Abstract
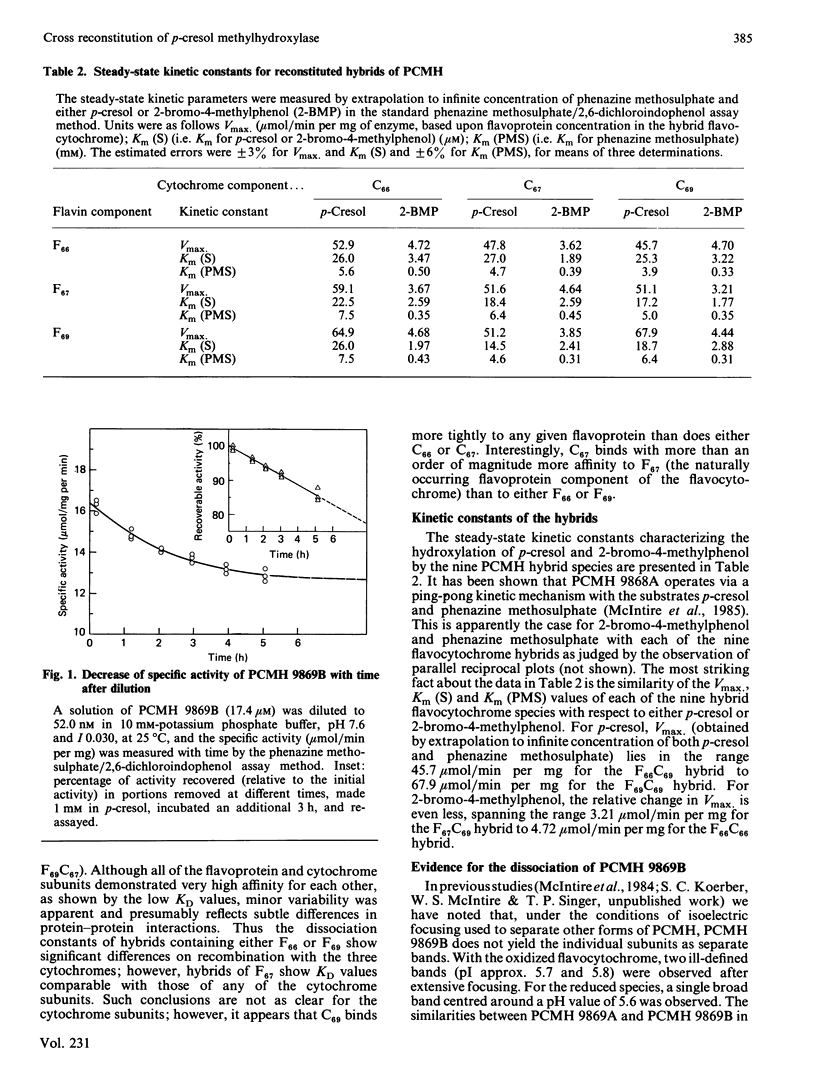
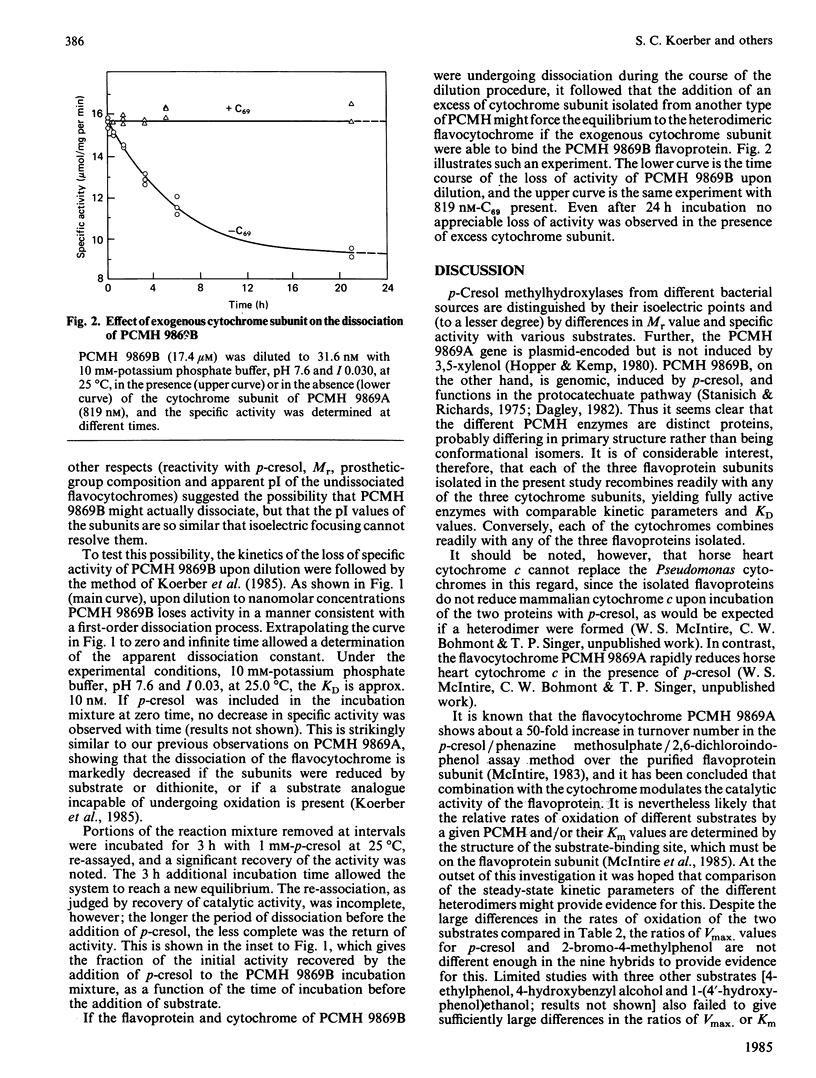
p-Cresol methylhydroxylases from four different pseudomonads differ in their isoelectric points and, to a lesser extent, in Mr values and substrate specificity. The enzymes from three species were isolated in homogeneous form, then resolved into their flavoprotein and cytochrome subunits, and the subunits were recombined to yield the nine possible hybrids (i.e. three intraspecies and six interspecies). The resulting flavocytochromes showed extensive similarities in steady-state kinetic parameters and in the dissociation constants of their subunits. Evidence is also presented that a fourth type of p-cresol methylhydroxylase, from Pseudomonas putida (N.C.I.B. 9869, form 'B'), the subunits of which cannot be isolated by the isoelectric focusing technique used to separate the subunits of the other flavocytochromes, nevertheless dissociates slowly at high dilution. The dissociation is reflected by a decline of catalytic activity with time. This process for the 'B' enzyme is prevented by the presence of substrate or an excess of a cytochrome subunit isolated from another enzyme species. Incubation of the dissociated subunits with p-cresol brings about extensive, albeit incomplete, re-association and regeneration of activity.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BRAY H. G., THORPE W. V. Analysis of phenolic compounds of interest in metabolism. Methods Biochem Anal. 1954;1:27–52. doi: 10.1002/9780470110171.ch2. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya A., Tollin G., McIntire W., Singer T. P. Laser-flash-photolysis studies of p-cresol methylhydroxylase. Electron-transfer properties of the flavin and haem components. Biochem J. 1985 Jun 1;228(2):337–345. doi: 10.1042/bj2280337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper D. J., Kemp P. D. Regulation of enzymes of the 3,5-xylenol-degradative pathway in Pseudomonas putida: evidence for a plasmid. J Bacteriol. 1980 Apr;142(1):21–26. doi: 10.1128/jb.142.1.21-26.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper D. J., Taylor D. G. The purification and properties of p-cresol-(acceptor) oxidoreductase (hydroxylating), a flavocytochrome from Pseudomonas putida. Biochem J. 1977 Oct 1;167(1):155–162. doi: 10.1042/bj1670155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keat M. J., Hopper D. J. P-cresol and 3,5-xylenol methylhydroxylases in Pseudomonas putida N.C.I.B. 9896. Biochem J. 1978 Nov 1;175(2):649–658. doi: 10.1042/bj1750649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntire W., Edmondson D. E., Hopper D. J., Singer T. P. 8 alpha-(O-Tyrosyl)flavin adenine dinucleotide, the prosthetic group of bacterial p-cresol methylhydroxylase. Biochemistry. 1981 May 26;20(11):3068–3075. doi: 10.1021/bi00514a013. [DOI] [PubMed] [Google Scholar]
- McIntire W., Hopper D. J., Craig J. C., Everhart E. T., Webster R. V., Causer M. J., Singer T. P. Stereochemistry of 1-(4'-hydroxyphenyl)ethanol produced by hydroxylation of 4-ethylphenol by p-cresol methylhydroxylase. Biochem J. 1984 Dec 1;224(2):617–621. doi: 10.1042/bj2240617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntire W., Hopper D. J., Singer T. P. p-Cresol methylhydroxylase. Assay and general properties. Biochem J. 1985 Jun 1;228(2):325–335. doi: 10.1042/bj2280325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamala N., Lim L. W., Mathews F. S., McIntire W., Singer T. P., Hopper D. J. Preliminary X-ray study of p-cresol methylhydroxylase (flavocytochrome c) from Pseudomonas putida N.C.I.B. 9869. J Mol Biol. 1985 Jun 5;183(3):517–518. doi: 10.1016/0022-2836(85)90020-8. [DOI] [PubMed] [Google Scholar]


