Abstract
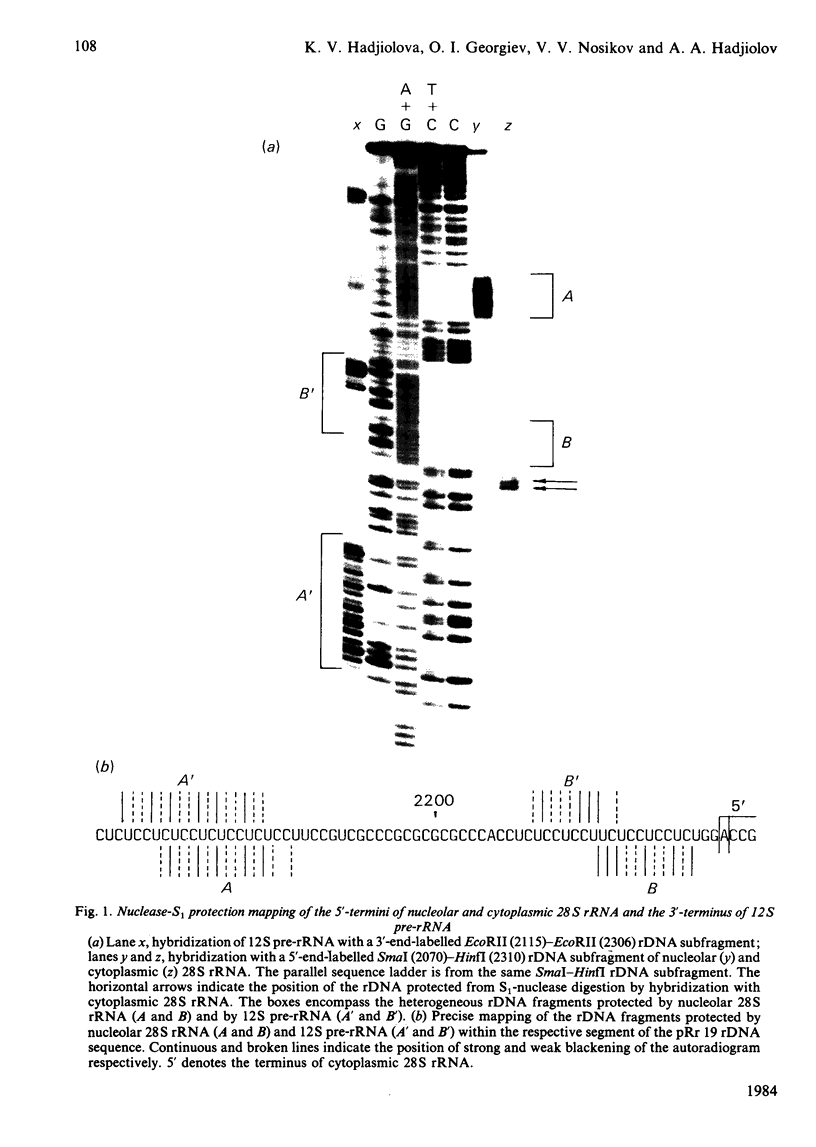
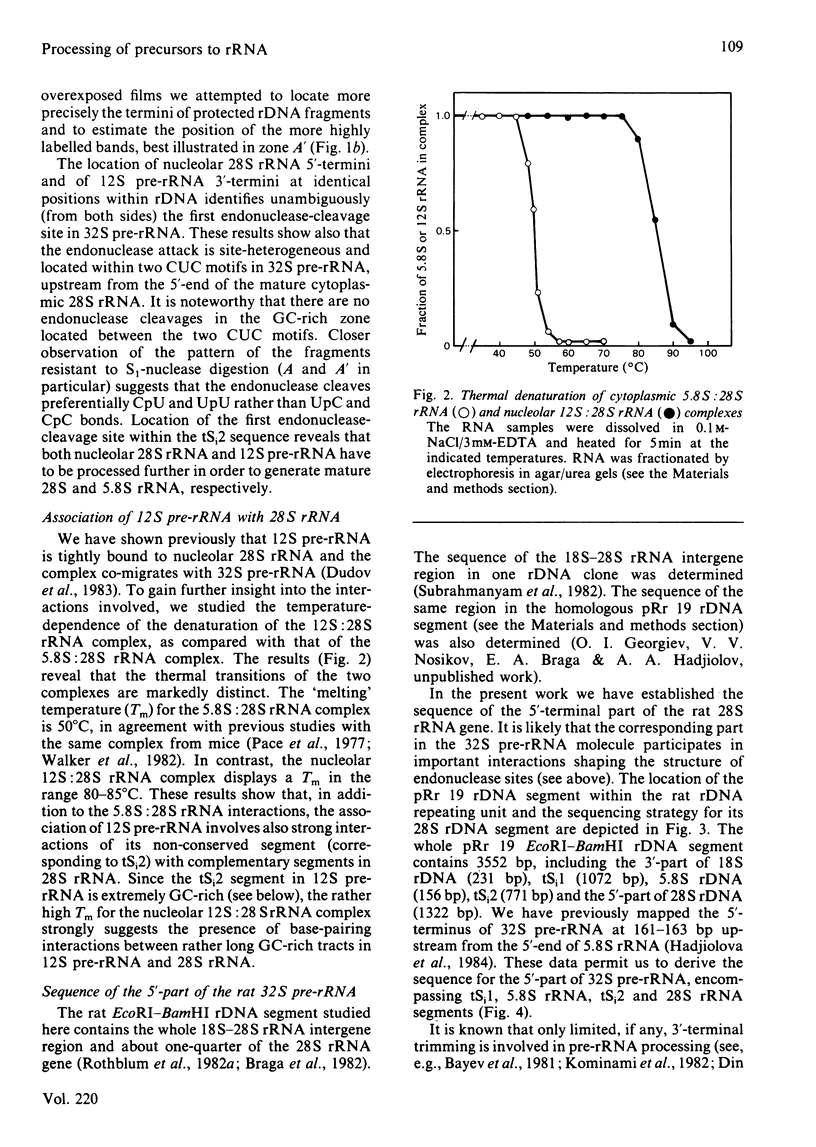
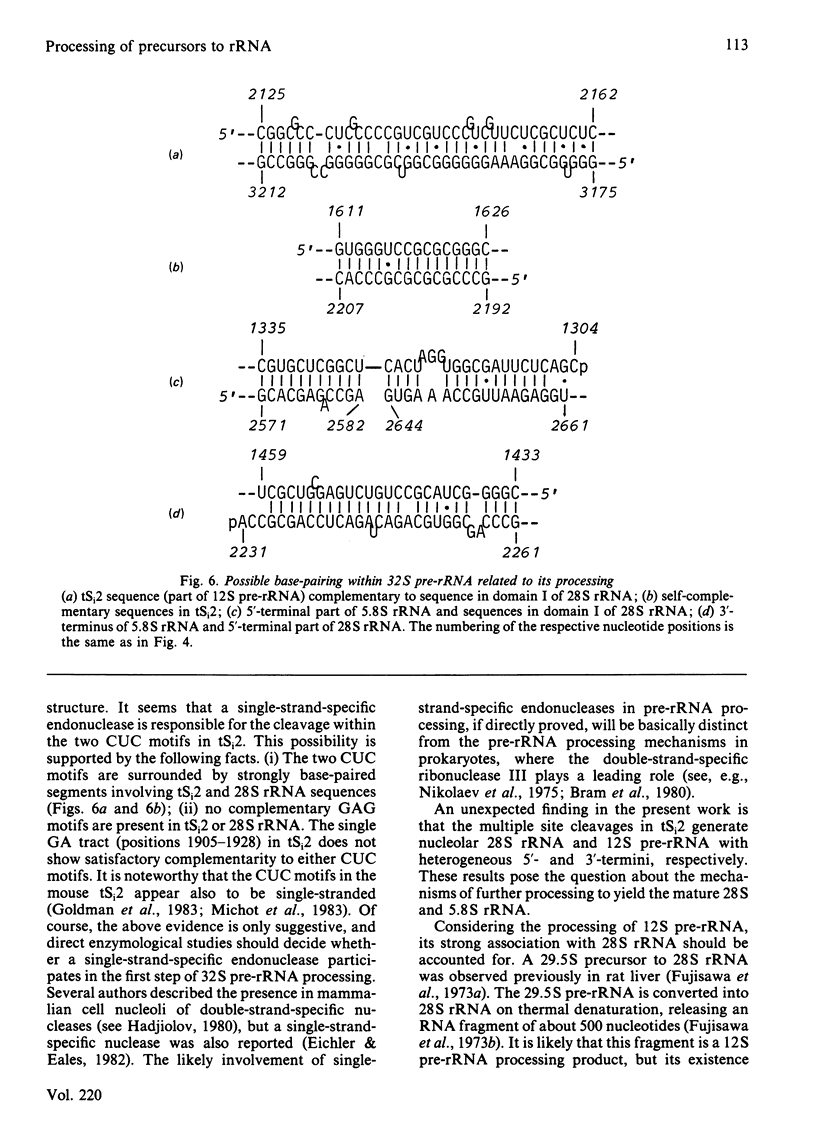
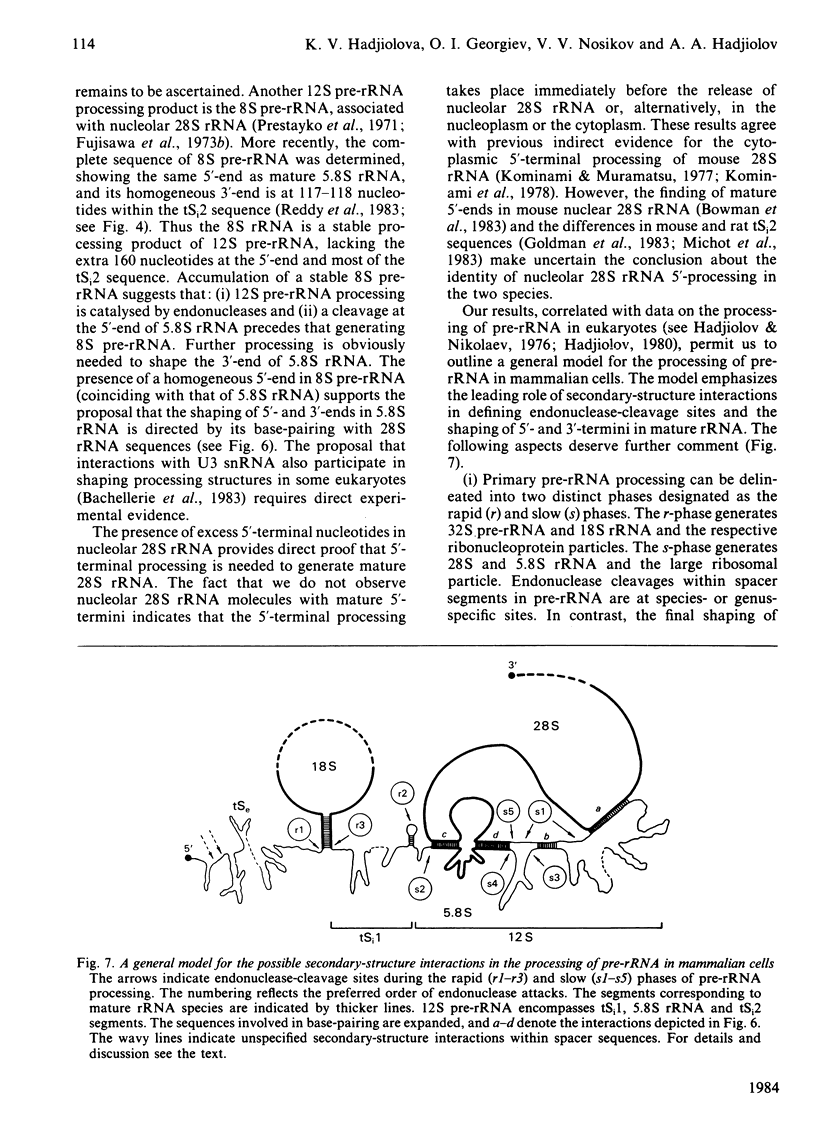
The initial endonuclease cleavage site in 32 S pre-rRNA (precursor to rRNA) is located within the rate rDNA sequence by S1-nuclease protection mapping of purified nucleolar 28 S rRNA and 12 S pre-rRNA. The heterogeneous 5'- and 3'-termini of these rRNA abut and map within two CTC motifs in tSi2 (internal transcribed spacer 2) located at 50-65 and 4-20 base-pairs upstream from the homogeneous 5'-end of the 28 S rRNA gene. These results show that multiple endonuclease cleavages occur at CUC sites in tSi2 to generate 28 S rRNA and 12 S pre-rRNA with heterogeneous 5'- and 3'-termini, respectively. These molecules have to be processed further to yield mature 28 S and 5.8 S rRNA. Thermal-denaturation studies revealed that the base-pairing association in the 12 S pre-rRNA:28 S rRNA complex is markedly stronger than that in the 5.8 S:28 S rRNA complex. The sequence of about one-quarter (1322 base-pairs) of the 5'-part of the rat 28 S rDNA was determined. A computer search reveals the possibility that the cleavage sites in the CUC motifs are single-stranded, flanked by strongly base-paired GC tracts, involving tSi2 and 28 S rRNA sequences. The subsequent nuclease cleavages, generating the termini of mature rRNA, seem to be directed by secondary-structure interactions between 5.8 S and 28 S rRNA segments in pre-rRNA. An analysis for base-pairing among evolutionarily conserved sequences in 32 S pre-rRNA suggests that the cleavages yielding mature 5.8 S and 28 S rRNA are directed by base-pairing between (i) the 3'-terminus of 5.8 S rRNA and the 5'-terminus of 28 S rRNA and (ii) the 5'-terminus of 5.8 S rRNA and internal sequences in domain I of 28 S rRNA. A general model for primary- and secondary-structure interactions in pre-rRNA processing is proposed, and its implications for ribosome biogenesis in eukaryotes are briefly discussed.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachellerie J. P., Michot B., Raynal F. Recognition signals for mouse pre-rRNA processing. A potential role for U3 nucleolar RNA. Mol Biol Rep. 1983 May;9(1-2):79–86. doi: 10.1007/BF00777477. [DOI] [PubMed] [Google Scholar]
- Bayev A., Georgiev O. I., Hadjiolov A. A., Nikolaev N., Skryabin K. G., Zakharyev V. M. The structure of the yeast ribosomal RNA genes. 3. Precise mapping of the 18 S and 25 S rRNA genes and structure of the adjacent regions. Nucleic Acids Res. 1981 Feb 25;9(4):789–799. doi: 10.1093/nar/9.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Bowman L. H., Goldman W. E., Goldberg G. I., Hebert M. B., Schlessinger D. Location of the initial cleavage sites in mouse pre-rRNA. Mol Cell Biol. 1983 Aug;3(8):1501–1510. doi: 10.1128/mcb.3.8.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman L. H., Rabin B., Schlessinger D. Multiple ribosomal RNA cleavage pathways in mammalian cells. Nucleic Acids Res. 1981 Oct 10;9(19):4951–4966. doi: 10.1093/nar/9.19.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braga E. A., Yussifov T. N., Nosikov V. V. Structural organization of rat ribosomal genes restriction endonuclease analysis of genomic and cloned ribosomal DNAs. Gene. 1982 Dec;20(2):145–156. doi: 10.1016/0378-1119(82)90033-6. [DOI] [PubMed] [Google Scholar]
- Bram R. J., Young R. A., Steitz J. A. The ribonuclease III site flanking 23S sequences in the 30S ribosomal precursor RNA of E. coli. Cell. 1980 Feb;19(2):393–401. doi: 10.1016/0092-8674(80)90513-9. [DOI] [PubMed] [Google Scholar]
- Dabeva M. D., Dudov K. P., Hadjiolov A. A., Emanuilov I., Todorov B. N. Intranuclear maturation pathways of rat liver ribosomal ribonucleic acids. Biochem J. 1976 Dec 15;160(3):495–503. doi: 10.1042/bj1600495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabeva M. D., Dudov K. P., Hadjiolov A. A., Stoykova A. S. Quantitative analysis of rat liver nucleolar and nucleoplasmic ribosomal ribonucleic acids. Biochem J. 1978 May 1;171(2):367–374. doi: 10.1042/bj1710367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Din N., Engberg J., Gall J. G. The nucleotide sequence at the transcription termination site of the ribosomal RNA gene in Tetrahymena thermophila. Nucleic Acids Res. 1982 Mar 11;10(5):1503–1513. doi: 10.1093/nar/10.5.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudov K. P., Dabeva M. D., Hadjiolov A. A. Simple agar--urea-gel electrophoretic fractionation of high molecular weight ribonucleic acids. Anal Biochem. 1976 Nov;76(50):250–258. doi: 10.1016/0003-2697(76)90283-9. [DOI] [PubMed] [Google Scholar]
- Dudov K. P., Dabeva M. D., Hadjiolov A. A., Todorov B. N. Processing and migration of ribosomal ribonculeic acids in the nucleolus and nucleoplasm of rat liver nuclei. Biochem J. 1978 May 1;171(2):375–383. doi: 10.1042/bj1710375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudov K. P., Dabeva M. D. Post-transcriptional regulation of ribosome formation in the nucleus of regenerating rat liver. Biochem J. 1983 Jan 15;210(1):183–192. doi: 10.1042/bj2100183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudov K. P., Hadjiolova K. V., Kermekchiev M. B., Stanchev B. S., Hadjiolov A. A. A 12 S precursor to 5.8 S rRNA associated with rat liver nucleolar 28 S rRNA. Biochim Biophys Acta. 1983 Jan 20;739(1):79–84. doi: 10.1016/0167-4781(83)90047-7. [DOI] [PubMed] [Google Scholar]
- Eichler D. C., Eales S. J. Isolation and characterization of a single-stranded specific endoribonuclease from Ehrlich cell nucleoli. J Biol Chem. 1982 Dec 10;257(23):14384–14389. [PubMed] [Google Scholar]
- Fujisawa T., Abe S., Kawada T., Satake M., Ogata K. Studies on the processing of 45-S RNA in rat liver nucleolus with specific reference to 29.5-S RNA. Biochim Biophys Acta. 1973 Oct 12;324(2):226–240. doi: 10.1016/0005-2787(73)90140-8. [DOI] [PubMed] [Google Scholar]
- Fujisawa T., Abe S., Satake M., Ogata K. Conversion of rat liver nucleolar 29.5-S RNA to 28-S RNA in vitro. Biochim Biophys Acta. 1973 Oct 12;324(2):241–253. doi: 10.1016/0005-2787(73)90141-x. [DOI] [PubMed] [Google Scholar]
- Georgiev O. I., Nikolaev N., Hadjiolov A. A., Skryabin K. G., Zakharyev V. M., Bayev A. A. The structure of the yeast ribosomal RNA genes. 4. Complete sequence of the 25 S rRNA gene from Saccharomyces cerevisae. Nucleic Acids Res. 1981 Dec 21;9(24):6953–6958. doi: 10.1093/nar/9.24.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman W. E., Goldberg G., Bowman L. H., Steinmetz D., Schlessinger D. Mouse rDNA: sequences and evolutionary analysis of spacer and mature RNA regions. Mol Cell Biol. 1983 Aug;3(8):1488–1500. doi: 10.1128/mcb.3.8.1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjiolov A. A. Biogenesis of ribosomes in eukaryotes. Subcell Biochem. 1980;7:1–80. doi: 10.1007/978-1-4615-7948-9_1. [DOI] [PubMed] [Google Scholar]
- Hadjiolov A. A., Nikolaev N. Maturation of ribosomal ribonucleic acids and the biogenesis of ribosomes. Prog Biophys Mol Biol. 1976;31(2):95–144. doi: 10.1016/0079-6107(78)90006-8. [DOI] [PubMed] [Google Scholar]
- Hadjiolova K. V., Golovinsky E. V., Hadjiolov A. A. The site of action of 5-fluoroorotic acid on the maturation of mouse liver ribonucleic acids. Biochim Biophys Acta. 1973 Sep 7;319(3):373–382. doi: 10.1016/0005-2787(73)90177-9. [DOI] [PubMed] [Google Scholar]
- Hall L. M., Maden B. E. Nucleotide sequence through the 18S-28S intergene region of a vertebrate ribosomal transcription unit. Nucleic Acids Res. 1980 Dec 20;8(24):5993–6005. doi: 10.1093/nar/8.24.5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M. S., Maden B. E. Maturation of 5.8 S RNA in HeLa cells. FEBS Lett. 1976 Jan 1;61(1):10–13. doi: 10.1016/0014-5793(76)80159-7. [DOI] [PubMed] [Google Scholar]
- Kominami R., Hamada H., Fujii-Kuriyama Y., Muramatsu M. 5'-Terminal processing of ribosomal 28S RNA. Biochemistry. 1978 Sep 19;17(19):3965–3970. doi: 10.1021/bi00612a014. [DOI] [PubMed] [Google Scholar]
- Kominami R., Mishima Y., Urano Y., Sakai M., Muramatsu M. Cloning and determination of the transcription termination site of ribosomal RNA gene of the mouse. Nucleic Acids Res. 1982 Mar 25;10(6):1963–1979. doi: 10.1093/nar/10.6.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kominami R., Muramatsu M. Heterogeneity of 5' -termini of nucleolar 45S, 32S and 28S RNA in mouse hepatoma. Nucleic Acids Res. 1977 Jan;4(1):229–240. doi: 10.1093/nar/4.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maden B. E., Moss M., Salim M. Nucleotide sequence of an external transcribed spacer in Xenopus laevis rDNA: sequences flanking the 5' and 3' ends of 18S rRNA are non-complementary. Nucleic Acids Res. 1982 Apr 10;10(7):2387–2398. doi: 10.1093/nar/10.7.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maden B. E., Salim M., Summers D. F. Maturation pathway for ribosomal RNA in the Hela cell nucleolus. Nat New Biol. 1972 May 3;237(70):5–9. doi: 10.1038/newbio237005a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Michot B., Bachellerie J. P., Raynal F. Sequence and secondary structure of mouse 28S rRNA 5'terminal domain. Organisation of the 5.8S-28S rRNA complex. Nucleic Acids Res. 1982 Sep 11;10(17):5273–5283. doi: 10.1093/nar/10.17.5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michot B., Bachellerie J. P., Raynal F. Structure of mouse rRNA precursors. Complete sequence and potential folding of the spacer regions between 18S and 28S rRNA. Nucleic Acids Res. 1983 May 25;11(10):3375–3391. doi: 10.1093/nar/11.10.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima Y., Yamamoto O., Kominami R., Muramatsu M. In vitro transcription of a cloned mouse ribosomal RNA gene. Nucleic Acids Res. 1981 Dec 21;9(24):6773–6785. doi: 10.1093/nar/9.24.6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaev N., Birge C. H., Gotoh S., Glazier K., Schlessinger D. Primary processing of high molecular weight preribosomal RNA in Escherichia coli and HeLa cells. Brookhaven Symp Biol. 1975 Jul;(26):175–193. [PubMed] [Google Scholar]
- Olsen G. J., Sogin M. L. Nucleotide sequence of Dictyostelium discoideum 5.8S ribosomal ribonucleic acid: evolutionary and secondary structural implications. Biochemistry. 1982 May 11;21(10):2335–2343. doi: 10.1021/bi00539a010. [DOI] [PubMed] [Google Scholar]
- Pace N. R., Walker T. A., Schroeder E. Structure of the 5.8S RNA component of the 5.8S-28S ribosomal RNA junction complex. Biochemistry. 1977 Nov 29;16(24):5321–5328. doi: 10.1021/bi00643a025. [DOI] [PubMed] [Google Scholar]
- Perry R. P. Processing of RNA. Annu Rev Biochem. 1976;45:605–629. doi: 10.1146/annurev.bi.45.070176.003133. [DOI] [PubMed] [Google Scholar]
- Peters M. A., Walker T. A., Pace N. R. Independent binding sites in mouse 5.8S ribosomal ribonucleic acid for 28S ribosomal ribonucleic acid. Biochemistry. 1982 May 11;21(10):2329–2335. doi: 10.1021/bi00539a009. [DOI] [PubMed] [Google Scholar]
- Prestayko A. W., Tonato M., Busch H. Low molecular weight RNA associated with 28 s nucleolar RNA. J Mol Biol. 1970 Feb 14;47(3):505–515. doi: 10.1016/0022-2836(70)90318-9. [DOI] [PubMed] [Google Scholar]
- Reddy R., Busch H. Small nuclear RNAs and RNA processing. Prog Nucleic Acid Res Mol Biol. 1983;30:127–162. doi: 10.1016/s0079-6603(08)60685-6. [DOI] [PubMed] [Google Scholar]
- Reddy R., Rothblum L. I., Subrahmanyam C. S., Liu M. H., Henning D., Cassidy B., Busch H. The nucleotide sequence of 8 S RNA bound to preribosomal RNA of Novikoff hepatoma. The 5'-end of 8 S RNA is 5.8 S RNA. J Biol Chem. 1983 Jan 10;258(1):584–589. [PubMed] [Google Scholar]
- Rothblum L. I., Parker D. L., Cassidy B. Isolation and characterization of rat ribosomal DNA clones. Gene. 1982 Jan;17(1):75–77. doi: 10.1016/0378-1119(82)90102-0. [DOI] [PubMed] [Google Scholar]
- Rothblum L. I., Reddy R., Cassidy B. Transcription initiation site of rat ribosomal DNA. Nucleic Acids Res. 1982 Nov 25;10(22):7345–7362. doi: 10.1093/nar/10.22.7345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skriabin K. G., Kraev A. S., Rubtsov P. M., Baev A. A. Polnaia posledovatel'nost' nukleotidov speisernoi oblasti, raspolozhennoi mezhdu genami 18S i 5.8S RNK drozhzhei. Dokl Akad Nauk SSSR. 1979;247(3):761–765. [PubMed] [Google Scholar]
- Subrahmanyam C. S., Cassidy B., Busch H., Rothblum L. I. Nucleotide sequence of the region between the 18S rRNA sequence and the 28S rRNA sequence of rat ribosomal DNA. Nucleic Acids Res. 1982 Jun 25;10(12):3667–3680. doi: 10.1093/nar/10.12.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldman G. M., Klootwijk J., de Regt V. C., Planta R. J., Branlant C., Krol A., Ebel J. P. The primary and secondary structure of yeast 26S rRNA. Nucleic Acids Res. 1981 Dec 21;9(24):6935–6952. doi: 10.1093/nar/9.24.6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldman G. M., Klootwijk J., van Heerikhuizen H., Planta R. J. The nucleotide sequence of the intergenic region between the 5.8S and 26S rRNA genes of the yeast ribosomal RNA operon. Possible implications for the interaction between 5.8S and 26S rRNA and the processing of the primary transcript. Nucleic Acids Res. 1981 Oct 10;9(19):4847–4862. doi: 10.1093/nar/9.19.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkov P. V., Hadjiolov A. A. Differential stability of 28s and 18s rat liver ribosomal ribonucleic acids. Biochem J. 1969 Oct;115(1):91–94. doi: 10.1042/bj1150091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker T. A., Endo Y., Wheat W. H., Wool I. G., Pace N. R. Location of 5.8 S rRNA contact sites in 28 S rRNA and the effect of alpha-sarcin on the association of 5.8 S rRNA with 28 S rRNA. J Biol Chem. 1983 Jan 10;258(1):333–338. [PubMed] [Google Scholar]
- Walker T. A., Johnson K. D., Olsen G. J., Peters M. A., Pace N. R. Enzymatic and chemical structure mapping of mouse 28S ribosomal ribonucleic acid contacts in 5.8S ribosomal ribonucleic acid. Biochemistry. 1982 May 11;21(10):2320–2329. doi: 10.1021/bi00539a008. [DOI] [PubMed] [Google Scholar]
- Walker T. A., Pace N. R. 5.8S ribosomal RNA. Cell. 1983 Jun;33(2):320–322. doi: 10.1016/0092-8674(83)90413-0. [DOI] [PubMed] [Google Scholar]
- Walseth T. F., Johnson R. A. The enzymatic preparation of [alpha-(32)P]nucleoside triphosphates, cyclic [32P] AMP, and cyclic [32P] GMP. Biochim Biophys Acta. 1979 Mar 28;562(1):11–31. doi: 10.1016/0005-2787(79)90122-9. [DOI] [PubMed] [Google Scholar]
- Weinberg R. A., Penman S. Processing of 45 s nucleolar RNA. J Mol Biol. 1970 Jan 28;47(2):169–178. doi: 10.1016/0022-2836(70)90337-2. [DOI] [PubMed] [Google Scholar]
- Winicov I. Alternate temporal order in ribosomal RNA maturation. J Mol Biol. 1976 Jan 15;100(2):141–155. doi: 10.1016/s0022-2836(76)80145-3. [DOI] [PubMed] [Google Scholar]



