for them. F.C. acknowledges support from NIH Early Independence Award (DP5, 1DP5OD024583), the NHGRI (R01, R01HG010647), the Burroughs Wellcome Fund CASI award, the Searle Scholars Foundation, the Harvard Stem Cell Institute, and the Merkin Institute. This research was supported by the NYSCF. FC is a New York Stem Cell Foundation – Robertson Investigator
Footnotes
DECLARATION OF INTERESTS
M.G.J. consults for and has equity in Vevo Therapeutics. K.E.Y. is a consultant for Cartography Biosciences. T.J. is a member of the Board of Directors of Amgen and Thermo Fisher Scientific, and a co-Founder of Dragonfly Therapeutics and T2 Biosystems. T.J. serves on the Scientific Advisory Board of Dragonfly Therapeutics, SQZ Biotech, and Skyhawk Therapeutics. T.J. is the President of Break Through Cancer. None of these affiliations represent a conflict of interest with respect to the design or execution of this study or interpretation of data presented in this manuscript. J.S.W. declares outside interest in 5 AM Venture, Amgen, Chroma Medicine, KSQ Therapeutics, Maze Therapeutics, Tenaya Therapeutics, Tessera Therapeutics, Ziada Therapeutics, DEM Biopharma, and Third Rock Ventures. D.Y. declares outside interest in DEM Biopharma.
DATA AND CODE AVAILABILITY
Custom code for the analysis of spatially-resolved lineage-tracing data is available on Github through Cassiopeia (https://github.com/YosefLab/Cassiopeia) and at https://github.com/mattjones315/KPSpatial-release. All raw and processed data will be made available on GEO and other public repositories.
REFERENCES
- 1.Nowell P. C. The clonal evolution of tumor cell populations. Science 194, 23–28 (1976). [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B. et al. Cancer genome landscapes. Science 339, 1546–1558 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Binnewies M. et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 24, 541–550 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Visser K. E. & Joyce J. A. The evolving tumor microenvironment: From cancer initiation to metastatic outgrowth. Cancer Cell 41, 374–403 (2023). [DOI] [PubMed] [Google Scholar]
- 5.Northey J. J., Przybyla L. & Weaver V. M. Tissue force programs cell fate and tumor aggression. Cancer Discov. 7, 1224–1237 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noble R. et al. Spatial structure governs the mode of tumour evolution. Nat. Ecol. Evol. 6, 207–217 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Derynck R., Turley S. J. & Akhurst R. J. TGFβ biology in cancer progression and immunotherapy. Nat. Rev. Clin. Oncol. 18, 9–34 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fang J. S., Gillies R. D. & Gatenby R. A. Adaptation to hypoxia and acidosis in carcinogenesis and tumor progression. Semin. Cancer Biol. 18, 330–337 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carmona-Fontaine C. et al. Emergence of spatial structure in the tumor microenvironment due to the Warburg effect. Proc. Natl. Acad. Sci. U. S. A. 110, 19402–19407 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Houlahan K. E. et al. Germline-mediated immunoediting sculpts breast cancer subtypes and metastatic proclivity. Science 384, (2024). [DOI] [PubMed] [Google Scholar]
- 11.McAllister S. S. & Weinberg R. A. The tumour-induced systemic environment as a critical regulator of cancer progression and metastasis. Nat. Cell Biol. 16, 717–727 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwartz R. & Schäffer A. A. The evolution of tumour phylogenetics: principles and practice. Nat. Rev. Genet. 18, 213–229 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones M. G., Yang D. & Weissman J. S. New tools for lineage tracing in cancer in vivo. Annu. Rev. Cancer Biol. 7, (2023). [Google Scholar]
- 14.McGranahan N. & Swanton C. Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future. Cell 168, 613–628 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Davis A., Gao R. & Navin N. Tumor evolution: Linear, branching, neutral or punctuated? Biochim. Biophys. Acta Rev. Cancer 1867, 151–161 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu Z. & Curtis C. Inferring tumor phylogenies from multi-region sequencing. Cell Syst. 3, 12–14 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Jones S. et al. Comparative lesion sequencing provides insights into tumor evolution. Proc. Natl. Acad. Sci. U. S. A. 105, 4283–4288 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerlinger M. et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 366, 883–892 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jamal-Hanjani M. et al. Tracking the evolution of non–small-cell lung cancer. N. Engl. J. Med. 376, 2109–2121 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Schwarz R. F. et al. Spatial and temporal heterogeneity in high-grade serous ovarian cancer: A phylogenetic analysis. PLoS Med. 12, e1001789 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sottoriva A. et al. A Big Bang model of human colorectal tumor growth. Nat. Genet. 47, 209–216 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao Y. et al. Selection of metastasis competent subclones in the tumour interior. Nat. Ecol. Evol. 5, 1033–1045 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turajlic S. et al. Tracking cancer evolution reveals constrained routes to metastases: TRACERx renal. Cell 173, 581–594.e12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Navin N. et al. Tumour evolution inferred by single-cell sequencing. Nature 472, 90–94 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao T. et al. Spatial genomics enables multi-modal study of clonal heterogeneity in tissues. Nature 601, 85–91 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Erickson A. et al. Spatially resolved clonal copy number alterations in benign and malignant tissue. Nature 608, 360–367 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lomakin A. et al. Spatial genomics maps the structure, nature and evolution of cancer clones. Nature 611, 594–602 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Househam J. et al. Phenotypic plasticity and genetic control in colorectal cancer evolution. Nature 611, 744–753 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heiser C. N. et al. Molecular cartography uncovers evolutionary and microenvironmental dynamics in sporadic colorectal tumors. Cell 186, 5620–5637.e16 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frieda K. L. et al. Synthetic recording and in situ readout of lineage information in single cells. Nature 541, 107–111 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chow K.-H. K. et al. Imaging cell lineage with a synthetic digital recording system. Science 372, (2021). [DOI] [PubMed] [Google Scholar]
- 32.Chan M. M. et al. Molecular recording of mammalian embryogenesis. Nature 570, 77–82 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McKenna A. et al. Whole-organism lineage tracing by combinatorial and cumulative genome editing. Science 353, aaf7907 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spanjaard B. et al. Simultaneous lineage tracing and cell-type identification using CRISPR–Cas9-induced genetic scars. Nat. Biotechnol. 36, 469–473 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He Z. et al. Lineage recording in human cerebral organoids. Nat. Methods 19, 90–99 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi J. et al. A time-resolved, multi-symbol molecular recorder via sequential genome editing. Nature 608, 98–107 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hwang B. et al. Lineage tracing using a Cas9-deaminase barcoding system targeting endogenous L1 elements. Nat. Commun. 10, 1–9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alemany A., Florescu M., Baron C. S., Peterson-Maduro J. & van Oudenaarden A. Whole-organism clone tracing using single-cell sequencing. Nature 556, 108–112 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Kalhor R., Mali P. & Church G. M. Rapidly evolving homing CRISPR barcodes. Nat. Methods 14, 195–200 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li L. et al. A mouse model with high clonal barcode diversity for joint lineage, transcriptomic, and epigenomic profiling in single cells. Cell 186, 5183–5199.e22 (2023). [DOI] [PubMed] [Google Scholar]
- 41.Jones M. G. et al. Inference of single-cell phylogenies from lineage tracing data using Cassiopeia. Genome Biol. 21, 92 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sashittal P., Schmidt H., Chan M. & Raphael B. J. Startle: A star homoplasy approach for CRISPR-Cas9 lineage tracing. Cell Syst. 14, 1113–1121.e9 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fang W. et al. Quantitative fate mapping: A general framework for analyzing progenitor state dynamics via retrospective lineage barcoding. Cell 185, 4604–4620.e32 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gong W. et al. Benchmarked approaches for reconstruction of in vitro cell lineages and in silico models of C. elegans and M. musculus developmental trees. Cell Syst 12, 810–826.e4 (2021). [DOI] [PubMed] [Google Scholar]
- 45.Pan X., Li H., Putta P. & Zhang X. LinRace: cell division history reconstruction of single cells using paired lineage barcode and gene expression data. Nat. Commun. 14, 1–15 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schiffman J. S. et al. Defining heritability, plasticity, and transition dynamics of cellular phenotypes in somatic evolution. Nat. Genet. 1–11 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Quinn J. J. et al. Single-cell lineages reveal the rates, routes, and drivers of metastasis in cancer xenografts. Science 371, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simeonov K. P. et al. Single-cell lineage tracing of metastatic cancer reveals selection of hybrid EMT states. Cancer Cell (2021) doi: 10.1016/j.ccell.2021.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang W. et al. The bone microenvironment invigorates metastatic seeds for further dissemination. Cell 184, 2471–2486.e20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang D. et al. Lineage tracing reveals the phylodynamics, plasticity, and paths of tumor evolution. Cell (2022) doi: 10.1016/j.cell.2022.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jackson E. L. et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 15, 3243–3248 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jackson E. L. et al. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res. 65, 10280–10288 (2005). [DOI] [PubMed] [Google Scholar]
- 53.Winslow M. M. et al. Suppression of lung adenocarcinoma progression by Nkx2-1. Nature 473, 101–104 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.LaFave L. M. et al. Epigenomic State Transitions Characterize Tumor Progression in Mouse Lung Adenocarcinoma. Cancer Cell 38, 212–228.e13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marjanovic N. D. et al. Emergence of a High-Plasticity Cell State during Lung Cancer Evolution. Cancer Cell 38, 229–246.e13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stickels R. R. et al. Highly sensitive spatial transcriptomics at near-cellular resolution with Slide-seqV2. Nat. Biotechnol. 39, 313–319 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rodriques S. G. et al. Slide-seq: A scalable technology for measuring genome-wide expression at high spatial resolution. Science 363, 1463–1467 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Russell A. J. C. et al. Slide-tags enables single-nucleus barcoding for multimodal spatial genomics. Nature 625, 101–109 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ståhl P. L. et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353, 78–82 (2016). [DOI] [PubMed] [Google Scholar]
- 60.Liu Y. et al. High-spatial-resolution multi-omics sequencing via deterministic barcoding in tissue. Cell 183, 1665–1681.e18 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sutherland K. D. et al. Multiple cells-of-origin of mutant K-Ras-induced mouse lung adenocarcinoma. Proc. Natl. Acad. Sci. U. S. A. 111, 4952–4957 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Arlauckas S. P. et al. Arg1 expression defines immunosuppressive subsets of tumor-associated macrophages. Theranostics 8, 5842–5854 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Saitou N. & Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 (1987). [DOI] [PubMed] [Google Scholar]
- 64.You Y. et al. Systematic comparison of sequencing-based spatial transcriptomic methods. Nat. Methods (2024) doi: 10.1038/s41592-024-02325-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chuang C.-H. et al. Molecular definition of a metastatic lung cancer state reveals a targetable CD109-Janus kinase-Stat axis. Nat. Med. 23, 291–300 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee J. Y. et al. Senescent fibroblasts in the tumor stroma rewire lung cancer metabolism and plasticity. bioRxivorg (2024) doi: 10.1101/2024.07.29.605645. [DOI] [Google Scholar]
- 67.Bill R. et al. CXCL9:SPP1 macrophage polarity identifies a network of cellular programs that control human cancers. Science 381, 515–524 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lewinsohn M. A., Bedford T., Müller N. F. & Feder A. F. State-dependent evolutionary models reveal modes of solid tumour growth. Nat. Ecol. Evol. 7, 581–596 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jones M. G., Rosen Y. & Yosef N. Interactive, integrated analysis of single-cell transcriptomic and phylogenetic data with PhyloVision. Cell Rep Methods 2, 100200 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moran P. A. P. Notes on continuous stochastic phenomena. Biometrika 37, 17–23 (1950). [PubMed] [Google Scholar]
- 71.Hayashi M. et al. Induction of glucose transporter 1 expression through hypoxia-inducible factor 1alpha under hypoxic conditions in trophoblast-derived cells. J. Endocrinol. 183, 145–154 (2004). [DOI] [PubMed] [Google Scholar]
- 72.Zhang J. Z., Behrooz A. & Ismail-Beigi F. Regulation of glucose transport by hypoxia. Am. J. Kidney Dis. 34, 189–202 (1999). [DOI] [PubMed] [Google Scholar]
- 73.Quail D. F. & Joyce J. A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 19, 1423–1437 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee J. H. et al. TGF-β and RAS jointly unmask primed enhancers to drive metastasis. Cell (2024) doi: 10.1016/j.cell.2024.08.014. [DOI] [PubMed] [Google Scholar]
- 75.McGinnis C. S. et al. The temporal progression of lung immune remodeling during breast cancer metastasis. Cancer Cell 42, 1018–1031.e6 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gong Z. et al. Lung fibroblasts facilitate pre-metastatic niche formation by remodeling the local immune microenvironment. Immunity 55, 1483–1500.e9 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kaczanowska S. et al. Genetically engineered myeloid cells rebalance the core immune suppression program in metastasis. Cell 184, 2033–2052.e21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Murdoch C., Muthana M. & Lewis C. E. Hypoxia regulates macrophage functions in inflammation. J. Immunol. 175, 6257–6263 (2005). [DOI] [PubMed] [Google Scholar]
- 79.Kugeratski F. G. et al. Hypoxic cancer-associated fibroblasts increase NCBP2-AS2/HIAR to promote endothelial sprouting through enhanced VEGF signaling. Sci. Signal. 12, eaan8247 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Corzo C. A. et al. HIF-1α regulates function and differentiation of myeloid-derived suppressor cells in the tumor microenvironment. J. Exp. Med. 207, 2439–2453 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Korbecki J. et al. Hypoxia alters the expression of CC chemokines and CC chemokine receptors in a tumor-A literature review. Int. J. Mol. Sci. 21, 5647 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chaturvedi P., Gilkes D. M., Takano N. & Semenza G. L. Hypoxia-inducible factor-dependent signaling between triple-negative breast cancer cells and mesenchymal stem cells promotes macrophage recruitment. Proc. Natl. Acad. Sci. U. S. A. 111, E2120–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.França G. S. et al. Cellular adaptation to cancer therapy along a resistance continuum. Nature 631, 876–883 (2024). [DOI] [PubMed] [Google Scholar]
- 84.Becker W. R. et al. Single-cell analyses define a continuum of cell state and composition changes in the malignant transformation of polyps to colorectal cancer. Nat. Genet. 54, 985–995 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hanahan D. Hallmarks of cancer: New dimensions. Cancer Discov. 12, 31–46 (2022). [DOI] [PubMed] [Google Scholar]
- 86.Kakani P. et al. Hypoxia-induced CTCF promotes EMT in breast cancer. Cell Rep. 43, 114367 (2024). [DOI] [PubMed] [Google Scholar]
- 87.Zhang L. et al. Hypoxia induces epithelial-mesenchymal transition via activation of SNAI1 by hypoxia-inducible factor −1α in hepatocellular carcinoma. BMC Cancer 13, 108 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rankin E. B. & Giaccia A. J. Hypoxic control of metastasis. Science 352, 175–180 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhao W. et al. A cellular and spatial atlas of TP53 -associated tissue remodeling in lung adenocarcinoma. bioRxivorg (2024) doi: 10.1101/2023.06.28.546977. [DOI] [Google Scholar]
- 90.De Zuani M. et al. Single-cell and spatial transcriptomics analysis of non-small cell lung cancer. Nat. Commun. 15, 4388 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Enfield K. S. S. et al. Spatial architecture of myeloid and T cells orchestrates immune evasion and clinical outcome in lung cancer. Cancer Discov. 14, 1018–1047 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Greenwald A. C. et al. Integrative spatial analysis reveals a multi-layered organization of glioblastoma. Cell 187, 2485–2501.e26 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen W. et al. Symbolic recording of signalling and cis-regulatory element activity to DNA. Nature 632, 1073–1081 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kempton H. R., Love K. S., Guo L. Y. & Qi L. S. Scalable biological signal recording in mammalian cells using Cas12a base editors. Nat. Chem. Biol. 18, 742–750 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Melsted P. et al. Modular, efficient and constant-memory single-cell RNA-seq preprocessing. Nat. Biotechnol. 39, 813–818 (2021). [DOI] [PubMed] [Google Scholar]
- 96.Fleming S. J. et al. Unsupervised removal of systematic background noise from droplet-based single-cell experiments using CellBender. Nat. Methods 20, 1323–1335 (2023). [DOI] [PubMed] [Google Scholar]
- 97.Wolf F. A., Angerer P. & Theis F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10 (2011). [Google Scholar]
- 99.Density-Based Algorithm for Discovering Clusters in Large Spatial Databases with Noise.
- 100.Gusfield D. Efficient algorithms for inferring evolutionary trees. Networks (N. Y.) 21, 19–28 (1991). [Google Scholar]
- 101.Xu C. et al. Probabilistic harmonization and annotation of single-cell transcriptomics data with deep generative models. Mol. Syst. Biol. 17, e9620 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lopez R., Regier J., Cole M. B., Jordan M. I. & Yosef N. Deep generative modeling for single-cell transcriptomics. Nat. Methods 15, 1053–1058 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gayoso A. et al. scvi-tools: a library for deep probabilistic analysis of single-cell omics data. bioRxiv 2021.04.28.441833 (2021) doi: 10.1101/2021.04.28.441833. [DOI] [Google Scholar]
- 104.Traag V. A., Waltman L. & van Eck N. J. From Louvain to Leiden: guaranteeing well-connected communities. Sci. Rep. 9, 5233 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.DeTomaso D. & Yosef N. Hotspot identifies informative gene modules across modalities of single-cell genomics. Cell Syst 12, 446–456.e9 (2021). [DOI] [PubMed] [Google Scholar]
- 106.Marconato L. et al. SpatialData: an open and universal data framework for spatial omics. Nat. Methods 1–5 (2024). [DOI] [PubMed] [Google Scholar]
- 107.Neher R. A., Russell C. A. & Shraiman B. I. Predicting evolution from the shape of genealogical trees. Elife 3, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Prillo S., Ravoor A., Yosef N. & Song Y. S. ConvexML: Scalable and accurate inference of single-cell chronograms from CRISPR/Cas9 lineage tracing data. bioRxivorg (2023) doi: 10.1101/2023.12.03.569785. [DOI] [Google Scholar]
- 109.Fang Z., Liu X. & Peltz G. GSEApy: a comprehensive package for performing gene set enrichment analysis in Python. Bioinformatics 39, btac757 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Fitness-neighborhood differential expression and GO Term analyses.
Data Availability Statement
Custom code for the analysis of spatially-resolved lineage-tracing data is available on Github through Cassiopeia (https://github.com/YosefLab/Cassiopeia) and at https://github.com/mattjones315/KPSpatial-release. All raw and processed data will be made available on GEO and other public repositories.