Abstract
Key Points
Patients with both COL4A3 and COL4A4 variants exhibited poor renal prognosis compared with those with autosomal dominant Alport syndrome.
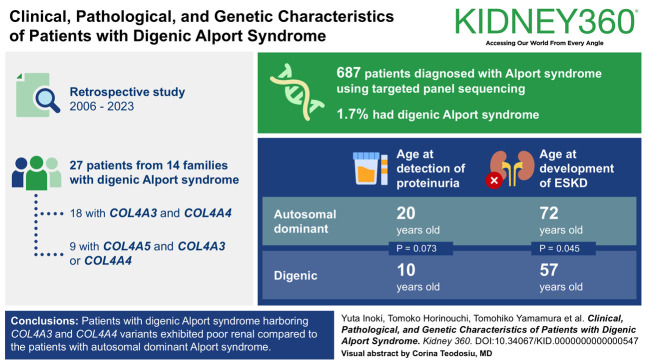
The proportion of patients with digenic Alport syndrome was 1.7% among all patients with Alport syndrome.
Background
Digenic Alport syndrome could be associated with poor renal prognosis. However, the characteristics of patients with digenic Alport syndrome remain ambiguous.
Methods
We retrospectively investigated the clinical symptoms, pathological findings, genetic variants, and proportions of patients with digenic Alport syndrome. The ages at detection of proteinuria and development of ESKD were compared between patients with digenic Alport syndrome with disease-causing variants in COL4A3 and COL4A4 and those with autosomal dominant Alport syndrome (ADAS) previously analyzed by our group.
Results
Eighteen patients from nine families with digenic variants in COL4A3 and COL4A4 and four male and five female patients with digenic variants in COL4A5 and COL4A3 or COL4A4 were enrolled in this study. Next-generation sequencing revealed that the proportion of patients with digenic Alport syndrome was 1.7% among all patients with Alport syndrome. In patients with digenic variants in COL4A3 and COL4A4, the median ages at detection of proteinuria and ESKD were 10.0 and 57.0 years, respectively. Compared with the patients with ADAS, the age at detection of proteinuria tended to be earlier (10.0 versus 20.0 years; P = 0.073) and that at development of ESKD was significantly earlier (57.0 versus 72.0 years; P = 0.045) in patients with digenic Alport syndrome.
Conclusions
Overall, patients with digenic Alport syndrome harboring COL4A3 and COL4A4 variants exhibited poor renal compared with the patients with ADAS. Therefore, timely identification of the two disease-causing variants is critical for the renal prognostic assessment and early treatment of patients with digenic Alport syndrome.
Keywords: Alport syndrome, ESKD, genetic renal disease
Visual Abstract
Introduction
Alport syndrome is an inherited kidney disease characterized by hematuria with proteinuria and renal dysfunction in progressive stages, hearing loss, and ocular abnormalities.1–3 Alport syndrome is caused by disease-causing variants in COL4A3, COL4A4, and COL4A5, which encode α3, α4, and α5 chains of type 4 collagen, respectively. The inheritance patterns of Alport syndrome include X-linked Alport syndrome (XLAS; OMIM: 301050), autosomal recessive Alport syndrome (ARAS; OMIM: 203780), and autosomal dominant Alport syndrome (ADAS; OMIM: 104200). We have previously revealed that renal prognosis is influenced by the inheritance pattern, and the median age at the development of ESKD was reported to be 35 years in men with XLAS,4 65 years in women with XLAS,5 21 years in ARAS,6 and 70 years in ADAS,7 respectively. In addition, two disease-causing variants of the COL4A3–COL4A5 genes have recently been recognized to cause digenic Alport syndrome.8,9
Many digenic variants may have been previously overlooked because laboratories often did not extend their analysis beyond identifying a single disease-causing variant, particularly before the widespread adoption of targeted panel sequencing technology.9 In recent years, the identification of digenic Alport syndrome cases has increased with the widespread use of targeted panel sequencing.7,8,10–14 Digenic Alport syndrome results from a disease-causing variant in COL4A5 combined with one in either COL4A3 or COL4A4 or disease-causing variants in COL4A3 and COL4A4. The renal prognosis in XLAS varies according to sex and variant type. Therefore, the clinical presentation, modes of inheritance, and frequency of digenic Alport syndrome should be considered separately according to the type of disease-causing variant: COL4A3 and COL4A4, COL4A5, and COL4A3 or COL4A4 in men and COL4A5 and COL4A3 or COL4A4 in women.9
Previous studies have suggested that digenic Alport syndrome may be associated with poor renal prognosis.9,14 However, limited data exist on the clinical presentation, pathology, and genotype-phenotype correlation in patients with digenic Alport syndrome. Furthermore, there are few reports on its prevalence in patients with Alport syndrome. In this study, we investigated the prevalence of digenic Alport syndrome in patients previously diagnosed with Alport syndrome through genetic analysis of large cohorts accumulated by our research group over several years. We also assessed the clinical characteristics, pathological findings, and variant patterns of patients with digenic Alport syndrome and their family members, to evaluate the potential association of a digenic disease-causing variant with a severe phenotype. Moreover, we conducted a comparative analysis of the age at the onset of proteinuria and renal prognosis between patients with digenic Alport syndrome (carrying two disease-causing variants in COL4A3 and COL4A4) and those with ADAS (carrying a single disease-causing variant in either COL4A3 or COL4A4) within the same cohort.
Methods
Diagnostic Criteria for Digenic Alport Syndrome
Digenic Alport syndrome in the study was defined as fulfilling one of the following criteria in accordance with previous study7,9: (1) Patients with clinical symptoms (hematuria, proteinuria, or renal dysfunction), along with renal pathology demonstrating abnormal glomerular basement membrane (GBM) and presence of two disease-causing variants located in different genes of COL4A3, COL4A4, or COL4A5; (2) patients with clinical symptoms, a family history of Alport syndrome or ESKD, and two disease-causing variants located in different genes of COL4A3, COL4A4, or COL4A5; and (3) among families of patients diagnosed using criteria (1) or (2), those who met the following subcategories: (1) symptomatic family members with two different disease-causing variants or (2) symptomatic family members of patients with disease-causing variants in COL4A3 and COL4A4 on the same allele (in cis) who have not undergone any genetic tests.
Study Design and Patient Population
In this retrospective observational study, patients with suspected Alport syndrome were referred to our hospital for genetic analysis between January 2006 and January 2023. A total of 27 patients from 14 families who met the diagnostic criteria for digenic Alport syndrome were included in this study.
Data Collection and Analysis
At the time of genetic analysis, the following data were extracted from medical records: (1) clinical data, including sex, initial symptoms, height, body wt, family history, age at detection of proteinuria, age at the development of ESKD, presence of hearing loss or eye lesions, and therapeutic interventions; (2) laboratory data, including serum creatinine levels, albumin levels, total cholesterol levels, urine protein-creatinine ratio, and urine red blood cell count; (3) pathological findings; and (4) family history details, including the age at the detection of proteinuria, age at the development of ESKD, and the presence of hearing loss or eye lesions. Age at ESKD onset was defined as the time of initiation of KRT. The age at detection of proteinuria was defined as the point when the protein/creatinine ratio exceeded 0.2 g/g Cre in the first morning urine. Cr-eGFR, measured in ml/min per 1.73 m2, was calculated using an equation designed for Japanese patients age between 2 and 18 years15 and a different equation for Japanese patients age 19 years and older.16
Patients were classified into three groups based on their genetic variants and sex: patients with disease-causing variants in COL4A3 and COL4A4, male patients with disease-causing variants in COL4A5 and COL4A3 or COL4A4, and female patients with disease-causing variants in COL4A5 and COL4A3 or COL4A4. We investigated the clinical symptoms, pathological findings, and genetic variants in these patients. In addition, the detection of proteinuria and development of ESKD were compared using the Kaplan–Meier method between patients with digenic Alport syndrome with disease-causing variants in COL4A3 and COL4A4 and previously reported patients with ADAS in our group.7 We also investigated the proportion of patients diagnosed with digenic Alport syndrome among all patients diagnosed with Alport syndrome via targeted panel sequencing analysis at our hospital.
Genetic Analysis
Genomic DNA was isolated from the peripheral blood leukocytes of patients using the Quick Gene Mini 80 System or QuickGene-Auto 12S (Kurabo Industries Ltd., Tokyo, Japan), according to the manufacturer's instructions. Mutational analyses of COL4A3, COL4A4, and COL4A5 were performed using Sanger sequencing from January 2006 to November 2015. Targeted panel sequencing, which was engineered custom panel, for genes associated with inherited kidney diseases, including COL4A3, COL4A4, and COL4A5 from November 2015 to January 2023 (Supplemental Table 1). If heterozygous variants were not identified via Sanger sequencing or targeted panel sequencing, additional confirmation was performed using either multiplex ligation–dependent probe amplification or RT-PCR analysis of mRNA. Sanger sequencing, targeted panel sequencing, multiplex ligation–dependent probe amplification, and PCR analysis of mRNA were performed as previously described.4
Variant Evaluation
Variants were classified according to the American College of Medical Genetics (ACMG) consensus guidelines.17,18 In this study, disease-causing variants were specifically defined based on the ACMG guidelines as pathogenic or likely pathogenic. To evaluate variant pathogenicity, in silico analyses were conducted using the following prediction tools: Sorting Intolerant from Tolerant (SIFT; http://sift.bii.a-star.edu.sg), PolyPhen2 (http://genetics.bwh.harvard.edu/pph2/), Mutation Taster (https://www.mutationtaster.org/), and Align GVGD (http://agvgd.iarc.fr). Each variant was also investigated to determine whether its heterozygous variant could cause urinary abnormality using the Human Gene Mutation Database variant database (https://digitalinsights.qiagen.com/products-overview/clinical-insights-portfolio/human-gene-mutation-database/).
Statistical Analyses
Data were presented as medians with interquartile ranges (IQRs) for continuous variables and percentages for categorical variables. The occurrence of events (age at detection of proteinuria and age at onset of ESKD) was analyzed using the Kaplan–Meier method. The log-rank test was used to calculate the P values. A P value of <0.05 was considered statistically significant. All statistical analyses were performed using JMP version 14.0 (SAS Institute Japan, Tokyo, Japan).
Ethical Considerations
All procedures in this study were conducted according to the ethical standards of the Institutional Review Board of Kobe University School of Medicine (approval number: 301), the principles of the Declaration of Helsinki, and ethical guidelines of the Japanese Ministry of Health, Labor, and Welfare. Informed consent was obtained from all patients and/or their parents.
Results
Patients' Characteristics
During the study period, 961 patients with Alport syndrome were identified, 687 of whom were diagnosed using targeted panel sequencing. The clinical features of 27 patients from 14 families diagnosed with digenic Alport syndrome are shown in Table 1. Among them, 18 patients from nine families carried disease-causing variants in COL4A3 and COL4A4, four male patients in COL4A5, and COL4A3 or COL4A4, and five female patients in COL4A5 and COL4A3 or COL4A4. The median age of genetic analysis was 34.0 years (IQR, 29.0–51.0 years old).
Table 1.
Clinical findings
| Patient ID | Age (yr) | Sex | Family Member | Height (cm) | Body Wt (kg) | Proteinuria Detected Age (yr) | ESKD Detected Age (yr) | Hearing Loss Detected Age (yr) | Ocular Lesion Detected Age (yr) | sCr, mg/dl (eGFR, ml/min per 1.73 m2) | sAlb (g/dl) | U-P/Cre (g/g) | Treatment | Variants | In Cis or In Trans | No. of Truncating Alleles |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| COL4A3 +COL4A4 | ||||||||||||||||
| 122 | 35 | M | 183 | 66 | 9 | — | — | — | 1.27 (53.8) | 4.3 | 0.45 | ARB | COL4A3+COL4A4 | In cis | 0 | |
| 122-1 | 39 | W | Sister | 156 | 51 | 2 | — | — | — | N/A | N/A | N/A | N/A | COL4A3+COL4A4 | In cis | 0 |
| 122-3 | 63 | M | Father | 163 | 60 | 15 | 57 | — | — | ESKD | ESKD | ESKD | ESKD | COL4A3+COL4A4 | In cis | 0 |
| 122-4 | Dead | W | Paternal sister | N/A | N/A | N/A | 20 | — | — | ESKD | ESKD | ESKD | ESKD | COL4A3+COL4A4 | In cis | 0 |
| 140 | 16 | M | 180.4 | 105.3 | 6 | — | — | — | 0.81 (80.9) | 4.2 | 0.92 | ACE-I | COL4A3+COL4A4 | In cis | 0 | |
| 140-1 | 57 | M | Father | 172 | 89.8 | › | 33 | — | — | ESKD | ESKD | ESKD | ESKD | COL4A3+COL4A4 | In cis | 0 |
| 140-2 | 70 | W | Paternal grandmother | N/A | N/A | N/A | 70 | N/A | N/A | ESKD | ESKD | ESKD | ESKD | COL4A3+COL4A4 | In cis | 0 |
| 503 | 51 | W | 162.2 | 58.5 | N/A | — | 33 | — | 1.43 (31.2) | 4.2 | 2.78 | ARB | COL4A3+COL4A4 | N/A | 0 | |
| 719 | 48 | M | N/A | N/A | 12 | — | — | N/A | 1.8 | 3.8 | 1.9 | ARB | COL4A3+COL4A4 | N/A | 1 | |
| 758 | 29 | W | 154 | 51 | — | — | — | — | 0.68 (83.2) | 4.2 | 0.03 | — | COL4A3+COL4A4 | In cis | 0 | |
| 758-1 | 34 | M | Brother | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | 0.54 | N/A | COL4A3+COL4A4 | In cis | 0 |
| 758-2 | 65 | M | Father | N/A | N/A | N/A | 40 | N/A | N/A | ESKD | ESKD | ESKD | ESKD | COL4A3+COL4A4 | In cis | 0 |
| 817 | 5 | W | 102.3 | 15.5 | — | — | — | — | 0.35 (100.5) | 4.5 | 0.13 | — | COL4A3+COL4A4 | In cis | 0 | |
| 817-1 | 30 | M | Father | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | COL4A3+COL4A4 | In cis | 0 |
| 817-2 | 71 | M | Paternal grandfather | N/A | N/A | N/A | 50 | N/A | N/A | ESKD | ESKD | ESKD | ESKD | COL4A3+COL4A4 | In cis | 0 |
| 1092 | 40 | M | 173 | 67 | 4 | — | — | — | 1.22 (54.1) | 4.1 | 0.67 | — | COL4A3+COL4A4 | N/A | 0 | |
| 1140 | 60 | W | 153.6 | 48 | 38 | — | — | — | 1.08 (40.7) | 3.7 | 1.59 | ARB | COL4A3+COL4A4 | N/A | 0 | |
| 1154 | 57 | W | 159 | 53 | 10 | — | — | — | 0.74 (62.5) | 3.9 | 2.14 | ARB | COL4A3+COL4A4 | in trans | 1 | |
| COL4A5 +COL4A3 or COL4A4 | ||||||||||||||||
| 967 | 35 | M | 168 | 58 | 4 | 27 | — | — | ESKD | ESKD | ESKD | ESKD | COL4A3+COL4A5 | |||
| 967-1 | 29 | M | Brother | 178 | 55 | 6 | — | — | — | 0.99 (74.6) | 3.4 | 0.3 | ACE-I | COL4A3+COL4A5 | ||
| 1062 | 26 | M | 170 | 62.8 | 11 | — | — | — | 0.90 (86) | 4.3 | 1.52 | ARB | COL4A4+COL4A5 | |||
| 1124 | 7 | M | 114 | 23.4 | 3 | — | — | — | 0.23 (165) | 4.8 | 0.12 | ACE-I | COL4A4+COL4A5 | |||
| 344 | 11 | W | 138 | 36.6 | 4.5 | — | — | — | 0.28 (171.1) | 3.5 | 0.84 | ARB | COL4A3+COL4A5 | |||
| 690 | 30 | W | 165 | 62.5 | N/A | — | — | — | 0.54 (106) | 4.2 | 0.33 | — | COL4A4+COL4A5 | |||
| 690-1 | N/A | W | Mother | N/A | N/A | N/A | 27 | + | —2 | N/A | N/A | N/A | N/A | COL4A4+COL4A5 | ||
| 1062-3 | 30 | W | Sister | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | COL4A4+COL4A5 | ||
| 1124-2 | 33 | W | Mother | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | COL4A4+COL4A5 |
In cis refers to two disease-causing variants on the same chromosome, whereas in trans refers to those on opposite chromosomes. ACE-I, angiotensin-converting enzyme inhibitor; ARB, angiotensin receptor blocker; ID, identification; M, man or boy; N/A, not applicable; sAlb, serum albumin; sCr, serum creatinine; U-P/Cre, urinary protein-to-creatinine ratio; W, woman or girl.
Among all patients diagnosed with Alport syndrome using targeted panel sequencing analysis at our hospital, the proportion of patients with digenic syndrome was 1.7% (12/687). Of 406 patients with a disease-causing variant in COL4A5, five patients (1.2%) had additional COL4A3 or COL4A4 disease-causing variants. Of 248 patients with pathogenic heterozygous variants in COL4A3 or COL4A4, seven patients (2.8%) had an additional COL4A3 or COL4A4 disease-causing variant.
Patients with Disease-Causing Variants in COL4A3 and COL4A4
The median age at proteinuria detection was 10.0 years. Varying degree of kidney dysfunction was observed. Six patients developed ESKD, with a median age that ESKD of 57.0 years. No patients exhibited hearing loss or eye lesions.
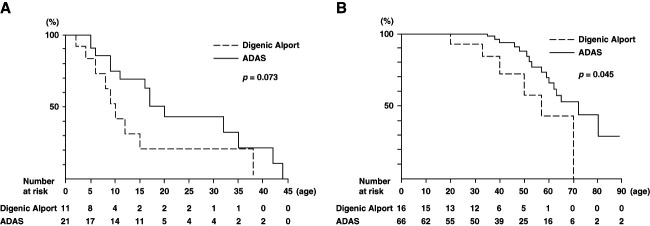
Among previously reported patients with ADAS in our group,7 two patients (patient identification; 120,141) had digenic variants and were consequently included in the digenic Alport syndrome group in the present investigation. Therefore, the ADAS group (referred to as the ADAS group) comprised 18 families. The age at detection of proteinuria in patients with a disease-causing variant in COL4A3 and COL4A4 tended to be earlier than patients of ADAS group, although there was no significant difference between two groups (10.0 versus 20.0 years; P = 0.073; Figure 1A). The age at the development of ESKD in patient with a disease-causing variant in COL4A3 and COL4A4 was significantly earlier than in the ADAS group (57.0 versus 72.0 years; P = 0.045; Figure 1B). In addition, we combined the data of our results and those of the recent review study of digenic Alport syndrome9 and analyzed the age at the development of ESKD compared with ADAS patients in our cohort. There was also a significant difference between the two groups (55.0 versus 72.0 years; P = 0.011; Supplemental Figure 1).
Figure 1.
Probability of each clinical symptom in Alport syndrome cases with digenic COL4A3 and COL4A4 variants compared with that in ADAS cases. (A) Probability of occurrence of proteinuria. No significant differences were observed between the digenic and ADAS cases (P = 0.073). (B) Probability of developing ESKD. Significant differences were observed between the digenic and ADAS cases (P = 0.045). ADAS, autosomal dominant Alport syndrome.
Patients with Disease-Causing Variants in COL4A5 and COL4A3 or COL4A4
The median age at detection of proteinuria was 5 years in men, whereas one female patient developed proteinuria at age 4.5 years. Two male patients developed ESKD at age 27 years. At the time of genetic analysis, a female patient with disease-causing variants in COL4A5 and COL4A3 or COL4A4 had hearing loss. No male patients exhibited hearing loss or eye lesions.
Pathological Findings
Table 2 shows the pathological findings of patients with digenic Alport syndrome for whom information was available. Renal biopsy was performed in seven patients with COL4A3 and COL4A4 variants and in four patients with COL4A5 and COL4A3 or COL4A4 variants. The median age at renal biopsy was 32.5 years (IQR, 18.3–40.0). Light microscopy revealed that three patients had FSGS, three had diffuse mesangial proliferation, and five had minimal glomerular changes. Electron microscopy revealed thin basement membrane in all 11 patients, whereas basket-weave changes (BWCs) and lamellation were observed in two patients. Immunohistochemical analysis of type 4 collagen α5 chain revealed normal expression in all nine patients who underwent examination.
Table 2.
Pathologic findings
| Patient ID | Digenic Variants | Age at Biopsy (yr) | Sex | Light Microscopy | Immunofluorescence Staining | Electron Microscopy | α5 Staining |
|---|---|---|---|---|---|---|---|
| 122 | COL4A3+COL4A4 | 35 | Man | DMP | Negative | TBM | Positive |
| 140 | COL4A3+COL4A4 | 16 | Man | FSGS | IgM | TBM | Positive |
| 503 | COL4A3+COL4A4 | 50 | Woman | MGA | IgA, IgM, C1q | TBM | Positive |
| 758 | COL4A3+COL4A4 | 28 | Woman | MGA | C3 | TBM | Positive |
| 1092 | COL4A3+COL4A4 | 40 | Man | FSGS | IgM, C3 | TBM, BWC, lamellation | Positive |
| 1140 | COL4A3+COL4A4 | 40 | Woman | MGA | Negative | TBM | N/A |
| 1154 | COL4A3+COL4A4 | 52 | Woman | MGA | Negative | TBM | Positive |
| 344 | COL4A3+COL4A5 | 11 | Woman | DMP | IgA, IgM | TBM | Positive |
| 690 | COL4A4+COL4A5 | 30 | Woman | DMP | Negative | TBM | Positive |
| 1062 | COL4A4+COL4A5 | 25 | Man | FSGS | IgA, IgG | TBM, BWC, lamellation | N/A |
| 1124 | COL4A4+COL4A5 | 6 | Man | MGA | Negative | TBM | Positive |
BWC, basket-weave changes; DMP, diffuse mesangial proliferation; MGA, minimal glomerular abnormality; TBM, thin basement membrane.
Detected Variants
The detected disease-causing variants are listed in Table 3. Six novel variants were identified in the five patients. Following the ACMG criteria modified by Alport experts,18 three variants were classified as pathogenic while the remaining 16 variants as likely pathogenic. All variants were located in the collagenic domain and all missense variants resulted in glycine substitutions.
Table 3.
Genetic findings
| Patient ID | Gene | Exon | Nucleotide Change | Amino Acid Change | Type | ACMG Classification |
|---|---|---|---|---|---|---|
| 122 | COL4A3 | 47 | c.4217G>A | p.Gly1406Glu | Missense | Likely pathogenic (PM1, PM2, PP2, PP3) |
| COL4A4 | 32 | c.2869G>A | p.Gly957Asp | Missense | Likely pathogenic (PM1, PM2, PP2, PP3) | |
| 140 | COL4A3 | 47 | c.4217G>A | p.Gly1406Glu | Missense | Likely pathogenic (PM1, PM2, PP2, PP3) |
| COL4A4 | 32 | c.2869G>A | p.Gly957Asp | Missense | Likely pathogenic (PM1, PM2, PP2, PP3) | |
| 503 | COL4A3 | 9 | c.469G>C | p.Gly157Arg | Missense | Likely pathogenic (PM1, PM2, PP2, PP3) |
| COL4A4 | 29 | c.2510G>C | p.Gly837ALa | Missense | Likely pathogenic (PM1, PM2, PP2, PP3, PP5) | |
| 719 | COL4A3 | 29-33 del | Large deletion | Pathogenic (PVS1, PM2, PP3) | ||
| COL4A4 | 36 | C.3307G>A | p.Gly1103Arg | Missense | Likely pathogenic (PM1, PM2, PP2, PP3) | |
| 758 | COL4A3 | 9 | c.469G>C | p.Gly157Arg | Missense | Likely pathogenic (PM1, PM2, PP2, PP3) |
| COL4A4 | 29 | c.2510G>C | p.Gly837ALa | Missense | Likely pathogenic (PM1, PM2, PP2, PP3, PP5) | |
| 817 | COL4A3 | 47 | c.4217G>A | p.Gly1406Glu | Missense | Likely pathogenic (PM1, PM2, PP2, PP3) |
| COL4A4 | 32 | c.2869G>A | p.Gly957Asp | Missense | Likely pathogenic (PM1, PM2, PP2, PP3) | |
| 1092 | COL4A3 | 35 | c.2962G>Ca | p.Gly988Arg | Missense | Likely pathogenic (PM1, PM2, PP2, PP3) |
| COL4A4 | 11 | c.677G>A | p.Gly226Glu | Missense | Likely pathogenic (PM1, PM2, PP2, PP3) | |
| 1140 | COL4A3 | 9 | c.469G>C | p.Gly157Arg | Missense | Likely pathogenic (PM1, PM2, PP2, PP3) |
| COL4A4 | 29 | c.2510G>C | p.Gly837ALa | Missense | Likely pathogenic (PM1, PM2, PP2, PP3, PP5) | |
| 1154 | COL4A3 | 40 | c.3427G>A | p.Gly1143Arg | Missense | Likely pathogenic (PM1, PM2, PP2, PP3) |
| COL4A4 | 37 | c3506-1G>Ca | Splicing | Pathogenic (PVS1, PM2, PP3) | ||
| 344 | COL4A5 | 32 | c.2732G>A | p.Gly911Glu | Missense | Likely pathogenic (PM1, PM2, PP2, PP3) |
| COL4A3 | 42 | c.3691G>Aa | p.Gly1231Ser | Missense | Likely pathogenic (PM1, PM2, PP2, PP3) | |
| 690 | COL4A5 | 41 | c.3764G>Aa | p.Gly1255Glu | Missense | Likely pathogenic (PM1, PM2, PP2, PP3) |
| COL4A4 | 46 | c.4440C>Aa | p.Cys1480a | Nonsense | Pathogenic (PVS1, PM2, PP5) | |
| 967 | COL4A5 | c.3107-4A>G | Splice-region | Likely pathogenic (PM2, PM4, PP3, PP5) | ||
| COL4A3 | 34 | c.2863G>A | p.Gly955Arg | Missense | Likely pathogenic (PM1, PM2, PP2, PP3) | |
| 1062 | COL4A5 | 47 | c.4351G>Ca | p.Gly1451Arg | Missense | Likely pathogenic (PM1, PM2, PP2, PP3) |
| COL4A4 | 14 | c.827G>C | p.Gly276Ala | Missense | Likely pathogenic (PM1, PM2, PP2, PP3) | |
| 1124 | COL4A5 | 19 | c1075G>C | p.Gly359Arg | Missense | Likely pathogenic (PM1, PM2, PP2, PP3, PP5) |
| COL4A4 | 14 | c.827G>C | p.Gly276Ala | Missense | Likely pathogenic (PM1, PM2, PP2, PP3) |
ACMG, American College of Medical Genetics.
Novel mutation.
Among the patients with disease-causing variants in COL4A3 and COL4A4, seven families had two missense variants, one had a splice site variant and a missense variant, and one had a large deletion and a missense variant. Three families had the same variant (c.4217G>A in COL4A3 and c.2869G>A in COL4A4), whereas the other three families had the same variant (c.469G>C in COL4A3 and c.2510G>C in COL4A4). Two disease-causing variants were detected on the same allele (in cis) in four families and on the opposite allele (in trans) in one family. Among patients with disease-causing variants in COL4A5 and COL4A3 or COL4A4, three families had two missense variants, one had a nonsense variant and a missense variant, and one had a splice-site variant and a missense variant. One female patient (patient identification: 344) had a somatic mosaic variant in COL4A5 with variant frequency 22.1%, as previously reported by our group.12
Genotype–Phenotype Correlation in Patients with Disease-Causing Variants in COL4A3 and COL4A4
Among the patients with disease-causing variants in COL4A3 and COL4A4, two families (719 and 1154) had one truncating variant, whereas the other families had no truncating variants. There was no significant difference in the age at proteinuria detection between patients with and without one truncating variant (P = 0.78). Two patients with one truncating variant did not develop ESKD at age 40 and 50 years, and no significant difference was observed with or without truncating variants (P = 0.53). Furthermore, there was no significant difference in the age of proteinuria detection and the development of ESKD between patients with two variants on the same allele (in cis) and those on the opposite allele (in trans) (P = 0.65 and 0.93, respectively).
Discussion
This study investigated the clinical presentation, pathological findings, and variant patterns in patients with digenic Alport syndrome. A total of 27 patients from 14 families were included in the study, comprising 18 patients from nine families with disease-causing variants in COL4A3 and COL4A4, four male patients in COL4A5 and COL4A3 or COL4A4, and five female patients in COL4A5 and COL4A3 or COL4A4. The proportion of digenic Alport syndrome among Alport syndrome was 1.7%. To our knowledge, this cohort represents one of the largest studies of patients with digenic Alport syndrome to date.7,8,10–14 Furthermore, this is the first study to elucidate the proportion of patients with digenic Alport syndrome, compare the renal prognosis of patients with COL4A3 and COL4A4 variants with those of ADAS patients within the same cohort, investigate the pathological findings in patients with COL4A3 and COL4A4 variants, and explore the genotype–phenotype relationship in patients with COL4A3 and COL4A4 variants.
In our study, using targeted panel sequencing analysis, another disease-causing variant was found in 1.2% of patients with XLAS and 2.8% of patients with ADAS. A recent study reported that approximately 1% (6/417) of patients with Alport syndrome were identified with disease-causing variants in COL4A5 and COL4A3 or COL4A4, which was consistent with our results.14 However, no reports have investigated the percentage of patients with XLAS with digenic variants or the percentage of patients with ADAS with digenic variants. A previous large population-based study showed that the frequency of disease-causing variants in COL4A5 was approximately one in 2000 and that for COL4A3 or COL4A4 was one in 100.19 Considering these frequencies, approximately 1% of patients with XLAS could have disease-causing variants in COL4A3 or COL4A4 if inheritance of each variant occurs independently. Similarly, considering a frequency of one in 200 for the COL4A3 variant and one in 200 for the COL4A4, approximately 0.5% of patients with ADAS could have the other variant of COL4A3 or COL4A4.9 Although our results for the frequency of COL4A5 and COL4A3 or COL4A4 variant were in line with this estimation, that of COL4A3 and COL4A4 was higher than the frequency estimated from population-based study. Our study included patients with urinary abnormalities, whereas a population-based study19 also included asymptomatic patients without hematuria. Thus, our study may have included patients with two variants resulting in urinary abnormalities, even if each disease-causing variant did not independently result in urinary abnormalities. In addition, owing to racial differences from a population-based study,19 it is possible that a higher percentage of Japanese patients, who are the patients in this study, have a variant in COL4A3 or COL4A4 classified as pathogenic or likely pathogenic by ACMG. The discrepancy in the prevalence may be due to the facts mentioned above, the nonindependent inheritance of two disease-causing variants on the same allele (in cis), potential random errors because of the small sample size, or collection bias via founder effects in our study.
In our study, patients with two heterozygous disease-causing variants in COL4A3 and COL4A4 tended to be detected proteinuria earlier and developed ESKD significantly earlier than patients with ADAS reported previously in our group.7 Although there have been no reports directly comparing the renal prognosis of digenic Alport syndrome and ADAS, Savige et al. summarized the characteristics of 32 patients with two heterozygous disease-causing variants in COL4A3 and COL4A4 from previous reports and showed that ESKD occurred earlier in patients with digenic Alport syndrome compared with those with ADAS (P = 0.01), which is consistent with our study.9 In Japan, an annual urine screening system contributes to accurate detection of the age at occurrence of proteinuria, which suggests that our data concerning the age of proteinuria detection was reliable. The reason for the poor renal prognosis in patients with digenic Alport syndrome seems to be that two disease-causing variants in COL4A3 and COL4A4 result in a loss of 75% of the collagen IV heterotrimers, whereas one disease-causing variant in COL4A3 or COL4A4 results in loss of 50% of these heterotrimers, as is mentioned in previous nice review papers.9,10
The phenotype can be more severe in patients with two variants in COL4A3 and COL4A4 on the opposite allele (in trans) compared with those on the same allele (in cis).9,14 However, in this study with a limited number of patients, there was no significant difference between these two groups. Further studies are required to assess whether the two variants of the opposite allele (in trans) result in more severe phenotypes. Identifying two variants located in trans or cis is crucial not only for assessing severity but also for correctly identifying and treating affected family members.
Our study also indicated that having one truncating variant may not necessarily influence the severity of digenic Alport syndrome. Whether the truncating variants are associated with the renal prognosis of patients with ARAS remains controversial.6,20–22 Large previous studies on patients with ADAS showed no association between the presence of truncating variants and renal prognosis.7,23 There have been no previous studies evaluating the impact of truncating alleles on severity in patients with digenic Alport syndrome, and further investigations are needed.
In this study, one male patient with a truncating variant in COL4A5 developed ESKD at age 27 years, which is consistent with previous studies on patients with XLAS with truncating variants.4,22,24 Our results support the hypothesis that an additional disease-causing variant in COL4A3 or COL4A4 may not affect the renal prognosis for patients with XLAS with a truncating variant because a truncating variant in COL4A5 leads to the destruction of the collagen Ⅳ α3 and α4 chain. 9,10 In addition, one female patient in the study developed ESKD at age 27 years, whereas her daughter with the same two variants had only mild proteinuria at the same age. This difference may be attributed to epigenetic factors, including skewed X inactivation, or environmental factors. 11 In addition, patients in our study with nontruncating variants in COL4A5 had mild renal dysfunction and proteinuria. None of the patients in this study, including the male patients with XLAS, had abnormal ocular lesions. One reason for this may be that ophthalmologic examinations were performed by local ophthalmologists at the time of genetic testing and abnormal findings may have been overlooked. Owing to the small sample size, further studies with larger cohorts could provide insights into the effect of an additional variant in COL4A3 or COL4A4 in patients with nontruncating variants in COL4A5.
To date, no comprehensive studies have reported α5 staining and electron microscopy findings in digenic Alport syndrome. In this study, normal expression of α5 in GBM was observed in all enrolled patients. Electron microscopy revealed thin basement membrane in all patients and BWC and lamellation in two adult male patients with renal dysfunction. Large-scale studies are needed to investigate whether GBM changes, including BWC and lamellation, are associated with age or disease severity in patients with digenic Alport syndrome.
This study has several limitations. First, owing to the retrospective study design, we could not collect information on confounders leading to renal dysfunction, such as hypertension, diabetes, and incidental glomerular disease, and use of drugs leading to renal dysfunction, such as anti-inflammatory drugs. Second, although the age at which ESKD develops could be affected by treatment, such as angiotensin-targeting drugs, we could not compare the patients with or without treatment owing to the small sample size. On the other hand, as angiotensin-targeting drugs were administered after proteinuria was detected in all cases, the use of these drugs did not affect the age at detection of proteinuria. Third, some patients with digenic Alport syndrome could have been overlooked in this study, even after targeted panel sequencing analysis.
In conclusion, our findings suggest that digenic Alport syndrome can account for approximately 1% of all Alport syndrome cases and that the number of patients with COL4A3 and COL4A4 variants may be higher than that of patients with COL4A5 and COL4A3 or COL4A4 variants. Moreover, patients with digenic Alport syndrome with COL4A3 and COL4A4 variants can exhibit poor renal prognosis. Therefore, identification of disease-causing variants is crucial for the renal prognosis assessment and treatment of patients with digenic Alport syndrome, as well as the detection of any affected family members.
Supplementary Material
Disclosures
Disclosure forms, as provided by each author, are available with the online version of the article at http://links.lww.com/KN9/A634.
Funding
K. Nozu: Japan Society for the Promotion of Science (KAKENHI 23K07698).
Author Contributions
Conceptualization: Tomoko Horinouchi, Yuta Inoki.
Data curation: Yuta Inoki.
Formal analysis: Yuta Inoki.
Investigation: Yuta Inoki.
Methodology: Tomoko Horinouchi.
Supervision: Tomoko Horinouchi, Kandai Nozu.
Validation: Tomoko Horinouchi.
Writing – original draft: Yuta Inoki.
Writing – review & editing: Tomoko Horinouchi, Yuta Ichikawa, Shingo Ishimori, Hideaki Kitakado, Atsushi Kondo, China Nagano, Kandai Nozu, Nana Sakakibara, Yu Tanaka, Chika Ueda, Tomohiko Yamamura.
Data Sharing Statement
The data underlying this article will be shared upon reasonable request by the corresponding authors.
Supplemental Material
This article contains the following supplemental material online at http://links.lww.com/KN9/A633.
Supplemental Table 1. List of 68 podocyte-related genes included in targeted sequencing analysis within a clinically approved gene panel test developed in our laboratory.
Supplemental Figure 1. Probability of developing ESKD in Alport syndrome cases with digenic COL4A3 and COL4A4 variants including previously reported1 cases compared with that in autosomal dominant Alport syndrome (ADAS) cases in our cohort (P = 0.011).
References
- 1.Alport AC. Hereditary familial congenital haemorrhagic nephritis. Br Med J. 1927;1(3454):504–506. doi: 10.1136/bmj.1.3454.504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kashtan CE. Alport syndrome and thin glomerular basement membrane disease. J Am Soc Nephrol. 1998;9(9):1736–1750. doi: 10.1681/ASN.V991736 [DOI] [PubMed] [Google Scholar]
- 3.Kruegel J, Rubel D, Gross O. Alport syndrome—insights from basic and clinical research. Nat Rev Nephrol. 2013;9(3):170–178. doi: 10.1038/nrneph.2012.259 [DOI] [PubMed] [Google Scholar]
- 4.Yamamura T Horinouchi T Nagano C, et al. Genotype-phenotype correlations influence the response to angiotensin-targeting drugs in Japanese patients with male X-linked Alport syndrome. Kidney Int. 2020;98(6):1605–1614. doi: 10.1016/j.kint.2020.06.038 [DOI] [PubMed] [Google Scholar]
- 5.Yamamura T Nozu K Fu XJ, et al. Natural history and genotype-phenotype correlation in female X-linked Alport syndrome. Kidney Int Rep. 2017;2(5):850–855. doi: 10.1016/j.ekir.2017.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oka M Nozu K Kaito H, et al. Natural history of genetically proven autosomal recessive Alport syndrome. Pediatr Nephrol. 2014;29(9):1535–1544. doi: 10.1007/s00467-014-2797-4 [DOI] [PubMed] [Google Scholar]
- 7.Kamiyoshi N Nozu K Fu XJ, et al. Genetic, clinical, and pathologic backgrounds of patients with autosomal dominant Alport syndrome. Clin J Am Soc Nephrol. 2016;11(8):1441–1449. doi: 10.2215/CJN.01000116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deltas C. Digenic inheritance and genetic modifiers. Clin Genet. 2018;93(3):429–438. doi: 10.1111/cge.13150 [DOI] [PubMed] [Google Scholar]
- 9.Savige J Renieri A Ars E, et al. Digenic Alport syndrome. Clin J Am Soc Nephrol. 2022;17(11):1697–1706. doi: 10.2215/CJN.03120322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mencarelli MA Heidet L Storey H, et al. Evidence of digenic inheritance in Alport syndrome. J Med Genet. 2015;52(3):163–174. doi: 10.1136/jmedgenet-2014-102822 [DOI] [PubMed] [Google Scholar]
- 11.Fallerini C Baldassarri M Trevisson E, et al. Alport syndrome: impact of digenic inheritance in patients management. Clin Genet. 2017;92(1):34–44. doi: 10.1111/cge.12919 [DOI] [PubMed] [Google Scholar]
- 12.Yokota K Nozu K Minamikawa S, et al. Female X-linked Alport syndrome with somatic mosaicism. Clin Exp Nephrol. 2017;21(5):877–883. doi: 10.1007/s10157-016-1352-y [DOI] [PubMed] [Google Scholar]
- 13.Choi M, Anistan YM, Eckardt KU, Gollasch M, Nickel P. Possible digenic disease in a Caucasian family with COL4A3 and COL4A5 mutations. Nephron. 2019;141(3):213–218. doi: 10.1159/000495764 [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y Ding J Zhang H, et al. Effect of heterozygous pathogenic COL4A3 or COL4A4 variants on patients with X-linked Alport syndrome. Mol Genet Genomic Med. 2019;7(5):e647. doi: 10.1002/mgg3.647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uemura O Nagai T Ishikura K, et al. Creatinine-based equation to estimate the glomerular filtration rate in Japanese children and adolescents with chronic kidney disease. Clin Exp Nephrol. 2014;18(4):626–633. doi: 10.1007/s10157-013-0856-y [DOI] [PubMed] [Google Scholar]
- 16.Matsuo S Imai E Horio M, et al. Revised equations for estimated GFR from serum creatinine in Japan. Am J Kidney Dis. 2009;53(6):982–992. doi: 10.1053/j.ajkd.2008.12.034 [DOI] [PubMed] [Google Scholar]
- 17.Richards S Aziz N Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Savige J Storey H Watson E, et al. Consensus statement on standards and guidelines for the molecular diagnostics of Alport syndrome: refining the ACMG criteria. Eur J Hum Genet. 2021;29(8):1186–1197. doi: 10.1038/s41431-021-00858-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gibson J Fieldhouse R Chan MMY, et al. Prevalence estimates of predicted pathogenic COL4A3-COL4A5 variants in a population sequencing database and their implications for Alport syndrome. J Am Soc Nephrol. 2021;32(9):2273–2290. doi: 10.1681/ASN.2020071065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Storey H, Savige J, Sivakumar V, Abbs S, Flinter FA. COL4A3/COL4A4 mutations and features in individuals with autosomal recessive Alport syndrome. J Am Soc Nephrol. 2013;24(12):1945–1954. doi: 10.1681/ASN.2012100985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Savige J Storey H Il Cheong H, et al. X-linked and autosomal recessive Alport syndrome: pathogenic variant features and further genotype-phenotype correlations. PLoS One. 2016;11(9):e0161802. doi: 10.1371/journal.pone.0161802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horinouchi T Nozu K Yamamura T, et al. Detection of splicing abnormalities and genotype-phenotype correlation in X-linked Alport syndrome. J Am Soc Nephrol. 2018;29(8):2244–2254. doi: 10.1681/ASN.2018030228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Furlano M Martínez V Pybus M, et al. Clinical and genetic features of autosomal dominant Alport syndrome: a cohort study. Am J Kidney Dis. 2021;78(4):560–570.e1. doi: 10.1053/j.ajkd.2021.02.326 [DOI] [PubMed] [Google Scholar]
- 24.Jais JP Knebelmann B Giatras I, et al. X-linked Alport syndrome: natural history in 195 families and genotype- phenotype correlations in males. J Am Soc Nephrol. 2000;11(4):649–657. doi: 10.1681/ASN.V114649 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article will be shared upon reasonable request by the corresponding authors.