Abstract
Alzheimer's disease (AD) is a gradual neurodegenerative ailment that lacks any disease-modifying intervention. Our objective was to pinpoint pharmacological targets with a focus on amyloid beta (Aβ) and tau to treat and prevent AD in the European population. A proteome-wide Mendelian randomization (MR) analysis was carried out to estimate the associations between proteins and cerebrospinal fluid (CSF) Aβ-42 and phosphorylated Tau (p-Tau). We utilized colocalization and MR analysis to investigate whether the identified proteins were associated with the risk of AD. Additionally, we expanded our investigation to include non-AD phenotypes by conducting a phenome-wide MR analysis of 1646 disease traits based on the FinnGen and UK Biobank databases to explore potential side effects. We identified 11 proteins that were genetically associated with both CSF Aβ-42 and p-Tau levels. The genetically predicted levels of three proteins, GAL3ST2, POLR1C, and BIN1, were found to be associated with an increased risk of AD with high colocalization. In the phenome-wide MR analysis, two out of the three biomarkers were associated with at least one disease, except for GAL3ST2, which was not associated with any disease under the threshold of FDR <0.1. POLR1C was found to be associated with the most disease traits, and all disease associations with genetically inhibited BIN1 were protective. The proteome-wide MR investigation revealed 11 proteins that were associated with the level of CSF Aβ-42 and p-Tau. Among them, GAL3ST2, POLR1C, and BIN1 were identified as potential therapeutic targets for AD and warrant further investigation.
Keywords: Alzheimer's disease, Mendelian randomization, Beta-amyloid, Phosphorylated tau
1. Introduction
Alzheimer's disease (AD) is becoming increasingly prevalent worldwide as the population ages. The current estimate is that the global population of individuals with AD dementia is 57.4 million, and this number is projected to increase to 152.8 million by the year 2050 [1]. The rise in AD has significant social, economic, family, and individual implications. There is an urgent need for therapeutic interventions that can prevent or delay the onset, slow down the progression, or improve the symptoms of AD.
While the molecular mechanisms of AD are not yet fully understood, there are two primary pathological features of the disease: the presence of extracellular beta-amyloid (Aβ) plaques and intracellular phosphorylated Tau (p-Tau) containing neurofibrillary tangles [2,3]. Aβ peptides are generated and released during regular synaptic activity in the brain through the catabolism of the amyloid precursor protein by secretases located in the neuronal membrane. In a healthy brain, these fragments would undergo degradation and subsequent elimination from the organism. Nevertheless, in AD, their breakdown is insufficient, leading to their accumulation and the subsequent formation of one of the distinctive features of the disease, namely amyloid plaques [4]. Aβ-42, one of the most pernicious and amyloidogenic variants of the peptide, plays a pivotal role in inducing cellular harm [5]. The Tau protein, an essential component of neuronal stability, actively contributes to the preservation of microtubules, which constitute a crucial element of the neuronal cytoskeleton [6]. Upon post-translational modification through hyperphosphorylation, the tau protein dissociates from microtubules, resulting in its self-aggregation and subsequent disassembly of the microtubule structure [7]. Consequently, amyloid β and tau emerge as the most auspicious therapeutic targets for modifying the progression of AD.
Basic research, preclinical studies, and clinical trials have elucidated the crucial role of amyloid β and tau in the pathogenesis of AD [8]. Despite this, the therapeutic targeting of amyloid β has not yet demonstrated convincing efficacy in the early stage [9]. Currently, Lecanemab, a humanized IgG1 monoclonal antibody that binds with high affinity to Aβ soluble protofibrils, has shown promise in reducing amyloid markers in early Alzheimer's disease. Over an 18-month period, it resulted in a moderate reduction in cognitive and functional decline compared to placebo. However, its use was also associated with adverse events [10]. The limited advancement in drug development could potentially be attributed to inadequate trial design, concerns about adverse events, and uncertainties surrounding the monitoring of cognitive function during follow-up assessments. Mendelian randomization (MR) is a well-established method in genetic epidemiology that enhances the ability to draw causal inferences about relationships between exposure and outcome. By leveraging genetic variants as instrumental variables, MR minimizes the impact of confounding factors and helps exclude potential reverse causality [11]. While MR has primarily been employed to investigate causal relationships between biomarkers and disease phenotypes, it also has an extension that leverages genotypes to verify drug targets [[12], [13], [14]].
Proteins are indisputably the paramount contributors to all biological processes, whether in the context of disease or health. The human plasma proteome comprises proteins that are either secreted or shed into circulation with the purpose of either executing their functions within the bloodstream or facilitating inter-tissue communication. The dysregulation of these proteins is a common occurrence in various diseases, rendering them pivotal targets for pharmacological intervention [15]. Consequently, in conjunction with the inherent accessibility of blood when compared to other tissues, circulating proteins emerge as an alluring reservoir for discerning molecular signatures of disease within expansive cohorts.
In this study, we aimed to identify the potentially causal circulating proteins and novel safety drug targets of Aβ-42 and p-Tau by means of an MR-based analytical framework, with the goal of providing insights into the prevention and treatment of AD.
2. Methods
2.1. Study design and ethics
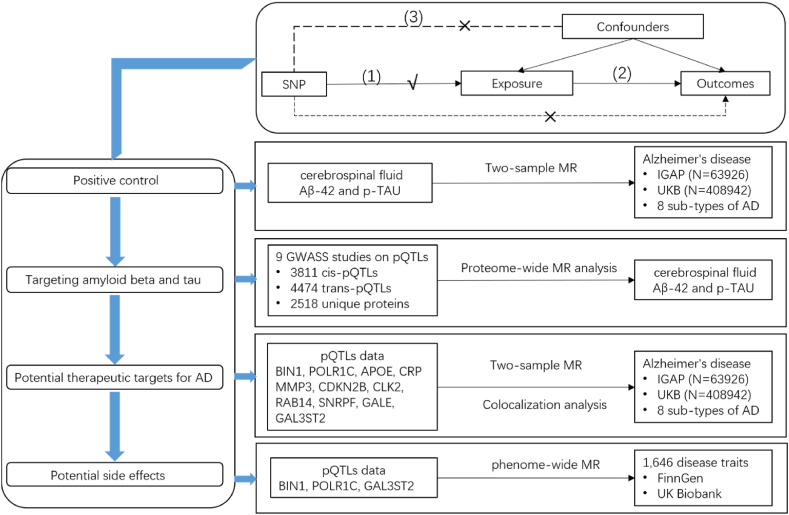
The study design is presented in Fig. 1. This genetic association study exclusively relied on published summary data from studies involving human participants who had provided written informed consent and received approval from their respective institutional ethics review committees.
Fig. 1.
Overview of the study design in our Mendelian randomization study. Three assumptions of MR analysis are as follows: (1) SNPs must be associated with the exposure, (2) SNPs not directly be associated with any outcomes, and (3) SNPs must not be associated with any confounding factor.
2.2. CSF Aβ-42 and p-tau data source
Genetic predictors of cerebrospinal fluid (CSF) Aβ-42 and p-Tau level were derived from the latest meta GWAS involving 8074 individuals from 16 European cohorts (Table S1) [16]. Aβ-42 was quantified with ELISA, Lumipulse or V-PLEX, while p-Tau was assessed using ELISA or Lumipulse. The normalized protein levels were employed as continuous phenotypes in the association analysis. Genetic associations were adjusted for gender, age, and the first 10 principal components.
To verify the GWASs of CSF Aβ-42 and p-Tau level used were causally associated with AD, we first investigate the association between genetically predicted CSF Aβ-42 and p-Tau level and AD as a positive control. Single nucleotide polymorphisms (SNPs) genome-wide significantly (P < 5 × 10−8) and independently (r2 < 0.01) associated with CSF Aβ-42 or p-Tau level were used as genetic instrumental variables. In order to verify the reliability of these selected SNPs as genetic instruments, we computed the F-statistic using a well-established formula, and all the SNPs exhibited F-statistics exceeding 10 (Table S2).
2.3. Proteomic data source
The variables investigated in this study consisted of genetically forecasted circulating proteins, procured from nine genome-wide association studies [15,[17], [18], [19], [20], [21], [22], [23], [24]]. Further information is presented in Table S1. For the selection of genetic instruments for each of the circulating proteins, genome-wide significant SNPs (p-value thresholds were based on the corresponding study) were extracted, and SNP harbouring the human leukocyte antigen (HLA) region (chr6:27,477,797–34,448,354, hg19/GRCh37) were removed. Clumping parameters were used to select independent protein quantitative trait loci (pQTLs) for each protein (clumping window of 5000 kb, LD r2 cutoff 0.01). Additionally, instruments linked to five or more proteins were omitted due to their high pleiotropic nature. In this study, we categorized instruments into cis-pQTLs (less than 500 kb) and trans-pQTLs (more than 500 kb) based on the window across the corresponding protein-coding sequences. Following the screening process, A total of 8285 pQTLs (3811 cis-pQTLs and 4474 trans-pQTLs) for 2518 unique proteins were retained as instrumental variables for the MR analysis.
2.4. AD data source
The data on Alzheimer's disease (AD) were derived from the International Genomics of Alzheimer's Project (IGAP) Genome-wide association studies (GWAS) Stage 1 result (N = 21,982 cases, 41,944 controls) [25]. The AD cases included in the study were either confirmed through autopsy or clinically validated using established criteria. The association analysis was adjusted for age, sex, and population substructure using principal components. Furthermore, we obtained an independent full summary level of Alzheimer's disease or family history of Alzheimer's disease data from the UK Biobank for validation, which consisted of 53,042 individuals and 355,900 controls [26]. AD cases were extracted from the UKB self-report ICD-10 (International Classification of Diseases, tenth revisions) diagnoses and ICD-10 cause of death data.
Furthermore, several subtypes of AD data were obtained in our study for sensitive analysis, including AD of early onset, AD of late-onset, family history of AD, paternal history of AD, maternal history of AD, illnesses of father: AD, illnesses of mother: AD and illnesses of siblings: AD. The details are presented in Table S3.
2.5. Sources of phenome-wide MR analysis of 1646 disease traits
To conduct the phenome-wide MR analysis, GWAS data on clinical health outcomes were procured from the FinnGen Consortium and UK Biobank, utilizing the MR-base database. In the FinnGen Consortium database, cases were defined based on electronic health records in accordance with ICD-9 or ICD-10 codes. We excluded the data of sex-specific outcomes and outcomes with fewer than 500 cases due to the unavailability of data and limited statistical power, respectively. The GWAS results were adjusted for age, sex, first ten principal components, and genotyping batch. For the UK biobank dataset, disease outcomes were extracted from the MR-base database based on ICD-10 diagnosis, and GWAS analysis was adjusted for age, sex, age∗sex, age∗age, age∗age∗sex and the first ten principal components. The study incorporated a comprehensive total of 1646 non-AD diseases in the final.
2.6. MR analysis
Following the extraction and harmonization of genetic instruments, we proceeded with the MR analysis. Once the eligible instrumental variables were selected, they were extracted from the outcome trait and harmonized across both the exposure and outcome GWAS. During this process, palindromic SNPs with intermediate allele frequencies were excluded to avoid ambiguity in allele assignment. Additionally, if a particular SNP was absent from the outcome GWAS, a proxy SNP in linkage disequilibrium with the target SNP (r2 ≥ 0.8, defined using 1000 Genomes European sample data) was identified and used as a substitute. After harmonizing the exposure and outcome datasets, MR analysis was conducted. The odds ratios (ORs) and their corresponding confidence intervals (CIs) on the associations between exposure and outcomes were calculated utilizing the inverse-variance weighted (IVW) and Wald ratios method. Additionally, we performed a sensitivity analysis using three additional MR methods. The weighted median method is an approach that is based on the median and can offer consistent estimates even when up to 50 % of the SNPs are invalid instrumental variables. MR-Egger introduces an intercept term to effectively control for the potential influence of invalid instrumental variables. The weighted mode categorizes the SNPs into groups based on their estimated causal effects and evaluates the evidence for causality using solely the largest set of SNPs. This approach essentially relaxes the assumptions of MR and possesses the capability to accurately identify the true effect even if a majority of instruments are invalid SNPs. Furthermore, we performed MR‐PRESSO and leave‐one‐out analyses to evaluate the robustness of the causality by removing potentially influential SNPs. All MR analyses were carried out using the “Mendelian Randomization” and “MR‐PRESSO” R packages. In the proteome-wide MR analysis, we considered a p-value of 0.05 and a Bonferroni-corrected p-value of May 0, 2396 = 2.09E-05 as significant. In the phenome-wide MR analysis, false discovery rate (FDR)-corrected p-values were calculated within each protein-outcome combination to correct for multiple testing, and we consider FDR <0.1 significant.
2.7. Colocalization analysis
Colocalization analyses are used to determine whether two genetic traits share a common causal variant at a particular locus. In the context of genomic studies, colocalization analyses are frequently applied to investigate the overlap between genetic signals associated with complex traits and molecular traits. This is especially useful in identifying whether the same genetic variant influences both the trait of interest and the molecular trait. In our study, we performed colocalization analysis using the coloc R package to evaluate the probability of AD risk loci and pQTL shared by the same causal signal. For each locus, the Bayesian method assessed the support for the following five exclusive hypotheses: (H0) no association with either trait; (H1) association with proteins only; (H2) association with AD only; (H3) both traits are associated, but distinct causal variants were for two traits; and (H4) both traits are associated, and the same shares causal variant for both traits. We assigned the default prior probabilities for an SNP being associated with stroke (p1 = 1 × 10−4), an SNP is a significant pQTL (p2 = 1 × 10−4) and an SNP being associated with both traits (p12 = 1 × 10−5). We used a filter of PP.H3+PP.H4 ≥ 0.8 to select potential drug targets with evidence of independent or colocalized AD GWAS and pQTL signals [27], and PP.H4 > 0.8 as strong evidence of colocalization.
3. Results
3.1. Genetically predicted CSF Aβ-42 and p-tau level were associated with the risk of AD
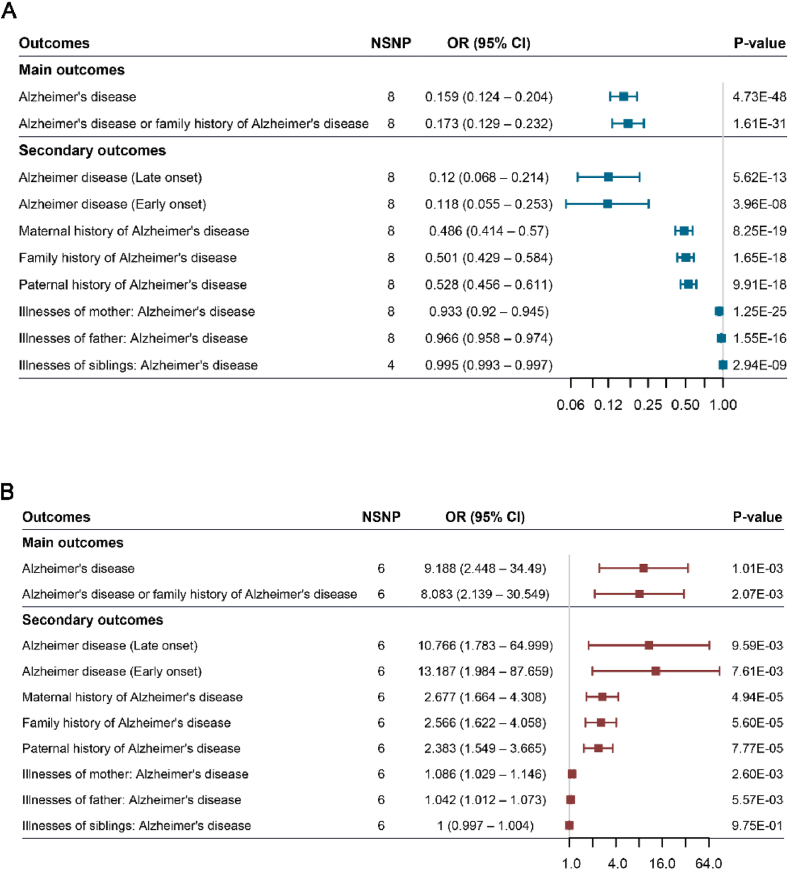
To validate the reliability of GWAS data of CSF Aβ-42 and p-Tau level, we designed a positive control study using the risk of AD as additional outcome traits in our MR models. We replicated the previous result that genetically predicted CSF Aβ-42 level was inversely associated with the risk of AD in two datasets (discovery cohort: OR = 0.159, 95 % CI: 0.124–0.204, p = 4.73E-48; validation cohort: OR = 0.173, 95 % CI: 0.129–0.232, p = 1.61E-31, Fig. 2A, Table S4). Additionally, genetically proxied CSF Aβ-42 level was negatively associated with subtypes of AD, including late onset of AD (OR = 0.12, 95 % CI: 0.068–0.214, p = 5.62E-13), early onset of AD (OR = 0.118, 95 % CI: 0.055–0.253, p = 3.96E-8), family history of AD (OR = 0.501, 95 % CI: 0.429–0.584, p = 1.65E-18).
Fig. 2.
Forest plot Mendelian randomization effect estimates and 95 % confidence intervals for the genetic proxied cerebrospinal fluid Aβ-42 (A), p-Tau (B) and the risk of Alzheimer's disease.
On the other hand, we also confirmed that higher CSF p-Tau were genetically related to a higher risk of AD (discovery cohort: OR = 9.188, 95 % CI: 2.448–34.49, p = 1.01E-03; validation cohort: OR = 8.083, 95 % CI: 2.139–30.55, p = 2.07E-03, Fig. 2B–Table S4) and some types of AD. As an example, the OR of disease per additional standard unit of genetically predicted CSF p-Tau concentration was 2.566 (95 % CI: 1.622–4.058, p = 5.60E-05) for family history of AD, 2.677 (95 % CI: 1.664–4.308, p = 4.94E-05) for maternal history of AD, and 2.383 (95 % CI: 1.549–3.665, p = 7.77E-05) for paternal history of AD.
Horizontal pleiotropy was not detected in this analysis (Tables S5–6, Figs. S1–2). Together, the presents confirm that the summary data of CSF Aβ-42 and p-Tau level are reliable and the result in the previous studies is reproducible.
3.2. Identification of proteins genetically associated with both CSF Aβ-42 and p-tau levels
We curated pQTLs from nine proteomic GWASs to construct genetic instruments. After the screening and harmonizing of the CSF GWAS data, a total of 6234 pQTLs for 2396 unique proteins were retained as instruments for MR analysis, including 3122 cis-pQTLs for 1491 unique proteins and 3112 cis-pQTLs for 1319 unique proteins.
The study examined the MR associations between 2396 proteins and the risk of CSF Aβ-42 and p-Tau level. The overview of the results of MR analysis is presented in Table 1. We identified 11 proteins genetically associated with both CSF Aβ-42 and p-Tau levels after Bonferroni-corrected.
Table 1.
The result of proteome-wide mendelian randomization analysis.
| Uniprot | Protein | Gene symbol | p-Tau |
Aβ42 |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Beta | S.e. | p-value | Beta | S.e. | p-value | cis/trans | |||
| O00499 | Myc box-dependent-interacting protein 1 | BIN1 | 0.468 | 0.107 | 1.25E-05 | −0.505 | 0.105 | 1.64E-06 | cis |
| O15160 | DNA-directed RNA polymerases I and III subunit RPAC1 | POLR1C | 1.399 | 0.071 | 1.58E-85 | −2.545 | 0.066 | 0 | trans |
| P02649 | Apolipoprotein E | APOE | 0.681 | 0.033 | 4.41E-94 | −1.280 | 0.031 | 0 | cis |
| P02741 | C-reactive protein | CRP | −1.231 | 0.107 | 8.62E-31 | 2.300 | 0.311 | 1.40E-13 | trans |
| P08254 | Stromelysin-1 | MMP3 | −1.174 | 0.085 | 1.38E-43 | 2.275 | 0.080 | ###### | trans |
| P42772 | Cyclin-dependent kinase 4 inhibitor B | CDKN2B | −1.877 | 0.341 | 3.78E-08 | Not available | trans | ||
| P49760 | CDC-Like Kinase 2 | CLK2 | 1.551 | 0.079 | 1.58E-85 | −2.820 | 0.074 | 0 | trans |
| P61106 | Ras-related protein Rab-14 | RAB14 | −0.624 | 0.045 | 1.38E-43 | 1.209 | 0.043 | ###### | trans |
| P62306 | Small nuclear ribonucleoprotein F | SNRPF | −0.656 | 0.032 | 4.41E-94 | 1.233 | 0.030 | 0 | trans |
| Q14376 | UDP-glucose 4-epimerase | GALE | −1.814 | 0.131 | 1.38E-43 | 3.516 | 0.124 | ###### | trans |
| Q9H3Q3 | Galactose-3-O-sulfotransferase 2 | GAL3ST2 | 1.425 | 0.088 | 2.31E-59 | −2.631 | 0.083 | ###### | trans |
Genetically predicted high level of Apolipoprotein E (APOE), Myc box-dependent-interacting protein 1 (BIN1), DNA-directed RNA polymerases I and III subunit RPAC1 (POLR1C), CDC-Like Kinase 2 (CLK2) and Galactose-3-O-sulfotransferase 2 (GAL3ST2) were significantly associated with an increased level of CSF p-Tau (APOE: OR = 1.98, 95 % CI: 1.851–2.108; BIN1: OR = 1.597, 95 % CI: 1.294–1.97; POLR1C: OR = 4.053, 95 % CI: 3.524–4.662; CLK2: OR = 4.715, 95 % CI: 4.037–5.506; GAL3ST2: OR = 4.16, 95 % CI: 3.502–4.94), but associated with a decreased level of CSF Aβ-42 (APOE: OR = 0.278, 95 % CI: 0.262–0.295; BIN1: OR = 0.604, 95 % CI: 0.491–0.742; POLR1C: OR = 0.078, 95 % CI: 0.069–0.089; CLK2: OR = 0.06, 95 % CI: 0.052–0.069; GAL3ST2: OR = 0.072, 95 % CI: 0.061–0.085).
In addition, genetically predicted higher levels of six circulating proteins including C-reactive protein (CRP), Ras-related protein rab-14 (RAB14), stromelysin-1 (MMP3), cyclin-dependent kinase 4 inhibitor B (CDKN2B), UDP-glucose 4-epimerase (GALE) and small nuclear ribonucleoprotein F (SNRPF) were significantly associated with a decreased level of CSF p-Tau, but associated with an increased level of CSF Aβ-42.
3.3. Associations between targeted proteins and the risk of AD
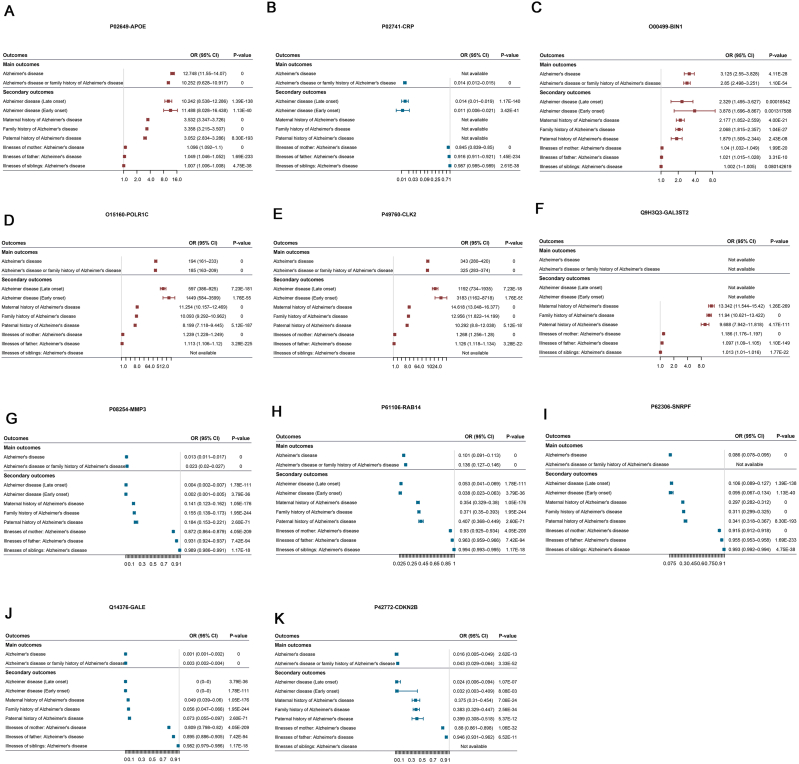
Using proteome-wide MR analysis, we identified 11 potential proteins targeting CSF Aβ-42 and p-Tau. We then tested whether these targets were related to the risk of AD. Interestingly, three of them had been reported to be associated with the risk AD previously, including APOE, CRP and BIN1, which had been replicated in our MR analysis (Fig. 3A–C and Table S7).
Fig. 3.
Forest plot Mendelian randomization effect estimates and 95 % confidence intervals for the genetic proxied identified circulating proteins and the risk of Alzheimer's disease. (A) APOE. (B) CRP. (C) BIN1. (D) POLR1C. (E) CLK2. (F) GAL3ST2. (G) MMP3. (H) RAB14. (I) SNRPF. (J) GLAE. (K) CDKN2B.
Of the rest eight novel proteins, we observed significant associations of higher genetically predicted levels of POLR1C, CLK2 and GAL3ST2 with increased risk of any AD (Fig. 3D–F, Table S7), of which direction was in accordance with proteome-wide MR analysis. Additionally, the negative associations of genetically predicted SNRPF, CDKN2B, RAB14, GALE and MMP3 levels with the risk of AD are shown in Fig. 3G–K.
We then performed colocalization analysis to examine whether the AD risk was controlled by the same genetic variants that influence protein levels. Genetic colocalization analysis on RAB14, MMP3 and GALE resulted in a large posterior of H3+H4 probability, indicating that multiple variants control AD susceptibility and the plasma protein levels (Table S8). Additionally, we found strong evidence of colocalization for GAL3ST2, POLR1C and BIN1 (PP.H4>0.95).
3.4. Associations between targeted proteins and the risk of 1646 disease traits
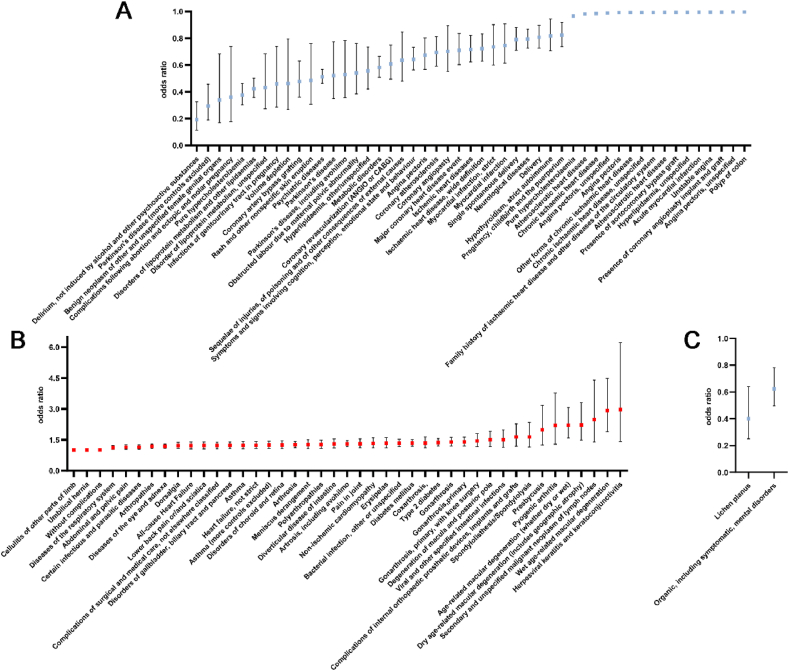
We have identified genetically predicted levels of three proteins including GAL3ST2, POLR1C and BIN1 associated with an increased risk of AD with high colocalization, indicating inhibitions of them could be therapeutic targets for AD. We then expanded the exploration of side effects to include non-AD phenotypes by performing an agnostic phenome-wide MR (Phe-MR) analysis of 1646 disease traits based on FinnGen and UK Biobank database, and the results are presented in Fig. 4A–C and Tables S9–10.
Fig. 4.
Forest plot shows the significant associations between genetical inhibitions of POLR1C (A, B), BIN1 (C) and numerous disease outcomes using phenome-wide MR analysis.
Two of three biomarkers were associated with at least one disease except for GAL3ST2, which was associated with none under the threshed of FDR <0.1. POLR1C was associated with the most disease traits (94, Figure A–B), and all disease associations with genetic inhibition of BIN1 were protective, including “organic, including symptomatic, mental disorders” (OR = 0.624, 95 % CI: 0.498–0.782, FDR = 0.07) and lichen planus (OR = 0.402, 95 % CI: 0.252–0.64, FDR = 0.099, Fig. 4C). The top three significant protective disease associations for POLR1C were psychiatric diseases (OR = 0.513, 95 % CI: 0.463–0.567, FDR = 9.23E-36), disorders of lipoprotein metabolism and other lipidaemia (OR = 0.423, 95 % CI: 0.358–0.501, FDR = 1.05E-21) and atherosclerotic heart disease (OR = 0.983, 95 % CI: 0.978–0.987, FDR = 1.63E-11, Fig. 4A). However, the most harmful disease associations for POLR1C could never be neglected, including Wet age-related macular degeneration (OR = 2.916, 95 % CI: 1.899–4.477, FDR = 8.9E-05), type 2 diabetes (OR = 1.372, 95 % CI: 1.205–1.562, FDR = 1.45E-04), and arthrosis (OR = 1.305, 95 % CI: 1.162–1.466, FDR = 3.91E-04, Fig. 4B).
4. Discussion
Our study employed an integrative approach combining proteome-wide MR and colocalization to evaluate the causal effects of thousands of plasma proteins from nine large proteomics GWAS on CSF Aβ-42 and p-Tau levels and to provide potential drug targets for AD. In summary, based on the MR analyses, we identified 11 distinct proteins that exhibited a causal association with CSF Aβ-42 and p-Tau level. Furthermore, by conducting colocalization analysis to account for the potential effects of linkage disequilibrium, we obtained compelling evidence indicating that genetically predicted elevated levels of GAL3ST2, POLR1C, and BIN1 were significantly associated with an increased risk of AD.
Our findings align with previous studies that have reported similar associations between APOE and BIN1 activities and AD. GWASs have identified the BIN1 and APOE genes as part of the most prevalent susceptibility locus for late-onset AD [28]. Most of the BIN1 variants associated with late-onset AD are situated several kilobases upstream of the BIN1 coding region. These variants have been postulated to elevate the risk of Alzheimer's disease by modulating the expression of cellular BIN1 within the cells [29,30]. In the brains affected by AD, elevated BIN1 mRNA expression levels have been linked to delayed onset of the disease and shorter duration of the disease [31]. Furthermore, neuronal loss was observed in BIN1 neuronal conditional knockout mice in the context of tauopathy [32], suggesting BIN1 may play a role in tau processing. The current study revealed that the attenuation of hippocampal tauopathy and neuroinflammation was observed following the loss of BIN1, which is akin to the findings reported on the modulation of tau pathology by the loss of expression of two genes associated with Alzheimer's disease, ApoE and Trem2 [33].
BIN1 has been extensively studied, and there is substantial evidence indicating its involvement in several AD-related pathways, including tau and amyloid pathologies. Emerging research has highlighted BIN1's role in Aβ generation, specifically its involvement in BACE1 proteolytic activity. BIN1 plays a critical role in the endocytic transport of BACE1 within endosomes, which is essential for regulating the amyloidogenic pathway of APP and BACE1. This pathway typically restricts the excessive production of Aβ. However, BIN1 knockdown in neurons disrupts the transport of BACE1 out of early endosomes, causing BACE1 to accumulate and increasing interactions between APP and BACE1, ultimately leading to elevated Aβ levels [29,34]. In addition to its role in amyloid pathology, BIN1 has been implicated in other pathways relevant to AD, such as inflammation, apoptosis, and calcium homeostasis [35,36]. Neuronal loss is a hallmark of AD pathology. Although research on BIN1-related apoptosis in AD remains limited, studies in oncology have shown that abnormal splicing or reduced levels of BIN1 are strongly associated with poor prognosis and increased tumor metastasis. Low levels of BIN1 enhance apoptosis resistance through the upregulation of c-FLIP. Thus, BIN1-mediated apoptosis, which is crucial in cancer biology, likely plays a significant role in AD as well.
POLR1C plays a role in rRNA synthesis, with the nucleolus being a critical site for this process. Research has shown that nucleolar dysfunction is closely associated with various neurodegenerative diseases, including AD. Neuronal damage in AD may be accompanied by nucleolar dysfunction, which affects protein synthesis capacity. Impaired POLR1C function could potentially lead to nucleolar dysfunction, exacerbating the degenerative changes observed in neurons [37,38]. In our study, we not only found the associations between PLOR1C level and psychiatric diseases, such as AD, delirium and Parkinson's disease but also a strong relationship with metabolic disorders, such as hypercholesterolemia, type 2 diabetes and heart diseases.
GAL3ST2 is involved in glycosylation and sulfation, processes that are essential for the proper functioning of proteins and lipids in the brain. These biochemical modifications play a role in maintaining the integrity of the extracellular matrix and facilitating cell-to-cell communication. Abnormal glycosylation and sulfation have been linked to the pathogenesis of neurodegenerative diseases. While the direct role of GAL3ST2 in AD is not yet fully understood, its involvement in sulfation, lipid metabolism, and inflammation suggests that it could influence AD progression in ways that warrant further investigation [39].
Several potential drug targets also need to be emphasized, though we did not find strong evidence of colocalization for AD. CLK2 belongs to the cdc2-like kinase (CLK) family. The CLKs are responsible for regulating the alternative splicing of microtubule-associated protein tau and are linked to frontotemporal dementia and Parkinson's disease [40]. A recent review has emphasized the role of CLKs in the pathophysiology of AD and the therapeutic potential of targeting CLK1, in particular, in AD drug discovery and development [41]. Similarly, CLK2 has also been suggested as a prospective drug target for AD [42]. Matrix metalloproteinases (MMPs) have been shown to have a significant role in the development of AD and other neurodegenerative conditions, and our study specifically focused on MMP3 as well. MMP-3 is upregulated in the dorsomedial prefrontal cortex throughout the preclinical and clinical stages of AD [43]. Dysregulated MMP-3 activity appears to exacerbate numerous neurodegenerative disorders by disrupting the integrity of the blood-brain barrier, promoting demyelination and apoptosis, and triggering additional inflammatory responses. In AD, MMPs have been implicated in the degradation of Aβ and tau, with MMP3 being one of the key enzymes involved in tau degradation. Additionally, several reports indicated that MMP3 is beneficial to the recovery process in the central nervous system [44]. Therefore, MMP3 could also serve as a potential therapeutic target for AD.
Mendelian randomization is an approach that uses the unique properties of genotypes to investigate causal associations. Since genetic variants are randomly assigned at conception, they are not influenced by reverse causation. In the absence of pleiotropy (ie, genetic variants being associated with the disease through alternative pathways) and population stratification, they can provide unconfounded estimates of disease risk. However, the MR study is essentially a genetic association study, and MR alone could not provide the full picture due to its inherent limitations, such as the potential for pleiotropy and population stratification. So, more evidence needs to be provided to support the claims, such as in silico experiments.
One of the strengths of this study is the utilization of Mendelian randomization (MR) and colocalization analyses, which leverage genetic variants to estimate potential drug targets for AD. The MR design helps to minimize biases stemming from confounding variables and reverse causality, thereby enhancing the reliability of causal inferences made in the study. Colocalization analysis has been demonstrated to be a powerful tool in identifying the pleiotropic effects of specific loci on multiple traits. Additionally, we employed more than one AD dataset to reinforce our findings.
It is important to note several limitations of our analysis. Firstly, horizontal pleiotropy cannot be entirely ruled out, despite the implementation of various sensitivity analyses to test the assumptions of MR analyses. Secondly, although we employed phenome-wide MR to investigate the potential side effects of the drug targets, clinical trials are necessary to assess their effectiveness and safety for the early management of AD. Thirdly, since our results were restricted to individuals of European ancestry, they may not be generalizable to other ethnic groups. Fourthly, pQTL studies typically focus on individual proteins rather than protein-protein interactions or complexes. Consequently, important pathways involving interactions between multiple proteins might be overlooked if the study design does not account for these collaborations.
5. Conclusion
In summary, this study utilized an integrated genetic approach to identify 11 proteins that are associated with the levels of CSF Aβ-42 and p-Tau. Among these proteins, GAL3ST2, POLR1C, and BIN1 were found to be significantly associated with an increased risk of AD and exhibited high colocalization. These findings prioritize them as potential drug targets for AD, which should be further validated in future clinical trials.
Ethics approval and consent to participate
This genetic association study exclusively relied on published summary data from studies involving human participants who had provided written informed consent and received approval from their respective institutional ethics review committees.
Consent for publication
All participants had signed informed written consent.
Availability of data and materials
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Funding
This work was supported by grants from the National Natural Science Foundation of China (82271234, 82060219); Jiangxi Province thousands of Plans (jxsq2019201023); Youth Team Project of the Second Affiliated Hospital of Nanchang University (2019YNTD12003).
CRediT authorship contribution statement
Xifeng Wang: Writing – original draft, Visualization, Software, Resources, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Huayu Yang: Software, Resources, Methodology, Data curation. Dengcheng Zhan: Visualization, Validation, Software, Data curation. Haiying Sun: Resources, Data curation. Qiang Huang: Validation, Software. Yiping Zhang: Software, Resources. Yue Lin: Resources, Methodology. Gen Wei: Software, Resources. Fuzhou Hua: Validation, Supervision, Investigation, Funding acquisition, Conceptualization. Li Liu: Visualization, Validation, Supervision, Investigation, Conceptualization. Shibiao Chen: Writing – review & editing, Validation, Supervision, Conceptualization.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:Fuzhou hua reports financial support was provided by National Natural Science Foundation of China. Fuzhou hua reports financial support was provided by Jiangxi Province thousands of Plans. Fuzhou hua reports financial support was provided by Youth Team Project of the Second Affiliated Hospital of Nanchang University. If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank the UK Biobank, MR-Base platform, FinnGen consortium, the participants, and all the investigators for making summary statistics publicly accessible for this analysis.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e39013.
Contributor Information
Li Liu, Email: 372808563@qq.com.
Shibiao Chen, Email: ndyfy00763@ncu.edu.cn.
Abbreviations
- AD
Alzheimer's disease
- APOE
Apolipoprotein E
- Aβ
amyloid beta
- BIN1
Myc box-dependent-interacting protein 1
- CDKN2B
cyclin-dependent kinase 4 inhibitor B
- CIs
confidence intervals
- CLK2
CDC-Like Kinase 2
- CRP
C-reactive protein
- CSF
cerebrospinal fluid
- FDR
false discovery rate
- GAL3ST2
Galactose-3-O-sulfotransferase 2
- GALE
UDP-glucose 4-epimerase
- GWAS
Genome-wide association study
- HLA
human leukocyte antigen
- IVW
inverse-variance weighted
- MR
mendelian randomization
- ORs
odds ratios
- POLR1C
DNA-directed RNA polymerases I and III subunit RPAC1
- pQTLs
protein quantitative trait loci
- p-Tau
phosphorylated Tau
- RAB14
Ras-related protein rab-14
- SNP
single nucleotide polymorphism
- SNRPF
small nuclear ribonucleoprotein F
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- 1.Collaborators G.B.D.D.F. Estimation of the global prevalence of dementia in 2019 and forecasted prevalence in 2050: an analysis for the Global Burden of Disease Study 2019. Lancet Public Health. 2022;7(2):e105–e125. doi: 10.1016/S2468-2667(21)00249-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kosik K.S., Joachim C.L., Selkoe D.J. Microtubule-associated protein tau (tau) is a major antigenic component of paired helical filaments in Alzheimer disease. Proc. Natl. Acad. Sci. U.S.A. 1986;83(11):4044–4048. doi: 10.1073/pnas.83.11.4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glenner G.G., Wong C.W. Alzheimer's disease: initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem. Biophys. Res. Commun. 1984;120(3):885–890. doi: 10.1016/s0006-291x(84)80190-4. [DOI] [PubMed] [Google Scholar]
- 4.Kang I.J., et al. Butanol extract of Ecklonia cava prevents production and aggregation of beta-amyloid, and reduces beta-amyloid mediated neuronal death. Food Chem. Toxicol. 2011;49(9):2252–2259. doi: 10.1016/j.fct.2011.06.023. [DOI] [PubMed] [Google Scholar]
- 5.Pijnenburg Y.A., Schoonenboom N.S., Scheltens P. Tau and Abeta42 protein in CSF of patients with frontotemporal degeneration. Neurology. 2003;60(2):353–354. doi: 10.1212/wnl.60.2.353-a. author reply 353-354. [DOI] [PubMed] [Google Scholar]
- 6.Avila J., et al. Role of tau protein in both physiological and pathological conditions. Physiol. Rev. 2004;84(2):361–384. doi: 10.1152/physrev.00024.2003. [DOI] [PubMed] [Google Scholar]
- 7.Arendt T., Stieler J.T., Holzer M. Tau and tauopathies. Brain Res. Bull. 2016;126(Pt 3):238–292. doi: 10.1016/j.brainresbull.2016.08.018. [DOI] [PubMed] [Google Scholar]
- 8.Sevigny J., et al. The antibody aducanumab reduces Abeta plaques in Alzheimer's disease. Nature. 2016;537(7618):50–56. doi: 10.1038/nature19323. [DOI] [PubMed] [Google Scholar]
- 9.Kuller L.H., Lopez O.L. ENGAGE and EMERGE: truth and consequences? Alzheimers Dement. 2021;17(4):692–695. doi: 10.1002/alz.12286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Dyck C.H., et al. Lecanemab in early Alzheimer's disease. N. Engl. J. Med. 2023;388(1):9–21. doi: 10.1056/NEJMoa2212948. [DOI] [PubMed] [Google Scholar]
- 11.Sheehan N.A., et al. Mendelian randomisation and causal inference in observational epidemiology. PLoS Med. 2008;5(8):e177. doi: 10.1371/journal.pmed.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y., et al. Genetic insights into therapeutic targets for aortic aneurysms: a Mendelian randomization study. EBioMedicine. 2022;83 doi: 10.1016/j.ebiom.2022.104199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Storm C.S., et al. Finding genetically-supported drug targets for Parkinson's disease using Mendelian randomization of the druggable genome. Nat. Commun. 2021;12(1):7342. doi: 10.1038/s41467-021-26280-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y., et al. Genetic insights into associations of multisite chronic pain with common diseases and biomarkers using data from the UK Biobank. Br. J. Anaesth. 2024;132(2):372–382. doi: 10.1016/j.bja.2023.11.007. [DOI] [PubMed] [Google Scholar]
- 15.Sun B.B., et al. Genomic atlas of the human plasma proteome. Nature. 2018;558(7708):73–79. doi: 10.1038/s41586-018-0175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jansen I.E., et al. Genome-wide meta-analysis for Alzheimer's disease cerebrospinal fluid biomarkers. Acta Neuropathol. 2022;144(5):821–842. doi: 10.1007/s00401-022-02454-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gudjonsson A., et al. A genome-wide association study of serum proteins reveals shared loci with common diseases. Nat. Commun. 2022;13(1):480. doi: 10.1038/s41467-021-27850-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferkingstad E., et al. Large-scale integration of the plasma proteome with genetics and disease. Nat. Genet. 2021;53(12):1712–1721. doi: 10.1038/s41588-021-00978-w. [DOI] [PubMed] [Google Scholar]
- 19.Suhre K., et al. Connecting genetic risk to disease end points through the human blood plasma proteome. Nat. Commun. 2017;8 doi: 10.1038/ncomms14357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hillary R.F., et al. Genome and epigenome wide studies of neurological protein biomarkers in the Lothian Birth Cohort 1936. Nat. Commun. 2019;10(1):3160. doi: 10.1038/s41467-019-11177-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pietzner M., et al. Genetic architecture of host proteins involved in SARS-CoV-2 infection. Nat. Commun. 2020;11(1):6397. doi: 10.1038/s41467-020-19996-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yao C., et al. Genome-wide mapping of plasma protein QTLs identifies putatively causal genes and pathways for cardiovascular disease. Nat. Commun. 2018;9(1):3268. doi: 10.1038/s41467-018-05512-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Folkersen L., et al. Mapping of 79 loci for 83 plasma protein biomarkers in cardiovascular disease. PLoS Genet. 2017;13(4) doi: 10.1371/journal.pgen.1006706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilly A., et al. Whole-genome sequencing analysis of the cardiometabolic proteome. Nat. Commun. 2020;11(1):6336. doi: 10.1038/s41467-020-20079-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kunkle B.W., et al. Genetic meta-analysis of diagnosed Alzheimer's disease identifies new risk loci and implicates Abeta, tau, immunity and lipid processing. Nat. Genet. 2019;51(3):414–430. doi: 10.1038/s41588-019-0358-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwartzentruber J., et al. Genome-wide meta-analysis, fine-mapping and integrative prioritization implicate new Alzheimer's disease risk genes. Nat. Genet. 2021;53(3):392–402. doi: 10.1038/s41588-020-00776-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Novikova G., et al. Integration of Alzheimer's disease genetics and myeloid genomics identifies disease risk regulatory elements and genes. Nat. Commun. 2021;12(1):1610. doi: 10.1038/s41467-021-21823-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karch C.M., Goate A.M. Alzheimer's disease risk genes and mechanisms of disease pathogenesis. Biol Psychiatry. 2015;77(1):43–51. doi: 10.1016/j.biopsych.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chapuis J., et al. Increased expression of BIN1 mediates Alzheimer genetic risk by modulating tau pathology. Mol Psychiatry. 2013;18(11):1225–1234. doi: 10.1038/mp.2013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martiskainen H., et al. Transcriptomics and mechanistic elucidation of Alzheimer's disease risk genes in the brain and in vitro models. Neurobiol. Aging. 2015;36(2):1221 e15–e28. doi: 10.1016/j.neurobiolaging.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 31.Karch C.M., et al. Expression of novel Alzheimer's disease risk genes in control and Alzheimer's disease brains. PLoS One. 2012;7(11) doi: 10.1371/journal.pone.0050976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McAvoy K.M., et al. Cell-autonomous and non-cell autonomous effects of neuronal BIN1 loss in vivo. PLoS One. 2019;14(8) doi: 10.1371/journal.pone.0220125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ponnusamy M., et al. Loss of forebrain BIN1 attenuates hippocampal pathology and neuroinflammation in a tauopathy model. Brain. 2023;146(4):1561–1579. doi: 10.1093/brain/awac318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crotti A., et al. BIN1 favors the spreading of Tau via extracellular vesicles. Sci. Rep. 2019;9(1):9477. doi: 10.1038/s41598-019-45676-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fugier C., et al. Misregulated alternative splicing of BIN1 is associated with T tubule alterations and muscle weakness in myotonic dystrophy. Nat Med. 2011;17(6):720–725. doi: 10.1038/nm.2374. [DOI] [PubMed] [Google Scholar]
- 36.Hong T., et al. Cardiac BIN1 folds T-tubule membrane, controlling ion flux and limiting arrhythmia. Nat Med. 2014;20(6):624–632. doi: 10.1038/nm.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nyhus C., et al. Evidence for nucleolar dysfunction in Alzheimer's disease. Rev. Neurosci. 2019;30(7):685–700. doi: 10.1515/revneuro-2018-0104. [DOI] [PubMed] [Google Scholar]
- 38.Brooks W.H. Polyamine dysregulation and nucleolar disruption in Alzheimer's disease. J Alzheimers Dis. 2024;98(3):837–857. doi: 10.3233/JAD-231184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ahmed T., et al. Cholinergic system and post-translational modifications: an insight on the role in Alzheimer's disease. Curr. Neuropharmacol. 2017;15(4):480–494. doi: 10.2174/1570159X14666160325121145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosenthal A.S., et al. Potent and selective small molecule inhibitors of specific isoforms of Cdc2-like kinases (Clk) and dual specificity tyrosine-phosphorylation-regulated kinases (Dyrk) Bioorg Med Chem Lett. 2011;21(10):3152–3158. doi: 10.1016/j.bmcl.2011.02.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jain P., et al. Human CDC2-like kinase 1 (CLK1): a novel target for Alzheimer's disease. Curr. Drug Targets. 2014;15(5):539–550. doi: 10.2174/1389450115666140226112321. [DOI] [PubMed] [Google Scholar]
- 42.Prak K., et al. Benzobisthiazoles represent a novel scaffold for kinase inhibitors of CLK family members. Biochemistry. 2016;55(3):608–617. doi: 10.1021/acs.biochem.5b01128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pentz R., et al. A new role for matrix metalloproteinase-3 in the NGF metabolic pathway: proteolysis of mature NGF and sex-specific differences in the continuum of Alzheimer's pathology. Neurobiol. Dis. 2021;148 doi: 10.1016/j.nbd.2020.105150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Hove I., et al. Matrix metalloproteinase-3 in the central nervous system: a look on the bright side. J. Neurochem. 2012;123(2):203–216. doi: 10.1111/j.1471-4159.2012.07900.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.