Abstract
Plant pathogens secrete nuclear effectors into the host nuclei to modulate the host immune system. Although several nuclear effectors of fungal pathogens have been recently reported, the molecular mechanism of NLS-associated transport vehicles of nuclear effectors and the roles of NLS in transcriptional reprogramming of host immunity genes remain enigmatic. We previously reported the MoHTR1, a nuclear effector of the rice blast fungus, Magnaporthe oryzae. MoHTR1 is translocated to rice nuclei but not in fungal nuclei. Here, we identify the core NLS (RxKK) responsible for MoHTR1’s nuclear localization. MoHTR1 is translocated in the host nucleus through interaction with rice importin α. MoHTR1 NLS empowers it to translocate the cytoplasmic effectors of M. oryzae into rice nuclei. Furthermore, other nuclear effector candidates of the blast pathogen and rice proteins which have RxKK also exhibit nuclear localization, highlighting the crucial role of RxKK in this process. We also unveil the importance of SUMOylation in the stability of MoHTR1 and translocation of MoHTR1 to host nuclei. Moreover, MoHTR1 NLS is essential for the pathogenicity of M. oryzae by reprogramming immunity-associated genes in the host. Our findings provide insights into the significance of plant-specific NLS on fungal nuclear effectors and its role in plant-pathogen interactions.
Subject terms: Protein translocation, Pathogens, Molecular biology
Nuclear effectors of plant pathogens modulate the host immunity. Here, Lim et al. unveiled the core sequence and mechanism for nuclear localization of the rice blast fungal effector, MoHTR1, revealing its role in regulating the host immune system.
Introduction
Effector of plant pathogens is a virulence determinant factor, which is secreted into the host and modulates the host environment to facilitate the infection of pathogens and successful colonization1. Depending on the effector’s secretion and functional localization, they can be categorized into three types; apoplastic, cytoplasmic, and nuclear effectors2. Apoplastic effectors interfere with Pathogen-associated molecular pattern (PAMP)-triggered immunity by recognizing plant immune receptors in the space between the pathogen and the host cell3. Cytoplasmic effectors and nuclear effectors are secreted into the host cell and localized in the cytoplasm or migrate to the host nucleus, and are involved in the manipulation of diverse host processes, including defense signaling, vesicle trafficking, reactive oxygen species (ROS) production, and signaling regulation4,5.
Nuclear effectors target host DNA and proteins to regulate the transcriptional reprogramming of immunity-associated genes, such as ROS and defense-related genes, hormone signaling, post-translational modification (PTM), and cytoplasmic mRNA transcription6–11. In order to migrate to the nucleus and regulate various nuclear processes of the host, nuclear effectors should traverse the nuclear envelope through nuclear pore complexes (NPCs)12. Small molecules (<40 kDa) are passively transported through NPC, while larger molecules require sorting factors to import through the NPC13. Nuclear localization sequence (NLS) is the most well-known sorting factor for nuclear transportation. It is classified into classical and non-classical NLS based on the abundance of basic residues, such as lysine and arginine, and the participation of importin α in the migration process12. The PTMs are essential biological processes that regulate the nuclear transport of various proteins12. Phosphorylation is the most comprehensively understood PTM regulating importation of proteins into the nucleus14,15. The others PTMs, such as methylation, acetylation, ubiquitination, and SUMOylation, are also crucial for accumulation of proteins in the nucleus and nucleocytoplasmic translocation16–19.
AvrBs3, a TAL effector targeting the host nucleus in a plant pathogenic bacterium, Xanthomonas campestris pv. vesicatoria, was first reported as a nuclear effector20. Nuclear effectors in plant pathogenic fungi, including Ustilago maydis, Melampsora larici-populina, Puccinia striiformis f.sp. tritici, and Verticillium dahliae, have also been recently reported3,9,11,21–24. However, so far, the core sequences of NLS for nuclear transportation have not yet been defined. Furthermore, the mechanism on how nuclear effectors migrate into the host nucleus and how this mechanism modulates the host immune response remain poorly understood.
The rice blast fungus, Magnaporthe oryzae, is a hemibiotroph plant pathogenic fungus that causes significant socioeconomic damage worldwide by invading rice and wheat25,26. M. oryzae has been extensively studied as a model organism for understanding the molecular mechanisms during host-pathogen interactions. This is due to the richness of genome and transcriptome analyses of both rice and the rice blast fungus27–30. During infection, M. oryzae secretes apoplastic effectors into the space between the host and the pathogen, known as the extra-invasive hyphal membrane31. Cytoplasmic and nuclear effectors are secreted into host cells through membrane-rich plant structure, biotrophy interfacial complex (BIC)25. In a previous study, two nuclear effectors of M. oryzae, MoHTR1 and MoHTR2, were found to modulate plant immunity by directly binding to the promoters of target genes and suppressing their expression. These two nuclear effectors are localized to the host nucleus without additional NLS but not in the fungal nucleus2. However, it is unknown how these nuclear effectors migrate to the host nuclei and what roles of NLS of the nuclear effectors in modulating the host immunity.
In this study, we found that MoHTR1 is localized in the host nucleus through interaction with rice importin α and confirmed that RxKK within MoHTR1 NLS is essential for nuclear localization. MoHTR1 NLS also plays a pivotal role in facilitating the migration of cytoplasmic effectors of M. oryzae into the host nucleus. Other nuclear effector candidates of M. oryzae bearing the RxKK are also predominantly localized in the host nucleus. Additionally, SUMOylation is crucial for translocation of MoHTR1 into the rice nucleus by contributing to the interaction with Osimpα and stability of MoHTR1. Furthermore, NLS of MoHTR1 is essential for fungal pathogenicity by regulating the transcription of host immunity-associated genes. Our findings will shed light on the plant-specific NLS and provide a novel perspective on the role of NLS in the nuclear effectors of plant-pathogenic fungi during host-pathogen interactions.
RESULTS
RxKK is core NLS of MoHTR1
MoHTR1, a nuclear effector, was predicted to possess a single nuclear NLS, PGRSKKE, using the WoLF PSORT program. To determine whether this NLS is indeed responsible for the nuclear localization of MoHTR1, we adopted two distinct strategies. Firstly, we deleted NLS in MoHTR1 (MoHTR1ΔNLS) (Fig. 1A). Secondly, to examine the key residues in the NLS, we substituted the lysine (Lys, K) residues with arginine (Arg, R) (NLS_K-R), and both K and R residues with alanine (Ala, A) (NLS_K,R-A) (Fig. 1A). Lysine and arginine, as basic amino acids, are essential for the functionality of NLS12. We cloned the coding sequences of MoHTR1, MoHTR1ΔNLS, NLS_K-R, and NLS_K, R-A into eGFP expression plasmid to confirm their subcellular localization in rice protoplasts. MoHTR1 was localized in 66% of rice protoplast nuclei. In contrast, the accumulation rate of MoHTR1ΔNLS decreased to 14%. These results indicate that PGRSKKE constitutes the native NLS of MoHTR1. Additionally, NLS_K-R and NLS_K,R-A were localized in 36% and 19% of rice protoplast nuclei, respectively (Fig. 1B, C). These results further suggest that RxKK motif is a core sequence of the NLS in MoHTR1.
Fig. 1. Roles of predicted nuclear localization sequence in the nuclear localization of MoHTR1.
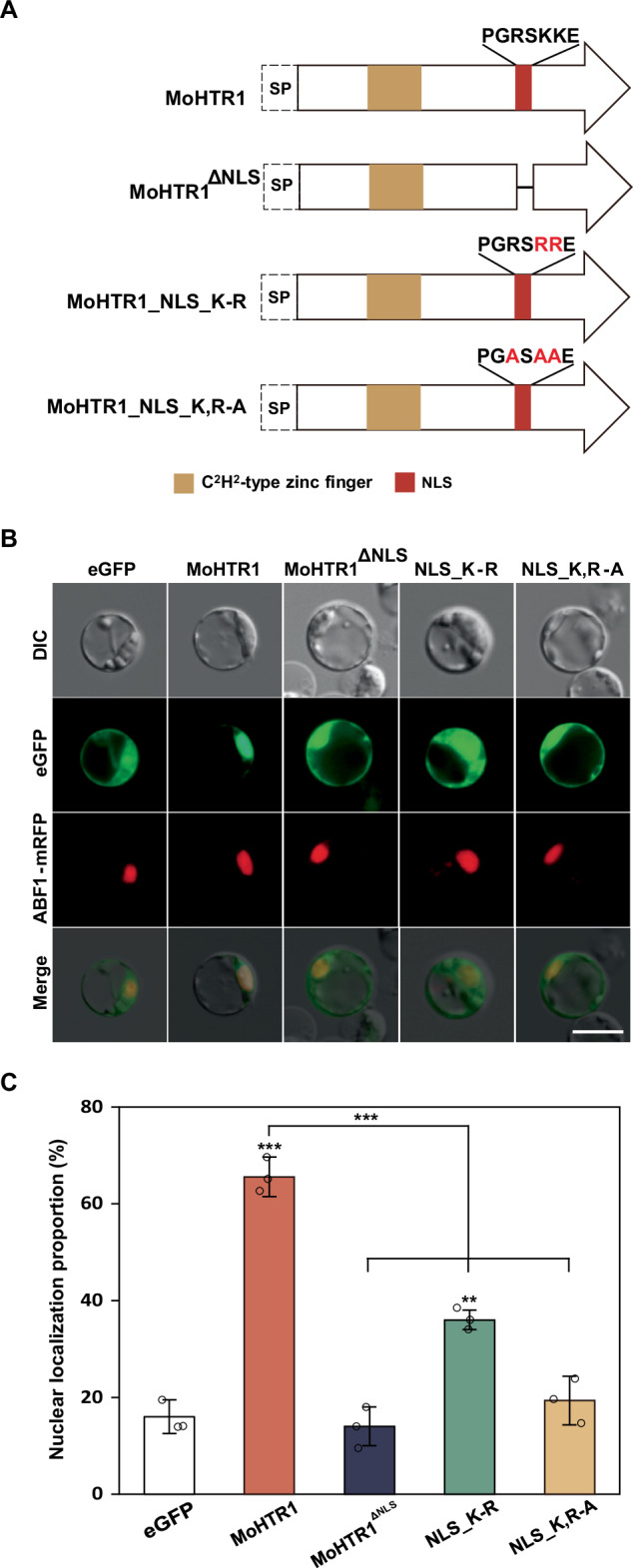
A Domain architectures, nuclear localization sequence (NLS), and substituted amino acids in MoHTR1 are represented. Red residues indicate point-mutation sites. B Subcellular localization of MoHTR1, MoHTR1∆NLS, and NLS site-directed mutants of MoHTR1 in the rice protoplasts. All protein coding sequences with eGFP were expressed under CaMV 35S promoter. ABF1:mRFP was used as a rice nuclei marker. Scale bar; 10 μm. C Nuclear localization proportion of MoHTR1, MoHTR1∆NLS, and NLS site-directed mutants of MoHTR1 in the rice protoplasts. The graph shows the percentage of nuclear localization in rice protoplasts. Mean ± SD, n = 3 independently transfected protoplasts, significance was determined by an unpaired two-tailed Student’s t-test (**p < 0.01 and ***p < 0.001). Representative data are shown from independently experiments and source data are provided as a Source Data file.
MoHTR1 is translocated into host nuclei by interacting with rice importin α
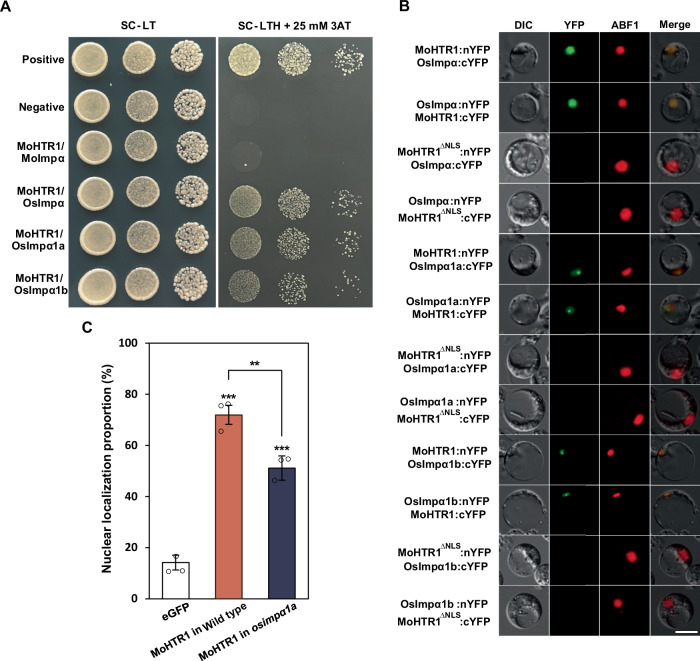
The most well-understood mechanism by which NLS-containing proteins are imported into the nucleus is the importin α-dependent pathway12. To confirm the interaction between MoHTR1 and importin α, we conducted a yeast two-hybrid assay. The coding gene for importin α in M. oryzae (MGG_15072, MoImpα) and three subunit genes of importin α in rice (Os01g24060, OsImpα; Os01g14950, OsImpα1a; Os05g06350, OsImpα1b) were cloned into prey vector, respectively, and MoHTR1 coding sequence was cloned into bait vector. As a result of the drop culture of serially diluted transformants on the selection medium (SC-LTH + 25 mM 3AT), we observed strong interactions between MoHTR1 and three rice importin αs. However, MoHTR1 did not interact with fungal importin α, MoImpα (Fig. 2A).
Fig. 2. Rice importin α-mediated transport of MoHTR1.
A Yeast two-hybrid assay of the MoHTR1 and importin α proteins of the rice blast fungus (Moimpα) and rice (Osimpα, Osimpα1a, and Osimpα1b). Interaction transformants were cultured with different dilutions on selection media. B In vivo interaction between MoHTR1 and rice importin α proteins. MoHTR1, NLS deleted MoHTR1 (MoHTR1∆NLS), and rice importin αs with split YFP were expressed under Ubiquitin promoter. BiFC assay was performed by co-transfecting of nYFP fused plasmid and cYFP fused plasmid in the rice protoplast. Scale bar; 10 μm. C Nuclear localization proportion of MoHTR1 in rice wild type cultivar and OsImpα1a T-DNA insertion mutant. Mean ± SD, n = 3 independently transfected protoplasts, significance was determined by an unpaired two-tailed Student’s t-test (**p < 0.01 and ***p < 0.001). Representative data are shown from independently experiments and source data are provided as a Source Data file.
To investigate the specificity of NLS interactions with fungal and rice importin α, we performed AlphaFold analysis using MoHTR1 NLS and MoHTR1 without both signal peptide and NLS (MoHTR1ΔNLS) with importin αs. The interface predicted template modeling (ipTM) scores, which indicate the accuracy of protein-protein interaction predictions, for MoHTR1 NLS with OsImpα, OsImpα1a, and OsImpα1b were 0.51, 0.49, and 0.49, respectively. In contrast, the ipTM scores for MoHTR1ΔNLS with each rice importin α decreased to 0.23, 0.23, and 0.22, respectively (Supplementary Table 1 and Supplementary Fig. 1). Similarly, the ipTM scores for MoHTR1 NLS with MoImpα was 0.6, which decreased to 0.29 for MoHTR1ΔNLS with MoImpα. This structural analysis did not provide insights into interaction specificity between rice and fungal importin αs. Therefore, to delve into the interaction specificity with importin α, we further observed the localization of eGFP by tagging the MoHTR1 NLS in the rice blast fungus. MoHTR1 NLS:eGFP was localized in the cytoplasm similar with eGFP, while the positive control, Simian virus 40 T antigen NLS (SV NLS):eGFP was localized in the nucleus (Supplementary Fig. 2).
In order to examine the in vivo interaction between MoHTR1 and importin αs of rice, we performed BiFC assay in the rice protoplasts. We generated split-YFP plasmids fused with coding sequences of MoHTR1, MoHTR1ΔNLS, OsImpα, OsImpα1a, and OsImpα1b, respectively. When these nYFP and cYFP fusion plasmids were co-transfected in the rice protoplasts, MoHTR1 was interacted with OsImpα in the host nucleus and interacted with OsImpα1a and OsImpα1b in the host nucleolus. However, NLS deleted MoHTR1 (MoHTR1ΔNLS) did not interact with three rice importin αs (Fig. 2B).
To further investigate whether rice importin α assists in the translocation of MoHTR1 into the rice nucleus, we examined the localization of MoHTR1 in a T-DNA insertion mutant line of rice importin α. Among the three rice importins αs, the OsImpα T-DNA mutant (osimpα1a) was identified in the Rice T-DNA insertion sequence database (http://signal.salk.edu/cgi-bin/RiceGE) (Supplementary Fig. 3). Through genotyping assay, we found homozygous and heterozygous T-DNA insertion lines, but homozygous T-DNA insertion line showed a lethal phenotype. Therefore, we observed the localization of MoHTR1 in the protoplasts of heterozygous T-DNA insertion line. In the wild type line, MoHTR1 was localized to the nucleus in 71% of the protoplasts. However, in the osimpα1a-hetero line, nuclear localization of MoHTR1 was decreased to 51% (Fig. 2C). These results indicated that MoHTR1 is imported in the host nucleus by interacting with rice importin α.
NLS of MoHTR1 escorts cytoplasmic effectors into host nuclei
The Simian virus 40 T antigen NLS (SV NLS), a most well-known classical NLS, has been widely used as a nuclear positioning marker in various previous studies32. To determine whether MoHTR1 NLS functions similarly to SV NLS in assisting nuclear import of proteins, we added MoHTR1 NLS and SV NLS to eGFP, respectively, and observed the nuclear localization. The nuclear localization rates were 84% and 65% for SV NLS and MoHTR1 NLS, respectively. There was no significant difference in the nuclear localization rates between MoHTR1 NLS alone and MoHTR1 (Supplementary Fig. 4). These results demonstrate that MoHTR1 NLS solely contributes to the nuclear localization of MoHTR1 and assists the translocation of proteins from cytoplasm to the nucleus.
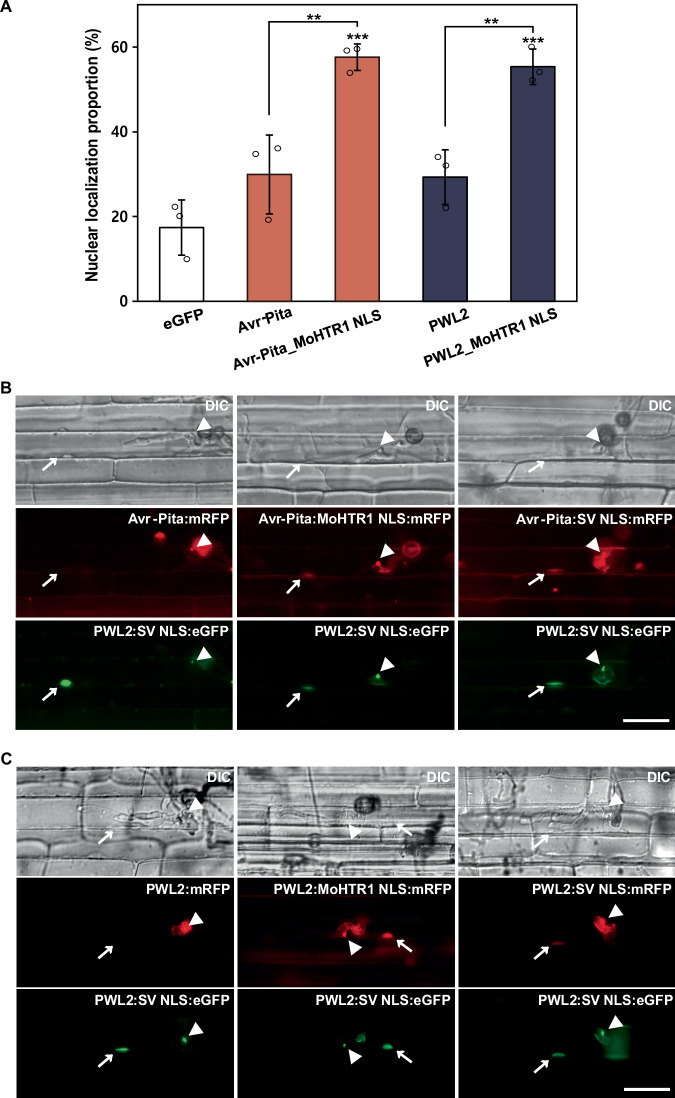
Avr-Pita and PWL2 are well-known cytoplasmic effectors of M. oryzae and are secreted into the host cytoplasm through BIC33. To investigate whether MoHTR1 NLS is able to translocate the cytoplasmic effectors of M. oryzae into the host nucleus, we observed the localization of two cytoplasmic effectors by tagging MoHTR1 NLS. Avr-Pita and PWL2 were nuclear-localized in 58% and 55% of rice protoplasts, respectively, by adding of the MoHTR1 NLS (PGRSKKE) (Fig. 3A). Similar increase of nuclear localization was also observed when two cytoplasmic effectors were tagged with the core NLS (RSKK) of MoHTR1 (Supplementary Fig. 5). To further determine whether nuclear translocation occurs by adding NLS during the M. oryzae-rice interactions, we observed nuclear localization in infected rice sheath cells. Avr-Pita was localized in 20% of rice nuclei in the infected cells, but this rate increased to 73% by adding MoHTR1 NLS. Similarly, PWL2 exhibited nuclear localization in 37% of infected cells, which increased to 83% by MoHTR1 NLS (Fig. 3B, C). These results strongly demonstrate that NLS of MoHTR1 is able to translocate cytoplasmic effectors into host nuclei.
Fig. 3. MoHTR1 NLS leading the nuclear transport of cytoplasmic effectors of M. oryzae in rice protoplasts and rice sheath cells.
A Intracellular localization of Avr-Pita and PWL2, two cytoplasmic effectors of M. oryzae, in rice protoplasts. Each cytoplasmic effector was fused with MoHTR1 NLS, and cloned into eGFP-expressing plasmid under CaMV 35S promoter. The proportion of these cytoplasmic effectors in nuclei was measured under a fluorescence microscope. B Localization of Avr-Pita and Avr-Pita tagged with a positive control NLS (SV NLS) and MoHTR1 NLS in rice sheath cells. Avr-Pita with mRFP and NLS were expressed under Avr-Pita native promoter. PWL2:eGFP:SV NLS was used as biotrophic interfacial complex (BIC) and rice nuclei marker. Arrowheads indicate BICs and arrows indicate rice nuclei. Scale bar; 20 μm. C Localization of PWL2 and PWL2 tagged with SV NLS and MoHTR1 NLS in rice sheath cells. PWL2 and PWL2 with SV NLS and MoHTR1 NLS were expressed under PWL2 native promoter and transformed into the wild type of M. oryzae. PWL2:eGFP:SV NLS was used as BIC and rice nuclei marker. The localization of Avr-Pita and PWL2 was observed at 30–34 hpi. Arrowheads indicate BICs and arrows indicate rice nuclei. Scale bar; 20 μm. Mean ± SD, n = 3 independently transfected protoplasts, significance was determined by an unpaired two-tailed Student’s t-test (**p < 0.01 and ***p < 0.001). Representative data are shown from independently experiments and source data are provided as a Source Data file.
RxKK is a core NLS for other nuclear effector candidates in M. oryzae
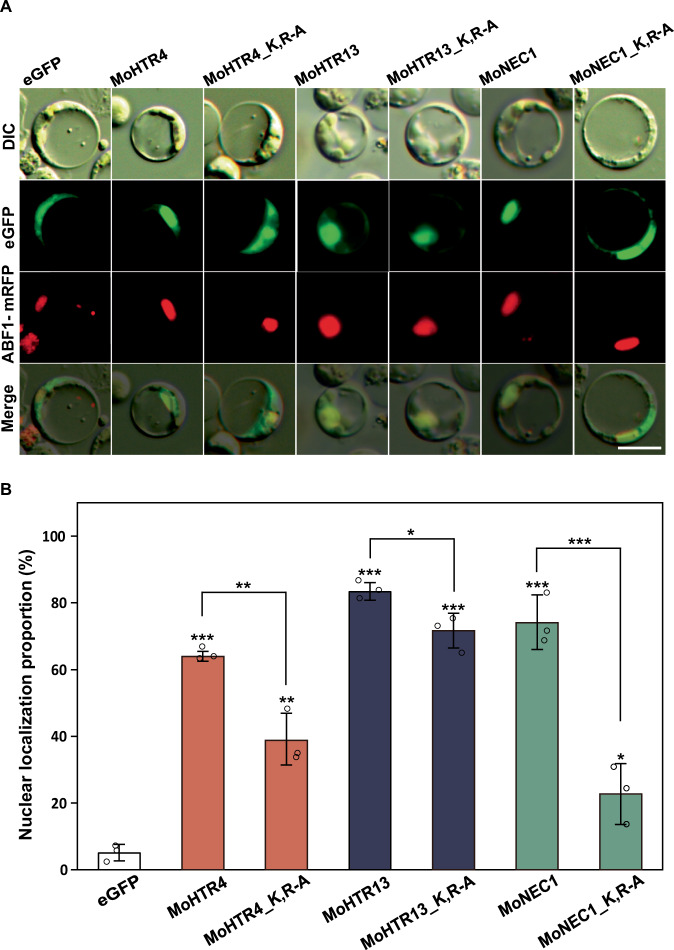
To investigate whether other nuclear effector candidates containing RxKK other than MoHTR1 also migrate to the rice nucleus, we observed the localization of these candidates. Among 1899 secretory proteins in M. oryzae, 440 proteins were predicted to have NLS by one or more NLS prediction tools. Out of 440 proteins, 23 proteins had RxKK within their predicted NLS. Twelve genes encoding these proteins were found to be expressed during rice infection from our previous report of in planta RNA-seq data30. We selected MoHTR4 (MGG_05518), MoHTR13 (MGG_13742), and MoNEC1 (nuclear effector candidates 1; MGG_14093), which showed the high expression levels during infection (Supplementary Fig. 6). We cloned coding sequences of MoHTR4, MoHTR13, and MoNEC1 without signal peptide into eGFP fusion vector and observed their nuclear localization in the rice protoplasts. MoHTR4, MoHTR13, and MoNEC1 were localized in 63%, 83%, and 73% of nuclei in the rice protoplasts, respectively. To further determine the RxKK is involved in the nuclear localization of these proteins, we mutated the K and R residues to A. Nuclear localization of each of the point-mutated proteins was decreased to 39%, 71%, and 23%, respectively (Fig. 4A, B). These results suggest that RxKK is required for nuclear localization of these nuclear effector candidates.
Fig. 4. Significance of RxKK for nuclear localization of nuclear effector candidates.
A Subcellular localization of three nuclear effector candidates of M. oryzae, including MoHTR4, MoHTR13, and MoNEC1. NLS site-directed mutants of each nuclear effector candidates were generated, and the localization of these NLS mutants and nuclear effector candidates were observed under fluorescence microscope. Scale bar; 10 μm. B The nuclear localization proportion of nuclear effector candidates and NLS site-directed mutant of nuclear effector candidates in rice protoplasts. The nuclear localization rate was counted under fluorescence microscope. Mean ± SD, n = 3 independently transfected protoplasts, significance was determined by an unpaired two-tailed Student’s t-test (*p < 0.05, **p < 0.01, and ***p < 0.001). Representative data are shown from independently experiments and source data are provided as a Source Data file.
Since MoHTR1 NLS is a plant-specific NLS, the significance of RxKK was evaluated in the nuclear localization of host proteins. We compiled a list of 870 rice proteins predicted to have NLS or to be nuclear-localized from Rice Seed Nuclear Protein DataBase (RSNP-DB) (https://pmb.du.ac.in/rsnpdb). Among them, 21 proteins were identified as the candidates containing only RxKK as monopartite NLS, and we randomly selected five proteins (Supplementary Data 1). Four out of five proteins were predominantly localized in the nucleus of rice protoplasts (Supplementary Fig. 7). These results further support that RxKK is also crucial for NLS function in the rice proteins.
SUMOylation is important for nuclear localization of MoHTR1
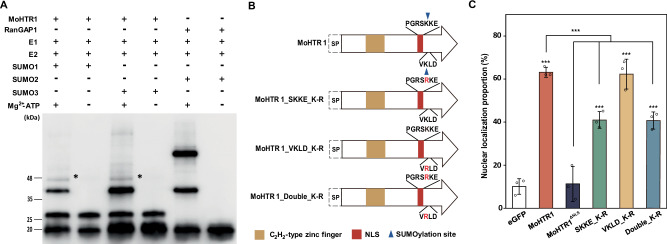
We identified two SUMOylation sites in MoHTR1 using GPS-SUMO software34. To confirm that MoHTR1 is a substrate for SUMOylation, we conducted in vitro SUMOylation assay. Initially, the MoHTR1 band was shown at 26 kDa, and it shifted to 46 kDa upon treatment with SUMO and ATP (Fig. 5A). These results indicate that MoHTR1 undergoes SUMO modification. One of the SUMOylation sites was located within the MoHTR1 NLS. To determine whether SUMOylation of the MoHTR1 NLS is involved in nuclear localization, we generated SUMOylation site-directed mutants of MoHTR1 and observed their localization in the rice protoplasts (Fig. 5B). Mutating the SUMOylation site in the non-NLS region (VKLD_K-R) did not show a significant difference with the nuclear localization of MoHTR1. However, compared to the nuclear localization rate of MoHTR1 (63%), the mutation of the SUMOylation site within the NLS region (SKKE_K-R) and mutation of both SUMOylation sites in the NLS region and non-NLS region (SKKE, VKLD_K-R) caused a decrease in the nuclear localization rate of MoHTR1 to 41% (Fig. 5C).
Fig. 5. SUMOylation is important for nuclear localization of MoHTR1.
A In vitro SUMOylation assay of MoHTR1. Purified MoHTR1 was incubated with E1, E2, SUMO, and ATP to perform in vitro SUMOylation assay. RanGAP1 was used as a positive control, and reaction without ATP served as a negative control for the SUMOylation assay. 4 ug of MoHTR1 was used for the SUMOylation assay, and Western blotting was performed using anti-His antibodies. E1, E1 SUMO activating enzyme; E2, E2 SUMO conjugating enzyme. B Domain architectures and substituted amino acids of predicted SUMOylation sites in MoHTR1. Red residues indicate point-mutation sites. Orange and red boxes represent C2H2-type zinc finger and NLS region, and blue inverted triangle represents SUMOylation site in MoHTR1. C The nuclear localization rates of SUMOylation site-directed mutants of MoHTR1 in rice protoplasts. Mean ± SD, n = 3 independently transfected protoplasts, significance was determined by an unpaired two-tailed Student’s t-test (***p < 0.001). Representative data are shown from independently experiments and source data are provided as a Source Data file.
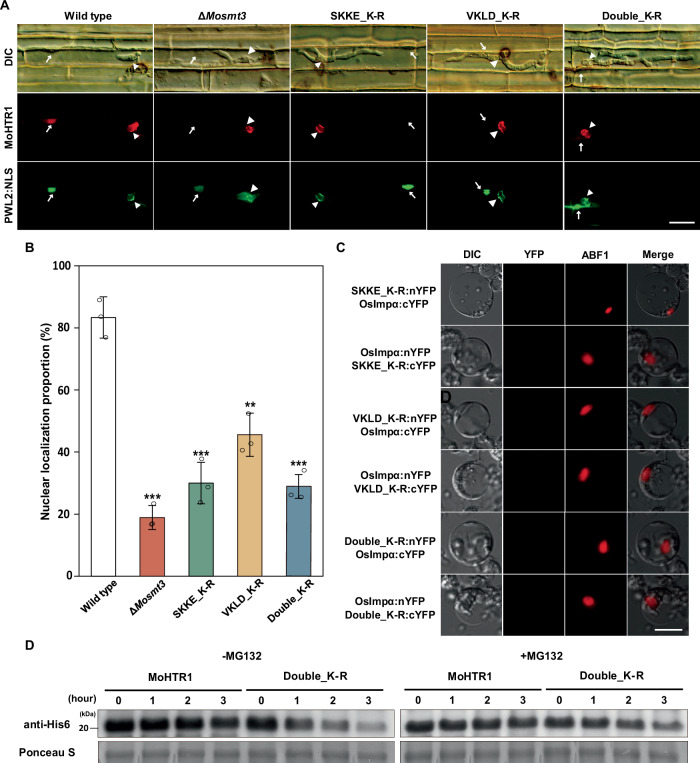
To ascertain whether SUMOylation of MoHTR1 is required not only for nuclear localization but also for secretion, we observed localization of MoHTR1 in infected rice sheath cells. In the SUMO deletion mutant (ΔMosmt3) infected rice cells, MoHTR1 was located in the BIC in 82% of inoculated cells, compared to wild type infected rice cells (98%). However, there was no significant difference in BIC localization proportion between SUMOylation site-mutated MoHTR1 and the wild type infected rice cells (Supplementary Fig. 8). In ΔMosmt3 infected rice cells, MoHTR1 was localized to the host nucleus in 19% of infected cells. Furthermore, the nuclear localization proportion of three SUMOylation site-mutated MoHTR1 variants (SKKE_K-R, VKLD_K-R, and SKKE, VKLD_K-R) were 30%, 46%, and 29% of infected cells, respectively, compared to the nuclear localization proportion of MoHTR1 (83%) (Fig. 6A, B). These results suggest that SUMOylation is essential for translocation of MoHTR1 into the host nucleus. To investigate the reason of decreased translocation of SUMOylation site-mutated MoHTR1 to the host nucleus, we examined the in vivo interaction between SUMOylation site-mutated MoHTR1 variants and rice importin αs, as well as the stability of SUMOylation site-mutated MoHTR1. In the BiFC assay, SUMOylation site-mutated MoHTR1 variants (SKKE_K-R, VKLD_K-R, and Double_K-R) interacted with OsImpα1a and OsImpα1b in the rice nucleus but did not interact with OsImpα (Fig. 6C and Supplementary Fig. 9). To assess the stability of MoHTR1 by SUMOylation, purified MoHTR1 and double SUMOylation site-mutated MoHTR1 (SKKE, VKLD_K-R) were treated with rice extracts. MG132, a 26S proteasome activity inhibitor, prevented the degradation of both MoHTR1 and SUMOylation site-mutated MoHTR1. However, without MG132 treatment, MoHTR1 level was retained throughout the time course experiment, whereas double SUMOylation site-mutated MoHTR1 was degraded depending on the incubation time (Fig. 6D). These results suggest that SUMOylation-mediated stability and interaction with OsImpα are essential for translocation of MoHTR1 into the host nucleus.
Fig. 6. SUMOylation is crucial for nuclear localization by interacting with Osimpα, and maintaing the stability of MoHTR1.
A Localization of MoHTR1 and SUMOylation site-directed mutants of MoHTR1 in rice sheath cells. MoHTR1 was transformed in the wild type and SUMO deletion mutant (ΔMosmt3), and SUMOylation site-directed mutants of MoHTR1 were transformed in the wild type. PWL2:eGFP:SV NLS was used as BIC and rice nuclei marker. The localization was observed in each mutant-infected cells at 30–34 hpi. Scale bar; 20 μm. B The nuclear localization percentage of SUMOylation site-directed mutants of MoHTR1 in rice sheath cells. C In vivo interaction between three SUMOylation site-mutated MoHTR1 and Osimpα. Site-muated MoHTR1 variants and Osimpα were cloned into split YFP expressed plasmids, respectively. BiFC assay was performed by co-transfecting of nYFP fused plasmid and cYFP fused plasmid in the rice protoplasts. Scale bar; 10 μm. D Protein stability assay of MoHTR1 and SUMOylation site-mutated MoHTR1. Purified MoHTR1 and double SUMOylation site-mutated MoHTR1 (Double_K-R) was incubated with rice extracts with or without proteasome inhibitor for 0, 1, 2, and 3 h. Western blotting was performed using anti-His antibodies. Mean ± SD, n = 3 independently inoculated rice sheath cells, significance was determined by an unpaired two-tailed Student’s t-test (**p < 0.01 and ***p < 0.001). Representative data are shown from independently experiments and source data are provided as a Source Data file.
MoHTR1 NLS has crucial roles for transcriptional reprogramming of host target genes
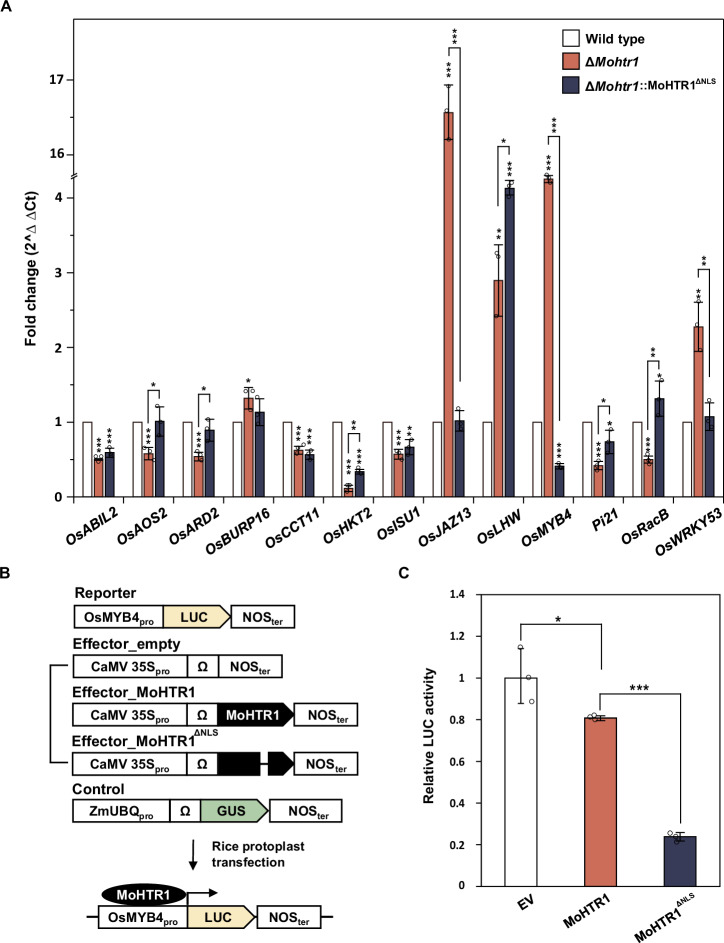
To investigate whether the nuclear localization of MoHTR1 is involved in the regulation of target gene expression, we conducted spray inoculation of the wild type, ΔMohtr1 and ΔMohtr1::MoHTR1ΔNLS on rice leaves. Total RNA samples were extracted from each infected rice and subjected to expression profiling of target genes. We performed qRT-PCR of 17 genes with promoters carrying effector binding element (EBE) of MoHTR1, and found that 13 genes showed differential expression depending on the presence of MoHTR1 (wild type vs. ΔMohtr1). When ΔMohtr1::MoHTR1ΔNLS was inoculated, the eight genes, including OsABIL2, OsCCT11, OsHKT2, OsISU1, OsLHW, OsMYB4, Pi21, and OsRacB were differentially expressed compare to the wild type-inoculated rice (Fig. 7A). These results indicate that the nuclear localization of MoHTR1 is important for the expression of host target genes. MoHTR1 is known to suppress OsMYB4 transcription by directly binding to its promoter2. To elucidate the transcriptional reprogramming of MoHTR1 target genes by NLS deletion, we assessed luciferase (LUC) activity expressed by the OsMYB4 promoter. We used a reporter vector containing LUC reporter gene under the OsMYB4 promoter and an effector vector containing MoHTR1 and NLS deleted MoHTR1 expressed genes under the CaMV 35S promoter, respectively (Fig. 7B). The expression of OsMYB4 was suppressed by MoHTR1, but the expression of OsMYB4 was even further suppressed by the deletion of the NLS (Fig. 7C).
Fig. 7. Crucial roles of MoHTR1 NLS in the regulation of host target gene expression.
A The transcripts abundance of rice target genes of MoHTR1, including OsMYB4. Each strain, including wild type, ΔMohtr1, and ΔMohtr1::MoHTR1ΔNLS, was inoculated onto 4-week-old rice seedlings, and infected rice was collected at 48 hpi. Transcription levels of host target genes of MoHTR1 were quantified using qRT-PCR, and gene abundance was normalized to actin encoding gene expression. B Features of the vectors used for the luciferase assay in rice protoplast. The LUC reporter gene was expressed under the OsMYB4 promoter, MoHTR1 was expressed under the CaMV 35S promoter, and GUS encoding gene was expressed under the ZmUBQ promoter. C Relative LUC activity in MoHTR1 and MoHTR1ΔNLS transfected rice protoplasts. Reporter, effector, and control vectors were transfected into rice protoplasts, and after 5 h, LUC activity was measured by luminometer. Relative LUC activity was calculated by comparing it to transfection with an empty effector vector. Mean ± SD, n = 3 independent RNA samples from infected rice leaves and independently transfected protoplasts, significance was determined by an unpaired two-tailed Student’s t-test (*p < 0.05, **p < 0.01 and ***p < 0.001). Representative data are shown from independently experiments and source data are provided as a Source Data file.
MoHTR1 NLS is essential for fungal virulence and host immune response
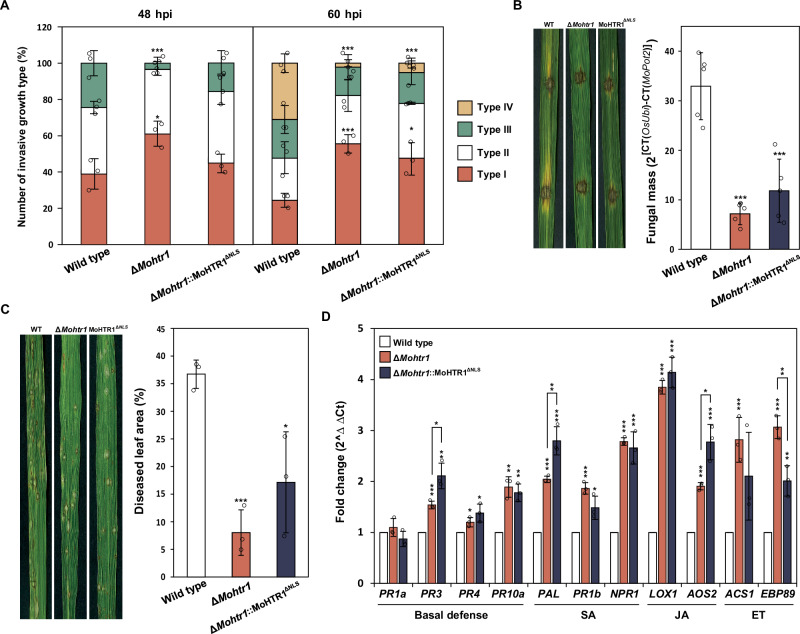
To investigate the roles of MoHTR1 NLS in the virulence of M. oryzae, we performed rice sheath inoculation with conidial suspensions of the wild type, ∆Mohtr1, and ∆Mohtr1::MoHTR1ΔNLS strains. After sheath inoculation for 48 h, the invasive growth of the wild type, ∆Mohtr1, and ∆Mohtr1::MoHTR1ΔNLS was exhibited 61%, 42%, and 55% of II and III types of virulence, respectively. At 60 h post inoculation (hpi), 76%, 44%, and 52% of the infection sites showed II, III, and IV types of virulence in the wild type, ∆Mohtr1, and ∆Mohtr1::MoHTR1ΔNLS, respectively (Fig. 8A). For more accurate virulence comparison, we measured the fungal mass in the infected rice. We performed drop inoculation assay using the wild type, ∆Mohtr1, and ΔMohtr1::MoHTR1ΔNLS strains on the 6-week-old rice leaves. Four days after inoculation, we extracted DNA from inoculated rice leaves and conducted DNA-based qPCR. The fungal mass of the wild-type-infected rice was 33, whereas the fungal mass was decreased in the ∆Mohtr1, and ΔMohtr1::MoHTR1ΔNLS-infected rice to 7 and 12, respectively (Fig. 8B). We also conducted spray inoculation on 4-week-old rice seedlings with the wild type, ∆Mohtr1, and ∆Mohtr1::MoHTR1ΔNLS strains. After 6 days post inoculation (dpi), we collected infected rice leaves and measured diseased leaf area (DLA). The percentage of DLA by ∆Mohtr1 and ∆Mohtr1::MoHTR1ΔNLS was decreased to 8% and 17%, respectively, compared to wild-type DLA (37%) (Fig. 8C). Since ∆Mohtr1::MoHTR1ΔNLS showed much less virulence compared to WT, we checked the expression of immunity-associated genes to confirm the roles of NLS in MoHTR1 in fungal virulence. These include basal defense-related genes (PR3, PR4, and PR10a), salicylic acid (SA) signaling pathway genes (PAL, PR1b, and NPR1), jasmonic acid (JA) signaling pathway genes (LOX1 and AOS2), and ethylene (ET) signaling pathway genes (ACS1 and EBP89). Expression of these genes was significantly increased not only in ∆Mohtr1-inoculated but also in ∆Mohtr1::MoHTR1ΔNLS-inoculated rice (Fig. 8D). These results indicate that MoHTR1 NLS is essential for the function of MoHTR1 in the regulation of host immune response and fungal virulence.
Fig. 8. Importance of MoHTR1 NLS in the pathogenicity of M. oryzae.
A Pathogenicity assay by sheath inoculation of wild type, ΔMohtr1, and ΔMohtr1::MoHTR1ΔNLS. Conidial suspensions (2 × 104 mL–1 for 48 hpi and 5 × 103 mL–1 for 60 hpi) were inoculated onto 6-week-old rice sheath cells. Invasive growth was observed, and the invasive growth type was counted using a microscope at 48 and 60 hpi. The criteria for the invasive growth types was described in Methods section. Scale bar, 50 μm. B Fungal mass in the wild type-, ΔMohtr1-, and ΔMohtr1::MoHTR1ΔNLS-infected rice leaves was quantified using DNA-based qPCR. The infected rice leaves were harvested at 4 dpi. C Pathogenicity assay by spray inoculation. Conidial suspensions (5 × 104 mL–1) were inoculated onto 4-week-old rice seedlings. The lesions were observed and collected at 6 dpi. Disease leaf area was measured using ImageJ software. D Transcription levels of defense-associated genes in wild type, ΔMohtr1, and ΔMohtr1::MoHTR1ΔNLS-infected rice. Infected rice leaves samples from the spray inoculation were collected at 48 hpi. The abundance of basal defense- and hormone signaling-associated genes in rice was quantified using qRT-PCR, and gene abundance was normalized with actin encoding gene expression. Mean ± SD, n = 5 independent drop inoculated rice leaves and n = 3 independent RNA samples from infected rice leaves and independently inoculated rice leaves and sheath cells, significance was determined by an unpaired two-tailed Student’s t-test (*p < 0.05, **p < 0.01 and ***p < 0.001). Representative data are shown from independently experiments and source data are provided as a Source Data file.
Since it was confirmed that MoHTR1 NLS affects the expression of host target genes and immunity-associated genes, we further investigated the role of NLS in transcriptional reprogramming of host genes at genome-wide level. We performed spray inoculation on 4-week-old rice seedlings using the wild type, ∆Mohtr1, and ∆Mohtr1::MoHTR1ΔNLS strains. Infected rice leaves were collected at 48 hpi and subjected to RNA extraction and RNA sequencing. Deletion of MoHTR1 or MoHTR1 NLS altered the expression of 1800 genes (967 up- and 833 downregulated) and 2836 genes (1204 up- and 1632 downregulated) compared to wild type, respectively. In addition, deletion of NLS altered the expression of 2255 genes (935 up- and 1320 downregulated) compared to ∆Mohtr1 (Supplementary Data 2). Defense response-related gene ontology (GO) terms were enriched in ∆Mohtr1 compared to the wild type as our previous study2 (Supplementary Data 2). The differentially expressed genes (DEGs) (p < 0.05) in ∆Mohtr1::MoHTR1ΔNLS -inoculated rice, compared to wild type-inoculated rice, were associated with 51 enriched GO terms. These include defense-related GO terms and several others, such as chloroplast (GO:0009507), cytoplasm (GO:0005737), nucleosome assembly (GO:0006334), response to hormone (GO:0009725), and response to endogenous stimulus (GO:0009719) compared to the wild type (Supplementary Data 2).
Discussion
Plant pathogens secrete effectors to manipulate the host immune system, thereby facilitating successful infection. Recently, among plant pathogenic effectors, there has been significant interest in nuclear effectors, which translocate into the host nucleus and modulate various biological processes by interacting with host genes or proteins. However, mechanisms for the translocation of nuclear effector into the host nucleus and the roles of these translocation factors on the fungal virulence remain poorly characterized.
Previously, a few nuclear effectors of plant pathogenic fungi have been reported and their NLS regions were predicted6,8,22–24,35. However, none of them identified the core NLS. In this study, we first identified the native core NLS, RxKK, essential for the translocation of the nuclear effector to the host nucleus through deletion and site-directed mutagenesis of NLS. This RxKK is unique and not present in NLS regions of other reported fungal nuclear effectors. (Supplementary Data 3). RxKK is required not only for the nuclear localization of fungal nuclear effectors but also for rice proteins. TAL effectors, the nuclear effectors of plant pathogenic bacteria, possess 2–3 conserved NLSs regardless of the bacterial species36,37. While reported NLSs of TAL effectors and fungal nuclear effectors contain abundant hydrophobic residues, NLS in nuclear effectors of plant pathogenic fungi are not well-conserved, and even among the reported nuclear effectors in the same species, such as MoHTR1, MoHTR2, and MoHTR3 in M. oryzae2,38.
Importin α is required for transporting cargo into the nucleus through NPC by directly binding to the classical NLS of the cargo and interacting with importin β12. However, the translocation mechanisms of the nuclear effector of plant pathogens have been remained elusive. More recently, it was demonstrated that rice importin α1b plays a crucial role in the nuclear import of TAL effectors, PthXo1 and TAL1 in X. oryzae pv. oryzae (Xoo) and X. oryzae pv. oryzicola (Xoc), respectively39. In this study, we also confirmed the significant roles of rice importin αs in the translocation of MoHTR1 and established that the MoHTR1 NLS is a classical NLS not only though yeast-two-hybrid experiment but also through experiments verifying in vivo interactions.
In the rice blast fungus, cytoplasmic effectors are accumulated into BIC via a Golgi-independent secretory pathway31. In more recent report, effectors are packaged by the rice plasma membrane component, clathrin, and delivered into plant cells from the BIC40. Many cytoplasmic effectors have been reported and their secretion and localization were identified by tagging with Simian virus 40 large T NLS peptide (SV NLS)33,41. In this study, MoHTR1 NLS translocated Avr-Pita and PWL2, cytoplasmic effectors of the rice blast fungus, into rice nuclei as much as those tagging with SV NLS. These data further support the functionality of MoHTR1 NLS in the translocation of proteins into the nucleus. Interestingly, when we observed the localization of these two cytoplasmic effectors, they were predominantly localized in the cytoplasm, but also in the nucleus without additional NLS. Avr-Pita and PWL2 were predicted to have protein molecular weights of 26 and 16 kDa, respectively, as determined using Compute pI/Mw tool (https://web.expasy.org/compute_pi/). Since small molecules below 40 kDa can diffuse into the nucleus through the NPC42, it is presumed that these cytoplasmic effectors might be passively translocated through the NPC. We further revealed that RxKK is crucial not only for nucleus localization of MoHTR1 and cytoplasmic effectors, but also of other nuclear effector candidates. Among three nuclear effector candidates tested, the nuclear localization rates of MoHTR4 and MoNEC1 were significantly decreased compared to that of MoHTR13 when RxKK was mutated. MoHTR4 and MoNEC1 have only one RxKK as their NLS, but MoHTR13 has three additional NLSs. Similar result was reported in SNF12, a nucleolar protein of Dictyostelium, a single-celled eukaryote. SNF12 possesses three NLSs (NLS 1, NLS2, and NLS3), but NLS1 and NLS2 are not involved in nuclear localization of this protein. However, the nuclear localization rate of NLS3-deleted mutant was significantly decreased43. Therefore, we may speculate that a modest reduction in the nuclear localization rate of RxKK mutant of MoHTR13 may be attributed to the presence of other NLSs with redundant functions.
Several studies have reported the roles of PTMs, such as phosphorylation, SUMOylation, and acetylation, on the function of plant pathogen’s effectors. AvrPto and AvrPtoB, type III secretion effectors of Pseudomonas syringae pv. tomato, are phosphorylated by host kinase (BAK1) and modulate pathogen’s virulence and plant immunity44,45. In addition, acetylation also affects the secretion of FolSvp1, an effector of Fusarium oxysporum f.sp. lycopersici46. In the rice blast fungus, the secretion of cytoplasmic effectors (Avr-Pita, PWL2, Avr-Piz-t, and Avr-Pia) and apoplastic effector (Slp1) was impaired in SUMO deletion mutant47,48. However, molecular mechanism on the roles of PTM in the translocation of nuclear effectors remains limited. In this study, we unraveled a novel translocation mechanism of nuclear effectors through in vitro SUMOylation assay, in vivo interaction assay with rice importin αs, protein stability assay, and site-directed mutagenesis of SUMOylation sites. We found that SUMOylation is necessary for the translocation of nuclear effectors into the host nucleus but indirectly affects the secretion of nuclear effectors. Our finding support that not only NLS and interaction with rice importin α but also SUMOylation play crucial roles in the translocation of fungal nuclear effectors to the host nucleus.
We demonstrated that MoHTR1 NLS is required for fungal virulence and modulates host immune response. These are supported by the facts that expression of basal defense-related genes and salicylic acid, jasmonic acid, and ethylene signaling pathway genes were increased in both ∆Mohtr1 and ∆Mohtr1::MoHTR1ΔNLS-inoculated rice compared to wild type-inoculated rice. On the other hand, we checked transcription levels of 17 biotic and abiotic stress response genes from previously reported 226 MoHTR1 host target candidates that harboring the EBE of MoHTR1 within their gene promoter. Although most genes showed differential expression, there was no significant difference in the expression of some target genes between the wild type and ∆Mohtr1::MoHTR1ΔNLS-inoculated rice. We may speculate the reasons how they showed similar expression patterns of host target genes in the absence of MoHTR1 NLS. First, deletion of MoHTR1 NLS is not completely removed MoHTR1 from the host nucleus, although it is a relatively small amount, MoHTR1 is present in the nucleus and regulate target gene expression by binding to their promoters. Second, interactions with cytoplasm-located host proteins may be involved in regulating the host genes expression. Pst_12806, an effector of Puccinia striiformis f. sp. tritici, interacts with photosynthesis-related protein and regulates the transcription of PAMP-triggered immunity (PTI)-associated genes49. Third, other mechanisms may be associated to regulate the amount of transcript, beyond direct binding to nucleic acid. Previously, involvement of nuclear effectors, VdSSR1 of V. dahlia and CSEP0064 of Blumeria graminis, in RNA processing has been reported, which plays a role in plant immune response and pathogenicity in plant fungal pathogens7,11.
In conclusion, we first identified the core NLS of MoHTR1 responsible for the translocation of nuclear effector to the host nucleus in the rice blast pathogen, M. oryzae. We further elucidated the importin α- and SUMOylation-mediated transportation of the nuclear effector and pivotal roles of the core NLS in the fungal virulence and host immune response. Our findings provide a novel perspective on the mechanism underlying the translocation of nuclear effectors during host-pathogen interactions and highlights their significant roles in the host immune response.
Methods
Prediction of NLS and strategy for selecting RxKK containing fungal nuclear effector candidates and rice proteins
We predicted presence of NLS in MoHTR1 using three prediction tools including WoLF-PSORT, cNLS mapper, and NLStradamus50–52. To identify RxKK-containing nuclear effector candidates, we listed-up the 440 secreted proteins predicted to have NLS using tools such as WoLF-PSORT, cNLS mapper, and NLStradamus from 1899 proteins with signal peptide in M. oryzae. After then, we found 23 proteins which were retained RxKK in predicted NLS. In addition, we referred previously studied in planta RNA-seq data, in planta stage specifically expressed 12 proteins were selected as nuclear effector candidates30. To observe the localization of RxKK containing rice proteins, we compiled a list of 870 rice proteins predicted to have NLS or to be nuclear-localized through subcellular localization prediction tools, such as WoLF-PSORT, YLoc, CELLO, and NucPred, from RSNP-DB (https://pmb.du.ac.in/rsnpdb). We found 21 proteins which were contained only RxKK as monopartite NLS, among them we randomly selected five proteins.
Yeast two-hybrid assay
The coding sequences of MoHTR1 without signal peptide and importin αs were amplified from cDNA of the M. oryzae KJ201 wild type and rice cv. Nakdongbyeo, respectively. We cloned the coding sequence of MoHTR1 into pDEST32 (BD vector, bait) and importins into pDEST22 (AD vector, prey). We performed co-transformation in the yeast strain, Mav203, using Pexp32:Krev1 as a bait and Pexp22:RalGDSwt as a prey for positive control, and using Pexp22:RalGDSm2 as a prey for negative control. The yeast two-hybrid (Y2H) assay was conducted using the ProQuest Two-Hybrid System kit (Invitrogen, USA) according to the supplier’s instructions. Transformants were cultured and isolated from selection media (SC-Leu/Trp (SC-LT) agar) and interaction transformants were selected on SC-Leu/Trp/His + 25 mM 3-amino-1,2,4-triazole (SC-LTH + 3AT) agar media.
AlphaFold analysis
We employed AlphaFold2 in its multimer mode on a local server to elucidate the protein-protein interactions (PPIs)53. The ipTM scores of PPIs were obtained by submitting the AlphaFold output into predicted aligned error (PAE) viewer54.
Bimolecular fluorescence complementation (BiFC) assay
We amplified the coding sequences without the signal peptide of MoHTR1 and without both signal peptide and NLS of MoHTR1 from the cDNA of the M. oryzae and the coding sequences of importin αs from the cDNA of O. sativa. Subsequently, we conducted a BP reaction using the amplified sequences and the donor vector (pENTR-TOPO), and then cloned them into pGA3574-GW-nYFP and pGA3574-GW-cYFP. Plasmids for transfection were prepared using PureLink™ HiPure Plasmid Midiprep Kit (Invitrogen, USA) in accordance with the supplier’s instructions. To isolate rice protoplasts, 12-day-old rice seedling was grown in 1/2 MS agar media and finely chopped rice tissues were incubated with lysing enzyme solution (containing 1.5% Cellulase and 0.75% Macerozyme R-10) for 3 h55. Following this, PEG-mediated co-transfection was performed using PEG solution (0.6 M Mannitol, 100 mM Calcium chloride, and 40% (w/v) PEG 4000) with 5 μg of each nYFP and cYFP plasmid in 100 μL of rice protoplast (2 × 106 cells mL–1). After transfection, protoplasts were incubated in W5 solution (154 mM Sodium chloride, 125 mM Calcium chloride, 5 mM Potassium chloride, and 2 mM MES) for overnight at RT. In vivo interaction was observed using a fluorescence microscope (Leica DM600 B).
Site directed mutagenesis
To generate the mutations of NLS and SUMOylation sites, we designed 30 bp primers containing desired mutation sequences (Supplementary Table 2). And then, target sequences were mutated using QuikChange Lightning Site-Directed Mutagenesis kit (Agilent) following the protocol of manufacturer.
Localization in rice protoplast
We isolated rice protoplasts as mentioned above. To observe the localization of MoHTR1, NLS mutated MoHTR1, nuclear effector candidates of the rice blast fungus, and RxKK containing rice proteins in rice protoplast, we amplified coding sequecnes (CDS) of these proteins from mycelial cDNA of M. oryzae and cDNA of the rice, and each CDS was cloned into donor vector, pENTR D-TOPO (Thermo Scientific™) (Supplementary Data 2). These entry vectors were cloned into eGFP fusion vector, p2GWF756 (destination vector) using the gateway system. And then, PEG-mediated co-transfected with ABF1:mRFP plasmid (marker for labeling rice nuclei) in rice protoplast. To confirm the function of the MoHTR1 NLS, localization was observed by adding the NLS of MoHTR1 (PGRSKKE) and classical NLS of SV40 large T antigen (SV) (PKKKRKV) to p2GWF7, Avr-pita cloned p2GWF7, and PWL2 cloned p2GWF7, respectively. Localization was observed at least 40 rice protoplasts using a fluorescence microscope (Leica DM600 B). These experiments were performed in triplicate.
Localization in fungal conidia and infected sheath cells
To determine the function of MoHTR1 NLS in fungal cells, MoHTR1 NLS and SV NLS were tagged to eGFP, respectively, and cloned with EF1α promoter and TrpC terminator in pCB1004. The EF1α pro:NLS:eGFP:TrpC term plasmids were transformed in wild type of M. oryzae protoplast. To determine the function of MoHTR1 NLS in rice infected cells, MoHTR1 NLS and SV NLS were tagged to C-term of two cytoplasmic effectors (Avr-Pita and PWL2) and cloned into pFPL-rh, respectively57. And then, they were co-transformed with PWL2:eGFP:SVNLS plasmid (marker for labeling BICs and rice nuclei) in wild type of M. oryzae protoplast. To observe the function of SUMOylation on MoHTR1, SUMOylation site mutated MoHTR1 was cloned into pFPL-rh and these plasmids were co-transformed with PWL2:eGFP:SVNLS plasmid (marker for labeling BICs and rice nuclei) in wild type and SUMO deletion mutant (∆Mosmt3) of M. oryzae47. To confirm the localization of these proteins tagged with fluorescent protein in rice sheath cells, we inoculated conidial suspensions (2 × 104 mL–1) on 6-week-old rice sheath and observed at least 40 infected sites using a fluorescence microscope (Leica DM600 B) at 30–32 h post inoculation (hpi). These experiments were performed in triplicate.
Protein purification and SUMOylation assay
His6 tagged MoHTR1 and SUMOylation site-mutated MoHTR1 without signal peptide were expressed using pET-28a (Novagen) in Escherichia coli strain BL21. To induce MoHTR1 expression, we treated with 0.1 mM IPTG and incubated at 37 °C for 4 h. The recombinant proteins were purified with His Mag Sepharose (Cytiva) following to the supplier’s instructions. After then, elution buffer was exchanged by dialysis to PBS (Phosphate-buffered saline). The protein concentration was measured using Bradford assay. In vitro, SUMOylation assay was conducted according to the supplier’s instructions using SUMOylation assay kit (Abcam). After incubation 4 μg of MoHTR1 with SUMOylation-associated components for 1 h at 37 °C, mixtures were loaded on 12% Tirs-glycine gel and separated by SDS-PAGE. And then, transferred to Immune-Blot LF PVDF membrane (BIO-RAD) and the membrane was probed with anti-SUMO1/2/3 (1:1000, Abcam, Rabbit polyclonal antibody, ab1394790) and anti-His6 (1:1000, Invitrogen, Mouse monoclonal antibody, MA1-21315) using Fast western blot kit, ECL substrate (Thermo Fisher). The probed membrane was developed using ChemiDoc MP Imaging System (BIO-RAD).
Protein stability analysis
We extracted total proteins from 10-day-old rice seedling using PBS buffer without protease inhibitors. Finely grinded rice tissues were sonicated and incubated on 4 °C for 1 h. After centrifugation, we obtained supernatant and measured concentration of proteins using Bradford assay. To assess the stability of 3 μg of MoHTR1, MoHTR1 and SUMOylation site-mutated MoHTR1 were incubated with 30 μg of rice protein extracts, respectively, at 25 °C for 0, 1, 2, and 3 h with or without 50 μM MG132, a 26S proteasome inhibitor. The incubated mixtures were heated at 95 °C for 5 min and loaded on 12% Tirs-glycine gel and separated by SDS-PAGE. And then, transferred to Immune-Blot LF PVDF membrane (BIO-RAD) and the membrane was probed with anti-His6 (1:1000, Invitrogen, Mouse monoclonal antibody, MA1-21315) using Fast western blot kit, ECL substrate (Thermo Fisher). To confirm the protein loading amount, we stained the membrane blot with Ponceau S. The probed and stained membrane were developed using ChemiDoc MP Imaging System (BIO-RAD).
RNA-seq, Go-term enrichment analysis, and qRT-PCR
To check how MoHTR1 NLS regulates the transcription levels of host genes during infection, we generated ΔMohtr1::MoHTR1ΔNLS strain in which NLS deleted MoHTR1 construct was complemented in ∆Mohtr12. Conidial suspensions (105 mL–1) of wild type (KJ201), ∆Mohtr1, and ΔMohtr1::MoHTR1ΔNLS were spray inoculated on 4-week-old rice seedlings, and infected leaves were harvested at 48 hpi. Total rice RNA was extracted using the Easy-spin total RNA extraction kit (iNtRON Biotechnology) and the quality of extracted RNA samples was assessed using Bioanalyzer 2100 (Agilent). RNA-Seq libraries were prepared using the TruSeq RNA Library Prep Kit (Illumina) and sequenced using HiSeq2500 (Illumina). Raw reads were processed to remove low-quality reads and trim adapter sequences using NGS QC Toolkit v2.3.3. The resulting reads were mapped against the Oryza sativa japonica reference genome (IRGSP-1.0, 2020-06-03) using HISAT2 v2.1.0. The transcriptome was assembled using the genome-guided method of StringTie v1.3.6 (Supplementary Table 3). We used fragments per kilobase of transcript per million mapped read pairs (FPKM) as the expression value. Genes with an absolute value of log2(fold-change) >1 were considered to be differentially expressed. DEGs were subjected to Gene Ontology term enrichment analysis at a 5% false discovery rate using g: Profiler.
To confirm the expression of defense-related genes and MoHTR1 target candidates, the extracted total RNA from infected rice was reverse transcribed to cDNA. And then we did qRT-PCR with 25 ng of cDNA, 3 μL of primers (10 pmole), and 5 μL of SYBR Green PCR Master Mix using Rotor-Gene Q 2plex (Qiagen)47 (Supplementary Data 2). Actin coding gene was used to normalize of rice genes expression.
Pathogenicity tests
We performed pathogenicity test using susceptible rice cultivar, Nakdongbyeo (Oryza sativa). To do spray and sheath inoculation assay, we used 5 × 104 mL–1 and 2 × 104 mL–1 conidia, respectively. We spray inoculated on 4-week-old rice and incubated at 25 °C for 1 day in humid and dark chamber (100% relative humidity) and then incubated at 28 °C for 5 days in growth chamber. We sheath inoculated on 6-week-old rice sheath and incubated at 25 °C for 48 and 60 hpi. After then, we collected infected leaves and observed invasive hyphal growth under a microscope (Leica DM600 B). Invasive hyphal growth in sheath cells was classified into four types. Type I for colonization occurred only in the penetrated cells, type II for invasive hyphae spread to neighboring cells from the penetrated cells, type III for invasive hyphae grow beyond the neighboring cells, and type IV for invasive hyphae extensive grow from the previous type. These experiments were performed triplicates. To measure the fungal mass in the infected rice leaves, we wounded drop inoculated on 6-week-old rice leaves and incubated at 25 °C for 4 days. We collected 4 cm of inoculated leaves and grinded using bead beater (MP Biomedical FastPrep Homogenizer). The grinded samples were incubated with CTAB buffer (100 mM Tris-HCl (pH 8.0), 20 mM EDTA (pH 8.0), 1.4 M NaCl, and 2% (w/v) Cetyltrimethyl ammonium bromide), proteinase K, and RNase A at 60 °C for 1 h. we added chloroform:isopropanol (24:1) to the supernatants and incubated at –20 °C for 30 min. After then, we suspended pellet in TE buffer.
Luciferase assay
To evaluate the transcriptional activity of MoHTR1 NLS on the target gene promoter, luciferase (LUC) reporter gene was expressed under OsMyb4 promoter. And the effector vector was designed CaMV 35Spro:MoHTR1-Δsp and CaMV 35Spro:MoHTR1-ΔspNLS. For the control vector, GUS gene was expressed under maize ubiquitin promoter. Rice protoplasts were generated from 2-week-old etiolated seedling as previously described53. Total 10 μg of these reporter, effector, and control vectors were PEG-mediated co-transfected into rice protoplast. After 5 h, we added 100 μl of cell lysis buffer and luciferase substrate of Luciferase assay system (Promega) for detecting LUC activity. To detect GUS activity, we added 100 μl of hypertonic solution (10 mM Tris-Hcl (pH 8.0), 2 mM MgCl2, 1 mM 4-Methylumbelliferyl-ß-D-glucuronide) and incubated for 2 h at 37 °C. After then, we added 1 mL of 0.2 M Na2CO3 to stop reaction. The LUC activity and GUS activity were detected by Promega GloMax 96 Microplate Luminometer (Promega) and Varioskan LUX Plate Reader (Thermo Fisher), respectively. The LUC activity was normalized by GUS activity. These experiments were performed triplicate.
Statistics & reproducibility
The statistical analyses were performed using SPSS 24.0. Statistical significance was determined by an unpaired two-tailed Student’s t-test. For gene ontology enrichment analysis, significance was assessed using the Hypergeometric test. Values are presented as mean ± SD, and the number of samples (n) and the statistical tests used are indicated in the figure legends. All experiments were repeated at least three times, with the main results verified by more than two investigators. Samples were observed by being randomly labeled with numbers.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of additional supplementary files
Source data
Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) grants funded by Ministry of Science and ICT (MSIT) (2018R1A5A1023599, RS-2023-00275965 to Y.-H.L. and RS-2023-00246565 to Y.-J.L.). We thank Dr. Jong-Seong Jeon for in planta expression plasmids and rice importin α T-DNA mutant line.
Author contributions
Y.-J.L. and Y.-H.L. designed the research. Y.-J.L. Y.-J.Y., H.L., and S.K. performed the experiments and analyzed the data. G.C., J.K., J.H.K., and K.-T.K. performed bioinformatics work. Y.-J.L. and Y.-H.L. wrote and revised the article. Y.-J.L. and Y.-H.L. acquired research grants.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
Data that support the findings of this study are available in the paper and its Supplementary information files. The datasets and fungal materials generated and analyzed during the current study are available from the corresponding author upon request. The RNA-seq data generated in this study has been deposited in the NCBI Sequence Read Archive under accession code PRJNA1103247. The processed RNA-seq data are available at NCBI Sequence Read Archive. The RNA-seq data generated in this study are provided in the Supplementary information. The RNA-seq data used in this study are available in the NCBI Sequence Read Archive under accession code PRJNA1103247. Source data for main figures and Supplementary Figs. are provided with this paper. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-54272-4.
References
- 1.Selin, C., de Kievit, T. R., Belmonte, M. F. & Fernando, W. G. Elucidating the role of effectors in plant-fungal interactions: progress and challenges. Front. Microbiol.7, 600 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim, S. et al. Two nuclear effectors of the rice blast fungus modulate host immunity via transcriptional reprogramming. Nat. Commun.11, 5845 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Wit, P. J. Apoplastic fungal effectors in historic perspective; a personal view. New Phytol.212, 805–813 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Figueroa, M., Ortiz, D. & Henningsen, E. C. Tactics of host manipulation by intracellular effectors from plant pathogenic fungi. Curr. Opin. Plant Biol.62, 102054 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Jwa, N.-S. & Hwang, B. K. Convergent evolution of pathogen effectors toward reactive oxygen species signaling networks in plants. Front. Plant Sci.8, 1687 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li, Q. et al. A Phytophthora capsici virulence effector associates with NPR1 and suppresses plant immune responses. Phytopathol. Res.1, 1–11 (2019).
- 7.Pennington, H. G. et al. The fungal ribonuclease-like effector protein CSEP0064/BEC1054 represses plant immunity and interferes with degradation of host ribosomal RNA. PLoS Pathog.15, e1007620 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qin, J. et al. The plant-specific transcription factors CBP60g and SARD1 are targeted by a Verticillium secretory protein VdSCP41 to modulate immunity. eLife7, e34902 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Redkar, A., Villajuana-Bonequi, M. & Doehlemann, G. Conservation of the Ustilago maydis effector See1 in related smuts. Plant Signal. Behav.10, e1086855 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh, S. K. et al. The nuclear effector ArPEC25 from the necrotrophic fungus Ascochyta rabiei targets the chickpea transcription factor CaβLIM1a and negatively modulates lignin biosynthesis for host susceptibility. Plant Cell35, 1134–1159 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu, C. et al. A fungal effector suppresses the nuclear export of AGO1–miRNA complex to promote infection in plants. PNAS119, e2114583119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu, J. et al. Types of nuclear localization signals and mechanisms of protein import into the nucleus. Cell Commun. Signal.19, 1–10 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cautain, B., Hill, R., de Pedro, N. & Link, W. Components and regulation of nuclear transport processes. FEBS J.282, 445–462 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harreman, M. T. et al. Regulation of nuclear import by phosphorylation adjacent to nuclear localization signals. J. Biol. Chem.279, 20613–20621 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Nardozzi, J. D., Lott, K. & Cingolani, G. Phosphorylation meets nuclear import: a review. Cell Commun. Signal.8, 1–17 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujimoto, H., Ikuta, T., Koike, A. & Koike, M. Acetylation of nuclear localization signal controls importin-mediated nuclear transport of Ku70. Biochem. Biophys. Rep.33, 101418 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hofweber, M. et al. Phase separation of FUS is suppressed by its nuclear import receptor and arginine methylation. Cell173, 706–719 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Park, I. S. et al. SUMOylation regulates nuclear localization and stability of TRAIP/RNF206. Biochem. Biophys. Res. Commun.470, 881–887 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Yao, F. et al. SKP2-and OTUD1-regulated non-proteolytic ubiquitination of YAP promotes YAP nuclear localization and activity. Nat. Commun.9, 2269 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van den Ackerveken, G., Marois, E. & Bonas, U. Recognition of the bacterial avirulence protein AvrBs3 occurs inside the host plant cell. Cell87, 1307–1316 (1996). [DOI] [PubMed] [Google Scholar]
- 21.Liu, L. et al. Verticillium dahliae secreted protein Vd424Y is required for full virulence, targets the nucleus of plant cells, and induces cell death. Mol. Plant Pathol.22, 1109–1120 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petre, B. et al. Heterologous expression screens in Nicotiana benthamiana identify a candidate effector of the wheat yellow rust pathogen that associates with processing bodies. PloS ONE11, e0149035 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang, L. et al. The Verticillium‐specific protein VdSCP7 localizes to the plant nucleus and modulates immunity to fungal infections. New Phytol.215, 368–381 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Ahmed, M. B. et al. A rust fungal effector binds plant DNA and modulates transcription. Sci. Rep.8, 14718 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fernandez, J. & Orth, K. Rise of a cereal killer: the biology of Magnaporthe oryzae biotrophic growth. Trends Microbiol.26, 582–597 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh, P. K. et al. Wheat blast: a disease spreading by intercontinental jumps and its management strategies. Front. Plant Sci. 12, 710707 (2021). [DOI] [PMC free article] [PubMed]
- 27.Dean, R. A. et al. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature434, 980–986 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Dong, Y. et al. Global genome and transcriptome analyses of Magnaporthe oryzae epidemic isolate 98-06 uncover novel effectors and pathogenicity-related genes, revealing gene gain and lose dynamics in genome evolution. PLoS Pathog.11, e1004801 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eckardt, N. A. Sequencing the rice genome. Plant Cell12, 2011–2017 (2000). (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeon, J. et al. Transcriptome profiling of the rice blast fungus Magnaporthe oryzae and its host Oryza sativa during infection. MPMI33, 141–144 (2020). [DOI] [PubMed] [Google Scholar]
- 31.Zhang, S. & Xu, J.-R. Effectors and effector delivery in Magnaporthe oryzae. PLoS Pathog.10, e1003826 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.You, H. S., Ok, Y. J., Lee, E. J., Kang, S. S. & Hyun, S. H. Development of a novel DsRed-NLS vector with a monopartite classical nuclear localization signal. 3 Biotech9, 1–10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khang, C. H. et al. Translocation of Magnaporthe oryzae effectors into rice cells and their subsequent cell-to-cell movement. Plant Cell22, 1388–1403 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao, Q. et al. GPS-SUMO: a tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res.42, W325–W330 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu, Z., Xiong, D., Han, Z. & Tian, C. A putative effector CcSp84 of Cytospora chrysosperma localizes to the plant nucleus to trigger plant immunity. Int. J. Mol. Sci.23, 1614 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roeschlin, R. A. et al. PthA4AT, a 7.5‐repeats transcription activator‐like (TAL) effector from Xanthomonas citri ssp. citri, triggers citrus canker resistance. Mol. Plant Pathol.20, 1394–1407 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ji, Z. et al. Interfering TAL effectors of Xanthomonas oryzae neutralize R-gene-mediated plant disease resistance. Nat. Commun.7, 13435 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee, S. et al. The nuclear effector MoHTR3 of Magnaporthe oryzae modulates host defence signalling in the biotrophic stage of rice infection. Mol. Plant Pathol.24, 602–615 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peng, J. et al. Editing of the rice importin gene IMPα1b results in sequestration of TAL effectors from plant cell nuclei. Phytopathol. Res.4, 1–16 (2022). [Google Scholar]
- 40.Oliveira-Garcia, E. et al. Clathrin-mediated endocytosis facilitates internalization of Magnaporthe oryzae effectors into rice cells. Plant Cell35, 2527–2551 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sharma, S. et al. Deployment of the Burkholderia glumae type III secretion system as an efficient tool for translocating pathogen effectors to monocot cells. Plant J.74, 701–712 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Jamali, T., Jamali, Y., Mehrbod, M. & Mofrad, M. Nuclear pore complex: biochemistry and biophysics of nucleocytoplasmic transport in health and disease. Int. Rev. Cell Mol. Biol.287, 233–286 (2011). [DOI] [PubMed] [Google Scholar]
- 43.Catalano, A. & O’Day, D. H. Nucleoplasmic/nucleolar translocation and identification of a nuclear localization signal (NLS) in Dictyostelium BAF60a/SMARCD1 homologue Snf12. Histochem. Cell Biol.138, 515–530 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Anderson, J. C., Pascuzzi, P. E., Xiao, F., Sessa, G. & Martin, G. B. Host-mediated phosphorylation of type III effector AvrPto promotes Pseudomonas virulence and avirulence in tomato. Plant Cell18, 502–514 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lei, L., Stevens, D. M. & Coaker, G. Phosphorylation of the Pseudomonas effector AvrPtoB by Arabidopsis SnRK2. 8 is required for bacterial virulence. Mol. Plant13, 1513–1522 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li, J. et al. Acetylation of a fungal effector that translocates host PR1 facilitates virulence. eLife11, e82628 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lim, Y.-J., Kim, K.-T. & Lee, Y.-H. SUMOylation is required for fungal development and pathogenicity in the rice blast fungus Magnaporthe oryzae. Mol. Plant Pathol.19, 2134–2148 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu, C. et al. Global analysis of sumoylation function reveals novel insights into development and appressorium‐mediated infection of the rice blast fungus. New Phytol.219, 1031–1047 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Xu, Q. et al. An effector protein of the wheat stripe rust fungus targets chloroplasts and suppresses chloroplast function. Nat. Commun.10, 5571 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Horton, P. et al. WoLF PSORT: protein localization predictor. Nucleic Acids Res.35, W585–W587 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kosugi, S., Hasebe, M., Tomita, M. & Yanagawa, H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. PNAS106, 10171–10176 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nguyen Ba, A. N., Pogoutse, A., Provart, N. & Moses, A. M. NLStradamus: a simple Hidden Markov Model for nuclear localization signal prediction. BMC Bioinform.10, 1–11 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Evans, R. et al. Protein complex prediction with AlphaFold-multimer. Preprint at bioRxivhttps://www.biorxiv.org/content/10.1101/2021.10.04.463034v2 (2021).
- 54.Elfmann, C. & Stülke, J. PAE viewer: a webserver for the interactive visualization of the predicted aligned error for multimer structure predictions and crosslinks. Nucleic Acids Res.51, W404–W410 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang, Y. et al. A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods7, 1–14 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Karimi, M., Inzé, D. & Depicker, A. GATEWAY™ vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci.7, 193–195 (2002). [DOI] [PubMed] [Google Scholar]
- 57.Gong, X. et al. pFPL vectors for high-throughput protein localization in fungi: detecting cytoplasmic accumulation of putative effector proteins. MPMI28, 107–121 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of additional supplementary files
Data Availability Statement
Data that support the findings of this study are available in the paper and its Supplementary information files. The datasets and fungal materials generated and analyzed during the current study are available from the corresponding author upon request. The RNA-seq data generated in this study has been deposited in the NCBI Sequence Read Archive under accession code PRJNA1103247. The processed RNA-seq data are available at NCBI Sequence Read Archive. The RNA-seq data generated in this study are provided in the Supplementary information. The RNA-seq data used in this study are available in the NCBI Sequence Read Archive under accession code PRJNA1103247. Source data for main figures and Supplementary Figs. are provided with this paper. Source data are provided with this paper.