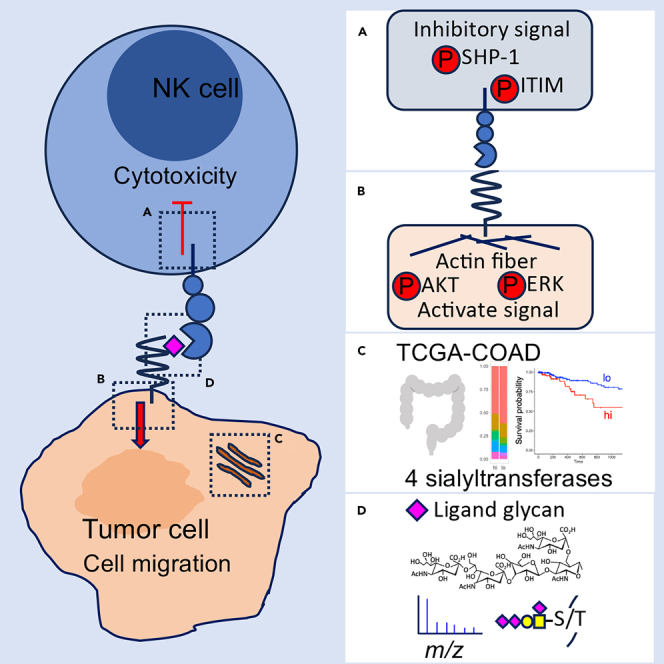
Summary
Siglec-7, an inhibitory receptor expressed on natural killer (NK) cells, recognizes sialic acid-containing glycans. However, the ligand glycan structures of Siglec-7 and its carrier proteins have not been comprehensively investigated. Here, we identified four sialyltransferases that are used for the synthesis of ligand glycans of Siglec-7 and two ligand O-glycan-carrier proteins, PODXL and MUC13, using a colon cancer line. Upon binding of these ligand glycans, Siglec-7-expressing immune cells showed reduced cytotoxic activity, whereas cancer cells expressing ligand glycans underwent signal activation, leading to enhanced invasion activity. To clarify the structure of the ligand glycan, podoplanin (PDPN) identified as a Siglec-7 ligand-carrier protein, was transfected into HEK293T cells using sialyltransferase cDNAs. Mass spectrometry of the products revealed a ligand glycan, tri-sialylated T antigen. These results indicate that Siglec-7 interaction with its ligand generates bidirectional signals in NK and cancer cells, leading to the efficient escape of cancers from host immune surveillance.
Subject areas: Cancer, Glycobiology, Immune response
Graphical abstract
Highlights
-
•
Siglec-7 inhibits cytotoxicity via the molecules identified as ligand glycan-carrier
-
•
The molecules enhance metastatic property of cancer cell by binding of Siglec-7
-
•
Sialyltransferase genes that synthesize the ligand relate a poor prognosis in cancer
-
•
The ligand structure is revealed to be a tri-sialyl T antigen
Cancer; Glycobiology; Immune response
Introduction
Siglecs, a family of sialic acid-recognizing lectins, are mainly expressed on the cell surface of immune cells.1 Most Siglecs have an immunoreceptor tyrosine-based inhibitory/switch motif (ITIM/ITSM) in the cytosol. These motifs undergo tyrosine phosphorylation by Src family kinase.2 Binding of the ligand glycans promotes receptor phosphorylation and transduction of an inhibitory signal into Siglec-expressing cells.3,4,5,6,7 To date, 15 family members of Siglecs have been reported in humans and 9 family members in mice. Siglecs belong to the immunoglobulin superfamily and are divided into the Siglec-1, 2, 4, and 15, which are widely conserved in mammals, and the Siglec-3 subfamily, the details of which vary depending on the species. Siglec-7 belongs to the human Siglec-3 subfamily. The extracellular domain is characterized by having a V-set domain that recognizes sialic acids.8
Although the Siglecs have been shown to be involved in regulation of the immune reaction, it has also been reported that they are used as tools for infection by bacteria and viruses. As for cancer cells, we already reported that Siglec-9 enhances motility and invasion of astrocytoma cells. Siglec-9 expressed on monocytes activates calpain in cancer cells that express ligand glycans, inactivates cell adhesion molecules, and induces adhesion dynamics.9 However, there are no definite reports on the escape of cancer cells from host immune surveillance based on the bidirectional signals, that is, inhibitory signals into immune cells and activation signals into cancer cells.
Siglec-7 is a membrane molecule that is mainly expressed in NK cells and monocytes. To date, GD3,10,11 di-sisialyl Gb5,12 di-sialyl Lewis a, and 6-sulfo-Lewis x13 have been reported as ligand glycans of Siglec-7 (Figure S1A). Recently, several groups reported that O-glycans14 are ligands for Siglec-7,15,16,17 in particular, the di-sialyl T was identified as a Siglec-7 ligand.7,16,18,19 Furthermore, structures in which the terminal galactose is sulfated by carbohydrate sulfotransferase 1 (6-O-sulfation) have also been reported as ligand glycans.20 However, these structures are inconsistent, and the common reactive structures are unclear. Here, we investigated the optimal sialyltransferases that promote the most efficient binding of Siglec-7 using all 20 types of sialyltransferase21 cDNAs defined to date, and identified the best combination of multiple cDNAs. We also identified previously unreported sialylated O-glycans on glycoproteins to which Siglec-7 strongly binds. Furthermore, upon interaction between those molecules, it was demonstrated that inhibitory signals were transduced into the immune cell side and activation signals were transduced into the cancer cell side, leading to increased invasion of cancer cells and the efficient escape of cancer cells from immuno-surveillance.
Results
Siglec-7 binds to O-glycans synthesized by 4 sialyltransferases on the cell surface of a colon adenocarcinoma cell line, DLD-1
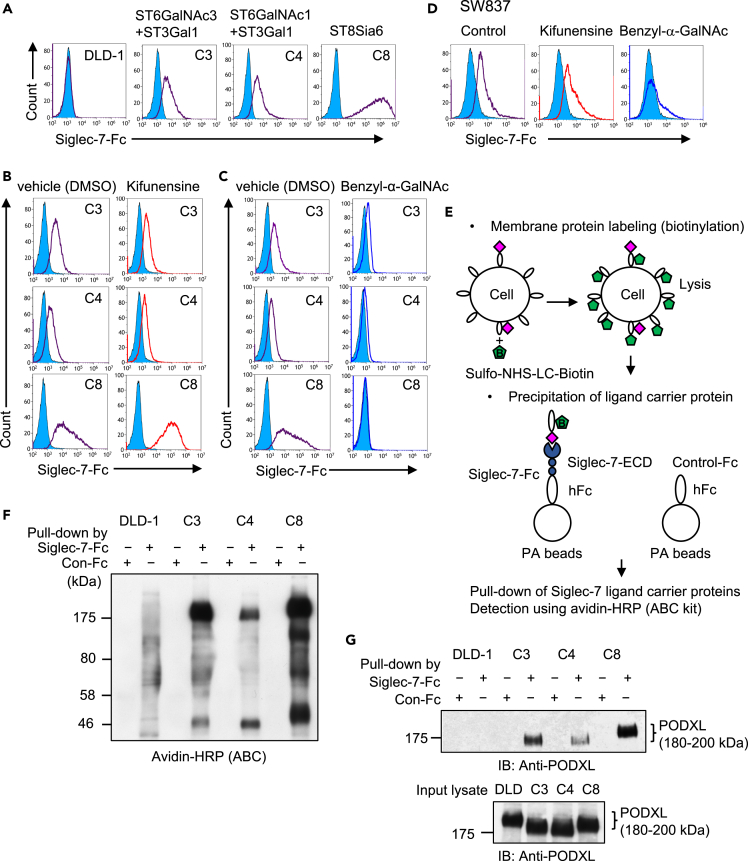
Because recombinant Siglec-7-Fc22 did not bind to DLD-1, we used DLD-1 to screen for sialyltransferase genes that could generate sialylated carbohydrates recognized by Siglec-7 using 20 sialyltransferase cDNAs (Table S1). Consequently, the expression of ST3Gal1,23 ST6GalNAc1,24,25 ST6GalNAc3,25 and ST8Sia626,27 induced Siglec-7 binding (Figure S2A). Previous studies have shown that at least two sialic acid residues are important for ligand-glycan recognition by Siglec-7 (Figure S1). Stable transfectant clones of these cDNAs were established and named C4 (ST3Gal1 and ST6GalNAc1), C3 (ST3Gal1 and ST6GalNAc3), and C8 (ST8Sia6) (Figure 1A). Siglec-7 did not bind to GD3 synthase (ST8Sia1) stably expressed ganglioside GD3 positive DLD-1 (Figure S2B),28 as described previously.11 Next, to clarify the properties of glycan structures recognized by Siglec-7, the three transfectants were treated with an N-type glycan synthesis inhibitor, kifunensine (Figure 1B),15,18 or an O-glycan synthesis inhibitor, benzyl-α-GalNAc (Figure 1C).29,30 While kifunesine had no effect on binding of Siglec-7-Fc, the transfectants treated by benzyl-α-GalNAc showed loss of the binding of Siglec-7-Fc. DLD-1 cells were peanut agglutinin (PNA) lectin-positive and O-glycans with Galβ1-3GalNac-structures were abundant (Figure S2C). This indicated that the substrate for the identified transferase was present on the DLD-1. Siglec-9 is a member of the CD33-related Siglec family and, like Siglec-7, is expressed on NK cells. The binding of Siglec-9 to DLD-1 and transfectants was assessed, and binding was also observed on DLD-1, in which the binding tendency of Siglec-9-Fc to DLD-1 was different from that of Siglec-7-Fc (Figure S2C). Consequently, we demonstrated that the ligand glycans recognized by Siglec-7 were O-type glycans. We investigated whether the four identified sialyltransferase genes are also expressed in colon cancer cell lines and other cancer cell lines (Figure S3A). hSt3gal1 and hSt6galnac1 were expressed at low levels in DLD-1. Only hSt6galnac3 expressed in Caco-2 cells, whose Siglec-7 binding was as negative as that of DLD-1 (data not shown). In contrast, HCT116 and SW837 cells expressed hSt3gal1, hSt6galnac3, and hSt8Sia6. In particular, SW837 cells with high expression of hSt8Sia6 also showed strong binding to Siglec-7-Fc (Figure 1D, left). hSt3gal1 and hSt8sia6 were expressed in MCF-7 cells. hSt3gal1, hSt6galnac1, and hSt8Sia6 were expressed in K562 cells. It has been reported that the O-type glycans of the disialyl T structure on K562 bind to Siglec-7-Fc.16,17 The expression levels of sialyltransferases transgenic for DLD-1 were compared with those of colon cancer and in other cell lines. Compared to SW837, St3gal1 was 2.44-fold, St6galnac1 was 143-fold, St6galnac3 was 0.48-fold and St8sia6 was 9.5-fold (Figure S3B). SW837 cells were treated with the kifunensine or benzyl-α-GalNAc. While kifunensine had no effect on binding of Siglec-7-Fc, SW837 treated by benzyl-α-GalNAc showed loss of the binding of Siglec-7-Fc (Figure 1D).
Figure 1.
Siglec-7 binding is enhanced by four sialyltransferases in colon cancer cell lines, and its ligand glycan is O-glycan on mucin-type proteins
(A) Flow cytometry analysis of Siglec-7-Fc binding to parent DLD-1 and stably expressing cells of sialyltransferases that have been identified as Siglec-7 ligand-synthase genes (Figure S2A). As a result, ST3Gal1, ST6GalNAc1, ST6GalNAc3, and ST8Sia6 were identified to be capable of the induction of Siglec-7 binding. DLD-1 co-transfected with ST3Gal1 and ST6GalNAc3 genes is C3. Co-transfectant DLD-1 of ST3Gal1 and ST6GalNAc1 genes is C4. ST8Sia6 gene transfectant DLD-1 is C8. Blue-filled lines indicate negative controls, and purple lines indicate Siglec-7-Fc binding.
(B and C) Effects of inhibition of N-glycosylation or O-glycosylation on the binding of Siglec-7. DLD-1 and the transfectants were treated with 20 nM kifunensine or DMSO as a negative control or 2 mM benzyl-α-GalNAc or DMSO as a negative control for 3 days, respectively. They were detached and incubated with Siglec-7-Fc (5 μg/100 μL) for 1 h and then with anti-human Fc IgG-FITC, and then analyzed using flow cytometry. (B) Kifunensine, an N-glycosylation inhibitor, showed no effect on the binding of Siglec-7. (C) Benzyl-α-GalNAc, an O-glycosylation synthesis inhibitor, led to a significant decrease in the binding of Siglec-7 in all three clones.
(D) N- and O-glycan synthesis inhibition was performed on the Siglec-7-binding cell colon adenocarcinoma cell line SW837 and its effects were evaluated.
(E) Biotinylated membrane proteins derived from DLD-1 and the ST transfectants were pulled down using Siglec-7-Fc or human IgG Fc only as a protein control Fc (Con-Fc) and protein A beads.
(F) They were electrophoresed and transferred onto a PVDF membrane. It was used for the avidin-biotin complex detection system (ABC kit) and ECL. Specific bands were detected at 175 kDa< and 50 kDa in ST-transfectants. A specific band detected at 120 kDa in C8 was Mucin-13 (MUC13) (Figure S2C).
(G) Immuno-blotting using anti-PODXL antibody. The upper panel indicates pull-down samples with Siglec-7-Fc or control-Fc. Lower panel indicates input lysates.
Siglec-7 binds to mucin-type proteins on the colon adenocarcinoma cell line DLD-1
To identify Siglec7 ligand glycan-carrier proteins, the cell surface membrane of these transfected cells, as well as DLD-1 cells, was biotinylated, and pull-down experiments of the cell lysates by Siglec-7-Fc were performed (Figure 1E). Bands higher than 175 kDa (180–200 kDa) and at 50 kDa were detected in C3, C4, and C8 using the avidin-biotin complex (ABC)-HRP kit. Another band was also observed at 120-kDa for C8 (Figure 1F). Mass spectrometry (MS) and immunoblotting (IB) revealed that the 175 kDa molecule was podocalyxin (PODXL)31,32,33 (Figures 1G and S4A) and the 120-kDa one was Mucin-13 (MUC13)34,35 (Figures S4A and S4B). The 50 kDa molecule is unknown.
Siglec-7 ligand O-glycans suppress NK cell cytotoxicity via inhibitory signal activation mediated by ITIM/ITSM
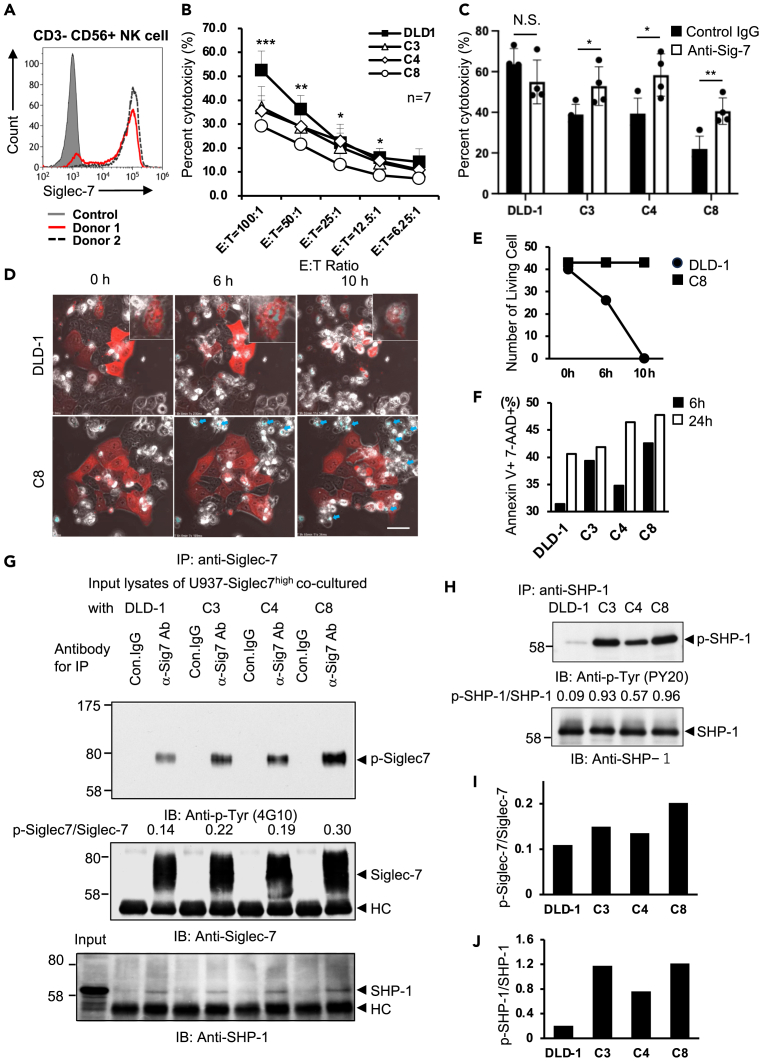
It has been reported that the expression pattern of Siglec-7 on NK cells varies between individuals.36,37 Siglec-7 was expressed in 88.08% and 98.28% of CD3− CD56+ NK cells and 100% of monocytes in human peripheral blood mononuclear cells (PBMCs) of donor 1 and donor 2, respectively (Figures 2A and S5A). NK cells exhibit cytotoxicity against cancer cells. Therefore, we examined the inhibitory effects of the interaction between ligand glycans and Siglec-7 on NK cell function. As a result of the co-culture of DLD-1 cells and transfectants with PBMCs from donor 1, the cytotoxic activity of NK cells was significantly suppressed by the transfectants (Figure 2B). As shown in Figure S2D, there was no difference in Siglec-9 binding between DLD-1 cells and transfectants. To further clarify whether the suppression of this cytotoxic activity was mediated by Siglec-7, we performed cytotoxic activity rescue experiments using blocking antibodies that inhibit the binding of Siglec-7 to the ligand.38 This antibody blocks the binding to the glycan ligand. The results showed a rescue effect on cytotoxic activity by the Siglec-7 blocking antibodies at C3, C4, and C8 (Figure 2C: donor 2; Figure S5B: donor 1). Using confocal microscopy, we analyzed the time-lapse images of what occurs when NK cells encounter cancer cells. An NK cell line was used to study the cytotoxic activity of NK cells. Because the human NK cell line KHYG-1 expressed Siglec-7 at low levels, we established a Siglec-7-highly-expressing clone, KHYG-1-Siglec-7high, from a Siglec-7-negative clone (Figure S3C). As a result of co-culture of DLD-1 cells with KHYG-1-Siglec-7high, almost all DLD-1 cells were killed after 10 h of co-culture. In contrast, most C8 cells survived during the co-culture with KHYG-1-Siglec-7high (Figure 2D and Video S1). KHYG-1 cells became DAPI-positive, indicating apoptosis (Figure 2C blue arrow). To determine whether binding to ligand glycans induced apoptosis in Siglec-7-expressing NK cells, NK cell apoptosis was assessed after co-culturing PBMCs with DLD transfectants. After 6 and 24 h of co-culture, PBMCs were collected and NK cell apoptosis was assessed by staining with Annexin V and 7-AAD, noting CD16 and CD56 positive NK cells within some gates (Gate:A, Figure S5D) in the forward scatter (FSC) and side scatter (SSC) plots. This gate is generally considered to have the gate with the highest number of dead cells; late apoptotic NK cells positive for both Annexin V and 7-AAD were found to have enhanced apoptosis when co-cultured with the transfectants (Figure 2F). (8% higher in C3, 3.4% in C4, and 11.2% in C8 at 6h) There was a slight apoptosis of NK cells in Gate: Lymphocyte. A few NK cells in Gate: A were classified as early apoptotic cells (Figure S5E). The proportion of late apoptosis in the gate: monocytes tended to be 3 to 4% higher in transfectants, but the difference was not significant (Figure S5F). These results suggest that the Siglec-7 signaling induced not only suppression of killing activity but also apoptosis of Siglec-7-expressing cells. To clarify the molecular mechanisms, we co-cultured here “non-cytotoxic” U937-Siglec7high cells instead of KHYG-1-Siglec-7high (Figure S6A) with DLD-1 and its transfectant cells of STs to analyze intracellular signals (Figure S6B). As a result, the ITIM/ITSM of Siglec-7 on U937-Siglec7high was phosphorylated in the co-culture with transfectants (Figure 2G upper panel). Simultaneously, the recruitment (binding) to ITIM/ITSM and phosphorylation of SHP-1 were observed (Figure 2G lower and 2H upper). The phosphorylation levels of Siglec7-ITIM/ITSM and SHP-1 were more than 2- and 10-fold higher, respectively, than those in parent DLD-1 cells (Figures 2I and 2J). In addition, when co-cultured with DLD GD3+, which is reported to be non-reactive with Siglec-7, U937-Siglec-7high did not activate of ITIM/ITSM signaling (Figure S6C).
Figure 2.
Siglec-7 ligand O-glycans suppress NK cell cytotoxity via inhibitory signal activation of ITIM/ITSM
(A) The gating information for picking up NK cells from PBMCs is in Figure S5B. The expression of Siglec-7 on NK cells from donor 1 and donor 2 was analyzed.
(B) Cytotoxicity assay using co-culture of DLD-1 or ST transfectants with PBMCs. Cytotoxicity was assessed by measuring lactate dehydrogenase (LDH) activity released into the medium from injured cells. They were measured using the LDH release detection kit. E:T refers to the ratio of effector cells (PBMCs) and target cells (DLD-1 and ST transfectants). Error bars indicate SEM. ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05, using one-way ANOVA. E:T = 100:1 p = 0.0003, E:T = 50:1 p = 0.0026, E:T = 25:1 p = 0.0166, E:T = 12.5:1 p = 0.0241. Tukey’s multiple comparison test, E:T = 100:1 DLD-1 vs. C3, p = 0.0129, DLD-1 vs. C4, p = 0.0059, DLD-1 vs. C8, p = 0.0002, E:T = 50:1 DLD-1 vs. C8, p = 0.0012, E:T = 25:1 DLD-1 vs. C8, p = 0.0346, E:T = 12.5:1 DLD-1 vs. C8, p = 0.0223, E:T = 6.25:1 DLD-1 vs. C8, p = 0.0314. Data are mean ± SD, n = 7 biological replicates.
(C) PBMCs were co-cultured with DLD at E:T = 100:1 after prior treatment with 10 μg/mL anti-Siglec-7 antibody or isotype control antibody. The cytotoxic activity was then assessed with the LDH release detection kit. DLD-1 Control IgG vs. DLD-1 Anti-Sig-7, p = 0.2328, C3 Control IgG vs. C3 Anti-Sig-7, p = 0.0404, C4 Control IgG vs. C4 Anti-Sig-7, p = 0.0260, C8 Control IgG vs. C8 Anti-Sig-7, p = 0.0062, Data are mean ± SD, n = 4 biological replicates.
(D and E) Co-culture of an NK cell line KHYG-1-Siglec-7high (Figure S3B) and DLD-1 or ST8Sia6-transfectant C8. DLD-1 and C8 were transiently transfected with a red fluoresceine protein gene for labeling of the cells. The medium contained DAPI, which stains dead cells. The magnified image in the upper right of the panel shows dead cells stained with DAPI. Dead KHYG-1-Siglec-7high stained using DAPI are indicated with blue arrows. Time-lapse imaging of co-cultured DLD-1 or C8 and KHYG-1-Siglec-7high was acquired for 10 h (Video S1). Scale bar indicates 50 μm.
(F) Assessment of NK apoptosis in PBMCs co-cultured with DLD; PBMCs were stained with anti-CD3-FITC and anti-CD16/CD56-PE after co-culturing. Further staining with Annexin-V-APC and 7-AAD comparing Annexin-V and 7-AAD co-positive populations in Gate:A (Appendix Gating Information).
(G) U937-Siglec-7high were co-cultured with DLD-1 or ST transfectants for 10 min at 37°C. The U937 cells were collected and lysed. Immunoprecipitation was performed using an anti-Siglec-7 antibody and subsequent immunoblotting with an anti-phospho-tyrosine antibody (4G10), anti-Siglec-7 antibody, or anti-SHP-1.
(H) Immunoprecipitation using anti-SHP-1 antibody and subsequent IB using anti-phosphotyrosine antibody (PY20) or anti-SHP-1 were performed. Phosphorylated Siglec-7 and SHP-1 were quantitated using ImageJ software (NIH) and the ratios of p-protein/total proteins in (G) and (H) are presented in (I) and (J), respectively.
(I and J) Summary of phosphorylation ratios for Siglec-7 ((I) p-Siglec-7/Siglec-7) and SHP-1 ((J) p-SHP-1/SHP-1). DLD-1 and ST transfectants vs. (A and B) PBMCs, (C) KHYG-1-Siglec-7high and (D–F) U937-Siglec-7high.
Siglec-7 binding transduces activation signals into cancer cells via the interaction with ligand O-glycans
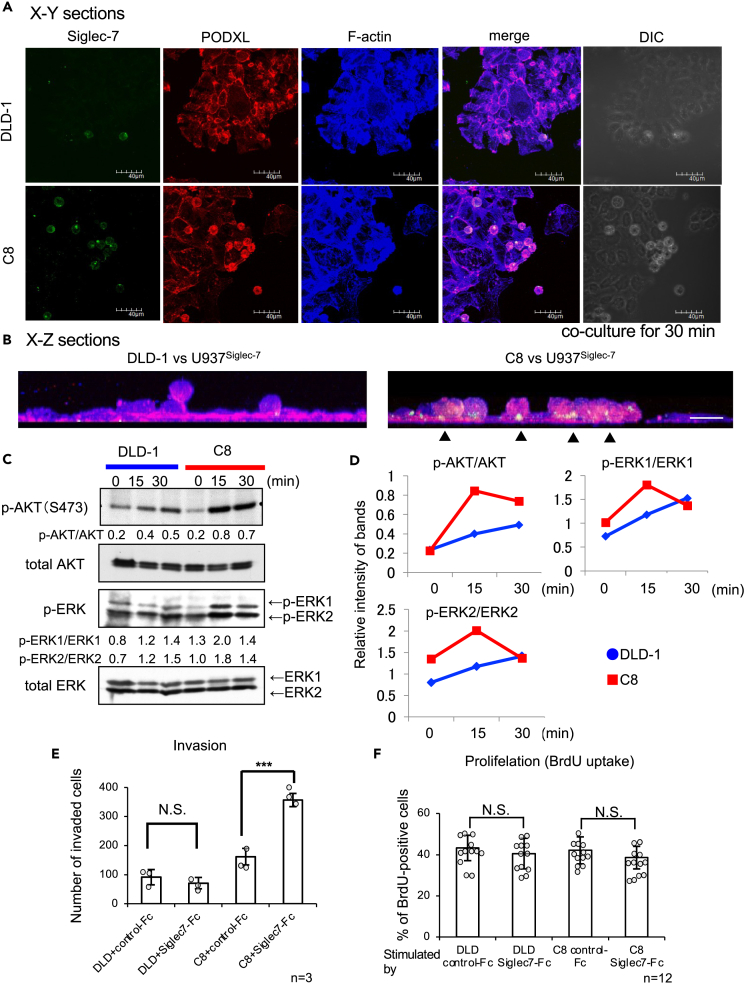
The following experiments were conducted to clarify how Siglec-7 expressed on lymphocytes recognizes and binds to cancer cells and what kind of signal is transmitted. From the results of the co-culture of DLD-1 and the transfected cells with U937-Siglec-7high, it was found that Siglec-7 co-localizes with the ligand glycan-carrier protein PODXL on the adhesive surface (Figure 3A). Furthermore, they co-localized with actin filaments that were present downstream of PODXL (Figure 3B). We analyzed the intracellular signaling changes that occurred along with the changes in actin localization. Upon co-culturing DLD-1 or C8 cells with U937-Siglec-7high, phosphorylation of AKT and ERK1/2 was observed in C8 cells (Figures 3C and 3D). Moreover, similar results were observed in the transfectants of STs when stimulated with the recombinant protein Siglec-7-Fc conjugated with protein A beads (Figures S7A–S7E). The results obtained from experiments using purified Siglec-7 signals were considered to exclude the possibility of other cellular signals. To elucidate the phenotypic changes in cancer cells induced by the Siglec-7-binding signals via ligand glycans, invasiveness was evaluated using the transwell invasion assay, and proliferation was evaluated using the BrdU cell proliferation assay. Invasion activity was enhanced in C8 cells, as shown by the transwell migration assay (Figure 3E). In contrast, the BrdU cell proliferation assay revealed no changes in the proliferation activity between DLD-1 and C8 cells (Figure 3F). These results suggest that Siglec-7 functions not only as a receptor that transmits inhibitory signals to its expressing cells (NK cells) but also as a ligand that transmits cytoskeletal activation signals to cancer cells via binding glycans, increasing the malignant phenotype.
Figure 3.
Siglec-7 transduces activation signals into cancer cells via ligand O-glycans
(A) Confocal microscopy analysis of Siglec-7, PODXL, and F-actin in DLD-1 and C8 cells during 30 min co-culture with U937-Siglec7high using confocal microscopy. FITC: Siglec-7, Alexa 568: PODXL, Alexa 650: F-actin (Phalloidin). Differential interference contrast microscope (DIC). In the co-culture of C8 and U937-Siglec7high, these three molecules were co-localized (white) in merge. Scale bar: 40 μm.
(B) Representative side view of confocal microscopy imaging. Arrow heads indicate co-localized point of Siglec-7, PODXL, and F-actin. Scale bar: 20 μm.
(C) The time-course of AKT and ERK1/2 activation in DLD-1 transfectants co-cultured with U937-Siglec7high. DLD-1 and C8 were co-cultured with U937-Siglec7high for indicated times, and then U937 cells were washed out. DLD-1 and C8 were lysed and analyzed using western blotting. Phosphorylated AKT and ERK1/2 were quantitated using ImageJ, and the ratios of p-protein/total protein were obtained as described in (D).
(D) Graphs summarizing the phosphorylation levels of AKT (p-AKT/AKT), ERK1 (p-ERK1/ERK1), and ERK2 (p-ERK2/ERK2).
(E) Transwell migration assay of DLD-1 and C8 stimulated using Siglec-7 binding. It showed increased migration of C8 cells when stimulated with Siglec-7-conjugated beads (Figure S7A). ∗∗∗p < 0.001, using Student’s t test. DLD+control-Fc vs. DLD+Siglec-7-Fc, p = 0.347. C8+control-Fc vs. C8+Siglec-7-Fc, p = 0.00095. Data are mean ± SD, n = 3 biological replicates.
(F) BrdU cell proliferation assay showed that there were no differences in DLD-1 and the transfectants either with no stimulation or when stimulated with Siglec-7. ∗∗∗p < 0.001, using Student’s t test. DLD+control-Fc vs. DLD+Siglec-7-Fc, p = 0.327. C8+control-Fc vs. C8+Siglec-7-Fc, p = 0.144. Data are mean ± SD, n = 12 biological replicates. DLD-1 or ST transfectant C8 vs. U937-Siglec7high (A-D) and Siglec-7-conjugated beads (E-F).
Expression analysis of sialyltransferase genes in human colon adenocarcinoma database
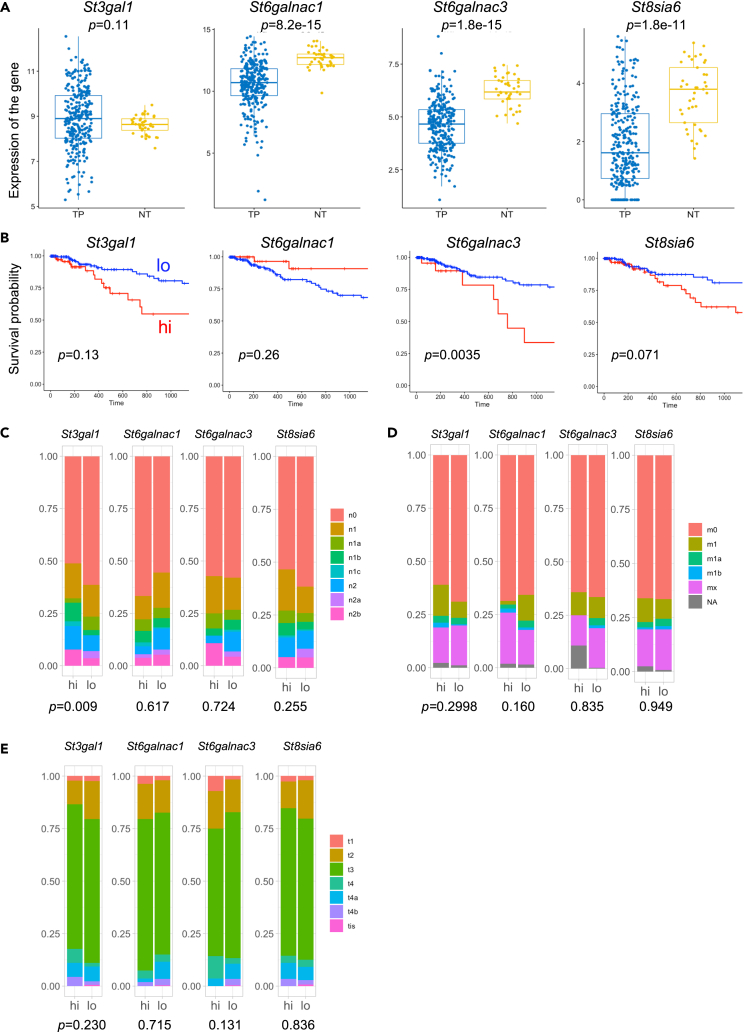
Four sialyltransferases that generate Siglec-7 ligand glycans on colon cancer cells have been identified. Ligand glycans suppress the cytotoxic activity of NK cells via Siglec-7 and further enhance their metastatic activity. Therefore, we analyzed the gene expression of these four sialyltransferases and their involvement in cancer grading using a database of human cancer cells. The expression levels of sialyltransferase genes were compared between cancerous and normal human colorectal cancer tissues in The Cancer Genome Atlas (TCGA) using Wilcoxon rank-sum test. St6galnac1, St6galnac3, and St8Sia6 were more highly expressed in normal tissues (NT) than in cancerous tissues (TP), whereas St3gal1 was more highly expressed in cancerous tissues (TP) (p = 0.11). (Figure 4A). In the survival time analysis, groups with high St3gal1, St6galnsc3, and St8sia6 expression tended to have lower survival rates, whereas the group with low expression of St6galnac1 tended to have a poorer prognosis (Figure 4B). In a comparative study of cancer metastasis, in the St3gal1 high group, n0 decreased and n1b, n1c, n2, and n2b increased to those in the low group (Figure 4C). A comparative analysis of cancer metastasis tendency using the chi-square test based on the expression of sialyltransferases showed that the high group had a high metastasis tendency in the St3gal1. In the St8sia6 group, the high group tended to metastasize easily. In addition, no significant differences were observed in the degree of metastasis, size, and/or extension of the main tumor (Figures 4D and 4E). However, because the expressions of St3gal1, St6galnac3, and St8sia6 tended to show malignant traits of cancer in any evaluation method, it may be necessary to evaluate the effects of the combination of multiple gene expressions on the poor prognosis of cancer patients. From the results described previously, it was shown that the sialyltransferase genes involved in the synthesis of the ligand glycan of Siglec-7 defined in this study were expressed (more or less) in human colon cancer tissues, and clinical cases with high expression of St3gal1 and St8sia6 clearly showed significantly lower survival rates, suggesting that the expression of these genes is involved in cancer metastasis. The “GlycoMaple” tool for visualizing the glycosyltransferase pathway for the biosynthesis of mucin-type O-glycans and the report by Huang et al.39 suggest that these four transferases may produce the tri-sialyl T structure. In order to clarify the predicted glycan, we next performed structural analysis of the glycan on the identified ligand-glycan-carrying proteins.
Figure 4.
Expression analysis of sialyltransferase genes in human colon cancer (COAD) in TCGA database
(A) Comparison of expression between cancer tissues and normal tissues. TP: primary solid tumor (285 samples). NT: solid tissue normal (41 samples). y axis is expression of the gene: log2 (transcripts per kilobase million+1). St3gal1 tended to be more abundant in cancer tissues (p = 0.11), but other sialyltransferase genes were significantly more expressed in normal tissues.
(B) Survival data from the COAD in TCGA database that had high (blue line) or low (red line) expression of the individual STs. The expression level of sialyltransferases in human colon cancer patients was divided into high and low groups. The low groups of St3gal1, St6galnac1, St6galnac3, and St8sia6 were those with expression values less than 650.0797, 4325.628, 58.5373, and 2.265, respectively. The groups with high St3gal1, high St6galnac3, and high St8sia6 tended to have lower survival rates (p = 0.013, 0.0035, and 0.071, respectively), whereas the group with low expression of St6galnac1 tended to have a poorer prognosis (p = 0.26).
(C) Analysis of COAD TNM staging in each gene expression. Lymph node metastasis analysis based on the difference of expression of sialyl transferase genes. “n0” means no metastasis. “1” means metastasis to the neighborhood, and “2” means metastasis to the distance. Symptoms become more serious from “A” to “C”.
(D) Metastasis analysis based on the difference of expression of sialyltransferase genes. “m0” means not spread to other regions. “1” means spread to other regions and b indicates a more serious condition than a. mx means “Metastasis cannot be measured”.
(E) Primary tumor status analysis for the difference in the expression levels of sialyltransferase genes. t1, t2, t3, and t4: Refers to the size and/or extent of the main tumor. The higher the number after t, the larger the tumor or the more it has grown into nearby tissues. t3 may be further divided to provide more details, such as t3a and t3b. A comparative analysis of cancer metastasis tendency based on the expression of sialyltransferases showed that the high group had a high metastatic tendency (p = 0.009) in the St3gal1. In the St8sia6 group, the high group tended to metastasize easily (p = 0.255). In addition, no significant difference was observed on evaluating the degree of the metastasis and size and/or extension of main tumor (Figures 3D and 3E). Statistics were performed in the following manner. (A) Differences between tumor and normal were analyzed using Wilcoxon rank-sum test and evaluated using p value. (B) Differences between ST-expression high and low group about survival rate were analyzed using Wilcoxon rank-sum test and evaluated using p value. (C–E) Differences in these groups were analyzed using the Chi square test and evaluated using p value. The p value indicates a difference in metastatic tendency between the high and low groups. ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05.
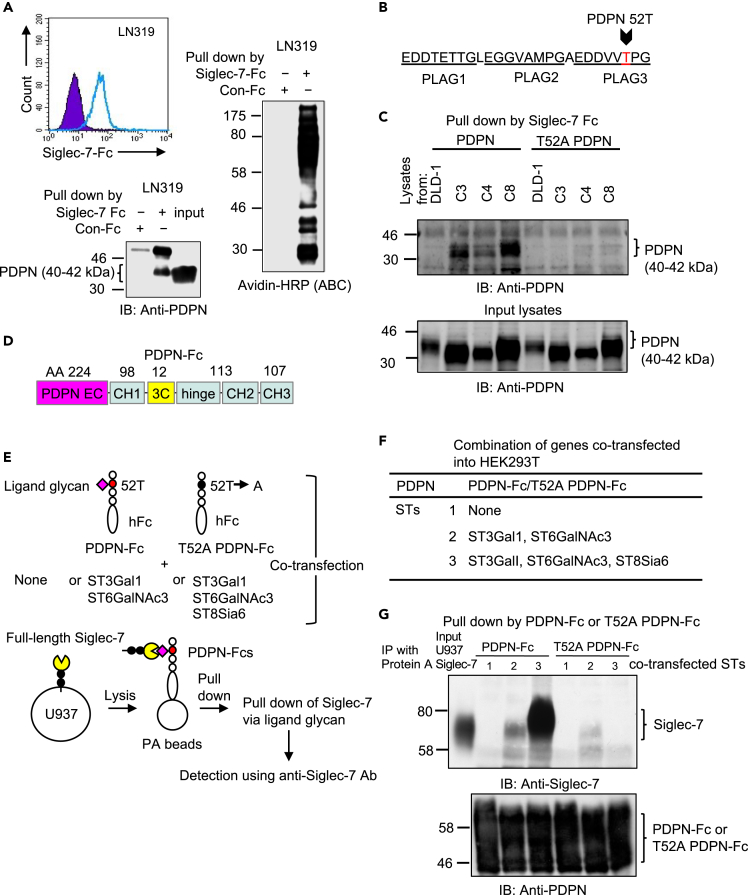
Siglec-7 binds to a glycan on the platelet aggregation-stimulating 3 domain of PDPN
It has been reported that PODXL and MUC13 have a large number of various O-glycans.31,34 Identifying the true ligand glycans on these molecules is challenging due to the large size of their backbone proteins and the abundance of glycan modifications. To simplify the experimental system, smaller glycan-carrying proteins were required. The glioblastoma cell line LN319 showed the binding of Siglec-7-Fc by flow cytometry (Figure 5A, upper panel). Pull-down Siglec-7-Fc of biotinylated membrane proteins revealed that LN319 contains several proteins that bind to Siglec-7 in the lower molecular weight region (Figure 5A, right). LN319 cells highly express podoplanin (PDPN, a mucin-type sialoglycoprotein).40,41,42,43 Furthermore, IB using pull-down products by Siglec-7-Fc from lysates of LN319 cells revealed that the 40 kDa protein is the ligand glycan-carrier protein PDPN (Figure 5A, lower). However, the amount of PDPN pulled down by Siglec-7-Fc was less than that pulled down by PDPN in the input lysate, suggesting that some PDPN binds to Siglec-7-Fc. As previously reported,41 the PDPN platelet aggregation-stimulating (PLAG) 3 domain in LN319 has a characteristic glycan di-sialyl-T structure attached at 52T, as shown in Figure 5B, suggesting that the Siglec-7 ligand glycan is attached to 52T of PDPN. PDPN is not expressed in DLD-1 cells. When PDPN was forcibly expressed in DLD-1 and ST transfectants, IB with an anti-PDPN antibody detected PDPN as a band around 40kDa in Siglec-7 pull-down samples from all ST clones, as expected (Figure 5C, 4 lanes on the left). It was detected more abundantly in clone 8 (ST8Sia6-transfected). In contrast, when the T52A mutant PDPN was forcibly expressed, Siglec-7 binding was not observed (Figure 5C, four lanes on the right). We wanted to clarify the structure of the Siglec-7 ligand-glycans. Therefore, generation of PDPN modified by the four sialyltransferases is required. HEK293T cells express PDPN, which does not bind to Siglec-7-Fc. When ST8Sia6 gene was transiently expressed, Siglec-7-Fc binding was enhanced (Figure S8A) and endogenous PDPN bound to Siglec-7-Fc in HEK293T (Figure S8B). Therefore, we generated an expression vector for the soluble fusion protein of the extracellular domain (ECD) of PDPN and human IgG1 Fc (Figure 5D), and the fusion proteins were expressed in HEK293T by transfection of this expression vector with sialyltransferases. To evaluate the binding properties of PDPN-Fc to full-length Siglec-7, a pull-down experiment with three types of PDPN-Fcs (Figure S8C) was performed using the U937-Siglec-7high cell lysate (Figure 5E). We also generated an expression vector for the PDPN-Fc fusion molecule in the T52A mutant. Together with these PDPN expression vectors, either expression vectors of ST3Gal1+ST6GalNAc3 or ST3Gal1+ST6GalNAc3+ST8Sia6 were co-transfected (Figure 5F) and analyzed by a pull-down assay. Although no binding of Siglec-7 was observed with PDPN-Fc alone (Figure 5G, 2nd lane), strong binding of Siglec-7 was noted in PDPN-Fc co-expressed with ST3Gal1 +ST6GalNAc3 +ST8Sia6 (Figure 5G, lane 4). However, none of the T52A point-mutated PDPN-Fc showed binding to Siglec-7 (Figure 5G, lanes 6 and 7). These results revealed that the ligand glycan structures of Siglec-7 were formed in the PLAG3 domain of PDPN.
Figure 5.
Siglec-7 ligand-glycan is present at 52T on PLAG3 domain of PDPN
(A) PDPN was identified as a Siglec-7 ligand glycan-carrier protein in glioblastoma LN319. Flow cytometry showed that LN319 expresses Siglec-7-Fc binding molecules (upper). A pull-down assay of biotinylated proteins revealed that there were various types of membrane proteins binding to Siglec-7 (right). One of them was identified as PDPN using IB (lower).
(B) Identification the of Siglec-7 ligand glycosylation site. Di-sialyl T antigen is present in glycosylation products at 52T on the PLAG3 domain of PDPN.
(C) The wild-type and a point mutant of PDPN-expression vectors were used to transfect DLD-1 and its ST transfectants. Their PDPNs were pulled-down using Siglec-7-Fc from the lysates of DLD-1 and its ST transfectants, and analyzed using IB, showing that WT PDPN derived from DLD-1 did not bind to Siglec-7-Fc. WT PDPN derived from ST transfectants bound to Siglec-7-Fc. T52A PDPN derived from either DLD-1 or ST transfectants did not bind to Siglec-7.
(D) Generation of fusion protein (PDPN-Fc) consisting of the WT and T52A PDPN extracellular (EC) domain and human IgG-Fc.
(E) Evaluation of Siglec-7 binding ability to PDPN-Fc co-transfected with various STs. WT and T52A PDPN-Fcs were prepared by co-expression with none or ST3Gal1 and ST6GalNAc3 or ST3Gal1, ST6GalNAc3 and ST8Sia6 in HEK293T cells, and purified using the protein A affinity column. U937 cells expressing full-length Siglec-7 were lysed and purified and PDPN-Fcs were added to the lysates. PDPN-Fcs were pulled-down with Protein A beads, and the amount of co-precipitated Siglec-7 was compared. A T52A point mutant of PDPN-Fc in the PLAG3 domain was also generated under the same conditions.
(F) A list of co-transfected PDPN-Fc and STs.
(G) Binding of WT full-length Siglec-7 and PDPN-Fcs was detected using IP with the Fc fusion proteins and subsequent IB with anti-Siglec-7 (top). Expression of PDPN-Fc or its mutant was confirmed using IB with an anti-PDPN antibody (bottom).
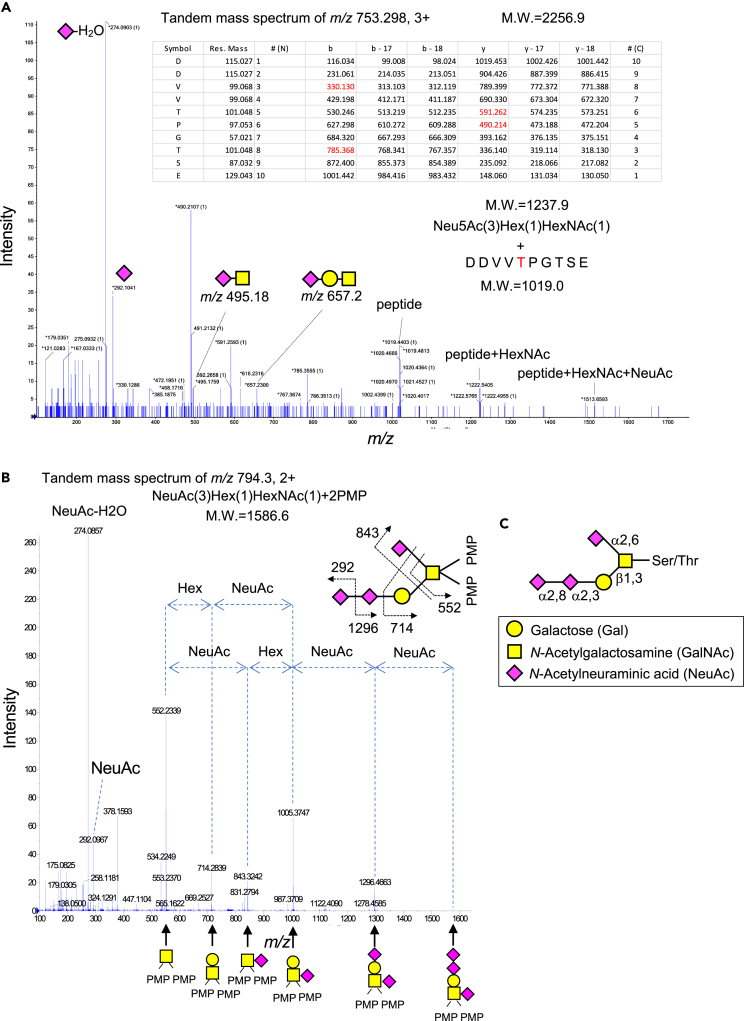
MS analysis of O-glycan structures recognized by Siglec-7
Glycopeptides and O-glycans derived from PDPN-Fcs were analyzed using MS. Glycopeptides were endoproteinase Asp-N digests. O-glycans was excised by the glycan release method (β-elimination and 1-phenyl-3-methyl-5-pyrazolone (PMP) labeling).44,45 These were derived from three types of PDPN-Fc (Figures 5F and 5G). Table S2 summarizes the m/z values of ions identified as amino acid sequences derived from PLAG3 based on the MS results of glycopeptides and the types of presumed glycan structures. Six peaks with m/z indicated were detected only for PDPN-Fc (ST3Gal1, ST6GalNAc3, ST8Sia6). m/z 753.3 (3+) indicated the Asp-N-digested PLAG3 domain (MW = 1019.0) and Neu5Ac(3)Hex(1)HexNAc(1) (MW = 1237.9) (Figure 6A; Tables S3 and S4). As a result of β-elimination of PDPNs, 4 types of O-glycans from PDPN-Fc (None), 3 types of O-glycans from PDPN-Fc (ST3Gal1, and ST6GalNAc3), and 5 types of O-glycans from PDPN-Fc (ST3Gal1, ST6GalNAc3, and ST8Sia6) were identified, respectively (Table S5). The tandem mass spectrum of m/z 794.3 the PMP-O-glycan from PDPN-Fc (ST3Gal1, ST6GalNAc3, and ST8Sia6) revealed that the ion contained one HexNAc, one Hex, and 3 Neu5Ac (Figure 6B). Thus, a tri-sialyl T structure (Figure 6C) exists in the PMP-labeled O-glycans derived from PDPN-Fc (ST3Gal1, ST6GalNAc3, and ST8Sia6), suggesting that the tri-sialyl T structure is characteristically formed in the PLAG3 domain. A quadrosialyl T structure was also found in peptides that were not derived from the PLAG3 domain (Figure S9A; Table S4) or PMP-labeled O-glycans (Figure S9B; Table S5). Since no binding to Siglec-7 was observed in the point mutant at 52T, and quadro-sialyl T was present outside the PLAG3 domain, it is considered that Siglec-7 does not bind to quadro-sialyl T. Table S5 summarizes the glycan structures predicted from the types and numbers of these glycans and the results of PMP-labeled cut-out glycans. Neu5Ac(2)HexNAc(1) “di-sialyl Tn” is the most abundant glycan structure. However, it cannot be a ligand glycan because our results suggest that α2-3, α2-6, and α2-8 Sia are important for Siglec-7 binding.
Figure 6.
Structure analysis of the Siglec-7 ligand glycan using MS
(A) MS/MS spectrum of PLAG3 domain-derived glycopeptide from Asp-N digested PDPN-Fc peptide. They were analyzed using LC/MS. Tandem mass spectrum of the precursor ion m/z 753.298 (3+) (Table S2) were analyzed using TOF MS/MS. The fragmentation table shows the PLAG domain 3 sequence and its detected product ions in red. The m/z 753.298 (3+) was identified to consist of a PLAG3 domain peptide and a HexNAc, a Hex, and 3 Neu5Ac, as identified using MS and MS/MS analysis. Then, the tandem mass spectrum indicated amino acid sequence of PLAG3 and NeuAc (m/z 274.1 is [NeuAc-H2O + H] and m/z 292.1 is [NeuAc+H]), NeuAc-HexNAc (m/z 495.2), and NeuAc-Hex-HexNAc (m/z 657.2).
(B) MS/MS spectrum of released and labeled tri-sialylated O-glycan. O-glycan was released by β-elimination and labeled with 1-phenyl-3-methyl-5-pyrazolone (PMP). Tandem mass spectrum of the precursor ion m/z 794.3 (2+) was analyzed using TOF MS/MS.
(C) Predicted glycan structure.
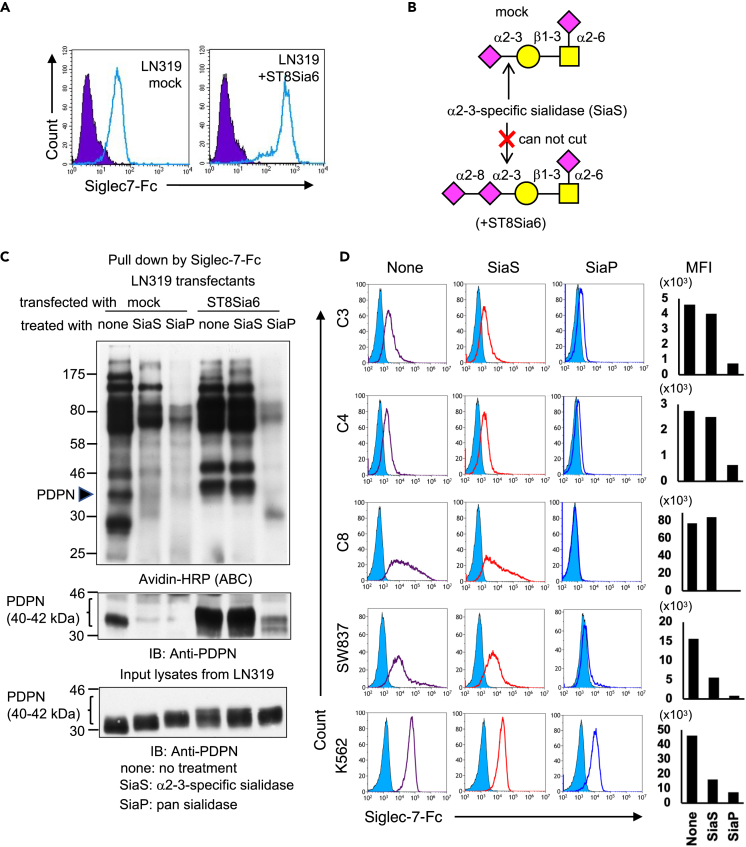
Sia α2-8 Sia α2-3 linkage, which cannot be removed by α2-3-linked Sia-specific sialidase, is essential for Siglec-7 binding
Using the α2-3-linked Sia-specific sialidase, it was clarified whether α2-8 Sia was bound to the end of α2-3 Sia or α2-6 Sia. Therefore, a stable transfectant, LN319, with high ST8Sia6 expression and markedly enhanced Siglec-7 binding was established (Figure 7A). We conducted a suppression experiment for Siglec-7 binding using a Sia-linkage-specific sialidase (Figure 7B). ST8Sia6-expressing LN319 cells showed more membrane proteins (biotinylated) bound to Siglec-7-Fc than mock cells. Arrowhead-indicated PDPN mainly increased in the pull-down assay using Siglec-7-Fc (Figure 7C, lanes 1 and 4), and the reactive protein was confirmed to be PDPN (Figure 7C, middle panel). Treatment with a regular sialidase (SiaP, lanes 3 and 6) suppressed the Siglec-7 binding, while α2-3 Sia-specific sialidase (SiaS, lanes 2 and 5)46 did not suppress binding in ST8Sia6-transfectant cells (Figure 7C). DLD-1 transfectants, SW837 and K562 cells were treated with SiaP and SiaS to assess their binding to Siglec-7. In contrast, SiaS treatment did not cause DLD-1 transfectants to lose their binding to Siglec-7, whereas SW837 and K562 cells showed reduced binding to Siglec-7 after SiaS treatment (Figure 7D). Since α2-3-linked Sia-specific sialidase cannot cleave Sia when α2-8 Sia is attached at the non-reducing end, it was concluded that α2-8-linked Sia is present at the non-reducing end of the glycan, being essential for the binding of Siglec-7. This result is consistent with the report that ST8Sia6 requires Neu5Acα2-3Galβ1-3GalNAc as a substrate47 and seems to provide evidence for the presence of a tri-sialyl T structure.
Figure 7.
Sia α2-8 Sia α2-3 structure is essential Siglec-7 binding to ligand glycans
(A) The glioblastoma cell line LN319 was transfected with pcDNA3.1 ST8Sia6 or pcDNA3.1 empty vector using Lipofectamine 2000. Those cells were selected using G418 and cloned using a limiting dilution method. The binding of Siglec-7-Fc to the LN319 ST8Sia6 gene transfectant and mock. Thus, ST8Sia6 further enhanced the binding of Siglec-7-Fc to the cell lines, as shown in colorectal carcinomas DLD-1.
(B) α2-3 specific sialidase SiaS can cut terminal α2-3-linked sialic acid, but it cannot cut non-terminal (inside) α2-3-linked sialic acid. However, pan sialidase (SiaP) removes all linkage modes of Sia.
(C) Biotinylated membrane proteins of LN319 and of ST8Sia6 transfectant LN319 were detected using avidin-HRP. Upper panel indicates biotinylated membrane proteins from mock and ST8Sia6-LN319 transfectants with pan sialidase (SiaP) or α2-3 Sia specific sialidase (SiaS) treatment and subsequent IB. Biotin-labeled products were applied for IP with recombinant Siglec-7-Fc and analyzed using IB with avidin-HRP (ABC) to detect all biotinylated molecules. Middle panel indicates IB of IP products with anti-PDPN antibody. Lower panel indicates PDPN molecular size changed due to sialylation and sialidases.
(D) DLD-1 and the ST transfectants, SW837 and K562 were treated with SiaP or SiaS for 1 h at 37°C. The binding of Siglec-7-Fc was analyzed using FACS and the mean fluorescence intensity is shown in the graph.
Discussion
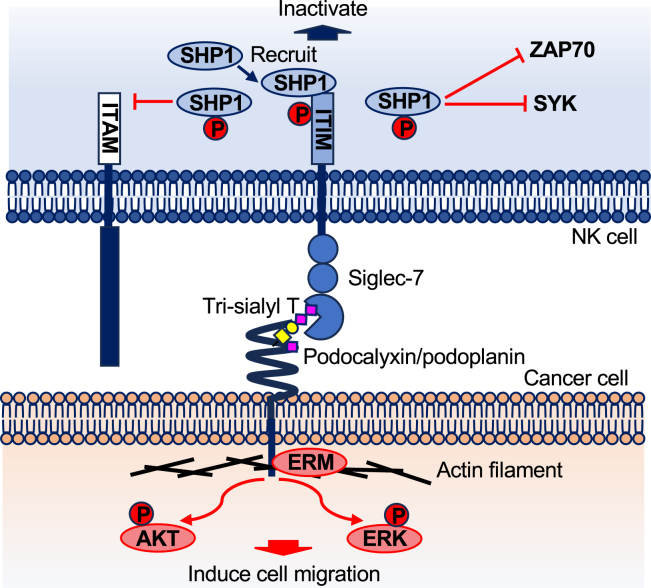
Although there have been several reports on the carbohydrates recognized by Siglec-7, no intensive studies on its ligand structures on the cell membrane and the resulting biological effects of their interactions have been performed to date. In this study, we revealed that Siglec-7 binds to O-glycans on PDPN, PODXL, and MUC13, and further identified the tri-sialyl T structure in O-glycans as a previously unreported ligand recognized by Siglec-7. Furthermore, this glycan structure induced a negative signal on the immune cell side by activating the phosphatase SHP-1, leading to the suppression of NK cytotoxic activity. In contrast, this interaction promotes actin filament recruitment to the ligand glycan carrier protein in cancer cells. Actually, it was shown that activation of AKT and ERK was induced, leading to the enhancement of cell invasion activity of cancer cells (Figure 8). When analyzing molecular signaling, we used transfectants expressing Siglec-7 or sialyltransferases. The expression levels of Siglec-7 in U937 transfectants are comparable to those in human NK cells (Figures S5A and S6A). Similarly, expression levels of sialyltransferases in DLD-1 transfectants were comparable to various cell lines (Figure S3B). Additionally, this ensures that our findings reflect natural cellular conditions and support the dual function hypothesis of Siglec-7 signaling.
Figure 8.
Model for pathway of bi-directional signaling via glyco-immune check point
Recently, it was reported that CD43 contains a cluster of disialylated O-glycans bound to Siglec-7 in leukemic cells.16,17 The disialyl T structure on CD45 and CD162 in chronic lymphocytic leukemia B cells has also been reported as a Siglec-7 interacting molecule.18 This study also showed that the formation of a high-density di-sialyl T structure is important for Siglec-7 binding. Our results suggest that the di-sialyl T structure of PDPN binds to Siglec-7. However, the tri-sialyl T structure identified in this study showed stronger binding strength than the di-sialyl T structure. Regarding the involvement of PLAG4 (Table S4),43,48 a peptide containing Hex(1), HexNAc(1), and Neu5Ac(3) was detected in MS of the glycopeptides. However, Siglec-7 ligand glycans were present only on PLAG3 as shown in the point mutation experiment (Figures 5B, 5C, and 5G). Thus, the tri-sialyl T structure was identified as a ligand glycan of Siglec-7 in this study. This glycan structure has been reported to be present on APP in the cerebrospinal fluid of patients with Alzheimer’s disease.49
Some Siglecs have been reported to be utilized during bacterial and viral infections.50,51,52,53 Pseudomonas aeruginosa takes up sialic acid from host body fluids and expresses it on its own membranes, where it interacts with Siglecs on immune cells and destroys the host immunity.53 However, the functions of these Siglecs in normal immune cells remains unclear. Siglecs help the immune system distinguish between self and non-self by recognizing sialic acid-containing glycans as ligands. The expression of sialic acid-containing glycans is restricted by the cell type and serves as a checkpoint for immune cell responses in human diseases such as cancer, asthma, allergies, neurodegeneration, and autoimmune diseases.5 It has been suggested that Siglec-7 may be used by cancer cells to escape from immune surveillance, similarly to immune checkpoint systems such as PD-1/PD-L1, which suppress immune responses. They are thought to maintain immune homeostasis and suppress excessive immune responses in normal tissues.
The activation signals transduced in cancer cells via Siglec-7 stimulation are shown in Figures 3C, 3D, and S7. We showed that Siglec-7 enhanced the phosphorylation levels of AKT and ERKs in cancer cells and promoted cancer invasion via ligand glycan-carrying molecules (Figure 3E). Experiments in which beads conjugated with recombinant Siglec-7-Fc were used also showed the activation of AKT and ERKs upon reaction with C8 (ST8Sia6) (Figures S5B–S5E). It is conceivable that the clustered Siglec-7 on both U937 cells and beads recognizes clustered ligand glycan carrier proteins on cancer cells and forms tight immunological synapses to transmit signals. Siglec-7 signaling on the cancer side promotes the interaction of PODXL with the cytoskeleton, leading to the activation of AKT and ERKs to enhance cell invasion. It has been reported that PDPN and PODXL regulate actin filaments through binding to Ezrin/Radixin/Moesin (ERM) and stimulate cancer cell invasion and metastasis through a variety of strategies.54 Thus, the binding of PDPN and PODXL to their ligands modulates signaling pathways that regulate proliferation, contractility, migration, epithelial-mesenchymal transition (EMT), and remodeling of the extracellular matrix.33,54 MUC13, a transmembrane (TM) mucin, has recently been implicated in cancer development and etiology. It may also be involved in cancers in a similar way.35 As described above, we observed similar signal reinforcing malignancy to the cancer side through ligand glycans by lectin stimulation.
ST3Gal1, ST6GalNAc1, ST6GalNAc3, and ST8Sia6 were identified as the enzymes involved in the synthesis of the tri-sialyl T structure. These genes have been reported to be involved in various malignancies. ST3GAL1 is involved in increased malignancy of ovarian cancer cells, such as cell growth.55 ST6GalNAc1 was highly expressed in colorectal cancer stem and cancer-initiating cells.56 ST8Sia6 expression inhibits the antitumor immune response, promotes tumor growth, and induces cancer-friendly tumor-associated macrophage.57 In that report, ST8Sia6 was reported to enhance the binding of mouse Siglec-E and human Siglec-7 and Siglec-9, and the same result as our findings. The results of our study suggest that the co-expression of three or four sialyltransferase genes contributes to highly enhanced cancer malignancy, and disturbance of the interaction between Siglecs and ligand glycans should lead to the suppression of cancer. Consequently, our study supports an immune checkpoint centered on the ligand glycans of a Siglec-member, leading to an unexplored area of antitumor immunity research. It has been reported that St3gal1,15 St3gal2,16 St6galnac1,16 and st6galnac418,19 sialylate T-antigen. Some of the results are consistent with ours and some are different depending on the tissue and cancer type, suggesting that the expression of carrier molecules may also be involved.
The web tool “GlycoMaple39” visualizes the synthetic pathways of glycan structures from comprehensive gene expression analysis data, such as RNA sequencing. In the paper presenting this study, we visualized the synthetic pathways of glycan structures based on gene expression data from normal colon tissues and colorectal cancers. Importantly, the sialyl transferase genes (ST3Gal1, ST6GalNAc1, ST6GalNAc3, and ST8Sia6) identified in our study synthesize tri-sialyl T structures during the biosynthesis of mucin-type O-glycans. This result suggests that sialyl-T is present in both normal and cancerous cells and may be involved in immunosuppression.
In a previous study, we reported that the interaction between Siglec-7 and GD3 was influenced by the differences in the structure of the lipid moiety of GD3.11 This finding suggests that GD3 clustering is essential for the binding of Siglec-7. The need for multiple tri-sialyl T epitopes to bind O-glycans to Siglec-7 remains unclear. In addition to the known ligand-binding region (containing R124), a new sialic acid-binding region (containing R67) has been reported by other research groups.58 An allosteric mechanism has been suggested for Siglec-7, in which the structure changes depending on the binding glycan and site, thereby affecting ligand glycan recognition at other sites. Whether quantitative and/or qualitative differences exist between gangliosides and O-glycans as ligand glycans, as recognized by Siglec-7, is an intriguing and an urgent issue to be examined. Transduced signals upon these interactions into immune cells and cancer cells should be investigated for individual Siglec-7-ligands, i.e., gangliosides and various O-glycan-carrying molecules. Whether competition or synergistic action occurs between glycosphingolipids and O-glycans in the interaction with Siglec-7 remains to be investigated in more sophisticated experimental systems.
Limitations of the study
This study had several limitations that require further investigation. The mechanism by which Siglec-7 interacts with ligand glycan carrier proteins requires further analysis. Previous studies have suggested that Siglec-7 recognizes multiple clusters of sialic acids.11 Sialic acid-binding sites have also been suggested to be present in domains other than the sialic acid-binding domain (V-set) of Siglec-7. More detailed investigation are needed to elucidate this; however, it is difficult to synthesize a highly purified protein with a Tri-sialyl T structure for NMR and X-ray crystallography. In addition, little information is available on the glycan structures identified in this study, and it is unclear how they behave under physiological conditions. Therefore, it is difficult to predict its behavior mathematically under physiological conditions. Siglec-7 is a human gene. It is said to correspond to Siglec-E in mice; however, there is little similarity in ligand glycan structures. Although there are some examples of human Siglec studies using humanized and transgenic mice in the immune system, they do not provide essential clarification of the issue in this study. However, we believe that using their use is necessary to elucidate glycan immune checkpoints, and we will discuss this issue in the future.
The potential effects of age, gender, and ethnicity were not considered in this study, and their influence remains unknown.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, (koichi@med.nagoya-u.ac.jp).
Material availability
Materials such as plasmids and cells generated in this study are available from the lead contact.
Data and code availability
-
•
The detailed data processing has been deposited on Mendeley Data and is publicly available as of the date of publication. The DOI is listed in the key resources table.
-
•
R-code has been deposited on Mendeley Data and is publicly available as of the date of publication. The DOI is listed in the key resources table.
-
•
Any additional information required to analyze the data reported will be shared by the lead contact upon request.
Acknowledgments
We thank P. R. Crocker at Dundee University for providing an expression vector pEE14-Siglec-7-Fc. We also thank T. Mizuno, Y. Nakayasu, Y. Mass spectrometry was performed at the Division for Medical Research Engineering, Nagoya University Graduate School of Medicine.
This work was supported by Grants-in-aids from the Ministry of Education, Culture, Sports, and Technology of Japan (MEXT, KAKENHI) grant number (26860320 [N.H.], 15K15080 [K.F.], 19K22518 [K.F.], 19K07393 [Y.O.], 20K08690 [N.H.], 21K06828 [K.F.], 23K06414 [N.H.]) and JST-CREST grant number JPMJCR17H2 (K.F.).
Author contributions
Conceptualization, N.H., A.Y., Y.O., T.O., Keiko F., Y.K., N.K., and Koichi F.; supervision, A.Y. and Y.O., Keiko F.., N.K., Koichi F.; investigation, N.H., S.I., A.H., A.T., and Y.M.; formal analysis, N.H., A.H., and Y.M.; software, N.H. and Y.M.; methodology, N.H., S.I., and A.H.; validation, A.H., A.T., Y.M., Y.K., and N.K., Koichi F.; data curation, A.H., A.T., Y.M., Y.K., and N.K., Koichi F.; writing-original draft preparation, N.H., Koichi F.; writing-review and editing, N.H., A.H., A.T., Y.M., Y.K., and N.K., Koichi F.; All authors have read and reviewed manuscript.
Declaration of interests
The authors declare no competing interests.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat anti-PDPN (NZ-1) | AngioBio | Cat# 11-009 |
| Cocktail Anti Hu CD3 FITC/(CD16 + 56) PE Cktl | BioLegend | Cat# 319101, RRID:AB_314999 |
| Anti-human CD3 PE/Cy7 | Thermo Fisher Scientific | Cat# 25-0037-42, RRID:AB_2573326 |
| Anti-human CD56 PE | Thermo Fisher Scientific | Cat# 12-0567-42, RRID:AB_10598200 |
| Human TruStain FcXTM (Fc Receptor Blocking Solution) | BioLegend | Cat# 422302, RRID:AB_2818986 |
| FITC Streptavidin | BioLegend | Cat#405202 |
| Anti-p-Tyr (PY20) HRP | BD Biosciences | Cat# 610012, RRID:AB_397433 |
| Hrizon V450 streptavidin | BD Biosciences | Cat#560797 |
| Rabbit Anti-AKT | Cell Signaling Technology | Cat# 9272, RRID:AB_329827 |
| Rabbit Anti-p-AKT | Cell Signaling Technology | Cat# 9271, RRID:AB_329825 |
| Rabbit Anti-p44/42 MAPK (ERK1/2) | Cell Signaling Technology | Cat# 4695, RRID:AB_390779 |
| Rabbit Anti-phospho ERK1/2 | Cell Signaling Technology | Cat# 4370, RRID:AB_2315112 |
| Goat Anti-rabbit IgG, HRP | Cell Signaling Technology | Cat# 7074, RRID:AB_2099233 |
| Anti-rabbit IgG Fab2 Alexa 555 | Cell Signaling Technology | Cat# 4413, RRID:AB_10694110 |
| Anti-mouse IgG Fab2 Alexa 647 | Cell Signaling Technology | Cat# 4410, RRID:AB_1904023 |
| Anti-Rat IgG-HRP | Proteintech | Cat# SA00001-15, RRID:AB_2864369 |
| Goat TrueBlot anti-goat Ig HRP | Rockland | Cat# 18-8814-31, RRID:AB_2610843 |
| Anti-human p75/AIRMI(Siglec-7)-biotin | Thermo Fisher Scientific | Cat# 13-5759-80, RRID:AB_763630 |
| Anti-mouse IgG HRP | Cytiva | Cat# NA931, RRID:AB_772210 |
| Rabbit Anti-MUC13 | IMGENEX | Cat# IMG-6250A, RRID:AB_1930301 |
| Anti-mouse IgG Alexa 488 | Thermo Fisher Scientific | Cat# A-21206, RRID:AB_2535792 |
| Phalloidin Alexa 647 | Invitrogen | Cat# A22287 |
| Anti-p-Tyr (4G10) HRP | Millipore | Cat# 16-105, RRID:AB_310779 |
| Rabbit Anti-SHP-1 | Millipore | Millipore Cat# 07-419, RRID:AB_310601 |
| Goat polyclonal Anti-Siglec-7/CD328 | R and D Systems | Cat# AF1138, RRID:AB_2189416 |
| Mouse Anti-PODXL (3D3) | Santa Cruz Biotechnology | Cat# sc-23904, RRID:AB_2166006 |
| 4′,6-diamidino-2-phenylindole (DAPI) | Sigma-Aldrich | Cat#D9542 |
| Mouse monoclonal Anti-β-Actin | Sigma-Aldrich | Cat# A1978, RRID:AB_476692 |
| Anti-human IgG (Fc specific) FITC | Sigma-Aldrich | Cat# F9512, RRID:AB_259808 |
| PE anti-human IgG Fc | BioLegend | Cat# 410708, RRID:AB_2565786 |
| Anti-mouse IgG FITC | Sigma-Aldrich | Cat# F0257, RRID:AB_259378 |
| ABC standard kit Elite | Vector | Cat#PK-6100 |
| Anti-GD3 monoclonal antibody | kindly provided by Dr. L. J. Old (Memorial Sloan Kettering Cancer Center) | N/A |
| Purified anti human CD328 (Siglec-7) (Blocking Antibody) | BioLegend | Cat# 347702, RRID:AB_2189411 |
| Mouse IgG1 Isotype Control | R and D Systems | Cat# MAB002, RRID:AB_357344 |
| Peanut Agglutinin (PNA), biotinylated | Sigma | Cat# L6135 |
| 7-AAD Viability Staining Solution | BioLegend | Cat# 420404 |
| APC Annexin V | BioLegend | Cat# 640920 |
| Biological samples | ||
| Human PBMC (Lot No. 22TL227189) | Lonza | Cat# CC-2704 |
| Chemicals, peptides, and recombinant proteins | ||
| ECL select | Amersham | Cat# RPN2235 |
| ITS Premix Universal Culture Supplement | Corning | Cat# 354351 |
| ITS Liquid Media Supplement (100x) | Sigma | Cat# I3146 |
| Matrigel | Corning | Cat# 356234 |
| transwell filter (8-μm pore) | Corning | Cat# 3422 |
| Ficoll-Paque PLUSTM | Cytiva | Cat# 17-1440-02 |
| EDTA3Na | DOJINDO | Cat# 342-01875 |
| Sodium pyruvate | Gibco | Cat# 11360070 |
| Opti-MEM | Gibco | Cat# 31985070 |
| GlutaMAX spplement | Gibco | Cat# 35-050-061 |
| Penicillin-Streptomycin | Gibco | Cat# 15140122 |
| nProtein A Sepharose 4 Fast Flow 5mL | GE | Cat# 17528001 |
| Lipofectamin 2000TM | Invitrogen | Cat# 11668019 |
| Lipofectamin 3000TM | Thermo Fisher | Cat# L3000001 |
| Acetonitrile LC/MS grade | Kanto Chemical | Cat# 01033-23 |
| Trifluoroacetic acid HPLC grade | Kanto Chemical | Cat# 40578-1B |
| Acetic acid | Kanto Chemical | Cat# 01021-96 |
| ammonium formate | Kanto Chemical | Cat# 01033-76 |
| 3-methyl-1-phenyl-5-pyrazolone | Kanto Chemical | Cat# 32148-30 |
| polyvinyldene fluoride (PVDF) membrane | Millipore | Cat# IPVH00010 |
| N-Acetylneuraminic Acid (Neu5Ac) | Nacarai | Cat# 08371-36 |
| Hind III | New England Biolabs Japan | Cat# R0104 |
| Bam HI | New England Biolabs Japan | Cat# R0136 |
| Trypsin | Promega | Cat# V5280 |
| IL-2 | PeproTech | Cat# 200-02 |
| Phenylmethylsulfonyl fluoride | Roche | Cat# 10837091001 |
| ammonium bicarbonate | Sigma | Cat# A6141 |
| GalNAc | Tokyo Chemical Industry | Cat# G0007 |
| Tris | WAKO | Cat# 103133 |
| G418 | WAKO | Cat# 076-05962 |
| BSA | WAKO | Cat# 011-27055 |
| Tunicamycin | WAKO | Cat# 202-08241 |
| Kifunensine | Sigma | Cat# K1140 |
| Benzyl-α-GalNAc | MCH | Cat# HY-129389 |
| Nonidet P-40 | WAKO | Cat# 21-3277 |
| sodium deoxycholate | WAKO | Cat# 190-08313 |
| sodium lauroylsarcosine | WAKO | Cat# 121-05502 |
| dithiothreitol | WAKO | Cat# 048-29224 |
| iodoacetamide | WAKO | Cat# 091-02153 |
| Triethylamine | WAKO | Cat# 202-02641 |
| Lys-C | WAKO | Cat# 125-05061 |
| D-Galactoce | WAKO | Cat# 071-00032 |
| Methanol | WAKO | Cat# 131-01826 |
| Neuraminidase (Vibrio cholerae) | Roche | Cat# 11080725001 |
| Neuraminidase (Arthrobacter ureafaciens) | Nacarai | Cat# 24229-74 |
| a2-3 Neuraminidase S | Prozyme | Cat# GK80020 |
| a2-3 Neuraminidase S | NEB | Cat# P0743S |
| Formic acid | WAKO | Cat# 063-04533 |
| Critical commercial assays | ||
| QuikChange II XLTM site-directed mutagenesis kit | Agilent | Cat# 200521 |
| Cell proliferation kit | Cytiva | Cat# RPN20 |
| CytoTox 96 Nonradioactive Cytotoxicity AssayTM kit | Promega | Cat# G1780 |
| EZ-Link Sulfo-NHS-LC-Biotin | Thermo | Cat# 21335 |
| Deposited data | ||
| Raw and analyzed data | This paper; Mendeley Data | https://doi.org/10.17632/r95br5ndzg.1 |
| Experimental models: cell lines | ||
| DLD-1 | ATCC | Cat# CCL-221, RRID:CVCL_0248 STR profile: https://doi.org/10.17632/r95br5ndzg.1 |
| Caco-2 | ATCC | Cat# HTB-37, RRID:CVCL_0025 |
| WiDr | ATCC | Cat# CCL-218, RRID:CVCL_2760 |
| HCT116 | RIKEN BRC | Cat# RCB2979, RRID:CVCL_0291 |
| SW837 | JCBR Cell Bank | JCBR9115 |
| MCF7 | ATCC | HTB-22, RRID:CVCL_0031 |
| K-562 | ATCC | CCL-243, RRID:CVCL_0004 |
| LN319 | Kindly provided by Dr. Kato (Tohoku Univ)42 | N/A |
| HEK293T | ATCC | Cat: RCRL-3216 RRID:CVCL_0063 |
| U937-mock | Kindly provided by Dr. Yamaji (Nat. Inst. Infections Diseases)39 | N/A |
| KHYG-1 | Kindly provided by Dr. Kimura (Nagoya Univ. Virology), JCRB Cell Bank59 | N/A |
| Oligonucleotides | ||
| PDPN extracellular cloning primer Forward 5′-ggaagcttcctccaaccccagattaaatgctgactccgctcgg-3′ |
See Construction of expression vector in this study | N/A |
| PDPN extracellular cloning primer Reverse 5′-ggggatccacttacctgtaactgttgtctgtgtgtctc-3′ |
See Construction of expression vector in this study | N/A |
| PDPN signal sequence cloning primer Forward 5′-ggaagcttcctccaaccccagattaaatgctgactccgctcgg-3′ |
See Construction of expression vector in this study | N/A |
| PDPN signal sequence cloning primer Reverse 5′-ggggatccacttacctgttccttctgccaggacccaga-3′ |
See Construction of expression vector in this study | N/A |
| hSt3gal1 qPCR primer forward, 5′- GGCACCATTTCCCACACCTA -3′ |
See Construction of expression vector in this study | N/A |
| hSt3gal1 qPCR primer reverse 5′- ACAAGTCCACCTCATCGCAG -3′ |
See Construction of expression vector in this study | N/A |
| hSt6galnac1 qPCR primer forward 5′- CAAAGCCTTCCAGGCATCAAC -3′ |
See Construction of expression vector in this study | N/A |
| hSt6galnac1 qPCR primer reverse 5′- GGCACTGGCTCTGCATAGAT -3′ |
See Construction of expression vector in this study | N/A |
| hSt6galnac3 qPCR primer forward 5′- GGCCTGCATCCTGAAGAGAAA -3′ |
See Construction of expression vector in this study | N/A |
| hSt6galnac3 qPCR primer reverse 5′- ACCAGGTTGTCCAAAGCAGT -3′ |
See Construction of expression vector in this study | N/A |
| hSt8sia6 qPCR primer forward 5′- TCTGAATGAGAAGTCGCTCCA-3′ |
See Construction of expression vector in this study | N/A |
| hSt8sia6 qPCR primer reverse 5′- GCATCACAGCAGGAAGCAAG -3′ |
See Construction of expression vector in this study | N/A |
| mSt3gal1 qPCR primer forward 5′- ATTGGAGGGAGGGGATGGGA -3′ |
See Construction of expression vector in this study | N/A |
| mSt3gal1 qPCR primer reverse 5′- ACGGTATCTTGCCCCTCTGC -3′ |
See Construction of expression vector in this study | N/A |
| mSt6galnac1 qPCR primer forward 5′- GGACCAGCCATCCACCATGA -3′ |
See Construction of expression vector in this study | N/A |
| mSt6galnac1 qPCR primer reverse 5′- CCTGGGCACTTGCGTCATTC -3′ |
See Construction of expression vector in this study | N/A |
| mSt6galnac3 qPCR primer forward 5′- ACCTCAGCACTGGCTGGTTT -3′ |
See Construction of expression vector in this study | N/A |
| mSt6galnac3 qPCR primer reverse 5′- CCCCATACGGGGCATGTTCA -3′ |
See Construction of expression vector in this study | N/A |
| mSt8sia6 qPCR primer forward 5′- ACTGGCTTCCTGTTGCGATG -3′ |
See Construction of expression vector in this study | N/A |
| mSt8sia6 qPCR primer reverse 5′- GTCCACAAAAGGCTGCGACA -3′ |
See Construction of expression vector in this study | N/A |
| Recombinant DNA | ||
| ST expression vectors | See Table S2 in this study. | N/A |
| pEE14-Siglec-7-Fc | Zang et al.21 | kindly provided by Dr. Crocker at the University of Dundee |
| pEE14-Siglec-9-Fc | Zang et al.21 | kindly provided by Dr. Crocker at the University of Dundee |
| pcDNA3.1 Siglec-7 | kindly provided by Dr. Yamaji (Nat. Inst. Infections Diseases)39 | N/A |
| PDPN (NM_001006624) Human Untagged Clone | OriGENE | Cat#SC301085 |
| pcDNA3.1 PDPN | This paper | N/A |
| pcDNA3.1 PDPN T52A | This paper | N/A |
| pEE14-PDPN-Fc | This paper | N/A |
| pEE14-PDPN T52A-Fc | This paper | N/A |
| pEE14-Fc | This paper | N/A |
| Software and algorithms | ||
| Analyst TF and LCMS Peptide Reconstruct software tool | Sciex | https://sciex.jp/products/software/analyst-tf-software |
| ImageJ | NIH | https://ImageJ.nih.gov/ij/ |
| R (4.2.0) | Ihaka, R., and R. Gentleman. 1996. | http://www.R-project.org. |
| Adobe Photoshop | Adobe Inc., 2023. Adobe Photoshop | https://www.adobe.com/products/photoshop.html |
| GraphPad Prism version 10.2.3 | GraphPad Soft Ware, LLC | https://www.graphpad.com/ |
| Excel for mac | Micro soft | https://www.microsoft.com/ja-jp/microsoft-365/mac/microsoft-365-for-mac |
| FLUOVIEW Viewer | Olympus | https://www.olympus-lifescience.com |
Experimental model and study participant details
Cells
Human colon adenocarcinoma cell lines DLD-1 [male], Caco-2 [male], WiDr [female], a human embryonic kidney cell line HEK 293T [female], MCF-7 [female] were obtained from ATCC. LN319 [male] was kindly provided by Dr. Kato (Tohoku Univ). A human colon adenocarcinoma cell line HCT116 [Male] was obtained from RIKEN BRC. They were cultured in DMEM (Nissui) including 7.5% FBS (Sigma-Aldrich) and Penicillin-Streptomycin (Gibco) in 5% CO2 at 37°C. A human colon adenocarcinoma cell line SW837 [male] was obtained from JCBR Cell bank. It was cultured in Lelbovitz’s L-15 medium (Gibco) including 7.5% FBS (Sigma-Aldrich) and Penicillin-Streptomycin (Gibco) in no CO2 applied at 37°C. K562 [female] were obtained from ATCC. U937 [male] (mock) and U937-Siglec-7high was kindly provided by Dr. Yamaji (Nat. Inst. Infections Diseases). KHYG-1 [female] was kindly provided by Dr. Kimura (Nagoya Univ. Virology). Human PBMCs of healthy donor origin were used as donor 1 [Male] and human PBMCs obtained from Lonza (CC-2704, Lot No. 22TL227189) were used as donor 2 [Male]. They were cultured in RPMI1640 (Sigma) including 10% FBS in 5% CO2 at 37°C. KHYG-1 was cultured with IL-2 (20U/mL, PeproTech) and GlutaMAX supplement (Gibco). No obvious differences by biological sex were detected, but the number of cell lines was apparently insufficient to conclude this unequivocally. DLD-1 was transiently transfected with expression vectors of 20 sialyltransferase cDNAs (see Table S1) by Lipofectamin 2000 (Invitrogen) and Opti-MEM (Gibco). Referring to the results of the transient transfection of single cDNA out of 20 STs (see Figure S2), ST3Gal1 and ST6GalNAc1, ST3Gal1 and ST6GalNAc3, or ST8Sia6 were stably transfected and selected by G418 (400 μg/mL, WAKO). Then, they were cloned by limiting dilution and checked for binding of Siglec-7-Fc. Human cells were used in accordance with the Ethics Committee and Clinical Research Review Bord of Nagoya University Graduate School of Medicine (1004, 1027 and 2012-0240-2) following the Declaration of Helsinki principles. DLD-1 are authenticated using Short Tandem Repeat (STR) profile by BEX Co., Ltd. (Tokyo, Japan). The results are available on mendeley data (see key resources table). Mycoplasma contamination test was performed by nested-PCR method. No issues were detected in either assessment.
Antibodies
The antibodies were Anti-β-Actin antibody (1:5000, Sigma), Anti-AKT (1:1000, Cell Signaling Technology), Anti-p-AKT (S473) (1:1000, Cell Signaling Technology), Anti-phospho ERK1/2 (1:1000, Cell Signaling Technology), Anti-p44/42 MAPK (ERK1/2) (1:1000, Cell Signaling Technology), Anti-rabbit IgG-HRP (1:1000, Cell Signaling Technology), Goat TrueBlot anti-goat Ig HRP (1:2000, ebioscience), Anti-Mouse IgG HRP-Linked Whole Ab (1:2000, GE HealthCare), Anti-MUC13 (1:1000, IMGENEX), Anti-p-Tyr (4G10) HEP conjugate (1:5000, Millipore), Anti-SHP-1 (1:1000, Millipore), Anti-Siglec-7/CD328 antibody (1:1000, R&D), Anti-PODXL (3D3) (1:1000, Santa Cruz), Anti-p-Tyr (PY20) HRP conjugate (1:2000, BD), Rat anti-PDPN (NZ-1) (1:1000, AngioBio Co), Anti-Rat IgG-HRP (1:2000, Cosmo bio), ABC standard kit Elite (5 μL/mL each, Vector) for immunoblotting, Anti-rabbit IgG Fab2 Alexa 555 (1:400, Cell Signaling Technology), Anti-mouse IgG Fab2 Alexa 647 (1:400, Cell Signaling Technology), Anti-mouse IgG Alexa 488 (1:400, Invitrogen), Phalloidin Alexa 647 (1:400, Invitrogen), DAPI (1:500, Sigma) for immunocyte chemistry. Cocktail Anti Hu CD3 FITC/(CD16 + 56) PE Cktl (20 μL/sample, BD), Hrizon V450 streptavidin (1:100, BD), Anti-human p75/AIRMI(Siglec-7)-biotin (1:100, ebioscience), Anti-human IgG (Fc specific) FITC (1:200, Sigma), Anti-mouse IgG FITC (1:200, Sigma) for flow cytometry.
Method details
Preparation of recombinant human Fc fusion proteins
The expression vector pEE14-Siglec-7/9-Fc was kindly provided by Dr. Crocker at the University of Dundee. Recombinant proteins Siglec-7/9-Fc and control Fc were expressed in serum-free medium containing ITS (Corning) by transfection of pEE14 expression vector with lipofectamine 2000 (Thermo Fisher) into HEK293T seeded to 1 × 106 cells in 10 cm dishes. The conditioned medium was affinity purified with Protein A Sepharose beads (GE) to obtain Siglec-7/9-Fc and control Fc.
Flow cytometry
Cell-surface expression of Siglec-7-ligand glycans and other surface glycans was analyzed by FCM, FACS Calibur (BD Bioscience) and CytoFlex (BECKMAN COULTER). Cells were detached by 0.01% trypsin and 0.53 mM EDTA/PBS. After counting, 5 × 105 cells were stained by Siglec-7/9-Fc, anti-GD3 (R24), and PNA lectin (none to control). FITC-rat anti-human Fc, FITC-goat anti-mouse IgG and FITC streptavidin was used for detection of binding. For FCM analysis of human PBMCs from healthy donor 1 and 2, PE-Cy7 conjugated anti-CD3, PE conjugated anti-CD56, biotinylated anti-Siglec-7 antibodies and FITC-streptavidin are used in 5 × 105 PBMCs. FCM analysis of desialylation using various sialidases and its effect on Siglec-7-binding. DMEM or RPMI 1640 containing 0.25% BSA and 2 mM CaCl2 was used as reaction medium for sialidase. DLD-1 and the transfectants, SW847 and K562 were cultured normally. DLD-1 and the transfectants and SW847 were detached by 0.01% trypsin and 0.53 mM EDTA/PBS. They were suspended in 1 × 105 cells/200 μL reaction medium. To this, 10 mU vibrio cholerae derived sialidase and 10 mU arthrobacter ureafaciens derived sialidase or 10 mU sialidase S (none to control) were added and allowed to react for 1.5 h at 37°C. The cells were then reacted with Siglec-7-Fc and the amount of binding was assessed using FACS. Apoptosis of NK cell was assessed in co-cultures of human NK cells and cancer cells using FACS. DLD-1 and the transfectants were seeded onto plates (1 × 105 cells/6-well plate) and culture for 24 h. Human PBMCs (from donor 1) were added at PBMCs: cancer = 10:1 and co-cultured for 6 and 24 h (control is none co-culture). PBMCs were collected by washing away and treated with Fc receptor blocking solution, followed by staining with CD3-FITC and CD16/CD56-PE. They were then suspended in annexin binding buffer of 10 mM HEPES pH 7.4, 140 mM NaCl and 2.5 mM CaCl2 and stained with Annexin V-APC and 7-AAD. Apoptosis of CD16+CD56+ NK cells was then assessed using FACS.
Real-time qPCR analysis
For gene expression analysis, total RNA was extracted using Isogen II from DLD-1 and transfectants, Caco-2, WiDr, HCT116, SW837, MCF7 and K562, seeded the previous day to 1 × 106 cells/6 cm dish. Total RNA was DNase treated, reacted with randam primer and treated with RNase OUT, then cDNA was synthesized using reverse transcriptase M-MLV. Real-time qPCR was performed using the Thunderbird qPCR mix and a standard curve with standard samples was used to quantify gene expression in each sample. To compare the expression levels of mouse-derived sialyltransferases with the corresponding human sialyltransferases, the following experiments were performed. Mouse and human primers were designed to have close PCR efficiency. After qPCR using the corresponding primer sets, the relative quantification ratio between DLD-1 transferant (DLD-1 ST) and various cancer cell lines was determined using the delta-delta Ct method with human gapdh as the housekeeping gene. A list of primers used can be found in the key resources table.
Inhibition of synthesis of N-glycan and O-glycan
For inhibition of N-glycan synthesis, 2 × 104 cells were seeded into 24-well plate with 20 nM kifunensine and culture for 1 day (vehicle control is DMSO). For inhibition of O-glycan synthesis, 1 × 104 cells were seeded into 24-well plate with 2 mM benzyl-α-GalNAc and cultured for 2 days (vehicle control is DMSO).
Siglec-7-Fc pull-down experiments
To biotinylate cell-surface proteins, Sulfo-NHS-Biotin (final conc. 2 mM, Thermo Fisher) was added to suspended DLD-1 and the transfectants (2 × 106 cells/sample) and incubated at room temperature for 30 min. Then, cells were washed with 100 mM glycine-containing PBS three times and lysed with a lysis buffer (1% Nonidet P-40 (Sigma), 50 mM Tris (WAKO)-HCl pH 7.5, 150 mM NaCl (WAKO) and 1 mM Phenylmethylsulfonyl fluoride (PMSF) (Roche)). The cell lysates were centrifuged at 20,000 × g for 30 min at 4°C and supernatants were collected. Ten μg Siglec-7-Fc or control Fc recombinant proteins were added to lysates and incubated overnight at 4°C with rotation. Siglec-7-Fc and ligand complexes were pulled-down with Protein A Sepharose 4 Fast Flow beads (GE Healthcare) from the lysates. These products were separated by SDS-PAGE and transferred onto polyvinyldene fluoride (PVDF) membranes (Millipore). These membranes were blocked with 3% BSA (WAKO)/PBS, and incubated with ABC reagent or individual antibodies according to the manufacturer’s protocol. HRP-labeled secondary antibody or avidin-HRP and ECL select were used for chemiluminescence, and then exposed to X-ray film. The band intensity was quantitatively analyzed using ImageJ software (NIH).
Identification of siglec-7-binding proteins by LC/MS
The pulled-down proteins by Siglec-7-Fc were dissolved in an MS sample buffer (12 mM sodium deoxycholate (190–08313, WAKO), 12 mM sodium lauroylsarcosine (WAKO), and 100 mM Tris-HCl, pH. 8.0), boiled at 95°C for 5 min, and centrifuged at 20,000 × g for 10 min. The supernatants underwent reduction with dithiothreitol (WAKO) and alkylation with iodoacetamide (WAKO). Then, the samples were 5-fold diluted with 50 mM ammonium bicarbonate, and digested by Lys-C (WAKO) for 3 h and then by trypsin (Promega) for 8 h at 37°C (protease-to-protein ratio of 1:25 (w/w)). They were desalted and concentrated with C18 StageTips. MS was performed using an Orbitrap fusion mass spectrometer (Thermo Fisher) system combined with a Dionex U3000 HPLC System (Thermo Fisher), as previously described.60 MS/MS data were submitted to the program Mascot 2.3 (Matrix Science) for the MS/MS ion search. Mascot was set up to search the Sprot_2011_12 database (selected for Homo sapiens, 20,249 entries) assuming the digestion enzyme as trypsin. Mascot was searched with a product ion mass tolerance of 0.80 Da, and a precursor ion tolerance of 10.0 PPM. The identified proteins were checked by Western blotting with specific antibodies.
Cytotoxicity assay with PBMCs
PBMCs were isolated by density gradient centrifugation using Ficoll-Paque PLUS (Cytiva) from healthy donors' peripheral blood. The cytotoxicity of NK cells (PBMCs) against Siglec-7-Fc-reactive or nonreactive DLD-1 cells was assessed using the CytoTox 96 Nonradioactive Cytotoxicity Assay kit (Promega) according to the manufacturer’s instruction. E/T indicates the ratio of effector (PBMCs) to target (DLD-1 and transfectants of STs). DLD-1 and the ST transfectants were plated in U-shaped 96-well plates (2,000 cells/well) and cultured for 24 h. Human PBMCs (2 × 105/well, Donor 1 and 2) were prepared, and anti-Siglec-7 blocking antibody (Biolegend) and control IgG (mouse IgG1k, R&D) at 10 μg/mL were added. The PBMCs were incubated in a CO2 incubator for 10 min before being applied to the plate. Each plate was then centrifuged at 250xg for 1 min. After 4–5 h, the supernatant was collected and analyzed using CytoTox96 (Promega). % of cytotoxicity was calculated. % Cytotoxicity = (Experimental − Effector Spontaneous − Target Spontaneous)/(Target Maximun − Target Spontaneous) ∗100.
Confocal microscopy
DLD-1 and C8 (1 × 105 cells/dish) co-cultured with U937-Siglec-7high (1 × 106 cells/dish) on 35-mm glass-bottom dishes (IWAKI) and washed with PBS to remove unattached cells were fixed in paraformaldehyde (4% in PBS) and were stained with biotinylated anti-Siglec-7 antibody and mouse anti-PODXL antibody in PBS containing 0.5% BSA for 60 min at room temperature. The cells were then stained with FITC-avidin, Alexa 567-conjugated goat anti-mouse IgG antibody, Alexa 647-conjugated phalloidin, and DAPI. The stained patterns were analyzed using a confocal microscope (Fluoview FV10i, Olympus). For live imaging analysis, red fluorescent protein was forced to be expressed to identify DLD-1 and C8 seeded in 3.5 cm dishes (1 × 105 cells/dish). The cells were then co-cultured with KHYG-1-Siglec-7high in medium containing DAPI (final 50 ng/mL) for 11 h and time-lapse analysis was performed using FV10i.
Invasion and proliferation assay
For the invasion assay, cells (0.5 × 106/well) were suspended in DMEM containing 7.5% FBS and seeded in the upper chamber of Matrigel Matrix (BD) -coated transwell filters (8-μm pore) (BD). DMEM containing 7.5% FBS was added to the lower chamber (6-well plate) and incubated at 37°C for 24 h. Non-invading cells remaining on the upper surface of the filter were removed, and the cells that appeared on the lower surface of the filter were fixed with 75% ethanol for 30 min and then stained with 0.025% Giemza and counted under a microscope.59 For the proliferation assay, cells grown on a 60-well plate were incubated in the presence of BrdU for 14 h according to the instructions of the cell proliferation kit (Amersham) and then fixed with acid-ethanol for 30 min. The cells were immunostained with anti-BrdU antibody and Alexa 546-conjugated anti-mouse antibody (Molecular Probes, Invitrogen). The BrdU-positive cells were observed by fluorescence microscopy (BX51, Olympus, Tokyo), and the percentage of BrdU-positive cells was calculated.61
Construction of expression vectors
Siglec-7-Fc is a fusion protein created through the joining of the extracellular domain of Siglec-7 and human IgG1 Fc region. PDPN cDNA (NM_001006624) Human Untagged Clone (ORIGNENE) was purchased and inserted into the pcDNA3.1 expression vector. The extracellular domain of PDPN was cloned using primers: forward: 5′-ggaagcttcctccaaccccagattaaatgctgactccgctcgg-3′ and reverse: 5′-ggggatccacttacctgtaactgttgtctgtgtgtctc-3′, and polymerase KOD-FX (Toyobo) and Taq polymerase (Promega). The products were inserted into the pCR2.1TM vector using the TA-cloning kit (Thermo Fisher). The plasmid vector was digested using Hind III (NEB) and Bam HI (NEB), and the PDPN cDNA sequence was inserted into pEE14-Fc vector, forming pEE14-PDPN-Fc. The PDPN signal sequence was cloned using primers: forward: 5′-ggaagcttcctccaaccccagattaaatgctgactccgctcgg-3′ and reverse: 5′-ggggatccacttacctgttccttctgccaggacccaga-3′, and KOD-FX. The product was digested using Hind III and Bam HI, and transferred into the pEE14-Fc vector. Their recombinant Fc-fusion proteins were expressed in HEK293T cells in DMEM containing ITS Premix Universal Culture Supplement (Corning) without FBS and purified using the protein A-Sepharose beads column.
Mutagenesis of disialyl O-glycosylation site 52T of PDPN and PDPN-Fc
The disialyl-core 1 (NeuAcα2-3Galβ1-3(NeuAcα2-6)GalNAcα1-O-Thr) glycan-substituted structure was modified at Thr52 of human PDPN (hPDPN)27. Thr52 was converted to alanine using the QuikChange II XL site-directed mutagenesis kit (Agilent), using sense primer 5′-gaagatgatgtggtggctccaggaaccagcg-3′, and antisense primer 5′-cgctggttcctggagccaccacatcatcttc-3’.
Structure analysis of released O-glycans on PDPN
PDPN-Fc was generated by co-transfection of pEE14-PDPN-Fc with either one of 3 conditions for sialyltransferase cDNAs to reinforce the binding of Siglec-7-Fc (1: PDPN-Fc only, 2: combined with ST3Gal1 and ST6GalNAc3, 3: combined with ST3Gal1, ST6GalNAc3, and ST8Sia6) in HEK293T. The culture medium for expression of these genes consisted of DMEM and 5 mM D-Galactoce (WAKO), 2.5 mM GalNAc (TCI), 5 mM Neu5Ac (Nakarai), 1 mM sodium pyruvate (Gibco), and ITS.
Releasing and PMP-labeling of O-glycans
For the analysis of O-glycans, the O-glycans were released from PDPN-Fcs by alkaline β-elimination with triethylamine (WAKO) and simultaneously derivatized with 3-methyl-1-phenyl-5-pyrazolone (PMP, Kanto Chemical) according to the published procedure wuth modifications.44 To 10 μL sample, was added 10 μL of water and 30 μL of 0.5 M PMP/methanol (WAKO) + triethylamine solution, and this was stirred well. The reaction was carried out at 60°C for 16 h. After cooling, 100 μL of water and 200 μL of chloroform were added, and the mixture was vigorously stirred by vortex. It was centrifuged at 10,000 rpm for 1 min. The aqueous phase was taken into another Eppendorf tube. The pH of the aqueous phase was made weakly acidic with 10% acetic acid. Chloroform (200 μL) was added again, and the mixture was vigorously stirred with a vortex, centrifuged at 10,000 rpm for 1 min, and the aqueous phase was collected. This operation was repeated once again. Six μL was used for analysis. MS was performed using a Triple TOF 6600 mass spectrometer system combined with a Ekspert nanoLC415 HPLC System (Sciex), as previously described44 (eksigent NanoLC column, 3C18-CL, 75 μm × 15 cm. A: 10 mM ammonium formate (WAKO) pH 6.0; B: 10 mM ammonium formate pH 6.0/90% acetonitrile (Kanto Chemical)). In the MS/MS spectrum, peaks with m/z 552 (HexNAc+2PMP) and m/z 292 (sialic acid) were all visually confirmed. The composition of the peak considered to be a glycan was estimated based on the mass information. The intensity of the peak was determined by the LCMS Peptide Reconstruct software tool (Sciex). MS analysis data were deposited in Mendeley Data. See key resources table for details.
O-glycosylated peptide analysis
For identification of the O-glycosylation site on PDPN, 0.1 mg PDPN-Fc and sialylated-PDPN-Fc were treated by oxidation and IAA, and digested by Asp-N. MS was performed using a Triple TOF 6600 mass spectrometer system combined with a Ekspert nanoLC415 HPLC System. The digests were purified and separated with the eksigent NanoLC column 3C18-CL 75 μm × 15 cm. Gradient conditions for the glycol-peptides were 300 nL/min; 0–25 min, 2–38% B, 25–26 min 38-90% B, 26–30 min 90% B (A: 0.1% formic acid (WAKO); B: 0.1% formic acid acetonitrile). Scan range of MS were m/z 400–2100 and m/z 700–2100. An scan range of MS/MS was m/z 100–2100. The top 10 peaks of MS intensity were applied for MS/MS analysis. Glycopeptide ions were selected by the presence of oxonium ions in the MS/MS spectrum, and the mass of the peptide moiety was deduced from the pattern of fragment ions. The composition of glycopeptides was estimated from the mass of the glycan portion.
TCGA data analysis
The analysis of four sialyltransferases in human colon cancers was performed using statistical software R (4.2.0). TCGA data (normalized expression levels of each gene and expression information in tumor and tumor free areas) were analyzed using the RTCGA packages (RTCGA.rnaseq and RTCGA.clinical) in R, and RNAseq data for COAD patients were obtained using COAD.rnaseq in the package. The code list of the analysis procedure of R is posted in the supplement. The Project ID used is TCGA-COAD. Wilcoxon was performed to compare expressions in the primary solid tumors and normal solid tissues. On analysis of the survival curve, cut-point analysis was performed for the expression level of each gene, and the group was divided into high and low groups. The cut-off points at which high and low groups were separated were as follows: ST3Gal1 was 650.0977, ST6GalNAc1 was 4325.628, ST6GalNAc3 was 58.5373, and ST8Sia6 was 2.8265. The R code is available on mendeley data (see key resources table, RAW and analyzed data, Figure 4/R code).
Quantification and statistical analysis
Statistics
Shapiro-Welk normality test was performed to confirm the normality of the data. The Bartlet test was then used to test for equal variances among the groups. After confirming normality and equal variances, we proceeded to the next test. For the comparison of the two groups, the student’s T test was performed. For comparisons between multiple groups, an analysis of variance was performed. Tukey’s multiple comparison test was performed as a post test to test for differences between groups. One-way ANOVA was performed to compare means of the percent cytotoxicity of human PBMCs toward Siglec-7-Fc negative parent DLD-1 and three transfectants. The above analyses were performed with the statistics software R (4.2.0), GraphPad Prism (10.2.3, GraphPad Software, MA, USA) and Excel for mac (16.66.1, Microsoft, WA, USA). The number of n and statistical tests carried out, as well as the p-values for these tests, are given in the legend of each figure. The actual values are shown in the Raw data of the repository Mendeley data (see key resources table, RAW and analyzed data). ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05, N.S. = not significant. All results of in vitro experiments were collected from at least 3 independent biological replicates.
Published: October 11, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.111139.
Supplemental information
References
- 1.Crocker P.R., Paulson J.C., Varki A. Siglecs and their roles in the immune system. Nat. Rev. Immunol. 2007;7:255–266. doi: 10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- 2.Tsubata T. Siglec cis-ligands and their roles in the immune system. Glycobiology. 2023;33:532–544. doi: 10.1093/glycob/cwad038. [DOI] [PubMed] [Google Scholar]
- 3.Fernandes R.A., Su L., Nishiga Y., Ren J., Bhuiyan A.M., Cheng N., Kuo C.J., Picton L.K., Ohtsuki S., Majzner R.G., et al. Immune receptor inhibition through enforced phosphatase recruitment. Nature. 2020;586:779–784. doi: 10.1038/s41586-020-2851-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Macauley M.S., Crocker P.R., Paulson J.C. Siglec-mediated regulation of immune cell function in disease. Nat. Rev. Immunol. 2014;14:653–666. doi: 10.1038/nri3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duan S., Paulson J.C. Siglecs as Immune Cell Checkpoints in Disease. Annu. Rev. Immunol. 2020;38:365–395. doi: 10.1146/annurev-immunol-102419-035900. [DOI] [PubMed] [Google Scholar]
- 6.Smith B.A.H., Bertozzi C.R. The clinical impact of glycobiology: targeting selectins, Siglecs and mammalian glycans. Nat. Rev. Drug Discov. 2021;20:217–243. doi: 10.1038/s41573-020-00093-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stewart N., Daly J., Drummond-Guy O., Krishnamoorthy V., Stark J.C., Riley N.M., Williams K.C., Bertozzi C.R., Wisnovsky S. The glycoimmune checkpoint receptor Siglec-7 interacts with T-cell ligands and regulates T-cell activation. J. Biol. Chem. 2024;300 doi: 10.1016/j.jbc.2023.105579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Varki A., Angata T. Siglecs--the major subfamily of I-type lectins. Glycobiology. 2006;16:1R–27R. doi: 10.1093/glycob/cwj008. [DOI] [PubMed] [Google Scholar]
- 9.Sabit I., Hashimoto N., Matsumoto Y., Yamaji T., Furukawa K., Furukawa K. Binding of a sialic acid-recognizing lectin Siglec-9 modulates adhesion dynamics of cancer cells via calpain-mediated protein degradation. J. Biol. Chem. 2013;288:35417–35427. doi: 10.1074/jbc.M113.513192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nicoll G., Avril T., Lock K., Furukawa K., Bovin N., Crocker P.R. Ganglioside GD3 expression on target cells can modulate NK cell cytotoxicity via siglec-7-dependent and -independent mechanisms. Eur. J. Immunol. 2003;33:1642–1648. doi: 10.1002/eji.200323693. [DOI] [PubMed] [Google Scholar]
- 11.Hashimoto N., Ito S., Tsuchida A., Bhuiyan R.H., Okajima T., Yamamoto A., Furukawa K., Ohmi Y., Furukawa K. The ceramide moiety of disialoganglioside (GD3) is essential for GD3 recognition by the sialic acid–binding lectin SIGLEC7 on the cell surface. J. Biol. Chem. 2019;294:10833–10845. doi: 10.1074/jbc.ra118.007083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawasaki Y., Ito A., Withers D.A., Taima T., Kakoi N., Saito S., Arai Y. Ganglioside DSGb5, preferred ligand for Siglec-7, inhibits NK cell cytotoxicity against renal cell carcinoma cells. Glycobiology. 2010;20:1373–1379. doi: 10.1093/glycob/cwq116. [DOI] [PubMed] [Google Scholar]
- 13.Miyazaki K., Sakuma K., Kawamura Y.I., Izawa M., Ohmori K., Mitsuki M., Yamaji T., Hashimoto Y., Suzuki A., Saito Y., et al. Colonic Epithelial Cells Express Specific Ligands for Mucosal Macrophage Immunosuppressive Receptors Siglec-7 and -9. J. Immunol. 2012;188:4690–4700. doi: 10.4049/jimmunol.1100605. [DOI] [PubMed] [Google Scholar]
- 14.Reily C., Stewart T.J., Renfrow M.B., Novak J. Glycosylation in health and disease. Nat. Rev. Nephrol. 2019;15:346–366. doi: 10.1038/s41581-019-0129-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodriguez E., Boelaars K., Brown K., Eveline Li R.J., Kruijssen L., Bruijns S.C.M., Van Ee T., Schetters S.T.T., Crommentuijn M.H.W., Van Der Horst J.C., et al. Sialic acids in pancreatic cancer cells drive tumour-associated macrophage differentiation via the Siglec receptors Siglec-7 and Siglec-9. Nat. Commun. 2021;12 doi: 10.1038/s41467-021-21550-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wisnovsky S., Möckl L., Malaker S.A., Pedram K., Hess G.T., Riley N.M., Gray M.A., Smith B.A.H., Bassik M.C., Moerner W.E., Bertozzi C.R. Genome-wide CRISPR screens reveal a specific ligand for the glycan-binding immune checkpoint receptor Siglec-7. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2015024118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoshimura A., Asahina Y., Chang L.-Y., Angata T., Tanaka H., Kitajima K., Sato C. Identification and functional characterization of a Siglec-7 counter-receptor on K562 cells. J. Biol. Chem. 2021;296 doi: 10.1016/j.jbc.2021.100477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang L.Y., Liang S.Y., Lu S.C., Tseng H.C., Tsai H.Y., Tang C.J., Sugata M., Chen Y.J., Chen Y.J., Wu S.J., et al. Molecular Basis and Role of Siglec-7 Ligand Expression on Chronic Lymphocytic Leukemia B Cells. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.840388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith B.A.H., Deutzmann A., Correa K.M., Delaveris C.S., Dhanasekaran R., Dove C.G., Sullivan D.K., Wisnovsky S., Stark J.C., Pluvinage J.V., et al. MYC-driven synthesis of Siglec ligands is a glycoimmune checkpoint. Proc. Natl. Acad. Sci. USA. 2023;120 doi: 10.1073/pnas.2215376120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Büll C., Nason R., Sun L., Van Coillie J., Madriz Sørensen D., Moons S.J., Yang Z., Arbitman S., Fernandes S.M., Furukawa S., et al. Probing the binding specificities of human Siglecs by cell-based glycan arrays. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2026102118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harduin-Lepers A., Vallejo-Ruiz V., Krzewinski-Recchi M.-A., Samyn-Petit B., Julien S., Delannoy P. The human sialyltransferase family. Biochimie. 2001;83:727–737. doi: 10.1016/s0300-9084(01)01301-3. [DOI] [PubMed] [Google Scholar]
- 22.Zhang J.Q., Nicoll G., Jones C., Crocker P.R. Siglec-9, a Novel Sialic Acid Binding Member of the Immunoglobulin Superfamily Expressed Broadly on Human Blood Leukocytes. J. Biol. Chem. 2000;275:22121–22126. doi: 10.1074/jbc.m002788200. [DOI] [PubMed] [Google Scholar]
- 23.Priatel J.J., Chui D., Hiraoka N., Simmons C.J., Richardson K.B., Page D.M., Fukuda M., Varki N.M., Marth J.D. The ST3Gal-I Sialyltransferase Controls CD8+ T Lymphocyte Homeostasis by Modulating O-Glycan Biosynthesis. Immunity. 2000;12:273–283. doi: 10.1016/s1074-7613(00)80180-6. [DOI] [PubMed] [Google Scholar]
- 24.Wu Z.L., Person A.D., Burton A.J., Singh R., Burroughs B., Fryxell D., Tatge T.J., Manning T., Wu G., Swift K.A.D., Kalabokis V. Direct fluorescent glycan labeling with recombinant sialyltransferases. Glycobiology. 2019;29:750–754. doi: 10.1093/glycob/cwz058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dimitroff C.J. Galectin-Binding O-Glycosylations as Regulators of Malignancy. Cancer Res. 2015;75:3195–3202. doi: 10.1158/0008-5472.can-15-0834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nairn A.V., Aoki K., Dela Rosa M., Porterfield M., Lim J.-M., Kulik M., Pierce J.M., Wells L., Dalton S., Tiemeyer M., Moremen K.W. Regulation of Glycan Structures in Murine Embryonic Stem Cells. J. Biol. Chem. 2012;287:37835–37856. doi: 10.1074/jbc.m112.405233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takashima S., Ishida H.K., Inazu T., Ando T., Ishida H., Kiso M., Tsuji S., Tsujimoto M. Molecular cloning and expression of a sixth type of alpha 2,8-sialyltransferase (ST8Sia VI) that sialylates O-glycans. J. Biol. Chem. 2002;277:24030–24038. doi: 10.1074/jbc.M112367200. [DOI] [PubMed] [Google Scholar]
- 28.Haraguchi M., Yamashiro S., Yamamoto A., Furukawa K., Takamiya K., Lloyd K.O., Shiku H., Furukawa K. Isolation of GD3 synthase gene by expression cloning of GM3 alpha-2,8-sialyltransferase cDNA using anti-GD2 monoclonal antibody. Proc. Natl. Acad. Sci. USA. 1994;91:10455–10459. doi: 10.1073/pnas.91.22.10455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huet G., Hennebicq-Reig S., De Bolos C., Ulloa F., Lesuffleur T., Barbat A., Carrière V., Kim I., Real F.X., Delannoy P., Zweibaum A. GalNAc-α-O-benzyl Inhibits NeuAcα2-3 Glycosylation and Blocks the Intracellular Transport of Apical Glycoproteins and Mucus in Differentiated HT-29 Cells. J. Cell Biol. 1998;141:1311–1322. doi: 10.1083/jcb.141.6.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kudelka M.R., Antonopoulos A., Wang Y., Duong D.M., Song X., Seyfried N.T., Dell A., Haslam S.M., Cummings R.D., Ju T. Cellular O-Glycome Reporter/Amplification to explore O-glycans of living cells. Nat. Methods. 2016;13:81–86. doi: 10.1038/nmeth.3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng H.-Y., Lin Y.-Y., Yu C.-Y., Chen J.-Y., Shen K.-F., Lin W.-L., Liao H.-K., Chen Y.-J., Liu C.-H., Pang V.F., Jou T.-S. Molecular Identification of Canine Podocalyxin-Like Protein 1 as a Renal Tubulogenic Regulator. J. Am. Soc. Nephrol. 2005;16:1612–1622. doi: 10.1681/asn.2004121145. [DOI] [PubMed] [Google Scholar]
- 32.Fröse J., Chen M.B., Hebron K.E., Reinhardt F., Hajal C., Zijlstra A., Kamm R.D., Weinberg R.A. Epithelial-Mesenchymal Transition Induces Podocalyxin to Promote Extravasation via Ezrin Signaling. Cell Rep. 2018;24:962–972. doi: 10.1016/j.celrep.2018.06.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nielsen J.S., McNagny K.M. The Role of Podocalyxin in Health and Disease. J. Am. Soc. Nephrol. 2009;20:1669–1676. doi: 10.1681/asn.2008070782. [DOI] [PubMed] [Google Scholar]
- 34.Shimamura T., Ito H., Shibahara J., Watanabe A., Hippo Y., Taniguchi H., Chen Y., Kashima T., Ohtomo T., Tanioka F., et al. Overexpression of MUC13 is associated with intestinal-type gastric cancer. Cancer Sci. 2005;96:265–273. doi: 10.1111/j.1349-7006.2005.00043.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gupta B.K., Maher D.M., Ebeling M.C., Stephenson P.D., Puumala S.E., Koch M.R., Aburatani H., Jaggi M., Chauhan S.C. Functions and regulation of MUC13 mucin in colon cancer cells. J. Gastroenterol. 2014;49:1378–1391. doi: 10.1007/s00535-013-0885-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shao J.Y., Yin W.W., Zhang Q.F., Liu Q., Peng M.L., Hu H.D., Hu P., Ren H., Zhang D.Z. Siglec-7 Defines a Highly Functional Natural Killer Cell Subset and Inhibits Cell-Mediated Activities. Scand. J. Immunol. 2016;84:182–190. doi: 10.1111/sji.12455. [DOI] [PubMed] [Google Scholar]
- 37.Bordoloi D., Kulkarni A.J., Adeniji O.S., Pampena M.B., Bhojnagarwala P.S., Zhao S., Ionescu C., Perales-Puchalt A., Parzych E.M., Zhu X., et al. Siglec-7 glyco-immune binding mAbs or NK cell engager biologics induce potent antitumor immunity against ovarian cancers. Sci. Adv. 2023;9 doi: 10.1126/sciadv.adh4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hudak J.E., Canham S.M., Bertozzi C.R. Glycocalyx engineering reveals a Siglec-based mechanism for NK cell immunoevasion. Nat. Chem. Biol. 2014;10:69–75. doi: 10.1038/nchembio.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang Y.F., Aoki K., Akase S., Ishihara M., Liu Y.S., Yang G., Kizuka Y., Mizumoto S., Tiemeyer M., Gao X.D., et al. Global mapping of glycosylation pathways in human-derived cells. Dev. Cell. 2021;56:1195–1209.e7. doi: 10.1016/j.devcel.2021.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Christou C.M., PEARCE A.C., Watson A.A., Mistry A.R., Pollitt A.Y., Fenton-May A.E., Johnson L.A., Jackson D.G., Watson S.P., O'Callaghan C.A. Renal cells activate the platelet receptor CLEC-2 through podoplanin. Biochem. J. 2008;411:133–140. doi: 10.1042/bj20071216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaneko M.K., Kato Y., Kameyama A., Ito H., Kuno A., Hirabayashi J., Kubota T., Amano K., Chiba Y., Hasegawa Y., et al. Functional glycosylation of human podoplanin: glycan structure of platelet aggregation-inducing factor. FEBS Lett. 2007;581:331–336. doi: 10.1016/j.febslet.2006.12.044. [DOI] [PubMed] [Google Scholar]
- 42.Kato Y., Kaneko M.K., Kunita A., Ito H., Kameyama A., Ogasawara S., Matsuura N., Hasegawa Y., Suzuki-Inoue K., Inoue O., et al. Molecular analysis of the pathophysiological binding of the platelet aggregation-inducing factor podoplanin to the C-type lectin-like receptor CLEC-2. Cancer Sci. 2008;99:54–61. doi: 10.1111/j.1349-7006.2007.00634.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suzuki H., Kaneko M.K., Kato Y. Roles of Podoplanin in Malignant Progression of Tumor. Cells. 2022;11:575. doi: 10.3390/cells11030575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang C., Fan W., Zhang P., Wang Z., Huang L. One-pot nonreductive O-glycan release and labeling with 1-phenyl-3-methyl-5-pyrazolone followed by ESI-MS analysis. Proteomics. 2011;11:4229–4242. doi: 10.1002/pmic.201000677. [DOI] [PubMed] [Google Scholar]
- 45.Wang C., Yuan J., Wang Z., Huang L. Separation of one-pot procedure released O-glycans as 1-phenyl-3-methyl-5-pyrazolone derivatives by hydrophilic interaction and reversed-phase liquid chromatography followed by identification using electrospray mass spectrometry and tandem mass spectrometry. J. Chromatogr. A. 2013;1274:107–117. doi: 10.1016/j.chroma.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 46.Hoyer L.L., Roggentin P., Schauer R., Vimr E.R. Purification and Properties of Cloned Salmonella typhimurium LT2 Sialidase with Virus-Typical Kinetic Preference for Sialyl α2→3 Linkages. J. Biochem. 1991;110:462–467. doi: 10.1093/oxfordjournals.jbchem.a123603. [DOI] [PubMed] [Google Scholar]
- 47.Teintenier-Lelievre M., Julien S., Juliant S., Guerardel Y., Duonor-Cerutti M., Delannoy P., Harduin-Lepers A. Molecular cloning and expression of a human hST8Sia VI (alpha2,8-sialyltransferase) responsible for the synthesis of the diSia motif on O-glycosylproteins. Biochem. J. 2005;392:665–674. doi: 10.1042/BJ20051120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sekiguchi T., Takemoto A., Takagi S., Takatori K., Sato S., Takami M., Fujita N. Targeting a novel domain in podoplanin for inhibiting platelet-mediated tumor metastasis. Oncotarget. 2016;7:3934–3946. doi: 10.18632/oncotarget.6598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Halim A., Brinkmalm G., Rüetschi U., Westman-Brinkmalm A., Portelius E., Zetterberg H., Blennow K., Larson G., Nilsson J. Site-specific characterization of threonine, serine, and tyrosine glycosylations of amyloid precursor protein/amyloid -peptides in human cerebrospinal fluid. Proc. Natl. Acad. Sci. USA. 2011;108:11848–11853. doi: 10.1073/pnas.1102664108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gummuluru S., Pina Ramirez N.G., Akiyama H. CD169-Dependent Cell-Associated HIV-1 Transmission: A Driver of Virus Dissemination. J. Infect. Dis. 2014;210:S641–S647. doi: 10.1093/infdis/jiu442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Izquierdo-Useros N., Lorizate M., Puertas M.C., Rodriguez-Plata M.T., Zangger N., Erikson E., Pino M., Erkizia I., Glass B., Clotet B., et al. Siglec-1 Is a Novel Dendritic Cell Receptor That Mediates HIV-1 Trans-Infection Through Recognition of Viral Membrane Gangliosides. PLoS Biol. 2012;10 doi: 10.1371/journal.pbio.1001448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Izquierdo-Useros N., Lorizate M., McLaren P.J., Telenti A., Kräusslich H.-G., Martinez-Picado J. HIV-1 Capture and Transmission by Dendritic Cells: The Role of Viral Glycolipids and the Cellular Receptor Siglec-1. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Khatua B., Roy S., Mandal C. Sialic acids siglec interaction: A unique strategy to circumvent innate immune response by pathogens. Indian J. Med. RES. 2013;138:648–662. [PMC free article] [PubMed] [Google Scholar]
- 54.Quintanilla M., Montero-Montero L., Renart J., Martín-Villar E. Podoplanin in Inflammation and Cancer. Int. J. Mol. Sci. 2019;20:707. doi: 10.3390/ijms20030707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu X., Zhao J., Ruan Y., Sun L., Xu C., Jiang H. Sialyltransferase ST3GAL1 promotes cell migration, invasion, and TGF-β1-induced EMT and confers paclitaxel resistance in ovarian cancer. Cell Death Dis. 2018;9 doi: 10.1038/s41419-018-1101-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ogawa T., Hirohashi Y., Murai A., Nishidate T., Okita K., Wang L., Ikehara Y., Satoyoshi T., Usui A., Kubo T., et al. ST6GALNAC1 plays important roles in enhancing cancer stem phenotypes of colorectal cancer via the Akt pathway. Oncotarget. 2017;8:112550–112564. doi: 10.18632/oncotarget.22545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Friedman D.J., Crotts S.B., Shapiro M.J., Rajcula M., McCue S., Liu X., Khazaie K., Dong H., Shapiro V.S. ST8Sia6 Promotes Tumor Growth in Mice by Inhibiting Immune Responses. Cancer Immunol. Res. 2021;9:952–966. doi: 10.1158/2326-6066.cir-20-0834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yamakawa N., Yasuda Y., Yoshimura A., Goshima A., Crocker P.R., Vergoten G., Nishiura Y., Takahashi T., Hanashima S., Matsumoto K., et al. Discovery of a new sialic acid binding region that regulates Siglec-7. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-64887-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Matsumoto Y., Zhang Q., Akita K., Nakada H., Hamamura K., Tsuchida A., Okajima T., Furukawa K., Urano T., Furukawa K. Trimeric Tn Antigen on Syndecan 1 Produced by ppGalNAc-T13 Enhances Cancer Metastasis via a Complex Formation with Integrin α5β1 and Matrix Metalloproteinase 9. J. Biol. Chem. 2013;288:24264–24276. doi: 10.1074/jbc.m113.455006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hashimoto N., Hamamura K., Kotani N., Furukawa K., Kaneko K., Honke K., Furukawa K. Proteomic analysis of ganglioside-associated membrane molecules: Substantial basis for molecular clustering. Proteomics. 2012;12:3154–3163. doi: 10.1002/pmic.201200279. [DOI] [PubMed] [Google Scholar]
- 61.Hamamura K., Tsuji M., Hotta H., Ohkawa Y., Takahashi M., Shibuya H., Nakashima H., Yamauchi Y., Hashimoto N., Hattori H., et al. Functional Activation of Src Family Kinase Yes Protein Is Essential for the Enhanced Malignant Properties of Human Melanoma Cells Expressing Ganglioside GD3. J. Biol. Chem. 2011;286:18526–18537. doi: 10.1074/jbc.m110.164798. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The detailed data processing has been deposited on Mendeley Data and is publicly available as of the date of publication. The DOI is listed in the key resources table.
-
•
R-code has been deposited on Mendeley Data and is publicly available as of the date of publication. The DOI is listed in the key resources table.
-
•
Any additional information required to analyze the data reported will be shared by the lead contact upon request.