Abstract
Natural products have proven themselves as a valuable resource for antibiotics. However, in view of increasing antimicrobial resistance, there is an urgent need for new, structurally diverse agents that have the potential to overcome resistance and treat Gram‐negative pathogens in particular. Historically, the search for new antibiotics was strongly focussed on the very successful Actinobacteria. On the other hand, other producer strains have been under‐sampled and their potential for the production of bioactive natural products has been underestimated. In this mini‐review, we highlight prominent examples of novel anti‐Gram negative natural products produced by Gram‐negative bacteria that are currently in lead optimisation or preclinical development. Furthermore, we will provide insights into the considerations and strategies behind the discovery of these agents and their putative applications.
In this mini‐review, we highlight prominent examples of novel anti‐Gram negative natural products produced by isolated Gram‐negative bacteria that are currently in pre‐clinical development. Furthermore, we will provide insights into the considerations and strategies behind the discovery of these agents and their putative applications.
INTRODUCTION – THE IMPORTANCE OF NATURAL PRODUCTS AS ANTIBIOTICS
Beginning with the development of salvarsan in the early 20th century (Ehrlich, 1913), small molecule antibiotics have become one of the pillars of modern medicine. These drugs gave humankind the ability to treat and cure a wide range of bacterial infections and paved the way for many modern medical procedures (Hutchings et al., 2019). However, this progress is jeopardised by the rise of antimicrobial resistances (AMR) against more and more classes of antibiotics, leading to increasing numbers of deaths that are associated with or directly attributed to AMR (Murray et al., 2022; O'Neil, 2014; Ventola, 2015). Therefore, the discovery and development of novel antibiotics with new chemical scaffolds and molecular targets are needed, in particular with activity against Gram‐negative bacteria (Walesch et al., 2022).
Although the first antibiotics were of synthetic nature (Ehrlich, 1913; Otten, 1986), the discoveries and subsequent use of penicillin and streptomycin shifted the focus of antibiotic discovery towards natural products from microbes (Abraham et al., 1941; Fleming, 1929; Waksman & Schatz, 1945; Walesch et al., 2022). The ongoing relevance of microbial natural products as antibiotics is attested by the fact that more than two thirds of antibiotics that were approved between 1981 and 2019 are natural products or derivatives thereof (Newman & Cragg, 2020). When taking into account antibiotics that are currently marketed in the United States for the systemic treatment of non‐mycobacterial infections, 18 out of 22 antibiotic classes are natural product‐based (Walesch et al., 2022). Compared to synthetic compounds, microbial natural products are believed optimised through evolution to facilitate microbial competition for nutrients and habitats. Therefore, their structural diversity and physico‐chemical properties are optimised to penetrate cell walls and selectively inhibit bacteria or other cells (Hutchings et al., 2019; Lakemeyer et al., 2018; Laraia & Waldmann, 2017; Wright, 2017).
Bacteria deserve a special place among microbial natural products producers. According to the latest version of the MIBiG database, the structural and biosynthetic diversity of bacterial natural products exceeds that of the compounds produced by fungi (Terlouw et al., 2023). Furthermore, the organisation of the genes responsible for the production of natural products are mostly clustered in biosynthetic gene clusters (BGCs) facilitating genetic manipulations to study and optimise the production of such compounds (Bode & Müller, 2005). Bacteria can be isolated from diverse habitats ranging from marine to soil and from permafrost to desert environments. In these complex ecosystems, bacteria may live independently or as symbionts and their natural products play a key role fulfilling a broad range of biological functions, e.g., cooperation and communication, defence mechanisms and predation (Figure 1). A vast majority – more than 99% – of bacteria have not been cultivated, thus providing an immense potential to discover novel natural products (Crits‐Christoph et al., 2018; Locey & Lennon, 2016).
FIGURE 1.
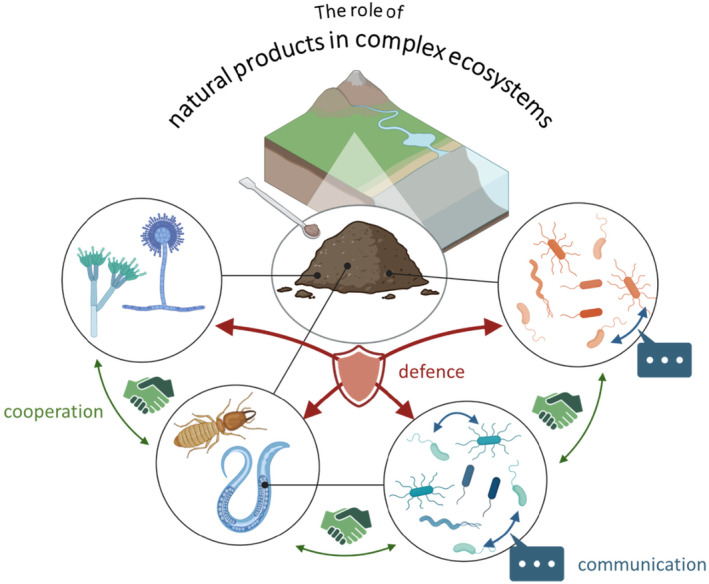
Microorganisms live in diverse habitats and ecosystems, as individuals, in competition or in symbiosis. Natural products are used as a means of communication, cooperation, inhibition, defence and predation. Exemplified by the cooperation of termites and fungi and of bacteria and nematodes, in defence of other microorganisms. Created in BioRender. Birkelbach, J. (2024) BioRender.com/x25z927
Hitherto, the majority of bacterial natural products listed in the NPAtlas database was discovered from Actinobacteria (Figure 2) (van Santen et al., 2022). Especially the genus Streptomyces was extremely fruitful for the discovery of clinically relevant antibiotics and anti‐cancer agents (Katz & Baltz, 2016; Newman & Cragg, 2020). However, the bias towards soil‐dwelling Actinobacteria and the resulting under‐sampling of other phyla underestimated the potential of other natural products producers (Hutchings et al., 2019). For example, the phyla Cyanobacteria, Proteobacteria, Firmicutes or Bacteriodetes are less explored than Actinobacteria and harbour more genera with less reported natural products (Figure 2) (van Santen et al., 2022; Walesch et al., 2022; Wright, 2017). In this mini‐review, we exemplarily highlight three different discovery approaches that were recently applied to generally underrepresented Gram‐negative producer strains, which yielded novel anti‐Gram negative antibiotics that are currently listed in preclinical development stages by the WHO, which comprise agents in lead optimisation, preclinical candidates applying Good Laboratory and Good Manufacturing Practices (GLP and GMP) and agents in Clinical Trial Application and Investigational New Drug‐enabling studies (CTA/IND) (Antimicrobial Resistance Division, 2024).
FIGURE 2.
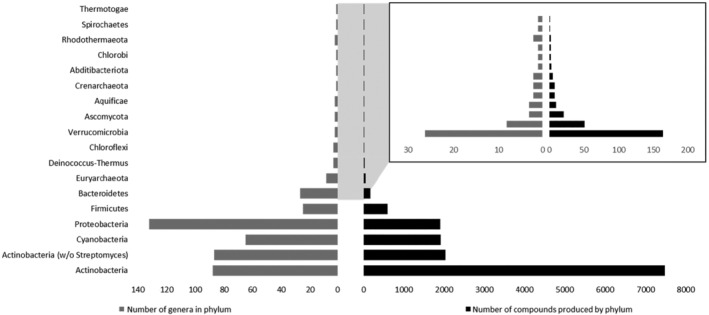
The number of bacterial natural products produced per phylum (left) versus the number of validly described genera per phylum (right) reported in the NPAtlas database (van Santen et al., 2022). Inset shows phyla that produce <200 reported natural products.
MAIN – GRAM‐NEGATIVE BACTERIA PRODUCING ANTI‐GRAM‐NEGATIVE ANTIBIOTICS – APPROACHES/ HABITATS
Predator approach
Myxococcota (or myxobacteria) are rod‐shaped Gram‐negative bacteria that grow in terrestrial and aquatic habitats all around the world (Mohr, 2018). They demonstrate a sophisticated and social life‐style including coordinated swarming on flat surfaces, cooperative predation of other microorganisms and the formation of multicellular fruiting bodies upon starvation (Munoz‐Dorado et al., 2016; Reichenbach, 1999). Furthermore, myxobacteria have the largest bacterial genomes with up to 16 Mbp (Han et al., 2013; Pal et al., 2021), harbouring a great potential to produce secondary metabolites (Garcia et al., 2024; Zaburannyi et al., 2016).
The potential value of myxobacteria as producers of antibiotics was first discussed in the middle of the past century, based on their ability to lyse other microbes (Oxford, 1947; Singh, 1947). Further research showed the ability of myxobacteria to lyse human pathogenic bacteria and to produce substances with antibacterial properties (Mathew & Dudani, 1955; Noren & Raper, 1962). Arguably, due to the comparably difficult (large scale) cultivation of most Myxococcota in the laboratory (Mohr, 2018), it took until the late 1970s before ambrucitin was isolated as the first natural product from a myxobacterium (Ringel et al., 1977). Since then, myxobacteria have proven themselves as a fruitful source of natural products with diverse chemical scaffolds displaying a wide range of biological activities (Herrmann et al., 2017). As of 2023, more than 800 natural products, belonging to ~170 chemical scaffolds were isolated from myxobacterial cultivation extracts (Wang et al., 2024). Among the many natural product scaffolds from myxobacteria with antibacterial activities, three compounds or compound classes are currently in preclinical development for their activities against Gram‐negative bacteria, corallopyronin A, cystobactamids and corramycins (Figure 3).
FIGURE 3.
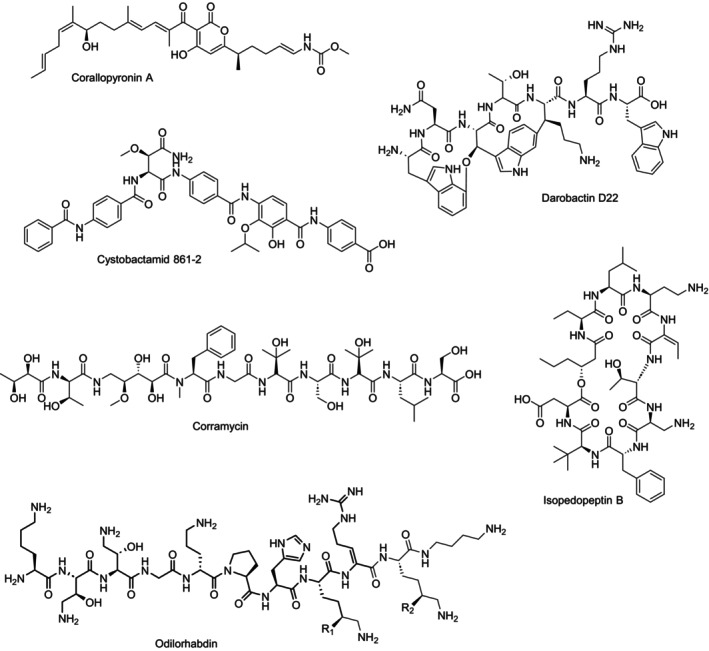
Chemical structures of corallopyronin A, cystobactamid 861–2, corramycin, isopedopeptin B, odilorhabdin and darobactin D22.
Although corallopyronin A (Figure 3) and its antibacterial activities against a range of Gram‐positive and ‐negative bacteria was already described in the 1980s (Irschik et al., 1985; Jansen et al., 1985), its development as antibiotic seemed unlikely, due to low production titres in the original producer Corallococcus coralloides and a poor yield in chemical synthesis (Krome et al., 2022). This changed with the implementation of a heterologous expression system of corallopyronin in Myxococcus xanthus, which increased the production of corallopyronin A greatly (Pogorevc et al., 2019; Sucipto et al., 2017). Corallopyronin A displays potent activity against Gram‐negative bacteria like Neisseria ghonorrhoeae, Chlamydia spp., Rickettsia spp. and Wolbachia spp. (Miethke et al., 2021). As a preclinical candidate it is currently developed for the treatment of filarial worm infections, by targeting their Wolbachia endosymbionts and has shown its efficacy in rodent infection models (Ehrens et al., 2022; Krome et al., 2022; Schiefer et al., 2020). The mode of action of corallopyronin A is the inhibition of the DNA‐dependent RNA polymerase (Mukhopadhyay et al., 2008). As its binding site at the ‘switch region’ of the enzyme is distinct from the binding sites of other RNA polymerase inhibitors, corallopyronin A does not show cross‐resistance with other antibiotics (Krome et al., 2022; Shima et al., 2018).
The antibacterial cystobactamids (Figure 3) were discovered in the cultivation extracts of Cystobacter velatus Cbv34 in the course of a screening campaign of a biodiverse collection of myxobacteria (Baumann et al., 2014; Herrmann et al., 2016). They are unusual peptides, featuring several para‐aminobenzoic acids and display activity against Gram‐positive and Gram‐negative bacteria (Baumann et al., 2014). The molecular target of cystobactamids, the bacterial type IIa topoisomerase was found through investigation of the self‐resistance mechanisms of the producing Cystobacter strain (Baumann et al., 2014). Obviously, cystobactamids target a different binding site in bacterial gyrases as the quinolone antibiotics, as they show a low to no cross‐resistance with this scaffold (Baumann et al., 2014; Hüttel et al., 2017). In the past years cystobactamids have progressed to the lead optimisation phase, as the implementation of a heterologous expression system and a total synthesis route have led to the development of derivatives with highly improved properties in vitro and in vivo (Elgaher et al., 2020; Groß et al., 2021; Moeller et al., 2019; Moreno et al., 2015; Testolin et al., 2020). Interestingly, the albicidins, originally isolated from the Gram‐negative bacterium Xanthomonas albilineans, have a similar scaffold and antibacterial activities to the cystobactamids (Cociancich et al., 2015). Due to their unique scaffold the cystobactamids and albicidins are currently in lead optimisation, which yielded derivatives with improved antibacterial properties and a better understanding of their modes of action and resistance mechanisms (Kleebauer et al., 2021; Michalczyk et al., 2023; Risch et al., 2024; Saathoff et al., 2023; Zborovsky et al., 2021).
Corramycins (Figure 3) are linear peptides with activity against E. coli that were first found in cultivation extracts of Corallococcus coralloides (Couturier et al., 2022). Lead optimisation by organic synthesis was used to develop a corramycin derivative with a more than 300‐fold increased activity against Gram‐negative bacteria, displaying promising activities against K. pneumoniae and A. baumannii (Renard et al., 2023). Inactivation of the warhead by phosphorylation was identified as a mechanism of self‐resistance in the producing myxobacteria (Adam et al., 2024). Although the mechanism of action of corramycins has not been elucidated yet, it can be assumed that it inhibits bacterial growth by a novel molecular target, as it shows no cross‐resistance with known antibiotic classes (Couturier et al., 2022; Renard et al., 2023). Moreover, corramycins have shown their in vivo efficacy in a range of rodent infection models (Couturier et al., 2022; Renard et al., 2023).
Resistance‐based isolation approach
Antibiotic producing bacteria obviously require self‐resistance mechanisms to protect themselves against the toxicity of the natural product they produced to affect their opponent.
To exploit self‐resistance mechanisms, Thaker et al. successfully developed the resistance‐guided cultivation approach selecting for glycolipopetide‐resistant actinomycetes to screen for the production of new glycolipopeptides (Thaker et al., 2013, 2014). Bjerketorp et al. adapted and extended this approach on the screening of soil isolates selecting for environmental multi drug resistance (MDR) bacteria (Bjerketorp et al., 2021). Therefore, the combination of several antibiotics from different chemical classes was used to screen for natural product producers with multiple self‐resistance mechanisms and thus the capacity to produce several antibacterial compounds. Furthermore, it enabled the isolation of low‐abundant underexplored natural product producers present in soil‐samples (Bjerketorp et al., 2021), which would have proven difficult without the selection bias.
This approach led to the isolation of several MDR Pedobacter spp., which are Gram‐negative bacteria belonging to the phylum Bacteroidota and the family Sphingobacteriaceae (Bjerketorp et al., 2021). Nord et al. discovered the lipodepsipeptide isopedopeptin scaffold, which shows low micro‐molar activity against WHO top priority pathogens including colistin resistant strains (Nord et al., 2020). Membrane disruption is proposed as one mode of action (MoA) of isopedopeptins, which is corroborated by the structural similarity to bacterial lipopolysaccharide (LPS)‐binding pedopeptins (Hirota‐Takahata et al., 2014; Kozuma et al., 2014). However, the discrepancy with the MIC suggests additional MoAs. Currently, isopedopeptin B (ULT3, Figure 3) is in CTA/IND studies showing good anti‐Gram‐negative activity and acceptable cytotoxicity. Although it is expected to discover similar antibiotics compared to the ones the isolates are resistant to, the extended resistance‐based approach facilitated the isolation of underrepresented natural products producers, thereby the discovery of novel antibacterial compounds.
Endosymbiont approach‐Xenorhabdus and Photorhabdus as a promising source of new anti‐Gram‐negatives
In addition to bacterial species that have long been studied for the discovery of novel antibiotics, such as Actinomyces or Myxococcota (Müller & Wink, 2014), entomopathogenic bacteria have become a focus of research interest due to their complex lifestyle (Chaston et al., 2011), which is mainly feasible due to the production of bioactive secondary metabolites that have a positive influence on persistence in the host (Cimen et al., 2022; Shi et al., 2022). In particular, studies have shown that the γ‐proteobacteria Xenorhabdus and Photorhabdus spp., that live in mutualistic symbiosis with nematodes, provide antibiotic candidates with promising anti‐Gram‐negative activity against difficult‐to‐treat pathogens (Chaston et al., 2011; Walesch et al., 2022). Xenorhabdus spp. primarily infect nematodes of the genus Steinema, whereas Photorhabdus spp. mainly infect Heterorhabditis spp. (Clarke, 2008; Forst, 2002; Poinar, 1966; Poinar & Thomas, 1967; Waterfield et al., 2001). Although they use functionally different approaches, both infect the intestinal tract of their host and are able to overcome or suppress the immune system of their hosts (Chaston et al., 2011; Clarke, 2008), or kill it by producing insecticides (Proschak et al., 2014; Sergeant et al., 2006). Furthermore, both are producing many antimicrobial compounds to prevent the growth of antagonistic microorganisms allowing the persistence in the intestinal tract of their nematodic host (Blackburn et al., 2016; Hu et al., 2006; Muangpat et al., 2017, 2020; Sajnaga & Kazimierczak, 2020; Wenski et al., 2020; Zhou et al., 2013). Recently, two promising novel antibacterial classes were discovered from these two endopathogenic bacteria: the odilorhabdins (Racine & Gualtieri, 2019) and the daropeptides (Ma et al., 2024), consisting of darobactins and dynobactin (Figure 3). Both classes exhibit strong anti‐Gram‐negative activity against, e.g., Escherichia coli, Pseudomonas aeruginosa, Klebsiella pneumoniae and Acinetobacter baumannii strains including clinical isolates (Chaston et al., 2011; Imai et al., 2019; Ma et al., 2024; Pantel et al., 2018).
Odilorhabdins
The discovery and development of odilorhabdins was largely driven by the start‐up Nosopharm (Racine & Gualtieri, 2019). In a comprehensive screening of various Xenorhabdus strains, the first odilorhabdins (Pantel et al., 2018) were isolated from X. nematophila using a traditional bioactivity‐guided approach, subsequently identifying the chemical structure and mode of action. Odilorhabdins represent a new class of broad‐spectrum antibiotics. These compounds target the 30S ribosomal subunit of Gram‐negative and Gram‐positive bacteria on a binding site not exploited by currently marketed antibiotics, thus they have a reduced risk of cross‐resistances (Pantel et al., 2018). Additionally, they conducted extensive derivatisation using chemical total synthesis to enhance antibacterial activity, particularly against E. coli and K. pneumoniae strains (Sarciaux et al., 2018). This led to the development of a frontrunner molecule, NOSO‐502 (Racine et al., 2018; Sarciaux et al., 2018), which is supposedly nearing clinical phase I studies for the treatment of critical urinary tract infections (UTIs) (Lanois‐Nouri et al., 2022; Präve et al., 2024; Racine et al., 2018; Racine & Gualtieri, 2019).
Darobactins
The first native darobactins were discovered after the bioactivity screening of Photorhabdus extracts (Imai et al., 2019). The bioactivity‐guided approach resulted in the isolation of darobactin A (DA) from P. khanii, a novel anti‐Gram‐negative agent that is a ribosomally synthesised and post‐translationally modified peptide (RiPP). Darobactins target the outer membrane protein BamA, part of the BamABCDE complex, preventing the incorporation and proper folding of outer membrane proteins into the cell membrane, thereby selectively killing Gram‐negative bacteria (Haysom et al., 2023; Imai et al., 2019; Kaur et al., 2021). Through binding on BamA, a novel antibacterial target, darobactins have a strongly reduced risk of cross‐resistance with currently used antibiotic classes.
Ongoing studies have focused on the heterologous production and total synthesis of native and artificial derivatives of darobactin A in E. coli by engineering the biosynthetic gene cluster (BGC) to produce optimised darobactin derivatives with modifications in the core peptide (Böhringer et al., 2021; Groß et al., 2021; Lin et al., 2022; Nesic et al., 2022; Seyfert et al., 2022; Wuisan et al., 2021). This has resulted in derivatives such as D22 (Figure 3) and D69, which exhibit up to 128‐fold enhanced in vitro anti‐Gram‐negative activity against pathogens classified as critical prioritised by the WHO, such as carbapenem‐resistant A. baumannii (CRAB), comparable to last‐resort antibiotics like colistin (Seyfert et al., 2022; Seyfert et al., 2023; World Health Organisation, 2018). Despite these advancements, both the total synthesis routes and the biotechnological production in alternative production hosts currently suffer from relatively low yields (Seyfert et al., 2023), which must be addressed to bring these highly promising agents into clinical development.
Outlook
The approaches described here, the predator approach, the resistance approach and the endosymbiont approach, led to promising anti‐Gram‐negative agents produced by Gram‐negative bacteria. Exemplary compounds such as cystobactamids, corramycins, corallopyoronin A, isopedopeptins, darobactins and odilorhabdins and their bioengineered or synthetic derivatives, respectively, are currently in development to prove their in vivo efficiency or have done so already (Couturier et al., 2022; Ehrens et al., 2022; Nord et al., 2020; Racine et al., 2018; Renard et al., 2023; Schiefer et al., 2020; Seyfert et al., 2023; Seyfert et al., 2023; Testolin et al., 2020). Those compounds, as well as other natural products in general, could be further optimised and their underlying biosynthesis investigated in more detail to allow for synthetic biology and evolution‐inspired bioengineering techniques (Bozhüyük et al., 2024; Präve et al., 2024). Moreover, novel bioinformatically guided tools could allow for the modification of the chemical structure, to, e.g., enhance target binding or alter the pharmaceutical properties (Ndagi et al., 2020; Wang et al., 2022). The discovery approaches described here, which aimed at identifying compounds with new target sites, unknown chemistry and exhibiting no cross‐resistances with marketed antibiotics, emphasise their potential for the development of novel molecules against Gram‐negative bacteria. Further approaches such as high‐throughput elicitor screening and the well‐established OSMAC approach enable the identification of additional antibacterial molecules. Those approaches can be used to investigate already cultivated but also uncultivated bacterial species, living in mostly underexplored habitats to discover novel chemistry to fight the AMR crisis (Bader et al., 2021; Claesen et al., 2020; Crits‐Christoph et al., 2018; Donia et al., 2014; Gavriilidou et al., 2022; Hegemann et al., 2023; Locey & Lennon, 2016; Nett et al., 2009; Nichols et al., 2010). As highlighted in Figure 2, e.g., Pseudomonas, Burkholderia, Cyanobacteria and Firmicutes species are further examples of underexplored yet promising natural products producers, which harbour the potential for the discovery of future antibiotic agents (Hegemann et al., 2023; van Santen et al., 2022; Walesch et al., 2022; Wright, 2017).
AUTHOR CONTRIBUTIONS
Joy Birkelbach: Conceptualization; writing – original draft; writing – review and editing; visualization. Carsten E. Seyfert: Conceptualization; writing – original draft; writing – review and editing. Sebastian Walesch: Conceptualization; writing – original draft; writing – review and editing. Rolf Müller: Conceptualization; writing – review and editing; project administration; supervision.
FUNDING INFORMATION
No funding information provided.
CONFLICT OF INTEREST STATEMENT
C.E.S. and R.M. are inventors of the patent application WO 2022/175443 A1.
Birkelbach, J. , Seyfert, C.E. , Walesch, S. & Müller, R. (2024) Harnessing Gram‐negative bacteria for novel anti‐Gram‐negative antibiotics. Microbial Biotechnology, 17, e70032. Available from: 10.1111/1751-7915.70032
Joy Birkelbach, Carsten E. Seyfert and Sebastian Walesch contributed equally to this work.
REFERENCES
- Abraham, E.P. , Chain, E. , Fletcher, C.M. , Gardner, A.D. , Heatley, N.G. , Jennings, M.A. et al. (1941) Further observations on penicillin. The Lancet, 238(6155), 177–189. [Google Scholar]
- Adam, S. , Fries, F. , Tesmar, A. , Rasheed, S. , Deckarm, S. , Sousa, C.F. et al. (2024) The peptide antibiotic Corramycin adopts a β‐hairpin‐like structure and is inactivated by the kinase ComG. Journal of the American Chemical Society, 146, 8981–8990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antimicrobial Resistance Division . (2024) 2023 Antibacterial agents in clinical and preclinical development: an overview and analysis. Geneva: World Health Organization. [Google Scholar]
- Bader, C.D. , Haack, P.A. , Panter, F. , Krug, D. & Müller, R. (2021) Expanding the scope of detectable microbial natural products by complementary analytical methods and cultivation systems. Journal of Natural Products, 84(2), 268–277. [DOI] [PubMed] [Google Scholar]
- Baumann, S. , Herrmann, J. , Raju, R. , Steinmetz, H. , Mohr, K.I. , Hüttel, S. et al. (2014) Cystobactamids: myxobacterial topoisomerase inhibitors exhibiting potent antibacterial activity. Angewandte Chemie, International Edition, 53(52), 14605–14609. [DOI] [PubMed] [Google Scholar]
- Bjerketorp, J. , Levenfors, J.J. , Nord, C. , Guss, B. , Öberg, B. & Broberg, A. (2021) Selective isolation of multidrug‐resistant Pedobacter spp., producers of novel antibacterial peptides. Frontiers in Microbiology, 12, 642829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn, D. , Wood, P.L. , Burk, T.J. , Crawford, B. , Wright, S.M. & Adams, B.J. (2016) Evolution of virulence in Photorhabdus spp., entomopathogenic nematode symbionts. Systematic and Applied Microbiology, 39(3), 173–179. [DOI] [PubMed] [Google Scholar]
- Bode, H.B. & Müller, R. (2005) The impact of bacterial genomics on natural product research. Angewandte Chemie, International Edition, 44(42), 6828–6846. [DOI] [PubMed] [Google Scholar]
- Böhringer, N. , Green, R. , Liu, Y. , Mettal, U. , Marner, M. , Modaresi, S.M. et al. (2021) Mutasynthetic production and antimicrobial characterization of Darobactin analogs. Microbiology Spectrum, 9(3), e0153521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozhüyük, K.A.J. , Präve, L. , Kegler, C. , Schenk, L. , Kaiser, S. , Schelhas, C. et al. (2024) Evolution‐inspired engineering of nonribosomal peptide synthetases. Science, 383(6689), eadg4320. [DOI] [PubMed] [Google Scholar]
- Chaston, J.M. , Suen, G. , Tucker, S.L. , Andersen, A.W. , Bhasin, A. , Bode, E. et al. (2011) The entomopathogenic bacterial endosymbionts Xenorhabdus and Photorhabdus: convergent lifestyles from divergent genomes. PLoS One, 6(11), e27909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimen, H. , Touray, M. , Gulsen, S.H. & Hazir, S. (2022) Natural products from Photorhabdus and Xenorhabdus: mechanisms and impacts. Applied Microbiology and Biotechnology, 106(12), 4387–4399. [DOI] [PubMed] [Google Scholar]
- Claesen, J. , Spagnolo, J.B. , Ramos, S.F. , Kurita, K.L. , Byrd, A.L. , Aksenov, A.A. et al. (2020) A Cutibacterium acnes antibiotic modulates human skin microbiota composition in hair follicles. Science Translational Medicine, 12(570), eaay5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, D.J. (2008) Photorhabdus: a model for the analysis of pathogenicity and mutualism. Cellular Microbiology, 10(11), 2159–2167. [DOI] [PubMed] [Google Scholar]
- Cociancich, S. , Pesic, A. , Petras, D. , Uhlmann, S. , Kretz, J. , Schubert, V. et al. (2015) The gyrase inhibitor albicidin consists of p‐aminobenzoic acids and cyanoalanine. Nature Chemical Biology, 11(3), 195–197. [DOI] [PubMed] [Google Scholar]
- Couturier, C. , Groß, S. , von Tesmar, A. , Hoffmann, J. , Deckarm, S. , Fievet, A. et al. (2022) Structure elucidation, total synthesis, antibacterial in vivo efficacy and biosynthesis proposal of Myxobacterial Corramycin. Angewandte Chemie, International Edition in English, 61, e202210747. Available from: 10.1002/anie.202210747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crits‐Christoph, A. , Diamond, S. , Butterfield, C.N. , Thomas, B.C. & Banfield, J.F. (2018) Novel soil bacteria possess diverse genes for secondary metabolite biosynthesis. Nature, 558(7710), 440–444. [DOI] [PubMed] [Google Scholar]
- Donia, M.S. , Cimermancic, P. , Schulze, C.J. , Wieland Brown, L.C. , Martin, J. , Mitreva, M. et al. (2014) A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell, 158(6), 1402–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrens, A. , Schiefer, A. , Krome, A.K. , Becker, T. , Rox, K. , Neufeld, H. et al. (2022) Pharmacology and early ADMET data of corallopyronin a, a natural product with macrofilaricidal anti‐wolbachial activity in filarial nematodes. Frontiers in Tropical Diseases, 39(9), 1705–1720. [Google Scholar]
- Ehrlich, P. (1913) Address in pathology, ON CHEMIOTHERAPY: delivered before the seventeenth international congress of medicine. British Medical Journal, 2(2746), 353–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgaher, W.A.M. , Hamed, M.M. , Baumann, S. , Herrmann, J. , Siebenbürger, L. , Krull, J. et al. (2020) Cystobactamid 507: concise synthesis, mode of action and optimization toward more potent antibiotics. Chemistry – A European Journal, 26, 7219–7225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming, A. (1929) On the antibacterial action of cultures of a Penicillium, with special reference to their use in the isolation of B. influenzæ . British Journal of Experimental Pathology, 10(3), 226–236. [Google Scholar]
- Forst, S. (2002) Bacteria‐nematode symbiosis. In: Entomopathogenic nematology, p. 57. Wallingford: CABI Publishing UK. [Google Scholar]
- Garcia, R. , Popoff, A. , Bader, C.D. , Löhr, J. , Walesch, S. , Walt, C. et al. (2024) Discovery of the Pendulisporaceae: an extremotolerant myxobacterial family with distinct sporulation behavior and prolific specialized metabolism. Chem, 10, 1–20. [Google Scholar]
- Gavriilidou, A. , Kautsar, S.A. , Zaburannyi, N. , Krug, D. , Müller, R. , Medema, M.H. et al. (2022) Compendium of specialized metabolite biosynthetic diversity encoded in bacterial genomes. Nature Microbiology, 7(5), 726–735. [DOI] [PubMed] [Google Scholar]
- Groß, S. , Panter, F. , Pogorevc, D. , Seyfert, C.E. , Deckarm, S. , Bader, C.D. et al. (2021) Improved broad‐spectrum antibiotics against gram‐negative pathogens via darobactin biosynthetic pathway engineering. Chemical Science, 12(35), 11882–11893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groß, S. , Schnell, B. , Haack, P.A. , Auerbach, D. & Müller, R. (2021) In vivo and in vitro reconstitution of unique key steps in cystobactamid antibiotic biosynthesis. Nature Communications, 12(1), 1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, K. , Li, Z.‐F. , Peng, R. , Zhu, L.‐P. , Zhou, T. , Wang, L. et al. (2013) Extraordinary expansion of a Sorangium cellulosum genome from an alkaline milieu. Scientific Reports, 3, 2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haysom, S.F. , Machin, J. , Whitehouse, J.M. , Horne, J.E. , Fenn, K. , Ma, Y. et al. (2023) Darobactin B stabilises a lateral‐closed conformation of the BAM complex in E. coli cells. Angewandte Chemie, 62(34), e202218783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegemann, J.D. , Birkelbach, J. , Walesch, S. & Müller, R. (2023) Current developments in antibiotic discovery: global microbial diversity as a source for evolutionary optimized anti‐bacterials: global microbial diversity as a source for evolutionary optimized anti‐bacterials. EMBO Reports, 24(1), e56184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann, J. , Fayad, A.A. & Müller, R. (2017) Natural products from myxobacteria: novel metabolites and bioactivities. Natural Product Reports, 34(2), 135–160. [DOI] [PubMed] [Google Scholar]
- Herrmann, J. , Lukezic, T. , Kling, A. , Baumann, S. , Hüttel, S. , Petkovic, H. et al. (2016) Strategies for the discovery and development of new antibiotics from natural products: three case studies. Current Topics in Microbiology and Immunology, 398, 339–363. [DOI] [PubMed] [Google Scholar]
- Hirota‐Takahata, Y. , Kozuma, S. , Kuraya, N. , Fukuda, D. , Nakajima, M. & Ando, O. (2014) Pedopeptins, novel inhibitors of LPS: taxonomy of producing organism, fermentation, isolation, physicochemical properties and structural elucidation. Journal of Antibiotics (Tokyo), 67(3), 243–251. [DOI] [PubMed] [Google Scholar]
- Hu, K.J. , Li, J.X. , Li, B. , Webster, J.M. & Chen, G.H. (2006) A novel antimicrobial epoxide isolated from larval galleria mellonella infected by the nematode symbiont, Photorhabdus luminescens (Enterobacteriaceae). Bioorganic & Medicinal Chemistry, 14(13), 4677–4681. [DOI] [PubMed] [Google Scholar]
- Hutchings, M.I. , Truman, A.W. & Wilkinson, B. (2019) Antibiotics: past, present and future. Current Opinion in Microbiology, 51, 72–80. [DOI] [PubMed] [Google Scholar]
- Hüttel, S. , Testolin, G. , Herrmann, J. , Planke, T. , Gille, F. , Moreno, M. et al. (2017) Discovery and total synthesis of natural Cystobactamid derivatives with superior activity against gram‐negative pathogens. Angewandte Chemie, International Edition, 56(41), 12760–12764. [DOI] [PubMed] [Google Scholar]
- Imai, Y. , Meyer, K.J. , Iinishi, A. , Favre‐Godal, Q. , Green, R. , Manuse, S. et al. (2019) A new antibiotic selectively kills gram‐negative pathogens. Nature, 576(7787), 459–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irschik, H. , Jansen, R. , Höfle, G. , Gerth, K. & Reichenbach, H. (1985) The corallopyronins, new inhibitors of bacterial RNA synthesis from Myxobacteria. The Journal of Antibiotics, 38(2), 145–152. [DOI] [PubMed] [Google Scholar]
- Jansen, R. , Höfle, G. , Irschik, H. & Reichenbach, H. (1985) Antibiotika aus Gleitenden Bakterien, XXIV. Corallopyronin a, B und C – drei neue Antibiotika aus Corallococcus coralloides cc c127 (Myxobacterales). Liebigs Annalen der Chemie, 1985(4), 822–836. [Google Scholar]
- Katz, L. & Baltz, R.H. (2016) Natural product discovery: past, present, and future. Journal of Industrial Microbiology & Biotechnology, 43(2–3), 155–176. [DOI] [PubMed] [Google Scholar]
- Kaur, H. , Jakob, R.P. , Marzinek, J.K. , Green, R. , Imai, Y. , Bolla, J.R. et al. (2021) The antibiotic darobactin mimics a β‐strand to inhibit outer membrane insertase. Nature, 593, 125–129. [DOI] [PubMed] [Google Scholar]
- Kleebauer, L. , Zborovsky, L. , Hommernick, K. , Seidel, M. , Weston, J.B. & Süssmuth, R.D. (2021) Overcoming AlbD protease resistance and improving potency: synthesis and bioactivity of antibacterial Albicidin analogues with amide Bond Isosteres. Organic Letters, 23(18), 7023–7027. [DOI] [PubMed] [Google Scholar]
- Kozuma, S. , Hirota‐Takahata, Y. , Fukuda, D. , Kuraya, N. , Nakajima, M. & Ando, O. (2014) Screening and biological activities of pedopeptins, novel inhibitors of LPS produced by soil bacteria. Journal of Antibiotics, 67(3), 237–242. [DOI] [PubMed] [Google Scholar]
- Krome, A.K. , Becker, T. , Kehraus, S. , Schiefer, A. , Gütschow, M. , Chaverra‐Muñoz, L. et al. (2022) Corallopyronin A: antimicrobial discovery to preclinical development. Natural Product Reports, 39(9), 1705–1720. [DOI] [PubMed] [Google Scholar]
- Lakemeyer, M. , Zhao, W. , Mandl, F.A. , Hammann, P. & Sieber, S.A. (2018) Thinking outside the box‐novel Antibacterials to tackle the resistance crisis. Angewandte Chemie, 57(44), 14440–14475. [DOI] [PubMed] [Google Scholar]
- Lanois‐Nouri, A. , Pantel, L. , Fu, J. , Houard, J. , Ogier, J.‐C. , Polikanov, Y.S. et al. (2022) The Odilorhabdin antibiotic biosynthetic cluster and acetyltransferase self‐resistance locus are niche and species specific. MBio, 13, e0282621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laraia, L. & Waldmann, H. (2017) Natural product inspired compound collections: evolutionary principle, chemical synthesis, phenotypic screening, and target identification. Drug Discovery Today: Technologies, 23, 75–82. [DOI] [PubMed] [Google Scholar]
- Lin, Y.‐C. , Schneider, F. , Eberle, K.J. , Chiodi, D. , Nakamura, H. , Reisberg, S.H. et al. (2022) Atroposelective total synthesis of Darobactin a. Journal of the American Chemical Society, 144(32), 14458–14462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locey, K.J. & Lennon, J.T. (2016) Scaling laws predict global microbial diversity. Proceedings of the National Academy of Sciences, USA, 113(21), 5970–5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, S. , Guo, S. , Ding, W. & Zhang, Q. (2024) Daropeptide natural products. Exploration of Drug Science, 2, 190–202. [Google Scholar]
- Mathew, S. & Dudani, A. (1955) Lysis of human pathogenic bacteria by myxobacteria. Nature, 175(4446), 125. [DOI] [PubMed] [Google Scholar]
- Michalczyk, E. , Hommernick, K. , Behroz, I. , Kulike, M. , Pakosz‐Stępień, Z. , Mazurek, L. et al. (2023) Molecular mechanism of topoisomerase poisoning by the peptide antibiotic albicidin. Nature Catalysis, 6(1), 52–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miethke, M. , Pieroni, M. , Weber, T. , Brönstrup, M. , Hammann, P. , Halby, L. et al. (2021) Towards the sustainable discovery and development of new antibiotics. Nature Reviews Chemistry, 5, 726–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller, M. , Norris, M.D. , Planke, T. , Cirnski, K. , Herrmann, J. , Müller, R. et al. (2019) Scalable syntheses of Methoxyaspartate and preparation of the antibiotic Cystobactamid 861‐2 and highly potent derivatives. Organic Letters, 21(20), 8369–8372. [DOI] [PubMed] [Google Scholar]
- Mohr, K.I. (2018) Diversity of Myxobacteria‐we only see the tip of the iceberg. Microorganisms, 6(3), 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno, M. , Elgaher, W. , Herrmann, J. , Schläger, N. , Hamed, M. , Baumann, S. et al. (2015) Synthesis and biological evaluation of cystobactamid 507: a bacterial topoisomerase inhibitor from Cystobacter sp. Synlett, 26(9), 1175–1178. [Google Scholar]
- Muangpat, P. , Suwannaroj, M. , Yimthin, T. , Fukruksa, C. , Sitthisak, S. , Chantratita, N. et al. (2020) Antibacterial activity of Xenorhabdus and Photorhabdus isolated from entomopathogenic nematodes against antibiotic‐resistant bacteria. PLoS One, 15(6), e0234129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muangpat, P. , Yooyangket, T. , Fukruksa, C. , Suwannaroj, M. , Yimthin, T. , Sitthisak, S. et al. (2017) Screening of the antimicrobial activity against drug resistant bacteria of Photorhabdus and Xenorhabdus associated with Entomopathogenic nematodes from Mae Wong National Park, Thailand. Frontiers in Microbiology, 8, 1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay, J. , Das, K. , Ismail, S. , Koppstein, D. , Jang, M. , Hudson, B. et al. (2008) The RNA polymerase “switch region” is a target of inhibitors. Cell, 135, 295–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, R. & Wink, J. (2014) Future potential for anti‐infectives from bacteria – how to exploit biodiversity and genomic potential. International Journal of Medical Microbiology, 304(1), 3–13. [DOI] [PubMed] [Google Scholar]
- Munoz‐Dorado, J. , Marcos‐Torres, F.J. , Garcia‐Bravo, E. , Moraleda‐Munoz, A. & Perez, J. (2016) Myxobacteria: moving, killing, feeding, and surviving together. Frontiers in Microbiology, 7, 781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray, C.J.L. , Ikuta, K.S. , Sharara, F. , Swetschinski, L. , Robles Aguilar, G. , Gray, A. et al. (2022) Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet, 399(10325), 629–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndagi, U. , Falaki, A.A. , Abdullahi, M. , Lawal, M.M. & Soliman, M.E. (2020) Antibiotic resistance: bioinformatics‐based understanding as a functional strategy for drug design. RSC Advances, 10(31), 18451–18468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesic, M. , Ryffel, D.B. , Maturano, J. , Shevlin, M. , Pollack, S.R. , Gauthier, D.R. et al. (2022) Total synthesis of Darobactin a. Journal of the American Chemical Society, 144(31), 14026–14030. [DOI] [PubMed] [Google Scholar]
- Nett, M. , Ikeda, H. & Moore, B.S. (2009) Genomic basis for natural product biosynthetic diversity in the actinomycetes. Natural Product Reports, 26(11), 1362–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman, D.J. & Cragg, G.M. (2020) Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. Journal of Natural Products, 83(3), 770–803. [DOI] [PubMed] [Google Scholar]
- Nichols, D. , Cahoon, N. , Trakhtenberg, E.M. , Pham, L. , Mehta, A. , Belanger, A. et al. (2010) Use of Ichip for high‐throughput in situ cultivation of “uncultivable” microbial species. Applied and Environmental Microbiology, 76(8), 2445–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nord, C. , Bjerketorp, J. , Levenfors, J.J. , Cao, S. , Strömstedt, A.A. , Guss, B. et al. (2020) Isopedopeptins A‐H: cationic cyclic Lipodepsipeptides from Pedobacter cryoconitis UP508 targeting WHO top‐priority Carbapenem‐resistant bacteria. ACS Chemical Biology, 15(11), 2937–2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noren, B. & Raper, K. (1962) Antibiotic activity of myxobacteria in relation to their bacteriolytic capacity. Journal of Bacteriology, 84(1), 157–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neil, J. (2014) Antimicrobial resistance: tackling a crisis for the health and wealth of nations.
- Otten, H. (1986) Domagk and the development of the sulphonamides. The Journal of Antimicrobial Chemotherapy, 17(6), 689–696. [DOI] [PubMed] [Google Scholar]
- Oxford, A. (1947) Observations concerning the growth and metabolic activities of myxococci in a simple protein‐free liquid medium. Journal of Bacteriology, 53, 129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal, S. , Sharma, G. & Subramanian, S. (2021) Complete genome sequence and identification of polyunsaturated fatty acid biosynthesis genes of the myxobacterium Minicystis rosea DSM 24000T. BMC Genetics, 22(1), 655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantel, L. , Florin, T. , Dobosz‐Bartoszek, M. , Racine, E. , Sarciaux, M. , Serri, M. et al. (2018) Odilorhabdins, antibacterial agents that cause miscoding by binding at a new ribosomal site. Molecular Cell, 70(1), 83–94.e7. [DOI] [PubMed] [Google Scholar]
- Pogorevc, D. , Panter, F. , Schillinger, C. , Jansen, R. , Wenzel, S.C. & Müller, R. (2019) Production optimization and biosynthesis revision of corallopyronin a, a potent anti‐filarial antibiotic. Metabolic Engineering, 55, 201–211. [DOI] [PubMed] [Google Scholar]
- Poinar, G.O. (1966) The presence of Achromobacter Nematophilus in the infective stage of a Neoaplectana Sp. (Steinernematidae: Nematoda). Nematology, 12(1), 105–108. [Google Scholar]
- Poinar, G.O. & Thomas, G.M. (1967) The nature of Achromobacter nematophilus as an insect pathogen. Journal of Invertebrate Pathology, 9(4), 510–514. [Google Scholar]
- Präve, L. , Seyfert, C.E. , Bozhüyük, K.A.J. , Racine, E. , Müller, R. & Bode, H.B. (2024) Investigation of the Odilorhabdin biosynthetic gene cluster using NRPS engineering. Angewandte Chemie, 63(33), e202406389. [DOI] [PubMed] [Google Scholar]
- Proschak, A. , Zhou, Q. , Schöner, T. , Thanwisai, A. , Kresovic, D. , Dowling, A. et al. (2014) Biosynthesis of the insecticidal xenocyloins in Xenorhabdus bovienii . Chembiochem, 15(3), 369–372. [DOI] [PubMed] [Google Scholar]
- Racine, E. & Gualtieri, M. (2019) From Worms to drug candidate: the story of Odilorhabdins, a new class of antimicrobial agents. Frontiers in Microbiology, 10, 2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racine, E. , Nordmann, P. , Pantel, L. , Sarciaux, M. , Serri, M. , Houard, J. et al. (2018) In vitro and in vivo characterization of NOSO‐502, a novel inhibitor of bacterial translation. Antimicrobial Agents and Chemotherapy, 62(9), e01016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichenbach, H. (1999) The ecology of the myxobacteria. Environmental Microbiology, 1(1), 15–21. [DOI] [PubMed] [Google Scholar]
- Renard, S. , Versluys, S. , Taillier, T. , Dubarry, N. , Leroi‐Geissler, C. , Rey, A. et al. (2023) Optimization of the antibacterial Spectrum and the Developability profile of the novel‐class natural product Corramycin. Journal of Medicinal Chemistry, 66(24), 16869–16887. [DOI] [PubMed] [Google Scholar]
- Ringel, S.M. , Greenough, R.C. , Roemer, S. , Connor, D. , Gutt, A.L. , Blair, B. et al. (1977) Ambruticin (W7783), a new antifungal antibiotic. The Journal of Antibiotics, 30(5), 371–375. [DOI] [PubMed] [Google Scholar]
- Risch, T. , Kolling, D. , Mostert, D. , Seedorf, T. , Heimann, D. , Kohnhäuser, D. et al. (2024) Point mutations in the ygiV promoter region lead to cystobactamid resistance and reduced virulence in E. coli . NPJ Antimicrobial Resistance. doi: 10.1038/s44259-024-00050-7 [DOI] [Google Scholar]
- Saathoff, M. , Kosol, S. , Semmler, T. , Tedin, K. , Dimos, N. , Kupke, J. et al. (2023) Gene amplifications cause high‐level resistance against albicidin in gram‐negative bacteria. PLoS Biology, 21(8), e3002186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajnaga, E. & Kazimierczak, W. (2020) Evolution and taxonomy of nematode‐associated entomopathogenic bacteria of the genera Xenorhabdus and Photorhabdus: an overview. Symbiosis, 80(1), 1–13. [Google Scholar]
- Sarciaux, M. , Pantel, L. , Midrier, C. , Serri, M. , Gerber, C. , Marcia de Figueiredo, R. et al. (2018) Total synthesis and structure‐activity relationships study of Odilorhabdins, a new class of peptides showing potent antibacterial activity. Journal of Medicinal and Pharmaceutical Chemistry, 61(17), 7814–7826. [DOI] [PubMed] [Google Scholar]
- Schiefer, A. , Hübner, M.P. , Krome, A. , Lämmer, C. , Ehrens, A. , Aden, T. et al. (2020) Corallopyronin a for short‐course anti‐wolbachial, macrofilaricidal treatment of filarial infections. PLoS Neglected Tropical Diseases, 14(12), e0008930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergeant, M. , Baxter, L. , Jarrett, P. , Shaw, E. , Ousley, M. , Winstanley, C. et al. (2006) Identification, typing, and insecticidal activity of Xenorhabdus isolates from entomopathogenic nematodes in United Kingdom soil and characterization of the xpt toxin loci. Applied and Environmental Microbiology, 72(9), 5895–5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyfert, C.E. , Müller, A.V. , Walsh, D.J. , Birkelbach, J. , Kany, A.M. , Porten, C. et al. (2023) New genetically engineered derivatives of antibacterial Darobactins underpin their potential for antibiotic development. Journal of Medicinal and Pharmaceutical Chemistry, 66, 16330–16341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyfert, C.E. , Porten, C. & Müller, R. (2023) Darobactine: eine neue Antibiotikaklasse in Entwicklung. Biospektrum, 29(5), 539–541. [Google Scholar]
- Seyfert, C.E. , Porten, C. , Yuan, B. , Deckarm, S. , Panter, F. , Bader, C. et al. (2022) Darobactins exhibiting superior antibiotic activity by Cryo‐EM structure guided biosynthetic engineering. Angewandte Chemie, International Edition, 62(2), e202214094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, Y.‐M. , Hirschmann, M. , Shi, Y.‐N. , Ahmed, S. , Abebew, D. , Tobias, N.J. et al. (2022) Global analysis of biosynthetic gene clusters reveals conserved and unique natural products in entomopathogenic nematode‐symbiotic bacteria. Nature Chemistry, 14, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima, K. , Ledig, S. , Loeper, N. , Schiefer, A. , Pfarr, K. , Hoerauf, A. et al. (2018) Effective inhibition of rifampicin‐resistant chlamydia trachomatis by the novel DNA‐dependent RNA polymerase inhibitor corallopyronin a. International Journal of Antimicrobial Agents, 52(4), 523–524. [DOI] [PubMed] [Google Scholar]
- Singh, B.N. (1947) Myxobacteria in soils and composts; their distribution, number and lytic action on bacteria. Journal of General Microbiology, 1(1), 1–10. [DOI] [PubMed] [Google Scholar]
- Sucipto, H. , Pogorevc, D. , Luxenburger, E. , Wenzel, S.C. & Müller, R. (2017) Heterologous production of myxobacterial α‐pyrone antibiotics in Myxococcus xanthus . Metabolic Engineering, 44, 160–170. [DOI] [PubMed] [Google Scholar]
- Terlouw, B.R. , Blin, K. , Navarro‐Muñoz, J.C. , Avalon, N.E. , Chevrette, M.G. , Egbert, S. et al. (2023) MIBiG 3.0: a community‐driven effort to annotate experimentally validated biosynthetic gene clusters. Nucleic Acids Research, 51(D1), D603–D610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testolin, G. , Cirnski, K. , Rox, K. , Prochnow, H. , Fetz, V. , Grandclaudon, C. et al. (2020) Synthetic studies of cystobactamids as antibiotics and bacterial imaging carriers lead to compounds with high in vivo efficacy. Chemical Science, 70, 3–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaker, M.N. , Waglechner, N. & Wright, G.D. (2014) Antibiotic resistance–mediated isolation of scaffold‐specific natural product producers. Nature Protocols, 9, 1469 EP. [DOI] [PubMed] [Google Scholar]
- Thaker, M.N. , Wang, W. , Spanogiannopoulos, P. , Waglechner, N. , King, A.M. , Medina, R. et al. (2013) Identifying producers of antibacterial compounds by screening for antibiotic resistance. Nature Biotechnology, 31(10), 922–927. [DOI] [PubMed] [Google Scholar]
- van Santen, J.A. , Poynton, E.F. , Iskakova, D. , McMann, E. , Alsup, T.A. , Clark, T.N. et al. (2022) The natural products atlas 2.0: a database of microbially‐derived natural products. Nucleic Acids Research, 50(D1), D1317–D1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventola, C.L. (2015) The antibiotic resistance crisis: part 1: causes and threats. P T, 40(4), 277–283. [PMC free article] [PubMed] [Google Scholar]
- Waksman, S.A. & Schatz, A. (1945) Streptomycin–origin, nature, and properties*††journal series paper of the Department of Microbiology of the New Jersey agricultural Experiment Station, Rutgers university. Journal of the American Pharmaceutical Association, 34(11), 273–291. [Google Scholar]
- Walesch, S. , Birkelbach, J. , Jézéquel, G. , Haeckl, F.P.J. , Hegemann, J.D. , Hesterkamp, T. et al. (2022) Fighting antibiotic resistance‐strategies and (pre)clinical developments to find new antibacterials. EMBO Reports, 24, e56033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C.‐Y. , Hu, J.‐Q. , Wang, D.‐G. , Li, Y.‐Z. & Wu, C. (2024) Recent advances in discovery and biosynthesis of natural products from myxobacteria: an overview from 2017 to 2023. Natural Product Reports, 41, 905–934. [DOI] [PubMed] [Google Scholar]
- Wang, Z. , Koirala, B. , Hernandez, Y. , Zimmerman, M. & Brady, S.F. (2022) Bioinformatic prospecting and synthesis of a bifunctional lipopeptide antibiotic that evades resistance. Science, 376(6596), 991–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterfield, N.R. , Bowen, D.J. , Fetherston, J.D. , Perry, R.D. & ffrench‐Constant, R.H. (2001) The tc genes of Photorhabdus: a growing family. Trends in Microbiology, 9(4), 185–191. [DOI] [PubMed] [Google Scholar]
- Wenski, S.L. , Cimen, H. , Berghaus, N. , Fuchs, S.W. , Hazir, S. & Bode, H.B. (2020) Fabclavine diversity in Xenorhabdus bacteria. Beilstein Journal of Organic Chemistry, 16, 956–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . (2018) Global Antimicrobial Surveillance System (GLASS) Report 2016‐2017. https://apps.who.int/iris/bitstream/handle/10665/279656/9789241515061‐eng.pdf?ua=1
- Wright, G.D. (2017) Opportunities for natural products in 21st century antibiotic discovery. Natural Product Reports, 34(7), 694–701. [DOI] [PubMed] [Google Scholar]
- Wuisan, Z.G. , Kresna, I.D.M. , Böhringer, N. , Lewis, K. & Schäberle, T.F. (2021) Optimization of heterologous Darobactin a expression and identification of the minimal biosynthetic gene cluster. Metabolic Engineering, 66, 123–136. [DOI] [PubMed] [Google Scholar]
- Zaburannyi, N. , Bunk, B. , Maier, J. , Overmann, J. & Muller, R. (2016) Genome analysis of the fruiting body‐forming Myxobacterium Chondromyces crocatus reveals high potential for natural product biosynthesis. Applied and Environmental Microbiology, 82(6), 1945–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zborovsky, L. , Kleebauer, L. , Seidel, M. , Kostenko, A. , Eckardstein, L. , Gombert, F.O. et al. (2021) Improvement of the antimicrobial potency, pharmacokinetic and pharmacodynamic properties of albicidin by incorporation of nitrogen atoms. Chemical Science, 12(43), 14606–14617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Q. , Grundmann, F. , Kaiser, M. , Schiell, M. , Gaudriault, S. , Batzer, A. et al. (2013) Structure and biosynthesis of xenoamicins from entomopathogenic Xenorhabdus . Chemistry – A European Journal, 19(49), 16772–16779. [DOI] [PubMed] [Google Scholar]


