Summary:
Animals exhibit sex-specific behaviors that are governed by sexually dimorphic circuits. One such behavior in male Drosophila melanogaster, courtship, is regulated by various sensory modalities, including olfaction. Here, we reveal how sexually dimorphic olfactory pathways in male flies converge at the third-order, onto lateral horn output neurons, to regulate courtship. To achieve this, we developed ds-Tango, a modified version of the monosynaptic tracing and manipulation tool trans-Tango. In ds-Tango, two distinct configurations of trans-Tango are positioned in series, thus providing selective genetic access not only to the monosynaptic partners of starter neurons but also to their disynaptic connections. Using ds-Tango, we identified a node of convergence for three sexually dimorphic olfactory pathways. Silencing this node results in deficits in sex recognition of potential partners. Our results identify lateral horn output neurons required for proper courtship behavior in male flies and establish ds-Tango as a tool for disynaptic circuit tracing.
Keywords: ds-Tango, disynaptic tracing, neural circuit, courtship, Drosophila, lateral horn
INTRODUCTION
Animals have evolved a variety of means to signal their suitability to potential mating partners. These signals relay information about the species, sex, and fitness of the animal via courtship rituals that consist of stereotyped sequences of behaviors, culminating in copulation. The courtship rituals of each species involve a specific array of sensory modalities that range from electroreception in electric fish1 to audition in bats2, from vision in birds of paradise3 to mechanosensation in spiders4.
In the fruit fly Drosophila melanogaster, courtship involves visual, auditory, tactile, gustatory, and olfactory signals5–9. Among the olfactory signals, volatile pheromones are used to correctly assess the suitability of a potential mate6,10,11, while environmental stimuli such as food odors affect the rate of copulation12–14. Since male and female fruit flies exhibit distinct mating behaviors15, they must respond differently to these olfactory stimuli. Indeed, several sexually dimorphic olfactory sensory pathways have been identified16–18.
Sexual dimorphism in the Drosophila nervous system is achieved by differential expression of sex-specific isoforms of the transcription factors fruitless (fru)19,20 and doublesex (dsx)21. Male-specific neurons express the male isoforms of these proteins (FruM and DsxM). In males, the FruM isoform is present in olfactory sensory neurons (OSNs) that express the odorant receptors Or47b or Or67d, or the ionotropic receptor Ir84a16–18. Hence, these three populations of OSNs likely play a role in male-specific behaviors. Indeed, these receptors detect distinct aspects of the environment relevant for courtship. Or47b detects two pheromones that are produced by both sexes, methyl laurate22 and palmitoleic acid23, and the activation of Or47b-OSNs promotes copulation in males24. Or67d, on the other hand, binds to a male-specific pheromone, 11-cis-vaccenyl acetate (cVA)25,26, which is an aphrodisiac for females and a suppressor of mating in males6. By contrast, the ligands of Ir84a are not pheromones but rather food-derived odors such as phenylacetic acid and phenylacetaldehyde12. The activation of Ir84a-OSNs enhances male courtship12.
The presence of the FruM isoform in these three olfactory circuits and their roles in male mating behaviors suggest that they might interact. This interaction could occur at the first-order level of the circuits (OSNs), the second-order level (local interneurons (LNs) or olfactory projection neurons (OPNs)), or at a higher level. Although OSNs affect the electrical properties of each other through ephaptic coupling27; this is unlikely to happen among Or47b-, Or67d-, and Ir84a-OSNs as they reside in different sensilla18,28. In addition, while Or47b-driven courtship is suppressed by cVA, this effect requires synaptic release from the Or67d-OSNs29. Alternatively, the interaction between these three pathways could happen via LNs that innervate all three glomeruli. This hypothesis is unlikely since the activity of the Ir84a circuit is less prone to lateral inhibition from other olfactory circuits30. However, an important caveat of this study is that the inhibitory effects of the activation of the Or47b and Or67d circuits were not directly examined30. We, therefore, expect minimal contribution from the first- and second-order neurons in these circuits to the integration of olfactory courtship cues.
Thus, olfactory courtship cues may be integrated in higher-order neurons within these three circuits. Indeed, the OPNs in these circuits innervate a common region in the lateral horn (LH)12,31, a brain area that acts as a hub of integration for olfactory stimuli governing innate behaviors31. Despite extensive mapping of the various cell types that make up the LH32,33, our understanding of how neuronal activity in the LH influences innate behaviors is limited. Individual sexually dimorphic neurons in the LH that respond to cVA have been identified34–36, but no such population is known to receive inputs from the other olfactory circuits affecting courtship. Further, while cVA negatively regulates courtship6, the other FruM+ circuits positively affect the courtship drive12,24. Thus, there remains a significant knowledge gap as to how olfactory information is routed through the brain to ultimately elicit male courtship behavior. We, therefore, sought to identify a potential node in the LH that translates neuronal activity in courtship-regulating olfactory circuits into the appropriate sexual behaviors.
To achieve this, we comprehensively mapped the FruM+ olfactory pathways starting from the OSNs through three layers of neural circuitry to the LH. Given that the published fly connectomes are from female brains37–39, we could not trace these pathways in the male brain. We, therefore, developed a new version of trans-Tango40 that selectively labels neurons across two consecutive synapses downstream from a starting population. We termed this new method ds-Tango for disynaptic Tango. Using ds-Tango, we revealed that Or47b, Or67d, and Ir84a pathways converge onto a group of LH neurons that are third-order to all three circuits. Silencing of these neurons lead to increased male-to-male courtship. Our study identifies a new circuit node important for proper copulation behavior in Drosophila melanogaster.
RESULTS
Tracing second-order projections within the sexually dimorphic olfactory circuits
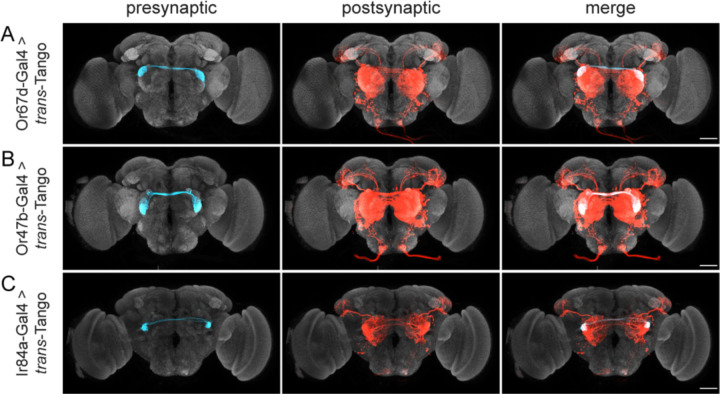
The antennal lobe (AL) is the first olfactory information processing center in the fly brain. In the AL, OSNs synapse onto OPNs that relay the olfactory information to higher-order brain regions, such as the mushroom body (MB) and the LH41. The MB is implicated in associative learning42 whereas the LH mostly mediates innate behaviors, including courtship43. To examine whether the three FruM+ olfactory circuits converge on a common node in the LH, we mapped the second-order neurons within these circuits via trans-Tango, a genetically encoded circuit tracing method that works monosynaptically in the anterograde direction40 (Figure 1). In all three cases, we observed strong post-synaptic signal in the LNs, as well as the characteristic projections of the OPNs to the MB and LH (Figure 1). The LH is subdivided into discrete regions for different classes of odorants, with food related odors being represented in the dorsal LH, and pheromone odors being represented in the ventral LH31,44. Accordingly, the post-synaptic signal in both Or67d and Or47b circuits was localized strictly in the ventral LH (Figure 1A and 1B). By contrast, the OPNs of the Ir84a circuit innervated both the ventral and dorsal compartments (Figure 1C), consistent with the ligands for Ir84a being courtship-promoting food odors12. Thus, the three FruM+ olfactory circuits may converge onto a common node in the ventral LH as this is a common projection target area. We, therefore, sought to map the circuits downstream of the OPNs to identify this putative node of convergence.
Figure 1. Tracing second-order projections within the sexually dimorphic olfactory circuits.
trans-Tango is used to trace the projections of the three FruM+ OSN populations: Or67d-OSNs (A), Or47b-OSNs (B), and Ir84a-OSNs (C). In the presynaptic channel (left column), only the OSNs are labeled (cyan). In the postsynaptic channel (center), both the LNs and PNs (red) are revealed. OPN axons target ventral LH in (A) and (B), and both ventral and dorsal LH in (C). Merge of both channels is shown in the rightmost column. Maximum intensity Z-stack projection of whole-mount brains are shown. Neuropil counterstain is shown in grey. Scale bars, 50μm.
Design of ds-Tango
To map the third-order neurons in the FruM+ olfactory circuits, we established a strategy for disynaptic anterograde tracing in the fly brain that we termed ds-Tango (For extended details on the design of ds-Tango, see supplemental text). In ds-Tango, two distinct versions of trans-Tango40 are configured in series, such that initiation of the first in the starter neurons leads to expression of the ligand of the second in the postsynaptic partners. This ligand, in turn, activates the second version of trans-Tango in third-order, disynaptic connections of the starter neurons. In this manner, ds-Tango enables selective genetic access into neurons mono- and disynaptic from the starter population.
The ds-Tango signaling pathway consists of five fusion proteins and three reporters (Figures 2A, S1, and S2). Three of the fusion proteins are expressed in all neurons: the human glucagon receptor tethered to a modified version of the transcription factor LexA45 via the cleavage site recognized by the tobacco etch virus N1a protease (hGCGR::TEVcs::LexA*), the human parathyroid hormone receptor tethered to the transcription factor QF by the same cleavage site (hPTHR::TEVcs::QF), and the human β-Arrestin2 fused to the N1a protease from the tobacco etch virus (hArr::TEV). The panneuronal expression of these components endows all neurons with the ability to react to both ds-Tango ligands, rendering the method flexible and versatile. The two remaining fusion proteins are the ligand constructs that are conditionally expressed in subsets of neurons. The first, a fusion between human glucagon and Drosophila Neurexin1 (hGCG::dNRXN1), is expressed in the starter neurons via the Gal4/UAS binary system. In the second, a modified version of human parathyroid hormone is similarly fused to Drosophila Neurexin1 (hPTH::dNRXN1). This fusion protein is expressed under the control of LexAop. To selectively label the three layers of the circuit, three reporter proteins are expressed under the control of the different binary systems. The starter neurons (shown in cyan) are labeled by CD2 under the control of Gal4/UAS, the monosynaptic partners (shown in green) are labeled by GFP under the control of LexA/LexAop, and the nuclei of the disynaptic connections (shown in magenta) are labeled by DsRed under the control of QF/QUAS46 (Figure 2A).
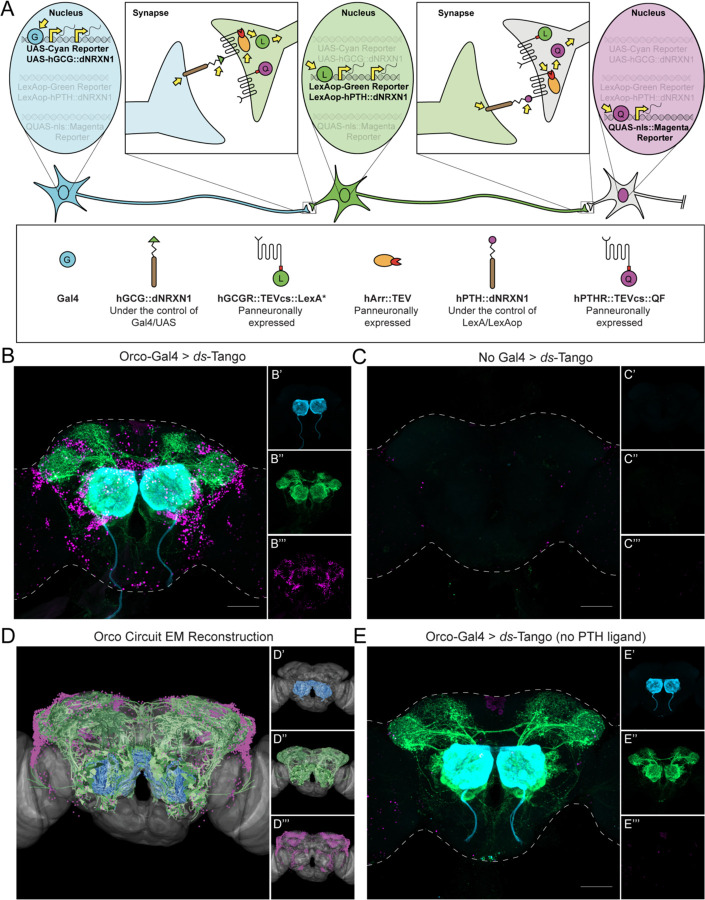
Figure 2. Design and implementation of ds-Tango in the olfactory system.
(A) Schematic and components of ds-Tango. In flies carrying a Gal4 driver, the presynaptic reporter (cyan) and the GCG ligand (hGCG::dNRXN1) are expressed in the starter neurons. The GCG ligand localizes to the presynaptic sites of the starter neurons and activates the GCGR Tango fusion (hGCGR::TEVcs::LexA*) across the synapse on the monosynaptic partners. Upon activation of the GCGR, the hArr::TEV fusion protein is recruited to it, TEV cleaves its recognition site (TEVcs) releasing LexA*. LexA* then translocates to the nucleus and initiates the expression of the PTH ligand (hPTH::dNRXN1) and of the monosynaptic reporter (green). The PTH ligand localizes to the presynaptic sites of the monosynaptic partners and activates the PTHR Tango fusion (hPTHR::TEVcs::QF) across the synapse on the disynaptic connections. Upon activation of the PTHR, the hArr::TEV fusion protein is recruited to it, TEV cleaves its recognition site (TEVcs), releasing QF. QF then translocates to the nucleus and initiates the expression of the nuclear disynaptic reporter (magenta). The various steps in the process are indicated by yellow arrows.
(B) Driving ds-Tango in the peripheral olfactory system using Orco-Gal4 labels OSNs (cyan, shown in B’), their monosynaptic partners LNs and OPNs (green, shown in B’’), and the nuclei of their disynaptic connections (magenta, shown in B’’’).
(C) A brain of a control fly bearing the ds-Tango components, but no Gal4 driver exhibits no background neurons in the presynaptic channel (cyan, shown in C’), virtually no background neurons in the monosynaptic channel (green, shown in C’’), and the nuclei of a few background neurons in the disynaptic channel (magenta, shown in C’’’).
(D) EM reconstruction of the Orco circuit projected on a template brain (gray) reveals OSNs (cyan, shown in D’), LNs and OPNs (green, shown in D’’), and the nuclei of third-order olfactory neurons (magenta, shown in D’’’).
(E) A brain of a control fly bearing Orco-Gal4 and a version of ds-Tango lacking the PTH ligand exhibits labeling in OSNs (cyan, shown in E’), LNs and OPNs (green, shown in E’’), but no staining of disynaptic partners except for the nuclei of a few background neurons (magenta, shown in E’’’).
Maximum intensity Z-stack projection of whole-mount brains are shown in B, D, and E. Dashed lines in B, D, and E depict the approximate outline of the fly brains. Scale bars = 50 μm.
The multilevel labeling scheme is initiated by Gal4 that, when expressed in a group of starter neurons, triggers the disynaptic ds-Tango signaling cascade (Figure 2A). In ds-Tango flies that do not carry a Gal4 driver, the GCG ligand is not expressed, and the ds-Tango cascade is not triggered. Thus, in all neurons, both QF and LexA* remain fused to their respective receptors and sequestered to the cell membrane, and none of the reporters are expressed. By contrast, in ds-Tango flies carrying a Gal4 driver, the CD2 reporter and the GCG ligand are expressed in all starter neurons. In these neurons, the GCG ligand construct targets to the presynaptic membrane, selectively initiating the GCGR::TEVcs::LexA* signaling cascade in their monosynaptic partners (Figure 2A). In the monosynaptic partners, the Arr::TEV fusion is recruited to the activated GCGR fusion, leading to cleavage of the TEVcs and releasing the transcription factor from the receptor. LexA* is now free to translocate to the nucleus where it initiates the expression of GFP and the PTH ligand construct. The PTH ligand construct is then targeted down the axon to the presynaptic membrane of the monosynaptic partners, allowing the ligand to bind its receptor on the disynaptic connections. This, in turn, initiates the PTHR::TEVcs::QF Tango signaling cascade in the disynaptic connections, culminating in translocation of QF to the nucleus and subsequent expression of nuclear DsRed. We used a nuclearly localized reporter as the final output for ds-Tango instead of a membrane-targeted reporter because the vast number of intermingled neurites labeled by the latter complicated analysis and troubleshooting. The modularity of ds-Tango enables one to readily replace the nuclearly localized reporter with other reporters, as shown below.
Implementation of ds-Tango in the olfactory circuits
To validate ds-Tango, we initiated disynaptic tracing from OSNs to visualize the expected first-, second-, and third-order neurons in the olfactory circuits. To this end, we initiated ds-Tango using orco-Gal4, a driver that expresses in most OSNs (Figure 2B). We observed the stereotypical innervation patterns in the AL, and the characteristic OPN projections to the MB, LH, and subesophageal zone (SEZ). This confirmed that the ds-Tango signal in the second-order neurons recapitulates previously published trans-Tango tracing experiments40 and connectome results47. We also observed canonical third-order nuclei labeling around the AL, MB, and LH, consistent with the known anatomy of the olfactory circuits. In addition, we revealed third-order nuclei surrounding the SEZ, the primary taste center in the fly brain (Figure 2B), likely due to the projections from the AL to the SEZ40. To confirm that the ds-Tango labeling that we observed is Gal4-dependent, we examined ds-Tango flies lacking a Gal4 driver. Indeed, no first- or second-order background noise was present in these flies. We did, however, observe few third-order background neurons that were mostly restricted to the optic lobes (Figure 2C).
To further validate that the neurons revealed by ds-Tango are bona fide third-order olfactory neurons, we simulated a disynaptic tracing experiment using the Drosophila EM hemibrain dataset37. In our EM reconstructions of the olfactory circuit, we observed similar first-, second-, and third- order signal as we did in our olfactory ds-Tango experiment (Figure 2D). To confirm that the GCG fusion does not activate PTHR and that the third-order signal is PTH-dependent, we generated flies bearing all the ds-Tango components except for the PTH ligand fusion (Figure S2B). As expected, when we initiated this deficient configuration of ds-Tango with orco-Gal4, we observed similar first- and second-order labeling of neurons as with ds-Tango, but without labeling of third-order neurons (Figure 2E). These experiments confirmed the successful implementation of ds-Tango for selective labeling of neurons within the first three layers of the olfactory circuit.
Specificity of ds-Tango
To examine the specificity of ds-Tango, we decided to initiate it from Or67d- and Or42a-OSNs, two populations we expect to have predominantly non-overlapping monosynaptic and disynaptic connections. Or67d is a pheromone receptor, and Or42a is broadly tuned to food odors48. Pheromones and food odors are represented in different compartments of the LH31. We, hence, sought to test whether ds-Tango would label distinct populations of LH neurons when initiated from these two OSN populations. Initially, we simulated ds-Tango experiments from the OSNs targeting the DA1 glomerulus (Or67d-OSNs) and OSNs targeting the VM7 glomerulus (Or42a-OSNs) using the Drosophila EM hemibrain dataset37 (Figures 3A and 3D). The EM reconstruction of the Or67d circuit revealed second-order projections to the ventral part of the LH and third-order neurons surrounding the LH (Figure 3A). Importantly, we noticed few to no cell bodies of the disynaptic connections surrounding the calyx of the MB. By contrast, the EM reconstruction of the Or42a circuit revealed second-order projections to the dorsal part of the LH, dense innervation of the MB, and numerous cell bodies of the disynaptic connections surrounding the calyx of the MB (Figure 3D). These differences serve as important benchmarks to evaluate the specificity of ds-Tango.
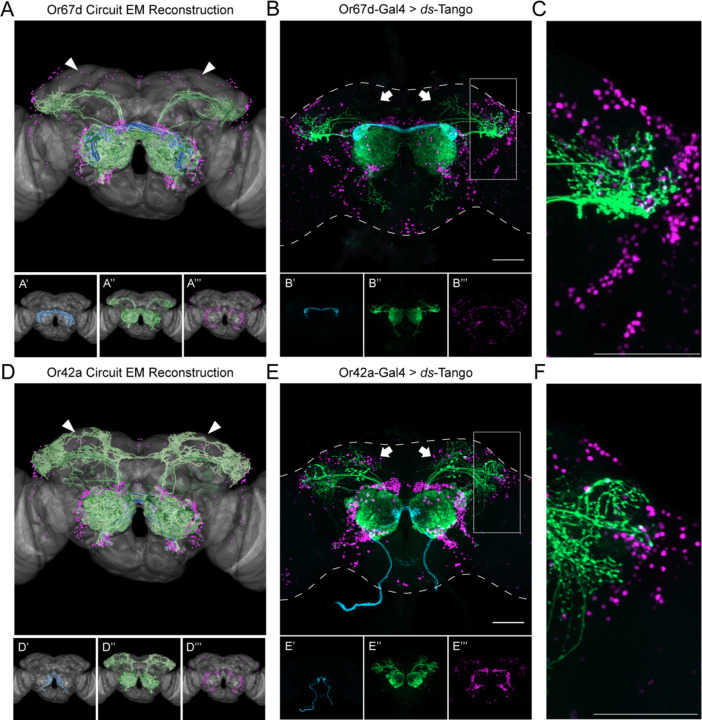
Figure 3. Specificity of ds-Tango.
(A) Or67d Circuit EM reconstruction projected on a template brain (gray) reveals the OSNs (cyan, shown in A’), LNs and OPNs (green, shown in A’’), and the nuclei of third-order neurons (magenta, shown in A’’’).
(B) Driving ds-Tango using Or67d-Gal4 labels Or67d-OSNs (cyan, shown in B’), their monosynaptic partner LNs and OPNs (green, shown in B’’), and the nuclei of their disynaptic connections (magenta, shown in B’’’).
(C) Higher magnification image of the gray inset in (B), depicting the lateral horn.
(D) Or42a Circuit EM reconstruction projected on a template brain (gray) reveals the OSNs (cyan, shown in D’), LNs and OPNs (green, shown in D’’), and the nuclei of third-order neurons (magenta, shown in D’’’).
(E) Driving ds-Tango using Or42a-Gal4 labels Or42a-OSNs (cyan, shown in E’), their monosynaptic partners LNs and OPNs (green, shown in E’’), and the nuclei of their disynaptic connections (magenta, shown in E’’’).
(F) Higher magnification image of the gray inset in (E), depicting the lateral horn.
Arrowheads in A and D highlight the absence and presence of third-order Kenyon cells in the MB, respectively. Arrows in B and E highlight the absence and presence of third-order Kenyon cells in the MB, respectively. Maximum intensity Z-stack projection of whole-mount brains are shown in B and E. Dashed lines in B and E depict the approximate outline of the fly brains. Scale bars = 50 μm.
We then initiated ds-Tango from Or67d-OSNs (Figure 3B and 3C) and Or42a-OSNs (Figures 3E and 3F) and observed labeling in anatomically distinct populations of monosynaptic partners and disynaptic connections. Notably, initiation of ds-Tango from Or67d-OSNs identified second-order OPNs that densely innervate the ventral part of the LH as well as previously described non-canonical PNs that target the SEZ40 (Figure 3B and 3C). It is worth noting that our EM simulations of the Or67d circuit did not reveal second-order PNs targeting the SEZ, or third-order neurons surrounding the SEZ, because this brain area has not yet been traced and thus is not included in the EM hemibrain dataset37. By contrast, initiating ds-Tango from Or42a-OSNs revealed second-order PNs that target predominantly the calyx of the MB and the dorsal LH (Figures 3E and 3F). Notably, the projection to the SEZ observed when driving ds-Tango from Or67d-OSNs is absent when ds-Tango is driven from Or42a-OSNs. Further, we observed more third-order neurons surrounding the LH in the Or67d circuit than in the Or42a circuit (Figures 3C and 3F). By contrast, ds-Tango labeled more cell bodies of disynaptic connections surrounding the MB, when initiated from Or42a-OSNs (Figures 3B and 3E). Thus, the differences between the Or67d and Or42a circuits predicted by the EM reconstruction were successfully recapitulated by ds-Tango in both monosynaptic partners and disynaptic connections. These results demonstrate the accuracy and specificity of ds-Tango.
While troubleshooting ds-Tango, we identified several variables that affect the signal, including the rearing temperature and age of dissected flies. We observed an inverse relationship between temperature and the signal in monosynaptic partners and disynaptic connections (Figure S3). We noticed an optimal signal-to-noise ratio for standard tracing at 21°C, but rearing flies at either higher or lower temperatures can also provide benefits in some experimental designs. Further, we observed a direct relationship between the age of flies and the third-order ds-Tango signal (Figure S4). Therefore, users should optimize the protocol of their ds-Tango experiments according to their driver lines and circuits of interest. In this study, we crossed and raised flies at 21°C and dissected them at 3–4 weeks of age, unless otherwise noted. Using these parameters, we noticed remarkably consistent ds-Tango signal in the brains of different flies when driving the system with Or67d-Gal4 (Figure S5). On average, we observed 39.75±7.21 monosynaptic partners and 533±73.70 disynaptic connections per hemisphere across eight brains.
These variables, and others, can impact the ds-Tango signal, and thus impact the interpretation of the experimental results. Bias can be introduced in both EM-simulated ds-Tango reconstructions (via synaptic thresholds) and ds-Tango tracing (via experimental parameters such as age or temperature). Therefore, we sought other ways to validate that the ds-Tango signal we observe accurately reflects the biological ground truth.
ds-Tango reveals sexual dimorphism in the lateral horn in the Or67d circuit
To further validate the specificity of ds-Tango in revealing differences between various neural pathways, we deployed it to explore sexually dimorphic circuits. Third order neurons in the Or67d circuit exhibit sexual dimorphism. While the FruM+ lateral cluster neurons in the LH (LC1 and LC2) are part of the Or67d circuit in both sexes, the FruM+ dorsal cluster neurons (DC1 and DC2) are postsynaptic to OPNs in males but not in females34,35. We wished to study whether ds-Tango is sufficiently specific to reveal this sexual dimorphism. To this end, we reasoned that a membrane bound reporter would enable easier identification of neurons, and thus, instead of the nuclearly localized one, we implemented a new reporter for the disynaptic connections, QUAS-mtdTomato49. In the absence of a Gal4 driver, this reporter exhibits little to no background in the central brain (Figures S6A and S6B). By contrast, when we initiated ds-Tango with this mtdTomato reporter from Or67d-OSNs, we observed dense disynaptic signal in the ventral portion of the LH in both males and females (Figures 4A and 4D). We also observed multiple distinct clusters of disynaptic neurons surrounding the LH in both sexes. Importantly, ds-Tango revealed sexual dimorphism in both the monosynaptic partners and disynaptic connections. We observed disynaptic LC1 and LC2 neurons in both sexes as expected34,35 (Figures 4A and 4D). By contrast, we noticed that the DC1 and DC2 neurons were strongly labeled in males; yet they were absent, or weakly labeled, in females (Figures 4A and 4D). To further confirm the identification of these neurons, we used an antibody against FruM alongside ds-Tango. We observed that among the disynaptic connections, not all neurons in the lateral clusters were FruM+. By contrast, we did not observe co-labeling of FruM within the dorsal clusters (Figure S7). This result contradicts previous reports that observed FruM immunoreactivity in dorsal cluster neurons35. A potential explanation for this discrepancy is that we analyzed flies at an old age (three to four-week-old). To resolve this discrepancy, we restricted the expression of the disynaptic reporter via an intersectional genetic approach using a FLP recombinase expressed from the fru locus (fru-FLP)50 together with our FLP-dependent reporter. Our results with this approach further confirmed that while ds-Tango reveals lateral cluster neurons to be disynaptic connections of Or67d-OSNs in both sexes, the dorsal cluster neurons appear to be part of the Or67d circuit in males only (Figures 4B, 4C, 4E, and 4F). Importantly, this intersectional approach yielded little to no background signal in the absence of a Gal4 driver (Figures S6C and S6D). Altogether, these experiments validate that ds-Tango is sufficiently specific to reveal differences in sexually dimorphic circuits.
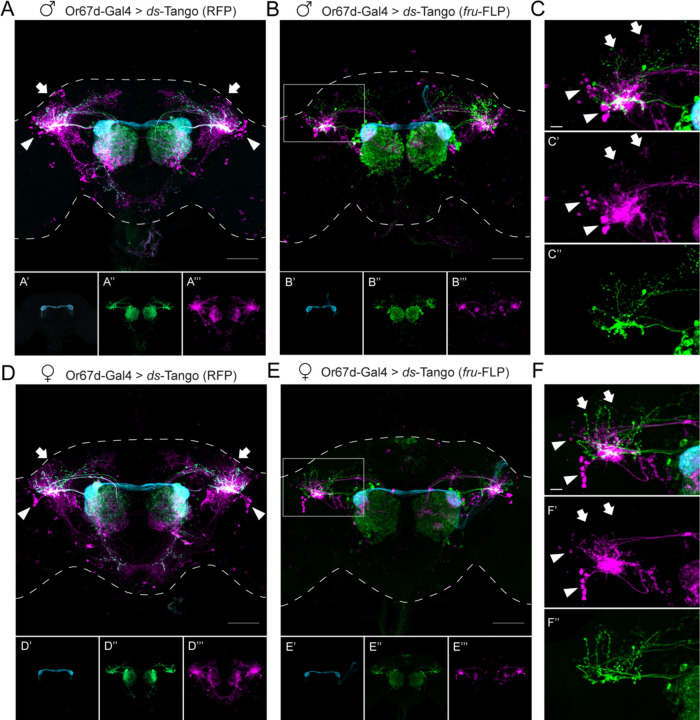
Figure 4. ds-Tango reveals sexual dimorphism in the lateral horn in the Or67d circuit.
(A) Driving ds-Tango with Or67d-Gal4 in males labels Or67d-OSNs (cyan, shown in A’), their monosynaptic partner LNs and OPNs (green, shown in A’’), and their disynaptic connections (magenta, shown in A’’’). Arrows indicate the dorsal cluster neurons; arrowheads indicate the lateral cluster neurons.
(B) Driving ds-Tango with Or67d-Gal4 and genetically restricting disynaptic reporter expression to fru-FLP+ neurons in males labels Or67d-OSNs (cyan, shown in B’), their monosynaptic partner LNs and OPNs (green, shown in B’’), and their fru-FLP+ disynaptic connections (magenta, shown in B’’’).
(C) A higher magnification image of the gray inset in (B) highlighting the left LH reveals PNs targeting the ventral region of the LH (green, shown in C’’), overlapping with the neurites of fru-FLP+ disynaptic connections (magenta, shown in C’). Note the presence of lateral cluster neurons (arrowheads) and the dorsal cluster neurons (arrows) in the male brain.
(D) Driving ds-Tango with Or67d-Gal4 in females labels Or67d-OSNs (cyan, shown in D’), their monosynaptic partner LNs and OPNs (green, shown in D’’), and their disynaptic connections (magenta, shown in D’’’). Arrowheads indicate the lateral cluster neurons; arrows indicate the location of dorsal cluster neurons.
Note the presence of lateral cluster neurons in both the male (arrowheads in A) and female (arrowheads in D) brains and the prominent dorsal cluster neurons in the male (arrows in A) brain that are less prominent or absent in the female (arrows in D) brain.
(E) Driving ds-Tango with Or67d-Gal4 and genetically restricting disynaptic reporter expression to fru-FLP+ neurons in females labels Or67d-OSNs (cyan, shown in E’), their monosynaptic partners LNs and OPNs (green, shown in E’’), and their fru-FLP+ disynaptic connections (magenta, shown in E’’’).
(F) A higher magnification image of the gray inset in (E) showing the left LH reveals PNs targeting the ventral region of the LH (green, shown in F’’), overlapping with the neurites of fru-FLP+ disynaptic connections (magenta, shown in F’). Note the presence of lateral cluster neurons (arrowheads) and the absence of dorsal cluster neurons (arrows) in the female brain.
Maximum intensity Z-stack projection of whole-mount brains are shown in A, B, D, and E. Dashed lines in A, B, D, and E depict the approximate outline of the fly brains. Scale bars, 50 μm.
A node of convergence for courtship-regulating olfactory circuits
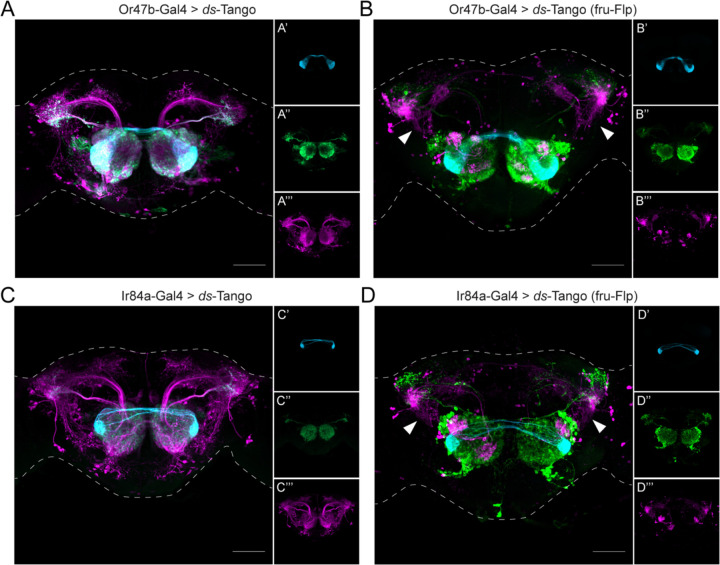
Having established ds-Tango as a specific tool for tracing disynaptic connections within a circuit, we turned to identifying nodes of convergence for courtship-regulating olfactory circuits. To this end, we sought to map the circuitry downstream from the two remaining FruM+ OSN populations. The use of the ds-Tango strategy allows us to identify common targets of the FruM+ OSNs in the LH. We first implemented ds-Tango to reveal all the third-order neurons in the Or47b and Ir84a circuits. We drove the configuration of ds-Tango in which the disynaptic reporter is mtdTomato from Or47b-OSNs and Ir84a-OSNs. We found that in each circuit the ds-Tango signal in the monosynaptic partners (Figures 5A and 5C) recapitulates the signal observed with trans-Tango (Figures 1B and 1C). Driving ds-Tango from Or47b-OSNs revealed innervation of the ventral compartment of the LH by the OPNs (Figure 5A). By contrast, driving ds-Tango from Ir84a-OSNs revealed OPNs that innervated the dorsal and ventral compartments of the LH (Figure 5C). Amongst the third-order neurons in both circuits, we observed ventral projections from the LH to the anterior ventrolateral protocerebrum (AVLP). However, when driving ds-Tango from Or47b-, or Ir84a-OSNs, the signal in the disynaptic connections was dense and hard to parse. We thus concluded that to identify a common node among the FruM+ olfactory circuits, a sparser ds-Tango signal in the third-order neurons would be necessary.
Figure 5. A node of convergence for courtship-regulating olfactory circuits.
(A) Driving ds-Tango with Or47b-Gal4 in males labels Or47b-OSNs (cyan, shown in A’), their monosynaptic partner LNs and OPNs (green, shown in A’’), and their disynaptic connections (magenta, shown in A’’’).
(B) Driving ds-Tango with Or47b-Gal4 and genetically restricting disynaptic reporter expression to fru-FLP+ neurons in males labels Or47b-OSNs (cyan, shown in B’), their monosynaptic partner LNs and OPNs (green, shown in B’’), and their fru-FLP+ disynaptic connections (magenta, shown in B’’’). Note the presence of ventral projections from the LH to the AVLP in the disynaptic connections characteristic of AV2b1/b2 neurons (arrowheads).
(C) Driving ds-Tango with Ir84a-Gal4 in males labels Ir84a-OSNs (cyan, shown in C’), their monosynaptic partners LNs and OPNs (green, shown in C’’), and their disynaptic connections (magenta, shown in C’’’).
(D) Driving ds-Tango with Ir84a-Gal4 and genetically restricting disynaptic reporter expression to fru-FLP+ neurons in males labels Ir84a-OSNs (cyan, shown in D’), their monosynaptic partners LNs and OPNs (green, shown in D’’), and their fru-FLP+ disynaptic connections (magenta, shown in D’’’). Note the presence of ventral projections from the LH to the AVLP in the disynaptic connections characteristic of AV2b1/b2 neurons (arrowheads).
Maximum intensity Z-stack projection of whole-mount brains are shown. Dashed lines depict the approximate outline of the fly brains. Scale bars, 50 μm.
We reasoned that a hypothetical LH neuron that would integrate information from the three FruM+ olfactory circuits would also express FruM. We, therefore, restricted the disynaptic ds-Tango signal using fru-FLP together with the FLP-dependent configuration of ds-Tango (Figure S2D). Driving this configuration of ds-Tango from Or47b- or Ir84a-OSNs labeled smaller populations of disynaptic connections (Figures 5B and 5D) than in the analogous experiments using the configuration of ds-Tango that does not require FLP expression (Figures 5A and 5C). When we compared the fru-FLP+ third-order neurons in the Or67d (Figure 4B), Or47b (Figure 5B), and Ir84a (Figure 5D) circuits, we noticed a common tract that projects ventrally from the LH into the AVLP. This projection is characteristic of AV2b1/b2 neurons33, a subset of the LC2 cluster34. Due to the distinct morphology of these neurons that resemble the meandering river Maiandros, we named them the Maiandros neurons. We reasoned that the Maiandros neurons could serve as a node of convergence that relays courtship-relevant olfactory cues to downstream processing centers to regulate courtship behaviors.
Silencing the Maiandros neurons leads to deficits in mate discrimination in males
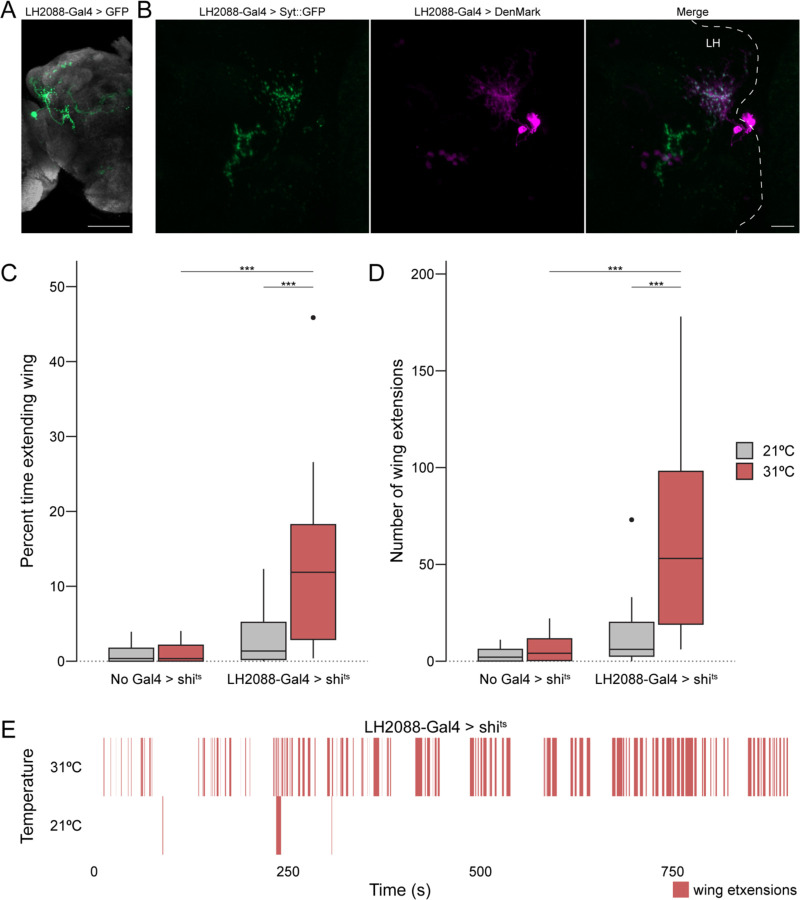
We next sought to characterize the Maiandros neurons through the use of a specific genetic driver line. We screened through the expression patterns of split-Gal4 drivers in a library that targets various cell types of the LH33. One such driver, LH2088-Gal433, provides weak access to the Maiandros neurons with only faint background in some of the Kenyon cells of the MB (Figure 6A). Using this driver, we sought to determine the polarity of the Maiandros neurons in males by expressing markers for presynaptic sites (synaptotagmin::GFP51) and dendrites (DenMark52). We reasoned that if the Maiandros neurons indeed relay information from the LH to downstream circuits, their dendrites should be localized in the LH, and their axons should be localized in the AVLP. Visualizing the neurite positions of the Maiandros neurons revealed that they have mixed axons and dendrites in the LH but only axons in the AVLP (Figure 6B). Because the neurites in the AVLP were strictly axonal, we conclude that the Maiandros neurons in males relay information from the LH to the AVLP as predicted for females53,54, indicating that they are indeed LH output neurons. In females, these neurons were determined to be both GABAergic and cholinergic through neurotransmitter staining33, yet only cholinergic by electron microscopy reconstructions53. To identify the neurotransmitters used by these neurons in males, we examined the expression of choline acetyltransferase (ChAT - cholinergic) and glutamic acid decarboxylase (GAD - GABAergic) drivers in these neurons. We observed that only the ChAT driver expressed in these neurons (Figure S8), indicating that in males these neurons are likely cholinergic.
Figure 6. Anatomical and functional analysis of the Maiandros neurons.
(A) Expression pattern of the LH2088-Gal4 driver line. LH2088-Gal4 drives UAS-GFP (green). The Maiandros neurons are clearly labeled along with minimal background in the MB. Neuropil counterstain is shown in grey.
(B) Locations of the Maiandros neuron axons and dendrites. LH2088-Gal4 drives synaptic vesicle marker Syt::GFP (green) and dendritic marker DenMark (magenta). Axons and dendrites are intermixed in the LH while ventral neurites in the AVLP are primarily axons. Dashed lines indicate the outline of the central brain. LH indicates lateral horn.
(C) Quantification of unilateral wing extension as indicator of attempted courtship when each experimental male fly is paired with a single naïve WT male. Boxplot depicting the percent of total trial time (15 min) that experimental male flies extended one wing during thermogenetic silencing using shits. Silencing of the Maiandros neurons in male flies tested at the restrictive temperature of 31°C results in increased time spent with one wing extended relative to genetic controls and to experimental flies tested at the permissive temperature of 21°C.
(D) Quantification of unilateral wing extension as indicator of attempted courtship when each experimental male fly is paired with a single naïve WT male. Boxplot depicting the total number of unilateral wing extensions exhibited by experimental male flies during thermogenetic silencing using shits. Silencing of the Maiandros neurons in male flies tested at the restrictive temperature of 31°C results in increases in total number of wing extensions relative to genetic controls and to experimental flies tested at the permissive temperature of 21°C.
(E) Rasterplot depicting bouts of unilateral wing extensions over time for representative trials of male flies bearing the LH2088-Gal4 and UAS-shits alleles in the thermogenetic silencing assay at the permissive temperature of 21°C and the restrictive temperature of 31°C. Unilateral wing extensions are sparse at 21°C but abundant throughout the 31°C trial.
Maximum intensity Z-stack projection of whole-mount brains are shown in (A) and (B). Scale bars, 50μm in (A) and 10μm in (B).
Because the Maiandros neurons receive synaptic inputs from the courtship-regulating olfactory circuits, we hypothesized that their activity would be necessary for proper control of courtship behavior in males. Under natural conditions, flies only exhibit minimal levels of intermale courtship due to pheromonal cues, such as cVA, that suppress courtship behaviors6,55. Thus, we tested whether silencing the Maiandros neurons would lead to intermale courtship. To achieve this, we used a temperature sensitive Dynamin allele (shits)56 that reversibly blocks neurotransmission at temperatures above 28°C. In each trial, we paired a single experimental male with a single naïve w1118 male in a circular behavior arena57 and used the unilateral wing extension behavior15 as a readout of attempted courtship. Predictably, control males that carried the shits allele but no driver exhibited only low levels of courtship towards a w1118 male at both 21°C and 31°C (Figures 6C and 6D). Flies expressing shits in the Maiandros neurons exhibited a slight, albeit insignificant, increase in courtship behavior at the permissive temperature of 21°C. At the restrictive temperature of 31°C, however, these flies exhibited dramatically increased levels of intermale courtship (Figures 6C and 6D). Experimental males typically pursued w1118 males throughout the duration of the 15-minute trial, indicating that the courtship pursuits persisted long after they would have presumably detected the pheromones on their targets (Figure 6E). Thus, silencing the Maiandros neurons elicits a prolonged arousal state that suppresses the effects of the anti-aphrodisiac pheromones present on the courtship targets. We interpret these data as evidence that activity in the Maiandros neurons is required for proper courtship behaviors in male flies. We conclude that the Maiandros neurons constitute a node of convergence for all three sexually dimorphic olfactory circuits and that their activity is required for proper identification of the sex of potential mates.
DISCUSSION
Control of innate behaviors, such as courtship, requires hardwired circuits to integrate a diverse array of sensory cues that are detected by multiple sensory systems. Even within a sensory modality, cues from various sources may be needed to convey diverse pieces of the information necessary for the proper execution of a certain behavior. For example, courtship in Drosophila melanogaster is regulated by sensory cues that are detected by at least three olfactory sensory channels. These cues may originate from conspecifics (e.g., the pheromones cVA and methyl laurate), or from the environment (e.g., the food odor phenylacetaldehyde). Regardless of their origin, these cues can either promote or inhibit courtship. Hence, the brain must integrate these distinct, and at times opposing, olfactory cues into a cohesive behavior. This integration may take place at a node of convergence for these three olfactory channels. Therefore, gaining a deeper understanding of how the brain transforms olfactory cues into courtship behaviors requires detailed mapping of the points of interaction between these channels. Since the LH is known to mediate innate behaviors, including courtship, it is well positioned to serve as the locus of this node of convergence.
The LH receives projections from the olfactory system that segregate into compartments based on the associated behaviors for the respective odors. Food odors are represented in the dorsal LH, and pheromones in the ventral LH31,58. However, whether this pattern of segregation of the OPN inputs into the LH is manifested as convergence onto a common node of output remained unknown. To address this, we developed ds-Tango, a genetic tool for tracing the disynaptic connections of starter neurons. Using ds-Tango, we traced the disynaptic connections of the three FruM+ OSN populations that detect courtship-regulating olfactory cues in males. Our analysis revealed the previously catalogued AV2b1/b2 neurons33, which we named the Maiandros neurons, as a common target receiving convergent inputs from the three courtship-regulating olfactory circuits. The anatomy of the Maiandros neurons indicates that they receive olfactory information from the OPNs projecting to the LH and relay this information to downstream targets in the AVLP. Thus, the AVLP is well positioned to inherit courtship-regulating olfactory signals from the LH and serve as a higher-order brain region for processing courtship-relevant sensory information into the appropriate behaviors. Consistent with this hypothesis, silencing the Maiandros neurons led to intermale courtship, likely due to increased arousal and impaired mate discrimination. The Maiandros neurons may, therefore, play a key role in processing courtship-relevant odor information to control arousal levels and decide whether to court or not to court. Since silencing of the Maiandros neurons in males leads to increases in intermale courtship, these neurons are likely inhibited by courtship promoting pheromones and food odors and activated by antiaphrodisiac pheromones. Indeed, in females these neurons have been described to receive inhibitory signals upon activation of the Or47b pathway59. Further work is required to determine how activity in the Maiandros neurons is affected in males by the three sexually dimorphic olfactory pathways. The Maiandros neurons may regulate levels of arousal in the male fly, perhaps through indirect inhibitory signals onto P1 neurons – a population that regulates arousal and the intensity of courtship behaviors60–63.
To identify third-order neurons within the olfactory circuits, we developed ds-Tango, a genetically encoded synthetic signaling pathway that allows differential genetic access into the monosynaptic partners and disynaptic connections of a given population of starter neurons. While we demonstrate the efficacy of ds-Tango by implementing it in the well-described Drosophila olfactory system, a major advantage of ds-Tango is its versatility. The signaling pathway that mediates ds-Tango labeling is expressed panneuronally. ds-Tango can, therefore, be deployed to achieve disynaptic tracing from any neuron for which genetic access is available via a Gal4 driver. While here the starter neurons are the first-order, sensory neurons of the olfactory circuits, ds-Tango can be initiated from any layer within any circuit. Additionally, the reporters that ds-Tango selectively expresses in the starter neurons, monosynaptic partners, and disynaptic connections are all genetically encoded. This design enables researchers to vary the types of effectors that are expressed in each layer of a circuit. Indeed, this modularity has enabled the implementation of various configurations of the precursor of ds-Tango, trans-Tango, for calcium imaging46,64 as well as optogenetic64,65 and chemogenetic66 manipulations. ds-Tango is, therefore, not limited to tracing experiments and can be extended to enable the recording or manipulation of neuronal activity via the extensive Drosophila genetic toolkit.
The success of a ds-Tango experiment, however, depends on the Gal4 driver line being used. Leaky or non-specific expression in a given Gal4 line will confound results by producing false positive ds-Tango signal in addition to bona fide monosynaptic partners and disynaptic connections. To minimize the false positive signal, we recommend selecting Gal4 driver lines that narrowly target the desired starting population, when possible. Importantly, all the alleles for the ds-Tango system can be carried by a single fly. Consequently, performing a ds-Tango experiment only requires researchers to cross flies bearing the Gal4 driver labeling their starter neurons of choice to flies carrying the ds-Tango alleles and analyze the progeny of this cross. This feature renders feasible the use of ds-Tango with intersectional genetic strategies such as split-Gal4s67. However, our attempts to trace the neurons downstream of the Maiandros neurons using ds-Tango were not successful, likely due to the low levels of expression induced by the LH2088 split-Gal4 driver. Therefore, obtaining a Gal4 driver line that is both sufficiently strong and specific to the population of interest is imperative when designing a ds-Tango experiment.
The recently completed connectome of a Drosophila brain37 and other EM volumes that are readily accessible via various databases, such as the Virtual Fly Brain68, have become the gold standard in profiling connectivity in the fly. Nevertheless, ds-Tango complements the use of these connectomic strategies in several ways. First, EM reconstruction of a connectome requires expertise and is costly, laborious, and time intensive. By contrast, ds-Tango is user-friendly and only requires basic maintenance of a fly line. Second, the information within each EM volume only represents a single fly of a single sex at a single timepoint raised in a single environment. Thus, the use of EM volumes does not allow for comparison between different group of individuals for the study of the effect of age, sex, environment, past experiences or genetic background. By contrast, the ease of use of ds-Tango and its inherent high throughput allow researchers to readily profile circuit connectivity across multiple individuals, sexes, or various other conditions. The utility of these features has been exemplified by the use of the other members of the Tango toolkit – trans-Tango and retro-Tango – to study the effects of environment69, sex49, and genetic background70,71 on brain wiring. Here, we demonstrate these features by deploying ds-Tango to map the courtship-regulating olfactory circuits in males – a study we would not have been able to conduct since the only currently available EM volumes are from female brains.
Mapping synaptic connections in the brain is an essential step towards understanding how sensory information is processed to elicit the appropriate behaviors to the environment. Our study introduces ds-Tango, a new tool that can be deployed to analyze various circuits in the fly brain. ds-Tango is the first transsynaptic labeling tool that provides selective genetic access to three layers within a given neural circuit and thus endows researchers with the capacity to differentially manipulate neurons within each layer. We demonstrate the utility of ds-Tango by implementing it in the olfactory circuits for courtship behaviors in the male. This study provides a step forward in our understanding of male courtship behavior in Drosophila melanogaster by unveiling a potential node of integration for courtship-regulating sensory cues. The underlying network structure we describe may generalize to other pathways in the LH, such as the ones underlying food seeking and aversion. Our experiments thus lay the groundwork for a deeper understanding of how the brain controls innate behaviors.
METHODS
Fly Strains
All Drosophila melanogaster lines used in this study were raised on cornmeal-agar media supplemented with the antifungal Tegosept. Flies were maintained in 70% humidity-controlled incubators with a 12-hour light/dark cycle. Flies were crossed and raised at a single temperature: 18°C, 21°C, or 25°C, unless otherwise stated. Temperature details for all experiments are available in Table S1. The publicly available fly lines used in this study are as follows: trans-Tango (BDSC #77123), fru-FLP50, Or42a-Gal4 (BDSC #9969), Or47b-Gal4 (BDSC #9983), Ir84a- Gal4 (BDSC #41750), Or67d-Gal46, Orco-Gal4 (Or83b-Gal4; BDSC #23292), QUAS-FRT-STOP-FRT-mCD8::GFP (BDSC #30134), QUAS-nlsRFP46, w1118 (BDSC #5905), LH2088-Gal4 (BDSC #86635), UAS-GFP (pJFRC81–10xUAS-Syn21-myr::GFP-p10)72 (Pfeiffer, Truman, and Rubin 2012), UAS-Sytaptotagmin::GFP (BDSC #6926), UAS-DenMark (BDSC #33061), UAS-Shibire(ts)56, SS25111-Gal4 (BDSC #86796), UAS-CsChrimson (BDSC #55135) and QUAS-tdTomato-HA49. The transgenic ds-Tango fly line that was generated for this study includes four new fly lines: elav-Arr::TEV_nSyb-PTHR::TEVcs::QF_gypsy insulator(INS)_nSyb-GCGR::TEVcs::LexA* (inserted at attP40), UAS-CD2_ gypsy insulator(INS)_UAS- GCG::dNRX1 (inserted at attP2), LexAop-tdTomato_ gypsy insulator(INS)_LexAop-PTH::dNRX1 (inserted at VK00027) and LexAop-GF_gypsy insulator(INS)_LexAop-PTH::dNRX1 (inserted at VK00027). Three other lines were also generated for use in this study: LexAop-GFP (inserted at VK00027), and LexAop-tdTomatoLexAop-PTH::dNRX1 (inserted at VK00027). More detailed information about the genetic components of ds-Tango flies and other flies used in this study are available in Figures S1 and S2. In addition, genotypes for flies used in all experiments are listed in Table S1.
Generation of Transgenic Fly Lines
All plasmids generated for this study were generated using standard molecular biology techniques: restriction digest of a vector plasmid, PCR amplification of DNA inserts with Gibson overhangs added to primers (NEB, Q5 High-Fidelity DNA Polymerase), and Gibson assembly of all components (NEB, NEBuilder HiFi DNA Assembly). Gibson assembly mix was transformed into competent cells (NEB, NEB Stable Competent E. coli), cells were spread on antibiotic resistant plates, colonies were screened and sequenced, and finally sent for injection and insertion into appropriate attP sites via PhiC31 integration (Rainbow Transgenic Flies Inc., Camarillo, CA). Detailed DNA sequence and integration site information and of injected plasmids is in Supplemental Figure and the Results section. Complete sequence map information of any constructs used in this study are available upon request. The vector backbone plasmid and the insert template plasmids used to generate flies in this study are listed below.
elav-Arr::TEV_nSyb-PTHR::TEVcs::QF_gypsy insulator(INS)_nSyb-GCGR::TEVcs::LexA* was generated using the trans-Tango plasmid40 as the vector backbone. The PTHR sequence was amplified from a synthesized DNA fragment containing human PTHR (GENEWIZ). The non-PTHR encoding sequences for the PTHR fusion protein were amplified from the trans-Tango plasmid40. The gypsy insulator sequence (INS) was amplified from the plasmid pJFRC10–10XUAS-IVS-mCD8::GFP, INS, 10XUAS-IVS-mCD8::GFP72. The nSyb- GCGR::TEVcs::LexA* sequence was partially amplified from the trans-Tango plasmid40. For the LexA* part, the LexADBD sequence was amplified from Addgene #46117 and and the QFAD* from #6131045. UAS-CD2, INS, UAS-GCG::dNRX1 was generated using the plasmid pJFRC10–10XUAS-IVS-mCD8::GFP, INS, 10XUAS-IVS-mCD8::GFP72 as the vector backbone. The UAS-CD2 sequence was amplified from the plasmid UAS- CD2_QUAS-mtdTomato(3xHA) (Scaplen et al. 2021). The UAS-GCG::dNRX1 sequence was amplified from the plasmid trans-Tango MkII73.
LexAop-GFP was generated using the pJFRC19–13XLexAop2-IVS-myr::GFP plasmid72 as the vector backbone and the “GFP” sequence was amplified from the pJFRC206–10XUAS-FRT>STOP>FRT-myr::smGFP-V5 plasmid74.
LexAop-GFP, INS, LexAop-PTH::dNRX1 was generated using the LexAop-GFP plasmid (this study) as the vector backbone. The INS sequence was amplified from plasmid UAS-CD2, INS, UAS-GCG::dNRX1 (this study). The LexAop-PTH::dNRX1 sequence was amplified from the following plasmids: LexAop-GFP plasmid (this study) — for the LexAop sequence and the sv40pA after the PTH ligand coding region — and the UAS-CD2, INS, UAS-GCG::dNRX1 plasmid (this study) — for the non-PTH encoding part of the PTH ligand. The PTH ligand sequence was generated using overhangs from Gibson primers. The PTH ligand sequence encodes all but the first amino acid from the propeptide and the entire mature PTH peptide: SVKKRSVSEIQLMHNLGKHLNSMERVEWLRKKLQDVHNFVALGAPLAPRDAGS QRPRKKEDNVLVESHEKSLGEADKADVNVLTKAKSQ We found that this specific peptide sequence gave a higher signal-to-noise ratio than the entire prepro-PTH peptide sequence, just the mature PTH peptide, or any other versions of the PTH peptide we generated (data not shown).
LexAop-tdTomato, INS, LexAop-PTH::dNRNX1 was generated using the LexAop-GFP, INS, LexAop-PTH::dNRX1 (this study) as the vector backbone. The tdTomato(3XHA) insert sequence was amplified from the QUAS-tdTomato(3XHA) plasmid49.
Immunohistochemistry and Tissue Processing
The age of each dissected experimental fly in this study is listed in Table 3-S1. Immunohistochemistry was performed on dissected adult fly brains as previously described40, with some adaptations. Briefly, adult flies were cold anesthetized on ice and dissected in 0.5% PBST. All following steps were performed while brains were nutating. Dissected brains were then fixed for 30 minutes at room temperature (RT) in 4% PFA/0.01% PBST, washed 4 times — each wash 15 minutes at RT in 0.5% PBST, and blocked for 90 minutes at RT in 5% heat-inactivated donkey serum (diluted in 0.5% PBST). Then, brains were incubated in primary antibodies (diluted in the donkey blocking solution) for two overnights. Following primary antibody incubation, brains were once again washed 4 times — each wash 15 minutes at RT in 0.5% PBST. Next, brains were incubated in secondary antibodies (diluted in the donkey blocking solution) for two overnights. Finally, brains were once again washed 4 times — each wash 15 minutes at RT in 0.5% PBST — and placed in 0.5% PBST for subsequent DPX clearing. The Janelia FlyLight Clearing Protocol “DPX Mounting” (https://www.janelia.org/sites/default/files/Project%20Teams/Fly%20Light/FL%20ProtocPr%20-%20DPX%20Mounting%202020-03-06.pdf) was followed exactly with a few modifications. First, we used Fisher Scientific coverslips for mounting brains (Cover glass 22×22 mm Square No. 1, Fisher Scientific #12–542B), as was done in the original DPX Mounting protocol, not Corning brand. As the protocol authors noted in the updated protocol, we also observed inconsistent and incomplete PLL coating using Fisher brand coverslips. For this reason, we made our second modification to the protocol, by increased the PLL coating of coverslips step to 5–10 seconds at RT. An abbreviated version of the DPX Mounting protocol is below including a few modifications.
Following wash of secondary antibodies, brains were postfixed for 4 hours at RT in 4% PFA/0.01% PBST. Brains were next washed 4 times — each wash 15 minutes at RT in 0.5% PBST — and then rinsed in PBS for 15 minutes at RT. Brains were then mounted on PLL-coated coverslips in PBS, quickly dipped in MilliQ water, and dehydrated for 10 minutes in successively higher concentration of ethanol baths. Finally, the brains were soaked in 3 xylene baths for 5 minutes each, DPX was applied, the coverslip was mounted on a slide and was left in the hood for at least 48 hours before imaging. A detailed list of antibodies used in each figure is in Table 3-S1. The primary antibodies used for this study are: Goat anti-GFP (Rockland #600–101-215, 1:1000), Guinea Pig anti-FruM (Gift from Michael Perry, UCSD, 1:100) (Wohl et al., 2020), Guinea Pig anti-RFP (Gift from Susan Brenner-Morton, Columbia University, 1:10,000), Mouse anti- CD2 (Bio-Rad #MCA154GA, 1:100), Rabbit anti-RFP (Abcam #ab62341, 1:500 – only used in Figure 4-S4), Rat anti-HA (Roche, 11867423001; 1:100). It is important to note that the Goat anti-GFP antibody works much better for second-order staining of all LexAop-GFP containing flies than any of the other GFP antibodies we tried and much better than any of the V5 epitope tag antibodies we tried (in LexAop-GFP, the GFP has a V5 epitope tag attached). Secondary antibodies were spun down at 15,000 RPM for 15 minutes before adding to blocking buffer. The secondary antibodies used for this study are: Donkey anti-goat 488 (ThermoFisher, #A32814), Donkey anti-Guinea Pig Cy3 550 (Jackson Immuno, #706–165-148), Donkey anti-Guinea Pig 647 (EMD Millipore, #AP193SA6), Donkey anti-mouse 647 (ThermoFisher, #A31571), Donkey anti-Rabbit 555 (ThermoFisher, #A32794), Donkey anti-Rat 555 (SouthernBiotech, #6430–32).
Thermogenetic silencing experiments
Thermogenetic silencing courtship experiments were performed in a custom behavior chamber that was based on the previously described Flybowl57. Flies that were assayed for our shibirets experiments were analyzed at either 21°C or 31°C by adjusting the temperature in our behavior chamber. Flies that were analyzed as part of these experiments were placed in 21°C temperature- and humidity-controlled incubators upon eclosion. Flies in the 31°C groups were placed in the chamber for at least 30 minutes before being assayed to allow flies to acclimate to the temperature increase. Flies were analyzed at age 5–7 days old. Each trial consisted of an experimental male and a w1118 male fly. w1118 males used as the courtship targets had a single wing clipped to allow differentiation between experimental fly and w1118 fly. Assays were performed under white light (~45 μW/cm2) with a polarizing filter. Courtship videos were manually annotated for frames when unilateral wing extensions occurred. Resulting data was then analyzed using custom R scripts. Code is available upon request.
Microscopy and Image Analysis
All images were taken on a confocal microscope (Zeiss, LSM800) with ZEN Blue software (version 2.3) using auto-Z brightness correction when appropriate, to maintain homogenous signal in the Z plane. Maximum intensity Z-stacks were created in ZEN and ultimately exported as TIFF files. All figures were generated in Adobe Illustrator (version 26.0.1).
Electron Microscopy Circuit Reconstructions
ds-Tango EM simulations were performed using the natverse suite of R packages75. The first order populations were queried using the “neuprint_search” function. The “neuprint_simple_connectivity” function was used to retrieve lists of postsynaptic partners that were annotated by the number of synapses between each neuron. We thresholded first-to-second and second-to-third synaptic partnerships using the total number of synapses each neuron received from the first order and second order populations, respectively. We used a threshold of 30 for first-to-second order connections and 50 for second-to-third connections. The “neuprint_read_skeletons” function was used to retrieve EM skeletonizations and the “mirror_brain” function was used to mirror skeletons between right and left hemispheres.
Supplementary Material
Acknowledgments:
We would like to thank Dr. Alexander Fleischmann and the members of the Barnea Laboratory for critical reading of the manuscript. We acknowledge Susan Brenner-Morton for sharing reagents and John Murphy from the Brown BioMed Machine Shop for aiding in the construction of our behavior chamber. This work was supported by NIH/NIDCD 5R01DC017146 (G.B.), Brown University Carney Institute for Brain Science, Suna Kıraç Fund for Brain Science (D.S.), Brown University Carney Institute for Brain Science, Graduate Award in Brain Science (D.S.) and NIH/NIDCD award F31DC019540 (A.M.C.). Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study.
Footnotes
Declaration of interests: Authors declare that they have no competing interests.
Additional information: All new fly strains will be deposited to Bloomington Drosophila Stock Center. Correspondence and requests for materials should be addressed to G.B.
References:
- 1.Wong R.Y., and Hopkins C.D. (2007). Electrical and behavioral courtship displays in the mormyrid fish Brienomyrus brachyistius. J Exp Biol 210, 2244–2252. 10.1242/jeb.003509. [DOI] [PubMed] [Google Scholar]
- 2.Bohn K.M., Schmidt-French B., Ma S.T., and Pollak G.D. (2008). Syllable acoustics, temporal patterns, and call composition vary with behavioral context in Mexican free-tailed bats. J Acoust Soc Am 124, 1838–1848. 10.1121/1.2953314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miles M.C., and Fuxjager M.J. (2018). Animal choreography of song and dance: a case study in the Montezuma oropendola,. Anim Behav 140, 99–107. 10.1016/j.anbehav.2018.04.006. [DOI] [Google Scholar]
- 4.Girard M.B., Elias D.O., and Kasumovic M.M. (2015). Female preference for multi-modal courtship: multiple signals are important for male mating success in peacock spiders. Proc Biol Sci 282, 20152222. 10.1098/rspb.2015.2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greenspan R.J., and Ferveur J.F. (2000). Courtship in Drosophila. Annu Rev Genet 34, 205–232. 10.1146/annurev.genet.34.1.205. [DOI] [PubMed] [Google Scholar]
- 6.Kurtovic A., Widmer A., and Dickson B.J. (2007). A single class of olfactory neurons mediates behavioural responses to a Drosophila sex pheromone. Nature 446, 542–546. 10.1038/nature05672. [DOI] [PubMed] [Google Scholar]
- 7.Thistle R., Cameron P., Ghorayshi A., Dennison L., and Scott K. (2012). Contact chemoreceptors mediate male-male repulsion and male-female attraction during Drosophila courtship. Cell 149, 1140–1151. 10.1016/j.cell.2012.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ejima A., and Griffith L.C. (2008). Courtship initiation is stimulated by acoustic signals in Drosophila melanogaster. PLoS One 3, e3246. 10.1371/journal.pone.0003246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Agrawal S., Safarik S., and Dickinson M. (2014). The relative roles of vision and chemosensation in mate recognition of Drosophila melanogaster. J Exp Biol 217, 2796–2805. 10.1242/jeb.105817. [DOI] [PubMed] [Google Scholar]
- 10.Ejima A., Smith B.P., Lucas C., van der Goes van Naters W., Miller C.J., Carlson J.R., Levine J.D., and Griffith L.C. (2007). Generalization of courtship learning in Drosophila is mediated by cis-vaccenyl acetate. Curr Biol 17, 599–605. 10.1016/j.cub.2007.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van der Goes van Naters W., and Carlson J.R. (2007). Receptors and neurons for fly odors in Drosophila. Curr Biol 17, 606–612. 10.1016/j.cub.2007.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grosjean Y., Rytz R., Farine J.P., Abuin L., Cortot J., Jefferis G.S., and Benton R. (2011). An olfactory receptor for food-derived odours promotes male courtship in Drosophila. Nature 478, 236–240. 10.1038/nature10428. [DOI] [PubMed] [Google Scholar]
- 13.Gorter J.A., Jagadeesh S., Gahr C., Boonekamp J.J., Levine J.D., and Billeter J.C. (2016). The nutritional and hedonic value of food modulate sexual receptivity in Drosophila melanogaster females. Sci Rep 6, 19441. 10.1038/srep19441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das S., Trona F., Khallaf M.A., Schuh E., Knaden M., Hansson B.S., and Sachse S. (2017). Electrical synapses mediate synergism between pheromone and food odors in Drosophila melanogaster. Proc Natl Acad Sci U S A 114, E9962–E9971. 10.1073/pnas.1712706114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall J.C. (1994). The mating of a fly. Science 264, 1702–1714. 10.1126/science.8209251. [DOI] [PubMed] [Google Scholar]
- 16.Stockinger P., Kvitsiani D., Rotkopf S., Tirian L., and Dickson B.J. (2005). Neural circuitry that governs Drosophila male courtship behavior. Cell 121, 795–807. 10.1016/j.cell.2005.04.026. [DOI] [PubMed] [Google Scholar]
- 17.Manoli D.S., Foss M., Villella A., Taylor B.J., Hall J.C., and Baker B.S. (2005). Male-specific fruitless specifies the neural substrates of Drosophila courtship behaviour. Nature 436, 395–400. 10.1038/nature03859. [DOI] [PubMed] [Google Scholar]
- 18.Silbering A.F., Rytz R., Grosjean Y., Abuin L., Ramdya P., Jefferis G.S., and Benton R. (2011). Complementary function and integrated wiring of the evolutionarily distinct Drosophila olfactory subsystems. J Neurosci 31, 13357–13375. 10.1523/JNEUROSCI.2360-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito H., Fujitani K., Usui K., Shimizu-Nishikawa K., Tanaka S., and Yamamoto D. (1996). Sexual orientation in Drosophila is altered by the satori mutation in the sex-determination gene fruitless that encodes a zinc finger protein with a BTB domain. Proc Natl Acad Sci U S A 93, 9687–9692. 10.1073/pnas.93.18.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryner L.C., Goodwin S.F., Castrillon D.H., Anand A., Villella A., Baker B.S., Hall J.C., Taylor B.J., and Wasserman S.A. (1996). Control of male sexual behavior and sexual orientation in Drosophila by the fruitless gene. Cell 87, 1079–1089. 10.1016/s0092-8674(00)81802-4. [DOI] [PubMed] [Google Scholar]
- 21.Villella A., and Hall J.C. (1996). Courtship anomalies caused by doublesex mutations in Drosophila melanogaster. Genetics 143, 331–344. 10.1093/genetics/143.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dweck H.K., Ebrahim S.A., Thoma M., Mohamed A.A., Keesey I.W., Trona F., Lavista-Llanos S., Svatos A., Sachse S., Knaden M., and Hansson B.S. (2015). Pheromones mediating copulation and attraction in Drosophila. Proc Natl Acad Sci U S A 112, E2829–2835. 10.1073/pnas.1504527112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin H.H., Cao D.S., Sethi S., Zeng Z., Chin J.S.R., Chakraborty T.S., Shepherd A.K., Nguyen C.A., Yew J.Y., Su C.Y., and Wang J.W. (2016). Hormonal Modulation of Pheromone Detection Enhances Male Courtship Success. Neuron 90, 1272–1285. 10.1016/j.neuron.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L., Han X., Mehren J., Hiroi M., Billeter J.C., Miyamoto T., Amrein H., Levine J.D., and Anderson D.J. (2011). Hierarchical chemosensory regulation of male-male social interactions in Drosophila. Nat Neurosci 14, 757–762. 10.1038/nn.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clyne P., Grant A., O’Connell R., and Carlson J.R. (1997). Odorant response of individual sensilla on the Drosophila antenna. Invert Neurosci 3, 127–135. 10.1007/BF02480367. [DOI] [PubMed] [Google Scholar]
- 26.Ha T.S., and Smith D.P. (2006). A pheromone receptor mediates 11-cis-vaccenyl acetate-induced responses in Drosophila. J Neurosci 26, 8727–8733. 10.1523/JNEUROSCI.0876-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Su C.Y., Menuz K., Reisert J., and Carlson J.R. (2012). Non-synaptic inhibition between grouped neurons in an olfactory circuit. Nature 492, 66–71. 10.1038/nature11712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Couto A., Alenius M., and Dickson B.J. (2005). Molecular, anatomical, and functional organization of the Drosophila olfactory system. Curr Biol 15, 1535–1547. 10.1016/j.cub.2005.07.034. [DOI] [PubMed] [Google Scholar]
- 29.Wu S.T., Chen J.Y., Martin V., Ng R., Zhang Y., Grover D., Greenspan R.J., Aljadeff J., and Su C.Y. (2022). Valence opponency in peripheral olfactory processing. Proc Natl Acad Sci U S A 119. 10.1073/pnas.2120134119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hong E.J., and Wilson R.I. (2015). Simultaneous encoding of odors by channels with diverse sensitivity to inhibition. Neuron 85, 573–589. 10.1016/j.neuron.2014.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jefferis G.S., Potter C.J., Chan A.M., Marin E.C., Rohlfing T., Maurer C.R. Jr., and Luo L. (2007). Comprehensive maps of Drosophila higher olfactory centers: spatially segregated fruit and pheromone representation. Cell 128, 1187–1203. 10.1016/j.cell.2007.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fisek M., and Wilson R.I. (2014). Stereotyped connectivity and computations in higher-order olfactory neurons. Nat Neurosci 17, 280–288. 10.1038/nn.3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dolan M.J., Frechter S., Bates A.S., Dan C., Huoviala P., Roberts R.J., Schlegel P., Dhawan S., Tabano R., Dionne H., et al. (2019). Neurogenetic dissection of the Drosophila lateral horn reveals major outputs, diverse behavioural functions, and interactions with the mushroom body. Elife 8. 10.7554/eLife.43079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruta V., Datta S.R., Vasconcelos M.L., Freeland J., Looger L.L., and Axel R. (2010). A dimorphic pheromone circuit in Drosophila from sensory input to descending output. Nature 468, 686–690. 10.1038/nature09554. [DOI] [PubMed] [Google Scholar]
- 35.Kohl J., Ostrovsky A.D., Frechter S., and Jefferis G.S. (2013). A bidirectional circuit switch reroutes pheromone signals in male and female brains. Cell 155, 1610–1623. 10.1016/j.cell.2013.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taisz I., Dona E., Munch D., Bailey S.N., Morris B.J., Meechan K.I., Stevens K.M., Varela-Martinez I., Gkantia M., Schlegel P., et al. (2023). Generating parallel representations of position and identity in the olfactory system. Cell 186, 2556–2573 e2522. 10.1016/j.cell.2023.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scheffer L.K., Xu C.S., Januszewski M., Lu Z., Takemura S.Y., Hayworth K.J., Huang G.B., Shinomiya K., Maitlin-Shepard J., Berg S., et al. (2020). A connectome and analysis of the adult Drosophila central brain. Elife 9. 10.7554/eLife.57443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dorkenwald S., Matsliah A., Sterling A.R., Schlegel P., Yu S.-c., McKellar C.E., Lin A., Costa M., Eichler K., Yin Y., et al. (2023). Neuronal wiring diagram of an adult brain. bioRxiv, 2023.2006.2027.546656. 10.1101/2023.06.27.546656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schlegel P., Yin Y., Bates A.S., Dorkenwald S., Eichler K., Brooks P., Han D.S., Gkantia M., dos Santos M., Munnelly E.J., et al. (2023). Whole-brain annotation and multi-connectome cell typing quantifies circuit stereotypy in Drosophila. bioRxiv, 2023.2006.2027.546055. 10.1101/2023.06.27.546055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Talay M., Richman E.B., Snell N.J., Hartmann G.G., Fisher J.D., Sorkac A., Santoyo J.F., Chou-Freed C., Nair N., Johnson M., et al. (2017). Transsynaptic Mapping of Second-Order Taste Neurons in Flies by trans-Tango. Neuron 96, 783–795 e784. 10.1016/j.neuron.2017.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stocker R.F. (1994). The organization of the chemosensory system in Drosophila melanogaster: a review. Cell Tissue Res 275, 3–26. 10.1007/BF00305372. [DOI] [PubMed] [Google Scholar]
- 42.de Belle J.S., and Heisenberg M. (1994). Associative odor learning in Drosophila abolished by chemical ablation of mushroom bodies. Science 263, 692–695. 10.1126/science.8303280. [DOI] [PubMed] [Google Scholar]
- 43.Heimbeck G., Bugnon V., Gendre N., Keller A., and Stocker R.F. (2001). A central neural circuit for experience-independent olfactory and courtship behavior in Drosophila melanogaster. Proc Natl Acad Sci U S A 98, 15336–15341. 10.1073/pnas.011314898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strutz A., Soelter J., Baschwitz A., Farhan A., Grabe V., Rybak J., Knaden M., Schmuker M., Hansson B.S., and Sachse S. (2014). Decoding odor quality and intensity in the Drosophila brain. Elife 3, e04147. 10.7554/eLife.04147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Riabinina O., Luginbuhl D., Marr E., Liu S., Wu M.N., Luo L., and Potter C.J. (2015). Improved and expanded Q-system reagents for genetic manipulations. Nat Methods 12, 219–222, 215 p following 222. 10.1038/nmeth.3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Snell N.J., Fisher J.D., Hartmann G.G., Zolyomi B., Talay M., and Barnea G. (2022). Complex representation of taste quality by second-order gustatory neurons in Drosophila. Curr Biol 32, 3758–3772 e3754. 10.1016/j.cub.2022.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schlegel P., Bates A.S., Sturner T., Jagannathan S.R., Drummond N., Hsu J., Serratosa Capdevila L., Javier A., Marin E.C., Barth-Maron A., et al. (2021). Information flow, cell types and stereotypy in a full olfactory connectome. Elife 10. 10.7554/eLife.66018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kreher S.A., Kwon J.Y., and Carlson J.R. (2005). The molecular basis of odor coding in the Drosophila larva. Neuron 46, 445–456. 10.1016/j.neuron.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 49.Sorkac A., Mosneanu R.A., Crown A.M., Savas D., Okoro A.M., Memis E., Talay M., and Barnea G. (2023). retro-Tango enables versatile retrograde circuit tracing in Drosophila. Elife 12. 10.7554/eLife.85041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu J.Y., Kanai M.I., Demir E., Jefferis G.S., and Dickson B.J. (2010). Cellular organization of the neural circuit that drives Drosophila courtship behavior. Curr Biol 20, 1602–1614. 10.1016/j.cub.2010.08.025. [DOI] [PubMed] [Google Scholar]
- 51.Zhang Y.Q., Rodesch C.K., and Broadie K. (2002). Living synaptic vesicle marker: synaptotagmin-GFP. Genesis 34, 142–145. 10.1002/gene.10144. [DOI] [PubMed] [Google Scholar]
- 52.Nicolai L.J., Ramaekers A., Raemaekers T., Drozdzecki A., Mauss A.S., Yan J., Landgraf M., Annaert W., and Hassan B.A. (2010). Genetically encoded dendritic marker sheds light on neuronal connectivity in Drosophila. Proc Natl Acad Sci U S A 107, 20553–20558. 10.1073/pnas.1010198107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bates A.S., Schlegel P., Roberts R.J.V., Drummond N., Tamimi I.F.M., Turnbull R., Zhao X., Marin E.C., Popovici P.D., Dhawan S., et al. (2020). Complete Connectomic Reconstruction of Olfactory Projection Neurons in the Fly Brain. Curr Biol 30, 3183–3199 e3186. 10.1016/j.cub.2020.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Frechter S., Bates A.S., Tootoonian S., Dolan M.J., Manton J., Jamasb A.R., Kohl J., Bock D., and Jefferis G. (2019). Functional and anatomical specificity in a higher olfactory centre. Elife 8. 10.7554/eLife.44590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Billeter J.C., Atallah J., Krupp J.J., Millar J.G., and Levine J.D. (2009). Specialized cells tag sexual and species identity in Drosophila melanogaster. Nature 461, 987–991. 10.1038/nature08495. [DOI] [PubMed] [Google Scholar]
- 56.Kitamoto T. (2001). Conditional modification of behavior in Drosophila by targeted expression of a temperature-sensitive shibire allele in defined neurons. J Neurobiol 47, 81–92. 10.1002/neu.1018. [DOI] [PubMed] [Google Scholar]
- 57.Simon J.C., and Dickinson M.H. (2010). A new chamber for studying the behavior of Drosophila. PLoS One 5, e8793. 10.1371/journal.pone.0008793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wong A.M., Wang J.W., and Axel R. (2002). Spatial representation of the glomerular map in the Drosophila protocerebrum. Cell 109, 229–241. 10.1016/s0092-8674(02)00707-9. [DOI] [PubMed] [Google Scholar]
- 59.Jeanne J.M., Fisek M., and Wilson R.I. (2018). The Organization of Projections from Olfactory Glomeruli onto Higher-Order Neurons. Neuron 98, 1198–1213 e1196. 10.1016/j.neuron.2018.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Clowney E.J., Iguchi S., Bussell J.J., Scheer E., and Ruta V. (2015). Multimodal Chemosensory Circuits Controlling Male Courtship in Drosophila. Neuron 87, 1036–1049. 10.1016/j.neuron.2015.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jung Y., Kennedy A., Chiu H., Mohammad F., Claridge-Chang A., and Anderson D.J. (2020). Neurons that Function within an Integrator to Promote a Persistent Behavioral State in Drosophila. Neuron 105, 322–333 e325. 10.1016/j.neuron.2019.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hindmarsh Sten T., Li R., Otopalik A., and Ruta V. (2021). Sexual arousal gates visual processing during Drosophila courtship. Nature 595, 549–553. 10.1038/s41586-021-03714-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hoopfer E.D., Jung Y., Inagaki H.K., Rubin G.M., and Anderson D.J. (2015). P1 interneurons promote a persistent internal state that enhances inter-male aggression in Drosophila. Elife 4. 10.7554/eLife.11346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.He Z., Luo Y., Shang X., Sun J.S., and Carlson J.R. (2019). Chemosensory sensilla of the Drosophila wing express a candidate ionotropic pheromone receptor. PLoS Biol 17, e2006619. 10.1371/journal.pbio.2006619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Feng K., Sen R., Minegishi R., Dubbert M., Bockemuhl T., Buschges A., and Dickson B.J. (2020). Distributed control of motor circuits for backward walking in Drosophila. Nat Commun 11, 6166. 10.1038/s41467-020-19936-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guo F., Holla M., Diaz M.M., and Rosbash M. (2018). A Circadian Output Circuit Controls Sleep-Wake Arousal in Drosophila. Neuron 100, 624–635 e624. 10.1016/j.neuron.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 67.Luan H., Peabody N.C., Vinson C.R., and White B.H. (2006). Refined spatial manipulation of neuronal function by combinatorial restriction of transgene expression. Neuron 52, 425–436. 10.1016/j.neuron.2006.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Court R., Costa M., Pilgrim C., Millburn G., Holmes A., McLachlan A., Larkin A., Matentzoglu N., Kir H., Parkinson H., et al. (2023). Virtual Fly Brain-An interactive atlas of the Drosophila nervous system. Front Physiol 14, 1076533. 10.3389/fphys.2023.1076533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kiral F.R., Dutta S.B., Linneweber G.A., Hilgert S., Poppa C., Duch C., von Kleist M., Hassan B.A., and Hiesinger P.R. (2021). Brain connectivity inversely scales with developmental temperature in Drosophila. Cell Rep 37, 110145. 10.1016/j.celrep.2021.110145. [DOI] [PubMed] [Google Scholar]
- 70.Chen Y.D., Park S.J., Joseph R.M., Ja W.W., and Dahanukar A.A. (2019). Combinatorial Pharyngeal Taste Coding for Feeding Avoidance in Adult Drosophila. Cell Rep 29, 961–973 e964. 10.1016/j.celrep.2019.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kiral F.R., Linneweber G.A., Mathejczyk T., Georgiev S.V., Wernet M.F., Hassan B.A., von Kleist M., and Hiesinger P.R. (2020). Autophagy-dependent filopodial kinetics restrict synaptic partner choice during Drosophila brain wiring. Nat Commun 11, 1325. 10.1038/s41467-020-14781-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pfeiffer B.D., Truman J.W., and Rubin G.M. (2012). Using translational enhancers to increase transgene expression in Drosophila. Proc Natl Acad Sci U S A 109, 6626–6631. 10.1073/pnas.1204520109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sorkac A., Savva Y.A., Savas D., Talay M., and Barnea G. (2022). Circuit analysis reveals a neural pathway for light avoidance in Drosophila larvae. Nat Commun 13, 5274. 10.1038/s41467-022-33059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nern A., Pfeiffer B.D., and Rubin G.M. (2015). Optimized tools for multicolor stochastic labeling reveal diverse stereotyped cell arrangements in the fly visual system. Proc Natl Acad Sci U S A 112, E2967–2976. 10.1073/pnas.1506763112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bates A.S., Manton J.D., Jagannathan S.R., Costa M., Schlegel P., Rohlfing T., and Jefferis G.S. (2020). The natverse, a versatile toolbox for combining and analysing neuroanatomical data. Elife 9. 10.7554/eLife.53350. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.