Abstract
Understanding the interplay between genetic drift, natural selection, gene flow, and demographic history in driving phenotypic and genomic differentiation of insular populations can help us gain insight into the speciation process. Comparing patterns across different insular taxa subjected to similar selective pressures upon colonizing oceanic islands provides the opportunity to study repeated evolution and identify shared patterns in their genomic landscapes of differentiation. We selected four species of passerine birds (Common Chaffinch Fringilla coelebs/canariensis, Red-billed Chough Pyrrhocorax pyrrhocorax, House Finch Haemorhous mexicanus and Dark-eyed/island Junco Junco hyemalis/insularis) that have both mainland and insular populations. Changes in body size between island and mainland populations were consistent with the island rule. For each species, we sequenced whole genomes from mainland and insular individuals to infer their demographic history, characterize their genomic differentiation, and identify the factors shaping them. We estimated the relative (Fst) and absolute (dxy) differentiation, nucleotide diversity (π), Tajima’s D, gene density and recombination rate. We also searched for selective sweeps and chromosomal inversions along the genome. All species shared a marked reduction in effective population size (Ne) upon island colonization. We found diverse patterns of differentiated genomic regions relative to the genome average in all four species, suggesting the role of selection in island-mainland differentiation, yet the lack of congruence in the location of these regions indicates that each species evolved differently in insular environments. Our results suggest that the genomic mechanisms involved in the divergence upon island colonization—such as chromosomal inversions, and historical factors like recurrent selection—differ in each species, despite the highly conserved structure of avian genomes and the similar selective factors involved. These differences are likely influenced by factors such as genetic drift, the polygenic nature of fitness traits and the action of case-specific selective pressures.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12862-024-02320-4.
Keywords: Comparative genomics, Island rule, Parallel evolution, Speciation
Introduction
The colonization of oceanic islands by mainland individuals has been a major engine of biological diversification, resulting in the evolution of thousands of new plant and animal species across the world [1–5]. These colonization events have also provided valuable research models to study processes like evolutionary divergence and local adaptation [6–8]. Upon colonization of oceanic islands, individuals across taxonomic groups have often been subjected to similar demographic and selective factors, like founder effects, population bottlenecks, strong selection for local adaptation, and reduced dispersal [9, 10], resulting in examples of repeated evolution (e.g., [11, 12]). The concept of repeated evolution involves the evolution of similar traits in response to similar environmental pressures, and it encompasses processes like parallel and convergent evolution. Parallel evolution occurs when the ancestral state prior to selection is similar in both populations or taxa, whereas convergent evolution occurs when the ancestral state is different [13].
The molecular basis of similar phenotypic traits across species could be entirely species-specific, or instead show evidence of repeated evolution among species. The degree of repeated evolution at the molecular level can range from sharing the same mutation on the same gene, to changes at different nucleotides within the same gene, to changes in different genes within the same pathway [14, 15]. The probability of repeated molecular evolution is determined by several factors, increasing when selective pressures are similar and genomic constraints such as demography and phylogenetic history are shared [16]. The genetic architecture of the phenotypic traits under selection is also important: single-locus traits have been often involved in repeated evolution (e.g., [17, 18]), yet for polygenic traits, which can be modified through multiple pathways, repeated molecular evolution becomes less likely [16, 19, 20] resulting instead in heterogeneous, species-specific patterns of genetic differentiation. Therefore, when dealing with polygenic traits, it is more likely to observe repeated phenotypic evolution achieved by modifying different loci that are involved in similar functions [21, 22]. In insular environments, the effect of genetic drift and founder effects that reduce genetic diversity randomly, likely limits the number of loci selection can act on [23]. This, added to the fact that a smaller effective population size reduces the efficacy of selection, results in insular populations generally having a reduced adaptive potential compared to their mainland counterparts, constraining the possibilities of observing parallel molecular evolution in insular environments [24].
Understanding the factors that generate heterogeneous patterns of differentiation across the genome is one of the main goals of population genomics [25–29]. When comparing differentiated populations, regions that are highly divergent relative to the genomic background are known as “islands of differentiation” [30, 31] and are usually detected as regions of high relative divergence (Fst [32]). Recent advances in sequencing technologies have allowed studying the genomic landscapes of variation, which show the distributional pattern of genomic variation across the entire genome [23, 31, 33, 34]. The main factors shaping differentiation patterns are drift and selection, but demographic history and genomic features such as gene content and recombination and mutation rate, also affect the distribution of the differentiated regions [27]. Early genome scans interpreted Fst peaks as signatures of strong selection surrounded by valleys homogenized by gene flow [35], where those Fst peaks were caused by marked differences in allele frequencies at locally adapted sites and the neutral loci linked to them [36, 37]. However, when considering patterns of absolute divergence (dxy) and within-population diversity (π) besides Fst, new interpretations of how these islands of differentiation originate have been put forward. Fst peaks could also appear when population diversity is low in either of the populations compared, while dxy is less affected by this pattern. Several processes such as positive and/or background selection can reduce within population nucleotide diversity and generate “islands” of relative divergence, while absolute divergence remains unchanged [25, 38, 39]. Four theoretical models have been proposed to explain the underlying cause of islands of differentiation [39, 40] and to identify each one it is crucial to understand the relationship between Fst, dxy and π [25, 39–41]. Two of those models account for speciation in the presence of gene flow (“Divergence-with-gene-flow” and “Sweep-before‐differentiation”) and the other two involve allopatric speciation (“Selection in allopatry” and “Recurrent selection”) [40]. In the island colonization scenario, both speciation in the presence of gene flow and speciation in allopatry could potentially occur, yet in oceanic islands allopatric models are more likely. Thus, to correctly interpret the genomic landscapes of differentiation it is important to understand the demographic and evolutionary history of the target taxa [27]. Moreover, variations in effective population size (Ne) can produce different genomic signatures. For instance, marked reductions in Ne such as those caused by population bottlenecks at founder events, can modify levels of background selection and therefore affect the baseline for the detection of outlier loci [24, 42].
Covariation of genomic patterns of differentiation among different avian species has been shown across broad evolutionary timescales [43–46] and the coincident location of differentiation peaks has been of special interest to understand the process of repeated molecular evolution where similar loci evolve independently in several species [47]. Bird genomes show high synteny [48], a relatively stable number of chromosomes [49], similar recombination landscapes [50, 51], and across species microchromosomes show higher density in gene content than macrochromosomes [50, 52]. The similarity in genomic landscapes of differentiation across closely related and recently diverged avian taxa could be attributed to a combination of factors. These include the non-random distribution of gene content across the genome and the coincidence of low recombination areas, along with linked selection, which may lead to the clustering of genes [44, 49]. Previous studies have demonstrated that the recombination landscape in birds can remain consistent across species over long evolutionary time periods [50].
To better understand the genomic underpinnings of phenotypic evolution, it is crucial to consider the role of chromosomal rearrangements, such as inversions. Inversions can keep sets of adaptive alleles together in strong linkage disequilibrium, promoting the maintenance of locally adapted genomic regions. Inversions can in some instances facilitate and accelerate the parallel adaptive process by making effective selection stronger [53]. The importance of inversions in repeated evolution has been demonstrated in various taxa (e.g., [54, 55]), and their role in avian evolution is receiving increased attention [56, 57], yet their role in the context of island colonization remains underexplored (but see [58]).
Repeated phenotypic changes on islands are often driven by similar selective pressures related to their unique features compared to mainland environments, such as simplified ecosystems, reduced trophic resources, the availability of new ecological niches, decreased predation (which can lead to increased intraspecific competition), and reduced interspecific competition [7, 59]. These insular selective pressures typically result in predictable changes, collectively known as the island syndrome [60, 61]. This syndrome includes modifications in body size [62], often attributed to the absence of predators and shifts in competition, as well as dietary changes to adapt to new trophic resources, leading to behavioral [63, 64], morphological [65, 66], and physiological adaptations [67, 68]. The consistent patterns of phenotypic evolution observed in insular populations across various taxonomic groups have given rise to general biogeographic rules, such as Foster’s rule, also known as the “island rule,” which posits that small animals tend to become larger and large animals tend to become smaller on islands [62, 69, 70].
These patterns suggest the potential for repeated evolutionary processes across species, providing an opportunity to investigate whether the selective mechanisms during island colonization are shared among species and whether selection targets the same or different genomic loci. However, newly established island populations often face significant genetic challenges due to founder effects and genetic drift, which can reduce their adaptive potential [24]. Small founding populations and random chance often lead to a substantial reduction in the genetic pool of colonizers compared to the source population [71]. The extent and persistence of this reduced diversity are influenced by several factors: the size of the founding population, the level of isolation, the number of colonization events, and the time elapsed since the initial migration, which allows for potential renewal of diversity through mutations and gene flow [72, 73]. Additionally, small effective population sizes, successive founder events, and frequent bottlenecks can further magnify the effects of genetic drift, leading to the rapid loss of genetic diversity [20, 74]. Understanding the genomic underpinnings of divergence in oceanic islands is complex due to the concurrent occurrence of multiple genomic processes. Nevertheless, recent advances in high-throughput DNA sequencing are enabling more studies to address this issue (reviewed in [15]). Both selection and drift play roles in influencing phenotypic changes in island populations, but patterns of repeated phenotypic change are more likely to be driven by selection rather than by random drift [16, 75].
Here we use a comparative approach to examine patterns of genome-wide differentiation in avian species that have colonized oceanic islands, with the goal of exploring if similar patterns of demographic history, time of divergence, and the effects of drift and directional selection in driving divergence could result in repeated molecular evolution upon island colonization. We selected four passerine species that have mainland populations and have also colonized oceanic islands; two species from mainland Europe that have colonized the island of La Palma in the Canary Islands, Atlantic Ocean, the Common Chaffinch (Fringilla coelebs/canariensis) and the Red-billed Chough (Pyrrhocorax pyrrhocorax), and two species from North America that have colonized Guadalupe Island on the Pacific Ocean, the House Finch (Haemorhous mexicanus) and the Dark-eyed Junco (Junco hyemalis/insularis). Given the considerable distance separating insular and mainland populations, and the fact that they are already recognized as separate subspecies or species, we can assume that the insular and mainland populations of these species are mainly allopatric with very restricted gene flow. The Red-billed Chough and the House Finch have diverged from mainland populations within the last 100,000 years, whereas the Common Chaffinch and the Junco have been separated from their mainland relatives for over 500,000 years [76–78]. Divergence times were estimated using mitochondrial DNA for all species, plus microsatellites for the Red-billed Chough and SNPs for the Common Chaffinch. Given that all four species have colonized oceanic islands and have been subjected to potentially similar selective pressures, we first analyzed if the differences in morphology between insular and mainland counterparts affected the same traits across species, as specific changes in morphological traits are expected upon colonization of the new insular environment [4, 10].
We also asked if the genomic landscapes of differentiation are similar among species with different divergence times between insular and mainland counterparts. We performed whole-genome resequencing of 9–12 individuals per treatment (i.e., island/mainland) per species to determine whether the four species showed similar patterns of differentiation in their genomic landscapes, and whether these patterns have been shaped by similar processes. We studied the demographic history and performed genomic scans of Fst, dxy, π, Tajima’s D, recombination rate, gene content and selective sweeps. We also scanned the genomes looking for putative chromosomal inversions, which have been shown to underlie repeated evolution in birds [79]. We detected regions under selection among insular and mainland counterparts as Fst outliers, and identified shared candidate genes among the four species. Comparing the genomic signatures of island colonization in four different species that have been exposed to similar selective pressures and that differ in colonization/divergence time (which can be considered as a proxy for different stages along the speciation continuum due to reproductive isolation in allopatry), can provide useful understanding for the mechanisms shaping the genomic landscapes through the divergence process over time.
Methods
Study area and fieldwork
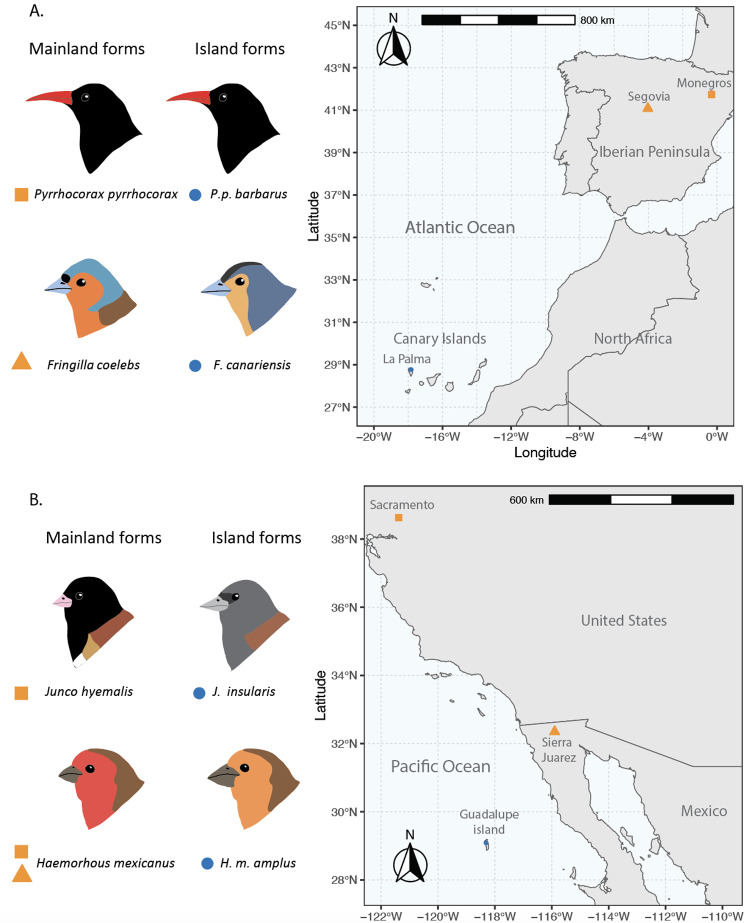
We sampled mainland populations of the Common Chaffinch (Fringillidae: Fringilla coelebs) and the Red-billed Chough (Corvidae: Pyrrhocorax pyrrhocorax) in the Iberian Peninsula at Segovia and Los Monegros, respectively (see [77, 78]). The insular populations from both species were sampled in La Palma, the most north-western island of the Canary Islands archipelago (Fig. 1A, Table S1). The Common Chaffinch lineage in the Canary Islands has recently been raised to species status [80], and we use its current name, Canary Islands Chaffinch (Fringilla canariensis). The mainland populations of the House Finch (Fringillidae: Haemorhous mexicanus) and Dark-eyed Junco (Passerellidae: Junco hyemalis oreganus) were sampled in California, and two House Finch individuals were sampled in Sierra Juarez (Baja California, Mexico). Insular populations for both species were sampled in Guadalupe Island, Mexico, in the Pacific Ocean (Fig. 1B, Table S1). The Junco on Guadalupe Island, until recently a subspecies of J. hyemalis, has been raised to species status, and we use its current name, Island Junco (Junco insularis).
Fig. 1.
Target taxa for comparative analysis. (A) Species that have colonized La Palma in the Atlantic Ocean: Red-billed Chough and Common Chaffinch. (B) Species that have colonized Guadalupe island in the Pacific Ocean: Dark-eyed Junco and House Finch. Nomenclature according to Clements et al., [178]. The shape and colors of markers next to the names correspond to the sampling locations on the map, with orange rectangles and triangles for the continental species and blue circles for the insular taxa
All individuals were captured in the field using mist nets and mesh traps in the case of Red-billed Choughs. All individuals were marked with uniquely numbered aluminum bands, sexed, aged and measured in the field. A blood sample was obtained by venipuncture of the sub-brachial vein and stored in absolute ethanol at -20 °C in the laboratory for DNA extraction. After processing, birds were released unharmed at the site of capture. As the red-billed chough was the only non-dimorphic species, we determined its sex by the amplification of the Chd1 gene following Griffiths et al. [81].
Morphological data and analysis
Morphological traits in adult males from mainland and insular populations were measured across all species. For the Common Chaffinch, the Junco and the House Finch a wing ruler was used to measure unflattened wing length to the nearest 0.5 mm, and dial callipers of 0.1-mm precision were used to measure tail length, tarsus length, bill culmen, total bill length, bill width and bill depth, following Milá et al. [82]. All measurements were taken by a single observer (BM). For the Red-billed Chough, the same traits were measured by a single observer (GB) following standard methods described previously [83]. We compared the morphological traits of adult males from mainland and insular populations for all species using principal components analysis (PCA) of all variables and univariate analysis of variance (ANOVA) to compare the means among treatments for each species. The PCA including all morphological variables was computed using the prcomp function in stats v. 3.6.2 R package.
Genome resequencing
Genomic DNA was extracted with a QIAGEN Blood and Tissue kit following the manufacturer’s protocol. Whole genome resequencing at 18x coverage of 24 individuals per species (12 per treatment, but only 9 for the mainland Common Chaffinch) was conducted on a SE50 Illumina™ platform at Novogene™. Reads were trimmed with Trim Galore [84] with default settings and a minimum length of 40 and then mapped to their respective reference genomes using BWA (Burrows-Wheeler Aligner [85]. For the Common Chaffinch and the House Finch we used the Common Chaffinch reference genome (GCA_015532645.2 [86]); for the Junco we used the Junco hyemalis reference genome (GCA_003829775.1 [87]); and for the Red-billed Chough we used the Corvus moneduloides reference genome (GCA_009650955.1, bCorMon1.pri). The Maximum likelihood genetic distance between the House Finch and the Common Chaffinch is 0.12 (16.5 Mya) [88] and between the Red-billed Chough and the Corvus moneduloides is around 5.9-8 Mya [89]. SNPs were called using BCFTOOLS v.1.3.1 [90] including invariant sites. Filtering was performed with VCFTOOLS v. 0.1.15 [91] separately for variant and invariant sites, using the following criteria for variant sites: (i) indels and sites with more than two alleles were removed; (ii) a number of reads per site between 10 and 40; (iii) a minimal genotype quality of 30; (iv) a minor allele count of 2 (--mac 2); and (v) 25% maximum missing data and for invariant sites a minimal genotype quality of 30. Variant and invariant sites were then merged using BCFTOOLS concat. The reference genomes from all four species were aligned to the zebra finch genome (Taeniopygia guttata, bTaeGut2.pat.W.v2) using nucmer from the MUMmer package (v.4.0, ‘-b 400’ and filtering with ‘delta-filter − 1’); [92] and chromosomes were numbered accordingly (see Table S2, Fig. S1).
Inference of demographic history
The change in effective population size (Ne) across time for each species was estimated using Pairwise Sequentially Markovian Coalescent (PSMC) analysis [93]. The PSMC model infers demographic history based on genome-wide heterozygous sequence data. We used SAMTOOLS [94] to obtain diploid consensus sequences from BAM files generated with BWA-mem [85]. Sites with sequencing depth lower than 10 and higher than 35 were removed. Because sex chromosomes can show different rates and patterns of evolution than autosomes (reviewed by [39, 95]), we focused our comparisons of differentiation statistics on autosomes only. We converted the diploid consensus sequence to PSMC input files (psmcfa) using the tool fq2psmcfa included in the PSMC software. Then, the program PSMC was used to infer the population history with the options ‘-N25 –t5 –r1 –p “4 + 30*2 + 4 + 6 + 10’, except for the mainland Common Chaffinch, and for both populations of the House Finch, where the upper time limit was set to 1 (-t1) to achieve convergence. We performed 100 bootstraps for one genome per species and treatment. The atomic time interval was set following Nadachowska-Brzyska et al., [96]. We used a mutation rate of 4.6 × 10− 9 mutations/site/generation [97], which has been used in other avian systems for PSMC analysis (e.g., [66, 98–100]). Generation time was set to two years for all species [101–103]. We also performed a test setting the generation time to three years for the Red-billed Chough and to one year for the Common Chaffinch, the Junco and the House Finch and to explore how this variability would affect divergence time estimates (Fig. S3).
Inference of recombination rate
To determine the effect of recombination rate on the genomic landscapes of differentiation, we estimated recombination rates across the genome for insular and continental populations for the four species using LDhat software [104]. First, we created a modified likelihood lookup table based on the LDhat precomputed tables using a sample size of 12 per treatment (9 for the continental Common Chaffinch) and a population mutation rate parameter estimate of 0.001. Then vcf files were split into chunks of 10,000 SNPs and converted to ldhat format using VCFTOOLS v. 0.1.15 [91]. The input files generated were used in LDhat “interval” to estimate the effective recombination rate by implementing a Bayesian MCMC sampling algorithm with five million iterations, sampling every 5,000 steps and a block penalty of 10. Finally, the results were summarized using the LDhat module “stat”, discarding 20% of the samples as burn-in.
Genome scans and detection of selective sweeps
In order to detect genomic signatures of selection among the island and mainland counterparts from the four different species, we estimated two different statistics, the fixation index (Fst [32]), and the cross-population extended haplotype homozygosity (XP-EHH) [105]. First, Fst, dxy and π using were calculated in non-overlapping windows of 10Kb using pixy v. 2 [106]. Pixy takes into account the invariant sites for π and dxy calculations, thus overcoming the problem of most programs that use VCF files to calculate those statistics but do not distinguish among invariant and missing sites, resulting in deflated estimates [106]. We also computed Tajima’s D [91, 107] in non-overlapping 10-Kb windows with VCFTOOLS [91, 108]. To detect outliers for all variables, first the averaged values of each variable were transformed to Z-scores using the scale command in R and then the p-value was calculated with the pnorm function to correct for multiple testing setting the false discovery rate (FDR) to 0.05 [108].
To detect selective sweeps, we computed the cross-population extended haplotype homozygosity (XP-EHH) [105], using the R package rehh v.3.2.2 [109]. First, we phased the vcf files containing only the variant sites in 50-Kb windows using Shapeit v2.r904 [110]. The XP-EHH is based on the comparison of haplotype lengths between populations and has most detection power when the selected haplotype is near fixation in one population and still polymorphic in the other. The genomic regions showing a –log10(p-value) ≥ 3 (i.e., p ≤ 0.001) were considered to be under selection. Then, we looked for overlapping regions between the Fst and the XP-EHH outliers. We generated Manhattan plots for all the statistics using the R package qqman [111] in R. All R analyses were performed in v. 3.6 [112].
Detecting putative chromosomal inversions
We examined how patterns of population structure varied along the genome to detect potential chromosomal inversions using the R package lostruct v.0.0.4 [113]. This method can help identify putative chromosomal inversions; however, it does not provide definitive evidence of their existence. SNP data for each species including only variant sites was converted to BCF format using BCFTOOLS version 1.9 [94]. We implemented the script provided by Huang et al., [114] dividing the genome into 1,000-SNP non-overlapping windows and applying a principal components analysis (PCA) to each window. Euclidian distances between the two first principal components (PCs) between windows were calculated and mapped using multidimensional scaling (MDS) into a 40-dimensional space to see the similarity of the relatedness patterns between windows. To identify genomic regions with extreme MDS values, windows with absolute values greater than 4 SD over the mean across all windows were selected for each MDS coordinate. We performed 1,000 permutations of windows over chromosomes to test if outlier regions were randomly distributed across chromosomes. The putative inversion coordinates were the start position of the first outlier window and the end position of the last outlier window. The script included additional analyses to check if the MDS outliers were detecting inversions or instead other processes such as linked selection. First, a PCA was performed using the SNPs from each putative inversion with SNPRelate [115]. Inversions in the PCA would split the samples into three different groups (i.e., the two orientations and the heterozygotes in an intermediate cluster). The R function kmeans with the Hartigan & Wong [116] method was used to identify the composition of groups of genotypes by performing clustering on the first PC, setting the initial cluster centers as the maximum, minimum and middle of the PC score range. Then, another test was performed averaging the individual heterozygosity per group detected by the k-means clustering. Inversions would show a pattern of higher heterozygosity of the central group relative to the other two groups. In addition, LD heatmaps of the putative inversion regions were constructed using the LDBlockShow v1.40 [117]. Finally, only MDS outlier regions that clustered into three groups in the PCA and showed higher heterozygosity in the middle group and high LD within the estimated regions were considered as putative inversions.
Candidate genes and GO-term enrichment analysis
We extracted the candidate genes of the genomic regions detected to be under selection by both methods separately (Fst and XP-EHH outliers) using bedtools intersect and the annotation of their respective reference genomes. We checked their functions in genecards [118]. We obtained the GO terms using the Zebra Finch dataset in biomaRt in R. We then performed a Gene ontology (GO) enrichment analysis for each set of outliers in the category “biological function” using the TopGO v.2.50.0 R package [119]. To estimate the statistical significance, we used the Fisher exact test implementing the weight01 method. As recommended by the TopGO authors, we did not implement corrections for multiple testing and presented raw p-values for the top-10 GO terms related to biological processes.
Results
Morphological differences
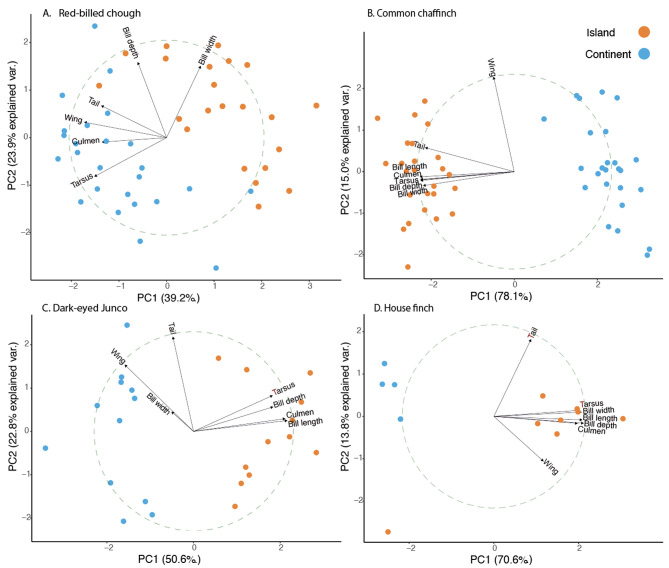
The morphological analysis revealed marked differences in most traits between insular and continental populations for all species. The threshold to consider small and large bird species in the context of the island rule has been shown to be around 60 g [62]. The smaller species (Common Chaffinch, Junco, and House Finch), shared a pattern of significantly larger values for most traits in the insular populations compared to mainland, except for the Junco wing length, which was longer in the continent (Table S3). We detected the opposite pattern in the larger sized Red-billed Chough, with significantly smaller values for most morphological traits in the insular populations, except for bill width which was smaller in the continent (Table S3). PC1 values showed marked separation of insular and mainland populations in all four species, explaining between 39 and 78% of the morphological variance (Fig. 2).
Fig. 2.
Principal Components Analysis (PCA) with morphological data per species A) Red-billed Chough, B) Common/Canary Islands Chaffinch, C) Dark-eyed/island Junco, D) House Finch. The variables included are wing, tail and tarsus length and bill culmen, bill depth, bill width and bill length (the latter is not included for the Red-billed Chough). The correlation circle with radius 1 show the loadings of each variable that are represented by the arrows. Red and blue markers correspond to insular and mainland individuals, respectively
Whole-genome resequencing
The total number of sites obtained in the variant calling was close to the length of the reference genomes. The number of variant sites (40–50 million) was similar for all species except for the Red-billed Chough, which was lower (~ 13 million), and the same pattern was maintained after filtering (Table S4). The lower number of variants of the Red-billed Chough is consistent with the lower ancestral effective population size compared to the other species [120].
Inference of demographic history
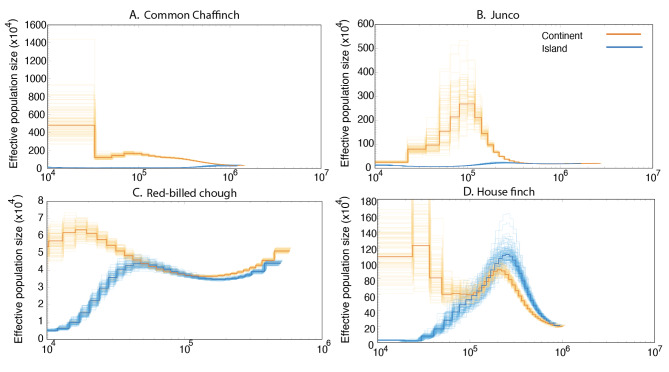
PSMC-based demographic inference revealed a consistent pattern for the four species, showing stable or growing effective population sizes for mainland populations and a sharp reduction in effective population size in insular populations following colonization. The island-mainland divergence time estimates obtained from the PSMC analysis are around 900,000 years for the Common Chaffinch, 100,000 years for the House Finch, 400,000 years for the Dark-eyed Junco, and 30,000 years for the Red-billed Chough (Fig. 3, S2). To account for the variability in generation time, we applied a generation time of three years for the chough and one year for the rest of the species, obtaining different split time estimates but still within the expected values: 70,000 years for the Red-billed Chough, 500,000 years for the Common Chaffinch, 200,000 years for the Dark-eyed Junco, and 80,000 years for the House Finch (Fig. S3). The continental population of the Red-billed Chough showed the smallest effective population size, and the smallest difference between the continental and insular populations among the study species.
Fig. 3.
Demographic history of insular and mainland populations. The analysis was performed using Pairwise Sequentially Markovian Coalescent (PSMC). Demographic inference for one individual per treatment and species, with the orange and blue dark lines corresponding to the continental and insular populations, respectively. Shown are PSMC plots for A) Common Chaffinch, B) Junco, C) Red-billed Chough and D) House Finch. The lighter orange and blue lines represent 100 bootstrap replicates. The point where both lines depart from each other corresponds to the time of colonization, which is around 40,000 y for the Red-billed Chough, 900,000 y for the Common Chaffinch, 100,000 y for the House Finch and 400,000 y for the Dark-eyed Junco. The mutation rate used was of 4.6e-9 mutation/site/generation for all species, and the generation time used in all cases was two years. See Fig. S2 for bootstrapped versions of the individual PSMC plots. See Fig. S3 for PSMC plots with generation time of one year for the Common Chaffinch, Dark-eyed Junco and House Finch and three years for the Red-billed Chough
Inferring parallel molecular evolution from genome-wide scans
Genome-wide scans of genetic differentiation showed high heterogeneity across the four target species. The Fst genomic landscapes varied strongly among species (Figs. 4, 5, 6 and 7). Mean Fst was higher in the Common Chaffinch, followed by the Dark-eyed Junco, as expected for relatively longer island-mainland divergence times. The Red-billed Chough showed a slightly higher mean Fst than the House Finch (Table 1). The Red-billed Chough’s absolute divergence and genetic diversity for insular and mainland populations were the lowest but comparable to the rest, mainly due to a specific region of about 3.4 Mb in chromosome 17 with exceptionally high dxy and genetic diversity. When this region is excluded, both dxy and π drop to an order of magnitude lower than those of the other species (Table 1). The continental Common Chaffinch population showed the highest diversity value (Table 1). All species showed consistently higher gene content and recombination rates at microchromosomes, and in general, recombination rates were higher at chromosome ends (Figs. 4, 5, 6 and 7).
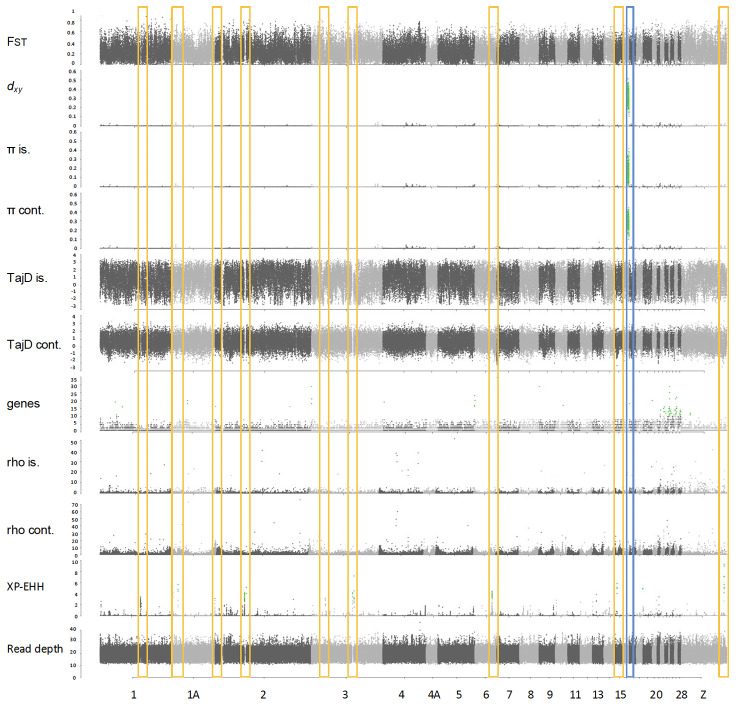
Fig. 4.
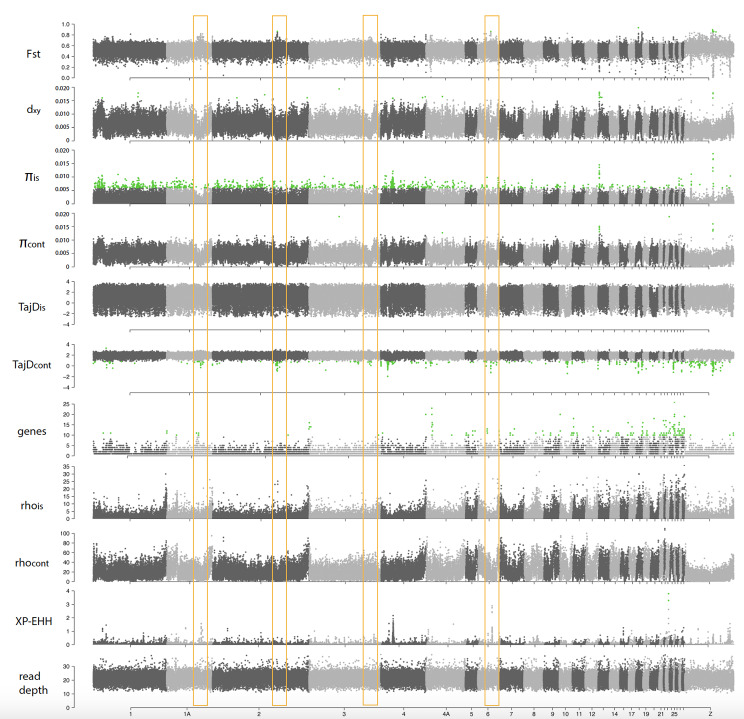
Genomic scans for several summary statistics for an island-mainland comparison in the Red-billed Chough (Pyrrhocorax pyrrhocorax). From top to bottom, fixation index (Fst), genomic divergence (dxy), genetic diversity for insular and continental populations (π), Tajima’s D for insular and continental populations (TajD), number of genes, recombination rates for insular and mainland populations (rho), cross-populations extended haplotype homozygosity (XP-EHH) and read depth. Chromosome numbers correspond to the Zebra Finch genome (Taeniopygia guttata). Green dots represent outliers with the false discovery rate (FDR) set at 0.05 after applying the Benjamini and Hochberg correction, except for the XP-EHH, where the threshold is set at –log10 (p-value) ≥ 3. The yellow boxes highlight the XP-EHH peaks coincident with regions with no coverage. The blue box correspond to the region with high absolute divergence (dxy ) and genetic diversity (π) in chromosome 17
Fig. 5.
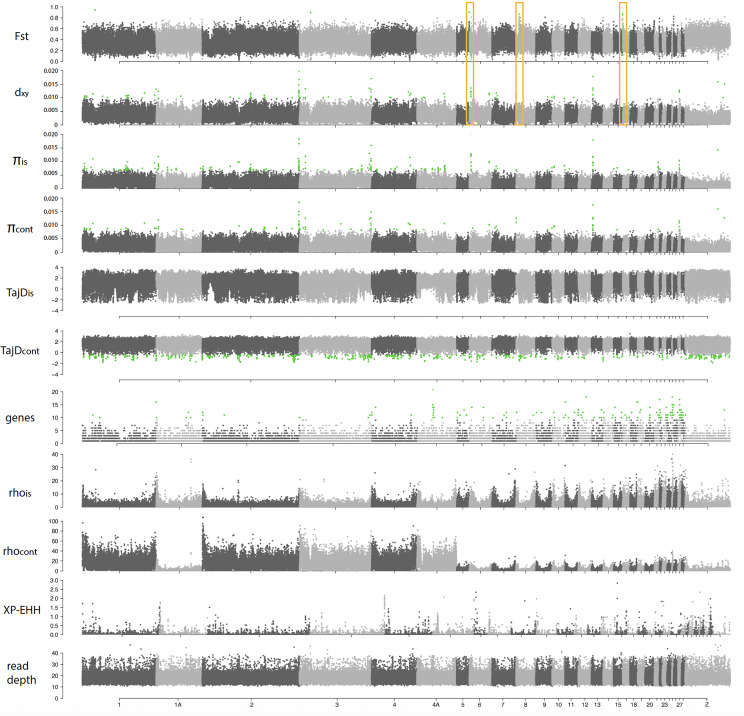
Genomic scans for several summary statistics for an island-mainland comparison in the Common Chaffinch (Fringilla coelebs). From top to bottom, fixation index (Fst), genomic divergence (dxy), genetic diversity for insular and continental populations (π), Tajima’s D for insular and continental populations (TajD), number of genes, recombination rates for insular and mainland populations (rho), cross-populations extended haplotype homozygosity (XP-EHH) and read depth. Chromosome numbers correspond to the Zebra Finch genome (Taeniopygia guttata). Green dots represent outliers with the false discovery rate (FDR) set at 0.05 after applying the Benjamini and Hochberg correction, except for the XP-EHH, where the threshold is set at –log10 (p-value) ≥ 3. The yellow boxes highlight the signatures of recurrent selection (Fst peaks coincident with drops in dxy and π). Some of them are also coincident with peaks in XP-EHH
Fig. 6.
Genomic scans for several summary statistics for an island-mainland comparison in the Dark-eyed Junco (Junco hyemalis). From top to bottom, fixation index (Fst), genomic divergence (dxy), genetic diversity for insular and continental populations (π), Tajima’s D for insular and continental populations (TajD), number of genes, recombination rates por insular and mainland populations (rho), cross-populations extended haplotype homozygosity (XP-EHH) and read depth. Chromosome numbers correspond to the Zebra Finch genome (Taeniopygia guttata). Green dots represent outliers with the false discovery rate (FDR) set at 0.05 after applying the Benjamini and Hochberg correction, except for the XP-EHH, where the threshold is set at –log10 (p-value) ≥ 3. The yellow boxes highlight the Fst peaks in the chromosome extremes
Fig. 7.
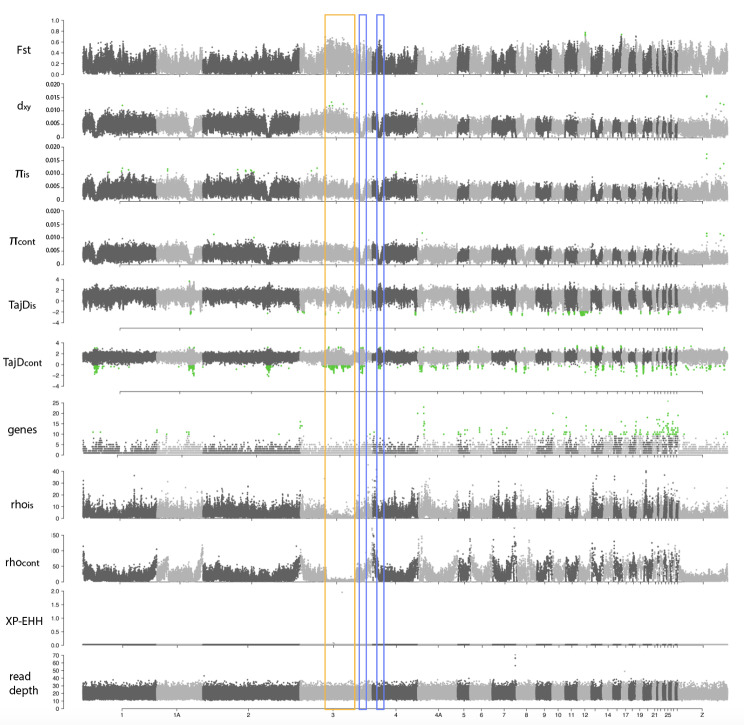
Genomic scans for several summary statistics for an island-mainland comparison in the House Finch (Haemorhous mexicanus). From top to bottom, fixation index (Fst), genomic divergence (dxy), genetic diversity for insular and continental populations (π), Tajima’s D for insular and continental populations (TajD), number of genes, recombination rates por insular and mainland populations (rho), cross-populations extended haplotype homozygosity (XP-EHH) and read depth. Chromosome numbers correspond to the Zebra Finch genome (Taeniopygia guttata). Green dots represent outliers with the false discovery rate (FDR) set at 0.05 after applying the Benjamini and Hochberg correction, except for the XP-EHH, where the threshold is set at –log10 (p-value) ≥ 3. The yellow box highlights the putative inversion in chromosome 3 (Fst peak that coincides with a drop in the recombination rate). The blue boxes highlight the signatures of recurrent selection (Fst peak coincident with drops in dxy and π)
Table 1.
Divergence and diversity across the genome. Mean values, standard deviation (sd) and range of genomic summary statistics for the four species including the Red-billed Chough without the region with high absolute divergence and genetic diversity. The table includes sample sizes for the continental and insular populations (Ncont and Nis), fixation index (Fst), absolute genomic divergence (dxy), and genetic diversity (π) for the insular and the continental populations
| Species | N cont | N is | Fst ± sd | range | dxy ± sd | range | πisland ± sd | range | πcontinent ± sd | range |
|---|---|---|---|---|---|---|---|---|---|---|
| Red-billed Chough | 12 | 12 | 0.24 ± 0.13 | [-0.062–0.89] | 0.002 ± 0.02 | [0–0.54] | 0.001 ± 0.01 | [0–0.42] | 0.002 ± 0.02 | [0–0.44] |
| Red-billed Chough_nopeakchr17 | 12 | 12 | 0.24 ± 0.13 | [-0.062–0.89] | 0.0009 ± 0.0009 | [0–0.065] | 0.0005 ± 0.0008 | [0–0.066] | 0.0008 ± 0.0009 | [0–0.068] |
| House Finch | 12 | 12 | 0.16 ± 0.10 | [-0.06–0.69] | 0.005 ± 0.002 | [0–0.017] | 0.004 ± 0.002 | [0–0.018] | 0.005 ± 0.002 | [0–0.016] |
| Dark-eyed Junco | 12 | 12 | 0.29 ± 0.08 | [-0.060–0.77] | 0.004 ± 0.002 | [0–0.023] | 0.002 ± 0.001 | [0–0.023] | 0.004 ± 0.002 | [0–0.022] |
| Common Chaffinch | 9 | 12 | 0.44 ± 0.05 | [-0.033–0.93] | 0.008 ± 0.003 | [0–0.021] | 0.002 ± 0.001 | [0–0.020] | 0.008 ± 0.003 | [0–0.022] |
The genomic landscape of the Red-billed Chough shows high levels of relative differentiation across the whole genome, with no clear outlier regions. Mean genetic diversity in both populations is generally very low throughout the genome, except for a highly differentiated region on chromosome 17. The XP-EHH analysis indicated evidence of selective sweeps, with a few distinct peaks along the genome (Fig. 4). However, most of these peaks are adjacent to regions of the genome with no coverage, probably corresponding to centromeric/high repeat content regions; and are therefore likely artifacts (Fig. S4).
The Common Chaffinch genome landscape is characterized by several Fst peaks that coincide with valleys in dxy and π, and negative peaks in Tajima’s D mainly in the continent (i.e., peaks in chromosomes 1,1 A, 2, 3, 4, 4 A, 6, Fig. 5). This pattern is consistent with the model of recurrent selection, which states that selection in the ancestral population prior to the mainland-island split generates a pattern of low dxy. Subsequent selection in those regions after divergence further reduces genetic diversity, generating Fst peaks. XP-EHH revealed peaks suggesting selective sweeps mostly concentrated in the microchromosomes and the Z chromosome and few of them coincided with the regions of recurrent selection.
The Dark-eyed Junco genomic landscape is highly differentiated across the entire genome, and there are few outlier genomic regions, which often coincide with chromosomal ends (Fig. 6). The XP-EHH scans did not detect significant selective sweeps across the genome, but there is again a pattern of peaks in the chromosomal ends.
The House Finch genomic landscape is characterized by a large, highly differentiated region in the middle of chromosome 3, representing 47 million base pairs, suggesting a large chromosomal inversion. It coincides with significant positive and negative peaks of Tajima’s D in the continental population and a region of low recombination, while dxy and π show regular values (Fig. 7). At the end of the same chromosome and at the beginning of chromosome 4, there are two Fst peaks that coincide with a valley in dxy and π, and a peak in Tajima’s D. This pattern is consistent with the recurrent selection model. In chromosomes 1, 1 A and 2 there is a similar pattern but, in these cases, there is no Fst peak. The microchromosomes show high relative differentiation along with high recombination rates and enriched gene content. The XP-EHH scan showed a relatively flat landscape with no evidence for significant selective sweeps.
Detecting putative chromosomal inversions
After combining all possible evidence, the analysis to detect inversions revealed that the Red-billed Chough genome has no putative inversions. The Dark-eyed Junco genome showed five possible inverted regions distributed in chromosomes 1, 3 and 5 (Table S5, Fig. S5) and the one in chromosome 1 coincided with an Fst outlier region. The Common Chaffinch genome showed two possible inversions in chromosomes 10 and 20, and the two of them coincided with Fst outlier regions (Table S5, Fig. S6). The House Finch genome revealed four putative inversions, with one particularly large in chromosome 3. (Table S5, Fig. S7). Only two putative inversions coincide with Fst outlier regions, the large inversion in chromosome 3 and a 21 Mb inversion in chromosome Z.
Detection of candidate genes and GO-term enrichment analysis
Sharing of candidate genes among species was limited (Table S10). There were only five genes putatively under selection that were shared between different species: the morc2 gene was shared between the House Finch and the Dark-eyed Junco; and the clic3, paxx, tmem141 and entp2 genes were shared between the House Finch and the Red-billed Chough dxy outliers. The morc2 gene is associated with Marie-Tooth disease, axonal, type 2z (CMT2Z) and developmental delay, impaired growth, dysmorphic facies, and axonal neuropathy (DIGFAN) diseases in humans. CMT2Z is characterized by distal lower limb muscle weakness and sensory impairment [121] and DIFGAN by impaired motor and intellectual development, poor overall growth, usually short body height and microcephaly and subtly dysmorphic facial features in humans [122, 123]. The clic3 gene has been associated with bone formation in humans [124]. The paxx and tmem141 genes are involved in neural development [125, 126]. The entp2 gene has been found to be related to taste transduction in core landbirds [127].
The number of genes included in the Fst outlier regions ranged from 2 to 84 and in all cases except for the Red-billed Chough were higher than the number of genes in the XP-EHH outlier regions (Table 2). Only in the Common Chaffinch there was an overlap of 1 gene between both methods (Table S10). In the Red-billed Chough, due to the high relative differentiation across the genome, the absence of clear Fst peaks, and the potential artifacts in selective sweep detection caused by regions with no coverage, focusing on the peak of absolute genomic differentiation could be a more effective approach to identify candidate regions in this species. For the dxy outliers, the top-10 GO terms are related to the regulation of signaling pathways, ion transport, and protein modifications (Table S6). In the Common Chaffinch, among the top-10 GO terms we found several involved in transmembrane transport, protein modification processes and organization of nuclear and cellular components (Table S7). In the Dark-eyed Junco, the top-10 GO terms revealed several involved in the regulation of cellular and molecular localization including two terms related to the centrosome and related to nervous system development and function (Table S8). Finally, in the House Finch, among the 84 genes identified under selection, 13 were clustered in the putative inversion in the middle region of chromosome 3 (Table S10). Within the top-10 significant GO terms (Table S9) we find terms related with development processes, morphogenesis and regulation of biochemical pathways.
Table 2.
Number of genes detected in the outlier regions detected with Fst and XP-EHH scans (and dxy for the Red-billed Chough) and their overlap. Number of genes available and feasible for the GO enrichment analysis and the number of outlier genes that were included in feasible genes (see Table S10 for genes IDs).
| Species | Fst / dxy Genes | XP-EHH Genes | Overlap | Available Genes | Feasible for GO |
Feasible outliers |
|---|---|---|---|---|---|---|
| Red-billed Chough | 2 / 96 | 8 | 0 | 20,580 | 8,788 | 4 |
| Common Chaffinch | 63 | 8 | 1 | 16,563 | 9,242 | 48 |
| Dark-eyed Junco | 15 | 0 | 0 | 17,038 | 9,410 | 9 |
| House Finch | 84 | 0 | 0 | 16,563 | 9,242 | 46 |
Discussion
Our comparative analysis of mainland and insular populations of four passerine species yielded shared patterns of demographic history and divergence of morphological traits consistent with the island rule, in contrast to species-specific patterns of genome-wide variation in the studied genomic variables. Relative to the mainland, all insular populations showed changes in body size, and suffered reductions in effective population size and genetic diversity, patterns that are consistent with previous findings [24, 62, 71]. While our focus was on body size, given its well-established association to island colonization, it’s important to note that there are other traits, such as plumage color or vocalizations, that showed noticeable differences between populations [76, 78]. The process of insular colonization is usually initiated by a small group of individuals, and the resulting genetic drift, combined with the small size of the island’s geographic area, leads to a small effective population size and low genetic diversity [24, 128]. Among the four species, the Red-billed Chough showed the smallest effective population size in both insular and mainland populations, which corresponds to the lowest levels of genetic diversity. In the mainland, this species has shown marked levels of genetic structure in the absence of geographic barriers, suggesting that social barriers due to complex behavioral interactions may constrain gene flow and thus the effective size of local populations [129]; the insular population is unlikely to be an exception [77].
While our focus was on body size—given its well-established connection to island colonization—it’s important to recognize that body size is a composite of multiple traits, each contributing differently across species, as observed in the varying PCA loadings. Additionally, other phenotypes, such as plumage divergence, also showed noticeable differences between populations. However, due to the complexity of comparing these traits across species, our primary investigation centered on body size, in line with previous findings of reduced effective population size and genetic diversity in insular populations [24, 60, 69].
Phenotypic divergence upon island colonization
Using PC1 and mean differences in tarsus length, as proxies for structural body size in birds [130–133], we found that the three smaller passerines increased in size and the larger species suffered a size reduction upon island colonization. This is consistent with the island rule, which posits that small birds evolve towards a larger size and large birds towards a smaller size upon island colonization [62, 70]. However, the difference in the House Finch tarsus length among insular and mainland populations was not significant probably due to the small sample size. Regarding beak size, we find that insular individuals from the small sized and short-billed species show longer bills than their mainland counterparts whereas the insular population of the long-billed chough species shows a reduction in bill length. All the species show also differences in at least other bill dimension; however, the Red-billed Chough is the only one in which the change is in the opposite direction, showing shorter but wider bills on the island. The beak is both a feeding and thermoregulatory structure with great evolutionary potential that allows birds to quickly adapt to new environmental conditions [134] and therefore plays a fundamental role in avian fitness [135–140].
Differing patterns of genomic divergence
Finding shared patterns of genomic variation and common regions of differentiation at the intra- or inter-specific levels has been of major interest to understand the mechanisms underlying divergence [38, 43, 44]. These shared divergent regions across taxa are particularly interesting when differentiation evolved independently in unrelated lineages [47]. Our comparative analysis of island-mainland populations in four passerine species showed a lack of parallelism in their respective genomic landscapes. In all four species, regions of higher divergence and genetic diversity are located in the microchromosomes, which have relatively higher recombination rates and higher gene content [141]. We also found highly differentiated genomic regions in all four species that were often associated with reduced genetic diversity, which could be the result of drift or/and selection either in the ancestral or the current populations. The lack of congruence in the location of these regions along the genome could indicate that drift likely played a role in the divergence, that different traits are under selection, or that due to the polygenic nature of traits the four species adapted to their respective insular environments in different ways, through genetic changes at different loci. Moreover, patterns of recombination rate in these regions suggest that the genomic mechanisms generating these patterns, which include chromosomal inversions, and historical factors like recurrent selection, differ in each of the four species.
Demographic history and functional genomic analysis
According to our demographic analysis, the divergence between Red-billed Choughs on La Palma and the Iberian Peninsula took place around 30,000 years ago, considering a generation time of two years. A previous study [77] estimated the divergence event in a similar time range, within the last 10,000 years using mitochondrial data and around 30,000 years using iMa2, however they used a generation time of 6 years based on mainland data. If we apply that value, the divergence time estimate changes to around 110,000 years. The Red-billed Chough also shows the smallest effective population size and lowest genetic diversity. This reduced genetic diversity also results in an inflated relative divergence [25, 142], causing a high baseline to detect outliers while the absolute divergence remains low. The scan for selective sweeps, which is more efficient in detecting recent divergence, revealed clear peaks along the genome, yet those peaks are near regions with no coverage and therefore we are considering them to be artifacts. The recent divergence of the Red-billed Chough is evident due to the low divergence across the genome, with a mean dxy value of 0.002. However, this value decreases drastically to 9·10⁻⁴ when the highly divergent region on chromosome 17 is excluded. We hypothesize that this region could represent a neo-sex chromosome because of preliminary sex-related differences found (unpublished data), though further research is required to confirm this. Neo-sex chromosomes have been observed in several bird species within the Sylvioidea superfamily, often resulting from fusions or translocations of autosomes and sex chromosomes [143]. Similarly, a neo-sex chromosome was recently discovered in the genus Zosterops, formed by the fusion of the W chromosome and chromosome 4 A [144]. Confirmation of a neo-sex chromosome in the Red-billed Chough would be important because, to date, no autosome-sex chromosome fusions have been observed within the Corvoidea superfamily. In this study, only males were included, which limits our ability to test this hypothesis further. Among the top ten GO terms of the genes within the absolute divergence peak we find two related with ubiquitination which has been found to be an important signaling mechanism controlling several physiological and pathological processes [145].
The Common Chaffinch of La Palma was found to have diverged from its mainland relatives around 0.8–0.9 my ago, which is in agreement with previous reconstructions of the species evolutionary history [78]. A study of the entire Common Chaffinch radiation across the Atlantic archipelagos revealed that it first colonized Azores, then Madeira and finally the Canary Islands [78]. This sequential colonization of isolated archipelagos has left a genomic signature of recurrent selection along the genome, leading to regions with low absolute divergence due to selection in the ancestor, that were subsequently selected in the daughter populations, reducing genetic diversity and generating Fst peaks [40]. This recurrent-selection model fits well with the known colonization history, as the first selective episode probably occurred upon colonization of the Azores, and then at every subsequent colonization step between islands, where successive selective events at the same genomic regions likely led to a loss of genetic diversity. Among the genes associated with outlier loci there were several involved in metabolism (i.e., kars1, nfrkb), four involved in pigmentation (ap3b1, hps6, ric1 and atrn) [146], and three related to singing (chrm5, mrps27, ube2d3) [147–151]. Within the top-ten significant GO terms we detected “positive regulation of endosome organization” and endosomes play an important role in neural development [151]. We also find the term “regulation of protein localization to adherens junction” and it has been shown that cell adhesion plays an important role in tissue morphogenesis [152].
In the Dark-eyed Junco, the demographic inference revealed that the insular population on Guadalupe diverged around 400,000 years ago, which is similar to previous estimates [76]. The differentiated regions were mainly distributed at the ends of chromosomes, coinciding with telocentric centromeres, as previously found in Swainson’s thrushes [153]. Consistent with this pattern, among the top-ten GO terms we identified several that were related to the centrosomes, increasingly recognized as signaling machines capable of regulating many cellular functions [154].
In the House Finch, the genomic landscape showed signatures of different processes. Despite the recent divergence time between mainland and Guadalupe Island populations, estimated at about 100,000 years before present, we did no detect signatures of significant selective sweeps. The large region showing high differentiation and very low recombination in chromosome 3 likely represents a major chromosomal inversion. Upon colonization of a new environment, such as an island, a chromosomal inversion can rapidly become fixed or prevalent within the population, especially if it provides an adaptive advantage [155]. If there is subsequent gene flow between island and mainland populations, the inversion can act as a barrier to genetic exchange by reducing recombination. This suppression of recombination facilitates the maintenance of distinct genomic regions that might be beneficial in the local environment, thus reinforcing divergence between the island and mainland populations [156]. Genomic islands of differentiation could be generated by chromosomal rearrangements that cluster highly differentiated loci together due to genomic hitchhiking [114, 157]. However, that could represent either a group of adaptive alleles or several neutral loci linked to a focal selected allele [157]. Several studies have found regions highly diverged within chromosomal inversions [114, 158–160]. In this case, 13 genes putatively under selection were found within the inversion. One of those genes (Fig. 4) is related to facial morphology and related disorders [161] and the gtf3c6 gene was found to be a candidate involved in sexual selection [162]. Another interesting candidate is the Iyd gene, which is also found within an inversion in chromosome 2 in the White-throated Sparrow (Zonotrichia albicollis) and has shown differences in expression between two morphs that differed in territorial aggression including song [163]. Within the top-ten significant GO terms, we found “growth plate cartilage chondrocyte morphogenesis”, which is involved in skeletal development and morphogenesis and regulated by multiple signaling pathways, including the bone morphogenetic proteins (BMP; [160, 164]) and Wingless/int.1 molecules (Wnt; [165]) that are known to be involved in facial development in different organisms including beak morphology in birds [166, 167].
Here we studied four cases of island-mainland divergence in passerine species that have colonized oceanic islands and share morphological modifications likely caused by similar selective pressures, and asked whether the genetic landscapes between species were also similar. Our general result in this respect is that the regions of the genome showing evidence of divergence under directional selection are lineage specific, suggesting that the genetic divergence is different in each case, so that evidence for repeated evolution at the genomic level appears to be lacking [43]. Even if the same regions had been detected as putatively under selection or with shared genomic features involved in genomic differentiation, such as the stable recombination landscape in avian lineages [50], it would be difficult to determine whether that pattern is generated by directional selection or by background and linked selection. Despite examples showing that few loci of large effect can drive adaptive divergence in complex traits, such as the bill (e.g., [168]), selection is likely to act on many loci of small effect due to the polygenic nature of most adaptive traits [169, 170]. Consequently, convergent phenotypes could in fact be due to divergent genotypes. Several examples to date show that phenotypic change in a given trait can be driven by different sets of genes, such as mouth morphology in cichlid fishes [171], or color pattern in mice [172, 173] and flies [174]. Even though the outlier genes differ among species, there could be common significant GO terms because different genes share functions and pathways. Interestingly, between the Common Chaffinch and the House Finch we found similar GO terms related to tRNA aminoacylation, and between the Red-billed Chough and the Common Chaffinch we also found the common term “protein autoubiquitination”. Remarkably, we found that in all four species, GO terms are mostly related to gene regulation, such as protein ubiquitination, transmembrane transport, and regulation of cellular localization, which are crucial for maintaining various physiological and developmental processes. Recently, Monroe et al. [175] reported that mutations occur less often in functional regions of the genome, and that epigenomic and physical chromosomal features account for the position of the mutations. In our case, most of the terms related to outlier loci are involved in regulatory and signaling pathways, suggesting that changes in gene regulation, instead of specific core genes, may be the main drivers of divergence. Currently, several models are being developed to understand the role of gene regulation in the evolution of complex traits [19, 176], implying that regulatory regions are disproportionately targeted by polygenic selection, highlighting the key role of gene regulatory networks in evolution [177].
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We are grateful to José Manuel González, Óscar Frías and Félix Medina for invaluable help in the field.
Author contributions
MR carried out the molecular lab work, carried out the data curation and analysis, participated in the design of the study, collected field data and drafted the manuscript; JM collected field data and critically revised the manuscript; GB conceived and designed the study, collected field data and critically revised the manuscript; BM conceived and designed the study, collected field data and critically revised the manuscript. All authors gave final approval for publication and agree to be held accountable for the work performed therein.
Funding
This work was supported by grants CGL-2015-66381P and PGC-2018-098897-B-I00 from Spain’s Ministry of Science and co-financed by the European Union’s Regional Development Fund (ERDF). MR was supported by a doctoral fellowship from Spain’s Ministry of Education, Culture, and Sport (FPU16/05724).
Data availability
Resequencing raw data is deposited at NCBI under the SRA data projects PRJNA661201 (for common chaffinch mainland population) with accession numbers SAMN16094451-SAMN16094459 and PRJNA1077913 with accession numbers SAMN39984864- SAMN39984947, for the common chaffinch insular population and both populations from the rest of the species, see Table S1 for details) and the datasets, are deposited in Figshare (https://doi.org/10.6084/m9.figshare.21590673).
Declarations
Ethics approval and consent to participate
Field sampling was carried out in compliance with ethical and animal welfare regulations under the corresponding permits, including: CSIC’s Ethics Committee approval of the bird handling protocol (Ref. 1415/2023), research and collecting permits from the Cabildo de La Palma, Canarian Regional Government (A/EST-004/2020 and A/EST-003/2021) and Madrid region (Ref: 10/090952.9/23) in Spain; research permit SGPA/DGVS/01190/14 for Guadalupe Island, Mexico and export permit Mexico-Spain (No. 34514); and scientific collecting permit for California, USA (D-0009344278-7) and export permit USA-Spain (MB079347-1). Bird banding by BM was conducted under permit No. 530531 in Spain and under Federal Bird Banding Permit No. 23402 in the USA.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
María Recuerda, Email: mariarecuerdacarrasco@gmail.com.
Borja Milá, Email: b.mila@csic.es.
References
- 1.Schluter D. The ecology of adaptive radiation. OUP Oxford; 2000.
- 2.Grant PR. Reconstructing the evolution of birds on islands: 100 years of research. Oikos. 2001;92(3):385–403. [Google Scholar]
- 3.Price T. Speciation in birds. Roberts and Company. greenwood Village, CO; 2008.
- 4.Warren BH, Simberloff D, Ricklefs RE, Aguilée R, Condamine FL, Gravel D, et al. Islands as model systems in ecology and evolution: prospects fifty years after MacArthur-Wilson. Ecol Lett. 2015;18(2):200–17. [DOI] [PubMed] [Google Scholar]
- 5.Gillespie RG, Bennett GM, De Meester L, Feder JL, Fleischer RC, Harmon LJ, et al. Comparing adaptive radiations across space, time, and taxa. J Hered. 2020;111(1):1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grant PR, Grant BR. Adaptive radiation of Darwin’s finches: recent data help explain how this famous group of Galapagos birds evolved, although gaps in our understanding remain. Am Sci. 2002;90(2):130–9. [Google Scholar]
- 7.Losos JB, Ricklefs RE. Adaptation and diversification on islands. Nature. 2009;457(7231):830–6. [DOI] [PubMed] [Google Scholar]
- 8.Brown RM, Siler CD, Oliveros CH, Esselstyn JA, Diesmos AC, Hosner PA, et al. Evolutionary processes of diversification in a model island archipelago. Annu Rev Ecol Evol Syst. 2013;44:411–35. [Google Scholar]
- 9.Woolfit M, Bromham L. Population size and molecular evolution on islands. Proc Royal Soc B: Biol Sci. 2005;272(1578):2277–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whittaker RJ, Fernández-Palacios JM, Matthews TJ, Borregaard MK, Triantis KA. Island biogeography: taking the long view of nature’s laboratories. Sci (1979). 2017;357(6354):eaam8326. [DOI] [PubMed] [Google Scholar]
- 11.Kavanagh PH, Burns KC. The repeated evolution of large seeds on islands. Proceedings of the Royal Society B: Biological Sciences. 2014;281(1786):20140675. [DOI] [PMC free article] [PubMed]
- 12.McCulloch GA. Digest: repeated body size evolution in island bats. Oxford University Press US; 2024. [DOI] [PubMed]
- 13.Cerca J. Understanding natural selection and similarity: convergent, parallel and repeated evolution. Mol Ecol. 2023;32(20):5451–62. [DOI] [PubMed] [Google Scholar]
- 14.Manceau M, Domingues VS, Linnen CR, Rosenblum EB, Hoekstra HE. Convergence in pigmentation at multiple levels: mutations, genes and function. Philosophical Trans Royal Soc B: Biol Sci. 2010;365(1552):2439–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sackton TB, Clark N. Convergent evolution in the genomics era: new insights and directions. Volume 374. Philosophical Transactions of the Royal Society B. The Royal Society; 2019. p. 20190102. [DOI] [PMC free article] [PubMed]
- 16.Rosenblum EB, Parent CE, Brandt EE. The molecular basis of phenotypic convergence. Annu Rev Ecol Evol Syst. 2014;45:203–26. [Google Scholar]
- 17.Reed RD, Papa R, Martin A, Hines HM, Counterman BA, Pardo-Diaz C et al. Optix drives the repeated convergent evolution of butterfly wing pattern mimicry. Science (1979). 2011;333(6046):1137–41. [DOI] [PubMed]
- 18.Colosimo PF, Hosemann KE, Balabhadra S, Villarreal G Jr, Dickson M, Grimwood J et al. Widespread parallel evolution in sticklebacks by repeated fixation of ectodysplasin alleles. Science (1979). 2005;307(5717):1928–33. [DOI] [PubMed]
- 19.Boyle EA, Li YI, Pritchard JK. An expanded view of complex traits: from polygenic to omnigenic. Cell. 2017;169(7):1177–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sendell-Price AT, Ruegg KC, Robertson BC, Clegg SM. An island‐hopping bird reveals how founder events shape genome‐wide divergence. Mol Ecol. 2021;30(11):2495–510. [DOI] [PubMed] [Google Scholar]
- 21.Martin CA, Sheppard EC, Ali HAA, Illera JC, Suh A, Spurgin LG et al. Genomic landscapes of divergence among island bird populations: evidence of parallel adaptation but at different loci? Mol Ecol. 2024;e17365. [DOI] [PubMed]
- 22.Corbett-Detig RB, Russell SL, Nielsen R, Losos J. Phenotypic convergence is not mirrored at the protein level in a lizard adaptive radiation. Mol Biol Evol. 2020;37(6):1604–14. [DOI] [PubMed] [Google Scholar]
- 23.Meier JI, Marques DA, Wagner CE, Excoffier L, Seehausen O. Genomics of parallel ecological speciation in Lake Victoria cichlids. Mol Biol Evol. 2018;35(6):1489–506. [DOI] [PubMed] [Google Scholar]
- 24.Leroy T, Rousselle M, Tilak MK, Caizergues AE, Scornavacca C, Recuerda M, et al. Island songbirds as windows into evolution in small populations. Curr Biol. 2021;31(6):1303–10. [DOI] [PubMed] [Google Scholar]
- 25.Cruickshank TE, Hahn MW. Reanalysis suggests that genomic islands of speciation are due to reduced diversity, not reduced gene flow. Mol Ecol. 2014;23(13):3133–57. [DOI] [PubMed] [Google Scholar]
- 26.Burri R. Interpreting differentiation landscapes in the light of long-term linked selection. Evol Lett. 2017;1(3):118–31. [Google Scholar]
- 27.Ravinet M, Faria R, Butlin RK, Galindo J, Bierne N, Rafajlović M, et al. Interpreting the genomic landscape of speciation: a road map for finding barriers to gene flow. J Evol Biol. 2017;30(8):1450–77. [DOI] [PubMed] [Google Scholar]
- 28.Feng S, Stiller J, Deng Y, Armstrong J, Fang QI, Reeve AH, et al. Dense sampling of bird diversity increases power of comparative genomics. Nature. 2020;587(7833):252–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chase MA, Ellegren H, Mugal CF. Positive selection plays a major role in shaping signatures of differentiation across the genomic landscape of two independent Ficedula flycatcher species pairs. Evol (N Y). 2021;75(9):2179–96. [DOI] [PubMed] [Google Scholar]
- 30.Turner TL, Hahn MW, Nuzhdin SV. Genomic islands of speciation in Anopheles gambiae. PLoS Biol. 2005;3(9):e285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ellegren H, Smeds L, Burri R, Olason PI, Backström N, Kawakami T, et al. The genomic landscape of species divergence in Ficedula flycatchers. Nature. 2012;491(7426):756–60. [DOI] [PubMed] [Google Scholar]
- 32.Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evol (N Y). 1984;1358–70. [DOI] [PubMed]
- 33.Nadeau NJ, Whibley A, Jones RT, Davey JW, Dasmahapatra KK, Baxter SW, et al. Genomic islands of divergence in hybridizing Heliconius butterflies identified by large-scale targeted sequencing. Philosophical Trans Royal Soc B: Biol Sci. 2012;367(1587):343–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poelstra JW, Vijay N, Bossu CM, Lantz H, Ryll B, Müller I, et al. The genomic landscape underlying phenotypic integrity in the face of gene flow in crows. Sci (1979). 2014;344(6190):1410–4. [DOI] [PubMed]
- 35.Nosil P, Funk DJ, Ortiz-Barrientos D. Divergent selection and heterogeneous genomic divergence. Mol Ecol. 2009;18(3):375–402. [DOI] [PubMed] [Google Scholar]
- 36.Zeng K, Corcoran P. The effects of background and interference selection on patterns of genetic variation in subdivided populations. Genetics. 2015;201(4):1539–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feder JL, Nosil P. The efficacy of divergence hitchhiking in generating genomic islands during ecological speciation. Evol (N Y). 2010;64(6):1729–47. [DOI] [PubMed] [Google Scholar]
- 38.Burri R, Nater A, Kawakami T, Mugal CF, Olason PI, Smeds L, et al. Linked selection and recombination rate variation drive the evolution of the genomic landscape of differentiation across the speciation continuum of Ficedula flycatchers. Genome Res. 2015;25(11):1656–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Irwin DE, Milá B, Toews DPL, Brelsford A, Kenyon HL, Porter AN, et al. A comparison of genomic islands of differentiation across three young avian species pairs. Mol Ecol. 2018;27(23):4839–55. [DOI] [PubMed] [Google Scholar]
- 40.Irwin DE, Alcaide M, Delmore KE, Irwin JH, Owens GL. Recurrent selection explains parallel evolution of genomic regions of high relative but low absolute differentiation in a ring species. Mol Ecol. 2016;25(18):4488–507. [DOI] [PubMed] [Google Scholar]
- 41.Han F, Lamichhaney S, Grant BR, Grant PR, Andersson L, Webster MT. Gene flow, ancient polymorphism, and ecological adaptation shape the genomic landscape of divergence among Darwin’s finches. Genome Res. 2017;27(6):1004–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferchaud A, Hansen MM. The impact of selection, gene flow and demographic history on heterogeneous genomic divergence: three-spine sticklebacks in divergent environments. Mol Ecol. 2016;25(1):238–59. [DOI] [PubMed] [Google Scholar]
- 43.Van Doren BM, Campagna L, Helm B, Illera JC, Lovette IJ, Liedvogel M. Correlated patterns of genetic diversity and differentiation across an avian family. Mol Ecol. 2017;26(15):3982–97. [DOI] [PubMed] [Google Scholar]
- 44.Delmore KE, Lugo Ramos JS, Van Doren BM, Lundberg M, Bensch S, Irwin DE, et al. Comparative analysis examining patterns of genomic differentiation across multiple episodes of population divergence in birds. Evol Lett. 2018;2(2):76–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vijay N, Weissensteiner M, Burri R, Kawakami T, Ellegren H, Wolf JBW. Genomewide patterns of variation in genetic diversity are shared among populations, species and higher-order taxa. Mol Ecol. 2017;26(16):4284–95. [DOI] [PubMed] [Google Scholar]
- 46.Carbeck K, Arcese P, Lovette I, Pruett C, Winker K, Walsh J. Candidate genes under selection in song sparrows co-vary with climate and body mass in support of Bergmann’s rule. Nat Commun. 2023;14(1):6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seehausen O, Butlin RK, Keller I, Wagner CE, Boughman JW, Hohenlohe PA, et al. Genomics and the origin of species. Nat Rev Genet. 2014;15(3):176–92. [DOI] [PubMed] [Google Scholar]
- 48.Zhang G, Li C, Li Q, Li B, Larkin DM, Lee C, et al. Comparative genomics reveals insights into avian genome evolution and adaptation. Sci (1979). 2014;346(6215):1311–20. [DOI] [PMC free article] [PubMed]
- 49.Ellegren H. Evolutionary stasis: the stable chromosomes of birds. Trends Ecol Evol. 2010;25(5):283–91. [DOI] [PubMed] [Google Scholar]
- 50.Singhal S, Leffler EM, Sannareddy K, Turner I, Venn O, Hooper DM et al. Stable recombination hotspots in birds. Science (1979). 2015;350(6263):928–32. [DOI] [PMC free article] [PubMed]
- 51.Kawakami T, Mugal CF, Suh A, Nater A, Burri R, Smeds L, et al. Whole-genome patterns of linkage disequilibrium across flycatcher populations clarify the causes and consequences of fine‐scale recombination rate variation in birds. Mol Ecol. 2017;26(16):4158–72. [DOI] [PubMed] [Google Scholar]
- 52.Dutoit L, Burri R, Nater A, Mugal CF, Ellegren H. Genomic distribution and estimation of nucleotide diversity in natural populations: perspectives from the collared flycatcher (Ficedula albicollis) genome. Mol Ecol Resour. 2017;17(4):586–97. [DOI] [PubMed] [Google Scholar]
- 53.Westram AM, Faria R, Johannesson K, Butlin R, Barton N. Inversions and parallel evolution. Philosophical Trans Royal Soc B. 2022;377(1856):20210203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koch EL, Morales HE, Larsson J, Westram AM, Faria R, Lemmon AR, et al. Genetic variation for adaptive traits is associated with polymorphic inversions in Littorina saxatilis. Evol Lett. 2021;5(3):196–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mérot C, Berdan EL, Cayuela H, Djambazian H, Ferchaud AL, Laporte M, et al. Locally adaptive inversions modulate genetic variation at different geographic scales in a seaweed fly. Mol Biol Evol. 2021;38(9):3953–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dunn PO, Sly ND, Freeman-Gallant CR, Henschen AE, Bossu CM, Ruegg KC et al. Sexually selected differences in warbler plumage are related to a putative inversion on the Z chromosome. Mol Ecol. 2024;e17525. [DOI] [PubMed]
- 57.Viitaniemi HM, Leder EH, Kauzál O, Stopková R, Stopka P, Lifjeld JT et al. Impact of Z chromosome inversions on gene expression in testis and liver tissues in the zebra finch. Mol Ecol. 2023. [DOI] [PMC free article] [PubMed]
- 58.Estandía A, Sendell-Price AT, Robertson BC, Clegg SM. Standing genetic variation and de novo mutations underlie parallel evolution of island bird phenotypes. bioRxiv. 2023;2023–6.
- 59.Blondel J. Evolution and ecology of birds on islands: trends and prospects. Vie et Milieu/Life & Environment. 2000;205–20.
- 60.Jezierski MT, Smith WJ, Clegg SM. The island syndrome in birds. J Biogeogr. 2023.
- 61.Adler GH, Levins R. The island syndrome in rodent populations. Q Rev Biol. 1994;69(4):473–90. [DOI] [PubMed] [Google Scholar]
- 62.Benítez-López A, Santini L, Gallego-Zamorano J, Milá B, Walkden P, Huijbregts MAJ, et al. The island rule explains consistent patterns of body size evolution in terrestrial vertebrates. Nat Ecol Evol. 2021;5(6):768–86. [DOI] [PubMed] [Google Scholar]
- 63.Sayol F, Downing PA, Iwaniuk AN, Maspons J, Sol D. Predictable evolution towards larger brains in birds colonizing oceanic islands. Nat Commun. 2018;9(1):2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lapiedra O, Sayol F, Garcia-Porta J, Sol D. Niche shifts after island colonization spurred adaptive diversification and speciation in a cosmopolitan bird clade. Proc Royal Soc B. 2021;288(1958):20211022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Glor RE, Gifford ME, Larson A, Losos JB, Schettino LR, Lara ARC, et al. Partial island submergence and speciation in an adaptive radiation: a multilocus analysis of the Cuban green anoles. Proc R Soc Lond B Biol Sci. 2004;271(1554):2257–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Campana MG, Corvelo A, Shelton J, Callicrate TE, Bunting KL, Riley-Gillis B, et al. Adaptive radiation genomics of two ecologically divergent Hawai ‘ian honeycreepers: the ‘akiapōlā ‘au and the Hawai ‘i ‘amakihi. J Hered. 2020;111(1):21–32. [DOI] [PubMed] [Google Scholar]
- 67.Blanco G, Laiolo P, Fargallo JA. Linking environmental stress, feeding-shifts and the ‘island syndrome’: a nutritional challenge hypothesis. Popul Ecol. 2014;56:203–16. [Google Scholar]
- 68.Tattersall GJ, Chaves JA, Danner RM. Thermoregulatory windows in Darwin’s finches. Funct Ecol. 2018;32(2):358–68. [Google Scholar]
- 69.Foster JB. Evolution of mammals on islands. Nature. 1964;202(4929):234–5. [Google Scholar]
- 70.Clegg SM, Owens PF. The ‘island rule’in birds: medium body size and its ecological explanation. Proc R Soc Lond B Biol Sci. 2002;269(1498):1359–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Frankham R. Do island populations have less genetic variation than mainland populations? Heredity (Edinb). 1997;78(3):311–27. [DOI] [PubMed] [Google Scholar]
- 72.Wang S, Zhu W, Gao XU, Li X, Yan S, Liu X, et al. Population size and time since island isolation determine genetic diversity loss in insular frog populations. Mol Ecol. 2014;23(3):637–48. [DOI] [PubMed] [Google Scholar]
- 73.Austerlitz F, Jung-Muller B, Godelle B, Gouyon PH. Evolution of coalescence times, genetic diversity and structure during colonization. Theor Popul Biol. 1997;51(2):148–64. [Google Scholar]
- 74.Clegg SM, Degnan SM, Kikkawa J, Moritz C, Estoup A, Owens IPF. Genetic consequences of sequential founder events by an island-colonizing bird. Proceedings of the National Academy of Sciences. 2002;99(12):8127–32. [DOI] [PMC free article] [PubMed]
- 75.Clegg S. Evolutionary changes following island colonization in birds. Theory Island Biogeogr Revisit. 2010;293–325.
- 76.Aleixandre P, Hernández Montoya J, Mila B. Speciation on oceanic islands: Rapid adaptive divergence vs. cryptic speciation in a Guadalupe Island Songbird (Aves: Junco). PLoS ONE. 2013;8(5):e63242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Morinha F, Milá B, Dávila JA, Fargallo JA, Potti J, Blanco G. The ghost of connections past: a role for mainland vicariance in the isolation of an insular population of the red-billed chough (Aves: Corvidae). J Biogeogr. 2020;47(12):2567–83. [Google Scholar]
- 78.Recuerda M, Illera JC, Blanco G, Zardoya R, Milá B. Sequential colonization of oceanic archipelagos led to a species-level radiation in the common chaffinch complex (Aves: Fringilla coelebs). Mol Phylogenet Evol. 2021;164:107291. [DOI] [PubMed] [Google Scholar]
- 79.Tuttle EM, Bergland AO, Korody ML, Brewer MS, Newhouse DJ, Minx P, et al. Divergence and functional degradation of a sex chromosome-like supergene. Curr Biol. 2016;26(3):344–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Billerman M, Keeney BK, Rodewald PG, Schulenberg TS. Birds of the World. Ithaca: Cornell Lab of Ornithology; 2022. [Google Scholar]
- 81.Griffiths R, Daan S, Dijkstra C. Sex identification in birds using two CHD genes. Proc R Soc Lond B Biol Sci. 1996;263(1374):1251–6. [DOI] [PubMed] [Google Scholar]
- 82.Milá B, Wayne RK, Smith TB. Ecomorphology of migratory and sedentary populations of the yellow-rumped warbler (Dendroica coronata). Condor. 2008;110(2):335–44. [Google Scholar]
- 83.Blanco G, Tella JL, Torre I. Age and sex determination of Monomorphic Non-breeding Choughs: a long-term study (Determinacion De La Edad Y El Sexo En Chovas Piquirrojas Pyrrhocorax pyrrhocorax no Reproductoras: Un Estudio a Largo Plazo). J Field Ornithol. 1996;428–33.
- 84.Krueger F, TrimGalore. A wrapper tool around Cutadapt and FastQC to consistently apply quality and adapter trimming to FastQ files. Babraham Bioinf. 2015.
- 85.Li H, Durbin R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics. 2010;26(5):589–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Recuerda M, Vizueta J, Cuevas-Caballé C, Blanco G, Rozas J, Milá B. Chromosome-level genome assembly of the common chaffinch (Aves: Fringilla coelebs): a valuable resource for evolutionary biology. Genome Biol Evol. 2021;13(4):evab034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Friis G, Vizueta J, Ketterson ED, Milá B. A high-quality genome assembly and annotation of the dark-eyed junco Junco Hyemalis, a recently diversified songbird. G3. 2022;12(6):jkac083. [DOI] [PMC free article] [PubMed]
- 88.Arnaiz-Villena A, Guillén J, Ruiz-del-Valle V, Lowy E, Zamora J, Varela P, et al. Phylogeography of crossbills, bullfinches, grosbeaks, and rosefinches. Cell Mol Life Sci. 2001;58:1159–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fernando SW, Peterson AT, Li SH. Reconstructing the geographic origin of the New World jays. Neotrop Biodivers. 2017;3(1):80–92. [Google Scholar]
- 90.Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, et al. Twelve years of SAMtools and BCFtools. Gigascience. 2021;10(2):giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, et al. The variant call format and VCFtools. Bioinformatics. 2011;27(15):2156–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Marçais G, Delcher AL, Phillippy AM, Coston R, Salzberg SL, Zimin A. MUMmer4: a fast and versatile genome alignment system. PLoS Comput Biol. 2018;14(1):e1005944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li H, Durbin R. Inference of human population history from individual whole-genome sequences. Nature. 2011;475(7357):493–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wright AE, Mank JE. The scope and strength of sex-specific selection in genome evolution. J Evol Biol. 2013;26(9):1841–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nadachowska-Brzyska K, Burri R, Smeds L, Ellegren H. PSMC analysis of effective population sizes in molecular ecology and its application to black‐and‐white Ficedula flycatchers. Mol Ecol. 2016;25(5):1058–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Smeds L, Qvarnström A, Ellegren H. Direct estimate of the rate of germline mutation in a bird. Genome Res. 2016;26(9):1211–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ericson PGP, Qu Y, Blom MPK, Johansson US, Irestedt M. A genomic perspective of the pink-headed duck Rhodonessa caryophyllacea suggests a long history of low effective population size. Sci Rep. 2017;7(1):16853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hanna ZR, Henderson JB, Wall JD, Emerling CA, Fuchs J, Runckel C, et al. Northern spotted owl (Strix occidentalis caurina) genome: divergence with the barred owl (Strix varia) and characterization of light-associated genes. Genome Biol Evol. 2017;9(10):2522–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sato Y, Ogden R, Kishida T, Nakajima N, Maeda T, Inoue-Murayama M. Population history of the golden eagle inferred from whole-genome sequencing of three of its subspecies. Biol J Linn Soc. 2020;130(4):826–38. [Google Scholar]
- 101.Baker AJ, Marshall HD. Population divergence in Chaffinches Fringilla coelebs assessed with control-region sequences. In: Proceedings XXII International Ornithological Congress (NJ Adams and RH Slotow, Eds) BirdLife South Africa, Durban. 1999. pp. 1899–913.
- 102.Reid JM, Bignal EM, Bignal S, McCracken DI, Monaghan P. Age-specific reproductive performance in red-billed choughs Pyrrhocorax pyrrhocorax: patterns and processes in a natural population. J Anim Ecol. 2003;765–76.
- 103.Friis G, Aleixandre P, Rodríguez-Estrella R, Navarro‐Sigüenza AG, Milá B. Rapid postglacial diversification and long‐term stasis within the songbird genus Junco: phylogeographic and phylogenomic evidence. Mol Ecol. 2016;25(24):6175–95. [DOI] [PubMed] [Google Scholar]
- 104.McVean G, Auton A. LDhat 2.1: a package for the population genetic analysis of recombination. Oxford, OX1 3TG, UK.: Department of Statistics; 2007. [Google Scholar]
- 105.Sabeti PC, Varilly P, Fry B, Lohmueller J, Hostetter E, Cotsapas C, et al. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449(7164):913–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Korunes KL, Samuk K. Pixy: unbiased estimation of nucleotide diversity and divergence in the presence of missing data. Mol Ecol Resour. 2021;21(4):1359–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123(3):585–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc: Ser B (Methodol). 1995;57(1):289–300. [Google Scholar]
- 109.Gautier M, Klassmann A, Vitalis R. Rehh 2.0: a reimplementation of the R package rehh to detect positive selection from haplotype structure. Mol Ecol Resour. 2017;17(1):78–90. [DOI] [PubMed] [Google Scholar]
- 110.Delaneau O, Zagury JF, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods. 2013;10(1):5–6. [DOI] [PubMed] [Google Scholar]
- 111.Turner SD. Qqman: an R package for visualizing GWAS results using QQ and manhattan plots. Biorxiv. 2014;005165.
- 112.Team RC, Team RC. R: a language and environment for statistical computing. R Found Stat Comput Vienna Austria. 2017.
- 113.Li H, Ralph P. Local PCA shows how the effect of population structure differs along the genome. Genetics. 2019;211(1):289–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Huang K, Andrew RL, Owens GL, Ostevik KL, Rieseberg LH. Multiple chromosomal inversions contribute to adaptive divergence of a dune sunflower ecotype. Mol Ecol. 2020;29(14):2535–49. [DOI] [PubMed] [Google Scholar]
- 115.Zheng X, Levine D, Shen J, Gogarten SM, Laurie C, Weir BS. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics. 2012;28(24):3326–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hartigan JA, Wong MA, Algorithm AS. A k-means clustering algorithm. J R Stat Soc Ser C Appl Stat. 1979;136(1):100–8. [Google Scholar]
- 117.Dong SS, He WM, Ji JJ, Zhang C, Guo Y, Yang TL. LDBlockShow: a fast and convenient tool for visualizing linkage disequilibrium and haplotype blocks based on variant call format files. Brief Bioinform. 2021;22(4):bbaa227. [DOI] [PubMed] [Google Scholar]
- 118.Rappaport N, Twik M, Plaschkes I, Nudel R, Iny Stein T, Levitt J, et al. MalaCards: an amalgamated human disease compendium with diverse clinical and genetic annotation and structured search. Nucleic Acids Res. 2017;45(D1):D877–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Alexa A, Rahnenfuhrer J. topGO: enrichment analysis for gene ontology. R Package Version. 2010;2(0):2010. [Google Scholar]
- 120.Tajima F. The effect of change in population size on DNA polymorphism. Genetics. 1989;123(3):597–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Vujovic D, Cornblath DR, Scherer SS. A recurrent MORC2 mutation causes Charcot-Marie‐tooth disease type 2Z. J Peripheral Nerv Syst. 2021;26(2):184–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sacoto MJG, Tchasovnikarova IA, Torti E, Forster C, Andrew EH, Anselm I, et al. De novo variants in the ATPase module of MORC2 cause a neurodevelopmental disorder with growth retardation and variable craniofacial dysmorphism. Am J Hum Genet. 2020;107(2):352–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lehti MS, Henriksson H, Rummukainen P, Wang F, Uusitalo-Kylmälä L, Kiviranta R, et al. Cilia-related protein SPEF2 regulates osteoblast differentiation. Sci Rep. 2018;8(1):859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.van der Brum AM, Schreuders Koedam M, Verhoeven J, Janssen M, Dekkers DHW, et al. Identification of chloride intracellular channel protein 3 as a novel gene affecting human bone formation. J Bone Mineral Res Plus. 2017;1(1):16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gago-Fuentes R, Oksenych V. Non-homologous end joining factors xlf, paxx and dna-pkcs maintain the neural stem and progenitor cell population. Biomolecules. 2020;11(1):20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sun L, Yang X, Khan A, Yu X, Zhang H, Han S, et al. Panoramic variation analysis of a family with neurodevelopmental disorders caused by biallelic loss-of-function variants in TMEM141, DDHD2, and LHFPL5. Front Med. 2024;18(1):81–97. [DOI] [PubMed] [Google Scholar]
- 127.Wu Y, Yan Y, Zhao Y, Gu L, Wang S, Johnson DH. Genomic bases underlying the adaptive radiation of core landbirds. BMC Ecol Evol. 2021;21:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Frankham R. Effective population size/adult population size ratios in wildlife: a review. Genet Res (Camb). 1995;66(2):95–107. [DOI] [PubMed] [Google Scholar]
- 129.Morinha F, Dávila JA, Bastos E, Cabral JA, Frías Ó, González JL, et al. Extreme genetic structure in a social bird species despite high dispersal capacity. Mol Ecol. 2017;26(10):2812–25. [DOI] [PubMed] [Google Scholar]
- 130.Rising JD, Somers KM. The measurement of overall body size in birds. Auk. 1989;106(4):666–74. [Google Scholar]
- 131.Jolicoeur P. 193. Note: the multivariate generalization of the allometry equation. Biometrics. 1963;19(3):497–9.
- 132.Freeman S, Jackson WM. Univariate metrics are not adequate to measure avian body size. Auk. 1990;107(1):69–74. [Google Scholar]
- 133.Senar JC, Pascual J. Keel and tarsus length may provide a good predictor of avian body size. ARDEA-WAGENINGEN-. 1997;85:269–74. [Google Scholar]
- 134.Grant PR, Grant BR. How and why species multiply: the radiation of Darwin’s finches. Princeton University Press; 2007.
- 135.Boag PT, Grant PR. Intense natural selection in a population of Darwin’s finches (Geospizinae) in the Galapagos. Science (1979). 1981;214(4516):82–5. [DOI] [PubMed]
- 136.Gibbs HL, Grant PR. Oscillating selection on Darwin’s finches. Nature. 1987;327(6122):511–3. [Google Scholar]
- 137.Tattersall GJ, Arnaout B, Symonds MRE. The evolution of the avian bill as a thermoregulatory organ. Biol Rev. 2017;92(3):1630–56. [DOI] [PubMed] [Google Scholar]
- 138.Gamboa MP, Ghalambor CK, Scott Sillett T, Morrison SA, Chris Funk W. Adaptive divergence in bill morphology and other thermoregulatory traits is facilitated by restricted gene flow in song sparrows on the California Channel Islands. Mol Ecol. 2022;31(2):603–19. [DOI] [PubMed] [Google Scholar]
- 139.Grant PR, EcologyandEvolutionofDarwin. sFinches・PrincetonUniversity. Princeton; 1986.
- 140.Price TD, Grant PR, Gibbs HL, Boag PT. Recurrent patterns of natural selection in a population of Darwin’s finches. Nature. 1984;309(5971):787–9. [DOI] [PubMed] [Google Scholar]
- 141.Burt DW. Origin and evolution of avian microchromosomes. Cytogenet Genome Res. 2002;96(1–4):97–112. [DOI] [PubMed] [Google Scholar]
- 142.Charlesworth B. Measures of divergence between populations and the effect of forces that reduce variability. Mol Biol Evol. 1998;15(5):538–43. [DOI] [PubMed] [Google Scholar]
- 143.Pala I, Naurin S, Stervander M, Hasselquist D, Bensch S, Hansson B. Evidence of a neo-sex chromosome in birds. Heredity (Edinb). 2012;108(3):264–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Leroy T, Anselmetti Y, Tilak MK, Bérard S, Csukonyi L, Gabrielli M et al. A bird’s white-eye view on avian sex chromosome evolution. Peer Community J. 2021;1.
- 145.Sun L, Chen ZJ. The novel functions of ubiquitination in signaling. Curr Opin Cell Biol. 2004;16(2):119–26. [DOI] [PubMed] [Google Scholar]
- 146.Ren S, Lyu G, Irwin DM, Liu X, Feng C, Luo R, et al. Pooled sequencing analysis of geese (Anser cygnoides) reveals genomic variations associated with feather color. Front Genet. 2021;12:650013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Qi LM, Mohr M, Wade J. Enhanced expression of tubulin-specific chaperone protein a, mitochondrial ribosomal protein S27, and the DNA excision repair protein XPACCH in the song system of juvenile male zebra finches. Dev Neurobiol. 2012;72(2):199–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Shi Z, Zhang Z, Schaffer L, Huang Z, Fu L, Head S, et al. Dynamic transcriptome landscape in the song nucleus HVC between juvenile and adult zebra finches. Adv Genet. 2021;2(1):e10035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lovell PV, Clayton DF, Replogle KL, Mello CV. Birdsong transcriptomics: neurochemical specializations of the oscine song system. PLoS ONE. 2008;3(10):e3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Oikkonen J, Onkamo P, Järvelä I, Kanduri C. Convergent evidence for the molecular basis of musical traits. Sci Rep. 2016;6(1):39707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yap CC, Winckler B. Harnessing the power of the endosome to regulate neural development. Neuron. 2012;74(3):440–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Harris TJC, Tepass U. Adherens junctions: from molecules to morphogenesis. Nat Rev Mol Cell Biol. 2010;11(7):502–14. [DOI] [PubMed] [Google Scholar]
- 153.Delmore KE, Hübner S, Kane NC, Schuster R, Andrew RL, Câmara F, et al. Genomic analysis of a migratory divide reveals candidate genes for migration and implicates selective sweeps in generating islands of differentiation. Mol Ecol. 2015;24(8):1873–88. [DOI] [PubMed] [Google Scholar]
- 154.Doxsey S, McCollum D, Theurkauf W. Centrosomes in cellular regulation. Annu Rev Cell Dev Biol. 2005;21:411–34. [DOI] [PubMed] [Google Scholar]
- 155.Faria R, Johannesson K, Butlin RK, Westram AM. Evolving inversions. Trends Ecol Evol. 2019;34(3):239–48. [DOI] [PubMed] [Google Scholar]
- 156.Wellenreuther M, Mérot C, Berdan E, Bernatchez L. Going beyond SNPs: the role of structural genomic variants in adaptive evolution and species diversification. Mol Ecol. 2019;28(6). [DOI] [PubMed]
- 157.Yeaman S. Genomic rearrangements and the evolution of clusters of locally adaptive loci. Proc Natl Acad Sci. 2013;110(19):E1743–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Hoffmann AA, Sgrò CM, Weeks AR. Chromosomal inversion polymorphisms and adaptation. Trends Ecol Evol. 2004;19(9):482–8. [DOI] [PubMed] [Google Scholar]
- 159.Ayala D, Ullastres A, González J. Adaptation through chromosomal inversions in Anopheles. Front Genet. 2014;5:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Christmas MJ, Wallberg A, Bunikis I, Olsson A, Wallerman O, Webster MT. Chromosomal inversions associated with environmental adaptation in honeybees. Mol Ecol. 2019;28(6):1358–74. [DOI] [PubMed] [Google Scholar]
- 161.Campeau PM, Lenk GM, Lu JT, Bae Y, Burrage L, Turnpenny P, et al. Yunis-Varon syndrome is caused by mutations in Fig. 4, encoding a phosphoinositide phosphatase. Am J Hum Genet. 2013;92(5):781–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Jaiswal SK, Gupta A, Shafer A, PK VP, Vijay N, Sharma VK. Genomic insights into the molecular basis of sexual selection in birds. Front Ecol Evol. 2021;2.
- 163.Zinzow-Kramer WM, Horton BM, McKee CD, Michaud JM, Tharp GK, Thomas JW, et al. Genes located in a chromosomal inversion are correlated with territorial song in white‐throated sparrows. Genes Brain Behav. 2015;14(8):641–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.De Luca F, Barnes KM, Uyeda JA, De-Levi S, Abad V, Palese T, et al. Regulation of growth plate chondrogenesis by bone morphogenetic protein-2. Endocrinology. 2001;142(1):430–6. [DOI] [PubMed] [Google Scholar]
- 165.Yang Y, Topol L, Lee H, Wu J. Wnt5a and Wnt5b exhibit distinct activities in coordinating chondrocyte proliferation and differentiation. 2003. [DOI] [PubMed]
- 166.Abzhanov A, Protas M, Grant BR, Grant PR, Tabin CJ. Bmp4 and morphological variation of beaks in Darwin’s finches. Science (1979). 2004;305(5689):1462–5. [DOI] [PubMed]
- 167.Brugmann SA, Powder KE, Young NM, Goodnough LH, Hahn SM, James AW, et al. Comparative gene expression analysis of avian embryonic facial structures reveals new candidates for human craniofacial disorders. Hum Mol Genet. 2010;19(5):920–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Enbody ED, Sendell-Price AT, Sprehn CG, Rubin CJ, Visscher PM, Grant BR et al. Community-wide genome sequencing reveals 30 years of Darwin’s finch evolution. Science (1979). 2023;381(6665):eadf6218. [DOI] [PubMed]
- 169.Pritchard JK, Di Rienzo A. Adaptation–not by sweeps alone. Nat Rev Genet. 2010;11(10):665–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Bosse M, Spurgin LG, Laine VN, Cole EF, Firth JA, Gienapp P et al. Recent natural selection causes adaptive evolution of an avian polygenic trait. Science (1979). 2017;358(6361):365–8. [DOI] [PubMed]
- 171.Elmer KR, Fan S, Kusche H, Luise Spreitzer M, Kautt AF, Franchini P, et al. Parallel evolution of Nicaraguan crater lake cichlid fishes via non-parallel routes. Nat Commun. 2014;5(1):5168. [DOI] [PubMed] [Google Scholar]
- 172.Hoekstra HE, Hirschmann RJ, Bundey RA, Insel PA, Crossland JP. A single amino acid mutation contributes to adaptive beach mouse color pattern. Science (1979). 2006;313(5783):101–4. [DOI] [PubMed]
- 173.Steiner CC, Römpler H, Boettger LM, Schöneberg T, Hoekstra HE. The genetic basis of phenotypic convergence in beach mice: similar pigment patterns but different genes. Mol Biol Evol. 2009;26(1):35–45. [DOI] [PubMed] [Google Scholar]
- 174.Wittkopp PJ, Williams BL, Selegue JE, Carroll SB. Drosophila pigmentation evolution: divergent genotypes underlying convergent phenotypes. Proceedings of the National Academy of Sciences. 2003;100(4):1808–13. [DOI] [PMC free article] [PubMed]
- 175.Monroe JG, Srikant T, Carbonell-Bejerano P, Becker C, Lensink M, Exposito-Alonso M, et al. Mutation bias reflects natural selection in Arabidopsis thaliana. Nature. 2022;602(7895):101–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Liu X, Li YI, Pritchard JK. Trans effects on gene expression can drive omnigenic inheritance. Cell. 2019;177(4):1022–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Fagny M, Austerlitz F. Polygenic adaptation: integrating population genetics and gene regulatory networks. Trends Genet. 2021;37(7):631–8. [DOI] [PubMed] [Google Scholar]
- 178.Clements JF, Schulenberg TS, Iliff MJ, Roberson D, Fredericks TA, Sullivan BL et al. The eBird/Clements checklist of birds of the world: v2019. 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Resequencing raw data is deposited at NCBI under the SRA data projects PRJNA661201 (for common chaffinch mainland population) with accession numbers SAMN16094451-SAMN16094459 and PRJNA1077913 with accession numbers SAMN39984864- SAMN39984947, for the common chaffinch insular population and both populations from the rest of the species, see Table S1 for details) and the datasets, are deposited in Figshare (https://doi.org/10.6084/m9.figshare.21590673).