Abstract
Background
The assembly of the rhizosphere community, even the diazotroph community, is mainly shaped by soil environmental factors (including soil climate and physiochemical characteristics) and plant selection. To better understand the driving forces on the active overall and nitrogen-fixing bacterial community compositions, we characterized the communities of tobacco rhizosphere soil collected from three sampling sites with a large geographic scale (> 600 km).
Results
The results indicate that the diversity and community composition of the overall bacterial and diazotroph communities are obviously differed according to the sampling sites. Still, no significant difference is found between the communities in rootzone and rhizosphere samples. Climate variables including mean annual precipitation (MAP) and mean annual temperature (MAT), soil physiochemical characteristics including available nitrogen (AN), available potassium (AK) and pH are main factors that affect the bacterial and diazotroph community structures in the three sampling sites. Furthermore, MAP and MAT, AN and available phosphorus (AP), total nitrogen (TN) and organic carbon (OC), AK and electrical conductivity (EC) showed similar effects, but pH showed independent effect on the composition of the overall bacteria and diazotroph communities. However, the alpha diversity indices of active overall and nitrogen-fixing bacteria in the rhizosphere are obviously higher than in the rootzone samples, and no significant differences are observed among different sampling sites. Proteobacteria is the predominant active phylum of all samples for overall and nitrogen-fixing bacteria. Escherichia-Shigella, Achromobacter, Streptomyces and Sphingomonas are the dominant active bacterial genera, and Bradyrhizobium, Skermanella and Extensimonas are dominant active nitrogen-fixing bacteria genera in rhizosphere. Furthermore, the high active abundance of Escherichia-Shigella but low abundance of Ralstonia in all three sampling sites indicate high root-knot nematode infection and low wilt disease endemic risk.
Conclusion
These results indicate that soil environmental factors contribute more to the tobacco rhizosphere bacterial community assemblage, but the rhizosphere contributes more to the diversity of active overall bacteria and nitrogen-fixing bacteria in the community. Our study provides novel knowledge for the assemble of rhizosphere bacterial and active bacteria communities across a large geographical scale.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12866-024-03611-y.
Keywords: Active nitrogen fixing, Bacterial community, Diazotroph, Rhizosphere
Introduction
The terrestrial plant lives in biogeochemically diverse soil and is associated with various soil microbiota [1]. Soil microbiome plays pivotal roles in plant nutrition, development, stress tolerance and immunity [1, 2]. The plant-associated microbiota usually comprises bacteria, archaea, fungi, etc. [3]. As the central hotspot region of plant-microorganism interaction, plant excretes more than 20–40% of the total fixed photosynthetic carbohydrate compound to the rhizosphere soil through the root, and the excreted compounds could be a stable carbon source for the rhizosphere microorganism [4, 5]. A stable isotope experiment indicating the excreta could be assimilated directly by the rhizosphere microorganism resulted in the microorganism community change and even could reshape the rhizosphere functional microbiota community [6]. In addition, special compounds such as flavonoids in excrete could mediate plant–microbe interactions and influence the overall microbiome assembly [7].
The soil and rhizosphere microbiomes are considered the most diverse ecosystems on earth, including hundreds of genotypes [8]. Soil environmental factors and plant selection are the two dominant forces shaping the rhizosphere microbiome [9]. The plant genetic background could determine the microbiome and was called a ‘plant genetic-dominated microbiome’ [9]. However, the core metabolome and root exudation varied from different plant species [10] and even different cultivars/varieties [11], which may also result in the change of rhizosphere bacterial community [12]. This dynamic community response to different plants could further highlight the reasonability of improving crop yields by exploiting beneficial plant–microbe interactions [6]. In addition to the host factor selection, the microbiome assemble could also be affected by soil physiochemical properties such as soil organic carbon [13], available nitrogen (AN) and available phosphorus (AP) [14], pH [15], even soil moisture [16] and climate variables such as mean annual precipitation (MAP) and mean annual temperature (MAT) [17]. According to the contribution of both factors to the rhizosphere microbiome assembly, two rhizosphere microbiomes were classified: environment-dominated and plant genetic-dominated, with the former accounting for the large proportion (96.5%) [9].
Biological nitrogen fixation plays a key role in nitrogen input in terrestrial ecosystems, contributing 40–100 trillion gram (Tg) fixed nitrogen per year [18]. The nitrogen fixation process is performed by bacteria or archaea with nitrogenase complex (mainly encoded by three genes: nifH, nifD and nifK) in free-living or symbiotic conditions [19, 20]. Unlike symbiotic nitrogen mainly happens between rhizobia and legumes, non-symbiotic nitrogen fixation does not rely on plant hosts [21]. The later contributes much more nitrogen to cereal crops than previously thought and could even qualifies 29–82% of maize nitrogen needs [22]. As a more ecological and friendly way to provide nitrogen for crops, diazotrophs have been widely isolated and utilized for various crops such as maize, potato, etc. [23, 24]. As part of the soil microbiome, the assemble of the rhizosphere diazotroph community could also be affected by soil physiochemical characteristics such as pH [25], organic matter [26], AN [27], and even soil management practices such as fertilization [28] also play essential roles in regulating diazotrophic community assemble process and the shifting of dominant taxa. However, it is still difficult to evaluate the contribution of nitrogen fixation for different members in the rhizosphere diazotroph community just based on the diazotroph community structure. Amplifying and analyzing nifH amplicon using cDNA extracted from rhizosphere soil to analyze the active nitrogen-fixing bacterial community provides insight into the contribution of nitrogen fixation by different diazotrophs for these plants [29]. Through such methods, active nitrogen-fixing diazotroph communities were uncovered from various plant rhizospheres, such as switch grass [30], sugarcane [31], and rice [32], and a unique community was observed for each plant.
Although culture independent technique proved to be effective methods to evaluate the overall bacterial (including diazotroph) and active (including nitrogen fixing) bacterial community, the correlation relationship between the two kinds of communities even the major determinant for each community still to be elucidated. Especially for diazotroph and active nitrogen fixing bacterial communities in tobacco rhizosphere had not been studied. To filling this knowledge gap, this study aimed to 1) elucidate the main factors that affect overall and diazotroph bacterial communities in different soil samples collected from a large geographic scale; 2) characterize the active bacterial community structures and formation; and 3) analyze the assembling mechanism of active overall and nitrogen-fixing bacteria community.
Material and methods
Study site and sampling process
To avoid the selection by different cultivars/varieties, in this study, the tobacco variety Yunyan 87 was selected to cultivate in the three sampling sites with a large geographic scale. The rhizosphere and root zone soil samples were collected from three sampling sites: Linqu (N36.16, E118.23), Biyang (N32.48, E113.25) and Bijie (N27.14, E106.00) (Fig. S1). For rhizosphere and rootzone soil sampling, five plants from the same field were randomly selected using the 5-point sampling method. For each plant, the uprooted plant was strongly shaken to remove the surface-attached soil; loose soil attached to roots (1–20 mm) was collected and snap-frozen as rootzone soil by shaking and kneading with a sterile glove [33]. Then, the thin, closely attached soil (0–1 mm) was removed from the smooth root using a sterilized brush, and the soil was preserved in a sterilized 50 mL tube as the rhizosphere soil. The tubes are quickly frozen by liquid nitrogen, and then frozen samples are transported to the lab and preserved at -80℃. For soil samples to determine the soil physiochemical analyses, both the rhizosphere and bulk soils around each plant were collected and mixed, then they were transferred to the lab and air-dried at room temperature.
Environmental variables
Each air-dried soil sample was sieved through a 2 mm mesh screen, and then they were analyzed according to the standard protocol [34]. Soil pH was measured by a pH meter (FE20 FiveEasy, Mettler Toledo, Germany) with the soil–water suspension ratio (1:2.5, w/v) using the potentiometric method. The available nitrogen (AN) was measured by detecting the concentration of alkali-hydrolysable nitrogen [35]. The available phosphorus (AP) of soil was determined using a colorimetry method [36]. The available potassium (AK) of the soil was determined using flame photometry [36]. The total nitrogen (TN) concentration of the soil was evaluated by the titration method [36]. The organic carbon (OC) was measured using the wet-oxidation method [36]. The electrical conductivity was detected by mixing the soil samples in distilled water with the soil: water ratio of 1:5 (w/v). And the EC was established for the soil filtrate using a WTW/LF-330 conductivity meter [37]. The climate variables, including mean annual precipitation (MAP), mean annual temperature (MAT) for each sampling sites were obtained from the WorldClim database (https://www.worldclim.org/). The temperature seasonality (TS) was calculated using the ratio of the standard deviation of the monthly mean temperatures to the mean of the monthly temperatures. And the precipitation seasonality (PS) was calculated using the ratio of the standard deviation of the monthly total precipitation to the mean monthly total precipitation [17].
Soil DNA and RNA extraction and PCR amplification
In this study, the rhizosphere overall bacterial and diazotroph community are analyzed through 16S rRNA gene and nifH amplicon based on the rhizosphere soil DNA, and the active overall bacteria and nitrogen-fixing bacteria are evaluated by analysis of the both gene sequences amplified using rootzone or rhizosphere soil cDNA as template. Soil DNA was extracted from 0.25 g of each soil sample using the ALFA Soil DNA Extraction Kit (Findrop, China) according to the manufacturer’s protocols. The concentration and content of all the DNA samples were evaluated by using agarose gel electrophoresis and NanoDrop One (Thermo, USA). The qualified DNA samples were then frozen and stored at − 80 °C. The V3-V4 region of bacterial 16S rRNA gene was amplified using the universal primer pair 338F (5′-ACTCCTACGGGAGGCAGCA-3′) and806R (5′-GGACTACHVGGGTWTCTAAT-3′) [14]. The nifH sequence was amplified using the bacterial universal primer pair nifHF (5′-AAAGGYGGWATCGGYAARTCCACCAC-3′) and nifHR (5′-TGSGCYTTGTCYTCRCGGATBGGCAT-3′) [38]. The PCR mixture contains 2 × premix Taq 25 μL, primer-F (10 μM) 1 μL, primer-R (10 μM) 1 μL, template DNA 50 ng, and ddH2O up to 50 μL. The PCR process under the following conditions: 95 ℃ for 3 min, 33 cycles of 95 ℃ for 10 s, 55℃ for 30 s, 72 ℃ for 30 s, followed by 72 ℃ for 1 min. The PCR products were evaluated and extracted from 1% agarose gel and purified using E.Z.N.A.® Gel Extraction Kit (Omega, USA).
Soil RNA was extracted from 0.25 g of each soil sample using the Mabio Soil RNA Extraction Mini Kit (Mabio, China) according to the manufacturer’s protocols. Qualified RNA samples were selected as templates for first-strand cDNA synthesis with the Superscript III system (Invitrogen Cat. no. 18080–051), using its random primer mix. To keep the data consistent, the bacterial 16S rRNA gene [14] and nifH [38] sequences were amplified using the same primer pairs with soil DNA amplification.
High-throughput sequencing and bioinformatics analysis
Based on the purified amplicons, we used NEBNext® Ultra™ II DNA Library Prep Kit for Illumina® (New England Biolabs, USA) for library construction according to the manufacturer’s protocol. The library quality was evaluated on a Qubit@ 2.0 Fluorometer (Thermo Fisher Scientific, USA) and Agilent Bioanalyzer 2100 system (Agilent Technologies, USA). The qualified library samples were sequenced on the Illumina NovaSeq PE250 platform (Illumina, San Diego, USA) using the standard protocols of Guangdong Magigene Biotechnology Co., Ltd. (Guangzhou, China). All the raw reads obtained in this study were submitted to the Genome Sequence Archive in National Genomics Data Center of China National Center for Bioinformation (Accession Numbers: CRA016587, CRA016608, CRA016612 and CRA016620) and the NCBI Sequence Read Archive (SRA) database (Accession Numbers: PRJNA1160359, PRJNA1160397, PRJNA1160409 and PRJNA1160160). The raw reads were demultiplexed, quality filtered by Fastp Version 0.23.4 [39], and merged by FLASH version 1.2.7 [30]. Then, sequences were quality filtered, denoised, and chimera was removed by using the UCHIME algorithm (www.drive5.com/usearch/manual/uchime_algo.html) [40]. The operational taxonomic units (OTU) were clustered with a 97% similarity threshold using UPARSE (www.drive5.com/uparse/) [41]. The OTU abundance for each sample was further normalized to the sample with the fewest sequences. The OTU representative sequences of the 16S rRNA gene were taxonomically annotated using the SILVA138 (http://www.arb-silva.de/) reference database [42]. And the OTU representative sequences of the nifH were taxonomically annotated using the Framebot tool in the RDP function gene pipeline (http://fungene.cme.msu.edu/FunGenePipeline/) and aligned with the nifH database (http://fungene.cme.msu.edu/FunGene) [43].
Statistical analyses
The alpha diversity for each sample, including Shannon diversity, Chao I, and Richness indices, were calculated by the vegan package in R (v. 4.3.2). One-way ANOVAs were selected to test the difference significance of diversity indices between different samples using SPSS 27. Nonmetric multidimensional scaling (NMDS) was applied using Bray–Curtis distance to identify the community composition using the vegdist function within the vegan v 2.6–4 R package [44]. The correlation between soil characteristics and bacterial community was also evaluated by using a vegan package in R (v. 4.3.2).
Results
Soil environmental variables
As is shown in Table 1, all the soil samples showed acid, and the pH values varied from 5.28 in Bijie to 5.84 in Linqu. The highest concentration of AN, AP, TN and OC occurred in Bijie, with the concentration of 172.20 mg/kg, 32.63 mg/kg, 2.20 g/kg and 35.49 g/kg, respectively, while the lowest corresponding values happened in Biyang with the concentration of 74.9 mg/kg, 5.51 mg/kg, 0.41 g/kg and 13.24 g/kg. However, Linqu has the highest AK concentration (230.90 mg/kg) and EC (624.30 μs/cm), and Biyang has the lowest AK concentration (115.01 mg/kg) and EC (1.7.90 μs/cm). The highest and lowest MAP happened in Bijie and Linqu, with the precipitation of 1080 and 758 mL, respectively. And the highest and lowest MAT occurred in Biyang and Linqu, with the annal temperature of 15.05 and 11.92 ℃, respectively. The PS ranges from 0.67 in Biyang to 1.12 in Linqu, and the TS varied from 0.51 in Bijie to 0.86 in Linqu.
Table 1.
Soil environmental variables for each sampling sites
| Sampling site | pH | AN (mg/kg) | AP (mg/kg) | AK (mg/kg) | TN (g/kg) | OC (g/kg) | EC (μs/cm) | MAP (mL) | MAT (℃) | PS | TS |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Linqu | 5.84 | 114.10 | 15.78 | 230.90 | 0.66 | 17.28 | 624.30 | 758 | 11.92 | 1.12 | 0.86 |
| Biyang | 5.58 | 74.90 | 5.51 | 115.01 | 0.41 | 13.24 | 137.90 | 935 | 15.05 | 0.67 | 0.61 |
| Bijie | 5.28 | 172.20 | 32.63 | 183.08 | 2.20 | 35.49 | 463.40 | 1080 | 13.67 | 0.72 | 0.51 |
Sequence information and OTU classification
For 16S rRNA gene amplicon based on soil DNA (16S-D), 3,777,582 reads were obtained, ranging from 120,019 to 131,749 reads were generated among the 30 samples from the three sampling sites. After quality control, chimeric filter, chloroplast and mitochondrion sequence deletion and singleton deletion, 2,694,350 high-quality reads were left and are further classified into 15,962 OTUs (Table S1). After each sample is normalized to the sample with the fewest sequences (56,809 sequences), the 15,458 OTUs belonging to Bacteria ranging from 2595 to 4961 OTUs were generated among the 30 samples. Furthermore, a total of 352 OTUs were found in all 30 samples (Fig. S2), 4164 OTUs were found in all three sampling sites, and 9692 OTUs were commonly shared by rootzone and rhizosphere soils (Fig. 1 A). Among the three sampling sites, Linqu has the most OTU numbers while Biyang has the least OTU numbers (Fig. 1A, Table S1). Furthermore, more OTUs are found in rootzone than in rhizosphere samples (Fig. 1E).
Fig. 1.
Schematic drawing of the common shared bacterial OTUs in different sampling sites of Biyang, Bijie and Linqu (A-D), rootzone and rhizosphere samples (E–H). A, E, data based on 16S-D; B, F, data based on nifH-D; C, G, data based on 16S-R; D, H, data based on nifH-R
For nifH amplicon amplified based on soil DNA (nifH-D), 3,612,041 raw reads are obtained, ranging from 88,625 to 131,537 reads generated among the 30 samples from three sampling sites. After quality control, chimeric filter and singleton deletion, 2,648,528 high-quality reads are left and are classified into 1432 OTUs (Table S2). After each sample was normalized to the sample with the fewest sequences (12,865 sequences), 1186 OTUs belonging to Bacteria ranging from 106 to 273 OTUs were generated among the 30 samples (Table S2). A total of 18 OTUs were found in all the samples, 225 OTUs were found in the three sampling sites, and 518 OTUs were commonly shared by root zone and rhizosphere soils (Fig. 1B, F and S2). Linqu has the most OTU numbers, and Bijie has the lowest OTU numbers. Furthermore, root zone samples contain more OTUs than rhizosphere soil (Fig. 1F).
For 16S rRNA gene amplicon based on soil cDNA (16S-R), a total of 3,767,978 raw reads are obtained, ranging from 120,106 to 130,826 reads generated from the 30 samples. After quality control, chimeric filter, and singleton deletion, 3,141,211 high-quality reads are left and are classified into 9021 OTUs (Table S1). After each sample is normalized to the sample with the fewest sequences (81,894 sequences), the 9008 OTUs belonging to Bacteria ranging from 584 to 3420 OTUs are generated among the 30 samples (Table S1). A total of 71 core OTUs are found in all 30 samples (Fig. S2). Furthermore, 1940 OTUs belonging to 285 genera are found in all three sampling sites (Fig. 1C), and 4413 OTUs (Fig. 1G) are found in both rootzone and rhizosphere soils. Linqu has the most out numbers, and the rhizosphere has more OTUs than the rootzone.
For nifH amplicon amplified based on soil cDNA (nifH-R), a total of 2,386,090 raw reads were obtained from 20 samples (14/15 of rhizosphere and 6/15 rootzone samples), ranging from 59,217 to 129,994 reads for each sample. After quality control, chimeric filter, and singleton deletion, 449,596 high-quality sequences were left and were classified into 58 OTUs (Table S2). However, no core OTUs are found in all the samples, even the three sampling sites (Fig. 1D, Fig. S2), but 18 core OTUs (Fig. 1H) are found in both rootzone and rhizosphere soils. Furthermore, Linqu, with the highest OTUs and rhizosphere, contains more OTUs than rootzone.
The (active) overall bacteria and diazotroph bacterial community composition
The overall and active bacterial communities
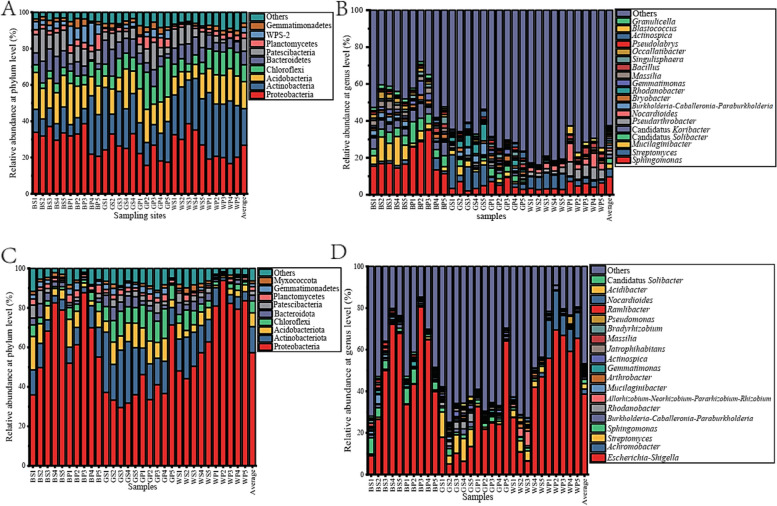
Based on 16S-D, all the OTUs were classified into 34 phyla and 529 genera (Table S3). The dominant phyla (> 10%) are Proteobacteria, Actinobacteria and Acidobacteria, and they are dominant in all three sampling sites (Fig. 2A). Proteobacteria showed the highest relative abundance in Biyang (31.20%), followed by Linqu (26.04%) and Bijie (24.22%). The abundance of Actinobacteria in Linqu is significantly higher than in Bijie and Biyang, and the candidate WPS-2 in Biyang is significantly higher than in Bijie and Linqu (Fig. S3). Furthermore, the abundance of Proteobacteria, Bacteroidetes and Patescibacteria in rhizosphere soil is significantly higher than in the root zone. However, the abundance of Chloroflexi, Planctomycetes, Gemmatimonadetes and Firmicutes is obviously higher than in the rootzone (Fig. S3). At the genus level, the dominant genus is Sphingomonas (> 5%) (Fig. 2B). A total of 109 core genera were found in each sample (Fig. S2), while 337 genera were found in the three sampling sites and 441 genera are simultaneously found in both rootzone and rhizosphere samples (Fig. S4). The bacterial community in Linqu includes 472 genera, which is higher than Bijie (441 genera) and Biyang (386 genera), and the rootzone bacterial community comprises more genera than rhizosphere (Fig. S4). Sphingomonas, Candidatus Solibacter, Candidatus Koribacter, Massilia, Gemmatimonas and Occallatibacter show the highest abundance in Biyang and significantly higher than in Bijie and Linqu. Rhodanobacter and Actinospica showed the highest abundance in Bijie and obviously higher than in Biyang and Linqu. Pseudarthrobacter, Nocardioides, Blastococcus and Ensifer in Linqu are significantly higher in Linqu than in Biyang and Bijie. Furthermore, the abundance of Streptomyces, Mucilaginibacter, Rhodanobacter, Allorhizobium-Neorhizobium-Pararhizobium-Rhizobium and Ensifer in rhizosphere samples are significantly higher than rootzone samples. However, Bacillus is obviously higher in rootzone than in rhizosphere samples (Fig. S3).
Fig. 2.
The overall bacterial (A, B) and active bacterial (C, D) community composition at phylum (A, C) or genus (B, D) level in different samples. A, B, data based on 16S-D; C, D, data based on 16S-R. BS1-BS5, rhizosphere soil samples of Biyang; BP1-BP5, rootzone soil samples of Biyang; GS1-GS5, rhizosphere soil samples of Bijie; GP1-GP5, rootzone soil samples of Bijie; WS1-WS5, rhizosphere soil samples of Linqu; WP1-WP5, rootzone soil samples of Linqu
All the active bacteria belong to 40 phyla and 511 genera (Table S3). The predominant phyla (> 10%) are Proteobacteria and Actinobacteriota (Fig. S4), and they are also dominant in all three sampling sites (Fig. 2C). Except the abundance of Proteobacteria is obviously higher in Linqu than in Bijie, Actinobacteriota and Chloroflexi significantly higher in Bijie than in Biyang, the other dominant (> 1%) phyla showed no obvious difference among the three sampling sites (Fig. S5). Although the expression of Proteobacteria and WPS-2 show higher in rhizosphere, Actinobacteriota, Acidobacteriota, Bacteroidota, Patescibacteria, and Myxococcota are higher in rootzone samples, but no obvious difference is observed (P > 0.30) (Fig. S5). The dominant genera are Escherichia-Shigella, Achromobacter, Streptomyces and Sphingomonas (Fig. 2D). Except for the relative expression level of Actinospica, Jatrophihabitans and Acidibacter are obviously higher in Bijie than in Biyang and Linqu, Rhodanobacter is significantly higher in Bijie than in Biyang, and Nocardioides in Linqu is obviously higher than Biyang, the other active bacteria genera shows no significantly difference (Fig. S5). Furthermore, the expression of the genera Escherichia-Shigella and Achromobacter is higher in the rootzone, Sphingomonas, Burkholderia-Caballeronia-Paraburkholderia, Rhodanobacter, Allorhizobium-Neorhizobium-Pararhizobium-Rhizobium, Mucilaginibacter and Arthrobacter is higher in rhizosphere samples, however, no significantly difference was observed (P > 0.65).
The diazotroph and active nitrogen-fixing bacterial communities
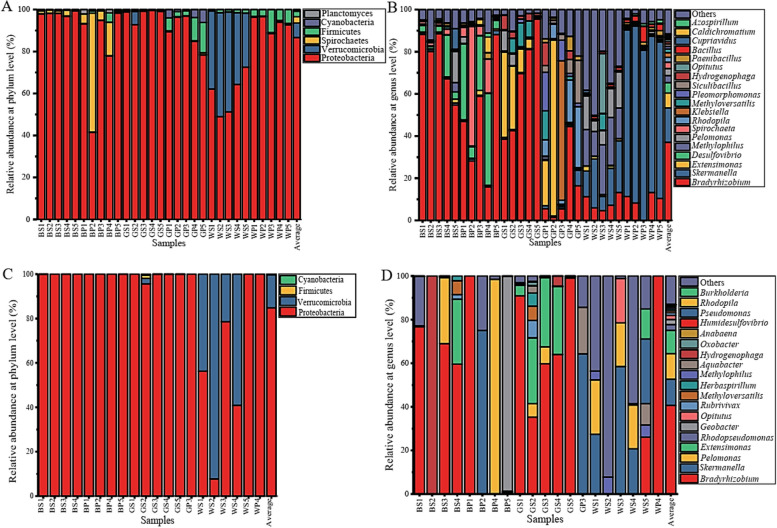
The diazotroph community comprises six phyla, with Proteobacteria as the only predominant phylum for each sampling site (> 10%, Fig. 3A). The abundance of Verrucomicrobia in Linqu is obviously higher than in Biyang, and Spirochaetes obviously higher in Biyang than in Liqu and Bijie (Fig. S6). Furthermore, the abundance of Firmicutes is obviously higher in rootzone than in rhizosphere soils. All the OTUs are further classified as 167 genera (Table S3), and 11 genera were found in all the samples (Fig. S2), with Bradyrhizobium, Skermanella, and Extensimonas are dominant genera (> 5%) (Fig. 3B). A total of 80 genera were commonly found in the three sampling sites, and 113 genera are found in both rootzone and rhizosphere soil (Fig. S4). The diazotroph in Linqu and Biyang belong to 121 genera, respectively, which is slightly higher than in Bijie (116 genera) (Fig. S4). The diazotroph belongs to genera Skermanella, Methylophilus and Bacillus, obviously enriched in Linqu, and the genera Desulfovibrio, Spirochaeta and Caldichromatium significantly enriched in Biyang and Rhodopila significantly enriched in Bijie (Fig. 3B). However, except Paenibacillus is obviously higher in the rhizosphere than in rootzone, the other genera showed no obvious different between the two conditions (Fig. S6).
Fig. 3.
The diazotroph (A, B) and active nitrogen-fixing (C, D) bacterial community composition at phylum (A, C) or genus (B, D) level in different samples. A, B, data based on nifH-D; C, D, data based on nifH-R. BS1-BS5, rhizosphere soil samples of Biyang; BP1-BP5, rootzone soil samples of Biyang; GS1-GS5, rhizosphere soil samples of Bijie; GP1-GP5, rootzone soil samples of Bijie; WS1-WS5, rhizosphere soil samples of Linqu; WP1-WP5, rootzone soil samples of Linqu
The active nitrogen-fixing bacteria belong to 4 phyla and 25 genera (Table S3). Proteobacteria (84.85%) and Verrucomicrobia (14.88%) are predominant phyla (> 10%, Fig. 3C). The abundance of Verrucomicrobia in Linqu is obviously higher than in Biyang and Bijie (Fig. S7). The highly expressed nifH belongs to the genera Bradyrhizobium, Skermanella, Pelomonas, Extensimonas, Rhodopseudomonas and Geobacter (> 2%, Fig. 3D). The predominant genera, Bradyrhizobium, is highly expressed in Biyang (65.88%) and Bijie (61.89%), and they are significantly higher than in Linqu (Fig. S7). While Skermanella, Pelomonas, Opitutus and Methylophilus expressed higher in Linqu. The nifH of Extensimonas was expressed significantly higher in Bijie than in Biyang and Linqu. And both Rhodopseudomonas and Geobacter expressed higher in Biyang than Bijie and Linqu. While the expression of Geobacter is mainly expressed in the rootzone, the other dominant expressed genera are mainly expressed in the rhizosphere (Fig. S7).
The alpha diversity of overall bacterial communities distributed in different sampling sites
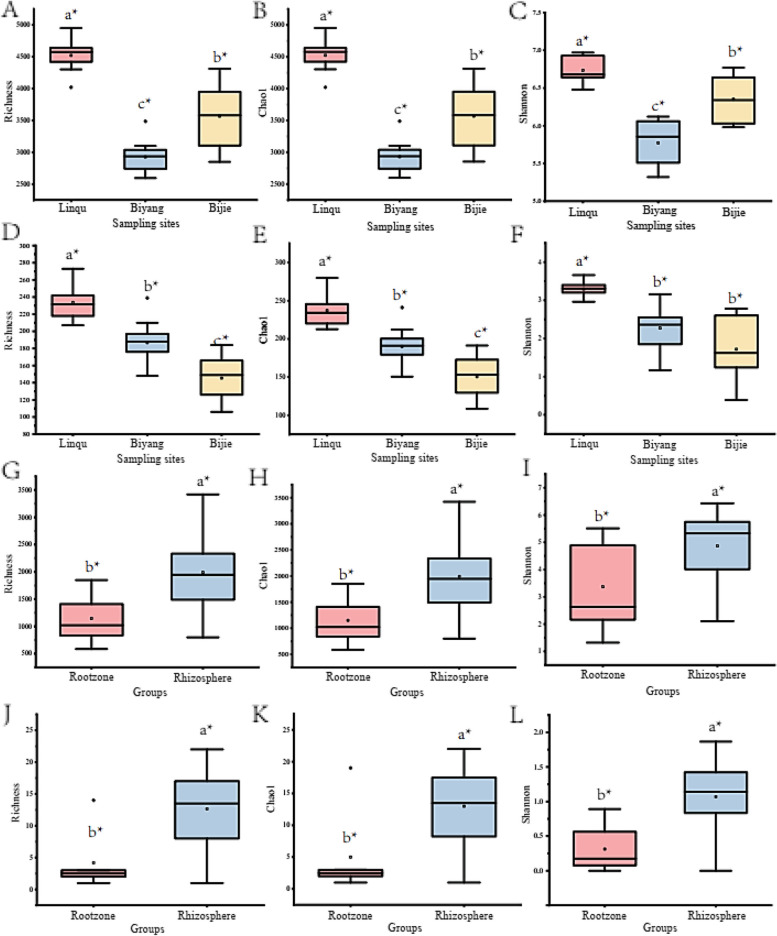
According to 16S-D, the overall bacterial community of Linqu with the highest and Biyang with the lowest indices, including richness, chao1 and Shannon indices (Fig. 4A, B and C). However, according to the results of nifH-D, the diazotroph community of Linqu with the highest indices and Bijie with the lowest indices (Fig. 4D, E and F). Except the Shannon indices between Biyang and Bijie of nifH-D showed no significant difference (Fig. 4F), each of the indexes of both 16S-D and nifH-D showed significant differences among the sampling sites (P < 0.03). The alpha diversity indices of Liqu are highest for both 16S-D and nifH-D. However, no significant difference is found between rootzone and rhizosphere samples (Fig. S8).
Fig. 4.
Species richness (A, D, G, J), Chao1 (B, E, H, K) and Shannon (C, F, I, L) indices of the overall (active) bacterial community, diazotroph and nitrogen-fixing bacteria collected from the three sampling sites (A-F), rootzone and rhizosphere (G-L) samples. A, B, C, data based on 16S-D; D, E, F, data based on nifH-D; G, H, I, data based on16S-R; J, K, L, data based on nifH-R. a*, b*, c* indicating the degree of significance
The highest indices of 16S-R happened in Bijie, and the lowest was found in Biyang (Fig. 4G-I). Except for the Shannon index of Bijie, obviously higher than in Linqu and Biyang (P < 0.05), no obvious differences are found for the three sampling sites for both 16S-R and nifH-R (P > 0.05) (Fig. S8). However, the three indices of both the 16S-R and nifH-R of the rhizosphere are significantly higher than rootzone samples (P < 0.01) (Fig. 4G-L).
Correlation between bacterial community compositions and environmental variables
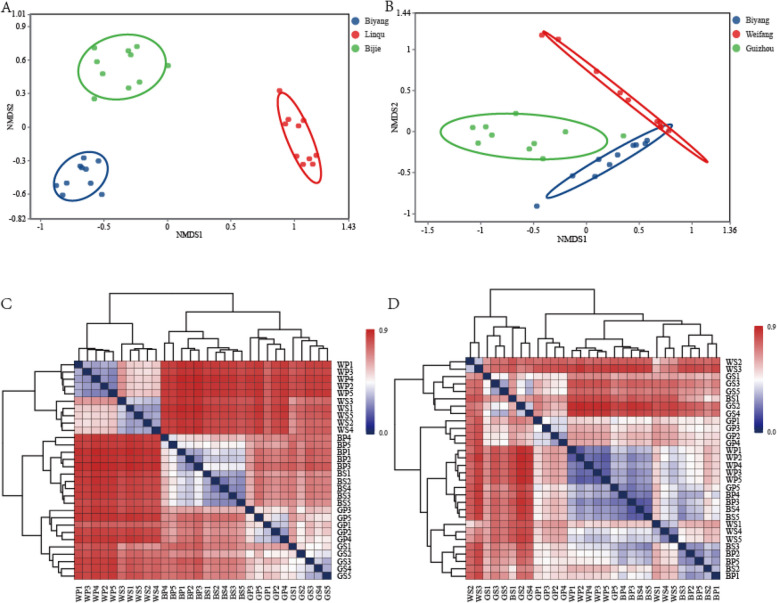
The beta diversity analyses based on Bray–Curtis distance showed that the overall bacteria and diazotroph bacteria community compositions significantly differed according to the sampling sites (the stress is 0.06 and 0.09 respectively) (Fig. 5A and Fig. S9), sticking to the soil environmental factors but not rhizosphere is the main factor determines the overall bacterial and diazotroph community structure. Although the overall active bacterial community sampling from the same site also clustered together (Fig. 5B), the distance between different sites is shorter than 16S-D (Fig. 5A) and nifH-D (Fig. S9).
Fig. 5.
Ordination plot of the NMDS based on the Bray–Curtis distance (A, B) and sample distance heatmap (C, D) of samples based on overall bacterial (A, C) and active bacterial (B, D) community. BS1-BS5, rhizosphere soil samples of Biyang; BP1-BP5, rootzone soil samples of Biyang; GS1-GS5, rhizosphere soil samples of Bijie; GP1-GP5, rootzone soil samples of Bijie; WS1-WS5, rhizosphere soil samples of Linqu; WP1-WP5, rootzone soil samples of Linqu
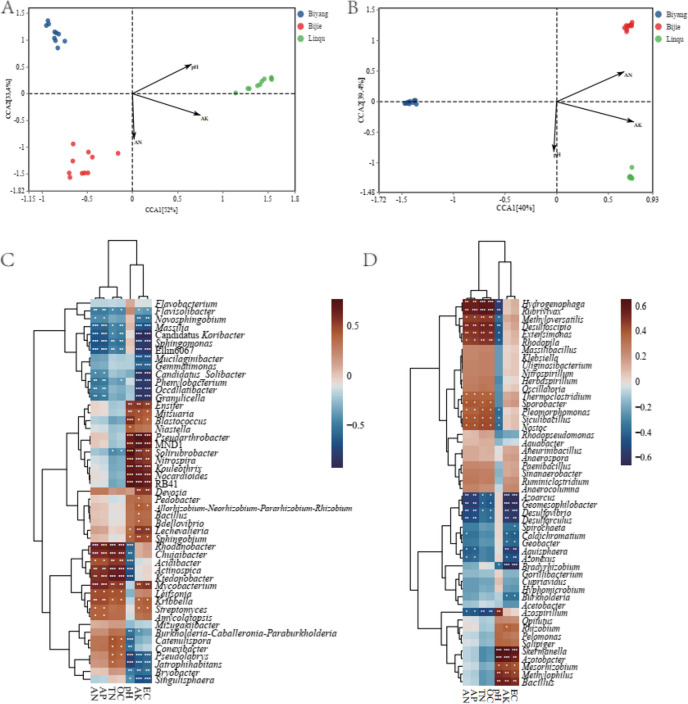
According to the first axis (3.35 for 16S-D and 6.49 for nifH-D), the CCA (canonical correspondence analysis) was selected to compare the community structure and the soil characteristics for the 16S-D and nifH-D (Fig. 6A and B). The AK, pH and AN (P < 0.05) are the main factors affected the community composition for both 16S-D and nifH-D, the first two axis contributed 85.4% and 79.4% of the total explanation rate for the two groups distributed in the three sampling sites. The overall bacterial and diazotroph communities of Linqu positively correlated with pH and AK, the communities of Biyang negatively correlated with AK and AN, and the communities of Bijie positively correlated with AN but negatively correlated with pH (Fig. 6A and B). The correlation analyses between the 50 most abundant genera and the environmental factors indicate AN and AP, TN and OC, AK and EC show similar effect on the enrichment of the genera (Fig. 6C and D), however, the effect of pH with independent effect for both 16S-D, and nifH-D (Fig. S10).
Fig. 6.
Correlation analyses between overall bacterial (A, C) or diazotroph bacterial (B, D) community and environment factors. A, B, CCA analyses between the overall bacterial (A) and diazotroph (B) community and soil physiochemical characteristics; C, D, correlation heatmap showing the top 50 dominant genera of overall bacterial (C) and diazotroph (D) community with soil physiochemical characteristics. The asterisk indicates a statistically significant t-test analysis. *, **, and *** represent P values < 0.05, 0.01, and 0.001, respectively
The CCA analyses between the bacterial community and climate variables indicating MAT and MAP are the main climate factors affect the overall bacterial and diazotroph community composition, and the first two axis contributed to 100% of the total explanation rate for the both groups (Fig. S10). The overall bacterial and diazotroph communities of Biyang positively correlated with MAT, the both communities of Bijie positively correlated with MAP, however the communities of Linqu negatively correlate with both MAT and MAP. Furthermore, the correlation between the 50 most abundant genera and the climate variables indicates MAT and MAP, PS and TS have similar effect on the enrichment of the genera, respectively (Fig. S10).
Discussion
The rhizosphere bacterial community composition and diversity are mainly determined by soil environmental factors
The soil microorganism community composition could be shaped by biotic factors such as host selection [5, 10] and soil environmental factors including soil physiochemical characteristics and climate variables [17, 45]. In this study, the rootzone and rhizosphere soil of the same tobacco cultivar (Yunyan 87) were collected from the three sampling sites with a large geographic scale. The bacterial community distributed with a large geographic scale usually differed significantly due to the affection environmental factors, such as climate variables MAT and MAP were predicted as the main factors affecting the rhizosphere bacterial community composition [17]. Consistent with it, in our study, the climate variables of MAP and MAT (Fig. S10) were also inferred as the main environmental factors affecting the overall bacterial and diazotroph community in this study. The soil in Bijie is the most fertile, but the soil in Biyang is the poorest, and the fertility of the soil in Liqu is in the middle (Table 1). As previous studies indicated, for bacterial community in fertile soils with higher diversity [46], but the soil with higher AN content, the nitrogen fixation could be weakened and with low diazotroph diversity [28]. The low diversity indices of 16S-D (Fig. 4A-C) but higher diversity indices of nifH-D (Fig. 4D-F) are consistent with the low AN content in Biyang soil. Soil characteristics including AK, AN and pH (Fig. 6) were inferred as the main environmental factors affecting the overall bacterial and diazotroph community in this study, consistent with previous studies they also as the main environmental factors affect the rhizosphere overall bacterial and diazotroph community composition [14, 15, 25, 27].
The overall bacterial and diazotroph community structure of both rootzone and rhizosphere samples within each sampling site are similar, indicating the stable community structure among the samples collected from each sampling site (Figs. 2 and 3). According to 16S-D, Linqu has the most and Biyang has the least OTUs and genera numbers (Fig. 1A), which is in accordance with Linqu with the highest and Biyang with the lowest alpha diversity indices (Fig. 4A-C). The alpha diversity indices of both 16S-D and nifH-D shows significant different between sampling sites but not between rootzone and rhizosphere (Fig. 4A-F, Fig. S9), indicating the soil environmental factors but not the rhizosphere selection determine both the overall bacterial and diazotroph community. The beta diversity generated by NMDS and sample distance heatmap indicates the unique overall bacterial and diazotroph community in each sampling site (Fig. 5A, B and Fig. S9). Thus, both the result of alpha and beta diversity analyses indicating the tobacco bacterial community structure in this study mainly determined by soil environmental factors, as previous studies found the bacterial community mainly determined by soil characteristics in farmland [47] and desert farming systems [48], and the community is mainly shaped by environmental factors and soil physiochemical characteristics [9].
Proteobacteria, Actinobacteria and Acidobacteria were the dominant phyla (> 10%) of tobacco rootzone and rhizosphere soil in this study, consistent with they are also dominant in tobacco rhizosphere soil collected from Guizhou and Yunnan provinces of China [49, 50]. Proteobacteria and Bacteroidota were obviously higher in the rhizosphere than rootzone samples, and Chloroflexi and Gemmatimonadetes are significantly enriched in rootzone soil than rhizosphere samples (Fig. S5) with the same distribution phenomenon was also observed in most of terrestrial ecosystems [51]. It indicating bacteria belong to the Proteobacteria and Bacteroidota may have more intimate relationships with the plants. Sphingomonas is the predominant genus in tobacco rhizosphere bacterial community in our study (Fig. 2B), which is commonly dominant in the tobacco rhizosphere and easier to isolate [49, 52]. Furthermore, Sphingomonas spp. are commonly considered dominant beneficial colonies for plants and could be used as effective plant growth-promoting bacteria (PGPB) to improve plant soil micro-ecology [53]. Interestingly, the tobacco bacterial wilt pathogen Ralstonia [50], with low-level abundance (0.02%) in our study, indicates the low-level risk of the wilt disease for tobacco in each sampling site.
Tobacco rhizosphere contributes more to active bacterial diversity
Although the OTU and genera numbers of 16S-D and nifH-D are more abundant in rootzone than in rhizosphere soil (Fig. 1E and F). However, the OTU numbers of 16S-R and nifH-R are higher in the rhizosphere than in rootzone soils (Fig. 1G and H, Fig. S4). Consistent with the α diversity indices of 16S-R and nifH-R are obviously higher in the rhizosphere than in rootzone, but no obvious difference among the three sampling sites (Fig. 4). Thus, the diversity of active rhizosphere bacterial and nitrogen fixing bacterial community are mainly regulated by tobacco plants but not soil environmental factors. Plants excreted a high proportion of fixed photosynthetic carbohydrate compounds to the soil by root [5], a higher concentration of carbon sources in the rhizosphere may result higher active bacterial diversity. Due to the unique bacteria and active bacterial communities are observed in the three sampling sites, the microorganisms involved in the same ecological functions may differ in different sampling sites, for the same ecological process function could be performed by different microorganisms [48]. The active bacteria in the rhizosphere may have a better interaction relationship with tobacco and the active dominant groups will be targeted to isolate and could be priorly used in subsequently synthetic community construction.
Tobacco is usually infected by the root-knot nematode in field, and the infection by nematode could result in the high abundance of Escherichia-Shigella in farmland soil [54]. The predominant high expression of Escherichia-Shigella in this study (Fig. 2D) may indicate the high risk of root-knot nematode outbreak in each sampling site. Interestingly, Escherichia-Shigella was also found to be the dominant genus of tobacco seed endophytic bacteria [55]. The higher active genera of Stretomyces, Shingomonas, Burkholderia-Caballeronia-Paraburkholderia, Rhodanobacter, Allorhizobium-Neorhizobium-Pararhizobium-Rhizobium, Mucilaginibacter and Arthrobacter in tobacco rhizosphere (Fig. 2B) consistent with many species from these genera usually enriched in the healthy tobacco rhizosphere [50, 56]. Furthermore, strains belong to those genera usually have beneficial agronomic utilization potential. Such as functional isolates belong to Streptomyces, Burkholderia were isolated from tobacco rhizosphere with abilities to prevent tobacco bacterial wilt [56, 57]. Shingomonas, Rhizobium and Arthrobacter were widely distributed in the tobacco rhizosphere with nicotine-degrading abilities [58]. Low expression level of active Ralstonia spp. in 16S-R (< 0.1%, data not shown) (Fig. 2D), again indicating a low level of bacterial wilt endemic risk in the three sampling sites.
The diazotroph community is mainly determined by soil environmental factors, and the diversity of active nitrogen-fixing bacteria is mainly selected by rhizosphere
The three sampling sites are far away from each other (600, 1000, and 1600 km) (Fig. S1), and the unique diazotroph bacterial community structure (Fig. S9) from each sampling site consistent with the geographic factor (MAT and MAP) could be one of the main factors shaping the diazotroph community structure [28]. It also consistent with soil characteristic change (such as AK concentration) with a greater effect on the diazotrophic community than the rhizosphere [59]. Thus, tobacco enrolls diazotrophs based on the local bacterial community to perform nitrogen fixation, and the functional taxa may differ from sampling sites. Due to only a small amount of nifH-R data being obtained from 4/15 rootzone samples, the significant statistical difference between rootzone and rhizosphere samples for each genus would be unscientific. Compared with the top 20 genera of nifH-D and niH-R, only 9/20 dominant genera in nifH-D are also observed in top 20 genera of nifH-R. Thus, the abundance of diazotroph taxa may not always correlate with nitrogen fixation contribution in the community. And the higher nifH expressing species could contribute more nitrogen to tobacco than lower expressing species. In our study, the 167 nifH-D genera are found in 16S-D, accounted for 10.51% of the total bacterial community (based on 16S-D). However, the 25 active nitrogen-fixing (nifH-R) genera only account for 2.08% of the overall active bacteria community (based on 16S-R) and for 1.29% of the overall bacteria community (based on 16S-D). The above indicates that nitrogen-fixing bacteria only account for a very small proportion of the bacterial community, and nitrogen fixation mainly happens in the rhizosphere. The reason may be that the nitrogen fixation process is a highly energy-consuming process [60], and massive nitrogen fixation could exceed the provision of carbon source in the rhizosphere.
The predominant of Bradyrhizobium in each sampling site and it enriched more in the rhizosphere, consistent with it also as the dominant diazotroph in many terrestrial environments, such as sugarcane rhizosphere [61], farmland [28], and forest [62]. Although most species in this genus perform symbiotic nitrogen fixation with diverse legume hosts such as peanut [63], non-symbiotic nitrogen fixation was also reported between Bradyrhizobium spp. and non-legume hosts such as sugarcane [64]. Thus, the dominant in the diazotroph community and highest nifH expression of Bradyrhizobium indicate species belong to this genus contributed more nitrogen for tobacco than other genera in the three sampling sites. The distribution of Bradyrhizobium shows an obviously negative correlation with soil pH and AK (Fig. 6D). It consistent with Bradyrhizobium dominant in many acid soils, such as acid rhizosphere soil of maize, peanut, and soybean [59]. However, the dominant (Fig. 3B) and high expression of nifH of Skermanella (Fig. 3D) in Linqu mainly correlates with its positive correlates with pH and AK (Fig. 6D). And the high expression of nifH belongs to Extensimonas in Bijie mainly due to it being positively correlated with AN, AP, TN, and OC and negatively correlated with pH (Fig. 6D). The high expression of Geobacter nifH in rootzone soil but not in rhizosphere is consistent with it also with high expression in farmland bulk soil [65]. In all, the tobacco rhizosphere active nitrogen-fixing bacteria are detected and analyzed in this study, and it would be interesting to evaluate the nitrogen-fixation bacteria belonging to the dominant diazotroph genera with non-active nitrogen-fixing genera under greenhouse or field conditions.
Conclusion
In this study, we collected tobacco rootzone and rhizosphere soil from three sampling sites. The overall bacteria and diazotroph community composition and diversity differed according to the sampling sites, but no obvious difference was observed between the rhizosphere and rootzone. Thus, the environment contributes more to the rhizosphere bacterial community assemble than rhizosphere selection. Furthermore, both the overall bacterial and diazotroph bacteria communities are mainly affected by climate variables including MAT and MAP, soil AK, pH and AN. However, the diversity of active overall bacteria and active nitrogen-fixing bacteria community shows obvious significant between rootzone and rhizosphere but not between different sampling sites. It indicating the rhizosphere contributes more to the active overall bacterial and nitrogen-fixing bacteria diversity than soil environmental factors. Importantly, our study suggests that active groups should be selected as the priority to construct a better and more stable synthetic community.
Supplementary Information
Acknowledgements
This study was financially supported by the Science and Technology Project of CNTC 110202201004(JY-04).
CRediT authorship contribution statement
Yalong Xu: Writing original draft, methodology. Jingjing: data analysis and soil characteristic determination. Chan Qiao: data analysis and figure drawing. Jinchu Yang: data analysis. Juan Li: prepare the soil samples and soil characteristic determination. Xueao Zheng: sample collecting and data analysis. Chen Wang: sample collection and data analyses. Peijian Cao: review and editing the manuscript. Yan Li: project design, sample collection, data analysis and writing draft manuscript. Qiansi Chen: project design and administration, supervision, and validation of the manuscript.
Abbreviations
- 16S-D
The 16S rRNA gene amplicons obtained by using soil DNA as a template
- 16S-R
The 16S rRNA gene amplicons obtained by using soil cDNA as a template
- nifH-D
The nifH amplicons obtained by using soil DNA as a template
- nifH-R
The nifH amplicons obtained by using soil cDNA as a template
- AN
Available nitrogen
- AK
Available potassium
- AP
Available phosphorus
- TN
Total nitrogen
- OC
Organic carbon
- MAP
Mean annual precipitation
- MAT
Mean annual temperature
- TS
Temperature seasonality
- PS
Precipitation seasonality
Authors’ contributions
The authors declare that they have no competing interests.
Data availability
All the raw reads obtained in this study were submitted to the Genome Sequence Archive in National Genomics Data Center of China National Center for Bioinformation (Accession Numbers: CRA016587, CRA016608, CRA016612 and CRA016620) and the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under the accession PRJNA1160359, PRJNA1160397, PRJNA1160409 and PRJNA1160160.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yalong Xu and Jingjing Li contributted equally to this work.
Contributor Information
Yan Li, Email: liyan0709@hotmail.com.
Qiansi Chen, Email: chen_qiansi@163.com.
References
- 1.Castrillo G, Teixeira PJPL, Paredes SH, Law TF, de Lorenzo L, Feltcher ME, Finkel OM, Breakfield NW, Mieczkowski P, Jones CD, Paz-Ares J, Dangl JL. Root microbiota drive direct integration of phosphate stress and immunity. Nature. 2017;543:513–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raaijmakers JM, Mazzola M. Soil immune responses. Science. 2016;352:1392–3. [DOI] [PubMed] [Google Scholar]
- 3.Bintarti AF, Wilson JK, Quintanilla-Tornel MA, Shade A. Biogeography and diversity of multi-trophic root zone microbiomes in Michigan apple orchards: analysis of rootstock, scion, and local growing region. Phytobiomes J. 2020;4:122–32. [Google Scholar]
- 4.Wang C, Kuzyakov Y. Rhizosphere engineering for soil carbon sequestration. Trends Plant Sci. 2024;29:447–68. [DOI] [PubMed] [Google Scholar]
- 5.Su Y, Wang J, Gao W, Wang R, Yang W, Zhang H, Huang L, Guo L. Dynamic metabolites: a bridge between plants and microbes. Sci Total Environ. 2023;899. [DOI] [PubMed] [Google Scholar]
- 6.Bakker PAHM, Pieterse CMJ, de Jonge R, Berendsen RL. The soil-borne legacy. Cell. 2018;172:1178–80. [DOI] [PubMed] [Google Scholar]
- 7.Wang L, Chen M, Lam P, Dini-Andreote F, Dai L, Wei Z. Multifaceted roles of flavonoids mediating plant-microbe interactions. Microbiome. 2022;10:233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bardgett R. The Biology of Soil: A community and ecosystem approach. UK: Oxford University Press; 2005. [Google Scholar]
- 9.Xun W, Liu Y, Ma A, Yan H, Miao Y, Shao J, Zhang N, Xu Z, Shen Q, Zhang R. Dissection of rhizosphere microbiome and exploiting strategies for sustainable agriculture. New Phytol. 2024;242(6):2401–10. [DOI] [PubMed] [Google Scholar]
- 10.Mclaughlin S, Zhalnina K, Kosina S, Northen TR, Sasse J. The core metabolome and root exudation dynamics of three phylogenetically distinct plant species. Nat Commun. 2023;14:1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng L, Karim MR, Hu Y, Shen R, Lan P. Greater morphological and primary metabolic adaptations in roots contribute to phosphate-deficiency tolerance in the bread wheat cultivar Kenong199. BMC Plant Biol. 2021;21:381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng Z, Zheng Q, Shi J, He Y, Yang X, Huang X, Wu L, Xu J. Metagenomic and machine learning-aided identification of biomarkers driving distinctive Cd accumulation features in the root-associated microbiome of two rice cultivars. ISME Commun. 2023;3(1):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xun W, Yan R, Ren Y, Jin D, Xiong W, Zhang G, Cui Z, Xin X, Zhang R. Grazing-induced microbiome alterations drive soil organic carbon turnover and productivity in meadow steppe. Microbiome. 2018;6:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang A, Zhang Y, Wang G, Zhang Z. Soil physicochemical properties and microorganisms jointly regulate the variations of soil carbon and nitrogen cycles along vegetation restoration on the Loess Plateau, China. Plant Soil. 2024;494:413–36. [Google Scholar]
- 15.Rousk J, Bååth E, Brookes PC, Lauber CL, Lozupone C, Caporaso JG, Knight R, Fierer N. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 2010;4:1340–51. [DOI] [PubMed] [Google Scholar]
- 16.Xu L, Naylor D, Dong Z, Simmons T, Pierroz G, Hixson KK, Kim Y, Zink EM, Engbrecht KM, Wang Y, Gao C, DeGraaf S, Madera MA, Sievert JA, Hollingsworth J, Birdseye D, Scheller HV, Hutmacher R, Dahlberg J, Jansson C, Taylor JW, Lemaux PG, Coleman-Derr D. Drought delays development of the sorghum root microbiome and enriches for monoderm bacteria. Proc Nat Acad Sci. 2018;115:E4284–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peng Z, Liu Y, Qi J, Gao H, Li X, Tian Q, Qian X, Wei G, Jiao S. The climate-driven distribution and response to global change of soil-borne pathogens in agroecosystems. Global Ecol Biogeogr. 2023;32:766–79. [Google Scholar]
- 18.Vitousek PM, Menge DNL, Reed SC, Cleveland CC. Biological nitrogen fixation: rates, patterns and ecological controls in terrestrial ecosystems. Philos T R Soc B: Biol Sci. 2013;368:20130119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuypers MMM, Marchant HK, Kartal B. The microbial nitrogen-cycling network. Nat Rev Microbiol. 2018;16:263–76. [DOI] [PubMed] [Google Scholar]
- 20.Dixon R, Kahn D. Genetic regulation of biological nitrogen fixation. Nat Rev Microbiol. 2004;2:621–31. [DOI] [PubMed] [Google Scholar]
- 21.Barron AR, Wurzburger N, Bellenger JP, Wright SJ, Kraepiel AML, Hedin LO. Molybdenum limitation of asymbiotic nitrogen fixation in tropical forest soils. Nat Geosci. 2009;2:42–5. [Google Scholar]
- 22.Van Deynze A, Zamora P, Delaux P, Heitmann C, Jayaraman D, Rajasekar S, Graham D, Maeda J, Gibson D, Schwartz KD, Berry AM, Bhatnagar S, Jospin G, Darling A, Jeannotte R, Lopez J, Weimer BC, Eisen JA, Shapiro HY, Ané JM, Bennett AB. Nitrogen fixation in a landrace of maize is supported by a mucilage-associated diazotrophic microbiota. PLoS Biol. 2018;16:e2006352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sivasakthivelan P, Saranraj P, Sayyed RZ, Arivukkarasu K, Kokila M, Manigandan M, Seifi S. Inoculant production and formulation of Azospirillum species. In: Mawar R, Sayyed RZ, Sharma SK, Sattiraju KS, editors. Plant growth promoting microorganisms of Arid Region. Singapore: Springer Nature Singapore; 2023. [Google Scholar]
- 24.Naqqash T, Malik KA, Imran A, Hameed S, Shahid M, Hanif MK, Majeed A, Arshad M, van Elsas JD. Isolation and characterization of Rhizobium from non-leguminous potato plants: new frontiers in Rhizobium research. Biol Fert Soils. 2024;60:307–25. [Google Scholar]
- 25.Fan K, Weisenhorn P, Gilbert JA, Shi Y, Bai Y, Chu H. Soil pH correlates with the co-occurrence and assemblage process of diazotrophic communities in rhizosphere and bulk soils of wheat fields. Soil Biol Biochem. 2018;121:185–92. [Google Scholar]
- 26.Gupta VVSR, Kroker SJ, Hicks M, Davoren CW, Descheemaeker K, Llewellyn R. Nitrogen cycling in summer active perennial grass systems in South Australia: non-symbiotic nitrogen fixation. Crop Pasture Sci. 2014;65(10):1044–56. [Google Scholar]
- 27.Wang C, Zheng M, Song W, Wen S, Wang B, Zhu C, Shen R. Impact of 25 years of inorganic fertilization on diazotrophic abundance and community structure in an acidic soil in southern China. Soil Biol Biochem. 2017;113:240–9. [Google Scholar]
- 28.Meng X, Liao H, Fan H, Zhang X, Li Y, Yao H, Razavi BS. The geographical scale dependence of diazotroph assembly and activity: effect of a decade fertilization. Geoderma. 2021;386. [Google Scholar]
- 29.Bahulikar RA, Chaluvadi SR, Torres-Jerez I, Mosali J, Bennetzen JL, Udvardi M. Nitrogen fertilization reduces nitrogen fixation activity of diverse diazotrophs in switchgrass roots. Phytobiomes J. 2020;5:80–7. [Google Scholar]
- 30.Magoc T, Salzberg S. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burbano CS, Liu Y, Rösner KL, Reis VM, Caballero-Mellado J, Reinhold-Hurek B, Hurek T. Predominant nifH transcript phylotypes related to Rhizobium rosettiformans in field-grown sugarcane plants and in Norway spruce. Env Microbiol Rep. 2011;3:383–9. [DOI] [PubMed] [Google Scholar]
- 32.Wang X, Bei Q, Yang W, Zhang H, Hao J, Qian L, Feng Y, Xie Z. Unveiling of active diazotrophs in a flooded rice soil by combination of NanoSIMS and 15N2-DNA-stable isotope probing. Biol Fert Soils. 2020;56:1189–99. [Google Scholar]
- 33.Xiao X, Chen W, Zong L, Yang J, Jiao S, Lin Y, Wang E, Wei G. Two cultivated legume plants reveal the enrichment process of the microbiome in the rhizocompartments. Mol Ecol. 2017;26:1641–51. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Z, Liu W, Shao S, Wang ET, Li Y. Diverse genomic backgrounds vs. highly conserved symbiotic genes in Sesbania-nodulating bacteria: shaping of the rhizobial community by host and soil properties. Microb Ecol. 2020;80:158–68. [DOI] [PubMed] [Google Scholar]
- 35.Shen J, Li R, Zhang F, Fan J, Tang C, Rengel Z. Crop yields, soil fertility and phosphorus fractions in response to long-term fertilization under the rice monoculture system on a calcareous soil. Field Crop Res. 2004;86:225–38. [Google Scholar]
- 36.Westerman RL. Soil testing and plant analysis. 3rd ed. Madison: Soil Science Society of America Inc; 1990.
- 37.Boudjabi S, Chenchouni H. Soil fertility indicators and soil stoichiometry in semi-arid steppe rangelands. CATENA. 2022;210. [Google Scholar]
- 38.Li C, Jia Z, Zhang S, Li T, Ma S, Cheng X, Chen M, Nie H, Zhai L, Zhang B, Liu X, Zhang J, Muller C. The positive effects of mineral-solubilizing microbial inoculants on asymbiotic nitrogen fixation of abandoned mine soils are driven by keystone phylotype. Sci Total Environ. 2023;882. [DOI] [PubMed] [Google Scholar]
- 39.Chen S. Ultrafast one-pass FASTQ data preprocessing, quality control, and deduplication using fastp. iMeta. 2023;2:e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–8. [DOI] [PubMed] [Google Scholar]
- 42.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2012;41:D590–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang J, Zheng M, Zhang Y, Wang J, Shen H, Lin Y, Tang X, Hui D, Lambers H, Sardans J, Peñuelas J, Liu Z. Soil phosphorus availability affects diazotroph communities during vegetation succession in lowland subtropical forests. Appl Soil Ecol. 2021;166. [Google Scholar]
- 44.Dixon P. VEGAN, a package of R functions for community ecology. J Veg Sci. 2003;14:927–30. [Google Scholar]
- 45.Liu Q, Wang S, Li K, Qiao J, Guo Y, Liu Z, Guo X. Responses of soil bacterial and fungal communities to the long-term monoculture of grapevine. Appl Microbiol Biot. 2021;105:7035–50. [DOI] [PubMed] [Google Scholar]
- 46.Deng J, Bai X, Zhou Y, Zhu W, Yin Y. Variations of soil microbial communities accompanied by different vegetation restoration in an open-cut iron mining area. Sci Total Environ. 2020;704. [DOI] [PubMed] [Google Scholar]
- 47.Martínez-García LB, Korthals G, Brussaard L, Jørgensen HB, De Deyn GB. Organic management and cover crop species steer soil microbial community structure and functionality along with soil organic matter properties. Agr Ecosyst Environ. 2018;263:7–17. [Google Scholar]
- 48.Lian W, Mohamad OAA, Dong L, Zhang L, Wang D, Liu L, Han M, Li S, Wang S, Antunes A, Fang B, Jiao J, Li W. Culturomics and metagenomics-based insights into the microbial community and function of rhizosphere soils in Sinai desert farming systems. Environ Microbiome. 2023;18:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gong B, He Y, Luo Z, Peng H, Cai H, Zhu Y, Bin J, Ding M. Response of rhizosphere soil physicochemical properties and microbial community structure to continuous cultivation of tobacco. Ann Microbiol. 2024;74:4. [Google Scholar]
- 50.Liang J, Wei C, Song X, Wang R, Shi H, Tan J, Cheng D, Wang W, Wang X. Bacterial wilt affects the structure and assembly of microbial communities along the soil-root continuum. Environ Microbiome. 2024;19:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ling N, Wang T, Kuzyakov Y. Rhizosphere bacteriome structure and functions. Nat Commun. 2022;13:836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou X, Mi Q, Yao J, Wu H, Liu X, Li Y, Duan Y, Chen J, Dang L, Mo M, Li X, Li W. Sphingomonas tabacisoli sp. nov., a member of the genus Sphingomonas, isolated from rhizosphere soil of Nicotiana tabacum L. Int J Syst Evol Microbiol. 2018;68:2574–9. [DOI] [PubMed] [Google Scholar]
- 53.Wang F, Wei Y, Yan T, Wang C, Chao Y, Jia M, An L, Sheng H. Sphingomonas sp. Hbc-6 alters physiological metabolism and recruits beneficial rhizosphere bacteria to improve plant growth and drought tolerance. Front Plant Sci. 2022;13:1002772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu R, Chen M, Liu B, Huang K, Mao Z, Li H, Zhao J. A root-knot nematode effector manipulates the rhizosphere microbiome for establishing parasitism relationship with hosts. Front Microbiol. 2023;14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xie H, Wang H, Cai L, Zhou H, Liu C, Lu N, Shi C, Wang X. Community structure and diversity of endophytic bacteria of tobacco seeds. Acta Microbiol Sin. 2020;60:601–16. [Google Scholar]
- 56.Tao J, Gu M, Yu S, Shi J, Cheng L, Jin J, Lu P, Zhang J, Li H, Cao P. The beneficial endophytic microbes enhanced tobacco defense system to resist bacterial wilt disease. Chem Biol Tech Agri. 2024;11:21. [Google Scholar]
- 57.Wang R, Li B, Cai S, Ding Y, Shi M, Jin T, Lin W, Liu P. Genetic diversity of Ralstonia solanacearum causes tobacco bacterial wilt in Fujian province and identification of biocontrol Streptomyces sp. Plant Dis. 2024;108(7):1946–58. [DOI] [PubMed] [Google Scholar]
- 58.Wang Y, Luo X, Chu P, Shi H, Wang R, Li J, Zheng S. Cultivation and application of nicotine-degrading bacteria and environmental functioning in tobacco planting soil. Bioresour Bioprocess. 2023;10:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang C, Zheng MM, Chen J, Shen RF. Land-use change has a greater effect on soil diazotrophic community structure than the plant rhizosphere in acidic ferralsols in southern China. Plant Soil. 2021;462:445–58. [Google Scholar]
- 60.Alleman AB, Costas AG, Mus F, Peters JW. Rnf and Fix have specific roles during aerobic nitrogen fixation in Azotobacter vinelandii. Appl Environ Microb. 2022;88:e1022–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.de Alencar Menezes Júnior I, Feitosa De Matos G, Moura De Freitas K, Da Conceição Jesus E, Rouws LFM. Occurrence of diverse Bradyrhizobium spp. in roots and rhizospheres of two commercial Brazilian sugarcane cultivars. Braz J Microbiol. 2019;50:759–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xiao D, He X, Zhang W, Cheng M, Hu P, Wang K. Diazotroph and arbuscular mycorrhizal fungal diversity and community composition responses to karst and non-karst soils. Appl Soil Ecol. 2022;170. [Google Scholar]
- 63.Shao S, Chen M, Liu W, Hu X, Wang ET, Yu S, Li Y. Long-term monoculture reduces the symbiotic rhizobial biodiversity of peanut. Syst Appl Microbiol. 2020;43. [DOI] [PubMed] [Google Scholar]
- 64.De Matos GF, Rouws L, Simoes-Araujo J, Baldani J. Evolution and function of nitrogen fixation gene clusters in sugarcane associated Bradyrhizobium strains. Environ Microbiol. 2021;23:6148–62. [DOI] [PubMed] [Google Scholar]
- 65.Calderoli PA, Collavino MM, Behrends KF, Morrás HJM, Aguilar OM. Analysis of nifH-RNA reveals phylotypes related to Geobacter and Cyanobacteria as important functional components of the N2-fixing community depending on depth and agricultural use of soil. Microbiol Open. 2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the raw reads obtained in this study were submitted to the Genome Sequence Archive in National Genomics Data Center of China National Center for Bioinformation (Accession Numbers: CRA016587, CRA016608, CRA016612 and CRA016620) and the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under the accession PRJNA1160359, PRJNA1160397, PRJNA1160409 and PRJNA1160160.