Abstract
Introduction
A better understanding of the physiological response of silage maize to a mild reduction in nitrogen (N) fertilization and the identification of predictive biochemical markers of N utilization efficiency could contribute to limit the detrimental effect of the overuse of N inputs.
Objectives
We integrated phenotypic and biochemical data to interpret the physiology of maize in response to a mild reduction in N fertilization under agronomic conditions and identify predictive leaf metabolic and proteic markers that could be used to pilot and rationalize N fertilization.
Methods
Eco-physiological, developmental and yield-related traits were measured and complemented with metabolomic and proteomic approaches performed on young leaves of a core panel of 29 European genetically diverse dent hybrids cultivated in the field under non-limiting and reduced N fertilization conditions.
Results
Metabolome and proteome data were analyzed either individually or in an integrated manner together with eco-physiological, developmental, phenotypic and yield-related traits. They allowed to identify (i) common N-responsive metabolites and proteins that could be used as predictive markers to monitor N fertilization, (ii) silage maize hybrids that exhibit improved agronomic performance when N fertilization is reduced.
Conclusions
Among the N-responsive metabolites and proteins identified, a cytosolic NADP-dependent malic enzyme and four metabolite signatures stand out as promising markers that could be used for both breeding and agronomic purposes.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11306-024-02186-z.
Keywords: Maize, Markers, Metabolome, Nitrogen nutrition, Proteome, Silage
Introduction
Maize (Zea mays L., also called corn) is cultivated worldwide in both temperate and tropical areas and is currently ranked first among cereals, representing more than 40% of the total world cereal production (http://www.worldagriculturalproduction.com/crops/corn.aspx). It is one of the major sources of food and feed for both animal and human consumption. Breeding strategies and sustainable agronomic practices have thus been developed to improve maize agronomic performance, either for silage or kernel production and to produce derived commercial products, such as bioethanol (Ranum et al., 2014). The challenge is to select new varieties adapted to a wide range of environmental constraints, including climate change, biotic and abiotic stresses while preserving the environment (Carena, 2021; Voss-Fels et al., 2019). An efficient use of nitrogen (N) fertilizers is required to attain maximal yield in most crops cultivated worldwide (Hirel & Krapp, 2020). However, owing to the detrimental effect of the overuse of N fertilizers on both marine and terrestrial ecosystems and on the release of greenhouse gases, several complementary actions have been conducted. They combine whole plant physiology, molecular genetics and breeding to identify the limiting steps of crop nitrogen use efficiency (NUE) (Sutton et al., 2020). Nitrogen use efficiency corresponds to the yield obtained per unit of N supplied by the soil in the form of mineral or organic N fertilizers (Beatty & Good, 2018).
Only nitrate transporters and the enzyme glutamine synthetase (GS) have proved to be involved in the control of crop productivity in general and maize in particular (Fortunato et al., 2023; Hirel & Krapp, 2020). Other studies have also shown that the undesirable side-effects of fertilization can be significantly decreased by improving agricultural practices (Giordano et al., 2021; Hirel et al., 2011), and by benefiting from the microbial rhizosphere (Dellagi et al., 2020; Porter et al., 2020). Until now, most diagnostic tools used in precision agriculture were sensors or chlorophyll-meters, measuring specific spectral indices that respond to variations in leaf area index or leaf chlorophyll concentration, either at plant or canopy level (Sahu et al., 2019). The information derived from these sensors can then be used to estimate fertilization requirements in a wide range of environments (Kumar & Ilango, 2018). The use of a core set of molecular and biochemical marker traits representative of the plant nutritional status has also been proposed both to monitor and pilot N fertilization (Sinha et al., 2020).
Importantly, our understanding of the complex control of NUE of maize has been greatly improved by the development of holistic approaches based on the use of these biological markers (Chowdhury et al., 2022; Simons et al., 2014a, 2014b). Although omics data collected from plants grown in the field and their integration remain scarce, especially in commercial maize hybrids, interesting perspectives have opened up concerning the use of metabolome-assisted breeding techniques for narrowing the genotype/phenotype gap of complex traits, such as yield and biomass production (Zhang et al., 2015).
Maize is a strategic species for the production of meat and milk, which are increasingly demanded worldwide (Henchion et al., 2017). It is one of the most high-yielding forage crops and it is generally cheaper to produce than other forage crops (Heuzé et al., 2017). Silage conservation guarantees an excellent feed and source of proteins for ruminants. This has led to the implementation of N management practices to significantly decrease pollutant emissions from maize cultivation (Piccini et al., 2016). However, further improvements in the nutrient management of silage maize are still required to adjust both the amount and timing of N fertilization and mitigate its potentially detrimental environmental effects (Adegbeye et al., 2020).
Therefore, to decipher the biochemical bases of N utilization and metabolism in relation to growth and productivity in silage maize, we combined a metabolomic and a proteomic approach exploiting the genetic diversity of 29 hybrids issued from the crossing of a core panel of 29 European dent lines with a flint tester.
Methods
Plant material, growth conditions and sampling
Twenty-nine genetically diverse Dent and Stiff Stalk maize inbred-lines (Zea mays ssp. mays, Table S1) structured into four admixture groups including European, Iodent, Lancaster and Stiff Stalk genotypes (Ganal et al., 2011; Rincent et al., 2014) were selected according to their diversity based on pedigree, genotyping and a narrow range of female flowering dates not exceeding two equivalent days at 20 °C. A dendrogram showing the genetic distances between the parental lines could be built as genotyping data of 26 of the 29 inbred-lines are open (Nicolas et al., 2020). SNP data, containing 978,134 genetic markers, were downloaded from Nicolas et al. (2020) and the 26 lines corresponding to the present study were selected. The dendrogram was created using R (version 4.3.3, R Core Team, 2022). A distance matrix was computed using the dist function with the Euclidean method. Then, clustering was performed with Ward’s method (ward.D) using the hclust function (Figure S1A). The inbred-lines were crossed with flint inbred-line UH007 (Univ. Hohenheim, Germany) developed to improve combining ability with Iodent and Stiff-Stalk lines for earliness, yield of kernel and stover, and sown as hybrids in a field located in Estrées-Mons (Northern France, 49°52′44″ N, 3°0′27″ E).
The soil was deep loam. N:P:K fertilizer and irrigation were applied. Plants were subjected to non-limiting and reduced N fertilization treatments (HN and LN, respectively). Before sowing, N fertilization was adjusted according to the amount of residual N already present in the soil (110 kgN.ha−1) to obtain a final yield of 100 quintals (q) for HN and 80 q for LN (147 kgN.ha−1 and 58 kgN.ha−1 for HN and LN plots, respectively). Air temperature, rainfall, air vapor pressure deficit and global radiation were recorded by a meteorological station located approximately 600 m from the field. Thermal times starting from emergence (equivalent to the number of equivalent days (EqD) at 20 °C) were calculated from air temperature (Parent et al., 2010) for each condition. The experimental design for each condition consisted in four individual rows of 40 plants per genotype, planted in three randomized blocks. Plants were sown on May 19th, 2014, and leaf samples were harvested on July 11th (20.1 EqD at 20 °C after emergence, d20°C). For each hybrid and block, the youngest ligulated leaf (usually from the third to the fifth leaf depending on the genotype) at the 8-visible-leaf plant stage was harvested between 10:00 am and 1:00 pm from 8 plants in each block. In each of the 8 leaves, a single central 5-cm section without the main vein was selected. The 8 leaf sections were pooled and immediately frozen in liquid nitrogen. The resulting powder was separated into two aliquots and stored at − 80 °C until further use. One of the two aliquots was lyophilized and stored in a dry environment at − 20 °C for metabolome analyses and targeted analyses of free amino acids, as well as total C and N analyses. The second aliquot was stored at − 80 °C for enzymatic activity measurements and proteomics. The youngest ligulated leaf was also harvested at flowering and ground as described above for total C and N analyses. The three biological replicates used for the different biochemical analyses corresponded to each block.
Measurement of eco-physiological, developmental and yield-related traits
In the 29 hybrids grown under HN and LN, eco-physiological, developmental and agronomic traits were measured at various developmental stages spanning from the four ligulated leaf (4LL) stage to maturity (kernel harvest) (Additional text, Annex 1, raw and Least Square Means [LSmeans] data in Table S2a,c).
Metabolome analyses
Metabolomic profiling based on proton nuclear magnetic resonance spectroscopy (1H-NMR) was performed on polar extracts obtained from 20 mg of lyophilized leaf powder, using hot ethanol–water extraction as described previously (Biais et al., 2009). The resulting extracts were prepared and analyzed according to Lamari et al. (Lamari et al., 2018) using a 500 MHz spectrometer. Metabolites were identified and quantified as described previously (Lamari et al., 2018). This resulted in the quantification of 28 metabolites expressed in μg gDW−1. The corresponding NMR data and metadata were deposited in the recherche.data.gouv.fr dataverse repository (10.57745/NZNMZH).
Liquid Chromatography Quadrupole Time-of-Flight Mass Spectrometry (LC–QTOF–MS) fingerprinting of semi-polar extracts was performed as in Lamari et al. (Lamari et al., 2018) with the modifications described in Additional text, Annex 2. This resulted in 2808 variables, or metabolite signatures, named using their nominal masses in Da and retention time in s (for instance M180T746 for a signature with a 180-Da nominal mass and a 746-s retention time). Annotation of 29 intense ions was based on previous studies (Lamari et al., 2018; Urrutia et al., 2021). Annotation with putative name assignments of the LC–QTOF–MS-based biomarkers highlighted by statistical analyses was performed by calculating chemical formulae using SmartFormula software (Bruker, Bremen, Germany) and comparing them with those available in the Dictionary of Natural Products (https://dnp.chemnetbase.com/). The LC–QTOF–MS data and metadata were deposited in the recherche.data.gouv.fr dataverse repository (10.57745/UPAO0X). Raw data and LSmeans for the identified metabolites and metabolite signatures in HN and LN fertilization conditions are presented in Table S3a and c, respectively. Annotations of the LC–MS–QTOF metabolite signatures are presented in Table S3d.
Starch, total protein and free amino acid quantification
After robotized ethanolic extraction, starch and total protein contents of the leaf samples were determined enzymatically or colorimetrically in the pellets, as previously described (Lamari et al., 2018). Starch and total proteins were expressed on a gDW−1 basis. Amino acids were quantified by Ultra High-Performance Liquid Chromatography (UHPLC) fluorimetry after derivatization with the AccQ-Tag method (Waters, Milford, MA), from a fourfold water dilution of the extraction performed for 1H-NMR analyses, following the supplier’s recommendations (Additional text, Annex 3). Final quantification was expressed in gDW−1.
Enzyme activity measurements
Soluble enzymes were extracted from fresh weight samples (20 ± 0.5 mg). Enzymatic activities for phosphoglucose isomerase (PGI), triose phosphate isomerase (TPI), NAD-glutamate dehydrogenase (GDH), glutamine synthetase (GS), alanine aminotransferase (AlaAT) and aspartate aminotransferase (AspAT) (Gibon et al., 2004; O'Neal & Joy, 1973) were measured following the procedure recommended by Bénard and Gibon (Bénard & Gibon, 2016) in a robotized Starlet platform (Hamilton, Villebon-sur-Yvette, France). Raw data and LSmeans for enzyme activities in HN and LN fertilization conditions are presented in Table S4a and c, respectively.
LC–MS/MS shotgun proteomics
Proteins were extracted from 50 mg of fresh material of leaf samples. Protein extraction and digestion were performed as described previously (Blein-Nicolas et al., 2020). LC–MS/MS analyses of protein digests (400 ng of peptides) were performed using an Ultimate 3000 RSLCnano System coupled with an Orbitrap Fusion™ Lumos™ Tribrid™ mass spectrometer (Thermo Electron, Waltham, MA) as described in Bednarz et al. (Bednarz et al., 2021) with minor modifications described in Additional text, Annex 4. Proteins represented by at least two reproducible and consistent peptides were quantified by summing their intensities, as in Balliau et al. (2018), in order to measure their relative abundance. Protein annotations and GO terms were extracted from maizegdb (https://www.maizegdb.org/, RRID:SCR_006600, (Portwood et al., 2018)) and complemented using Mercator V4.0 (https://www.plabipd.de/portal/mercator4) and enzymatic data included in CornCyc v8.0 (https://www.plantcyc.org/databases/corncyc/8.0). The mass spectrometry proteomics data were deposited in the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2021) partner repository with dataset identifier PXD034145. Raw data and LSmeans for the identified proteins in HN and LN are presented in Table S5a and c, respectively.
Data analyses
Univariate and multivariate data analyses (ANOVA, principal component analysis [PCA], hierarchical clustering analysis [HCA]) were conducted as described in Annex 5. Eco-physiological, developmental and yield-related traits were combined with the metabolite and proteome data, using multiblock sparse partial least-squares discriminant analysis (PLS-DA) on LSmeans data (calculated for each N fertilization treatment) and correlation networks. For each variable affected by the N treatment according to a two-way ANOVA, the “susceptibility” value per genotype was calculated as follows: susceptibility = (LSmeans (HN) − LSmeans (LN))/LSmeans (HN), to search for the less-susceptible hybrids.
Results
Reduced N fertilization had a significant impact on eco-physiological, developmental and agronomic traits
In the 29 hybrids, a two-way ANOVA analysis (P < 0.05 after FDR correction) showed that in the LN condition, a significant decrease was observed in 17 out of the 25 eco-physiological traits (Table 1). An increase was observed in only one of them (Sen80) under reduced N fertilization. A greater impact of reduced N fertilization tended to be observed at flowering. On average, at flowering, a 35% and 30% decrease were observed for QN_Flo and NNI, respectively. In addition, the rate of leaf senescence (Sen80) increased by 6% at the latest stages of plant development. When N fertilization was reduced, there was a lower than the 20% expected yet significant decrease in kernel production (KW) at maturity (6%) and in total plant biomass production (8% to 10%), both at the vegetative stage (TDW_4DL) and at maturity (TDW), (Figure S1B).
Table 1.
Variability of eco-physiological, developmental and yield-related traits measured in a panel of 29 maize hybrids under two N treatments
| Trait | Trait abbreviation | Variation (%) |
|---|---|---|
| Foliar surface (1 to 8 FDL) | Surf_i | − 9.67 |
| Foliar surface (12 to 14 FDL) | Surf_o | − 9.69 |
| Total foliar surface (FDL) | Surf_tot | − 10.05 |
| Leaf emergence vigour at 10 EqD | EV | − 2.99 |
| Leaf emergence rate (VL) | LER_V | ns |
| Leaf emergence rate (FDL) | LER_D | ns |
| Senescence at 40 EqD | Sen40 | − 5.99 |
| Senescence at 80 EqD | Sen80 | 6.27 |
| Ear leaf rank | Rk | ns |
| Male flowering date | FloM | ns |
| Female flowering date | FloF | ns |
| Total nitrogen at 4FDL | QN_4FDL | − 13.45 |
| Total carbon at 4FDL | QC_4FDL | − 8.77 |
| Nitrogen nutrition index at 4FDL | NNI_4FDL | − 4.09 |
| Total nitrogen at flowering | QN_Flo | − 34.72 |
| Total carbon at flowering | QC_Flo | − 8.74 |
| Nitrogen Nutrition Index at flowering | NNI_Flo | − 30.52 |
| Relative growth rate (until 11 VL) | Rgr_Veg | ns |
| Relative growth rate (until flowering) | Rgr_Flo | ns |
| Thousand kernel weight | TKW | − 1.87 |
| Kernel number per ear | KN | − 4.42 |
| Total kernel dry weight | KW | − 6.33 |
| Total dry weight at 4FDL | TDW_4FDL | − 9.73 |
| Total dry weight at harvest | TDW | − 8.49 |
| Kernel nitrogen percent | K%N | − 12.95 |
Variations were calculated using the mean of 3 replicates corresponding to three different blocks. Each replicate is composed of a pool of 8 individual plants. Details of the analysis and statistics are presented in Table S2
ns not significant
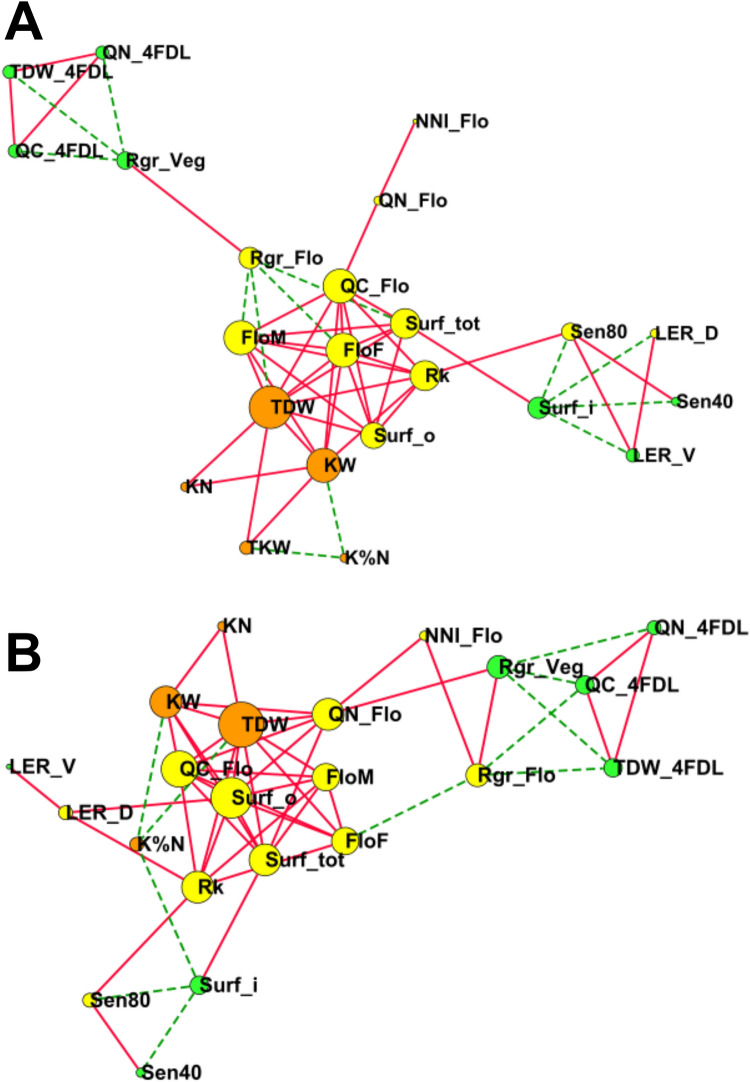
When the LS-means of all these traits were used, we found that most of the correlations were significant (P < 0.05 after FDR correction), irrespective of the level of N fertilization. The correlation coefficients are shown for the HN and LN conditions in Fig. 1A and B, respectively, and their values are presented in Table S2d. All the traits exhibiting a positive or negative correlation could be grouped into three main clusters, irrespective of the level of N fertilization. One of these three clusters contained most of the measured traits, including the kernel and shoot-biomass yield components measured at harvest (KN, KW, TDW, K%N) together with the traits related to the leaf surface (Surf_o and Surf_tot) and to status of plant C and N at flowering (QN_Flo, NNI_Flo, QC_Flo). All these traits were the most strongly and positively correlated. In this cluster, only the kernel N content (K%N) was negatively correlated to KW in both HN and LN conditions.
Fig. 1.
Network diagram showing the main correlations between the different eco-physiological, developmental and yield-related traits. Traits were measured in the panel of 29 maize hybrids grown under reduced (LN) and non-limiting (HN) fertilization conditions. Network diagrams show significant correlations with FDR correction (P < 0.05) between trait LSmeans (n = 29) based on positive and negative Pearson coefficients in HN A and LN B. Traits with a larger number of significant correlations are represented by larger dots. Traits with a smaller number of significant correlations are represented by smaller dots. Green dots correspond to the vegetative stage, yellow dots to the flowering stage and orange dots to plant maturity at harvest. Lines represent a significant correlation between two traits. Full red lines represent positive correlations. Dashed green lines represent negative correlations. See Table 1 for definitions of abbreviations and Table S2d for Pearson coefficients and corresponding statistical analyses
Two other small clusters of traits were also identified, including those related to the plant physiological status during vegetative growth at 4DL and to leaf senescence. Within these two clusters, negative correlations were observed between Rgr_Veg and the other traits measured at the vegetative stage of plant development (TDW_4FDL, QN_4FDL, GC_4FDL), and a positive correlation was observed between the latter three traits. In both HN and LN conditions, the second minor cluster encompassed the two traits used to monitor leaf senescence (Sen40 and Sen80) and Surf_i, which were negatively correlated.
For the key marker enzymes involved in primary C and N metabolism (Table S4), the most interesting result was a positive correlation between total leaf glutamine synthetase (GS) activity and the two main yield components represented by KW and TDW when N fertilization was reduced (Table S4d). For this enzyme, both the level of N fertilization and its interaction with the genotypic effect were highly significant, with 65% variation for the former (two-way ANOVA, corrected P < 0.05, Table S4c).
Strong genotypic variability for tolerance to N-limiting conditions
A PCA was conducted using as variables the different phenotypic and physiological traits measured in the 29 hybrids that were significantly different between the HN and LN conditions (Fig. 2). Along the first two components of the PCA (PC1 and PC2), which accounted for 27% and 19% of the variance respectively, a clear separation between HN and LN could be observed for most hybrids along PC1. The Euclidian distance (Ed) between the two N fertilization conditions in the PC1 x PC2 plan depended on the hybrids and roughly defined their responsiveness to N fertilization. For example, B84 (Ed = 6.5) exhibited the greatest difference between HN and LN. This difference was the lowest for F618 (Ed = 1.3), whereas an intermediate N responsiveness (Ed spanning from 2 to 5) was obtained for FR19 (Ed = 4.1).
Fig. 2.
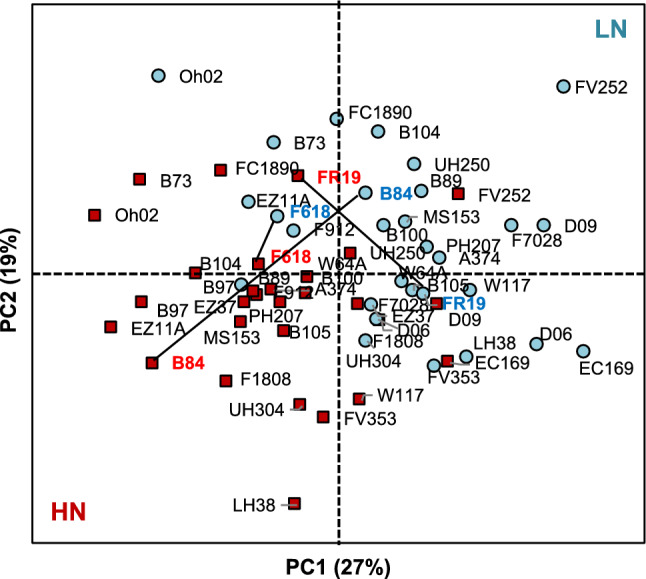
Ordination diagram of the PCA analysis for the 29 maize hybrids based on the different eco-physiological, developmental and yield-related traits. Traits were measured in the panel of 29 maize hybrids grown under reduced (LN) and non-limiting (HN) fertilization conditions. Diagrams are defined by the first two PCs of the PCA of the different variables using their LSmeans: PC1 (27% of variance explained) and PC2 (19% of variance explained). LN: reduced N fertilization (blue circles). HN: non-limiting N fertilization (red squares). Black lines between LN and HN represent the Euclidian distance between the three hybrids (B84, FR19 and F618) exhibiting the most contrasted difference with respect to their morphological and physiological responsiveness to N fertilization
Identification of N-responsive metabolite and protein markers
PCAs were performed to obtain an overview of the metabolite (Figure S2) and protein (Figure S3) composition in the 29 hybrids, together with a visual representation of their distribution in the HN and LN conditions. The score plots show that the two levels of N fertilization were clearly separated along the first principal component (PC1), accounting for 13 and 18% of total variability for metabolites and proteins, respectively. The 29 hybrids were separated along the PC2 axis, which reflects the genetic variability for their metabolite and protein contents. Moreover, the distribution of the 29 hybrids along the PC2 axis was not always similar in the HN and LN conditions, thus indicating that a genotype/level of N fertilization interaction probably occurred in some of them.
For the metabolites (Figure S2), several carbohydrates, an organic acid and amino acid glutamate were more abundant in the LN condition. Several other amino acids and choline were more abundant in the HN condition. Higher amounts of precursors of lignin biosynthesis such as quinic acids and derivates, flavonoids, coumaric acid ester and several benzoxazinoids were detected in the HN condition. A two-way ANOVA (N fertilization and genotype, corrected p-value < 0.05, Table S3b) confirmed the trends observed in the PCA presented in Figure S2. In the LN condition, among the 1800 metabolites or metabolic signatures exhibiting changes in their level of accumulation (both according to the genotype and to the level of N fertilization), 658 were significantly up-regulated and 1142 were significantly down-regulated. Higher amounts of starch, sucrose, inositol, malate, trans-aconitate, glutamate, HBOA-glucoside, DIM2BOA-glucoside and three flavonoids were detected in LN, whereas eight free amino acids, trigonelline, choline, HMBOA-glucoside, DIMBOA-glucoside and 13 flavonoids including three caffeoyl-quinates and two N-acyl amines were less abundant. Among the 40 metabolites exhibiting a change in their level of accumulation according to the level of N fertilization yet not affected by genotype, 20 were present in higher amounts (including rhamnose) and 20 in lower amounts (including tyrosine).
The proteome PCA loadings plot (Figure S3) showed that most of the proteins involved in the control of biosynthesis and homeostasis were more abundant in the LN condition. In contrast, proteins involved in the control of cellular respiration were more abundant in the HN condition. The two-way ANOVA confirmed the results of the PCA (corrected p-value < 0.05, Table S5b). Among the 2846 identified proteins, 873 exhibited differences in their level of accumulation, according to both the genotype and the level of N fertilization. The level of accumulation of 140 proteins was modified only according to the N fertilization regime. Among these 140 N-responsive proteins, the two predominant functional categories up-regulated in LN were proteins involved in the control of their biosynthesis and in photosynthesis, and those down-regulated in LN were proteins involved in photosynthesis and in solute transport (Table S5d).
Identification of coregulations between metabolites, proteins, eco-physiological and agronomic traits
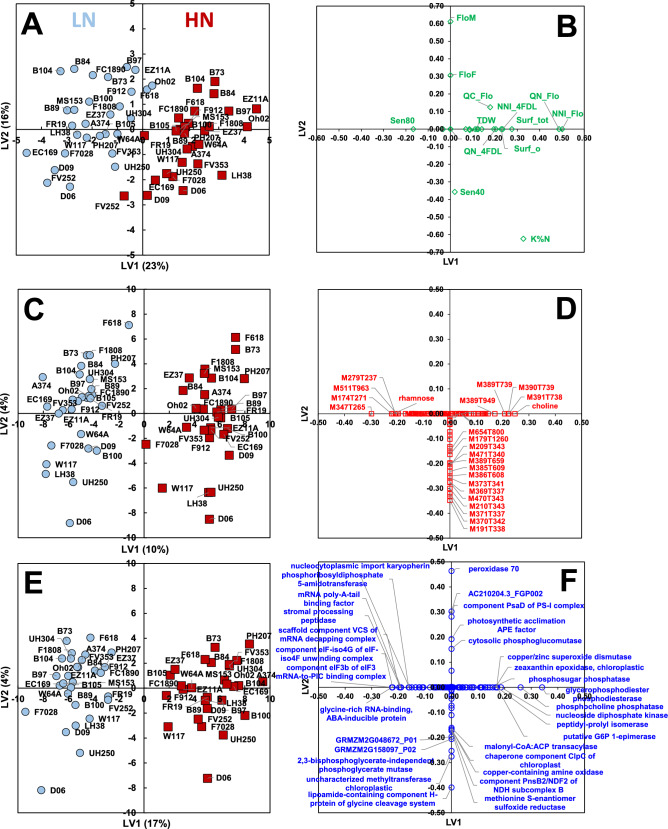
A multiblock sparse Partial-Least-Squares Discriminant Analysis (sPLS-DA) was first performed using their LSmeans values (Tables S2c, S3c and S5c). This analysis helped to select 100 metabolites and proteins that discriminated the two fertilization regimes and covaried across the three sets of traits (Fig. 3A–F). The first latent variable (LV1) separated the two N fertilization regimes as expected (Fig. 3A, C and E). The loading values of the variables selected on LV1 are presented in Table S6. On the positive side of LV1, NNI_Flo, QN_Flo, Surf_tot, NNI_4FDL, Surf_o and QN_4FDL were higher in HN, covaried positively with choline, with four LC–QTOF–MS metabolic signatures (M391T738, M390T739, M389T739, M389T949), and for proteins with a glycerophosphodiester phosphodiesterase, a phosphocholine phosphatase, a nucleoside diphosphate kinase, a putative glucose-6-phosphate 1-epimerase, a peptidyl-prolyl isomerase, a copper/zinc superoxide dismutase, a chloroplastic zeaxanthin epoxidase and a phosphosugar phosphatase. On the negative side of LV1, Sen80 was higher in LN, covaried positively with M347T265, M174T271, rhamnose, M279T237 and M511T963, and with a component eIF-iso4G of eIF-iso4F unwinding complex, a component eIF3b of eIF3 mRNA-to-PIC binding complex and a glycine-rich RNA-binding ABA-inducible protein. A separation of genotypes could be observed along LV2. On the positive side of LV2, FloF and FloM, were delayed in several hybrids, including B97, B84, B104, B73 and F618. These two developmental traits covaried positively with a peroxidase 70, AC210204.3_FGP002 and a component PsaD of PS-I complex. On the negative side of LV2, Sen40 occurred later in hybrids D09, FV252 and D06. This marker of leaf senescence covaried positively with a range of LC–QTOF–MS metabolic signatures, including M191T338, M370T342, M371T337 and M210T343, and for proteins with a lipoamide-containing component H-protein of glycine cleavage system, an uncharacterized chloroplastic methyltransferase and a 2,3-bisphosphoglycerate-independent phosphoglycerate mutase.
Fig. 3.
Multiblock sparse PLS-DA of the LSmeans per condition and hybrid combining the eco-physiological, developmental and yield-related traits with metabolome data and proteome data measured in the panel of 29 maize hybrids grown under reduced (LN, blue circles) and non-limiting (HN, red squares) fertilization conditions. A model with two latent variables (LV) was chosen. A Scores plot for the eco-physiological, developmental and yield-related data block. B Scores plot for the metabolome data block. C Scores plot for the proteome data block. Hybrid codes are indicated on each scores plot. D Loadings plot for the eco-physiological, developmental and yield-related traits. E Loadings plot for the metabolome. F Loadings plot for the proteome. On each loadings plot, variables with a loading value higher than 0.15 on at least one latent variable are annotated. See Table S3 for details on metabolite annotations and Table S6 for the highest loading values on LV1
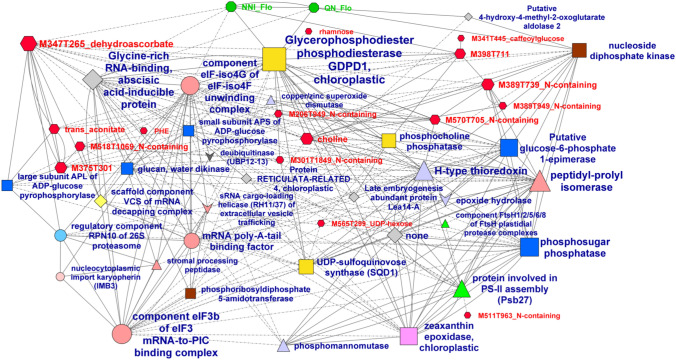
A correlation network between the variables exhibiting an absolute loading value higher than 0.1 on LV1 was then constructed if the Spearman correlation values between the variables were above 0.75 or below − 0.75. Before reconstructing the network, redundancies within the selected LC–QTOF–MS signatures of the metabolite data set were discarded. The selected LC–QTOF–MS variables were clustered manually according to their chromatographic profiles and putative chemical formulas. For each cluster, only the predominant metabolic signatures were selected. A tentative annotation of these metabolite signatures led to the putative annotation of three metabolites: dehydroascorbate, a caffeoyl-glucose and a UDP-hexose (Table S3e). The resulting network presented in Fig. 4 contained two phenotypic traits NNI_Flo and QN_Flo, 16 metabolites and 31 proteins. Metabolite signatures and proteins directly correlated with NNI_Flo and QN_Flo are listed in Table S7. To illustrate the effect of environmental conditions (here the two fertilization regimes) on the correlations between phenotypic traits and metabolites or proteins, the plots of the relationships with NNI_Flo and QN_Flo listed in Table S7 are presented in Figure S4.
Fig. 4.
Correlation network between the variables contributing to separate the two N treatments selected based on the multiblock sparse PLS-DA presented in Fig. 3 and having an absolute loading value higher than 0.1 on LV1. Spearman correlations with an absolute value higher than 0.75 are shown in a network built with Cytoscape. Only the subnetwork comprising variables of the three datasets is shown. Node size is proportional to the number of connections. For edges, a solid line means a positive correlation and a dashed line means a negative correlation. Node symbols are represented by green octagons for eco-physiological, developmental and yield-related traits and red hexagons for metabolites. For proteins, different symbols and colors are used according to Mercator categories
Within this network, five proteins were involved in carbohydrate metabolism, three in lipid metabolism, three in protein biosynthesis and three in redox homeostasis. NNI_Flo was negatively correlated to M347T265_dehydroascorbate, a glycine-rich RNA-binding ABA-inducible protein, and component eIF-iso4G of eIF-iso4F unwinding complex. NNI_Flo was positively correlated to M398T711. NNI_Flo and QN_Flo were both positively correlated to chloroplastic glycerophosphodiester phosphodiesterase GDPD1. QN_Flo was also positively correlated to a nucleoside diphosphate kinase and a 4-hydroxy-4-methyl-2-oxoglutarate aldolase. The three metabolite nodes exhibiting the highest number of connections were M347T265_dehydroascorbate, choline and M389T739 (an unidentified N-containing compound), with at least 12 connections each. The three protein nodes exhibiting the highest number of connections were chloroplastic glycerophosphodiester phosphodiesterase GDPD1, the glycine-rich RNA-binding ABA-inducible protein and a peptidyl-prolyl isomerase, with at least 26 connections each.
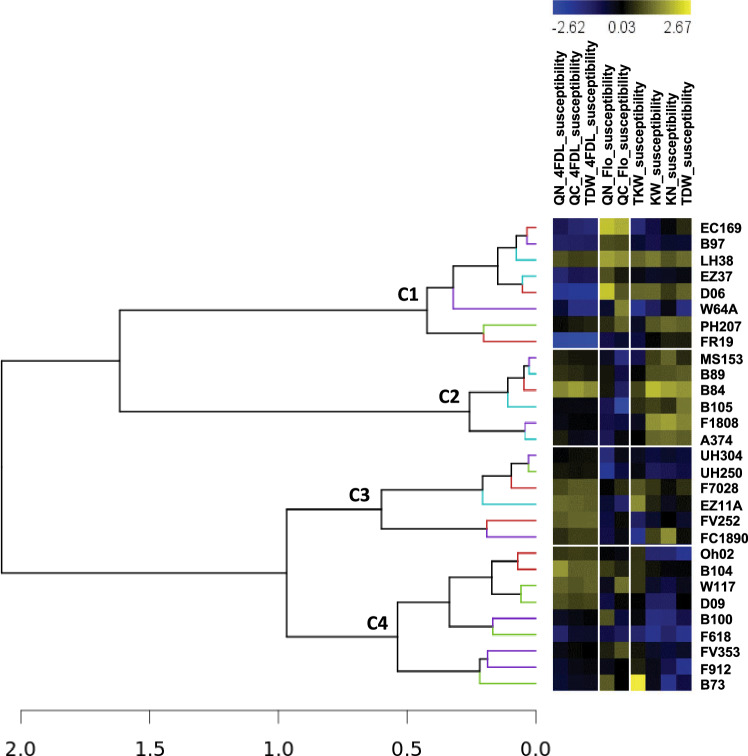
Metabolites and proteins linked to N-susceptibility
When the susceptibility to LN was calculated using QN_4FDL, QC_4FDL, QN_Flo, QC_Flo, TKW, KN, KW, TDW_4FDL and TDW LS-means data, an HCA identified four distinct groups of hybrids (Fig. 5). The first group (cluster C1) was characterized by low susceptibility to reduced N fertilization at the early stage of plant development (4DL), including shoot biomass production (TDW_4FDL). All these hybrids were less susceptible to the LN condition, not only in terms of their C and N contents at flowering but also for all yield-related traits, especially TDW at maturity. In contrast, the fourth group (cluster C4) contained hybrids that were more susceptible to reduced N fertilization at 4DL and less susceptible at maturity, especially regarding the traits related to kernel and total biomass production. The other two clusters C2 and C3 were mostly characterized by low susceptibility in the LN condition, mostly for their C and N contents at flowering. N-susceptibility for all other traits was high, except for the yield-related traits in C3.
Fig. 5.
Hierarchical classification of the 29 maize hybrids based on the susceptibility of key physiological and yield-related traits to reduced N fertilization (LN). The level of susceptibility to LN for each trait was calculated using its LSmean value as follows: [(LS-means (HN)−LS-means (LN)]/LS-means (HN). The different clusters identified for the 29 hybrids were named C1, C2, C3 and C4
A one-way ANOVA was then performed to determine which metabolites and proteins differentiated the hybrids grouped in cluster C4, compared to all other hybrids (Figure S5). For metabolites in this cluster, susceptibility to the LN condition was characterized by an increase in metabolic signatures M519T1858, M385T279 and M468T733 and a decrease in M343T742 (P < 0.01). M519T1858 was tentatively identified as hypatulin B, and M343T742 as a dimethoxybenzoic acid glucoside. All these metabolite signatures had a mean signal-over-noise ratio above 60. Among these metabolite signatures, M468T733 had the highest peak intensity (Table S3). For proteins, susceptibility to the LN condition was characterized by an increase in GRMZM2G018074, a regulatory protein (GCN4) of RIN4, and a decrease in GRMZM5G886257, a cytosolic NADP-dependent malic enzyme. The later was an abundant protein, whereas the former was present in a much lower amount (Table S5).
Discussion
Predictive value of eco-physiological markers for yield and its components
Across the 29 hybrids and irrespective of the N fertilization regime, there was a high positive correlation between KW and KN, as expected (Bertin & Gallais, 2000). Total plant dry matter production at harvest (TDW) was also strongly correlated with kernel yield-related traits, thus indicating that the hybrids producing more vegetative biomass are also the most productive in terms of kernel yield. Consequently, in these hybrids, it was logical to find that the leaf surface and the plant C and N contents at flowering were also correlated with their ability to produce more harvestable material for silage production. This is likely because they absorbed more N and had higher photosynthetic capacity at the same time. In line with these findings, the flowering time (FloM and FloF) was also correlated with TDW, thus indicating that before kernel filling, the most productive hybrids were also able to accumulate more C and N assimilates. In contrast, although correlated together, the various traits measured at the very early stage of plant development (e.g. QN_4FDL, QC_4FDL, TDW_4FDL) were not correlated with yield-related traits. It is therefore unlikely that they can be used as predictors for plant agronomic performance.
Irrespective of the level of N fertilization, a negative relationship was observed between KN and KW. Such a result agrees with the negative relationship previously observed between kernel yield and the amount of proteins present in the kernels (Caballero-Rothar et al., 2019). As previously proposed by Cañas et al., (Cañas et al., 2017), most agronomic traits measured at flowering and at harvest, can be used to identify predictive biochemical traits related to kernel and shoot biomass production.
Correlation studies performed between eco-physiological, developmental and yield-related traits and the activity of several key enzymes involved in primary C and N assimilation revealed that leaf GS activity was positively correlated (> 0.5) with TDW and KW only with reduced N fertilization. Under this N feeding condition, leaf GS activity was 65% higher on average. Although we did not identify which GS isoform was involved at this stage of the study, this result confirms that the enzyme plays a key role in the control of kernel production both in lines (Martin et al., 2006) and in hybrids (Amiour et al., 2021).
N-responsive metabolite and protein markers
As already observed (Amiour et al., 2012; Schlüter et al., 2013), an accumulation of starch and sucrose occurred in plants grown under N-limiting conditions. The accumulation of several organic acids involved in the TCA cycle, such as trans-aconitate and malate, was characteristic in N-deficient conditions (Amiour et al., 2012), as observed in the present study. In addition, rhamnose and glutamate levels were higher. These two molecules are known to be markers of stress conditions (Jiang et al., 2021; Qiu et al., 2020). However, a decrease in glutamate was generally observed in other studies, which could be explained by the fact that only a single genotype was examined (line B73) and that the level of the N-stress was much higher (Amiour et al., 2012; Schlüter et al., 2013). In these two studies, a decrease in the most abundant amino acids was also a characteristic of plant N deficiency. In the present investigation, higher amounts of several other amino acids e.g. methionine and phenylalanine in the HN condition suggest that in silage hybrids they could be markers representative of non-limiting N feeding conditions.
Several other molecules represented mostly by metabolites belonging to the phenylpropanoid or benzoxazinoid families were found to accumulate when N fertilization was not reduced. The accumulation of phenolics in plant tissues is considered as an adaptive response of plants to adverse environmental conditions (Akhi et al., 2021). Under N-limiting conditions, a low chlorogenate content in the leaves alters lignin biosynthesis (Amiour et al., 2012). These specialized metabolites also appear to be important markers that can be used to select maize lines producing larger kernels (Cañas et al., 2017). In the present study, the level of ferruloylhydroxycitrate and caffeoylhydroxycitrate was higher under N-limiting conditions, while under non-limiting N feeding conditions, the level of several quinate derivatives and rutin was higher. In addition to biotic stresses induced by pathogen infection, benzoxazinoid accumulation can also be regulated by abiotic stresses (Niemeyer, 2009). However, the molecular mechanisms underlying the physiological function of these metabolites synthesized only by grasses (Medeiros et al., 2021) needs to be further investigated, as we observed that their N-dependent level of accumulation also seems to be compound-dependent. This, exclude the possibility of using them as markers for NUE.
The classes of proteins involved in the control of their biosynthesis were relatively more abundant in the LN condition. This result is surprising at first, as their amount was found to increase (mostly ribosomal proteins) when N was provided to the plant (Prinsi & Espen, 2018). However, protein synthesis is a crucial multi-faceted process in plant adaptation to N availability (Prinsi & Espen, 2018). In maize hybrids subjected to a mild N deficit, a reprogramming of protein synthesis may thus occur, irrespective of the genetic background. Such reprogramming has been observed in Arabidopsis upon N starvation, leading to an increased amount of proteins involved in the translation machinery and amino acid degradation (Zhu et al., 2018). In HN, most other classes of proteins involved in amino acid biosynthesis, lipid metabolism and nutrient uptake were more abundant. This is in line with previously reported results in maize (Simons et al., 2014a) and other crop species (Zhu et al., 2018). Proteins involved in respiration were down-regulated in the LN condition, fitting with the hypothesis that mitochondrial metabolism is tightly linked to NUE and its improvement (Foyer et al., 2011).
Choline was identified among the various metabolites covarying positively with most of the agronomic traits and present in higher amounts in HN only. Choline is known to protect plants against oxidative damage and it is also a marker when the phosphorus (P) supply is optimal (Salinas et al., 2013). Our results suggest that choline could also be a marker of non-limiting N fertilization. The possibility of such a relationship between P and N metabolisms is strengthened by the finding that among the proteins covarying positively with the increase in most agronomic traits in HN, a number of them (glycerophosphodiester phosphodiesterase, glucose-6-phosphate-1-epimerase, phosphosugar phosphatase, phosphocholine phosphatase) are known to be involved in optimizing P use efficiency (Nakamura, 2021; Wang & Lambers, 2020). The other identified proteins (nucleoside diphosphate kinase, peptidyl-prolyl isomerase, copper/zinc superoxide dismutase, chloroplastic zeaxanthin epoxidase) were related to a general stress response of the plant (Eskling et al., 2001; Li et al., 2019), thus suggesting that these proteins are up-regulated in maize hybrids under non-limiting N fertilization. Although the exact nature of the four LC–QTOF–MS metabolic signatures covarying positively with the agronomic traits remains unknown, we were able to determine that they are all N-containing compounds. They could represent predictive markers of the agronomic performance of the plant.
When N fertilization was reduced, rhamnose was the only identified metabolic marker covarying with only one phenotypic trait related to leaf senescence at a late stage of plant development (Sen80). This is in line with the finding that an increase in several carbohydrates originating from cell wall degradation occurs during leaf aging (Quirino et al., 2001) and when N fertilization is reduced (Amiour et al., 2012; Bassi et al., 2018). It may therefore be suggested that rhamnose is a possible marker for N deficiency in maize hybrids, at least from the flowering period.
Among the proteins covarying with leaf senescence, a component eIF3b of eIF3 mRNA-to-PIC binding complex, a component eIF-iso4G of eIF-iso4F unwinding complex, scaffold component VCS of mRNA decapping complex and a stromal processing peptidase were identified. One may hypothesize that three of these proteins are involved in the regulation of translation, probably involving a decay in mRNA during certain phases of plant development (Belostotsky & Sieburth, 2009), including leaf protein degradation. The role of the stroma processing peptidase is more difficult to interpret in the context of leaf senescence as it is involved in the import of proteins into the chloroplast (Day & Theg, 2018). However, such peptidase could be involved in chloroplast remodeling during certain phases of leaf senescence during which chloroplast degradation occurs (Nishimura et al., 2017).
The susceptibility of the hybrids to reduced N fertilization helped to propose a maize ideotype mainly characterized by a high GS activity, an accumulation of amino acids and a low content in phenolics. At least the latter first two markers could be used for monitoring N fertilization and for the selection of productive hybrids. In the group of hybrids that were less susceptible to N deficiency with respect to yield, the most interesting result was to find a cytosolic NADP-dependent malic enzyme and a GCN4 regulatory protein, which are both known to be involved in a variety of biotic and abiotic stresses (Chen et al., 2019; Toruño et al., 2019). Both proteins thus appear to be putative markers for the selection of the most productive silage hybrids under mild N stress. Although the use of protein markers to pilot N fertilization remains to be developed, their use for breeding purposes appears to be feasible if we consider that fast and efficient throughput techniques have been recently developed for proteomics in medical research (Ivanov et al., 2020; Messner et al., 2020). As the two proteins were also detected in maize plants grown in a controlled-environment chamber (Balliau & Zivy, 2020; Urrutia et al., 2021), it seems therefore possible to test genotypes under various growth conditions.
Four metabolite signatures were also observed in this group of hybrids, two of which were tentatively identified. Among these four metabolite signatures, only M519T1858 was also quantified in the controlled-environment experiment using an untargeted approach (Urrutia et al., 2020, 2021). Thus, a targeted LC–HRMS or LC–MS/MS strategy should be prefered to screen a larger quantity of genotypes for these markers under various environmental conditions. Better agronomic performance in LN seems to be linked to higher susceptibility of hypatulin terpene, and lower susceptibility of dimethoxybenzoic acid glucoside. The role of these two specialized metabolites in maize needs to be further assessed, although as natural products present in certain plant species (Tanaka & Kashiwada, 2021) and in cereals (Van Hung, 2016), they have an antioxidant function in human nutrition.
Conclusions
Predictive leaf metabolic and proteic markers that could be used to pilot and rationalize N fertilization at the very early stage of plant development to match the plant demand were identified, e.g. higher amounts in LN of starch, sucrose, inositol, malate, trans-aconitate, glutamate, HBOA-glucoside, DIM2BOA-glucoside and three flavonoids and of most of the proteins involved in the control of their biosynthesis and homeostasis. Susceptibility to the LN condition was characterized by an increase in several LC–MS based metabolic signatures including a tentatively identified hypatulin B and a dimethoxybenzoic acid glucoside and in a regulatory protein of RIN4, and by a decrease in a cytosolic NADP-dependent malic enzyme. These markers could be used to select high-yielding commercial maize hybrids for silage production requiring less N fertilizer inputs.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file1 (DOCX 32 KB)—Detailed methods for plant phenotyping and biochemical and data analyses are presented in Annexes1 to 5.
Acknowledgements
We thank the GCIE team for plant culture and sample harvest, the Saint-Martin de Hinx INRAE Experimental Unit for providing the seeds, and Emilie Cooke-Martageix for copyediting the manuscript. The IJPB benefited from the support of Saclay Plant Sciences-SPS. Proteomics analyses were performed on the PAPPSO platform (http://pappso.inra.fr), which is funded by INRAE (http://www.inrae.fr), the Ile-de-France regional council (https://www.iledefrance.fr/education-recherche), IBiSA (https://www.ibisa.net) and Saclay Plant Sciences-SPS (ANR-17-EUR-0007).
Abbreviations
- ANOVA
Analysis of variance
- 1H-NMR
Proton nuclear magnetic resonance spectroscopy
- GS
Glutamine synthetase
- LC–QTOF–MS
Liquid chromatography quadrupole time-of-flight mass spectrometry
- HCA
Hierarchical clustering analysis
- HN
Non-limiting fertilization treatment
- LN
Reduced N fertilization treatment
- LSmeans
Least square means
- LV
Latent variable
- PLS-DA
Partial least-squares discriminant analysis
- PC
Principal component
- PCA
Principal component analysis
- UHPLC
Ultra-high-performance liquid chromatography
Author contributions
The author contributions were as follows: CG, BH, AM and MZ designed the study. DR coordinated phenotyping and sample preparation. MU, OF, SB, MM, CD, CB and IQ performed the metabolite analyses and other biochemical or chemical analyses. MZ, MBN and TB performed the proteome analyses. SP reannotated the proteins. AM, MZ and DJ analysed the data. AM, BH and MZ wrote the initial version of the manuscript. MU and YG edited the manuscript. BH and AM contributed equally to this work. All authors read and approved the final manuscript.
Funding
We thank the French Research National Agency (ANR), MetaboHUB (ANR-11-INBS-0010), PHENOME (ANR-11-INBS-0012) and AMAIZING (ANR-10-BTBR-01) projects and FranceAgrimer for financing.
Data availability
The proteomics data were deposited in the ProteomeXchange Consortium via the PRIDE partner repository (PXD034145 identifier). The NMR and LC–QTOF–MS data were deposited in the recherche.data.gouv.fr repository (10.57745/NZNMZH and 10.57745/UPAO0X).
Declarations
Conflict of interest
The authors have no competing interests to declare that are relevant to the content of this article.
Ethical approval
Our experimental studies on plants compiled with relevant institutional, national and international guidelines and registration.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Adegbeye, M. J., Ravi Kanth Reddy, P., Obaisi, A. I., Elghandour, M. M. M. Y., Oyebamiji, K. J., Salem, A. Z. M., Morakinyo-Fasipe, O. T., Cipriano-Salazar, M., & Camacho-Díaz, L. M. (2020). Sustainable agriculture options for production, greenhouse gasses and pollution alleviation, and nutrient recycling in emerging and transitional nations—An overview. Journal of Cleaner Production,242, 118319. 10.1016/j.jclepro.2019.118319 [Google Scholar]
- Akhi, M. Z., Haque, M. M., & Biswas, M. S. (2021). Role of secondary metabolites to attenuate stress damages in plants. In W. Viduranga (Ed.), Antioxidants (p. 27). IntechOpen. [Google Scholar]
- Amiour, N., Décousset, L., Rouster, J., Quenard, N., Buet, C., Dubreuil, P., Quilleré, I., Brulé, L., Cukier, C., Dinant, S., Sallaud, C., Dubois, F., Limami, A. M., Lea, P. J., & Hirel, B. (2021). Impacts of environmental conditions, and allelic variation of cytosolic glutamine synthetase on maize hybrid kernel production. Communications Biology,4(1), 1095. 10.1038/s42003-021-02598-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiour, N., Imbaud, S., Clément, G., Agier, N., Zivy, M., Valot, B., Balliau, T., Armengaud, P., Quilleré, I., Cañas, R., Tercet-Laforgue, T., & Hirel, B. (2012). The use of metabolomics integrated with transcriptomic and proteomic studies for identifying key steps involved in the control of nitrogen metabolism in crops such as maize. Journal of Experimental Botany,63(14), 5017–5033. 10.1093/jxb/ers186 [DOI] [PubMed] [Google Scholar]
- Balliau, T., Blein-Nicolas, M., & Zivy, M. (2018). Evaluation of optimized tube-gel methods of sample preparation for large-scale plant proteomics. Proteomes,6(1), 6. 10.3390/proteomes6010006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balliau, T., & Zivy, M. (2020). Genetic variation of maize responses to low temperatures in controlled conditions, PRIDE, Retrieved October 1, 2024, from https://www.ebi.ac.uk/pride/archive/projects/PXD021790
- Bassi, D., Menossi, M., & Mattiello, L. (2018). Nitrogen supply influences photosynthesis establishment along the sugarcane leaf. Scientific Reports,8(1), 2327. 10.1038/s41598-018-20653-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatty, P. H., & Good, A. G. (2018). Improving nitrogen use efficient in crop plants using biotechnology approaches. In A. Shrawat, A. Zayed, & D. A. Lightfoot (Eds.), Engineering nitrogen utilization in crop plants (pp. 15–35). Springer. [Google Scholar]
- Bednarz, B., Millan-Oropeza, A., Kotowska, M., Świat, M., Quispe Haro, J. J., Henry, C., & Pawlik, K. (2021). Coelimycin synthesis activatory proteins are key regulators of specialized metabolism and precursor flux in Streptomyces coelicolor A3(2). Frontiers in Microbiology. 10.3389/fmicb.2021.616050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belostotsky, D. A., & Sieburth, L. E. (2009). Kill the messenger: mRNA decay and plant development. Current Opinion in Plant Biology,12(1), 96–102. 10.1016/j.pbi.2008.09.003 [DOI] [PubMed] [Google Scholar]
- Bénard, C., & Gibon, Y. (2016). Measurement of enzyme activities and optimization of continuous and discontinuous assays. Current Protocols in Plant Biology,1(2), 247–262. 10.1002/cppb.20003 [DOI] [PubMed] [Google Scholar]
- Bertin, P., & Gallais, A. (2000). Physiological and genetic basis of nitrogen use efficiency in maize. I. Agrophysiological results. Maydica,45, 53–66. [Google Scholar]
- Biais, B., Allwood, J. W., Deborde, C., Xu, Y., Maucourt, M., Beauvoit, B., Dunn, W. B., Jacob, D., Goodacre, R., Rolin, D., & Moing, A. (2009). 1H NMR, GC−EI-TOFMS, and data set correlation for fruit metabolomics: Application to spatial metabolite analysis in melon. Analytical Chemistry,81(8), 2884–2894. 10.1021/ac9001996 [DOI] [PubMed] [Google Scholar]
- Blein-Nicolas, M., Negro, S. S., Balliau, T., Welcker, C., Cabrera-Bosquet, L., Nicolas, S. D., Charcosset, A., & Zivy, M. (2020). A systems genetics approach reveals environment-dependent associations between SNPs, protein coexpression, and drought-related traits in maize. Genome Research,30(11), 1593–1604. 10.1101/gr.255224.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero-Rothar, N. N., Abdala, L. J., Borrás, L., & Gerde, J. A. (2019). Role of yield genetic progress on the biochemical determinants of maize kernel hardness. Journal of Cereal Science,87, 301–310. 10.1016/j.jcs.2019.04.019 [Google Scholar]
- Cañas, R. A., Yesbergenova-Cuny, Z., Simons, M., Chardon, F., Armengaud, P., Quilleré, I., Cukier, C., Gibon, Y., Limami, A. M., Nicolas, S., Brulé, L., Lea, P. J., Maranas, C. D., & Hirel, B. (2017). Exploiting the genetic diversity of maize using a combined metabolomic, enzyme activity profiling, and metabolic modeling approach to link leaf physiology to kernel yield. The Plant Cell,29(5), 919–943. 10.1105/tpc.16.00613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carena, M. J. (2021). Germplasm enhancement and cultivar development: The need for sustainable breeding. Crop Breeding and Applied Biotechnology. 2, e385621S4. 10.1590/1984-70332021v21Sa17 [Google Scholar]
- Chen, Q., Wang, B., Ding, H., Zhang, J., & Li, S. (2019). Review: The role of NADP-malic enzyme in plants under stress. Plant Science,281, 206–212. 10.1016/j.plantsci.2019.01.010 [DOI] [PubMed] [Google Scholar]
- Chowdhury, N. B., Schroeder, W. L., Sarkar, D., Amiour, N., Quillere, I., Hirel, B., Maranas, C. D., & Saha, R. (2022). Dissecting the metabolic reprogramming of maize root under nitrogen-deficient stress conditions. Journal of Experimental Botany,73(1), 275–291. 10.1093/jxb/erab435 [DOI] [PubMed] [Google Scholar]
- Day, P. M., & Theg, S. M. (2018). Evolution of protein transport to the chloroplast envelope membranes. Photosynthesis Research,138(3), 315–326. 10.1007/s11120-018-0540-x [DOI] [PubMed] [Google Scholar]
- Dellagi, A., Quillere, I., & Hirel, B. (2020). Beneficial soil-borne bacteria and fungi: A promising way to improve plant nitrogen acquisition. Journal of Experimental Botany,71(15), 4469–4479. 10.1093/jxb/eraa112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskling, M., Emanuelsson, A., & Åkerlund, H.-E. (2001). Enzymes and mechanisms for violaxanthin-zeaxanthin conversion. In E.-M. Aro & B. Andersson (Eds.), Regulation of photosynthesis (pp. 433–452). Springer. [Google Scholar]
- Fortunato, S., Nigro, D., Lasorella, C., Marcotuli, I., Gadaleta, A., & de Pinto, M. C. (2023). The Role of glutamine synthetase (GS) and glutamate synthase (GOGAT) in the improvement of nitrogen use efficiency in cereals. Biomolecules,13(12), 1771. 10.3390/biom13121771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer, C. H., Noctor, G., & Hodges, M. (2011). Respiration and nitrogen assimilation: Targeting mitochondria-associated metabolism as a means to enhance nitrogen use efficiency. Journal of Experimental Botany,62(4), 1467–1482. 10.1093/jxb/erq453 [DOI] [PubMed] [Google Scholar]
- Ganal, M. W., Durstewitz, G., Polley, A., Bérard, A., Buckler, E. S., Charcosset, A., Clarke, J. D., Graner, E.-M., Hansen, M., Joets, J., Le Paslier, M.-C., McMullen, M. D., Montalent, P., Rose, M., Schön, C.-C., Sun, Q., Walter, H., Martin, O. C., & Falque, M. (2011). A large maize (Zea mays L.) SNP genotyping array: Development and germplasm genotyping, and genetic mapping to compare with the B73 reference genome. PLoS ONE,6(12), e28334. 10.1371/journal.pone.0028334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon, Y., Blaesing, O. E., Hannemann, J., Carillo, P., Höhne, M., Hendriks, J. H. M., Palacios, N., Cross, J., Selbig, J., & Stitt, M. (2004). A robot-based platform to measure multiple enzyme activities in Arabidopsis using a set of cycling assays: Comparison of changes of enzyme activities and transcript levels during diurnal cycles and in prolonged darkness. The Plant Cell,16(12), 3304–3325. 10.1105/tpc.104.025973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano, M., Petropoulos, S. A., & Rouphael, Y. (2021). The fate of nitrogen from soil to plants: Influence of agricultural practices in modern agriculture. Agriculture,11(10), 944. 10.3390/agriculture11100944 [Google Scholar]
- Henchion, M., Hayes, M., Mullen, A. M., Fenelon, M., & Tiwari, B. (2017). Future protein supply and demand: Strategies and factors influencing a sustainable equilibrium. Foods,6(7), 53. 10.3390/foods6070053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuzé, V., Tran, G., Edouard, N., & Lebas, F. (2017). Maize silage. Feedipedia, a programme by INRAE, CIRAD, AFZ and FAO. (14), 24
- Hirel, B., & Krapp, A. (2020). Nitrogen utilization in plants I biological and agronomic importance. Encyclopedia of Biochemistry. 3rd Edition (Reference Module in Life Sciences): Elsevier
- Hirel, B., Tétu, T., Lea, P. J., & Dubois, F. (2011). Improving nitrogen use efficiency in crops for sustainable agriculture. Sustainability,3(9), 1452–1485. 10.3390/su3091452 [Google Scholar]
- Ivanov, M. V., Bubis, J. A., Gorshkov, V., Tarasova, I. A., Levitsky, L. I., Lobas, A. A., Solovyeva, E. M., Pridatchenko, M. L., Kjeldsen, F., & Gorshkov, M. V. (2020). DirectMS1: MS/MS-free identification of 1000 proteins of cellular proteomes in 5 minutes. Analytical Chemistry,92(6), 4326–4333. 10.1021/acs.analchem.9b05095 [DOI] [PubMed] [Google Scholar]
- Jiang, N., Dillon, F. M., Silva, A., Gomez-Cano, L., & Grotewold, E. (2021). Rhamnose in plants - from biosynthesis to diverse functions. Plant Science,302, 110687. 10.1016/j.plantsci.2020.110687 [DOI] [PubMed] [Google Scholar]
- Kumar, S. A., & Ilango, P. (2018). The impact of wireless sensor network in the field of precision agriculture: A review. Wireless Personal Communications,98(1), 685–698. 10.1007/s11277-017-4890-z [Google Scholar]
- Lamari, N., Zhendre, V., Urrutia, M., Bernillon, S., Maucourt, M., Deborde, C., Prodhomme, D., Jacob, D., Ballias, P., Rolin, D., Sellier, H., Rabier, D., Gibon, Y., Giauffret, C., & Moing, A. (2018). Metabotyping of 30 maize hybrids under early-sowing conditions reveals potential marker-metabolites for breeding. Metabolomics,14(10), 132. 10.1007/s11306-018-1427-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., Cui, J., Zhao, Q., Yang, Y., Wei, L., Yang, M., Liang, F., Ding, S., & Wang, T. (2019). Physiology and proteomics of two maize genotypes with different drought resistance. Biologia Plantarum,63, 519–528. 10.32615/bp.2019.085 [Google Scholar]
- Martin, A., Lee, J., Kichey, T., Gerentes, D., Zivy, M., Tatout, C., Du-bois, F., Balliau, T., Valot, B., Davanture, M., Tercé-Laforgue, T., Quilleré, I., Coque, M., Gallais, A., Gonzalez-Moro, M.-B., Bethencourt, L., Habash, D. Z., Lea, P. J., Charcosset, A., … Hirel, B. (2006). Two cytosolic glutamine synthetase isoforms of maize are specifically involved in the control of grain production. The Plant Cell,18(11), 3252–3274. 10.1105/tpc.106.042689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros, D. B., Brotman, Y., & Fernie, A. R. (2021). The utility of metabolomics as a tool to inform maize biology. Plant Communications,2(4), 100187. 10.1016/j.xplc.2021.100187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messner, C. B., Demichev, V., Wendisch, D., Michalick, L., White, M., Freiwald, A., Textoris-Taube, K., Vernardis, S. I., Egger, A.-S., Kreidl, M., Ludwig, D., Kilian, C., Agostini, F., Zelezniak, A., Thibeault, C., Pfeiffer, M., Hippenstiel, S., Hocke, A., von Kalle, C., … Ralser, M. (2020). Ultra-high-throughput clinical proteomics reveals classifiers of COVID-19 infection. Cell Systems,11(1), 11-24.e14. 10.1016/j.cels.2020.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, Y. (2021). Headgroup biosynthesis of phosphatidylcholine and phosphatidylethanolamine in seed plants. Progress in Lipid Research,82, 101091. 10.1016/j.plipres.2021.101091 [DOI] [PubMed] [Google Scholar]
- Nicolas, S., Negro, S., Madur, D., Clipet, C., Combes, V., Bauland, C., Tardieu, F., Charcosset, A., & Moreau, L. (2020). Amaizing Dent Panel Genotyping Dataset (354 Public Lines), recherche.data.gouv, Retrieved September 27, 2024, from 10.15454/GAHEU0
- Niemeyer, H. M. (2009). Hydroxamic acids derived from 2-hydroxy-2H-1,4-benzoxazin-3(4H)-one: Key defense chemicals of cereals. Journal of Agricultural and Food Chemistry,57(5), 1677–1696. 10.1021/jf8034034 [DOI] [PubMed] [Google Scholar]
- Nishimura, K., Kato, Y., & Sakamoto, W. (2017). Essentials of proteolytic machineries in chloroplasts. Molecular Plant,10(1), 4–19. 10.1016/j.molp.2016.08.005 [DOI] [PubMed] [Google Scholar]
- O’Neal, D., & Joy, K. W. (1973). Glutamine synthetase of pea leaves. I. Purification, stabilization, and pH optima. Archives of Biochemistry and Biophysics,159(1), 113–122. 10.1016/0003-9861(73)90435-9 [DOI] [PubMed] [Google Scholar]
- Parent, B., Turc, O., Gibon, Y., Stitt, M., & Tardieu, F. (2010). Modelling temperature-compensated physiological rates, based on the co-ordination of responses to temperature of developmental processes. Journal of Experimental Botany,61(8), 2057–2069. 10.1093/jxb/erq003 [DOI] [PubMed] [Google Scholar]
- Perez-Riverol, Y., Bai, J., Bandla, C., García-Seisdedos, D., Hewapathirana, S., Kamatchinathan, S., Kundu, D. J., Prakash, A., Frericks-Zipper, A., Eisenacher, M., Walzer, M., Wang, S., Brazma, A., & Vizcaíno, J. A. (2021). The PRIDE database resources in 2022: A hub for mass spectrometry-based proteomics evidences. Nucleic Acids Research,50(D1), D543–D552. 10.1093/nar/gkab1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccini, C., Di Bene, C., Farina, R., Pennelli, B., & Napoli, R. (2016). Assessing nitrogen use efficiency and nitrogen loss in a forage-based system using a modeling approach. Agronomy,6(2), 23. 10.3390/agronomy6020023 [Google Scholar]
- Porter, S. S., Bantay, R., Friel, C. A., Garoutte, A., Gdanetz, K., Ibarreta, K., Moore, B. M., Shetty, P., Siler, E., & Friesen, M. L. (2020). Beneficial microbes ameliorate abiotic and biotic sources of stress on plants. Functional Ecology,34(10), 2075–2086. 10.1111/1365-2435.13499 [Google Scholar]
- Portwood, J. L., II., Woodhouse, M. R., Cannon, E. K., Gardiner, J. M., Harper, L. C., Schaeffer, M. L., Walsh, J. R., Sen, T. Z., Cho, K. T., Schott, D. A., Braun, B. L., Dietze, M., Dunfee, B., Elsik, C. G., Manchanda, N., Coe, E., Sachs, M., Stinard, P., Tolbert, J., … Andorf, C. M. (2018). MaizeGDB 2018: The maize multi-genome genetics and genomics database. Nucleic Acids Research,47(D1), D1146–D1154. 10.1093/nar/gky1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinsi, B., & Espen, L. (2018). Time-course of metabolic and proteomic responses to different nitrate/ammonium availabilities in roots and leaves of maize. International Journal of Molecular Sciences,19(8), 2202. 10.3390/ijms19082202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, X.-M., Sun, Y.-Y., Ye, X.-Y., & Li, Z.-G. (2020). Signaling role of glutamate in plants. Frontiers in Plant Science. 10.3389/fpls.2019.01743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirino, B. F., Reiter, W.-D., & Amasino, R. D. (2001). One of two tandem Arabidopsis genes homologous to monosaccharide transporters is senescence-associated. Plant Molecular Biology,46(4), 447–457. 10.1023/A:1010639015959 [DOI] [PubMed] [Google Scholar]
- R Core Team (2022). R: A language and environment for statistical computing. R Foundation for Statistical Computing, from https://www.R-project.org
- Ranum, P., Peña-Rosas, J. P., & Garcia-Casal, M. N. (2014). Global maize production, utilization, and consumption. Annals of the New York Academy of Sciences,1312(1), 105–112. 10.1111/nyas.12396 [DOI] [PubMed] [Google Scholar]
- Rincent, R., Nicolas, S., Bouchet, S., Altmann, T., Brunel, D., Revilla, P., Malvar, R. A., Moreno-Gonzalez, J., Campo, L., Melchinger, A. E., Schipprack, W., Bauer, E., Schoen, C. C., Meyer, N., Ouzunova, M., Dubreuil, P., Giauffret, C., Madur, D., Combes, V., … Charcosset, A. (2014). Dent and flint maize diversity panels reveal important genetic potential for increasing biomass production. Theoretical and Applied Genetics,127(11), 2313–2331. 10.1007/s00122-014-2379-7 [DOI] [PubMed] [Google Scholar]
- Sahu, B., Chatterjee, S., Mukherjee, S., & Sharma, C. (2019). Tools of precision agriculture: A review. International Journal of Chemical Studies,7(6), 2692–2697. [Google Scholar]
- Salinas, R., Sánchez, E., Ruíz, J. M., Lao, M. T., & Romero, L. (2013). Proline, betaine, and choline responses to different phosphorus levels in green bean. Communications in Soil Science and Plant Analysis,44(1–4), 465–472. 10.1080/00103624.2013.744146 [Google Scholar]
- Schlüter, U., Colmsee, C., Scholz, U., Bräutigam, A., Weber, A. P. M., Zellerhoff, N., Bucher, M., Fahnenstich, H., & Sonnewald, U. (2013). Adaptation of maize source leaf metabolism to stress related disturbances in carbon, nitrogen and phosphorus balance. BMC Genomics,14(1), 442. 10.1186/1471-2164-14-442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons, M., Saha, R., Amiour, N., Kumar, A., Guillard, L., Clément, G., Miquel, M., Li, Z., Mouille, G., Lea, P. J., Hirel, B., & Maranas, C. D. (2014a). Assessing the metabolic impact of nitrogen availability using a compartmentalized maize leaf genome-scale model. Plant Physiology,166(3), 1659–1674. 10.1104/pp.114.245787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons, M., Saha, R., Guillard, L., Clément, G., Armengaud, P., Cañas, R., Maranas, C. D., Lea, P. J., & Hirel, B. (2014b). Nitrogen-use efficiency in maize (Zea mays L.): from ‘omics’ studies to metabolic modelling. Journal of Experimental Botany,65(19), 5657–5671. 10.1093/jxb/eru227 [DOI] [PubMed] [Google Scholar]
- Sinha, V. B., Jangam, A. P., & Raghuram, N., et al. (2020). Biological determinants of crop nitrogen use efficiency and biotechnological avenues for improvement. In M. A. Sutton, K. E. Mason, A. Bleeker, W. K. Hicks, C. Masso, & N. Raghuram (Eds.), Just enough nitrogen: Perspectives on how to get there for regions with too much and too little nitrogen (pp. 157–171). Springer. [Google Scholar]
- Sutton, M. A., Mason, K. E., Bleeker, A., Hicks, W. K., Masso, C., Raghuram, N., Reis, S., & Bekunda, M., et al. (2020). Just enough nitrogen: Summary and synthesis of outcomes. In M. A. Sutton, K. E. Mason, A. Bleeker, W. K. Hicks, C. Masso, & N. Raghuram (Eds.), Just enough nitrogen: Perspectives on how to get there for regions with too much and too little nitrogen (pp. 1–25). Springer. [Google Scholar]
- Tanaka, N., & Kashiwada, Y. (2021). Characteristic metabolites of Hypericum plants: Their chemical structures and biological activities. Journal of Natural Medicines,75(3), 423–433. 10.1007/s11418-021-01489-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toruño, T. Y., Shen, M., Coaker, G., & Mackey, D. (2019). Regulated disorder: Posttranslational modifications control the RIN4 plant immune signaling hub. Molecular Plant-Microbe Interactions,32(1), 56–64. 10.1094/MPMI-07-18-0212-FI [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urrutia, M., Bernillon, S., & Moing, A. (2020). LC–MS metabolomic analyses of maize young leaf cultivated in a growth chamber, recherche.data.gouv, Retrieved September, 30, 2024 from 10.15454/J9KO72
- Urrutia, M., Blein-Nicolas, M., Prigent, S., Bernillon, S., Deborde, C., Balliau, T., Maucourt, M., Jacob, D., Ballias, P., Bénard, C., Sellier, H., Gibon, Y., Giauffret, C., Zivy, M., & Moing, A. (2021). Maize metabolome and proteome responses to controlled cold stress partly mimic early-sowing effects in the field and differ from those of Arabidopsis. Plant, Cell & Environment,44(5), 1504–1521. 10.1111/pce.13993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hung, P. (2016). Phenolic compounds of cereals and their antioxidant capacity. Critical Reviews in Food Science and Nutrition,56(1), 25–35. 10.1080/10408398.2012.708909 [DOI] [PubMed] [Google Scholar]
- Voss-Fels, K. P., Stahl, A., & Hickey, L. T. (2019). Q&A: Modern crop breeding for future food security. BMC Biology,17(1), 18. 10.1186/s12915-019-0638-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., & Lambers, H. (2020). Root-released organic anions in response to low phosphorus availability: Recent progress, challenges and future perspectives. Plant and Soil,447(1), 135–156. 10.1007/s11104-019-03972-8 [Google Scholar]
- Zhang, N., Gibon, Y., Wallace, J. G., Lepak, N., Li, P., Dedow, L., Chen, C., So, Y.-S., Kremling, K., Bradbury, P. J., Brutnell, T., Stitt, M., & Buckler, E. S. (2015). Genome-wide association of carbon and nitrogen metabolism in the maize nested association mapping population. Plant Physiology,168(2), 575–583. 10.1104/pp.15.00025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, F.-Y., Chen, M.-X., Chan, W.-L., Yang, F., Tian, Y., Song, T., Xie, L.-J., Zhou, Y., Xiao, S., Zhang, J., & Lo, C. (2018). SWATH-MS quantitative proteomic investigation of nitrogen starvation in Arabidopsis reveals new aspects of plant nitrogen stress responses. Journal of Proteomics,187, 161–170. 10.1016/j.jprot.2018.07.014 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file1 (DOCX 32 KB)—Detailed methods for plant phenotyping and biochemical and data analyses are presented in Annexes1 to 5.
Data Availability Statement
The proteomics data were deposited in the ProteomeXchange Consortium via the PRIDE partner repository (PXD034145 identifier). The NMR and LC–QTOF–MS data were deposited in the recherche.data.gouv.fr repository (10.57745/NZNMZH and 10.57745/UPAO0X).