Abstract
The nucleotide sequences of two approx. 4 kilobase pair segments of the bovine genome are presented. One segment contains a coding region for bovine pancreatic trypsin inhibitor (BPTI) and the other segment contains a coding region for a BPTI homologue. The two 4 kilobase pair sequences are strikingly similar over approx. 3.4 kilobase pairs of their sequence, including putative intron sequences, suggesting that they have evolved from a gene duplication event.
Full text
PDF



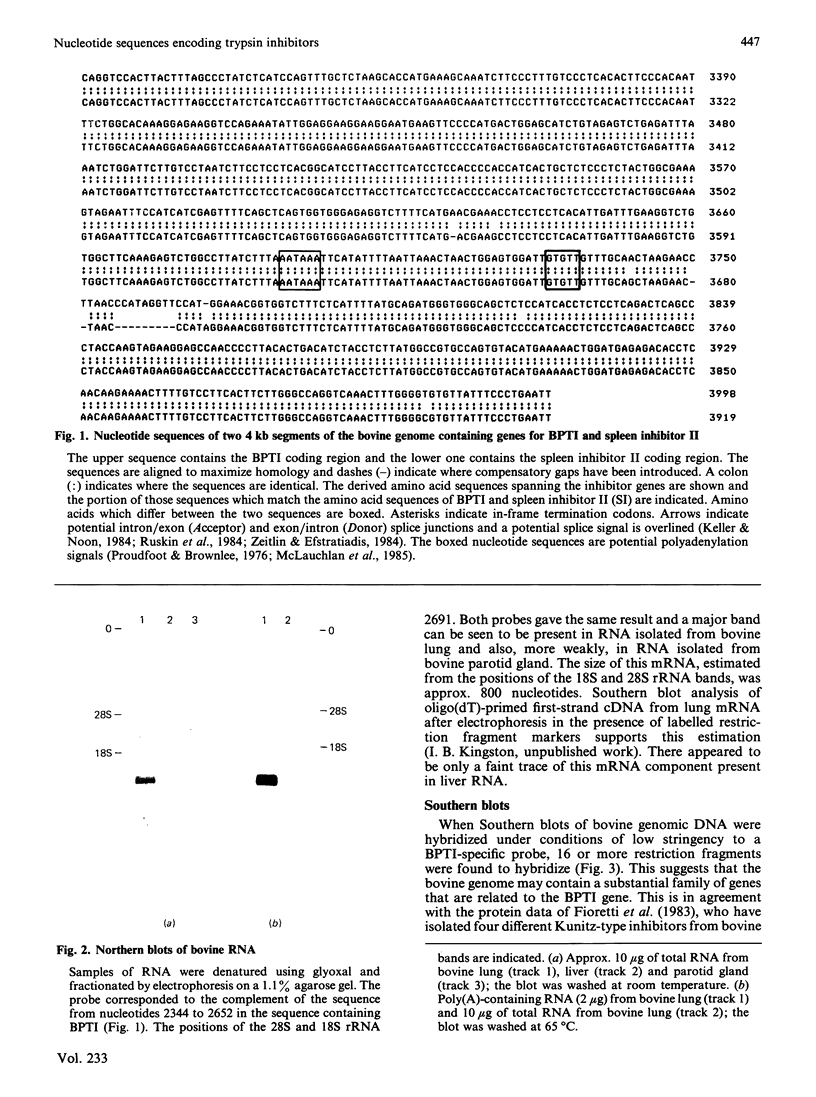



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson S., Kingston I. B. Isolation of a genomic clone for bovine pancreatic trypsin inhibitor by using a unique-sequence synthetic DNA probe. Proc Natl Acad Sci U S A. 1983 Nov;80(22):6838–6842. doi: 10.1073/pnas.80.22.6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard O., Hozumi N., Tonegawa S. Sequences of mouse immunoglobulin light chain genes before and after somatic changes. Cell. 1978 Dec;15(4):1133–1144. doi: 10.1016/0092-8674(78)90041-7. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brégégère F., Abastado J. P., Kvist S., Rask L., Lalanne J. L., Garoff H., Cami B., Wiman K., Larhammar D., Peterson P. A. Structure of C-terminal half of two H-2 antigens from cloned mRNA. Nature. 1981 Jul 2;292(5818):78–81. doi: 10.1038/292078a0. [DOI] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Creighton T. E. Experimental studies of protein folding and unfolding. Prog Biophys Mol Biol. 1978;33(3):231–297. doi: 10.1016/0079-6107(79)90030-0. [DOI] [PubMed] [Google Scholar]
- Creighton T. E. Homology of protein structures: proteinase inhibitors. Nature. 1975 Jun 26;255(5511):743–745. doi: 10.1038/255743a0. [DOI] [PubMed] [Google Scholar]
- Deininger P. L. Random subcloning of sonicated DNA: application to shotgun DNA sequence analysis. Anal Biochem. 1983 Feb 15;129(1):216–223. doi: 10.1016/0003-2697(83)90072-6. [DOI] [PubMed] [Google Scholar]
- Doolittle R. F. Similar amino acid sequences: chance or common ancestry? Science. 1981 Oct 9;214(4517):149–159. doi: 10.1126/science.7280687. [DOI] [PubMed] [Google Scholar]
- Efstratiadis A., Kafatos F. C., Maniatis T. The primary structure of rabbit beta-globin mRNA as determined from cloned DNA. Cell. 1977 Apr;10(4):571–585. doi: 10.1016/0092-8674(77)90090-3. [DOI] [PubMed] [Google Scholar]
- Empie M. W., Laskowski M., Jr Thermodynamics and kinetics of single residue replacements in avian ovomucoid third domains: effect on inhibitor interactions with serine proteinases. Biochemistry. 1982 May 11;21(10):2274–2284. doi: 10.1021/bi00539a002. [DOI] [PubMed] [Google Scholar]
- Fioretti E., Binotti I., Barra D., Citro G., Ascoli F., Antonini E. Heterogeneity of the basic pancreatic inhibitor (Kunitz) in various bovine organs. Eur J Biochem. 1983 Jan 17;130(1):13–18. doi: 10.1111/j.1432-1033.1983.tb07110.x. [DOI] [PubMed] [Google Scholar]
- Fritz H., Fink E., Truscheit E. Kallikrein inhibitors. Fed Proc. 1979 Dec;38(13):2753–2759. [PubMed] [Google Scholar]
- Girvitz S. C., Bacchetti S., Rainbow A. J., Graham F. L. A rapid and efficient procedure for the purification of DNA from agarose gels. Anal Biochem. 1980 Aug;106(2):492–496. doi: 10.1016/0003-2697(80)90553-9. [DOI] [PubMed] [Google Scholar]
- Hardison R. C., Butler E. T., 3rd, Lacy E., Maniatis T., Rosenthal N., Efstratiadis A. The structure and transcription of four linked rabbit beta-like globin genes. Cell. 1979 Dec;18(4):1285–1297. doi: 10.1016/0092-8674(79)90239-3. [DOI] [PubMed] [Google Scholar]
- Hochstrasser K., Wachter E., Bretzel G. Identification of the acid-stable trypsin-plasma inhibitors of the human and bovine serum as Kunitz-type inhibitors. Preliminary sequencing data. Hoppe Seylers Z Physiol Chem. 1976 Nov;357(11):1659–1661. [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Huber R., Bode W., Kukla D., Kohl U., Ryan C. A. The structure of the complex formed by bovine trypsin and bovine pancreatic trypsin inhibitor III. Structure of the anhydro-trypsin-inhibitor complex. Biophys Struct Mech. 1975 May 30;1(3):189–201. doi: 10.1007/BF00535756. [DOI] [PubMed] [Google Scholar]
- Huber R., Kukla D., Bode W., Schwager P., Bartels K., Deisenhofer J., Steigemann W. Structure of the complex formed by bovine trypsin and bovine pancreatic trypsin inhibitor. II. Crystallographic refinement at 1.9 A resolution. J Mol Biol. 1974 Oct 15;89(1):73–101. doi: 10.1016/0022-2836(74)90163-6. [DOI] [PubMed] [Google Scholar]
- KASSELL B., RADICEVIC M., ANSFIELD M. J., LASKOWSKI M., Sr THE BASIC TRYPSIN INHIBITOR OF BOVINE PANCREAS. IV. THE LINEAR SEQUENCE OF THE 58 AMINO ACIDS. Biochem Biophys Res Commun. 1965 Jan 18;18:255–258. doi: 10.1016/0006-291x(65)90749-7. [DOI] [PubMed] [Google Scholar]
- Karn J., Brenner S., Barnett L., Cesareni G. Novel bacteriophage lambda cloning vector. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5172–5176. doi: 10.1073/pnas.77.9.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus M., McCammon J. A. The internal dynamics of globular proteins. CRC Crit Rev Biochem. 1981;9(4):293–349. doi: 10.3109/10409238109105437. [DOI] [PubMed] [Google Scholar]
- Keller E. B., Noon W. A. Intron splicing: a conserved internal signal in introns of animal pre-mRNAs. Proc Natl Acad Sci U S A. 1984 Dec;81(23):7417–7420. doi: 10.1073/pnas.81.23.7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LASKOWSKI M., MARS P. H., LASKOWSKI M. Comparison of trypsin inhibitor from colostrum with other crystalline trypsin inhibitors. J Biol Chem. 1952 Oct;198(2):745–752. [PubMed] [Google Scholar]
- Laskowski M., Jr, Kato I. Protein inhibitors of proteinases. Annu Rev Biochem. 1980;49:593–626. doi: 10.1146/annurev.bi.49.070180.003113. [DOI] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Mason A. J., Evans B. A., Cox D. R., Shine J., Richards R. I. Structure of mouse kallikrein gene family suggests a role in specific processing of biologically active peptides. Nature. 1983 May 26;303(5915):300–307. doi: 10.1038/303300a0. [DOI] [PubMed] [Google Scholar]
- McLauchlan J., Gaffney D., Whitton J. L., Clements J. B. The consensus sequence YGTGTTYY located downstream from the AATAAA signal is required for efficient formation of mRNA 3' termini. Nucleic Acids Res. 1985 Feb 25;13(4):1347–1368. doi: 10.1093/nar/13.4.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T., Yasunaga T., Nishida T. Nucleotide sequence divergence and functional constraint in mRNA evolution. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7328–7332. doi: 10.1073/pnas.77.12.7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perler F., Efstratiadis A., Lomedico P., Gilbert W., Kolodner R., Dodgson J. The evolution of genes: the chicken preproinsulin gene. Cell. 1980 Jun;20(2):555–566. doi: 10.1016/0092-8674(80)90641-8. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. J., Brownlee G. G. 3' non-coding region sequences in eukaryotic messenger RNA. Nature. 1976 Sep 16;263(5574):211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Krainer A. R., Maniatis T., Green M. R. Excision of an intact intron as a novel lariat structure during pre-mRNA splicing in vitro. Cell. 1984 Aug;38(1):317–331. doi: 10.1016/0092-8674(84)90553-1. [DOI] [PubMed] [Google Scholar]
- Rühlmann A., Kukla D., Schwager P., Bartels K., Huber R. Structure of the complex formed by bovine trypsin and bovine pancreatic trypsin inhibitor. Crystal structure determination and stereochemistry of the contact region. J Mol Biol. 1973 Jul 5;77(3):417–436. doi: 10.1016/0022-2836(73)90448-8. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S. H., Slightom J. L., Smithies O. A history of the human fetal globin gene duplication. Cell. 1981 Oct;26(2 Pt 2):191–203. doi: 10.1016/0092-8674(81)90302-0. [DOI] [PubMed] [Google Scholar]
- Slightom J. L., Blechl A. E., Smithies O. Human fetal G gamma- and A gamma-globin genes: complete nucleotide sequences suggest that DNA can be exchanged between these duplicated genes. Cell. 1980 Oct;21(3):627–638. doi: 10.1016/0092-8674(80)90426-2. [DOI] [PubMed] [Google Scholar]
- Staden R. An interactive graphics program for comparing and aligning nucleic acid and amino acid sequences. Nucleic Acids Res. 1982 May 11;10(9):2951–2961. doi: 10.1093/nar/10.9.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Automation of the computer handling of gel reading data produced by the shotgun method of DNA sequencing. Nucleic Acids Res. 1982 Aug 11;10(15):4731–4751. doi: 10.1093/nar/10.15.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Graphic methods to determine the function of nucleic acid sequences. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):521–538. doi: 10.1093/nar/12.1part2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Measurements of the effects that coding for a protein has on a DNA sequence and their use for finding genes. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):551–567. doi: 10.1093/nar/12.1part2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talmadge K., Vamvakopoulos N. C., Fiddes J. C. Evolution of the genes for the beta subunits of human chorionic gonadotropin and luteinizing hormone. Nature. 1984 Jan 5;307(5946):37–40. doi: 10.1038/307037a0. [DOI] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA transferred or dotted nitrocellulose paper. Methods Enzymol. 1983;100:255–266. doi: 10.1016/0076-6879(83)00060-9. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Wachter E., Deppner K., Hochstrasser K., Lempart K., Geiger R. A new Kunitz-type inhibitor from bovine serum amino acid sequence determination. FEBS Lett. 1980 Sep 22;119(1):58–62. doi: 10.1016/0014-5793(80)80997-5. [DOI] [PubMed] [Google Scholar]
- Wachter E., Hochstrasser K. Kunitz-type proteinase inhibitors derived by limited proteolysis of the inter-alpha-trypsin inhibitor, III. Sequence of the two Kunitz-type domains inside the native inter-alpha-trypsin inhibitor, its biological aspects and also of its cleavage products. Hoppe Seylers Z Physiol Chem. 1979 Sep;360(9):1305–1311. doi: 10.1515/bchm2.1979.360.2.1305. [DOI] [PubMed] [Google Scholar]
- Wagner G., Wüthrich K. Sequential resonance assignments in protein 1H nuclear magnetic resonance spectra. Basic pancreatic trypsin inhibitor. J Mol Biol. 1982 Mar 5;155(3):347–366. doi: 10.1016/0022-2836(82)90009-2. [DOI] [PubMed] [Google Scholar]
- Watanabe Y., Tsukada T., Notake M., Nakanishi S., Numa S. Structural analysis of repetitive DNA sequences in the bovine corticotropin-beta-lipotropin precursor gene region. Nucleic Acids Res. 1982 Mar 11;10(5):1459–1469. doi: 10.1093/nar/10.5.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbur W. J., Lipman D. J. Rapid similarity searches of nucleic acid and protein data banks. Proc Natl Acad Sci U S A. 1983 Feb;80(3):726–730. doi: 10.1073/pnas.80.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlin S., Efstratiadis A. In vivo splicing products of the rabbit beta-globin pre-mRNA. Cell. 1984 Dec;39(3 Pt 2):589–602. doi: 10.1016/0092-8674(84)90466-5. [DOI] [PubMed] [Google Scholar]
- van Ooyen A., van den Berg J., Mantei N., Weissmann C. Comparison of total sequence of a cloned rabbit beta-globin gene and its flanking regions with a homologous mouse sequence. Science. 1979 Oct 19;206(4416):337–344. doi: 10.1126/science.482942. [DOI] [PubMed] [Google Scholar]