Abstract
Mammalian wounds leave visible scars, and there are no methods for complete regeneration. However, mouse fetuses regenerate their skin, including epidermal and dermal structures, up to embryonic day (E)13. This regeneration pattern requires the formation of actin cables in the wound margin epithelium; however, the molecular mechanisms are not fully understood. Rac1 alters actin in cells and is involved in the formation of filopodia. We investigated whether actin remodeling and skin regeneration patterns can be reproduced through the regulation of Rac1 signaling. Rac1 expression was downregulated in E13 wounds and upregulated after E15 when scars remained. NSC23766, a Rac1-specific inhibitor, altered actin dynamics at the cell margin from filopodia formation to cable formation and inhibited the migration of mouse epidermal keratinocyte, PAM212, by Rac1 signaling suppression. NSC23766 suppressed Rac1 activity and completely regenerated the fetal mouse wounds, even at E14, by changing actin dynamics. Knocked-out Rac1 transgenic mice experienced delayed epithelialization of wounds with suppressed epidermal migration in adults; however, in fetuses, complete wound regeneration via Rac1 signal suppression was observed. Therefore, Rac1 suppression in the wound epidermis can achieve regenerative wound healing in fetuses and may be a potential candidate for healing scars.
Keywords: Rac1, Skin regeneration, Scar, Epidermis, Actin cable
Subject terms: Cell migration, Cell signalling, Mechanisms of disease
Introduction
In adult mammals, wound healing results in the formation of scar tissue with fibrosis and a lack of skin appendages. Although scar formation can fulfill the requirements of the skin’s basic function of preventing infection and dehydration, it has a distinctly different appearance from that of the original intact skin. Thus, scars formed as a result of injuries or burns can have devastating cosmetic and psychological consequences and reduce an individual’s quality of life1. In contrast, mammals can fully regenerate wounds up to a certain developmental stage. For example, fetal lamb wounds before 120 days of gestation were first shown to heal without scar formation in 19712. Similarly, human fetal skin injuries in early gestation have been shown to heal and form normal skin tissue without scar formation3. Engineering approaches that mimic the process of fetal wound healing may enable technologies that minimize or eliminate scar formation after injury.
The characteristics of the skin that forms scar tissue can be divided into four categories: (1) fibrosis of the dermis, (2) loss of skin texture, (3) loss of skin appendages, and (4) change in color tone4. Therefore, scarless wound healing can only be achieved by focusing on the regeneration mechanism of each of these parameters. In our previous study, we found that conventional histological regeneration occurred between embryonic day (E)16 and E17, whereas epidermal regeneration, including skin texture, occurred between E13 and E145. A necessary condition for this regeneration is the presence of actin fibers in the wound margin epidermis, which was first discovered during chicken embryo regeneration6. This occurs because of the polymerization and contraction of actomyosin in the limb of a single epidermal cell at the wound margin7. In contrast, during the non-regenerative period of the epidermis, healing occurs through epidermal cell migration over the exposed connective tissue8. The actin cables allow the epidermis to cover the wound surface later than the dermis; the defect is repaired by dermal-derived fibroblasts, and the wound regenerates without scar tissue formation through the retention of epidermal–dermal interactions9. Therefore, epidermal regeneration may be achieved by altering actin dynamics, thereby disrupting epidermal cell migration patterns and inducing actin cable formation. However, the signaling mechanisms and factors that regulate this function remain poorly understood.
One of the few intracellular signaling molecules with established non-overlapping functions in wound re-epithelialization is the Rho family GTP-binding protein Rac110. The small GTPase Rac1 is ubiquitously expressed and exerts important functions in the epidermis.
Although an essential function of Rac in establishing intraepidermal cell–cell contacts has been suggested11,12, deletion of Rac1 in epidermal keratinocytes does not necessarily cause destruction of epidermal keratinocytes, indicating destruction of hair follicles, with no detectable changes in the interfollicular epidermis13. Additionally, Rac1 is involved in the maintenance of epidermal stem cell populations14, and its expression in the epidermis is required for adult animal-type wound healing. In addition, Rac1 contributes to foliar pseudopodia formation via phosphorylation of S418 in the ARP3 subunit of the ARP2/3 complex and promotes cell migration15. In this context, it has been suggested that Rac1 repression may be involved in fetal wound healing patterns during early mouse development; however, the mechanism is unclear.
We hypothesized that Rac1 inhibition in acute fetal mouse wounds and wound-healing patterns in transgenic mice with epidermal-specific Rac1 inhibition would alter actin dynamics, resulting in skin regeneration or scar inhibition. Using an originally developed mouse fetal wound healing model, we administered NSC23766, a Rac1-specific inhibitor, to fetal wounds when the skin had not regenerated and observed its effect on wound healing16. Wounds in mouse fetuses heal within 48–72 h; therefore, we focused on the 24–48-h post-injury time point6. Our finding indicate that Rac1 suppression leads to epidermal regenerative wound healing via changes in actin dynamics and has the potential for the development of scarless skin regeneration therapies.
Results
Rac1 expression is attenuated in fully regenerating wounds in E13 fetuses
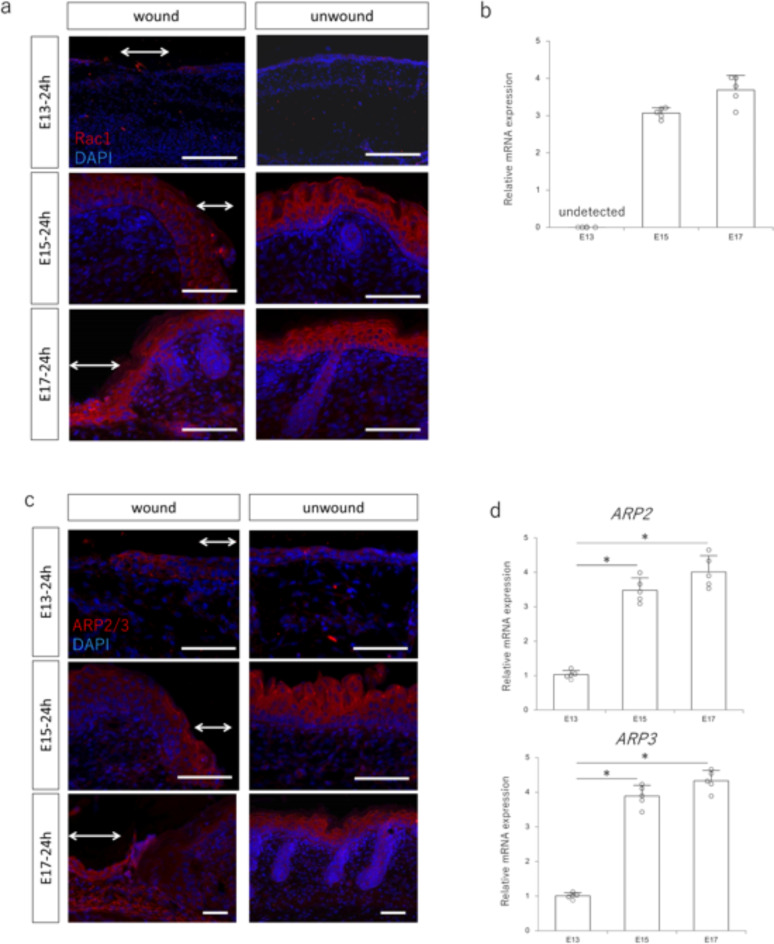
First, we evaluated Rac1 expression in mouse fetuses to investigate the changes in Rac1 dynamics during the transition from complete scarless regeneration to scar formation. We found that Rac1 expression was relatively weak in the epidermis at E13, which regenerates without scarring, and Rac1 was expressed in the epidermis from E15 onward when scarring occurred (Fig. 1a). RT–PCR using RNA collected from the wound area also showed that Rac1 expression in the wound area was lower at E13 than that at E15 (Fig. 1b). Furthermore, the ARP2/3 complex, a downstream signaling molecule of Rac1, was weakly expressed in the wound epidermis at E13, with expression observed from E15 onward (Fig. 1c). Consistent with this finding, ARP2/3 expression in the wound was lower at E13 than that at E15 and beyond (Fig. 1d). These results indicate a switch between complete wound regeneration and no wound regeneration, and altered expression of Rac1 and its downstream signaling molecules, suggesting that epidermal Rac1 repression may be involved in wound regeneration.
Fig. 1.
Expression of Rac1 signaling in the wound during embryonic mouse development. (a) Immunostaining of Rac1 in embryonic day (E)13, E15, and E17 mouse fetuses 24 h after wounding. Rac1 is weakly expressed at E13, but is highly expressed in the wound epidermis after E15. Arrows: area of wound. Red: Rac1; blue: DAPI (nuclei). Scale bar = 100 μm. (b) Quantitative evaluation of Rac1 gene expression in E13, E15, and E17 mouse fetuses 24 h after wounding. (c) Immunostaining of ARP2/3 in E13, E15, and E17 mouse fetuses 24 h after wounding. Weak ARP2/3 expression is visible on E13, whereas high expression is visible in the wound epidermis after E15. Arrows: area of wound. Red: ARP2/3; blue: DAPI (nuclei). Scale bar = 100 μm. (d) Quantitative evaluation of ARP2 and ARP3 gene expression in E13, E15, and E17 mouse fetuses 24 h after wounding. *P < 0.05.
Inhibition of Rac1 decreases epidermal cell migration
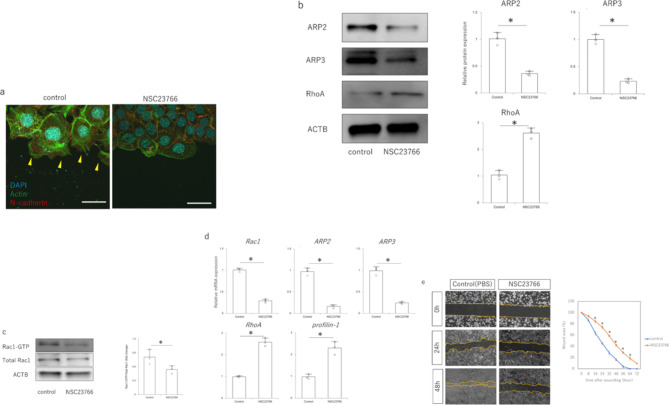
Based on changes in Rac1 expression in fetal wound healing, we investigated the effects of Rac1 inhibition in epidermal cells in vitro. In the wound-mimicking scratch assay, actin-induced filopodia were formed at the margins of the epidermal cells. However, when NSC23766 was administered, filopodia did not form, the cells were held together by N-cadherin and actin, and a cable-like actin structure was observed on the cell membrane at the wound edge (Fig. 2a). In addition, treatment with NSC23766 decreased the expression of ARP2 and ARP3, which are proteins downstream of Rac1 signaling, and increased the expression of RhoA, which is complementary to Rac1 (Fig. 2b). In the Rac1activation assay, NSC23766 treatment decreased Rac1-GTP expression compared with that in the controls (Fig. 2c). Consistent with protein expression, RT–PCR also showed decreased expression of Rac1, ARP2, and ARP3 mRNA and increased expression of RhoA and its downstream profilin-1 (Fig. 2d). In the scratch assay, NSC23766 treatment suppressed PAM212 cell migration (Fig. 2e). Thus, suppression of Rac1 reduced the migration ability by changing actin dynamics from filopodia formation to cytoskeletal formation.
Fig. 2.
Effect of NSC23766 treatment on mouse epidermal keratinocyte PAM212. (a) Actin dynamics at the PAM212 cell margin in a scratch assay (immunocytochemistry). Scale bar = 10 μm. Red: E-cadherin; green: actin; blue: DAPI (nuclei). Yellow arrowheads: filopodia. (b) Expression of Rac1 signaling-related molecules in response to NSC23766 treatment. *P < 0.05. (c) Changes in Rac1 activity in response to NSC23766 treatment (Rac1 activation assay). *P < 0.05. (d) Quantitative comparison of Rac1 signaling gene expression after NSC23766 treatment. *P < 0.05. (e) Effect of NSC23766 treatment on cell migration capacity in the scratch assay. The area covered by migrating cells was measured as the area of migration relative to the area of the gap due to scratching. *P < 0.05.
NSC23766 regenerates wounds in mouse fetuses via Rac1 signaling repression
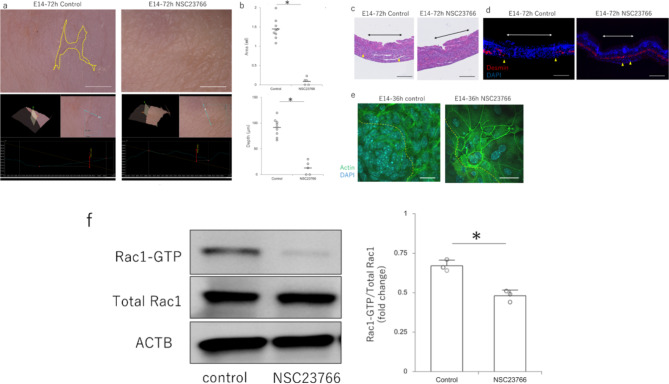
To observe the effect of NSC23766 in vivo, the drug was administered to the amniotic fluid of mice and the wound healing pattern of the fetuses was observed. At E14, which normally leaves visible mark, the controls had visible marks, whereas the wounds had regenerated, including the skin texture, in the NSC23766-treated group (Fig. 3a). When all samples were analyzed, the wounds remained significantly shallower, smaller, and regenerated in the NSC23766-treated group compared with those of the control (Fig. 3b). Histologically, after 72 h, all structures, including the panniculus carnosus muscle, were completely regenerated in the NSC23766-treated group, whereas disruption of the panniculus carnosus muscle was observed in the control group (Fig. 3c,d). Actin dynamics in the epidermis at the wound margin showed a random pattern of filopodia on the cell surface in the controls, whereas NSC23766 caused actin to be strongly expressed on the cell surface and connected to neighboring cells in a cable-like fashion (Fig. 3e). The expression of Rac1–GTP, which indicates Rac1 activity at the wound margin, was significantly decreased by NSC23766 (Fig. 3f). Thus, Rac1 suppression at the wound site by NSC23766 alters actin dynamics in the epidermis at the wound margin.
Fig. 3.
Effect of NSC23766 on wound healing in mouse fetuses. (a) Macroscopic analysis of the scar 72 h after the administration of NSC23766 to embryonic day (E)14. The upper panel shows the analysis of 2D photographs and the lower panel shows the analysis of 3D images. Yellow dotted line: area of visible mark. Scale bar = 1 mm. (b) Quantitative comparison of wound depth and area 72 h after injury following NSC23766 administration at E14. *P < 0.05. (c) Hematoxylin & eosin-stained images of the scar 72 h after the administration of NSC23766 to embryonic day (E)14. Skin structures completely regenerated in the NSC23766-treated group. Black arrows: area of the wound; yellow triangles: tips of the panniculus carnosus muscle. Scale bar = 200 μm. (d) Regeneration of panniculus carnosus muscle 72 h after the administration of NSC23766 to embryonic day (E)14 (fluorescent staining of desmin). Red: desmin; blue: DAPI (nuclei); white arrows: area of the wound; yellow triangles: tips of the panniculus carnosus muscle. Scale bar = 200 μm. (e) Immunostaining for actin in the wound epidermis at E14 and 36 h after NSC23766 administration. Green: actin; blue: DAPI (nuclei); yellow dotted line: wound margins. Scale bar = 20 μm. (f) Evaluation of Rac1 activity in the wound epidermis at E14 at 36 h after NSC23766 administration and wounding. *P < 0.05.
Epidermal-specific Rac1-knockout mice have prolonged epithelialization
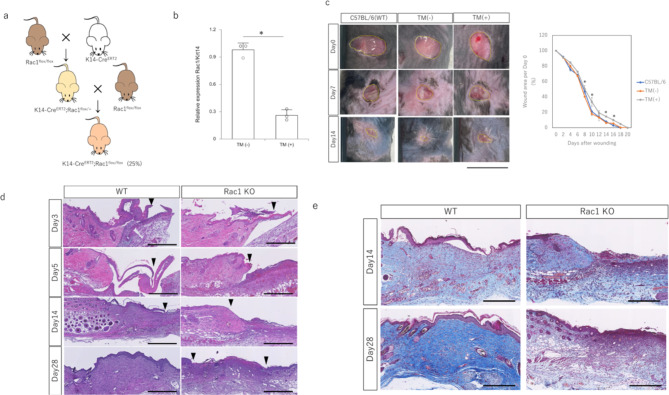
To investigate the effect of Rac1 inhibition on wound healing in vivo, we generated epidermal-specific Rac1-knockout mice. We crossed STOCK Tg(krt14-cre/ER(T2))20Efu, which expresses Cre recombinase in epidermis-specific krt14-positive cells under tamoxifen (TM) treatment, with STOCK Rac1tm1Djk/J (Rac1flox), which has a loxP site in the exon encoding Rac1 and can knock out Rac1 in cells expressing Cre recombinase (Fig. 4a). Homozygous (homo) and heterozygous (hetero) mutant offspring were obtained at the predicted Mendelian ratios. Rac1 expression in the epidermis was significantly suppressed in homozygotes after sequential administration of TM (Fig. 4b). The wound healing pattern of transgenic mice showed significantly more prolonged epithelialization than that of wild-type (WT) and non-TM-treated mice (Fig. 4c). Histologically, TM-treated epidermal-specific Rac1-knockout mice showed prolonged epidermal migration and reduced thickening of epidermis (Fig. 4d). Numerous cell infiltrations were observed at the base of the wound, indicating enhanced granulation (Fig. 4e). Thus, the epidermis-specific inhibition of Rac1 caused prolonged epidermal migration.
Fig. 4.
Wound healing in adult epidermis-specific Rac1-knockout mice (K14-CreERT2;Rac1flox/flox). (a) Shema of K14-CreERT2;Rac1flox/flox mouse crosses. (b) Evaluation of Rac1 expression in the epidermis. TM: Tamoxifen. *P < 0.05. (c) Progression of wound healing in epidermis-specific Rac1-knockout mice. Tamoxifen treatment significantly prolongs epithelialization in epidermis-specific Rac1-suppressed mice. Scale bar = 1 cm. *P < 0.05. (d) Hematoxylin & eosin stained images of wound healing process in epidermis-specific Rac1-knockout mice. Scale bar = 100 μm. (e) Masson’s trichrome stained image of the wound healing process in epidermis-specific Rac1-knockout mice. Scale bar = 100 μm.
Epidermis-specific Rac1-knockout mouse fetuses completely regenerate wounds
Wound healing patterns were observed in fetal epidermis-specific Rac1-knockout mice. Hetero crosses yielded homo and hetero mutant offspring with the predicted Mendelian ratios. Both hetero and homo fetuses survived after surgery, with survival rates comparable to those of WT fetuses.
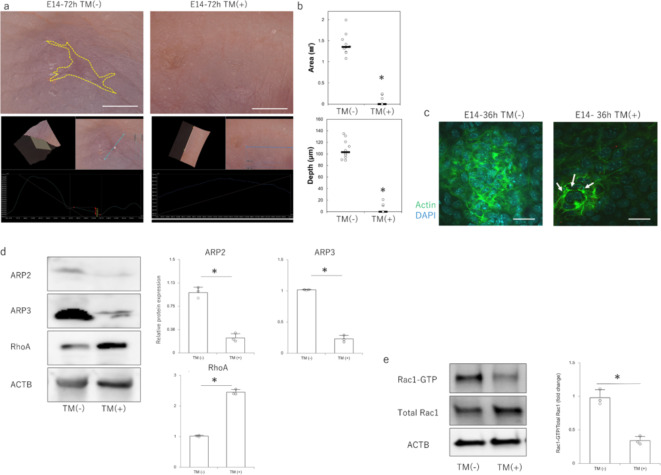
At E14, when visible makers normally remained, homozygotes in the non-TM-treated group had scars similar to those of the WT, but homozygotes in the TM-treated group had visible marks that disappeared, and the scars regenerated completely (Fig. 5a). In all recovered fetuses, the wound area was significantly smaller and shallower compared with that of the non-TM-treated group (Fig. 5b). At 24 h after the injury, during the wound healing process, actin was randomly expressed around the cells and mobilized to form filopodia in the non-TM-treated group, whereas cable-like structures were observed on the cell surface in the TM-treated group (Fig. 5c). In terms of Rac1 signaling-related molecules in the epidermis at the wound margin, TM treatment decreased the expression of ARP2 and ARP3, which are downstream proteins of Rac1 signaling, and increased the expression of RhoA, which is complementary to Rac1 (Fig. 5d). In the Rac1activation assay, the expression of Rac1–GTP decreased in the TM-treated group compared with that in the non-TM-treated group (Fig. 5e). Taken together, these results indicate that epidermis-specific Rac1 suppression contributes to the complete regeneration of fetal skin by suppressing Rac1 signaling.
Fig. 5.
Wound healing in fetuses of epidermis-specific Rac1-knockout mice (K14-CreERT2;Rac1flox/flox). (a) Macroscopic analysis of scars in embryonic day (E)14 fetuses 72 h after wounding. The wound was completely regenerated in the tamoxifen group. Yellow dotted line: extent of visible mark. Scale bar = 1 mm. (b) Quantitative comparison of wound depth and area 72 h after wounding at E14. *P < 0.05. (c) Immunostaining for actin in the wound epidermis at E14 and 36 h after wounding. Green: actin; blue: DAPI (nuclei); arrows: areas where actin is expressed in a cable-like structure. Scale bar = 20 μm. (d) Protein expression of Rac1-signaling-related molecules in E14 embryos from epidermis-specific Rac1-knockout mice. *P < 0.05. (e) Changes in Rac1 activity in fetal E14 epidermis-specific Rac1-knockout mice (Rac1 activation assay). *P < 0.05.
Discussion
In vitro suppression of Rac1 signaling by NSC23766 resulted in the inhibition of filopodia formation and cell migration at the cell edge in a wound-mimicking scratch assay. Furthermore, at E14, when the wound persisted, NSC23766 treatment suppressed Rac1 and altered actin dynamics, resulting in complete wound regeneration. Through observation of epidermis-specific Rac1-knockout transgenic mice, accelerated granulation and prolonged epithelialization occurred in adult animals, whereas for fetuses, NSC23766 administration resulted in altered actin dynamics and complete wound regeneration.
Although the potential function of Rac1 in wound healing has been predicted from previous in vitro studies and loss-of-function or inhibition experiments in Drosophila17–20, the complexity of the mammalian wound-healing process cannot be accurately represented by these models. This later reported expression of Rac1 in mammalian wound healing is a key factor in the early closure of wounds, as regulation of both RhoA and Rac1 activity contributes to bronchial epithelial wound repair mechanisms21, and activation of Rac1 by bFGF administration promotes fibroblast migration and wound healing22, which are necessary for early wound closure. Similar to the present study, to evaluate the effect of Rac1 activity in the epidermis, a report investigating the wound-healing phenotype of mice expressing a dominant-negative version of Rac1 as a transgene in epidermal keratinocytes and mice lacking epidermis-specific Rac1 showed that Rac1 inhibition was associated with prolonged wound epithelialization and suppression of keratinocyte proliferation and migration23. However, all these studies focused only on the rapidity of epithelialization in the scarring pattern of wound healing in adult animals and insufficiently discussed the relationship with scarless wound healing, including epidermal texture and regeneration of dermal structures. We established a mouse model of epidermis-specific Rac1-knockout mice by crossing K14CreERT2;Rac1flox/flox mice with JXSTOCK Rac1tm1Djk/J mice (Rac1flox/flox). In a previous report, time-lapse imaging of epithelial-conditioned Rac1-/- mice (K14-CreER; Rac1-/-; K14-H2BGFP) showed that during re-epithelialization in vivo, when compared with Rac1+/- littermates, skin epidermal cell migration and division are indeed impaired and wound-induced stratification is inhibited24. This suggests that the rate and direction of cell migration influences epidermal differentiation, promoting wound-induced stratification and regulating cell movement to form progressively thicker epidermis closer to the wound. Consistent with previously reported results, epidermal thinning (that is, reduced proliferation and migration) was observed. In contrast, Rac1 suppression resulted in complete skin regeneration in the fetus via changes in actin dynamics in epidermal cells. The difference is that the structure of the epidermis and dermis becomes more complex and less mobile than in early embryonic stages, as tissue differentiation proceeds through the developmental stages25,26. Thus, the interaction between the epidermis and dermis may not be maintained in the embryonic form by epidermal control alone. In the future, reproduction of the skin regeneration pattern in late embryonic and adult animals will require a combination of other factors that contribute to scarring, such as degradation of the extracellular matrix of the dermis and removal of the stratum corneum27,28.
A limitation of the study is that actin cables form only in the epithelium as far as has been reported, and the effects of Rac1 inhibition on regeneration and repair of dermal and fascial structures have not been fully examined and require further investigation. In addition, the structures of mouse and human skin are different, and additional experiments are needed to determine whether the effects of Rac1 shown in this study can be applied to adult mice or humans, as well as the possibility of adverse effects. Furthermore, fetal observations did not distinguish between sexes, and further experiments are needed to determine the differences in wound healing patterns between sexes.
In conclusion, we found that the suppression of Rac1 activity in the epidermis leads to wound regeneration in mouse fetuses via changes in the actin dynamics of epidermal cells. These findings may lead to the development of new therapeutic modalities for wound regeneration.
Methods
Ethical consideration
The research protocol was reviewed and approved by the Institutional Animal Care and Use Committee of Keio University School of Medicine (approval number: 20170914). All experiments were conducted in accordance with the institutional guidelines for animal experiments at Keio University. This study was conducted in accordance with the Reporting of In Vivo Experiments on Animals (ARRIVE) guidelines.
Mouse
Jcl: ICR mice were obtained from Sankyo Laboratory Services, Inc. (Osaka, Japan). STOCK Rac1tm1Djk/J (Rac1flox, #005550) and Tg(KRT14-cre/ERT)20Efu/J (KRT14ERT, #005107) mice were purchased from Jackson Laboratories (Bar Harbor, ME, USA). C57BL/6J mice (Sankyo Laboratory Services) were used as WT mice. Hetero crosses were performed, and the offspring were maintained. The phenotypes of adult animals were determined by genotyping earlobe tissue. Fetal phenotypes were confirmed by genotyping performed after the fetuses were operated on and the wound tissue was recovered.
Cell assay
Mouse epidermal keratinocytes, PAM212, were purchased from Thermo Fisher Scientific (Waltham, MA, USA). Cells were plated in 96-well plates (5 × 103 cells per well, n = 4) and maintained in 100 µL of Dulbecco’s modified Eagle’s medium (DMEM, Wako Pure Chemical, Osaka, Japan). Cells were treated with mitomycin C (NAKARAI TESQUE, INC, Kyoto, Japan; 10 µg/mL, 3 h at 37 °C) to remove proliferative effects before assay. The cells were grown to confluence on a plastic dish and 500 μm was scratched with a pipette tip. The Rac1–GTP inhibitor, NSC23766, (Tocris Bioscience, Bristol, UK) was then dissolved in phosphate buffered saline (PBS) and 100 µL was administered to each well to a final concentration of 100 µM. An equal volume of PBS was used as the control. Cells were cultured in DMEM at 37 °C, 5% CO2, and 95% relative humidity for up to 48 h. Cell migration from the abraded edge was visualized under a microscope (BZ-X800; KEYENCE, Osaka, Japan). The area covered by migrating cells was measured as the area of migration relative to the area of the gap due to scratching. The experiment was repeated three times.
Immunocytochemistry
Cells were fixed in 4% paraformaldehyde for 10 min at room temperature (20℃–25℃) and then washed once with PBS for 5 min. They were blocked with 3% bovine serum albumin (BSA)/PBS solution for 1 h at room temperature. After blocking, the primary antibody was diluted in blocking solution (3% BSA/PBS) and the sample was incubated overnight at 4 °C in antibody solution with the following antibody: rabbit anti-N-cadherin antibody (1: 200, ab76011, Abcam, Cambridge, UK). After washing three times with PBS, samples were reacted with secondary antibody Alexa Fluor 555 (Thermo Fisher Scientific, 1:200), diluted in PBS for 1 h each at room temperature. After washing three times with PBS, actin and nuclei were stained with Acti-stain 488 phalloidin (PHDG1-A; Cytoskeleton, Inc., Denver, CO, USA; 1:200) and diamidino-2-phenylindole (DAPI) solution (Thermo Fisher Scientific, 1:500) for 1 h at room temperature. The sections were sealed on glass slides using ProLong Gold (Thermo Fisher Scientific). All slides were viewed under a confocal laser-scanning microscope (FLUO-VIEW FV3000; Olympus Co., Ltd., Osaka, Japan). The experiment was repeated thrice.
Adult wounding procedure
Tamoxifen (Sigma-Aldrich, St. Louis, MO, USA) was dissolved in corn oil to a final concentration of 10 mg/mL. TM solution (100 µL of 10 mg/mL) was administered intraperitoneally to male K14-CreERT2;Rac1flox/flox mice aged 8–10 weeks for 4 consecutive days. The control group was administered the same amount of corn oil. The mice were anesthetized using 3% isoflurane inhalation, and an 8 mm dermo-punch was placed on the shaved back skin to form two 8 mm full-layer wounds per animal on both sides of the chest. Eight mice in each group were wounded. The wounds were covered with a film to avoid external irritation; a silicone adnexa, non-absorbable sutures, and surgical glue were applied to prevent skin contracture. Laceration sutures were replaced as needed, and care was taken to avoid wound destruction. Wounds were photographed weekly, and the wound area was calculated using the ImageJ software (version 1.53p (NIH, Bethesda, MD, USA). Healed skin samples were collected at 3, 5, 14, and 28 days after wounding for histological and molecular biological analyses.
Fetal wounding procedure
Pregnant Jcl: ICR mice were used for the NSC23766 administration experiment. K14CreERT2;Rac1flox/flox mice were mated overnight with JXSTOCK Rac1tm1Djk/J (Rac1flox/flox), and plug checks were performed the next day. Vaginal plugs were checked twice daily. When a plug was observed, the fetus was designated as E0, and 100 µL of 10 mg/mL TM solution was administered intraperitoneally from E10 to the mother. As a control group, female C57BL/6jjcl mice were purchased on day 10 of gestation, and fetuses were obtained by administering TM in the same manner.
Surgery was performed on five pregnant mice from each group at E13, E14, E15, and E17. Pregnant mice were anesthetized with 3% isoflurane and the abdominal wall was incised to expose the uterus. The myometrium and amnion/yolk sac were excised under an operating microscope. Next, using surgical microscissors, a full-layer incision, approximately 2 mm long, was made in the lateral thoracic region of the fetus. During the NSC23766 experiment, the control group received 100 µL of saline and the treated group received 100 µL of NSC23766 (100 mM) dissolved in the same volume of saline in the amniotic fluid at the same time as wound surgery. The treated group received 100 µL of NSC23766 dissolved in the same volume of saline. At E13 and E14, the amnion and yolk sac were sutured with 9 − 0 nylon, but the myometrium was left open and unsutured. Therefore, the fetus was returned to the abdominal cavity with the amnion and yolk sac covered, but the myometrium uncovered, and the abdomen was closed; in E15 and E17, after making fetal wounds, the myometrium was sutured with 9 − 0 nylon, the uterus was returned to the abdominal cavity, and the abdomen was closed. To mark and visualize the wound site, the wound was labeled with 0.25% 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate (DiI) dissolved in 1% ethanol in PBS after wound preparation. Then, just before the closure of both wounds, a uterine relaxant, 1 µg/gbw of ritodrine hydrochloride (FUJIFILM Wako Pure Chemical), was intraperitoneally administered. The peritoneum and skin were sutured using a 5 − 0 nylon thread. Maternal mice were euthanized by cervical dislocation and fetuses were harvested 24, 36, and 72 h after wounding. At each time point, wounds were induced in at least four fetuses. DiI fluorescence was confirmed with a stereomicroscope (SZX16, Olympus Corporation, Tokyo, Japan) to identify the site of the wound. Images of the wound were captured using an ultra-high precision digital microscope VHX-8000 (Keyence Corporation, Osaka, Japan) and reconstructed in three-dimensions (3D). Morphological images of the wound were imported into ImageJ, wound area was measured, and area ratios were calculated. For wound depth, the deepest point of the cross-sectional view across the center of the wound in the 3D reconstructed image was measured. Fetal bodies were fixed in 4% paraformaldehyde for 24 h, wound tissue was cut out with a No. 11 scalpel and microsurgical scissors, and fixed tissue was embedded in paraffin for analysis.
Immunohistochemistry
Paraffin-embedded specimens were sliced into 7 μm-thick sections and mounted on glass slides. After drying overnight at room temperature to allow the specimens to adhere to the slides, the slides were deparaffinized by changing xylene twice at room temperature (5 min each). Slides were transferred twice to 100% ethanol (3 min each), once to 95%, 70%, and 50% ethanol (3 min each), and rehydrated at room temperature. After antigen activation by heat, the samples were incubated with 2% goat serum in PBS for 30 min at room temperature to block nonspecific binding sites. Samples were then incubated with anti-Rac1 antibody (1:100, GTX100761, GeneTex, Irvine, CA, USA), anti-ARP2/3 antibody (1:100, MABT95, Sigma-Aldrich), and anti-desmin antibody (1:1000, MA122150, Thermo Fisher Scientific) diluted with PBS at 4 °C overnight. After washing three times with PBS, they were reacted with secondary antibody Alexa Fluor 555 (1:2000, Thermo Fisher Scientific) diluted with PBS for 1 h each at room temperature. Nuclei were washed thrice with PBS and stained with DAPI solution (Thermo Fisher Scientific, 1:500) for 1 h at room temperature. Slides sealed on glass slides by ProLong® Gold (Thermo Fisher Scientific) were mounted on an integrated stereomicroscope (BZ-X800; KEYENCE). The experiment was repeated thrice.
Western blotting
Total protein was extracted from cells and tissues using RIPA buffer (Santa Cruz Biotechnology, Santa Cruz, CA, USA). Each sample (30 µg) was electrophoresed on 10% polyacrylamide gels Mini-PROTEAN® TGX™ Precast Gels (Bio-Rad Laboratories, Inc., CA, USA) and transferred to a Trans-Blot Turbo Transfer System (Bio-Rad Laboratories, Inc.). The cells were transferred to polyvinylidene difluoride membranes using the Trans-Blot Turbo Transfer System (Bio-Rad Laboratories, Inc.). After blocking with 3% nonfat milk at room temperature for 1 h, primary antibodies diluted in blocking solution, ARP2 (#4738, 1:100, Cell Signalling Technology, Danvers, MA, USA), ARP3 (ab49671, 1:100, Abcam, Cambridge UK), RhoA (#2117, 1:100, Cell Signalling Technology), and GAPDH (1:2000; Santa Cruz Biotechnology) were incubated overnight at 4 °C. The next day, incubation was conducted with the following secondary antibodies at 37 °C for 1 h: donkey anti-goat IgG H&L (HRP; ab6885; Abcam) and goat anti-rabbit IgG H&L (HRP; ab205718; Abcam). After washing three times with Tris-buffered saline (TBS), the immunoreactive protein bands were visualized using an electrochemiluminescence detection kit (Pierce Biotechnology, Rockford, IL, USA). A chemiluminescence imager (ImageQuant LAS4000 mini; GE Healthcare, Chicago, IL, USA) was used to image the bands. Image analysis was performed using the ImageJ software. Each experiment was repeated thrice.
Laser micro dissection (LMD)
LMD was performed using a PALM MicroBeam (Carl Zeiss, Germany). The manufacturer’s recommended slides and collection tubes (AdhesiveCap 500 opaque; Carl Zeiss) were set up and the tissue was carefully cut after adjusting the aperture and intensity using a 20× magnification objective lens. The tube caps were filled with Buffer RLT (RNeasy Micro Kit; Qiagen, Germany) with β-mercaptoethanol to allow separation of intact RNA.
RNA isolation and reverse transcription
Total RNA was extracted from the cells using the RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Total RNA was mixed with random primers, reverse transcriptase, and dNTP mixture (Takara Bio, Shiga, Japan). The mixture was incubated in a T100TM thermal cycler (Bio-Rad Laboratories, Inc.) at 25 °C for 5 min, 55 °C for 10 min, and 80 °C for 10 min to heat inactivate the reverse transcriptase and produce cDNA.
Quantitative real-time PCR
RT–PCR was performed using the Applied Biosystems 7500 Fast Real-Time PCR System (Thermo Fisher Scientific). A total of 40 cycles were performed, and the fluorescence of each sample was measured at the end of each cycle. The PCR reaction was performed in two major steps: holding the reagent at 95 °C for 3 s (denaturation) and at 60 °C for 30 s (annealing and extension). In the subsequent melting curve analysis phase, the temperature was increased from 60 °C to 95 °C and fluorescence was measured continuously. Rac1 (Mm00488845_m1), ARP2 (Mm07300461_g1), ARP3 (Mm02342769_g1), RhoA (Mm00834507_g1), and profilin-1 (Mm00726691_s1) were used as the primers (Thermo Fisher Scientific). The PCR master mix (4352042; Applied Biosystems, Foster City, CA, USA) was used according to the manufacturer’s instructions, and ACTB (Mm02619580_g1) was used as a control gene for normalization. Gene expression levels at normal sites were used as baseline and fold-change values were determined using the 2-ΔΔCt method.
Rac1 activation assay
Proteins extracted using the method described above were used to assess Rac1 activation using the G-LISA Rac1 Activation Assay Biochem Kit (Cytoskeleton Inc., Denver, CO, USA) according to the manufacturer’s protocol. Briefly, 50 µL of protein adjusted to 0.5 mg/mL was added to a special 96-well plate of the kit and incubated in a cold orbital microplate shaker (Biosan, Riga, Latvia) for 30 min at 4 °C. Antigen presenting buffer was added for 2 min, and a 1:300 dilution of anti-Rac1 antibody in the kit was added and allowed to react at room temperature for 60 min. After washing with TBS, HRP-conjugated secondary antibody diluted at 1:100 was added and allowed to react at room temperature for 60 min. After washing with TBS, HRP detection reagent was added, and absorbance was detected at 490 nm using a microplate reader (SpectraMax Paradigm, Molecular Devices, San Jose, CA, USA). Each experiment was repeated thrice.
Statistical analysis
The Mann–Whitney U test was performed using the Statistica software (version 9.0; StatSoft, Tulsa, OK, USA) to determine the significance of differences in gene expression. Descriptive statistics are presented as the mean ± standard deviation. Statistical significance was set at P < 0.05. Each experiment was performed in triplicate.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors thank the laboratory members for their technical assistance with the experiments. This study was funded by JSPS KAKENHI (grant number: JP 23K24511).
Author contributions
Conceptualization: K.T., Y.I., and K.K. Data curation: K.T., Y.I., and N.A-H. and K. K.; formal analysis, K. K. and Y. I.; funding acquisition, K. K.; investigation, K. K., Y. I., S. S., and K. K.; methodology, K. K.; project administration, K. O. and K. K.; and resources, N.A-H. K. K., Q. W., and K. K.; supervision, K. K.; validation, K. T. and K. K.; visualization, K. T. ; writing—original draft preparation, K. T.; writing—review and editing, K. K.
Data availability
The data presented in this study are available on request from the corresponding author.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-78395-2.
References
- 1.Erickson, J. R. & Echeverri, K. Learning from regeneration research organisms: the circuitous road to scar free wound healing. Dev. Biol.433, 144–154 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burrington, J. D. Wound healing in the fetal lamb. J. Pediatr. Surg.6, 523–528 (1971). [DOI] [PubMed] [Google Scholar]
- 3.Rowlatt, U. Intrauterine wound healing in a 20 week human fetus. Virchows Arch. Pathol. Anat. Histol.381, 353–361 (1979). [DOI] [PubMed] [Google Scholar]
- 4.Ishii, T. et al. Skin wound healing of the adult newt, Cynops pyrrhogaster: a unique re-epithelialization and scarless model. Biomedicines9, 1892 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takaya, K. et al. Actin cable formation and epidermis-dermis positional relationship during complete skin regeneration. Sci. Rep.12, 15913 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin, P. & Lewis, J. Actin cables and epidermal movement in embryonic wound healing. Nature360, 179–183 (1992). [DOI] [PubMed] [Google Scholar]
- 7.Henson, J. H. et al. Wound closure in the lamellipodia of single cells: mediation by actin polymerization in the absence of an actomyosin purse string. Mol. Biol. Cell.13, 1001–1014 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yumura, S. et al. Dynamics of actin cytoskeleton and their signaling pathways during cellular wound repair. Cells11, 3166 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kishi, K., Okabe, K., Shimizu, R. & Kubota, Y. Fetal skin possesses the ability to regenerate completely: complete regeneration of skin. Keio J. Med.61, 101–108 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Simon, A. R. et al. Regulation of STAT3 by direct binding to the Rac1 GTPase. Science290, 144–147 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Braga, V. M., Betson, M., Li, X. & Lamarche-Vane, N. Activation of the small GTPase Rac is sufficient to disrupt cadherin-dependent cell-cell adhesion in normal human keratinocytes. Mol. Biol. Cell.11, 3703–3721 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mertens, A. E., Rygiel, T. P., Olivo, C., van der Kammen, R. & Collard, J. G. The rac activator Tiam1 controls tight junction biogenesis in keratinocytes through binding to and activation of the Par polarity complex. J. Cell. Biol.170, 1029–1037 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chrostek, A. et al. Rac1 is crucial for hair follicle integrity but is not essential for maintenance of the epidermis. Mol. Cell. Biol.26, 6957–6970 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DiPersio, C. M. Double duty for Rac1 in epidermal wound healing. Sci. STKE pe33 (2007). [DOI] [PubMed]
- 15.Bogucka-Janczi, K. et al. ERK3/MAPK6 dictates CDC42/RAC1 activity and ARP2/3-dependent actin polymerization. eLife12, e85167 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang, C. et al. Inhibition of Rac1 activity by NSC23766 prevents cartilage endplate degeneration via Wnt/β-catenin pathway. J. Cell. Mol. Med.24, 3582–3592 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farooqui, R. & Fenteany, G. Multiple rows of cells behind an epithelial wound edge extend cryptic lamellipodia to collectively drive cell-sheet movement. J. Cell. Sci.118, 51–63 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Fenteany, G., Janmey, P. A. & Stossel, T. P. Signaling pathways and cell mechanics involved in wound closure by epithelial cell sheets. Curr. Biol.10, 831–838 (2000). [DOI] [PubMed] [Google Scholar]
- 19.Hakeda-Suzuki, S. et al. Rac function and regulation during Drosophila development. Nature416, 438–442 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Zhou, S. et al. Two Rac1 pools integrate the direction and coordination of collective cell migration. Nat. Commun.13, 6014 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Desai, L. P., Aryal, A. M., Ceacareanu, B., Hassid, A. & Waters, C. M. RhoA and Rac1 are both required for efficient wound closure of airway epithelial cells. Am. J. Physiol. Lung Cell. Mol. Physiol.287, L1134–L1144 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Kanazawa, S. et al. bFGF regulates PI3-kinase-Rac1-JNK pathway and promotes fibroblast migration in wound healing. PLoS One5, e12228 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tscharntke, M. et al. Impaired epidermal wound healing in vivo upon inhibition or deletion of Rac1. J. Cell. Sci.120, 1480–1490 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Park, S. et al. Tissue-scale coordination of cellular behaviour promotes epidermal wound repair in live mice. Nat. Cell. Biol.19, 155–163 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cass, D. L. et al. Wound size and gestational age modulate scar formation in fetal wound repair. J. Pediatr. Surg.32, 411–415 (1997). [DOI] [PubMed] [Google Scholar]
- 26.Longaker, M. T. & Adzick, N. S. The biology of fetal wound healing: a review. Plast. Reconstr. Surg.87, 788–798 (1991). [DOI] [PubMed] [Google Scholar]
- 27.Jiang, D. et al. Injury triggers fascia fibroblast collective cell migration to drive scar formation through N-cadherin. Nat. Commun.11, 5653 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, C. et al. Establishment of an in vitro scab model for investigating different phases of wound healing. Bioeng. (Basel)9, 191 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available on request from the corresponding author.