Abstract
Background
Gut microbes are important to the health and fitness of many animals. Many factors have been shown to affect gut microbial communities including diet, lifestyle, and age. Most animals have very complex physiologies, lifestyles, and microbiomes, making it virtually impossible to disentangle what factors have the largest impact on microbiota composition. Honeybees are an excellent model to study host-microbe interactions due to their relatively simple gut microbiota, experimental tractability, and eusociality. Worker honey bees have distinct gut microbiota from their queen mothers despite being close genetic relatives and living in the same environment. Queens and workers differ in numerous ways including development, physiology, pheromone production, diet, and behavior. In the prolonged absence of a queen or Queen Mandibular Pheromones (QMP), some but not all workers will develop ovaries and become “queen-like”. Using this inducible developmental change, we aimed to determine if diet and/or reproductive development impacts the gut microbiota of honey bee workers.
Results
Microbiota-depleted newly emerged workers were inoculated with a mixture of queen and worker gut homogenates and reared under four conditions varying in diet and pheromone exposure. Three weeks post-emergence, workers were evaluated for ovary development and their gut microbiota communities were characterized. The proportion of workers with developed ovaries was increased in the absence of QMP but also when fed a queen diet (royal jelly). Overall, we found that diet, rather than reproductive development or pheromone exposure, led to more “queen-like” microbiota in workers. However, we revealed that diet alone cannot explain the microbiota composition of workers.
Conclusion
The hypothesis that reproductive development explains microbiota differences between queens and workers was rejected. We found evidence that diet is one of the main drivers of differences between the gut microbial community compositions of queens and workers but cannot fully explain the distinct microbiota of queens. Thus, we predict that behavioral and other physiological differences dictate microbiota composition in workers and queens. Our findings not only contribute to our understanding of the factors affecting the honey bee microbiota, which is important for bee health, but also illustrate the versatility and benefits of utilizing honeybees as a model system to study host-microbe interactions.
Supplementary Information
The online version contains supplementary material available at 10.1186/s42523-024-00350-3.
Keywords: Honey bees (Apis mellifera), Gut microbiota, Queen Mandibular Pheromone (QMP), Royal jelly, Ovary development, Diet, Queen-worker dimorphism, Caste differentiation
Introduction
Gut microbes provide a multitude of functions for their hosts, including metabolizing nutrients, removing toxins, modulating immune function, stimulating growth and development, and protecting against pathogens [1]. A disrupted or altered gut microbiota can have impacts on host health, therefore, determining what and how different factors impact microbial community structure and function has been a major goal of microbiome studies [2–5]. Differences in diet, geography, age, genetics, physiology, and lifestyle can result in substantial variation in microbiota composition across individuals from the same species [6–8]. Intraspecific variation across microbial communities makes it difficult to understand the significance of changes in microbiota structure and hinders our ability to define a “healthy” microbiome [7, 9]. Moreover, in hosts with complex physiologies, lifestyles, and microbiomes, such as mammals, it is virtually impossible to disentangle how and to what extent different host and environmental factors impact the microbiota, mostly due to numerous confounding variables. Thus, simpler, more tractable model systems are needed to address fundamental questions about host-associated microbial communities [8, 10, 11].
Honey bees (Apis mellifera) constitute an excellent model system for studying host-microbe dynamics because their gut microbiota is relatively simple but displays many parallels to the microbiota of mammals [11–14]. The honey bee worker gut microbiota consists of five core bacterial genera: Lactobacillus, Bombilactobacillus, Gilliamella, Snodgrassella, and Bifidobacterium [12, 15–18]. In addition, three other non-core bacteria (Frischella, Bartonella, and Commensalibacter) are often detected in workers [12]. Together, these eight bacterial genera account for > 90% of the diversity within the honey bee gut microbiota [12]. Honey bees acquire their characteristic gut microbiota after emerging from their pupal state via contact with nestmates and hive material [19, 20]. Although the composition of the core microbiota is stable across honey bee workers, differences among individuals can be seen in the relative frequency of the core species and the presence and abundance of atypical (transient or opportunistic) bacteria [12, 15, 21]. Moreover, there is a high degree of strain-level variation within individuals and across bee gut microbial communities, which has been shown to correspond to differences in functional capabilities [22–28]. Additionally, the honey bee gut microbiota has been associated with many components of health, including metabolism, pathogen resistance, and immunity [15, 23, 28–39].
Female workers make up the majority of the honey bee colony population (> 90%) followed by a small male drone population (0 to 10%, depending on the season), and a singular queen which is the sole reproductive female in the colony [40]. Despite being immediate relatives that are exposed to the same environment, the gut microbiota of queens is very distinct from workers [41, 42]. The queen gut microbiota is variable across individuals but is typically dominated by only four bacteria, Lactobacillus spp., Bombella apis, Apilactobacillus kunkeei, and Commensalibacter sp., of which only Lactobacillus is consistently present in all worker guts [43–46]. The root of this difference is unclear but could be due to biological or dietary differences between honey bee queens and workers. A diploid honey bee egg has the potential to develop into a queen or a worker depending on the diet they are provided during development. Larvae that will develop into queens are fed royal jelly, a protein-rich secretion produced by the hypopharyngeal glands of nurse bees, during development and throughout their entire life [40, 47]. Larvae destined to become workers are fed royal jelly for ∼ 3.5 days, after which their main source of protein becomes worker jelly [48, 49]. Following emergence, the main protein source of workers is bee bread and pollen [40, 47]. Queens are also much larger than workers, lack worker morphological characteristics (e.g., notched mandibles, hypopharyngeal and wax glands, barbed stingers, and pollen baskets), differentially express genes, particularly vitellogenin (vg) and the major royal jelly protein 1 (mrjp1), and have fully developed reproductive organs (ovaries with ovarioles and a spermatheca) [50–52]. Additionally, queens secrete Queen Mandibular Pheromones (QMP) which suppress the development of ovaries, egg production, and other queen-like morphological and physiological characteristics in workers [53, 54]. However, in the prolonged absence of QMP, some adult workers will develop ovaries, become “queen-like” in both morphology and physiology, and have the ability to lay haploid (unfertilized) drone eggs [55, 56]. A recent study has also shown that even in the presence of QMP, workers can develop ovaries if they are provided a royal jelly diet [57].
We hypothesized that workers with developed ovaries would possess gut microbial communities that more closely resemble the queen gut microbiota (i.e., be dominated by Lactobacillus spp., Bombella apis, Apilactobacillus kunkeei, and Commensalibacter sp). To test our hypothesis, we inoculated microbiota-depleted newly emerged workers (NEWs) with a cocktail of queen and worker microbes and then provided them with either royal jelly or pollen as a sole protein source, both in the presence or absence of QMP. After approximately three and a half weeks, we evaluated ovary development and sampled the guts of individual worker bees to characterize their gut microbial communities. We confirmed that workers not exposed to QMP and/or fed a royal jelly diet had increased ovary activation. However, reproductive organ development appeared to have no effect on microbiota composition. Instead, we found that a royal jelly diet led to increased abundance of queen-associated microbes but cannot fully explain the differences between the queen and worker gut microbial communities. Our findings not only shed light on the factors that drive microbial community structure in honey bees but also emphasize the benefit of using the honey bee as a model system to disentangle how different factors impact host-associated microbial communities.
Results
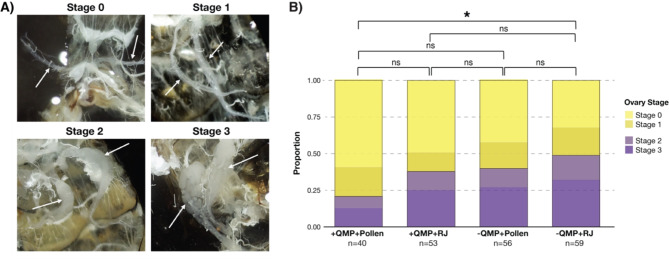
To determine whether diet, exposure to QMP, or reproductive (ovary) development explains the variation between the gut microbiota of queens and workers, late-stage worker pupae were aseptically removed from a brood frame and reared under sterile conditions in the lab. Upon emergence, approximately 240 microbiota-depleted newly emerged workers (NEWs) were inoculated with a mixture of queen and worker gut homogenates. After inoculation, NEWs were split into four experimental groups: (1) QMP with a royal jelly diet (+ QMP + RJ), (2) QMP with a pollen diet + QMP + Pollen), (3) no QMP and a royal jelly diet (-QMP + RJ), and (4) no QMP and a pollen diet (-QMP + Pollen). Approximately three and a half weeks after the start of the experiment, bees were dissected, their guts were aseptically removed, and the level of ovary activation for each bee was determined based on a scale of 0–3 [58, 59], with Stage 0 indicating no ovary development and Stage 3 representing highly developed ovaries (Fig. 1A). All worker honey bees possess ovary organs but they are typically undeveloped and nonfunctional [60]. Thus, we considered workers to have developed ovaries if they were ranked Stage 2 or 3 (Fig. 1A). Only 20% of the + QMP + Pollen exposed workers exhibited developed ovaries, whereas 49% of workers that were not exposed to QMP and given a royal jelly diet (-QMP + RJ) presented developed ovaries (Fig. 1B; P = 0.009, Chi-Squared Test). Consistent with previous studies [57, 61], we found that ovary development occurred more frequently in the absence of QMP (39–49%), but in the presence of QMP, a royal jelly diet (+ QMP + RJ) also led to increased ovary development in workers (38% of workers; Fig. 1B). Variation in ovary development was observed across replicate cup cages within each experimental group, with the exception of the -QMP + RJ group in which all replicates had ∼ 50% of workers with developed (Stage 2–3) ovaries (Figure S1), indicating that ovary activation more consistently occurs in royal jelly fed workers in the absence of QMP.
Fig. 1.
Ovary development in worker bees. (A) Microscope photographs demonstrating the four ovary developmental stages (0–3) of workers. Arrows point to the location of the ovaries on the photographs. Ovaries classified as stage 0 were indistinguishable from typical queen-right worker morphology and exhibited no visible thickening or oocyte development. Stage 1 ovaries demonstrated visible thickening but no constriction of oocyte development. Stage 2 ovaries were characterized by both visible thickening and constriction around at least one developed oocyte. Ovaries with multiple maturing eggs and across several ovarioles were categorized as 3. (B) Proportion of workers assigned to each ovary development stage from the four experimental groups. For statistical testing Stages 0–1 were considered to have undeveloped ovaries and Stages 2–3 were considered to have developed ovaries (*=P < 0.005, Chi-Squared Test)
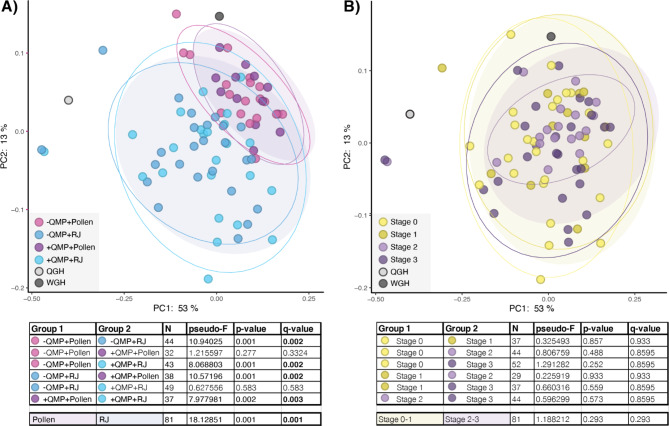
We compared the microbiota composition of workers from each experimental group and each ovary developmental stage. We found no significant difference between workers that were fed the same diet and reared in the presence or absence of QMP (Q > 0.3, Pairwise PERMANOVA). However, workers fed pollen possessed significantly different microbiota compositions than workers fed royal jelly, regardless of QMP exposure (Fig. 2A; Q < 0.003, Pairwise PERMANOVA). No differences in microbiota composition were observed based on ovary developmental stage (Fig. 2B; Q > 0.8, Pairwise PERMANOVA). These findings indicate that diet plays a much larger role in shaping microbiota composition in workers than reproductive development or exposure to QMP.
Fig. 2.
Beta diversity in the gut microbiota of experimentally reared workers. Principle Coordinate Analysis using weighted UniFrac comparing workers from (A) our four experimental groups and (B) workers at different ovary developmental stages. Significance was tested using PERMANOVA with 999 permutations followed by Benjamini–Hochberg FDR correction. Ellipses represent the 95% confidence intervals
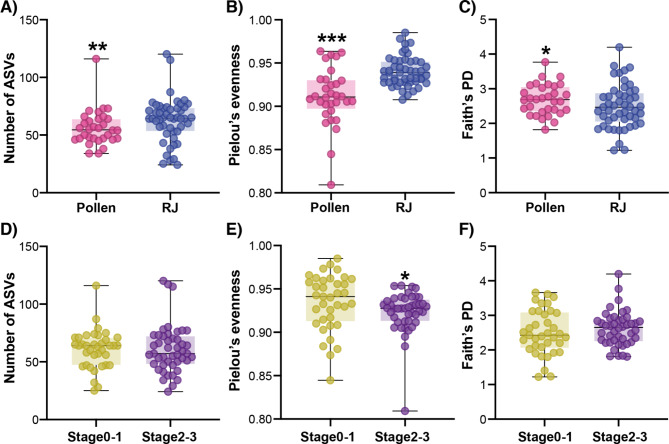
Alpha diversity also differed depending on diet, with pollen-fed workers having less rich (Fig. 3A; P = 0.009, Mann Whitney Test) and less even (Fig. 3B: P < 0.0001, Mann Whitney Test) but more phylogenetically diverse (Fig. 3C; P = 0.04, Mann Whitney Test) microbial communities than workers fed royal jelly. Very few differences in microbiota alpha diversity were observed based on ovary development stage (Fig. 3D-F); only evenness differed between workers with undeveloped (Stages 0–1) and developed (Stages 2–3) ovaries (Fig. 3E; P = 0.01, Mann Whitney Test). These results provide further evidence that diet –rather than ovary activation or QMP– impacts the microbiota composition of workers.
Fig. 3.
Alpha diversity in the worker gut microbiota compared across diets (A-C) and ovary developmental stages (D-F). Richness was measured based on the number of ASVs (A and D). Pielou’s evenness index was used to calculate microbiota evenness (B and E) and phylogenetic diversity was measured using Faith’s PD (C and F). Significance was tested using the Mann-Whitney test. *= P < 0.05, **= P < 0.005, ***= P < 0.0005
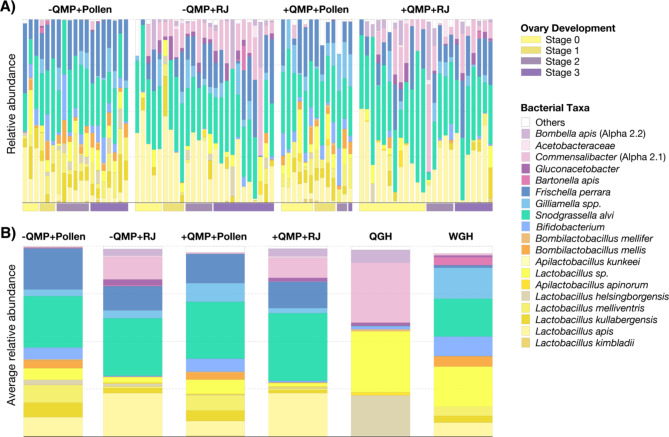
To evaluate taxonomic differences across experimental groups, we analyzed the relative abundance of each taxon within individual bees (Fig. 4A, Dataset S1). Again, we found that gut microbiota composition was more similar across bees that were fed the same diet, regardless of their ovary developmental stage or exposure to QMP (Fig. 4A). Workers fed pollen possessed all of the core taxa of the worker gut microbiota, i.e., Lactobacillus, Bombilactobacillus, Bifidobacterium, Snodgrassella, and Gilliamella, and had a high diversity of Lactobacillus species (Fig. 4A). Conversely, nearly all workers that were fed royal jelly lacked Bifidobacterium and Bombilactobacillus and were mainly dominated by a single Lactobacillus species, L. apis (Fig. 4A). Virtually all the royal jelly-fed workers also contained a high abundance of two queen-associated bacteria, Commensalibacter sp. and Bombella apis, which were very rarely found in workers that were fed a pollen diet (Fig. 4A) and are not consistently observed in conventional honey bee worker guts [46].
Fig. 4.
Taxonomic classification of bacteria present in honey bee guts. (A) Relative abundance of each bacterial taxon within individual worker bees from each of the four experimental groups (-QMP + Pollen, -QMP + RJ, +QMP + Pollen, +QMP + RJ). Vertical bars represent the relative abundance of the bacterial taxa and horizontal bars along the bottom of the graph indicate the ovary development stage (0–3) of the workers. (B) Average relative abundance of bacterial taxa in the workers from each experimental group compared to the queen (QGH) and worker (WGH) gut homogenates that were used to inoculate all experimental bees in our study. See Dataset S1 for raw relative abundance data
All NEWs in this study were inoculated with a mixture of queen (QGH) and worker (WGH) gut homogenates, which was created from two randomly sampled conventional workers and two randomly sampled conventional queens. Thus, we also compared the bacterial taxa present in the WGH and QGH to the average relative abundance of the bacterial taxa identified in the guts of workers from each of our four experimental groups (Fig. 4B, Dataset S1). Although the workers in our study did not directly mirror the microbiota composition of the QGH or WGH used to inoculate them, we found that workers that were fed royal jelly possessed a higher abundance of queen-associated microbes whereas workers that were given pollen displayed gut microbiota more similar to conventional worker honey bees (Fig. 4B). However, royal jelly fed workers still retained most of the core worker gut bacteria. Taken together, our results suggest that diet plays a significant role in driving microbiota composition in queen and worker honey bees but cannot fully explain the differences between queen and worker gut communities.
Discussion
Physiological changes, such as organ development during insect metamorphosis have been correlated with different microbial communities in insects [62], suggesting a potential relationship between reproductive physiology and gut microbiota in honey bees. Diet can also have major impacts on the gut microbiota of many animals, including honey bees [21, 63–66]. Female honey bee workers have undeveloped nonfunctional ovaries and eat pollen as their main protein source. Conversely, honey bee queens have developed ovaries and are fed royal jelly as their sole protein source. Queens also secrete pheromones (QMP) that suppress ovary activation and regulate the development and behavior of workers [53, 54]. In many animals, microbes have been demonstrated to play a role in pheromone production [67–69], but it is unclear if pheromones impact gut microbial communities. The main goal of this study was to investigate if ovary development could explain the naturally occurring differences in microbiota composition between queens and workers [41, 42]. Consistent with a recent study [57], we found that workers not exposed to QMP and/or fed a diet of royal jelly exhibited increased ovary development when compared to workers kept in the presence of QMP and fed a pollen diet. Because not all workers within each experimental group developed ovaries, even in the absence of QMP, we were also able to investigate if diet or QMP exposure (regardless of ovary activation) affects worker microbiota composition. We found no evidence that ovary development or QMP impacts the gut microbiota of worker honey bees. Notably, we revealed that diet, in part, explains the differences between queen and worker microbiota composition, as workers fed a royal jelly diet displayed more “queen-like” gut microbial communities than workers fed pollen.
Although workers fed royal jelly possessed a higher abundance of queen-associated microbes, they still maintained many of the characteristic gut microbes of conventional workers, e.g., Snodgrassella, Gilliamella and Frischella, which are typically not present in queens [41, 43, 45, 46, 70]. This finding suggests that another factor, aside from diet, ovary development, or exposure to QMP, dictates bacterial community composition in workers. Many other physiological, developmental, and morphological differences exist between workers and queens [71]. For example, workers have notched mandibles, hypopharyngeal and wax glands, barbed stingers, and pollen baskets and they produce several pheromones and chemicals that queens do not produce or secrete at different levels (e.g., alarm pheromone, Nasonov pheromone, 2-heptanone, ethyl oleate) [50–52, 54]. Unlike queens, workers undergo a caste transition from nurse to forager which is accompanied by changes in gene expression, physiology, chemical production, and behavior [50–52, 72, 73]. Age and caste are generally coupled in workers, with younger bees being nurses and older bees being foragers [40, 74]. However, worker caste transitioning can occur independent of age, depending on colony needs, and the transition is reversible [75]. In fact, a recent study showed that age-controlled NEWs kept in the lab often existed in both caste states (nurses and foragers), which corresponded to differences in cuticular hydrocarbons (CHCs), body and gut weight, and hypopharyngeal gland size [76], demonstrating the extreme plasticity of honey bee workers even when kept in a controlled lab setting. Moreover, queens have a shorter developmental period than workers and are exclusively fed royal jelly as a protein source during development and throughout their lives [40]. Thus, there are numerous differences between workers and queens that were not specifically investigated in our study, which could contribute to shaping their microbiota composition.
Social interactions and behaviors may also explain the higher microbiota diversity in workers when compared to queens. Honey bee workers take care of all hive maintenance which includes building, cleaning, repairing, and guarding the hive, foraging for food and water, and feeding and caring for all developing larvae (workers, drones, and queens), adult queens, and drones [40, 71]. Queens do not feed themselves and are fed royal jelly directly from the hypopharyngeal glands of workers [71]. Workers also constantly groom and clean the queen, but she does not reciprocate these behaviors [77]. It is well-established that workers acquire their microbes via interactions with other workers and hive material and through consuming pollen [16, 19, 20]. Although coprophagy has never been reported to occur in honey bees, exposure to fecal particles could occur when workers groom and clean one another or through contaminated food and hive materials [19, 78]. As queens do not clean or groom workers or participate in hive maintenance, we predict that queens have less exposure to worker-associated microbes and mainly acquire their microbes from workers during feeding. In fact, queen-associated gut microbes, such as Bombella apis, Apilactobacillus kunkeei and Commensalibacter sp., have been found in the worker mouth parts, hypopharyngeal glands, and royal jelly [41, 45, 46, 70, 79], but are rarely present in the guts of workers [79–83]. Early colonization and niche occupancy by these bacterial taxa in the queen gut could prevent colonization by worker-associated microbes that queens are undoubtably exposed to but potentially later and to a lesser degree.
In this study we exposed microbiota-depleted NEWs to a cocktail of both worker and queen gut homogenates, predicting that different conditions (e.g., diet, QMP, and/or ovary activation) would select for a queen versus worker microbiota composition. However, even though workers fed royal jelly as their sole protein source had more “queen-like” gut microbial communities, they still harbored most of the typical worker-associated microbes [41, 44–46, 70]. Thus, we hypothesize that workers are exposed to a larger diversity of microbes than queens due to their exclusive role in maintaining the hive (e.g., building, cleaning, grooming, feeding, foraging, and storing) and that the unique low diversity microbiota of queens is only partially due to diet. This hypothesis could be tested in future studies by rearing microbiota-depleted queens in the lab and exposing them to both worker and queen microbes. Overall, our results indicate that diet plays a role in governing the differences between worker and queen gut microbiota but does not explain why workers possess more diverse and conserved gut microbial communities.
Conclusions
Due to the complexity of most animal microbiomes and the inability to control for confounding physiological and environmental variables, we still have a poor understanding of the factors driving the composition of gut microbiota. Using the honey bee, a tractable model system for host-microbiome studies, we investigated how differences in reproductive status, diet, and pheromones affect gut microbiota composition and structure. We demonstrated that diet plays a significant role in driving the naturally observed differences between the gut microbial community compositions of honey bee queens and workers, whereas ovary activation and/or exposure to queen pheromones appeared to have no effect on the microbiota. Our results also suggest that although diet can have a major impact on the microbiota, it is not the only factor governing microbial community composition in honey bees. We predict that exposure to microbes via social interactions and behavioral traits plays a major role in dictating microbiota composition. Diet, social interactions, and behavior are all factors that have been shown to affect the microbiota composition of many animals, including humans [84–87]. Thus, our findings not only further our understanding of the honey bee microbiome, which is important for bee health [13, 15, 29], but they also reinforce the significance of using the honey bee as a model system to address fundamental questions about host-microbe interactions.
Methods
Experimental design
Approximately 240 microbiota-depleted newly emerged workers (NEWs) were obtained by extracting 16–17-day-old pupae from a brood frame and incubating them at 35 °C and 80% RH in sterile rearing containers until emergence. Upon emergence, NEWs were immobilized at 4 °C and randomly placed into 16 different 50 mL conical tubes (∼ 15 bees per tube). Two “healthy” mated queens were provided to us from the North Carolina State University (NCSU) Queen and Disease Clinic, which were used to create a queen gut homogenate (QGH). Because we were only able to obtain two queens for this study and we wanted to have equal representation of queen and worker guts, we sampled two worker bees from a single colony from our apiary at NCSU to create a worker gut homogenate (WGH). In brief, the guts of the queens and workers were aseptically extracted and homogenized to create a single queen gut homogenate QGH and a single worker gut homogenate WGH (total volume for each ∼ 200 μL). 50 μL of the QGH and 50 μl of the WGH were then combined and diluted 1:100 in 0.5 M filter-sterilized sucrose syrup and immediately fed to the microbiota-depleted NEWs. The remaining gut WGH (150 μL) and QGH (150 μL) were suspended in 20% glycerol and preserved at -80 °C for future use. The conical tubes containing 15 bees were then inoculated with 150 μL of the QGH/WGH sucrose solution via the immersion method [30, 88] and subsequently placed into cup cages [89]. All cup cages were supplied with a 10 mL tube feeder containing filter-sterilized 0.5 M sucrose syrup and half of the cup cages were supplied with USDA certified organic royal jelly (+ RJ) and the other half with irradiated pollen (+ Pollen). The organic royal jelly used (Greenbow® Royal Jelly) was tested for microbial contamination via plating on Luria-Bertani (LB) agar, De Man–Rogosa–Sharpe agar (MRS) agar, Columbia agar with 5% sheep blood, and Brain Heart Infusion (BHI) agar with 5% sheep blood and incubated at 37 C in an aerobic incubator and also in a 5% CO2 incubator. No microbial growth was seen on any of the plates after five days. Synthetic queen mandibular pheromone (QMP), manufactured by TempQueen, was cut into thirds and one piece was placed into the respective treatment cages (+ QMP + RJ and + QMP + Pollen). Cages containing QMP were kept in a separate incubator. All bees were kept in incubators at 35 °C and 80% RH throughout the experiment. Cages were censused regularly during which dead bees were removed and recorded. We found no significant differences in probability of survival across experimental groups (Figure S2). Ten days after the start of the experiment, 150 μl of the same queen/worker gut homogenate suspended in sterile sucrose (1:100) was pipetted onto a cotton ball placed in each cup cage. Food and sugar syrup were replenished as needed.
Dissections and ovary activation assignments
After a period of ∼ 3.5 weeks, surviving workers were immobilized at 4 °C and then randomly distributed into numbered 1.5 mL tubes with the experimental group of the bee being unknown to the dissector (blinded dissection). Bees were pinned to a dissection plate dorsal side up and their abdomens were cut directly under the thorax and then laterally along each side. The abdomen was then pulled open, pinned, and the gut was carefully removed, placed in 500 μl of 100% molecular-grade ethanol, and stored at 4 °C for further analysis. Ovaries were visually assessed for development using a dissection microscope and were graded on a four-point scale (0–3) adapted from [58, 59]. Ovaries classified as stage 0 were indistinguishable from typical queen-right worker morphology and exhibited no visible thickening or oocyte development. Stage 1 ovaries demonstrated visible thickening but no constriction of oocyte development. Stage 2 ovaries were characterized by both visible thickening and constriction around at least one developed oocyte. Ovaries with multiple maturing eggs and across several ovarioles were categorized as 3. Ovaries scored 0–1 were deemed undeveloped, and those graded 2–3 were considered developed. Ovaries scored 0–1 were deemed undeveloped, and those graded 2–3 were considered developed. We ranked ovary development in 53 + QMP + RJ, 40 + QMP + Pollen, 59 -QMP + RJ, and 55 -QMP + Pollen workers.
DNA extractions and amplicon sequencing
We then extracted DNA from 22 to 25 individual gut tissue samples from each experimental group as well as the queen (QGH) and worker (WGH) gut homogenates used to inoculate the NEWs. Dissected guts were homogenized and a phenol-chloroform DNA extraction with bead cleaning was performed on each sample individually; this protocol as described in [19]. Following DNA extraction, PCR amplification of the V4 region of the 16 S rRNA gene was performed using the universal primers 515 F and 806R with Illumina-specific adapters: Hyb515F_rRNA: 5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGGTGYCAGCMGCCGCGGTA − 3′ and Hyb806R_rRNA: 5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGGACTACHVGGGTWTCTAAT-3′. The PCR cycling conditions were: 98 °C for 30s followed by 30 cycles of 98 °C (10s), 58 °C (30s), 72 °C (30s), with a final extension at 72 °C for 7 m and a hold at 4 °C. The PCR amplicons were cleaned using the Axygen AxyPrep Mag PCR Clean-up Kit at 0.8x bead concentration. Samples were then indexed with the Illumina Nextera XT Index kit v2 set A. The PCR cycling conditions for indexing were 98 °C for 2 m followed by 15 cycles of 98 °C (10s), 55 °C (30s), 72 °C (30s), with a final extension at 72 °C for 7 m and a hold at 4 °C. The indexed PCR products were cleaned again with the Axygen AxyPrep Mag PCR Clean-up Kit at 0.8x bead concentration and then quantified using a Qubit3.0 (Life Technologies) with the Qubit dsDNA HS Assay kit. Our negative controls did not contain enough DNA to quantify and thus were excluded from the sequencing run. All samples were pooled into equal concentrations and sequenced on an Illumina iSeq 100 (2 × 150 bp). A 30% PhiX spike-in was included in the final sample pool to increase the diversity on the run. See Dataset S2 for sample metadata.
16 S sequencing analysis
Forward and reverse raw sequencing reads were merged using FLASH v1.2.10 [90] before being imported into Qiime2 v2023.7 [91]. The DADA2 [92] pipeline in Qiime2 v2023.7 was used to denoise paired reads (qiime dada2 denoise-single). After denoising, the data was filtered to remove reads assigned to Mitochondria, Chloroplast, or Unassigned; additionally reads at less than 1% frequency were removed. All downstream analysis was performed on Qiime2 at a sequencing depth of 4000; this sequencing depth allowed us to retain the most samples while still maintaining enough reads to capture the diversity present in the dataset. After filtering and rarefaction, a total of 82 samples were retained for our analysis: 24 + QMP + RJ, 13 + QMP + Pollen, 19 -QMP + Pollen, 25 -QMP + RJ, 1 QGH, and 1 WGH.
The Qiime2 script “qiime diversity core-metrics-phylogenetic” was used to perform all alpha and beta diversity analyses. Taxonomy was assigned to the sequences using “qiime feature-classifier classify-sklearn” using a classifier trained on the SILVA 16 S v138 [93] reference database. GraphPad Prism v10.2.1 was used to plot alpha diversity results and test for significance (Mann-Whitney Tests). Beta diversity analyses were performed in Qiime2 based on weighted UniFrac [94, 95] and tested using a PERMANOVA (999 permutations) with Benjamini-Hochberg FDR correction. PCoA plots with 95% confidence intervals (stats_ellipse) were made using Qiime2R [96].
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We would like to thank Dr. Bradley Metz for providing us with queens (from the NCSU Queen Clinic), training on ovary identification and classification, lending us dissection tools, and giving feedback on our experimental design and results. We would like to thank Dr. David Tarpy for obtaining funding for the NCSU BeeMORE REEU program, allowing us to use his dissection microscope, and providing feedback on our results. We would also like to thank Caroline Stott and Dr. Meng-Jia Lau for assisting with beekeeping, bee maintenance, and experimental blinding and Dr. Louis-Marie Bobay for reading and providing feedback on the manuscript.
Author contributions
AZ-C performed the experiments, collected the data, and analyzed the results. PG helped with experiments, data collection, and data analysis. KR designed and funded the study and helped perform experiments, collect data, and analyze the results. AZ-C, PG, and KR wrote and edited the manuscript. All authors read and approved the final manuscript.
Funding
This work was funded by the National Science Foundation under grant DEB- 2344788 to KR, the United States Department of Agriculture (USDA), the National Institute of Food and Agriculture (NIFA), Agriculture and Food Research Initiative (AFRI) under grant 2022-67013-42296 to KR. AZC was supported by the BeeMORE program funded by USDA-NIFA Education and Workforce Development grant 2021-67037-34626.
Data availability
The raw sequencing files generated and analyzed during the current study are available on the NCBI SRA repository under BioProject PRJNA1157463. All other data generated or analyzed during this study are included in this published article and its supplementary information files.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ma L, Zhao H, Wu LB, Cheng Z, Liu C. Impact of the microbiome on human, animal, and environmental health from a one health perspective. Sci One Health. 2023;2:100037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hou K, Wu Z-X, Chen X-Y, Wang J-Q, Zhang D, Xiao C, et al. Microbiota in health and diseases. Sig Transduct Target Ther. 2022;7:1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang J, Wu J, Li Y, Zhang Y, Cho WC, Ju X, et al. Gut bacteria formation and influencing factors. FEMS Microbiol Ecol. 2021;97:fiab043. [DOI] [PubMed] [Google Scholar]
- 4.Scepanovic P, Hodel F, Mondot S, Partula V, Byrd A, Hammer C, et al. A comprehensive assessment of demographic, environmental, and host genetic associations with gut microbiome diversity in healthy individuals. Microbiome. 2019;7:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Das B, Nair GB. Homeostasis and dysbiosis of the gut microbiome in health and disease. J Biosci. 2019;44:117. [PubMed] [Google Scholar]
- 6.Division on Earth and Life Studies; Board on Life Sciences. Board on Environmental Studies and Toxicology; Committee on advancing understanding of the implications of environmental-chemical interactions with the human microbiome. Microbiome variation. Environmental chemicals, the human microbiome, and health risk: a research strategy. National Academies Press (US); 2017. [PubMed]
- 7.Anderson BD, Bisanz JE. Challenges and opportunities of strain diversity in gut microbiome research. Front Microbiol. 2023;14. [DOI] [PMC free article] [PubMed]
- 8.Bobay L-M, Raymann K. Population genetics of host-associated microbiomes. Curr Mol Bio Rep. 2019;5:128–39. [Google Scholar]
- 9.Shanahan F, Ghosh TS, O’Toole PW. The healthy microbiome—what is the definition of a healthy gut microbiome? Gastroenterology. 2021;160:483–94. [DOI] [PubMed] [Google Scholar]
- 10.Douglas AE. Simple animal models for microbiome research. Nat Rev Microbiol. 2019;17:764–75. [DOI] [PubMed] [Google Scholar]
- 11.Zheng H, Steele MI, Leonard SP, Motta EVS, Moran NA. Honey bees as models for gut microbiota research. Lab Anim. 2018;47:317–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwong WK, Moran NA. Gut microbial communities of social bees. Nat Rev Microbiol. 2016;14:374–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engel P, Kwong WK, McFrederick Q, Anderson KE, Barribeau SM, Chandler JA et al. The bee microbiome: impact on bee health and model for evolution and ecology of host-microbe interactions. mBio. 2016;7. [DOI] [PMC free article] [PubMed]
- 14.Kwong WK, Moran NA. Evolution of host specialization in gut microbes: the bee gut as a model. Gut Microbes. 2015;6:214–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raymann K, Moran NA. The role of the gut microbiome in health and disease of adult honey bee workers. Curr Opin Insect Sci. 2018;26:97–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinson VG, Moy J, Moran NA. Establishment of characteristic gut bacteria during development of the honeybee worker. Appl Environ Microbiol. 2012;78:2830–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moran NA, Hansen AK, Powell JE, Sabree ZL. Distinctive gut microbiota of honey bees assessed using deep sampling from individual worker bees. PLoS ONE. 2012;7:e36393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martinson VG, Danforth BN, Minckley RL, Rueppell O, Tingek S, Moran NA. A simple and distinctive microbiota associated with honey bees and bumble bees. Mol Ecol. 2011;20:619–28. [DOI] [PubMed] [Google Scholar]
- 19.Powell JE, Martinson VG, Urban-Mead K, Moran NA. Routes of acquisition of the gut microbiota of the honey bee Apis mellifera. Appl Environ Microbiol. 2014;80:7378–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson KE, Ricigliano VA, Copeland DC, Mott BM, Maes P. Social interaction is unnecessary for hindgut microbiome transmission in honey bees: the effect of diet and social exposure on tissue-specific microbiome assembly. Microb Ecol. 2023;85:1498–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kešnerová L, Emery O, Troilo M, Liberti J, Erkosar B, Engel P. Gut microbiota structure differs between honeybees in winter and summer. ISME J. 2020;14:801–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ellegaard KM, Suenami S, Miyazaki R, Engel P. Vast differences in strain-level diversity in the gut microbiota of two closely related honey bee species. Curr Biol. 2020;30:2520–e25317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng H, Nishida A, Kwong WK, Koch H, Engel P, Steele MI et al. Metabolism of toxic sugars by strains of the bee gut symbiont Gilliamella Apicola. mBio. 2016;7. [DOI] [PMC free article] [PubMed]
- 24.Baud GLC, Prasad A, Ellegaard KM, Engel P. Turnover of strain-level diversity modulates functional traits in the honeybee gut microbiome between nurses and foragers. Genome Biol. 2023;24:283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ellegaard KM, Engel P. Genomic diversity landscape of the honey bee gut microbiota. Nat Commun. 2019;10:446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bobay L-M, Wissel EF, Raymann K. Strain structure and dynamics revealed by targeted deep sequencing of the honey bee gut microbiome. mSphere. 2020;5. [DOI] [PMC free article] [PubMed]
- 27.Steele MI, Kwong WK, Whiteley M, Moran NA. Diversification of type VI secretion system toxins reveals ancient antagonism among bee gut microbes. mBio. 2017;8. [DOI] [PMC free article] [PubMed]
- 28.Zheng H, Perreau J, Powell JE, Han B, Zhang Z, Kwong WK, et al. Division of labor in honey bee gut microbiota for plant polysaccharide digestion. Proc Natl Acad Sci USA. 2019;116:25909–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Motta EVS, Moran NA. The honeybee microbiota and its impact on health and disease. Nat Rev Microbiol. 2024;22:122–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raymann K, Shaffer Z, Moran NA. Antibiotic exposure perturbs the gut microbiota and elevates mortality in honeybees. PLoS Biol. 2017;15:e2001861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng H, Powell JE, Steele MI, Dietrich C, Moran NA. Honeybee gut microbiota promotes host weight gain via bacterial metabolism and hormonal signaling. Proc Natl Acad Sci USA. 2017;114:4775–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kwong WK, Mancenido AL, Moran NA. Immune system stimulation by the native gut microbiota of honey bees. R Soc open sci. 2017;4:170003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horak RD, Leonard SP, Moran NA. Symbionts shape host innate immunity in honeybees. Proc Biol Sci. 2020;287:20201184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steele MI, Motta EVS, Gattu T, Martinez D, Moran NA. The gut microbiota protects bees from invasion by a bacterial pathogen. Microbiol Spectr. 2021;9:e00394–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller DL, Smith EA, Newton ILG. A bacterial symbiont protects honey bees from fungal disease. mBio. 2021;12:e00503–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang J, Lang H, Zhang W, Zhai Y, Zheng L, Chen H, et al. Stably transmitted defined microbial community in honeybees preserves Hafnia alvei inhibition by regulating the immune system. Front Microbiol. 2022;13:1074153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lang H, Duan H, Wang J, Zhang W, Guo J, Zhang X et al. Specific strains of honeybee gut Lactobacillus stimulate host immune system to protect against pathogenic Hafnia alvei. Microbiol Spectr. 2022;10:e01896–21. [DOI] [PMC free article] [PubMed]
- 38.Kešnerová L, Mars RAT, Ellegaard KM, Troilo M, Sauer U, Engel P. Disentangling metabolic functions of bacteria in the honey bee gut. PLoS Biol. 2017;15:e2003467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bonilla-Rosso G, Engel P. Functional roles and metabolic niches in the honey bee gut microbiota. Curr Opin Microbiol. 2018;43:69–76. [DOI] [PubMed] [Google Scholar]
- 40.Winston ML. The biology of the honey bee. Cambridge, MA: Harvard University Press; 1987. [Google Scholar]
- 41.Tarpy DR, Mattila HR, Newton ILG. Development of the honey bee gut microbiome throughout the queen-rearing process. Appl Environ Microbiol. 2015;81:3182–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kapheim KM, Rao VD, Yeoman CJ, Wilson BA, White BA, Goldenfeld N, et al. Caste-specific differences in hindgut microbial communities of honey bees (Apis mellifera). PLoS ONE. 2015;10:e0123911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Copeland DC, Ricigliano VA, Mott BM, Kortenkamp OL, Erickson RJ, Gorrochategui-Ortega J, et al. A longitudinal study of queen health in honey bees reveals tissue specific response to seasonal changes and pathogen pressure. Sci Rep. 2024;14:8963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Copeland DC, Anderson KE, Mott BM. Early queen development in honey bees: social context and queen breeder source affect gut microbiota and associated metabolism. Microbiol Spectr. 2022;10:e00383–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anderson KE, Ricigliano VA, Mott BM, Copeland DC, Floyd AS, Maes P. The queen’s gut refines with age: longevity phenotypes in a social insect model. Microbiome. 2018;6:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Caesar L, Rice DW, McAfee A, Underwood R, Ganote C, Tarpy DR, et al. Metagenomic analysis of the honey bee queen microbiome reveals low bacterial diversity and Caudoviricetes phages. mSystems. 2024;9:e01182–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Snodgrass RE. Anatomy of the honey bee. Cornell University Press; 1956.
- 48.Shi YY, Huang ZY, Zeng ZJ, Wang ZL, Wu XB, Yan WY. Diet and cell size both affect queen-worker differentiation through DNA methylation in honey bees (Apis mellifera, Apidae). PLoS ONE. 2011;6:e18808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Crailsheim K. The protein balance of the honey bee worker. Apidologie. 1990;21:417–29. [Google Scholar]
- 50.De Souza DA, Kaftanoglu O, De Jong D, Page RE, Amdam GV, Wang Y. Differences in the morphology, physiology and gene expression of honey bee queens and workers reared in vitro versus in situ. Biol Open. 2018;7:bio036616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Linksvayer TA, Kaftanoglu O, Akyol E, Blatch S, Amdam GV, Page RE Jr. Larval and nurse worker control of developmental plasticity and the evolution of honey bee queen–worker dimorphism. J Evol Biol. 2011;24:1939–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Souza DAD, Wang Y, Kaftanoglu O, Jong DD, Amdam GV, Gonçalves LS, et al. Morphometric identification of queens, workers and intermediates in in vitro reared honey bees (Apis mellifera). PLoS ONE. 2015;10:e0123663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jarriault D, Mercer AR. Queen mandibular pheromone: questions that remain to be resolved. Apidologie. 2012;43:292–307. [Google Scholar]
- 54.Bortolotti L, Costa C. Chemical communication in the honey bee society. In: Mucignat-Caretta C, editor. Neurobiology of Chemical Communication. Boca Raton (FL): CRC Press/Taylor & Francis; 2014. Chapter 5. [PubMed] [Google Scholar]
- 55.Butler CG, Fairey EM. The role of the queen in preventing oogenesis in worker honeybees. J Apic Res. 1963;2:14–8. [Google Scholar]
- 56.Hoover SER, Keeling CI, Winston ML, Slessor KN. The effect of queen pheromones on worker honey bee ovary development. Naturwissenschaften. 2003;90:477–80. [DOI] [PubMed] [Google Scholar]
- 57.Cardoso-Júnior C a M, Oldroyd BP, Ronai I. Vitellogenin expression in the ovaries of adult honeybee workers provides insights into the evolution of reproductive and social traits. Insect Mol Biol. 2021;30:277–86. [DOI] [PubMed] [Google Scholar]
- 58.Ronai I, Oldroyd BP, Vergoz V. Queen pheromone regulates programmed cell death in the honey bee worker ovary. Insect Mol Biol. 2016;25:646–52. [DOI] [PubMed] [Google Scholar]
- 59.Duncan EJ, Hyink O, Dearden PK. Notch signalling mediates reproductive constraint in the adult worker honeybee. Nat Commun. 2016;7:12427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aamidor SE, Cardoso-Júnior CAM, Harianto J, Nowell CJ, Cole L, Oldroyd BP, et al. Reproductive plasticity and oogenesis in the queen honey bee (Apis mellifera). J Insect Physiol. 2022;136:104347. [DOI] [PubMed] [Google Scholar]
- 61.Lin H, Winston ML. The role of nutrition and temperature in the ovarian development of the worker honey bee (Apis mellifera). Can Entomol. 1998;130:883–91. [Google Scholar]
- 62.Girard M, Luis P, Valiente Moro C, Minard G. Crosstalk between the microbiota and insect postembryonic development. Trends Microbiol. 2023;31:181–96. [DOI] [PubMed] [Google Scholar]
- 63.Powell JE, Lau P, Rangel J, Arnott R, Jong TD, Moran NA. The microbiome and gene expression of honey bee workers are affected by a diet containing pollen substitutes. PLoS ONE. 2023;18:e0286070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Singh RK, Chang H-W, Yan D, Lee KM, Ucmak D, Wong K, et al. Influence of diet on the gut microbiome and implications for human health. J Transl Med. 2017;15:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Youngblut ND, Reischer GH, Walters W, Schuster N, Walzer C, Stalder G, et al. Host diet and evolutionary history explain different aspects of gut microbiome diversity among vertebrate clades. Nat Commun. 2019;10:2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang H, Liu C, Liu Z, Wang Y, Ma L, Xu B. The different dietary sugars modulate the composition of the gut microbiota in honeybee during overwintering. BMC Microbiol. 2020;20:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Engl T, Kaltenpoth M. Influence of microbial symbionts on insect pheromones. Nat Prod Rep. 2018;35:386–97. [DOI] [PubMed] [Google Scholar]
- 68.Sarkar A, Harty S, Johnson KV-A, Moeller AH, Carmody RN, Lehto SM, et al. The role of the microbiome in the neurobiology of social behaviour. Biol Rev Camb Philos Soc. 2020;95:1131–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ezenwa VO, Williams AE. Microbes and animal olfactory communication: where do we go from here? BioEssays. 2014;36:847–54. [DOI] [PubMed] [Google Scholar]
- 70.Powell JE, Eiri D, Moran NA, Rangel J. Modulation of the honey bee queen microbiota: effects of early social contact. PLoS ONE. 2018;13:e0200527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yadav S, Kumar Y, Jat BL. Honeybee: diversity, castes and life cycle. In: Omkar, editor. Industrial Entomology. Singapore: Springer; 2017. pp. 5–34. [Google Scholar]
- 72.Severson DW, Williamson JL, Aiken JM. Caste-specific transcription in the female honey bee. Insect Biochem. 1989;19:215–20. [Google Scholar]
- 73.Chen X, Hu Y, Zheng H, Cao L, Niu D, Yu D, et al. Transcriptome comparison between honey bee queen- and worker-destined larvae. Insect Biochem Mol Biol. 2012;42:665–73. [DOI] [PubMed] [Google Scholar]
- 74.Huang Z-Y, Robinson GE. Regulation of honey bee division of labor by colony age demography. Behav Ecol Sociobiol. 1996;39:147–58. [Google Scholar]
- 75.Herb BR, Wolschin F, Hansen KD, Aryee MJ, Langmead B, Irizarry R, et al. Reversible switching between epigenetic states in honeybee behavioral subcastes. Nat Neurosci. 2012;15:1371–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liberti J, Frank ET, Kay T, Kesner L, Monié--Ibanes M, Quinn A, et al. Gut microbiota influences onset of foraging-related behavior but not physiological hallmarks of division of labor in honeybees. mBio. 2024;0:e01034–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pankiw T, Winston ML, Slessor KN. Queen attendance behavior of worker honey bees (Apis mellifera L.) that are high and low responding to queen mandibular pheromone. Ins Soc. 1995;42:371–8. [Google Scholar]
- 78.Cabirol A, Chhun A, Liberti J, Kesner L, Neuschwander N, Schaerli Y, et al. Fecal transplant allows transmission of the gut microbiota in honey bees. mSphere. 2024;0:e00262–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Anderson KE, Copeland DC. The honey bee hive microbiota: meta-analysis reveals a native and aerobic microbiota prevalent throughout the social resource niche. Front Bee Sci. 2024;2.
- 80.Anderson KE, Maes P. Social microbiota and social gland gene expression of worker honey bees by age and climate. Sci Rep. 2022;12:10690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dalenberg H, Maes P, Mott B, Anderson KE, Spivak M. Propolis envelope promotes beneficial bacteria in the honey bee (Apis mellifera) mouthpart microbiome. Insects. 2020;11:453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Asama T, Arima T-H, Gomi T, Keishi T, Tani H, Kimura Y, et al. Lactobacillus kunkeei YB38 from honeybee products enhances IgA production in healthy adults. J Appl Microbiol. 2015;119:818–26. [DOI] [PubMed] [Google Scholar]
- 83.Corby-Harris V, Snyder LA, Schwan MR, Maes P, McFrederick QS, Anderson KE. Origin and effect of alpha 2.2 acetobacteraceae in honey bee larvae and description of parasaccharibacter apium gen. nov., sp. nov. Appl Environ Microbiol. 2014;80:7460–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dong TS, Gupta A. Influence of early life, diet, and the environment on the microbiome. Clin Gastroenterol Hepatol. 2019;17:231–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mallott EK, Amato KR. Host specificity of the gut microbiome. Nat Rev Microbiol. 2021;19:639–53. [DOI] [PubMed] [Google Scholar]
- 86.Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Archie EA, Tung J. Social behavior and the microbiome. Curr Opin Behav Sci. 2015;6:28–34. [Google Scholar]
- 88.Raymann K, Coon KL, Shaffer Z, Salisbury S, Moran NA. Pathogenicity of serratia marcescens strains in honey bees. mBio. 2018;9. [DOI] [PMC free article] [PubMed]
- 89.Evans JD, Schwarz RS, Chen YP, Budge G, Cornman RS, De La Rua P, et al. Standard methods for molecular research in Apis mellifera. J Apic Res. 2013;52:1–54. [Google Scholar]
- 90.Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37:852–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: high-resolution sample inference from illumina amplicon data. Nat Methods. 2016;13:581–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R. UniFrac: an effective distance metric for microbial community comparison. ISME J. 2011;5:169–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bisanz JE. qiime2R: importing QIIME2 artifacts and associated data into R sessions. Version 099. 2018;13.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequencing files generated and analyzed during the current study are available on the NCBI SRA repository under BioProject PRJNA1157463. All other data generated or analyzed during this study are included in this published article and its supplementary information files.