Abstract
The field of bile acid microbiology in the gastrointestinal tract is going through a current rebirth after a peak of activity in the late 1970s and early 1980s. This renewed activity is a result of many factors including the discovery near the turn of the century that bile acids are potent signaling molecules, technological advances in next-generation sequencing, computation, culturomics, gnotobiology, and metabolomics. We describe the current state of the field with particular emphasis on questions that have remained unanswered for many decades in both bile acid synthesis by the host, and metabolism by the gut microbiota. Current knowledge of established enzymatic pathways including bile salt hydrolase, hydroxysteroid dehydrogenases involved in the oxidation and epimerization of bile acid hydroxy groups, the Hylemon–Bjӧrkhem Pathway of bile acid C7-dehydroxylation, and the formation of secondary allo-bile acids are described. We cover aspects of bile acid conjugation and esterification, and evidence for bile acid C3 and C12-dehydroxylation that are less well understood, but potentially critical for our understanding of bile acid metabolism in the human gut. The physiological consequences of bile acid metabolism for human health, important caveats and cautionary notes on experimental design and interpretation of bile acid metabolism are also explored.
Introduction
Intestinal digestion in all vertebrates is a complex physicochemical process whereby an animal obtains energy and nutrients to sustain life. Lipid digestion in the aqueous environment of the small intestine is a special problem due to the intrinsic insolubility of dietary lipids. Natural selection solved this problem in vertebrates through the development of amphipathic detergents known as bile acids formed via the enzymatic ring-system and side-chain oxidation of cholesterol in the liver1–5. These detergent molecules also activate complex cellular signaling pathways that regulate bile acid and cholesterol homeostasis6, glucose and lipid metabolism in the liver7, energy homeostasis8, and inflammation9.
Bile has long been known to have a critical role in lipid absorption as impairment of bile flow into the intestine results in malabsorption of fat (steatorrhea), and malabsorption of lipid-soluble vitamins10. The function of bile acids as detergents is implicit in their chemical structures. The two-faced Roman god Janus represents transitions, dualities, and boundaries between territories. In general, primary bile acids are ‘Janus-like’ in that they possess two faces, structural dualities, and boundaries: one hydrophobic, the other hydrophilic (FIG. 1; Table 1). The hydrophilic face (α-face) operates largely at the surface of mixed micelles, facing the aqueous milieu of the intestinal lumen, whilst their hydrophobic face (β-face) points inwards towards interior lipids11,12. Evidence early in the 20th century began to establish that intestinal bacteria modify the structures and functions of bile acids produced by the host liver, even blurring the boundary between their ‘faces’ (e.g., ursodeoxycholic acid [UDCA], which derives from ‘flipping’ the C7α hydroxyl group from CDCA to the β-face). This knowledge has catalyzed a major revolution that is rediscovering fundamental research during the later decades of the 20th century, which lacked the technologies available today.
Fig. 1|. Bile acid structure and function.
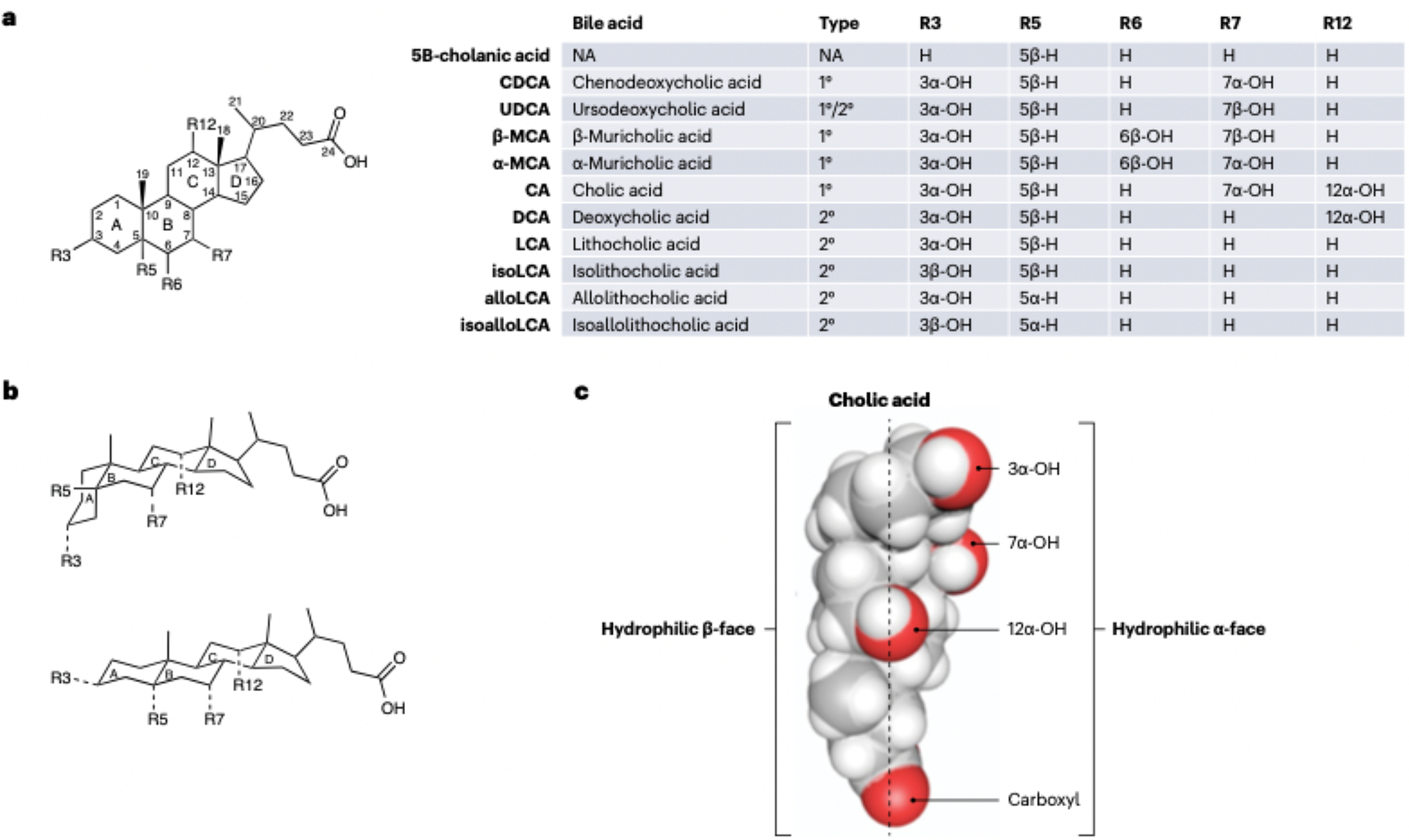
a, Generic form of a bile acid with functional groups (R) defined for select bile acids listed in the corresponding insert table. b, Structural comparisons of 5β-bile acids (R5 = 5β-H), whose A/B rings are cis. and 5α-bile acids (R5 = 5α-H), in which A/B rings are trans. c, Bile acids have a ‘Janus-like’ quality in that their faces have distinct properties that lend to detergent function, 1°, primary; 2°, secondary; NA. not applicable.
Table 1|.
Bacterial taxa contributing to bile acid metabolism in the human colon
| Enzymatic | Bacterial taxon | Substrate | Product | Refs. |
|---|---|---|---|---|
| Bile salt hydrolase or conjugase | Clostridia, Bacteroides, Enterococcuc. Bifidobacteria, Lactobacilli, methanogenic archaea | Conjugated bile acide or free bile acids | Unconjugated bile acide, MCBAs | 66–72 |
| C3-oxidation or epimerization (iso-pathway) | Ruminococcus spp., Eggerthella lenta, Baccillus coagulans, Clostridium inoculum, Lachnospira pectinoschiza, Peptoniphilus harei, Catenibacterium. Lactobacillus, Aldercreutzia equolifaciens, Dorea formicigenerans, Blautia hydrogenotrophica | CA, CDCA, DCA, LCA | 3-OxoCA, isoCA, 3-oxoCDCA, isoCDCA, 3-oxoDCA, isoDCA, 3-oxoLCA, isoLCA | 169 |
| C7-oxidation or epimerization (urso-pathway) | Escherichia coli, Clostridium sardiniense, Ruminococcus gnavus. Ruminococcus torques, Collinsella aerofaciens, Clostridium scindens, Bacteroides spp. | CDCA, CA | UDCA (from CDCA), UCA (from CA) | 205 |
| C12-oxidation or epimerization (epi-pathway) | C. scindens, Clostridium hytemonae, Peptacetobacter hiranonis, Clostridium leptum, Holdemania filiformis, Anaerostipes hadrus. Coprococcus comes. Bacteroides pectinophilus, Methanobrevibacter, Methanosphaera. Dorea sp., Clostridium paraputrificum, Eisenbergiella. Olsenella, Collinsella spp., Ruminococcus lactaris. E. lenta | CA, DCA | 12-OxoCDCA, 12-oxoLCA, epiCA, epiDCA (lagoDCA) | 169,181,182 |
| C7-dehydroxylation (Hylemon-Bjorkhem pathway) | C. scindens, C. hylemonae, P. hiranonis, Extibacter muris, Dorea sp., Proteocatella sphenisci, Faecalicatena contorta, C. leptum, Clostridium sordellii | CDCA, UDCA, CA | DCA, LCA | 37,129,136,143,206,207,214 |
| Bile acid fatty acid-conjugating enzymes | Unknown (mixed faecal bacteria) | 118 | ||
| Bile acid sulfatase | Clostridium spp., Pseudomonas aeruginosa | 3-Sulfo bile acids (for example, 3-sulfoCA) | Unconjugated bile acids (for example, CA) | 102,208,209 |
| C3-dehydroxylation | Clostridium sporogenes, Clostridium perfringens, C. paraputrificum, Clostridium sordellii | 3-SulfoLCA | 5β-cholanic acid, Δ3-cholenic acid, isoLCA | 157 |
| C12-dehydroxylation | Bacteroides spp., R. gnavus | CA, DCA | CDCA from CA, LCA from DCA | 168,169 |
| Allo-secondary bile acid pathways | C. scindens, C. hylemonae, Parabacteroides spp., Bacteroides spp., Holdmania hathewayi, Odoribacter sp., Alistipes spp. | DCA, LCA, isoDCA, isoLCA | alloDCA, alloLCA, isoDCA, isoLCA | 57,59 |
CA, cholic acid; CDCA, chenodeoxycholic acid; DCA, deoxycholic acid; LCA, lithocholic acid; MCBAs, microbially conjugated bile acids; UDCA, ursodeoxycholic acid.
Having been regarded for much of their history as mere detergents (or before this as acrimonious laxatives)12 and, as a consequence, interest in the chemistry and biology of bile acids largely waned in the 1990s as gallstone dissolution gave way to laparoscopic cholecystectomy12. However, interest in the carcinogenic role of deoxycholic acid (DCA) in the gastrointestinal tract13–16, and indeed the role of DCA in the formation of certain types of cholesterol gallstones persisted16. Today, there is renewed interest in bile acids because of the discoveries, starting in 1999, that bile acids act as powerful nutrient signaling hormones, whose structures are negotiated through the actions of eukaryotic, bacterial, and archaeal enzymes17.
A structure–function relationship is evident in the ability of distinct bile acids to differentially act as ligands for a complex array of nuclear receptors such as the farnesoid X receptor (FXR, also known as NR1H4)18–20, pregnane X receptor (PXR, or NR1I2)21, vitamin D3 receptor (VDR)22, and the constitutive androstane receptor (CAR, or NR1I3)23. Bile acids are also ligands for G protein-coupled receptors, such as TGR5 (GPBAR1)24,25, muscarinic receptors (CHRM2, CHRM3)25, sphingosine-1-phosphate receptor 2 (S1PR2)26, retinoic acid receptor gamma T (RORɣT)27,28, nuclear receptor 4A1 (NR4A1)29, and downstream effectors have emerged in multiple digestive organs to sense and respond to nutrient and metabolic status by sampling bile acids7,30, and thereby influence intestinal inflammation and gastrointestinal cancers31,32.
Strikingly, derivatives of hydrophobic secondary bile acid products of microbial metabolism, such as DCA and lithocholic acid (LCA), are in general preferred ligands for many of these host receptors over the primary bile acids from which they derive30. What has emerged in the past few decades is that the ‘Western lifestyle’ of inactivity, diets low in fiber and high in processed carbohydrates and saturated fats, increase both the amount of bile entering the gastrointestinal tract and the hydrophobicity of the bile acid pool, thereby increasing the risk of hepatobiliary and gastrointestinal cancers in humans32,33. In their own way, the microorganisms are ‘talking’, and we have developed the analytical tools to ‘listen’, and hopefully will learn to act accordingly to modulate the numerous human disorders and diseases affected by microbial bile acid metabolism.
Humans are composed of roughly a trillion mammalian cells and harbor an estimated equivalent number of microorganisms that dwarf us in gene content34,35. It is now apparent from comparisons between conventional and germ-free rodents that the host genome and the microbiome (collective microbial genomes) together contribute substantially to the complex repertoire of small molecules in biological tissues and fluids collectively known as the metabolome36,37. Such studies established early on, and beyond doubt, that the gut microbiota deconstruct host-derived primary bile acids into a myriad of metabolites known as secondary bile acids during enterohepatic circulation (FIG. 2)38. Indeed, bile acid modification has been identified as one of the major bidirectional modes by which microorganisms communicate with their host, and how the host senses, responds to, and shapes the composition of its symbionts7,37. In this Review, we focus on the production of and modifications to the secondary bile acids, DCA and LCA. In addition, we describe lesser-known bile acid dehydroxylation reactions, how these reactions might alter the fecal bile acid pool, and experiments that will be needed in the future to confirm and quantify these novel microbial metabolites. We also summarize the latest findings on how secondary bile acid derivatives affect host immune function.
Fig. 2|. The enterohepatlc circulation or bile acids.
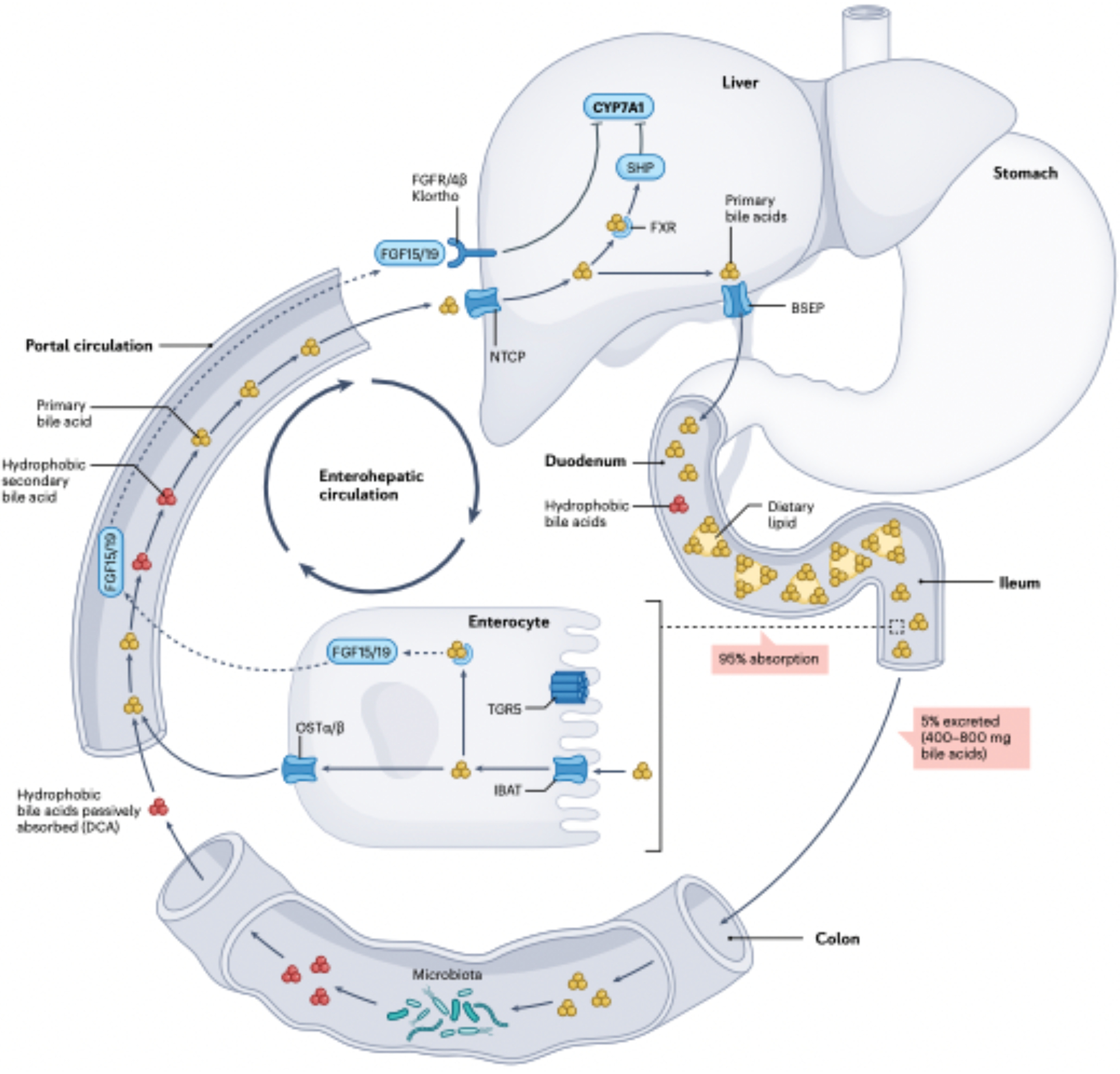
Primary bile acids (yellow) are synthesized de novo in hepatocytes from cholesterol and secreted into bile through transport protein BSEP. During a meal, the gallbladder contracts, releasing bile into the duodenum, where mixed micelles composed of phosphdipods. fatty acids, cholesterol and lipid-soluble vitamins form and are surrounded by amphipathic conjugated bile acids. When conjugated bile acids reach the terminal ileum, they are transposed into enterocytes by the illeal sodium-bile acid cotransporter (JBAT), bound to FABP6 and transported into portal circulation via OSTα and OSTβ expressed basolaterally on enterocytes. As part of the negative feedback function of bile acid synthesis, intracellular bile acids activate the nuclear farnesoild X receptor (FXR) in enterocytes, resulting in upregulation in the synthesis and secretion of the protein FGF15/19 into the portal circulation. FGF15/19 binds to fibroblast growth factor receptor FGFR4/β-Klortho receptor-dependent manner resulting in inhibition of CYP7AL the rate-limiting enzyme in bile acid biosynthesis in the liver. Bile acids returning to the liver are transported by NTCP. Activation of FXR in hepatocytes represses CYP7A1 expression dependent on small heterodimer partner (SHP) and liver-related homologue 1(LRHI). This process allows bile acid levels to remain in steady state. TCRS activation in intestinal stem cells promotes regeneration of enterocytes20. Roughly 5% of bile acids (400–800 mg per day) escape fieal transport and enter the large Intestine, which is the major route by which cholesterol is removed from the body. In the large intestine, bile acid structure and function are diversified by the gut microbiota. Part of this diversification is increasing the hydrophobicity of bile acids in the large intestine, allowing passive absorption into colonocytes and entry into the portal circulation, where secondary bile acids (mainly decoycholic acid (DCA)) accumulate to roughly one-quarter of the bile acid pool in healthy humans.
Bile acid synthesis and composition
Liver metabolism is primarily conjugative and oxidative. Chenodeoxycholic acid (CDCA) is considered the root of all C24 bile acids in vertebrates (FIG. 1, Table 1)12. In humans, bile acid synthesis yields CDCA and cholic acid (CA) in roughly equal amounts. The enzymology and regulation of genes involved in the formation of the primary bile acids CA and CDCA are well established6,39. Most of the total bile acid pool is produced in the liver by the classical or neutral pathway, whereas the alternative (or acidic pathway) contributes, mostly in the form of CDCA, a smaller portion to the pool (≤10%)6,40. In both humans and rodents, there are sex and age differences in the rate of bile acid synthesis and the composition of the biliary pool41. Important species differences exist between humans and the model organisms we approximate to our physiology. For example, in mice and rats, the biliary pool is composed of the primary bile acids CA and CDCA, as well as muricholic acids, primarily β-muricholic acid (β-MCA), and small quantities of the C7 epimer of CDCA, UDCA6. The enzymes involved in the formation of MCAs have been identified and their disruption through gene knockout has provided researchers with mouse models that better approximate the human biliary pool (synthesis of CDCA and CA) although there are reported trade-offs as rodents are not well adapted to the more hydrophobic biliary pool of humans42,43.
Bile acids are amidated to the amino acids glycine or taurine in the human liver. In humans, glycine predominates by an order of magnitude over taurine on a plant-based diet, but this feature can be reversed by a diet high in animal protein44. In rodents, bile acids are conjugated almost exclusively to taurine, irrespective of diet45. During the enterohepatic circulation, conjugated bile acids are secreted into the small intestine where they form mixed micelles with dietary lipids enabling absorption, before being transported back to the liver through the portal circulation via high-affinity transporters on the apical and basolateral sides of enterocytes (FIG. 2). Each day, several hundred milligrams of conjugated bile acids escape enterohepatic circulation, and enter the large intestine, where they are rapidly deconjugated by bacterial bile salt hydrolases (BSH), releasing taurine or glycine and free bile acids46. Several pathways for taurine utilization exist in the gastrointestinal tract47, however, microbial respiration of taurine by anaerobes results in the formation of hydrogen sulfide, excess formation of which is associated with colorectal cancer (CRC)48,49.
Bile acids are among the most structurally diverse biomolecules in nature2. Bile acid structure varies with respect to side-chain length (C27 versus C24), side-chain carboxylation (bile acids) versus hydroxylation (bile alcohols), side-chain conjugation (amino acids versus sulfate), A/B ring cis (5β-H; non-planar ring system) versus trans (5ɑ-H; planar ring system) stereochemistry, and hydroxylation patterns and hydroxyl stereochemistry (α, β, and Ω)2. Animal bile has a long history in ethnopharmacology, which Western medicine has been slow to examine critically4. Notable exceptions include naturally occurring bile acids such as ursodeoxycholic acid (UDCA) that has wide usage in the treatment of biliary disorders50, and avicholic acid, which has therapeutic potential51. The extensive work of Haselwood1,3 and Hofmann and Hagey2,52 provided extensive surveys of biliary bile acid composition across vertebrates, suggesting the diversity and pattern of bile acid structure has evolutionary significance2. Strikingly, no clear pattern exists between dietary strategy (such as carnivore, herbivore or omnivore) and bile acid composition, suggesting other evolutionary forces are driving bile acid diversification (e.g., interspecies and intraspecies communication and developmental and cellular signaling)52. Comprehensive fecal bile acid profiles reflecting bacterial bile acid metabolism across vertebrates are beginning to be reported53. It is known in at least some species that the fecal bile acid profiles differ little from the liver, indicating selection for microbiota that leave bile acids intact54.
A substantial gap in our knowledge still exists with respect to the synthesis of bile acid A/B-ring trans-isomers known as allo-bile acids. Primary allo-bile acids are produced in miniscule quantities in healthy adults, whereas they are produced at more substantial amounts in neonates, after liver transplantation in adults, and during proliferation associated with hepatocellular carcinoma55,56. By contrast, allo-bile acids are primary bile acids in agnathan fishes (lampreys and hagfish) and cyprinid fishes (such as zebrafish and carp), as well as some reptiles, and ancestral mammals such as the Afrotheria (such as elephants, manatees, and tenrecs) and rhinoceros2,57. It is predicted that SDR5A1 and/or SDR5A2 are responsible for bile acid 5α-reduction in humans, and orthologues of these genes have been found in zebrafish52. Presumably, the production of allo-bile acids in humans activates developmental signaling pathways associated with hepatic growth; however, research in this area is currently lacking. Work published in 2021 has indicated that the gut microbiome is capable of producing small quantities of allo-secondary bile acids (such as isoallolithocholic acid [isoalloLCA]) that are enriched in Japanese centenarians58. Whether the source of planar secondary bile acids such as isoalloLCA derived mainly from primary bile acids (for example, CDCA) or from primary allo-bile acids (for example, allochenodeoxycholic acid [alloCDCA]), or both, remains unclear, and will be a topic discussed at length later in this Review58–60.
Bile acid metabolism by gut microbiome
Bile acid deconjugation or conjugation
The earliest evidence for microbial metabolism of host primary bile acids was the deconjugation of conjugated bile acids by mixed fecal bacteria and microbial isolates61. BSH (E.C.3.5.1.24) is a member of the choloylglycine hydrolase family along with penicillin V amidase of the Ntn-hydrolase superfamily of proteins46. BSH enzymes are some of the most studied microbial bile acid-metabolizing enzymes and have been extensively reviewed62–66, so our comments highlight key points and latest advances. BSH enzymes are widely represented in a variety of species distributed across most phyla in the gut microbiome. BSH activity has been characterized in Gram-positive commensal bacteria including Lactobacillus67,68, Bifidobacterium69, and Enterococcus70. BSH activity is also represented in Gram-negative bacteria, especially widely distributed among species of the genus Bacteroides71,72. Archaea common to the mammalian gastrointestinal tract have also been reported to express BSH73. Phylogenetic analysis indicates horizontal gene transfer of bsh genes from members of the Bacillota to gut methanogens73. BSH is necessary for gastrointestinal colonization by pathogens including Brucella abortis74,75 and Listeria monocytogenes76–78. The sensing of conjugated bile acids provides an important environmental signal of gastrointestinal colonization, distinct from the secondary bile acids common in soil. It is hypothesized that bsh genes are colonization factors that provide some combination of: amino acid sources of carbon, nitrogen, sulfur, and energy; protection against the detergent properties of conjugated bile acids allowing gastrointestinal colonization and persistence; and incorporation of bile acids into bacterial membranes, which increases tensile strength, fluidity, and charge, thus protecting bacteria against host immune function65,79. Caution should be taken against generalization of bsh function across taxa, and where possible, isogenic bsh mutants should be generated to determine both bacterial physiological function and consequences of bsh loss on host physiology65,71,73.
In healthy individuals, the deconjugation of bile acids is a substrate-limiting reaction and goes essentially to completion. This feature is assumed to be true in most vertebrates, with notable exceptions such as the cyprinid fishes (such as zebrafish), whose side-chain sulfate is retained during intestinal transit54,80–82. The fecal composition of conjugated bile acids increases substantially in mammals that are taking broad-spectrum antibiotics83. By contrast, BSH activity in the small bowel generates unconjugated bile acids that are less polar than conjugated bile acids, and inefficiently transported by ileal sodium/bile acid cotransporter (IBAT or SLC10A2) in the ileum leading to increased fecal excretion84,85. Increased fecal excretion of bile acids leads to more de novo bile acid synthesis from cholesterol or reverse cholesterol transport to the liver, reducing serum cholesterol. There is evidence that BSH activity leads to cholesterol lowering through reduction in micellar lipid and cholesterol reabsorption and loss of bile acids in feces, increasing cholesterol conversion to bile acids64,86–88.
Conversely, prior studies in poultry indicated that sub-therapeutic levels of antibiotics promote growth, at least in part, through the reduction in intestinal BSH activity resulting in an improvement in lipid digestion89,90. Thus, targeting BSH activity in malnourished populations might improve weight gain partially through enhanced lipid absorption. Indeed, a longitudinal study in children supports an inverse association between BSH and weight gain caused by macrolide antibiotic use91. By contrast, other studies suggest increased BSH activity leads to enhanced weight gain92. The latest ‘omics’ applications reveal that the physiological consequences of BSH activity to the host might be mediated more through cellular signaling in the intestine and liver than through mere detergent actions and fat absorption. BSH activity affects host gastrointestinal maturation93, alters liver and intestinal gene expression relating to circadian rhythm, glucose and lipid homeostasis in the liver, and immune function71,94. As BSH enzymes differ in substrate specificity relating both to the amino acid conjugate (glycine versus taurine) and the sterol nucleus, targeting subsets of BSH enzymes is likely necessary to achieve a clinical outcome of interest. Indeed, large scale metagenomic surveys reveals associations between the pattern of bsh genes in both human populations95 and chronic diseases96 suggesting further functional characterization is warranted. Specific inhibitors targeting BSH enzymes have been developed, and further refinements in pharmacological inhibition of different subsets of these enzymes might be therapeutically useful both in human disease and animal production97,98.
Metabolism of tertiary bile acids
Gut microbial products are capable of modulating hepatic conjugation of bile acids, with potential for therapeutic benefits. The term tertiary bile acid has been applied to describe unique bile acids formed from hepatic metabolism of secondary bile acids12. Phase II metabolism of the toxic secondary bile acid LCA yields the tertiary bile acid 3-sulfo-LCA99. The microbial formation and ileal absorption of LCA can also lead to sulfation of other bile acids in the liver. Indeed, a study published in 2021 observed LCA-induced enrichment of cholic acid-7-sulfate (CA7S) in the feces of humans and mice subjected to partial sleeve gastrectomy100. CA7S is a gut-specific apical activator of TGR5 expression, which can lead to secretion of glucagon-like peptide 1 (GLP-1) conferring anti-diabetic effects100.
Gut microorganisms are also capable of removing the sulfate from tertiary bile acids through expression of aryl sulfatase enzymes101–103. Desulfation activity is associated with Peptococcus, Clostridium, Pseudomonas, and Fusobacterium103–106. However, the microbial sulfatases involved have yet to be identified. The gut microbiota might also be capable of bile acid sulfation in the gut, a function that has historically been assumed to be host-enzyme dependent107. If confirmed, sulfation of LCA by bacteria would blur the distinction between so-called secondary and tertiary bile acids. In the absence of a final consensus relating to host versus microbial biochemical potential, bile acid sulfation by gut microprganisms would further support the suggestion to abandon the term “tertiary bile acid” altogether108.
MCBAs and bile acid polyesters
The advent of next generation sequencing coupled with computational progress has driven a renaissance in gut microbiology led largely by bioinformatics109,110. Powerful advances in untargeted metabolomics have paved the way for discovery of novel microbial metabolites by so-called cheminformaticians111. Indeed, this cheminformatics approach has led to the startling identification of microbially conjugated bile acids (MCBAs) in which non-canonical amino acids (such as amino acids other than glycine or taurine) are amidated to free bile acids by gut bacteria such as Enterocloster boltaea112. It seems that the BSH enzymes are responsible for generation of MCBAs, and that patterns are emerging between BSH amino acid sequences and amino acid conjugation specificity113. Although it is widely understood that microbial biotransformations in the gut are limited by fecal analysis, new technologies are emerging that promise to enable clinicians and microbiome scientists to sample along the gastrointestinal tract using pill-like sampling devices that can be swallowed by individuals114. Indeed, this approach revealed that MCBAs are generated largely in the small bowel where BSH enzymes are active114. Evidence indicates that MCBAs can signal through PXR and FXR115; however, their physiological relevance is only beginning to be understood.
Bacteria in the gastrointestinal tract can also esterify bile acids with alcohols, short-chain fatty acids (SCFAs), and long-chain fatty acids. Methyl esters of DCA are formed by human fecal isolates, and whilst the mechanism is not known, it was hypothesized that the reaction is dependent on C1-transfer (methyl or methoxyl group) to the C24 carboxyl group rather than methanol as a substrate116. By contrast, esterification of bile acids associated with Lactobacillus, Eubacterium, and Bacteroides was reported to be dependent on the addition of ethanol117. Bacteria also generate bile acid fatty acid esters in which long-chain fatty acids (C16 and C18-fatty acids) as well as SCFAs such as acetate are linked to C3 of isoDCA and isoLCA118. Several reports describe the oligomerization of C-24 carboxyl group of one molecule of DCA to the 3α-hydroxy group of the another to form a polyester chain119. It is presumed that these reactions are a detoxification strategy to precipitate bile acid esters, therefore reducing the concentration of both hydrophobic secondary bile acids and toxic fatty acids and alcohols from fecal water120.
Despite progress in LC/MS approaches to bile acid profiling, bacteria generate a number of bile acid structural and stereoisomers (such as DCA and alloDCA, isoCDCA and isoDCA), making separation and detection of some bile acids challenging. By contrast, bile acid esters are rarely measured in fecal samples118. Comparison of stool samples from healthy individuals with or without strong alkaline hydrolysis revealed that between 10–30% of total bile acids (primarily isoDCA and isoLCA) are esterified118. The diversity and quantity of bacterial bile acid conjugates therefoee represents an important consideration when designing fecal bile acid extraction protocols to address particular clinical and research questions. Methodological advances can now be harnessed to identify novel bile acid metabolites, discover new bile acid metabolizing enzymes, and alter bile acid metabolism through targeting microbial strains and biochemical pathways (FIG. 3).
Fig. 3|. Targeting microbiota-bile acid interactions as potential therapeutic approaches for gastrointestinal and metabolic diseases.
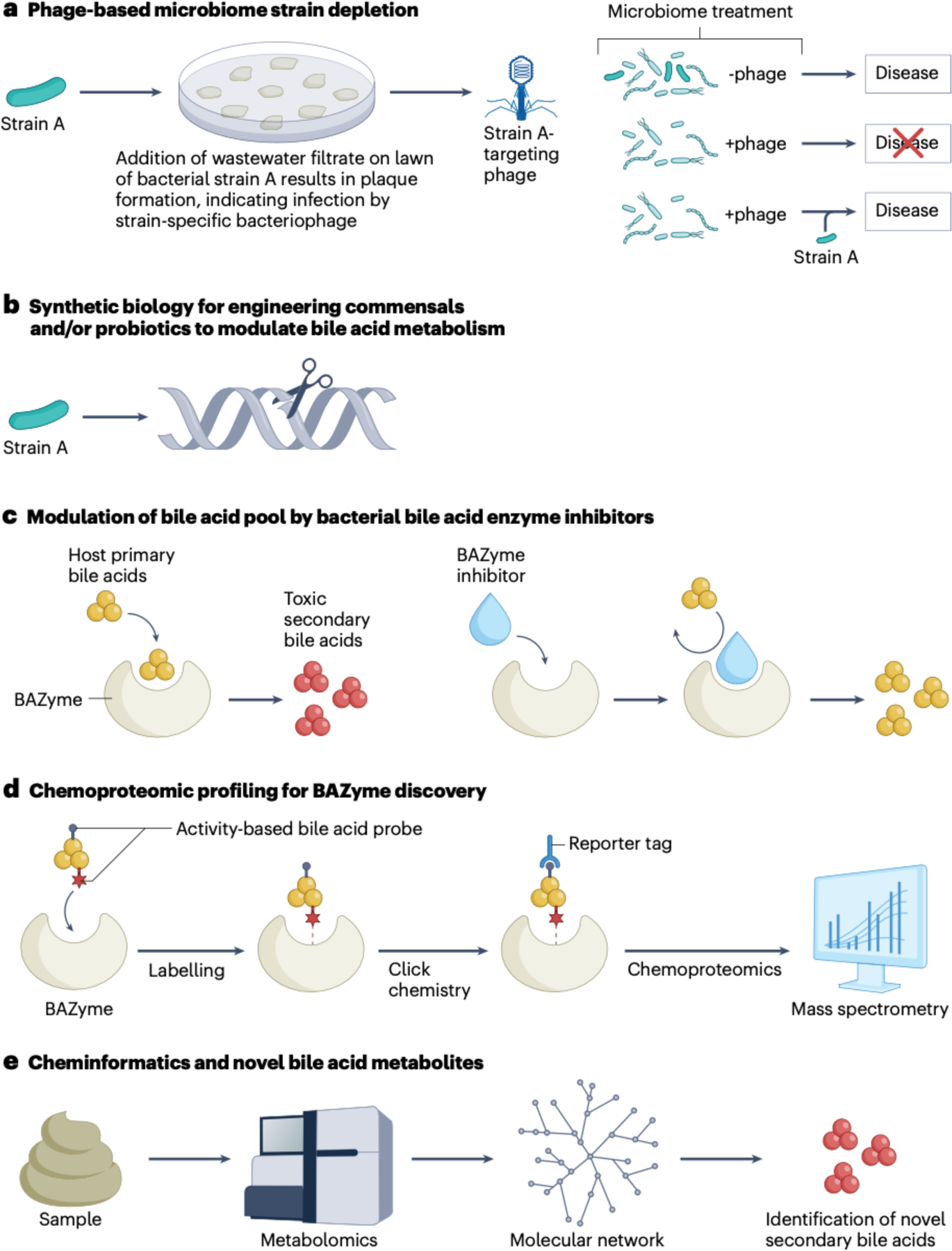
a, Studies have demonstrated the potential utility of selecting for bacterial strain-dependent bacteriophages to remove microbial strains that have a causal rolle in diseases such as inflammatory bowel disease12. b, Synthetic biology offers the potential to rationally design commensal or probiotic bacteria to modulate bile acid metabolism in vivo129. c, The development of specific inhibitors against the microblome is expected to provide therapeutic potential22. The development of bile salt hydrolase (BSH) enayme inhibitors has allowed interrogation of the effects of altering bile acid metabolism71,91,10; other studies indicate that inhibitors against bar enzymes might also be therapeutically Important11. d, Chemoproteomic profiling using click chemistry bile acid probes allows the discovery of novel bacterial enzymes involved in bile acid metabolism23. After the bile acid probe covalently bonds to a bile acid binding enzyme (BAZyme), proteomic mass spectrometry allows the identification of gene sequence candidates. e, Cheminformatics couples metabolomics with computation to obtain metabolite networks in which some nodes represent novel metabolites that repeal previously unknown bacterial metabolism121. BAZyme, bile acid enzyme.
The Hylemon–Bjӧrkhem Pathway
Historically, bile acids designated primary were defined as possessing a 7ɑ-hydroxyl group, and removal of the 7ɑ-hydroxyl group by the action of microbial enzymes defined secondary bile acids38. Today, secondary bile acids are regarded as microbial reaction products (with the exception of C24 deconjugation) of host primary bile acids, and include products of microbial oxidation, epimerization, and dehydroxylation products of CA and CDCA in humans (FIG. 2) and expand to other products of primary bile acids in rodents (e.g., UDCA and muricholic acid metabolites), and pigs (e.g., hyocholic acid metabolites). This diversification of bile acid structure indicates that numerous enzymatic pathways exist, distributed across bacterial and archaeal phyla inhabiting the gastrointestinal tract. The collective genes involved in bile acid and sterol biotransformations have been termed the sterolbiome (BOX 1)121,122.
Box 1. Introduction to the importance of strain variation in the gut sterolbiome.
The field of microbial endocrinology218 approaches gut microbial metabolism of sterols as an integral yet overlooked part of the human endocrine system218–220. We previously defined the term ‘sterolbiome’ to describe the genetic potential of the gut microbiota to produce endocrine molecules from endogenous and exogenous sterols in the gastrointestinal tract121,122. An intersection exists between sex hormones and the regulation of bile acid synthesis40. Both bile acids and sex hormones alter the gut microbiome, although the mechanisms remain poorly understood122. Strain level variation with respect to bile acid and steroid metabolizing genes has been known for some time with the few isolates historically available but is now becoming clearer at the molecular level with the advent of shotgun metagenome sequencing, which enables the integration of taxonomic information with gene content. This advance is evident in species such as Eggerthella lenta and Clostridium scindens. which vary at the strain level in their capacity to metabolize bile acids, steroid hormones and steroid drugs178,182,220–223. indeed, some strains of these species convert cortisol to androgens224 and corticosterone to progesterone225. C. scindens can convert prednisone to an androgenic product that promotes the growth of prostate cancer cells in vitro221. The sterolbiome is therefore an important dimension to consider with the advent of live biotherapeutic products to treat recurrent Clostridiodes difficile infection and potentially other diseases such as Crohn’s disease, ulcerative colitis and metabolic dysfunction-associated steatohepatitis (formerly non-alcoholic steatohepatitis)226–228.
Excess hydrophobic secondary bile acids have been long associated with cancers of the gastrointestinal tract31 and cholesterol gallstone formation15. For this reason, determining which gut bacteria are responsible for forming DCA and LCA, the bile acid intermediates formed during this conversion, the genes encoding enzymes catalyzing these biotransformations, and the mechanism(s) by which each reaction is catalyzed are important in interpreting microbiome data and devising future interventions to modulate the bile acid metabolome to treat disease.
Two models have been proposed for bile acid 7-dehydroxylation. The first was a two-step mechanism we termed the Samuelsson–Bergstrom model that was proposed in 1960123. This model, arrived at by a series of elegant and rigorous in vivo experiments, proposed a diaxial trans-elimination of the 7α-hydroxyl group of CA yielding a 𝚫6-intermediate (3α,12α-dihydroxy-5β-chol-6-enoic acid), followed by trans-hydrogenation resulting in DCA. Synthesis of 3α,12α-dihydroxy-5β-chol-6-enoic acid and incubation with cultures of Clostridium bifermentans124 and Clostridium scindens125 resulted in conversion to DCA providing some early support for the model. However, in vitro and in vivo studies during the 1980s and 1990s demonstrated that while there is indeed a 𝚫6-reduction step (catalyzed by BaiH and BaiN), the pathway is far more complex than initially thought38. The bile acid intermediates in a multi-step bifurcating pathway from CA to DCA and its ‘flat’ isomer, alloDCA have been identified in cell extracts of C. scindens VPI 12708126. A reverse genetic approach facilitated cloning of the ~12 kB bile acid inducible (bai) operon from C. scindens VPI 12708127. Subsequent work with purified and recombinant Bai enzymes over several decades128 led to identification of Bai enzymes catalyzing each step of what we have termed the Hylemon–Bjӧrkhem Pathway38. Elegant confirmation of the bai operon catalyzing the conversion of CA to DCA both in vivo and in vitro has been reported129. The measurement of bai genes in human stool samples is now becoming standard as a marker for bile acid dysbiosis in the case of IBD130, antibiotic treatment131, or excess in the case of cancers of the gastrointestinal tract132.
Several key aspects of bile acid 7-dehydroxylation have emerged over the course of study in this area. First, bile acid C24 amides (conjugated bile acids) are not substrates133–135. Thus, bile acid hydrolysis is a prerequisite for bile acid 7-dehydroxylation. This is important to note as BSH inhibitors have indeed been observed to enrich the host in primary bile acids98. Second, bile acid 7-dehydroxylation seems to be relegated to relatively small populations (103-107 CFU per gram wet weight feces)15 of species within the Bacillota (Ruminococcaceae, Peptostreptococcaceae, Lachnospiraceae, and Oscillospiraceae)136–138. Third, these populations of species have been separated into two groups (low activity versus high activity) by their relative rates of conversion of CA to DCA, which differs ~100 fold (Table 1; Box 2)139. Fourth, despite these small populations, defined communities of microorganisms with complexities ranging from a handful up to 100 members demonstrates so far that organisms with the bai operon are necessary for DCA and LCA formation140–143. Fifth, microbial bai pathway enzymes have evolved to recognize the bile acids endogenously produced by the hosts they are adapted to. Thus, although rodent gut microbial isolates can convert β-muricholic acid (β-MCA) to murideoxycholic acid (MDCA), human gut microbiota that are able to convert CA to DCA and CDCA to LCA are unable to metabolize β-MCA when colonized in germ-free mice140,144. The only exception is UDCA, which can be 7β-dehydroxylated to LCA (FIG. 4)38,121. Sixth, the bai pathway is a redox process yielding a net 2-electron reduction (that is, bile acids as an electron acceptor) that provides an important proximal cause for its evolution. In the large intestine (a highly reductive, anaerobic environment), microorganisms must dispose of reducing equivalents, and reducing unsaturated bile acids during dehydroxylation accomplishes this feat to a degree. However, there might be underlying ultimate causes of equal or greater importance, which include but are not limited to: elimination of microbial competition for key nutrients by increasing toxic bile acids, and interdomain-signaling with the host to improve the fitness of DCA producers in the gut.
Box 2. A cautionary tale in gut microbiology studies.
Much confusion still exists in the literature relating to which bacteria generate deoxycholic acid and lithocholic acid37. This confusion is partly a product of uncertain taxonomic placement early in the field that has since been clarified by increasingly sensitive nucleic acid sequencing techniques37. Conversely, the advent of highly sensitive metabolomics platforms has led to some false-positive detection of bile acid metabolism by gut bacteria. Animal-based microbiological medium components (for example, tryptone, meat extract and brain heart infusions) contain measurable quantities of bile acids. Thus, care should be taken to differentiate background bile acids in culture media from the accumulation over time of increasing quantities of a bioconversion product indicative of bona fide biological activity. Alternatively, plant-based protein sources (trypticase soy) might be used to eliminate background bile acids.
In addition, the search for sterolbiome genes in stool metagenomes should be accompanied by biochemical confirmation of function. The identification of a ‘bai-like’ operon in Eggerthella lenta178, whilst not originally claimed to encode enzymes that catalyse C7-dehydroxylation, has nonetheless been cited to incorrectly list E. lenta among species capable of forming deoxycholic acid and lithocholic acid in other articles. Indeed, there is ample evidence to the contrary, including a study that identified the ‘bai-like’ operon178 Although E. lenta is capable of extensive oxidation and epimerization of bile acid hydroxyl groups, removing the C7 hydroxyl has not been demonstrated despite extensive characterization of many strains independently in several labs over four decades175,182,229–231. By contrast, a report of ‘bai-like genes’ in the Oscillisporaceae in human gut metagenome sequences further expands the diversity of potential bile acid 7-dehydroxylating bacteria because the authors performed functional confirmation of bile acid metabolism by characterizing the recombinant Bai enzymes136. To clarify, Table 1 lists species currently confirmed to express bile acid 7-dehydroxylation activity.
Fig. 4|. Bile acid biotransformations in the human large intestine.
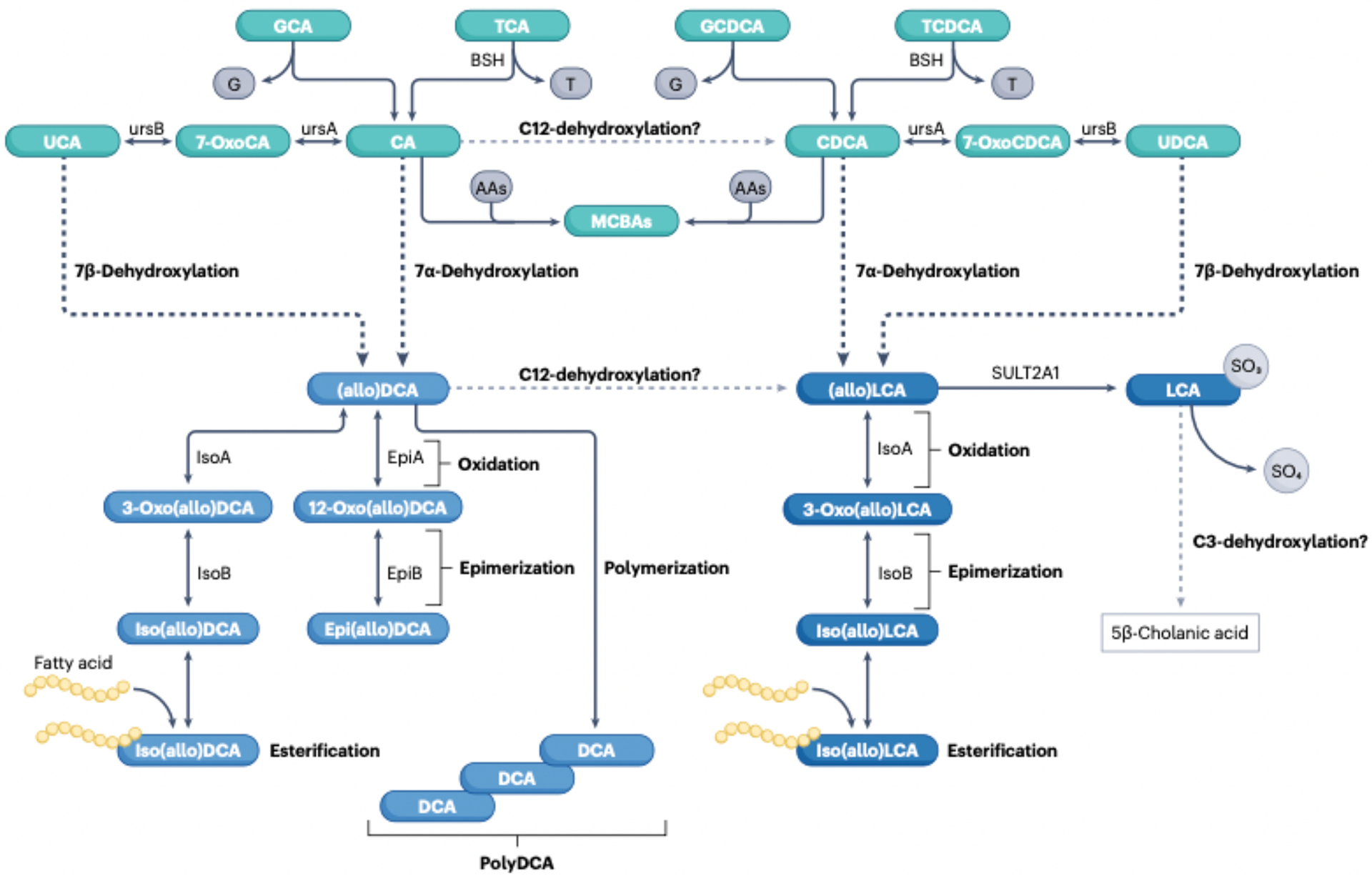
Conjugated primary bile acids (taurocholic acid (TCA), glycocholic acid (GCA), taurochenodeoxycholic acid (TCDCA), glycochenodeoxycholic acid (GCDCA)) enter the large intestineand are deconjugated by the enzyme bile salt hydrolase (BSID by diverse gut bacterial taxa (Table 1). BSH is also reported to reconjugate free bile acids with a wide range of amino acids (AAs) yielding microbially conjugated bile acids (MCBAs). The primary bile acids cholic acid (CA) and chenodeoxycholic acid i.CDCA) can be oxidized to 7-oxoCA and 7-oxoCDCA, respectively, by the enzyme 7α hydroxysteroid dehydrogenase(ursA) (Table 1) and epimerized to ursochollc acid (UCA) and ursodeoxycholic acid (UDCA), respectively, by the enzyme 7β-hydroxysteroid dehydrogenase(ursB). CA and CDCA are also converted to deoxycholic acid (DCA) and lithocholic acid (LCA), respectively, through the multipstep Hylemon-Björkhem pathway of bile acid 7α-dehydroxylation by intestinal Clostridia (Table 1). UCA and UDCA can be directly 7β-dehydroxylated through the Hylemon-Björkhem pathway or, alternatively, epimerized back to CA and CDCA and 7α-dehydroxylated to DCA and LCA. The Hylemon-Björkhem pathway also yields A/B-ring epimers that are planar, known as ‘allo’ bile acids. We have designated both epimers of DCA and alloDCAas (allo)DCA as well as (allo)LCA and their derivatives. (allo)DCA has two hydroxyl groups that can be oxidized to 3-oxoDCA by 3α-hydroxysteroid dehydrogenase (isoA) and/or 12-oxoDCA by 12ahydroxysteroid dehydrogenase (epiA), and epimerized by 3β-hydroxysteroid dehydrogenase (isoB) and/or 12β-hydroxysteroid dehydrogenase (epiB) to iso(allo)DCA and epi(allo)DCA, respectively. (allo)LCA is monohydroxylated at C3 and can yield 3-oxo(allo)LCA and lso(allo)LCAonly. Additionally, the host sulfates lca to yield 3-sulfoLCA. It is reported that 3 sulfoLCA can be C3-dehydroxylated to the non bile acid, 5β-cholanic acid. isoDCA and isoLCA are also esterified to short-chain and long-chain fatty acids.and DCA can be polymerized through esterification between C3-OH and C24-COOH. There is some support for the C12-dehydroxylation of CA to CDCA and of DCA to LCA.
Although hydrophobic secondary bile acids such as DCA and LCA are strongly implicated in cancers of the gastrointestinal tract, their production has important physiological functions to host immune function27,28,145,146, serotonin production147, cellular signaling16,30, prevention of Clostridioides difficile colonization and vegetative growth131, and enteric viral infection148. By adopting diets lower in animal protein and fat, and higher in complex carbohydrates and fiber, intestinal bile acid levels can be lowered such that the benefits of hydrophobic bile acids can be maintained whilst reducing the risks associated with elevated fecal levels and enrichment of the bile with DCA32.
Patients with cirrhosis have a reduced bile acid pool relative to control, substantial reduction in the abundance of bile acid 7α-dehydroxylating bacteria with reduced fecal DCA and LCA, and there is a concomitant dysbiosis characterized by toxic Gram-negative gut microbiota149,150. When patients with cirrhosis receive a liver transplant, an increase in bile acid secretion, increased fecal secondary bile acids, and diversification of the gut microbiome along with reduced systemic inflammation are observed151. Faecal microbiota transplant (FMT) restored cognitive function and improved inflammation with concomitant increase in Gram-positive bacteria associated with secondary bile acid formation, along with increases in DCA and LCA in stool152. Patients with poor outcomes were observed to have markedlt lower levels of secondary bile acids in serum and feces and diminished bacterial genes associated with secondary bile acid formation152. These results indicate an important role for the maintenance of basal levels of hydrophobic secondary bile acids and the liver–gut axis.
Formation of allo-secondary bile acids
Two pathways to allo-secondary bile acids have been elucidated. The first pathway we have termed the ‘direct pathway’ in which primary bile acids are converted to alloDCA or alloLCA through the Hylemon–Bjӧrkhem Pathway38. After the rate limiting 7α-dehydration step (catalyzed by BaiE), a 3-oxo-4-DCA or 3-oxo-4-LCA intermediate is formed. In the Hylemon–Bjӧrkhem Pathway, the conversion of 3-oxo-4-DCA to DCA proceeds via a reduction catalyzed by BaiCD (bile acid 5β-reductase) and BaiA (3α-HSDH)38,60. Alternatively, 3-oxo-4-DCA can be converted to alloDCA through reduction by BaiP or BaiJ (bile acid 5α-reductase) and BaiA60. The second pathway is what we have termed the indirect pathway and first requires a bile acid 7-dehydroxylating bacterium to produce DCA or LCA. In this scheme, members of the gut microbiome expressing 3α-HSDH, bile acid 5β-reductase, and bile acid 5α-reductase generate allo-secondary bile acids through a metabolic equilibrium: DCA, 3-oxo-DCA, 3-oxo-4-DCA, 3-oxo-alloDCA, alloDCA58,153. The relative contribution of the direct and indirect pathways to the formation of secondary allo-bile acids is currently unclear and may exhibit both intra- and inter-individual variation.
Bile acid C3-dehydroxylation
Lithocholic acid, a microbial product of C7-dehydroxylation of CDCA and UDCA, is mono-hydroxylated and the most hydrophobic of the prominent bile acids detected in vertebrates. LCA is a suspected carcinogen, generating reactive oxygen species and DNA adducts154. LCA acts as a tumor-promoter through the inhibition of DNA repair enzymes that drives the proliferation of apoptosis resistant cells154. Studies of LCA metabolism in humans during CDCA or UDCA treatment of gallstones revealed extensive sulfation of LCA yielding 3-sulfo-LCA155,156. It is now well understood that LCA is a strong ligand for the vitamin D receptor (VDR), which induces sulfotransferase SULT2A1 expression99. The sulfation of LCA, during phase II metabolism, generates a hydrophilic derivative that is not readily absorbed by the intestine, facilitating excretion. As noted earlier, gut bacteria express aryl sulfatases that deconjugate 3-sulfo-LCA leading to release of LCA. Thus, there is a predicted ‘back and forth’ between host phase II metabolism and microbial deconjugation. Yet, there is evidence for an alternative microbial pathway out of this cycle (described later), yielding products that are no longer defined as bile acids.
Although the focus of bile acid dehydroxylation is principally with respect to the Hylemon–Bjӧrkhem Pathway, there are reports of other bile acid dehydroxylation reactions, which includes C3-dehydroxylation of bile acids. Bile acid C3-dehydroxylation is particularly interesting in that removal of C3 changes the designation from bile acid to derivatives of 5β-cholanic acid (FIG. 4). This process presents a particular difficulty with respect to bile acid metabolic profiling as cholanic acids are almost never measured during bile acid analysis and their origins from microbial bile acid metabolism cannot be made certain without proper stable isotope labeling in vivo to establish that fecal cholanic acids derive from bile acids.
Studies from independent labs report that human fecal suspensions convert 3-sulfo-LCA to isoLCA, Δ3-cholenic acid, and 5β-cholanic acid157,158. The addition of vancomycin to fecal suspensions inhibited 3-sulfo-LCA metabolism; however, methods to select for Gram-positive spore-forming bacteria (such as heat and alcohol treatment of feces) enriched for 3-sulfo-LCA metabolism157. Pure cultures of clostridia were able to generate isoLCA, Δ3-cholenic, and 5β-cholanic acid from 3-sulfo-LCA (FIG. 4)157. The authors speculated that C-O bond cleavage with inversion, indicative of aryl sulfatases, results in formation of isoLCA157. Notably, this process represents a potential alternative route for isoLCA formation distinct from oxidation and epimerization of LCA (described later)157. In this scheme, isoLCA is dehydroxylated to Δ3-cholenic acid and reduced to 5β-cholanic acid. Alternatively, it is possible that trans-elimination of the sulfo-ester yields Δ3-cholenic acid, which is reduced to 5β-cholanic acid. Intriguingly, a study reported that CDCA is converted to 7α-hydroxy-5β-cholan-24-oic acid in human fecal suspensions159, suggesting that the Hylemon–Bjӧrkhem pathway, yielding LCA from CDCA, need not precede C3-dehydroxylation. Further work is needed to firmly establish C3-dehydroxylation, its mechanism(s), and determine the substrate range for bile acid C3-dehydroxylation.
The physiological consequences of 5β-cholanic acid are unclear, but enhancement of bile acid C3-dehydroxylation might represent a strategy to reduce bile acid concentration in the gastrointestinal tract to prevent gastrointestinal cancers, similar to the proposed enhancement of cholesterol conversion to coprostanol by gut bacteria to reduce serum cholesterol in the prevention of cardiovascular disease160,161. In support of this theory, a series of 5β-cholanic acid derivatives were shown to activate FXR more potently than hydroxylated bile acids in a reporter gene assay162. A later pharmacological screen of a small library of 5β-cholanic acid derivatives identified for the first time potent FXR agonists or TGR5 (GP-BAR1) antagonists derived from this steroid backbone163. Notably in the context of hepatogastroenterological disorders, use of an orthotopic mouse model of hepatocellular carcinoma demonstrated that administration of the FXR agonist obeticholic acid, to mimic primary bile acids, together with the TGR5 antagonist 5β-cholanic acid, to block the downstream signaling of secondary bile acids, exhibited substantial tumor suppressive effects through interactions between liver sinusoidal endothelial cells and natural killer T cells mediated by paracrine chemokine signaling164. Clearly, the extent to which host and/or microbial-derived 5β-cholane derivatives are physiologically relevant should be defined, with accurate measurements of such being a first step.
Bile acid C12-dehydroxylation
The removal of the C12 hydroxyl group from bile acids would blur the line between the current distinction between CA metabolites (such as derivatives of DCA) and CDCA or UDCA metabolites (such as derivatives of LCA). Few studies have measured or confirmed the occurrence of C12-dehydroxylation and its prevalence in the human population. There are data demonstrating that the ratio of CA:CDCA metabolites in serum is variable in the population165. A higher ratio of 12α-hydroxylated bile acids (CA, DCA) versus non-12α-hydroxylated bile acids (CDCA and LCA) is associated with insulin resistance in humans166, and the gut microbiomes of individuals with insulin resistance differ from those without167. It is unclear if this altered ratio reflects regulation in bile acid synthesis in the liver (neutral pathway) and extrahepatic tissues (acidic pathway)40, C12-dehydroxylation by the gut microbiome, or a combination of these factors. It was reported that eight human fecal Bacteroides isolates were capable of converting CA to CDCA through C12-dehydroxylation (FIG. 4)168. Unfortunately, however, serial transfers of the bacterial culture resulted in loss of dehydroxylation activity168. Yet, this aspect was also common in the history of isolation of bile acid 7-dehydroxylating strains38. Other studies, including another report further support the possibility that C12-dehydroxylation occurs in the human gut169.
An intermediate expected in this pathway is the formation of an 11,12-unsaturated bile acid, which would be subsequently reduced. Studies assessing the stability of [11,12-3H]CDCA and [11,12-3H]LCA as tracers for isotope dilution studies during enterohepatic circulation in humans inadvertently provided evidence that bacteria can indeed (de)hydrogenate the C11–C12 bond170. Comprehensive and carefully designed studies are needed to resolve the uncertainty regarding C12-dehydroxylation in the human gut. In vitro and in vivo tracing of the fate of isotopically labeled CA is essential in resolving this important question in the field of bile acid microbiology.
Oxidation and epimerization of bile acids
The oxidation and epimerization of bile acid hydroxyl groups greatly expands the diversity of bile acid metabolites as each hydroxyl toggles between three stable positions (such as 3α-OH, 3-oxo, and 3β-OH) (FIG. 4). These reactions are catalyzed by regiospecific and stereospecific pyridine nucleotide-dependent hydroxysteroid dehydrogenases (HSDHs)171. Early work on these enzymes was important for identifying Eggerthella lenta, Blautia producta, Clostridium absonum, Clostridium perfringens, Clostridium paraputrificum, Escherichia coli, Bacteroides fragilis, and Ruminococcus gnavus as species capable of oxidation and reduction of bile acids (Table 2)172–177. The hsd genes encoding these enzymes were later identified and characterized in strains of these species178–182. The latest efforts are identifying a slate of other bacterial isolates capable of oxido-reduction of bile acids28,169.
Important physiological consequences of oxo-bile acids and bile acid epimers have been identified. 7-oxo-CDCA, a product of microbial 7α-HSDH, competitively inhibits hepatic 11β-HSD2, therefore altering glucocorticoid metabolism183. Intriguingly, disruption in the activity of 11β-HSD1 isoform through genetic knockout markedly alters gut microbiome structure and function184. Secondary oxo-bile acids (such as 3-oxo-LCA) inhibit the development of T helper cells that express IL-17 (T helper 17 (TH17) cells) in the gastrointestinal tract27,29. Three bacterial HSDH pathways exist for the metabolism of human bile acids, the genes of which are widely distributed among gut microbial species. CDCA and CA are reversibly oxidized and epimerized at C7 to yield urso-bile acids172,180. UDCA has a long history in treating biliary and gastrointestinal disorders and is a current maintenance therapy for primary sclerosing cholangitis185. CA and CDCA (as well as DCA and LCA) are also epimerized at C3 to form iso-bile acids178,179,186.
Iso-bile acids, particularly isoDCA and isoLCA, are some of the most predominant secondary bile acids in feces, and the fraction returned to the liver in portal circulation are ‘repaired’ to the 3α-hydroxy orientation187. Iso-DCA has been shown to be less toxic to Bacteroides spp.178, and ‘flat’ secondary allo-bile acids (discussed later) such as isoalloLCA modulate colonic regulatory T (Treg) cell function27. Only CA derivatives can be epimerized to so-called lago bile acids (Ancient Greek lagos [λαγώς, “hare”] also known as 12-epi bile acids)117,181. LagoDCA (3α,12β-dihydroxy) is predicted to have similar hydrophilicity to UDCA (3α,7β-dihydroxy), and was tested for decreased toxicity relative to its epimer DCA (3α,12α-dihydroxy) in a rabbit model188. Indeed, whilst lagoDCA was far less toxic than DCA, it was progressively epimerized in the gut to DCA, which was enriched in the bile188. Unlike rats, the liver of rabbits is incapable of converting DCA to CA, so DCA accumulates in rabbit bile, as is also observed in humans. Similar epimerization of UDCA to CDCA occurs in patients with gallstones, but unlike DCA, CDCA enrichment desaturates bile188. Bacterial enzymes in the epi-bile acid pathway have been reported (Table 2), reviewed elsewhere171,179,181,182,189.
Effects of bile acids on host immune cells
Secondary bile acids have long been associated with gastrointestinal disorders associated with chronic inflammation including inflammatory bowel disease (IBD) and colorectal cancer (CRC) with a vast literature providing data consistent with multiple mechanisms of action including: direct cytotoxicity; direct DNA damage; inflammation associated with NF-κB activation; perturbation of cellular redox poise due to reactive oxygen species induction; and enhanced cell proliferation through activation of various cell cycle and inflammatory signaling pathways31,32,190–193. It is generally accepted that these effects reflect to varying degrees both the hydrophobic nature of secondary bile acids resulting in membrane damage to host cells and their activation of numerous cell signaling cascades through interaction with both cell surface and nuclear receptors. There are also numerous reports that secondary bile acids exert anti-inflammatory, immunosuppressive responses in ex vivo and in vitro systems as was expertly reviewed in Jia et al (2018)31, and Cai et al (2022)9.
As already discussed, it is now clear that through numerous enzymatic pathways, the colonic microbiota is capable of generating a highly diverse secondary bile acid metabolome with numerous derivatives rarely being measured due to both inadequate analytical techniques and the lack of chemical standards for less abundant secondary bile acids. It is this diverse secondary bile acid metabolome, as a whole, that likely contributes to setting the inflammatory tone and regulation of tumor cell growth in the colon. Much additional work and novel tissue and cell engineering approaches are needed to gain a more complete and accurate understanding of how the secondary bile acid metabolome contributes to local inflammation and growth control. With such new knowledge, it should become possible to identify a range of somewhat innocuous approaches for manipulating the co-metabolism of secondary bile acids by microbial and host cells to prevent local inflammation or restore normal growth control. Nevertheless, by using gnotobiotic mouse, microbial engineering, and various omics-based approaches, novel insight is emerging regarding contributions of several previously overlooked bile acid derivatives that seem to modulate balance between pathogenic TH17 inflammation and Treg cells with anti-inflammatory properties and a brief summary follows.
With the advantage of looking back, it is now obvious that gnotobiotic studies attempting to identify gut bacterial taxa that influence the development of intestinal CD4+ Treg cells provided an early clue of the likely importance of secondary bile acids when it was observed that the induction of colonic Treg cells was specific to Clostridium-colonized gnotobiotic mice194. This research group later expanded the search for particular clostridial strains capable of inducing CD41+FOXP3+ Treg cells by screening gnotobiotic mice colonized by human microbiota195. This approach identified 17 strains within Clostridia clusters XIVa, IV and XVIII with potent Treg cell induction capabilities. After testing each of the strains individually as well as randomly selected combinations of 3–5 strains, it was concluded that the 17 clostridial strains likely act synergistically to amplify the induction of Treg cells in a microbial-community-dependent manner195. Additional studies designed to pinpoint mechanisms, led to the conclusion that “the 17 strains provide…SCFAs, bacterial antigens and probably other factors, which together contribute to differentiation, expansion and colonic homing of Treg cells” in the mouse196. The potential that the necessity of clostridia for colonic Treg cell development related to their production of secondary bile acids was not acknowledged at that time, although one of the strains was identified as C. scindens196.
A later study, however, by Hang et al. (2019)27 identified two distinct derivatives of LCA, 3-oxoLCA and isoalloLCA, as key regulators of naive CD4+ T cell differentiation in mice after screening a library of 30 primary and secondary bile acid metabolites in in vitro assays under either TH17 cell or Treg cell differentiation conditions. Specifically, 3-oxoLCA inhibited TH17 cell differentiation, as shown by reduced expression of IL-17a, and isoalloLCA enhanced induction of Treg cells, as shown by increased FOXP3 expression. It was further demonstrated that 3-oxoLCA inhibited the differentiation of TH17 cells by directly binding to the key transcription factor retinoid-related orphan receptor-γt (RORγt) and that isoalloLCA enhanced the differentiation of Treg cells through the production of mitochondrial reactive oxygen species, which led to increased expression of FOXP3. Prior evidence that a variety of oxysterols were known to interact with the somewhat promiscuous RORγt transcription factor197,198 provided precedence for the observation of 3-OxoLCA inhibiting TH17 differentiation through direct binding to RORγt. Additional studies published around the same time provided additional support for the importance of secondary bile acids as important modulators of TH17 and Treg cell differentiation. Song et al. (2020)145 reported evidence that LCA and 3-oxoLCA modulate FOXP3+ Treg cells expressing RORγ+ through interactions with the nuclear receptor VDR. By screening the major species of deconjugated bile acids found in mice and humans for their ability to enhance Foxp3 induction in vitro, Campbell et al. (2020)146 found that isoDCA increased Foxp3 induction by acting on dendritic cells (DCs) to diminish their immunostimulatory properties.
Potentiation of Treg cell generation by isoDCA required FXR expression in DCs providing evidence for involvement of an isoDCA-FXR interaction in cells of the myeloid lineage also possibly contributing to the induction of peripherally induced Treg (pTreg) in the mouse intestine (FIG. 5). Li et al. (2021)29 demonstrated that the planar secondary bile acid isoalloLCA enhances Treg cell differentiation through interactions with the nuclear hormone receptor NR4A1 leading to activation of Foxp3 gene transcription and identified a biosynthetic gene cluster in gut Bacteroidota that converts 3-oxoLCA to isoalloLCA as discussed earlier. These authors provided additional compelling evidence through examination of longitudinal metabolomic and metagenomic profiles of stool samples from individuals in the HMP2 IBDMDB cohort199 (Crohn’s disease, ulcerative colitis versus non-IBD controls) and found that isoalloLCA and its biosynthetic genes are substantially reduced in patients with IBD. Intriguingly, the fold change in isoalloLCA in patients with Crohn’s disease and ulcerative colitis compared with controls was the largest among all identified bile acids in the metabolomics data from the HMP2 cohort.
Fig. 5|. Modulation of inflammation and immune cell differentiation and function by secondary bile acid derivatives in the gastrointestinal tract.
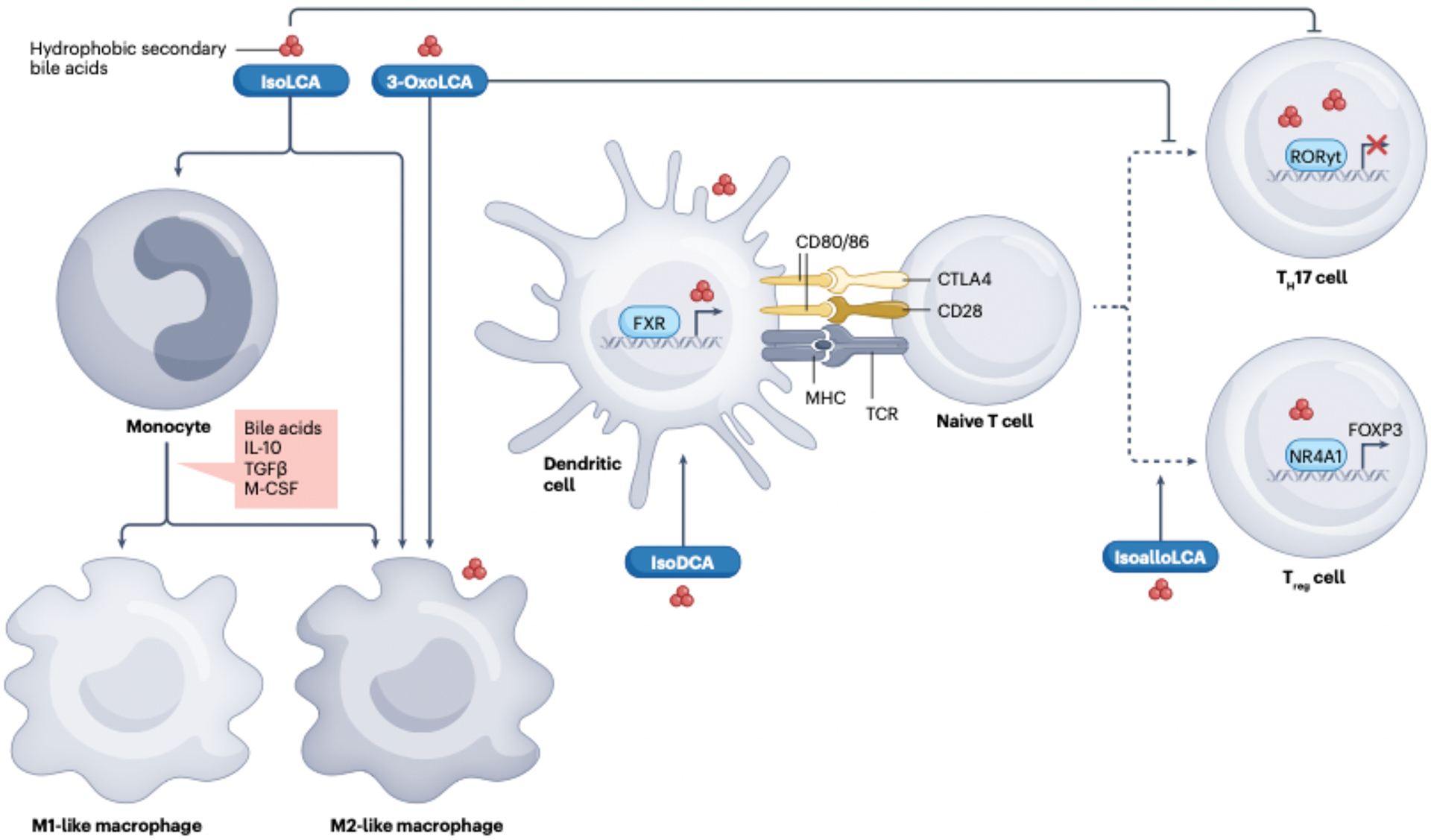
The illustration summarizes the latest observations on the effects of rarely measured and hence understudied microbially derived secondary bile acid derivatives on host immune cells. In general, the data indicate that particular secondary bile acid derivatives exert distinct effects on the differentiation of macrophage progenitors as well as dendritic cell antigen presentation and naive CD4+ T cell differentiation, thereby purportedly influencing the inflammatory tone in the gastrointestinal tract. For specific details on the findings illustrated, readers are referred to the original research papers26–28,57,59,145,146,178,194–196,198, which are summarized in the body of the present Review and subsequent review articles9,29,30,55,216. Briefly, iso-lithocholic acid (isoLCA) and 3-oxoLCA were shown to modulate macrophage polarization states217, iso-deoxycholic acid (isoDCA) induced FOXP3 expression in dendritic cells to diminish their immunostimulatory properties146, the planar secondary bile acid isoalloLCA enhanced regulatory T (Treg) cell differentiation through interactions with the nuclear hormone receptor NR4A1, leading to activation of FOXP3gene transcription28 .and 3-oxoLCA inhibited T helper 17 (TH17) cell differentiation26,27. Similar to 3-oxoLCA, isoLCA suppressed TH17 cell differentiation by inhibiting the canonical transcription factor retinoid-related orphan receptor γt (RORγt+)27. FXR, farnesoid X receptor; M-CSF, macrophage colony-stimulating factor. TCR, T cell receptor.
Further evidence for an apparent beneficial role of certain secondary bile acid derivatives was provided by Sato et al. (2021)58, who showed that the gut microbiome of Japanese centenarians was enriched in bacteria capable of generating isoforms of LCA including isoLCA, 3-oxoLCA, alloLCA, 3-oxoalloLCA, and isoalloLCA. These researchers further identified a biosynthetic pathway for isoalloLCA production by Odoribacteraceae strains and demonstrated that this planar bile acid wielded potent antimicrobial effects against Gram-positive (but not Gram-negative) multidrug-resistant pathogens, including C. difficile and Enterococcus faecium. Paik et al. (2022)28 demonstrated that similar to 3-oxoLCA, isoLCA suppressed TH17 cell differentiation by inhibiting the canonical transcription factor RORγ+ and that fecal concentrations of both 3-oxoLCA and isoLCA and the 3α-HSDH genes required for their biosynthesis were markedly reduced and inversely correlated with the expression of TH17-cell-associated genes in patients with IBD (FIG. 5).
Thus, a series of studies have provided considerable evidence that a subset of secondary bile acid derivatives exerts potent effects on setting inflammatory tone in the intestine through modulation of the balance between TH17-mediated inflammation and immunosuppressive Treg cells. However, a clear mechanistic perspective does not emerge from a stereochemical perspective. Namely, LCA, 3-oxoLCA, isoLCA, alloLCA, isoalloLCA, and isoDCA vary both qualitatively and quantitatively in individual stool samples. Also, in addition to bile acids, the host mucosa is exposed to gradients of a myriad of microbial metabolites that affect immune and stromal cell functions. It is also likely that the bile acid metabolome and the other microbial metabolites vary substantially longitudinally along the gastrointestinal tract, and novel sampling approaches promise to shed light on this aspect in the future114.
Given that bile acids are involved in the regulation of glucose, lipid and energy metabolism, it is not surprising that they often underlie the association between the gut microbiota and metabolic diseases including obesity, diabetes and metabolic dysfunction-associated steatotic liver disease (formerly nonalcoholic fatty liver disease). Relevant mechanisms defined to date involving bile acid signaling through both FXR and TGR5 are summarized in a number of excellent reviews on the topic of bile acids and metabolic disorders95,200–204. A short list of crucial questions that remain largely unanswered include: the extent of heterogeneity in the carriage of bsh and bai genes in the microbiome across human populations in both healthy individuals and those presenting with various metabolic disorders95; the range of responsiveness to macronutrients and micronutrients in diet among taxa harboring bsh and bai genes; the effect of stereoisomers of secondary bile acids and MCBAs on metabolic processes; and the effectiveness of phage-based approaches and engineered commensal microorganisms to generate bile acid pools that optimize host metabolic function (FIG. 3).
Conclusions
This Review has attempted to update key advances in classical bile acid biotransformation pathways by the gut microbiome such as bile acid hydrolysis, oxidation and epimerization, and C7-dehydroxylation. The link between the products of these microbial biotransformations and human health and disease, particularly immune function, are highlighted. We also critically reviewed the evidence for lesser-known microbial reactions such as bile acid esterification, C3-dehydroxylation and C12-dehydroxylation, for which future work is needed to firmly establish these functions, identify the bacteria responsible, and the enzymes catalyzing these reactions. With advances in microbiome science, and the rebirth in interest in bile acid microbiology, the time has arrived to settle outstanding questions in bile acid (micro)biology. The knowledge gained is anticipated to lead to novel microbiome-based interventions aimed at modulating the bile acid pool in the prevention and treatment of gastrointestinal diseases.
Key Points.
Co-metabolism of bile acids is among the most studied aspects of host–microbiota interactions important for human health, although many mechanistic questions remain unanswered.
A substantial gap in our knowledge still exists with respect to the host synthesis of bile acid A/B-ring trans-isomers known as allo-bile acids.
Untargeted metabolomics identified microbially conjugated bile acids, which seem to be generated via bile salt hydrolase enzymes and can signal through PXR and FXR, although their physiological relevance is not fully understood.
Much of the biochemistry and enzymology of microbial bile acid 7-dehydroxylation is established, however, the enzymology of C3 and C12-dehydroxylation requires additional work, as does host responses to these products.
The oxidation and epimerization of bile acid hydroxyl groups greatly expands the diversity of bile acid metabolites as each hydroxyl toggles between three stable positions (e.g., 3α-OH, 3-oxo, and 3β-OH).
Secondary bile acid epimers that have not been measured historically are emerging as potent modulators of the balance between T-helper-17-mediated inflammation and immunosuppressive regulatory T cells in the intestine.
This Review discusses the synthesis and metabolism of bile acids and the role of the gut microbiome in bile acid metabolism. Insights into how secondary bile acid derivatives influence host immune function are also described.
Acknowledgements
We express sincere appreciation to Steven Daniel for his assistance in editing and offering constructive comments on this review, and to Hanchu Dai for providing Figure 3. We acknowledge financial support from National Institutes of Health grants (R01 CA204808-01 [J.M.R., H.R.G.], R01 GM134423-01A1 [J.M.R.], R01 GM145920-01 [J.M.R], R03 AI147127-01A1 [J.M.R.]) as well as UIUC Department of Animal Sciences Matchstick grant and Hatch ILLU-538-916 (J.M.R.).
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Haslewood GA The biological significance of chemical differences in bile salts. Biol Rev Camb Philos Soc 39, 537–574, doi: 10.1111/j.1469-185x.1964.tb01170.x (1964). [DOI] [PubMed] [Google Scholar]
- 2.Hofmann AF, Hagey LR & Krasowski MD Bile salts of vertebrates: structural variation and possible evolutionary significance. J Lipid Res 51, 226–246, doi: 10.1194/jlr.R000042 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haslewood GA Bile salt evolution. J Lipid Res 8, 535–550 (1967). [PubMed] [Google Scholar]
- 4.Wang DQ & Carey MC Therapeutic uses of animal biles in traditional Chinese medicine: An ethnopharmacological, biophysical chemical and medicinal review. World J Gastroenterol 20, 9952–9975, doi: 10.3748/wjg.v20.i29.9952 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frisch K & Alstrup AKO On the evolution of bile salts and the farnesoid X receptor in vertebrates. Physiol Biochem Zool 91, 797–813, doi: 10.1086/695810 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Russell DW The enzymes, regulation, and genetics of bile acid synthesis. Annu Rev Biochem 72, 137–174, doi: 10.1146/annurev.biochem.72.121801.161712 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Wahlstrom A, Sayin SI, Marschall HU & Backhed F Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab 24, 41–50, doi: 10.1016/j.cmet.2016.05.005 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Watanabe M et al. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature 439, 484–489, doi: 10.1038/nature04330 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Cai J, Sun L & Gonzalez FJ Gut microbiota-derived bile acids in intestinal immunity, inflammation, and tumorigenesis. Cell Host Microbe 30, 289–300, doi: 10.1016/j.chom.2022.02.004 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dam H Medical aspects of vitamin K. Journal-Lancet 63, 353 (1943). [Google Scholar]
- 11.Hofmann AF Bile acids: Trying to understand their chemistry and biology with the hope of helping patients. Hepatology 49, 1403–1418, doi: 10.1002/hep.22789 (2009). [DOI] [PubMed] [Google Scholar]
- 12.Hofmann AF & Hagey LR Key discoveries in bile acid chemistry and biology and their clinical applications: History of the last eight decades. J Lipid Res 55, 1553–1595, doi: 10.1194/jlr.R049437 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Faassen A et al. Plasma deoxycholic acid is related to deoxycholic acid in faecal water. Cancer Lett 114, 293–294, doi: 10.1016/s0304-3835(97)04683-1 (1997). [DOI] [PubMed] [Google Scholar]
- 14.Bayerdorffer E et al. Unconjugated secondary bile acids in the serum of patients with colorectal adenomas. Gut 36, 268–273, doi: 10.1136/gut.36.2.268 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berr F, Kullak-Ublick GA, Paumgartner G, Munzing W & Hylemon PB 7α-dehydroxylating bacteria enhance deoxycholic acid input and cholesterol saturation of bile in patients with gallstones. Gastroenterology 111, 1611–1620, doi: 10.1016/s0016-5085(96)70024-0 (1996). [DOI] [PubMed] [Google Scholar]
- 16.Zhou H & Hylemon PB Bile acids are nutrient signaling hormones. Steroids 86, 62–68, doi: 10.1016/j.steroids.2014.04.016 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Makishima M et al. Identification of a nuclear receptor for bile acids. Science 284, 1362–1365, doi: 10.1126/science.284.5418.1362 (1999). [DOI] [PubMed] [Google Scholar]
- 18.Wang H, Chen J, Hollister K, Sowers LC & Forman BM Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol Cell 3, 543–553, doi: 10.1016/s1097-2765(00)80348-2 (1999). [DOI] [PubMed] [Google Scholar]
- 19.Parks DJ et al. Bile acids: Natural ligands for an orphan nuclear receptor. Science 284, 1365–1368, doi: 10.1126/science.284.5418.1365 (1999). [DOI] [PubMed] [Google Scholar]
- 20.Staudinger JL et al. The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc Natl Acad Sci U S A 98, 3369–3374, doi: 10.1073/pnas.051551698 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Makishima M et al. Vitamin D receptor as an intestinal bile acid sensor. Science 296, 1313–1316, doi: 10.1126/science.1070477 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Guo GL et al. Complementary roles of farnesoid X receptor, pregnane X receptor, and constitutive androstane receptor in protection against bile acid toxicity. J Biol Chem 278, 45062–45071, doi: 10.1074/jbc.M307145200 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Maruyama T et al. Identification of membrane-type receptor for bile acids (M-BAR). Biochem Biophys Res Commun 298, 714–719, doi: 10.1016/s0006-291x(02)02550-0 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Kawamata Y et al. A G protein-coupled receptor responsive to bile acids. J Biol Chem 278, 9435–9440, doi: 10.1074/jbc.M209706200 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Raufman JP, Cheng K & Zimniak P Activation of muscarinic receptor signaling by bile acids: Physiological and medical implications. Dig Dis Sci 48, 1431–1444, doi: 10.1023/a:1024733500950 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Nagahashi M et al. The roles of bile acids and sphingosine-1-phosphate signaling in the hepatobiliary diseases. J Lipid Res 57, 1636–1643, doi: 10.1194/jlr.R069286 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hang S et al. Bile acid metabolites control TH17 and Treg cell differentiation. Nature 576, 143–148, doi: 10.1038/s41586-019-1785-z (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paik D et al. Human gut bacteria produce TH17-modulating bile acid metabolites. Nature 603, 907–912, doi: 10.1038/s41586-022-04480-z (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li W et al. A bacterial bile acid metabolite modulates Treg activity through the nuclear hormone receptor NR4A1. Cell Host Microbe 29, 1366–1377 e1369, doi: 10.1016/j.chom.2021.07.013 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fuchs CD & Trauner M Role of bile acids and their receptors in gastrointestinal and hepatic pathophysiology. Nat Rev Gastroenterol Hepatol 19, 432–450, doi: 10.1038/s41575-021-00566-7 (2022). [DOI] [PubMed] [Google Scholar]
- 31.Jia W, Xie G & Jia W Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat Rev Gastroenterol Hepatol 15, 111–128, doi: 10.1038/nrgastro.2017.119 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Keefe SJ Diet, microorganisms and their metabolites, and colon cancer. Nat Rev Gastroenterol Hepatol 13, 691–706, doi: 10.1038/nrgastro.2016.165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ocvirk S & O’Keefe SJD Dietary fat, bile acid metabolism and colorectal cancer. Semin Cancer Biol 73, 347–355, doi: 10.1016/j.semcancer.2020.10.003 (2021). [DOI] [PubMed] [Google Scholar]
- 34.Sender R, Fuchs S & Milo R Revised estimates for the number of human and bacteria cells in the body. PLoS Biol 14, e1002533, doi: 10.1371/journal.pbio.1002533 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qin J et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65, doi: 10.1038/nature08821 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swann JR et al. Systemic gut microbial modulation of bile acid metabolism in host tissue compartments. Proc Natl Acad Sci U S A 108 Suppl 1, 4523–4530, doi: 10.1073/pnas.1006734107 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sayin SI et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab 17, 225–235, doi: 10.1016/j.cmet.2013.01.003 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Ridlon JM, Daniel SL & Gaskins HR The Hylemon-Bjorkhem pathway of bile acid 7-dehydroxylation: History, biochemistry, and microbiology. J Lipid Res 64, 100392, doi: 10.1016/j.jlr.2023.100392 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Russell DW Fifty years of advances in bile acid synthesis and metabolism. J Lipid Res 50 Suppl, S120–125, doi: 10.1194/jlr.R800026-JLR200 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pandak WM & Kakiyama G The acidic pathway of bile acid synthesis: Not just an alternative pathway. Liver Res 3, 88–98, doi: 10.1016/j.livres.2019.05.001 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Phelps T, Snyder E, Rodriguez E, Child H & Harvey P The influence of biological sex and sex hormones on bile acid synthesis and cholesterol homeostasis. Biol Sex Differ 10, 52, doi: 10.1186/s13293-019-0265-3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sjoland W et al. Absence of gut microbiota reduces neonatal survival and exacerbates liver disease in Cyp2c70-deficient mice with a human-like bile acid composition. Clin Sci (Lond) 137, 995–1011, doi: 10.1042/CS20230413 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Truong JK et al. Ileal bile acid transporter inhibition in Cyp2c70 KO mice ameliorates cholestatic liver injury. J Lipid Res 63, 100261, doi: 10.1016/j.jlr.2022.100261 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ridlon JM, Wolf PG & Gaskins HR Taurocholic acid metabolism by gut microbes and colon cancer. Gut Microbes 7, 201–215, doi: 10.1080/19490976.2016.1150414 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li J & Dawson PA Animal models to study bile acid metabolism. Biochim Biophys Acta Mol Basis Dis 1865, 895–911, doi: 10.1016/j.bbadis.2018.05.011 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Daly JW, Keely SJ & Gahan CG M. Functional and phylogenetic diversity of BSH and PVA enzymes. Microorganisms 9, doi: 10.3390/microorganisms9040732 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wolf PG et al. Diversity and distribution of sulfur metabolic genes in the human gut microbiome and their association with colorectal cancer. Microbiome 10, 64, doi: 10.1186/s40168-022-01242-x (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hale VL et al. Distinct microbes, metabolites, and ecologies define the microbiome in deficient and proficient mismatch repair colorectal cancers. Genome Med 10, 78, doi: 10.1186/s13073-018-0586-6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yazici C et al. Race-dependent association of sulfidogenic bacteria with colorectal cancer. Gut 66, 1983–1994, doi: 10.1136/gutjnl-2016-313321 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goossens JF & Bailly C Ursodeoxycholic acid and cancer: From chemoprevention to chemotherapy. Pharmacol Ther 203, 107396, doi: 10.1016/j.pharmthera.2019.107396 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Pellicciari R et al. Avicholic acid: A lead compound from birds on the route to potent tgr5 modulators. ACS Med Chem Lett 3, 273–277, doi: 10.1021/ml200256d (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hagey LR, Vidal N, Hofmann AF & Krasowski MD Evolutionary diversity of bile salts in reptiles and mammals, including analysis of ancient human and extinct giant ground sloth coprolites. BMC Evol Biol 10, 133, doi: 10.1186/1471-2148-10-133 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gregor R et al. Mammalian gut metabolomes mirror microbiome composition and host phylogeny. ISME J 16, 1262–1274, doi: 10.1038/s41396-021-01152-0 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wen J et al. Fxr signaling and microbial metabolism of bile salts in the zebrafish intestine. Sci Adv 7, doi: 10.1126/sciadv.abg1371 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shiffka SJ et al. Quantification of common and planar bile acids in tissues and cultured cells. J Lipid Res 61, 1524–1535, doi: 10.1194/jlr.D120000726 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shiffka SJ, Kane MA & Swaan PW Planar bile acids in health and disease. Biochim Biophys Acta Biomembr 1859, 2269–2276, doi: 10.1016/j.bbamem.2017.08.019 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hagey LR, Moller PR, Hofmann AF & Krasowski MD Diversity of bile salts in fish and amphibians: Evolution of a complex biochemical pathway. Physiol Biochem Zool 83, 308–321, doi: 10.1086/649966 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sato Y et al. Novel bile acid biosynthetic pathways are enriched in the microbiome of centenarians. Nature 599, 458–464, doi: 10.1038/s41586-021-03832-5 (2021). [DOI] [PubMed] [Google Scholar]
- 59.Bokkenheuser V, Hoshita T & Mosbach EH Bacterial 7-dehydroxylation of cholic acid and allocholic acid. J Lipid Res 10, 421–426 (1969). [PubMed] [Google Scholar]
- 60.Lee JW et al. Formation of secondary allo-bile acids by novel enzymes from gut Firmicutes. Gut Microbes 14, 2132903, doi: 10.1080/19490976.2022.2132903 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Frankel M The biological splitting of conjugated bile acids. Biochem J 30, 2111–2116, doi: 10.1042/bj0302111 (1936). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dong Z & Lee BH Bile salt hydrolases: Structure and function, substrate preference, and inhibitor development. Protein Sci 27, 1742–1754, doi: 10.1002/pro.3484 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chand D et al. Molecular features of bile salt hydrolases and relevance in human health. Biochim Biophys Acta Gen Subj 1861, 2981–2991, doi: 10.1016/j.bbagen.2016.09.024 (2017). [DOI] [PubMed] [Google Scholar]
- 64.Joyce SA, Shanahan F, Hill C & Gahan CG Bacterial bile salt hydrolase in host metabolism: Potential for influencing gastrointestinal microbe-host crosstalk. Gut Microbes 5, 669–674, doi: 10.4161/19490976.2014.969986 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Begley M, Hill C & Gahan CG Bile salt hydrolase activity in probiotics. Appl Environ Microbiol 72, 1729–1738, doi: 10.1128/AEM.72.3.1729-1738.2006 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Foley MH, O’Flaherty S, Barrangou R & Theriot CM Bile salt hydrolases: Gatekeepers of bile acid metabolism and host-microbiome crosstalk in the gastrointestinal tract. PLoS Pathog 15, e1007581, doi: 10.1371/journal.ppat.1007581 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Foley MH et al. Bile salt hydrolases shape the bile acid landscape and restrict Clostridioides difficile growth in the murine gut. Nat Microbiol 8, 611–628, doi: 10.1038/s41564-023-01337-7 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Foley MH et al. Lactobacillus bile salt hydrolase substrate specificity governs bacterial fitness and host colonization. Proc Natl Acad Sci U S A 118, doi: 10.1073/pnas.2017709118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tanaka H, Hashiba H, Kok J & Mierau I Bile salt hydrolase of Bifidobacterium longum-biochemical and genetic characterization. Appl Environ Microbiol 66, 2502–2512, doi: 10.1128/aem.66.6.2502-2512.2000 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chand D, Panigrahi P, Varshney N, Ramasamy S & Suresh CG Structure and function of a highly active bile salt hydrolase (BSH) from Enterococcus faecalis and post-translational processing of BSH enzymes. Biochim Biophys Acta Proteins Proteom 1866, 507–518, doi: 10.1016/j.bbapap.2018.01.003 (2018). [DOI] [PubMed] [Google Scholar]
- 71.Yao L et al. A selective gut bacterial bile salt hydrolase alters host metabolism. Elife 7, doi: 10.7554/eLife.37182 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sun L et al. Bile salt hydrolase in non-enterotoxigenic Bacteroides potentiates colorectal cancer. Nat Commun 14, 755, doi: 10.1038/s41467-023-36089-9 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jones BV, Begley M, Hill C, Gahan CG & Marchesi JR Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc Natl Acad Sci U S A 105, 13580–13585, doi: 10.1073/pnas.0804437105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Marchesini MI et al. Brucella abortus choloylglycine hydrolase affects cell envelope composition and host cell internalization. PLoS One 6, e28480, doi: 10.1371/journal.pone.0028480 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Delpino MV et al. A bile salt hydrolase of Brucella abortus contributes to the establishment of a successful infection through the oral route in mice. Infect Immun 75, 299–305, doi: 10.1128/iai.00952-06 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gahan CG & Hill C Listeria monocytogenes: survival and adaptation in the gastrointestinal tract. Front Cell Infect Microbiol 4, 9, doi: 10.3389/fcimb.2014.00009 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dussurget O et al. Listeria monocytogenes bile salt hydrolase is a PrfA-regulated virulence factor involved in the intestinal and hepatic phases of listeriosis. Mol Microbiol 45, 1095–1106, doi: 10.1046/j.1365-2958.2002.03080.x (2002). [DOI] [PubMed] [Google Scholar]
- 78.Begley M, Sleator RD, Gahan CG & Hill C Contribution of three bile-associated loci, bsh, pva, and btlB, to gastrointestinal persistence and bile tolerance of Listeria monocytogenes. Infect Immun 73, 894–904, doi: 10.1128/iai.73.2.894-904.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Begley M, Gahan CG & Hill C The interaction between bacteria and bile. FEMS Microbiol Rev 29, 625–651, doi: 10.1016/j.femsre.2004.09.003 (2005). [DOI] [PubMed] [Google Scholar]
- 80.Buchinger TJ, Li W & Johnson NS Bile salts as semiochemicals in fish. Chem Senses 39, 647–654, doi: 10.1093/chemse/bju039 (2014). [DOI] [PubMed] [Google Scholar]
- 81.Hahn MA, Effertz C, Bigler L & von Elert E 5α-cyprinol sulfate, a bile salt from fish, induces diel vertical migration in Daphnia. Elife 8, doi: 10.7554/eLife.44791 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Goto T et al. Physicochemical and physiological properties of 5α-cyprinol sulfate, the toxic bile salt of cyprinid fish. J Lipid Res 44, 1643–1651, doi: 10.1194/jlr.M300155-JLR200 (2003). [DOI] [PubMed] [Google Scholar]
- 83.Abt MC, McKenney PT & Pamer EG Clostridium difficile colitis: Pathogenesis and host defence. Nat Rev Microbiol 14, 609–620, doi: 10.1038/nrmicro.2016.108 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Craddock AL et al. Expression and transport properties of the human ileal and renal sodium-dependent bile acid transporter. Am J Physiol 274, G157–169, doi: 10.1152/ajpgi.1998.274.1.G157 (1998). [DOI] [PubMed] [Google Scholar]
- 85.Dietschy JM & Turley SD Control of cholesterol turnover in the mouse. J Biol Chem 277, 3801–3804, doi: 10.1074/jbc.R100057200 (2002). [DOI] [PubMed] [Google Scholar]
- 86.De Smet I, De Boever P & Verstraete W Cholesterol lowering in pigs through enhanced bacterial bile salt hydrolase activity. Br J Nutr 79, 185–194, doi: 10.1079/bjn19980030 (1998). [DOI] [PubMed] [Google Scholar]
- 87.Damodharan K, Lee YS, Palaniyandi SA, Yang SH & Suh JW Preliminary probiotic and technological characterization of Pediococcus pentosaceus strain KID7 and in vivo assessment of its cholesterol-lowering activity. Front Microbiol 6, 768, doi: 10.3389/fmicb.2015.00768 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ooi LG & Liong MT Cholesterol-lowering effects of probiotics and prebiotics: a review of in vivo and in vitro findings. Int J Mol Sci 11, 2499–2522, doi: 10.3390/ijms11062499 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Feighner SD & Dashkevicz MP Subtherapeutic levels of antibiotics in poultry feeds and their effects on weight gain, feed efficiency, and bacterial cholyltaurine hydrolase activity. Appl Environ Microbiol 53, 331–336, doi: 10.1128/aem.53.2.331-336.1987 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Geng W et al. Evaluation of bile salt hydrolase inhibitor efficacy for modulating host bile profile and physiology using a chicken model system. Sci Rep 10, 4941, doi: 10.1038/s41598-020-61723-7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Korpela K et al. Intestinal microbiome is related to lifetime antibiotic use in Finnish pre-school children. Nat Commun 7, 10410, doi: 10.1038/ncomms10410 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Long SL, Gahan CG & Joyce SA Interactions between gut bacteria and bile in health and disease. Mol Aspects Med 56, 54–65 (2017). [DOI] [PubMed] [Google Scholar]
- 93.Nunez-Sanchez MA et al. Microbial bile salt hydrolase activity influences gene expression profiles and gastrointestinal maturation in infant mice. Gut Microbes 14, 2149023, doi: 10.1080/19490976.2022.2149023 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Joyce SA et al. Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut. Proc Natl Acad Sci U S A 111, 7421–7426, doi: 10.1073/pnas.1323599111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Song Z et al. Taxonomic profiling and populational patterns of bacterial bile salt hydrolase (BSH) genes based on worldwide human gut microbiome. Microbiome 7, 9, doi: 10.1186/s40168-019-0628-3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jia B, Park D, Hahn Y & Jeon CO Metagenomic analysis of the human microbiome reveals the association between the abundance of gut bile salt hydrolases and host health. Gut Microbes 11, 1300–1313, doi: 10.1080/19490976.2020.1748261 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Adhikari AA et al. A gut-restricted lithocholic acid analog as an inhibitor of gut bacterial bile salt hydrolases. ACS Chem Biol 16, 1401–1412, doi: 10.1021/acschembio.1c00192 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Adhikari AA et al. Development of a covalent inhibitor of gut bacterial bile salt hydrolases. Nat Chem Biol 16, 318–326, doi: 10.1038/s41589-020-0467-3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chatterjee B, Echchgadda I & Song CS Vitamin D receptor regulation of the steroid/bile acid sulfotransferase SULT2A1. Methods Enzymol 400, 165–191, doi: 10.1016/s0076-6879(05)00010-8 (2005). [DOI] [PubMed] [Google Scholar]
- 100.Chaudhari SN et al. A microbial metabolite remodels the gut-liver axis following bariatric surgery. Cell Host Microbe 29, 408–424 e407, doi: 10.1016/j.chom.2020.12.004 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Eyssen HJ, Parmentier GG & Mertens JA Sulfate bile acids in germ-free and conventional mice. Eur J Biochem 66, 507–514, doi: 10.1111/j.1432-1033.1976.tb10576.x (1976). [DOI] [PubMed] [Google Scholar]
- 102.Robben J, Caenepeel P, Van Eldere J & Eyssen H Effects of intestinal microbial bile salt sulfatase activity on bile salt kinetics in gnotobiotic rats. Gastroenterology 94, 494–502, doi: 10.1016/0016-5085(88)90443-x (1988). [DOI] [PubMed] [Google Scholar]
- 103.Robben J, Parmentier G & Eyssen H Isolation of a rat intestinal Clostridium strain producing 5α- and 5β-bile salt 3α-sulfatase activity. Appl Environ Microbiol 51, 32–38, doi: 10.1128/aem.51.1.32-38.1986 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Huijghebaert SM & Eyssen HJ Specificity of bile salt sulfatase activity from Clostridium sp. strains S1. Appl Environ Microbiol 44, 1030–1034, doi: 10.1128/aem.44.5.1030-1034.1982 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Eyssen H, Van Eldere J, Parmentier G, Huijghebaert S & Mertens J Influence of microbial bile salt desulfation upon the fecal excretion of bile salts in gnotobiotic rats. J Steroid Biochem 22, 547–554, doi: 10.1016/0022-4731(85)90176-1 (1985). [DOI] [PubMed] [Google Scholar]
- 106.Van Eldere J, Robben J, De Pauw G, Merckx R & Eyssen H Isolation and identification of intestinal steroid-desulfating bacteria from rats and humans. Appl Environ Microbiol 54, 2112–2117, doi: 10.1128/aem.54.8.2112-2117.1988 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yao L et al. A biosynthetic pathway for the selective sulfonation of steroidal metabolites by human gut bacteria. Nat Microbiol 7, 1404–1418, doi: 10.1038/s41564-022-01176-y (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hofmann AF et al. A proposed nomenclature for bile acids. J Lipid Res 33, 599–604 (1992). [PubMed] [Google Scholar]
- 109.Morgan XC, Segata N & Huttenhower C Biodiversity and functional genomics in the human microbiome. Trends Genet 29, 51–58, doi: 10.1016/j.tig.2012.09.005 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Consortium HMP A framework for human microbiome research. Nature 486, 215–221, doi: 10.1038/nature11209 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Quinn RA et al. Bridging the gap between analytical and microbial sciences in microbiome research. mSystems 6, e0058521, doi: 10.1128/mSystems.00585-21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Quinn RA et al. Global chemical effects of the microbiome include new bile-acid conjugations. Nature 579, 123–129, doi: 10.1038/s41586-020-2047-9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Guzior D et al. Bile salt hydrolase/aminoacyltransferase shapes the microbiome. Not Published (2022). [Google Scholar]
- 114.Shalon D et al. Profiling the human intestinal environment under physiological conditions. Nature 617, 581–591, doi: 10.1038/s41586-023-05989-7 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dorrestein P et al. A synthesis-based reverse metabolomics approach for the discovery of chemical structures from humans and animals. Not Published (2021). [Google Scholar]
- 116.Edenharder R & Hammann R Deoxycholic acid methyl ester — a novel bacterial metabolite of cholic acid. Syst Appl Microbiol 6, 18–22, doi: 10.1016/s0723-2020(85)80005-9 (1985). [DOI] [Google Scholar]
- 117.Edenharder R & Schneider J 12β-dehydrogenation of bile acids by Clostridium paraputrificum, C. tertium, and C. difficile and epimerization at carbon-12 of deoxycholic acid by cocultivation with 12α-dehydrogenating Eubacterium lentum. Appl Environ Microbiol 49, 964–968, doi: 10.1128/aem.49.4.964-968.1985 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Takei H et al. Characterization of long-chain fatty acid-linked bile acids: A major conjugation form of 3β-hydroxy bile acids in feces. J Lipid Res 63, 100275, doi: 10.1016/j.jlr.2022.100275 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Benson GM et al. Polydeoxycholate in human and hamster feces: a major product of cholate metabolism. J Lipid Res 34, 2121–2134 (1993). [PubMed] [Google Scholar]
- 120.Schoeler M & Caesar R Dietary lipids, gut microbiota and lipid metabolism. Rev Endocr Metab Disord 20, 461–472, doi: 10.1007/s11154-019-09512-0 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ridlon JM & Bajaj JS The human gut sterolbiome: Bile acid-microbiome endocrine aspects and therapeutics. Acta Pharm Sin B 5, 99–105, doi: 10.1016/j.apsb.2015.01.006 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ridlon JM Conceptualizing the vertebrate sterolbiome. Appl Environ Microbiol 86, doi: 10.1128/AEM.00641-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Samuelsson B Bile acids and steroids: 96. On the mechanism of the biological formation of deoxycholic acid from cholic acid. J Biol Chem 235, 361–366, doi: 10.1016/s0021-9258(18)69529-8 (1960). [DOI] [Google Scholar]
- 124.Ferrari A & Beretta L Activity on bile acids of a Clostridium bifermentans cell-free extract. FEBS Lett 75, 163–165, doi: 10.1016/0014-5793(77)80076-8 (1977). [DOI] [PubMed] [Google Scholar]
- 125.White BA et al. Cofactor requirements for 7α-dehydroxylation of cholic and chenodeoxycholic acid in cell extracts of the intestinal anaerobic bacterium, Eubacterium species V.P.I. 12708. J. Lipid Res 22, 891–898 (1981). [PubMed] [Google Scholar]
- 126.Hylemon PB, Melone PD, Franklund CV, Lund E & Björkhem I Mechanism of intestinal 7α-dehydroxylation of cholic acid: Evidence that allo-deoxycholic acid is an inducible side-product. J. Lipid Res 32, 89–96 (1991). [PubMed] [Google Scholar]
- 127.Mallonee DH, White WB & Hylemon PB Cloning and sequencing of a bile acid-inducible operon from Eubacterium sp. strain VPI 12708. J Bacteriol 172, 7011–7019, doi: 10.1128/jb.172.12.7011-7019.1990 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wise JL & Cummings BP The 7-α-dehydroxylation pathway: An integral component of gut bacterial bile acid metabolism and potential therapeutic target. Front Microbiol 13, 1093420, doi: 10.3389/fmicb.2022.1093420 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Funabashi M et al. A metabolic pathway for bile acid dehydroxylation by the gut microbiome. Nature 582, 566–570, doi: 10.1038/s41586-020-2396-4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Heinken A et al. Systematic assessment of secondary bile acid metabolism in gut microbes reveals distinct metabolic capabilities in inflammatory bowel disease. Microbiome 7, 75, doi: 10.1186/s40168-019-0689-3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Buffie CG et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517, 205–208, doi: 10.1038/nature13828 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wirbel J et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat Med 25, 679–689, doi: 10.1038/s41591-019-0406-6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Schmassmann A et al. Cholylsarcosine, a new bile acid analogue: Metabolism and effect on biliary secretion in humans. Gastroenterology 104, 1171–1181, doi: 10.1016/0016-5085(93)90289-o (1993). [DOI] [PubMed] [Google Scholar]
- 134.Batta AK et al. Side chain conjugation prevents bacterial 7-dehydroxylation of bile acids. J Biol Chem 265, 10925–10928 (1990). [PubMed] [Google Scholar]
- 135.White BA, Lipsky RL, Fricke RJ & Hylemon PB Bile acid induction specificity of 7α-dehydroxylase activity in an intestinal Eubacterium species. Steroids 35, 103–109, doi: 10.1016/0039-128x(80)90115-4 (1980). [DOI] [PubMed] [Google Scholar]
- 136.Kim KH et al. Identification and characterization of major bile acid 7 α-dehydroxylating bacteria in the human gut. mSystems 7, e0045522, doi: 10.1128/msystems.00455-22 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Devendran S et al. Clostridium scindens ATCC 35704: Integration of nutritional requirements, the complete genome sequence, and global transcriptional responses to bile acids. Appl Environ Microbiol 85, doi: 10.1128/AEM.00052-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ridlon JM, Harris SC, Bhowmik S, Kang DJ & Hylemon PB Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes 7, 22–39, doi: 10.1080/19490976.2015.1127483 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Doerner KC, Takamine F, LaVoie CP, Mallonee DH & Hylemon PB Assessment of fecal bacteria with bile acid 7α-dehydroxylating activity for the presence of bai-like genes. Appl Environ Microbiol 63, 1185–1188, doi: 10.1128/aem.63.3.1185-1188.1997 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Marion S et al. Biogeography of microbial bile acid transformations along the murine gut. J Lipid Res 61, 1450–1463, doi: 10.1194/jlr.RA120001021 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Studer N et al. Functional intestinal bile acid 7α-dehydroxylation by Clostridium scindens associated with protection from Clostridium difficile infection in a gnotobiotic mouse model. Front Cell Infect Microbiol 6, 191, doi: 10.3389/fcimb.2016.00191 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wang M et al. Strain dropouts reveal interactions that govern the metabolic output of the gut microbiome. Cell 186, 2839–2852. e2821 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ridlon JM et al. The ‘in vivo lifestyle’of bile acid 7α-dehydroxylating bacteria: Comparative genomics, metatranscriptomic, and bile acid metabolomics analysis of a defined microbial community in gnotobiotic mice. Gut microbes 11, 381–404 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Narushima S et al. Deoxycholic acid formation in gnotobiotic mice associated with human intestinal bacteria. Lipids 41, 835–843, doi: 10.1007/s11745-006-5038-1 (2006). [DOI] [PubMed] [Google Scholar]
- 145.Song X et al. Microbial bile acid metabolites modulate gut RORγ+ regulatory T cell homeostasis. Nature 577, 410–415, doi: 10.1038/s41586-019-1865-0 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Campbell C et al. Bacterial metabolism of bile acids promotes generation of peripheral regulatory T cells. Nature 581, 475–479, doi: 10.1038/s41586-020-2193-0 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Yano JM et al. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 161, 264–276, doi: 10.1016/j.cell.2015.02.047 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Grau KR et al. The intestinal regionalization of acute norovirus infection is regulated by the microbiota via bile acid-mediated priming of type III interferon. Nat Microbiol 5, 84–92, doi: 10.1038/s41564-019-0602-7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ridlon JM, Kang DJ, Hylemon PB & Bajaj JS Gut microbiota, cirrhosis, and alcohol regulate bile acid metabolism in the gut. Dig Dis 33, 338–345, doi: 10.1159/000371678 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kakiyama G et al. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J Hepatol 58, 949–955, doi: 10.1016/j.jhep.2013.01.003 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hylemon PB, Su L, Zheng PC, Bajaj JS & Zhou H Bile acids, gut microbiome and the road to fatty liver disease. Compr Physiol 12, 2719–2730, doi: 10.1002/cphy.c210024 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Bajaj JS et al. Antibiotic-associated disruption of microbiota composition and function in cirrhosis is restored by fecal transplant. Hepatology 68, 1549–1558, doi: 10.1002/hep.30037 (2018). [DOI] [PubMed] [Google Scholar]
- 153.Kallner A The transformation of deoxycholic acid into allodeoxycholic acid in the rat. Bile acids and steroids.174. Acta Chem Scand 21, 87–92, doi: 10.3891/acta.chem.scand.21-0087 (1967). [DOI] [PubMed] [Google Scholar]
- 154.Bernstein H, Bernstein C, Payne CM & Dvorak K Bile acids as endogenous etiologic agents in gastrointestinal cancer. World J Gastroenterol 15, 3329–3340, doi: 10.3748/wjg.15.3329 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Allan RN, Thistle JL, Hofmann AF, Carter JA & Yu PY Lithocholate metabolism during chenotherapy for gallstone dissolution. 1. Serum levels of sulphated and unsulphated lithocholates. Gut 17, 405–412, doi: 10.1136/gut.17.6.405 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hofmann AF Detoxification of lithocholic acid, a toxic bile acid: Relevance to drug hepatotoxicity. Drug Metab Rev 36, 703–722, doi: 10.1081/dmr-200033475 (2004). [DOI] [PubMed] [Google Scholar]
- 157.Borriello SP & Owen RW The metabolism of lithocholic acid and lithocholic acid-3-α-sulfate by human fecal bacteria. Lipids 17, 477–482, doi: 10.1007/BF02535328 (1982). [DOI] [PubMed] [Google Scholar]
- 158.Kelsey MI, Molina JE, Huang SK & Hwang KK The identification of microbial metabolites of sulfolithocholic acid. J Lipid Res 21, 751–759 (1980). [PubMed] [Google Scholar]
- 159.Garcia CJ, Kosek V, Beltran D, Tomas-Barberan FA & Hajslova J Production of new microbially conjugated bile acids by human gut microbiota. Biomolecules 12, doi: 10.3390/biom12050687 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Juste C & Gérard P Cholesterol-to-coprostanol conversion by the gut microbiota: What we know, suspect, and ignore. Microorganisms 9, doi: 10.3390/microorganisms9091881 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kenny DJ et al. Cholesterol metabolism by uncultured human gut bacteria influences host cholesterol level. Cell Host Microbe 28, 245–257.e246, doi: 10.1016/j.chom.2020.05.013 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Fukuchi J, Song C, Dai Q, Hiipakka RA & Liao S 5β-Cholane activators of the farnesol X receptor. J Steroid Biochem Mol Biol 94, 311–318, doi: 10.1016/j.jsbmb.2004.11.012 (2005). [DOI] [PubMed] [Google Scholar]
- 163.Sepe V et al. Investigation on bile acid receptor regulators. Discovery of cholanoic acid derivatives with dual G-protein coupled bile acid receptor 1 (GPBAR1) antagonistic and farnesoid X receptor (FXR) modulatory activity. Steroids 105, 59–67, doi: 10.1016/j.steroids.2015.11.003 (2016). [DOI] [PubMed] [Google Scholar]
- 164.Gou H et al. Obeticholic acid and 5β-cholanic acid 3 exhibit anti-tumor effects on liver cancer through CXCL16/CXCR6 pathway. Front Immunol 13, 1095915, doi: 10.3389/fimmu.2022.1095915 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wang D et al. Characterization of gut microbial structural variations as determinants of human bile acid metabolism. Cell Host Microbe 29, 1802–1814.e1805, doi: 10.1016/j.chom.2021.11.003 (2021). [DOI] [PubMed] [Google Scholar]
- 166.Haeusler RA, Astiarraga B, Camastra S, Accili D & Ferrannini E Human insulin resistance is associated with increased plasma levels of 12α-hydroxylated bile acids. Diabetes 62, 4184–4191, doi: 10.2337/db13-0639 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Mancera-Hurtado Y et al. A dysregulated bile acids pool is associated with metabolic syndrome and gut microbial dysbiosis in early adolescence. Obesity (Silver Spring) 31, 2129–2138, doi: 10.1002/oby.23797 (2023). [DOI] [PubMed] [Google Scholar]
- 168.Edenharder R Dehydroxylation of cholic acid at C12 and epimerization at C5 and C7 by Bacteroides species. J Steroid Biochem 21, 413–420, doi: 10.1016/0022-4731(84)90304-2 (1984). [DOI] [PubMed] [Google Scholar]
- 169.Lucas LN et al. Dominant bacterial phyla from the human gut show widespread ability to transform and conjugate bile acids. mSystems, e0080521, doi: 10.1128/mSystems.00805-21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ng PY, Allan RN & Hofmann AF Suitability of [11, 12-3H2]chenodeoxycholic acid and [11, 12-3H2]lithocholic acid for isotope dilution studies of bile acid metabolism in man. J Lipid Res 18, 753–758 (1977). [PubMed] [Google Scholar]
- 171.Doden HL & Ridlon JM Microbial hydroxysteroid dehydrogenases: From alpha to omega. Microorganisms 9, doi: 10.3390/microorganisms9030469 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Macdonald IA, White BA & Hylemon PB Separation of 7α- and 7β-hydroxysteroid dehydrogenase activities from Clostridium absonum ATCC# 27555 and cellular response of this organism to bile acid inducers. J. Lipid Res 24, 1119–1126 (1983). [PubMed] [Google Scholar]
- 173.Macdonald IA & Sutherland JD Further studies on the bile salt induction of 7α- and 7β-hydroxysteroid dehydrogenases in Clostridium absonum. Biochim Biophys Acta 750, 397–403, doi: 10.1016/0005-2760(83)90045-0 (1983). [DOI] [PubMed] [Google Scholar]
- 174.Macdonald IA et al. Metabolism of primary bile acids by Clostridium perfringens. J Steroid Biochem 18, 97–104, doi: 10.1016/0022-4731(83)90336-9 (1983). [DOI] [PubMed] [Google Scholar]
- 175.Macdonald IA, Jellett JF, Mahony DE & Holdeman LV Bile salt 3α- and 12α-hydroxysteroid dehydrogenases from Eubacterium lentum and related organisms. Appl Environ Microbiol 37, 992–1000, doi: 10.1128/aem.37.5.992-1000.1979 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Macdonald IA, Williams CN, Mahony DE & Christie WM NAD- and NADP-dependent 7α-hydroxysteroid dehydrogenases from Bacteroides fragilis. Biochim Biophys Acta 384, 12–24, doi: 10.1016/0005-2744(75)90091-1 (1975). [DOI] [PubMed] [Google Scholar]
- 177.Hirano S & Masuda N Characterization of NADP-dependent 7β-hydroxysteroid dehydrogenases from Peptostreptococcus productus and Eubacterium aerofaciens. Appl Environ Microbiol 43, 1057–1063, doi: 10.1128/aem.43.5.1057-1063.1982 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Devlin AS & Fischbach MA A biosynthetic pathway for a prominent class of microbiota-derived bile acids. Nat Chem Biol 11, 685–690, doi: 10.1038/nchembio.1864 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Mythen SM, Devendran S, Mendez-Garcia C, Cann I & Ridlon JM Targeted synthesis and characterization of a gene cluster encoding NAD(P)H-dependent 3α-, 3β-, and 12α-hydroxysteroid dehydrogenases from Eggerthella CAG:298, a gut metagenomic sequence. Appl Environ Microbiol 84, doi: 10.1128/AEM.02475-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Lee JY et al. Contribution of the 7β-hydroxysteroid dehydrogenase from Ruminococcus gnavus N53 to ursodeoxycholic acid formation in the human colon. J Lipid Res 54, 3062–3069, doi: 10.1194/jlr.M039834 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Doden HL et al. Completion of the gut microbial epi-bile acid pathway. Gut Microbes 13, 1–20, doi: 10.1080/19490976.2021.1907271 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Harris SC et al. Bile acid oxidation by Eggerthella lenta strains C592 and DSM 2243T. Gut Microbes 9, 523–539, doi: 10.1080/19490976.2018.1458180 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Odermatt A et al. Hepatic reduction of the secondary bile acid 7-oxolithocholic acid is mediated by 11β-hydroxysteroid dehydrogenase 1. Biochem J 436, 621–629, doi: 10.1042/BJ20110022 (2011). [DOI] [PubMed] [Google Scholar]
- 184.Johnson JS et al. 11β-hydroxysteroid dehydrogenase-1 deficiency alters the gut microbiome response to Western diet. J Endocrinol 232, 273–283, doi: 10.1530/joe-16-0578 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Dyson JK et al. Novel therapeutic targets in primary biliary cirrhosis. Nat Rev Gastroenterol Hepatol 12, 147–158, doi: 10.1038/nrgastro.2015.12 (2015). [DOI] [PubMed] [Google Scholar]
- 186.Edenharder R, Pfutzner A & Hammann R Characterization of NAD-dependent 3α- and 3β-hydroxysteroid dehydrogenase and of NADP-dependent 7β-hydroxysteroid dehydrogenase from Peptostreptococcus productus. Biochim Biophys Acta 1004, 230–238, doi: 10.1016/0005-2760(89)90272-5 (1989). [DOI] [PubMed] [Google Scholar]
- 187.Marschall HU et al. Human liver class I alcohol dehydrogenase γγ isozyme: The sole cytosolic 3β-hydroxysteroid dehydrogenase of iso bile acids. Hepatology 31, 990–996, doi: 10.1053/he.2000.5720 (2000). [DOI] [PubMed] [Google Scholar]
- 188.Schmassmann A et al. Transport, metabolism, and effect of chronic feeding of lagodeoxycholic acid. A new, natural bile acid. Gastroenterology 99, 1092–1104, doi: 10.1016/0016-5085(90)90630-j (1990). [DOI] [PubMed] [Google Scholar]
- 189.Doden H et al. Metabolism of oxo-bile acids and characterization of recombinant 12α-hydroxysteroid dehydrogenases from bile acid 7α-dehydroxylating human gut bacteria. Appl Environ Microbiol 84, doi: 10.1128/AEM.00235-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Bernstein H, Bernstein C, Payne CM, Dvorakova K & Garewal H Bile acids as carcinogens in human gastrointestinal cancers. Mutat Res 589, 47–65, doi: 10.1016/j.mrrev.2004.08.001 (2005). [DOI] [PubMed] [Google Scholar]
- 191.Bernstein H & Bernstein C Bile acids as carcinogens in the colon and at other sites in the gastrointestinal system. Exp Biol Med (Maywood) 248, 79–89, doi: 10.1177/15353702221131858 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Thibaut MM & Bindels LB Crosstalk between bile acid-activated receptors and microbiome in entero-hepatic inflammation. Trends Mol Med 28, 223–236, doi: 10.1016/j.molmed.2021.12.006 (2022). [DOI] [PubMed] [Google Scholar]
- 193.Bayerdörffer E et al. Increased serum deoxycholic acid levels in men with colorectal adenomas. Gastroenterology 104, 145–151, doi: 10.1016/0016-5085(93)90846-5 (1993). [DOI] [PubMed] [Google Scholar]
- 194.Atarashi K et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331, 337–341, doi: 10.1126/science.1198469 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Atarashi K et al. Treg induction by a rationally selected mixture of clostridia strains from the human microbiota. Nature 500, 232–236, doi: 10.1038/nature12331 (2013). [DOI] [PubMed] [Google Scholar]
- 196.Narushima S et al. Characterization of the 17 strains of regulatory T cell-inducing human-derived Clostridia. Gut Microbes 5, 333–339, doi: 10.4161/gmic.28572 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Soroosh P et al. Oxysterols are agonist ligands of RORγt and drive Th17 cell differentiation. Proc Natl Acad Sci U S A 111, 12163–12168, doi: 10.1073/pnas.1322807111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Hu X et al. Sterol metabolism controls TH17 differentiation by generating endogenous RORγ agonists. Nat Chem Biol 11, 141–147, doi: 10.1038/nchembio.1714 (2015). [DOI] [PubMed] [Google Scholar]
- 199.Lloyd-Price J et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 569, 655–662, doi: 10.1038/s41586-019-1237-9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.He YJ & You CG The Potential Role of Gut Microbiota in the Prevention and Treatment of Lipid Metabolism Disorders. Int J Endocrinol 2020, 8601796, doi: 10.1155/2020/8601796 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Gonzalez FJ, Jiang C & Patterson AD An Intestinal Microbiota-Farnesoid X Receptor Axis Modulates Metabolic Disease. Gastroenterology 151, 845–859, doi: 10.1053/j.gastro.2016.08.057 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Gonzalez FJ, Jiang C, Xie C & Patterson AD Intestinal Farnesoid X Receptor Signaling Modulates Metabolic Disease. Dig Dis 35, 178–184, doi: 10.1159/000450908 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Francis GA, Fayard E, Picard F & Auwerx J Nuclear receptors and the control of metabolism. Annu Rev Physiol 65, 261–311, doi: 10.1146/annurev.physiol.65.092101.142528 (2003). [DOI] [PubMed] [Google Scholar]
- 204.Lefebvre P, Cariou B, Lien F, Kuipers F & Staels B Role of bile acids and bile acid receptors in metabolic regulation. Physiol Rev 89, 147–191, doi: 10.1152/physrev.00010.2008 (2009). [DOI] [PubMed] [Google Scholar]
- 205.Sorrentino G et al. Bile Acids Signal via TGR5 to Activate Intestinal Stem Cells and Epithelial Regeneration. Gastroenterology 159, 956–968.e958, doi: 10.1053/j.gastro.2020.05.067 (2020). [DOI] [PubMed] [Google Scholar]
- 206.Federici S et al. Targeted suppression of human IBD-associated gut microbiota commensals by phage consortia for treatment of intestinal inflammation. Cell 185, 2879–2898.e2824, doi: 10.1016/j.cell.2022.07.003 (2022). [DOI] [PubMed] [Google Scholar]
- 207.Woo AYM et al. Targeting the human gut microbiome with small-molecule inhibitors. Nat Rev Chem 7, 319–339, doi: 10.1038/s41570-023-00471-4 (2023). [DOI] [PubMed] [Google Scholar]
- 208.Li DK et al. Inhibition of microbial deconjugation of micellar bile acids protects against intestinal permeability and liver injury. Sci Adv 8, eabo2794, doi: 10.1126/sciadv.abo2794 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Jin WB et al. Genetic manipulation of gut microbes enables single-gene interrogation in a complex microbiome. Cell 185, 547–562 e522, doi: 10.1016/j.cell.2021.12.035 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Parasar B et al. Chemoproteomic Profiling of Gut Microbiota-Associated Bile Salt Hydrolase Activity. ACS Cent Sci 5, 867–873, doi: 10.1021/acscentsci.9b00147 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Ivanov II, Tuganbaev T, Skelly AN & Honda K T Cell Responses to the microbiota. Ann Rev Immunol 40, 559–587, doi: 10.1146/annurev-immunol-101320-011829 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Neuman H, Debelius JW, Knight R & Koren O Microbial endocrinology: The interplay between the microbiota and the endocrine system. FEMS Microbiol Rev 39, 509–521, doi: 10.1093/femsre/fuu010 (2015). [DOI] [PubMed] [Google Scholar]
- 213.Plottel CS & Blaser MJ Microbiome and malignancy. Cell Host Microbe 10, 324–335, doi: 10.1016/j.chom.2011.10.003 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Ly LK, Doden HL & Ridlon JM Gut feelings about bacterial steroid-17,20-desmolase. Mol Cell Endocrinol 525, 111174, doi: 10.1016/j.mce.2021.111174 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Ly LK et al. Bacterial steroid-17,20-desmolase is a taxonomically rare enzymatic pathway that converts prednisone to 1,4-androstanediene-3,11,17-trione, a metabolite that causes proliferation of prostate cancer cells. J Steroid Biochem Mol Biol 199, 105567, doi: 10.1016/j.jsbmb.2019.105567 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Haiser HJ et al. Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science 341, 295–298, doi: 10.1126/science.1235872 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Zimmermann M, Zimmermann-Kogadeeva M, Wegmann R & Goodman AL Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature 570, 462–467, doi: 10.1038/s41586-019-1291-3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Ridlon JM et al. Clostridium scindens: A human gut microbe with a high potential to convert glucocorticoids into androgens. J Lipid Res 54, 2437–2449, doi: 10.1194/jlr.M038869 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Feighner SD, Bokkenheuser VD, Winter J & Hylemon PB Characterization of a C21 neutral steroid hormone transforming enzyme, 21-dehydroxylase, in crude cell extracts of Eubacterium lentum. Biochim Biophys Acta 574, 154–163, doi: 10.1016/0005-2760(79)90094-8 (1979). [DOI] [PubMed] [Google Scholar]
- 220.Oliveira RA & Pamer EG Assembling symbiotic bacterial species into live therapeutic consortia that reconstitute microbiome functions. Cell Host Microbe 31, 472–484, doi: 10.1016/j.chom.2023.03.002 (2023). [DOI] [PubMed] [Google Scholar]
- 221.Louie T et al. VE303, a defined bacterial consortium, for prevention of recurrent Clostridioides difficile infection: A randomized clinical trial. JAMA 329, 1356–1366, doi: 10.1001/jama.2023.4314 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Britton GJ & Faith JJ Causative microbes in host-microbiome interactions. Annu Rev Microbiol 75, 223–242, doi: 10.1146/annurev-micro-041321-042402 (2021). [DOI] [PubMed] [Google Scholar]
- 223.Hirano S & Masuda N Transformation of bile acids by Eubacterium lentum. Appl Environ Microbiol 42, 912–915, doi: 10.1128/aem.42.5.912-915.1981 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Pedersen KJ et al. Eggerthella lenta DSM 2243 alleviates bile acid stress response in Clostridium ramosum and Anaerostipes caccae by transformation of bile acids. Microorganisms 10, 2025, doi: 10.3390/microorganisms10102025 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Bokkenheuser VD et al. New markers for Eubacterium lentum. Appl Environ Microbiol 37, 1001–1006, doi: 10.1128/aem.37.5.1001-1006.1979 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Song C, Wang B, Tan J, Zhu L & Lou D Discovery of tauroursodeoxycholic acid biotransformation enzymes from the gut microbiome of black bears using metagenomics. Sci Rep 7, 45495, doi: 10.1038/srep45495 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Vital M, Rud T, Rath S, Pieper DH & Schluter D Diversity of bacteria exhibiting bile acid-inducible 7α-dehydroxylation genes in the human gut. Comput Struct Biotechnol J 17, 1016–1019, doi: 10.1016/j.csbj.2019.07.012 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Wells JE & Hylemon PB Identification and characterization of a bile acid 7α-dehydroxylation operon in Clostridium sp. strain TO-931, a highly active 7α-dehydroxylating strain isolated from human feces. Appl Environ Microbiol 66, 1107–1113, doi: 10.1128/AEM.66.3.1107-1113.2000 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Huijghebaert SM, Mertens JA & Eyssen HJ Isolation of a bile salt sulfatase-producing Clostridium strain from rat intestinal microflora. Appl Environ Microbiol 43, 185–192, doi: 10.1128/aem.43.1.185-192.1982 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Imperato TJ, Wong CG, Chen LJ & Bolt RJ Hydrolysis of lithocholate sulfate by Pseudomonas aeruginosa. J Bacteriol 130, 545–547, doi: 10.1128/jb.130.1.545-547.1977 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]


