Abstract
Shoot stem cells, harbored in the shoot apical meristem (SAM), play key roles during post-embryonic development of Arabidopsis and function as the origin of plant aerial tissues. Multiple transcription factors are involved in the sophisticated transcriptional regulation of stem cell homeostasis, with the WUSCHEL (WUS)/CLAVATA3 (CLV3) negative feedback loop playing a central role. WUS acts as a master regulator in maintaining stem cells through its transcriptional regulatory activity including repressive and activating abilities. Although the interaction between WUS and TOPLESS confers the repressive activity of WUS in transcriptional control, the mechanism by which WUS activates gene expression remains elusive. Here, we showed that DORNRÖSCHEN competitively interacts with WUS and disturbs the WUS homodimer, which recruits BRAHMA to activate CLV3 expression via nucleosome depletion for maintaining the stem cell pool.
DRN and WUS are known to upregulate the transcription of CLV3 in the shoot apical meristem of Arabidopsis, but how remains unclear. In this study, DRN is shown to interfere with WUS dimerization and chromatin remodeling to activate CLV3 transcription, thus contributing to the maintenance of the shoot stem cell pool.
Introduction
Aboveground organs in plants, such as true leaves, flowers, and stems, originate from stem cells embedded in the shoot apical meristem (SAM) [1]. Stem cells also contribute to the developmental plasticity of plants to adapt to the ever-changing environment [2]. In Arabidopsis, the shoot stem cells, embedded in the central zone (CZ) of the SAM, divide slowly [3]. Some daughter cells retain the undifferentiated state of stem cells, while others differentiate into organ primordia through the periphery zone (PZ).
Previous studies have demonstrated that the WUSCHEL (WUS)/CLAVATA3 (CLV3) negative feedback loop is a key hub of multiple regulatory networks underlying stem cell maintenance [4–7]. WUS transcribes in the organizing center (OC) beneath stem cells in the SAM. However, WUS proteins migrate to stem cells through the plasmodesmata, which maintain stem cells and directly activate the expression of CLV3 [8–10]. CLV3 encodes a peptide that acts as a signal to suppress WUS at the transcriptional and posttranslational levels via membrane receptors, CLAVATA1 (CLV1)/CLAVATA2 (CLV2)/CORYNE (CRN)/BARELY ANY MERISTEMS (BAMs)/RECEPTORLIKE PROTEIN KINASE 2 (RPK2)/CLAVATA3 INSENSITIVE RECEPTOR KINASES (CIKs), for sustaining the stable pool of stem cells [11–14]. The robust activity of stem cells depends on the ability of WUS to positively and negatively regulate gene transcription, which relies on multiple domains of WUS [15–19]. A homeodomain (HD) of WUS can bind to DNA; a middle region is responsible for homodimerization; an acidic region and a WUS box are required for retention in the nucleus; and an EAR domain is related to nuclear export [9,20,21]. TOPLESS (TPL), a transcriptional corepressor, interacts with the WUS box and EAR domain, helping WUS to suppress gene expression [22,23]. However, the mechanism by which WUS activates downstream genes is unclear.
Interestingly, the ability of WUS to regulate gene expression seems to be closely related to the WUS protein concentration. Low levels of WUS proteins can activate CLV3 expression, while high levels of WUS proteins function in the opposite way [24]. One mechanism underlying these effects could be that the WUS dimer and monomer, depending on the WUS concentration, have opposite effects on the regulation of CLV3 expression. In this process, DNA cis-elements associated with WUS determine the threshold of WUS level for activating or repressing CLV3 transcription [25]. Alternatively, WUS heterodimerization with co-regulators may play opposite roles in regulating downstream genes. Multiple WUS-interacting proteins have been identified in diverse developmental processes. During floral development, KNUCKLES (KNU) competitively interacts with WUS to expel it from the CLV3 promoter, which ultimately inhibits CLV3 expression and terminates the floral meristem (FM) [17]. In SAMs, the HAIRYMERISTEM (HAM) family proteins are expressed in the rib meristem (RM) and associated with WUS, which is important for stem cell homeostasis [26,27]. SHOOT MERISTEMLESS (STM) has also been reported to interact with WUS and bind to the CLV3 promoter for stabilizing the DNA-protein complex to activate CLV3 expression in SAMs [16]. Yet, how WUS initiates CLV3 transcription is largely unknown.
Previous studies have shown that DORNRÖSCHEN (DRN) is mainly expressed in the CZ and directly up-regulates CLV3 transcription independent of its DNA-binding activity [28,29], which suggests that DRN can interact with other proteins to jointly facilitate CLV3 expression.
Here, we depicted a mechanistic framework for modulating CLV3 expression by a protein complex including WUS, DRN, and BRAHMA (BRM). Chromatin remodeling processes by this complex positively regulate CLV3 expression through specific nucleosome depletion, which depends on the unlocking of WUS-DRN anchored chromatin by SWItch/Sucrose Non-Fermentable (SWI/SNF) ATPases. Upon the specific relaxed DNA instead of nucleosome occupation, related transcription factors (TFs) including RNA polymerases are able to access the DNA and commence CLV3 transcription to limit the stem cell population. This model provides a clear mechanism following the association between DNA and TFs, which is required for the positive transcriptional regulatory activity of WUS.
Results
DRN physically interacts with WUS and competitively inhibits WUS homodimerization
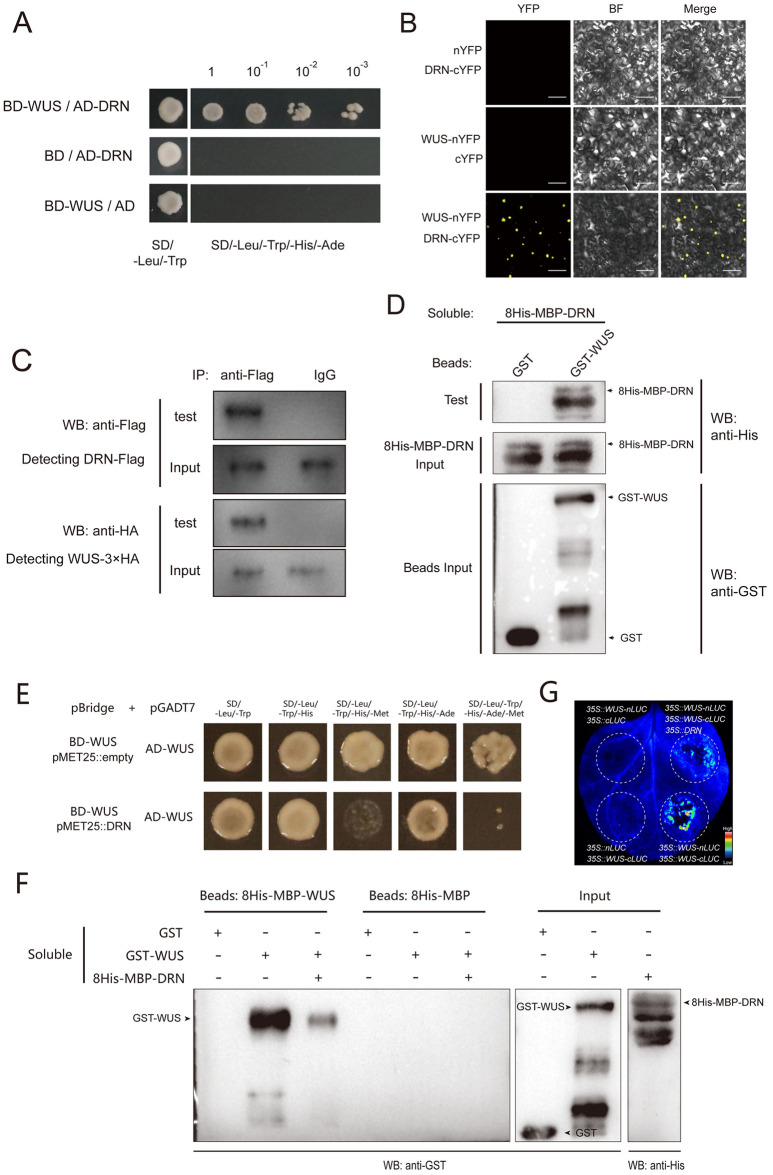
WUS proteins are able to sustain stem cells and bind to the CLV3 promoter to activate its transcription, which maintains stem cell homeostasis [8,9]. DRN can directly activate CLV3 expression independent of its DNA-binding activity [29]. Furthermore, WUS and DRN are both required for shoot regeneration in tissue culture [30]. These findings suggest that WUS may interact with DRN to jointly regulate CLV3 transcription and sustain stem cells. To test this hypothesis, we performed yeast two-hybrid (Y2H) assays and observed that WUS interacted with DRN in yeast cells with the positive control WUS-TPL and the negative control WUS-ARABIDOPSIS RESPONSE REGULATOR 7 (ARR7) (Figs 1A and S1). Additionally, bimolecular fluorescence complementation (BiFC) was also applied in tobacco (Nicotiana benthamiana) leaves, and we observed that WUS and DRN interacted with each other in vivo (Figs 1B and S2). To test this interaction in SAMs, we conducted BiFC assays in Arabidopsis SAMs using split YFP, driven by the native promoters of WUS and DRN, respectively. The fluorescent positive cells indicated that the WUS-DRN interactions occurred in SAMs (S3 Fig). Co-immunoprecipitation (co-IP) assays were also performed, and the immunoblot analysis showed that WUS-3×HA was co-immunoprecipitated with DRN-Flag using the anti-Flag antibody for IP, indicating the interaction of WUS and DRN in Arabidopsis (Fig 1C). To test whether the WUS-DRN interaction was direct, we performed pull-down assays with purified glutathione-S-transferase (GST)-WUS and 8His-maltose binding protein (MBP)-DRN expressed in Escherichia coli (E. coli), which confirmed our hypothesis (Fig 1D). Together, these data demonstrated that DRN physically interacts with WUS and may function as a co-activator in regulating CLV3 expression.
Fig 1. DRN competitively interacts with WUS.
(A) Y2H assays exhibiting the interaction of WUS-DRN. BD and AD empty vectors were used as negative controls. Yeast cells were grown on the selective medium (SD/−Leu/−Trp/−His/−Ade) in a series of dilutions of 10−1, 10−2, and 10−3. Two independent experiments were performed with similar results. (B) BiFC exhibiting that the interaction of WUS-DRN occurs in tobacco leaves. BF, bright field. Scale bars, 100 μm. Five leaves were analyzed for each group. Two independent experiments were performed with similar results. (C) Co-IP showing the interaction of WUS and DRN in Arabidopsis. 35S::DRN-Flag and 35S::WUS-3×HA were transformed into Arabidopsis protoplasts. The anti-Flag antibody was used for IP. Instead of the anti-Flag antibody, IgG was used for IP as the negative control. Two independent experiments were performed with similar results. (D) Pull-down assays showing the interaction of WUS and DRN directly. Recombinant proteins were expressed in E. coli. Anti-His and anti-GST antibodies were used for immunoblot analysis. Two independent experiments were performed with similar results. (E) WUS–WUS interaction is disrupted by DRN in Y3H. Selective media (SD/−Leu/−Trp/−His) and (SD/−Leu/−Trp/−His/−Ade) were used to test the interaction of WUS-WUS. Selective media (SD/−Leu/−Trp/−His/−Met) and (SD/−Leu/−Trp/−His/−Ade/−Met) were used to test the competitive binding of WUS-DRN. Two independent experiments were performed with similar results. (F) Pull-down assays were used to determine the interruption of WUS homodimer by DRN. The binding group demonstrates the interaction of WUS-WUS, which is compromised by introducing 8His-MBP-DRN. Two independent experiments were performed with similar results. (G) Split-luciferase complementation assays in tobacco leaves were conducted to test that DRN competitively interacts with WUS. Five leaves were analyzed in each independent replicate. Two independent experiments were performed with similar results. BiFC, bimolecular fluorescence complementation; Y2H, yeast two-hybrid; Y3H, yeast three-hybrid.
Given that WUS contains multiple domains, we employed Y2H to determine which domain is responsible for the WUS-DRN interaction. The results exhibited that the middle region of WUS is required for the interaction with DRN (S4 Fig). With respect to DRN, the N-terminal part containing an AP2 domain (DNA-binding domain) accounts for the WUS-DRN interaction, but not the C-terminus (S5 Fig).
Previous studies have shown that WUS proteins can form homodimers via their middle region [9,24], which was also confirmed in our BiFC and Y2H assays (S6 Fig). The fact that the middle region of WUS contributes to both the WUS-WUS homodimers and WUS-DRN heterodimers suggests a hypothesis that the WUS-DRN interaction may compete with WUS homodimerization. To test this possibility, we carried out yeast three-hybrid (Y3H) assays. BD-WUS and AD-WUS were introduced into yeast along with the inducible DRN proteins driven by a conditional methionine promoter (pMET25). In the absence of methionine, pMET25 promoter transcription occurred, and the induction of DRN proteins greatly impaired the WUS-WUS interaction (Fig 1E). These results indicated that DRN is able to disturb the WUS homodimerization in yeast. To exclude the effects of unrelated proteins in yeast cells, we performed pull-down assays with purified GST-WUS, 8His-MBP-WUS, and 8His-MBP-DRN, expressed in E. coli. These results, agreeing with those of Y3H, displayed that WUS dimerization was largely interrupted in the presence of DRN proteins through competitive binding in vitro (Fig 1F). In the competitive group of this pull-down experiment, the agarose beads with attached 8His-MBP-WUS proteins had been incubated with GST-WUS soluble proteins for 6 h before adding the 8His-MBP-DRN soluble proteins, which ruled out the possibility that the amount of free agarose beads loaded with 8His-MBP-WUS would decrease in the presence of 8His-MBP-DRN. In addition, split-luciferase complementation assays showed that DRN disrupted WUS homodimerization in vivo (Fig 1G). Collectively, our data demonstrated that DRN competitively interacts with WUS, which suggests that the WUS-DRN complex activates CLV3 expression in the CZ.
Direct activation of CLV3 expression by WUS-DRN interactions
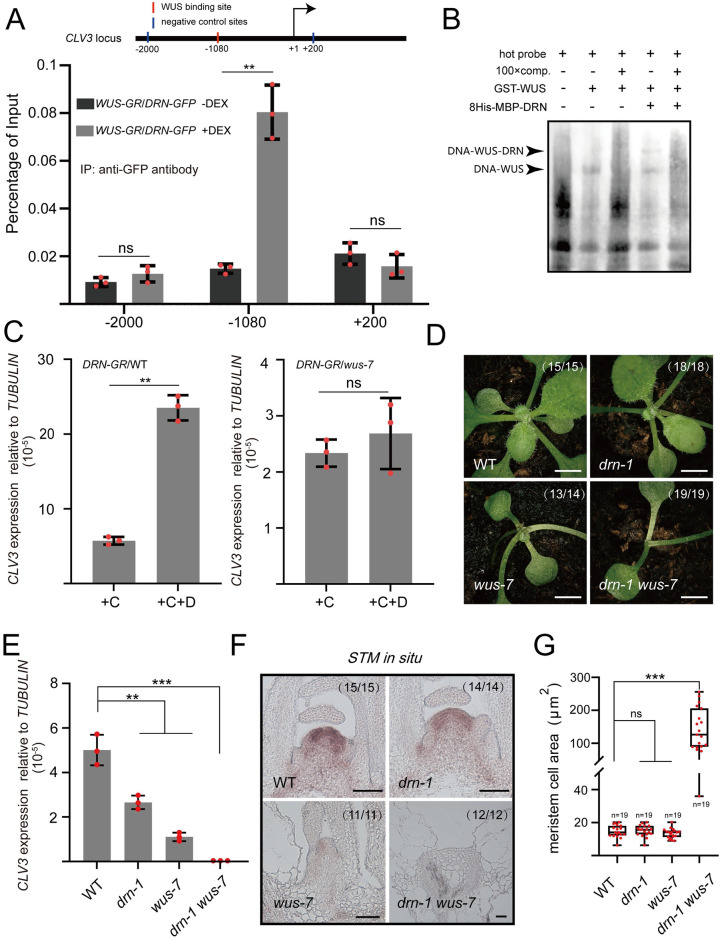
To further explore the regulation of CLV3 expression by the WUS-DRN complex, we generated transgenic plants, carrying UBQ10::DRN-GFP and UBQ10::mCherry-WUS-GR that produce dexamethasone (DEX) inducible WUS-GR. Chromatin immunoprecipitation (ChIP) was employed to determine the associations between DRN proteins and CLV3 chromatin with or without DEX induction. The results showed that, upon nuclear localization of WUS-GR after DEX treatment, DRN strongly enriched the fragment of the CLV3 promoter containing the WUS-binding site (TAAT, upstream from −1,082 to −1079), using the anti-GFP antibody for IP (Fig 2A). In contrast, DRN-GFP failed to effectively associate with the CLV3 promoter without DEX induction (Fig 2A). However, why did the ChIP assays fail to detect the association of CLV3 promoter with DRN in our previous study [29]? We reasoned that DRN::DRN-GFP lines, used for ChIP assays in the previous study [29], were in shortage of cells expressing WUS and DRN, and thus, extremely low abundance of DNA-WUS-DRN were immunoprecipitated. In the present study, we used materials with abundant DRN and inducible WUS, UBQ10::mCherry-WUS-GR/UBQ10::DRN-GFP lines, and succeeded in detecting the association of CLV3 promoter by immunoprecipitation of DRN-GFP. These data indicated that DRN can access the CLV3 promoter, which relies on WUS proteins in vivo. Previous studies have reported that multiple additional WUS-binding sites exist in downstream of CLV3, called the “cis-regulatory module” (CRM), and are involved in modulating CLV3 expression [24,25]. We also performed ChIP to check the association of CRM DNA with the WUS-DRN complex. The results showed that CRM DNA cannot be co-immunoprecipitated with DRN-GFP in the presence of inducible WUS-GR (S7 Fig). We considered that WUS-DRN fails to associate with CRM DNA, which may be attributed to the low threshold of WUS level for WUS dimers in the CRM region [25]. To further investigate whether the WUS-DRN complex recognizes the fragment of the CLV3 promoter with a WUS-binding site (upstream from −1,082 to −1,079), electrophoretic mobility shift assays (EMSAs) were conducted using the fragment of CLV3 promoter (upstream from −1,090 to −1,064) as a probe. We observed a shift band in the presence of WUS, and an additional super-shift in the presence of both WUS and DRN (Fig 2B). Given that DRN cannot associate with the CLV3 promoter alone [29], the ChIP and EMSA data in the present study suggested a regulatory model in which the activation of CLV3 expression by DRN depends on WUS. To test this model genetically, we generated CLV3::DRN-GR/WT transgenic lines and introduced CLV3::DRN-GR into wus-7, a weak mutant allele of WUS, by crossing. Since the wus null mutants, in the absence of SAMs, fail to illustrate the genetic interaction on regulating SAM development, the weak mutant wus-7 was selected for analysis (S8A Fig) [26,31]. After DEX induction, the CLV3 expression of CLV3::DRN-GR/WT was significantly up-regulated compared with the control group (Fig 2C). However, the same DEX induction had no effect on CLV3 transcription in CLV3::DRN-GR/wus-7 (Fig 2C), which indicated that the activation of CLV3 transcription by DRN-GR relies on WUS.
Fig 2. WUS-DRN complex associates with CLV3 promoter to regulate CLV3 expression.
(A) The 14-day-old seedlings were used for ChIP assays. Upon DEX induction, the nuclear localization of WUS-GR enables the association of CLV3 promoter (WUS-binding site, from −1,082 to −1,079) with DRN-GFP, using anti-GFP antibodies for IP. The upstream −2,000 bp site and downstream +200 site acted as negative control loci (no binding site). Two independent experiments were performed with similar results. (B) EMSAs results showing the direct binding of WUS-DRN and the CLV3 promoter fragment enriched in ChIP. The black arrows indicate protein-DNA complexes. Two independent experiments were performed with similar results. (C) qRT-PCR was applied to test the relative transcript level of CLV3 in CLV3::DRN-GR/WT and CLV3::DRN-GR/wus-7 after DEX induction compared with the mock, using 14-day-old seedlings. +C, cycloheximide; +D, dexamethasone. Two independent experiments were performed with similar results. (D) Phenotypes of 14-day-old WT, drn-1, wus-7, and drn-1 wus-7. Scale bars, 1 mm. Two independent experiments were performed with similar results. (E) qRT-PCR was applied to test the relative CLV3 expression in drn-1, wus-7, and drn-1 wus-7 compared with WT, using 14-day-old seedlings. Two independent experiments were performed with similar results. (F) STM expression patterns were checked in SAMs of WT, drn-1, wus-7, and drn-1 wus-7 by RNA in situ hybridization, using 14-day-old seedlings. Scale bars, 50 μm. Two independent experiments were performed with similar results. (G) Meristematic cell sizes of WT, drn-1, wus-7, and drn-1 wus-7 in F were measured by image J software. Black bars, highest and lowest values; box, median 50%; black line in the box, median. Two independent experiments were performed with similar results. ***P < 0.001; **P < 0.01; ns, no significant difference; Student’s t test in A, C, E, and G. Data represent means ± SDs from 3 biological replicates in A, C, and E. The data underlying this figure can be found in S1 Data. ChIP, chromatin immunoprecipitation; DEX, dexamethasone; EMSA, electrophoretic mobility shift assay; SAM, shoot apical meristem.
To further explore the biological significance of the WUS-DRN interaction, we analyzed drn-1, wus-7, and drn-1 wus-7 mutants. The drn-1 single mutants showed no obvious phenotypic defect in post-embryonic development, whereas the wus-7 mutants grew slowly accompanied by a decrease of true leaves (Figs 2D and S8B–S8D). Interestingly, drn-1 wus-7 double mutants showed severely arrested SAMs and fewer true leaves than wus-7 single mutants, similar to wus-8, a null mutant allele of WUS (Figs 2D, S8A, S8E and S8F) [32]. The inflorescence meristem was also analyzed, and the drn-1 wus-7 double mutants completely lacked their meristem, more severe than drn-1 and wus-7 single mutant (S9A and S9B Fig). The transcript level of CLV3 significantly declined in the drn-1 and wus-7 single mutants, consistent with the activating roles of DRN and WUS (Figs 2E and S8J–S8L). However, the CLV3 expression was barely detected in the drn-1 wus-7 double mutant, similar to the wus-8 null mutants (Figs 2E, S8M and S8N). Given the severe defects in true leaf formation in drn-1 wus-7 double mutants, we speculated that the SAMs of the double mutants are completely dysfunctional. To test this hypothesis, RNA in situ hybridization was performed to examine the STM expression pattern in wild-type, drn-1, wus-7, and drn-1 wus-7 plants, as STM is a well-recognized marker gene of the SAM [33]. The results showed that there was no significant difference in the region of STM expression in SAMs between drn-1 and the wild type, whereas a decrease in wus-7, and STM RNA was barely detectable in drn-1 wus-7 (Fig 2F). Additionally, it was exhibited that cells in the meristem region of drn-1 wus-7 were much larger than those of wild-type, drn-1, and wus-7 plants, indicating that these cells in drn-1 wus-7 are highly differentiated rather than normally smaller meristematic cells (Fig 2F and 2G). We also generated drn-1 wus-8 double mutants, showing similar phenotype to drn-1 wus-7 and wus-8 (S9A and S9B Fig), suggesting the role of DRN-WUS interaction in sustaining stem cells.
Since no obvious phenotypic defect in drn-1 was observed, we also crossed the drn-1 drnl-2 double mutant with wus-7. DORNRÖSCHEN-LIKE (DRNL) is a homolog of DRN, and they have been reported to redundantly modulate shoot stem cell homeostasis and organ initiation [29,34]. The drn-1 drnl-2 double mutant showed an enlarged SAM, a decrease in the CLV3 expression region (S8B, S8C, S8G, S8H, S8J, S8K, S8O and S8P Fig), as well as a pin-like inflorescence in the reproductive stage (S9A–S9C Fig) [29,34], whereas the SAM was absolutely absent in drn-1 drnl-2 wus-7 triple mutants without subsequent development (S8I and S8Q Fig). Moreover, we generated drn-1 clv3-7 and wus-7 clv3-7 double mutants using clv3-7, a null allele of CLV3 [5]. We found that there was no significant difference in SAM size between drn-1 clv3-7 and clv3-7 (S10A and S10B Fig). The SAM size of wus-7 clv3-7 decreased obviously compared with clv3-7, and increased obviously compared with wus-7, indicating that CLV3 negatively regulates WUS expression to maintain stem cell activity (S10A and S10B Fig). Together with the severe defects of SAMs in drn-1 wus-7, these data suggested that the WUS-DRN complex is not only involved in the regulation of CLV3 expression, but also extensively participates in sustaining stem cells, and WUS seems to act as a master regulator in this process.
DRN physically interacts with BRM
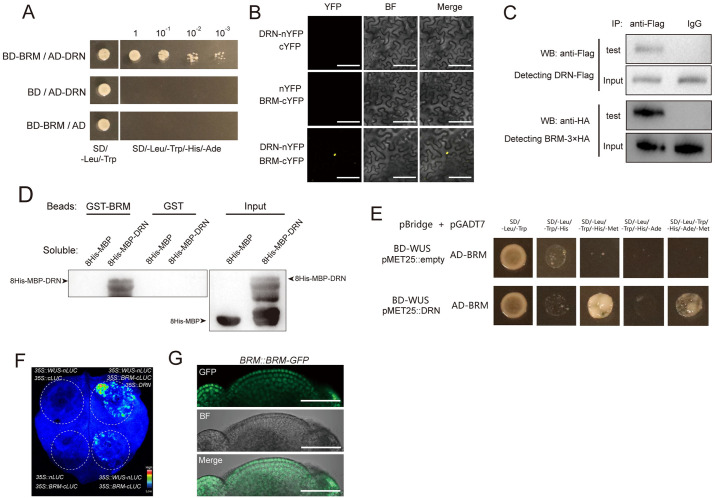
To further explore the mechanism underlying the activation of CLV3 by WUS-DRN, we hypothesized that the WUS-DRN complex recruits additional proteins to facilitate the transcription of CLV3. To test this possibility, Y2H screening was employed to identify candidate proteins using BD-DRN as the bait. The results showed that BRM was identified as a candidate protein interacting with DRN and was independently isolated 3 times among 512 sequenced clones. BRM is a vital factor involved in the chromatin remodeling process to enable gene transcription [35–37], and mutating BRM leads to a decrease in SAM size [38]. Y2H assays were subsequently used to confirm the DRN-BRM interaction using the full-length coding sequences of DRN and BRM (Fig 3A). We also identified the N-terminus of BRM (amino acids 1–976), which is responsible for the interaction with DRN, using truncated BRM in Y2H assays (S11 Fig). We further adopted BiFC assays to verify DRN-BRM interactions in tobacco leaves (Figs 3B and S12). BiFC assays were also performed in Arabidopsis using DRN::DRN-nYFP and BRM::BRM-cYFP transgenic plants, which showed positive signals in the SAM, indicating that DRN-BRM interactions can occur in meristematic cells (S13 Fig). In addition, co-IP was applied to test this DRN-BRM interaction in Arabidopsis (Fig 3C). To further determine whether the DRN-BRM interaction is direct, pull-down assays were conducted, and the results showed a direct interaction between DRN and BRM in vitro (Fig 3D).
Fig 3. BRM interacts with DRN mediating WUS-DRN-BRM complex.
(A) Y2H shows the interaction of DRN-BRM. The combinations with BD and AD empty vectors were introduced as negative controls. Yeast cells in a series of dilutions were grown on the selective medium (SD/−Leu/−Trp/−His/−Ade). Two independent experiments were performed with similar results. (B) BiFC exhibiting the interaction of DRN-BRM in tobacco leaves. YFP was split into N-terminus and C-terminus, which were fused to DRN and BRM, respectively. BF, bright field. Scale bars, 100 μm. Two independent experiments were performed with similar results, and 5 leaves were analyzed for each group. (C) Co-IP showing the interaction of DRN and BRM in Arabidopsis. 35S::DRN-Flag and 35S::BRM-3×HA were transformed into Arabidopsis protoplasts. The anti-Flag antibody was used for IP. Instead of the anti-Flag antibody, IgG was used for IP as the negative control. Two independent experiments were performed with similar results. (D) Pull-down assay showing the interaction of DRN and BRM directly. Recombinant proteins were expressed in E. coli and the anti-His antibody was used for immunoblot analysis. Two independent experiments were performed with similar results. (E) Y3H results showing that WUS fails to directly bind to BRM (upper lane) and that introducing DRN produces the WUS-DRN-BRM complex (bottom lane). SD/−Leu/−Trp/−His and SD/−Leu/−Trp/−His/−Ade were used for selective media. Lacking Met in addition allowed the transcription of pMET25 promoter to produce DRN in the bottom lane but not in the upper lane. Two independent experiments were performed with similar results. (F) Split-luciferase complementation assays were used to test the association between WUS and BRM mediated by DRN in tobacco leaves. Two independent experiments were performed with similar results. Six leaves were analyzed for each independent replicate. (G) BRM::BRM-GFP lines were used to detect the distribution of BRM-GFP in SAMs. Green, GFP signal; BF, bright field. Scale bars, 50 μm. Eight apices were analyzed. BiFC, bimolecular fluorescence complementation; SAM, shoot apical meristem; Y2H, yeast two-hybrid; Y3H, yeast three-hybrid.
To test which region of the DRN protein is responsible for the interaction with BRM, truncated DRN proteins were used to perform Y2H assays. We observed that the C-terminus of DRN interacts with BRM (S14A and S14B Fig). Given that the DRN N-terminus interacts with WUS (S5 Fig), these data suggested that DRN may bridge the association between WUS and BRM and that the WUS-DRN-BRM protein complex may represent the molecular basis for WUS to activate CLV3 expression. To further verify the protein interaction module of WUS-DRN-BRM, Y3H assays were performed using pBridge (BD-WUS, pMET25::DRN) and pGADT7 (AD-BRM) constructs. In the presence of methionine, the pMET25 promoter was inhibited and WUS alone failed to interact with BRM (Fig 3E). When DRN proteins were expressed in yeast cells without methionine, the yeast reporter genes HIS3 and ADE2 were activated, indicating that the association between WUS and BRM relies on DRN (Fig 3E). In addition, split-luciferase assays showed that DRN functioned as a bridge connecting WUS and BRM in vivo (Fig 3F).
WUS and DRN proteins have been reported to be located and function in shoot stem cells [8,9,28,29]. DRN::DRN-GFP/drn-1 and WUS::WUS-GFP/wus-8 rescue lines were used to determine the distribution of DRN and WUS proteins in SAMs. The results showed that DRN and WUS proteins were present in shoot stem cells, whereas low levels of WUS were detected in L1 and L2 cell layers, and CLV3::GFP/WT lines were used to mark the shoot stem cells, as described in previous studies (S15A–S15C Fig) [5,8,10,29]. To carefully examine the expression pattern of BRM in the SAM, BRM::BRM-GFP lines were used and the results showed that BRM-GFPs occupied the whole SAM including the central zone, in line with a previous study [35] (Fig 3G). Specifically, abundant BRM-GFP proteins were located within the L1 cell layer of SAMs (S15D Fig). RNA in situ hybridization was also used to detect BRM mRNAs, which showed that BRM transcripts exist throughout the SAM, including in stem cells, but no signal in the sense probe control (S15E and S15F Fig). The presence of WUS, DRN, and BRM proteins in stem cells suggested that they may function in maintaining stem cells.
Collectively, DRN interacts with WUS and BRM proteins, forming a large protein complex mediated by DRN, which suggests a mechanism of regulating CLV3 transcription.
DRN and BRM jointly regulate CLV3 transcription
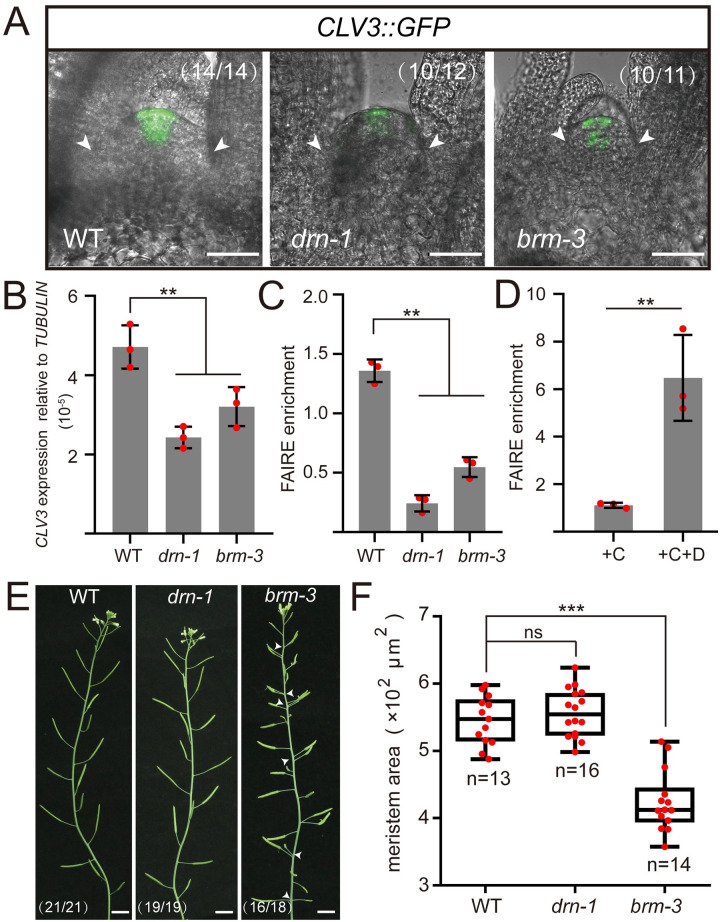
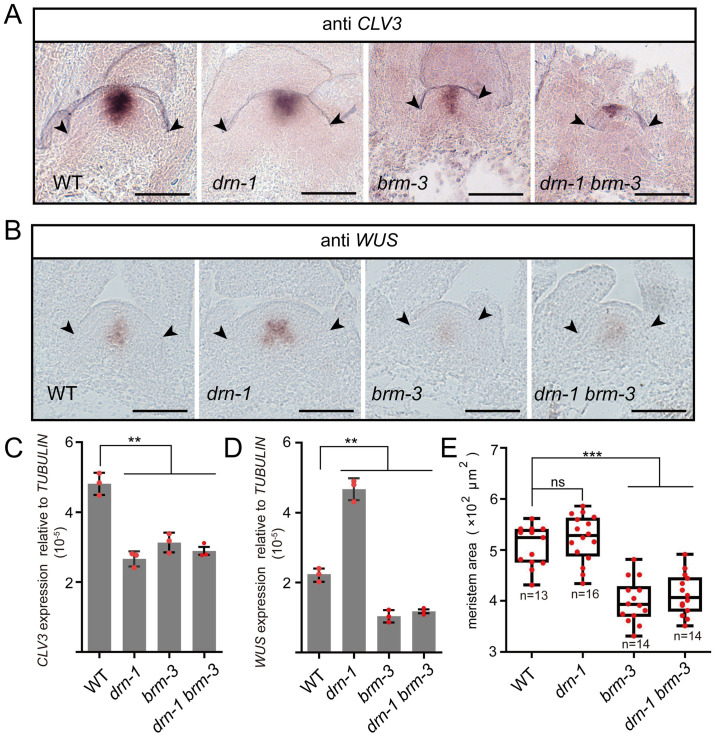
To further investigate the function of the WUS-DRN-BRM complex in regulating CLV3 transcription, we analyzed CLV3 expression in wild-type, drn-1, and brm-3 plants by introducing CLV3::GFP. We observed that the transcriptional activity of CLV3 promoter declined in drn-1 and brm-3 mutants compared with wild-type plants (Fig 4A). Consistently, qRT-PCR was also used to check CLV3 transcripts, which demonstrated the markedly reduced CLV3 expression in the drn-1 and brm-3 mutants compared with wild-type plants (Fig 4B).
Fig 4. BRM contributes to CLV3 expression and SAM maintenance.
(A) CLV3 expression was checked in WT, drn-1, and brm-3 introduced with CLV3::GFP, using 14-day-old seedlings. The white arrows indicate the boundaries of SAMs. Scale bars, 50 μm. Two independent experiments were performed with similar results. (B) qRT-PCR was applied to test the relative CLV3 transcript levels of drn-1 and brm-3 compared with WT, using 14-day-old seedlings. Two independent experiments were performed with similar results. (C) DNA accessibility at the CLV3 locus was evaluated by FAIRE experiments in WT, drn-1, and brm-3, using 10-day-old seedlings. The ratio of FAIRE enrichment at the CLV3 promoter (WUS-binding site −1,080) was normalized to the Ta3 retrotransposon. Two independent experiments were performed with similar results. (D) DNA accessibility at the CLV3 locus (WUS-binding site −1,080) was checked by FAIRE assays in 35S::DRN-GR/WT after DEX treatment, using 10-day-old seedlings. The ratio of FAIRE enrichment at the CLV3 promoter (WUS-binding site −1,080) was normalized to the Ta3 retrotransposon. +C, cycloheximide; +D, dexamethasone. Two independent experiments were performed with similar results. (E) The inflorescences of WT, drn-1, and brm-3 are shown. The white arrows indicate phyllotaxy defects. Scale bars, 1 cm. (F) The areas of SAMs in A were measured by image J and statistically analyzed. Black bars, highest and lowest values; box, median 50%; black line in the box, median. Two independent experiments were performed with similar results. ***P < 0.001; **P < 0.01; ns, no significant difference; Student’s t test in B, C, D, and F. Data represent means ± SDs from 3 biological replicates in B, C, and D. The data underlying this figure can be found in S1 Data. DEX, dexamethasone; FAIRE, formaldehyde-assisted isolation of regulatory elements; SAM, shoot apical meristem.
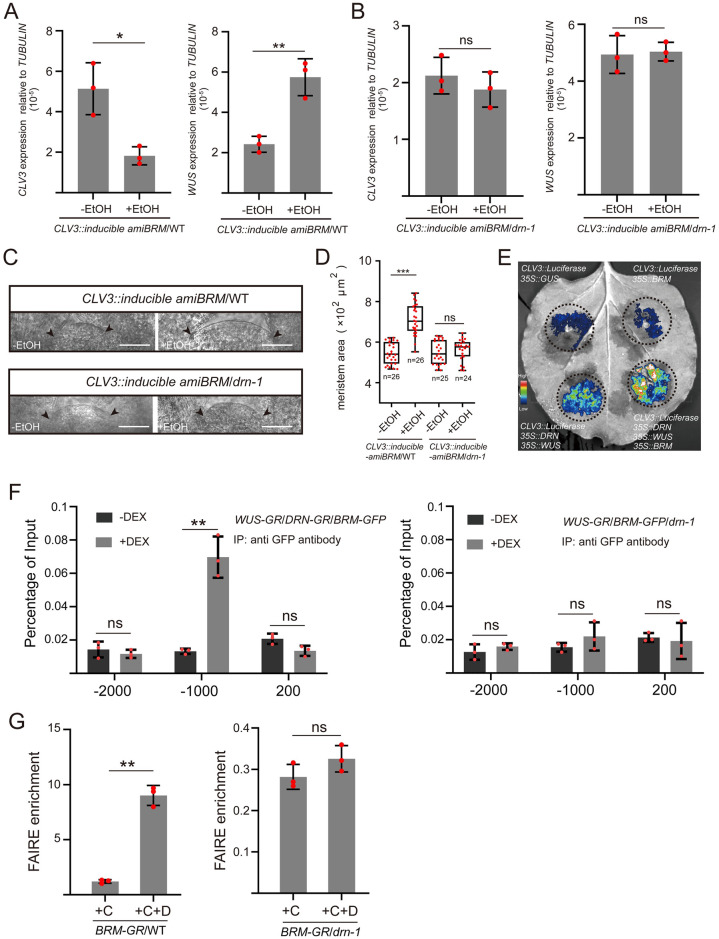
To further confirm the regulation of CLV3 expression by BRM, ethanol-inducible artificial microRNA-BRM (amiBRM) was introduced into wild-type plants. CLV3 transcripts were subsequently examined by qRT-PCR in ethanol-inducible amiBRM lines, and the results showed that CLV3 transcription was markedly reduced after 1% ethanol induction for 24 h, accompanied by the partial silencing of BRM transcripts (S16A and S16B Fig). These data indicated that both DRN and BRM contribute to the regulation of CLV3 transcription.
Many studies have reported that chromatin remodeling complexes, such as SWI/SNF, are able to disrupt nucleosome structure and produce relaxed DNA, enabling the access of TFs, which eventually initiates gene transcription [35,37]. Considering that BRM is referred to as a key component in the SWI/SNF machinery in regulating gene transcription [36], we speculated that the WUS-DRN-BRM interactions might recruit SWI/SNF to the CLV3 promoter, allowing specific nucleosome depletion to initiate CLV3 transcription. To test this hypothesis, we performed formaldehyde-assisted isolation of regulatory elements (FAIRE) assays to examine the nucleosome-depleted regions (NDRs) in the CLV3 promoter including the WUS-binding site (TAAT, from −1,082 to −1,079) in drn-1 and brm-3 mutants. In line with the reduced CLV3 expression in these mutants, we observed the decline of NDRs enrichment in drn-1 and brm-3 mutants compared with the wild type, whereas no significant change was detected in −2,000 upstream site, as a negative control, indicating reduced DNA accessibility in these mutants (Figs 4C and S17A). In addition, FAIRE assays were also applied to 35S::DRN-GR/WT lines. Upon DEX induction, we observed a higher level of NDRs located at the CLV3 promoter, including WUS-binding site (TAAT, from −1,082 to −1,079), agreeing with the activation of CLV3 expression by DEX treatment (Figs 2C and 4D, and S17B), but not found in CLV3 downstream CRM region (S18 Fig). These data demonstrated that the WUS-DRN-BRM complex participates in nucleosome destabilization of the CLV3 promoter, eventually allowing its transcription. In addition to disturbed CLV3 expression, we also observed a decrease of SAM size and severely disturbed phyllotaxis in brm-3 mutants but no significant change in drn-1 mutants (Fig 4A, 4E, and 4F), suggesting additional functions of BRM in regulating SAM activity.
To determine the genetic interaction between DRN and BRM, we generated drn-1 brm-3 double mutants by crossing. Similar to single mutants, CLV3 expression in drn-1 brm-3 double mutants was decreased significantly compared with wild-type plants (Figs 5A, 5C, and S19). Despite decreased CLV3 expression, WUS mRNAs were surprisingly reduced in brm-3 and drn-1 brm-3, examined by RNA in situ hybridization, qRT-PCR, and the florescent reporter (Figs 5B, 5D, and S19), which agreed with the reduced SAM sizes of brm-3 and drn-1 brm-3 in both vegetative and reproductive development (Figs 5E, S20A and S20B). To exclude the possibility that overall developmental defects in brm-3 affect SAM activity, we checked the number of true leaves of 14-day-old seedlings, showing no significant difference between the wild type and mutants (S21A and S21B Fig). Although WUS expression in drn-1 increased moderately, that in drn-1 brm-3 still decreased, similar to brm-3, indicating that BRM is epistatic to DRN in regulating WUS transcription (Fig 5D). In addition, we generated brm-3 clv3-7 double mutants, which showed a decreased SAM size compared to clv3-7 (S10A and S10B Fig). Together, we concluded that BRM not only regulates CLV3 expression, but may also participate in additional signaling pathways to sustain SAM activity.
Fig 5. BRM is epistatic to DRN in regulating WUS transcription.
(A) CLV3 mRNAs were checked in SAMs of WT, drn-1, brm-3, and drn-1 brm-3 by RNA in situ hybridization, using 14-day-old seedlings. Scale bars, 50 μm. Two independent experiments were performed with similar results, and 10 samples of each mutant were analyzed. (B) WUS mRNAs were checked in SAMs of WT, drn-1, brm-3, and drn-1 brm-3 by RNA in situ hybridization, using 14-day-old seedlings. Scale bars, 50 μm. Two independent experiments were performed with similar results, and 10 samples of each mutant were analyzed. (C) qRT-PCR was applied to test the relative CLV3 transcript levels of drn-1, brm-3, and drn-1 brm-3 compared with WT, using 14-day-old seedlings. Two independent experiments were performed with similar results. (D) qRT-PCR was applied to test the relative WUS transcript levels of drn-1, brm-3, and drn-1 brm-3 compared with WT, using 14-day-old seedlings. Two independent experiments were performed with similar results. (E) The areas of SAMs in A and B were measured by Image J software. Black bars, highest and lowest values; box, median 50%; black line in the box, median. Two independent experiments were performed with similar results. Black arrows indicate the boundaries of SAMs in A and B. **P < 0.01; ***P < 0.001; ns, no significant difference; Student’s t test in C, D, and E. Data represent means ± SDs from 3 biological replicates in C and D. The data underlying this figure can be found in S1 Data. SAM, shoot apical meristem.
Given the decrease of WUS expression in the brm-3 mutant, we fail to rule out the likelihood that the decline in CLV3 expression is a consequence of the reduced WUS levels in brm-3. To address this issue, we generated transgenic plants with ethanol-inducible amiBRM driven by the CLV3 promoter (CLV3::inducible amiBRM) to specifically knock down BRM mRNAs in shoot stem cells. Upon ethanol induction for 24 h, RNA in situ hybridization results showed that BRM transcripts faded away in stem cells, but not in the OC and inner cells (S22 Fig). In CLV3::inducible amiBRM/WT plants, knocking down BRM in stem cells by ethanol induction for 24 h resulted into a decrease of CLV3 transcription and, subsequently, an increase of WUS transcription (Fig 6A), whereas not occurring in the ethanol-inducible GUS control (S23A and S23B Fig). However, the CLV3 and WUS transcription were not altered in CLV3::inducible amiBRM/drn-1 plants (Fig 6B). Agreeing with the alteration of CLV3 and WUS transcription, the SAM size noticeably increased in CLV3::inducible amiBRM/WT plants after ethanol treatment for 72 h, which was not found in CLV3::inducible amiBRM/drn-1 plants and GUS negative control (Figs 6C, 6D, S23C and S23D). These data indicated that BRM regulates stem cell activity via modulating CLV3 expression, which requires DRN. Furthermore, transient transfection of tobacco leaves was used to illustrate the synergistic effects of WUS, DRN, and BRM in regulating CLV3 transcription. The results showed that the presence of WUS and DRN notably activated the activity of the CLV3 promoter, whereas the luciferase signal of the group with BRM alone was comparable to the GUS control (Fig 6E). Introducing WUS, DRN, and BRM simultaneously resulted in much more intense signals (Fig 6E), indicating that the activation of CLV3 expression by BRM relies on DRN and WUS.
Fig 6. The positive regulation of CLV3 by BRM relies on DRN.
(A) qRT-PCR was applied to test the relative transcript levels of CLV3 and WUS after ethanol (EtOH) induction for 24 h in CLV3::inducible amiBRM/WT transgenic seedlings (14-day-old). Two independent experiments were performed with similar results. (B) qRT-PCR was applied to test the relative transcript levels of CLV3 and WUS after ethanol (EtOH) induction for 24 h in CLV3::amiBRM/drn-1 transgenic seedlings (14-day-old). Two independent experiments were performed with similar results. (C) SAMs of the mock group and EtOH treated group (72 h) in CLV3::inducible amiBRM/WT and CLV3::inducible amiBRM/drn-1 transgenic seedlings (14-day-old). Black arrows indicate the boundaries of SAMs. Bars, 50 μm. Two independent experiments were performed with similar results. (D) Areas of SAMs in C were measured by Image J software. Two independent experiments were performed with similar results. (E) Different combinations of vectors, including CLV3::Luciferase, 35S::GUS, 35S::WUS, 35S::DRN, and 35S::BRM were transformed into tobacco leaves via Agrobacterium. The intensity of luciferase signal represents the activity of CLV3 promoter. The regions where Agrobacterium transformed were circled by dotted lines. Two independent experiments were performed with similar results. (F) UBQ10::mCherry-WUS-GR/35S::DRN-GR/UBQ10::BRM-GFP lines (14-day-old seedlings) were used for ChIP assays. Upon DEX induction, the nuclear localization of WUS-GR enables the association of the CLV3 promoter (WUS-binding site, from −1,082 to −1,079) by BRM-GFP, using the anti-GFP antibody for IP, but not found in UBQ10::mCherry-WUS-GR/UBQ10::BRM-GFP/drn-1 lines. The upstream −2,000 bp site and downstream +200 site acted as negative control loci (no binding site). Two independent experiments were performed with similar results. (G) DNA accessibility at the CLV3 locus (WUS-binding site −1,080) was checked by FAIRE assays after DEX treatment, using UBQ10::BRM-GR/WT and UBQ10::BRM-GR/drn-1 lines (10-day-old seedlings). The ratio of FAIRE enrichment at the CLV3 promoter (WUS-binding site −1,080) was normalized to Ta3 retrotransposon. +C, cycloheximide; +D, dexamethasone. Two independent experiments were performed with similar results. *P < 0.05; **P < 0.01; ***P < 0.001; ns, no significant difference; Student’s t test in A, B, D, F, and G. Data represent means ± SDs from 3 biological replicates in A, B, F, and G. The data underlying this figure can be found in S1 Data. ChIP, chromatin immunoprecipitation; DEX, dexamethasone; FAIRE, formaldehyde-assisted isolation of regulatory elements; SAM, shoot apical meristem.
In order to test the association between CLV3 and BRM, we conducted ChIP assays using UBQ10::WUS-GR/35S::DRN-GR/UBQ10::BRM-GFP lines and UBQ10::WUS-GR/UBQ10::BRM-GFP/drn-1 lines. We observed that BRM was able to bind to the CLV3 promoter (−1,080 site) in the presence of DRN-GR and WUS-GR induced by DEX, which was not found in drn-1 background (Fig 6F). We also performed FAIRE assays to test NDR levels using UBQ10::BRM-GR and UBQ10::BRM-GR/drn-1 lines. The results showed that BRM overexpression could up-regulate the NDR level of CLV3 promoter, but not in drn-1 background, indicating that the modification of CLV3 promoter chromatin by BRM requires DRN (Fig 6G). These data genetically demonstrated that BRM modulates CLV3 expression by altering the chromatin state to tune WUS/CLV3 feedback loop and maintain stem cell activity, which depends on WUS and DRN.
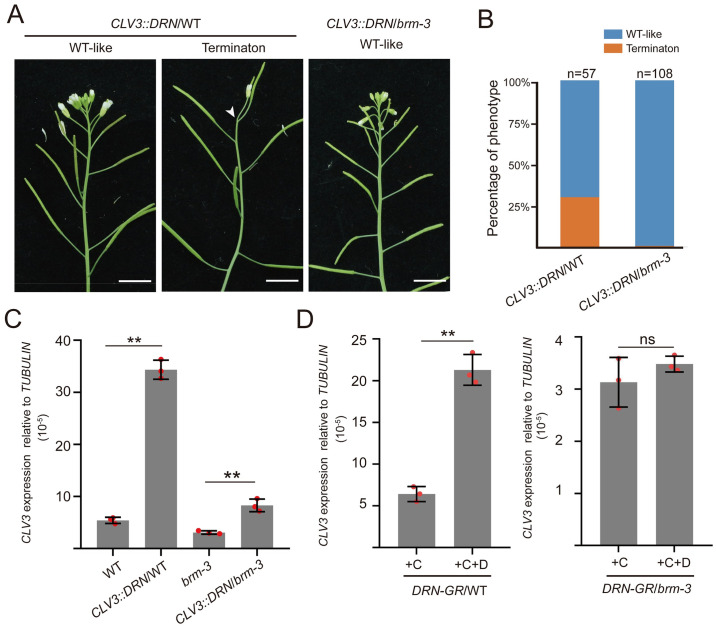
To conversely determine whether the activation of CLV3 expression by DRN required BRM, we generated 2 transgenic plants, CLV3::DRN/WT and CLV3::DRN/brm-3. Consistent with our previous study [29], CLV3::DRN/WT lines failed to produce SAMs, which was not found in CLV3::DRN/brm-3 lines (Fig 7A and 7B). Moreover, qRT-PCR experiments were also performed on the 14-day-old seedlings of these 2 transgenic plants, and the results exhibited a dramatic increase in CLV3 expression in CLV3::DRN/WT lines compared with wild-type plants (Fig 7C). However, the marked increase of CLV3 transcription caused by CLV3::DRN was compromised in the CLV3::DRN/brm-3 lines (Fig 7C), agreeing with the phenotypes of these 2 transgenic lines (Fig 7A and 7B), which suggests that the positive regulation of CLV3 expression by DRN requires BRM.
Fig 7. The activation of CLV3 expression by DRN requires BRM.
(A, B) The phenotypes of CLV3::DRN transgenic plants in WT and brm-3 backgrounds are shown and statistically analyzed. The plants in A were analyzed in B. Two independent experiments were performed with similar results. (C) qRT-PCR was used to test the relative transcript level of CLV3 in CLV3::DRN/WT, brm-3, and CLV3::DRN/brm-3 lines, using 14-day-old seedlings. Two independent experiments were performed with similar results. (D) qRT-PCR was applied to test the relative CLV3 transcript level of CLV3::DRN-GR in the wild-type background or brm-3 mutant background after DEX induction compared with the mock group, using 14-day-old seedlings. +C, cycloheximide; +D, dexamethasone. Two independent experiments were performed with similar results. **P < 0.01; ns, no significant difference; Student’s t test in C, and D. Data represent means ± SDs from three biological replicates in C and D. The data underlying this figure can be found in S1 Data. DEX, dexamethasone.
To test whether the direct regulation of CLV3 expression by DRN requires BRM, we introduced CLV3::DRN-GR into wild-type and brm-3 mutant plants. After DEX induction, CLV3 expression was significantly activated compared with the mock group in CLV3::DRN-GR/WT plants, whereas the brm-3 mutation attenuated the up-regulation of CLV3 expression induced by DEX in CLV3::DRN-GR/brm-3 plants (Fig 7D). Taken together, our data demonstrated that DRN interacts with BRM and synergistically regulates CLV3 expression depending on each other.
Discussion
Stem cells embedded in the SAM generate almost all aerial organs of plants, which is critical for the success of plant life cycles and human sustenance. The WUS/CLV3 feedback loop, known as a classical circuit in controlling the stem cell pool, has been extensively studied, and WUS has dual functions of modulating gene expression for stem cell activity [15]. Here, we showed that DRN competitively interacts with WUS and recruits BRM, located at the CLV3 promoter, to activate its transcription via nucleosome depletion. This finding explains the mechanism by which WUS positively regulates gene transcription, whereas the negative transcriptional activity of WUS relies on the interaction with TPL [22,23]. Moreover, previous studies have reported that a dose-dependent manner confers the transcriptional regulatory ability of WUS [24]. WUS monomers, in the low levels of WUS proteins, tend to activate CLV3 expression, whereas high levels of WUS result in WUS dimers to negatively regulate CLV3 expression [24]. Our results showed that DRN competitively interacts with WUS to disturb the WUS dimer and recruits BRM to form the WUS-DRN-BRM complex, which uncovers the molecular base of WUS monomer activating CLV3 expression. Yet, how the WUS dimer, with its homodimerization threshold is affected by CRM, represses CLV3 expression remains elusive [24].
CLV3, acting as the marker gene of shoot stem cells, specifically transcribes in the CZ [5], which is under the precise control of multiple gene regulatory networks. WUS transcribes in the OC, and a fraction of WUS proteins move to stem cells to activate CLV3 expression. However, a large portion of WUS proteins are retained in the OC and accumulate at high concentrations, which allow WUS homodimerization to repress CLV3 expression [24]. Additionally, HAMs have been reported to be expressed in deep cell layers beneath stem cells and to repress CLV3 expression [27]. Both WUS and HAMs are considered as the signals from the stem cell niche to modulate CLV3 expression. Here, we also found that the transcription factor DRN, mainly located in stem cells, can positively regulate CLV3 expression together with WUS, describing a new mechanism of restricting CLV3 expression in shoot stem cells. Proper CLV3 expression requires the synergetic regulation of signals from stem cells and stem cell niche. Furthermore, despite the reduced CLV3 expression in drn-1 and drn-1 drnl-2 (Figs 4B, S8J, S8K, and S8P) [29], the SAM size of drn-1 was comparable to the wild type, and drn-1 drnl-2 does not have extremely enlarged SAMs like clv3-7 (S9 and S10 Figs) [29], suggesting that additional unknown factors are involved in activating CLV3 in stem cells.
WUS proteins have 2 functions in regulating shoot stem cell activity: activating CLV3 and sustaining stem cell fate [4,6,8]. WUS interacts with DRN to activate CLV3 expression to limit a stable stem cell pool. Our data also showed that the drn-1 wus-7 double mutants completely lost their functional SAMs, more serious than either of the single mutants (Figs 2D, S9A and S9B), indicating the positive regulation of WUS-DRN in sustaining SAMs. Given that WUS is necessary for initiating shoot regeneration in tissue culture [39–41], our data may explain why DRN overexpression can also enhance shoot regeneration [42]. Similar to DRN and WUS, BRM also seems to have 2 opposite effects on modulating stem cells, activating CLV3 and sustaining SAM activity. The brm-3 clv3-7 mutants showed smaller SAMs than clv3-7 (S10 Fig), whereas silencing BRM transcripts in stem cells led to decreased CLV3 expression and increased SAM size (Fig 6A, 6C, and 6D). These results suggested that the WUS-DRN-BRM machinery may support stem cell activity via targeting additional unknown downstream genes, as well as limiting the stem cell number via activating CLV3.
We also found reduced WUS expression in brm-3, whereas CLV3 expression likewise declined in brm-3 (Figs 5A–5D and S19). These seemingly paradoxical results suggested that BRM not only regulates stem cell activity, but also sustains stem cell niche. The reduction in CLV3 expression of brm-3 is not sufficient to affect SAM size, as observed in the drn-1 single mutant, and the decrease in WUS expression is predominantly responsible for the smaller SAM size in brm-3 (Fig 5C–5E). Although BRM proteins are located in the whole SAM (Fig 3G), DRN proteins were not detected in OC (S15A Fig), indicating that the regulation of WUS expression by BRM does not rely on DRN.
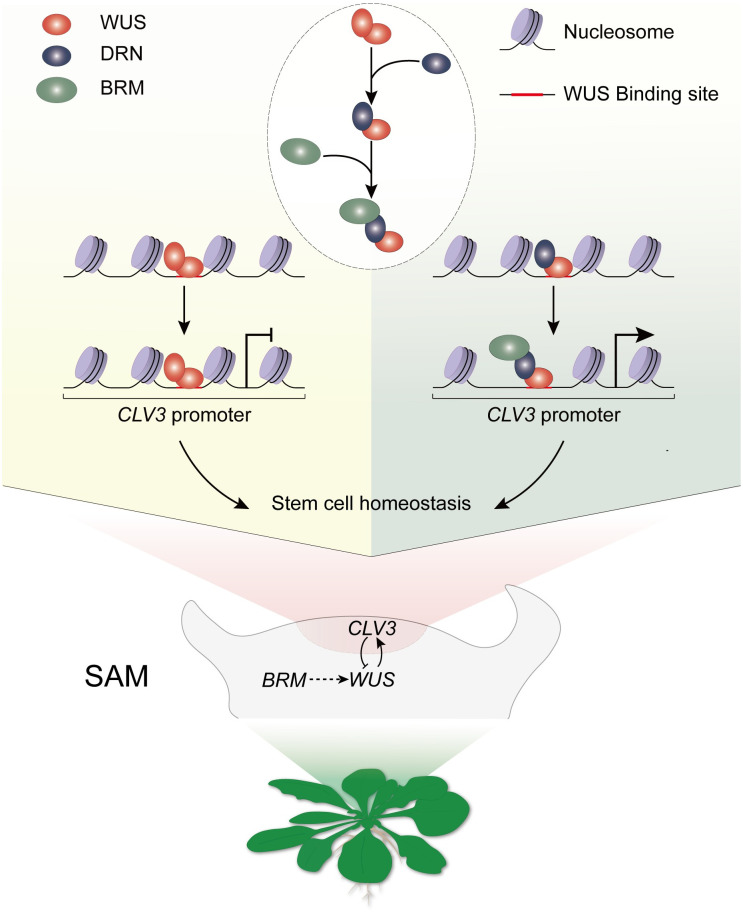
Overall, we illustrated a mechanism underlying the positive transcriptional regulatory activity of WUS, which relies on the WUS-DRN-BRM complex (Fig 8). This ternary complex plays a key role in regulating the CLV3 transcription via chromatin remodeling process to maintain the stem cell pool (Fig 8). Further investigation is required to determine (1) the additional unknown downstream genes of the WUS-DRN-BRM complex in the maintenance of stem cell activity; and (2) the molecular basis underlying BRM regulates the stem cell niche.
Fig 8. Schematic model of the WUS-DRN-BRM complex maintaining stem cell homeostasis.
SAM, shoot apical meristem.
Methods
Plant materials and treatments
All plants are Columbia-0 background. The wus-8 (CS349353) is from ABRC [32]. The wus-7 is a weak mutant allele of WUS [26,31] and was backcrossed with Columbia-0 for 5 times. The seeds of brm-3 and BRM::BRM-GFP lines were kindly provided by Prof. Doris Wagner and Prof. Jun Xiao [35,37]. clv3-7 is a null allele [5]. All seeds, except those used for the ethanol induction experiments, were sterilized in 70% ethanol and 0.5% Tween 20 for 10 min, and then washed twice with 95% ethanol. In the ethanol induction experiments, 0.16 M NaClO was used for sterilizing seeds, followed by several washes with water. Plants were grown under long-day conditions at 21°C. Half-strength Murashige and Skoog medium with 0.8% agar was used for cultivating the seedlings. For ethanol induction, we collected the inflorescence apices (35S::AlcR AlcA::amiBRM) and 14-day-old seedlings (CLV3::AlcR AlcA::amiBRM) with the induction condition of 1% ethanol for 24 h to analyze gene expression, for 72 h to analyze SAM size. In the DEX induction assays for gene expression analysis, 14-day-old seedlings were treated with 15 μM DEX and 50 μM cycloheximide in 1/2 MS medium for 2 h. For all induction assays, 3 biological replicates were performed.
Plasmid construction
The 1.4 kb promoter and 1.3 kb terminator of the CLV3 were used, respectively. CLV3::DRN and CLV3::DRN-GR were described in our previous study [29]. In Y2H assays, the full-length and truncated coding sequences of WUS, DRN, and BRM with stop codons were introduced into pGADT7 and pGBKT7 using EcoRI and BamHI sites. In pull-down assays, BRM1-976 and full-length coding sequence of WUS and DRN with stop codons were introduced into modified pET28b with an 8His-MBP tag using SalI and NotI sites. BRM1-976 and full-length coding sequence of WUS were introduced into pGEX6p-1 with a GST tag using BamHI and NotI sites. In co-IP assays, BRM1-976 and the full-length coding sequence of WUS and DRN without the stop codon were introduced into pUC19 with the Flag or 3×HA tag using SalI or BcuI site. We applied the gateway system to construct the plasmids used for BiFC assays in tobacco leaves. BRM1-976 and full-length coding sequence of WUS, and DRN without stop codon were introduced into entry vector PJLBlue, and were recombined into pGreen with the split YFP terminus by LR reaction. The 5.6 kb promoter and 1.2 kb terminator of the WUS [8], the 4.8 kb promoter and 1.5 kb terminator of the DRN [29], the 1.2 kb promoter and 0.5 kb terminator of the BRM [35], the full-length of WUS, DRN, and BRM CDS were used for BiFC in SAMs. The full-length of BRM CDS was used in UBQ10::BRM-GR. The target sequence of ethanol-inducible amiBRM was referred in Wu and colleagues [37]. Pre-amiRNA was assembled by PCR using the RS300 plasmid as templates. Subsequently, the PCR fragments were cloned after the AlcA promoter (35S::AlcR AlcA::amiBRM, CLV3::AlcR AlcA::amiBRM) in pGreen using gateway system. The primer sequences used in plasmid construction are listed in S1 Table.
Chromatin immunoprecipitation (ChIP)
ChIP was performed as described in previous studies [29,43,44]. The entire seedlings (14-day-old) were inducted with 15 μM DEX for 2 h and fixed in fixation buffer (100 mM Na3PO4, 50 mM NaCl, 0.1 M sucrose, and 1% formaldehyde (pH 7)) under vacuum conditions 3 times for 10 min at room temperature. Subsequently, use glycine under vacuum for 5 min to end the fixation. The tissues were ground in liquid nitrogen. Nuclei were isolated from the tissues and resuspended with sonication buffer (10 mM Na3PO4, 100 mM NaCl, 0.5% sarkosyl, 10 mM EDTA, 1 mM PMSF, one complete protease inhibitor cocktail tablet per 10 ml (pH 7)). The chromatin was interrupted into fragments with the average DNA size of 0.2 to 1.0 kb using Bioruptor UCD-200 for sonication (30 s on, 30 s off, medium level, 15 min duration). The lysate was precleared by an incubation with 15 μl protein A beads (catalog no. 26162, Thermo Fisher) for 1 h and was incubated with the anti-GFP antibody (catalog no. Ab290, Abcam). The bound DNA was purified and analyzed by qPCR. The primers used for qPCR are shown in S1 Table.
Co-immunoprecipitation (co-IP)
Plasmids were introduced into wild-type protoplasts. Total proteins were extracted from the protoplasts with the PEN-140 buffer (140 mM NaCl, 2.7 mM KCl, 25 mM Na2HPO4, 1.5 mM KH2PO4, 1 mM EDTA, 0.05% NP-40, 0.5 mM PMSF) and then incubated with Flag-Trap beads (catalog no. M20038, Abmart) at 4°C for 4 h (IgG group as a negative control), followed by 5 times of washing with PEN-140 buffer. Western blotting was performed to analyze the immunoprecipitated proteins using the anti-HA antibody (catalog no. M20003, Abmart) and the anti-Flag antibody (catalog no. M20008, Abmart).
Pull-down
Recombinant GST-WUS, GST-BRM, 8His-MBP-WUS, and 8His-MBP-DRN proteins were expressed in E. coli BL21 (DE3) and purified; 1 ml well-cultured cells was added to 500 ml fresh lysogeny broth (LB) medium and cultured at 37°C until the optical density at 600 nm (OD600) reached 0.8 to 1. Proteins were induced by adding isopropyl-β-D-thiogalactopyranoside (IPTG) to a final concentration of 0.5 mM at 16°C for 16 h. The bacteria were harvested by centrifuging at 12,000 rpm. The cell precipitate was washed with buffer (100 mM NaCl, 50 mM Tris-HCl (pH 7.4)) 3 times and resuspended with buffer (100 mM NaCl, 50 mM Tris-HCl, 1 mM PMSF (pH 7.4)). After sonication (1 s on, 2 s off, at 30% amplitude for 10 min), the expressed GST-WUS and GST-BRM proteins were purified using glutathione resin (catalog no. P2253, Beyotime); 8His-MBP-WUS and 8His-MBP-DRN proteins were purified using Ni resin (catalog no. P2233, Beyotime). Beads loaded with proteins and soluble proteins were incubated by rotating at 4°C for 4 h in pull-down buffer (200 mM NaCl, 1 mM EDTA, 20 mM Tris-HCl, 0.15 mM PMSF (pH 8.0)). After being washed 3 times with pull-down buffer, the resin beads were boiled and processed for western blotting analyses using anti-GST (catalog no. M20025, Abmart) and anti-His (catalog no. M20020, Abmart) antibodies.
Yeast two-hybrid (Y2H)
The pGBKT7 (BD) and pGADT7 (AD) vectors (Clontech) were used for plasmid construction. The combinations of AD and BD were transformed into the yeast strain Y2H Gold, which was cultured on complete medium lacking leucine and tryptophan (SD/−Leu/−Trp). The protein interactions were tested on complete medium lacking leucine, tryptophan, histidine, and adenine (SD/−Leu/−Trp/−His/−Ade). The empty BD and empty AD were used as negative controls.
Yeast three-hybrid (Y3H)
The vectors of pBridge and pGADT7 were used for constructing plasmids. The full-length coding sequence of WUS and DRN were cloned behind the BD and pMET25 promoter of pBridge, respectively. WUS and BRM full-length coding sequence were cloned into pGADT7 vector, respectively. The designed combinations of pBridge vectors and pGADT7 vectors were cotransformed into the yeast strain Y2H Gold. The interactions relationship among the 3 proteins were measured on selective medium (SD/-Leu/-Trp/-His, SD/-Leu/-Trp/-His/-Met SD/-Leu/-Trp/-His/-Ade, and SD/-Leu/-Trp/-His/-Ade/-Met).
Bimolecular fluorescence complementation (BiFC)
The constructs were transformed into the Agrobacterium. The Agrobacterium were cultured, collected, and resuspended in infiltration buffer (10 mM MES, 10 mM MgCl2, 0.15 mM acetosyringone (pH 5.8)). Equal volumes of different Agrobacterium with YFP N-terminus or C-terminus fused proteins were mixed and incubated for 2 h at room temperature, subsequently injected into N. benthamiana leaves. The plants with injected leaves were grown in the green house for 48 h. The DRN::DRN-nYFP/WUS::WUS-cYFP and DRN::DRN-nYFP/BRM::BRM-cYFP lines were generated by crossing. YFP fluorescence was detected by confocal laser-scanning microscope (ZEISS, LSM710).
Split-luciferase complementary assays
The constructs were transformed into the Agrobacterium. The Agrobacterium was cultured, collected, and resuspended in infiltration buffer (10 mM MES, 10 mM MgCl2, 0.15 mM acetosyringone (pH 5.8)) and adjusted to the OD = 1. Equal volumes of different Agrobacterium with luciferase N-terminus or C-terminus fused proteins were mixed and incubated for 2 h at room temperature, and subsequently injected into N. benthamiana leaves. The injected plants were cultured under normal conditions for 48 h; 1 mM D-Luciferin (catalog no. 40902ES01, YEASEN) was infiltrated into leaves, which were subsequently incubated for 3 min after injection. The luciferase activity was detected by an imaging system (Tanon, 5200).
Transient activation assays in tobacco
The Agrobacterium with constructs were cultured, collected, and resuspended in infiltration buffer (10 mM MES, 10 mM MgCl2, 0.15 mM acetosyringone (pH 5.8)) and adjusted to OD = 1. The Agrobacterium carrying different constructs were injected into N. benthamiana leaves with identical Agrobacterium concentrations in all groups. The injected plants were grown under normal conditions for 48 h. Subsequently, 1 mM D-Luciferin (catalog no. 40902ES01, YEASEN) was injected into leaves, which were subsequently incubated for 3 min after injection. The luciferase activity was detected by an imaging system (Tanon, 5200).
Electrophoretic mobility shift assay (EMSA)
The GST-WUS and 8His-MBP-DRN proteins were expressed in E. coli BL21 (DE3) and purified using Ni resin (catalog no. P2233, Beyotime). Size-exclusion chromatography was used to yield proteins with the desired molecular weight. The biotin-labeled probes were synthesized by Tsingke Biotechnology Co. The experiments were performed using the EMSA kit (catalog no. 20148, Thermo).
Quantitative reverse transcription PCR (qRT-PCR)
The leaves and roots of 14-day-old seedlings were removed. Seedlings (0.05 g) were ground into powder in liquid nitrogen. RNA isolater Total RNA Extraction Reagent (catalog no. R401-01, Vazyme) was used to extract total RNA from the plant samples. The HiScript III RT SuperMix Kit (catalog no. R323-01, Vazyme) was used for cDNA synthesis. The sequences of primers used for qRT-PCR are listed in S1 Table. qRT-PCR was performed using a Roche LightCycler 96 instrument with SYBR qPCR Master Mix (catalog no. Q711-02, Vazyme) and the following PCR program: Step 1, 95°C for 5 min; Step 2, 40 cycles of 95°C for 10 s followed by 62°C for 30 s; and Step 3, 20°C for 10 s. TUBULIN was used for normalization. Three biological replicates were performed.
Confocal microscopy
For SAM images of the CLV3::GFP, WUS::GFP, WUS::WUS-GFP, DRN::DRN-GFP, and BRM::BRM-GFP lines, the tissues were fixed with agarose and sectioned into 50 μm slices. Afterwards, the sections were visualized by a confocal microscope (ZEISS, LSM710). The top view of the BRM::BRM-GFP lines was performed by the confocal microscope (Olympus, FV3000), after the lateral floral organs were dissected and stained with propidium iodide (PI).
RNA in situ hybridization
Templates of RNA probes were amplified from cDNA using gene-specific primers containing T7 promoter sequences at the 5′ end. The primer sequences are listed in S1 Table. The RNA probes with DIG-labeled UTP were synthesized by T7 RNA polymerase, and RNA in situ hybridization was performed according to standard protocols [29]. The RNA probes were detected by the anti-DIG antibody (catalog no. 11093274910, Roche).
Formaldehyde-assisted isolation of regulatory elements (FAIRE) assays
FAIRE was performed as described [45]. For each replicate, 0.1 g of 10-day-old seedlings (removed leaves and roots) were crosslinked with crosslinking buffer (1% formaldehyde, 400 mM sucrose,10 mM Tris-HCl, 5 mM β-mercaptoethanol, 0.1 mM PMSF (pH 8.0)) under vacuum for 5 min, and 0.1 g materials was under vacuum for 5 min in crosslinking buffer without formaldehyde. The isolated DNA fragments were purified with Phenol/Chloroform/Isoamyl alcohol (25:24:1) for 2 times. The purified DNA was used as templates for qPCR. Primer sequences are listed in S1 Table. The Ta3 retrotransposon (At1g37110) was used as a reference for normalization [46–48]. The fold enrichment of a locus was obtained by normalization to Ta3 in crosslinked samples over that in uncrosslinked samples.
Supporting information
(DOCX)
Yeast cells were grown on the selective medium (SD/−Leu/−Trp/−His/−Ade) in a series of dilutions of 10–1, 10–2, and 10–3. ARR7 and TPL served as the negative and positive controls, respectively. Two independent experiments were performed with similar results.
(TIF)
ARF1 was used as the negative controls in BiFC. YFP was split into the N-terminus and C-terminus, fused to WUS, ARF1, and DRN. Scale bars, 100 μm.
(TIF)
The DRN::DRN-nYFP/WUS::WUS-cYFP transgenic plants were used to detect DRN-WUS interactions in inflorescence SAMs. DRN::nYFP and WUS::cYFP were introduced as negative controls. Scale bars, 50 μm. Two independent experiments were performed with similar results.
(TIF)
(A) Diagram of the WUS coding sequence. (B) Truncated WUS and full-length DRN were used for Y2H. BD and AD empty vectors were introduced as negative controls. Yeast cells were grown on the selective medium (SD/−Leu/−Trp/−His/−Ade) in a series of dilutions of 10–1, 10–2, and 10–3. The experiments were independently performed two times with similar results.
(TIF)
(A) Diagram of DRN coding sequence. (B) The full-length WUS, truncated and full-length DRN were used for Y2H. BD and AD empty vectors were used as negative controls. Yeast cells were grown on the selective medium (SD/−Leu/−Trp/−His/−Ade) in a series of dilutions of 10–1, 10–2, and 10–3. The experiments were independently performed 2 times with similar results.
(TIF)
(A) BiFC exhibiting that the interaction of WUS-WUS in tobacco leaves. YFP was split into the N-terminus and C-terminus, fused to WUS, respectively. Scale bars, 100 μm. (B) The full-length and truncated WUS were used for Y2H. Yeast cells were grown on the selective medium (SD/−Leu/−Trp/−His/−Ade) in a series of dilutions of 10–1, 10–2, and 10–3. All experiments were independently performed 2 times with similar results.
(TIF)
UBQ10::mCherry-WUS-GR/UBQ10::DRN-GFP lines (14-day-old seedlings) were used for ChIP assays. The nuclear localization of WUS-GR induced by DEX failed to confer the association of CRM with DRN-GFP, using the anti-GFP antibody for IP. The upstream -2,000 bp site acted as the negative control (no binding site). The experiments were independently performed 2 times with similar results. The data underlying this figure can be found in S1 Data.
(TIF)
(A) The diagram showing the point mutation of wus-7 mutants, and the T-DNA insertion of wus-8 mutants. (B–I) The images of 10-day-old seedlings including WT, drn-1, wus-7, drn-1 wus-7, wus-8, drnl-2, drn-1 drnl-2, and drn-1 drnl-2 wus-7. The dotted circle indicates the meristem in H. Bars in B–G, 1 mm. Bars in H and I, 100 μm. (J–O) The in situ hybridization was performed in the wild-type and mutants (10-day-old seedlings) to check CLV3 expression. Bars in J–L, O, and P, 30 μm. Bars in M and N, 100 μm. Bar in Q, 15 μm. All experiments were independently performed 2 times with similar results.
(TIF)
(A) The phenotypes of indicated mutants, including the shoots and SAMs, are shown. A portion of drn-1 wus-7, wus-8, and drn-1 wus-8 mutants failed to produce shoots. Black scale bars in WT, drn-1, wus-7, and drn-1 drnl-2, 100 μm. Black scale bars in drn-1 wus-7, wus-8, and drn-1 wus-8, 1 mm. White scale bars, 5 mm. (B) The percentage of phenotypic plants in A was analyzed. (C) The SAM sizes of the plants in A were analyzed. Black bars, highest and lowest values; box, median 50%; black line in the box, median. ***P < 0.001; **P < 0.01; ns, no significant difference; Student’s t test. The experiments were independently performed 2 times with similar results. The data underlying this figure can be found in S1 Data.
(TIF)
(A) The SAMs of indicated mutants are shown. The white arrows indicate the diameter of SAMs. Scale bars, 100 μm. (B) The SAM sizes of plants in A were analyzed. Black bars, highest and lowest values; box, median 50%; black line in the box, median. ***P < 0.001; ns, no significant difference; Student’s t test. The experiments were independently performed 2 times with similar results. The data underlying this figure can be found in S1 Data.
(TIF)
(A) Diagram of the BRM coding sequence. (B) The full-length of DRN and truncated BRM were used for Y2H. BD and AD empty vectors were used as negative controls. Yeast cells were grown on the selective medium (SD/−Leu/−Trp/−His/−Ade) in a series of dilutions of 10–1, 10–2, and 10–3. The experiments were independently performed 2 times with similar results.
(TIF)
ARF1 was used as the negative control in BiFC. YFP was split into the N-terminus and C-terminus, fused to ARF1, BRM, and DRN. Scale bars, 100 μm.
(TIF)
The DRN::DRN-nYFP/BRM::BRM-cYFP transgenic plants were used to detect DRN-BRM interactions in inflorescence SAMs. DRN::nYFP and BRM::cYFP were introduced as negative controls. Scale bars, 50 μm. The experiments were independently performed 2 times with similar results.
(TIF)
(A) Diagram of the DRN coding sequence. (B) The full-length BRM, truncation and full-length DRN were used for Y2H. BD and AD empty vectors were used as negative controls. Yeast cells were grown on the selective medium (SD/−Leu/−Trp/−His/−Ade) in a series of dilutions of 10–1, 10–2, and 10–3. The experiments were independently performed 2 times with similar results.
(TIF)
(A) The distribution of DRN proteins in the SAM was checked using DRN::DRN-GFP/drn-1 rescue lines during the reproductive stage, and 8 apices were analyzed. (B) The distribution of WUS proteins in the SAM was checked using WUS::WUS-GFP/wus-8 rescue lines during the reproductive stage. Ten apices were analyzed. The red arrows indicate WUS-GFP signals in the L1 and L2 cell layers. (C) CLV3 expression pattern was checked in CLV3::GFP/WT lines during the reproductive stage. Six apices were analyzed. (D) The top view of the SAM of BRM::BRM-GFP represents the L1 cell layer. Five apices were analyzed. Green, BRM-GFP signals; red, propidium iodide (PI) signals. (E) BRM mRNAs were detected by RNA in situ hybridization in the SAMs of wild-type plants. Seven apices were analyzed. (F) The BRM sense probe was used as a negative control. Five apices were analyzed. Scale bars in A–F, 50 μm. Bright field, BF, in A–C. All experiments were independently performed 2 times with similar results.
(TIF)
qRT-PCR was applied to check the relative transcript levels of BRM and CLV3 in 35S::inducible amiBRM/WT transgenic plants after 1% ethanol (EtOH) induction for 24 h using 14-day-old seedlings. Data represent means ± SDs from 3 biological replicates. *P < 0.1; **P < 0.01; Student’s t test. The experiments were independently performed 2 times with similar results. The data underlying this figure can be found in S1 Data.
(TIF)
The −2,000 site upstream of CLV3 was selected to be the negative control of FAIRE assays in Fig 4C and 4D, using 10-day-old seedlings. Data represent means ± SDs from 3 biological replicates. ns, no significant difference; Student’s t test. The experiments were independently performed 2 times with similar results. The data underlying this figure can be found in S1 Data.
(TIF)
FAIRE assays were performed, using 35S::DRN-GR lines (10-day-old seedlings) inducted with 15 μM DEX for 3 h, to check the chromatin state at CRM region, downstream of CLV3. The upstream −2,000 site was used as the negative control. Data represent means ± SDs from 3 biological replicates. ns, no significant difference; Student’s t test. The experiments were independently performed 2 times with similar results. The data underlying this figure can be found in S1 Data.
(TIF)
CLV3::GFP and WUS::GFP lines were crossed with drn-1, brm-3, and drn-1 brm-3, respectively. CLV3 and WUS expression was checked in these mutants by fluorescence. Six apices were analyzed in each line. Scale bars, 50 μm. The experiments were independently performed 2 times with similar results.
(TIF)
(A) The phenotypes of indicated mutants, including the shoots and SAMs, are shown. The white arrows indicate the diameter of SAMs. Black scale bars, 100 μm. White scale bars, 5 mm. (B) The SAM sizes of plants in A were analyzed. Black bars, highest and lowest values; box, median 50%; black line in the box, median. ***P < 0.001; ns, no significant difference; Student’s t test. The experiments were independently performed 2 times with similar results. The data underlying this figure can be found in S1 Data.
(TIF)
The number of true leaves was analyzed in WT, drn-1, brm-3, and drn-1 brm-3, using 14-day-old seedlings. Black bars, highest and lowest values; box, median 50%. ns, no significant difference; Student’s t test. The experiments were independently performed 2 times with similar results. The data underlying this figure can be found in S1 Data.
(TIF)
BRM mRNAs were detected by RNA in situ hybridization in SAMs of CLV3::inducible amiBRM plants after 1% ethanol induction for 24 h during the reproductive stage. Scale bars, 50 μm. Ten apices were analyzed in each group. The experiments were independently performed 2 times with similar results.
(TIF)
(A and B) CLV3::inducible GUS/WT lines (14-day-old seedlings), as the negative control of Fig 6A and 6B, were used for EtOH induction. qRT-PCR was performed to test CLV3 and WUS expression. Data represent means ± SDs from 3 biological replicates. (C and D) The SAM sizes of CLV3::inducible GUS/WT lines (14-day-old seedlings) with EtOH induction were analyzed, as the negative control of Fig 6C and 6D. The black arrows indicate the boundaries of SAMs. Scale bars, 50 μm. Black bars, highest and lowest values; box, median 50%; black line in the box, median. ns, no significant difference; Student’s t test. All experiments were independently performed 2 times with similar results. The data underlying this figure can be found in S1 Data.
(TIF)
Values of histograms and box plots in main figures and supplemental figures.
(XLSX)
Raw gels of co-IP, pull-down, and EMSA assays in this study. The gels used in figures and the repetitive results are included in this file.
(PDF)
Acknowledgments
The authors thank Prof. Wolfgang Werr, Prof. Doris Wagner, and Prof. Jun Xiao for sharing the mutants.
Abbreviations
- BiFC
bimolecular fluorescence complementation
- ChIP
chromatin immunoprecipitation
- CRM
cis-regulatory module
- CZ
central zone
- DEX
dexamethasone
- EMSA
electrophoretic mobility shift assay
- FAIRE
formaldehyde-assisted isolation of regulatory elements
- FM
floral meristem
- HD
homeodomain
- LB
lysogeny broth
- MBP
maltose binding protein
- NDR
nucleosome-depleted region
- OC
organizing center
- PI
propidium iodide
- PZ
periphery zone
- RM
rib meristem
- SAM
shoot apical meristem
- TF
transcription factor
- Y2H
yeast two-hybrid
- Y3H
yeast three-hybrid
Data Availability
All relevant data are within the paper and its Supporting information files.
Funding Statement
The study design, data collection, and analysis were funded by the National Natural Science Foundation of China (https://www.nsfc.gov.cn/) with grants 31900624 to L. Luo, 32071270 to G. Z., and 32130009 to Z. Z.. Additionally, the Natural Science Foundation of Anhui Province of China (http://kjt.ah.gov.cn/) provided support for study design, data collection, and analysis with grant 1908085QC91 to L. Luo. Data collection and analysis were also supported by the Major Science and Technology Projects in Anhui Province (http://kjt.ah.gov.cn/) with grant 202003a06020009 to G. Z.. The preparation and publication fees of the manuscript were supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (https://www.cas.cn/) with grant XDB27030105 to Z. Z., and by the Outstanding Innovative Research Team for Molecular Enzymology and Detection in Anhui Provincial Universities (https://www.ahnu.edu.cn/) with grant 2022AH010012 to G. Z.. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Aichinger E, Kornet N, Friedrich T, Laux T. Plant Stem Cell Niches. Annu Rev Plant Biol. 2012;63:615–636. doi: 10.1146/annurev-arplant-042811-105555 [DOI] [PubMed] [Google Scholar]
- 2.Pfeiffer A, Wenzl C, Lohmann JU. Beyond flexibility: controlling stem cells in an ever changing environment. Curr Opin Plant Biol. 2017;35:117–123. doi: 10.1016/j.pbi.2016.11.014 [DOI] [PubMed] [Google Scholar]
- 3.Reddy GV, Heisler MG, Ehrhardt DW, Meyerowitz EM. Real-time lineage analysis reveals oriented cell divisions associated with morphogenesis at the shoot apex of Arabidopsis thaliana. Development. 2004;131:4225–4237. doi: 10.1242/dev.01261 [DOI] [PubMed] [Google Scholar]
- 4.Mayer KFX, Schoof H, Haecker A, Lenhard M, Jurgens G, et al. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell. 1998;95:805–815. doi: 10.1016/s0092-8674(00)81703-1 [DOI] [PubMed] [Google Scholar]
- 5.Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science. 1999;283:1911–1914. doi: 10.1126/science.283.5409.1911 [DOI] [PubMed] [Google Scholar]
- 6.Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science. 2000;289:617–619. doi: 10.1126/science.289.5479.617 [DOI] [PubMed] [Google Scholar]
- 7.Schoof H, Lenhard M, Haecker A, Mayer KF, Jurgens G, et al. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 2000;100:635–644. doi: 10.1016/s0092-8674(00)80700-x [DOI] [PubMed] [Google Scholar]
- 8.Yadav RK, Perales M, Gruel J, Girke T, Jonsson H, et al. WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes Dev. 2011;25:2025–2030. doi: 10.1101/gad.17258511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daum G, Medzihradszky A, Suzaki T, Lohmann JU. A mechanistic framework for noncell autonomous stem cell induction in Arabidopsis. Proc Natl Acad Sci U S A. 2014;111:14619–14624. doi: 10.1073/pnas.1406446111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma Y, Miotk A, Šutiković Z, Ermakova O, Wenzl C, et al. WUSCHEL acts as an auxin response rheostat to maintain apical stem cells in Arabidopsis. Nat Commun. 2019;10:5093. doi: 10.1038/s41467-019-13074-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kondo T, Sawa S, Kinoshita A, Mizuno S, Kakimoto T, et al. A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science. 2006;313:845–848. doi: 10.1126/science.1128439 [DOI] [PubMed] [Google Scholar]
- 12.Ohyama K, Shinohara H, Ogawa-Ohnishi M, Matsubayashi Y. A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nat Chem Biol. 2009;5:578–580. doi: 10.1038/nchembio.182 [DOI] [PubMed] [Google Scholar]
- 13.Hu C, Zhu YF, Cui YW, Cheng KL, Liang W, et al. A group of receptor kinases are essential for CLAVATA signalling to maintain stem cell homeostasis. Nat Plants. 2018;4:205–211. doi: 10.1038/s41477-018-0123-z [DOI] [PubMed] [Google Scholar]
- 14.Plong A, Rodriguez K, Alber M, Chen WT, Reddy GV. CLAVATA3 mediated simultaneous control of transcriptional and post-translational processes provides robustness to the WUSCHEL gradient. Nat Commun. 2021;12:6361. doi: 10.1038/s41467-021-26586-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han H, Liu X, Zhou Y. Transcriptional circuits in control of shoot stem cell homeostasis. Curr Opin Plant Biol. 2020;53:50–56. doi: 10.1016/j.pbi.2019.10.004 [DOI] [PubMed] [Google Scholar]
- 16.Su YH, Zhou C, Li YJ, Yu Y, Tang LP, et al. Integration of pluripotency pathways regulates stem cell maintenance in the Arabidopsis shoot meristem. Proc Natl Acad Sci U S A. 2020;117:22561–22571. doi: 10.1073/pnas.2015248117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shang EL, Wang X, Li TH, Guo FF, Ito T, et al. Robust control of floral meristem determinacy by position-specific multifunctions of KNUCKLES. Proc Natl Acad Sci U S A. 2021;118:e2102826118. doi: 10.1073/pnas.2102826118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu HJ, Qu XY, Dong ZC, Luo LJ, Shao C, et al. WUSCHEL triggers innate antiviral immunity in plant stem cells. Science. 2020;370:227–231. doi: 10.1126/science.abb7360 [DOI] [PubMed] [Google Scholar]
- 19.Ikeda M, Mitsuda N, Ohme-Takagi M. Arabidopsis WUSCHEL Is a Bifunctional Transcription Factor That Acts as a Repressor in Stem Cell Regulation and as an Activator in Floral Patterning. Plant Cell. 2009;21:3493–3505. doi: 10.1105/tpc.109.069997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodriguez K, Perales M, Snipes S, Yadav RK, Diaz-Mendoza M, et al. DNA-dependent homodimerization, sub-cellular partitioning, and protein destabilization control WUSCHEL levels and spatial patterning. Proc Natl Acad Sci U S A. 2016;113:E6307–E6315. doi: 10.1073/pnas.1607673113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sloan J, Hakenjos JP, Gebert M, Ermakova O, Gumiero A, et al. Structural basis for the complex DNA binding behavior of the plant stem cell regulator WUSCHEL. Nat Commun. 2020;11:2223. doi: 10.1038/s41467-020-16024-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kieffer M, Stern Y, Cook H, Clerici E, Maulbetsch C, et al. Analysis of the transcription factor WUSCHEL and its functional homologue in Antirrhinum reveals a potential mechanism for their roles in meristem maintenance. Plant Cell. 2006;18:560–573. doi: 10.1105/tpc.105.039107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dolzblasz A, Nardmann J, Clerici E, Causier B, van der Graaff E, et al. Stem Cell Regulation by Arabidopsis WOX Genes. Mol Plant. 2016;9:1028–1039. doi: 10.1016/j.molp.2016.04.007 [DOI] [PubMed] [Google Scholar]
- 24.Perales M, Rodriguez K, Snipes S, Yadav RK, Diaz-Mendoza M, et al. Threshold-dependent transcriptional discrimination underlies stem cell homeostasis. Proc Natl Acad Sci U S A. 2016;113:E6298–E6306. doi: 10.1073/pnas.1607669113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodriguez K, Do A, Senay-Aras B, Perales M, Alber M, et al. Concentration-dependent transcriptional switching through a collective action of cis-elements. Sci Adv. 2022;8:eabo6157. doi: 10.1126/sciadv.abo6157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou Y, Liu X, Engstrom EM, Nimchuk ZL, Pruneda-Paz JL, et al. Control of plant stem cell function by conserved interacting transcriptional regulators. Nature. 2015;517:377–380. doi: 10.1038/nature13853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou Y, Yan A, Han H, Li T, Geng Y, et al. HAIRY MERISTEM with WUSCHEL confines CLAVATA3 expression to the outer apical meristem layers. Science. 2018;361:502–506. doi: 10.1126/science.aar8638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kirch T, Simon R, Grunewald M, Werr W. The DORNROSCHEN/ENHANCER OF SHOOT REGENERATION1 gene of Arabidopsis acts in the control of meristem cell fate and lateral organ development. Plant Cell. 2003;15:694–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo LJ, Zeng J, Wu HJ, Tian ZX, Zhao Z. A Molecular Framework for Auxin-Controlled Homeostasis of Shoot Stem Cells in Arabidopsis. Mol Plant. 2018;11:899–913. doi: 10.1016/j.molp.2018.04.006 [DOI] [PubMed] [Google Scholar]
- 30.Xu MX, Du QW, Tian CH, Wang Y, Jiao YL. Stochastic gene expression drives mesophyll protoplast regeneration. Science. Adv Ther. 2021;7:eabg8466. doi: 10.1126/sciadv.abg8466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Graf P, Dolzblasz A, Würschum T, Lenhard M, Pfreundt U, et al. MGOUN1 Encodes an Arabidopsis Type IB DNA Topoisomerase Required in Stem Cell Regulation and to Maintain Developmentally Regulated Gene Silencing. Plant Cell. 2010;22:716–728. doi: 10.1105/tpc.109.068296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen L, Liang Z, Gu X, Chen Y, Teo Zhi Wei N, et al. N6-Methyladenosine RNA Modification Regulates Shoot Stem Cell Fate in Arabidopsis. Dev Cell. 2016;38:186–200. doi: 10.1016/j.devcel.2016.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hake S, Smith HMS, Holtan H, Magnani E, Mele G, et al. The role of knox genes in plant development. Annu Rev Cell Dev Biol. 2004;20:125–151. doi: 10.1146/annurev.cellbio.20.031803.093824 [DOI] [PubMed] [Google Scholar]
- 34.Dai YQ, Luo LJ, Zhao Z. Genetic robustness control of auxin output in priming organ initiation. Proc Natl Acad Sci U S A. 2023;120:e2221606120. doi: 10.1073/pnas.2221606120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu MF, Sang Y, Bezhani S, Yamaguchi N, Han SK, et al. SWI2/SNF2 chromatin remodeling ATPases overcome polycomb repression and control floral organ identity with the LEAFY and SEPALLATA3 transcription factors. Proc Natl Acad Sci U S A. 2012;109:3576–3581. doi: 10.1073/pnas.1113409109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han SK, Wu MF, Cui SJ, Wagner D. Roles and activities of chromatin remodeling ATPases in plants. Plant J. 2015;83:62–77. doi: 10.1111/tpj.12877 [DOI] [PubMed] [Google Scholar]
- 37.Wu MF, Yamaguchi N, Xiao J, Bargmann B, Estelle M, et al. Auxin-regulated chromatin switch directs acquisition of flower primordium founder fate. Elife. 2015;4:e09269. doi: 10.7554/eLife.09269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Farrona S, Hurtado L, Bowman JL, Reyes JC. The Arabidopsis thaliana SNF2 homolog AtBRM controls shoot development and flowering. Development. 2004;131:4965–4975. doi: 10.1242/dev.01363 [DOI] [PubMed] [Google Scholar]
- 39.Zhang TQ, Lian H, Zhou CM, Xu L, Jiao Y, et al. A Two-Step Model for de Novo Activation of WUSCHEL during Plant Shoot Regeneration. Plant Cell. 2017;29:1073–1087. doi: 10.1105/tpc.16.00863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meng WJ, Cheng ZJ, Sang YL, Zhang MM, Rong XF, et al. Type-B ARABIDOPSIS RESPONSE REGULATORs Specify the Shoot Stem Cell Niche by Dual Regulation of WUSCHEL. Plant Cell. 2017;29:1357–1372. doi: 10.1105/tpc.16.00640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dai X, Liu Z, Qiao M, Li J, Li S, et al. ARR12 promotes de novo shoot regeneration in Arabidopsis thaliana via activation of WUSCHEL expression. J Integr Plant Biol. 2017;59:747–758. doi: 10.1111/jipb.12567 [DOI] [PubMed] [Google Scholar]
- 42.Banno H, Ikeda Y, Niu QW, Chua NH. Overexpression of Arabidopsis ESR1 induces initiation of shoot regeneration. Plant Cell. 2001;13:2609–2618. doi: 10.1105/tpc.010234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leibfried A, To JPC, Busch W, Stehling S, Kehle A, et al. WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature. 2005;438:1172–1175. doi: 10.1038/nature04270 [DOI] [PubMed] [Google Scholar]
- 44.Dahl JA, Collas P. A rapid micro chromatin immunoprecipitation assay (ChIP). Nat Protoc. 2008;3:1032–1045. [DOI] [PubMed] [Google Scholar]
- 45.Omidbakhshfard MA, Winck FV, Arvidsson S, Riano-Pachon DM, Mueller-Roeber B. A step-by-step protocol for formaldehyde-assisted isolation of regulatory elements from Arabidopsis thaliana. J Integr Plant Biol. 2014;56:527–538. doi: 10.1111/jipb.12151 [DOI] [PubMed] [Google Scholar]
- 46.Johnson LM, Cao XF, Jacobsen SE. Interplay between two epigenetic marks: DNA methylation and histone H3 lysine 9 methylation. Curr Biol. 2002;12:1360–1367. [DOI] [PubMed] [Google Scholar]
- 47.Yang L, Wang Z, Hua J. Multiple chromatin-associated modules regulate expression of an intracellular immune receptor gene in Arabidopsis. New Phytol. 2023;237:2284–2297. doi: 10.1111/nph.18672 [DOI] [PubMed] [Google Scholar]
- 48.Jing Y, Guo Q, Zha P, Lin R. The chromatin-remodelling factor PICKLE interacts with CONSTANS to promote flowering in Arabidopsis. Plant Cell Environ. 2019;42:2495–2507. doi: 10.1111/pce.13557 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Yeast cells were grown on the selective medium (SD/−Leu/−Trp/−His/−Ade) in a series of dilutions of 10–1, 10–2, and 10–3. ARR7 and TPL served as the negative and positive controls, respectively. Two independent experiments were performed with similar results.
(TIF)
ARF1 was used as the negative controls in BiFC. YFP was split into the N-terminus and C-terminus, fused to WUS, ARF1, and DRN. Scale bars, 100 μm.
(TIF)
The DRN::DRN-nYFP/WUS::WUS-cYFP transgenic plants were used to detect DRN-WUS interactions in inflorescence SAMs. DRN::nYFP and WUS::cYFP were introduced as negative controls. Scale bars, 50 μm. Two independent experiments were performed with similar results.
(TIF)
(A) Diagram of the WUS coding sequence. (B) Truncated WUS and full-length DRN were used for Y2H. BD and AD empty vectors were introduced as negative controls. Yeast cells were grown on the selective medium (SD/−Leu/−Trp/−His/−Ade) in a series of dilutions of 10–1, 10–2, and 10–3. The experiments were independently performed two times with similar results.
(TIF)
(A) Diagram of DRN coding sequence. (B) The full-length WUS, truncated and full-length DRN were used for Y2H. BD and AD empty vectors were used as negative controls. Yeast cells were grown on the selective medium (SD/−Leu/−Trp/−His/−Ade) in a series of dilutions of 10–1, 10–2, and 10–3. The experiments were independently performed 2 times with similar results.
(TIF)
(A) BiFC exhibiting that the interaction of WUS-WUS in tobacco leaves. YFP was split into the N-terminus and C-terminus, fused to WUS, respectively. Scale bars, 100 μm. (B) The full-length and truncated WUS were used for Y2H. Yeast cells were grown on the selective medium (SD/−Leu/−Trp/−His/−Ade) in a series of dilutions of 10–1, 10–2, and 10–3. All experiments were independently performed 2 times with similar results.
(TIF)
UBQ10::mCherry-WUS-GR/UBQ10::DRN-GFP lines (14-day-old seedlings) were used for ChIP assays. The nuclear localization of WUS-GR induced by DEX failed to confer the association of CRM with DRN-GFP, using the anti-GFP antibody for IP. The upstream -2,000 bp site acted as the negative control (no binding site). The experiments were independently performed 2 times with similar results. The data underlying this figure can be found in S1 Data.
(TIF)
(A) The diagram showing the point mutation of wus-7 mutants, and the T-DNA insertion of wus-8 mutants. (B–I) The images of 10-day-old seedlings including WT, drn-1, wus-7, drn-1 wus-7, wus-8, drnl-2, drn-1 drnl-2, and drn-1 drnl-2 wus-7. The dotted circle indicates the meristem in H. Bars in B–G, 1 mm. Bars in H and I, 100 μm. (J–O) The in situ hybridization was performed in the wild-type and mutants (10-day-old seedlings) to check CLV3 expression. Bars in J–L, O, and P, 30 μm. Bars in M and N, 100 μm. Bar in Q, 15 μm. All experiments were independently performed 2 times with similar results.
(TIF)
(A) The phenotypes of indicated mutants, including the shoots and SAMs, are shown. A portion of drn-1 wus-7, wus-8, and drn-1 wus-8 mutants failed to produce shoots. Black scale bars in WT, drn-1, wus-7, and drn-1 drnl-2, 100 μm. Black scale bars in drn-1 wus-7, wus-8, and drn-1 wus-8, 1 mm. White scale bars, 5 mm. (B) The percentage of phenotypic plants in A was analyzed. (C) The SAM sizes of the plants in A were analyzed. Black bars, highest and lowest values; box, median 50%; black line in the box, median. ***P < 0.001; **P < 0.01; ns, no significant difference; Student’s t test. The experiments were independently performed 2 times with similar results. The data underlying this figure can be found in S1 Data.
(TIF)
(A) The SAMs of indicated mutants are shown. The white arrows indicate the diameter of SAMs. Scale bars, 100 μm. (B) The SAM sizes of plants in A were analyzed. Black bars, highest and lowest values; box, median 50%; black line in the box, median. ***P < 0.001; ns, no significant difference; Student’s t test. The experiments were independently performed 2 times with similar results. The data underlying this figure can be found in S1 Data.
(TIF)
(A) Diagram of the BRM coding sequence. (B) The full-length of DRN and truncated BRM were used for Y2H. BD and AD empty vectors were used as negative controls. Yeast cells were grown on the selective medium (SD/−Leu/−Trp/−His/−Ade) in a series of dilutions of 10–1, 10–2, and 10–3. The experiments were independently performed 2 times with similar results.
(TIF)
ARF1 was used as the negative control in BiFC. YFP was split into the N-terminus and C-terminus, fused to ARF1, BRM, and DRN. Scale bars, 100 μm.
(TIF)
The DRN::DRN-nYFP/BRM::BRM-cYFP transgenic plants were used to detect DRN-BRM interactions in inflorescence SAMs. DRN::nYFP and BRM::cYFP were introduced as negative controls. Scale bars, 50 μm. The experiments were independently performed 2 times with similar results.
(TIF)
(A) Diagram of the DRN coding sequence. (B) The full-length BRM, truncation and full-length DRN were used for Y2H. BD and AD empty vectors were used as negative controls. Yeast cells were grown on the selective medium (SD/−Leu/−Trp/−His/−Ade) in a series of dilutions of 10–1, 10–2, and 10–3. The experiments were independently performed 2 times with similar results.
(TIF)
(A) The distribution of DRN proteins in the SAM was checked using DRN::DRN-GFP/drn-1 rescue lines during the reproductive stage, and 8 apices were analyzed. (B) The distribution of WUS proteins in the SAM was checked using WUS::WUS-GFP/wus-8 rescue lines during the reproductive stage. Ten apices were analyzed. The red arrows indicate WUS-GFP signals in the L1 and L2 cell layers. (C) CLV3 expression pattern was checked in CLV3::GFP/WT lines during the reproductive stage. Six apices were analyzed. (D) The top view of the SAM of BRM::BRM-GFP represents the L1 cell layer. Five apices were analyzed. Green, BRM-GFP signals; red, propidium iodide (PI) signals. (E) BRM mRNAs were detected by RNA in situ hybridization in the SAMs of wild-type plants. Seven apices were analyzed. (F) The BRM sense probe was used as a negative control. Five apices were analyzed. Scale bars in A–F, 50 μm. Bright field, BF, in A–C. All experiments were independently performed 2 times with similar results.
(TIF)
qRT-PCR was applied to check the relative transcript levels of BRM and CLV3 in 35S::inducible amiBRM/WT transgenic plants after 1% ethanol (EtOH) induction for 24 h using 14-day-old seedlings. Data represent means ± SDs from 3 biological replicates. *P < 0.1; **P < 0.01; Student’s t test. The experiments were independently performed 2 times with similar results. The data underlying this figure can be found in S1 Data.
(TIF)
The −2,000 site upstream of CLV3 was selected to be the negative control of FAIRE assays in Fig 4C and 4D, using 10-day-old seedlings. Data represent means ± SDs from 3 biological replicates. ns, no significant difference; Student’s t test. The experiments were independently performed 2 times with similar results. The data underlying this figure can be found in S1 Data.
(TIF)
FAIRE assays were performed, using 35S::DRN-GR lines (10-day-old seedlings) inducted with 15 μM DEX for 3 h, to check the chromatin state at CRM region, downstream of CLV3. The upstream −2,000 site was used as the negative control. Data represent means ± SDs from 3 biological replicates. ns, no significant difference; Student’s t test. The experiments were independently performed 2 times with similar results. The data underlying this figure can be found in S1 Data.
(TIF)
CLV3::GFP and WUS::GFP lines were crossed with drn-1, brm-3, and drn-1 brm-3, respectively. CLV3 and WUS expression was checked in these mutants by fluorescence. Six apices were analyzed in each line. Scale bars, 50 μm. The experiments were independently performed 2 times with similar results.
(TIF)
(A) The phenotypes of indicated mutants, including the shoots and SAMs, are shown. The white arrows indicate the diameter of SAMs. Black scale bars, 100 μm. White scale bars, 5 mm. (B) The SAM sizes of plants in A were analyzed. Black bars, highest and lowest values; box, median 50%; black line in the box, median. ***P < 0.001; ns, no significant difference; Student’s t test. The experiments were independently performed 2 times with similar results. The data underlying this figure can be found in S1 Data.
(TIF)
The number of true leaves was analyzed in WT, drn-1, brm-3, and drn-1 brm-3, using 14-day-old seedlings. Black bars, highest and lowest values; box, median 50%. ns, no significant difference; Student’s t test. The experiments were independently performed 2 times with similar results. The data underlying this figure can be found in S1 Data.
(TIF)
BRM mRNAs were detected by RNA in situ hybridization in SAMs of CLV3::inducible amiBRM plants after 1% ethanol induction for 24 h during the reproductive stage. Scale bars, 50 μm. Ten apices were analyzed in each group. The experiments were independently performed 2 times with similar results.
(TIF)
(A and B) CLV3::inducible GUS/WT lines (14-day-old seedlings), as the negative control of Fig 6A and 6B, were used for EtOH induction. qRT-PCR was performed to test CLV3 and WUS expression. Data represent means ± SDs from 3 biological replicates. (C and D) The SAM sizes of CLV3::inducible GUS/WT lines (14-day-old seedlings) with EtOH induction were analyzed, as the negative control of Fig 6C and 6D. The black arrows indicate the boundaries of SAMs. Scale bars, 50 μm. Black bars, highest and lowest values; box, median 50%; black line in the box, median. ns, no significant difference; Student’s t test. All experiments were independently performed 2 times with similar results. The data underlying this figure can be found in S1 Data.
(TIF)
Values of histograms and box plots in main figures and supplemental figures.
(XLSX)
Raw gels of co-IP, pull-down, and EMSA assays in this study. The gels used in figures and the repetitive results are included in this file.
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting information files.