Abstract
Identification of tissue-specific gene regulatory networks can yield insights into the molecular basis of a tissue's development, function and pathology. Here, we present a computational approach designed to identify potential regulatory target genes of photoreceptor cell-specific transcription factors (TFs). The approach is based on the hypothesis that genes related to the retina in terms of expression, disease and/or function are more likely to be the targets of retina-specific TFs than other genes. A list of genes that are preferentially expressed in retina was obtained by integrating expressed sequence tag, SAGE and microarray datasets. The regulatory targets of retina-specific TFs are enriched in this set of retina-related genes. A Bayesian approach was employed to integrate information about binding site location relative to a gene's transcription start site. Our method was applied to three retina-specific TFs, CRX, NRL and NR2E3, and a number of potential targets were predicted. To experimentally assess the validity of the bioinformatic predictions, mobility shift, transient transfection and chromatin immunoprecipitation assays were performed with five predicted CRX targets, and the results were suggestive of CRX regulation in 5/5, 3/5 and 4/5 cases, respectively. Together, these experiments strongly suggest that RP1, GUCY2D, ABCA4 are novel targets of CRX.
INTRODUCTION
Understanding of the regulatory networks controlling retinal gene expression will probably provide insights into the molecular basis of retinal development, function and disease. Development of network models requires knowledge about the transcription factors (TFs) involved, the target genes that are regulated by these factors, and the interactions of the products of these genes with other downstream and upstream genes. Traditionally, the identity and nature of TF-DNA regulatory element interactions have been studied by wet-lab-based approaches, usually analyzing one gene at a time. Among the approaches that have been employed are affinity chromatography and related protein purification methods, yeast one-hybrid cloning, electrophoretic mobility shift assays (EMSAs), protein–DNA cross-linking studies, DNase I footprint analysis and chromatin immunoprecipitation (ChIP). More recently, a method termed ChIP-chip, which combines techniques of ChIP and microarray (chip), has been developed to determine TF binding locations on a genomic scale (1,2). Although advances have certainly been made in using these approaches to identify retinal regulatory factors and elements (3,4) and a number of TF mutations associated with retinal disease have been identified (5–12), our overall knowledge of retinal regulatory networks is still rather limited.
With the goal of ultimately developing more comprehensive and accurate models of retinal regulatory networks, we have been trying to apply and further develop computational approaches to the analysis of retinal gene expression datasets. As a specific model, we have so far focused on identification of the regulatory targets of CRX (13,14), and to a lesser extent on NRL (15–17) and NR2E3 (10,18,19). These TFs are predominantly retina-specific and play an important role in retinal development, function and pathology. We have concentrated on CRX, not only because of its biological importance but also because significant experimental data is already available related to its regulatory targets. For example, microarray and SAGE analysis have been performed comparing gene expressions between Crx null (−/−) and wild-type mice (20,21).
Increasing efforts are being made to utilize bioinformatics to complement laboratory-based methods in the analysis of transcriptional regulatory networks. Due to the relative simplicity of its genome, many of these efforts have focused on yeast (1,22–24), but some have also explored mammalian systems (25–28). The difficulty of predicting regulatory targets based on TF binding sites is largely due to the fact that TF binding sequences are short and often degenerate. The short sequences of binding motifs by themselves do not appear to be sufficient for appropriate and specific protein–DNA recognition in vivo. A full understanding of recognition mechanisms is likely to require information on protein–protein interactions and chromatin structure. Two widely used computational methods that can increase prediction specificity are phylogenetic footprinting and identifying cis-regulatory module. Phylogenetic footprinting is based on the observation that functional binding motifs are more often located on evolutionarily conserved regions (26,29–31). The method of identification of cis-regulatory modules assumes that clusters of binding motifs of related TFs are more likely to be functional than a solitary binding motif (25,32–36).
Here, we propose a complementary method to enrich for potential target genes of a tissue-specific TF. The method is based on the reasonable hypothesis that most genes that are regulated by retina-specific TFs are related to the retina in terms of expression, function or disease. Instead of searching for TF targets from the entire genome, we have concentrated on the subset of genes that are retina-related. This idea is actually quite intuitive. When researchers experimentally hunt for target genes of tissue-specific TFs, the genes relevant to the tissue are often the good candidates to be examined. Like other computational methods for target enrichment, this approach will miss some true positives since some targets of retina-specific TF may not be specifically expressed in retina, or may not have a known retinal function. However, the important question here is how much we gain in specificity by losing a certain amount of sensitivity. Based on the results from our computational and experimental work, our approach seems to provide a reasonable balance.
Identification of a set of retina-related genes, however, is not trivial. A large proportion of retina-related genes are retina-enriched genes that are preferentially expressed in the retina compared to other tissues. A number of groups have utilized a variety of approaches to identify such retina-enriched genes (37). These studies have been somewhat successful, in that they have identified interesting retina-specific genes (21,38–44), but they have also been limited by technical and interpretive problems (45). One manifestation of these problems is that only a surprisingly small portion of the identified retina-enriched genes overlaps across the studies, suggesting significant error rates in at least some of the individual studies (46). One approach to reducing the overall error rate is to integrate data across the independent studies. In this paper, we proposed a score-based integration approach to identify retina-enriched genes.
This identification of genes preferentially expressed in retina turned out to be useful in enriching for the targets of retina-specific TFs. We identified 591 retina-related genes, which is ∼35-fold reduction in prediction space from the entire human genome (20 000–25 000 genes) (47). Among the 591 retina-related genes, we identified 169, 166 and 97 putative targets of CRX, NRL and NR2E3, respectively. A significant fraction of known targets was recovered in our predictions. Furthermore, we applied a Bayesian approach to rank these targets for prioritizing the experimental validation. Finally, we performed a set of experiments (EMSA, transient transfection and ChIP) on five genes which were predicted as novel targets of CRX. Three of them yielded positive results in all experiments, strongly suggesting that they are indeed novel targets of CRX, and that the inclusion of expression data into TF target predictions can yield reasonable specificity.
MATERIALS AND METHODS
EST, SAGE and microarray datasets
Expressed sequence tag (EST) data were obtained from NCBI's UniGene dataset (http://www.ncbi.nlm.nih.gov/UniGene). SAGE data was obtained from NCBI GEO (Gene Expression Omnibus) website (http://www.ncbi.nlm.nih.gov/GEO). Microarray data was from Chowers et al. (46). Additional EST and SAGE data was extracted from public domain cDNA libraries (NCBI). Only non-normalized libraries from normal tissues were included in the analysis (‘non-normalized’ and ‘normal’ were used as key words for library searching). Two sets of reference libraries were constructed to compare with the retina libraries. One represented libraries from normal brain tissues (including different subregions such as cortex, pineal gland and cerebellum), and the other represented ‘pooled’ libraries from a variety of normal tissues including liver, kidney and brain. The library numbers of (retina, brain, ‘pooled’) for EST are (3,14,74), for SAGE (4,8,32) and for microarray (5,2,4). The detailed description of each library can be found in the Supplementary Materials. The gene expression levels for the EST and SAGE data sets were normalized by the library size. The numbers of genes in the three sets are 16 569, 32 435 and 6098 for the EST, SAGE and microarray studies, respectively. Only the genes found in all three studies were considered in the next stage.
Genome sequences and alignments
The human and mouse alignments were obtained from the UCSC genome web browser (48) using blastz (49). The alignments were filtered so that only the best alignment for any given region of the human genome was left. The alignment file we used is axtBest. The human assembly we used is build 33 (or hg16), and the mouse assembly is MGSCv4 (or mm3).
Promoter sequences
In order to reduce the complexity of our analysis, we restricted the regions of interest to sequences from 2000 bp upstream to 200 bp downstream relative to each gene's transcriptional start site (TSS). To identify the upstream sequence of a gene, however, can be non-trivial. Most of the cDNA information stored in current databases is incomplete in the sense that they lack the precise information TSSs. To address this limitation, we combined data from the database of Eukaryotic Promoter Database (EPD) (50,51) and the DataBase of human Transcriptional Start Sites (DBTSSs) (52) to obtain a set of experimentally determined TSSs. A total of 1871 human promoter sequences were obtained from EPD and ∼9000 full-length 5′-untranslated region sequences were obtained from DBTSS.
Bayesian approach for motif location constrain
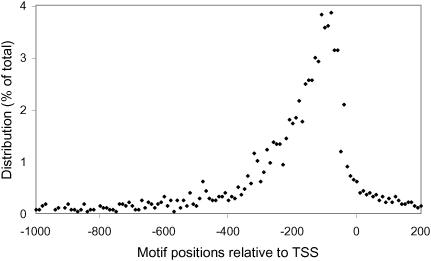
We used Bayes' rule for update: p(motif|dis) = p(dis|motif) * p(motif)/p(dis), where p(dis|motif) is the probability of a given distance (dis) for motif. We obtained the distribution from TRANSFAC (53,54) (see Figure 3). p(motif) is the prior probability that a hit is a regulatory motif and p(dis) is the distance distribution for all hits of the motifs. p(motif) was estimated from the hit score and defined as the ratio of the number of positive examples versus the total number of hits in a certain score range.
Figure 3.
Distribution of the positions of binding sites relative to the TSS. Negative values represent upstream regions. Approximately 2100 eukaryotic binding sites, extracted from the TRANSFAC database, were used for the calculation.
False discovery rate (FDR)
First, permutations were performed. The tissue labels were randomly assigned to retina and other tissues. Since values of gene expression in various studies usually have different scale, the permutations were performed only within each study; label randomization did not cross the different studies. For each permutation, the t scores for each individual study and the summary score for integrated data set were calculated. Average t scores and average summary score for permutations were obtained. Then, we compared the score distributions in original data sets and those from permutation. For a given threshold x, the FDR was calculated as np/n, where np is the average number of genes whose summary scores are larger than x after permutation, and n is the same number in the original data. Also, np and n denote the numbers of falsely significant genes and genes called significant, respectively. The genes called significant include both true and false significant genes. With a series of threshold x's, we obtained the number of falsely significant genes as a function of the number of genes called significant. The calculation was performed for three studies and integrated data set.
Hypergeometric probability
To determine if the number of overlapped target genes between two factors is over-represented or by chance, we calculated the hypergeometric probability by the formula:
where N is the total number of retina-enriched genes (N = 617), t1 and t2 are the numbers of target genes of two factors and x is the number of shared targets of these factors. Notice it is not symmetric for exchanging t1 and t2 in the formula. We chose the larger one as the P-value.
EMSAs
Assays were carried out essentially as previously described (13), with the exception of using p32-α-dGTP as the radioactive nucleotide for probe labeling. The radioactive probes were purified through G25-columns according to the manufacturer's protocol (Amersham Pharmacia Biotech 27-5325-01). Approximately 10 000 c.p.m. of probe and 20 ng of CRX-HD-GST protein were used for each assay. The DNA oligomer pairs used to generate the target probes were listed in Supplementary Material.
Generation of luciferase reporter constructs
The promoter regions of RP1, GUCY2D, ABCA4, ARR3 or BBS4 were amplified from human genomic DNA by PCR, using primers containing XhoI (5′ end) and HindIII (3′ end) restriction sites. Promoter–luciferase reporter constructs were then generated by directionally cloning the PCR products into the XhoI and HindIII sites of the pGL2-Basic vector containing firefly luciferase gene (Promega). Construct sequences were confirmed by sequencing. DNA used for transient transfection was prepared using Qiagen plasmid maxi-prep according to the manufacturer's protocol. The primers used for PCR cloning were shown in Supplementary Material.
Transient transfection and luciferase assay
Transient transfections were performed using a modification of our previously described procedure (13). Lipofectamine 2000 (Invitrogen) was used instead of calcium phosphate, and six-well culture plates were used for culturing GripTite 293 MSR cells (Invitrogen). Transfections were performed using 80–90% confluent cells and a 1:2.5 ratio of DNA (μg)/Lipofectamine (μl). A total of 2.2 μg of DNA was used for each transfection, including 0.2 μg of reporter construct, different amounts (0, 0.2, 1 or 2 μg) of pcDNA3.1/HisC-bovin Crx expression construct and 2 ng of Renilla luciferase reporter (pRL-CMV, Promega) as an internal control for transfection efficiency. Luciferase assays were performed using the Dual Luciferase Reporter Assay System (Promega) as described by the manufacturer. Each construct was transfected in triplicate per experiment and three independent experiments were performed. Since we noted that increasing amounts of the CRX expression construct consistently led to decreasing amounts of Renilla luciferase activity, which would have led to artifactually high CRX transactivation values, we performed a second normalization based on Renilla luciferase-normalized firefly luciferase values obtained with empty pGL2-Basic vector.
ChIP
Primers were designed to amplify 150–250 bp fragments of the promoter regions containing predicted CRX binding site(s) of mouse Rp1, Gucy2d, Bbs4, Abca4 and Arr3. The promoter regions of Rho and Alb were also analyzed as positive and negative control, respectively. ChIP assays were performed using adult mouse retina as described previously (55,56), with minor modifications. Intact retinas harvested from 8 week old BALB/cJ mice (The Jackson Laboratory) were treated with 1% formaldehyde in PBS at room temperature for 15 min and then homogenized with a Dounce homogenizer. One and a half mouse retinas were used for each ChIP reaction. Chromatin complexes were sheared in SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris–HCl, pH 8.1, 1 mM PMSF, 1 μg/ml aprotinin and 1 μg/ml pepstatin A) to an average length of ∼500 bp by 3 repeats of 10 s sonication at 100% duty cycle and 1.5 power output using a Branson Sonifier 250. After diluting the SDS concentration, immunoprecipitation was performed with an anti-CRX antibody (p261, a gift from Dr Shiming Chen, Washington University) followed by Protein A agarose (Upstate Biotechnology). After washing and eluting the DNA–protein complexes with 300 μl of elution buffer (1% SDS and 0.1 M NaHCO3), cross-links were reversed by heating at 65°C for 4 h. The precipitated DNA was purified by phenol/chloroform extraction and ethanol precipitation, resuspended in 30 μl TE buffer and 1 μl of the resultant solution was analyzed by PCR using gene-specific primers. The same procedures with no antibody were performed in parallel as negative control. The primers used for analysis were listed in Supplementary Material.
RESULTS
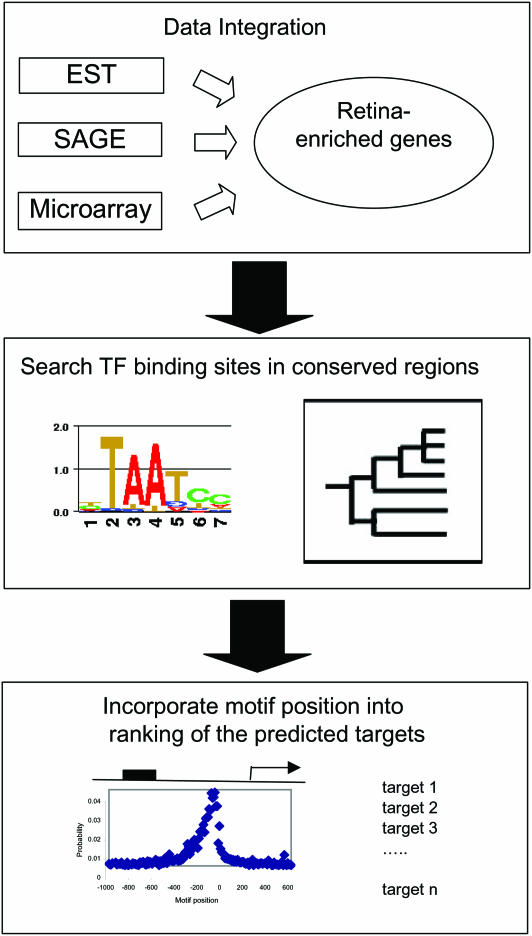
We utilized a multiple-step approach to predict the regulatory targets of retina-specific TFs (see Figure 1). First, we used a statistical approach to identify a set of retina-enriched genes, which we hypothesize to be more likely to be the target genes of retina-specific TFs than a set of random genes. Then, we searched for the presence of the binding sites of these TFs in the promoter regions of the retina-enriched genes using a phylogenetic footprinting approach. Finally, the information of binding site position relative to TSS was incorporated and the predicted targets were prioritized based on a probability score.
Figure 1.
Schematic view of our approach to identification of regulatory target genes of retina-specific TFs.
Positive controls
As a guide to assess the sensitivity and specificity of target prediction, we chose a set of positive control genes that have already been reported, based on experimental data, to be regulatory targets of CRX (13,14,56). The genes chosen were rhodopsin (RHO), arrestin (SAG), S-cone opsin (OPN1SW), M-cone opsin (OPN1MW), phosphodiesterase 6B (PDE6B) and guanine nucleotide binding protein (GNAT1), and they are referred to below as positive control set I. Although the experimental data supporting control set I is strong, the set is biased by the genes that researchers have happened to choose as their genes of interest. Most of them are expressed specifically in photoreceptor cells. Using this set of genes as positive controls is likely to over-estimate the sensitivity in our study. We therefore chose another set, which is from a SAGE analysis comparing retinal gene expression in Crx null compared to that in wild-type mice (21). The genes that were identified as significantly down-regulated in the Crx (−/−) animals are potential CRX target genes. Of the 122 differentially expressed murine genes, we identified 45 human orthologs. Among these 45 genes, 27 genes contain at least one CRX binding site in their promoter regions. This set of 27 genes is defined as positive control set II. We did not combine sets I and II because they represent two different approaches. Compared with set II, set I might have a higher confidence level, but on the other hand, it is biased to retina-related genes.
Prediction of CRX target genes
We attempted to predict CRX target genes in a subset of the human genome by incorporating tissue-specificity information. The rationale for this was that since the expression of CRX is largely retina-specific, it seemed reasonable that most of its target genes would be relevant to retina in terms of expression, function or disease. This set of retina-related genes is expected to be enriched for CRX targets. First, we identified a set of genes that are preferentially expressed in the retina compared to other tissues.
Identification of retina-enriched genes
We sought to compile a reliable list of genes that were preferentially expressed in the retina by integrating EST, SAGE and microarray datasets. Since it was not clear a priori which of the available lists were more accurate, we used a statistical approach to combine the datasets, reasoning that a set combining the information from the different experimental approaches would more closely approximate the ‘true’ list of retina-enriched genes.
Statistical testing was performed based on the null hypothesis that there is no gene that is preferentially expressed in retina compared to other tissues. A statistical t-test score for each gene was calculated for each study (i.e. EST, SAGE and microarray). The t-test score for gene i is defined as
where and are the average expression levels for retinal and non-ocular libraries, respectively, Vr and Vn are defined as , ; and n1 and n2 are the numbers of libraries. For the genes that were present in all three studies, a summary score was calculated as the average of the three scores from these individual studies, i.e. t = (tEST + tSAGE + tarray)/3. The use of the average function to combine t scores is based on the assumption that these studies yield data of equal quality. An alternative way for integration is using effect size (57,58) instead of t score. In fact, the results obtained by effect size and t score are similar in this particular case. The correlation coefficient of the gene rankings from the two integration approaches is 0.998. A more sophisticated integration approach might be to assign a weight to each study based on its data quality and then use a Bayesian method for the integration. However, since there are only a few known retina-enriched genes available, it is not statistically sound to assess the quality of each data set based on a limited group of known retina-enriched genes. Furthermore, we checked the distributions of t values. They are comparable for three studies and thus justify the simple averaging.
By comparing gene expression from retina libraries with that from ‘pooled’ tissues, summary scores, which reflect the confidence level of a gene being preferentially expressed in retina, were calculated. By ranking the summary score, we obtained a corresponding list of retina-enriched gene. Table 1 shows the top 20 genes from this list, and the whole list can be found in the Supplementary Material. The list can be classified into three types of genes: (i) genes already known to be retina-enriched, such as guanine nucleotide binding protein (GNAT1) and arrestin (ARR3); (ii) genes previously not known to be retina enriched, such as WNT inhibitory factor 1 (WIF1) and frizzled-related protein (FRZB) and (iii) unknown genes, such as EST clusters.
Table 1.
Retina-enriched genes by integrating EST, SAGE and microarray data
| Rank | UniGene | Gene name |
|---|---|---|
| 1 | Hs.51147 | Guanine nucleotide binding protein (G protein), (GNAT1), mRNA |
| 2 | Hs.261204 | 17b8 Homo sapiens cDNA |
| 3 | Hs.32721 | S-antigen; retina and pineal gland (arrestin) (SAG), mRNA |
| 4 | Hs.13768 | mRNA; cDNA DKFZp434I1216 (from clone DKFZp434I1216) |
| 5 | Hs.416707 | ATP-binding cassette, sub-family A (ABC1), member 4 (ABCA4) |
| 6 | Hs.308 | Arrestin 3, retinal (X-arrestin) (ARR3), mRNA |
| 7 | Hs.92858 | Guanylate cyclase activator 1A (retina) (GUCA1A), mRNA |
| 8 | Hs.128453 | Frizzled-related protein (FRZB), mRNA |
| 9 | Hs.284122 | WNT inhibitory factor 1 (WIF1), mRNA |
| 10 | Hs.247565 | Rhodopsin (opsin 2, rod pigment) (RHO), mRNA |
| 11 | Hs.281564 | Retinal outer segment membrane protein 1 (ROM1), mRNA |
| 12 | Hs.129882 | Interphotoreceptor matrix proteoglycan 1 (IMPG1), mRNA |
| 13 | Hs.110080 | mRNA; cDNA DKFZp434C0631 (from clone DKFZp434C0631) |
| 14 | Hs.410455 | unc-119 homolog (Caenorhabditis elegans) (UNC119), transcript variant 2 |
| 15 | Hs.89606 | Neural retina leucine zipper (NRL), mRNA |
| 16 | Hs.154131 | Voltage-gated potassium channel Kv11.1 (Kv11.1), mRNA |
| 17 | Hs.857 | Retinol binding protein 3, interstitial (RBP3), mRNA |
| 18 | Hs.135058 | tc57d10.x1 Homo sapiens cDNA, 3′-end |
| 19 | Hs.433923 | Transferrin (TF), mRNA |
| 20 | Hs.93828 | AGENCOURT_6543695 Homo sapiens cDNA, 5′-end |
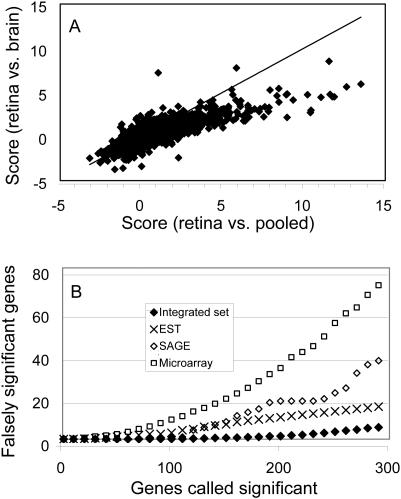
To check if the results are sensitive to the choice of reference dataset, we also compared gene expression from retina libraries with that from brain tissues. The two lists (retina versus ‘pooled’ and retina versus brain) are similar, but with slight differences in ranking. The difference can be attributed to technical variation (e.g. library sampling) and/or biological variation (e.g. expression variation between the brain tissues and the ‘pooled’ tissues). To compare the two lists globally, we plotted the summary scores from the two comparisons as shown in Figure 2A. Each point in the figure corresponds to one gene. The scatter plot displays a good correlation with a correlation coefficient of 0.82. However, the summary scores from the ‘retina versus pooled’ comparison tend to be larger than those from the ‘retina versus brain’ comparison. This observation probably reflects the greater similarity of retina to brain than to the pooled tissues.
Figure 2.
(A) Correlation of summary scores between the comparisons of retina versus brain and retina versus ‘pooled’; x-axis is the score from the comparison of retina versus ‘pooled’ and the y-axis is from retina versus brain. The line is for perfect correlation and only used for eye guide. (B) False discovery rates for EST, SGE, microarray and integrated data sets; x-axis is the number of significant genes and y-axis is the genes falsely called significant.
Statistical validation of retina-enriched genes
To assess the validity of the list of differentially expressed genes, it would be desirable to compare the obtained list with positive and negative controls. Due to limited knowledge on retina-enriched genes, we utilized an FDR calculation to evaluate statistical significance (59–61). The FDR is the expected proportion of false positives among the significant tests. In practice, we used an empirical Bayes method to calculate the FDR as described by Efron and Tibshirani (62) (see Materials and Methods for details). Since FDR is a ratio of expected false positives and overall significant genes, for a given number of significant genes, FDR is proportional to the number of falsely significant genes. Figure 2B illustrates the number of false positive genes in function of number of significant genes. As a comparison, we also calculated the corresponding rates for each individual study. The FDR for the integrated set is significantly lower than those for each of the individual studies. For example, after the data integration, with 200 genes called significant, there are four falsely significant genes, leading to an FDR of 2%. In contrast, the corresponding FDRs are 18, 10 and 7% for the microarray, SAGE and EST, respectively. Consequently, with the data integration procedure, it appears we can obtain a more reliable list of retina-enriched genes. We chose to use the top 500 genes on the list for further prediction, which from the above analysis has an FDR of 5%.
Additional potential target genes
One caveat in the analyses described above is that genes that are potentially important to retina function are not necessarily retina-enriched. For instance, mutation in the pre-mRNA splicing factor gene PRPC8 is associated with the disease retinitis pigmentosa (RP13 locus), but it is a ubiquitously expressed gene (63). Some genes are retina-specific, but their gene expression levels are so low that our approach does not recognize them as significantly retina-enriched. OPN1SW is one such example. OPN1SW is included as a positive control in the study and is well known to be retina-specific (64). The corresponding UniGene cluster (Hs.102119), however, contains only 10 EST sequences. Of these, two sequences are from an optic nerve library, one from an eye library and the rest are from other libraries. This cluster would not be considered as a significantly retina-enriched gene by our EST criteria, even though it is believed to be retina-specific and very likely a target of CRX.
To address this problem, we compiled an additional list of genes related to the retina in terms of disease or function. The list was based on information from two sources: (i) RetNet (http://www.sph.uth.tmc.edu/Retnet/), which, at the time of the analysis, consisted of 94 retinal disease genes and (ii) key word search of LocusLink (65) summary descriptions. Sixty-nine genes contain either ‘retina’ or ‘visual’ in their LocusLink's summary description. Combined with the 500 retina-enriched genes, we had overall 591 retina-related genes for prediction at this stage.
Enrichment of CRX targets in the retina-related gene list
To assess the effect of reducing the prediction space from the whole genome (20 000–25 000 genes) (47) to the 591 retina-related genes, we first examined the retention of positive control genes in the reduced set. All positive control genes from positive control set I were retained, while for positive set II, 6 of 27 were present in the 591 gene list, yielding sensitivities of 100 and 22%, respectively. For this 6 positive genes, 4 of them can be found in retina-enriched gene list, while all of them are retina disease genes. As mentioned earlier, the sensitivity based on positive set I is likely to be an overestimate due to its bias toward photoreceptor genes. On the other hand, since positive set II is derived from gene expression data instead of a direct measure of CRX binding, and thus probably includes indirectly regulated genes, the sensitivity obtained from this control is likely to be an underestimate. More accurate sensitivity assessment will be possible only when a more reliable and larger set of positive controls is available.
Searching for CRX targets in the retina-related gene set
We next searched the retina-related set for genes containing sequences resembling the CRX binding site (see Figure 4 for CRX binding motif). A position-specific score matrix was constructed for CRX binding sites based on previously published data and alignments (13). This was used to search the promoter sequences of the 591 retina-related genes using the program Patser (66). We used −log(P) as the score, where ‘P’ is the P-value provided by the program. The score for known binding sites ranged from 6.54 to 9.63. So as to include most potential regulatory motifs, while realizing that the resultant set probably contained many more false than true positives, we defined a score cut-off of 6 for further analysis. We restricted the search domain to sequences from 2000 bp upstream to 200 bp downstream relative to known or predicted TSSs of all RefSeq genes (see Materials and Methods for TSS).
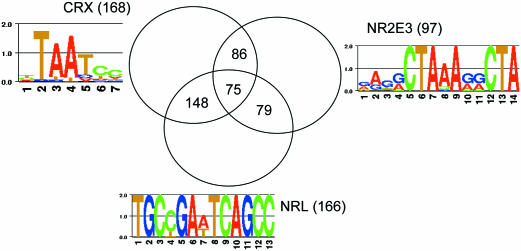
Figure 4.
Venn diagram for the target genes for CRX, NRL and NR2E3. The binding motif logos for each factor are shown. The numbers in the parentheses represent the total number of predicted targets for each factor.
We applied a phylogenetic footprinting approach to improve specificity. Only CRX binding sequences within conserved regions between the human and mouse genomes were taken into account (see Materials and Methods for details). About one-third of the hits remain after the phylogenetic footprint-based filtering. Consistent with the finding that regulatory regions tend to be evolutionarily conserved, those positive controls among the 591 retina-related genes still remain after application of the phylogenetic footprinting filter (6 of 6 positive control genes from positive set I and 6 of 27 from positive set II). Besides the positive controls, our analysis predicted as CRX targets a number of genes not previously implicated as being regulated by CRX. In total, among the 591 retina-related genes, 169 of them contain at least one CRX binding site in their promoter regions.
Bayesian approach to ranking the list of putative targets
We next sought to take advantage of transcription binding site localization information to help rank the 169 predicted CRX targets for prioritizing the follow-up experimental tests. Although eukaryotic TFs can bind many thousand of base pairs away from their target genes, the distribution of their binding sites is non-random. In order to explore this issue quantitatively, we extracted 2100 eukaryotic binding sites from the TRANSFAC database (53,54) and calculated the distribution of their positions relative to their corresponding TSSs. The peak density of binding sites was found between 100 and 200 bp upstream (Figure 3). In order to incorporate this spatial information into our target prediction algorithm, we utilized a Bayesian approach (see Materials and Methods).
A list of putative CRX target genes ranked by confidence level was obtained after we applied the Bayesian analysis. The top 25 putative target genes, with the positive controls marked, are displayed in Table 2. As evidence of the efficacy of the Bayesian approach, four of the positive controls were ranked within the top 10 (OPN1SW-ranked 1, SAG-ranked 3, PDE6B-ranked 6 and RHO-ranked 9). The average ranking of the six positive control genes is 20.2, while the random expectation of average ranking is 84.5 (= 169/2). The P-value of the observed average ranking is <0.0002 according to a random simulation, indicating that the target genes are further enriched in the top-ranking positions in the predicted list.
Table 2.
Predicted CRX target genes
| Ranking | RefSeq ID | Chromosome | Gene name | EMSAa | ChIP | Transfection |
|---|---|---|---|---|---|---|
| 1b | NM_001708 | chr7 | Opsin 1 (cone pigments), short-wave-sensitive (OPN1SW) | |||
| 2 | NM_001297 | chr16 | Cyclic nucleotide-gated channel beta 1 (CNGB1) | |||
| 3 | NM_000541 | chr2 | S-Antigen; retina and pineal gland (arrestin) (SAG) | |||
| 4c | NM_033028 | chr15 | Bardet–Biedl syndrome 4 (BBS4) | + | ||
| 5 | NM_000326 | chr15 | Retinaldehyde binding protein 1 (RLBP1) | |||
| 6 | NM_000283 | chr4 | Phosphodiesterase 6B, rod, beta (PDE6B) | |||
| 7 | NM_012265 | chr22 | Chromosome 22 open reading frame 3 (C22orf3) | |||
| 8 | NM_000180 | chr17 | Guanylate cyclase 2D, membrane (retina-specific) (GUCY2D) | + | + | + |
| 9 | NM_000539 | chr3 | Rhodopsin (opsin 2, rod pigment) (RHO) | |||
| 10 | NM_000330 | chrX | Retinoschisis (X-linked, juvenile) 1 (RS1) | |||
| 11 | NM_001604 | chr11 | Paired box gene 6 (aniridia, keratitis) (PAX6) | |||
| 12 | NM_002900 | chr10 | Retinol binding protein 3, interstitial (RBP3) | |||
| 13 | NM_006269 | chr8 | Retinitis pigmentosa 1 (autosomal dominant) (RP1) | + | + | + |
| 14d | NM_000440 | chr5 | Phosphodiesterase 6A, cGMP-specific, rod, alpha (PDE6A) | |||
| 15 | NM_000350 | chr1 | ATP-binding cassette, sub-family A (ABC1), member 4 (ABCA4) | + | + | + |
| 16 | NM_004312 | chrX | Arrestin 3, retinal (X-arrestin) (ARR3) | + | + | |
| 17 | NM_014848 | chr15 | Synaptic vesicle protein 2B homolog (SV2B) | |||
| 18 | NM_007123 | chr1 | Usher syndrome 2A (autosomal recessive, mild) (USH2A) | |||
| 19 | NM_006493 | chr13 | Ceroid-lipofuscinosis, neuronal 5 (CLN5) | |||
| 20 | NM_022567 | chrX | Nyctalopin (NYX) | |||
| 21 | NM_005272 | chr1 | Guanine nucleotide binding protein (G protein), (GNAT2) | |||
| 22 | NM_002574 | chr1 | Peroxiredoxin 1 (PRDX1) | |||
| 23 | NM_005316 | chr11 | General transcription factor IIH, polypeptide 1 (GTF2H1) | |||
| 24 | NM_000253 | chr4 | Microsomal triglyceride transfer protein (MTP) | |||
| 25 | NM_000409 | chr6 | Guanylate cyclase activator 1A (retina) (GUCA1A) |
aPositive results from each experiment are marked with ‘+’.
bThe positive controls are highlighted by italics.
cThe genes selected for experimental validation are in bold font.
dThis gene was not selected as positive control in our analysis, but has been found to be CRX target recently (52).
Other retina-specific transcription factors (NRL, NR2E3)
The same approach was also applied to two other retina-specific TFs, NRL and NR2E3. NRL is a basic motif-leucine zipper TF that is preferentially expressed in rod photoreceptors and is involved in regulating photoreceptor development (15–17,67). NRL interacts with CRX and the two work synergistically to activate rhodopsin expression (17,68). NR2E3, also known as PNR, is a retinal nuclear receptor that is a presumed ligand-dependent TF that functions as a regulator of photoreceptor gene expression (10,18,19). Using the binding sites of NRL and NR2E3 (shown in Figure 4), we applied the techniques of phylogenetic footprinting and binding location constraint to the 591 retina-related genes. This resulted in the prediction of 166 and 97 putative targets of NRL and NR2E3, respectively. The lists of the predicted target genes can be found in the Supplementary Material.
Recently, a microarray analysis of Nrl null mice has been employed to identify NRL targets (69). Eighteen putative NRL targets were identified from a follow-up experiment of ChIP analysis. Five of them are predicted by our bioinformatic approach, with two of them being the top two in our list (RHO, ROM1). For the targets of NR2E3, it has been found that NR2E3 activates transcription of rod-specific genes and represses cone-specific genes (70,71). Among 14 genes with aberrant expression patterns in Nr2e3 mutant mice, five of them are predicted by us as putative targets.
We also examined the combinatorial regulation of these TFs. Figure 4 is a Venn diagram for the putative targets of the three factors. The overlap between these targets is much larger than would be expected from random assortment. The respective P-values obtained from a hypergeometric probability (see Materials and Methods) are 9.9 × 10−62, 1.3 × 10−21 and 5.1 × 10−26 for the overlap between the targets of CRX and NRL, NRL and NR2E3, NR2E3 and CRX, respectively (note that the P-values were adjusted for multiple testing). Thus, the occurrences of binding motifs of these factors are correlated. This observation corroborates the finding that these three factors form a TF complex that co-regulates rod photoreceptor genes (19,71).
Experimental assessment of target predictions
To assess the validity of our bioinformatic predictions, we have selected a sample of five genes predicted as novel targets of CRX. They were analyzed by EMSA, transient transfection and ChIP. The selected genes, chosen as representative well-characterized retinal genes, were Bardet–Biedl syndrome-4 (BBS4; rank 4, see Table 2), rod outer segment membrane guanylate cyclase (GUCY2D; rank 8), retinitis pigmentosa 1 (RP1; rank 13), ATP binding cassette transporter retina-specific (ABCA4; rank 15) and X-arrestin (arrestin 3, cone arrestin; ARR3; rank 16).
By EMSA, affinity-purified human CRX homeodomain GST fusion protein (CRX-HD-GST) bound to DNA oligomers containing predicted CRX binding sites for all five genes (Figure 5, lanes 2, 4, 6, 8, 10, 12 and 14). The finding of multiple shifted bands with GUCY2D and ARR3 (lanes 4 and 14) suggests that CRX may bind to multiple sites within these probes or may bind as a multimer. The fraction of probe shifted also varied with the different probes, particularly with ABCA4 (compare lanes 10 and 12). These results indicate that CRX-HD can show preference in selecting its binding targets in vitro, which presumably is determined by the sequences flanking the core TAAT/ATTA target sequence.
Figure 5.
EMSA analysis of predicted CRX targets genes. Lanes 1, 3, 5, 7, 9, 11, 13 show the indicated free probes without CRX homeodomain (CRX-HD). Lanes 2, 4, 6, 8, 10, 12, 14 contain the indicated probe plus 20 ng of CRX-HD. Mobility shifts are evident for all the genes.
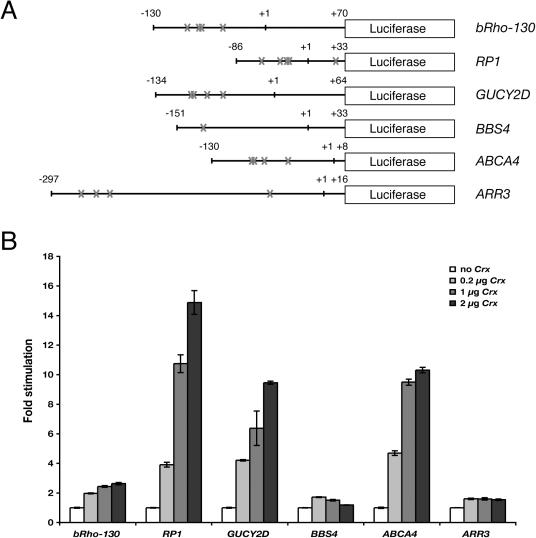
We next used transient transfection assays to test whether the predicted target genes could in fact be transactivated by CRX. Three of the genes (RP1, GUCY2D and ABCA4) showed levels of transactivation that were higher than that seen with a known CRX target, rhodopsin (BRho130) (Figure 6). Significant activation was not seen with either ARR3 or BBS4. Interestingly, and perhaps of significance, the basal activity of the genes that did not demonstrate transactivation (ARR3 and BBS4) was significantly higher than the genes that did (data not shown).
Figure 6.
CRX transactivates RP1, GUCY2D and ABCA4 in transient transfection assays. (A) Schematic diagram showing the luciferase reporter constructs carrying upstream regions of RP1 (−86 to +33), GUCY2D (−134 to +64), ARR3 (−297 to +16), BBS4 (−151 to +33) and ABCA4 (−130 to +8) in the pGL2-basic vector. The positions of CRX core binding sites (TAAT) are labeled by crosses. (B) Transient transfection assays. GripTite 293 MSR cells were transfected with 0.2 μg of the indicated luciferase reporter construct shown in (A) and increasing amounts (0, 0.2, 1 or 2 μg) of the CRX expression vector pcDNA3.1/HisC-Crx. The fold stimulation was calculated relative to control transfections without pcDNA3.1/HisC-Crx. Error bars show the standard error, n = 3.
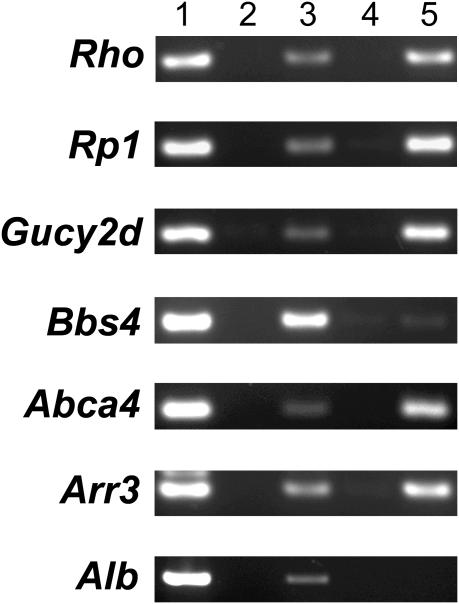
In order to determine whether the promoters of the predicted target genes were in fact bound by CRX in vivo, ChIP was performed (Figure 7). This is important because the genome contains far more potential TF binding sites than are actually occupied in vivo, and the finding of DNA binding and transactivation activity in vitro does not necessarily prove that a gene is a transcriptional target in vivo. Because of the relative ease of obtaining fresh murine retina compared to fresh human retina, the immunoprecipitates were prepared from mouse retina. Consistent with previously published work (56), in the ChIP assay the positive control Rho showed a clearly positive band (lane 5) that was absent in the no antibody control (lane 4), and the negative control Albumin (Alb) did not show any evidence of a positive signal (lane 5). Of the five predicted genes tested, Rp1, Gucy2d, Abca4 and Arr3 all showed reproducible signal that was present with the anti-CRX antibody (lane 5) but absent in the no antibody control (lane 4). Bbs4 did not show a consistent clear signal, although in some experiments a faint band was obtained.
Figure 7.
The promoter regions of Rp1, Gucy2d, Abca4 and Arr3 are occupied by CRX in vivo. ChIP analysis was performed on fresh murine retina using oligomer PCR primers corresponding to the upstream regions of the indicated genes. Lane 1, genomic DNA template; lane 2, no DNA control; lane 3, input DNA pre-immunoprecipitation; lane 4, immunoprecipitation with no antibody; lane 5, immunoprecipitation with anti-CRX antibody.
DISCUSSION
The prediction of regulatory target genes of a TF, especially in eukaryotic systems, is notoriously difficult. This is, in part, due to the challenge of identifying limited size DNA binding sites in a sea of largely random sequences. The TF binding sequence is not a sufficient condition for protein–DNA interaction. Therefore, prediction of TF targets based solely on short binding sequences yields poor specificity. Many methods have been proposed to enrich the target genes and improve the prediction specificity. The approach of identifying cis-regulatory modules is particularly useful when a set of interacting TFs is known. For instance, Wasserman and colleagues successfully applied this method to liver- and muscle-specific expression (25,72). Berman et al. (32) used this method to exploit TF binding sites involved in pattern formation in Drosophila. However, information on which TFs work cooperatively is not always available.
In this paper, we proposed an alternative method for enrichment of the targets of tissue-specific TFs. The assumption is that the genes controlled by tissue-specific TFs are likely to be related to the tissue. We used three retina-specific TFs as model systems. Instead of searching their regulatory target genes in the entire genome, we focused on the genes that are retina-related. Undoubtedly, prediction on this subset of the genome will miss some true positive because some targets may not have known retinal function, or may not be preferentially expressed in the retina. From our computational and experimental analysis, however, we demonstrated that the loss of a certain amount of sensitivity seems to be worth the benefit of a significant gain in prediction specificity.
There is of course much room for improvement in our method. One possible approach is to combine tissue specificity information with other relevant information. For example, it has been found that the genes sharing the same TFs are likely to have similar expression patterns (73–75). If we had available more information about cell-type-specific expression patterns in the retina, and more information about how expression patterns change with various stress and related conditions, subgroups of similarly expressed genes could be extracted that would be more likely to be regulated by the same TFs.
Several lines of evidence suggest that our combined approach generated reasonable results. As one piece of evidence, since the completion of our analysis a number of papers have appeared in the literature that provide experimental evidence that several of the novel predicted targets are in fact regulated by CRX. Pickrell et al. (76) showed, using transient transfection and a Xenopus expression system, that mutation of two putative CRX binding sites in cone arrestin (rank 16, Table 2) leads to significantly decreased expression. Pittler et al. (77) using a combination of approaches, implicated CRX in the regulation of cGMP phosphodiesterase type 6 alpha (PDE6A) (rank 14, Table 2). Chen et al. (56) performed ChIP on mouse retina and found evidence for CRX binding to Rho, L/M cone opsin, S-opsin and beta-PDE (rank 9, 48/49, 1 and 6, respectively, Table 2).
In addition to these published studies, we experimentally tested an additional five predicted target genes, using a combination of EMSA, transient transfection and ChIP studies. The EMSA results indicated that the promoters of all five genes could be bound in vitro by CRX. This finding is perhaps not surprising given that the binding site of CRX is well defined and an important criteria in the bioinformatics analysis was the presence of a good consensus sequence.
However, the presence of a consensus binding sequence does not always lead to strong protein–DNA interaction, as in the case of BBS4. In the more stringent transfection assays, RP1, GUCY2D and ABCA4 all showed significant activation, from 9- to 15-fold, which was higher than that observed with Rho, the prototypic CRX target. It should be noted that although highly suggestive, the finding of transactivation by CRX in such an assay does not necessarily mean that the activated gene is a CRX target in vivo, because in transient transfection studies the transfected TF is generally overexpressed compared to the in vivo situation and 293 cells almost certainly differ from photoreceptor cells in terms of chromatin structure and the availability of other TFs and coregulators. Likewise, a negative result, such as observed with ARR3 and BBS4, does not preclude these genes as CRX targets in vivo, since we may have not chosen the proper upstream fragment for the luciferase assay, or a required cofactor might not be present in the host 293 cells.
Of the three assays we employed, probably the best predictor of in vivo significance was the ChIP study. These studies were clearly positive with Rp1, Gucy2d, Abca4 and Arr3. A weak and non-reproducible band was observed with Bbs4, making it hard to interpret. As powerful as ChIP studies are, however, it should be kept in mind that it is theoretically possible that a TF could bind to a promoter region in vivo without actually altering its activity, perhaps because the gene is already maximally activated, a required cofactor is missing, or because the local chromatin structure is not in the required state. Despite these caveats, taking the data from the three assays together, it seems likely that RP1, GUCY2D and ABCA4 are indeed bona fide targets of CRX in vivo.
Identification of targets of TFs is a difficult task, both computationally and experimentally. The combination of the recent published data cited above and our experimental data suggests that our bioinformatic predictions of CRX target genes are reasonable. Additional work will be necessary to further improve the sensitivity and accuracy of the method, and to broaden it to include other retinal TFs. Hopefully, integration of developing bioinformatic approaches with increasing experimental data will yield new insights into the complex networks regulating retinal gene expression.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
The authors wish to thank Dr Shiming Chen (Washington University School of Medicine) for generously providing antibody against CRX, Drs Nicholas Katsanis, Giovanni Parmigiani, Dongmei Liu (Johns Hopkins University) for stimulating discussions and Dr George Hanson (Invitrogen) for providing the GripTite 293 MSR cell line. The research is supported in part by grants from the National Eye Institute (EY015684, J.Q. and EY009769, D.J.Z.) and from the Foundation Fighting Blindness, and by a generous gift from Mr Robert Smith and Mrs Clarice Smith. This work was also supported by the National Eye Institute Core Grant P30 EY001765-29. D.J.Z. is the Guerrieri Professor of Genetic Engineering and Molecular Ophthalmology, and the recipient of a Research to Prevent Blindness Senior Investigator Award. Funding to pay the Open Access publication charges for this article was provided by National Eye Institute.
Conflict of interest statement. None declared.
REFERENCES
- 1.Iyer V.R., Horak C.E., Scafe C.S., Botstein D., Snyder M., Brown P.O. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- 2.Lee T.I., Rinaldi N.J., Robert F., Odom D.T., Bar-Joseph Z., Gerber G.K., Hannett N.M., Harbison C.T., Thompson C.M., Simon I., et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- 3.Freund C., Horsford D.J., McInnes R.R. Transcription factor genes and the developing eye: a genetic perspective. Hum. Mol. Genet. 1996;5:1471–1488. doi: 10.1093/hmg/5.supplement_1.1471. [DOI] [PubMed] [Google Scholar]
- 4.Mu X., Klein W.H. A gene regulatory hierarchy for retinal ganglion cell specification and differentiation. Semin. Cell Dev. Biol. 2004;15:115–123. doi: 10.1016/j.semcdb.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 5.Freund C.L., Wang Q.L., Chen S., Muskat B.L., Wiles C.D., Sheffield V.C., Jacobson S.G., McInnes R.R., Zack D.J., Stone E.M. De novo mutations in the CRX homeobox gene associated with Leber congenital amaurosis. Nature Genet. 1998;18:311–312. doi: 10.1038/ng0498-311. [DOI] [PubMed] [Google Scholar]
- 6.Freund C.L., Gregory-Evans C.Y., Furukawa T., Papaioannou M., Looser J., Ploder L., Bellingham J., Ng D., Herbrick J.A., Duncan A., et al. Cone-rod dystrophy due to mutations in a novel photoreceptor-specific homeobox gene (CRX) essential for maintenance of the photoreceptor. Cell. 1997;91:543–553. doi: 10.1016/s0092-8674(00)80440-7. [DOI] [PubMed] [Google Scholar]
- 7.Bessant D.A., Payne A.M., Mitton K.P., Wang Q.L., Swain P.K., Plant C., Bird A.C., Zack D.J., Swaroop A., Bhattacharya S.S. A mutation in NRL is associated with autosomal dominant retinitis pigmentosa. Nature Genet. 1999;21:355–356. doi: 10.1038/7678. [DOI] [PubMed] [Google Scholar]
- 8.Wang Q.L., Chen S., Esumi N., Swain P.K., Haines H.S., Peng G., Melia B.M., McIntosh I., Heckenlively J.R., Jacobson S.G., et al. QRX, a novel homeobox gene, modulates photoreceptor gene expression. Hum. Mol. Genet. 2004;13:1025–1040. doi: 10.1093/hmg/ddh117. [DOI] [PubMed] [Google Scholar]
- 9.Sharon D., Sandberg M.A., Caruso R.C., Berson E.L., Dryja T.P. Shared mutations in NR2E3 in enhanced S-cone syndrome, Goldmann–Favre syndrome, and many cases of clumped pigmentary retinal degeneration. Arch. Ophthalmol. 2003;121:1316–1323. doi: 10.1001/archopht.121.9.1316. [DOI] [PubMed] [Google Scholar]
- 10.Milam A.H., Rose L., Cideciyan A.V., Barakat M.R., Tang W.X., Gupta N., Aleman T.S., Wright A.F., Stone E.M., Sheffield V.C., et al. The nuclear receptor NR2E3 plays a role in human retinal photoreceptor differentiation and degeneration. Proc. Natl Acad. Sci. USA. 2002;99:473–478. doi: 10.1073/pnas.022533099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haider N.B., Jacobson S.G., Cideciyan A.V., Swiderski R., Streb L.M., Searby C., Beck G., Hockey R., Hanna D.B., Gorman S., et al. Mutation of a nuclear receptor gene, NR2E3, causes enhanced S cone syndrome, a disorder of retinal cell fate. Nature Genet. 2000;24:127–131. doi: 10.1038/72777. [DOI] [PubMed] [Google Scholar]
- 12.Swain P.K., Chen S., Wang Q.L., Affatigato L.M., Coats C.L., Brady K.D., Fishman G.A., Jacobson S.G., Swaroop A., Stone E., et al. Mutations in the cone-rod homeobox gene are associated with the cone-rod dystrophy photoreceptor degeneration. Neuron. 1997;19:1329–1336. doi: 10.1016/s0896-6273(00)80423-7. [DOI] [PubMed] [Google Scholar]
- 13.Chen S., Wang Q.L., Nie Z., Sun H., Lennon G., Copeland N.G., Gilbert D.J., Jenkins N.A., Zack D.J. Crx, a novel Otx-like paired-homeodomain protein, binds to and transactivates photoreceptor cell-specific genes. Neuron. 1997;19:1017–1030. doi: 10.1016/s0896-6273(00)80394-3. [DOI] [PubMed] [Google Scholar]
- 14.Furukawa T., Morrow E.M., Cepko C.L. Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell. 1997;91:531–541. doi: 10.1016/s0092-8674(00)80439-0. [DOI] [PubMed] [Google Scholar]
- 15.Swaroop A., Xu J.Z., Pawar H., Jackson A., Skolnick C., Agarwal N. A conserved retina-specific gene encodes a basic motif/leucine zipper domain. Proc. Natl Acad. Sci. USA. 1992;89:266–270. doi: 10.1073/pnas.89.1.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farjo Q., Jackson A., Pieke-Dahl S., Scott K., Kimberling W.J., Sieving P.A., Richards J.E., Swaroop A. Human bZIP transcription factor gene NRL: structure, genomic sequence, and fine linkage mapping at 14q11.2 and negative mutation analysis in patients with retinal degeneration. Genomics. 1997;45:395–401. doi: 10.1006/geno.1997.4964. [DOI] [PubMed] [Google Scholar]
- 17.Kumar R., Chen S., Scheurer D., Wang Q.L., Duh E., Sung C.H., Rehemtulla A., Swaroop A., Adler R., Zack D.J. The bZIP transcription factor Nrl stimulates rhodopsin promoter activity in primary retinal cell cultures. J. Biol. Chem. 1996;271:29612–29618. doi: 10.1074/jbc.271.47.29612. [DOI] [PubMed] [Google Scholar]
- 18.Kobayashi M., Takezawa S., Hara K., Yu R.T., Umesono Y., Agata K., Taniwaki M., Yasuda K., Umesono K. Identification of a photoreceptor cell-specific nuclear receptor. Proc. Natl Acad. Sci. USA. 1999;96:4814–4819. doi: 10.1073/pnas.96.9.4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng H., Khanna H., Oh E.C., Hicks D., Mitton K.P., Swaroop A. Photoreceptor-specific nuclear receptor NR2E3 functions as a transcriptional activator in rod photoreceptors. Hum. Mol. Genet. 2004;13:1563–1575. doi: 10.1093/hmg/ddh173. [DOI] [PubMed] [Google Scholar]
- 20.Livesey F.J., Furukawa T., Steffen M.A., Church G.M., Cepko C.L. Microarray analysis of the transcriptional network controlled by the photoreceptor homeobox gene Crx. Curr. Biol. 2000;10:301–310. doi: 10.1016/s0960-9822(00)00379-1. [DOI] [PubMed] [Google Scholar]
- 21.Blackshaw S., Fraioli R.E., Furukawa T., Cepko C.L. Comprehensive analysis of photoreceptor gene expression and the identification of candidate retinal disease genes. Cell. 2001;107:579–589. doi: 10.1016/s0092-8674(01)00574-8. [DOI] [PubMed] [Google Scholar]
- 22.Cliften P., Sudarsanam P., Desikan A., Fulton L., Fulton B., Majors J., Waterston R., Cohen B.A., Johnston M. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science. 2003;301:71–76. doi: 10.1126/science.1084337. [DOI] [PubMed] [Google Scholar]
- 23.Roth F.P., Hughes J.D., Estep P.W., Church G.M. Finding DNA regulatory motifs within unaligned noncoding sequences clustered by whole-genome mRNA quantitation. Nat. Biotechnol. 1998;16:939–945. doi: 10.1038/nbt1098-939. [DOI] [PubMed] [Google Scholar]
- 24.Beer M.A., Tavazoie S. Predicting gene expression from sequence. Cell. 2004;117:185–198. doi: 10.1016/s0092-8674(04)00304-6. [DOI] [PubMed] [Google Scholar]
- 25.Wasserman W.W., Fickett J.W. Identification of regulatory regions which confer muscle-specific gene expression. J. Mol. Biol. 1998;278:167–181. doi: 10.1006/jmbi.1998.1700. [DOI] [PubMed] [Google Scholar]
- 26.Wasserman W.W., Palumbo M., Thompson W., Fickett J.W., Lawrence C.E. Human–mouse genome comparisons to locate regulatory sites. Nature Genet. 2000;26:225–228. doi: 10.1038/79965. [DOI] [PubMed] [Google Scholar]
- 27.Hardison R.C., Oeltjen J., Miller W. Long human–mouse sequence alignments reveal novel regulatory elements: a reason to sequence the mouse genome. Genome Res. 1997;7:959–966. doi: 10.1101/gr.7.10.959. [DOI] [PubMed] [Google Scholar]
- 28.Conkright M.D., Guzman E., Flechner L., Su A.I., Hogenesch J.B., Montminy M. Genome-wide analysis of CREB target genes reveals a core promoter requirement for cAMP responsiveness. Mol. Cell. 2003;11:1101–1108. doi: 10.1016/s1097-2765(03)00134-5. [DOI] [PubMed] [Google Scholar]
- 29.Hoglund A., Kohlbacher O. From sequence to structure and back again: approaches for predicting protein–DNA binding. Proteome Sci. 2004;2:3. doi: 10.1186/1477-5956-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCue L., Thompson W., Carmack C., Ryan M.P., Liu J.S., Derbyshire V., Lawrence C.E. Phylogenetic footprinting of transcription factor binding sites in proteobacterial genomes. Nucleic Acids Res. 2001;29:774–782. doi: 10.1093/nar/29.3.774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boffelli D., McAuliffe J., Ovcharenko D., Lewis K.D., Ovcharenko I., Pachter L., Rubin E.M. Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science. 2003;299:1391–1394. doi: 10.1126/science.1081331. [DOI] [PubMed] [Google Scholar]
- 32.Berman B.P., Nibu Y., Pfeiffer B.D., Tomancak P., Celniker S.E., Levine M., Rubin G.M., Eisen M.B. Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc. Natl Acad. Sci. USA. 2002;99:757–762. doi: 10.1073/pnas.231608898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu Z., Pilpel Y., Church G.M. Computational identification of transcription factor binding sites via a transcription-factor-centric clustering (TFCC) algorithm. J. Mol. Biol. 2002;318:71–81. doi: 10.1016/S0022-2836(02)00026-8. [DOI] [PubMed] [Google Scholar]
- 34.Banerjee N., Zhang M.Q. Identifying cooperativity among transcription factors controlling the cell cycle in yeast. Nucleic Acids Res. 2003;31:7024–7031. doi: 10.1093/nar/gkg894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sinha S., Van Nimwegen E., Siggia E.D. A probabilistic method to detect regulatory modules. Bioinformatics. 2003;19:i292–i301. doi: 10.1093/bioinformatics/btg1040. [DOI] [PubMed] [Google Scholar]
- 36.Halfon M.S., Grad Y., Church G.M., Michelson A.M. Computation-based discovery of related transcriptional regulatory modules and motifs using an experimentally validated combinatorial model. Genome Res. 2002;12:1019–1028. doi: 10.1101/gr.228902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swaroop A., Zack D.J. Transcriptome analysis of the retina. Genome Biol. 2002;3:Reviews1022. doi: 10.1186/gb-2002-3-8-reviews1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharon D., Blackshaw S., Cepko C.L., Dryja T.P. Profile of the genes expressed in the human peripheral retina, macula and retinal pigment epithelium determined through serial analysis of gene expression (SAGE) Proc. Natl Acad. Sci. USA. 2002;99:315–320. doi: 10.1073/pnas.012582799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Katsanis N., Worley K.C., Gonzalez G., Ansley S.J., Lupski J.R. A computational/functional genomics approach for the enrichment of the retinal transcriptome and the identification of positional candidate retinopathy genes. Proc. Natl Acad. Sci. USA. 2002;99:14326–14331. doi: 10.1073/pnas.222409099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hackam A.S., Bradford R.L., Bakhru R.N., Shah R.M., Farkas R., Zack D.J., Adler R. Gene discovery in the embryonic chick retina. Mol. Vis. 2003;9:262–276. [PubMed] [Google Scholar]
- 41.Mu X., Zhao S., Pershad R., Hsieh T.F., Scarpa A., Wang S.W., White R.A., Beremand P.D., Thomas T.L., Gan L., et al. Gene expression in the developing mouse retina by EST sequencing and microarray analysis. Nucleic Acids Res. 2001;29:4983–4993. doi: 10.1093/nar/29.24.4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Diaz E., Yang Y.H., Ferreira T., Loh K.C., Okazaki Y., Hayashizaki Y., Tessier-Lavigne M., Speed T.P., Ngai J. Analysis of gene expression in the developing mouse retina. Proc. Natl Acad. Sci. USA. 2003;100:5491–5496. doi: 10.1073/pnas.0831080100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wistow G., Bernstein S.L., Wyatt M.K., Ray S., Behal A., Touchman J.W., Bouffard G., Smith D., Peterson K. Expressed sequence tag analysis of human retina for the NEIBank Project: retbindin, an abundant, novel retinal cDNA and alternative splicing of other retina-preferred gene transcripts. Mol. Vis. 2002;8:196–204. [PubMed] [Google Scholar]
- 44.Schulz H.L., Rahman F.A., Fadl El., Moula F.M., Stojic J., Gehrig A., Weber B.H. Identifying differentially expressed genes in the mammalian retina and the retinal pigment epithelium by suppression subtractive hybridization. Cytogenet Genome Res. 2004;106:74–81. doi: 10.1159/000078564. [DOI] [PubMed] [Google Scholar]
- 45.Haverty P.M., Hsiao L.L., Gullans S.R., Hansen U., Weng Z. Limited agreement among three global gene expression methods highlights the requirement for non-global validation. Bioinformatics. 2004;20:3431–3441. doi: 10.1093/bioinformatics/bth421. [DOI] [PubMed] [Google Scholar]
- 46.Chowers I., Gunatilaka T.L., Farkas R.H., Qian J., Hackam A.S., Duh E., Kageyama M., Wang C., Vora A., Campochiaro P.A., et al. Identification of novel genes preferentially expressed in the retina using a custom human retina cDNA microarray. Invest. Ophthal. Mol. Vis. Sci. 2003;44:3732–3741. doi: 10.1167/iovs.02-1080. [DOI] [PubMed] [Google Scholar]
- 47.International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 48.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schwartz S., Kent W.J., Smit A., Zhang Z., Baertsch R., Hardison R.C., Haussler D., Miller W. Human–mouse alignments with BLASTZ. Genome Res. 2003;13:103–107. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cavin Perier R., Junier T., Bucher P. The Eukaryotic Promoter Database EPD. Nucleic Acids Res. 1998;26:353–357. doi: 10.1093/nar/26.1.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmid C.D., Praz V., Delorenzi M., Perier R., Bucher P. The Eukaryotic Promoter Database EPD: the impact of in silico primer extension. Nucleic Acids Res. 2004;32:D82–D85. doi: 10.1093/nar/gkh122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suzuki Y., Yamashita R., Nakai K., Sugano S. DBTSS: DataBase of human Transcriptional Start Sites and full-length cDNAs. Nucleic Acids Res. 2002;30:328–331. doi: 10.1093/nar/30.1.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wingender E., Chen X., Hehl R., Karas H., Liebich I., Matys V., Meinhardt T., Pruss M., Reuter I., Schacherer F. TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Res. 2000;28:316–319. doi: 10.1093/nar/28.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wingender E., Chen X., Fricke E., Geffers R., Hehl R., Liebich I., Krull M., Matys V., Michael H., Ohnhauser R., et al. The TRANSFAC system on gene expression regulation. Nucleic Acids Res. 2001;29:281–283. doi: 10.1093/nar/29.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weinmann A.S., Bartley S.M., Zhang T., Zhang M.Q., Farnham P.J. Use of chromatin immunoprecipitation to clone novel E2F target promoters. Mol. Cell Biol. 2001;21:6820–6832. doi: 10.1128/MCB.21.20.6820-6832.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen S., Peng G.H., Wang X., Smith A.C., Grote S.K., Sopher B.L., Spada A.R. Interference of Crx-dependent transcription by ataxin-7 involves interaction between the glutamine regions and requires the ataxin-7 carboxy-terminal region for nuclear localization. Hum. Mol. Genet. 2004;13:53–67. doi: 10.1093/hmg/ddh005. [DOI] [PubMed] [Google Scholar]
- 57.Choi J.K., Yu U., Kim S., Yoo O.J. Combining multiple microarray studies and modeling interstudy variation. Bioinformatics. 2003;19:i84–i90. doi: 10.1093/bioinformatics/btg1010. [DOI] [PubMed] [Google Scholar]
- 58.Hedges L.V., Olkin I. Statistical Methods for Meta-analysis. Orlando: Academic Press; 1985. [Google Scholar]
- 59.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Statist. Soc. B. 1995;57:289–300. [Google Scholar]
- 60.Storey J.D. A direct approach to false discovery rates. J. R. Statist. Soc. B. 2002;64:479–498. [Google Scholar]
- 61.Tusher V.G., Tibshirani R., Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl Acad. Sci. USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Efron B., Tibshirani R. Empirical Bayes methods and false discovery rates for microarrays. Genet. Epidemiol. 2002;23:70–86. doi: 10.1002/gepi.1124. [DOI] [PubMed] [Google Scholar]
- 63.McKie A.B., McHale J.C., Keen T.J., Tarttelin E.E., Goliath R., van Lith-Verhoeven J.J., Greenberg J., Ramesar R.S., Hoyng C.B., Cremers F.P., et al. Mutations in the pre-mRNA splicing factor gene PRPC8 in autosomal dominant retinitis pigmentosa (RP13) Hum. Mol. Genet. 2001;10:1555–1562. doi: 10.1093/hmg/10.15.1555. [DOI] [PubMed] [Google Scholar]
- 64.Nathans J., Merbs S.L., Sung C.H., Weitz C.J., Wang Y. Molecular genetics of human visual pigments. Annu. Rev. Genet. 1992;26:403–424. doi: 10.1146/annurev.ge.26.120192.002155. [DOI] [PubMed] [Google Scholar]
- 65.Pruitt K.D., Maglott D.R. RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res. 2001;29:137–140. doi: 10.1093/nar/29.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hertz G.Z., Stormo G.D. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics. 1999;15:563–577. doi: 10.1093/bioinformatics/15.7.563. [DOI] [PubMed] [Google Scholar]
- 67.Mears A.J., Kondo M., Swain P.K., Takada Y., Bush R.A., Saunders T.L., Sieving P.A., Swaroop A. Nrl is required for rod photoreceptor development. Nat. Genet. 2001;29:447–452. doi: 10.1038/ng774. [DOI] [PubMed] [Google Scholar]
- 68.Rehemtulla A., Warwar R., Kumar R., Ji X., Zack D.J., Swaroop A. The basic motif-leucine zipper transcription factor Nrl can positively regulate rhodopsin gene expression. Proc. Natl Acad. Sci. USA. 1996;93:191–195. doi: 10.1073/pnas.93.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yoshida S., Mears A.J., Friedman J.S., Carter T., He S., Oh E., Jing Y., Farjo R., Fleury G., Barlow C., et al. Expression profiling of the developing and mature Nrl−/− mouse retina: identification of retinal disease candidates and transcriptional regulatory targets of Nrl. Hum. Mol. Genet. 2004;13:1487–1503. doi: 10.1093/hmg/ddh160. [DOI] [PubMed] [Google Scholar]
- 70.Chen J., Rattner A., Nathans J. The rod photoreceptor-specific nuclear receptor Nr2e3 represses transcription of multiple cone-specific genes. J. Neurosci. 2005;25:118–129. doi: 10.1523/JNEUROSCI.3571-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Peng G.H., Ahmad O., Ahmad F., Liu J., Chen S. The photoreceptor-specific nuclear receptor Nr2e3 interacts with Crx and exerts opposing effects on the transcription of rod versus cone genes. Hum. Mol. Genet. 2005;14:747–764. doi: 10.1093/hmg/ddi070. [DOI] [PubMed] [Google Scholar]
- 72.Krivan W., Wasserman W.W. A predictive model for regulatory sequences directing liver-specific transcription. Genome Res. 2001;11:1559–1566. doi: 10.1101/gr.180601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yu H., Luscombe N.M., Qian J., Gerstein M. Genomic analysis of gene expression relationships in transcriptional regulatory networks. Trends Genet. 2003;19:422–427. doi: 10.1016/S0168-9525(03)00175-6. [DOI] [PubMed] [Google Scholar]
- 74.Stuart J.M., Segal E., Koller D., Kim S.K. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302:249–255. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- 75.Lee H.K., Hsu A.K., Sajdak J., Qin J., Pavlidis P. Coexpression analysis of human genes across many microarray data sets. Genome Res. 2004;14:1085–1094. doi: 10.1101/gr.1910904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pickrell S.W., Zhu X., Wang X., Craft C.M. Deciphering the contribution of known cis-elements in the mouse cone arrestin gene to its cone-specific expression. Invest. Ophthal Mol. Vis. Sci. 2004;45:3877–3884. doi: 10.1167/iovs.04-0663. [DOI] [PubMed] [Google Scholar]
- 77.Pittler S.J., Zhang Y., Chen S., Mears A.J., Zack D.J., Ren Z., Swain P.K., Yao S., Swaroop A., White J.B. Functional analysis of the rod photoreceptor cGMP phosphodiesterase alpha-subunit gene promoter: Nrl and Crx are required for full transcriptional activity. J. Biol. Chem. 2004;279:19800–19807. doi: 10.1074/jbc.M401864200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.