Abstract
Background
Panarthropods, a major group of invertebrate animals comprised of arthropods, onychophorans, and tardigrades, are the only limb-bearing members of Ecdysozoa. The complexity and versatility of panarthropod paired limbs has prompted great interest in their development to better understand the formation of these structures and the genes involved in this process. However, studies of limb patterning and development are overwhelmingly focused on arthropods, followed by select work on onychophorans but almost entirely lacking for tardigrades. This model organism bias is inherently limited and precludes a comparative analysis of how panarthropod legs originated, have evolved, and the likely limb patterning genes present in the earliest panarthropod ancestors. In this study, we investigated tardigrade homologs of seven arthropod distal limb patterning genes (apterous, aristaless, BarH1, clawless, Lim1, rotund, and spineless) to better characterize tardigrade limb development in a comparative context.
Results
We detected homologs of all seven genes in the eutardigrade Hypsibius exemplaris and heterotardigrade Echiniscoides cf. sigismundi suggesting their conservation in both tardigrade lineages. Hybridization chain reaction experiments in H. exemplaris reveal a regionalized expression pattern for the genes aristaless, BarH1, clawless, rotund and spineless.
Conclusion
The observed regionalized expression of the distal limb patterning genes in H. exemplaris might reflect the external morphological features of tardigrade legs, such as the distal claws, sensory organs in the proximal region, and specific muscle attachment sites. The comparison between the expression of these limb patterning genes in H. exemplaris relative to other panarthropods suggests their conserved role in the last common panarthropod ancestor, such as establishing the distal limb end and the distribution of sensory structures. Our results support the hypothesis that tardigrade legs are homologous to the distal region of other panarthropod limbs, as suggested by previous work on the expression of leg gap genes in H. exemplaris.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13227-024-00235-1.
Keywords: Tardigrada, Panarthropoda, Distal limb patterning genes, Regionalization, Body plan
Introduction
Panarthropoda is a major animal group composed of arthropods (e.g. arachnids, myriapods and pancrustaceans), onychophorans (velvet worms), and tardigrades (water bears) [1]. The panarthropods are the only limb-bearing ecdysozoans (i.e., moulting invertebrates), featuring diverse sets of paired segmental appendages that can either be sclerotized and jointed (i.e., arthropods) or lobopodous (i.e., onychophorans and tardigrades). The Cambrian fossil record of panarthropods [2–4], coupled with molecular phylogenomic analyses supporting the sister-group position of tardigrades relative to onychophorans and arthropods [5, 6], indicate that the lobopodous legs are ancestral whereas jointed legs are derived and unique to arthropods. However, paired limbs are complex morphological features, and so numerous studies have addressed the question of how panarthropod legs first originated, how they have evolved, and what genes were responsible for limb patterning in the earliest panarthropod ancestors. Arthropods have been the subject of numerous developmental genetics studies to answer these questions (e.g. [7–11]), and more recent studies on onychophorans have revealed both similarities and differences compared to arthropod models [12, 13]. However, a comprehensive understanding of panarthropod limb evolution is hindered by the paucity of developmental work on tardigrades, with only one recent study available that investigated genes involved in tardigrade limb development in the eutardigrade Hypsibius exemplaris [14].
Tardigrades are a phylum of microinvertebrates that feature four pairs of lobopodous legs that typically bear terminal claws [15]. As a clade, tardigrades can be broadly subdivided into the heterotardigrades, which are morphologically variable and showcase dorsal cuticular specializations, and the comparatively simpler eutardigrades, which have a more conservative overall appearance with minimal or lacking external cuticular specializations [15]. Heterotardigrades feature pronounced differences in limb morphology, including distal finger-like digits where claws are attached, while some lack these structures [16], or they may also display sensory organs of different shapes and sizes around the proximal or medial part of the legs [16, 17]. The structural components of both the claws and sensory organs are secreted by cells [15, 18], which are most likely regulated by leg patterning genes to define their cellular identity, determine which structures they produce, or mark the location where they develop. Some heterotardigrades even have telescopic limbs which show pseudo-segmentation and whose sections can be differentiated from each other [16]. Even the superficially simple legs of eutardigrades have internal musculature with proximodistal differences, such as their attachment site [19]. Collectively, these phenotypes suggest that there are genes involved in patterning leg regionalization in tardigrades akin to those observed in the more morphologically variable legs of arthropods.
Understanding the genetic underpinning of panarthropod limbs is fundamental for explaining the substantial proximodistal (PD) specialization of paired appendages in these versatile organisms. For instance, gene interactions in the fruit fly Drosophila melanogaster result in the emergence of gene expression domains involved in patterning the PD axis of the legs [20]. Differential expression of the leg gap genes, such as homothorax [hth], dachsund [dac], and Distal-less [Dll] broadly divides the developing adult walking leg into three regions—proximal, medial, and distal, respectively (reviewed in [21]). One role of Dll in D. melanogaster is to activate EGFR signaling, which leads to the activation of distal limb patterning genes [22, 23]. The interactions of these genes result in the regionalization and specialization of the distal end of the legs, namely the tarsus and pretarsus. This second set of genes includes aristaless (al), clawless (cll), and Lim1, which are involved in the development of the pretarsus [24–27], Bar (BarH1) and apterous (ap) that control distal tarsi development [25, 28], and rotund (rn) and spineless (ss) which affect the development of the proximal tarsi [29–31]. Despite the profound morphological differences between arthropod and onychophoran legs, studies on the velvet worm Euperipatoides kanangrensis have shown that most homologs of the leg gap genes and distal limb patterning genes have similar expression patterns in the developing embryonic walking limbs of onychophorans [12, 13], hinting at a conserved gene patterning network shared between these organisms. A recent study on the eutardigrade Hypsibius exemplaris, however, showed a different expression pattern for the leg gap genes [14], particularly that the gene that specifies the medial part (i.e., dac) is absent in tardigrades. The gene that is normally expressed in the proximal (i.e., hth and extradenticle [exd]) and distal (i.e., Dll) regions are expressed across the entire first three pairs of limbs, while only Dll is expressed across the hind limbs. These results suggest that either the hind limbs have a different PD identity compared to other front legs, or all tardigrade legs have a distal identity (i.e., homologous to the distal region) relative to other panarthropod legs. Furthermore, the broad expression of leg gap genes across the entire legs of H. exemplaris suggest that these genes are not necessarily the direct cause for the observed regionalized tardigrade legs. Since Dll is required for activating the distal limb patterning genes in D. melanogaster [22, 23] and these genes also showed regionalized expression in the distal limb region of arthropods and onychophorans, their homologs in tardigrades are good candidates to test whether they are expressed in a regionalized manner and potentially involved in tardigrade limb regionalization.
In this study, we investigated homologs of arthropod distal limb patterning genes in two tardigrade species. We were able to identify homologs of seven D. melanogaster distal limb patterning genes—ap, al, BarH1, cll, Lim1, rn, and ss in the eutardigrade H. exemplaris and the heterotardigrade Echiniscoides cf. sigismundi. We then determined the expression patterns of these putative homologs during limb development in embryos of the emerging tardigrade model H. exemplaris to facilitate direct comparisons across all three panarthropod phyla.
Materials and methods
Gene mining of distal-limb patterning genes
Tardigrade homologs of seven Drosophila melanogaster distal limb-patterning genes—apterous (ap), aristaless (al), Bar (BarH1), clawless (cll), Lim1, rotund (rn), and spineless (ss)—were identified from predicted protein sequences of the eutardigrade Hypsibius exemplaris (https://www.ncbi.nlm.nih.gov/datasets/genome/GCA_002082055.1/). Gene mining was done using a modified method from [32] (File S1 for the commands). For each gene of interest, one D. melanogaster protein isoform obtained from Flybase (https://flybase.org/; Table S1) was used for building profile hidden Markov models (HMMs) using HMMER v3 [33] which were then used as queries against the H. exemplaris predicted protein sequences as database to obtain the first set of candidate homologs using the Easel application of HMMER. Then, a reciprocal hit search using BLAST v2.6 [34] was done by using the same D. melanogaster isoform for building HMMs as query (Table S1) in a BLASTp search against the first set of candidate genes as database. Hits with an e-value less than 10–6 were retained and formed the second set of candidate genes. This set was then used as query in an online BLASTp search (https://blast.ncbi.nlm.nih.gov/Blast.cgi) against the D. melanogaster protein database (taxid:7227). The third set of candidate genes were composed of genes that showed the specific D. melanogaster gene as the top hit and had a max score greater than 80, a percent identity greater than 20%, and e-value less than 10–6. After obtaining the final set of candidate homologs via reciprocal BLASTp searches, their protein domains were determined using the NCBI Batch Conserved Domain tool (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi) to check whether they contain the domains found in the D. melanogaster genes. As a final step of validation, this final set of candidates were used as queries in BLASTp searches against the D. melanogaster annotated proteins using the Flybase BLAST tool to check if the top hit corresponds to their putative gene homolog. The same process was done to identify tardigrade homologs of the heterotardigrade Echiniscoides cf. sigismundi. These homologs were identified from translated sequences of a previously assembled transcriptome (https://doi.org/10.7910/DVN/CFNUGF) [32]. Gene sequences are listed in the supplementary material (Table S2–S3).
Phylogenetic analyses
As a separate test for homology, phylogenetic analyses were done with the identified tardigrade homologs and homologs from other organisms to assess how they cluster together. Non-tardigrade homologs were obtained from Flybase and OrthoDB 11 [35]. All the homeobox-containing genes (i.e., ap, al, BarH1, cll, and Lim1) were run in the same analysis, while rn and ss were run with homologs of glass (gl) and clock (clk), respectively. The sequences were aligned using MAFFT 7 [36] using the L-INS-i algorithm (File S2-S4). The alignments were subjected to maximum likelihood (ML) and Bayesian inference (BI) analyses. The ML trees were reconstructed using IQTree 1.6 [37] with the best model based on the Akaike information criterion (AIC) and selected by the program (VT + F + R5 for the homeobox-containing genes, DCMut + F + I + G4 for rn-gl, and JTT + F + I + G4 for ss-clk). Bootstrap analysis was done using 500 replicates, and the consensus tree was obtained using the default setting. The BI trees were reconstructed using MrBayes 3.2 [38] using the best models (DAYHOFF + I + G for the homeobox-containing genes and rn-gl, and VT + I + G for ss-clk) obtained using Partitionfinder 2 [39] based on the AIC. The analyses were run for at least 1,000,000 generations, sampling every 500 generations and with a 25% burn-in frequency. Two runs were done simultaneously, each with one cold and three heated chains. Convergence was assessed by checking that the average deviation of split frequencies of the two runs was less than 0.01, effective sample size values were greater than 200, and the potential scale reduction factor was approximately = 1. A 50% majority rule consensus tree was then obtained to summarize the resulting analysis (File S5 for p and t files).
In situ hybridization and imaging
To determine expression patterns of the genes of interest during limb development, we focused on H. exemplaris, whose embryogenesis is well-documented [40] and became a model organism for studying tardigrade development [41]. We focused on three stages of embryonic development [40]—(i) stage 14, when limb bud formation is first easily observed; (ii) stage 16, when the limbs are more elongated, but claws have not yet developed; and (iii) stage 17, when claws start to appear. In our cultures, which are incubated at 23 °C, these stages were observed at 35 h, 45 h, and 55 h post laying (hpl), respectively.
In situ hybridization was done using the hybridization chain reaction (HCR) method following the protocol by Smith et al. [42]. To aid in identifying the legs, all samples were counterstained with Dll (Fig S1). HCR probe sets were synthesized by Molecular Instruments based on the predicted RNA sequences of the identified H. exemplaris homologs of the genes of interest (Table S2). After the final wash, embryos were then mounted in DAPI Flouromount-G (SouthernBiotech). Images were collected using the LSM 980 Confocal Microscope with Airyscan2 (Zeiss). The 405 nm, 488 nm, 561 nm, and 639 nm lasers were used to visualize the DAPI-stained nucleus, B1 probe, B2 probe, and B3 probe-bound RNA, respectively. Different parts of the optical sections were obtained to create the final image to highlight specific parts (e.g., leg I or leg IV) using the ‘maximum intensity’ projection type of the Z-stack feature in ImageJ 2.3. Levels were adjusted using ImageJ. Figures were assembled using Adobe Illustrator 26.5.
For transmitted light microscopy, slides of tardigrades were imaged using an Axioscope 5 compound microscope (Zeiss) with Axiocam 208 color camera (Zeiss). Different optical sections were obtained, and the “auto-blend” function of Adobe Photoshop 23.5 was used to create the final image. Figures were assembled using Adobe Illustrator 26.5.
Despite finding homologs in Echiniscoides cf. sigismundi, we did not investigate their expression patterns since laboratory techniques for this species are currently unavailable and culturing them is methodologically challenging because of their marine habitat.
Results
Identification of tardigrade gene homologs
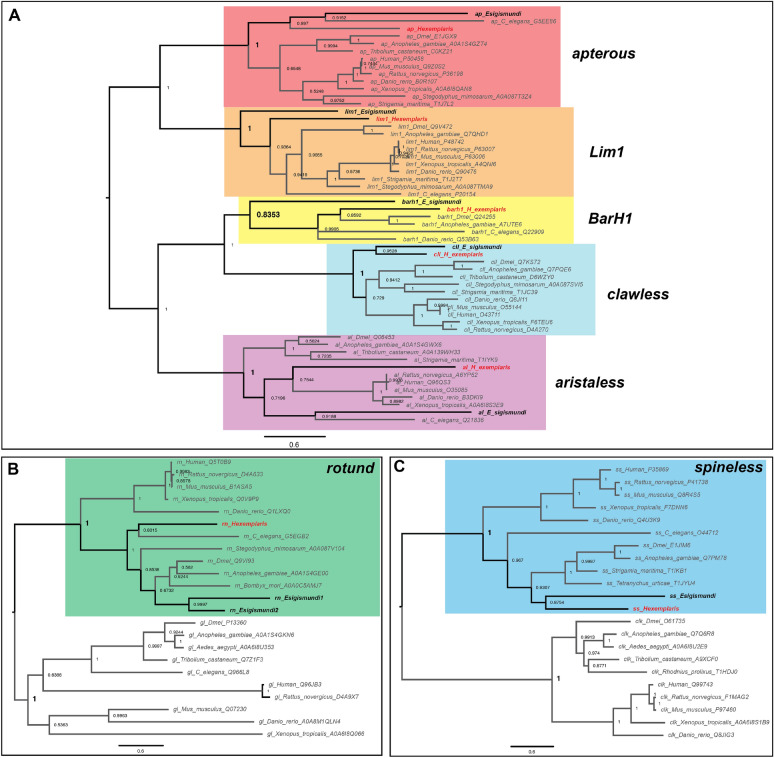
Using our gene mining strategy, we were able to identify homologs of all seven genes of interest—apterous (ap), aristaless (al), Bar (BarH1), clawless (cll), Lim1, rotund (rn), and spineless (ss)—in Hypsibius exemplaris (He), with single gene copies for each (Table S2–S3). The reciprocal BLASTp against the Drosophila melanogaster protein predictions showed that the top hits for the tardigrade homolog query were their corresponding D. melanogaster homologs (Table S4). The homologs also possess protein domains similar to D. melanogaster homologs (Table S5). Our phylogenetic analyses showed that the identified H. exemplaris genes clustered with other putative homologs from different organisms (Fig. 1, S2). Our results suggest that the identified genes in H. exemplaris are the true homologs of the D. melanogaster distal limb patterning genes.
Fig. 1.
Phylogenetic result of the Bayesian inference using MrBayes. A Homeobox-containing genes dataset. B rotund-glass genes dataset. C spineless-clock genes dataset. Values on the node represent the posterior probability values. Tardigrade taxa are highlighted in bold
We were also able to identify homologs in Echiniscoides cf. sigismundi (Es) (Table S6). Similar to H. exemplaris, single copies for each gene were identified, except for rotund, in which two sequences were identified. The BLASTp search (Table S7), protein domain inspection (Table S5), and phylogenetic analyses (Fig. 1, S2) all support the identified candidates as true homologs of the seven genes of interest.
Expression patterns of Hypsibius exemplaris homologs of arthropod distal limb patterning genes
Since D. melanogaster was used as a reference in the homolog search, the expression patterns of the tardigrade homologs are discussed below relative to how these genes are involved in the development of fruit fly limbs from the distal to proximal podomeres (i.e., al, cll, and Lim1 for pretarsus, ap and BarH1 for distal tarsus, and rn and ss for proximal tarsus). The spatial positioning of the embryonic limbs follows Chavarria et al. [43] (Fig. 2).
Fig. 2.
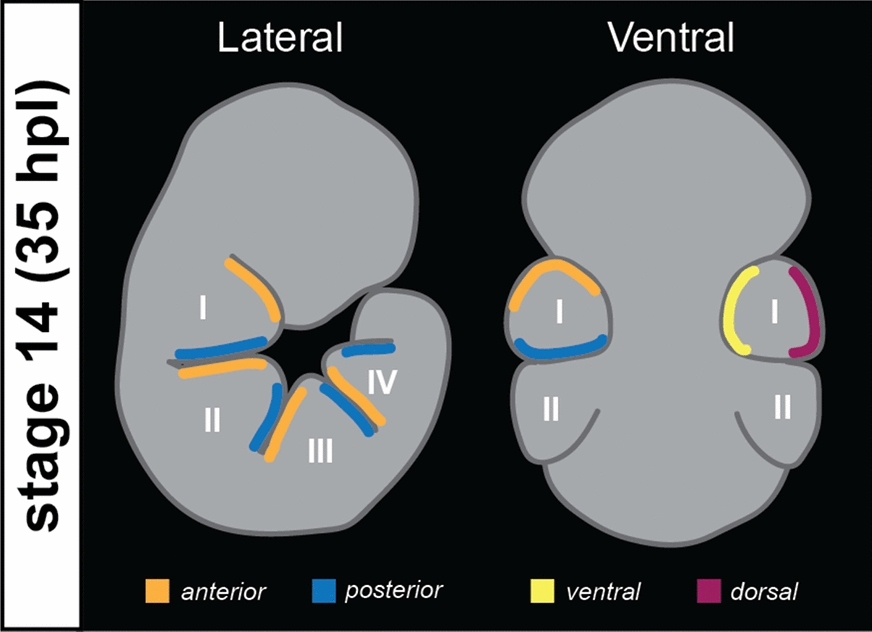
Schematic drawing of the tardigrade embryo at stage 14 with the relative leg regions based on [43]. The same regions correspond to legs in stage 16 and 17
He-al, He-cll, and He-Lim1 were expressed in the legs during H. exemplaris embryogenesis (Fig. 2, 4A–C, 5, S3, S4, S5A-C, S6). At stage 14, He-al was expressed mostly at the posterior side of the legs (Fig S3A,B,B*, S4, S6A,B,B*). As the legs extended at stage 16, He-al expression was seen at a more interior position (i.e., not touching the posterior border of the limbs) (Fig S3C,E’, 6C,D,D*). These observations resemble the He-al expression pattern observed in another study that used a different (i.e., chromogenic) in situ hybridization method [14]. At stage 17, a clear expression pattern of He-al was not detected (Fig S3D, F’, F**, S6E, F). Unlike He-al, He-cll was expressed broadly in the legs at stage 14 (Fig. 3A, B, B*). At stage 16, the expression was mostly concentrated at the posterior side of the legs with a slight overlap with He-al (Fig. 3C, C’, S3E,E”). By the time the claws started to appear at stage 17, He-cll expression was barely detectable (Fig. 3D, S3F,F*). Outside the leg region, He-cll was also visibly expressed as a wide band along the dorsal side of the embryo at stage 14 (Fig. 3B, B*). This pattern started to disappear at stage 16, but a clear, concentrated expression at the dorsal side of the head was seen (Fig. 3C, S3E). He-Lim1 was expressed broadly across the legs, as well as the head region, at all three stages, but its expression was noticeably reduced at stage 17 (Fig. 4A–C, S5A-C).
Fig. 4.
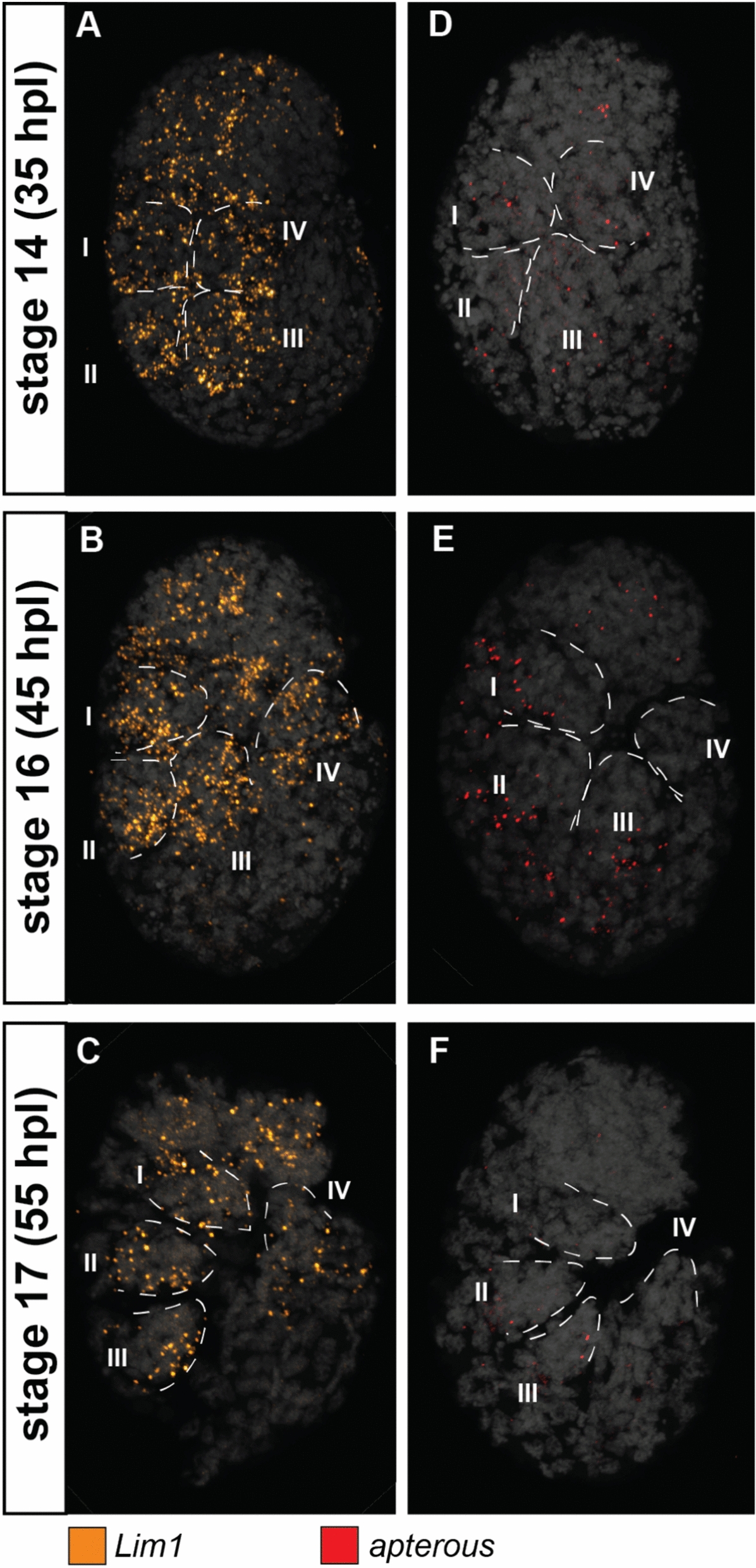
Expression patterns of apterous (ap) and Lim1 at different stages of the eutardigrade Hypsibius exemplaris limb development. A, D Stage 14. B, E Stage 16. (C,F) Stage 17. A–C Lim1 expression. D–F ap expression. Embryos are in lateral view and facing right in all panels. Roman numeral number indicates leg number (e.g., I—1st pair of legs). Nuclei are labeled with DAPI (Gray)
Fig. 5.
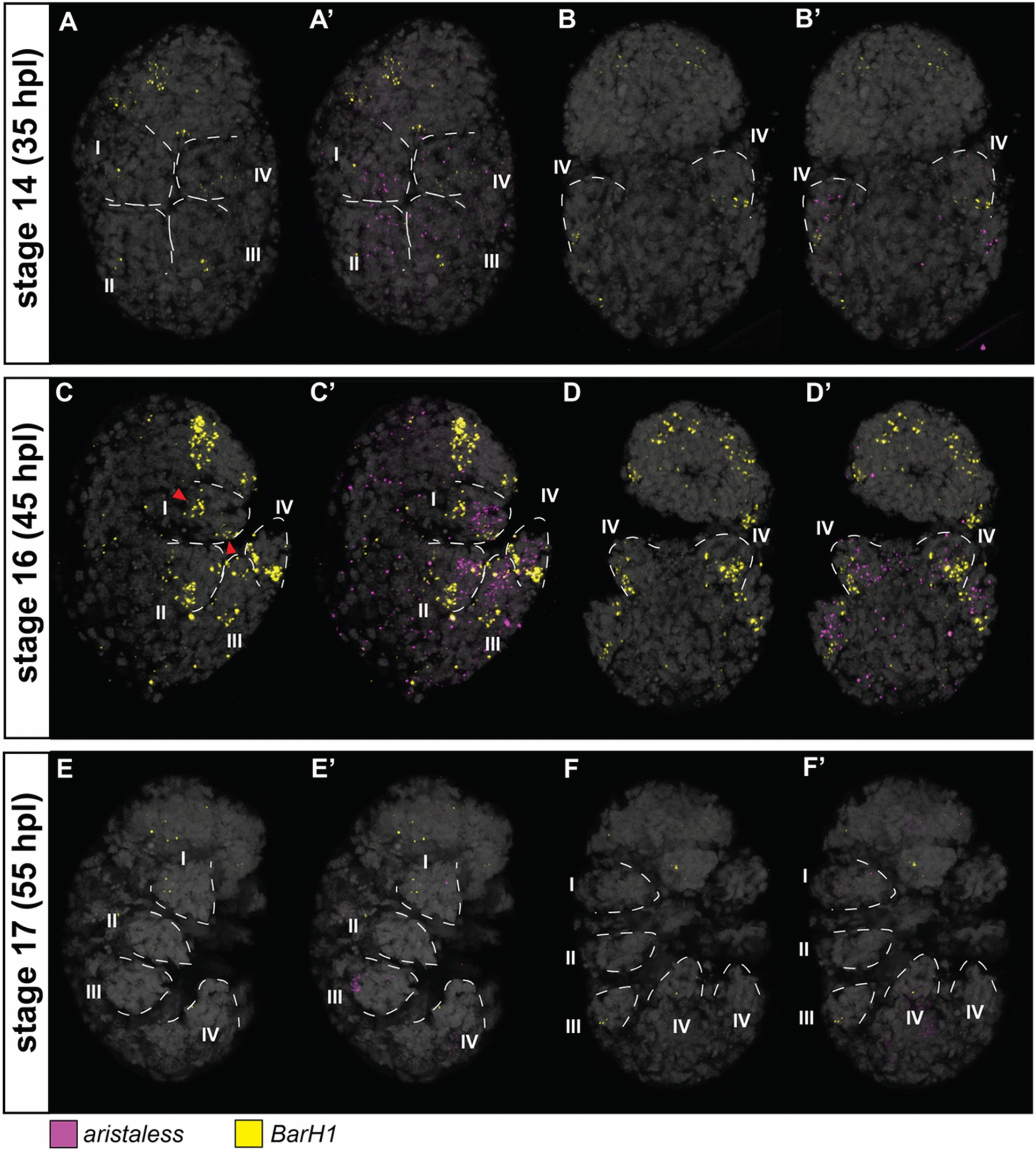
Expression patterns of aristaless (al) and BarH1 at different stages of the eutardigrade Hypsibius exemplaris limb development. A, B Stage 14. C, D Stage 16. E, F Stage 17. A–F BarH1 expression. A’–F’ al and BarH1 expression. Figures with the same letter indicates the same embryo viewed in similar optical sections. A, C, E Embryos in lateral view and facing right. B, D, F Embryos in dorsoventral mount. Roman numeral number indicates leg number (e.g., I—1st pair of legs). Nuclei are labeled with DAPI (Gray). Red arrowheads indicate the two patches of BarH1 expression observed at stage 16
Fig. 3.
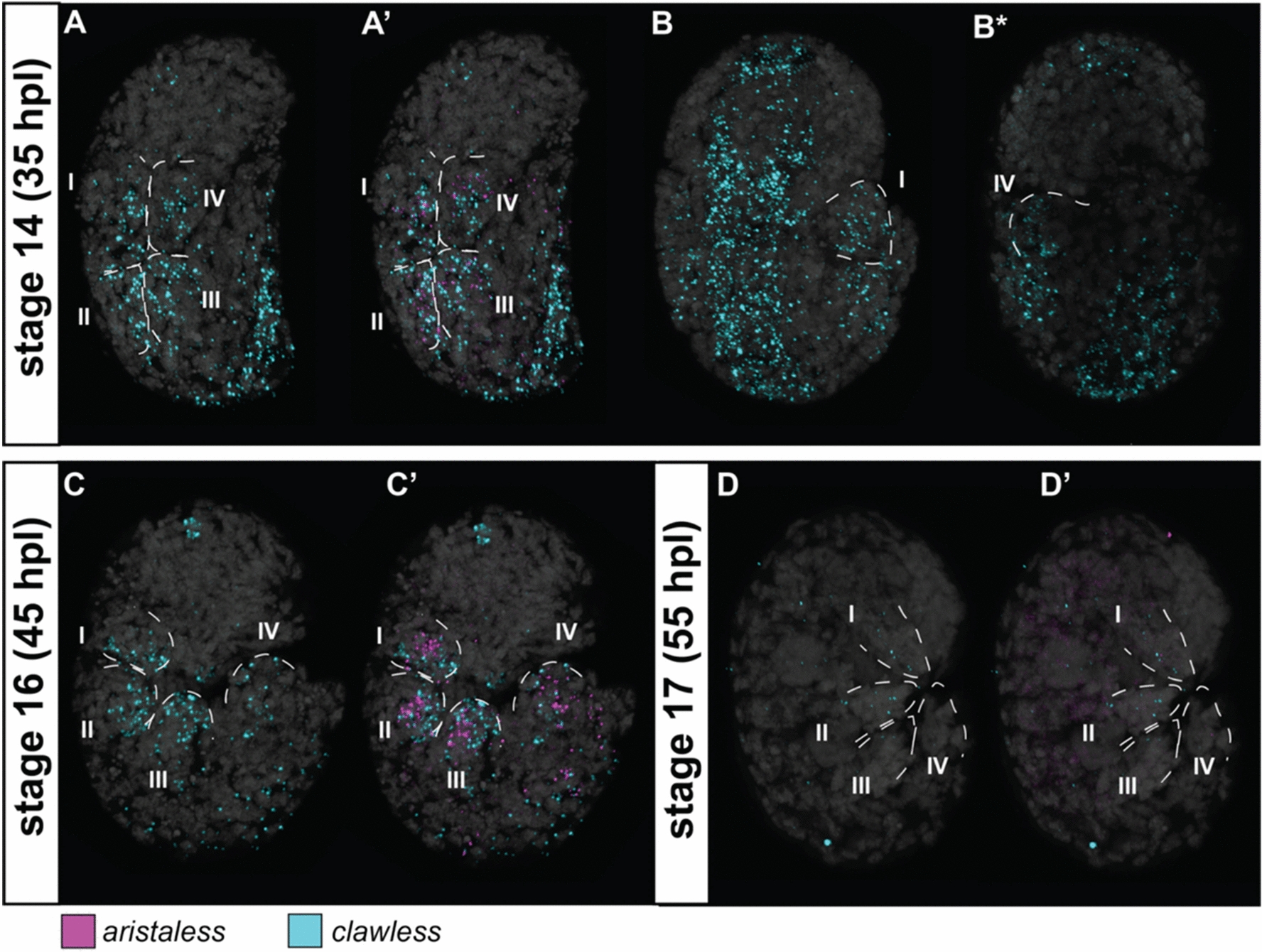
Expression patterns of aristaless (al) and clawless (cll) at different stages of the eutardigrade Hypsibius exemplaris limb development. A, B Stage 14. C Stage 16. D Stage 17. A, B, B*, C, D cll expression. A’, C’, D’ al and cll expression. Figures with the same letter indicates the same embryo viewed in similar optical sections; with asterisk (*) indicate the same embryo but viewed at different optical sections. Embryos are in lateral view and facing right in all panels, except for B and B*, which are in dorsoventral mount. Roman numeral number indicates leg number (e.g., I—1st pair of legs). Nuclei are labeled with DAPI (Gray)
At all three stages, He-ap expression was sparse in the legs and across the entire embryo (Fig. 4D–F, S5D-F). He-BarH1 showed a different expression pattern than He-ap. At stage 14, expression was observed at the proximal side of the developing limb bud (Fig. 5A, B). The signal became stronger at stage 16, and two patches of expression were observed in the elongated leg (Fig. 5C, D red arrowheads). The first was observed at the proximal side of the leg and expressed proximally relative to He-al (Fig. 5C’, D’, S6D***). The other was observed at the tip of the leg around the postero-ventral side (Fig. 5C). This second location of He-BarH1 expression was observed in the first three limbs but not in the last limb. At stage 17, the expression of He-BarH1 was no longer detectable (Fig. 5E, F). A notable expression pattern of He-BarH1 was also seen in the head region at stages 14 and 16 (Fig. 5A, C). This pattern was visible on either side of the head as seen from a ventral view (Fig S6B**,D**).
At stage 14, He-rn showed a strong expression on one side of the leg (Fig. 6A’, S7A’), similar to how the expression of He-al. He-rn was observed in the most distal part of the leg at stage 16 (Fig. 6B’, S7B’,B**) and continued to be expressed at the same region at stage 17 (Fig. 6C’, S7C’,C**). Outside the legs and at all three stages, He-rn was strongly expressed at the dorsal side of the body region containing the third pair of legs (Fig S8). At stage 14, He-ss was strongly expressed at a region without He-rn (Fig. 6A, S7A) and only showed a slight overlap with this gene (Fig. 6A”, S7A”). The expression of He-ss at the proximal region of the legs became evident at stage 16 (Fig. 6B, S7B,B*) and was detected at the postero-ventral side, similar to He-BarH1. A slight overlap with He-rn was also observed at this stage (Fig. 6B”, S7B”,B***). The same expression pattern was observed at stage 17 (Fig. 6C, S7C,C*). Outside the legs, a notable expression of He-ss was seen in a head region at all three embryonic stages (Fig. 6A, B, C, S7A, B, C), similar to the He-BarH1 expression in the head.
Fig. 6.
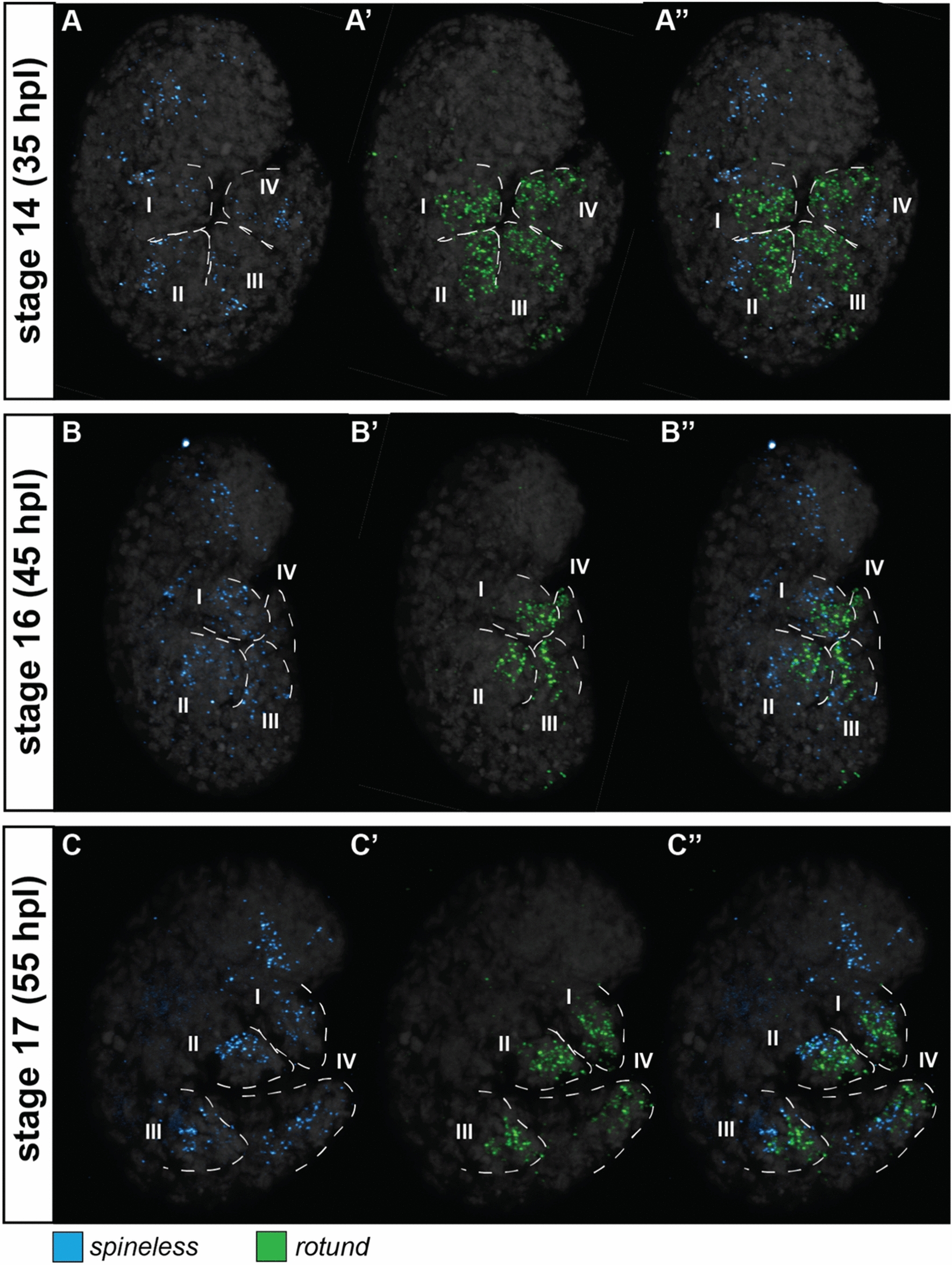
Expression patterns of rotund (rn) and spineless (ss) at different stages of the eutardigrade Hypsibius exemplaris limb development. A Stage 14. B Stage 16. C Stage 17. A–C ss expression. A’-C’ rn expression. A”–C” ss and rn expression. Figures with the same letter indicates the same embryo viewed in similar optical sections. Embryos are in lateral view and facing right in all panels. Roman numeral number indicates leg number (e.g., I—1st pair of legs). Nuclei are labeled with DAPI (Gray)
Discussion
Tardigrade homologs of arthropod distal limb patterning genes
Our results show that the eutardigrade Hypsibius exemplaris (He) and heterotardigrade Echiniscoides cf. sigismundi (Es) have homologs of the seven arthropod distal limb patterning genes, apterous (ap), aristaless (al), Bar (BarH1), clawless (cll), Lim1, rotund (rn), and spineless (ss). All genes appear to have single copies in each tardigrade species, except for Es-rn where two transcripts were identified. Since the domains found for each Es-rn copy are different from each other, and each corresponds to different domains of the Drosophila melanogaster rotund gene (Table S5), the two copies could be parts of one single transcript. Given that these two species belong to the two major classes of tardigrades, positive detection of these homologs suggests that these genes are broadly conserved in the phylum.
Limb patterning in tardigrades
Six of the seven genes we investigated (i.e., all but He-ap) appear to be involved in limb development of H. exemplaris embryos. The expression of He-al and He-cll at the distal end of the leg at stage 16 (45 hpl) suggests their involvement in claw production, most likely by either marking the location of claw glands or directly regulating the secretion of claws. This is further supported by their lack of expression at stage 17 (55 hpl), when claws have already formed, which would be similar to insects where functional studies of al and cll revealed that these genes are involved in patterning the pretarsus (or claws)—the distal part of the walking leg [24, 27, 44]. He-rn also showed a regionalized expression pattern similar to He-al at stage 14, suggesting it could also be involved in claw production. However, unlike the He-al, its expression persisted until stage 17, when claws have already been produced.
The expression of He-BarH1 and He-ss is evident in the proximal region of the legs. In heterotardigrades, this region is where sensory cirri are located (Fig. 7A, B) [16, 17]. Interestingly, these genes are involved in the development of sensory structures in arthropods, such as the antenna and bristles [30, 45–47]. Therefore, we hypothesize that BarH1 and ss could be involved in the development of sensory structures in tardigrades, and that this could be tested by gene expression experiments in heterotardigrades. Even though eutardigrades lack sensory structures on their limbs (Fig. 7C, D), the expression of He-BarH1 and He-ss at the proximal side could indicate that there are sensory regions at this area that are related to the sensory organs in heterotardigrades. Indeed, sensory fields have been found in the cephalic regions of eutardigrades despite the lack of flagellate or papillate-like sensory organs, which are present on the head of heterotardigrades [48–51]. A recent neuroanatomical study based on innervation patterns on the head of the heterotardigrade Echiniscus testudo suggested that the area where the cephalic cirri and clava are can be homologized to the cephalic sensory fields in eutardigrades that are devoid of cuticular extensions [52]. He-BarH1 and He-ss were also observed to be expressed at the lateral side of the embryonic head (Figs. 5A, C, 6A, B, C, S6B**,D**, S7A, B, C), which spatially corresponds to the region of the heterotardigrade head where cephalic cirri (i.e., cirri A) and the primary clava are found. The expression patterns of He-BarH1 and He-ss suggest that the lack of sensory structures does not indicate a lack of sensory area on the legs of eutardigrades. This also suggests that the ancestral tardigrade may have had sensory organs in this area. Additionally, He-BarH1 could be marking the leg ganglia since these are found around the proximal side of the H. exemplaris legs [53, 54]. Since this gene is also expressed in the ventral ganglia (Fig S6D**), it could be marking neurons or other cell types that are present in ganglia.
Fig. 7.
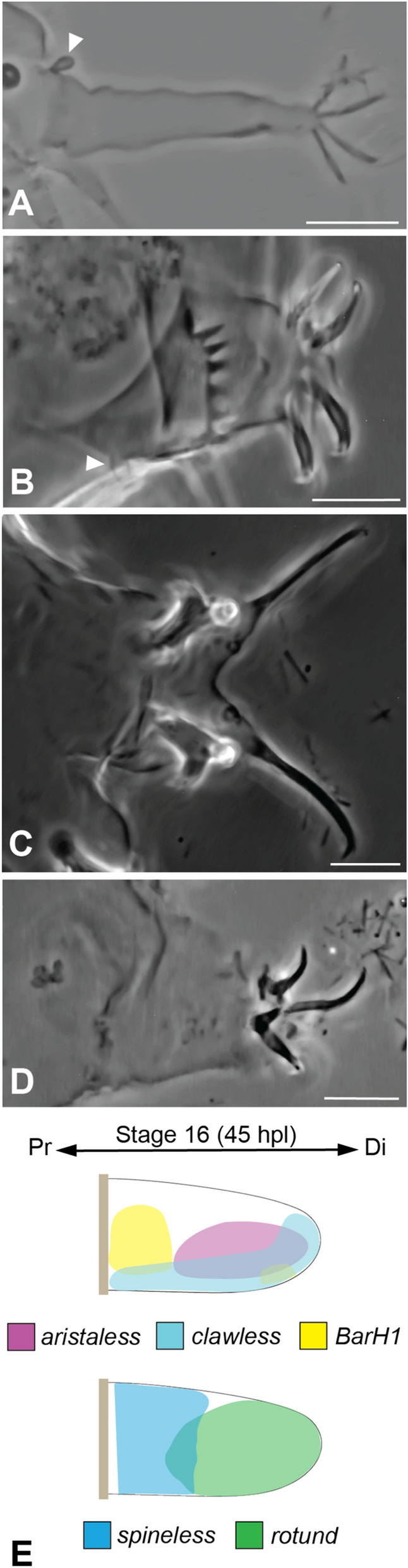
Comparison of limb patterning genes in the eutardigrade Hypsibius exemplaris relative to the morphology of the fourth leg pair in different tardigrade groups. A Heterotardigrade Stygarctus leg. B Heterotardigrade echiniscid leg. C Eutardigrade Milnesium leg. D Eutardigrade H. exemplaris leg. E Expression pattern at stage 16 of H. exemplaris development. Scale bar = 10 μm. White arrowheads indicate leg sensory organs
He-Lim1 and He-ap did not show regionalized expression patterns. He-Lim1 is expressed across the entire limb in all the observed embryonic stages, which suggests that it is involved in leg development, but it is not possible to identify a specific role because it lacks any distinct patterns. By contrast, in situ experiments for He-ap lack noticeable expression in the legs during development, which suggests that it might not be involved in this process or is not active in the studied embryonic stages.
Our results show that homologs of the arthropod distal limb patterning genes have regionalized expression patterns in eutardigrade legs (Fig. 6E). These regionalized expression patterns may be important for forming different structures during development, such as claws and sensory organs at the distal and proximal regions of the legs, respectively. Since all these genes are also found in the heterotardigrade E. cf. sigismundi, determining their expression patterns in this species during limb development would help confirm if this regionalization is present in both tardigrade groups and, thus, present in the tardigrade ancestor. It will also help test whether some of these genes are truly associated with sensory structures, which are known to be present in heterotardigrade legs. The function of these genes could then be experimentally disrupted to test their functions and determine if altering them influences tardigrade leg morphology.
Distal limb patterning genes in Panarthropoda
The new data on the eutardigrade H. exemplaris allows us to compare the expression patterns of the seven distal limb-patterning genes across representatives of Panarthropoda (Fig S9). In the insects Drosophila melanogaster, Tribolium castaneum, and Gryllus bimaculatus, al is expressed at the distal tip of the legs and at the putative segmental boundaries that form the podomeres [44, 55, 56]. However, functional studies on D. melanogaster and T. castaneum show that al mutants and knockdowns caused the loss of the pretarsus [27, 44], suggesting that this gene is only involved in patterning the distalmost part of the leg. Furthermore, since al and Dll are proposed to be co-expressed ancestrally [57], the other proximal expression domains of al that do not overlap with Dll are most likely derived in insects. Since al is also expressed at the distal tip of the embryonic legs of the myriapod Glomeris marginata [10], the onychophoran Euperipatoides kanangrensis [12], and the eutardigrade H. exemplaris (Fig. 7E), we conclude that the role of this gene for patterning the distalmost part of the legs is most likely conserved across extant panarthropods, and was likely also expressed in the distalmost part of the leg in the panarthropod ancestor. The cll gene also shows distal expression in the legs of disparate panarthropods [10, 12, 24, 58], which suggest that it has a similarly conserved role.
In D. melanogaster, positive regulatory feedback between al and cll, and their antagonistic relationship with BarH1, result in the more proximal expression of BarH1 relative to the more distal expression of the other two genes [24, 59]. A similar proximal BarH1 expression pattern is observed in G. marginata, and E. kanangrensis [10], as well as in H. exemplaris (Fig. 7E). However, H. exemplaris showed a different pattern from other panarthropods since BarH1 is also expressed at the distal region of the leg where al and cll are also expressed at stage 16 (Fig. 7E). Even in onychophorans, the congeners E. kanangrensis and E. rowelli showed different BarH1 expression patterns [10, 60]. Given these different observations, the role of BarH1 in the development of the limbs of the panarthropod ancestors remain uncertain.
The ancestral role of Lim1 is also uncertain given its different expression patterns across panarthropods (Fig S9). In D. melanogaster and T. castaneum, Lim1 has three expression domains—proximal, medial, and distal leg regions [25, 61]. In G. marginata, however, Lim1 is expressed across the entire leg [10]. Lim1 is only expressed at the tip of the leg in E. kanangrensis [12], while it is broadly expressed in H. exemplaris (Fig. 4B). Thus, whether Lim1 ancestrally has an expression pattern that was broad or more regionalized remains unknown.
Like Lim1, rn expression also varies across Panarthropoda (Fig S9). For example, rn is one of the most proximally expressed distal limb patterning genes in D. melanogaster [62], but it is expressed across the entire leg in G. marginata [10]. In non-arthropods, rn is only expressed at the distal tip in E. kanangrensis [12] and the distal part of H. exemplaris. Expression data for T. castaneum is not available, but RNAi experiments on rn produced abnormalities on the tarsus and pretarsus—the distal part of the legs [63]. This suggests that rn is involved in patterning the distal end of the leg of T. castaneum, unlike in D. melanogaster wherein this gene is only involved in the development of the proximal tarsi [29, 31]. We hypothesize that rn could be involved in the development of the distal end of the panarthropod ancestral leg, while the patterns observed in D. melanogaster and G. marginata are the derived state.
Similar to rn, ss is also expressed in the proximal tarsi in D. melanogaster and is involved in patterning this region [30], but has a more variable expression pattern across the panarthropods (Fig S9). In T. castaneum, ss is expressed at the proximal end of the thoracic legs of T. castaneum embryos [45] but is still involved in the proper development of the proximal tarsi [63]. This gene is expressed in the trunk appendages of the malacostracan Parhyale hawaiensis, but no expression is observed in the walking legs of the insect Oncopeltus fasciatus and myriapod Lithobius atkinsoni [45, 47]. The chelicerates Parasteatoda tepidariorum and Phalangium opilio showed distal expression of ss in the walking legs, but these are not limited to the tarsal region [47]. A similar distal expression is observed in E. kanangrensis [12], but not in H. exemplaris wherein it is seen more at the proximal end of the legs. We hypothesize that ss is not involved in patterning specific regions in the walking legs of the panarthropod ancestor, but instead, it was probably involved in patterning areas with sensory functions. This is supported by the fact that ss is involved in the proper development of sensory organs, such as antennae and bristles [30, 45–47]. Additionally, this gene is expressed in the antennal limb buds of mandibulates and the protocerebral antennae of E. kanangrensis, both of which have sensory functions [12, 47]. Expression of ss, therefore, could indicate the presence of sensory fields, and the role of this gene in tarsal development could have been derived in the ancestor of D. melanogaster and T. castaneum. It has also been suggested that this role in tarsal development has been independently acquired in arachnids [47].
Distal nature of tardigrade legs
Investigation of the expression of leg gap genes in the eutardigrade H. exemplaris [14] revealed that Dll, the gene that specifies the distal region in arthropods and onychophorans, was broadly expressed across all the legs. The genes specifying the proximal region of the leg, hth and exd, were only expressed in the first three pairs of limbs in H. exemplaris. These results broadly concur with the hypothesis that either the tardigrade hind limbs have a different PD identity compared to the three anterior leg pairs or that all tardigrade legs have a distal identity relative to other panarthropod legs based on their gene expression [14].
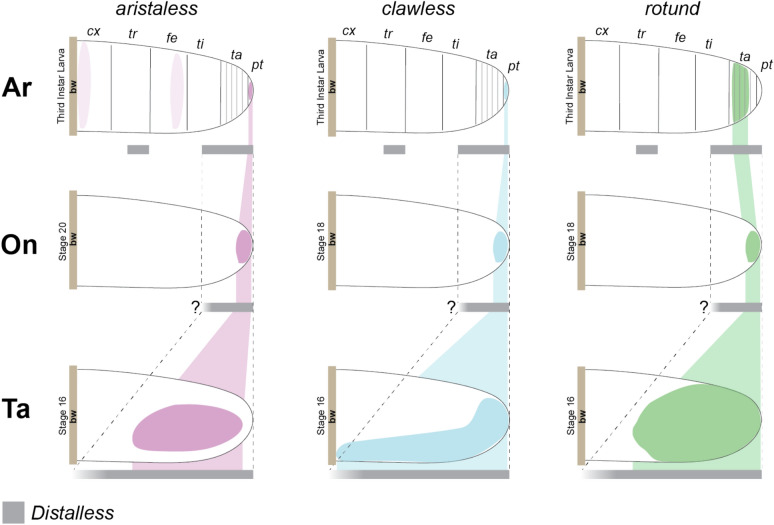
Our results highlight the wide area of expression of He-rn in H. exemplaris compared to other panarthropods (Fig. 8). He-rn is expressed distally, encompassing almost half the length of the H. exemplaris legs compared to onychophorans and D. melanogaster where rn expression only covers a small area at the distal tip of the legs (Fig S9). The same observation can be made when comparing He-al and He-cll to other panarthropods. If the expression patterns of He-al, He-cll, and He-rn are superimposed to other panarthropods, the entire tardigrade leg would correspond to the distal ends of the onychophoran and arthropod legs. We propose that these observations support the tardigrade leg having a distal identity and, thus, are homologous to the distal region of other panarthropod appendages.
Fig. 8.
Expression patterns of aristaless (al), clawless (cll), and rotund (rn) in the walking legs of different panarthropod phyla relative to Distalless (Dll) expression. Ar—Arthropoda (Drosophila melanogaster), On—Onychophora (Euperipatoides kanangrensis), Ta—Tardigrada (Hypsibius exemplaris). bw—body wall. Stages when the illustrated expression patterns appeared are indicated. Lighter shades of al in D. melanogaster denotes positive expression, but the gene is not involved in patterning the specific leg region (Campbell & Tomlinson, 1998). The “?” in E. kanangrensis denotes the uncertainty of the extent of proximal expression of Dll relative to the other genes. Expression references: H. exemplaris (this study, Game & Smith, 2020); E. kanangrensis (Janssen et al. 2010, Oliveira et al. 2014); D. melanogaster (Campbell et al. 1993, Kojima et al. 2005, Natori et al. 2012, Jockusch & Smith 2015)
Conclusion
We identified homologs of seven D. melanogaster distal limb patterning genes in tardigrades and showed that at least six genes (al, BarH1, cll, Lim1, rn, and ss) are expressed during H. exemplaris limb development. We detected regionalized expression of some of these genes (i.e., al, BarH1, cll, rn, and ss), which could reflect their role in patterning different structures in the legs, such as claws and sensory organs. Our data supports the hypothesis that developmental regionalization predates the external morphological segmentation and differentiation of panarthropod limbs [13]. By comparing the eutardigrade H. exemplaris to representatives of Onychophora and Arthropoda, we could identify both conserved and divergent expression patterns across disparate members of Panarthropoda (Fig S9). Our findings allow us to hypothesize possible roles of these limb patterning genes in the panarthropod ancestor and lead us to propose that tardigrade legs have a distal identity relative to the limbs of other panarthropods. These results will serve as a reference point for future functional studies aimed to elucidate further the role of these genes in tardigrade limb development.
Supplementary Information
Acknowledgements
We thank the Harvard Center for Biological Imaging (HCBI) for infrastructure and support in microscope imaging. We thank the anonymous reviewers for their valuable comments and suggestions that significantly improved the quality of the manuscript. Published by a grant from the Wetmore Colles Fund.
Author contributions
MAM, FWS, and JOH designed the study; MAM and MG performed wet lab work; MAM performed bioinformatic analyses, collected confocal data, and wrote the original draft. All authors read and edited the manuscript and approved the final version.
Funding
This material is based in part upon work supported by the National Science Foundation under Grant No. 1951257 to FWS.
Availability of data and materials
All sequence data analyzed in this study are publicly available. Sequences obtained via the homology search and alignments used for the phylogenetic analyses are available in the supplementary materials.
Declarations
Ethics approval and consent to participate
Studies of tardigrades do not require ethics approval or consent to participate.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Frank W. Smith, Email: frank.smith@unf.edu
Javier Ortega-Hernández, Email: jortegahernandez@fas.harvard.edu.
References
- 1.Nielsen C. Panarthropoda. In: Nielsen C, editor. Animal evolution: interrelationships of the living phyla. New York: Oxford University Press; 2012. p. 240–1. [Google Scholar]
- 2.Ortega-Hernández J. Lobopodians. Curr Biol. 2015;25:R873–5. 10.1016/j.cub.2015.07.028. [DOI] [PubMed] [Google Scholar]
- 3.Yang J, Ortega-Hernández J, Gerber S, Butterfield NJ, Hou JB, Lan T, Zhang XG. A superarmored lobopodian from the Cambrian of China and early disparity in the evolution of Onychophora. Proc Natl Acad Sci U S A. 2015;112:8678–83. 10.1073/pnas.1505596112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ortega-Hernández J. Making sense of ‘lower’ and ‘upper’ stem-group Euarthropoda, with comments on the strict use of the name Arthropoda von Siebold, 1848. Biol Rev. 2016;91:255–73. 10.1111/brv.12168. [DOI] [PubMed] [Google Scholar]
- 5.Campbell LI, et al. MicroRNAs and phylogenomics resolve the relationships of Tardigrada and suggest that velvet worms are the sister group of Arthropoda. Proc Natl Acad Sci. 2011;108:15920–4. 10.1073/pnas.1105499108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laumer CE, et al. Revisiting metazoan phylogeny with genomic sampling of all phyla. Proc R Soc B Biol Sci. 2019. 10.1098/rspb.2019.0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jockusch EL. Developmental and evolutionary perspectives on the origin and diversification of arthropod appendages. Integr Comp Biol. 2017;57:533–45. 10.1093/icb/icx063. [DOI] [PubMed] [Google Scholar]
- 8.Jockusch EL, Smith FW. 2015 Hexapoda: Comparative aspects of later embryogenesis and metamorphosis. In Evolutionary Developmental Biology of Invertebrates 5: Ecdysozoa III: Hexapoda (ed A Wanninger), Springer. pp. 111–208. 10.1007/978-3-7091-1868-9
- 9.Bruce HS, Patel NH. Knockout of crustacean leg patterning genes suggests that insect wings and body walls evolved from ancient leg segments. Nat Ecol Evol. 2020;4:1703–12. 10.1038/s41559-020-01349-0. [DOI] [PubMed] [Google Scholar]
- 10.Janssen R. Gene expression reveals evidence for EGFR-dependent proximal-distal limb patterning in a myriapod. Evol Dev. 2017;19:124–35. 10.1111/ede.12222. [DOI] [PubMed] [Google Scholar]
- 11.Pechmann M, Khadjeh S, Sprenger F, Prpic NM. Patterning mechanisms and morphological diversity of spider appendages and their importance for spider evolution. Arthropod Struct Dev. 2010;39:453–67. 10.1016/j.asd.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 12.Oliveira MB, et al. Expression of arthropod distal limb-patterning genes in the onychophoran Euperipatoides kanangrensis. Dev Genes Evol. 2014;224:87–96. 10.1007/s00427-014-0466-z. [DOI] [PubMed] [Google Scholar]
- 13.Janssen R, Eriksson BJ, Budd GE, Akam M, Prpic NM. Gene expression patterns in an onychophoran reveal that regionalization predates limb segmentation in pan-arthropods. Evol Dev. 2010;12:363–72. 10.1111/j.1525-142X.2010.00423.x. [DOI] [PubMed] [Google Scholar]
- 14.Game M, Smith FW. Loss of intermediate regions of perpendicular body axes contributed to miniaturization of tardigrades. Proc R Soc B Biol Sci. 2020. 10.1098/rspb.2020.1135rspb20201135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Møbjerg N, Jørgensen A, Kristensen RM, Neves RC. Morphology and functional anatomy. In: Schill RO, editor. Water bears: the biology of tardigrades. Cham, Switzerland: Springer Nature Switzerland; 2018. p. 57–94. [Google Scholar]
- 16.Fontoura P, Bartels PJ, Jørgensen A, Kristensen RM, Hansen JG. A dichotomous key to the genera of the Marine Heterotardigrades (Tardigrada). Zootaxa. 2017;4294:1–45. 10.11646/zootaxa.4294.1.1. [Google Scholar]
- 17.Kristensen RM. Generic revision of the Echiniscidae (Heterotardigrada), with a discussion of the origin of the family. In: Bertolani R, editor. Biology of tardigrades. Modena, Italy: Mucchi Editore; 1987. p. 261–335. [Google Scholar]
- 18.Kristensen RM. Sense organs of two marine arthrotardigrades (Heterotardigrada, Tardigrada). Acta Zool. 1981;62:27–41. [Google Scholar]
- 19.Gross V, Mayer G. Cellular morphology of leg musculature in the water bear Hypsibius exemplaris (Tardigrada) unravels serial homologies. R Soc Open Sci. 2019. 10.1098/rsos.191159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Estella C, Voutev R, Mann RS. A dynamic network of morphogens and transcription factors patterns the fly leg. Curr Top Dev Biol. 2012;98:173–98. 10.1016/B978-0-12-386499-4.00007-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kojima T. The mechanism of Drosophila leg development along the proximodistal axis. Dev Growth Differ. 2004;46:115–29. 10.1111/j.1440-169X.2004.00735.x. [DOI] [PubMed] [Google Scholar]
- 22.Galindo MI, Bishop SA, Greig S, Couso JP. Leg patterning driven by proximal-distal interactions and EGFR signaling. Science (80-). 2002;297:256–9. 10.1126/science.1072311. [DOI] [PubMed] [Google Scholar]
- 23.Galindo MI, Bishop SA, Couso JP. Dynamic EGFR-Ras signalling in Drosophila leg development. Dev Dyn. 2005;233:1496–508. 10.1002/dvdy.20452. [DOI] [PubMed] [Google Scholar]
- 24.Kojima T, Tsuji T, Saigo K. A concerted action of a paired-type homeobox gene, aristaless, and a homolog of Hox11/tlx homeobox gene, clawless, is essential for the distal tip development of the Drosophila leg. Dev Biol. 2005;279:434–45. 10.1016/j.ydbio.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 25.Pueyo JI, Galindo MI, Bishop SA, Couso JP. Proximal-distal leg development in Drosophila requires the apterous gene and the Lim1 homologue dlim1. Development. 2000;127:5391–402. 10.1242/dev.127.24.5391. [DOI] [PubMed] [Google Scholar]
- 26.Tsuji T, Sato A, Hiratani I, Taira M, Saigo K, Kojima T. Requirements of Lim1, a Drosophila LIM-homeobox gene, for normal leg and antennal development. Development. 2000;127:4315–23. 10.1242/dev.127.20.4315. [DOI] [PubMed] [Google Scholar]
- 27.Campbell G, Tomlinson A. The roles of the homeobox genes aristaless and Distal-less in patterning the legs and wings of Drosophila. Development. 1998;125:4483–93. 10.1242/dev.125.22.4483. [DOI] [PubMed] [Google Scholar]
- 28.Kojima T, Sato M, Saigo K. Formation and specification of distal leg segments in Drosophila by dual Bar homeobox genes, BarH1 and BarH2. Development. 2000;127:769–78. 10.1242/dev.127.4.769. [DOI] [PubMed] [Google Scholar]
- 29.St. Pierre SE, Galindo MI, Couso JP, Thor S. Control of Drosophila imaginal disc development by rotund and roughened eye: Differentially expressed transcripts of the same gene encoding functionally distinct zinc finger proteins. Development. 2002;129:1273–81. 10.1242/dev.129.5.1273. [DOI] [PubMed] [Google Scholar]
- 30.Duncan DM, Burgess EA, Duncan I. Control of distal antennal identity and tarsal development in Drosophila by spineless-aristapedia, a homolog of the mammalian dioxin receptor. Genes Dev. 1998;12:1290–303. 10.1101/gad.12.9.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pueyo JI, Couso JP. The 11-aminoacid long Tarsal-less peptides trigger a cell signal in Drosophila leg development. Dev Biol. 2008;324:192–201. 10.1016/j.ydbio.2008.08.025. [DOI] [PubMed] [Google Scholar]
- 32.Mapalo MA, Arakawa K, Baker CM, Persson DK, Mirano-Bascos D, Giribet G. The unique antimicrobial recognition and signaling pathways in tardigrades with a comparison across ecdysozoa. G3 Genes Genomes Genet. 2020;10:1137–48. 10.1534/g3.119.400734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mistry J, Finn RD, Eddy SR, Bateman A, Punta M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 2013;41: e121. 10.1093/nar/gkt263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:1–9. 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuznetsov D, Tegenfeldt F, Manni M, Seppey M, Berkeley M, Kriventseva EV, Zdobnov EM. OrthoDB v11: annotation of orthologs in the widest sampling of organismal diversity. Nucleic Acids Res. 2023;51:D445–51. 10.1093/nar/gkac998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–80. 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–74. 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ronquist F, et al. Mrbayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–42. 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. Partitionfinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 2017;34:772–3. 10.1093/molbev/msw260. [DOI] [PubMed] [Google Scholar]
- 40.Gabriel WN, McNuff R, Patel SK, Gregory TR, Jeck WR, Jones CD, Goldstein B. The tardigrade Hypsibius dujardini, a new model for studying the evolution of development. Dev Biol. 2007;312:545–59. 10.1016/j.ydbio.2007.09.055. [DOI] [PubMed] [Google Scholar]
- 41.Goldstein B. The emergence of the tardigrade hypsibius exemplaris as a model system. Cold Spring Harb Protoc. 2018;2018:859–66. 10.1101/pdb.emo102301. [DOI] [PubMed] [Google Scholar]
- 42.Smith FW, Game M, Mapalo MA, Chavarria RA, Harrison TR, Janssen R. Developmental and genomic insight into the origin of the tardigrade body plan. Evol Dev. 2023. 10.1111/ede.12457. [DOI] [PubMed] [Google Scholar]
- 43.Chavarria RA, Game M, Arbelaez B, Ramnarine C, Snow ZK, Smith FW. Extensive loss of Wnt genes in Tardigrada. BMC Ecol Evol. 2021;21:1–21. 10.1186/s12862-021-01954-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beermann A, Schröder R. Functional stability of the aristaless gene in appendage tip formation during evolution. Dev Genes Evol. 2004;214:303–8. 10.1007/s00427-004-0411-7. [DOI] [PubMed] [Google Scholar]
- 45.Toegel JP, Wimmer EA, Prpic NM. Loss of spineless function transforms the Tribolium antenna into a thoracic leg with pretarsal, tibiotarsal, and femoral identity. Dev Genes Evol. 2009;219:53–8. 10.1007/s00427-008-0265-5. [DOI] [PubMed] [Google Scholar]
- 46.Sato M, Kojima T, Michiue T, Saigo K. Bar homeobox genes are latitudinal prepattern genes in the developing Drosophila notum whose expression is regulated by the concerted functions of decapentaplegic and wingless. Development. 1999;126:1457–66. 10.1242/dev.126.7.1457. [DOI] [PubMed] [Google Scholar]
- 47.Setton EVW, March LE, Nolan ED, Jones TE, Cho H, Wheeler WC, Extavour CG, Sharma pp. Expression and function of spineless orthologs correlate with distal deutocerebral appendage morphology across Arthropoda. Dev Biol. 2017;430:224–36. 10.1016/j.ydbio.2017.07.016. [DOI] [PubMed] [Google Scholar]
- 48.Walz B. Electron microscopic investigation of cephalic sense organs of the tardigrade Macrobiotus hufelandi C.A.S. Schultze Zoomorphologie. 1978;89:1–19. [Google Scholar]
- 49.Biserova NM, Kuznetsova KG. Head sensory organs of Halobiotus stenostomus (Eutardigrada, Hypsibiidae). Biol Bull. 2012;39:579–89. 10.1134/S1062359012070035. [Google Scholar]
- 50.Persson DK, Halberg KA, Jørgensen A, Møbjerg N, Kristensen RM. Neuroanatomy of Halobiotus crispae (Eutardigrada: Hypsibiidae): tardigrade brain structure supports the clade panarthropoda. J Morphol. 2012;273:1227–45. 10.1002/jmor.20054. [DOI] [PubMed] [Google Scholar]
- 51.Kihm JH, Zawierucha K, Rho HS, Park TYS. Homology of the head sensory structures between Heterotardigrada and Eutardigrada supported in a new species of water bear (Ramazzottiidae: Ramazzottius). Zool Lett. 2023;9:1–17. 10.1186/s40851-023-00221-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gross V, Epple L, Mayer G. Organization of the central nervous system and innervation of cephalic sensory structures in the water bear Echiniscus testudo (Tardigrada: Heterotardigrada) revisited. J Morphol. 2021;282:1298–312. 10.1002/jmor.21386. [DOI] [PubMed] [Google Scholar]
- 53.Gross V, Mayer G. Neural development in the tardigrade Hypsibius dujardini based on anti-acetylated α-tubulin immunolabeling. EvoDevo. 2015. 10.1186/s13227-015-0008-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martin C, Gross V, Hering L, Tepper B, Jahn H, de Sena OI, Stevenson PA, Mayer G. The nervous and visual systems of onychophorans and tardigrades: learning about arthropod evolution from their closest relatives. J Comp Physiol A Neuroethol Sensory Neural Behav Physiol. 2017;203:565–90. 10.1007/s00359-017-1186-4. [DOI] [PubMed] [Google Scholar]
- 55.Miyawaki K, Inoue Y, Mito T, Fujimoto T, Matsushima K, Shinmyo Y, Ohuchi H, Noji S. Expression patterns of aristaless in developing appendages of Gryllus bimaculatus (cricket). Mech Dev. 2002;113:181–4. 10.1016/S0925-4773(02)00020-5. [DOI] [PubMed] [Google Scholar]
- 56.Campbell G, Weaver T, Tomlinson A. Axis specification in the developing Drosophila appendage: the role of wingless, decapentaplegic, and the homeobox gene aristaless. Cell. 1993;74:1113–23. 10.1016/0092-8674(93)90732-6. [DOI] [PubMed] [Google Scholar]
- 57.Lemons D, Fritzenwanker JH, Gerhart J, Lowe CJ, McGinnis W. Co-option of an anteroposterior head axis patterning system for proximodistal patterning of appendages in early bilaterian evolution. Dev Biol. 2010;344:358–62. 10.1016/j.ydbio.2010.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grossmann D, Prpic NM. Egfr signaling regulates distal as well as medial fate in the embryonic leg of Tribolium castaneum. Dev Biol. 2012;370:264–72. 10.1016/j.ydbio.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 59.Campbell G. Regulation of gene expression in the distal region of the Drosophila leg by the Hox11 homolog, C15. Dev Biol. 2005;278:607–18. 10.1016/j.ydbio.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 60.Treffkorn S, Mayer G. Expression of NK genes that are not part of the NK cluster in the onychophoran Euperipatoides rowelli (Peripatopsidae). BMC Dev Biol. 2019;19:1–22. 10.1186/s12861-019-0185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Beermann A, Prühs R, Lutz R, Schröder R. A context-dependent combination of Wnt receptors controls axis elongation and leg development in a short germ insect. Development. 2011;138:2793–805. 10.1242/dev.063644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Natori K, Tajiri R, Furukawa S, Kojima T. Progressive tarsal patterning in the Drosophila by temporally dynamic regulation of transcription factor genes. Dev Biol. 2012;361:450–62. 10.1016/j.ydbio.2011.10.031. [DOI] [PubMed] [Google Scholar]
- 63.Angelini DR, Smith FW, Jockusch EL. Extent with modification: leg patterning in the beetle Tribolium castaneum and the evolution of serial homologs. G3 Genes Genomes Genet. 2012;2:235–48. 10.1534/g3.111.001537. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequence data analyzed in this study are publicly available. Sequences obtained via the homology search and alignments used for the phylogenetic analyses are available in the supplementary materials.