Abstract
In mammalian cells, repair of the most abundant endogenous premutagenic lesion in DNA, 7,8-dihydro-8-oxoguanine (8-oxoG), is initiated by the bifunctional DNA glycosylase OGG1. By using purified human proteins, we have reconstituted repair of 8-oxoG lesions in DNA in vitro on a plasmid DNA substrate containing a single 8-oxoG residue. It is shown that efficient and complete repair requires only hOGG1, the AP endonuclease HAP1, DNA polymerase (Pol) β and DNA ligase I. After glycosylase base removal, repair occurred through the AP lyase step of hOGG1 followed by removal of the 3′-terminal sugar phosphate by the 3′-diesterase activity of HAP1. Addition of PCNA had a slight stimulatory effect on repair. Fen1 or high concentrations of Pol β were required to induce strand displacement DNA synthesis at incised 8-oxoG in the absence of DNA ligase. Fen1 induced Pol β strand displacement DNA synthesis at HAP1-cleaved AP sites differently from that at gaps introduced by hOGG1/HAP1 at 8-oxoG sites. In the presence of DNA ligase I, the repair reaction at 8-oxoG was confined to 1 nt replacement, even in the presence of high levels of Pol β and Fen1. Thus, the assembly of all the core proteins for 8-oxoG repair catalyses one major pathway that involves single nucleotide repair patches.
INTRODUCTION
In all organisms the mutagenic effects of 7,8-dihydro-8-oxoguanine (8-oxoG) lesions, caused by DNA oxidation, are prevented by base excision repair (BER). This repair mechanism is initiated by a specific DNA glycosylase that recognises and removes the damaged base through N-glycosylic bond hydrolysis. The human enzyme hOGG1 is a bifunctional DNA glycosylase endowed with an AP lyase activity with specificity for cleavage at 8-oxoG opposite C (1,2). The generated AP site can be repaired in human cells by two alternative pathways which involve either the replacement of one (short patch BER) or more nucleotides (long patch BER) at the lesion site (reviewed in 3). Short patch BER requires three proteins: the major mammalian AP endonuclease HAP1, DNA polymerase (Pol) β and the DNA ligase III/XRCC1 heterodimer or DNA ligase I (4,5). Long patch BER involves, besides HAP1, proliferating cell nuclear antigen (PCNA), structure-specific nuclease Flap endonuclease 1 (Fen1), Pol δ/ɛ (6,7) or Pol β (8,9) and DNA ligase I (10). It has been hypothesised (reviewed in 3) that the type of termini generated by different enzymatic cleavage of AP sites is a major determinant for selection of the BER pathway. Single nucleotide BER might be favoured by the presence of a break with an intact nucleotide residue at the 3′ side such as that generated by the AP lyase activity of a bifunctional DNA glycosylase. Conversely, if a terminal deoxyribose phosphate (dRP) is produced 3′ to the break by the sequential action of a monofunctional DNA glycosylase and HAP1, this frayed 5′ end can be excised either as a monomer by Pol β or as a part of an oligomer by Fen1. In agreement with this model, mammalian cell extracts repair DNA containing 8-oxoG (11,12) and thymine glycol (which is a substrate for the DNA glycosylase/AP lyase hNTH1) (13) predominantly by Pol β-catalysed resynthesis of a single nucleotide residue, whereas both short and long repair patches are observed during repair of DNA containing uracil (14) or hypoxathine (11).
We have reconstituted in vitro the BER pathway for 8-oxoG by using a core of four proteins: hOGG1, HAP1, Pol β and DNA ligase I. We show that, in the absence of DNA ligase I, long patch repair synthesis occurs in the presence of Fen1 or of high levels of Pol β. The pattern of repair intermediates is strongly affected by the interaction between Fen1 and Pol β and by the nature of the gap. However, in the presence of DNA ligase I, long patch BER synthesis at 8-oxoG is completely abolished and the repair reaction involves exclusively the replacement of single nucleotide residues.
MATERIALS AND METHODS
Chemicals
[γ-32P]ATP (3000 Ci/mmol), [α-32P]dCTP (3000 Ci/mmol), [α-32P]dGTP (3000 Ci/mmol) and [α-32P]dTTP (3000 Ci/mmol) were from Amersham Pharmacia Biotech (Little Chalfont, UK) and unlabelled dNTPs from Roche Molecular Biochemicals (Mannheim, Germany). All other reagents were of analytical grade and purchased from Merck or Fluka-Sigma-Aldrich (St Louis, MO).
Nucleic acid substrates
The oligonucleotides were synthesized on a Biosearch Expedite 8909, by the phosphoramidite method, at the 1 µmol scale. 8-oxodG phosphoramidite was prepared according to the literature (15). Cleavage from the solid support (1 h at room temperature) and deprotection (17 h at 55°C) were performed with ammonium hydroxide and 0.25 M 2-mercaptoethanol. Oligonucleotides containing a single 8-oxoG were purified first by HPLC on a C-4 column, according to standard procedures, and then by preparative PAGE. Oligonucleotides containing a single uracil were purchased from M-Medical.
Closed circular DNA molecules containing a single 8-oxoG or abasic site were used as substrates in the BER assays. These single lesion-containing plasmids were produced as previously described (16) by priming single-stranded (+) pGEM-3Zf DNA (Promega) with a 30-fold molar excess of single lesion-containing oligonucleotide 5′-GAT CCT CTA 8-oxoG/U TCG ACC TGC AGG CAT GCA-3′, followed by in vitro DNA synthesis with T4 DNA polymerase holoenzyme, single-stranded DNA-binding protein and T4 DNA ligase (Boehringer Mannheim). Closed circular DNA duplex molecules were purified by cesium chloride equilibrium centrifugation. In the case of the uracil-containing plasmids a further digestion with Escherichia coli uracil-DNA glycosylase was required to produce a single abasic site.
The incision assay was carried out with slightly modified circular molecules obtained by priming single-stranded phagemid DNA with an oligonucleotide containing a 5′-terminal 8-oxoG residue, 5′-8-oxoG TCGACCTGCAGGCATGCAAGCT-3′. This oligonucleotide was 5′-end-labelled with [γ-32P]ATP prior to in vitro DNA synthesis.
Enzymes and proteins
Restriction enzymes were from Biolabs. Recombinant hOGG1 was purified from E.coli as previously described (1); recombinant human AP endonuclease (HAP1) was purified from E.coli as described in Rothwell et al. (17). Human PCNA (18), replication factor C (RF-C) (19) and DNA ligase I (20) were purified according to published procedures. Rat Pol β was purified according to Date et al. (21). Escherichia coli UDG and NTH were a gift from J. Laval.
Enzymatic assays
Incision assay. To measure hOGG1 and HAP1 activities the incision reactions were performed in a final volume of 50 µl containing 1 nM plasmid DNA, 40 mM HEPES–KOH, pH 7.9, 75 mM KCl, 5 mM MgCl2, 0.5 mM DTT, 2 mM ATP, 40 mM phosphocreatine, 18 µg BSA and purified enzymes as described. Reactions were incubated at 30°C for different periods of time followed by phenol/chloroform extraction and ethanol precipitation. Incised products were separated by 20% denaturing PAGE and visualised by autoradiography. Following incision reactions, the DNA samples were digested with BamHI and HindIII restriction enzymes to release the fragment originally containing the 8-oxoG residue.
BER assay. The repair reactions were carried out essentially as described (16), with minor modifications, by using highly purified components. Briefly, reaction mixtures (50 µl) contained 1 nM plasmid DNA containing a single 8-oxoG residue or a single AP site, 40 mM HEPES–KOH, pH 7.9, 75 mM KCl, 5 mM MgCl2, 0.5 mM DTT, 50 µM each dNTP, 2 µCi [α-32P]dNTPs as indicated (the concentration of the corresponding cold dNTP was lowered to 5 µM, unless differently specified), 2 mM ATP, 40 mM phosphocreatine, 2.5 µg creatine phosphokinase and 18 µg BSA. Reactions contained various combinations of purified enzymes as described. After 60 min at 30°C the plasmid DNA was recovered and digested with the appropriate restriction enzymes. The digestion products were resolved by denaturing 20% PAGE. The BER products were visualised by autoradiography.
RESULTS
HAP1 stimulates OGG1 activity on plasmid DNA and exerts a 3′-diesterase activity on the hOGG1 β-elimination products
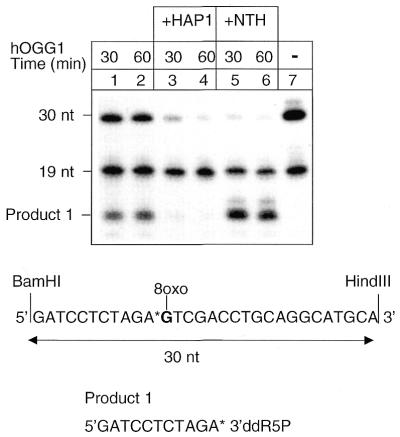
hOGG1 is a bifunctional DNA glycosylase endowed with an AP lyase activity (reviewed in 22). However, this activity is inefficient compared with the base removing activity, even on the biologically correct substrate (1,2,23). As shown in Figure 1 (lanes 1 and 2), when circular plasmid molecules containing a single 5′-end-labelled 8-oxoG moiety were incubated with high concentrations (10-fold molar excess relative to the substrate) of hOGG1 alone and then digested with the restriction enzymes BamHI and HindIII, the full-length 30 nt restriction fragment was still visible on the gel, implying that cleavage by hOGG1 at the lesion site was incomplete. The presence of cleavage products (product 1) that co-migrate with the products of the AP lyase activity of E.coli NTH (lanes 5 and 6) confirms that hOGG1 DNA strand cleavage is by β-elimination. This reaction plateaued in 30 min (compare lane 1 with lane 2). Complete cleavage of the 30 nt restriction fragment upon incubation with NTH (lanes 5 and 6) demonstrates that OGG1 DNA glycosylase activity has worked to completion leaving preformed AP sites amenable to cleavage by the AP lyase acitivity of NTH. These data confirm that the DNA glycosylase and AP lyase activities of hOGG1 are uncoupled. The additional 19 nt fragment visible in all the lanes is due to inefficient ligation during construction of the substrate (data not shown). This fragment is present in similar amounts in the control DNA sample (lane 7) and in the repaired samples (lanes 1–6) and will not be further discussed.
Figure 1.
HAP1 stimulates OGG1 activity and exerts a 3′-diesterase activity on the hOGG1 β-elimination products. Autoradiograph of a denaturing polyacrylamide gel. Incision reactions were performed on circular duplex plasmids containing a single 5′-end-labelled 8-oxoG moiety. Incubation at 30°C was conducted with various combinations of proteins for different periods of time (30–60 min) as indicated. Proteins used in this experiment were hOGG1 (13 nM) and HAP1 (10 nM). The plasmid DNAs were then digested with BamHI and HindIII to release the incised products. The cleavage product of E.coli NTH is shown for comparison. The sequence of the 8-oxoG-containing oligonucleotide used as a primer for in vitro DNA synthesis and of the cleaved products are reported. 3′ddR5P, 2,3-didehydro-2,3-dideoxyribose-5-phosphate.
With the DNA substrate constructed for this assay, elimination by HAP1 of the 3′-terminal sugar phosphate relative to the AP lyase break site can be monitored by disappearance of the radioactivity associated with the cleavage product. When the plasmids were incubated with HAP1 along with hOGG1 (lanes 3 and 4) the lesions were incised to completion (as shown by the disappearance of the 30 nt fragment) and the labelled cleavage products were no longer detectable.
These results show that HAP1 plays an important role in the first steps of 8-oxoG repair by stimulating hOGG1 to cleave all the AP sites by its AP lyase activity, thus allowing complete removal of the radiolabelled phosphates through the 3′-diesterase activity of HAP1.
Efficient repair of 8-oxoG DNA by hOGG1, HAP1, Pol β and DNA ligase I
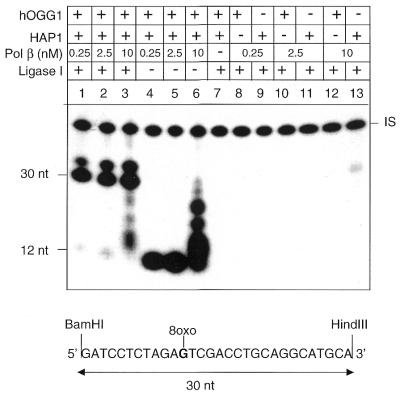
The excision/synthesis steps were reconstituted using human purified proteins and 8-oxoG-containing circular duplex molecules as substrate. The single lesion-containing plasmids were designed to allow identification of the repair intermediates and patches at the lesion site by restriction digestions and/or by using different labelled dNTPs in the repair reaction. The repair reaction in the presence of radiolabelled dGTP followed by BamHI and HindIII restriction of the repaired molecules allows identification of the length of the repair intermediates. One nucleotide replacement reactions are expected to produce 12 nt fragments and long patch BER intermediates will measure >13 nt, whereas complete repair products will co-migrate with the full-length BamHI–HindIII restriction fragment (30 nt). In some experiments the full-length fragment migrated as a double band. This is due to inefficient protein extraction before DNA loading. A 5′-end-labelled 60mer (indicated as internal standard, IS) was added to all reaction tubes in order to correct the repair incorporation values for DNA recovery. As shown in Figure 2 (lanes 1–3), efficient repair was observed in the presence of hOGG1, HAP1, Pol β and DNA ligase I. At the highest Pol β concentration (10 nM) unligated repair intermediates >1 nt are visible on the gel (lane 3). When DNA ligase I was omitted from the repair reaction (lanes 4–6) repair intermediates strictly confined to 1 nt replacement events were observed in the presence of Pol β up to 2.5 nM (lanes 4 and 5). At higher concentrations of Pol β (10 nM) a significant amount of strand displacement DNA synthesis was observed (lane 6). As expected, following hOGG1 cleavage, Pol β could not employ the blocked 3′ termini as a primer (lanes 8, 10 and 12) and in the absence of hOGG1 no repair synthesis was detected (lanes 9, 11 and 13). These data show that repair of 8-oxoG requires hOGG1, HAP1, Pol β and DNA ligase I.
Figure 2.
Efficient repair of 8-oxoG by hOGG1, HAP1, Pol β and DNA ligase I. Autoradiograph of a denaturing polyacrylamide gel. Repair replication was performed for 1 h in the presence of [α-32P]dGTP at 30°C on circular duplex plasmids containing a single 8-oxoG residue. The plasmids were then digested with BamHI and HindIII to release the 30 bp fragment originally containing the lesion. Repair reactions were conducted with various combinations of proteins. Proteins used in these experiments were hOGG1 (13 nM), HAP1 (10 nM), Pol β (0.25–10 nM) and DNA ligase I (50 nM). IS, internal standard.
Fen1, RF-C and PCNA are dispensable for Pol β-dependent repair of 8-oxoG DNA
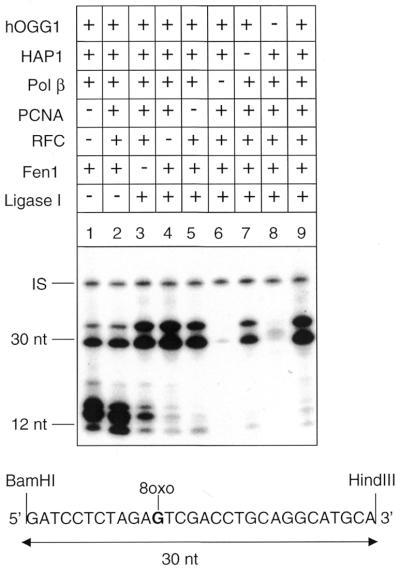
The ability of Pol β to mediate strand displacement at the incised 8-oxoG was further investigated in the presence of long patch BER components like Fen1, PCNA and RF-C. Fen1 has been shown to stimulate Pol β-mediated long patch BER (9), PCNA stimulates excision of the flap structures by Fen1 (24) and RF-C is required for the loading of PCNA onto circular DNA substrates. Figure 3 shows the repair products obtained when dGTP is used as labelled precursor in a 1 h repair reaction. PCNA (2.8 nM) and RF-C (0.1 U) were dispensable for Pol β-mediated repair of 8-oxoG (compare lanes 4 and 5 with lane 9) although in the presence of PCNA the yield of the repaired fragment was slightly increased. The type and amount of repair intermediates detected in the absence of DNA ligase I (mostly 1–5 nt in length) are also unaffected by the presence of PCNA/RF-C in the repair mixture (compare lane 1 with lane 2). The radioactive 30 nt products (lanes 1 and 2) represent elongation by Pol β beyond the HindIII restriction site converted into 30 nt fragments by digestion with BamHI and HindIII. The flap endonuclease Fen1 (10 nm) was also dispensable for efficient repair of the oxidised base (lane 3) although, when missing, some repair intermediates >1 nt were left unligated. As expected, in the absence of Pol β (lane 6) or hOGG1 (lane 8) no repair incorporation was observed. In the absence of HAP1 (lane 7), some repair synthesis was detected, most likely originating from a nick generated by spontaneous hydrolysis of the hOGG1-induced abasic site. The AP site-containing vector is in fact fully converted into nicked forms following 1 h incubation in the repair synthesis buffer (data not shown). These results indicate that PCNA, RF-C and Fen1 are dispensable for repair at concentrations of Pol β (10 nM) that will induce strand displacement at the incised 8-oxoG.
Figure 3.
Fen1, RF-C and PCNA are dispensable for Pol β-dependent repair of 8-oxoG. Autoradiograph of a denaturing polyacrylamide gel. Repair replication was performed for 1 h in the presence of [α-32P]dGTP at 30°C on circular duplex plasmids containing a single 8-oxoG residue. The plasmids were then digested with BamHI and HindIII to release the 30 bp fragment originally containing the lesion. Repair reactions were conducted with various combinations of proteins. Proteins used in these experiments were hOGG1 (13 nM), HAP1 (10 nM), Pol β (10 nM), PCNA (2.8 nM), RF-C (0.10 U), Fen1 (10 nM) and DNA ligase I (50 nM). IS, internal standard.
Fen1 controls strand displacement synthesis by Pol β, which varies depending on the structure of the 5′-terminal moiety
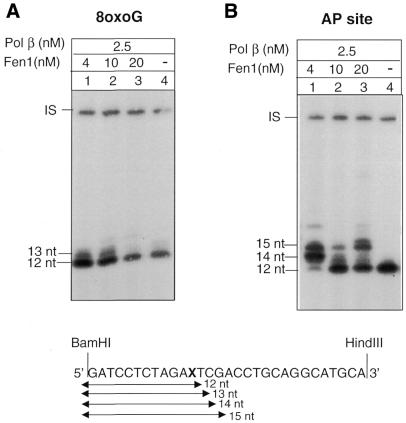
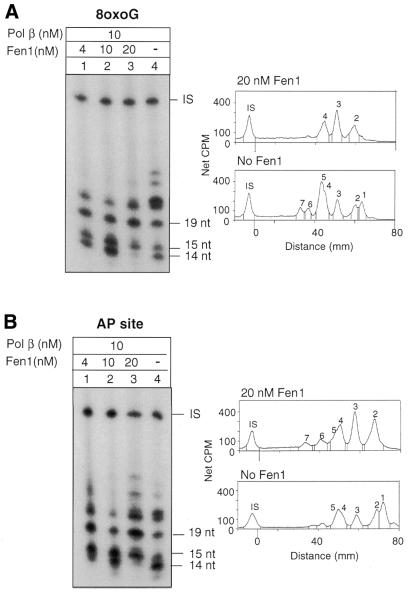
The role of Fen1 in BER of 8-oxoG DNA was further analysed. Since Fen1 is required for Pol β-mediated long patch BER at AP sites the experiments were run in parallel on circular molecules containing either a single 8-oxoG or an abasic site located at the same nucleotide position of the plasmid molecule. DNA ligase I was omitted and dGTP was used as labelled precursor in 1 h repair reactions. 8-oxoG DNA was cleaved with hOGG1/HAP1 and the AP DNA with HAP1, thus generating 5′ ends with an intact nucleotide residue or a terminal dRP residue, respectively. In the absence of Fen1 and with low Pol β concentrations (2.5 nM) repair intermediates almost exclusively confined to 1 nt replacement events were observed with both substrates (Fig. 4A and B, lanes 4). Strand displacement DNA synthesis by Pol β was dependent on the addition of Fen1. In the case of 8-oxoG the strand displacement induced by Fen1 was limited to very short patches (mostly 2 nt) (Fig. 4A, lanes 1–3). In the case of the AP site, Fen1 determined a more robust strand displacement with a pattern of repair intermediates that was dependent on Fen1 concentration. At a low concentration of Fen 1 (4 nM) (Fig. 4B, lane 1) the major intermediates corresponded to 3–4 nt replacement reactions. At higher concentrations of Fen1 (10 and 20 nM) the repair intermediates were almost exclusively 1 and 4 nt in length (Fig. 4B, lanes 2 and 3).
Figure 4.
Effect of Fen1 on strand displacement DNA synthesis by low concentrations of Pol β: dependence on the structure of the 5′-terminal moiety. Autoradiograph of a denaturing polyacrylamide gel. Repair replication was performed for 1 h in the presence of [α-32P]dGTP at 30°C. The plasmids were then digested with BamHI and HindIII to release the 30 bp fragment originally containing the lesion. (A) Repair replication was performed on circular duplex plasmids containing a single 8-oxoG residue with hOGG1 (13 nM), HAP1 (10 nM), Pol β (2.5 nM) and different concentrations of Fen1 (0–20 nM). (B) Repair replication was performed on circular duplex plasmids containing a single AP site with HAP1 (1.25 nM), Pol β (2.5 nM) and different concentrations of Fen1 (0–20 nM). IS, internal standard.
At high concentrations of Pol β (10 nM) Fen1 was dispensable for strand displacement (Fig. 5). DNA synthesis beyond 1 nt replacement was observed either at the 8-oxoG or the AP site in the absence of Fen1 (Fig. 5A and B, lanes 4). In the presence of high concentrations of Fen1 (20 nM) strand displacement was increased, with the disappearance of the 3 nt replacement reactions (Fig. 5A and B, lanes 3). The repair intermediate profile was slightly but consistently different when comparing the two types of gaps (Fig. 5, right).
Figure 5.
Effect of Fen1 on strand displacement DNA synthesis by high concentrations of Pol β: dependence on the structure of the 5′-terminal moiety. Autoradiograph of a denaturing polyacrylamide gel. Repair replication was performed for 1 h in the presence of [α-32P]dGTP at 30°C. The plasmid was then digested with BamHI and HindIII to release the 30 bp fragment originally containing the lesion. (A) Repair replication was performed on circular duplex plasmids containing a single 8-oxoG residue with hOGG1 (13 nM), HAP1 (10 nM), Pol β (10 nM) and different concentrations of Fen1 (0–20 nM). (B) Repair replication was performed on circular duplex plasmids containing a single AP site with HAP1 (1.25 nM), Pol β (10 nM) and different concentrations of Fen1 (0–20 nM). To the right of (A) and (B) are shown the distribution and levels of radioactivity in the repair intermediates measured by electronic autoradiography and corrected for DNA recovery (Instant Imager, Packard). IS, internal standard. To allow a comparison between the two types of gaps the numbers on the peaks identify the same repair intermediate.
These data confirm that a functional interaction between Fen1 and Pol β may control the size of the repair patches (9). Fen1 is able to alter the DNA synthesis activity of Pol β with a mode that is dependent on the structure of the 5′ termini.
DNA ligase I acts as a patch size determinator in Pol β-mediated BER of 8-oxoG
In a previous study, we have shown that DNA ligase I tightly controls the repair patch size when Pol δ/ɛ are involved in the resynthesis step (6). Here, the effect of DNA ligase I was explored in Pol β-mediated repair at 8-oxoG. The fine mapping of the repair patches was performed under high Pol β concentrations that induced significant strand displacement reactions. The repair reactions were performed in the presence of different radiolabelled dNTPs: dGTP to monitor incorporation at the lesion site and dTTP and dCTP to determine whether resynthesis extended beyond the lesion site. As shown in Figure 6, the incorporation of dGMP (lane 1), but not of dTMP (lane 2) and dCMP (lane 3), into the BamHI–HincII restriction fragment clearly showed that, in the presence of DNA ligase I, repair reactions were exclusively confined to 1 nt replacement. This result was also confirmed by analysis of repair incorporation in restriction fragments located 5′ and 3′ to the lesion site (data not shown). Pol β-mediated strand displacement is inhibited in the presence of DNA ligase I. The assemby of all the proteins required to complete 8-oxoG repair drives the repair reaction to the single nucleotide BER mode.
Figure 6.
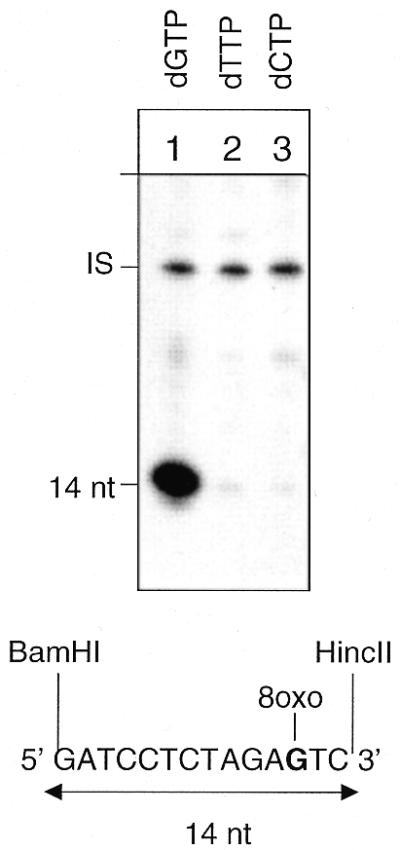
DNA ligase I acts as a patch size determinator in Pol β-mediated BER of 8-oxoG. Autoradiograph of a denaturing polyacrylamide gel. Repair replication was performed for 1 h in the presence of [α-32P]dGTP, [α-32P]dTTP or [α-32P]dCTP at 30°C on circular duplex plasmids containing a single 8-oxoG residue. The plasmids were then digested with BamHI and HincII to release the 14 bp fragment originally containing the lesion. Proteins used in this experiment were hOGG1 (13 nM), HAP1 (10 nM), Pol β (10 nM), PCNA (2.8 nM), RF-C (0.1 U), Fen1 (10 nM) and DNA ligase I (50 nM). IS, internal standard.
DISCUSSION
BER is initiated by the action of a DNA glycosylase that removes the modified base. Two general pathways for BER exist in mammalian cells, short and long patch BER, which involve different sets of proteins. Several studies indicate that mammalian cells repair 8-oxoG predominantly by short patch BER and that DNA Pol β is responsible for the resynthesis step (11,12). Since Pol β has been shown to conduct gap filling in either BER pathway (4,8) we have now reconstituted BER in vitro on 8-oxoG-containing DNA in the presence and absence of the long patch BER proteins Fen1, PCNA and RF-C. Fen1 or high levels of Pol β induced strand displacement DNA synthesis. However, the recruitment of DNA ligase I at the 5′-phosphate terminal moiety, created by the combined activities of hOGG1 and HAP1, abolishes the Pol β/Fen1-induced DNA synthesis and confines the repair patches to 1 nt replacement. In a previous study (6), we reported that DNA ligase I is responsible for limiting the Pol δ/ɛ/PCNA-dependent repair at incised AP sites to 2–4 nt. The mechanism behind these two apparently similar phenomena is likely to be different and might have important implications for the distinction between short and long patch BER. Both the main repair DNA ligases, DNA ligases I and III, interact with Pol β: via XRCC1 in the case of DNA ligase III/XRCC1 heterodimer (4,25) and directly in the case of DNA ligase I (26). These interactions might force the replacement of only 1 nt and will promote short patch BER. Conversely, no direct interaction between Pol δ/ɛ and DNA ligase I has been observed. Long patch BER is initiated by the loading of PCNA at the lesion site through its multiple interactions with Fen1, DNA ligase I and Pol δ/ɛ that leads to the formation of the 5′-flap structures excised by Fen1 (reviewed in 27). Accordingly, the excision step in Pol δ/ɛ-catalysed long patch BER does not require strand displacement DNA synthesis (28).
Additionally, we show the important role played by the nature of the 5′-terminal moiety in processing of the damaged strand by the BER machinery. In the absence of DNA ligase I, Fen1 ‘senses’ the nature of the gap ahead of the polymerase. The stimulation of strand displacement is stronger when the downstream nucleotide is represented by a 5′-dRP as compared with an ordinary 5′-phosphate and the profile of the repair intermediate appears to be gap-specific (Figs 4 and 5). We confirm (9) that the addition of Fen1 to Pol β changes the 1 nt filling mode to synthesis of longer patches and in the case of the incised AP sites to the production of a predominant 4 nt (primer extension) product. Formation of this product requires the 5′-terminal dRP since it is not observed with the 8-oxoG 5′-phosphate termini. As suggested by the crystal structure of Fen1 from Pyrococcus furiosus (29), the plasticity in this enzyme structure would allow Fen1 to slide down the single-stranded DNA leading to formation of a Fen1–DNA complex. The positioning of this complex should be very precise to lead to the excision of a 5′-dRP trinucleotide independently of the sequence context (this study; 9). When Pol β is present in excess it is sufficient per se to induce strand displacement. These results indicate that Pol β concentration and its interaction with Fen1 determine the strand displacement DNA synthesis.
These findings, although interesting from a mechanistic point of view, are likely to have a minor impact, if any, on the repair synthesis mode in cells that contain ample amounts of DNA ligase. This is shown by the strict control of the repair patches at 8-oxoG (this study) and at AP sites (6,7) in in vitro reconstituted repair systems. It has also been suggested from the profile of the repair patches at abasic sites observed by using whole cell extracts from mouse cells (30). We have reported that the repair patches were mainly confined to 1 nt replacement reactions (70%), with 30% long patch BER. A large fraction (40%) of these long patch repair events did not exceed 2 nt, which is the minimum size for release by Fen1 of the 5′-terminal AP site (31). The lack of predominant 4 nt repair patches, the hallmark of Fen1/Pol β processing of AP sites, or of longer repair patches such as those synthesised at AP sites by Pol δ/ɛ (6) in the absence of DNA ligase I, suggests that cells are provided with tight control of the resynthesis step beyond the lesion site. The recruitment of DNA ligase I at the lesion site via its direct interaction with the repair polymerase or with the scaffolding protein PCNA (32) would prevent the introduction of errors caused by the repair polymerases.
The reconstitution in vitro of the first steps of 8-oxoG repair also deserves some comments. We have confirmed the uncoupling between the DNA glycosylase and AP lyase activities of hOGG1 (1,2) and the stimulation by HAP1 of its catalytic activity. Stimulation of the DNA glycosylase activity of hOGG1 by HAP1 has been recently described (33,34). It has been hypothesized that HAP1 exerts its function by preventing hOGG1 reassociation to the AP site. In this model, HAP1 would preclude the AP lyase reaction of hOGG1 (34). The recent finding that the dRP lyase activity of Pol β is required in repair of oxidative base lesions by mammalian cell extracts supports this mechanism (35). We observe that HAP1 can stimulate the AP lyase activity of hOGG1. When hOGG1 is stoichiometrically in excess, as in our experiments, HAP1 might promote a second distinct interaction of hOGG1 with the AP site, thus giving a new chance for AP lyase cleavage of the DNA strand (M.Bjoras, B.Pascucci, E.Dogliotti and E.Seeberg, manuscript in preparation).
In conclusion, we have shown that there are multiple interactions between BER enzymes both at early and late steps during repair. Most importantly, ligase plays a crucial role in determining and minimising the repair patch size. Small repair patch size is important to limit the possibility of the formation of double-strand breaks caused by the repair of closely spaced lesions in opposite strands. Clustering of lesions will frequently occur after cell exposure to ionising radiation (36).
Acknowledgments
ACKNOWLEDGEMENTS
B.P. was partially supported by EU/ISS contract ‘Microsatellite instability and cancer’ and by a FIRC fellowship. The work performed in Hübscher’s laboratory by G.M. and U.H. was supported by the Kanton of Zürich. The work of G.V. was supported by the Association pour le Recherche sur le Cancer.
REFERENCES
- 1.Bjoras M., Luna,L., Johnsen,B., Hoff,E., Haug,T., Rognes,T. and Seeberg,E. (1997) Opposite base-dependent reactions of a human base excision repair enzyme on DNA containing 7,8-dihydro-8-oxoguanine and abasic sites. EMBO J., 16, 6314–6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zharkov D.O., Rosenquist,T.A., Gerchman,S.E. and Grollman,A.P. (2000) Substrate specificity and reaction mechanism of murine 8-oxoguanine-DNA glycosylase. J. Biol. Chem., 275, 28607–28617. [DOI] [PubMed] [Google Scholar]
- 3.Dogliotti E., Fortini,P., Pascucci,B. and Parlanti,E. (2001) Multiple pathways for DNA base excision repair. The mechanism of switching among multiple BER pathways. Prog. Nucleic Acid Res. Mol. Biol., 68, 1–28. [DOI] [PubMed] [Google Scholar]
- 4.Kubota Y., Nash,R., Klungland,A., Schar,P., Barnes,D. and Lindahl,T. (1996) Reconstitution repair of DNA base excision-repair with purified human proteins: interaction between DNA polymerase beta and the XRCC1 protein. EMBO J., 15, 6662–6670. [PMC free article] [PubMed] [Google Scholar]
- 5.Srivastava D.K., Vande,B.J., Prasad,R., Molina,J.T., Beard,W.A., Tomkinson,A.E. and Wilson,S.H. (1998) Mammalian base excision repair. Identification of the reaction sequence and rate-determining steps. J. Biol. Chem., 273, 21203–21209. [DOI] [PubMed] [Google Scholar]
- 6.Pascucci B., Stucki,M., Jonsson,Z.O., Dogliotti,E. and Hubscher,U. (1999) Long patch base excision repair with purified human proteins. J. Biol. Chem., 274, 33696–33702. [DOI] [PubMed] [Google Scholar]
- 7.Matsumoto Y., Kim,K., Hurwitz,J., Gary,R., Levin,D.S., Tomkinson,A.E. and Park,M.S. (1999) Reconstitution of proliferating cell nuclear antigen-dependent repair of apurinic/apyrimidinic sites with purified human proteins. J. Biol. Chem., 274, 33703–33708. [DOI] [PubMed] [Google Scholar]
- 8.Dianov G., Prasad,R., Wilson,S. and Bohr,V. (1999) Role of DNA polymerase β in the excision step of long patch mammalian base excision repair. J. Biol. Chem., 274, 13741–13743. [DOI] [PubMed] [Google Scholar]
- 9.Prasad R., Dianov,G.L., Bohr,V.A. and Wilson,S.H. (2000) FEN1 stimulation of DNA polymerase β mediates an excision step in mammalian long patch base excision repair. J. Biol. Chem., 275, 4460–4466. [DOI] [PubMed] [Google Scholar]
- 10.Cappelli E., Taylor,R., Cevasco,M., Abbondandolo,A., Caldecott,K. and Frosina,G. (1997) Involvement of XRCC1 and DNA ligase III gene products in DNA base excision repair. J. Biol. Chem., 272, 23970–23975. [DOI] [PubMed] [Google Scholar]
- 11.Fortini P., Parlanti,E., Sidorkina,O.M., Laval,J. and Dogliotti,E. (1999) The type of DNA glycosylase determines the base excision repair pathway in mammalian cells. J. Biol. Chem., 274, 15230–15236. [DOI] [PubMed] [Google Scholar]
- 12.Dianov G., Bischoff,C., Piotrowski,J. and Bohr,V.A. (1998) Repair pathways for processing of 8-oxoguanine in DNA by mammalian cell extracts. J. Biol. Chem., 278, 33511–33516. [DOI] [PubMed] [Google Scholar]
- 13.Dianov G.L., Thybo,T., Dianova,I.I., Lipinski,L.J. and Bohr,V.A. (2000) Single nucleotide patch base excision repair is the major pathway for removal of thymine glycol from DNA in human cell extracts. J. Biol. Chem., 275, 11809–11813. [DOI] [PubMed] [Google Scholar]
- 14.Dianov G., Price,A. and Lindahl,T. (1992) Generation of single-nucleotide repair patches following excision of uracil residues from DNA. Mol. Cell. Biol., 12, 1605–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bodepudi V., Shibutani,S. and Johnson,F. (1992) Synthesis of 2′-deoxy-7,8-dihydro-8-oxoguanosine and 2′-deoxy-7,8- dihydro-8-oxoadenosine and their incorporation into oligomeric DNA. Chem. Res. Toxicol., 5, 608–617. [DOI] [PubMed] [Google Scholar]
- 16.Frosina G., Fortini,P., Rossi,O., Carrozzino,F., Raspaglio,G., Cox,L.S., Lane,D.P., Abbondandolo,A. and Dogliotti,E. (1996) Two pathways for base excision repair in mammalian cells. J. Biol. Chem., 271, 9573–9578. [DOI] [PubMed] [Google Scholar]
- 17.Rothwell D.G., Hang,B., Gorman,M.A., Freemont,P.S., Singer,B. and Hickson,I.D. (2000) Substitution of Asp-210 in HAP1 (APE/Ref-1) eliminates endonuclease activity but stabilises substrate binding. Nucleic Acids Res., 28, 2207–2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schurtenberger P., Egelhaaf,S.U., Hindges,R., Maga,G., Jonsson,Z.O., May,R.P., Glatter,O. and Hubscher,U. (1998) The solution structure of functionally active human proliferating cell nuclear antigen determined by small-angle neutron scattering. J. Mol. Biol., 275, 123–132. [DOI] [PubMed] [Google Scholar]
- 19.Hübscher U., Mossi,R., Ferrari,E., Stucki,M. and Jonsson,Z.O. (1999) Functional analysis of DNA replication accessory proteins. In Cotterill,S. (ed.), Eukaryotic DNA Replication: A Practical Approach. Oxford University Press, pp. 119–137.
- 20.Jonsson Z.O., Hindges,R. and Hubscher,U. (1998) Regulation of DNA replication and repair proteins through interaction with the front side of proliferating cell nuclear antigen. EMBO J., 17, 2412–2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Date T., Yamaguchi,M., Hirose,F., Nishimoto,Y., Tanihara,K. and Matsukage,A. (1998) Expression of active rat DNA polymerase β in Escherichia coli. Biochemistry, 27, 2983–2990. [DOI] [PubMed] [Google Scholar]
- 22.Boiteux S. and Radicella,J.P. (2000) The human OGG1 gene: structure, functions and its implication in the process of carcinogenesis. Arch. Biochem. Biophys., 377, 1–8. [DOI] [PubMed] [Google Scholar]
- 23.Asagoshi K., Yamada,T., Terato,H., Ohyama,Y., Monden,Y., Arai,T., Nishimura,S., Aburatani,H., Lindahl,T. and Ide,H. (2000) Distinct repair activities of human 7,8-dihydro-8-oxoguanine DNA glycosylase and formamidopyrimidine DNA glycosylase for formamidopyrimidine and 7,8-dihydro-8-oxoguanine. J. Biol. Chem., 275, 4956–4964. [DOI] [PubMed] [Google Scholar]
- 24.Tom S., Henricksen,L.A. and Bambara,R.A. (2000) Mechanism whereby proliferating cell nuclear antigen stimulates flap endonuclease 1. J. Biol. Chem., 275, 10498–10505. [DOI] [PubMed] [Google Scholar]
- 25.Caldecott K.W., Aoufouchi,S., Johnson,P. and Shall,S. (1996) XRCC1 polypeptide interacts with DNA polymerase β and possibly poly(ADP-ribose) polymerase and DNA ligase III is a novel molecular ‘nick-sensor’ in vitro. Nucleic Acids Res., 24, 4387–4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prasad R., Singhal,R.K., Srivastava,D.K., Molina,J.T., Tomkinson,A.E. and Wilson,S.H. (1996) Specific interaction of DNA polymerase β and DNA ligase I in a multiprotein base excision repair complex from bovine testis. J. Biol. Chem., 271, 16000–16007. [DOI] [PubMed] [Google Scholar]
- 27.Caldecott K.W. (2001) Mammalian DNA single-strand break repair: an X-ra(y)ted affair. Bioessays, 23, 447–455. [DOI] [PubMed] [Google Scholar]
- 28.Gary R., Kim,K., Cornelius,H.L., Park,M.S. and Matsumoto,Y. (1999) Proliferating cell nuclear antigen facilitates excision in long-patch base excision repair. J. Biol. Chem., 274, 4354–4363. [DOI] [PubMed] [Google Scholar]
- 29.Hosfield D.J., Mol,C.D., Shen,B. and Tainer,J.A. (1998) Structure of the DNA repair and replication endonuclease and exonuclease FEN-1: coupling DNA and PCNA binding to FEN-1 activity. Cell, 95, 135–146. [DOI] [PubMed] [Google Scholar]
- 30.Fortini P., Pascucci,B., Belisario,F. and Dogliotti,E. (2000) DNA polymerase β is required for efficient DNA strand break repair induced by methyl methanesulfonate but not by hydrogen peroxide. Nucleic Acids Res., 28, 3040–3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeMott M.S., Shen,B., Park,M.S., Bambara,R.A. and Zigman,S. (1996) Human RAD2 homolog 1 5′- to 3′-exo/endonuclease can efficiently excise a displaced DNA fragment containing a 5′ terminal abasic lesion by endonuclease activity. J. Biol. Chem., 271, 30068–30076. [DOI] [PubMed] [Google Scholar]
- 32.Tom S., Henricksen,L.A., Park,M.S. and Bambara,R.A. (2001) DNA ligase I and proliferating cell nuclear antigen form a functional complex. J. Biol. Chem., 276, 24817–24825. [DOI] [PubMed] [Google Scholar]
- 33.Hill J.W., Hazra,T.K., Izumi,T. and Mitra,S. (2001) Stimulation of human 8-oxoguanine-DNA glycosylase by AP-endonuclease: potential coordination of the initial steps in base excision repair. Nucleic Acids Res., 29, 430–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vidal A.E., Hickson,I.D., Boiteux,S. and Radicella,J.P. (2001) Mechanism of stimulation of the DNA glycosylase activity of hOGG1 by the major human AP endonuclease: bypass of the AP lyase activity step. Nucleic Acids Res., 29, 1285–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Allinson S.L., Dianova,I.I. and Dianov,G.L. (2001) DNA polymerase β is the major dRP lyase involved in repair of oxidative base lesions in DNA by mammalian cell extracts. EMBO J., 20, 6919–6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sutherland B.M., Bennett,P.V., Sidorkina,O. and Laval,J. (2000) Clustered damages and total lesions induced in DNA by ionizing radiation: oxidized bases and strand breaks. Biochemistry, 39, 8026–8031. [DOI] [PubMed] [Google Scholar]