Abstract
Loop-mediated isothermal amplification (LAMP), a novel nucleic acid amplification method, was developed for the rapid detection of the major periodontal pathogens Porphyromonas gingivalis, Tannerella forsythia, and Treponema denticola. The LAMP method amplifies DNA with high specificity, efficiency, and rapidity under isothermal conditions using a set of four specially designed primers and a DNA polymerase with strand displacement activity. In this study, we initially designed the primers for LAMP assays to detect these bacteria and evaluated the specificity and sensitivity of these assays. The specificities of the primers for these bacteria were examined using various oral bacteria and various reaction times. The lower detection limits of the 60-min LAMP reaction without loop primers were 1 μg/tube for P. gingivalis, 10 fg/tube for T. forsythia, and 1 ng/tube for T. denticola. Addition of the loop primers for each bacterium improved the detection specificities and sensitivities by several magnitudes. Furthermore, LAMP assays were applied to the rapid detection of these periodontal pathogens in clinical specimens, and the results were compared with those of conventional PCR detection. The results of the LAMP assays corresponded to those of conventional PCR assays. These results indicate that the LAMP assay is an extremely rapid, highly sensitive, specific method. This method is very useful for the rapid detection of periodontopathic bacteria and the diagnosis of periodontal disease.
Periodontitis is an infectious disease caused by periodontopathic bacteria that bring about destructive changes leading to the loss of bone and connective tissue attachment (40, 46). Two periodontopathic bacteria, Porphyromonas gingivalis and Tannerella forsythia formerly Bacteroides forsythus and Tannerella forsythensis (24, 34), which are black-pigmented, gram-negative anaerobic rods, have been strongly implicated as major pathogens in the etiology of this disease (13, 39, 51). T. forsythia is frequently isolated together with P. gingivalis, indicating an ecological relationship between these organisms (41). Treponema denticola, which is a helical oral spirochete, has also been implicated as a major pathogen in periodontitis (15). A previous study found a strong correlation between mixed infections by P. gingivalis, T. forsythia, and T. denticola and adult periodontitis (18, 38). In addition, these organisms are strongly implicated in the development of oral malodor (20, 31, 32).
Genetic analyses of infectious diseases have been developed to obtain detailed genetic information on the virulence and antibiotic resistance of microbes (8). Of the various methods used to diagnose infectious disease (3, 6, 19, 37), molecular-based methods are often used (13, 44); of these, the PCR is one of the most widely used techniques (4, 42, 45). PCR-based detection of bacteria is sensitive and specific, and some PCR-derived methods, such as nested PCR (7, 9) and PCR-restriction fragment length polymorphism (36), have been developed. However, PCR-based detection methods require equipment such as thermal cyclers and several operations.
Recently, Eiken Chemical Co., Ltd., developed loop-mediated isothermal amplification (LAMP), which constitutes a novel nucleic acid amplification method (28, 29). The LAMP reaction requires a DNA polymerase with strand displacement activity and a set of four specially designed primers, termed inner and outer primers. First, a stem-loop DNA structure, in which the sequences of both DNA ends are derived from the inner primer, is constructed (28, 29). Subsequently, self-primed DNA synthesis rapidly occurs at the 3′ terminus of the stem-loop DNA structure, and one inner primer hybridizes to the loop on the product in the LAMP cycle and initiates strand displacement DNA synthesis, yielding the original stem-loop DNA and new stem-loop DNA with a stem twice as long as the original was. The final products are stem-loop DNA of the target DNA. LAMP is a novel approach for nucleic acid amplification with high specificity, selectivity, and rapidity. The primary characteristic of the LAMP method is its ability to amplify nucleic acids under isothermal conditions at temperatures between 60 and 65°C (28, 29). Importantly, this method does not require denaturation of a DNA template (28). The second characteristic of this method is that it has high specificity. The LAMP reaction requires a DNA polymerase with strand displacement activity and a set of four specially designed primers to improve specificity. Furthermore, the amplification efficiency of the LAMP method is extremely high because there is no time loss for thermal change, since the reaction is isothermal.
Therefore, the LAMP assay has emerged as a powerful tool to facilitate genetic testing for the rapid diagnosis of infectious diseases (10, 11, 17, 26, 27, 33, 49). In this study, we developed and evaluated a LAMP method for the rapid detection of three major periodontopathic bacteria. This is the first report of a LAMP assay for oral bacteria.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains used in this study are listed in Table 1. The P. gingivalis strains and T. forsythia were grown in GAM (Nissui Medical Co., Tokyo, Japan) broth supplemented with 5 μg of hemin per ml, 1.0 μg of menadion per ml, 1.0% l-cysteine, and 15 μg N-acetylmuramic acid per ml (for T. forsythia) at 37°C under anaerobic conditions, as reported previously (2, 50). T. denticola and other oral spirochete strains were cultured in TYGVS medium at 37°C under anaerobic conditions, as described previously (15). The purities of the bacterial cultures used are confirmed as follows: P. gingivalis and T. forsythia were inoculated on blood agar plates (containing 2% agar), and single colonies were picked and inoculated into GAM broth supplemented with 0.2 mg/ml gentamicin. T. denticola were inoculated in TYGVS medium, and the culture was mixed with TYGVS medium containing 0.8% agar. After incubation, individual colonies were isolated with a capillary pipette and reinoculated into TYGVS medium. All bacteria were confirmed by microscopy.
TABLE 1.
Strains and amplification results
| Strain | Amplification with the LAMP primers for:
|
||
|---|---|---|---|
| P. gingivalis | T. forsythia | T. denticola | |
| Porphyromonas gingivalis W83 | + | − | − |
| Porphyromonas gingivalis W50 | + | − | − |
| Porphyromonas gingivalis 381 | + | − | − |
| Tannerella forsythia ATCC 43037 | − | + | − |
| Treponema denticola ATCC 35404 | − | − | + |
| Treponema denticola ATCC 35405 | − | − | + |
| Treponema medium ATCC 700293 | − | − | − |
| Treponema vincentii ATCC 35580 | − | − | − |
| Treponema socranskii subsp. paredis ATCC 35535 | − | − | − |
| Actinobacillus actinomycetemcomitans ATCC 29522 | − | − | − |
| Actinobacillus actinomycetemcomitans ATCC 29524 | − | − | − |
| Actinobacillus actinomycetemcomitans ATCC 43718 | − | − | − |
| Actinobacillus actinomycetemcomitans ATCC 43719 | − | − | − |
| Actinobacillus actinomycetemcomitans OMZ534 | − | − | − |
| Actinobacillus actinomycetemcomitans OMZ541 | − | − | − |
| Actinobacillus actinomycetemcomitans OMZ546 | − | − | − |
| Actinobacillus actinomycetemcomitans SUNYaB67 | − | − | − |
| Actinobacillus actinomycetemcomitans SUNYaB75 | − | − | − |
| Actinobacillus actinomycetemcomitans NCTC9709 | − | − | − |
| Actinobacillus actinomycetemcomitans NCTC9710 | − | − | − |
| Actinobacillus actinomycetemcomitans TN-1 | − | − | − |
| Actinobacillus actinomycetemcomitans Y4 | − | − | − |
| Actinobacillus actinomycetemcomitans JP2 | − | − | − |
| Prevotella intermedia ATCC 25611 | − | − | − |
| Prevotella melaninogenica ATCC 25845 | − | − | − |
| Prevotella nigrescens ATCC 25261 | − | − | − |
| Prevotella denticola ATCC 33185 | − | − | − |
| Prevotella loescheii ATCC 15930 | − | − | − |
| Prevotella corporis ATCC 33547 | − | − | − |
| Prevotella bivia ATCC 29303 | − | − | − |
| Prevotella pallens ATCC 700821 | − | − | − |
| Prevotella oralis ATCC 33322 | − | − | − |
| Prevotella veroralis ATCC 33779 | − | − | − |
| Fusobacterium nucleatum ATCC 10953 | − | − | − |
| Haemophilus aphrophilus NCTC5980 | − | − | − |
| Streptococcus mutans Xc | − | − | − |
| Streptococcus mutans OMZ175 | − | − | − |
| Streptococcus mutans MT703R | − | − | − |
| Streptococcus mutans MT8148 | − | − | − |
| Streptococcus sobrinus OU8 | − | − | − |
| Streptococcus sobrinus OMZ176 | − | − | − |
| Streptococcus sobrinus 6715 | − | − | − |
| Streptococcus sobrinus AHT-K | − | − | − |
| Streptococcus sobrinus MT8145 | − | − | − |
| Streptococcus mitis 903 | − | − | − |
| Streptococcus sanguinis ATCC 10556 | − | − | − |
| Streptococcus sanguinis OMZ9 | − | − | − |
| Streptococcus sanguinis 556 | − | − | − |
| Streptococcus gordonii DL1 | − | − | − |
| Streptococcus oralis ATCC 10557 | − | − | − |
| Streptococcus salivarius HT9R | − | − | − |
| Escherichia coli DH5α | − | − | − |
DNA techniques.
Routine molecular biology techniques were performed as described by Sambrook et al. (35). Chromosomal DNA was isolated from the bacteria listed in Table 1 with an IsoQuick Nucleic Acid Extraction kit (ORCA Research, Inc., Bothell, WA) or a PureGene DNA Isolation kit (Gentra Systems, Minneapolis, Minn.).
Primer design for LAMP.
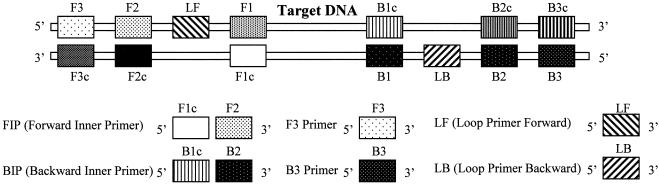
The oligonucleotide primers used in this study are listed in Table 2. The LAMP method requires a set of four specially designed primers (F3, B3, the forward inner primer [FIP], and backward inner primer [BIP]) that recognize a total of six distinct sequences (F1, F2, F3, B1, B2, and B3) in the target DNA (Fig. 1). The two inner primers, FIP and BIP, contain two distinct sequences corresponding to the sense and antisense sequences of the DNA, one for priming in the first stage and the other for self priming in later stages. FIP consists of complementary sequence F1 (F1c) and direct sequence F2 (F2). BIP consists of complementary sequence B1 (B1c) and direct sequence B2 (B2). The two outer primers, F3 and B3c (the sequence complementary to B3), are located outside the F2-B2 region. To increase amplification efficacy, two loop primers, the forward loop primer (LF) and backward loop primer (LB), were designed using Primer Explorer software, version 2.0 (Fujitsu Co., Ltd., Tokyo, Japan) as shown in Fig. 1. The primers for P. gingivalis, T. forsythia, and T. denticola were designed from the pepO, cct, and opdB genes, respectively, and encode P. gingivalis endopeptidase (1, 5), the putative cytocidal toxin of T. forsythia (2), and a trypsin-like peptidase of T. denticola (12). The specificities of the designed primers were initially confirmed using BLAST on the National Center for Biotechnology Information server (http://www.ncbi.nlm.nih.gov/) and then confirmed by LAMP (Table 1).
TABLE 2.
Oligonucleotide primers for LAMP
| Primer | Type | Sequencea | Amplicon size (bp) | Target | Source (Strain) |
|---|---|---|---|---|---|
| Porphyromonas gingivalis | |||||
| Pg-F3 | F3 | 5′-GGC AGT AAT CGG CGC ATT-3′ | 225 | pepO | 381 |
| Pg-B3 | B3 | 5′-TCG TGC AGG ATG TCG AAT G-3′ | |||
| Pg-FIP | FIP | 5′-ACT GAG GTC GAT GGC CGG TAG CTG CAA TGG CAA TAA GGG T-3′ | |||
| Pg-BIP | BIP | 5′-CCG CAG GAC GAC TTT TAT CGC TAG CCG TAG CGA CTA TAA GCA-3′ | |||
| Pg-LF | LF | 5′-GCT TCC TGT CAG TAT CGT TAG TCT G-3′ | |||
| Pg-LB | LB | 5′-ACT GCA ACG GCA ATT GGA TG-3′ | |||
| Tannerella forsythia | |||||
| Tf-F3 | F3 | 5′-GCA ACC AAG ATT GCC AGA GA-3′ | 219 | cct | ATCC 43037 |
| Tf-B3 | B3 | 5′-AAC AGC GAC TGC AAC GAA-3′ | |||
| Tf-FIP | FIP | 5′-GGC ACC ACA CAG GAA CGA GTT AGG GAA TTG CCA AGG ATG TCA-3′ | |||
| Tf-BIP | BIP | 5′-GGG TAA GCC AAC GGT AGA GAC CGG AAT CTG CAT TCA CAC-3′ | |||
| Tf-LF | LF | 5′-ACG TAGTTG CCG GTG TCA-3′ | |||
| Tf-LB | LB | 5′-TCA GTT CCG CCA AGT CAA TG-3′ | |||
| Treponema denticola | |||||
| Td-F3 | F3 | 5′-AAA GGC TTT GGG CGA CAG-3′ | 240 | opdB | ATCC 35405 |
| Td-B3 | B3 | 5′-TCC CGT CCT CAT ACC ACT TT-3′ | |||
| Td-FIP | FIP | 5′-GGT GAG GAC CCG TCC TTT ACC ACG GAG TGA AGG TGC CTA TG-3′ | |||
| Td-BIP | BIP | 5′-GCT CCG ATG CGT TTT TCA GTC CGC CCC TGA TTT GAG CAA CA-3′ | |||
| Td-LF | LF | 5′-AAC CTT TTT TGT AAA CGG CAG C-3′ | |||
| Td-LB | LB | 5′-GAG TGT TTA CAG CCT TGT AGA GAG G-3′ |
FIG. 1.
Double-stranded target DNA and the design of the LAMP primers. The LAMP inner (FIP and BIP), outer (F3 and B3), and loop (LF and LB) primer pairs are shown.
LAMP.
The LAMP reaction was carried out in a 25-μl volume containing 1.6 μM each FIP and BIP, 0.2 μM each F3 and B3, 0.8 μM each LF and LB, 1.4 mM each deoxynucleoside triphosphate, 0.8 M betaine (Sigma, St. Louis, Mo.), 20 mM Tris-HCl (pH 8.8), 10 mM KCl, 10 mM (NH4)2SO4, 8 mM MgSO4, 0.2% Tween 20, 8 U of the Bst DNA polymerase large fragment (New England Biolabs, Beverly, MA), and 5 μl of target DNA. The mixture was incubated at 65°C using a conventional heating block and was heated at >80°C for 2 min to terminate the reaction.
Detection of the LAMP products.
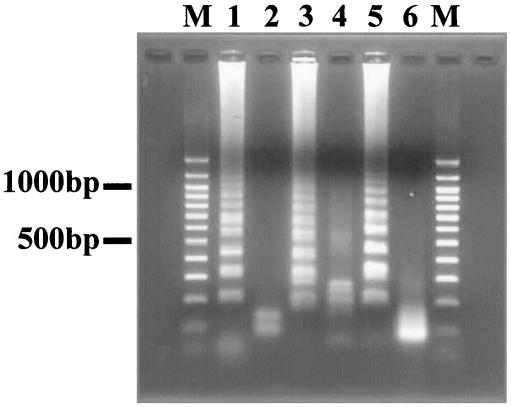
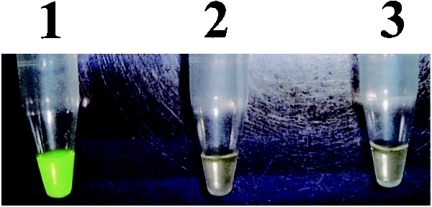
LAMP amplicons in the reaction mixture were detected directly by the naked eye on addition of 1.0 μl of 1/10-diluted original SYBR Green I (Molecular Probes, Inc., Eugene, OR) to the mixture and observation of the solution color. The solution turned green in the presence of a LAMP amplicon, while it remained orange with no amplification. Otherwise, the turbidity derived from the white precipitate of magnesium pyrophosphate in the mixture was detected by the naked eye (25). Furthermore, the amplified products were subjected to agarose gel electrophoresis. The LAMP products were digested with the appropriate restriction enzymes (NcoI for P. gingivalis amplicons, SnaBI for T. forsythia amplicons, and AluI for T. denticola amplicons) and electrophoresed in 2% agarose gels (Fig. 3). The sensitivities of the LAMP assays were confirmed using serially diluted chromosomal DNA.
FIG. 3.
Restriction analysis of LAMP products. Lanes: M, 100-bp DNA ladder (Promega); 1, amplified products of P. gingivalis pepO; 2, P. gingivalis pepO digested with NcoI; 3, amplified products of T. forsythia cct; 4, T. forsythia cct digested with SnaBI; 5, amplified products of T. denticola opdB; 6, T. denticola opdB digested with AluI.
Preparation of subgingival plaque.
Human subgingival plaque was prepared as follows. Subgingival plaque samples were obtained by inserting a sterile endodontic paperpoint into the subgingival site for 10 s. The paperpoint was transferred into 200 μl of phosphate-buffered saline and centrifuged at 12,000 × g for 5 min. The cells were resuspended in 100 μl of cell lysis buffer and boiled at 100°C for 5 min, and the supernatant served as the template (42).
RESULTS
Specificity of LAMP for periodontal bacteria.
The specificities of the LAMP assay for detecting P. gingivalis, T. forsythia, and T. denticola were confirmed by checking the reactivity with various oral bacteria DNA samples (Table 1). For this purpose, the LAMP reaction was performed at 65°C for 60 min without the loop primer. The primers used in this assay did not react with the other bacterial DNA. Initially, the LAMP products were subjected to agarose gel electrophoresis, and a characteristic ladder of multiple bands was seen (Fig. 3). This ladder pattern is characteristic of the LAMP amplification and indicates that stem-loop DNA with inverted repeats of the target sequence was produced. LAMP amplified extremely large amounts of target DNA and produced magnesium pyrophosphate as a by-product. The magnesium pyrophosphate production was confirmed as white turbidity (data not shown). Furthermore, the existence of an amplicon in the LAMP reaction mixture was confirmed using SYBR Green I. A mixture containing an amplicon turned green in the presence of SYBR Green I (Fig. 2), allowing the confirmation of LAMP products by the naked eye. The specificity of the amplification was confirmed by restriction endonuclease digestion with NcoI (for the P. gingivalis amplicon), SnaBI (for the T. forsythia amplicon), and AluI (for the T. denticola amplicon). Each amplicon digested with restriction endonuclease was subjected to agarose gel electrophoresis (Fig. 3).
FIG. 2.
Visual inspection of LAMP products detected using SYBR Green I. The P. gingivalis primers were used for this assay. Tubes: 1, P. gingivalis positive; 2, P. gingivalis negative (T. forsythia and T. denticola positive); 3, no DNA (water only).
Sensitivity of LAMP.
The sensitivity of this assay for each periodontopathic bacteria was evaluated. A serial dilution of the chromosomal DNA of each periodontopathic bacteria was used to evaluate the lower detection limit. Using chromosomal DNA, the P. gingivalis primer set without the loop primer set had a detection limit of 1 μg/tube for a 60-min reaction (Table 3). By contrast, the P. gingivalis primer set with the pair of loop primers was faster; the detection limit was 1 μg/tube for chromosomal DNA in a 30-min reaction (Table 3). Similarly, the detection limits of the T. forsythia primer set were 10 fg/tube for a 40-min reaction (without the loop primers) and 10 fg/tube for a 20-min reaction (with the loop primers) and those of the T. denticola primer set were 100 ng/tube for a 50-min reaction (without the loop primers) and 10 μg/tube for a 20-min reaction (with the loop primers) (Table 3).
TABLE 3.
Detection sensitivities of LAMP
| Primer set (min) | Detection of genomic DNA (fg/tube) witha:
|
|||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No loop primers
|
Loop primers
|
|||||||||||||||||||||
| 0 | 101 | 102 | 103 | 104 | 105 | 106 | 107 | 108 | 109 | 1010 | 0 | 101 | 102 | 103 | 104 | 105 | 106 | 107 | 108 | 109 | 1010 | |
| P. gingivalis | ||||||||||||||||||||||
| 0 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 10 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 20 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 30 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | ± | ± | ± | ± | ± | ± | + | + |
| 40 | − | − | − | − | − | − | − | − | − | − | − | − | + | + | + | + | + | + | + | + | + | + |
| 50 | − | − | − | − | − | − | − | − | − | − | ± | − | + | + | + | + | + | + | + | + | + | + |
| 60 | − | − | − | − | − | ± | ± | ± | ± | + | + | − | + | + | + | + | + | + | + | + | + | + |
| T. forsythia | ||||||||||||||||||||||
| 0 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 10 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 20 | − | − | − | − | − | − | − | − | − | − | − | − | + | + | + | + | + | + | + | + | + | + |
| 30 | − | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | − | + | + | + | + | + | + | + | + | + | + |
| 40 | − | + | + | + | + | + | + | + | + | + | + | − | + | + | + | + | + | + | + | + | + | + |
| 50 | − | + | + | + | + | + | + | + | + | + | + | − | + | + | + | + | + | + | + | + | + | + |
| 60 | − | + | + | + | + | + | + | + | + | + | + | − | + | + | + | + | + | + | + | + | + | + |
| T. denticola | ||||||||||||||||||||||
| 0 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 10 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 20 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | ± | + |
| 30 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | + | + | + | + |
| 40 | − | − | − | − | − | − | − | − | − | ± | ± | − | − | − | − | − | + | + | + | + | + | + |
| 50 | − | − | − | − | − | − | − | − | + | + | + | − | − | − | + | + | + | + | + | + | + | + |
| 60 | − | − | − | − | − | − | + | + | + | + | + | − | − | + | + | + | + | + | + | + | + | + |
+, clearly visible; ±, visible but not clear; −, not visible.
LAMP-based rapid detection of the periodontopathic bacteria in subgingival plaque.
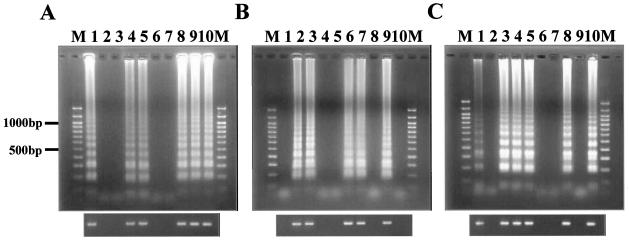
We initially confirmed the inhibitory effects of the oral specimens on LAMP. The presence of LAMP inhibitors in subgingival plaque was assessed using lysates spiked with P. gingivalis-, T. forsythia-, and T. denticola-negative saliva (1 μl per mixture) and dental plaque (ca. 1 μg [wet weight] per mixture, mimicking subgingival plaque) and showed negligible inhibition (data not shown). Therefore, we applied the assays to the rapid detection of P. gingivalis, T. forsythia, and T. denticola in subgingival plaque from 10 individuals (Fig. 4 and Table 4). As shown in Fig. 4 and Table 4, various detection patterns were observed.
FIG. 4.
Electrophoretic analysis of LAMP products in subgingival plaque. Detection of P. gingivalis (A), T. forsythia (B), and T. denticola (C). Lanes 1 to 10 correspond to patient numbers 1 to 10 in Table 4; lane M, 100-bp DNA ladder (Promega). The lower figures are electrophoretic data from the PCR analysis.
TABLE 4.
Patient characteristics and result of LAMPa with loop primer detected in subgingival plaque samples
| Patient no. | Age (yr) | Genderb | Pocket probing depth (mm) | BOPc | Detection of:
|
||
|---|---|---|---|---|---|---|---|
| P. gingivalis | T. forsythia | T. denticola | |||||
| 1 | 41 | M | 3 | − | + | − | + |
| 2 | 35 | M | 4 | + | − | + | − |
| 3 | 38 | F | 6 | − | − | + | + |
| 4 | 63 | F | 3 | − | + | − | + |
| 5 | 58 | M | 8 | + | + | − | + |
| 6 | 43 | F | 2 | − | − | + | − |
| 7 | 24 | M | 4 | + | − | + | − |
| 8 | 61 | M | 3 | − | + | − | + |
| 9 | 41 | F | 4 | + | + | + | − |
| 10 | 41 | M | 4 | − | + | − | + |
A result from a 40-min reaction is shown.
F, female; M, male.
BOP, bleeding on probing.
DISCUSSION
Nucleic acid amplification is one of the most valuable methods for research in the life sciences; the new technique has particularly benefited amplicon-oriented sciences, including studies concerned with the diagnosis of infectious diseases, genetic disorders, and genetic traits in clinical medicine (29). Of the nucleic acid amplification methods, PCR-based amplification methods are widely used for the diagnosis of various diseases. Periodontitis is a common infectious disease, and a PCR-based diagnosis system for periodontitis has been developed (4, 42, 45). Despite their simplicity and accuracy, PCR-based diagnosis methods are not widely used in private clinics as routine diagnostic tools, due to the need for a thermal cycler. By contrast, the LAMP method needs only a conventional heating block. An accurate and rapid diagnosis system for periodontitis is essential for periodontal treatment. Several recent reports have demonstrated the usefulness of the LAMP method (10, 11, 17, 26, 27, 33, 49). Therefore, we focused on the LAMP method for the rapid detection of periodontopathic bacteria. The principle of LAMP involves autocycling strand displacement DNA synthesis using a DNA polymerase with high strand displacement activity and a set of two specially designed inner and two outer primers. This novel nucleic acid amplification method was developed by Notomi et al. (28, 29). This report is the first application of the LAMP method for the diagnosis of oral disease.
Previously, Kasuga et al. reported that mixed infection with P. gingivalis, T. forsythia, and T. denticola in periodontal sites is strongly correlated with the severity of adult periodontitis (18). Furthermore, they suggested that the detection of these organisms provides essential information on the severity of periodontitis. There has been a recent focus on periodontal bacteria and periodontitis, due to the latter's relationship with cardiovascular disease (14, 22, 23) and atherosclerosis (16, 30). Our technique could be useful for evaluating periodontal conditions in relation to these general health conditions. Therefore, we focused on the LAMP method for the rapid detection of these three organisms. Initially, we evaluated the specificities of the LAMP assays for these organisms. LAMP is highly specific for the target sequence. This is attributed to recognition of the target sequence by six independent sequences in the initial stage and by four independent sequences during the later stages of the reaction (29). We confirmed the specificities of the bacteria-specific primers using various oral bacteria (Table 1). This is very important in the detection of oral bacteria because over 500 species or phylotypes have been detected in subgingival plaque (20, 21).
To develop a rapid detection system for bacteria, the assay sensitivity is also essential. LAMP amplifies DNA with high efficiency under isothermal conditions without a significant influence of the copresence of nontarget DNA (29). This characteristic is also suited to the detection of oral bacteria. In this study, we evaluated the detection limits of these assays using serially diluted chromosomal DNA. The lower detection limits with loop primers in a 60-min reaction are 10 fg/tube for P. gingivalis and T. forsythia and 100 fg/tube for T. denticola. These sensitivities are consistent with previous studies (10, 11).
Using this assay system, we detected these periodontal bacteria in subgingival plaque from 10 individuals (Table 4). In this experiment, various detection patterns were observed, and we confirmed that LAMP-based detection is applicable to oral specimens. Furthermore, we show the clinical characteristics of the patients and summarize the detection of the three periodontopathic bacteria listed in Table 4. We were unable to identify any relationship between the clinical characteristics and the detection of these bacteria from this result. A comparison of the relationship between the clinical characteristics and the symbiotic relationship of these bacteria could be the subject of a follow-up study.
We reported a novel rapid detection system for pathogenic bacteria in periodontitis. This novel method constitutes an extremely rapid qualitative system. We showed that LAMP is suitable for rapid screening of oral bacteria and chairside diagnosis. Quantitative analysis of infectious disease pathogens is essential for accurate, detailed diagnosis (43, 47, 48). LAMP technology possesses the potential for quantitative analysis (25). For a quantitative determination of the amount of DNA in a clinical specimen, a kinetic analysis of the time-related changes in turbidity due to the precipitation of magnesium pyrophosphate is possible (25). The development of an extremely rapid quantitative genetic detection system for periodontal pathogen using the LAMP method is now under way.
Acknowledgments
This investigation was supported by a Grant-in-Aid for the Encouragement of Scientists 13771265 (to T.T.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, by Research Fellowships from the Japan Society for the Promotion of Science for Young Scientists (to S.N.), and by a research grant from the Clinical Research Foundation (to A.Y.).
REFERENCES
- 1.Ansai, T., W. Yu, S. Urnowey, S. Barik, and T. Takehara. 2003. Construction of a pepO gene-deficient mutant of Porphyromonas gingivalis: potential role of endopeptidase O in the invasion of host cells. Oral Microbiol. Immunol. 18:398-400. [DOI] [PubMed] [Google Scholar]
- 2.Arakawa, S., T. Nakajima, H. Ishikura, S. Ichinose, I. Ishikawa, and N. Tsuchida. 2000. Novel apoptosis-inducing activity in Bacteroides forsythus: a comparative study with three serotypes of Actinobacillus actinomycetemcomitans. Infect. Immun. 68:4611-4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armitage, G. C., W. R. Dickinson, R. S. Jenderseck, S. M. Levine, and D. W. Chambers. 1982. Relationship between the percentage of subgingival spirochetes and the severity of periodontal disease. J. Periodontol. 53:550-556. [DOI] [PubMed] [Google Scholar]
- 4.Ashimoto, A., C. Chen, I. Bakker, and J. Slots. 1996. Polymerase chain reaction detection of 8 putative periodontal pathogens in subgingival plaque of gingivitis and advanced periodontitis lesions. Oral Microbiol. Immunol. 11:266-273. [DOI] [PubMed] [Google Scholar]
- 5.Awano, S., T. Ansai, H. Mochizuki, W. Yu, K. Tanzawa, A. J. Turner, and T. Takehara. 1999. Sequencing, expression and biochemical characterization of the Porphyromonas gingivalis pepO gene encoding a protein homologous to human endothelin-converting enzyme. FEBS Lett. 460:139-144. [DOI] [PubMed] [Google Scholar]
- 6.Bretz, W. A., D. E. Lopatin, and W. J. Loesche. 1990. Benzoyl-arginine naphthylamide (BANA) hydrolysis by Treponema denticola and/or Bacteroides gingivalis in periodontal plaques. Oral Microbiol. Immunol. 5:275-279. [DOI] [PubMed] [Google Scholar]
- 7.Carroll, N. M., E. E. Jaeger, S. Choudhury, A. A. Dunlop, M. M. Matheson, P. Adamson, N. Okhravi, and S. Lightman. 2000. Detection of and discrimination between gram-positive and gram-negative bacteria in intraocular samples by using nested PCR. J. Clin. Microbiol. 38:1753-1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Del Vecchio, V. G., J. M. Petroziello, M. J. Gress, F. K. McCleskey, G. P. Melcher, H. K. Crouch, and J. R. Lupski. 1995. Molecular genotyping of methicillin-resistant Staphylococcus aureus via fluorophore-enhanced repetitive-sequence PCR. J. Clin. Microbiol. 33:2141-2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.el Fantroussi, S., J. Mahillon, H. Naveau, and S. N. Agathos. 1997. Introduction of anaerobic dechlorinating bacteria into soil slurry microcosms and nested-PCR monitoring. Appl. Environ. Microbiol. 63:806-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Endo, S., T. Komori, G. Ricci, A. Sano, K. Yokoyama, A. Ohori, K. Kamei, M. Franco, M. Miyaji, and K. Nishimura. 2004. Detection of gp43 of Paracoccidioides brasiliensis by the loop-mediated isothermal amplification (LAMP) method. FEMS Microbiol. Lett. 234:93-97. [DOI] [PubMed] [Google Scholar]
- 11.Enosawa, M., S. Kageyama, K. Sawai, K. Watanabe, T. Notomi, S. Onoe, Y. Mori, and Y. Yokomizo. 2003. Use of loop-mediated isothermal amplification of the IS900 sequence for rapid detection of cultured Mycobacterium avium subsp. paratuberculosis. J. Clin. Microbiol. 41:4359-4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fenno, J. C., S. Y. Lee, C. H. Bayer, and Y. Ning. 2001. The opdB locus encodes the trypsin-like peptidase activity of Treponema denticola. Infect. Immun. 69:6193-6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gersdorf, H., A. Meissner, K. Pelz, G. Krekeler, and U. B. Gobel. 1993. Identification of Bacteroides forsythus in subgingival plaque from patients with advanced periodontitis. J. Clin. Microbiol. 31:941-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herzberg, M. C., and M. W. Weyer. 1998. Dental plaque, platelets, and cardiovascular diseases. Ann. Periodontol. 3:151-160. [DOI] [PubMed] [Google Scholar]
- 15.Ishihara, K., and H. K. Kuramitsu. 1995. Cloning and expression of a neutral phosphatase gene from Treponema denticola. Infect. Immun. 63:1147-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishihara, K., A. Nabuchi, R. Ito, K. Miyachi, H. K. Kuramitsu, and K. Okuda. 2004. Correlation between detection rates of periodontopathic bacterial DNA in carotid coronary stenotic artery plaque and in dental plaque samples. J. Clin. Microbiol. 42:1313-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iwamoto, T., T. Sonobe, and K. Hayashi. 2003. Loop-mediated isothermal amplification for direct detection of Mycobacterium tuberculosis complex, M. avium, and M. intracellulare in sputum samples. J. Clin. Microbiol. 41:2616-2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kasuga, Y., K. Ishihara, and K. Okuda. 2000. Significance of detection of Porphyromonas gingivalis, Bacteroides forsythus and Treponema denticola in periodontal pockets. Bull. Tokyo Dent. Coll. 41:109-117. [DOI] [PubMed] [Google Scholar]
- 19.Kazor, C., G. W. Taylor, and W. J. Loesche. 1999. The prevalence of BANA-hydrolyzing periodontopathic bacteria in smokers. J. Clin. Periodontol. 26:814-821. [DOI] [PubMed] [Google Scholar]
- 20.Kazor, C. E., P. M. Mitchell, A. M. Lee, L. N. Stokes, W. J. Loesche, F. E. Dewhirst, and B. J. Paster. 2003. Diversity of bacterial populations on the tongue dorsa of patients with halitosis and healthy patients. J. Clin. Microbiol. 41:558-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kolenbrander, P. E. 2000. Oral microbial communities: biofilms, interactions, and genetic systems. Annu. Rev. Microbiol. 54:413-437. [DOI] [PubMed] [Google Scholar]
- 22.Kuramitsu, H. K., I. C. Kang, and M. Qi. 2003. Interactions of Porphyromonas gingivalis with host cells: implications for cardiovascular diseases. J. Periodontol. 74:85-89. [DOI] [PubMed] [Google Scholar]
- 23.Kuramitsu, H. K., M. Qi, I. C. Kang, and W. Chen. 2001. Role for periodontal bacteria in cardiovascular diseases. Ann. Periodontol. 6:41-47. [DOI] [PubMed] [Google Scholar]
- 24.Maiden, M. F., P. Cohee, and A. C. Tanner. 2003. Proposal to conserve the adjectival form of the specific epithet in the reclassification of Bacteroides forsythus Tanner et al. 1986 to the genus Tannerella Sakamoto et al. 2002 as Tannerella forsythia corrig., gen. nov., comb. nov. Request for an opinion. Int. J. Syst. Evol. Microbiol. 53:2111-2112. [DOI] [PubMed] [Google Scholar]
- 25.Mori, Y., K. Nagamine, N. Tomita, and T. Notomi. 2001. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 289:150-154. [DOI] [PubMed] [Google Scholar]
- 26.Nagamine, K., T. Hase, and T. Notomi. 2002. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell. Probes 16:223-229. [DOI] [PubMed] [Google Scholar]
- 27.Nagamine, K., Y. Kuzuhara, and T. Notomi. 2002. Isolation of single-stranded DNA from loop-mediated isothermal amplification products. Biochem. Biophys. Res. Commun. 290:1195-1198. [DOI] [PubMed] [Google Scholar]
- 28.Nagamine, K., K. Watanabe, K. Ohtsuka, T. Hase, and T. Notomi. 2001. Loop-mediated isothermal amplification reaction using a nondenatured template. Clin. Chem. 47:1742-1743. [PubMed] [Google Scholar]
- 29.Notomi, T., H. Okayama, H. Masubuchi, T. Yonekawa, K. Watanabe, N. Amino, and T. Hase. 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28:E63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okuda, K., K. Ishihara, T. Nakagawa, A. Hirayama, and Y. Inayama. 2001. Detection of Treponema denticola in atherosclerotic lesions. J. Clin. Microbiol. 39:1114-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Persson, S., R. Claesson, and J. Carlsson. 1989. The capacity of subgingival microbiotas to produce volatile sulfur compounds in human serum. Oral Microbiol. Immunol. 4:169-172. [DOI] [PubMed] [Google Scholar]
- 32.Persson, S., M. B. Edlund, R. Claesson, and J. Carlsson. 1990. The formation of hydrogen sulfide and methyl mercaptan by oral bacteria. Oral Microbiol. Immunol. 5:195-201. [DOI] [PubMed] [Google Scholar]
- 33.Poon, L. L., C. S. Leung, M. Tashiro, K. H. Chan, B. W. Wong, K. Y. Yuen, Y. Guan, and J. S. Peiris. 2004. Rapid detection of the severe acute respiratory syndrome (SARS) coronavirus by a loop-mediated isothermal amplification assay. Clin. Chem. 50:1050-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sakamoto, M., M. Suzuki, M. Umeda, L. Ishikawa, and Y. Benno. 2002. Reclassification of Bacteroides forsythus (Tanner et al. 1986) as Tannerella forsythensis corrig., gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 52:841-849. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Sato, T., and H. K. Kuramitsu. 1999. Restriction fragment-length polymorphism analysis of 16S ribosomal RNA genes amplified by polymerase chain reaction for rapid identification of cultivable oral treponemes. Oral Microbiol. Immunol. 14:117-121. [DOI] [PubMed] [Google Scholar]
- 37.Simonson, L. G., C. H. Goodman, J. J. Bial, and H. E. Morton. 1988. Quantitative relationship of Treponema denticola to severity of periodontal disease. Infect. Immun. 56:726-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simonson, L. G., P. J. Robinson, R. J. Pranger, M. E. Cohen, and H. E. Morton. 1992. Treponema denticola and Porphyromonas gingivalis as prognostic markers following periodontal treatment. J. Periodontol. 63:270-273. [DOI] [PubMed] [Google Scholar]
- 39.Slots, J., and R. J. Genco. 1984. Black-pigmented Bacteroides species, Capnocytophaga species, and Actinobacillus actinomycetemcomitans in human periodontal disease: virulence factors in colonization, survival, and tissue destruction. J. Dent. Res. 63:412-421. [DOI] [PubMed] [Google Scholar]
- 40.Socransky, S. S., and A. D. Haffajee. 1992. The bacterial etiology of destructive periodontal disease: current concepts. J. Periodontol. 63:322-331. [DOI] [PubMed] [Google Scholar]
- 41.Socransky, S. S., A. D. Haffajee, M. A. Cugini, C. Smith, and R. L. Kent, Jr. 1998. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25:134-144. [DOI] [PubMed] [Google Scholar]
- 42.Suzuki, N., Y. Nakano, Y. Yoshida, D. Ikeda, and T. Koga. 2001. Identification of Actinobacillus actinomycetemcomitans serotypes by multiplex PCR. J. Clin. Microbiol. 39:2002-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suzuki, N., A. Yoshida, T. Saito, M. Kawada, and Y. Nakano. 2004. Quantitative microbiological study of subgingival plaque by real-time PCR shows correlation between levels of Tannerella forsythensis and Fusobacterium spp. J. Clin. Microbiol. 42:2255-2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tanner, A. C., M. F. Maiden, J. J. Zambon, G. S. Thoren, and R. L. Kent, Jr. 1998. Rapid chair-side DNA probe assay of Bacteroides forsythus and Porphyromonas gingivalis. J. Periodont. Res. 33:105-117. [DOI] [PubMed] [Google Scholar]
- 45.Watanabe, K., and T. O. Frommel. 1993. Detection of Porphyromonas gingivalis in oral plaque samples by use of the polymerase chain reaction. J. Dent. Res. 72:1040-1044. [DOI] [PubMed] [Google Scholar]
- 46.Williams, R. C. 1990. Periodontal disease. N. Engl. J. Med. 322:373-382. [DOI] [PubMed] [Google Scholar]
- 47.Yoshida, A., M. Kawada, N. Suzuki, Y. Nakano, T. Oho, T. Saito, and Y. Yamashita. 2004. TaqMan real-time polymerase chain reaction assay for the correlation of Treponema denticola numbers with the severity of periodontal disease. Oral Microbiol. Immunol. 19:196-200. [DOI] [PubMed] [Google Scholar]
- 48.Yoshida, A., N. Suzuki, Y. Nakano, T. Oho, M. Kawada, and T. Koga. 2003. Development of a 5′ fluorogenic nuclease-based real-time PCR assay for quantitative detection of Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis. J. Clin. Microbiol. 41:863-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoshikawa, T., M. Ihira, S. Akimoto, C. Usui, F. Miyake, S. Suga, Y. Enomoto, R. Suzuki, Y. Nishiyama, and Y. Asano. 2004. Detection of human herpesvirus 7 DNA by loop-mediated isothermal amplification. J. Clin. Microbiol. 42:1348-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoshimura, M., Y. Nakano, Y. Yamashita, T. Oho, T. Saito, and T. Koga. 2000. Formation of methyl mercaptan from L-methionine by Porphyromonas gingivalis. Infect. Immun. 68:6912-6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zambon, J. J., H. S. Reynolds, and J. Slots. 1981. Black-pigmented Bacteroides spp. in the human oral cavity. Infect. Immun. 32:198-203. [DOI] [PMC free article] [PubMed] [Google Scholar]