Abstract
Plasmodium falciparum intraerythrocytic development is a complex process. Development proceeds rapidly from the trophozoite phase of nutrient acquisition and growth through to the synthetic and reproductive schizont phase, which ends with production of new invasive merozoites. During this process, the malaria parasite must express a series of different gene products, depending on its metabolic and synthetic needs. We are particularly interested in the development of the merozoite’s organelles in the apical complex, which form during the later schizont stages. We have used quantitative real-time RT–PCR fluorogenic 5′ nuclease assays (TaqMan®) for the first time on malaria parasites for analysis of erythrocytic stage-specific gene expression. We analyzed transcripts of the P.falciparum eba-175 and other erythrocyte binding-like (eb l) family genes in temperature-synchronized parasites and found ebl genes have tightly controlled, stage-specific transcription. As expected, eba-175 transcripts were abundant only at the end of schizont development in a pattern most common among ebl, including baebl, pebl and jesebl. The maebl transcript pattern was unique, peaking at mid-late trophozoite stage, but absent in late-stage schizonts. ebl-1 demonstrated another pattern of expression, which peaked during mid-schizont stage and then significantly diminished in late-stage schizonts. Our analysis demonstrates that using real-time RT–PCR fluorogenic 5′ nuclease assays is a sensitive, quantitative method for analysis of Plasmodium transcripts.
INTRODUCTION
Malaria causes 300–500 million cases of clinical disease and 2–3 million deaths every year. Unfortunately, relatively little is known about the basic biology of this pathogenic protozoan. Recently, much new information and significant insights have been achieved by sequencing the Plasmodium falciparum genome. This initial phase of genomic discovery has led to identification of previously unknown gene families and novel metabolic pathways. New technologies need to be applied in malaria research to take full advantage of this wealth of genetic information and its relevance to the parasite’s unique biology. Many of the traditional analytical methods for gene activity lack sensitivity (e.g. northern blot hybridization) and are not adaptable to high-throughput analysis requiring large-scale preparations of parasite materials. Other more sensitive methods that rely on PCR amplification of genes or their transcripts are only semi-quantitative and results often vary among laboratories.
Real-time quantitative RT–PCR transcript analysis, utilizing fluorogenic 5′ nuclease assays, or TaqMan® chemistry, has the potential to overcome many of the limitations of currently used methods. The real-time RT–PCR method allows less inherent potential variation as it is standardized to stringent instrumental specifications and has rigorous internal standardization for each set of reactions. The TaqMan® chemistry enhances the specificity and the sensitivity of detection of the real-time PCR products, since the TaqMan® probe only reacts with a specific nucleotide target sequence. In other eukaryote systems TaqMan® real-time PCR assays are extremely robust, as they can accurately quantify nucleotide abundance over a wide range of concentrations. Previous studies have been successful in using TaqMan® real-time PCR on P.falciparum to determine total parasite numbers in extra-erythrocytic development using the abundant 18S rRNA as a target molecule (1,2). We were curious in testing whether this method can quantify P.falciparum expressed gene transcripts from the erythrocytic stages.
A major interest of our research group is the study of molecules important for P.falciparum erythrocytic growth and development. The ability of malaria parasites to cause disease is dependent on erythrocytic growth so this stage includes important vaccine targets. Merozoites utilize proteins sequestered in organelles of the apical complex to mediate invasion into host erythrocytes via specific receptor/ligand interactions (3). Host cell invasion by merozoites is a multi-step process thought to require numerous interactions between these apical complex proteins and erythrocyte receptors. Erythrocyte binding antigen-175 (EBA-175) is a P.falciparum ligand that binds to sialic acid dependent determinants on glycophorin A of host erythrocytes (4). The P.falciparum EBA-175 is a member of the Duffy binding-like erythrocyte-binding protein (DBL-EBP) family homologous to the Plasmodium knowlesi/Plasmodium vivax Duffy binding proteins (DBPs). The DBL-EBPs represent the functionally important products of the erythrocyte binding-like (ebl) gene family, which now in P.falciparum contains six members: baebl, eba-175, ebl-1, jesebl, maebl and pebl (5). The ebl family represents one of the best-characterized molecular families of malaria parasites. Common features of these ebls include a multi-exon gene structure, conserved exon/intron splicing boundaries, and cysteine-rich (cys-rich) domains (5,6). Expression of EBA-175, BAEBL and DBP is stage specific, occurring only at the end of erythrocytic-stage development, and the proteins are targeted to the microneme apical organelle (7–9). We developed a method for real-time quantitative RT–PCR transcript analysis, utilizing fluorogenic 5′ nuclease assays, or TaqMan® chemistry, on the P.falciparum ebl members to more precisely determine the time of asexual stage-specific expression. To facilitate our analysis we utilized temperature-synchronized parasite cultures. The data presented validates real-time RT–PCR for accurate quantitative analysis of stage-specific gene activity in malaria parasites.
MATERIALS AND METHODS
Parasites, cell culture and nucleic acid extraction
Clones of P.falciparum 3D7 were used for all studies unless otherwise stated. Cultures were initiated from cryopreserved stocks at the Naval Medical Research Center or were obtained from Dr Kim Williamson, Loyola University, Chicago. Additional clones of Dd2, NM2, HB3 and FVO were obtained from frozen stocks maintained at the National Institutes of Health, (Bethesda, MD). Routine maintenance of parasite cultures was done using standard methods at 37°C and gassing (5% O2, 5% CO2, nitrogen balanced) in standard RPMI media supplemented with Albumax I (Gibco BRL) or human AB sera. In addition, for the time course analysis, continuous synchronization of parasite cultures was achieved using a custom temperature-cycling incubator as described and presented later (10). After temperature synchronization, parasites were returned to a 37°C incubator for periodic time point collection. Parasites were lysed in 0.15% saponin in TSE (50 mM Tris, pH 8.0, 50 mM EDTA and 100 mM NaCl) and genomic DNA was extracted using 0.3 M sodium acetate and ethanol precipitation followed by standard chloroform/phenol methods. Total RNA was isolated using TriReagent (Molecular Research Center, Inc.) RNA isolation protocol. NM2 DNA was provided by Dr Stephen Dolan.
ebl gene identification
The six genes of the P.falciparum ebl family were all identified within the preliminary contigs of the P.falciparum malaria genome project. Four of the genes, baebl, eba-175, pebl and ebl-1, are at least partially characterized, while all six have been placed in the ebl family (4,5,7,11–14). Using a PCR-based approach our laboratory has independently sequenced P.falciparum 3D7 baebl and maebl in their entirety. Additionally, the gene-specific TaqMan® primer pair (see below) amplified PCR products were also cloned and sequenced to verify each gene and assure specificity (GenBank accession nos AF461093–AF461098 and AY090552). PCR conditions were identical to those used for the TaqMan® assays below and PCRs were performed in a Perkin Elmer 9600 Thermocycler.
Southern blot analysis
Plasmodium falciparum 3D7, Dd2 and HB3 genomic DNA (2 µg) was digested with restriction enzymes EcoRI or NsiI, separated by agarose electrophoresis, fragmented in 0.25 M HCl, denatured in 0.5 M NaOH/1.5 M NaCl, and blotted as previously described (8,15). The blots were probed with gene-specific fragments amplified from 3D7 gDNA for each of the P.falciparum ebl genes. The probes were created from cloned (pCR 2.1) products of the following primer pair sets (F, R): baebl, JA 419–479 (5′-TAAGGGAGGAAATAATTAAATTATCGAAGCA-3′, 5′-GTACTCGGAATAATCTGAAATGTTTGTCTC-3′); eba-175, JA 349–350 (5′-ataggatccTTTAATAATATTCCAAGTAG-3′, 5′-atagaattcTTGAAAAAGCCTCCTTTCTGAAAC-3′); jesebl, JA 421–480 (5′-ATAGAGAAGAAATATTGAATAGGTCAAACAC-3′, 5′-CCAATCAAACATCCAGAAAATTAAAAATG-3′); maebl, JA 302–310 (5′-AATCCTCAAGCCGAATATATGGATAGGTTTGATAT-3′, 5′-CTTTTTATTTATAAGACTTTTGCATTTTCC); pebl, JA 420–481 (5′-TAGCTGATGATATCATGAGATCTTATAAAAG-3′, 5′-CCAGCAAAGAAAATAAAAACAATACAGAG-3′); and ebl-1, JA 571–572 (5′-ataggatccATACCAGAACATCATAATAAGAGT-3′, 5′-atagaattcCTAACTTGTCATATCTGTTTGAAACAT-3′). The cloned fragments were each confirmed by sequencing. The blots were washed in 2× SSC/0.5% SDS (1× SSC: 0.15 M sodium chloride/0.015 M sodium citrate, pH 7.0) for 30 min before exposure to DuPont NEF-496 film for 1 day at –80°C. All probes were radiolabeled using the Klenow fragment of DNA polymerase (Random Primer Labeling Kit, Gibco-BRL).
Real-time quantitative transcript analysis
Synchronized P.falciparum 3D7 parasite cultures were maintained in a temperature-cycling incubator (Forma) with a periodicity of 48 h (10). Before transcript analysis, cultures were moved to a constant 37°C incubator and harvested at seven time points spanning most of the estimated 40-h cycle at 37°C. Total RNA (665 ng for a final concentration of 50 ng/reaction) was isolated as previously described and treated with DNase (Gibco BRL catalog no. 18068-015). The RNA for each time point was converted to cDNA using the RNA PCR Core kit (Perkin Elmer catalog no. N808-0143) as described by the manufacturer. For real-time transcript quantification, the cDNA was used in a fluorogenic 5′ nuclease assay utilizing the chemistry of the TaqMan® system on the ABI Prism 7700 Sequence Detector (Applied Biosystems). Gene-specific TaqMan® primer (F, R) and probe (P) sets were created using the Primer Express Software v.1.5 (Applied Biosystems) for the six known ebls: baebl (F, 5′-GCAAAATAAATGCAACAATGAATA-3′; R, 5′-AACAAGGACCCGGTGAACTA-3′; P, 5′-CCATGGAATATTGTACCTATTCTGACGAAAGG-3′), eba-175 (F, 5′-AATTTCTGTAAAATATTGTGACCATATG-3′; R, 5′-GATACTGCACAACACAGATTTCTTG-3′; P, 5′-ATGAAGAAATCCCATTAAAAACATGCACTAAAGA-3′), ebl-1 (F, 5′-CAGAAGTAGACGATGGAGTGGA; R, 5′-TCATGTTTCACATCTTTCTCTTCTT-3′; P, 5′-AAAAGAAGAATGACCCAAAACCATCCAGG-3′), jesebl (F, 5′-GCGGGTAGTACAATATTAGATGATTC-3′; R, 5′-TGTTGTGTGCTAAAA- TTATGTTCTTG-3′; P, 5′-AAATGACAGAAGGTAGCGAAAGTGATGTTGGAG-3′), maebl (F, 5′-TCAAAATATTGTGATTATATGAAGGATAA-3′; R, 5′-TAGACAAAAATCAGAAATGGAACAAC-3′; P, 5′-TTCATCAGGTACATGTTCTAATGAAGAAAGAAAAAGT-3′) and pebl (F, 5′-ATTAAATCGTACATCACATACGCA-3′; R, 5′-ACGCCCATCATGCACATT-3′; P, 5′-ATAGAAACAACAGCTGAGAACAATATAGGTGGTTTAAGT-3′); for one P.falciparum gene expressed in liver stages: liver stage antigen-1 (lsa-1) (F, CATGGAGATGTATTAGCAGAGGATTTAT; R, CCCTCTGTTGTCCTGAGGTAAAGA; P, TGGTCGTTTAGAAATACCAGCTATAGAACTTCCATCA) and for an 18S rRNA control (F, 5′-GCTGACTACGTCCCTGCCC-3′; R, 5′-ACAATTCATCATATCTTTCAATCGGTA-3′; P, 5′-TTGTACACACCGCCCGTCGCTC-3′). All ebl probes and the lsa-1 probe were labeled with 6-carboxy-fluorescein (FAM), and the 18S rRNA probe labeled with VIC and used in single reporter assays. TAMRA was used as the 3′ quencher in all cases. Experimental PCRs included 300 nM of each primer, 100 nM probe, 1× TaqMan® buffer A (10×: 500 mM KCl, 100 mM Tris–HCl, 0.1 mM EDTA), 5 mM MgCl2, dATP (200 µM), dCTP (200 µM), dGTP (200 µM), dUTP (400 µM), 0.5 U AmpErase UNG and 2.5 U AmpliTaq Gold DNA Polymerase. The real-time PCR consisted of one cycle of each 50°C for 2 min and 95°C for 10 min, followed by 50 cycles of 95°C for 15 s and 60°C for 1 min. Two ebl genes analyzing all seven time points in triplicate could be analyzed on a single 96-well plate including all necessary controls (18S rRNA) and gDNA standards (10-fold dilutions from 6 µg/reaction to 600 pg/reaction). A P.falciparum gene not normally expressed in the erythrocytic cycle, lsa-1, was examined to account for evidence of relaxed transcriptional control. Serial dilutions of gDNA were used as the standard reference control after determining each ebl is present as a single-copy gene. Experiments on independent 96-well plates were duplicated matching a different ebl pair from the initial run. Data analysis utilized the Sequence Detector software (version 1.6.3 and 1.7) to determine threshold cycle, Ct, and reduced normalized fluorescence values, ΔRn, for each amplified product for each time point.
RESULTS
ebl gene identification
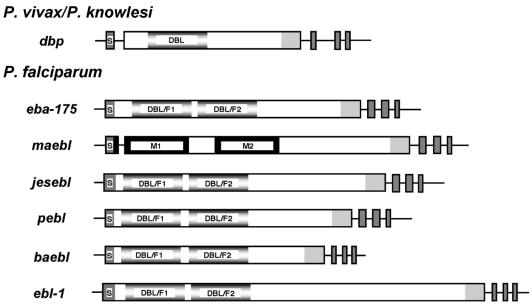
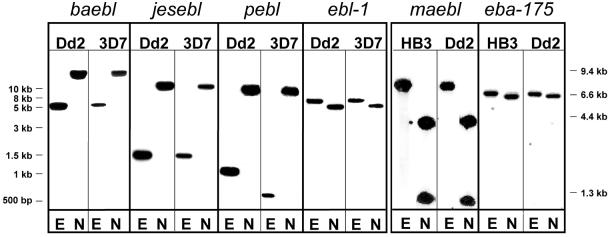
Sequence data generated by the P.falciparum genome project and in our laboratory, together with previous reports, verified the presence of six ebl family members in the P.falciparum genome (4,5,7,11–13). Each ebl was directly identified by sequencing RT–PCR products which spanned the final three exons and elucidated intron/exon splicing boundaries (data not shown). Each gene had the ebl consensus multi-exon gene structure encoding type I transmembrane proteins with the conserved cys-rich domains, except maebl which had a unique amino cys-rich domain as described for the MAEBL from Plasmodium yoelii (Fig. 1) (16). Southern blot hybridizations produced unique banding patterns and determined that the ebl genes are single-copy genes (Fig. 2). The significance of the RFLP of pebl is not yet known.
Figure 1.
Schematic representations defining the multi-exon gene structure for P.falciparum ebl. Boxed regions denote exons and horizontal lines signify flanking untranslated regions and introns. Regions of similarity are designated and/or shaded similarly: putative signal sequence (S), dbp and eba-175 DBL ligand domains (fading gray), maebl M1/M2 domains and ama-1 subdomains 1 and 2 (black), c-cys domain (light gray), 3′ exons encoding putative transmembrane domain and two cytoplasmic tail domains, respectively (dark gray).
Figure 2.
Southern blot hybridization confirms the ebl are single-copy genes. Plasmodium falciparum gDNA (2 µg) was digested with either EcoRI (E) or NsiI (N) restriction enzymes, separated by standard electrophoresis and blotted. Each blot was probed with an ebl gene-specific probe (see Materials and Methods). Left, P.falciparum gDNA derived from clones Dd2 and 3D7 on independent blots probed for baebl, jesebl, pebl and ebl-1. Right, P.falciparum HB3 and Dd2 gDNA on a single blot probed first for maebl, stripped, and probed for eba-175. Similar hybridization results were found for maebl and eba-175 using 3D7 gDNA. DNA standard markers are listed in the margins adjacent to each panel.
ebl PCR amplification
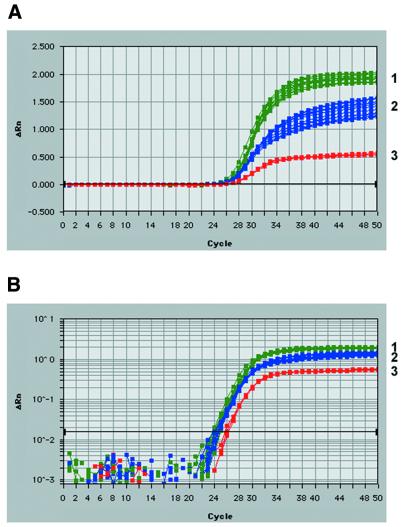
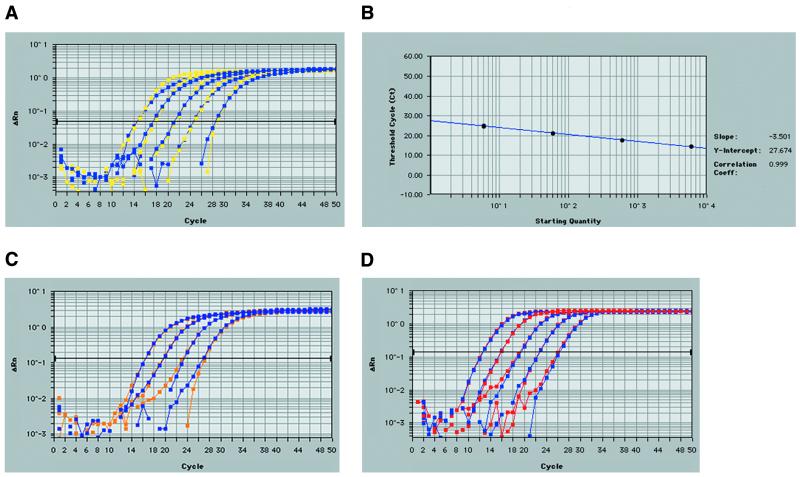
Real-time quantitative RT–PCR analysis of ebl transcripts, using fluorogenic 5′ nuclease assays or TaqMan® chemistry, was developed as an exceptionally sensitive and reproducible means to determine transcript profiles of each P.falciparum ebl gene. The Primer Express Software provided optimal TaqMan® primer and probe positions. The criteria for gene-specific primer/probe design included the production of short amplicons (50–150 bp) without probe overlap, a primer Tm of 58–60°C, a probe Tm of 68–70°C, and limited GC content. The positions of each of the ebl primer pairs were targeted toward the carboxyl cys-rich region 3′ end of each gene for efficient cDNA synthesis by oligo-dT priming. PCR amplification of the six specific P.falciparum ebl genes using TaqMan® primer sets and thermocycler conditions consistent with the TaqMan® 5′ nuclease assay yielded unique bands of the predicted size. Sequencing of these cloned products verified gene specificity. Experimental primer and probe concentrations were optimized using manufacturer’s recommendations. Briefly, for primer sets, an optimal PCR was achieved by choosing concentrations producing the lowest threshold cycle (Ct) and highest reduced normalized fluorescence values (ΔRn). Therefore, varying concentrations (50, 300 and 900 nM) of each primer (F, R) were used in all combinations in a real-time PCR on a set template of P.falciparum 3D7 genomic DNA (6 ng) (Fig. 3). Primer concentrations were optimized for each of the six ebl genes and controls and found to be 300 nM in all cases. Primer sets which involved the 50 nM concentrations always resulted in the highest Ct and lowest ΔRn, while the 300 nM often was equivalent (in Ct and ΔRn) with the seemingly excess 900 nM concentration. Likewise, real-time PCR probe optimization was achieved using various probe concentrations (50, 100, 150, 200 and 250 nM) on a fixed DNA (6 ng) and primer concentration (300 nM). For each ebl gene, only the 50 nM concentration gave lower Ct values (∼0.5 cycle) compared with the higher concentrations tested. Since gene expression quantification directly depends on Ct (and not ΔRn), the 50 nM probe concentration could not be used. Therefore, based on cost effectiveness, the 100 nM probe concentration was utilized for each TaqMan® reaction. Real-time PCR amplification of 3D7 gDNA in 10-fold dilutions (6 µg to 600 pg) resulted in equivalent profiles for each of the single-copy ebl genes, proving that each ebl-specific primer/probe set could accurately measure each ebl over their potential range of its transcript concentrations (Fig. 4A). Real-time amplifications of multiple gDNA dilutions were used to construct standard curves for setting quantitative expression levels in each assay (Fig. 4B). Correlation coefficients for the resulting standard curves never varied <0.995, and often were 1.000. Therefore, each TaqMan® primer/probe set using the FAM reporter dye provided optimal, equivalent amplification of each ebl (baebl, eba-175, ebl-1, jesebl, maebl and pebl) could be used in subsequent analysis of relative transcript abundance. Sensitivity was tested successfully to template levels to 6 pg of gDNA without attempting lower concentrations. Similarly, the P.falciparum lsa-1 gene was successfully amplified from gDNA dilutions (Fig. 4C). TaqMan® assays on the 18S rRNA control primer/probe set using the VIC reporter dye had similar and highly reproducible results (Fig. 4D).
Figure 3.
Preliminary TaqMan® 5′ nuclease assays determine optimal ebl gene-specific primer concentrations. ebl forward and reverse TaqMan® primers were tested in real-time 5′ exonuclease reactions at three concentrations (50, 300 and 900 nM) in all combinations with a constant probe concentration (100 nM). The plots relate PCR cycle number to change of detected fluorescence with background removed (ΔRn) on a linear (A) and logarithmic scale (B). Three isolated groups based on total ΔRn resulted: (1) reactions containing 300 or 900 nM of each primer (green), (2) reactions containing 300 or 900 nM of one primer and 50 nM of the other (blue), and (3) reactions containing 50 nM of each primer (red). Therefore, 300 nM of each primer was the optimal concentration achieving highest fluroscence (ΔRn) and lowest threshold cycle (Ct) while being the lowest concentration in group 1. Amplification plots show the results for varying eba-175 primer set concentrations only. The remaining ebl genes demonstrated similar results. Images were taken from data analysis provided by the Sequence Detection Software (version 1.6.3 or 1.7) for the ABI Prism 7700. The optimization strategy was followed from manufacturer’s recommendations (PE Biosystems).
Figure 4.
Validation of TaqMan® real-time RT–PCR for quantitative detection of P.falciparum ebl. (A) Amplification plot of P.falciparum 3D7 gDNA with eba-175 and maebl TaqMan® primer/probe sets. maebl (yellow) and eba-175 (blue)-specific primer probes sets were used in the 5′ nuclease TaqMan® assay with varying gDNA concentrations (10-fold dilutions from 6 µg to 600 pg/reaction). The darkened horizontal bar represents the chosen threshold cycle (Ct) during the linear phase of the PCR. PCRs with higher initial gDNA concentrations cross the Ct at an earlier cycle number with subsequent dilutions crossing the Ct in later cycles. Due to the stringent and similar design for TaqMan® primer and probe sets, maebl and eba-175-specific products produced near exact amplification plots. These results verified an optimized PCR, achieving the theoretical gap of ~3.33 cycles per 10-fold dilution of gDNA. The primer/probe sets for baebl, ebl-1, jesebl and pebl showed similar and reproducible results. These and similar plots validated the use of each ebl primer and probe set as well as the use of gDNA to construct a standard curve. (B) A representative standard curve constructed from the eba-175 profile given in (A) with its replicate. The logarithmic plot relates starting DNA concentration (ng) versus Ct for the gDNA dilutions. Replicate values almost perfectly overlay each other (circles). A similar curve was created for each of the ebls for every TaqMan® experiment. (C) The lsa-1 (violet) and replicate (orange) show similar amplification profiles using four dilutions of gDNA. (D) As in (A), five dilutions (10-fold dilutions from 6 µg to 600 pg/reaction) of genomic DNA validate real-time quantitative RT–PCR using TaqMan® chemistry for the 18S rRNA control primer and probe set using the VIC reporter dye. The replicates shown (red and blue) almost perfectly overlay with one another indicating reproducibility. These images were taken from data analysis provided by the Sequence Detection Software (version 1.6.3 or 1.7) for the ABI Prism 7700.
Temperature-synchronized parasite cultures
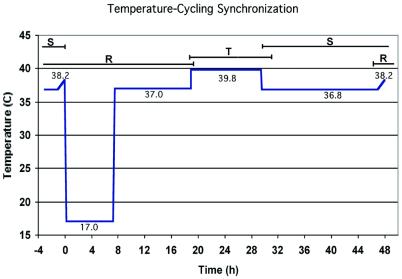
For accurate measurements of P.falciparum transcript levels spanning erythrocytic development, it was essential to use highly synchronized parasite cultures. We utilized a custom-designed temperature cycling incubator (Forma) in which P.falciparum 3D7 parasites were synchronized through five temperature changes during a programmed 48-h cycle (10) (Fig. 5). Briefly, just after merozoite invasion, the temperature was lowered to 17°C (for 7 h 15 min, including ramping) postponing the maturation of early ring stages to more mature forms and likely destroying residual schizonts. Upon raising the temperature to 37°C (11 h 40 min) parasites resumed typical ring-form development. An increase in temperature to 39.8°C (10 h 35 min) followed, which allowed the development of mature trophozoites but inhibited schizont development. This mimicked P.falciparum cyclic fevers thought to influence parasite synchronization in vivo (17,18). Lowering the temperature to 36.8°C (17 h 30 min) allowed for completion of schizont development including ultimate rupture and merozoite release. A final continuous ramp (1 h) to 38.2°C may coax lagging parasites to invade and concluded the adjusted 48-h cycle. Figure 5 illustrates the scheme of the temperature cycling incubator.
Figure 5.
The timings for temperature-cycling synchronization of clone 3D7 P.falciparum malaria parasites. The approximate times are indicated for ring-form parasites (R), trophozoites (T) and schizonts (S). Invasion occurs during the overlap between S and R. During the time at 17°C, the ring form parasites have minimal development (they are in suspended animation), and during the time at 39.8°C, the trophozoites are prevented from maturing into schizonts. Before sampling for transcription analysis, the cultures were moved to a constant temperature 37°C incubator for uninterrupted development, during which the total cycle time was ∼40 h. Sampling at seven time points at 6 h intervals covered development from early rings to late schizonts at 37°C. Synchronization of parasites was ∼90% pure as determined from examining Giemsa-stained blood smears. A detailed description of the cycling conditions is given in the Materials and Methods and Haynes and Moch (10).
ebl stage-specific transcript profiles
Real-time quantitative RT–PCR was successfully performed on 3D7 total RNA extracted from periodic time points (6 h) of temperature-synchronized P.falciparum blood-stage parasites. Standard deviation of the Cts for each ebl on a particular plate never reached >0.40. The data were exported into Excel (Microsoft) and a standard curve was generated using known gDNA concentrations and resulting Cts. Standard curves were determined for each TaqMan® plate and ebls were only compared with the standard curve associated with their own primer/probe sets. Correlation coefficients for the standard curves were always >0.995. Using the Ct for each ebl sample a genome equivalent for each sample and time point was determined by interpolating from the standard curve using the Excel GROWTH algorithm. The ebl genome equivalents were then normalized by dividing by the 18S rRNA genome equivalents. This adjustment controlled for efficiency in the cDNA synthesis across experimental time point samples and acted as a loading control by dividing through with expression values from a constitutively expressed gene. This normalized genome equivalent was then transformed by a factor of 10 000. Stage-specific patterns of ebl expression were maintained for each TaqMan® plate.
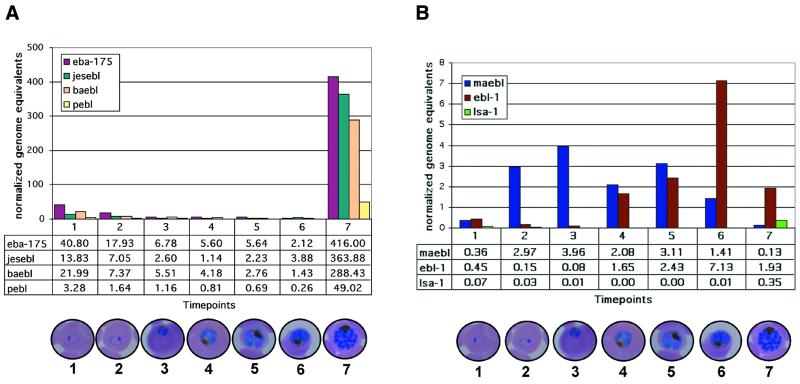
ebl transcripts were detected in a tightly-controlled stage-specific manner (Fig. 6), revealing three distinct patterns of their transcription during development of the blood-stage P.falciparum. eba-175, baebl, jesebl and pebl had peak transcript abundance in the final time point of late segmenting schizonts (time point 7 in Fig. 6A) just prior to merozoite release. eba-175, baebl and jesebl displayed similar high levels (each >250 normalized genome equivalents) at this time point. Although pebl was significantly lower relative to these other late-stage ebl, at only 49 normalized genome equivalents, this was still significantly higher than its preceding time points. Transcript levels for these late-stage ebl genes were significantly lower in all other time points. Transcripts detected in the early stages for these four late-stage ebls were attributed to contaminating late-stage parasites used to initiate the cultures or those that were asynchronous or lagging in their development. In comparison, ebl-1 transcript abundance plateaued at time point 6, which is considered the mid-schizont stage of the temperature-synchronized growth cycle. maebl had an extended duration compared with the other ebls. Its transcripts peaked early at the third time point, considered from mid to late trophozoites, with a broad range extending to the fifth time point (early schizonts), but maebl transcripts were always completely absent from the late-stage schizonts (Fig. 6B). Transcript abundance given in normalized genome equivalents were consistently lower for maebl and ebl-1 relative to the other ebls, but well above lsa-1 and within the sensitivity of the TaqMan® chemistry and fluorescence detection of the ABI Prism 7700.
Figure 6.
Real-time quantitative RT–PCR using TaqMan® chemistry identified ebl stage-specific transcripts from synchronized blood-stage P.falciparum. Periodic (∼6 h) time points taken from synchronized 3D7 cultures spanning the erythrocytic cycle are represented on the abscissa with merozoite invasion occurring just before the first time point. Ordinate values of the transcript levels are given in normalized genome equivalents (see Materials and Methods). Actual normalized genome equivalent values are given in the data table beneath each graph. Standard deviation of the Cts for each ebl on a particular plate never reached >0.40. (A) eba-175, jesebl, baebl and pebl are expressed at high levels in late-stage schizonts (time point 7). (B) maebl transcript levels peaked during the late trophozoites (time point 3) and ebl-1 were most abundant in mid-stage schizonts (time point 6). The abundance of lsa-1 transcripts was negligible and indicates the level of background detection. Digital images taken from Giemsa-stained thin blood smears show a representative parasite stage for each time point.
DISCUSSION
We found transcription of the P.falciparum ebl genes was highly regulated during blood-stage development. This study is the first use of quantitative real-time RT–PCR for the analysis of P.falciparum stage-specific gene transcription during erythrocytic development. The method proved to be a very successful way to compare the relative temporal dynamics of transcriptional regulation for members of the ebl gene family. Quantitative real-time RT–PCR will greatly improve the speed and accuracy with which transcription of individual genes can be characterized.
The observed transcript profiles of eba-175 and maebl match the expected pattern based on their protein expression. The temporal expression pattern for P.falciparum MAEBL contrasts markedly with that characterized for BAEBL and EBA-175, which were expressed later in the merozoites of segmenting schizonts precisely when we detected their peak transcription (7,19,20). maebl transcripts appeared earlier in the erythrocytic cycle and were absent when peak transcription occurred for all of the other ebls (Fig. 6). Detection of maebl transcripts, which peaked during mid to late trophozoites, is consistent with the expression pattern observed previously for the P.yoelii YM MAEBL (21). In YM blood-stage parasites, a burst of MAEBL expression was concentrated at the onset of schizogony, during an estimated 30 min time period, in which the full-length protein could be detected in the endoplasmic reticulum concurrent with the reorganization of this and the other compartments of the secretory pathway.
We presume that temporal control of gene expression is related to specific apical organelle development and targeting of proteins to these different subcellular compartments. A previous study, using transgene expression from an episomal element, demonstrated that the erythrocytic, stage-specific time of Plasmodium gene expression determined correct subcellular targeting (22). Apical membrane antigen-1 (AMA-1), another protein product thought to be involved in malaria merozoite invasion of erythrocytes, normally localizes to the rhoptries in only late-stage schizonts (23,24). Heterologous transgene expression of P.falciparum AMA-1 (PfAMA-1) in Plasmodium berghei, when under the control of a dhfr-ts promoter expressed throughout the asexual cycle as well as in gametes (22). Expression at times other than the late schizont led to abnormal localizations in the parasites. Transgene expression under the control of the pbama-1 promoter achieved characteristic and genuine targeting of PfAMA-1 to the rhoptries. These results suggest a promoter-driven mechanism for stage-specific expression for malaria parasites and therefore might provide insight into the means of regulated transcription described for the ebl genes.
Transcript analysis in malaria parasites may be complicated by changes in mRNA stability as parasites proceed through development and by transcriptional activity that does not translate into protein expression. The relatively low levels of maebl and ebl-1 transcripts in trophozoites and early schizonts may be explained by a rapid RNA turnover rate associated with a brief intense period of translation, as these developing stages undergo an extensive succession of transcriptional activity and processing. During the final stages of intraerythrocytic development, as the abundant synthesis of the major merozoite proteins is completed, there is a general termination of gene expression; therefore, it is likely that the rapid degradation of the late-stage transcripts is not as necessary as the parasite’s priority turns to preparing for merozoite release and the ensuing invasion rather than RNA maintenance. This model can explain the apparent heightened transcript abundance for parasite ligands such as baebl, eba-175, jesebl and pebl relative to maebl and ebl-1. Consistent with this interpretation is the persistence of y235 transcripts, coding for the HMW 235 kDa rhoptry proteins, which persist in P.yoelii from schizogony through to mature invasive merozoites (25). Some transcriptionally active genes, including pebl, are proposed to be pseudogenes (13,26), while some multi-copy genes exhibit a relaxed transcriptional control early in intraerythrocytic development (27,28). The reduced transcript levels of pebl at late schizont stages compared with baebl, jesebl and eba-175 might be explained by the presence of upstream frame-shift mutations thereby potentially impeding full-length pebl transcription (13). We anticipate that the pattern of apparent high transcript levels will be observed for other late-stage schizont proteins, expressed during final differentiation of merozoites. Precise synchronization of parasite growth and narrow time point intervals will be required to accurately determine relative transcript abundance when comparing widely separated times during parasite development along with confirmation of protein expression.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Heidi Rottschafer, Kelli Shannon and Jorge Maciel for their assistance in the analysis of ebl transcripts. This work was supported by the National Institutes of Health (R29/R01 AI33656) and the UNDP/World Bank/WHO Special Program for Research and Training in Tropical Diseases. J.H.A. is a Burroughs Wellcome Fund New Investigator in Molecular Parasitology. P.L.B. was supported by a Predoctoral Fellowship (training grant) in Experimental Parasitology and Vector Biology (T32 AI0703018) from the National Institute of Allergy and Infectious Diseases. We wish to thank the scientists and funding agencies comprising the international Malaria Genome Project for making sequence data from the genome of P.falciparum (3D7) public prior to publication of the completed sequence. The Sanger Centre (UK) provided sequence for chromosomes 1, 3–9 and 13, with financial support from the Wellcome Trust. A consortium composed of The Institute for Genome Research, along with the Naval Medical Research Center (USA), sequenced chromosomes 2, 10, 11 and 14, with support from NIAID/NIH, the Burroughs Wellcome Fund, and the Department of Defense. The Stanford Genome Technology Center (USA) sequenced chromosome 12, with support from the Burroughs Wellcome Fund. The Plasmodium Genome Database is a collaborative effort of investigators at the University of Pennsylvania (USA) and Monash University (Melbourne, Australia), supported by the Burroughs Wellcome Fund. The opinions expressed are those of the authors and do not reflect the official policy of the Department of the Navy, Department of Defense, or the US government.
DDBJ/EMBL/GenBank accession nos+ To whom correspondence should be addressed. Tel: +1 574 631 8676; Fax: +1 574 631 7413; Email: adams.20@nd.edu AF461093–AF461098, AY090552
REFERENCES
- 1.Witney A.A., Doolan,D.L., Anthony,R.M., Weiss,W.R., Hoffman,S.L. and Carucci,D.J. (2001) Determining liver stage parasite burden by real time quantitative PCR as a method for evaluating pre-erythrocytic malaria vaccine efficacy. Mol. Biochem. Parasitol., 118, 233–245. [DOI] [PubMed] [Google Scholar]
- 2.Hermsen C., Telgt,D., Linders,E., van de Locht,L., Eling,W., Mensink,E. and Sauerwein,R. (2001) Detection of Plasmodium falciparum malaria parasites in vivo by real-time quantitative PCR. Mol. Biochem. Parasitol., 118, 247–251. [DOI] [PubMed] [Google Scholar]
- 3.Ward G.E., Chitnis,C.E. and Miller,L.H. (1994) The invasion of erythrocytes by malarial merozoites. In Russell,D.G. (ed.), Clinical Infectious Diseases. Bailliere Tindall, London, Vol. 1, pp. 155–187.
- 4.Sim B.K.L., Chitnis,C.E., Wasniowska,T.J., Hadley,T.J. and Miller,L.H. (1994) Receptor and ligand domains for invasion of erythrocytes by Plasmodium falciparum. Science, 264, 1941–1944. [DOI] [PubMed] [Google Scholar]
- 5.Adams J.H., Blair,P.L., Kaneko,O. and Peterson,D.S. (2001) An expanding ebl family of Plasmodium falciparum. Trends Parasitol., 17, 297–299. [DOI] [PubMed] [Google Scholar]
- 6.Adams J.H., Sim,B.K.L., Dolan,S.A., Fang,X., Kaslow,D.C. and Miller,L.H. (1992) A family of erythrocyte binding proteins of malaria parasites. Proc. Natl Acad. Sci. USA, 89, 7085–7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mayer D.C., Kaneko,O., Hudson-Taylor,D.E., Reid,M.E. and Miller,L.H. (2001) Characterization of a Plasmodium falciparum erythrocyte-binding protein paralogous to EBA-175. Proc. Natl Acad. Sci. USA, 98, 5222–5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adams J.H., Hudson,D.E., Torii,M., Ward,G.E., Wellems,T.E., Aikawa,M. and Miller,L.H. (1990) The Duffy receptor family of Plasmodium knowlesi is located within the micronemes of invasive malaria merozoites. Cell, 63, 141–153. [DOI] [PubMed] [Google Scholar]
- 9.Sim B.K., Toyoshima,T., Haynes,J.D. and Aikawa,M. (1992) Localization of the 175-kilodalton erythrocyte binding antigen in micronemes of Plasmodium falciparum merozoites. Mol. Biochem. Parasitol., 51, 157–159. [DOI] [PubMed] [Google Scholar]
- 10.Haynes J.D. and Moch,J.K. (2002) Automated synchronization of P. falciparum malaria parasites by culture in a temperature-cycling incubator. In Doolan,D.L. (ed.), Malaria Methods and Protocols. Humana Press, Totowa, NJ, in press. [DOI] [PubMed]
- 11.Peterson D.S. and Wellems,T.E. (2000) EBL-1, a putative erythrocyte binding protein of Plasmodium falciparum, maps within a favored linkage group in two genetic crosses. Mol. Biochem. Parasitol., 105, 105–113. [DOI] [PubMed] [Google Scholar]
- 12.Thompson J.K., Triglia,T., Reed,M.B. and Cowman,A.F. (2001) A novel ligand from Plasmodium falciparum that binds to a sialic acid-containing receptor on the surface of human erythrocytes. Mol. Microbiol., 41, 47–58. [DOI] [PubMed] [Google Scholar]
- 13.Triglia T., Thompson,J.K. and Cowman,A.F. (2001) An EBA175 homologue which is transcribed but not translated in erythrocytic stages of Plasmodium falciparum. Mol. Biochem. Parasitol., 116, 55–63. [DOI] [PubMed] [Google Scholar]
- 14.Narum D.L., Fuhrmann,S.R., Luu,T. and Sim,B.K. (2002) A novel Plasmodium falciparum erythrocyte binding protein-2 (EBP2/BAEBL) involved in erythrocyte receptor binding. Mol. Biochem. Parasitol., 119, 159–168. [DOI] [PubMed] [Google Scholar]
- 15.Kappe S.H.I., Curley,G.P., Noe,A.R., Dalton,J.P. and Adams,J.H. (1997) Erythrocyte binding protein homologues of rodent malaria parasites. Mol. Biochem. Parasitol., 89, 137–148. [DOI] [PubMed] [Google Scholar]
- 16.Kappe S.H.I., Noe,A.R., Fraser,T.S., Blair,P.L. and Adams,J.H. (1998) A family of chimeric erythrocyte binding proteins of malaria parasites. Proc. Natl Acad. Sci. USA, 95, 1230–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gravenor M.B. and Kwiatkowski,D. (1998) An analysis of the temperature effects of fever on the intra-host population dynamics of Plasmodium falciparum. Parasitology, 117, 97–105. [DOI] [PubMed] [Google Scholar]
- 18.Kwiatkowski D. (1989) Febrile temperatures can synchronize the growth of Plasmodium falciparum in vitro. J. Exp. Med., 169, 357–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaneko O., Fidock,D.A., Schwartz,O.M. and Miller,L.H. (2000) Disruption of the C-terminal region of EBA-175 in the Dd2/Nm clone of Plasmodium falciparum does not affect erythrocyte invasion. Mol. Biochem. Parasitol., 110, 135–146. [DOI] [PubMed] [Google Scholar]
- 20.Reed M.B., Caruana,S.R., Batchelor,A.H., Thompson,J.K., Crabb,B.S. and Cowman,A.F. (2000) Targeted disruption of an erythrocyte binding antigen in Plasmodium falciparum is associated with a switch toward a sialic acid-independent pathway of invasion. Proc. Natl Acad. Sci. USA, 97, 7509–7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noe A.R. and Adams,J.H. (1998) Plasmodium yoelii YM MAEBL protein is coexpressed and colocalizes with rhoptry proteins. Mol. Biochem. Parasitol., 96, 27–35. [DOI] [PubMed] [Google Scholar]
- 22.Kocken C.H.M., van der Wel,A.M., Dubbeld,M.A., Narum,D.L., van de Rijke,F.M., van Gemert,G.J., van der Linde,X., Bannister,L.H., Janse,C., Waters,A.P. et al. (1998) Precise timing of expression of a Plasmodium falciparum-derived transgene in Plasmodium berghei is a critical determinant of subsequent subcellular localization. J. Biol. Chem., 273, 15119–15124. [DOI] [PubMed] [Google Scholar]
- 23.Narum D.L. and Thomas,A.W. (1994) Differential localization of full length and processed forms of Pf83/AMA-1 an apical membrane antigen of Plasmodium falciparum merozoites. Mol. Biochem. Parasitol., 67, 59–68. [DOI] [PubMed] [Google Scholar]
- 24.Thomas A.W., Deans,J.A., Mitchell,G.H., Alderson,T. and Cohen,S. (1984) The Fab fragments of monoclonal IgG to a merozoite surface antigen inhibit Plasmodium knowlesi invasion of erythrocytes. Mol. Biochem. Parasitol., 13, 187–199. [DOI] [PubMed] [Google Scholar]
- 25.Preiser P.R., Jarra,W., Capiod,T. and Snounou,G. (1999) A rhoptry-protein-associated mechanism of clonal phenotypic variation in rodent malaria. Nature, 398, 618–622. [DOI] [PubMed] [Google Scholar]
- 26.Taylor H.M., Triglia,T., Thompson,J., Sajid,M., Fowler,R., Wickham,M.E., Cowman,A.F. and Holder,A.A. (2001) Plasmodium falciparum homologue of the genes for Plasmodium vivax and Plasmodium yoelii adhesive proteins, which is transcribed but not translated. Infect. Immun., 69, 3635–3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scherf A., Hernandez-Rivas,R., Buffet,P., Bottius,E., Benatar,C., Pouvelle,B., Gysin,J. and Lanzer,M. (1998) Antigenic variation in malaria: in situ switching, relaxed and mutually exclusive transcription of var genes during intra-erythrocytic development in Plasmodium falciparum. EMBO J., 17, 5418–5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Q., Fernandez,V., Sundstrom,A., Schlichtherle,M., Datta,S., Hagblom,P. and Wahlgren,M. (1998) Developmental selection of var gene expression in Plasmodium falciparum. Nature, 394, 392–395. [DOI] [PubMed] [Google Scholar]