Abstract
Proteins containing both intrinsically disordered regions (IDRs) and RNA binding domains (RBDs) can phase separate in vitro, forming bodies similar to cellular biomolecular condensates. However, how IDR and RBD domains contribute to in vivo recruitment of proteins to biomolecular condensates remains poorly understood. Here, we analyzed the roles of IDRs and RBDs in L-bodies, biomolecular condensates present in Xenopus oocytes. We show that a cytoplasmic isoform of hnRNPAB, which contains two RBDs and an IDR, is highly enriched in L-bodies. While both of these domains contribute to hnRNPAB self-association and phase separation in vitro and mediate enrichment into L-bodies in oocytes, neither the RBDs nor the IDR replicate the localization of full-length hnRNPAB. Our results suggest a model where the combined effects of the IDR and RBDs regulate hnRNPAB partitioning into L-bodies. This model likely has widespread applications as proteins containing RBD and IDR domains are common biomolecular condensate residents.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-79409-9.
Subject terms: RNA, Cell biology, Molecular biology
Introduction
Biomolecular condensates have emerged as a key feature of cellular architecture, serving to organize biomolecules such as RNAs and proteins into discreet bodies without a lipid membrane1–3. Biomolecular condensates are found in both the cytoplasm and the nucleus, and display a range of functions and biophysical properties4. The biophysical behavior of a biomolecular condensate tends to align with its function: transient structures like stress granules are more liquid-like and dynamic5–7, while biomolecular condensates that persist on the order of days to months, such as L-bodies, P-granules and germline granules, are less dynamic and more hydrogel- or solid-like in their physical state8–11.
In vitro studies performed with a variety of proteins and RNAs found in biomolecular condensates have elucidated many of their governing principles12–14. Prominent among these is the prevalence of intrinsically disordered regions (IDRs) in biomolecular condensate constituent proteins13,15,16. IDRs are regions of a protein that display low degrees of sequence complexity, and consequently are unable to form stable secondary or tertiary structures, remaining more or less unfolded5. Many studies have shown that proteins containing IDRs phase separate in vitro via protein-protein interactions, forming bodies that display properties similar to biomolecular condensates7,12,17–19. Many IDR-containing proteins also have RNA binding domains (RBDs). Studies have consistently shown that addition of RNA to RNA binding proteins (RBPs) in vitro facilitates the formation of phase separated bodies, indicating a role for RNA-protein interactions in their formation in addition to IDR interactions19–21. This is notable given that many biomolecular condensates found in cells are composed of RNA and protein, suggesting that both protein-protein and RNA-protein interactions occur in in vivo phase-separated bodies22. However, while the mechanics of in vitro phase separated bodies have been well-studied, how these principles translate to more complex heterogeneous biomolecular condensates in cells remains less clear.
The Xenopus laevis oocyte organizes its cytoplasm in part through RNA localization, which specifies germ layer patterning during embryogenesis23,24. mRNAs encoding morphogens such as the TGF-β growth factor vg1 are transported to the vegetal cortex during oogenesis via large cytoplasmic granules containing both RNA and protein25. Work from our laboratory discovered that these RNP granules are novel cytoplasmic biomolecular condensates termed Localization bodies, or L-bodies26. Localizing RNAs are highly enriched in L-bodies and form a non-dynamic scaffold within them, in contrast to L-body constituent proteins, which display a range of dynamics, from moderate to high26. As with several other biomolecular condensates, the L-body proteome is enriched for proteins containing RBDs and IDRs, with many proteins containing both26. While RBDs and IDRs have been shown to be integral to phase separation in vitro, their relative importance to protein enrichment and dynamics in biomolecular condensates in vivo remains less clear, particularly in the context of proteins that contain both types of domains.
L-bodies present a unique opportunity to study how proteins associate with long-lived biomolecular condensates, as other experimentally tractable biomolecular condensates such as stress granules tend to be short-lived6. As such, previous studies have often focused on protein recruitment via de novo assembly of biomolecular condensate16,27,28, whereas L-bodies allow us to investigate how proteins are recruited to extant biomolecular condensates.
In this work we investigated how RBD and IDR domains regulate protein enrichment and dynamics within L-bodies. We focused on the Xenopus RBP heterogenous nuclear ribonucleoprotein AB (hnRNPAB) due to its known localization to L-bodies, association with L-body constituent mRNA vg1, and structure, which consists of two RBDs and C-terminal IDR29,30. We found that hnRNPAB X2, a novel cytoplasmic splice isoform of hnRNPAB, lacks the C-terminal nuclear localization signal found in the canonical form of hnRNPAB and is highly enriched in L-bodies. While both the hnRNPAB X2 RBDs and IDR are sufficient individually for L-bodies enrichment, enrichment was lower than for the full-length protein, indicating that the two domains together are necessary to replicate enrichment of hnRNPAB X2 in phase separated L-bodies. Consistent with this, both the RBD and the IDR domains phase separate in vitro and the dynamic behavior of hnRNPAB in vivo is dependent on both domains. While hnRNPAB X2 is highly dynamic within L-bodies, the RBD and IDR of hnRNPAB X2 each diffuse more rapidly than the full-length protein. Moreover, the addition of the hnRNPAB X2 IDR to PTBP3, an L-body RBP that contains four RBDs and lacks an IDR, slows translational diffusion of the chimeric protein in L-bodies. Taken together, our results suggest that interactions mediated by both the RBD and IDR domains act to regulate protein enrichment and dynamics within biomolecular condensates such as L-bodies.
Results
hnRNPAB X2 is a novel cytoplasmic isoform of hnRNPAB
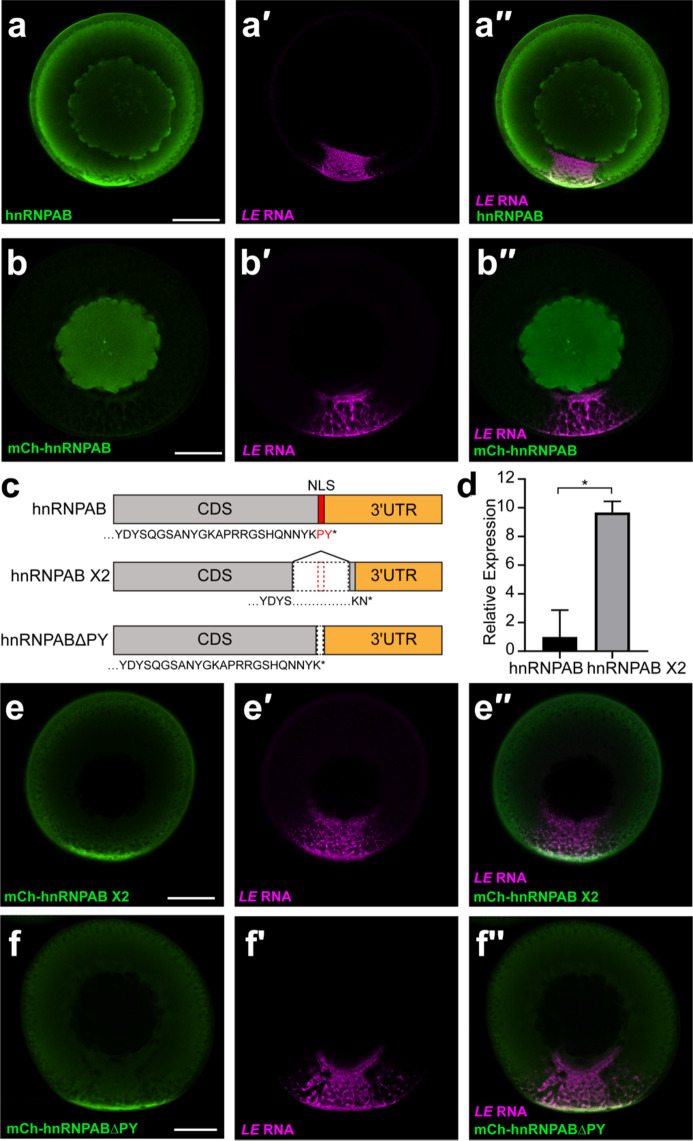
To investigate how RBDs and IDRs regulate protein enrichment and dynamics in biomolecular condensates we focused on hnRNPAB, a constituent of L-bodies that contains two RBDs and an IDR (Figure S1a). vg1 mRNA is transported to the vegetal cortex of Xenopus oocytes during stages II-III of oogenesis in L-bodies in a process that can be visualized by imaging a fluorescently-labeled localization element (LE) derived from sequences from the 3′ UTR of vg1 mRNA31,32. Analysis of the distribution of endogenous hnRNPAB by immunostaining in stage II-III oocytes showed that hnRNPAB is present in both the nucleus and cytoplasm, with notable enrichment to the vegetal cortex and L-bodies as demonstrated by colocalization with LE RNA (Fig. 1a). However, expression of mCherry-tagged hnRNPAB in stage II-III oocytes following microinjection of a cloned, tagged mRNA showed predominantly nuclear localization (Fig. 1b). This led us to ask whether an endogenous hnRNPAB splice isoform represented the cytoplasmic protein seen for the endogenous protein by immunofluorescence (IF). We identified a computationally predicted splice isoform, hnRNPAB X2 (Fig. 1c), and successfully cloned a cDNA from stage II oocyte RNA corresponding to the predicted sequence. IF of mCherry-tagged hnRNPAB X2 (Fig. 1e) showed exclusively cytoplasmic signal with enrichment in L-bodies at the vegetal cortex similar to that observed in the cytoplasm by IF for endogenous hnRNPAB (Fig. 1a). These results indicate that the distribution of endogenous hnRNPAB observed by IF (Fig. 1a) represents the combined distribution of two isoforms, with the hnRNPAB X2 splice isoform localized in the cytoplasm and canonical hnRNPAB in the nucleus.
Fig. 1.
hnRNPAB X2 is a novel cytoplasmic splice isoform of hnRNPAB that is enriched in L-bodies. (a) Stage II oocytes were microinjected with Cy5-labeled LE RNA (magenta, a′) and immunostained for endogenous hnRNPAB (green, a) using antibodies raised against Xenopus hnRNPAB30. The overlap is shown in a′′. (b) Cy5-labeled LE RNA (magenta, b′) was microinjected into stage II oocytes expressing mCherry-tagged canonical hnRNPAB, as detected by immunostaining with anti-RFP (green, b). The overlap is shown in b′′. (c) Schematics of canonical hnRNPAB, hnRNPAB X2 and hnRNPABΔPY. C-terminal sequence is shown below each, with the PY NLS indicated in red, the protein coding sequence (CDS) in gray, and the 3′UTR shown in orange. (d) RNA isolated from stage II oocytes was used to measure the relative expression of hnRNPAB X2 compared to canonical hnRNPAB by qPCR. ΔCt values were calculated normalizing to refence gene vg1 (* indicates p < 0.05 by T-test). Error bars represent standard deviation of the mean, n = 3. (e) Cy5-labeled LE RNA (magenta, e′) was microinjected into stage II oocytes expressing mCherry-tagged hnRNPAB X2, as detected by immunostaining with anti-RFP (green, e). The overlap is shown in e′′. (f) Cy5-labeled LE RNA (magenta, e′) was microinjected into stage II oocytes expressing mCherry-tagged hnRNPABΔPY, as detected by immunostaining with anti-RFP (green, e). The overlap is shown in e′′. Confocal sections (a-b, e-f) are shown with the vegetal hemisphere at the bottom; scale bars = 100 μm.
Pre-mRNA processing of the hnRNPAB X2 isoform splices out nucleotides encoding the C-terminal region of the protein (Fig. 1c) which contains a putative PY nuclear localization sequence (NLS)33; related proteins such as JKTBP and hnRNPD have a NLS in this region, which we hypothesized might be ablated in hnRNPAB X2. To test this, we ablated the putative PY NLS in canonical hnRNPAB (construct hnRNPABΔPY; Fig. 1c) by deleting the two most C-terminal amino acids (PY). Expression and imaging of this construct (Fig. 1f) showed that deletion of the C-terminal PY residues is sufficient to recapitulate hnRNPAB X2-like cytoplasmic localization (Fig. 1e-f), suggesting that hnRNPAB X2 represents an isoform of hnRNPAB that is targeted to the cytoplasm through the loss of a C-terminal NLS. Indeed, qPCR carried out with primers specific to canonical hnRNPAB or hnRNPAB X2 show that hnRNPAB X2 is ~ 10-fold more highly expressed than the canonical splice isoform in stage II-III oocytes (Fig. 1d).
Both the RBDs and the IDR contribute to hnRNPAB X2 L-body localization
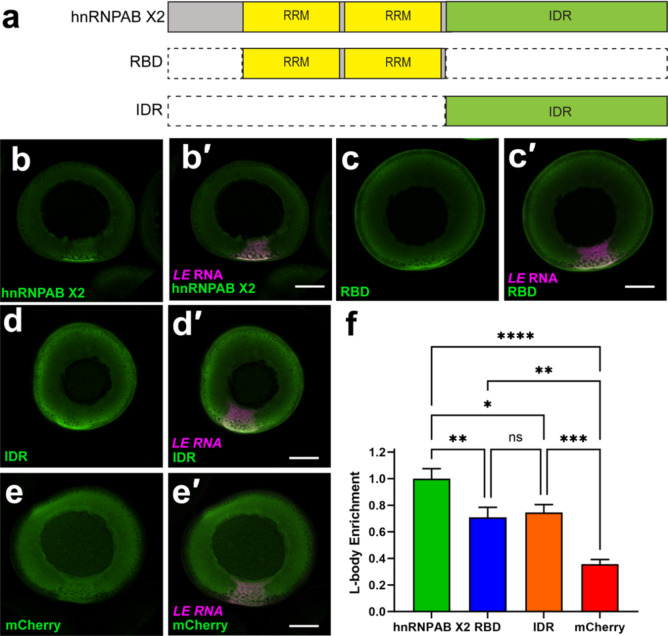
We next investigated the roles of the RBD and IDR domains of hnRNPAB X2 in its enrichment in L-bodies. We first analyzed hnRNPAB X2 via Prion Like Amino Acid Composition (PLAAC)34 and found that the IDR has significant predicted Prion-like character (Figure S1a). Based on the PLAAC analysis and structured domain annotation, we generated two mCherry (mCh) tagged constructs termed RBD and IDR, which each isolated their respective functional domains (Fig. 2a). The RBD construct contains two RNA recognition motif (RRM) class RNA-binding domains, and the IDR construct contains the prion-like IDR (Fig. 2a). In the case of stress granule localization of the human RBP Fused in Sarcoma (FUS), that contains a prion-like domain (very much like that of hnRNPAB) and RNA-binding domains (RRM, zinc finger, as well as disordered RGG motifs in the case of FUS), the RBDs but not the prion-like domains are sufficient for, and contribute to, incorporation in stress granules35. Therefore, we hypothesized that the hnRNPAB X2 RBDs are required for recruitment to L-bodies and the IDR is dispensable. Expression of the mCh tagged RBD and IDR constructs in oocytes co-microinjected with fluorescent LE RNA indicated that both were enriched in L-bodies at the vegetal cortex relative to free mCh (Fig. 2b-e), and were expressed at levels similar to full-length hnRNPAB X2 (Figure S2). To quantitate L-body enrichment, the fraction of total protein fluorescence at the vegetal cortex was measured for mCh-hnRNPAB X2, mCh-IDR, mCh-RBD, and mCh (Fig. 2f). While mCh-RBD and mCh-IDR were significantly enriched in L-bodies relative to free mCh, hnRNPAB X2 was significantly more enriched than either of the isolated domains, indicating that neither domain alone is sufficient to recapitulate hnRNPAB X2 localization. Therefore, it is likely that hnRNPAB X2 localizes to L-bodies due to a combination of interactions by both the RBD and IDR domains.
Fig. 2.
hnRNPAB X2 enriches in L-bodies through its RBD and IDR. (a) Schematics of domain constructs. (b–e) Stage II oocytes expressing (b) mCh-hnRNPAB X2, (c) mCh-RBD, (d) mCh-IDR, or (e) free mCh (green; detected by anti-RFP IF) were co-microinjected with Cy5 LE RNA (magenta, b′-e′) to label L-bodies. Colocalization (white) is shown in the merged confocal images (b′-e′). Scale bars = 100 μm. (f) L-body enrichment was quantitated by measuring the fraction of total protein fluorescence localized to the vegetal cortical region. Shown are relative levels of L-body enrichment for hnRNPAB X2 (green, set to 1.0 ± 0.075), RBD (blue, 0.71 ± 0.076), IDR (orange, 0.75 ± 0.059), and mCherry (red, 0.38 ± 0.034). n = 24 oocytes from four biological replicates; error bars represent standard error of the mean, **** indicates p < 0.0001, *** indicates p < 0.001, ** indicates p < 0.01, * indicates p < 0.05, and ns indicates not significant (p > 0.05). Statistics shown are an ordinary one-way ANOVA followed by Tukey’s multiple comparisons.
The hnRNPAB X2 and the IDR and RBD domains self-assemble and phase separate in vitro
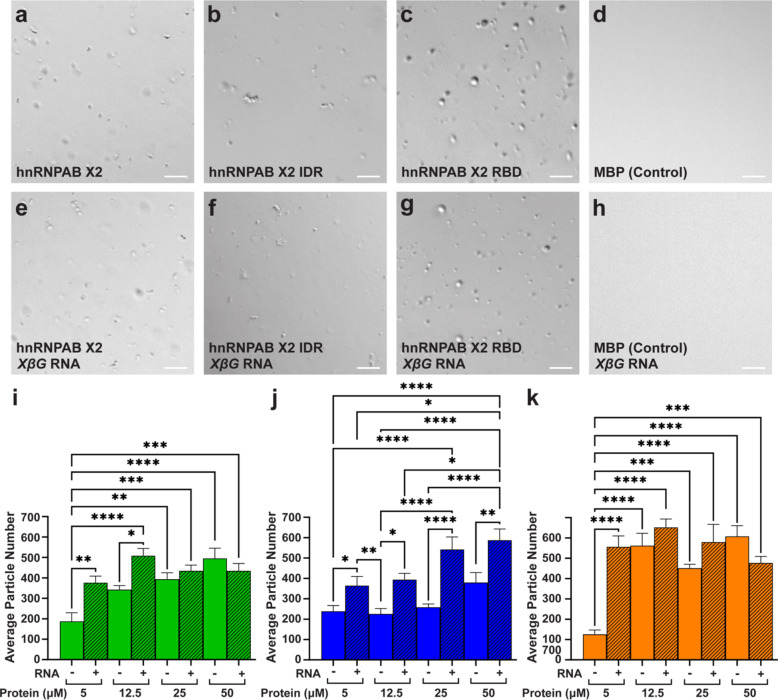
Biomolecular condensates have been characterized to form through phase separation36. We used an in vitro approach to test if hnRNPAB X2 or its domains phase separate. Differential interference microscopy was used to analyze phase separation on samples of recombinant hnRNPAB X2, and the IDR and RBD domains purified as fusions with maltose binding protein (MBP) solubility tags. After cleavage of the MBP tag by TEV protease and in the presence of crowding agent (10% PEG), full-length hnRNPAB X2, the IDR domain, and the RBD all formed round condensates in the absence (Fig. 3a-c, Figure S3a-c) or presence of RNA (Fig. 3e-g, Figure S3d-f). A control protein (MBP alone) did not exhibit phase separation (Fig. 3d, h). Quantitation of condensate number for hnRNPAB X2 and its IDR and RBD domains (Fig. 3i-k), at protein concentrations between 5 µM and 50 µM with and without Xenopus β-globin RNA, revealed that condensates are formed at all protein concentrations tested in the presence and absence of RNA. Both the full-length protein and the RBD domain form fewer condensates at 5 µM protein in the absence of RNA, while at higher protein concentrations (12.5–50 µM), RNA had little effect (Fig. 3i, k). For IDR domain (Fig. 3j), the presence of RNA increased the number of condensates at all protein concentrations tested and the individual condensates were smaller in the presence of RNA (Figure S3k). The size of the condensates formed by the RBD domain increased with increasing protein concentration (Figure S3l), and RNA showed an effect on condensate size only at the lowest protein concentration (5 µM) where the average size was higher with RNA. The size of the full length hnRNPAB X2 condensates was not strongly affected by varying protein concentration or addition of RNA (Figure S3j). Although the condensates formed by hnRNPAB X2 and its IDR and RBD domains appear as round structures (Fig. 3a-c, e-g) that exhibit high levels of circularity (Figure S3g-i), only the condensates formed by the RBD domain can be observed to fuse and wet the slide (Videos S1-2), a property consistent with liquid-like condensates. By contrast, the condensates formed by the full-length protein (Videos S3-4) and the IDR domain (Videos S5-6) cluster together and do not fuse, which may be an indication of more static-like structures. Taken together, these results indicate that with and without RNA full-length hnRNPAB X2 is able to phase separate in vitro and that both the RBD and IDR domains alone are sufficient in vitro to form phase separated condensates.
Fig. 3.
hnRNPAB X2 and its domains self-assemble and phase separate in vitro. (a–h) DIC micrographs of (a,e) full-length hnRNPA2 X2, (b,f) hnRNPAB X2 IDR, (c,g) hnRNPAB X2 RBD, and (d,h) MBP (control) proteins at 12.5 µM in 20 mM NaPi (pH 7.4), 150 mM NaCl, and 10% PEG, in the absence (a–d) or the presence (e–h) of 0.25 mg/mL Xenopus β-globin (XβG) RNA. Images are representative from three or more biological replicates (with independently expressed and purified protein) and three or more technical replicates. Scale bars = 10 μm. (i–k) Condensate formation was quantitated for fluorescently labeled condensates by determining the average particle number per field of view for (i) full-length hnRNPA2 X2 (green), (j) hnRNPAB X2 IDR (blue), and (k) hnRNPAB X2 RBD (orange) at 5, 12.5, 25, and 50 µM protein concentrations in the presence (+, hatched bars) and absence (-, open bars) of 0.25 mg/mL Xenopus β-globin RNA. Statistics shown are an ordinary two-way ANOVA followed by Tukey’s multiple comparisons. Error bars represent standard error of the mean, **** indicates p < 0.0001, *** indicates p < 0.001, ** indicates p < 0.01, * indicates p < 0.05, and all brackets not shown are not significant (p > 0.05).
hnRNPAB X2 is highly dynamic in L-bodies
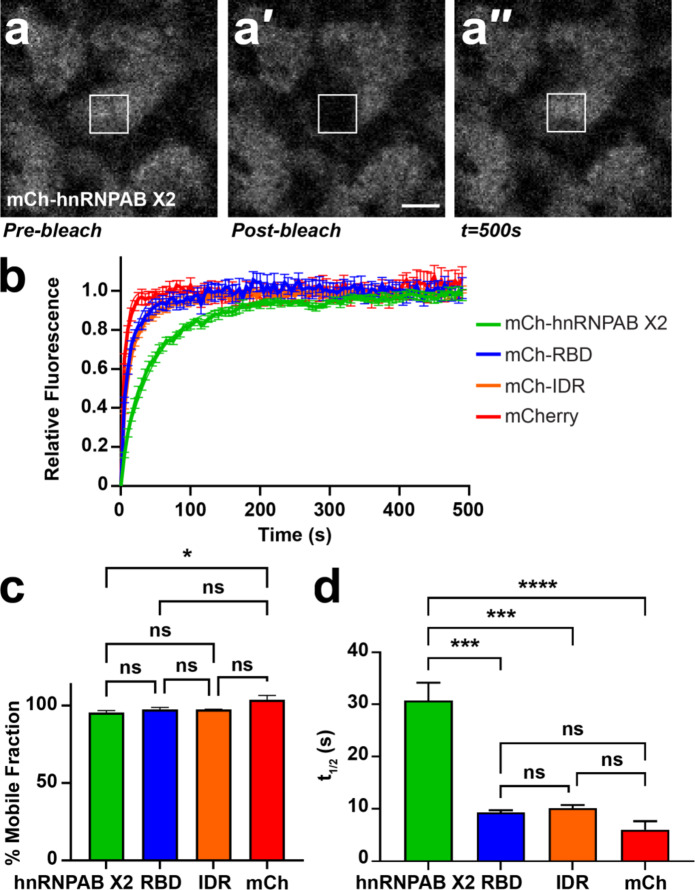
We next sought to determine the dynamics of hnRNPAB X2 in L-bodies and how the IDR and RBD domains regulate hnRNPAB X2 dynamics in L-bodies in vivo. Proteins containing IDRs, including hnRNPAB, have been shown to dynamically associate with biomolecular condensates like L-bodies37. Therefore, we hypothesized that the RBDs may form stable RNA interactions and therefore be less dynamic than the full-length protein and that the IDR modulates the dynamic physical association of hnRNPAB X2 within L-bodies. We assessed the dynamics of mCh-tagged hnRNPAB X2 by fluorescence recovery after photobleaching (FRAP), defining the regions of interest (ROIs) such that L-bodies were only partially bleached, to allow for recovery both from within the L-body and from the surrounding cytoplasm (Fig. 4a). Despite its enrichment to L-bodies, hnRNPAB X2 is highly dynamic within them, with a mobile fraction of 96% (Fig. 4b,c). We then tested whether the dynamics of hnRNPAB X2 were governed by the IDR domain, reasoning that the RBDs alone might display less dynamic behavior, due to potential interactions with the non-dynamic RNA phase. We tested the mCh-RBD and mCh-IDR proteins by FRAP and found that both the RBD and the IDR display mobile fractions of 98%, which is not statistically different from the mobile fractions observed for mCherry (104%) or full-length hnRNPAB X2 (Fig. 4c), contradicting the idea that the RBD would display lower dynamics. Notably, both mCh-RBD and mCh-IDR display significantly shorter t1/2 measurements (9.3 and 10.1 s, respectively) than the full-length protein (30.8 s) (Fig. 4d), indicating that each of the domains diffuse more rapidly than the full-length hnRNPAB X2 protein does in L-bodies. These FRAP experiments suggest that, similar to protein enrichment in L-bodies, the dynamic behavior of hnRNPAB X2 is influenced by both the IDR and RBD domains, and that neither type of domain alone is sufficient to recapitulate the behavior of the full-length protein.
Fig. 4.
hnRNPAB X2 dynamically associates with L-bodies via its RBD and IDR. (a) Shown is an image of the vegetal cytoplasm of a stage II oocyte expressing mCh-hnRNPAB X2. FRAP was conducted such that an individual L-body was partially bleached (a′), to allow for recovery (a′′) both from within the L-body and from its environment. The 10 μm2 ROI is indicated by a white box; scale bar = 10 μm. (b) Stage II oocytes were microinjected with mCherry (mCh), mCh-hnRNPAB X2, mCh-RBD or mCh-IDR RNA to express the mCh-tagged proteins, along with Cy5 LE RNA to mark L-bodies. Normalized FRAP recovery curves are shown (n = 21 oocytes per fusion protein); error bars represent standard error of the mean. Measurements were taken at 5 s intervals over 100 iterations. (c) Plateau values (% mobile fraction) are shown for mCh-hnRNPAB X2 (green, 96%±0.013), mCh-RBD (blue, 98%±0.014), mCh-IDR (orange, 98%±0.002), and mCherry (red, 104%±0.29). Error bars represent standard error of the mean. Statistics shown are an Ordinary one-way ANOVA with Tukey’s multiple comparisons; ns indicates not significant (p > 0.05) and * indicates p = 0.04. (d) T1/2 measurements are shown for mCh-hnRNPAB X2 (green, 30.1s ± 3.4), mCh-RBD (blue, 9.3s ± 0.4), mCh-IDR (orange, 10.1s ± 0.6), and mCherry (red, 6.0s ± 1.6), measured as the average time at which the protein recovers half of its plateau value. Error bars represent standard error of the mean. Statistics shown are an Ordinary one-way ANOVA with Tukey’s multiple comparisons; ns indicates not significant (p > 0.05), *** indicates p < 0.001, **** indicates p < 0.0001.
The hnRNPAB X2 IDR can reduce protein dynamics in L-bodies
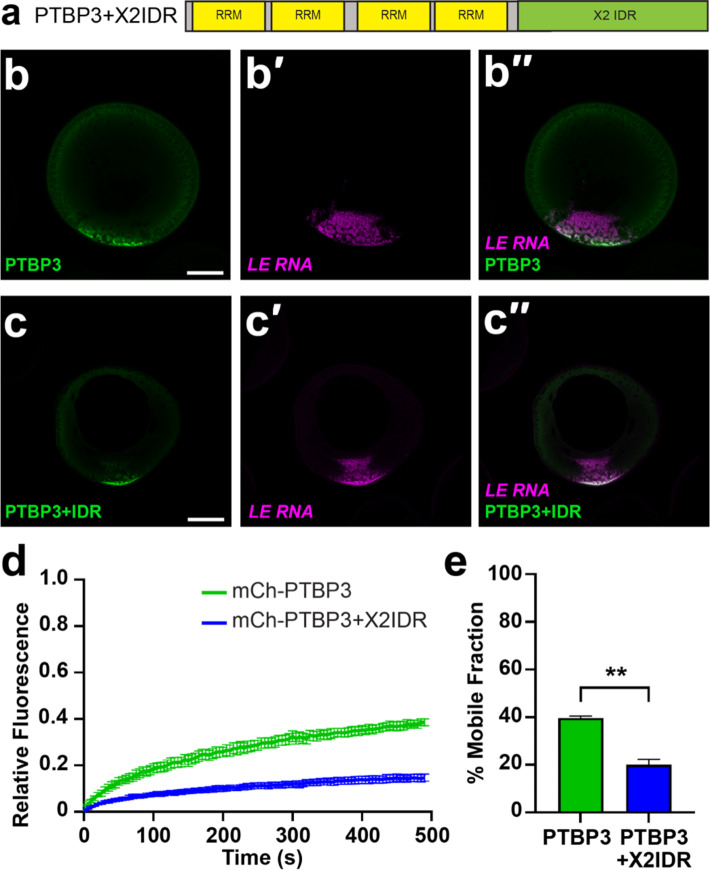
Given that the hnRNPAB X2 and the IDR alone are highly dynamic within L-bodies, we wondered whether IDR interactions, either homo-typic or with other L-body proteins might be responsible for the high mobility of hnRNPAB X2 in L-bodies. Thus, we sought to ask if the dynamic behavior of hnRNPAB X2 could be conferred to a less dynamic L-body protein by the hnRNPAB X2 IDR. For this, we used the RBP PTBP3, an L-body protein with four well-folded RRM domains but no discernable IDR (Figure S1B), which is moderately dynamic (mobile fraction ~ 40–50%) in L-bodies38. Work from our laboratory has previously shown that the dynamic behavior of PTBP3 in vivo is due to multivalent interactions between two of the PTBP3 RRM domains and the non-dynamic localized RNAs in L-bodies38. If addition of the hnRNPAB IDR to PTBP3 causes an increase in interactions between PTBP3 and other dynamic proteins in L-bodies, such as hnRNPAB, and decreases interactions between PTBP3 and the static RNA component of L-bodies, we would expect the dynamics of the chimeric protein to be increased relative to that of PTBP3. To test this, we generated a mCh-tagged construct of PTBP3 fused with the hnRNPAB X2 IDR, termed PTBP3+IDR (Fig. 5a). Expression and imaging of mCh-PTBP3+IDR showed localization to L-bodies similar to wild type PTBP3 (Fig. 5b-c)38. However, upon assaying PTBP3+IDR by FRAP, we found that the addition of the hnRNPAB X2 IDR decreased the dynamics of PTBP3 in L-bodies, as the chimeric protein displayed a mobile fraction of 20%, compared to a mobile fraction of 40% observed for PTBP3 (Fig. 5d-e). These results suggest that the hnRNPAB X2 IDR domain can act to slow translational diffusion of PTBP3 within L-bodies, likely by providing an interface for protein-protein interactions.
Fig. 5.
The hnRNPAB X2 IDR acts to stabilize proteins in L-bodies. (a) The hnRNPAB X2 IDR was fused to the C-terminus of PTBP3 (PTBP3+X2IDR) and tagged with mCherry (mCh). (b) Stage II oocytes were microinjected with mCh-PTBP3 RNA to express the encoded protein (green, detected by anti-RFP IF) along with Cy5 LE RNA to label L-bodies (magenta, b′). The merge is shown in b′′; scale bar = 100 μm. (c) Stage II oocytes were microinjected with mCh-PTBP3+IDR to express the encoded protein (green, detected by anti-RFP IF) along with Cy5 LE RNA (magenta, c′) to label L-bodies. The merge is shown in c′′; scale bar = 100 μm. (d) Stage II oocytes were microinjected with RNA encoding PTBP3 or PTBP3+X2IDR, along with Cy5 LE RNA to label L-bodies. Normalized FRAP recovery curves are shown (n = 21 oocytes); error bars represent standard error of the mean. Measurements were taken at 5 s intervals over 100 iterations. (e) Average percent mobile fractions are shown for mCh-PTBP3 (green, 40%±0.88) and mCh-PTBP3+X2IDR (blue, 20%±2.31). Error bars represent standard error of the mean and ** indicates p < 0.01. Statistics shown are an Ordinary one-way ANOVA with Tukey’s multiple comparisons.
Discussion
We investigated how a protein containing RBDs and an IDR localizes to a biomolecular condensate. The RBD- and IDR-containing protein hnRNPAB X2 is both highly enriched in L-body biomolecular condensates and highly dynamic within them. Interestingly, the nuclear form of hnRNPAB, which contains the same architecture with the addition of only a short nuclear localization sequence, is not assembled into granules. This difference may be due to the high concentration of RNA throughout the nucleus which can prevent phase separation of RBPs36 or due to the lack of L-body-specific components that would be needed to recruit hnRNPAB to other nuclear bodies. We also probed the contribution of each domain to hnRNPAB X2 behavior within L-bodies. The isolated hnRNPAB X2 RBD and IDR domains are sufficient for localization to L-bodies, but to a lesser degree than the full-length protein. In vitro, full-length hnRNPAB X2 protein forms condensates, as do both the IDR and RBD domains. Hence, both domains contribute to hnRNPAB X2 self-interactions and phase separation in vitro. Furthermore, this self-interaction does not require RNA, similar to other RBPs such as FUS, which can phase separate readily without RNA and whose disordered domain phase separates but does not interact with RNA39. Both the RBD and IDR domains of hnRNPAB X2 also affect protein dynamics in L-bodies: as assessed by FRAP both the RBD and IDR displayed shorter recovery half-times than the full-length protein, indicating that interactions anchoring the domains to L-bodies are reduced with the removal of the other domain.
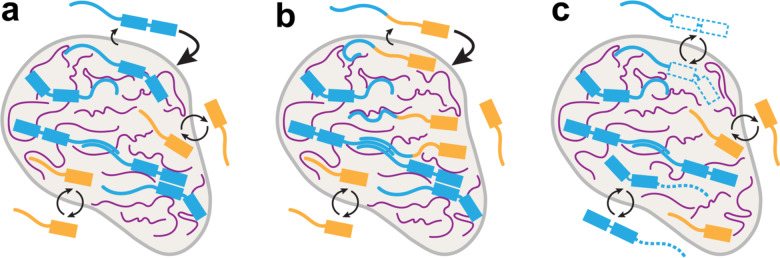
Taken together, our results suggest a model (Fig. 6) in which in vivo enrichment of RBPs like hnRNPAB X2 that contain phase separation prone RBDs and IDRs is determined by a combination of its interactions with L-body constituents, independent of the types of forces governing those interactions. Similarly, this model also suggests that RBP dynamics within L-bodies is determined by the combined effects of the interaction domains found in a protein. We had hypothesized that increasing an RBP’s association with the dynamic protein layer of L-bodies (such as by adding the hnRNPAB X2 IDR to PTBP3, which is known to bind directly to LE RNA) would cause it to be more dynamic, but found instead that adding the IDR causes the protein to become less dynamic and more stably associated with L-bodies. This indicates that protein enrichment and dynamics are likely agnostic of the manner in which a protein interacts with L-bodies; whether it binds the stable RNA directly as PTBP3 does or interacts with the dynamic protein layer as the hnRNPAB X2 IDR might, these interactions serve to enrich and stabilize the protein in L-bodies.
Fig. 6.
Model for how RBDs and IDRs together determine a protein’s localization (hnRNPAB) to and dynamics within L-bodies. (a) While RNA (magenta) is highly stable within L-bodies (grey), L-body associated proteins (blue, gold) dynamically associate with the RNA and other proteins. The strength of these associations determines the degree of their enrichment in L-bodies: proteins that interact strongly with L-bodies (blue) show a greater degree of enrichment to L-bodies and lower degrees of dynamic activity, while proteins that interact more weakly (gold) are less enriched and more dynamic. (b–c) The association of a protein with L-bodies can be tuned by modifying its interaction domains. Additional interaction domains (b) may stabilize a protein within L-bodies, while removal of interaction domains (c) may serve to destabilize a protein within L-bodies. This principle holds true for both RBDs (represented by boxes) and IDRs (represented by lines).
Indeed, it is possible that this model extends beyond proteins and that the stable RNA phase we observe in L-bodies10,38 is in fact an extreme end of this sliding scale of interaction strength. We previously showed38 that mutating LE RNA such that it could no longer bind two sequence-specific RBPs abolished its ability to localize to L-bodies and caused a sharp increase in its percent mobile fraction (from ~ 5% to ~ 60%). This linkage in LE RNA between a reduction in interaction strength, reduced localization, and increased dynamics is similar to what we have found with hnRNPAB X2 and illustrates what may be fundamental principles underlying the behavior of L-body constituents. In addition, our results, which indicate that IDRs play a role in protein recruitment to stable, long-lived biomolecular condensates, mirror those found in other systems37, suggesting that while a biomolecular condensate may mature over time, the fundamental physical properties governing protein recruitment to condensates are likely to remain constant.
Methods
Oocyte isolation and culture
All animal experiments were approved by the Brown University Institutional Animal Care and Use Committee. All experiments were conducted in accordance with relevant guidelines, and the study complied with the ARRIVE guidelines. Oocytes were harvested from wild type Xenopus laevis females (Nasco, catalog # LM00535MX). Oocytes were enzymatically defolliculated in 3 mg/ml collagenase (Sigma) followed by washes in MBSH (88mM NaCl, 1mM KCl, 2.4mM NaHCO3, 0.82mM MgSO4, 0.33mM CaCl2, 0.33mM Ca(NO3)2, 10mM HEPES pH7.6). Stage II-III oocytes were cultured at 18 °C in XOCM [50% Leibovitz L-15 (Thermofisher), 15mM HEPES (pH 7.6), 1 mg/mL insulin, 50U/mL nystatin, 100U/mL penicillin/streptomycin, 0.1 mg/mL gentamicin].
Cloning
Total RNA from stage II-III oocytes was extracted by TRIZOL. cDNA was generated via the iScript cDNA synthesis kit. Primers specific to hnRNPAB X2 (Table S3) were used to amplify hnRNPAB X2 from cDNA using Phusion PCR master mix which was then cloned into pSP64TNRLMCS:mCh10 using Gibson Assembly master mix (New England Biolabs) to generate pSP64:mCh-hnRNPAB-X2. Table S3 shows the primer sequences used to generate pSP64:mCh-RBD-X2, pSP64:mCh-IDR-X2, and pSP64:mCh-hnRNPABΔPY using pSP64:mCh-hnRNPAB-X2 as a template for PCR, followed by cloning into pSP64TNRLMCS:mCh as described above. Plasmids for expression of hnRNPAB and its isolated domains with a N-terminal maltose-binding protein tag were generated by PCR using primers (Table S3) specific to the appropriate sequence region with NdeI and XhoI sites for subcloning into the pTHMT vector39.
RNA transcription and microinjection
Fluorescently labeled vg1 LE RNA (Gautreau et al., 1997) was generated from linearized pSP73-2×135 using the MEGAscript T7 transcription kit (Ambion) in the presence of 250 nM Cy5-UTP (Fisher Scientific, PA55026). mCherry (mCh)-tagged protein-coding mRNAs were generated using the mMessage Machine transcription kit from the following linearized plasmids: pSP64:mCh-hnRNPAB-X2, pSP64:mCh-RBD-X2, pSP64:mCh-IDR-X2, pSP64:mCh, pSP64:mCh-hnRNPAB-FL, pSP64:mCh-hnRNPABΔPY, pSP64:mCh-PTBP3 and pSP64:mCh-PTBP3 + IDR. mRNAs and Cy-labeled RNAs were co-microinjected at 500 nM at a volume of 2 nL per oocyte. Following injection, oocytes were cultured for ~ 48 h at 18 °C in XOCM. Unlabeled Xenopus β-globin RNA was transcribed with the MEGAscript T7 kit (ThermoFisher) using a PCR product generated from pSP64:XBM40 (see Table S3 for primers).
Whole-mount immunofluorescence
Oocytes were washed in PBS to remove XOCM and then fixed for 1 h at room temperature in PEMT-FA (80 mM PIPES pH6.8, 1 mM MgCl2, 5 mM EGTA, 0.2% Triton X-100, 3.7% formaldehyde). Fixed oocytes were washed in PBT (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, 0.2% BSA, 0.1% Triton X-100) 3 times for 15 min each, then blocked in PBT+ (PBT supplemented with 2% normal goat serum, 2% BSA) for 2 h at room temperature. Oocytes were then incubated overnight at 4° C in a 1:500 dilution of primary antibody (listed in Table S2) in PBT+. Following incubation, oocytes were washed three times for two hours each in PBT, then incubated overnight at 4° C in a 1:1000 dilution of secondary antibody (Thermofisher goat anti-rabbit AF546, 11010) in PBT+. Oocytes were then washed three times for two h each in PBT. Following the washes, oocytes were dehydrated in anhydrous methanol and stored at -20° C until imaging. Immediately prior to imaging, oocytes were cleared in BABB solution (1:2 benzyl alcohol: benzyl benzoate). Oocytes were imaged on an inverted Olympus FV3000 confocal microscope using 20× UPlan Super Apochromat objective (air, NA = 0.75) and 30× UPlan Super Apochromat objective (silicon oil, NA = 1.05) using GaAsP detectors. To quantitate protein enrichment in L-bodies, image files were converted to channel-separated greyscale TIFs with ImageJ software and subjected to background subtraction with a rolling ball radius of 50.0. In the mCh channel (570–620 nm), to quantitate the fluorescence for hnRNPAB X2, RBD, IDR, or mCherry proteins, a circular region of interest (ROI) was drawn surrounding the oocyte and the RawIntDen was calculated as a measure of total fluorescence in the oocyte. In the Cy5 channel (650–750 nm), representing the Cy5-labeled LE signal, a second ROI was drawn surrounding the LE fluorescence at the vegetal cortex, this second ROI was copied to the mCh channel and the RawIntDen was recorded as a measure of vegetal cortical IF signal (fluorescence was only measured in the mCh channel, not the Cy5 LE channel). The fraction of fluorescence present in the cortex for each oocyte was calculated by dividing the vegetal cortex fluorescence by total oocyte fluorescence. Six images from each group were analyzed for each of four biological replicates (n = 24 images total per group). The data were analyzed by ordinary one-way ANOVA followed by Tukey’s multiple comparison tests with Prism software.
In vitro expression and purification of hnRNPAB X2 full length protein, IDR, and RBD domains
N-terminally MBP tagged (pTHMT) hnRNPAB X2 full-length isoform, IDR and RBD domains, and MBP (control) were expressed in E. coli BL21 star (DE3) cells (C600003; Thermofisher Scientific). Bacterial cultures were grown to an optical density at 600 nm of 0.7–0.9 before induction with 1 mM isopropyl-β-D-1-thiogalactopyranoside (IPTG) for 4 h at 37° C. Cell pellets were harvested by centrifugation and stored at -80 °C. Cells were resuspended in ~ 40 mL of 20 mM Tris, 500 mM NaCl, 10 mM Imidazole (pH 8.0), with one Roche EDTA-free protease inhibitor tablet (11697498001; Sigma Aldrich) per ~ 4 g cell pellet, and lysed using the Avestin Emulsiflex C3 (Ottawa, Ontario, Canada). The lysate was cleared by centrifugation at 47,850 ×g (20,000 rpm) for 50 min at 4 °C, filtered using a 0.2 μm syringe filter, and loaded onto a HisTrap HP 5 ml column (17524701; Cytiva, Marlborough, MA, USA). The protein was eluted with a gradient from 10 to 300 mM imidazole in 20 mM Tris, 500 mM NaCl, pH 8.0. Fractions containing MBP-tagged hnRNPAB X2 full length isoform were loaded onto a HiLoad 26/600 Superdex 200 pg column (28-9893-36; Cytiva) equilibrated in 20 mM Tris, 150 mM NaCl, and 1 mM dithiothreitol (DTT). Fractions containing MBP-tagged hnRNPAB X2, hnRNPAB X2-IDR, hnRNPAB X2-RBD, or MBP alone were loaded onto a HiLoad 26/600 Superdex 75 pg (28-9893-34; Cytiva) equilibrated in 20 mM Tris and 150 mM NaCl. Fractions with high purity were identified by SDS-PAGE and concentrated using a centrifugation filter with a 10 kDa cutoff (ACS501024; MilliporeSigma, Burlington, MA, USA). MBP-tagged hnRNPAB X2 full length isoform, the IDR and RBD domains, and MBP were flash frozen in liquid nitrogen in 20 mM Tris and 150 mM NaCl and stored at -80 °C.
In vitro phase separation
Purified MBP-tagged hnRNPAB X2 full length isoform, and the IDR and RBD domains or, as a control, MBP alone (including TEV cleavage artifacts and all residues present in MBP-tagged hnRNPAB X2 proteins) in 20 mM NaPi, pH 7.4, 150 mM NaCl were incubated with 0.03 mg/mL TEV protease for 20 min to cleave the MBP tag from the protein. Protein concentrations of 5 µM, 12.5 µM. 25 µM and 50 µM were tested in the absence and presence of 0.25 mg/mL Xenopus β-globin RNA, with the addition of 10% PEG to induce phase separation, analogous to recent protocols for other RNA-binding proteins41. After inducing phase separation in microcentrifuge tubes, the samples were incubated at room temperature for ~ 20 min before imaging.
Condensate imaging
All condensate imaging was performed on a Nikon Ti2-E Fluorescence microscope using a 60× objective. Samples were spotted onto a glass coverslip and condensate formation was evaluated by imaging with differential interference contrast (DIC), acquired with a Prime 95B Mono camera. For DIC time-lapse videos, frames were collected every 0.6 s for 20 s. For fluorescence imaging, in vitro phase separation was carried out in the presence of 25 µM Thioflavin T (2390-54-7, Sigma Aldrich). Fluorescence images were quantified with ImageJ software. Images were subjected to background subtraction with a 50.0 pixel rolling ball radius and manual thresholding. The Analyze Particles plugin was then used to measure condensate number per field of view (294.8 × 294.8 μm), size (in µm), and circularity (on a scale of 0–1.0, with 1.0 representing a perfect circle), excluding particles on the edges. Six to nine images were analyzed per group for each of 3 replicates. Results were analyzed by ordinary two-way ANOVA followed by Tukey’s multiple comparison tests for hnRNPAB X2 and the IDR and RBD domains.
FRAP
Stage II oocytes were microinjected with 2 nL of 500 nM mRNA encoding mCherry-tagged constructs and 250 nM Cy5-UTP labeled LE RNA to mark L-bodies. Oocytes were cultured in XOCM at 18° C for ~ 48 h post-injection. A 10 μm2 ROI was bleached using a 488 nm laser at 100% for 2 s. Recovery of the fluorescence was measured at 5 s increments over 100 iterations. Seven oocytes per biological replicate (21 oocytes total per protein construct) were analyzed. Fluorescence measurements were corrected based on background fluorescence and nonspecific photobleaching, as previously described38,42,43. Data were analyzed via one phase non-linear regression analysis using GraphPad Prism 9. Statistics shown in the main text for the percent immobile fractions are an ordinary one-way ANOVA with Tukey’s multiple comparison of the one phase association data.
qPCR
One hundred stage II oocytes per biological replicate were washed in PBS and homogenized in Trizol to extract total oocyte RNA. Total oocyte RNA was reverse transcribed using the iScript cDNA synthesis kit (BioRad). qPCR was performed using SybrGreen Powerup Master mix (ThermoFisher, A25742) per the manufacturer’s protocol for vg1, canonical hnRNPAB and hnRNPAB X2 RNAs, using the primers shown in Table S3. ΔCt values were calculated44 normalizing to vg1 as a refence gene, and compared by T-test.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Juan Alfonzo for comments on the manuscript. We thank Kevin Czaplinski for gifting the 40-LoVe (Xenopus hnRNPAB) antibody. This work was funded by R01GM071049 from the NIH to KLM and by R01GM147677 from the NIH to NLF.
Author contributions
LCO and KLM conceptualized the study and contributed to the writing and editing of the manuscript. KLM carried out supervision, acquired funding, contributed to visualization, writing and editing of the manuscript. LCO carried out methodology development, investigation, formal analysis, visualization, and writing of the original draft. VJ carried out investigation, formal analysis, and visualization. JPO carried out investigation, formal analysis, visualization, and editing of the manuscript. AKH carried out investigation, formal analysis, and visualization. ACM carried out methodology development and investigation. MCL carried out methodology development and investigation. S-HW carried out investigation. NLF carried out supervision, acquired funding, contributed to visualization, and editing of the manuscript.
Data availability
All data supporting the findings of this study are available within the paper and its Supplementary Information.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Banani, S. F., Lee, H. O., Hyman, A. A. & Rosen, M. K. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol.18, 285–298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fare, C. M., Villani, A., Drake, L. E. & Shorter, J. Higher-order organization of biomolecular condensates. Open. Biology11 (2021). [DOI] [PMC free article] [PubMed]
- 3.Banani, S. F. et al. Compositional control of phase-separated Cellular bodies. Cell166, 651–663 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swain, P. & Weber, S. C. Dissecting the complexity of biomolecular condensates. Biochem. Soc. Trans.48, 2591–2602 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Protter, D. S. W. & Parker, R. Principles and properties of stress granules. Trends Cell. Biol.26, 668 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hofmann, S., Kedersha, N., Anderson, P. & Ivanov, P. Molecular mechanisms of stress granule assembly and disassembly. Biochim. et Biophys. Acta - Mol. Cell. Res.1868 (2021). [DOI] [PMC free article] [PubMed]
- 7.Molliex, A. et al. Phase separation by low complexity domains promotes stress Granule Assembly and drives pathological fibrillization. Cell163, 123–133 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bose, M., Lampe, M., Mahamid, J. & Ephrussi, A. Liquid-to-solid phase transition of oskar ribonucleoprotein granules is essential for their function in Drosophila embryonic development. Cell185, 1308–1324e23 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boke, E. et al. Amyloid-like self-assembly of a cellular compartment. Cell166, 637–650 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neil, C. R. et al. L-bodies are RNA-protein condensates driving RNA transport in Xenopus oocytes. Mol. Biol. Cell ar27 (2021). [DOI] [PMC free article] [PubMed]
- 11.Putnam, A., Cassani, M., Smith, J. & Seydoux, G. A gel phase promotes condensation of liquid P granules in Caenorhabditis elegans embryos. Nat. Struct. Mol. Biol.26, (2019). [DOI] [PMC free article] [PubMed]
- 12.Fawzi, N. L., Parekh, S. H. & Mittal, J. Biophysical studies of phase separation integrating experimental and computational methods. Curr. Opin. Struct. Biol.70, 78–86 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chong, S. & Mir, M. Towards decoding the sequence-based Grammar governing the functions of intrinsically disordered protein regions. J. Mol. Biol. 433 (2021). [DOI] [PubMed]
- 14.Murthy, A. C. et al. Molecular interactions underlying liquid – liquid phase separation of the FUS low-complexity domain. Nat. Struct. Mol. Biol.26, 637–648 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin, Y., Currie, S. L. & Rosen, M. K. Intrinsically disordered sequences enable modulation of protein phase separation through distributed tyrosine motifs. J. Biol. Chem.292, 19110–19120 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang, P. et al. G3BP1 is a tunable switch that triggers phase separation to assemble stress granules. Cell181, 325–345e28 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Monahan, Z. et al. Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity. EMBO J.36, 2951–2967 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elbaum-Garfinkle, S. et al. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl. Acad. Sci. U S A112, 7189–7194 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maharana, S. et al. RNA buffers the phase separation behavior of prion-like RNA binding proteins. Sci. (1979)360, 918–921 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roden, C. & Gladfelter, A. S. RNA contributions to the form and function of biomolecular condensates. Nat. Rev. Mol. Cell Biol.22, 183–195 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zimyanin, V. L. et al. In vivo imaging of oskar mRNA transport reveals the mechanism of posterior localization. Cell134, 843–853 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wiedner, H. J. & Giudice, J. It’s not just a phase: function and characteristics of RNA-binding proteins in phase separation. Nat. Struct. Mol. Biology28, 465–473 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Medioni, C., Mowry, K. & Besse, F. Principles and roles of mRNA localization in animal development. Dev. (Cambridge)139, 3263–3276 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cabral, S. E. & Mowry, K. L. Organizing the oocyte: RNA localization meets phase separation. Curr. Top. Dev. Biol.140, 87–118 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O’Connell, L. C. & Mowry, K. L. Regulation of spatially restricted gene expression: linking RNA localization and phase separation. Biochem. Soc. Trans.49, 2591–2600 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neil, C. R. et al. L-bodies are RNA–protein condensates driving RNA localization in Xenopus oocytes. Mol. Biol. Cell.32, (2021). [DOI] [PMC free article] [PubMed]
- 27.Mittag, T. & Parker, R. Multiple modes of protein–protein interactions promote RNP granule assembly. J. Mol. Biol.430, 4636–4649 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verma, A., Sumi, S. & Seervi, M. Heat shock proteins-driven stress granule dynamics: yet another avenue for cell survival. Apoptosis (2021). [DOI] [PubMed]
- 29.Czaplinski, K. & Mattaj, I. W. 40LoVe interacts with Vg1RBP/Vera and hnRNP I in binding the Vg1-localization element. RNA 12, 213–222 (2006). [DOI] [PMC free article] [PubMed]
- 30.Czaplinski, K. et al. Identification of 40LoVe, a xenopus hnRNP D family protein involved in localizing a TGF-β-related mRNA during oogenesis. Dev. Cell.8, 505–515 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Lewis, R. A. et al. Conserved and clustered RNA recognition sequences are a critical feature of signals directing RNA localization in Xenopus oocytes. Mech. Dev.121, 101–109 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Gautreau, D., Cote, C. A. & Mowry, K. L. Two copies of a subelement from the Vg1 RNA localization sequence are sufficient to direct vegetal localization in Xenopus oocytes. Development124, 5013–5020 (1997). [DOI] [PubMed] [Google Scholar]
- 33.Lee, B. J. et al. Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell126, 543–558 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lancaster, A. K., Nutter-Upham, A., Lindquist, S. & King, O. D. PLAAC: a web and command-line application to identify proteins with prion-like amino acid composition. Bioinformatics30, 2501–2502 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bentmann, E. et al. Requirements for stress granule recruitment of fused in sarcoma (FUS) and TAR DNA-binding protein of 43 kDa (TDP-43). J. Biol. Chem.287, 23079–23094 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boeynaems, S. et al. Protein phase separation: a new phase in cell biology. Trends Cell. Biol.28, 420–435 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ditlev, J. A., Case, L. B. & Rosen, M. K. Who’s in and who’s out—compositional control of Biomolecular condensates. J. Mol. Biol.430, 4666–4684 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cabral, S. E., Otis, J. P. & Mowry, K. L. Multivalent interactions with RNA drive recruitment and dynamics in biomolecular condensates in Xenopus oocytes. iScience 104811 (2022). [DOI] [PMC free article] [PubMed]
- 39.Burke, K. A., Janke, A. M., Rhine, C. L. & Fawzi, N. L. Residue-by-residue view of in vitro FUS granules that bind the C-terminal domain of RNA polymerase II. Mol. Cell.60, 231–241 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krieg, P. A. & Melton, D. A. Functional messenger RNAs are produced by SP6 in vitro transcription of cloned cDNAs. Nucleic Acids Res.12, 7057–7070 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hallegger, M. et al. TDP-43 condensation properties specify its RNA-binding and regulatory repertoire. Cell184, 4680–4696e22 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Powrie, E. A. et al. Using in vivo imaging to measure RNA mobility in Xenopus laevis oocytes. Methods98, 60–65 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gagnon, J. A., Kreiling, J. A., Powrie, E. A., Wood, T. R. & Mowry, K. L. Directional transport is mediated by a dynein-dependent step in an RNA localization pathway. PLoS Biol.11, (2013). [DOI] [PMC free article] [PubMed]
- 44.Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available within the paper and its Supplementary Information.